- 1Department of Hepatobiliary and Pancreatic Surgery, the Second Hospital of Jilin University, Changchun, China
- 2Department of Radiotherapy, the Second Hospital of Jilin University, Changchun, China
Hepatocellular carcinoma (HCC) is the most common primary liver cancer. Surgery has been the major treatment method for HCC owing to HCC’s poor sensitivity to radiotherapy and chemotherapy. However, its effectiveness is limited by postoperative tumour recurrence and metastasis. Systemic therapy is applied to eliminate postoperative residual tumour cells and improve the survival of patients with advanced HCC. Recently, the emergence of various novel targeted and immunotherapeutic drugs has significantly improved the prognosis of advanced HCC. However, targeted and immunological therapies may not always produce complete and long-lasting anti-tumour responses because of tumour heterogeneity and drug resistance. Traditional and patient-derived cell lines or animal models are used to investigate the drug resistance mechanisms of HCC and identify drugs that could reverse the resistance. This study comprehensively reviewed the established methods and applications of in-vivo and in-vitro HCC drug resistance models to further understand the resistance mechanisms in HCC treatment and provide a model basis for possible individualised therapy.
1 Introduction
Primary liver cancer is a common digestive system malignancy with extremely high rates of incidence and mortality, ranking sixth and fourth, respectively (1). Primary liver cancer includes hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma and combined hepatocellular cholangiocarcinoma, with HCC as the most common, accounting for almost 90% of cases (1). Surgery remained the most important treatment method for HCC over the years owing to HCC’s poor sensitivity to chemotherapy. However, postoperative patients show a high risk of recurrence or metastasis. Moreover, many HCC patients are diagnosed with advanced tumours and have lost the opportunity for surgery. In recent years, with the gradual popularisation of new treatment techniques, such as radiofrequency, microwave, freezing and TACE, and the development of numerous targeted and immunotherapeutic drugs, the progression-free and overall survival rates of HCC have greatly improved. However, primary and acquired drug resistance to these medications remains the most critical challenge in HCC treatment. This study reviewed original articles about drug resistance of HCC published in the last 5 years. The drug resistance models that were employed are presented, as well as a detailed introduction to some of the major drug resistance mechanisms that were discovered utilising the drug resistance models.
2 Traditional in vitro and in vivo drug resistance models
2.1 Establishment methods of traditional drug resistance models
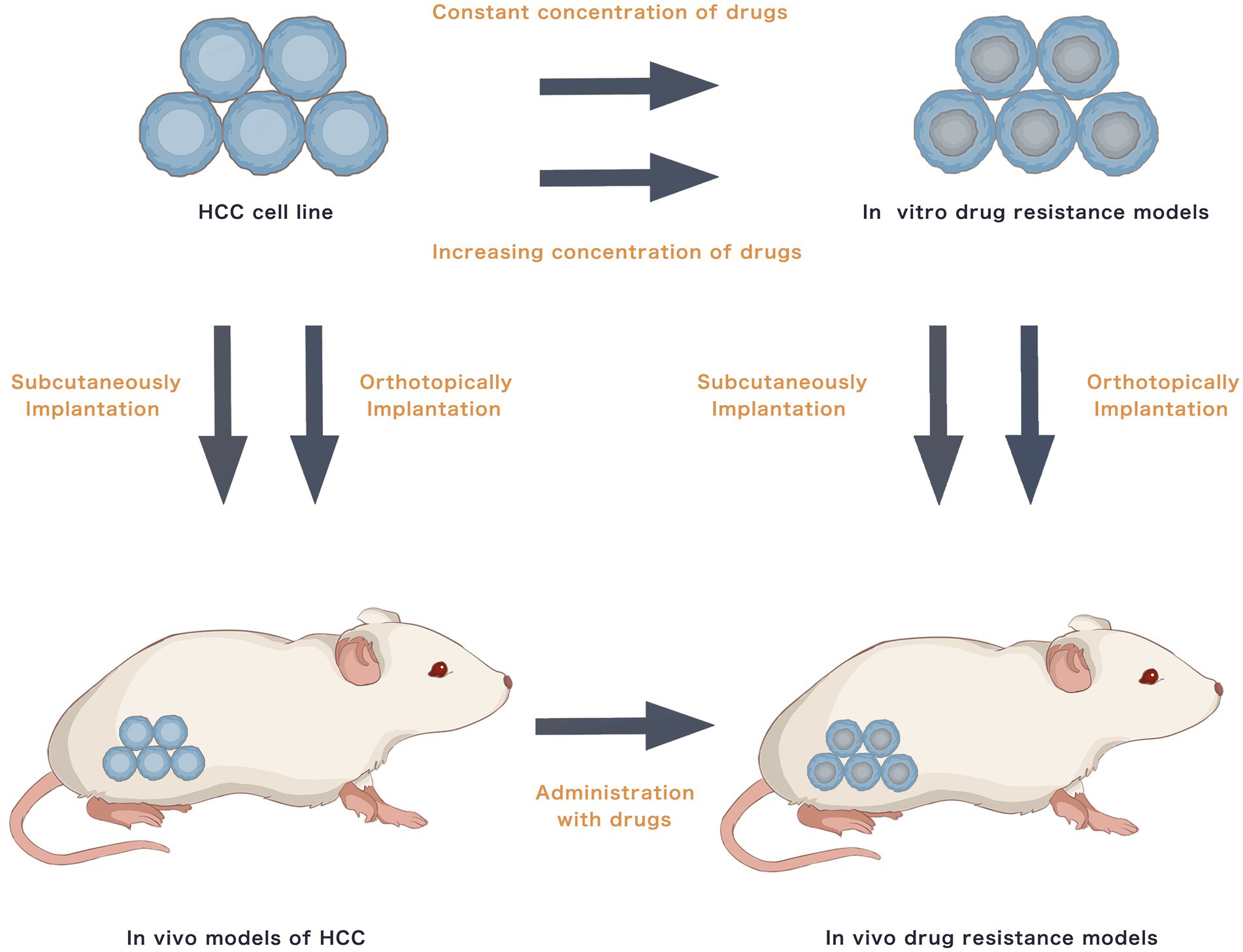
Most studies have used traditional commercial HCC cell lines to establish drug resistance models. In-vitro models involve exposing parental HCC cells to medications at either a continuous high or a progressively increasing concentration for 3–6 months or 20–30 generations for the cells to develop drug-specific resistance. For in-vivo drug resistance models, drug-resistant RCC cells can directly be implanted subcutaneously or orthotopically into nude mice. Furthermore, parental RCC cells may be injected subcutaneously or orthotopically into nude mice, followed by prolonged oral drug feeding to acquire drug resistance. Figure 1 presents the establishment methods of traditional drug resistance models. Table 1 presents studies on HCC drug resistance using traditional in-vitro and in-vivo models. Briefly, HepG2, Huh7, SMMC-7721, MHCC97H, MHCC97L, Hep3B, BEL-7402, PLC/PRF/5 and SK-Hep-1 are commonly used to induce acquired drug resistance. Traditional chemotherapy drugs, such as cisplatin, oxaliplatin, 5-FU and doxorubicin, and TKIs, including sorafenib, lenvatinib and regorafenib, are major research topics. Numerous signalling pathways may be involved in resistance to a single agent. A specific drug that could reverse the drug resistance-associated signalling may present its potential as an alternative or combined treatment.
2.2 Drug-resistant mechanisms based on traditional models
Li et al. (3) established sorafenib-resistant HCC (HepG2 and Huh7) cell lines and mouse models. To help Huh7-SR cells maintain their sorafenib-resistant ability, mice injected with Huh7-SR cells subcutaneously underwent daily treatment of sorafenib at a dose of 10 mg/kg. SR-HCC cells showed higher levels of lncRNA SNHG1 expression, miR-21 expression and Akt pathway activation than parental cells. SNHG1 activates the Akt pathway by regulating SLC3A2. Akt pathway inactivation induced by SNHG1 inhibition significantly increased the sensitivity of SR-HCC cells to sorafenib. Additionally, sorafenib induced the transfer of miR-21 to the nucleus, and miR-21 could continue to induce SNHG1 expression. Using the in-vitro and in-vivo sorafenib-resistant models, they found that LncRNA SNHG1 caused sorafenib resistance through activation of the Akt pathway. Reiter et al. (8) created a sorafenib-resistant HCC cell line to investigate the effect of ribociclib on the treatment of sorafenib-resistant HCC. HepG2 cells that were continuously incubated with sorafenib at escalating concentrations for 8 months, up to a final sorafenib concentration of 4 μM, were used as sorafenib-resistant cells and maintained in the medium with 4 μM sorafenib. They discovered that ribociclib reduced Rb expression and induced G1 cell cycle arrest in SR-HepG2 cells with Rb-high/p16-low protein expression profiles, indicating that ribociclib may be a good choice for the treatment of certain sorafenib-resistant HCC. Qiu et al. (13) established sorafenib-resistant HuH7 cells and xenograft mouse models and examined a group of patients with advanced, recurrent HCC to evaluate the clinical significance of sorafenib therapy. They found that high 14-3-3η expression and low miR-16 expression were related to sorafenib resistance and poor prognosis of HCC. Moreover, Chen et al. (36) initiated secondary sorafenib-resistant HuH-7 cells and xenograft mouse models. EphA2 was recognised as a crucial molecule in sorafenib resistance by quantitative phosphoproteomic analysis. It has been confirmed in in vivo animal models that sorafenib resistance can be successfully treated by concurrently inhibiting EphA2. Yang et al. (63) established sorafenib-resistant HepG2 cells and analysed the expression differences of circRNAs between sorafenib-sensitive and sorafenib-resistant HepG2 cells. CircFN1 was upregulated in sorafenib-resistant HepG2 cells and induced sorafenib resistance through the miR-1205/E2F1 signalling pathway. Zhao et al. (70) created sorafenib-resistant in-vitro and in-vivo models to explore the role of CBX4 in sorafenib resistance of HCC. CBX4 was upregulated in sorafenib-resistant Huh7 and PLC cells. Tumour growth could be suppressed by CA3- and UNC3866-mediated YAP1 and CBX4 inhibition in vivo. Younis et al. (71) designed an ultra-small lipid nanoparticle encapsulating sorafenib and midkine-siRNA and examined the effect of the new nanoparticles on treating sorafenib-resistant HCC with sorafenib-resistant cell-derived (HepG2) xenograft mouse models. The tumours in the xenograft models were eradicated by 70% using the nanoparticles, demonstrating the potential of this new approach in HCC treatment with sorafenib resistance. Xu et al. (79) constructed sorafenib-resistant in-vitro and in-vivo models to explore the role of UBQLN1 in HCC sorafenib resistance. For the sorafenib-resistant HCC mouse model, 100 million HCCLM3 cells were initially implanted into the flank of a BALB/c mouse. The formed tumours were re-implanted into 4-week-old BALB/c nude mice and fed with sorafenib (30 mg/kg/day). The mice that survived after 8 weeks of treatment were regarded as sorafenib-resistant mice. The ROS levels decreased in sorafenib-resistant HCC cells. However, sorafenib-resistant cells have better mitochondrial function and integrity with less mitochondrial content and respiratory capacity. Mechanically, these phenomena may be achieved by UBQLN1 through PGC1β inhibition in HCC. Fang et al. (96) established radiation-resistant HCC cell lines and xenograft mouse models to investigate the resistant mechanism of HCC to radiotherapy. MHCC97L cells were exposed to 8 Gy IR every 2 days for 5 fractions, and MHCC97H cells were exposed to 2 Gy IR daily for 25 fractions. After 4 weeks of recovery time, cells were again exposed to 10 Gy IR. MHCC97L IR-R cells were xenografted into nude mice and exposed to IR (8 Gy × 2 F) to establish radiation-resistant in-vivo models. Subsequent functional experiments demonstrated that the integration of glucose and cardiolipin anabolism was crucial for the radiation resistance of HCC. Zhou et al. (97) performed single-cell RNA sequencing in parental and sorafenib-resistant Huh7 cells using the 10X Genomic Chromium System. Huh7-R cells presented upregulations of stemness markers, EMT-related genes and Notch signalling-related genes, indicating that Notch signalling activation may be crucial for the induction of tumour stemness/EMT traits and acquired sorafenib resistance. Moreover, ZNFX1 antisense RNA 1 (ZFAS1), a new regulator IncRNA, had the highest upregulation in Huh7-R cells. Mechanically, the knockdown of ZFAS1 caused the downregulation of various mRNAs related to stemness and notch signalling pathways, indicating the critical role of this noncoding RNA in HCC sorafenib resistance. Huang et al. (112) created lenvatinib-resistant HCC cell lines and cell-derived xenograft mouse models. The two essential parts of the tRNA N7-methylguanosine (m7G) methyltransferase complex—methyltransferase-like protein-1 (METTL1) and WD repeat domain 4 protein (WDR4)—were significantly increased in lenvatinib-resistant cells. The crucial role that METTL1/WDR4-mediated m7G tRNA modification plays in developing lenvatinib resistance in vivo was further elucidated by xenograft mice models. Mechanically, METTL1 genes triggered drug resistance by EGFR pathway activation in HCC. Yang et al. (113) established the Pt-resistant HCC cell line 7404DDP to evaluate the antitumour effect of a cascade targeted and mitochondrion-dysfunctional nanomedicine (Gal-NP@TPt). When compared to cisplatin, Gal-NP@TPt caused a 9-fold increase in Pt accumulation in 7404DDP cells. Moreover, Gal-NP@TPt caused significant DNA damage in 7404DDP cells. Furthermore, Gal-NP@TPt may mitigate platinum resistance as the ratio of IC50 for 7404DDP to that for 7,404 dropped from 6.34 for cisplatin to 0.71 for Gal-NP@TPt. Kim et al. (133) established secondary sorafenib-resistant HCC cells and cell-derived mouse models to investigate the role of SIRT7 in sorafenib resistance. In Huh7SR and SK-Hep1SR cells, hyperactivated pERK1/2 was seen in conjunction with increased SIRT7 expression. In-vivo tumour development was suppressed by inhibiting SIRT7. Mechanically, SIRT7 inhibition eliminates sorafenib resistance by decreasing ERK1/2 phosphorylation via the DDX3X-mediated NLRP3 inflammasome in HCC.
3 Patient-derived drug resistance models
3.1 Establishment methods of patient-derived drug resistance models
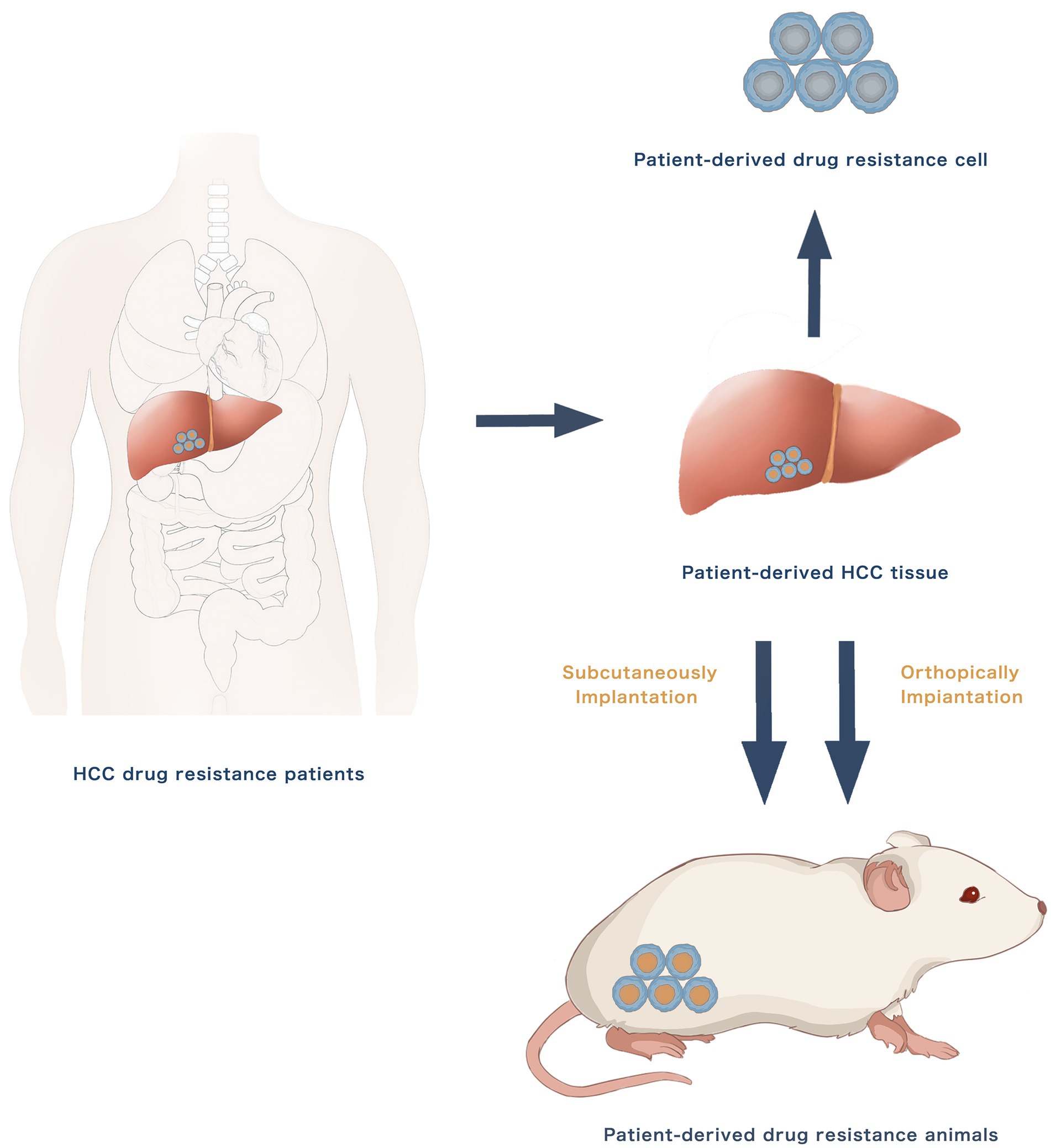
The use of patient-derived HCC cell lines and xenograft mouse models has increased in the study of HCC drug resistance. New drug-resistant cell lines may be created using primary cultures of HCC tissues from patients who have developed drug resistance. The resected tumour tissue samples may endure tissue digestion, cell separation and purification and primary cell culture to form a stable cell line. Patient-derived xenografts (PDXs) from patients with drug resistance could be directly implanted in nude mice subcutaneously or orthotopically to establish in-vivo drug resistance models. Figure 2 presents the establishment methods of patient-derived drug resistance models. The patient-derived drug resistance models better reserve individual molecular signature, including DNA copy number alterations, mutations and gene expression levels. The patient-derived drug resistance models may have additional advantages over the traditional models in the investigation of drug resistance mechanisms and individualised treatment as they can more accurately imitate the pathophysiological features of individual patients.
3.2 Drug-resistant mechanisms based on patient-derived models
Hu et al. (139) established sorafenib-resistant and sorafenib-sensitive PDX mouse models and examined the expression pattern differences between them. KPNA3 was found overexpressed in the sorafenib-resistant PDX models and was further confirmed to induce EMT and sorafenib resistance of HCC cells through KPNA3-AKT-ERK-TWIST signalling. Wang et al. (140) used HCC patient-derived organoid models to investigate the functions of Hedgehog signalling and CD44 in sorafenib resistance. In CD44-positive HCC, GANT61 dramatically reduced Hedgehog signalling to reverse sorafenib resistance, demonstrating that sorafenib with Hedgehog signalling inhibitors may be a useful therapeutic strategy for HCC patients with elevated CD44 levels. Hashiba et al. (33) revealed a patient-derived sorafenib-resistant HCC cell line (HCC-SR). The single-cell suspensions obtained from post-sorafenib HCC tissues were subcutaneously injected in nonobese diabetic, severe combined immunodeficient (NOD/SCID) mice, and the subcutaneous tumours were harvested to establish the new cell line. Sorafenib treatment induced higher cell proliferation of HCC-SR than other HCC cell lines, showing the resistance of HCC-SR to sorafenib. CIC S1595P missense mutation was detected in HCC-SR by whole-exome sequence analysis, revealing the potential function of this gene on HCC sorafenib resistance. Prawira et al. (141) established infigratinib-resistant PDX mouse models to explore the function of ribociclib in overcoming infigratinib resistance in HCC. SCID mice were subcutaneously implanted with four HCC PDX tumours with high FGFR1-4 expression, administered infigratinib (15 mg/kg per day) and sacrificed when tumours were 1800 mm3. Infigratinib-resistant tumours were re-implanted into SCID mice, and nine treatment cycles were required to maintain infigratinib resistance. Then, the infigratinib-resistant PDX mouse models were treated with either infigratinib alone or in combination with ribociclib and sacrificed for subsequent examination. The results showed that the combined inhibition of FGFR/CDK4/6 pathways is efficient in overcoming infigratinib resistance. Xu et al. (62) established four sorafenib-resistant HCC cell lines (HepG2-SR, LM3-SR, Huh7-SR, and SKhep1-SR) and cell-derived and patient-derived sorafenib-resistant xenograft mouse models to explore the mechanisms of sorafenib resistance in HCC. They found that circRNA-SORE was overexpressed in sorafenib-resistant HCC cells, induced sorafenib resistance and activated the Wnt/β-catenin pathway by sponging miR-660-3p and miR-103a-2-5p. Moreover, The cytoplasmic binding of circRNA-SORE to the master oncogenic protein YBX1 inhibits PRP19-mediated YBX1 degradation by blocking YBX1’s nuclear interaction with the E3 ubiquitin ligase PRP19 (142). Moreover, sorafenib resistance could be spread via exosomal circRNA-SORE transport (142). A local injection of circRNA-SORE shRNA lentivirus could significantly enhance the sensitivity of sorafenib treatment in mouse models. Liao et al. (143) investigated the potential of 17-AAG in overcoming sorafenib resistance using secondary sorafenib-resistant PDX mouse models. The HCC tissues from patients were transplanted into the armpit of severe NSG immunodeficient mice. Sorafenib (80 mg/kg) was administered orally to the mice once a day when the tumour size reached 100 mm3. The tumours were significantly resistant to sorafenib in the fourth generation. Additionally, 17-AAG suppressed HSP90α and reversed sorafenib resistance in vivo, indicating the potential of 17-AAG in overcoming sorafenib resistance. Leung et al. (144) created two sorafenib-resistant PDX mouse models, which resembled the emergence of sorafenib-induced acquired resistance in patients with HCC. The tumours from two HCC patients were xenografted into the immunodeficient mice, which were then administered with several cycles of sorafenib treatment to acquire sorafenib resistance. Two sorafenib-resistant HCC cell lines were further developed from the above two PDX models, with stronger self-renewal and tumourigenicity. RNA-sequencing recognised EPH receptor B2 (EPHB2) as the most significantly upregulated kinase in sorafenib-resistant PDXs. Functional experiments demonstrated that EPHB2 may increase tumour stemness and induce sorafenib resistance through the EPHB2/β-catenin/TCF1 positive feedback loop. Prawira et al. (145) established seven acquired infigratinib-resistant PDX models and developed a new infigratinib-resistant HCC cell line from one of these PDXs. Infigratinib-resistant tumours presented higher p-ErbB2, p-ErbB3 and EZH2 levels. Mechanically, EZH2 may promote infigratinib resistance by upregulating the ErbB family. Gao et al. (84) found high USP29, HIF1α and GLUT1 levels in sorafenib-resistant PDX tumours, and follow-up research revealed that USP29-induced sorafenib resistance by mediating HIF1α stabilisation and upregulated glycolysis. Mok et al. (146) used two drug-resistant tumour xenografts derived from HCC patients to investigate driving resistance and CSC repopulation in HCC. The xenografts mimicked the development of acquired resistance to sorafenib or lenvatinib treatment observed in HCC patients. RNA sequencing showed that cholesterol production was most frequently elevated in the drug-resistant xenografts. Mechanically, the drug resistance in HCC is driven by caspase-3-induced SREBP2 activation, which promotes cholesterol biosynthesis. Tao et al. (147) constructed six PDX models from HCC patients. The organoids from different patients showed different IC50 values of sorafenib treatment, indicating their sensitivity differences to sorafenib. BBOX1-AS1 was significantly upregulated in the organoids with the highest IC50 value, indicating that BBOX1-AS1 may be associated with sorafenib resistance. Ruan et al. (148) established a sorafenib-resistant PDX mouse model to investigate the function of a circular RNA, cDCBLD2. Tumour tissues from sorafenib-resistant HCC patients were implanted into the livers of NOD/SCID mice. Four weeks later, the mice were administered sorafenib at 30 mg/kg/d by gavage for 8 weeks to sustain sorafenib resistance. Then, the tissues were cut into equal pieces and re-implanted into the armpits of 4-week-old BALB/c nude mice, which were reared for subsequent in-vivo experiments. When in vivo grade cholesterol-conjugated si-cDCBLD2 was locally injected around the PDX implantation site, the sensitivity to sorafenib treatment was much higher than when a control siRNA was injected. In-vitro experiments further discovered that cDCBLD2-mediated sorafenib resistance was achieved by sponging miR-345-5p binding to the TOP2A coding sequence. Zhang et al. (149) found that YTHDF1 enhanced CSC renewal and resistance to the multiple tyrosine kinase inhibitors lenvatinib and sorafenib by upregulating NOTCH1 in patient-derived organoids and HCC cell lines. Additionally, Leung et al. (150) established sorafenib-resistant HCC cell lines and sorafenib-resistant PDX mouse models to evaluate the combined therapeutic effect of sorafenib and Src homology 2 domain-containing phosphatase 2 (SHP2) inhibitor on sorafenib-resistant HCC. NOD/SCID mice bearing PDX were orally administered sorafenib at 100 mg/kg/day for 25 days to acquire sorafenib resistance. SHP2 was significantly upregulated in sorafenib-resistant HCC cell lines and PDXs. Sorafenib combined with SHP2 inhibitor SHP099 showed high treatment efficacy in sorafenib-resistant PDX mice.
4 Direct detection of clinical drug-resistant samples from HCC patients
Peripheral blood and tumour tissues from HCC patients may be directly detected for gene expression levels using quantitative or semi-quantitative techniques such as qPCR, western blot or immunohistochemistry. In drug-resistant HCC samples, highly expressed genes are generally more likely to perform as drug resistance genes, whereas suppressed genes may be able to withstand drug resistance. Table 2 presents research on the direct detection of clinical drug-resistant samples from HCC patients.
Circulating tumour DNA (ctDNA) sequencing is a minimally invasive method that enables the collection of repeat samples. Hatlen et al. (19) sequenced the ctDNA of acquired fisogatinib-resistant HCC patients in a fisogatinib phase I trial. Two patients with disease progression had mutations in the gatekeeper and hinge-1 residues in the FGFR4 kinase domain. Further, subsequent experiments using in-vivo and in-vitro secondary fisogatinib-resistant models demonstrated that acquired fisogatinib resistance was related to FGFR4 kinase domain mutations. Yu et al. (67) examined the HCC tissues of patients who experienced recurrence after primary HCC resection and sequential sorafenib treatment and found an inverse expression between CYP1A2 and NF-κB p65 in the sorafenib-naïve primary HCC compared with its paired sorafenib-experienced recurrence. Moreover, Weng et al. (73) divided patients who received two cycles of sorafenib treatment into the sorafenib-sensitive and sorafenib-resistant groups. RNA sequencing showed higher circFOXM1 levels in sorafenib-resistant HCC tissues. Functional experiments revealed circFOXM1-induced sorafenib resistance by upregulating MECP2 expression via sponging miR-1324. Ma et al. (87) discovered that transcriptional factor myocyte enhancer factor 2D (MEF2D) was overexpressed in sorafenib-resistant HCC cell lines and HCC specimens, indicating a poor prognosis for sorafenib-treated HCC patients. Mechanically, coupling HDAC4 with MEF2D may activate ERK by inhibiting SPRY4, causing sorafenib resistance of HCC. Wang et al. (160) examined the compared tumour tissues (pretreatment and post-progression samples) of one HCC patient with acquired resistance to a combined treatment of atezolizumab and bevacizumab who subsequently underwent surgical resection of the tumour. The number of CD8+ T cells in the tumour area and PD-L1 level in tumour-infiltrating immune cells were decreased in the drug-resistant HCC tissue. Additionally, the drug-resistant tissue presented more progenitor/hepatoblast features in the gene expression profile. The abovementioned results show that the acquired resistance to the combined treatment may be caused by the immune-excluded tumour microenvironment and tumour dedifferentiation. Lu et al. (161) found that circTMEM181 expression was upregulated in puncture biopsies of HCC tissues from anti-PD1 antibody-resistant patients compared to those from anti-PD1-sensitive patients. Furthermore, a high exosomal circTMEM181 level was associated with immunosuppression of microenvironment and anti-PD1 resistance in HCC. Mechanistically, exosomal circTMEM181 promoted CD39 expression by sponging miR-488-3p in macrophages. They further created macrophage-specific CD39 knockout mice and discovered that CD73 expression in HCC cells and CD39 expression in macrophages could impair the function of CD8 + T cells and induce anti-PD1 resistance by activating the eATP-adenosine pathway. Meng et al. (170) discovered that CD10 + ALPL+ neutrophils were more abundant in the tumour tissues of anti-PD-1-resistant patients than in those of anti-PD-1-sensitive patients. Mechanically, tumour cells secreted NAMPT, which reprogrammed CD10 + ALPL+neutrophils via NTRK1, keeping them immature and preventing their maturation and activation. This was how CD10 + ALPL+neutrophils were generated. Immunosuppressive CD10 + ALPL+ neutrophils further mediated ongoing T-cell exhaustion, which increased resistance to anti-PD-1 treatment in HCC.
5 Transgenic drug resistance models
5.1 Establishment methods of transgenic drug resistance models
Gene-editing techniques could be used in studies on mechanisms of drug resistance by direct insertion or knockout of drug-resistant genes in cells or animal models. An RNA-guided DNA endonuclease derived from the type II CRISPR bacterial immune system, namely clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9), has been extensively employed as a highly effective gene-editing method because of its capacity to target novel genes through the simple modification of single-guide RNA (sgRNA) sequences (176). The precise Watson–Crick base pairing between Cas9’s guide RNA and the target DNA region and a direct contact between Cas9 and a short DNA protospacer adjacent motif are critical for the targeted-sequence specificity of Cas9 (177–180). The two Cas9 nuclease domains—HNH and RuvC—catalytically cleave the double-stranded DNA (179, 180). A shift in the reading frame caused by random insertions or deletions may result in mutations at the targeted locations (176). Homologous recombination with an introduced homologous donor DNA may be used to complete homology-directed repair (181, 182).
5.2 Drug-resistant mechanisms based on transgenic models
The transcription factor called hepatic leukaemia factor (HLF) is a member of the proline and acidic amino acid-rich family, which are known to regulate circadian rhythms (183). Xiang et al. (184) constructed an HLF-knockout mouse model (HLFPB/PB mouse) and found that sorafenib resistance requires HLF-mediated c-Jun activation, demonstrating that the HLF-c-Jun axis may control the haepatoma’s response to sorafenib. A piggyBac transposon-encoding CAG-RFP (a red fluorescent protein sequence under the CAG promoter) was inserted into the HLF gene to create the HLFPB/PB mice. Diethylnitrosamine was injected into the abdomen of mice to induce haepatoma development. The mice were then euthanised 7, 8 or 9 months later. Haepatoma cells that overexpressed HLF became resistant to growth inhibition and cell death caused by sorafenib. The c-Jun interference eliminated sorafenib resistance in haepatoma cells overexpressing HLF. Furthermore, HLF interference weakened c-Jun activation due to sorafenib and made haepatoma cells more susceptible to sorafenib. Wei et al. (185) found phosphoglycerate dehydrogenase (PHGDH) to be a critical sorafenib-resistance gene by genome-wide CRISPR/Cas9 library screening. The Human GeCKOv2A CRISPR knockout pooled library contains 65,386 sgRNAs targeting 19,052 human genes and 1864 miRNAs. They constructed a stable Cas9-expressing HCC cell line (MHCC97L-Cas9) by lentiviral transfection and transfected it with the GeCKOv2A library. Mutant cells were selected by puromycin and treated with sorafenib and DMSO for a week and were collected. The sgRNAs were amplified by PCR for subsequent high-throughput sequencing and bioinformatic analysis. PHGDH targeting sgRNAs were negatively selected after sorafenib treatment. They further established CRISPR/Cas9 knockout and RNAi knockdown cell models and found that inhibition of PHGDH promotes HCC apoptosis by suppressing the synthesis pathway upon sorafenib treatment. Sueangoen et al. (186) developed transgenic cell models to evaluate the functions of seven HCC-derived EGFR mutations on erlotinib resistance. Seven missense mutations (K757E, N808S, R831C, V897A, P937L, T940A and M947T) in the kinase domain of EGFR were recognised and retrovirally transducted into NIH-3 T3 cells. T790M and L858R were used as erlotinib-resistant and erlotinib-sensitive mutant controls, respectively. In vitro experiments showed that the seven EGFR mutants are dependent on EGF and resistant to erlotinib. Sofer et al. (187) performed a genome-wide CRISPR/Cas9 activation screen in Huh7 and recognised hexokinase 1 as a critical factor in promoting regorafenib resistance in HCC. Moreover, Chen et al. recognised FGF21 as a sorafenib-resistant gene in HCC by CRISPR/CAS9 genome library screening. Mechanically, FGF21 induces sorafenib resistance by directly combining with NRF2 to prevent NRF2 ubiquitination degradation. Huang et al. (188) used CRISPR/CAS9 genome library screening and discovered that lenvatinib resistance was mediated by DUSP4 deficiency through the activation of MAPK/ERK signalling. Another gene associated with lenvatinib resistance, LAPTM5, was identified by Pan et al. (189). MiR-3689a-3p was found to be the most upregulated miRNA in sorafenib-sensitive HCC by CRISPR/Cas9 screens (190). Mechanically, miR-3689a-3p may suppress CCS/SOD1-dependent mitochondrial oxidative stress to regulate sorafenib resistance. Zhu et al. (191) used a transgenic HCC mouse model driven by MYC overexpression and beta-catenin (encoded by CTNNB1) activation (termed MYC-lucOS; CTNNB1) (192), which shows resistance to anti-PD-1 immunotherapy, to evaluate the effect of the combination of anti-PD-L1 immunotherapy and anti-VEGF. After 4 weeks of the combined treatment, a significant survival improvement and a decline in the proportion of mice with tumours were observed in this model, indicating the potential of the anti-PD-L1/anti-VEGF dual treatment in overcoming resistance to either of a single agent. Martin et al. (193) used CRISPR/Cas9 to directionally knock out tumour suppressor genes in HCC cells. The p53-knockout HepG2 cells performed increased malignant properties and multidrug resistance to cisplatin, regorafenib, sorafenib and doxorubicin. Alb-R26Met mice carry a conditional mouse–human chimeric Met transgene into the Rosa26 locus. The Alb-R26Met HCC is resistant to sorafenib. ADAMTSL5 overexpression was noted at the early stages of liver carcinogenesis, and its upregulation was reproduced in the Alb-R26Met HCC model. Various oncogenic inputs associated with HCC decreased as a result of ADAMTSL5 abrogation, such as MET, EGFR, PDGFRβ, IGF1Rβ and FGFR4 receptor tyrosine kinases, which were all expressed and/or phosphorylated to a lesser extent, showing the potential role of ADAMTSL5 in HCC drug resistance (194).
6 Summary
To further understand drug resistance mechanisms in HCC, useful models must be developed. Different kinds of models have their own advantages and disadvantages. Traditional drug resistance models are easier to establish. Meanwhile, the corresponding experimental results have higher stability and reproducibility. However, individual differences in HCC gene expression could not be reflected in a single cell line. Patient-derived models maintain more individual traits, which are essential for studying the various drug resistance pathways related to distinct clinical subtypes of cancer. Despite the difficulty of the construction process, patient-derived models may be a better choice to investigate the drug resistance mechanisms of various malignancies. Sometimes a direct detection of clinical drug-resistant samples may be a simpler method to screen for drug-resistant genes. Gene-editing methodologies can be employed to generate genetically engineered cell lines or animal models that exhibit resistance to particular pharmaceutical agents. Theoretically, a gene-editing cell line may be an ideal model to investigate the function of a specific gene on drug resistance. It has to be admitted that none of the aforementioned techniques is flawless. In the actual research, we should choose the appropriate drug resistance model according to the research purpose and the realistic research environment.
Author contributions
YX: Writing – original draft. JW: Writing – original draft. HQ: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Llovet, JM, Kelley, RK, Villanueva, A, Singal, AG, Pikarsky, E, Roayaie, S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. (2021) 7:6. doi: 10.1038/s41572-020-00240-3
2. Chowdhury, KD, Sarkar, A, Chatterjee, S, Patra, D, Sengupta, D, Banerjee, S, et al. Cathepsin B mediated scramblase activation triggers cytotoxicity and cell cycle arrest by andrographolide to overcome cellular resistance in cisplatin resistant human hepatocellular carcinoma HepG2 cells. Environ Toxicol Pharmacol. (2019) 68:120–32. doi: 10.1016/j.etap.2019.03.003
3. Li, W, Dong, X, He, C, Tan, G, Li, Z, Zhai, B, et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. (2019) 38:183. doi: 10.1186/s13046-019-1177-0
4. Dong, N, Shi, X, Wang, S, Gao, Y, Kuang, Z, Xie, Q, et al. M2 macrophages mediate sorafenib resistance by secreting HGF in a feed-forward manner in hepatocellular carcinoma. Br J Cancer. (2019) 121:22–33. doi: 10.1038/s41416-019-0482-x
5. Sui, C, Dong, Z, Yang, C, Zhang, M, Dai, B, Geng, L, et al. LncRNA FOXD2-AS1 AS a competitive endogenous RNA against miR-150-5p reverses resistance to sorafenib in hepatocellular carcinoma. J Cell Mol Med. (2019) 23:6024–33. doi: 10.1111/jcmm.14465
6. Yang, Y, Yao, JH, Du, QY, Zhou, YC, Yao, TJ, Wu, Q, et al. Connexin 32 downregulation is critical for chemoresistance in oxaliplatin-resistant HCC cells associated with EMT. Cancer Manag Res. (2019) 11:5133–46. doi: 10.2147/CMAR.S203656
7. Liao, X, Bu, Y, Jiang, S, Chang, F, Jia, F, Xiao, X, et al. CCN2-MAPK-Id-1 loop feedback amplification is involved in maintaining stemness in oxaliplatin-resistant hepatocellular carcinoma. Hepatol Int. (2019) 13:440–53. doi: 10.1007/s12072-019-09960-5
8. Reiter, FP, Denk, G, Ziesch, A, Ofner, A, Wimmer, R, Hohenester, S, et al. Predictors of ribociclib-mediated antitumour effects in native and sorafenib-resistant human hepatocellular carcinoma cells. Cell Oncol (Dordr). (2019) 42:705–15. doi: 10.1007/s13402-019-00458-8
9. Liu, YS, Kuo, WW, Chen, MC, Hsu, HH, Tu, CC, Yeh, YL, et al. Inhibition of protein phosphatase 1 stimulates noncanonical ER stress eIF2α activation to enhance fisetin-induced chemosensitivity in HDAC inhibitor-resistant hepatocellular carcinoma cells. Cancers (Basel). (2019) 11:918. doi: 10.3390/cancers11070918
10. Wu, J, Qu, J, Cao, H, Jing, C, Wang, Z, Xu, H, et al. Monoclonal antibody AC10364 inhibits cell proliferation in 5-fluorouracil resistant hepatocellular carcinoma via apoptotic pathways. Onco Targets Ther. (2019) 12:5053–67. doi: 10.2147/OTT.S206517
11. Singh, MP, Cho, HJ, Kim, JT, Baek, KE, Lee, HG, and Kang, SC. Morin hydrate reverses cisplatin resistance by impairing PARP1/HMGB1-dependent autophagy in hepatocellular carcinoma. Cancers (Basel). (2019) 11:986. doi: 10.3390/cancers11070986
12. Zhou, Y, Wang, Y, Zhou, W, Chen, T, Wu, Q, Chutturghoon, VK, et al. YAP promotes multi-drug resistance and inhibits autophagy-related cell death in hepatocellular carcinoma via the RAC1-ROS-mTOR pathway. Cancer Cell Int. (2019) 19:179. doi: 10.1186/s12935-019-0898-7
13. Qiu, Y, Shan, W, Yang, Y, Jin, M, Dai, Y, Yang, H, et al. Reversal of sorafenib resistance in hepatocellular carcinoma: epigenetically regulated disruption of 14-3-3η/hypoxia-inducible factor-1α. Cell Death Discov. (2019) 5:120. doi: 10.1038/s41420-019-0200-8
14. Provvisiero, DP, Negri, M, de Angelis, C, Di Gennaro, G, Patalano, R, Simeoli, C, et al. Vitamin D reverts resistance to the mTOR inhibitor everolimus in hepatocellular carcinoma through the activation of a miR-375/oncogenes circuit. Sci Rep. (2019) 9:11695. doi: 10.1038/s41598-019-48081-9
15. Liu, Z, Wang, Y, Yao, Y, Fang, Z, Miao, QR, and Ye, M. Quantitative proteomic and phosphoproteomic studies reveal novel 5-fluorouracil resistant targets in hepatocellular carcinoma. J Proteome. (2019) 208:103501. doi: 10.1016/j.jprot.2019.103501
16. Pang, L, Xu, L, Yuan, C, Li, X, Zhang, X, Wang, W, et al. Activation of EGFR-KLF4 positive feedback loop results in acquired resistance to sorafenib in hepatocellular carcinoma. Mol Carcinog. (2019) 58:2118–26. doi: 10.1002/mc.23102
17. Ding, Y, Li, S, Ge, W, Liu, Z, Zhang, X, Wang, M, et al. Design and synthesis of parthenolide and 5-fluorouracil conjugates as potential anticancer agents against drug resistant hepatocellular carcinoma. Eur J Med Chem. (2019) 183:111706. doi: 10.1016/j.ejmech.2019.111706
18. Makol, A, Kaur, H, Sharma, S, Kanthaje, S, Kaur, R, and Chakraborti, A. Vimentin as a potential therapeutic target in sorafenib resistant HepG2, a HCC model cell line. Clin Mol Hepatol. (2020) 26:45–53. doi: 10.3350/cmh.2019.0031
19. Hatlen, MA, Schmidt-Kittler, O, Sherwin, CA, Rozsahegyi, E, Rubin, N, Sheets, MP, et al. Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discov. (2019) 9:1686–95. doi: 10.1158/2159-8290.CD-19-0367
20. Sanchez-Carranza, JN, González-Maya, L, Razo-Hernández, RS, Salas-Vidal, E, Nolasco-Quintana, NY, Clemente-Soto, AF, et al. Achillin increases Chemosensitivity to paclitaxel, overcoming resistance and enhancing apoptosis in human hepatocellular carcinoma cell line resistant to paclitaxel (Hep3B/PTX). Pharmaceutics. (2019) 11:512. doi: 10.3390/pharmaceutics11100512
21. Saraswati, S, Alhaider, A, Abdelgadir, AM, Tanwer, P, and Korashy, HM. Phloretin attenuates STAT-3 activity and overcomes sorafenib resistance targeting SHP-1-mediated inhibition of STAT3 and Akt/VEGFR2 pathway in hepatocellular carcinoma. Cell Commun Signal. (2019) 17:127. doi: 10.1186/s12964-019-0430-7
22. Zan, Y, Dai, Z, Liang, L, Deng, Y, and Dong, L. Co-delivery of plantamajoside and sorafenib by a multi-functional nanoparticle to combat the drug resistance of hepatocellular carcinoma through reprograming the tumor hypoxic microenvironment. Drug Deliv. (2019) 26:1080–91. doi: 10.1080/10717544.2019.1654040
23. Suk, FM, Liu, CL, Hsu, MH, Chuang, YT, Wang, JP, and Liao, YJ. Treatment with a new benzimidazole derivative bearing a pyrrolidine side chain overcomes sorafenib resistance in hepatocellular carcinoma. Sci Rep. (2019) 9:17259. doi: 10.1038/s41598-019-53863-2
24. Lai, SC, Su, YT, Chi, CC, Kuo, YC, Lee, KF, Wu, YC, et al. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J Exp Clin Cancer Res. (2019) 38:474. doi: 10.1186/s13046-019-1442-2
25. Liao, X, Bu, Y, Chang, F, Jia, F, Song, G, Xiao, X, et al. Remodeling of hepatic stellate cells orchestrated the stroma-derived oxaliplatin-resistance through CCN3 paracrine in hepatocellular carcinoma. BMC Cancer. (2019) 19:1192. doi: 10.1186/s12885-019-6362-1
26. Shen, JH, Chen, PH, Liu, HD, Huang, DA, Li, MM, and Guo, K. HSF1/AMPKα2 mediated alteration of metabolic phenotypes confers increased oxaliplatin resistance in HCC cells. Am J Cancer Res. (2019) 9:2349–63.
27. Liao, X, Song, G, Xu, Z, Bu, Y, Chang, F, Jia, F, et al. Oxaliplatin resistance is enhanced by saracatinib via upregulation Wnt-ABCG1 signaling in hepatocellular carcinoma. BMC Cancer. (2020) 20:31. doi: 10.1186/s12885-019-6480-9
28. Wu, B, Li, A, Zhang, Y, Liu, X, Zhou, S, Gan, H, et al. Resistance of hepatocellular carcinoma to sorafenib can be overcome with co-delivery of PI3K/mTOR inhibitor BEZ235 and sorafenib in nanoparticles. Expert Opin Drug Deliv. (2020) 17:573–87. doi: 10.1080/17425247.2020.1730809
29. Wang, S, Cai, L, Zhang, F, Shang, X, Xiao, R, and Zhou, H. Inhibition of EZH2 attenuates Sorafenib resistance by targeting NOTCH1 activation-dependent liver Cancer stem cells via NOTCH1-related MicroRNAs in hepatocellular carcinoma. Transl Oncol. (2020) 13:100741. doi: 10.1016/j.tranon.2020.01.002
30. Liu, Y, Chen, L, Yuan, H, Guo, S, and Wu, G. LncRNA DANCR promotes Sorafenib resistance via activation of IL-6/STAT3 signaling in hepatocellular carcinoma cells. Onco Targets Ther. (2020) 13:1145–57. doi: 10.2147/OTT.S229957
31. Yu, X, Gao, X, Mao, X, Shi, Z, Zhu, B, Xie, L, et al. Knockdown of FOXO6 inhibits glycolysis and reduces cell resistance to paclitaxel in HCC cells via PI3K/Akt signaling pathway. Onco Targets Ther. (2020) 13:1545–56. doi: 10.2147/OTT.S233031
32. Wu, XL, Chen, Y, Kong, WC, and Zhao, ZQ. Amyloid precursor protein regulates 5-fluorouracil resistance in human hepatocellular carcinoma cells by inhibiting the mitochondrial apoptotic pathway. J Zhejiang Univ Sci B. (2020) 21:234–45. doi: 10.1631/jzus.B1900413
33. Hashiba, T, Yamashita, T, Okada, H, Nio, K, Hayashi, T, Asahina, Y, et al. Inactivation of transcriptional repressor Capicua confers Sorafenib resistance in human hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. (2020) 10:269–85. doi: 10.1016/j.jcmgh.2020.02.009
34. Bamodu, OA, Chang, HL, Ong, JR, Lee, WH, Yeh, CT, and Tsai, JT. Elevated PDK1 expression drives PI3K/AKT/MTOR signaling promotes radiation-resistant and dedifferentiated phenotype of hepatocellular carcinoma. Cells. (2020) 9:746. doi: 10.3390/cells9030746
35. Liu, Z, Wang, Q, Mao, J, Wang, K, Fang, Z, Miao, QR, et al. Comparative proteomic analysis of protein methylation provides insight into the resistance of hepatocellular carcinoma to 5-fluorouracil. J Proteome. (2020) 219:103738. doi: 10.1016/j.jprot.2020.103738
36. Chen, CT, Liao, LZ, Lu, CH, Huang, YH, Lin, YK, Lin, JH, et al. Quantitative phosphoproteomic analysis identifies the potential therapeutic target EphA2 for overcoming sorafenib resistance in hepatocellular carcinoma cells. Exp Mol Med. (2020) 52:497–513. doi: 10.1038/s12276-020-0404-2
37. Shi, Y, Yang, X, Xue, X, Sun, D, Cai, P, Song, Q, et al. HANR enhances autophagy-associated Sorafenib resistance through miR-29b/ATG9A Axis in hepatocellular carcinoma. Onco Targets Ther. (2020) 13:2127–37. doi: 10.2147/OTT.S229913
38. Fan, L, Huang, X, Chen, J, Zhang, K, Gu, YH, Sun, J, et al. Long noncoding RNA MALAT1 contributes to Sorafenib resistance by targeting miR-140-5p/Aurora-a signaling in hepatocellular carcinoma. Mol Cancer Ther. (2020) 19:1197–209. doi: 10.1158/1535-7163.MCT-19-0203
39. Li, J, Qin, X, Wu, R, Wan, L, Zhang, L, and Liu, R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med. (2020) 24:5152–61. doi: 10.1111/jcmm.15162
40. Wu, MY, Tang, YP, Liu, JJ, Liang, R, and Luo, XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer. (2020) 11:2993–3001. doi: 10.7150/jca.39854
41. Cai, J, Chen, J, Wu, T, Cheng, Z, Tian, Y, Pu, C, et al. Genome-scale CRISPR activation screening identifies a role of LRP8 in Sorafenib resistance in hepatocellular carcinoma. Biochem Biophys Res Commun. (2020) 526:1170–6. doi: 10.1016/j.bbrc.2020.04.040
42. Ji, L, Lin, Z, Wan, Z, Xia, S, Jiang, S, Cen, D, et al. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. (2020) 11:250. doi: 10.1038/s41419-020-2413-4
43. Jing, Z, Ye, X, Ma, X, Hu, X, Yang, W, Shi, J, et al. SNGH16 regulates cell autophagy to promote Sorafenib resistance through suppressing miR-23b-3p via sponging EGR1 in hepatocellular carcinoma. Cancer Med. (2020) 9:4324–38. doi: 10.1002/cam4.3020
44. Li, R, Dong, C, Jiang, K, Sun, R, Zhou, Y, Yin, Z, et al. Rab27B enhances drug resistance in hepatocellular carcinoma by promoting exosome-mediated drug efflux. Carcinogenesis. (2020) 41:1583–91. doi: 10.1093/carcin/bgaa029
45. Zhu, Y, Xu, J, Hu, W, Wang, F, Zhou, Y, Xu, W, et al. TFAM depletion overcomes hepatocellular carcinoma resistance to doxorubicin and sorafenib through AMPK activation and mitochondrial dysfunction. Gene. (2020) 753:144807. doi: 10.1016/j.gene.2020.144807
46. Liao, YJ, Hsu, SM, Chien, CY, Wang, YH, Hsu, MH, and Suk, FM. Treatment with a new Barbituric acid derivative exerts Antiproliferative and Antimigratory effects against Sorafenib resistance in hepatocellular carcinoma. Molecules. (2020) 25:2856. doi: 10.3390/molecules25122856
47. Huang, W, Huang, F, and Feng, C. CircFoxo3 promotes Adriamycin resistance through regulation of miR-199a-5p/ATP binding cassette subfamily C member 1 Axis in hepatocellular carcinoma. Onco Targets Ther. (2020) 13:5113–22. doi: 10.2147/OTT.S243571
48. Yang, Y, Liao, Y, Gui, YP, Zhao, L, and Guo, LB. GL-V9 reverses adriamycin resistance in hepatocellular carcinoma cells by affecting JNK2-related autophagy. Chin J Nat Med. (2020) 18:491–9. doi: 10.1016/S1875-5364(20)30059-5
49. Meßner, M, Schmitt, S, Ardelt, MA, Fröhlich, T, Müller, M, Pein, H, et al. Metabolic implication of tigecycline as an efficacious second-line treatment for sorafenib-resistant hepatocellular carcinoma. FASEB J. (2020) 34:11860–82. doi: 10.1096/fj.202001128R
50. Shi, W, Zhang, S, Ma, D, Yan, D, Zhang, G, Cao, Y, et al. Targeting SphK2 reverses acquired resistance of Regorafenib in hepatocellular carcinoma. Front Oncol. (2020) 10:694. doi: 10.3389/fonc.2020.00694
51. Liu, J, Yang, X, Liang, Q, Yu, Y, Shen, X, and Sun, G. Valproic acid overcomes sorafenib resistance by reducing the migration of Jagged2-mediated Notch1 signaling pathway in hepatocellular carcinoma cells. Int J Biochem Cell Biol. (2020) 126:105820. doi: 10.1016/j.biocel.2020.105820
52. Xiao, L, Hou, Y, He, H, Cheng, S, Hou, Y, Jin, H, et al. A novel targeted delivery system for drug-resistant hepatocellular carcinoma therapy. Nanoscale. (2020) 12:17029–44. doi: 10.1039/D0NR01908A
53. Shen, Q, Jiang, S, Wu, M, Zhang, L, Su, X, and Zhao, D. LncRNA HEIH confers cell Sorafenib resistance in hepatocellular carcinoma by regulating miR-98-5p/PI3K/AKT pathway. Cancer Manag Res. (2020) 12:6585–95. doi: 10.2147/CMAR.S241383
54. Lv, S, Wang, X, Bai, X, Ning, H, Li, Y, Wen, H, et al. Mesenchymal epithelial transition factor regulates tumor necrosis factor-related apoptotic induction ligand resistance in hepatocellular carcinoma cells through down-regulation of cyclin B1. Int J Biochem Cell Biol. (2020) 128:105844. doi: 10.1016/j.biocel.2020.105844
55. Wang, W, Ding, B, Lou, W, and Lin, S. Promoter Hypomethylation and miR-145-5p downregulation- mediated HDAC11 overexpression promotes Sorafenib resistance and metastasis of hepatocellular carcinoma cells. Front Cell Dev Biol. (2020) 8:724. doi: 10.3389/fcell.2020.00724
56. Regan-Fendt, K, Li, D, Reyes, R, Yu, L, Wani, NA, Hu, P, et al. Transcriptomics-based drug repurposing approach identifies novel drugs against Sorafenib-resistant hepatocellular carcinoma. Cancers (Basel). (2020) 12:2730. doi: 10.3390/cancers12102730
57. Zhang, J, Zhao, X, Ma, X, Yuan, Z, and Hu, M. KCNQ1OT1 contributes to sorafenib resistance and programmed death-ligand-1-mediated immune escape via sponging miR-506 in hepatocellular carcinoma cells. Int J Mol Med. (2020) 46:1794–804. doi: 10.3892/ijmm.2020.4710
58. Hu, YT, Shu, ZY, Jiang, JH, Xie, QF, and Zheng, SS. Torin2 overcomes sorafenib resistance via suppressing mTORC2-AKT-BAD pathway in hepatocellular carcinoma cells. Hepatobiliary Pancreat Dis Int. (2020) 19:547–54. doi: 10.1016/j.hbpd.2020.09.010
59. Zhang, Z, Tan, X, Luo, J, Yao, H, Si, Z, and Tong, JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis. (2020) 11:902. doi: 10.1038/s41419-020-03123-3
60. Xu, GL, Ni, CF, Liang, HS, Xu, YH, Wang, WS, Shen, J, et al. Upregulation of PD-L1 expression promotes epithelial-to-mesenchymal transition in sorafenib-resistant hepatocellular carcinoma cells. Gastroenterol Rep (Oxf). (2020) 8:390–8. doi: 10.1093/gastro/goaa049
61. Zhu, Q, Ren, H, Li, X, Qian, B, Fan, S, Hu, F, et al. Silencing KIF14 reverses acquired resistance to sorafenib in hepatocellular carcinoma. Aging (Albany NY). (2020) 12:22975–3003. doi: 10.18632/aging.104028
62. Xu, J, Wan, Z, Tang, M, Lin, Z, Jiang, S, Ji, L, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. (2020) 19:163. doi: 10.1186/s12943-020-01281-8
63. Yang, C, Dong, Z, Hong, H, Dai, B, Song, F, Geng, L, et al. circFN1 mediates Sorafenib resistance of hepatocellular carcinoma cells by sponging miR-1205 and regulating E2F1 expression. Mol Ther Nucleic Acids. (2020) 22:421–33. doi: 10.1016/j.omtn.2020.08.039
64. Leung, HW, Lau, EYT, Leung, CON, Lei, MML, Mok, EHK, Ma, VWS, et al. NRF2/SHH signaling cascade promotes tumor-initiating cell lineage and drug resistance in hepatocellular carcinoma. Cancer Lett. (2020) 476:48–56. doi: 10.1016/j.canlet.2020.02.008
65. Zhang, X, He, B, Chen, E, Lu, J, Wang, J, Cao, H, et al. The aryl hydrocarbon receptor ligand ITE inhibits cell proliferation and migration and enhances sensitivity to drug-resistance in hepatocellular carcinoma. J Cell Physiol. (2021) 236:178–92. doi: 10.1002/jcp.29832
66. Cao, Y, Zhang, F, Wang, H, Bi, C, Cui, J, Liu, F, et al. LncRNA MALAT1 mediates doxorubicin resistance of hepatocellular carcinoma by regulating miR-3129-5p/Nova1 axis. Mol Cell Biochem. (2021) 476:279–92. doi: 10.1007/s11010-020-03904-6
67. Yu, J, Wang, N, Gong, Z, Liu, L, Yang, S, Chen, GG, et al. Cytochrome P450 1A2 overcomes nuclear factor kappa B-mediated sorafenib resistance in hepatocellular carcinoma. Oncogene. (2021) 40:492–507. doi: 10.1038/s41388-020-01545-z
68. Zhao, Z, Zhang, D, Wu, F, Tu, J, Song, J, Xu, M, et al. Sophoridine suppresses lenvatinib-resistant hepatocellular carcinoma growth by inhibiting RAS/MEK/ERK axis via decreasing VEGFR2 expression. J Cell Mol Med. (2021) 25:549–60. doi: 10.1111/jcmm.16108
69. Hsu, WC, Ramesh, S, Shibu, MA, Chen, MC, Wang, TF, Day, CH, et al. Platycodin D reverses histone deacetylase inhibitor resistance in hepatocellular carcinoma cells by repressing ERK1/2-mediated cofilin-1 phosphorylation. Phytomedicine. (2021) 82:153442. doi: 10.1016/j.phymed.2020.153442
70. Zhao, W, Ma, B, Tian, Z, Han, H, Tang, J, Dong, B, et al. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer. (2021) 124:1237–48. doi: 10.1038/s41416-020-01240-6
71. Younis, MA, Khalil, IA, Elewa, YHA, Kon, Y, and Harashima, H. Ultra-small lipid nanoparticles encapsulating sorafenib and midkine-siRNA selectively-eradicate sorafenib-resistant hepatocellular carcinoma in vivo. J Control Release. (2021) 331:335–49. doi: 10.1016/j.jconrel.2021.01.021
72. Miao, J, Meng, C, Wu, H, Shan, W, Wang, H, Ling, C, et al. Novel hybrid CHC from β-carboline and N-Hydroxyacrylamide overcomes drug-resistant hepatocellular carcinoma by promoting apoptosis, DNA damage, and cell cycle arrest. Front Pharmacol. (2020) 11:626065. doi: 10.3389/fphar.2020.626065
73. Weng, H, Zeng, L, Cao, L, Chen, T, Li, Y, Xu, Y, et al. circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Mol Ther Nucleic Acids. (2021) 23:811–20. doi: 10.1016/j.omtn.2020.12.019
74. Sun, T, Mao, W, Peng, H, Wang, Q, and Jiao, L. YAP promotes sorafenib resistance in hepatocellular carcinoma by upregulating survivin. Cell Oncol (Dordr). (2021) 44:689–99. doi: 10.1007/s13402-021-00595-z
75. He, W, Huang, X, Berges, BK, Wang, Y, An, N, Su, R, et al. Artesunate regulates neurite outgrowth inhibitor protein B receptor to overcome resistance to Sorafenib in hepatocellular carcinoma cells. Front Pharmacol. (2021) 12:615889. doi: 10.3389/fphar.2021.615889
76. Lee, ACK, Lau, PM, Kwan, YW, and Kong, SK. Mitochondrial fuel dependence on glutamine drives chemo-resistance in the Cancer stem cells of hepatocellular carcinoma. Int J Mol Sci. (2021) 22:3315. doi: 10.3390/ijms22073315
77. Xia, S, Ji, L, Tao, L, Pan, Y, Lin, Z, Wan, Z, et al. TAK1 is a novel target in hepatocellular carcinoma and contributes to Sorafenib resistance. Cell Mol Gastroenterol Hepatol. (2021) 12:1121–43. doi: 10.1016/j.jcmgh.2021.04.016
78. Yu, T, Yu, J, Lu, L, Zhang, Y, Zhou, Y, Zhou, Y, et al. MT1JP-mediated miR-24-3p/BCL2L2 axis promotes Lenvatinib resistance in hepatocellular carcinoma cells by inhibiting apoptosis. Cell Oncol (Dordr). (2021) 44:821–34. doi: 10.1007/s13402-021-00605-0
79. Xu, J, Ji, L, Ruan, Y, Wan, Z, Lin, Z, Xia, S, et al. UBQLN1 mediates sorafenib resistance through regulating mitochondrial biogenesis and ROS homeostasis by targeting PGC1β in hepatocellular carcinoma. Signal Transduct Target Ther. (2021) 6:190. doi: 10.1038/s41392-021-00594-4
80. Lin, JC, Yang, PM, and Liu, TP. PERK/ATF4-dependent ZFAS1 upregulation is associated with Sorafenib resistance in hepatocellular carcinoma cells. Int J Mol Sci. (2021) 22:5848. doi: 10.3390/ijms22115848
81. Zhu, Y, Xu, J, Hu, W, Wang, F, Zhou, Y, Gong, W, et al. Inhibiting USP8 overcomes hepatocellular carcinoma resistance via suppressing receptor tyrosine kinases. Aging (Albany NY). (2021) 13:14999–5012. doi: 10.18632/aging.203061
82. Gao, L, Morine, Y, Yamada, S, Saito, Y, Ikemoto, T, Tokuda, K, et al. The BAFF/NFκB axis is crucial to interactions between sorafenib-resistant HCC cells and cancer-associated fibroblasts. Cancer Sci. (2021) 112:3545–54. doi: 10.1111/cas.15041
83. Xia, P, Zhang, H, Xu, K, Jiang, X, Gao, M, Wang, G, et al. MYC-targeted WDR4 promotes proliferation, metastasis, and sorafenib resistance by inducing CCNB1 translation in hepatocellular carcinoma. Cell Death Dis. (2021) 12:691. doi: 10.1038/s41419-021-03973-5
84. Gao, R, Buechel, D, Kalathur, RKR, Morini, MF, Coto-Llerena, M, Ercan, C, et al. USP29-mediated HIF1α stabilization is associated with Sorafenib resistance of hepatocellular carcinoma cells by upregulating glycolysis. Oncogenesis. (2021) 10:52. doi: 10.1038/s41389-021-00338-7
85. Du, Z, Mao, Y, Zhang, P, Hu, J, Fu, J, You, Q, et al. TPGS-galactose-modified Polydopamine co-delivery nanoparticles of nitric oxide donor and doxorubicin for targeted chemo-Photothermal therapy against drug-resistant hepatocellular carcinoma. ACS Appl Mater Interfaces. (2021) 13:35518–32. doi: 10.1021/acsami.1c09610
86. Ma, L, Xu, A, Kang, L, Cong, R, Fan, Z, Zhu, X, et al. LSD1-Demethylated LINC01134 confers Oxaliplatin resistance through SP1-induced p62 transcription in HCC. Hepatology. (2021) 74:3213–34. doi: 10.1002/hep.32079
87. Ma, Q, Xu, Q, Zhao, J, Zhang, W, Wang, Q, Fang, J, et al. Coupling HDAC4 with transcriptional factor MEF2D abrogates SPRY4-mediated suppression of ERK activation and elicits hepatocellular carcinoma drug resistance. Cancer Lett. (2021) 520:243–54. doi: 10.1016/j.canlet.2021.07.049
88. Ngo, MT, Peng, SW, Kuo, YC, Lin, CY, Wu, MH, Chuang, CH, et al. A yes-associated protein (YAP) and insulin-like growth factor 1 receptor (IGF-1R) signaling loop is involved in Sorafenib resistance in hepatocellular carcinoma. Cancers (Basel). (2021) 13:3812. doi: 10.3390/cancers13153812
89. Guan, X, Wu, Y, Zhang, S, Liu, Z, Fan, Q, Fang, S, et al. Activation of FcRn mediates a primary resistance response to Sorafenib in hepatocellular carcinoma by single-cell RNA sequencing. Front Pharmacol. (2021) 12:709343. doi: 10.3389/fphar.2021.709343
90. Karabicici, M, Azbazdar, Y, Ozhan, G, Senturk, S, Firtina Karagonlar, Z, and Erdal, E. Changes in Wnt and TGF-β signaling mediate the development of Regorafenib resistance in hepatocellular carcinoma cell line HuH7. Front Cell Dev Biol. (2021) 9:639779. doi: 10.3389/fcell.2021.639779
91. Chung, JY, Chan, MK, Tang, PC, Chan, AS, Chung, JS, Meng, XM, et al. AANG: a natural compound formula for overcoming multidrug resistance via synergistic rebalancing the TGF-β/Smad signalling in hepatocellular carcinoma. J Cell Mol Med. (2021) 25:9805–13. doi: 10.1111/jcmm.16928
92. Wang, JW, Ma, L, Liang, Y, Yang, XJ, Wei, S, Peng, H, et al. RCN1 induces sorafenib resistance and malignancy in hepatocellular carcinoma by activating c-MYC signaling via the IRE1α-XBP1s pathway. Cell Death Discov. (2021) 7:298. doi: 10.1038/s41420-021-00696-6
93. Gao, R, Kalathur, RKR, Coto-Llerena, M, Ercan, C, Buechel, D, Shuang, S, et al. YAP/TAZ and ATF4 drive resistance to Sorafenib in hepatocellular carcinoma by preventing ferroptosis. EMBO Mol Med. (2021) 13:e14351. doi: 10.15252/emmm.202114351
94. Pandit, SK, Sandrini, G, Merulla, J, Nobili, V, Wang, X, Zangari, A, et al. Mitochondrial plasticity promotes resistance to Sorafenib and vulnerability to STAT3 inhibition in human hepatocellular carcinoma. Cancers (Basel). (2021) 13:6029. doi: 10.3390/cancers13236029
95. Zhang, L, Zhou, L, Zhang, H, Zhang, Y, Li, L, Xie, T, et al. Development of a DNA aptamer against multidrug-resistant hepatocellular carcinoma for in vivo imaging. ACS Appl Mater Interfaces. (2021) 13:54656–64. doi: 10.1021/acsami.1c12391
96. Fang, Y, Zhan, Y, Xie, Y, Du, S, Chen, Y, Zeng, Z, et al. Integration of glucose and cardiolipin anabolism confers radiation resistance of HCC. Hepatology. (2022) 75:1386–401. doi: 10.1002/hep.32177
97. Zhou, K, Nguyen, R, Qiao, L, and George, J. Single cell RNA-seq analysis identifies a noncoding RNA mediating resistance to sorafenib treatment in HCC. Mol Cancer. (2022) 21:6. doi: 10.1186/s12943-021-01473-w
98. Lu, Y, Chan, YT, Tan, HY, Zhang, C, Guo, W, Xu, Y, et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J Exp Clin Cancer Res. (2022) 41:3. doi: 10.1186/s13046-021-02208-x
99. Lai, YL, Wang, KH, Hsieh, HP, and Yen, WC. Novel FLT3/AURK multikinase inhibitor is efficacious against sorafenib-refractory and sorafenib-resistant hepatocellular carcinoma. J Biomed Sci. (2022) 29:5. doi: 10.1186/s12929-022-00788-0
100. Chang, YS, Su, CW, Chen, SC, Chen, YY, Liang, YJ, and Wu, JC. Upregulation of USP22 and ABCC1 during Sorafenib treatment of hepatocellular carcinoma contribute to development of resistance. Cells. (2022) 11:634. doi: 10.3390/cells11040634
101. Shao, YY, Chen, PS, Lin, LI, Lee, BS, Ling, A, Cheng, AL, et al. Low miR-10b-3p associated with sorafenib resistance in hepatocellular carcinoma. Br J Cancer. (2022) 126:1806–14. doi: 10.1038/s41416-022-01759-w
102. Li, D, Yao, Y, Rao, Y, Huang, X, Wei, L, You, Z, et al. Cholesterol sensor SCAP contributes to sorafenib resistance by regulating autophagy in hepatocellular carcinoma. J Exp Clin Cancer Res. (2022) 41:116. doi: 10.1186/s13046-022-02306-4
103. Alhamad, DW, Elgendy, SM, Hersi, F, El-Seedi, HR, and Omar, HA. The inhibition of autophagy by spautin boosts the anticancer activity of fingolimod in multidrug-resistant hepatocellular carcinoma. Life Sci. (2022) 304:120699. doi: 10.1016/j.lfs.2022.120699
104. Vishnoi, K, Ke, R, Viswakarma, N, Srivastava, P, Kumar, S, Das, S, et al. Ets1 mediates sorafenib resistance by regulating mitochondrial ROS pathway in hepatocellular carcinoma. Cell Death Dis. (2022) 13:581. doi: 10.1038/s41419-022-05022-1
105. Luo, J, Li, L, Zhu, Z, Chang, B, Deng, F, Wang, D, et al. Fucoidan inhibits EGFR redistribution and potentiates sorafenib to overcome sorafenib-resistant hepatocellular carcinoma. Biomed Pharmacother. (2022) 154:113602. doi: 10.1016/j.biopha.2022.113602
106. Hu, B, Zou, T, Qin, W, Shen, X, Su, Y, Li, J, et al. Inhibition of EGFR overcomes acquired Lenvatinib resistance driven by STAT3-ABCB1 signaling in hepatocellular carcinoma. Cancer Res. (2022) 82:3845–57. doi: 10.1158/0008-5472.CAN-21-4140
107. Lai, Y, Lu, N, Luo, S, Wang, H, and Zhang, P. A Photoactivated Sorafenib-ruthenium(II) prodrug for resistant hepatocellular carcinoma therapy through Ferroptosis and purine metabolism disruption. J Med Chem. (2022) 65:13041–51. doi: 10.1021/acs.jmedchem.2c00880
108. Zhou, X, Luo, J, Xie, H, Wei, Z, Li, T, Liu, J, et al. MCM2 promotes the stemness and sorafenib resistance of hepatocellular carcinoma cells via hippo signaling. Cell Death Discov. (2022) 8:418. doi: 10.1038/s41420-022-01201-3
109. Yuan, J, Lv, T, Yang, J, Wu, Z, Yan, L, Yang, J, et al. HDLBP promotes hepatocellular carcinoma proliferation and Sorafenib resistance by suppressing Trim71-dependent RAF1 degradation. Cell Mol Gastroenterol Hepatol. (2023) 15:307–25. doi: 10.1016/j.jcmgh.2022.10.005
110. Zhang, F, Hu, K, Liu, W, Quan, B, Li, M, Lu, S, et al. Oxaliplatin-resistant hepatocellular carcinoma drives immune evasion through PD-L1 up-regulation and PMN-singular recruitment. Cell Mol Gastroenterol Hepatol. (2023) 15:573–91. doi: 10.1016/j.jcmgh.2022.12.002
111. Deng, J, and Ke, H. Overcoming the resistance of hepatocellular carcinoma to PD-1/PD-L1 inhibitor and the resultant immunosuppression by CD38 siRNA-loaded extracellular vesicles. Onco Targets Ther. (2023) 12:2152635. doi: 10.1080/2162402X.2022.2152635
112. Huang, M, Long, J, Yao, Z, Zhao, Y, Zhao, Y, Liao, J, et al. METTL1-mediated m7G tRNA modification promotes Lenvatinib resistance in hepatocellular carcinoma. Cancer Res. (2023) 83:89–102. doi: 10.1158/0008-5472.CAN-22-0963
113. Yang, Y, Mai, Z, Zhang, Y, Yu, Z, Li, W, Zhang, Y, et al. A Cascade targeted and mitochondrion-dysfunctional nanomedicine capable of overcoming drug resistance in hepatocellular carcinoma. ACS Nano. (2023) 17:1275–86. doi: 10.1021/acsnano.2c09342
114. Guo, XJ, Huang, XY, Yang, X, Lu, JC, Wei, CY, Gao, C, et al. Loss of 5-hydroxymethylcytosine induces chemotherapy resistance in hepatocellular carcinoma via the 5-hmC/PCAF/AKT axis. Cell Death Dis. (2023) 14:79. doi: 10.1038/s41419-022-05406-3
115. Wang, Z, Pan, B, Yao, Y, Qiu, J, Zhang, X, Wu, X, et al. XPO1 intensifies sorafenib resistance by stabilizing acetylation of NPM1 and enhancing epithelial-mesenchymal transition in hepatocellular carcinoma. Biomed Pharmacother. (2023) 160:114402. doi: 10.1016/j.biopha.2023.114402
116. Miyazaki, K, Morine, Y, Xu, C, Nakasu, C, Wada, Y, Teraoku, H, et al. Curcumin-mediated resistance to Lenvatinib via EGFR signaling pathway in hepatocellular carcinoma. Cells. (2023) 12:612. doi: 10.3390/cells12040612
117. Wang, L, Yang, Q, Zhou, Q, Fang, F, Lei, K, Liu, Z, et al. METTL3-m(6)A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. (2023) 559:216122. doi: 10.1016/j.canlet.2023.216122
118. Huang, Y, Kou, Q, Su, Y, Lu, L, Li, X, Jiang, H, et al. Combination therapy based on dual-target biomimetic nano-delivery system for overcoming cisplatin resistance in hepatocellular carcinoma. J Nanobiotechnology. (2023) 21:89. doi: 10.1186/s12951-023-01840-3
119. Eun, JW, Yoon, JH, Ahn, HR, Kim, S, Kim, YB, Lim, SB, et al. Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib. Cancer Commun (Lond). (2023) 43:455–79. doi: 10.1002/cac2.12414
120. Zhou, S, Liu, Y, Zhang, Q, Xu, H, Fang, Y, Chen, X, et al. Human menstrual blood-derived stem cells reverse sorafenib resistance in hepatocellular carcinoma cells through the hyperactivation of mitophagy. Stem Cell Res Ther. (2023) 14:58. doi: 10.1186/s13287-023-03278-8
121. Gu, L, Jin, X, Liang, H, Yang, C, and Zhang, Y. Upregulation of CSNK1A1 induced by ITGB5 confers to hepatocellular carcinoma resistance to sorafenib in vivo by disrupting the EPS15/EGFR complex. Pharmacol Res. (2023) 192:106789. doi: 10.1016/j.phrs.2023.106789
122. Sha, Y, Pan, M, Chen, Y, Qiao, L, Zhou, H, Liu, D, et al. PLEKHG5 is stabilized by HDAC2-related deacetylation and confers sorafenib resistance in hepatocellular carcinoma. Cell Death Discov. (2023) 9:176. doi: 10.1038/s41420-023-01469-z
123. Fang, S, Zheng, L, Chen, X, Guo, X, Ding, Y, Ma, J, et al. MEX3A determines in vivo hepatocellular carcinoma progression and induces resistance to sorafenib in a hippo-dependent way. Hepatol Int. (2023) 17:1500–18. doi: 10.1007/s12072-023-10565-2
124. Zheng, Y, Zhan, Y, Zhang, Y, Zhang, Y, Liu, Y, Xie, Y, et al. Hexokinase 2 confers radio-resistance in hepatocellular carcinoma by promoting autophagy-dependent degradation of AIMP2. Cell Death Dis. (2023) 14:488. doi: 10.1038/s41419-023-06009-2
125. Suk, FM, Wu, CY, Fang, CC, Chen, TL, and Liao, YJ. β-HB treatment reverses sorafenib resistance by shifting glycolysis-lactate metabolism in HCC. Biomed Pharmacother. (2023) 166:115293. doi: 10.1016/j.biopha.2023.115293
126. Sun, J, Liu, Q, Jiang, Y, Cai, Z, Liu, H, and Zuo, H. Engineered small extracellular vesicles loaded with miR-654-5p promote ferroptosis by targeting HSPB1 to alleviate sorafenib resistance in hepatocellular carcinoma. Cell Death Discov. (2023) 9:362. doi: 10.1038/s41420-023-01660-2
127. He, K, An, S, Liu, F, Chen, Y, Xiang, G, and Wang, H. Integrative analysis of multi-omics data reveals inhibition of RB1 signaling promotes apatinib resistance of hepatocellular carcinoma. Int J Biol Sci. (2023) 19:4511–24. doi: 10.7150/ijbs.83862
128. Leung, CON, Yang, Y, Leung, RWH, So, KKH, Guo, HJ, Lei, MML, et al. Broad-spectrum kinome profiling identifies CDK6 upregulation as a driver of lenvatinib resistance in hepatocellular carcinoma. Nat Commun. (2023) 14:6699. doi: 10.1038/s41467-023-42360-w
129. Liu, W, Zhang, F, Quan, B, Yao, F, Chen, R, Ren, Z, et al. NLRP3/IL-1β induced myeloid-derived suppressor cells recruitment and PD-L1 upregulation promotes oxaliplatin resistance of hepatocellular carcinoma. MedComm (2020). (2023) 4:e447. doi: 10.1002/mco2.447
130. Xu, W, Yang, M, Zhang, W, Jia, W, Zhang, H, and Zhang, Y. Tumor microenvironment responsive nano-platform for overcoming sorafenib resistance of hepatocellular carcinoma. Mater Today Bio. (2024) 24:100902. doi: 10.1016/j.mtbio.2023.100902
131. Hsu, TW, Su, YH, Chen, HA, Liao, PH, Shen, SC, Tsai, KY, et al. Galectin-1-mediated MET/AXL signaling enhances sorafenib resistance in hepatocellular carcinoma by escaping ferroptosis. Aging (Albany NY). (2023) 15:6503–25. doi: 10.18632/aging.204867
132. Wang, S, You, X, Liu, X, Fengwei, Z, Zhou, H, Shang, X, et al. SMYD3 induces sorafenib resistance by activating SMAD2/3-mediated epithelial-mesenchymal transition in hepatocellular carcinoma. iScience. (2023) 26:106994. doi: 10.1016/j.isci.2023.106994
133. Kim, Y, Jung, KY, Kim, YH, Xu, P, Kang, BE, Jo, Y, et al. Inhibition of SIRT7 overcomes sorafenib acquired resistance by suppressing ERK1/2 phosphorylation via the DDX3X-mediated NLRP3 inflammasome in hepatocellular carcinoma. Drug Resist Updat. (2024) 73:101054. doi: 10.1016/j.drup.2024.101054
134. Xie, P, Yu, M, Zhang, B, Yu, Q, Zhao, Y, Wu, M, et al. CRKL dictates anti-PD-1 resistance by mediating tumor-associated neutrophil infiltration in hepatocellular carcinoma. J Hepatol. (2024) 81:93–107. doi: 10.1016/j.jhep.2024.02.009
135. Chan, YT, Wu, J, Lu, Y, Li, Q, Feng, Z, Xu, L, et al. Loss of lncRNA LINC01056 leads to sorafenib resistance in HCC. Mol Cancer. (2024) 23:74. doi: 10.1186/s12943-024-01988-y
136. Zhang, QY, Ding, W, Mo, JS, Ou-Yang, SM, Lin, ZY, Peng, KR, et al. Novel STAT3 oligonucleotide compounds suppress tumor growth and overcome the acquired resistance to sorafenib in hepatocellular carcinoma. Acta Pharmacol Sin. (2024). doi: 10.1038/s41401-024-01261-4 [Epub ahead of print].
137. Huang, X, Li, G, Li, H, Zhong, W, Jiang, G, Cai, J, et al. Glycyrrhetinic acid as a hepatocyte targeting ligand-functionalized platinum(IV) complexes for hepatocellular carcinoma therapy and overcoming multidrug resistance. J Med Chem. (2024) 67:8020–42. doi: 10.1021/acs.jmedchem.4c00144
138. Rodrigo, MAM, Michalkova, H, Jimenez, AMJ, Petrlak, F, Do, T, Sivak, L, et al. Metallothionein-3 is a multifunctional driver that modulates the development of sorafenib-resistant phenotype in hepatocellular carcinoma cells. Biomark Res. (2024) 12:38. doi: 10.1186/s40364-024-00584-y
139. Hu, B, Cheng, JW, Hu, JW, Li, H, Ma, XL, Tang, WG, et al. KPNA3 confers Sorafenib resistance to advanced hepatocellular carcinoma via TWIST regulated epithelial-mesenchymal transition. J Cancer. (2019) 10:3914–25. doi: 10.7150/jca.31448
140. Wang, S, Wang, Y, Xun, X, Zhang, C, Xiang, X, Cheng, Q, et al. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J Exp Clin Cancer Res. (2020) 39:22. doi: 10.1186/s13046-020-1523-2
141. Prawira, A, Le, TBU, Vu, TC, and Huynh, H. Ribociclib enhances infigratinib-induced cancer cell differentiation and delays resistance in FGFR-driven hepatocellular carcinoma. Liver Int. (2021) 41:608–20. doi: 10.1111/liv.14728
142. Xu, J, Ji, L, Liang, Y, Wan, Z, Zheng, W, Song, X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. (2020) 5:298. doi: 10.1038/s41392-020-00375-5
143. Liao, Y, Yang, Y, Pan, D, Ding, Y, Zhang, H, Ye, Y, et al. HSP90α mediates Sorafenib resistance in human hepatocellular carcinoma by necroptosis inhibition under hypoxia. Cancers (Basel). (2021) 13:243. doi: 10.3390/cancers13020243
144. Leung, HW, Leung, CON, Lau, EY, Chung, KPS, Mok, EH, Lei, MML, et al. EPHB2 activates β-catenin to enhance Cancer stem cell properties and drive Sorafenib resistance in hepatocellular carcinoma. Cancer Res. (2021) 81:3229–40. doi: 10.1158/0008-5472.CAN-21-0184
145. Prawira, A, Le, TBU, Ho, RZW, and Huynh, H. Upregulation of the ErbB family by EZH2 in hepatocellular carcinoma confers resistance to FGFR inhibitor. J Cancer Res Clin Oncol. (2021) 147:2955–68. doi: 10.1007/s00432-021-03703-6
146. Mok, EHK, Leung, CON, Zhou, L, Lei, MML, Leung, HW, Tong, M, et al. Caspase-3-induced activation of SREBP2 drives drug resistance via promotion of cholesterol biosynthesis in hepatocellular carcinoma. Cancer Res. (2022) 82:3102–15. doi: 10.1158/0008-5472.CAN-21-2934
147. Tao, H, Zhang, Y, Li, J, Liu, J, Yuan, T, Wang, W, et al. Oncogenic lncRNA BBOX1-AS1 promotes PHF8-mediated autophagy and elicits sorafenib resistance in hepatocellular carcinoma. Mol Ther Oncolytics. (2023) 28:88–103. doi: 10.1016/j.omto.2022.12.005
148. Ruan, Y, Chen, T, Zheng, L, Cai, J, Zhao, H, Wang, Y, et al. cDCBLD2 mediates sorafenib resistance in hepatocellular carcinoma by sponging miR-345-5p binding to the TOP2A coding sequence. Int J Biol Sci. (2023) 19:4608–26. doi: 10.7150/ijbs.86227
149. Zhang, X, Su, T, Wu, Y, Cai, Y, Wang, L, Liang, C, et al. N6-Methyladenosine reader YTHDF1 promotes Stemness and therapeutic resistance in hepatocellular carcinoma by enhancing NOTCH1 expression. Cancer Res. (2024) 84:827–40. doi: 10.1158/0008-5472.CAN-23-1916
150. Leung, CON, Tong, M, Chung, KPS, Zhou, L, Che, N, Tang, KH, et al. Overriding adaptive resistance to Sorafenib through combination therapy with Src homology 2 domain-containing phosphatase 2 blockade in hepatocellular carcinoma. Hepatology. (2020) 72:155–68. doi: 10.1002/hep.30989
151. Gramantieri, L, Pollutri, D, Gagliardi, M, Giovannini, C, Quarta, S, Ferracin, M, et al. MiR-30e-3p influences tumor phenotype through MDM2/TP53 Axis and predicts Sorafenib resistance in hepatocellular carcinoma. Cancer Res. (2020) 80:1720–34. doi: 10.1158/0008-5472.CAN-19-0472
152. Zhang, Y, Huang, G, Miao, H, Song, Z, Zhang, X, Fan, W, et al. Apatinib treatment may improve survival outcomes of patients with hepatitis B virus-related sorafenib-resistant hepatocellular carcinoma. Ther Adv Med Oncol. (2020) 12:175883592093742. doi: 10.1177/1758835920937422
153. von Felden, J, Craig, AJ, Garcia-Lezana, T, Labgaa, I, Haber, PK, D'Avola, D, et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene. (2021) 40:140–51. doi: 10.1038/s41388-020-01519-1
154. Wei, X, Zhao, L, Ren, R, Ji, F, Xue, S, Zhang, J, et al. MiR-125b loss activated HIF1α/pAKT loop, leading to Transarterial chemoembolization resistance in hepatocellular carcinoma. Hepatology. (2021) 73:1381–98. doi: 10.1002/hep.31448
155. Huang, G, Li, L, Liang, C, Yu, F, Teng, C, Pang, Y, et al. Upregulated UCA1 contributes to oxaliplatin resistance of hepatocellular carcinoma through inhibition of miR-138-5p and activation of AKT/mTOR signaling pathway. Pharmacol Res Perspect. (2021) 9:e00720. doi: 10.1002/prp2.720
156. Liu, Y, Zhuang, H, Cao, F, Li, J, Guo, Y, Zhang, J, et al. Shc3 promotes hepatocellular carcinoma stemness and drug resistance by interacting with β-catenin to inhibit its ubiquitin degradation pathway. Cell Death Dis. (2021) 12:278. doi: 10.1038/s41419-021-03560-8
157. Zhao, G, Zhang, A, Sun, S, and Ding, Y. Long non-coding RNA LINC00173 enhances cisplatin resistance in hepatocellular carcinoma via the microRNA-641/RAB14 axis. Oncol Lett. (2021) 21:371. doi: 10.3892/ol.2021.12632
158. Liu, QQ, Liu, YW, Xie, YK, Zhang, JH, Song, CX, Wang, JZ, et al. Amplification of DDR2 mediates sorafenib resistance through NF-κB/c-Rel signaling in hepatocellular carcinoma. Cell Biol Int. (2021) 45:1906–16. doi: 10.1002/cbin.11625
159. Jiang, Q, Ma, Y, Han, J, Chu, J, Ma, X, Shen, L, et al. MDM2 binding protein induces the resistance of hepatocellular carcinoma cells to molecular targeting agents via enhancing the transcription factor activity of the Pregnane X receptor. Front Oncol. (2021) 11:715193. doi: 10.3389/fonc.2021.715193
160. Wang, Y, Lu, LC, Guan, Y, Ho, MC, Lu, S, Spahn, J, et al. Atezolizumab plus bevacizumab combination enables an unresectable hepatocellular carcinoma resectable and links immune exclusion and tumor dedifferentiation to acquired resistance. Exp Hematol Oncol. (2021) 10:45. doi: 10.1186/s40164-021-00237-y
161. Lu, JC, Zhang, PF, Huang, XY, Guo, XJ, Gao, C, Zeng, HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. (2021) 14:200. doi: 10.1186/s13045-021-01207-x
162. Liu, S, Bu, X, Kan, A, Luo, L, Xu, Y, Chen, H, et al. SP1-induced lncRNA DUBR promotes stemness and oxaliplatin resistance of hepatocellular carcinoma via E2F1-CIP2A feedback. Cancer Lett. (2022) 528:16–30. doi: 10.1016/j.canlet.2021.12.026
163. Zhou, J, Shao, Q, Lu, Y, Li, Y, Xu, Z, Zhou, B, et al. Monocarboxylate transporter upregulation in induced regulatory T cells promotes resistance to anti-PD-1 therapy in hepatocellular carcinoma patients. Front Oncol. (2022) 12:960066. doi: 10.3389/fonc.2022.960066
164. Wang, HC, Haung, LY, Wang, CJ, Chao, YJ, Hou, YC, Yen, CJ, et al. Tumor-associated macrophages promote resistance of hepatocellular carcinoma cells against sorafenib by activating CXCR2 signaling. J Biomed Sci. (2022) 29:99. doi: 10.1186/s12929-022-00881-4
165. Wei, CY, Zhu, MX, Zhang, PF, Huang, XY, Wan, JK, Yao, XZ, et al. PKCα/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti-PD1 resistance in hepatocellular carcinoma. J Hepatol. (2022) 77:163–76. doi: 10.1016/j.jhep.2022.02.019
166. Tan, Z, Chiu, MS, Yang, X, Yue, M, Cheung, TT, Zhou, D, et al. Isoformic PD-1-mediated immunosuppression underlies resistance to PD-1 blockade in hepatocellular carcinoma patients. Gut. (2023) 72:1568–80. doi: 10.1136/gutjnl-2022-327133
167. Yang, Z, Suda, G, Maehara, O, Ohara, M, Yoda, T, Sasaki, T, et al. Changes in serum growth factors during resistance to Atezolizumab plus bevacizumab treatment in patients with Unresectable hepatocellular carcinoma. Cancers (Basel). (2023) 15:593. doi: 10.3390/cancers15030593
168. Zou, X, Xu, Q, You, R, and Yin, G. Efficacy and safety of TACE combined with Regorafenib plus PD-1 inhibitor in the treatment of hepatocellular carcinoma after Sorafenib resistance. J Hepatocell Carcinoma. (2023) 10:267–79. doi: 10.2147/JHC.S399874
169. Zheng, R, Weng, S, Xu, J, Li, Z, Wang, Y, Aizimuaji, Z, et al. Autophagy and biotransformation affect sorafenib resistance in hepatocellular carcinoma. Comput Struct Biotechnol J. (2023) 21:3564–74. doi: 10.1016/j.csbj.2023.07.005
170. Meng, Y, Ye, F, Nie, P, Zhao, Q, An, L, Wang, W, et al. Immunosuppressive CD10(+)ALPL(+) neutrophils promote resistance to anti-PD-1 therapy in HCC by mediating irreversible exhaustion of T cells. J Hepatol. (2023) 79:1435–49. doi: 10.1016/j.jhep.2023.08.024
171. Zhou, C, Sun, BY, Zhou, PY, Yang, ZF, Wang, ZT, Liu, G, et al. MAIT cells confer resistance to Lenvatinib plus anti-PD1 antibodies in hepatocellular carcinoma through TNF-TNFRSF1B pathway. Clin Immunol. (2023) 256:109770. doi: 10.1016/j.clim.2023.109770
172. Zhang, S, Yuan, L, Danilova, L, Mo, G, Zhu, Q, Deshpande, A, et al. Spatial transcriptomics analysis of neoadjuvant cabozantinib and nivolumab in advanced hepatocellular carcinoma identifies independent mechanisms of resistance and recurrence. Genome Med. (2023) 15:72. doi: 10.1186/s13073-023-01218-y
173. Hao, SH, Ma, XD, Xu, L, Xie, JD, Feng, ZH, Chen, JW, et al. Dual specific phosphatase 4 suppresses ferroptosis and enhances sorafenib resistance in hepatocellular carcinoma. Drug Resist Updat. (2024) 73:101052. doi: 10.1016/j.drup.2024.101052
174. He, M, Liu, Y, Chen, S, Deng, H, Feng, C, Qiao, S, et al. Serum amyloid a promotes glycolysis of neutrophils during PD-1 blockade resistance in hepatocellular carcinoma. Nat Commun. (2024) 15:1754. doi: 10.1038/s41467-024-46118-w
175. Tu, X, Chen, L, Zheng, Y, Mu, C, Zhang, Z, Wang, F, et al. S100A9(+)CD14(+) monocytes contribute to anti-PD-1 immunotherapy resistance in advanced hepatocellular carcinoma by attenuating T cell-mediated antitumor function. J Exp Clin Cancer Res. (2024) 43:72. doi: 10.1186/s13046-024-02985-1
176. Wang, H, La Russa, M, and Qi, LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. (2016) 85:227–64. doi: 10.1146/annurev-biochem-060815-014607
177. Marraffini, LA, and Sontheimer, EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. (2008) 322:1843–5. doi: 10.1126/science.1165771
178. Garneau, JE, Dupuis, M, Villion, M, Romero, DA, Barrangou, R, Boyaval, P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. (2010) 468:67–71. doi: 10.1038/nature09523
179. Gasiunas, G, Barrangou, R, Horvath, P, and Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. (2012) 109:E2579–86. doi: 10.1073/pnas.1208507109
180. Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA, and Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. (2012) 337:816–21. doi: 10.1126/science.1225829
181. Rudin, N, Sugarman, E, and Haber, JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. (1989) 122:519–34. doi: 10.1093/genetics/122.3.519
182. Choulika, A, Perrin, A, Dujon, B, and Nicolas, JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. (1995) 15:1968–73. doi: 10.1128/MCB.15.4.1968
183. Inaba, T, Shapiro, LH, Funabiki, T, Sinclair, AE, Jones, BG, Ashmun, RA, et al. DNA-binding specificity and trans-activating potential of the leukemia-associated E2A-hepatic leukemia factor fusion protein. Mol Cell Biol. (1994) 14:3403–13.
184. Xiang, DM, Sun, W, Zhou, T, Zhang, C, Cheng, Z, Li, SC, et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. (2019) 68:1858–71. doi: 10.1136/gutjnl-2018-317440
185. Wei, L, Lee, D, Law, CT, Zhang, MS, Shen, J, Chin, DW, et al. Genome-wide CRISPR/Cas9 library screening identified PHGDH as a critical driver for Sorafenib resistance in HCC. Nat Commun. (2019) 10:4681. doi: 10.1038/s41467-019-12606-7
186. Sueangoen, N, Tantiwetrueangdet, A, and Panvichian, R. HCC-derived EGFR mutants are functioning, EGF-dependent, and erlotinib-resistant. Cell Biosci. (2020) 10:41. doi: 10.1186/s13578-020-00407-1
187. Sofer, S, Lamkiewicz, K, Armoza Eilat, S, Partouche, S, Marz, M, Moskovits, N, et al. A genome-wide CRISPR activation screen reveals hexokinase 1 as a critical factor in promoting resistance to multi-kinase inhibitors in hepatocellular carcinoma cells. FASEB J. (2022) 36:e22191. doi: 10.1096/fj.202101507RR
188. Huang, S, Ma, Z, Zhou, Q, Wang, A, Gong, Y, Li, Z, et al. Genome-wide CRISPR/Cas9 library screening identified that DUSP4 deficiency induces Lenvatinib resistance in hepatocellular carcinoma. Int J Biol Sci. (2022) 18:4357–71. doi: 10.7150/ijbs.69969
189. Pan, J, Zhang, M, Dong, L, Ji, S, Zhang, J, Zhang, S, et al. Genome-scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma. Autophagy. (2023) 19:1184–98. doi: 10.1080/15548627.2022.2117893
190. Lu, Y, Chan, YT, Wu, J, Feng, Z, Yuan, H, Li, Q, et al. CRISPR/Cas9 screens unravel miR-3689a-3p regulating sorafenib resistance in hepatocellular carcinoma via suppressing CCS/SOD1-dependent mitochondrial oxidative stress. Drug Resist Updat. (2023) 71:101015. doi: 10.1016/j.drup.2023.101015
191. Zhu, AX, Abbas, AR, de Galarreta, MR, Guan, Y, Lu, S, Koeppen, H, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med. (2022) 28:1599–611. doi: 10.1038/s41591-022-01868-2
192. Ruiz de Galarreta, M, Bresnahan, E, Molina-Sánchez, P, Lindblad, KE, Maier, B, Sia, D, et al. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. (2019) 9:1124–41. doi: 10.1158/2159-8290.CD-19-0074
193. Sanchez-Martin, A, Sanchon-Sanchez, P, Romero, MR, Marin, JJG, and Briz, O. Impact of tumor suppressor genes inactivation on the multidrug resistance phenotype of hepatocellular carcinoma cells. Biomed Pharmacother. (2023) 165:115209. doi: 10.1016/j.biopha.2023.115209
Keywords: hepatocellular cell carcinoma, in vitro model, in vivo model, drug resistance, patient-derived xenograft
Citation: Xiang Y, Wu J and Qin H (2024) Advances in hepatocellular carcinoma drug resistance models. Front. Med. 11:1437226. doi: 10.3389/fmed.2024.1437226
Edited by:
Kai Qu, The First Affiliated Hospital of Xi’an Jiaotong University, ChinaReviewed by:
Yu Bai, Jilin Medical University, ChinaHongquan Wang, Peking University Aerospace School of Clinical Medicine, China
Copyright © 2024 Xiang, Wu and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanjiao Qin, cWluaGFuamlhb0BqbHUuZWR1LmNu
 Yien Xiang
Yien Xiang Jun Wu
Jun Wu Hanjiao Qin
Hanjiao Qin