- 1Department of ICU, Ningbo Municipal Hospital of Traditional Chinese Medicine (TCM), Affiliated Hospital of Zhejiang Chinese Medical University, Ningbo, China
- 2WillingMed Technology (Beijing) Co., Ltd, Beijing, China
- 3Department of Nephrology, Ningbo Municipal Hospital of Traditional Chinese Medicine (TCM), Affiliated Hospital of Zhejiang Chinese Medical University, Ningbo, China
Background: Cases of severe pneumonia complicated by empyema due to normal anaerobic flora from the oral cavity are infrequent. Diagnosing anaerobic infections through conventional microbiological test (CMT) is often challenging.
Case presentation: This study describes the case of a 67-year-old man, bedridden long-term, who developed severe pneumonia with empyema caused by multiple anaerobic bacterial infections. The patient was hospitalized with a 5-day history of cough, sputum and fever, accompanied by a 2-day history of dyspnea. Despite CMT, the specific etiology remained elusive. However, metagenomic next-generation sequencing (mNGS) identified various anaerobic bacteria in bronchoalveolar lavage fluid (BALF), blood and pleural effusion. The patient was diagnosed with a polymicrobial infection involving multiple anaerobic bacteria. Following treatment with metronidazole and moxifloxacin, the patient’s pulmonary symptoms improved.
Conclusion: mNGS serves as a valuable adjunctive tool for diagnosting and managing patients whose etiology remains unidentified following CMT.
1 Introduction
Empyema refers to the accumulation of purulent exudate in the pleural cavity due to invasion by pathogenic bacteria, a common complication of pneumonia. In recent years, there has been a global rise in the incidence of pneumonia complicated by empyema. Common causative agents include aerobic bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, and Klebsiella pneumoniae, as well as anaerobic bacteria like Bacteroides and Peptostreptococcus (1, 2). Conversely, normal oral flora rarely lead to pneumonia or empyema and are challenging to identify through conventional microbiological test (CMT). However, isolated cases of severe pneumonia with empyema caused by single or dual oral anaerobic bacteria have been increasing reported (3–5), while infections involving multiple anaerobic species remain uncommon (6). Here, we report a case of pneumonia complicated by empyema in a patient admitted with a 5-day history of cough, sputum production, and fever, alongside 2 days of dyspnea. Metagenomic next generation sequencing (mNGS) confirmed a complex oral anaerobic infection as the etiology. This case underscores the critical role of advanced detection techniques in clinical practice.
2 Case presentation
A 67-year-old male patient was admitted to the hospital on February 26, 2024, presenting with a 5 day history of cough, sputum production, and fever, along with 2 days of dyspnea. Since Feb-22, the patient had developed an unprovoked cough, sputum and fever peaking at 38°C, without other notable symptoms. Administration of antipyretic drugs initially reduced the fever, which subsequently fluctuated between 37.3 and 37.5°C. By Feb-24, the patient began experiencing chest tightness and exacerbated dyspnea, especially at rest. These symptoms worsened by Feb-26, necessitating hospitalization at a local facility, Followed by transfer to our emergency department due to the severity of his condition. On admission, the patient presented with apathy, decreased peripheral blood oxygen level, and required emergency tracheal intubation before transfer to the intensive care unit (ICU). In 2005, the patient suffered a brainstem hemorrhage from a traffic accident, resulting in left limb hemiplegia and 2 years of bed rest. He had been in long-term nursing home care before hospitalization, suggesting nursing-home acquired pneumonia (NHAP). However, no definitive etiological examination results were provided by family members, nor was there information on a history of recurrent aspiration.
Upon admission to our hospital, the patient exhibited a temperature of 37.7°C, heart rate of 111 times/min, respiratory rate was 22 breaths/min, blood pressure was 91/61 mmHg, and oxygen saturation (SpO2%) of 95%. The patient was in a shallow coma, intubated and mechanically ventilated. Physical examination revealed moist rales in the right lung, diminished breath sounds in the left lung, normal heart rhythm, distended abdomen, soft without edema in the limbs, cool, moist skin on the left lower limb, intact left dorsal foot artery, and negative bilateral Babinski’s sign. Laboratory findings included elevated white blood cell (WBC, 18.2 × 109/L), neutrophil percentage (N%, 94.6%), C-reactive protein (CRP, 293.9 mg/L), procalcitonin (PCT, 28.42 ng/mL), and creatinine (155 μmol/L), with significantly decreased albumin (21.1 g/L). There were no evidence of HIV or other immunosuppressive conditions. Abdominal ultrasound revealed dense liver echogenicity, gallbladder polyp, right kidney cyst, and multiple stones. Chest CT showed left pleural effusion, pneumothorax, left lung atelectasis, focal consolidation, and infection in the right lung with a small right-sided pleural effusion (Figures 1A–C). The initial diagnosis was severe pneumonia with empyema, with a SOFA score of 17. Detailed SOFA scores throughout the clinical course were listed in Supplementary Table 1. Upon ICU admission, a closed drainage puncture of the left chest was performed for purulent drainage. Empirically, meropenem (1.0 g, q8h+) and tigecycline (50 mg, q12h, ivgtt) was given for anti-infection treatment, as well as methylprednisolone (40 mg, q12h) for intensive anti-inflammatory therapy and stress relief.
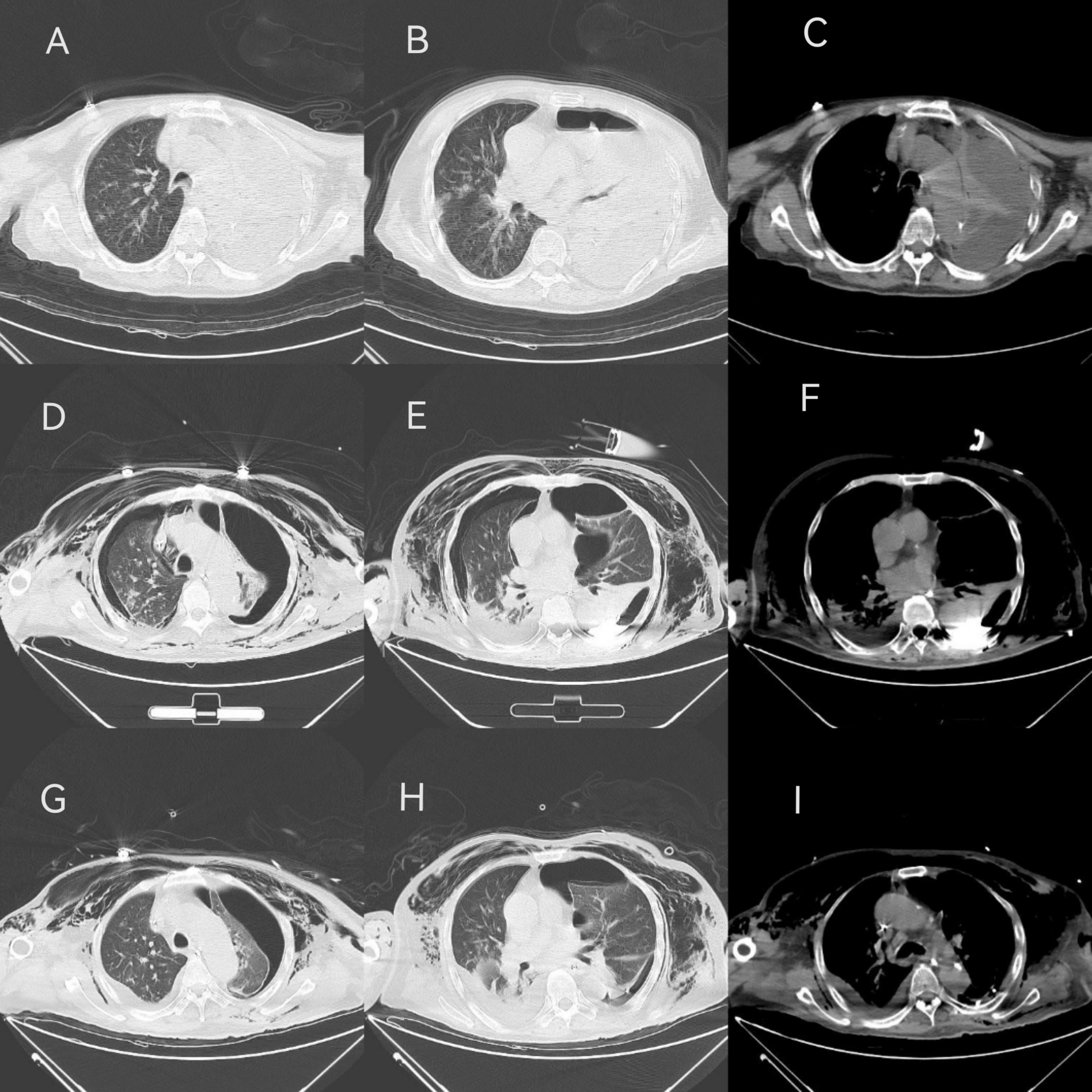
Figure 1. CT findings in different periods. (A–C) On admiration, the patient showed left pleural effusion, air accumulation, left lung atelectasis, local compactness, right lung infection, a small amount of pleural effusion on the right. (D–F) On Mar-01, the CT scan indicated bilateral liquid pneumothorax, local fluid accumulation on the left side showed a high-density shadow, two lungs atelectasis, local consolidation, two lungs infection, left lung consolidation and atelectasis improved. (G–I) On Mar-05, bilateral liquid pneumothorax still can be found, but left attasis slightly improved.
Further etiological tests included antibody and nucleic acid testing for COVID-19, respiratory virus antibody panels (respiratory syncytial virus, adenovirus, parainfluenza virus), influenza A + B virus antigen, routine peripheral venous blood culture, midstream urine culture, thoracic drainage fluid culture, (1, 3)-β-D-glucan (G) and galactomannan (GM) test, all yielding negative results. Sputum and BALF cultures identified Candida albicans, and methicillin-susceptible Staphylococcus aureus, which did not fully correspond with the clinical symptoms.
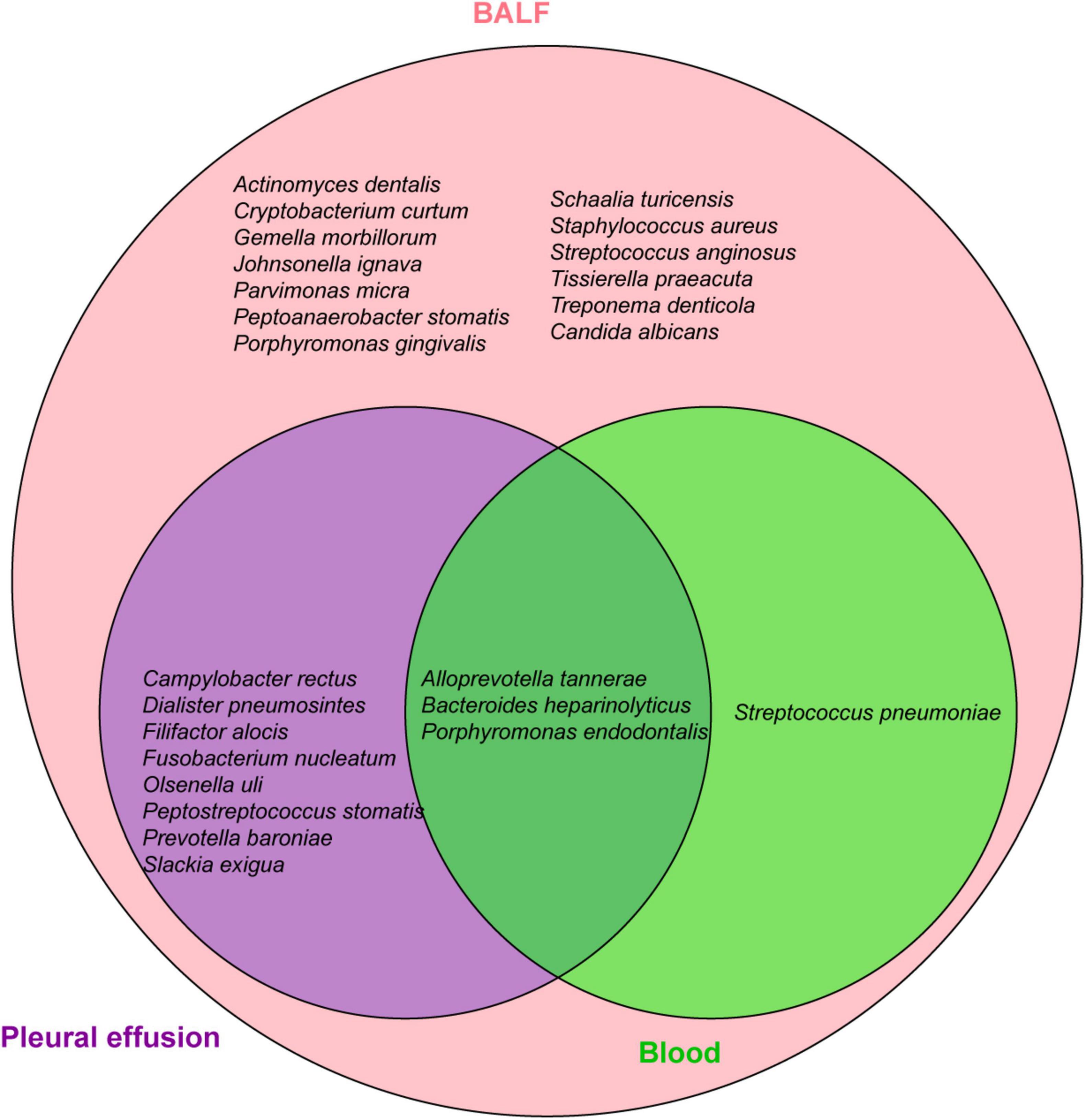
To identify the etiology, mNGS of blood, pleural effusion and bronchoalveolar lavage fluid (BALF) were performed on Feb-28 following family consent. The mNGS procedure followed previous report (7, 8). Both DNA and RNA were extracted from the samples, and libraries were prepared for sequencing using a 50 bp single-end sequencing kit on the MGISEQ-200 platform (MGI Technology). Raw FASTQ-format data underwent Fastq for quality control and evaluation. High-quality sequencing reads were aligned against the human reference genome GRCh37 (hg19) using Bowtie2 v2.4.3 to remove human host sequences. The remaining sequences were then compared against the NCBI GenBank database using Kraken2 v2.1.0 to annotate pathogen genomes and identify pathogens present in the samples. Pathogens were identified based on the specific reads per ten million (RPTM value). For virus detection, an RPTM value ≥ 3 was used as the threshold, while bacteria and fungi required an RPTM ≥ 8 for positive identification. Anaerobic bacteria, primarily oral colonizers, including Parvimomas micra, Peptostreptococcus stomatis, Olsenella uli, and Slackia exigua, were identified on Mar-01 (Figure 2). Relative pathogen abundance is detailed in Supplementary Table 2. Detection of the same pathogen in multiple sites indicates potential disseminated infection, and combined with the highest abundance of the anaerobic bacteria in BALF, pleural fluid, and blood, an anaerobic infection is strongly suspected in this patient. Thus, the antibiotic regimen was adjusted to metronidazole (500 mg, q6h+) and moxifloxacin (0.4 g, qd). Subsequent CT scans demonstrated bilateral pleural effusions, denser left sided effusion, atelectasis in both lungs, focal consolidation, and improved left lung consolidation and atelectasis compared to the previous one (Figures 1D–F).
Following antibiotic adjustment, WBC, CRP, and PCT levels declined significantly, accompanied by gradual reduction in vasopressor requirements, indicating clinical improvement. On Mar-04, following infectious disease expert consultation, meropenem was substituted with imipenem for anaerobic bacteria. By Mar-05, drainage from the left lung was satisfactory, with improved atelectasis compared to previous scans (Figures 1G–I). However, subsequent fever fluctuations and increased WBC and CRP levels prompted further adjustments to the antibiotic regimen: imipenem (1 g, q8h), metronidazole (500 mg, q6h), and subsequently polymyxin B (750000 U, q12h) and aerosolized colistin (250000 U, q12h), due to suspected bloodstream infection. Despite ongoing treatment, family decision led to discharge on Mar-22 with tracheotomy and ventilator support. The patient’s treatment course is detailed in Figure 3.
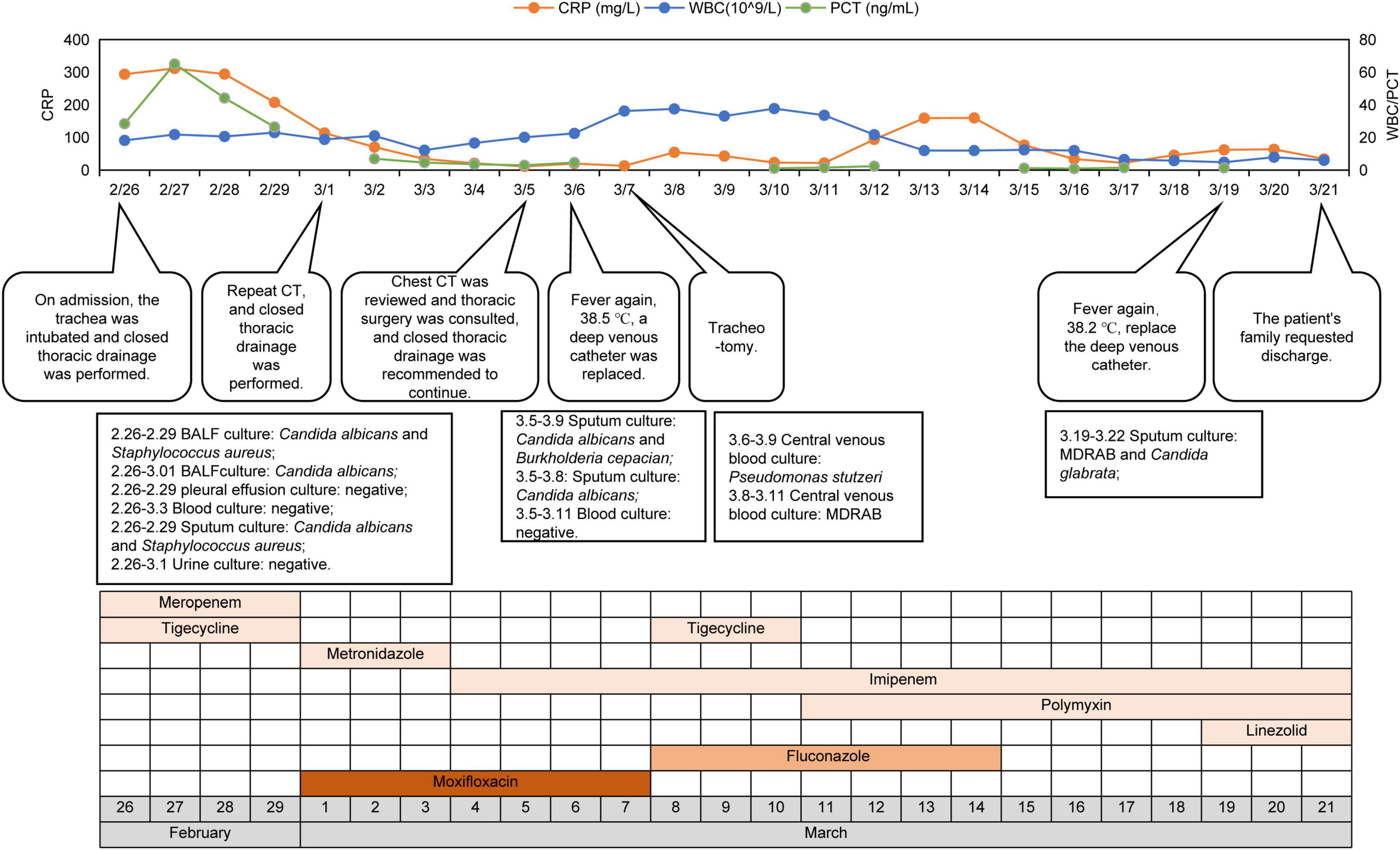
Figure 3. The clinical course of the patient. CRP, C-reactive protein; WBC, white blood cell; MDRAB, multidrug-resistant Acinetobacter baumannii.
3 Discussion
The majority of complex empyema cases arise subsequent to pneumonia. During pathogenesis, pathogens must breach pulmonary defense mechanisms, spread via the respiratory tract and alveoli, and ultimately reach the pleura. The high oxygen tension in the respiratory tract effectively inhibits anaerobic bacterial growth, through it does not entirely prevent it; infection typically occurs through accidental inhalation of oral or gastric contents (9). At present, empyema caused by oral or gastrointestinal anaerobe is common, but cases involving rare or multiple anaerobic infections, or severe infections, are infrequent. Etiological detection and culture of rare anaerobic bacteria pose challenges, often necessitating mNGS or 16s rRNA analysis.
Out study presents a case of severe pneumonia with empyema caused by various oral anaerobes identified by mNGS in a long-term bedridden patient. A PubMed search using keywords “Pleural Empyema” and “anaerobic” yielded clinical case reports of pneumonia and anaerobic thoracic infections (Supplementary Table 3). Most cases involved male smokers, some with documented oral infections, and a few with anatomical anomalies such as pleural fistula, none had significant immunosuppression. Most patients presented solely with empyema, rarely concurrent with pneumothorax. Treatment primarily involved closed chest drainage of pus, with some undergoing thoracoscopic surgery or pleural dissection; mechanical ventilation was seldom required, and prognosis was generally favorable.
The patient in this study had no history of smoking or alcohol abuse, no did he have clear oral infection history or related anatomical abnormalities. The patient suffered from extensive and severe lung infection, predominantly affecting the entire left lung. This led to the accumulation of a significant amount of pleural pus, resulting in left lung atelectasis and compression of the right lung, necessitating endotracheal tube, mechanical ventilation and vasoactive agents to maintain vital signs. The patient also displayed signs of systemic infection, prompting the use of vasoactive medications and endotracheal intubation for ventilator support. Throughout the disease course, the patient developed severe pneumothorax and subcutaneous emphysema. In addition to drainage of pleural fluid and two closed chest drains to relieve pneumothorax symptoms, the patient lung re-expansion was suboptimal, and recurrent fever persisted. The possibility of bloodstream infections was considered, and surgical interventions were deliberated but ultimately withheld due to the patient’s poor overall condition and the family’s wishes. The prognosis was grim, and efforts were made to minimize sedation and analgesia to facilitate awakening before discharge. However, the patient remained unconsciousness and unable to breathe spontaneously off the ventilator.
Anaerobic bacterial infections mainly involve single or dual types of oral or intestinal anaerobes, including Parvimonas micra, Fusobacterium, Staphylococcus, Actinomyces, and Peptostreptococcus (Supplementary Table 3). The anaerobic profile in our patient were complex; mNGS identified distinct bacteria in blood, BALF, and pleural effusion, predominantly oral anaerobes and opportunistic pathogens. Blood reads were minimal, whereas BALF and pleural effusion were dominated by Peptostreptococcus stomatis, Parvimonas micra, Olsenella uli, particularly Slackia exigua with more than 10,000 reads, and P. stomatis with over 50,000 reads in the pleural effusion (Figure 2). Apart from P. micra, implicated in prior anaerobic empyema cases (10, 11), reports on P. stomatis, O. uli, and S. exigua are scarce. P. micra, a component of the normal flora in the oral cavity, and the gastrointestinal tract mucous, skin, and female reproductive tract (3), rarely causes pneumonia or empyema (12), but can lead to bacteremia (13). P. stomatis, widely distributed in the human oral cavity, upper respiratory tract, gastrointestinal tract, female genitourinary system, and skin, is part of the normal flora. No reports of P. stomatis-associated empyema have been reported, although other Peptostreptococcus species have caused empyema with atypical infection symptoms and possible chest pain (14–16). O. uli is a common member of the microbiota associated with primary endodontic infections and can also be isolated from patients with local oral or gastrointestinal infections (17–19), but cases of lung infections are rare. Yan et al. reported the first case of O. uli lung infection (4). The patient had a 30-year history of smoking but had quit 10 years prior. The patient developed a cough with bloody sputum without an obvious cause. CT findings indicated bilateral pneumonia, pyogenic necrosis of the right lower lobe, empyema of the right thoracic capsule, and bilateral emphysema. Treatment with ornidazole combined with ceftazidime resulted in significant improvement in lung lesions and pleural effusion. S. exigua is an obligate anaerobic bacterium associated with dental infections, but rarely causes extraneous infections. However, cases of pulmonary and bloodstream infections have been reported in recent years. Roingeard et al. performed non-invasive ventilation tracheotomy on a 29-year-old man with a history of severe neurological deficit due to Ravine syndrome, who was hospitalized with fever (20). The patient was found to have a large abscess in his left lung, and a localized effusion in the left pectoral muscle. Pulmonary abscess culture confirmed the presence of S. exigua, which responded to treatment with amoxicillin/clavulanic acid. Three cases of S. exigua bacteremia were reported, including one community-acquired bacteremia associated with pleural empyema and a postoperative intraperitoneal abscess in a 73-year-old man with primary intestinal diffuse large B-cell lymphoma (Supplementary Table 3), and one case associated with pyometra (21). These studies indicate that anaerobic bacteria can not only cause lung infections but also lead to bloodstream infections in patients with disease progression.
In this case, the patient had obvious pulmonary symptoms initially. Following the diagnosis of anaerobic bacterial infection, targeted medication led to improvement in the pulmonary symptoms. However, the patient’s condition subsequently deteriorated, and multidrug-resistant Acinetobacter baumannii (MDRAB) was detected in the blood, raising suspicion of bloodstream infection. The discontinuation of mNGS testing prevented accurate determination of whether anaerobic bloodstream infection was complicating the patient’s condition. This case underscores the importance of preventing anaerobic bacteria from spreading beyond the pulmonary infection site when it becomes severe.
In our current clinical practice, microbial diagnosis still heavily relies on conventional bacterial culture (22). However, bacteriological study of lung abscesses encounters significant challenges in sampling and culturing, particularly in avoiding oral contamination and exposure to oxygen. Moreover, bacterial growth conditions in vivo differ markedly from those in laboratory media, make it difficult to replicate the in vivo environment accurately. These factors, coupled with pathogen complexity and empirical antibiotics use, result in a lower sensitivity of conventional culture (23, 24). Consequently, the proportion of culture-negative samples exceeds that in actual situations. Additionally, 16S rRNA sequencing and Matrix-Assisted Laser Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) were also important tools for pathogen identification. While 16S rRNA sequencing rapidly and accurately identifies microorganisms at the genus level, its ability to differentiate between species or subspecies is limited (25). MALDI-TOF MS, a spectroscopic method reliant on a comprehensive database, offers rapid (less than 1 h) identification and high discriminatory power, particularly valuable for characterizing rare bacteria traditionally challenging to identify using routine methods (26). However, the database limitations of MALDI-TOF MS in anaerobic organism identification significantly impact MS sequencing data analysis, particularly in discerning mixed strains. Additionally, MALDI-TOF MS identification relies on bacteria culture, which as mentioned earlier, exhibits lower sensitivity. In our case, routine peripheral blood and pleural effusion culture yielded negative results, while Candida albicans and Staphylococcus aureus detected by sputum and BALF culture failed to explain the patient’s severe empyema. Despite obtaining cultures prior to antibiotic administration, the results were unsatisfactory. In such scenarios, mNGS, free from culture dependence and capable of detecting all nucleic acids without bias, appears optimal for identifying etiology (27). Although mNGS has limitations, such as generating a large number of sequences matching multiple microorganisms, resulting in false negatives and positives, and its high cost (28), its use remain crucial for etiological diagnosis in cases of culture-negative specimens post–antibiotic treatment, suspected uncommon pathogens infections.
In conclusion, pathogen detection in pneumonia and empyema patients infected by anaerobic bacteria remains challenging, even with pre-antibiotic culture samples. When suspicion of rare pathogen infection arises or when clinical etiological test results are negative or incongruent with symptoms, mNGS proves invaluable for timely and accurate pathogen identification, enabling targeted antimicrobial treatment in clinical practice.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Ethics committee of Ningbo Municipal Hospital of T.C.M. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
FY: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review and editing. XZ: Formal analysis, Visualization, Writing – original draft, Writing – review and editing. YL: Writing – original draft, Writing – review and editing. WG: Writing – review and editing. YZ: Writing – review and editing. XC: Project administration, Visualization, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
XZ, WG, and YZ were employed by Willingmed Technology (Beijing) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1435823/full#supplementary-material
References
1. Addala D, Bedawi E, Rahman N. Parapneumonic effusion and empyema. Clin Chest Med. (2021) 42:637–47. doi: 10.1016/j.ccm.2021.08.001
2. Kuryłek A, Stasiak M, Kern-Zdanowicz I. Virulence factors of Streptococcus anginosus - a molecular perspective. Front Microbiol. (2022) 13:1025136. doi: 10.3389/fmicb.2022.1025136
3. Yu Q, Sun L, Xu Z, Fan L, Du Y. Severe pneumonia caused by Parvimonas micra: A case report. BMC Infect Dis. (2021) 21:364. doi: 10.1186/s12879-021-06058-y
4. Yan Y, Li H, Li S, Liu S, Jia N, Liu Y, et al. Olsenella uli-induced pneumonia: A case report. Ann Clin Microbiol Antimicrob. (2022) 21:9. doi: 10.1186/s12941-022-00499-2
5. Duan Y, Feng W, Shen Y, Li Y, Li N, Chen X, et al. Severe pneumonia with empyema caused by Parvimonas micra and Streptococcus constellatus co-infection: A case report. J Int Med Res. (2023) 51:3000605231210657. doi: 10.1177/03000605231210657
6. Duport P, Miltgen G, Kebbabi C, Belmonte O, Coolen-Allou N, Allyn J, et al. First case of pleural empyema and pulmonary abscess caused by Eggerthia catenaformis. Anaerobe. (2018) 50:9–11. doi: 10.1016/j.anaerobe.2018.01.006
7. Chen H, Zheng Y, Zhang X, Liu S, Yin Y, Guo Y, et al. Clinical Evaluation of cell-free and cellular metagenomic next-generation sequencing of infected body fluids. J Adv Res. (2023) 55:119–29. doi: 10.1016/j.jare.2023.02.018
8. Wu C, Yu X, Gai W, Liu Y, Qi Y, Zheng Y, et al. Diagnostic value of plasma and blood cells metagenomic next-generation sequencing in patients with sepsis. Biochem Biophys Res Commun. (2023) 683:149079. doi: 10.1016/j.bbrc.2023.10.011
9. Allewelt M. Aspiration pneumonia and primary lung abscess: Diagnosis and therapy of an aerobic or an anaerobic infection? Expert Rev Respir Med. (2007) 1:111–9. doi: 10.1586/17476348.1.1.111
10. Gumbs S, Kwentoh I, Atiku E, Gikunda W, Safavi A. Parvimonas micra: A rare cause of pleural empyema with Covid-19 Co-infection. Cureus. (2024) 16:e51998. doi: 10.7759/cureus.51998
11. Vilcarromero S, Small M, Lizarzaburu A, Rivadeneyra-Rodriguez A. Pleural empyema by Parvimonas micra in an immunocompetent patient: A case report. Rev Peru Med Exp Salud Publica. (2023) 40:99–104. doi: 10.17843/rpmesp.2023.401.11956
12. Shimizu K, Horinishi Y, Sano C, Ohta R. Infection route of Parvimonas micra: A case report and systematic review. Healthcare (Basel). (2022) 10:1727. doi: 10.3390/healthcare10091727
13. Yamada K, Taniguchi J, Kubota N, Kawai T, Idemitsu R, Inoshima N, et al. Empyema and bacteremia caused by Parvimonas micra: A case report. Respir Med Case Rep. (2023) 45:101892. doi: 10.1016/j.rmcr.2023.101892
14. Gülmez D, Alp S, Topeli Iskit A, Akova M, Hasçelik G. [Pneumonia caused by Fusobacterium necrophorum: Is lemierre syndrome still current?]. Mikrobiyol Bul. (2011) 45:729–34.
15. Kikuchi N, Nomura A, Endo T, Sekizawa K. Anaerobic bacterial empyema accompanying intrathoracic gas formation in anorexia nervosa. Int J Eat Disord. (2006) 39:621–3. doi: 10.1002/eat.20275
16. Martinez K, Mangat G, Sherwani N, Glover D, Silver M. Veillonella intrapulmonary abscess with empyema. Cureus. (2023) 15:e45210. doi: 10.7759/cureus.45210
17. Dewhirst F, Paster B, Tzellas N, Coleman B, Downes J, Spratt D, et al. Characterization of novel human oral isolates and cloned 16s RDNA sequences that fall in the family coriobacteriaceae: Description of Olsenella Gen. Nov., Reclassification of Lactobacillus uli as Olsenella uli Comb. Nov. and description of Olsenella profusa Sp. Nov. Int J Syst Evol Microbiol. (2001) 51:1797–804. doi: 10.1099/00207713-51-5-1797
18. Lau S, Woo P, Fung A, Chan K, Woo G, Yuen K. Anaerobic, non-sporulating, gram-positive bacilli bacteraemia characterized by 16s RRNA gene sequencing. J Med Microbiol. (2004) 53:1247–53. doi: 10.1099/jmm.0.45803-0
19. Bahrani-Mougeot F, Paster B, Coleman S, Ashar J, Barbuto S, Lockhart P. Diverse and novel oral bacterial species in blood following dental procedures. J Clin Microbiol. (2008) 46:2129–32. doi: 10.1128/jcm.02004-07
20. Roingeard C, Jaubert J, Guilleminault L. A large and unusual lung abscess with positive culture to Slackia exigua. Int J Infect Dis. (2015) 40:37–8. doi: 10.1016/j.ijid.2015.09.015
21. Lim K, Son J, Moon SY. A case of Slackia exigua bacteremia associated with pyometra in a patient with poor dentition. Anaerobe. (2022) 73:102477. doi: 10.1016/j.anaerobe.2021.102477
22. Lasken R, McLean J. Recent advances in genomic DNA sequencing of microbial species from single cells. Nat Rev Genet. (2014) 15:577–84. doi: 10.1038/nrg3785
23. Saglani S, Harris K, Wallis C, Hartley J. Empyema: The use of broad range 16s RDNA PCR for pathogen detection. Arch Dis Child. (2005) 90:70–3. doi: 10.1136/adc.2003.042176
24. Le Monnier A, Carbonnelle E, Zahar J, Le Bourgeois M, Abachin E, Quesne G, et al. Microbiological diagnosis of empyema in children: Comparative evaluations by culture, polymerase chain reaction, and pneumococcal antigen detection in pleural fluids. Clin Infect Dis. (2006) 42:1135–40. doi: 10.1086/502680
25. Church D, Cerutti L, Gürtler A, Griener T, Zelazny A, Emler S. Performance and application of 16s RRNA gene cycle sequencing for routine identification of bacteria in the clinical microbiology laboratory. Clin Microbiol Rev. (2020) 33:e53–19. doi: 10.1128/cmr.00053-19
26. Fernández Vecilla D, Roche Matheus M, Calvo Muro F, Iglesias Hidalgo G, Díaz de Tuesta Del Arco JL. Identification of curved gram-negative rods by maldi-tof mass spectrometer in a patient with fournier’S gangrene. A bacteremia caused by desulfovibrio desulfuricans and Escherichia Coli. Rev Esp Quimioter. (2023) 36:629–31. doi: 10.37201/req/026.2023
27. Duan H, Li X, Mei A, Li P, Liu Y, Li X, et al. The diagnostic value of metagenomic next-generation sequencing in infectious diseases. BMC Infect Dis. (2021) 21:62. doi: 10.1186/s12879-020-05746-5
Keywords: severe pneumonia, empyema, anaerobic, mixed infection, metagenomic next generation sequencing
Citation: Yu F, Zhang X, Li Y, Gai W, Zheng Y and Cai X (2024) Severe pneumonia with empyema due to multiple anaerobic infections: case report and literature review. Front. Med. 11:1435823. doi: 10.3389/fmed.2024.1435823
Received: 24 May 2024; Accepted: 29 July 2024;
Published: 14 August 2024.
Edited by:
George Grant, University of Aberdeen (Retired) / now Independent Researcher, Aberdeen, United KingdomReviewed by:
Katerina Tsilipounidaki, University of Thessaly, GreeceDharmendra Kumar Soni, Uniformed Services University of the Health Sciences, United States
Domingo Fernández Vecilla, Osakidetza Basque Health Service, Spain
Haopu Yang, University of Pittsburgh, United States
Takuichi Sato, Niigata University, Japan
Copyright © 2024 Yu, Zhang, Li, Gai, Zheng and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xudong Cai, MTM3NzcxMjQ4NzdAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Fangyu Yu
Fangyu Yu Xiaojing Zhang
Xiaojing Zhang Yujiao Li1
Yujiao Li1 Yafeng Zheng
Yafeng Zheng