- 1Department of Dermatology, West China Hospital, Sichuan University, Chengdu, China
- 2Laboratory of Dermatology, Clinical Institute of Inflammation and Immunology, Frontiers Science Center for Disease-related Molecular Network, West China Hospital, Sichuan University, Chengdu, China
- 3Med-X Center for Informatics, Sichuan University, Chengdu, China
- 4Research Institute of Tissue Engineering and Stem Cells, Nanchong, China
- 5Department of Burns and Plastic Surgery, West China Hospital, Sichuan University, Chengdu, China
Background: Pathological scars, including keloids and hypertrophic scars, represent a significant dermatological challenge, and emerging evidence suggests a potential role for the gut microbiota in this process.
Methods: Utilizing a two-sample Mendelian randomization (MR) methodology, this study meticulously analyzed data from genome-wide association studies (GWASs) relevant to the gut microbiota, keloids, and hypertrophic scars. The integrity and reliability of the results were rigorously evaluated through sensitivity, heterogeneity, pleiotropy, and directionality analyses.
Results: By employing inverse variance weighted (IVW) method, our findings revealed a causal influence of five bacterial taxa on keloid formation: class Melainabacteria, class Negativicutes, order Selenomonadales, family XIII, and genus Coprococcus2. Seven gut microbiota have been identified as having causal relationships with hypertrophic scars: class Alphaproteobacteria, family Clostridiaceae1, family Desulfovibrionaceae, genus Eubacterium coprostanoligenes group, genus Eubacterium fissicatena group, genus Erysipelotrichaceae UCG003 and genus Subdoligranulum. Additional sensitivity analyses further validated the robustness of the associations above.
Conclusion: Overall, our MR analysis supports the hypothesis that gut microbiota is causally linked to pathological scar formation, providing pivotal insights for future mechanistic and clinical research in this domain.
Introduction
Wound healing is a multifaceted and dynamic process, governed by a balance of numerous regulatory pathways. A deviation from this balance can lead to pathological scars, chiefly hypertrophic scars and keloids, characterized by excessive extracellular matrix deposition and abnormal fibroblast behavior (1, 2). The profound impact of pathological scars extends beyond their physical appearance, encompassing cosmetic concerns, functional impairments such as contractures, and subjective symptoms including pruritus and pain (3). Collectively, these aspects highlight the far-reaching effects of pathological scars, with significantly influencing various facets of patient well-being (3).
The intricate mechanisms underlying the formation of these scars involve both local and systemic factors, including genetic predispositions (4). Recent advancements in dermatological research have highlighted the potential influence of the gut microbiota on skin health, particularly in the context of inflammatory disorders (5). This has led to the exploration of the “gut-skin axis,” a concept describing the complex interactions between the gut microbiota and skin health (5). Alterations in the gut microbiota have been linked to skin inflammation and the manifestation of various skin diseases, including rosacea, psoriasis, and atopic dermatitis (6–8). The gut-skin axis involves complex interactions where the gut microbiome impacts skin health through immune modulation, metabolic functions, and hormonal pathways. Gut bacteria influence immune responses, producing anti-inflammatory mediators that reduce skin inflammation (9). They also metabolize dietary components into bioactive metabolites like short-chain fatty acids, enhancing skin barrier function and hydration (10). Additionally, the gut microbiome affects nutrient absorption, essential for skin health (5). Hormonal pathways are influenced as gut bacteria modulate hormones such as cortisol and serotonin, impacting conditions like acne and psoriasis (11). Therapeutic approaches include probiotics and dietary modifications to restore gut microbiome balance, offering potential for improved skin health (12). Notably, a recent study revealed distinct differences in the gut microbiota composition between individuals with normal scars and those with pathological scars (13). This emerging evidence suggests that the gut microbiota may also play a role in the development of pathological scars, adding a new dimension to our understanding of skin disorders. A pivotal question that arises from these observations is whether the relationship between gut microbiota imbalance and the onset or exacerbation of pathological scars is merely observational or indicative of a direct causal influence. Addressing this question is challenging due to the multifaceted nature of gut microbiota, which is influenced by a range of factors such as age, sex, body mass index, and even environmental factors. These variables not only affect the composition of the gut microbiota but also complicate the task of assembling a diverse and representative sample for research. Furthermore, ethical considerations and practical limitations pose significant hurdles in conducting clinical trials to investigate the causal links between the gut microbiota alterations and pathological scar development.
To address these challenges, our study employs a two-sample Mendelian randomization (MR) approach. MR utilizes genetic variants as instrumental variables to establish causal relationships between exposures (such as variations in the gut microbiota) and outcomes (such as pathological scars) (14). This method offers a strategic advantage over traditional randomized controlled trials (RCTs) and observational studies, as it reduces the confounding biases and ethical concerns associated with direct manipulation of the gut microbiota in human subjects (15). By leveraging this approach, our study aims to dissect the potential causal relationship between gut microbiota and the development of pathological scars. The findings from this research could significantly enhance our understanding of the gut-skin axis and its implications for skin health, potentially leading to novel therapeutic strategies for managing pathological scars.
Methods
Study design
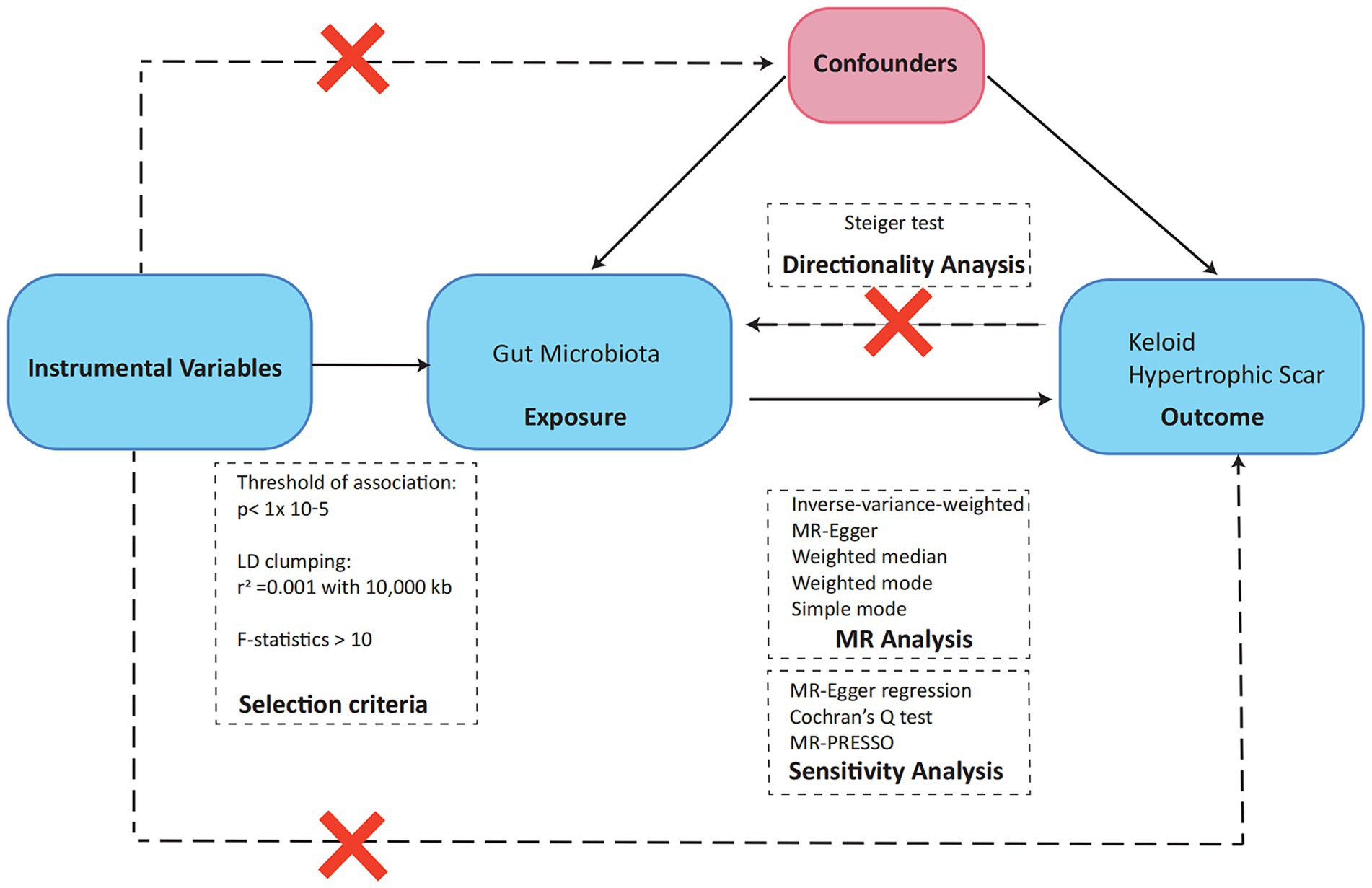
Our study’s analytical framework, depicted in Figure 1, employs a two-sample MR design to investigate the potential causal relationships between gut microbiota and pathological scar formation. This approach is underpinned by three fundamental hypotheses, each aligning with core MR principles and collectively crucial in elucidating the connection between the gut microbiota and pathological scars. The initial hypothesis centers on the representativeness of the genetic instruments utilized in our MR analysis. These instruments must accurately reflect the exposure of interest, which in this case is the gut microbiota. This is essential for reliably measuring the impact of gut microbiota variations on the development of pathological scars. Our second hypothesis pertains to the independence of these genetic instruments from confounding factors. This aspect is critical for mitigating the risk of spurious associations, thereby bolstering the validity of our causal inferences. By ensuring this independence, we strengthened the credibility of our findings in linking gut microbiota alterations to pathological scars. Finally, the third hypothesis, known as the exclusion restriction hypothesis, plays a pivotal role in establishing a direct causal pathway between changes in gut microbiota and the manifestation of pathological scars. This hypothesis is integral to reinforcing the causal interpretation derived from our MR analysis, as it asserts that the observed associations are not influenced by external factors unrelated to the gut microbiota-pathological scar nexus (16).
Data source
In this study, genetic variants for the gut microbiota were derived from an extensive genome-wide association study (GWAS) meta-analysis (17). This analysis included a predominantly European cohort of 18,340 participants across 24 separate cohorts. Post-imputation, the analysis covered 5,717,754 SNPs. In terms of the gut microbiota, the original study categorized it into 257 taxa across six taxonomic levels: phylum [p], class [c], order [o], family [f], and genus [g]. For our microbial quantitative trait locus (mbQTL) mapping analysis, this number was reduced due to quality control and filtering, excluding taxa with low prevalence or abundance. Out of the remaining 211 taxa, we excluded 15 unknown taxa and ultimately included 196 taxa. This selection comprised 9 phyla, 16 classes, 20 orders, 32 families, and 119 genera.
Additionally, GWAS summary statistics for hypertrophic scars were obtained from the latest R10 release data of the FinnGen consortium. This dataset comprises 1,641 documented hypertrophic scar cases, juxtaposed with a control cohort of 385,559 individuals. The GWAS summary statistics for keloids were extracted from a meta-analysis conducted by Sakaue et al. (18). The present study included 668 documented keloid cases with 481,244 control cases and identified a total of 24,197,210 SNPs to explore their potential associations sdwith keloids. The utilization of these diverse and comprehensive sources of data will undoubtedly facilitate a meticulous and thorough investigation into delineating the potential causal relationships that exist between the gut microbiota and pathological scars. No obvious overlap between exposures and outcomes.
Instrumental variable selection
To identify potential instrumental variables (IVs) for our study, we first concentrated on single nucleotide polymorphisms (SNPs) associated with gut bacterial taxa, adhering to the genome-wide significance benchmark of p < 1.0 × 10–5. The selection of appropriate IVs hinged on meeting stringent quality control standards, which included establishing a linkage disequilibrium (LD) threshold for clumping at r2 < 0.001 within a 10,000 kb window, essential for minimizing LD impact and ensuring SNP independence. Additionally, we aligned the effect estimates of exposure and outcome variants, discarded SNPs with incompatible alleles or palindromic characteristics, and selected only SNPs present for all evaluated traits as IVs, avoiding the use of proxies for absent traits. Furthermore, we conducted comprehensive examinations of each SNP via PhenoScanner V21 to determine whether the SNPs influenced outcomes exclusively through their exposure. Finally, we evaluated the robustness of our chosen instruments by calculating the F statistic using the formula: F = (β/SE)2, where β represents the effect size and SE the standard error, adhering to an F > 10 criterion to mitigate bias toward weak IVs (19).
MR analysis
Our foundational analysis incorporated a comprehensive array of MR techniques to establish causal effects. Central to this was the inverse-variance weighted (IVW) method, which formed the core of our analysis, and was augmented by additional methods, including the simple model, weighted model, weighted median, and MR-Egger methods. These factors collectively enabled a thorough evaluation of causal relationships. To address the potential bias due to pleiotropy, we focused on the intercept term of MR-Egger regression. A near-zero intercept term suggests the absence of horizontal pleiotropy in our bidirectional MR approach, a crucial aspect in determining the validity of the SNP under investigation (20). To further investigate horizontal pleiotropy, where a single genetic variant may affect multiple traits and complicate causal inference, we employed the MR-PRESSO global test (21). We also meticulously assessed heterogeneity within the IVW method using Cochran’s Q statistics and funnel plots, tools that illuminate the consistency and reliability of our results. Given the extensive scope of hypothesis testing, the Benjamini-Hochberg (BH) procedure was rigorously applied to adjust for multiple comparisons in our analysis. The results with a false discovery rate p value (PFDR) below 0.1 were considered significant, setting a high bar was used to indicate statistical significance. However, outcomes with a p value less than 0.05 but a PFDR above 0.1, while not meeting this stringent criterion, were still noted for their nominal significance, indicating potential emerging trends meriting further investigation. Finally, the MR Steiger test was employed to rigorously determine the direction of causality between the exposure and the outcome, offering essential insight into the causative pathways involved in our study. All analyses were performed using the package “Two-Sample-MR” (version 0.5.6), “MR-PRESSO” (version 1.0) in R (version 4.3.0).
Results
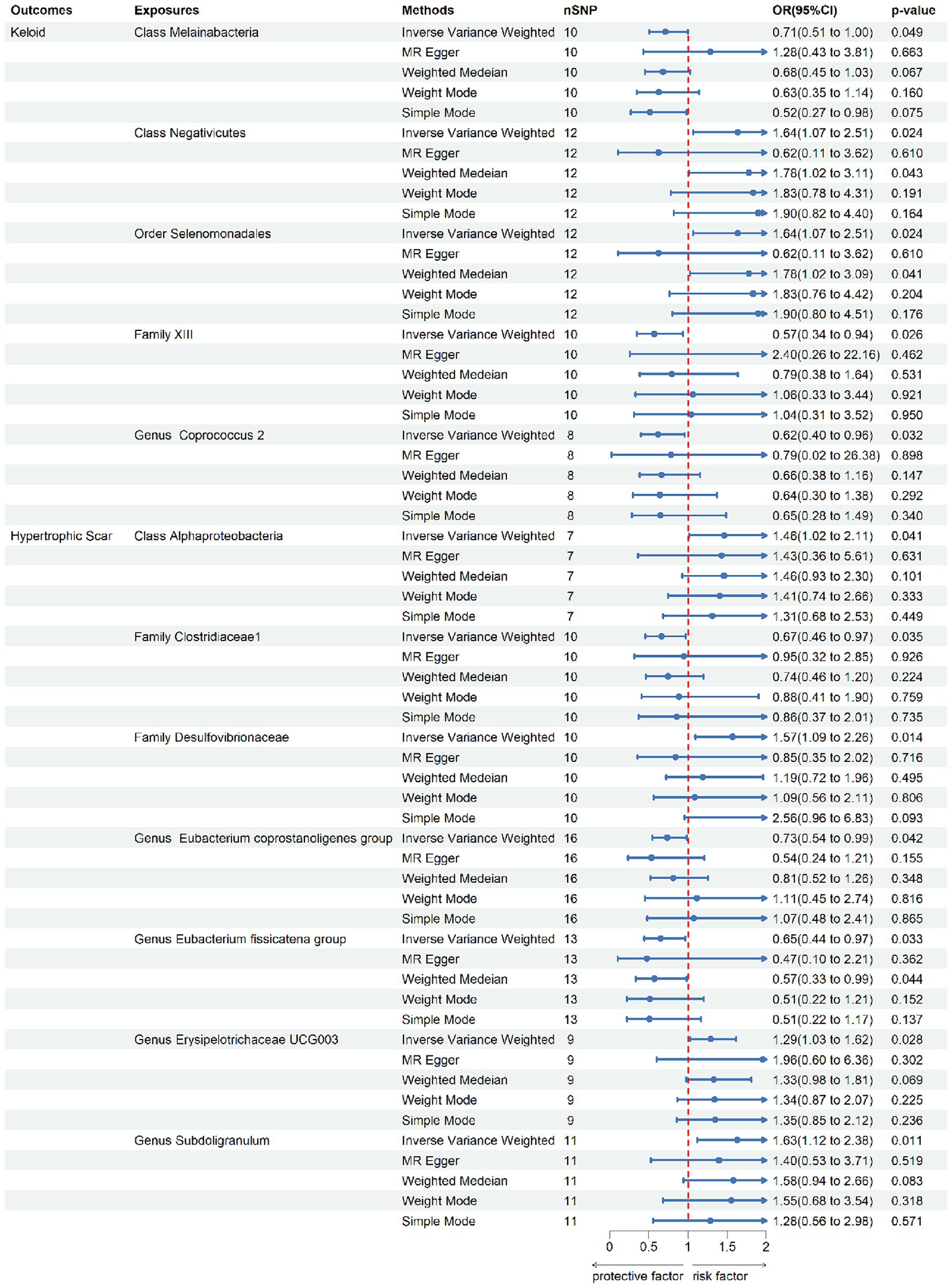
According to the IV selection, a total of 2078 SNPs were used as IVs for SNPs associated with the gut microbiota. We explored the association between the gut microbiota and pathological scars using five MR methods (Figure 2; Supplementary Tables S1, S2). All the F-statistics of the IVs were greater than 10, which indicated that weak instrument bias was unlikely. The IVW methods highlighted five bacterial genera: class Melainabacteria (OR = 0.71, 95% CI: 0.51–1.00, p = 0.046), class Negativicutes (OR = 1.64, 95% CI: 1.07–2.51, p = 0.024), order Selenomonadales (OR = 1.64, 95% CI: 1.07–2.51, p = 0.024), family XIII (OR = 0.57, 95% CI: 0.35–0.94, p = 0.026) and genus Coprococcus2 (OR = 0.62, 95% CI: 0.40–0.96, p = 0.032) (Figure 3). These factors play a protective role by preventing the onset of keloids. Additionally, our examination revealed seven gut microbiota taxa exerting significant causal effects on hypertrophic scars, and a range of taxa from broad phyla to specific genera was identified: class Alphaproteobacteria (OR = 1.46, 95% CI: 1.02–2.11, p = 0.041), family Clostridiaceae1 (OR = 0.67, 95% CI: 0.46–0.97, p = 0.035), family Desulfovibrionaceae (OR = 1.57, 95% CI: 1.09–2.26, p = 0.014), genus Eubacterium coprostanoligenes group (OR = 0.73, 95% CI: 0.55–0.99, p = 0.042), genus Eubacterium fissicatena group (OR = 0.65, 95% CI: 0.44–0.97, p = 0.033), genus Erysipelotrichaceae UCG003 (OR = 1.29, 95% CI: 1.03–1.62, p = 0.028) and genus Subdoligranulum (OR = 1.64, 95%CI: 1.07–2.51, p = 0.024) (Figure 3). The above results are depicted by scatter plots in Supplementary Figure S1 and illustrated through forest plots for causal effects of gut microbiota on keloids and hypertrophic scars with individual SNPs in Supplementary Figure S2. Despite the observed associations, no MR outcomes met the FDR correction threshold for multiple testing (QFDR < 0.1). Nevertheless, with p-values < 0.05, these results hold nominally significant (Supplementary Table S1).
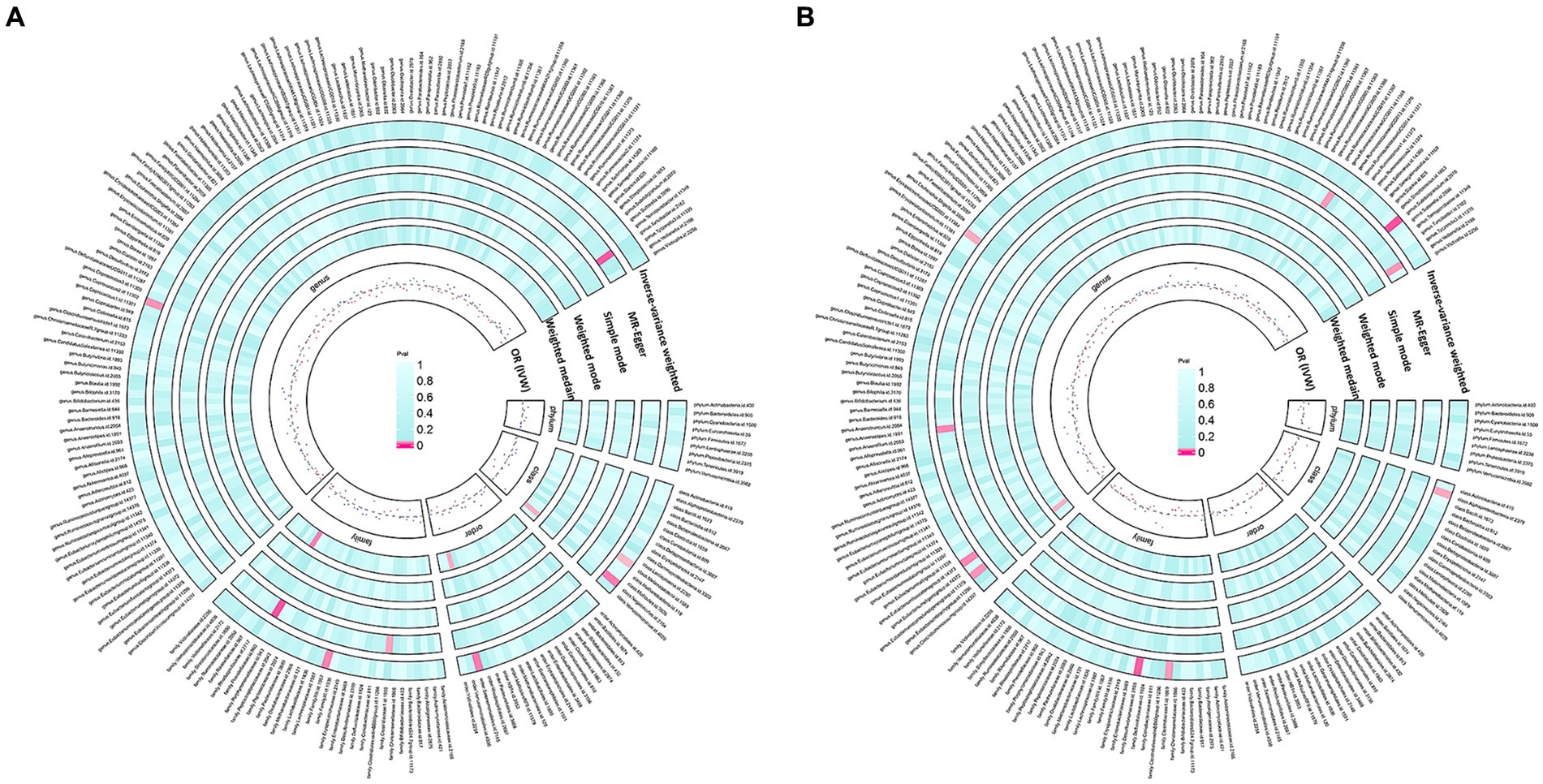
Figure 2. Preliminary MR estimates of gut microbiota’s association with keloids (A) and hypertrophic scars (B).
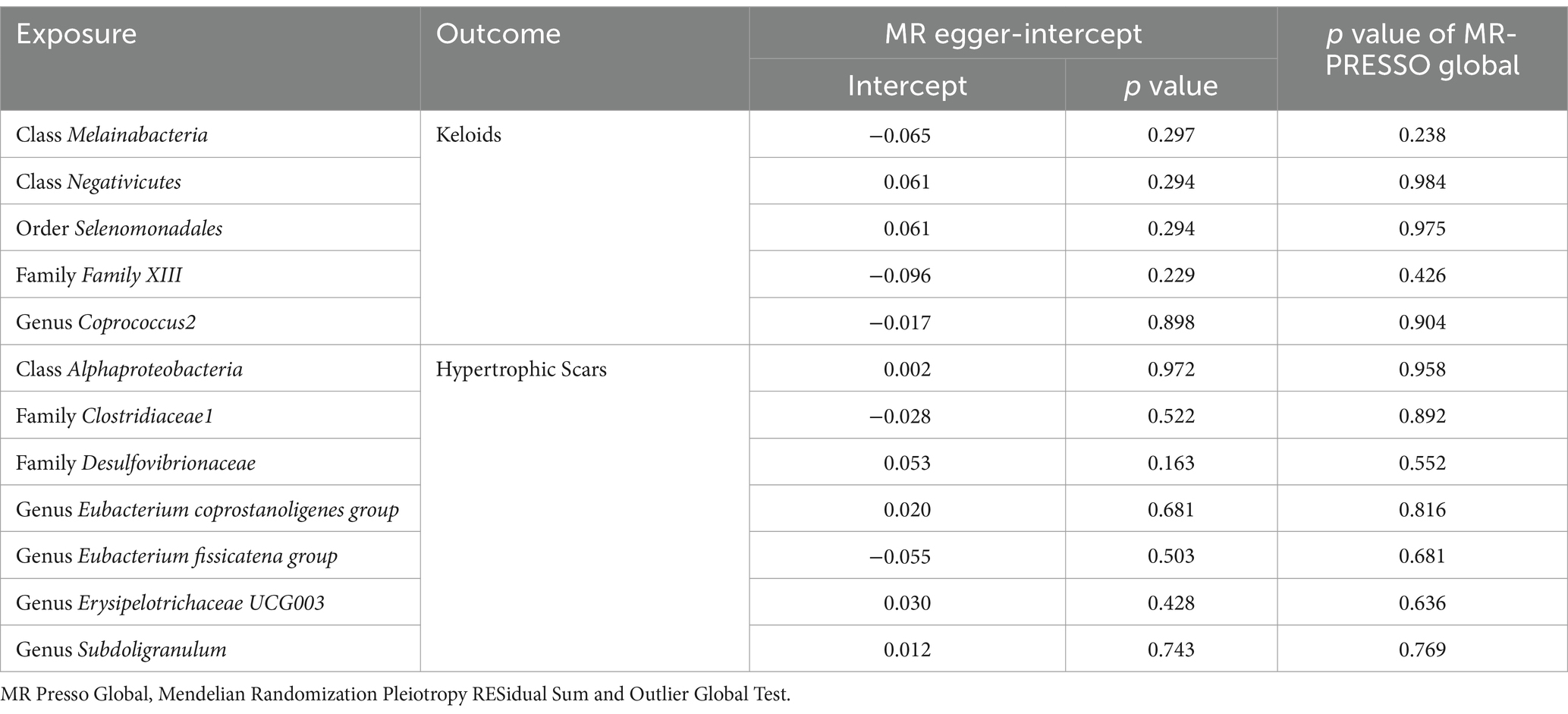
In our rigorous analysis of data homogeneity, we utilized Cochrane’s Q test and observed no significant heterogeneity among the selected single nucleotide polymorphisms (SNPs), with p-values exceeding 0.05, as detailed in Table 1. This lack of heterogeneity underscores the consistency of the SNP selection for our study. Additionally, we conducted comprehensive pleiotropy assessments using both the MR Egger test and the MR-PRESSO analysis. Pleiotropy occurs when a genetic variant influences multiple traits, which can bias the results of a Mendelian Randomization study. The MR Egger intercept provides evidence of directional pleiotropy if the intercept significantly deviates from zero. MR-PRESSO identifies and corrects for outliers that may bias the MR estimates. These tests consistently indicated the absence of pleiotropy, with all p values again exceeding the 0.05 threshold (Table 2). Crucially, our study employed MR Steiger directionality tests, which uniformly indicated a strong causal relationship running from the gut microbiota to pathological scars across all evaluated outcomes. This finding, detailed in Supplementary Table S3, solidifies the direction of the causal pathway in our research. This consistent demonstration of a causal direction not only reinforces the robustness of our findings but also highlights the potential impact of the gut microbiota on the formation of pathological scars.
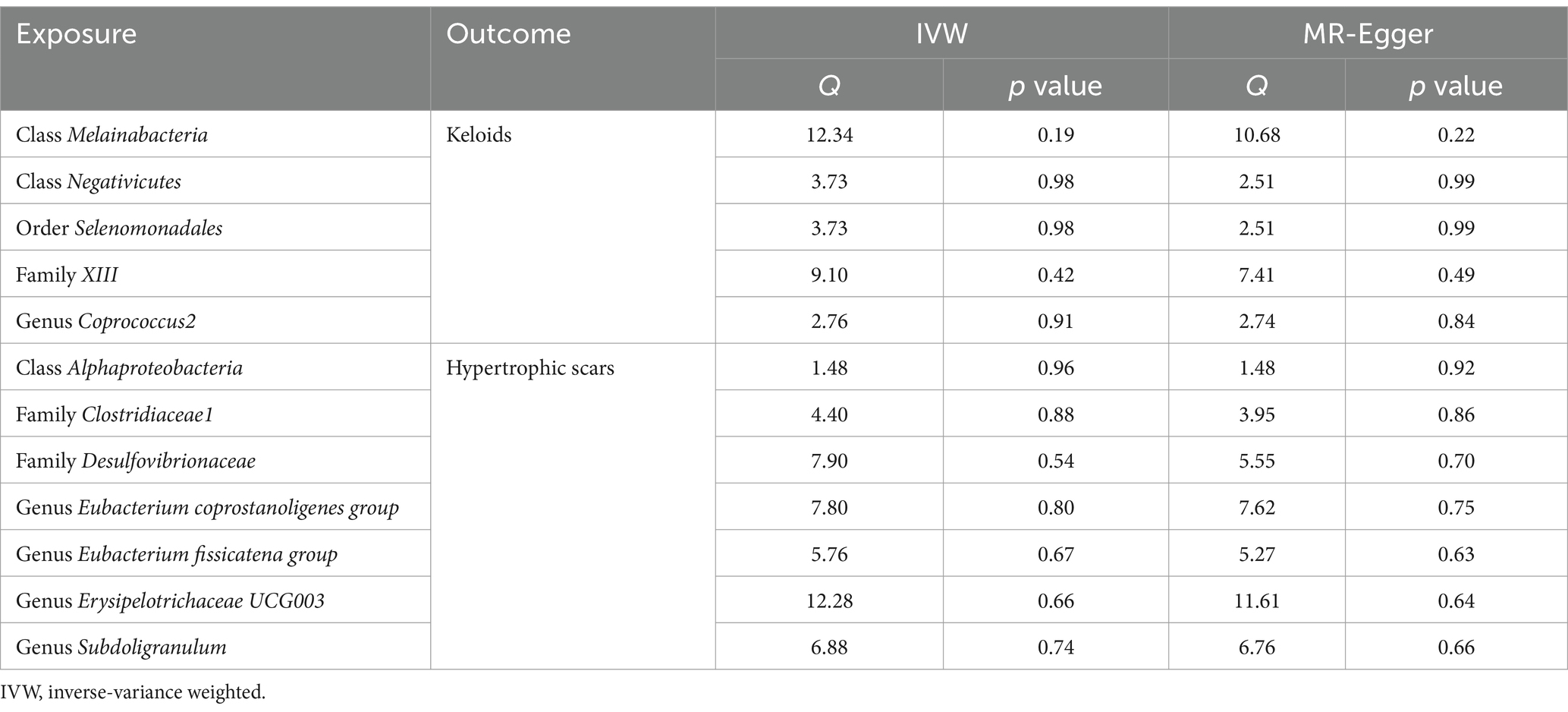
Table 1. Results of heterogeneity analysis using Cochran’s Q test for causal associations between gut microbiota and pathological scars.
Discussion
The complex interplay between the gut microbiota and various health outcomes has been a focal point of numerous investigations in recent years. Our study focused on this ever-evolving frontier to delineate the causal relationships between the gut microbiota and pathological scars. We identified five and seven distinct gut microbiota taxa causally linked to keloids and hypertrophic scar formations, respectively. Our findings underscore a promising direction for understanding the etiopathogenesis of pathological scars and open new vistas for potential therapeutic interventions.
The complex interplay between the gastrointestinal tract and skin, known as the gut-skin axis, is pivotal for sustaining skin homeostasis (22). Central to this interaction is the role of the gut microbiome in modulating both systemic and local inflammation through immune system engagement. In the gut, microbial communities maintain barrier integrity by metabolizing complex polysaccharides into vital vitamins and short-chain fatty acids (SCFAs), such as butyrate and propionate (23, 24). These substances are integral to reinforcing intestinal barrier strength and minimizing permeability (24). Furthermore, the presentation of commensal antigens by dendritic cells (DCs) facilitates the differentiation of gut commensal bacteria-specific Tregs, IgA-producing B cells, and Th17 cells (22). This process is vital for maintaining the equilibrium of the gut microbiota and preventing bacterial entry into the bloodstream, thereby contributing to skin homeostasis. Skin inflammation can be influenced by minor alterations in specific bacterial species of the intestinal microbiome, potentially leading to disorders such as acne, alopecia areata, atopic dermatitis and psoriasis (5). For instance, Faecalibacterium prausnitzii helps protect against psoriasis by competitively inhibiting the colonization of pathogenic skin flora and producing SCFAs, which modulate inflammatory responses (25), and Lactobacillus casei helps reduce skin inflammation by decreasing the number of cytotoxic CD8+ T cells, thereby modulating the immune response and promoting skin health (26). In this study, it was discovered that the microbiota identified as potentially protective against pathological scars may exert its protective effects on pathological scars through similar pathways. The Coprococcus 2 genus, classified as a gram-positive bacteria, is instrumental in producing butyrate, a compound with anti-inflammatory properties that notably inhibits nuclear factor κB, mitigates reactive oxygen species, and fosters both cell differentiation and intestinal health (25). A decrease in Coprococcus 2 levels, leading to a consequential decrease in butyrate within the gastrointestinal tract, may adversely affect immune functionality, potentially precipitating the development of keloids. Erysipelotrichaceae UCG-003, another butyrate-producing bacterium, is essential for colonic epithelial integrity, and its abundance is notably lower in patients with neurological disorders and lung cancer than in healthy individuals (27–29). This study also revealed that other gut microbiota taxa can influence the formation of pathological scars as protective factors or risk factors, but their specific mechanisms require further investigation.
Emerging evidence increasingly underscores a significant correlation between pathological scars and the microbiota. Regarding the skin microbiota, studies have identified dysbiosis predominantly characterized by S. aureus, which is linked to chronic inflammation and subsequent hypertrophic scar development (30). The topical application of probiotics may also represent a promising approach for the prevention of pathological scars. Lombardi et al.’s research revealed that S. thermophilus lysate markedly reduces the primary mediators and activities involved in the abnormal activation of myofibroblasts triggered by TGF-β1 under fibrotic conditions. This treatment notably decreased the expression of α-SMA, fibronectin, and collagen-I, while also diminishing the collagen contraction ability of activated dermal fibroblasts (31). Focusing on the gut microbiota, Li et al. utilized 16S rRNA gene sequencing to investigate generalizable microbial signatures associated with pathological scars and their relationship with the gut microbiota. Their findings highlighted distinctions in alpha and beta diversity at the phylum and genus levels between patients with physiological and pathological scars, indicating a dysregulated gut microbiota in individuals with pathological scars (13). Notably, a significantly greater presence of Subdoligranulum was observed in the gut microbiota of patients with pathological scars than in that of patients with physiological scars. Our research further corroborates this finding, establishing a causal link between increased Subdoligranulum abundance and the development of hypertrophic scars, thus offering a mechanistic insight into previous observations. However, our findings present some divergences from those reported in Li′s study. This disparity may arise from several factors. First, Li et al. did not distinguish keloids from hypertrophic scars within pathological scars, and our study results indicate that the impact of the gut microbiota on keloids and hypertrophic scars is not identical. Second, our research sample comprised individuals of European descent, in contrast to the East Asian population in Li′s study. Third, the methodological strengths of our MR study include its capacity to explore causal relationships between exposure and outcomes while effectively controlling for confounders, which pose significant challenges in observational settings. Additionally, we have noted recent studies exploring the causal relationship between hypertrophic scars and gut microbiota by MR (32). Our research uniquely investigates the causal relationships between gut microbiota and both hypertrophic scars and keloids. Interestingly, we found distinct causal relationships for these two types of pathological scars with gut microbiota, offering new perspectives on their pathogenesis.
Our MR analysis revealed a subset of the gut microbiota with protective effects against pathological scars, most of which are directly or indirectly related to the production of SCFAs. For instance, the genus Coprococcus, a classic butyrate-producing microbial group, has been shown to have protective effects against a variety of diseases, including atopic disease and Parkinson’s disease (33, 34). Within the genus Eubacterium, groups such as Eubacterium coprostanoligenes and Eubacterium fissicatena are involved in cholesterol metabolism and are capable of producing SCFAs during the degradation of cholesterol (35). Additionally, certain microbial groups within family XIII and family Clostridiaceae 1, such as Clostridium, are well-known producers of butyric acid and other SCFAs (36, 37). The relationships among the gut microbiota, SCFAs, and the formation of pathological scars, such as keloids and hypertrophic scars, have emerged as a focal point of interest in understanding the complex mechanisms of fibrosis. The lipid hypothesis of pathological scar pathogenesis has emphasized the role of triglycerides, cholesterol, and unsaturated fatty acids, while investigations into SCFAs, such as butyric acid, isobutyric acid, malonic acid, isovaleric acid, and valeric acid, have been comparatively limited. Recent studies underscore the significance of these SCFAs, which are produced by the anaerobic fermentation of dietary fibers by colonic bacteria, and play crucial roles in various cellular processes, including apoptosis, proliferation, and differentiation, largely through their histone deacetylase inhibitor activity (38). SCFAs are present at lower concentrations in keloid tissues than in normal skin, suggesting disrupted metabolic equilibrium (39, 40). This reduction, particularly of butyric acid, suggests a compromised antifibrotic defense mechanism, potentially exacerbating scar pathogenesis through the deregulation of keloid-derived fibroblast activity. Such deregulation could culminate in unchecked cellular proliferation and excessive collagen deposition. Indeed, in vitro experiments have shown that butyrate can attenuate fibroblast proliferation and modulate inflammatory responses, in a concentration-dependent manner: lower concentrations of butyrate promote cellular proliferation, while higher concentrations of butyrate promote apoptosis (41, 42). Pathological scars are also intricately associated with inflammation and the connection between SCFAs and inflammation is highlighted by their regulatory effect on mast cells and the production of inflammatory mediators. Specifically, butyrate and propionate have been shown to inhibit the activation of mast cells, potentially mitigating inflammation-driven scar formation (43). In summary, the relationships among the gut microbiota, SCFAs, and pathological scars present a complex yet logically coherent narrative that encompasses metabolic, immunological, and cellular perspectives. Our investigation, through a bioinformatics approach, contributes novel insights into this intricate relationship, enriching the existing body of knowledge with additional reference points for further understanding the underlying mechanisms involved.
This study, while insightful, is subject to several limitations. First, it’s important to acknowledge that the results did not adhere to the stringent FDR correction threshold. Despite not meeting this rigorous standard, the nominal significance of the findings warrants consideration. Second, our analysis did not account for sex differences, a factor that could have influenced the outcomes, particularly given known sex-specific variations in the skin microbiome of burn scars (44). Third, the genetic studies we referenced were limited to populations of European ancestry, potentially introducing stratification bias. This is especially relevant considering the higher incidence of pathological scars among African and Asian individuals, which may limit the applicability of our results to these groups. Fourthly, our study has only validated the causal relationship between gut microbiota and pathological scars at a bioinformatics level; these relationships still lack validation through molecular biology and clinical trials. Finally, our analysis was limited by the taxonomic resolution of the exposure dataset, which was limited to the genus level, thereby restricting our ability to investigate species-level relationships.
Conclusion
In this study, we conducted a comprehensive two-sample MR analysis utilizing publicly accessible GWAS summary-level data, to evaluate the causal influence of the gut microbiota on the development of pathological scars. Our MR analysis robustly supports the hypothesis that the gut microbiota is causally linked to pathological scar formation, providing pivotal insights for future mechanistic and clinical research in this domain.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
In accordance with local legislation and institutional requirements, ethical review and approval for the study involving human participants was not necessary. This article does not include any potentially identifiable images or data.
Author contributions
HS: Validation, Methodology, Conceptualization, Writing – review & editing, Writing – original draft. JL: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Visualization. Y-lL: Visualization, Writing – review & editing, Writing – original draft. XC: Validation, Methodology, Data curation, Conceptualization, Writing – review & editing, Writing – original draft. XJ: Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was conducted with the generous financial support from the Med-X Center for Informatics funding project at Sichuan University (YGJC-003), the National Natural Science Foundation of China (NSFC grants 82273559 and 82073473), and the 1.3.5 Project for Disciplines of Excellence at West China Hospital, Sichuan University (ZYJC21036).
Acknowledgments
The authors wish to express their profound appreciation to the FinnGen biobank and MiBioGen consortium for their invaluable contribution to this research. By granting public access to their comprehensive statistical data, they have significantly facilitated our Mendelian randomization analyses using the original datasets.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1405097/full#supplementary-material
Footnotes
References
1. Ghazawi, FM, Zargham, R, Gilardino, MS, Sasseville, D, and Jafarian, F. Insights into the pathophysiology of hypertrophic scars and keloids: how Do they differ? Adv Skin Wound Care. (2018) 31:582–95. doi: 10.1097/01.ASW.0000527576.27489.0f
2. Lee, HJ, and Jang, YJ. Recent understandings of biology, prophylaxis and treatment strategies for hypertrophic scars and keloids. Int J Mol Sci. (2018) 19:711. doi: 10.3390/ijms19030711
3. Bijlard, E, Kouwenberg, CA, Timman, R, Hovius, SE, Busschbach, JJ, and Mureau, MA. Burden of keloid disease: a cross-sectional health-related quality of life assessment. Acta Derm Venereol. (2017) 97:225–9. doi: 10.2340/00015555-2498
4. Limandjaja, GC, Niessen, FB, Scheper, RJ, and Gibbs, S. The keloid disorder: heterogeneity, histopathology, mechanisms and models. Front Cell Dev Biol. (2020) 8:360. doi: 10.3389/fcell.2020.00360
5. Mahmud, MR, Akter, S, Tamanna, SK, Mazumder, L, Esti, IZ, Banerjee, S, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. (2022) 14:2096995. doi: 10.1080/19490976.2022.2096995
6. Joura, MI, Brunner, A, Nemes-Nikodém, É, Sárdy, M, and Ostorházi, E. Interactions between immune system and the microbiome of skin, blood and gut in pathogenesis of rosacea. Acta Microbiol Immunol Hung. (2021) 68:1–6. doi: 10.1556/030.2021.01366
7. Buhaș, MC, Gavrila, LI, Candrea, R, Cătinean, A, Mocan, A, Miere, D, et al. Gut Microbiota in Psoriasis. Nutrients. (2022) 14:14. doi: 10.3390/nu14142970
8. Liu, X, Xu, J, Wang, Z, Xu, X, Wen, H, Su, H, et al. Differential changes in the gut microbiota between extrinsic and intrinsic atopic dermatitis. J Autoimmun. (2023) 141:103096. doi: 10.1016/j.jaut.2023.103096
9. Salem, I, Ramser, A, Isham, N, and Ghannoum, MA. The gut microbiome as a major regulator of the gut-skin Axis. Front Microbiol. (2018) 9:1459. doi: 10.3389/fmicb.2018.01459
10. O'Neill, CA, Monteleone, G, McLaughlin, JT, and Paus, R. The gut-skin axis in health and disease: a paradigm with therapeutic implications. Bioessays. (2016) 38:1167–76. doi: 10.1002/bies.201600008
11. Chen, G, Chen, ZM, Fan, XY, Jin, YL, Li, X, Wu, SR, et al. Gut-brain-skin Axis in psoriasis: a review. Dermatol Ther (Heidelb). (2021) 11:25–38. doi: 10.1007/s13555-020-00466-9
12. Szántó, M, Dózsa, A, Antal, D, Szabó, K, Kemény, L, and Bai, P. Targeting the gut-skin axis-probiotics as new tools for skin disorder management? Exp Dermatol. (2019) 28:1210–8. doi: 10.1111/exd.14016
13. Li, M, Li, M, Dai, Y, Li, D, Yu, H, Liu, J, et al. 16S rRNA gene sequencing reveals the correlation between the gut microbiota and the susceptibility to pathological scars. Front Microbiol. (2023) 14:1215884. doi: 10.3389/fmicb.2023.1215884
14. Richmond, RC, and Davey, SG. Mendelian randomization: concepts and scope. Cold Spring Harb Perspect Med. (2022) 12:a040501. doi: 10.1101/cshperspect.a040501
15. Burgess, S, Daniel, RM, Butterworth, AS, and Thompson, SG. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. (2015) 44:484–95. doi: 10.1093/ije/dyu176
16. Davey Smith, G, Holmes, MV, Davies, NM, and Ebrahim, S. Mendel's laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol. (2020) 35:99–111. doi: 10.1007/s10654-020-00622-7
17. Kurilshikov, A, Medina-Gomez, C, Bacigalupe, R, Radjabzadeh, D, Wang, J, Demirkan, A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53:156–65. doi: 10.1038/s41588-020-00763-1
18. Sakaue, S, Kanai, M, Tanigawa, Y, Karjalainen, J, Kurki, M, Koshiba, S, et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. (2021) 53:1415–24. doi: 10.1038/s41588-021-00931-x
19. Burgess, S, and Thompson, SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
20. Burgess, S, and Thompson, SG. Interpreting findings from Mendelian randomization using the MR-egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
21. Verbanck, M, Chen, CY, Neale, B, and Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
22. De Pessemier, B, Grine, L, Debaere, M, Maes, A, Paetzold, B, and Callewaert, C. Gut-skin Axis: current knowledge of the interrelationship between microbial Dysbiosis and skin conditions. Microorganisms. (2021) 9:353. doi: 10.3390/microorganisms9020353
23. LeBlanc, JG, Milani, C, de Giori, GS, Sesma, F, van Sinderen, D, and Ventura, M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. (2013) 24:160–8. doi: 10.1016/j.copbio.2012.08.005
24. Merra, G, Noce, A, Marrone, G, Cintoni, M, Tarsitano, MG, Capacci, A, et al. Influence of Mediterranean diet on human gut microbiota. Nutrients. (2020) 13:7. doi: 10.3390/nu13010007
25. Scher, JU, Ubeda, C, Artacho, A, Attur, M, Isaac, S, Reddy, SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol (Hoboken, NJ). (2015) 67:128–39. doi: 10.1002/art.38892
26. Chen, L, Li, J, Zhu, W, Kuang, Y, Liu, T, Zhang, W, et al. Skin and gut microbiome in psoriasis: gaining insight into the pathophysiology of it and finding novel therapeutic strategies. Front Microbiol. (2020) 11:589726. doi: 10.3389/fmicb.2020.589726
27. Liu, S, Li, E, Sun, Z, Fu, D, Duan, G, Jiang, M, et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci Rep. (2019) 9:287. doi: 10.1038/s41598-018-36430-z
28. Zhao, F, An, R, Wang, L, Shan, J, and Wang, X. Specific gut microbiome and serum metabolome changes in lung Cancer patients. Front Cell Infect Microbiol. (2021) 11:725284. doi: 10.3389/fcimb.2021.725284
29. Kaiyrlykyzy, A, Kozhakhmetov, S, Babenko, D, Zholdasbekova, G, Alzhanova, D, Olzhayev, F, et al. Study of gut microbiota alterations in Alzheimer's dementia patients from Kazakhstan. Sci Rep. (2022) 12:15115. doi: 10.1038/s41598-022-19393-0
30. Yu, J, Mao, Z, Zhou, Z, Yuan, B, and Wang, X. Microbiome dysbiosis occurred in hypertrophic scars is dominated by S. aureus colonization. Front Immunol. (2023) 14:1227024. doi: 10.3389/fimmu.2023.1227024
31. Lombardi, F, Augello, FR, Artone, S, Bahiti, B, Sheldon, JM, Giuliani, M, et al. Efficacy of probiotic Streptococcus thermophilus in counteracting TGF-beta1-induced fibrotic response in normal human dermal fibroblasts. J Inflamm (Lond). (2022) 19:27. doi: 10.1186/s12950-022-00324-9
32. Xue, K, Zhang, G, Li, Z, Zeng, X, Li, Z, Wang, F, et al. Dissecting the association between gut microbiota and hypertrophic scarring: a bidirectional Mendelian randomization study. Front Microbiol. (2024) 15:1345717. doi: 10.3389/fmicb.2024.1345717
33. Keshavarzian, A, Green, SJ, Engen, PA, Voigt, RM, Naqib, A, Forsyth, CB, et al. Colonic bacterial composition in Parkinson's disease. Mov Disord. (2015) 30:1351–60. doi: 10.1002/mds.26307
34. Nylund, L, Nermes, M, Isolauri, E, Salminen, S, de Vos, WM, and Satokari, R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. (2015) 70:241–4. doi: 10.1111/all.12549
35. Blaak, EE, Canfora, EE, Theis, S, Frost, G, Groen, AK, Mithieux, G, et al. Short chain fatty acids in human gut and metabolic health. Benefic Microbes. (2020) 11:411–55. doi: 10.3920/BM2020.0057
36. Hwang, J, Yoo, W, Shin, SC, Kim, KK, Kim, HW, Do, H, et al. Structural and biochemical insights into Bis(2-hydroxyethyl) terephthalate degrading carboxylesterase isolated from Psychrotrophic bacterium Exiguobacterium antarcticum. Int J Mol Sci. (2023) 24:12022. doi: 10.3390/ijms241512022
37. Stoeva, MK, Garcia-So, J, Justice, N, Myers, J, Tyagi, S, Nemchek, M, et al. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes. (2021) 13:1–28. doi: 10.1080/19490976.2021.1907272
38. Li, CY, Xie, RX, Zhang, SW, Yun, J, Zhong, A, Cen, Y, et al. Metabolism, fibrosis, and apoptosis: the effect of lipids and their derivatives on keloid formation. Int Wound J. (2024) 21:e14733. doi: 10.1111/iwj.14733
39. Yang, J, Chen, M, and He, L. To explore ideas from the altered metabolites: the metabolomics of pathological scar. J Craniofac Surg. (2022) 33:1619–25. doi: 10.1097/SCS.0000000000008470
40. Yang, JX, Li, SY, Chen, ML, and He, LR. The role of altered fatty acid in pathological scars and their dermal fibroblasts. Chin J Traumatol. (2022) 25:218–23. doi: 10.1016/j.cjtee.2022.03.006
41. Torii, K, Maeshige, N, Aoyama-Ishikawa, M, Miyoshi, M, Terashi, H, and Usami, M. Combination therapy with butyrate and docosahexaenoic acid for keloid fibrogenesis: an in vitro study. An Bras Dermatol. (2017) 92:184–90. doi: 10.1590/abd1806-4841.20176198
42. Maeshige, N, Torii, K, Tabuchi, H, Imai, M, Koga, Y, Uemura, M, et al. Inhibitory effects of short-chain fatty acids and ω-3 polyunsaturated fatty acids on Profibrotic factors in dermal fibroblasts. Eplasty. (2019) 19:e4
43. Wang, CC, Wu, H, Lin, FH, Gong, R, Xie, F, Peng, Y, et al. Sodium butyrate enhances intestinal integrity, inhibits mast cell activation, inflammatory mediator production and JNK signaling pathway in weaned pigs. Innate Immun. (2018) 24:40–6. doi: 10.1177/1753425917741970
Keywords: Mendelian randomization, gut microbiota, keloids, hypertrophic scars, pathological scars
Citation: Shucheng H, Li J, Liu Y-l, Chen X and Jiang X (2024) Causal relationship between gut microbiota and pathological scars: a two-sample Mendelian randomization study. Front. Med. 11:1405097. doi: 10.3389/fmed.2024.1405097
Edited by:
Jurica Zucko, University of Zagreb, CroatiaReviewed by:
Shikha Sharma, National Institutes of Health (NIH), United StatesXiuzu Song, Third People’s Hospital of Hangzhou, China
Copyright © 2024 Shucheng, Li, Liu, Chen and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian Jiang, amlhbmd4aWFuQHNjdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Huidi Shucheng1,2,3
†
Huidi Shucheng1,2,3
† Jiaqi Li
Jiaqi Li Xinghan Chen
Xinghan Chen