
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 02 July 2024
Sec. Gastroenterology
Volume 11 - 2024 | https://doi.org/10.3389/fmed.2024.1379335
This article is part of the Research Topic The Role of Gut Microbiota-Gut-Brain Axis in Inflammatory Bowel Disease View all 5 articles
 Jeannie Devereaux1
Jeannie Devereaux1 Ainsley M. Robinson1,2,3
Ainsley M. Robinson1,2,3 Rhian Stavely1,4
Rhian Stavely1,4 Majid Davidson1
Majid Davidson1 Narges Dargahi1
Narges Dargahi1 Ramya Ephraim1
Ramya Ephraim1 Dimitros Kiatos1
Dimitros Kiatos1 Vasso Apostolopoulos1,3,5
Vasso Apostolopoulos1,3,5 Kulmira Nurgali1,3,6*
Kulmira Nurgali1,3,6*Background: Inflammatory bowel disease is an incurable and idiopathic disease characterized by recurrent gastrointestinal tract inflammation. Tryptophan metabolism in mammalian cells and some gut microbes comprise intricate chemical networks facilitated by catalytic enzymes that affect the downstream metabolic pathways of de novo nicotinamide adenine dinucleotide (NAD+) synthesis. It is hypothesized that a correlation exists between tryptophan de novo NAD+ synthesis and chronic intestinal inflammation.
Methods: Transcriptome analysis was performed using high-throughput sequencing of mRNA extracted from the distal colon and brain tissue of Winnie mice with spontaneous chronic colitis and C57BL/6 littermates. Metabolites were assessed using ultra-fast liquid chromatography to determine differences in concentrations of tryptophan metabolites. To evaluate the relative abundance of gut microbial genera involved in tryptophan and nicotinamide metabolism, we performed 16S rRNA gene amplicon sequencing of fecal samples from C57BL/6 and Winnie mice.
Results: Tryptophan and nicotinamide metabolism-associated gene expression was altered in distal colons and brains of Winnie mice with chronic intestinal inflammation. Changes in these metabolic pathways were reflected by increases in colon tryptophan metabolites and decreases in brain tryptophan metabolites in Winnie mice. Furthermore, dysbiosis of gut microbiota involved in tryptophan and nicotinamide metabolism was evident in fecal samples from Winnie mice. Our findings shed light on the physiological alterations in tryptophan metabolism, specifically, its diversion from the serotonergic pathway toward the kynurenine pathway and consequential effects on de novo NAD+ synthesis in chronic intestinal inflammation.
Conclusion: The results of this study reveal differential expression of tryptophan and nicotinamide metabolism-associated genes in the distal colon and brain in Winnie mice with chronic intestinal inflammation. These data provide evidence supporting the role of tryptophan metabolism and de novo NAD+ synthesis in IBD pathophysiology.
Accumulating evidence has shown that disruption of the gut-brain axis, a bidirectional communication system that involves neural, hormonal, metabolic, immunological, and microbial signals, may be related to the occurrence and development of gastrointestinal (GI) disorders, such as inflammatory bowel disease (IBD) (1). IBD is a chronic inflammatory disorder of the GI tract that can manifest as Crohn's disease (CD) or ulcerative colitis (UC) (2). Typical clinical manifestations of IBD include diarrhea, abdominal pain, weight loss, fatigue, tenesmus, perianal fissures, and bloody stool (2, 3). Progression of the disease may lead to complications, including fistulas, strictures, rectal bleeding, abscesses, bowel perforations, toxic megacolon, and an increased risk of colorectal cancer (4–6). Additionally, IBD is often complicated by psychological comorbidities, such as anxiety and depression, which affect patient quality of life and disease management (7). It is widely considered that the gut-brain axis disruption is a key element underlying the association between psychological distress and IBD (8, 9). While the etiology of IBD is yet to be fully elucidated, it is considered to be multifactorial, involving complex interactions between host genetic susceptibility, environmental exposures, gut dysbiosis, aberrant immune responses, and enteric nervous system (ENS) dysfunction (10).
IBD symptoms have been associated with dysregulation of 5-hydroxytryptamine (5-HT, serotonin), a tryptophan derivative, which is an essential amino acid and vital ENS signaling molecule (11, 12). 5-HT regulates energy metabolism, which is necessary for maintaining gut homeostasis (13, 14). In response to numerous stimuli, including nutrients, toxins, or neuromodulatory agents, enterochromaffin cells release 5-HT, which regulates gut motility, glucose absorption, and fluid balance (11). Recent evidence indicates that peripheral 5-HT is involved in multiple metabolic control pathways (13), including links to the brain relating to behavioral changes, anxiety, and depression (15). Intestinal tryptophan metabolism involves complex interactions between host genetic, microbial, and dietary factors. Tryptophan metabolites are produced via three major metabolic pathways: kynurenine, serotonin, and indole pathways, which have essential roles in the regulation of intestinal inflammation by acting directly or indirectly on pro/anti-inflammatory cytokines, immune cell function, intestinal microbial composition, and gut homeostasis (16). Results of studies investigating the connection between the gut-brain axis and intestinal inflammation suggest that dysregulated tryptophan metabolism contributes to disease severity and depressive symptoms in IBD patients and experimental models of colitis; however, these studies have limited their focus to only a few tryptophan metabolites (15, 17–20).
Gut microbial dysbiosis has been implicated in the pathogenesis of IBD and numerous studies mention it as a contributory factor for the development and progression of the disease (21, 22). Commensal gut microbes have diverse impacts on host tryptophan availability, directly and indirectly controlling tryptophan metabolism and contributing to the synthesis of neuroactive molecules, such as 5-HT, kynurenines, tryptamine, and indolic compounds which participate in the microbiota-gut-brain communication (23–25). Dysbiosis can be associated with imbalanced activation of the three pathways of tryptophan metabolism (26, 27). Furthermore, variations in tryptophan metabolism can negatively influence microbial proliferation and diversity; thus, the gut microbiota is considered a driving force affecting the tryptophan metabolism in the gut (24).
Despite the association between chronic intestinal inflammation and dysregulated tryptophan metabolism, few studies have investigated the intrinsic molecules involved in tryptophan metabolism that may distinguish the downstream physiological alterations in IBD. Approximately 90%−95% of tryptophan degradation occurs through the kynurenine pathway leading to de novo NAD+ biosynthesis (28), which depends on expressions of interferon (IFN)-γ, tumor necrosis factor (TNF)-α, bacterial lipopolysaccharides, and the pro-inflammatory cytokine interleukin (IL)-6, all of which are key pathogenic factors in IBD (28). Deviations from NAD+ homeostasis distort NAD-dependent enzyme levels and impact DNA repair, neuron regeneration, and inflammation resistance, contributing to the development of a spectrum of diseases (29). However, studies investigating NAD+ biosynthesis have been restricted to the comparison of just one or two genes within these pathways (30). Indole, or the aryl hydrocarbon receptor (AhR) pathway, metabolizes 4%−6% of tryptophan into indole derivatives via gut microbiota, including Lactobacillus, Bifidobacterium, Clostridium, and Bacteroides, activating AhR, which is important in the modulation of intestinal inflammation (24, 31–33). About 1%−2% of tryptophan enters the serotonergic pathway, which processes tryptophan mainly via tryptophan hydroxylase 1 (TPH1) within enterochromaffin cells in the gut or TPH2 in the brain into 5-HT and melatonin (34). Enterochromaffin cell hyperplasia and downregulation of serotonin reuptake transporter lead to elevated levels of 5-HT within the colonic mucosa, contributing to the onset and progression of chronic intestinal inflammation, shown in the Winnie mouse model of chronic colitis (35).
Winnie mice, an experimental model of spontaneous chronic colitis, are affected by a missense mutation of the Muc2 gene, resulting in aberrant biosynthesis of mucin within the goblet cells (36). Consequently, progressive chronic intestinal inflammation develops, with signs appearing from 6 weeks of age, progressing to severe colitis by 12–16 weeks of age (37). Although previous studies have demonstrated inflammation-induced structural and functional alterations in the Winnie mouse colon, establishing this experimental model to be highly representative of IBD (36–42), no studies have evaluated tryptophan metabolism and de novo NAD+ biosynthesis in Winnie mice. Since IBD development is closely related to gene interactions within diseased tissue (43), this study aimed to determine changes in the tryptophan de novo NAD+ synthesis. Subsequently, we aimed to investigate correlation between dysregulated tryptophan metabolism in the colon and the brain and fecal microbiota involved in tryptophan metabolism using the Winnie mouse model. Enhancing understanding of changes in the tryptophan de novo NAD+ synthesis may potentially reveal novel insights into IBD pathophysiology.
Winnie (males and females, aged 14 weeks, n = 17) and C57BL/6 mice (males and females, aged 14 weeks, n = 18) were obtained from the same breeding colony at the Victoria University Werribee Animal Facility (Melbourne, Victoria, Australia) and delivered to the Animal Holding Facility at the Western Centre for Health, Research and Education (Melbourne, Victoria, Australia), where experiments were performed. All mice were permitted ad libitum access to food and water and housed in a temperature-controlled environment with a 12-h day/night cycle. Animals were acclimatized for 1 week prior to euthanasia via administration of pentobarbitone (1:16 dilution, 30 g, 100 μl/20 g, Lethabarb, Virbac, Australia). The distal colon tissues and brains were excised and used for subsequent experiments. All experimental procedures adhered to the Australian National Health and Medical Research Council (NHMRC) guidelines and were approved by the Victoria University Animal Experimentation Ethics Committee (AEC-17-016).
Total RNA was extracted from the distal colons and brains of C57BL/6 (n = 8) and Winnie (n = 6) mice using the RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany), as per manufacturer instructions. The extracted RNA underwent a quality assessment on a 2100 Bioanalyzer using the RNA 6000 Nano Kit (Agilent Technologies, USA) to confirm that the samples were devoid of genomic DNA contamination and 16S ribosomal RNA from bacteria. RNA concentration was then measured by a Nanodrop spectrophotometer (Denovix, Melbourne, Australia), with 260/280 nm and 260/230 nm ratios exceeding 1.8, indicating purity. The mRNA samples were submitted to Micromon Genomics (Monash University, Melbourne, Australia) for RNA-sequencing (RNA-Seq) using the next-generation sequencing (NGS) platform. Micromon Genomics performed RNA quality control with Invitrogen Qubit RNA BR assay (Gene Target Solutions, Sydney, Australia), as well as the polyA purification of mRNA from total RNA samples and RNA-Seq library construction. All samples quantified and analyzed with the Qubit and Bio-analyzer passed the QNA quality control (QC) standard. Micromon Genomics conducted the bioinformatics and performed the sequencing on MGITech MGISEQ2000RS hardware (MGISEQ-2000RS High-throughput Sequencing Set). The MGIEasy V2 chemistry set was used to generate the library, with at least 400 m raw reads per lane and paired-end 100b reads for accuracy and processing to compressed FASTQ files. Raw FASTQ files were analyzed using the RNAsik pipeline version 1.5.4 (44) and the Mus musculus reference genome GRCm38 (GenBank accession GCA_000001635.2) (45). Raw read counts were quantified using the FeatureCounts program (46) and analyzed with Degust for counts per million (CPM) library size normalization (47). Differentially expressed genes (DEGs) of the colon and brain were identified using EdgeR with a false discovery rate (FDR) of < 0.001, applying the Benjamini–Hochberg correction (48). Genes with log2 fold change (FC) value of ±0.585 (FC ± 1.5) were selected as the cut-off. To analyze differential gene expression based on overrepresentation analysis, software for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was used (49). Sequencing data are deposited at the Gene Expression Omnibus (GEO) repository, with the accession numbers GSE244558 (colon) and GSE264317 (brain).
Gene lists for tryptophan and nicotinamide metabolism-associated gene expressions were generated by merging related gene lists from multiple databases accessed via the molecular signature database (MSigDB). Heat maps of tryptophan and nicotinamide metabolism-associated gene expression values for each colon sample were visualized using the Morpheus web-based tool and presented as Z-score distributions (across samples) of the CPM values. Data are hierarchically clustered by Euclidian distance. Visualization of selected pathways was performed using the R package, Pathview, using default parameters (50). Input data included significance scores of gene expression values between Winnie and C57BL/6 mice computed as the sign Log2FC × Log10P and represented in a KEGG graph.
The homology of genes in Winnie mice and IBD patients was determined as previously described (51). Briefly, the National Center for Biotechnology Information (NCBI) GEO public data source was used to gather gene expression data on the transcriptome of human IBD patients (52, 53) to establish homology to tryptophan and nicotinamide metabolism DEGs identified in the Winnie mouse colon in this study. High-throughput sequencing was used to create the expression profile of the inflamed intestine in IBD patients and the healthy colon regions in uninflamed controls undergoing resection of non-obstructive colorectal adenocarcinoma. Peters et al. (54) assembled these data as reads per kilobase of transcript per million mapped reads. Similar to our methods, polyA-purified mRNA was used in this study from RNA isolated using TRIzol. These data are available at https://www.ncbi.nlm.nih.gov/geo/ under the GEO series accession number GSE83687. This dataset included CD patients (n = 42), UC patients (n = 31), and controls (n = 60).
Reference standards: L-tryptophan; 5-hydroxyindole-acetic acid (5-HIAA), kynurenic acid (KYNA), L-kynurenine 98%, 2,3-pyridinedicarboxylic acid [quinolinic acid (QUIN)], nicotinamide, and 2-picolinic acid (PIC; all purchased from Sigma–Aldrich, Castle Hill, Australia) were accurately weighed using the calculation of normality and subsequently dissolved individually into the mobile phase composition (15% acetonitrile, 0.2% trifluoroacetic acid (TFA), and 84.8% Milli-Q purified water) degassed under vacuum. The eluents were filtered through a Millipore Membrane Filter (type HA, pore size 0.45 μm, Billerica, MA, USA); 0.5 ml of eluent was aliquoted into a 2 ml dark-colored glass bottle and injected into the ultra-fast liquid chromatograph (UFLC) for method validation (55, 56).
The Shimadzu Prominence UFLC (Shimadzu Corporation, Kyoto, Japan) consisted of variable wavelength UV-visible (UV-Vis) detectors. The high-precision six-port valve achieves 0.3% or less area repeatability in 100,000 cycle endurance tests. The measurements for simultaneous tryptophan compounds were obtained via the isocratic, ion-pair, reversed-phased C18 (250 mm length × 4.6 mm internal diameter) analytical column in the aforementioned mobile phase composition. The chromatogram was run under multistep gradient conditions with a pump flow rate of 0.8 ml/min, a column temperature of 25°C and an injection volume of 10 μl/min. The observed analysis column pump pressure fluctuated around 2500 pounds per square inch (psi). Elution was monitored by the photodiode array detector (collecting UV–Vis spectra from 190 to 800 nm). Standard retention times were monitored using the 254 nm wavelength.
The UFLC analytical method for measuring tryptophan metabolite concentration was validated for the parameters of selectivity, specificity, linearity, precision (robustness and ruggedness), accuracy (% recovery), sensitivity, limits of detection (LOD), and limits of quantification (LOQ) in calibrating the solutions. The linearity graph was plotted as ‘concentration vs. peak area response' via the Lab Solutions calibration curve function, with a correlation coefficient ranging from 0.9995 to 1.000 for all compounds. The intra-day reproducibility was assessed using five solutions daily, while the inter-day reproducibility was determined by measuring the same solutions on three consecutive days. Intra-day accuracy and precision were calculated from the % bias [% (measured–theoretical)/theoretical concentration] and relative standard deviation (RSD) [%RSD = % standard deviation/mean] for the five replicates of each QC point. LOD and LOQ values were calculated by expressing the noise-to-signal ratio of the lowest known concentration of linearity samples in μg/ml or ppm, then converted to percentages (mg/L). Following analysis, the output chromatogram displayed the constituents of standard solutions as a peak against the retention time. The retention times were utilized for quantitative analysis and detection of unknown concentrations in the biological samples (57). Our method is consistent with the profiles observed in preceding studies involving tryptophan metabolites (58, 59).
Biological samples of the distal colon and brains from C57BL/6 (n = 5–8) and Winnie mice (n = 6–8) were weighed and placed into safe-lock tubes, containing ice-cold 0.2 M HCIO4 solution (10 μl/mg of tissue). Homogenization of the tissues was performed in the pre-cooled homogenizing bead beater (TissueLyser LT, Qiagen, Melbourne, Australia). The homogenates were retained at 5°C for 15 min to aid the precipitation of proteins, cell debris, and microscopic tissues. Next, the homogenates were centrifuged at 12,000 revolutions per minute at 4°C for 15 min to extract the metabolites from the matrix. The supernatants were immediately separated from the precipitates and transferred into a sterile microtube, adjusted to a ratio of 4:1 v/v with ice-cold 0.1 M HClO4, vortexed for 30 s, and filtered into a new tube for subsequent experiments (60).
Prepared distal colon and brain samples from C57BL/6 and Winnie mice were injected separately into the UFLC. The concentrations of each tryptophan metabolite were determined by analyzing the peak heights of individual analytes using the inbuilt Shimadzu software. The metabolite concentrations recorded in the Shimadzu acquired “American Standard Code for Information Interchange” text file analyzed by Origin Pro, Version 2021b (OriginLab Corporation, Northampton, MA, USA). All peak fitting, surface fitting, and signal processing capacities reached chi-square tolerance values of 1E-6.
Fecal samples were collected from C57BL/6 and Winnie mice (n = 11 for both) and stored at −80°C until microbial DNA extraction. Extraction of DNA was performed as previously described (40). Briefly, the MoBio PowerFecal DNA Isolation Kit (GeneWorks, Thebarton, South Australia) was used to extract DNA from 0.25 g of each fecal sample according to the manufacturer's instructions. Homogenization and lysis of cells were performed with a FastPrep-24 instrument (MP Biomedicals, Seven Hills, NSW, Australia). The DNA was cleaned with an ethanol-based wash solution to remove residual salt and other contaminants. Subsequently, a sterile elution buffer (10 mM Tris) released the DNA from the spin column filter, yielding DNA that was ready for downstream applications. Centrifugation steps were performed for 1 min at 13,000 g. PCR amplicons spanning the 16S rRNA V3–V4 hypervariable region (forward primer: CCTAYGGGRBGCASCAG, reverse primer GGACTACNNGGGTATCTAAT) underwent high-throughput sequencing on the Illumina MiSeq platform using a 300 bp paired end protocol (600 cycles, 0.04–0.10 GB per run) at the Australian Genome Research Facility (University of Queensland, Brisbane, Australia). All microbiota data have been uploaded to the GitHub repository. The data are accessible via https://github.com/Nurgali-lab/C57-Win-APX-16SV3-4.git. Bioinformatics analysis involved demultiplexing, QC, Amplicon Sequence Variant (ASV) calling, and taxonomic classification. Briefly, diversity profiling analysis was performed with QIIME 2 2019.7. The demultiplexed raw reads were primer trimmed and quality filtered using the cutadapt plugin followed by denoising with DADA2 (via q2-dada2). Taxonomy was assigned to ASVs using the q2-featureclassifier classify-sklearn naïve Bayes taxonomy classifier. Evaluations present at the genus taxonomic level, including percentage compilations, represent all sequences resolved to their primary identification or their closest relative. The relative abundance of genera in fecal samples from C57BL/6 and Winnie mice was calculated by the number of ASVs relative to the total number of ASVs per sample and means compared between groups in this study. Genera involved in tryptophan and nicotinamide metabolism were identified from previous reports, including some studies in IBD and colitis (23, 24, 61–67). Alpha diversity was performed for species richness, Chao1, Shannon–Wiener's diversity index and Simpson's index. Beta diversity was obtained by calculating the Bray-Curtis dissimilarity index. Calculations were performed using Microsoft Excel.
GraphPad Prism v9 (GraphPad Software, San Diego, CA, USA) was used to determine statistical differences using an unpaired t-test with Welch's correction. Data are presented as the mean ± standard error of the mean (SEM). Differences were considered statistically significant when p < 0.05.
Gene lists for tryptophan and nicotinamide metabolism-associated gene expressions were generated by merging related gene lists from multiple databases accessed via the MSigDB (Supplementary Figure S1). Subsequently, RNA-Seq was performed to determine changes in these gene expressions in the distal colon of Winnie mice (n = 6) vs. age-matched C57BL/6 littermates (n = 8). Heat maps were used to visualize DEGs associated with tryptophan and nicotinamide metabolism and are shown as individual sample Z-scores from the colons of C57BL/6 and Winnie mice (Figure 1). In total, six DEGs associated with tryptophan metabolism were observed in colons from Winnie when compared to samples from C57BL/6 mice (Figure 1A), while 24 DEGs associated with nicotinamide metabolism were noted (Figure 1B).
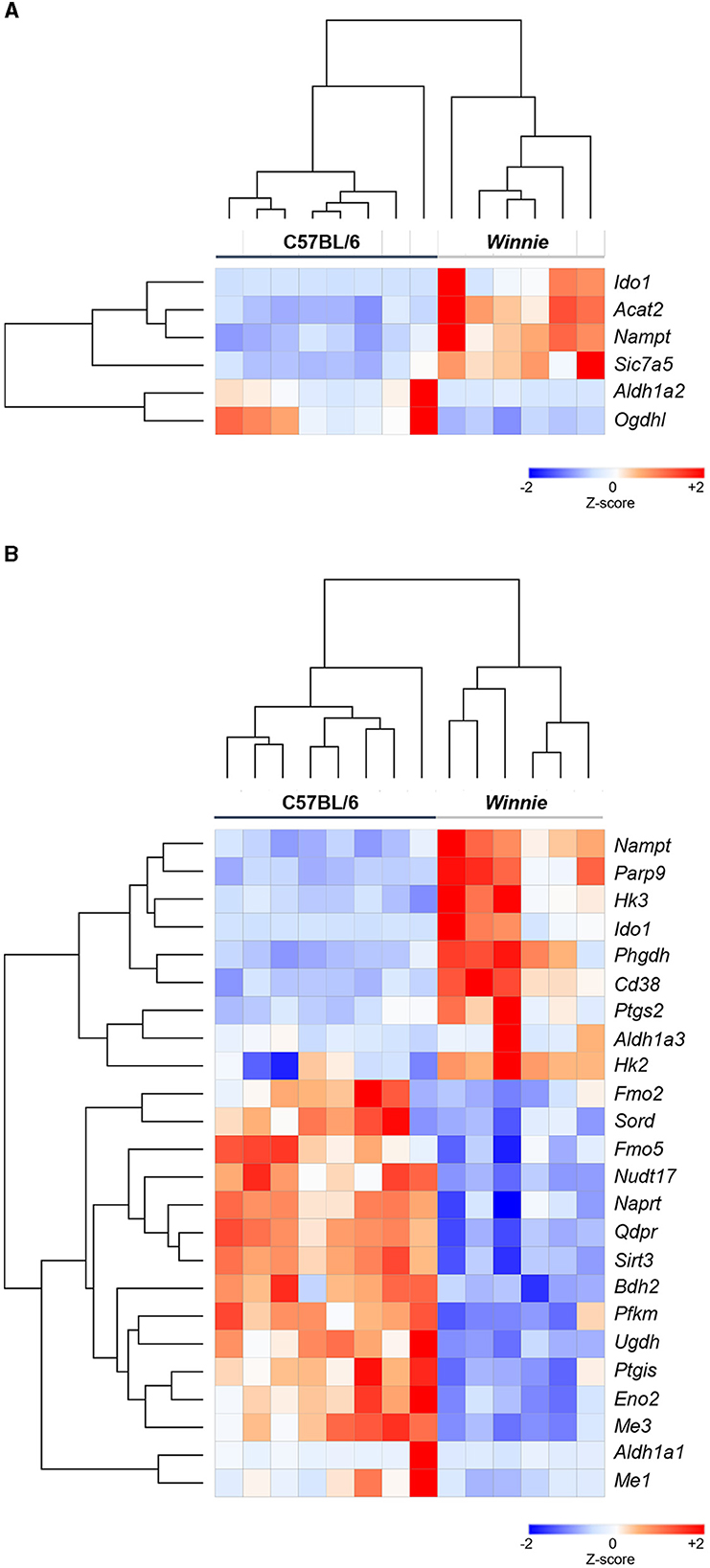
Figure 1. Heatmap representation of DEGs associated with tryptophan and nicotinamide metabolism in the Winnie mouse colon Heat map representations of tryptophan (A) and nicotinamide (B) metabolism-associated DEGs in colons from Winnie mice (n = 6) compared to colons from C57BL/6 mice (n = 8). Individual colon samples from C57BL/6 and Winnie mice are visualized in columns and DEGs are represented in rows. Plotted data presented as Z-score distributions (−2 to +2 across samples) of the counts per million values indicate gene expression values from DEGs analysis. Red represents upregulated gene expression, while blue represents downregulated gene expression.
Visualization of the KEGG tryptophan and nicotinamide metabolism pathways underscored changes to the regulation of key enzymes involved in these pathways in the Winnie mouse colon compared to colon samples from C57BL/6 mice (Figure 2). In colons from Winnie mice, analysis revealed a total of 11 DEGs involved in the tryptophan metabolic pathway (Figure 2A). In the Winnie mouse colon, the expression of genes involved in the catabolic process of tryptophan to kynurenine and the de novo NAD+ biosynthesis [kynureninase (Kynu), indoleamine 2,3-dioxygenase1 (Ido1)] were upregulated. Increased expression of Kynu, Ido1, and acetyl-CoA acetyltransferase 1 (Acat1) is associated with driving cellular amino acid catabolic processes and nucleotide biosynthetic processes. Additionally, upregulation of Kynu and Acat1 implicate the acetyl-CoA metabolic process, potentially indicating heightened energy metabolism. An increase in the expression of Kynu, Ido1, Acat1, and monoamine oxidase-b (Mao-b) suggests enhanced cellular catabolic processes, while downregulation in glutaryl-CoA dehydrogenase (Gcdh) and aryl-formamidase (Afmid) suggests reduced substrate availability for tryptophan metabolism. There was a substantial reduction in the genes involved in the cellular metabolism of biogenic amines, indole-containing compounds, and cellular amino acid metabolic processes.
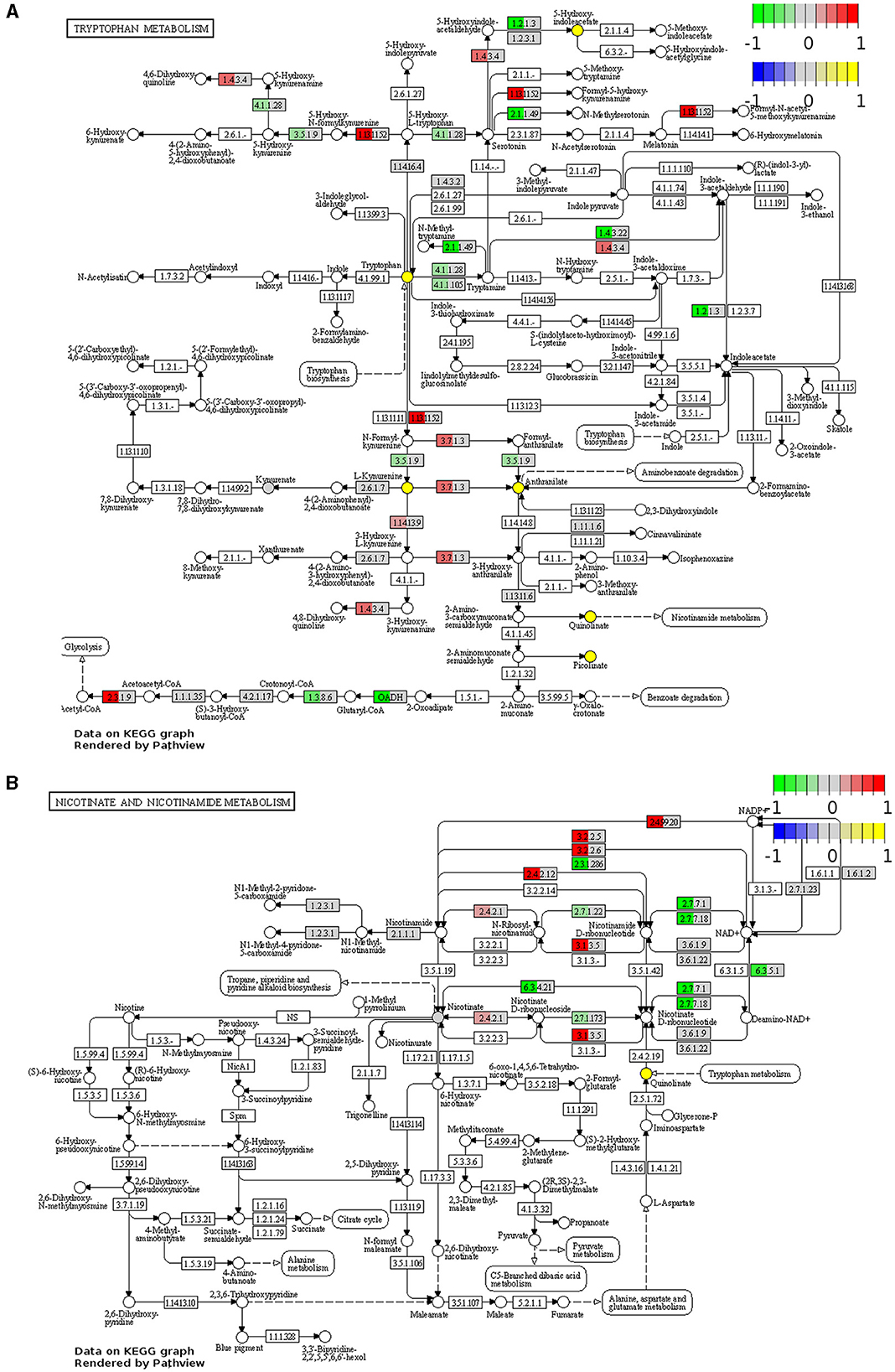
Figure 2. Functional analysis identifies upregulation and downregulation of key elements of tryptophan and nicotinamide metabolism in the Winnie mouse colon. Tryptophan (A) and nicotinamide (B) metabolism KEGG pathway analysis of distal colon samples from Winnie mice (n = 6) vs. C57BL/6 littermates (n = 8). DEGs of the tryptophan and nicotinamide metabolism KEGG pathways are colored according to their sign Log2FC × Log10P value changes. Rectangular nodes indicate gene expression data by RNA-seq gene expression data. Positive values (red) indicate genes that are upregulated (tryptophan—EC:1.4.3.4, Mao-b; EC:1.13.11.52, Ido1; EC:3.7.1.3, Kynu; EC:2.3.1.9, Acat1, nicotinamide—EC:2.4.99.20, Cd38; EC:3.2.2.6, Cd38; EC:3.2.2.5, Art2a; EC:2.4.2.12, Nampt1; EC:3.1.3.5, Cd73; EC:2.4.2.1, Pnp) and negative values (green) indicate genes that are downregulated [tryptophan—(EC):3.5.1.9, Afmid; EC:4.1.1.28 and EC:4.1.1.105, Ddc; EC:2.1.1.49, SAMe; EC:1.4.3.22, Aoc1; EC:1.2.1.3, Aldh2; OADH, Dhtkd1; EC:1.3.8.6, Gcdh, nicotinamide—EC:2.3.1.286, Sirt2; EC:2.7.7.1, Nmnat2; EC:2.7.7.18, Nmnat3; EC:2.7.1.22, Nmrk1; EC:2.7.1.173, Nmrk2; EC:6.3.5.1, Nadsyn1; EC:6.3.4.21, Naprt] in the Winnie mouse colon relative to colons from C57BL/6 mice.
There were 12 DEGs associated with the nicotinamide metabolism pathway in the Winnie mouse colon when compared to colons from C57BL/6 mice (Figure 2B). Upregulation of ecto-5′-nucleotidase (Cd73) and purine-nucleoside phosphorylase (Pnp) suggests adenosine monophosphate (AMP) catabolic activity, adenosine metabolic processes, and the biosynthetic process for purine ribonucleosides. Nicotinamide phosphoribosyltransferase (Nampt) and Pnp upregulation can be associated with the metabolic processing of pyridine-containing compounds, while cluster of differentiation 38 (Cd38) plays a significant role in broader metabolic processes. The downregulation in nicotinamide ribose kinases (Nmrk), nicotinamide mononucleotide adenylyl transferases (Nmnat) nicotinamide adenine dinucleotide synthetase 1 (Nadsyn1), nicotinate phosphoribosyltransferase (Naprt), and silent information regulator 2 (Sirt2) are known to affect biological processes of NAD+ biosynthesis, catalytic activity, and adenosine triphosphate (ATP) binding.
The homology in the expression of target genes involved in tryptophan and nicotinamide metabolism between colon samples from Winnie mice and IBD patients was determined respectively to their uninflamed controls (Figure 3). We found 81% concordance in the expression of genes associated with tryptophan metabolism in Winnie mice with both UC and CD patients (Figure 3A). Evaluation of homology for nicotinamide metabolism-associated genes in Winnie mice colons revealed a 64% concordance with UC patients and 45% with CD patients (Figure 3B). These findings further underscore the relevance and significance of the Winnie mouse as a model for exploring the pathophysiology of IBD.
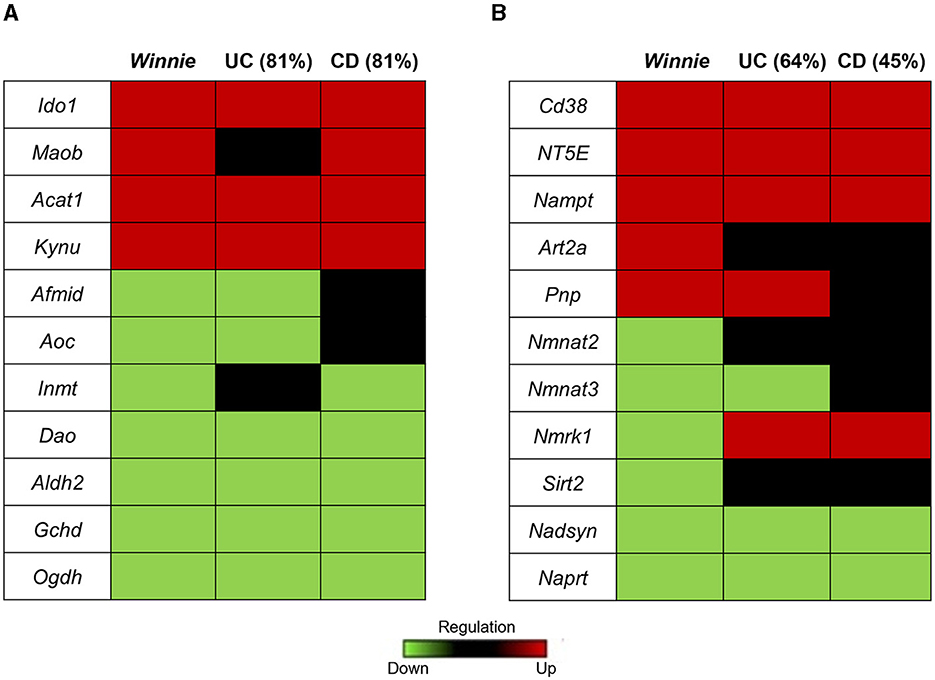
Figure 3. Homology of tryptophan and nicotinamide gene expression in colon samples from Winnie mice and IBD patients. Heat map representations of homology of tryptophan (A) and nicotinamide (B) metabolism associated genes in Winnie mice and IBD patients. Upregulated gene expression levels are marked red, low expression levels are marked green, and neutral expression levels are marked black.
Metabolomics analysis was employed to quantify tryptophan metabolites, including tryptophan, 5-HIAA, KYNA, kynurenine, QUIN, nicotinamide, and PIC, in the distal colons of Winnie mice and compared to colon samples from C57BL/6 littermates (n = 6–8 for both, Table 1, Figure 4). In Winnie mice colons, quantification of tryptophan metabolites revealed higher concentrations of tryptophan (p < 0.05), 5-HIAA (p < 0.05), kynurenine (p < 0.001), QUIN (p < 0.001), and PIC (p < 0.05) when compared to colons samples from C57BL/6 mice. No differences in concentrations of KYNA and nicotinamide were observed between groups (Table 1, Figure 4).
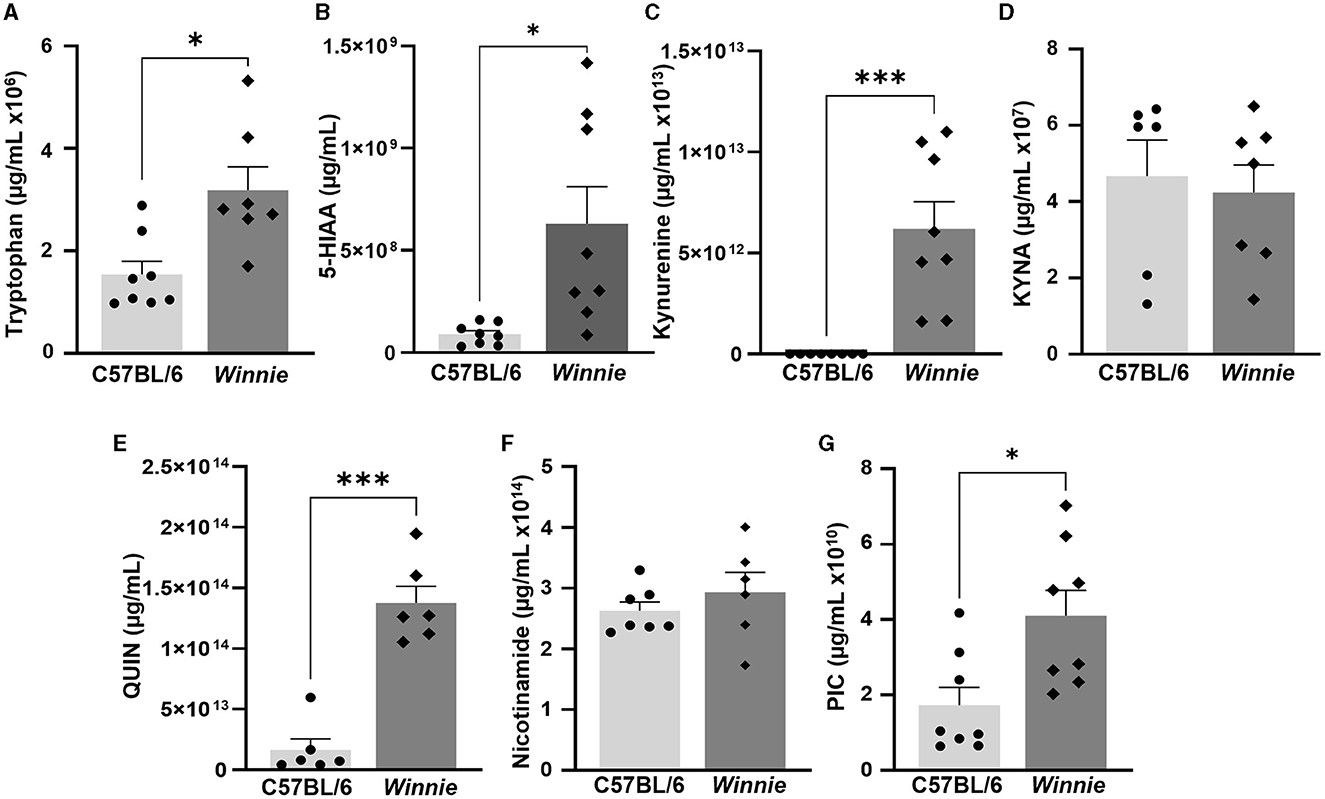
Figure 4. Changes in tryptophan metabolite concentrations in the Winnie mouse colon. Quantification of tryptophan metabolites (μg/ml) in Winnie mice distal colons compared to colons from C57BL/6 littermates (n = 6–8) as measured by UFLC: (A) Tryptophan, (B) 5-HIAA, (C) Kynurenine, (D) KYNA, (E) QUIN, (F) Nicotinamide, and (G) PIC. Data are expressed as mean ± SEM. *p < 0.05, ***p < 0.001 when compared to C57BL/6 mice.
Since gut microbes contribute to the synthesis of neuroactive molecules and indolic compounds (23–25) and dysbiosis can be associated with disrupted tryptophan metabolism (26, 27), we next investigated whether changes in tryptophan metabolite concentrations in Winnie mouse colons corresponded to changes in the abundance of gut microbes involved in tryptophan and nicotinamide metabolism. To evaluate the relative abundance of gut microbial genera involved in tryptophan and nicotinamide metabolism, we performed 16S rRNA gene sequencing of fecal samples from C57BL/6 and Winnie mice (n = 11 for both). α-diversity was assessed based on community richness (Chao1), diversity (Shannon–Wiener diversity index), and evenness (Simpson index). Community richness (p = 0.06) did not differ significantly between groups, however, diversity and evenness were increased in Winnie (p < 0.001 for both) when compared to C57BL/6 mice (Figures 5A–C). For β-diversity metrics, the Bray-Curtis dissimilarity was calculated as 0.37, indicating a 37% difference in the relative abundance of genera between fecal samples from C57BL/6 and Winnie mice. At the genus level, the abundance of microbiota in samples from Winnie mice was distinctly different to samples from C57BL/6 mice, including substantial increases (FC > 10) in Turicibacter, unknown Christensenellaceae, unknown Clostridiaceae, Blautia, Butyrivibrio, Clostridium (Lachnospiraceae family), Ruminococcus (Lachnospiraceae family), unknown Desulfovibrionaceae, unknown ML615J-28, and Akkermansia, as well as decreased Butyricicoccus (Table 2). Since this study aimed to investigate whether there is a correlation between changes in tryptophan metabolites and changes in intestinal microbiota involved in tryptophan and nicotinamide metabolism in Winnie mice, we identified genus level tryptophan and nicotinamide metabolism-associated microbiota from previous studies, including studies in IBD and colitis (23, 24, 61–67). Analysis revealed dysbiosis of microbiota involved in tryptophan and nicotinamide metabolism in fecal samples from Winnie mice compared to samples from C57BL/6 mice (Table 2, Figure 5D). Key changes comprised large-scale decreases in the abundance of Lactobacillus (p < 0.001) and Bifidobacterium (p < 0.05), as well as increased abundance of Akkermansia (p < 0.001) in Winnie mice feces, demonstrating clear alterations in the composition of bacteria that contribute to the synthesis and/or degradation of tryptophan metabolites. We also observed increased abundance of certain indole-producing bacteria, including Bacteroides (p < 0.001), Prevotella (p < 0.01), Ruminococcus (Ruminococcaceae family) (p < 0.05), and Oscillospira (p < 0.05) in samples from Winnie mice, whereas the abundance of others, such as Enterococcus (p < 0.05), and Butyricicoccus (p < 0.01) decreased. Similarly, there were changes in the abundance of indole-3-lactic acid (ILA), indole-3-propionic acid (IPA), and indole-3-lactic acid (IAA) synthesizing bacteria in Winnie mice feces, including increases in Parabacteroides (p < 0.001) and Coprococcus (p < 0.05), while the abundance of [Prevotella] (p < 0.01) was reduced. Streptococcus, a 5-HT-producing microbe was decreased in samples from Winnie mice, while Turicibacter, a bacterium that directly imports 5-HT through a mechanism similar to a mammalian serotonin transporter, was significantly increased (p < 0.05 for both). Furthermore, the abundance of tryptamine-synthesizing Blautia (p < 0.05) and Ruminococcus (Lachnospiraceae family) (p < 0.01) were increased in samples from Winnie mice when compared to samples from C57BL/6 mice. These results highlight colitis-associated changes in tryptophan and nicotinamide metabolism in the microbiome; however, it is difficult to ascertain whether microbiota involved in these pathways are more affected than those involved in other pathways considering genera may have unknown functions participating in tryptophan metabolism that are yet to be reported in the literature.
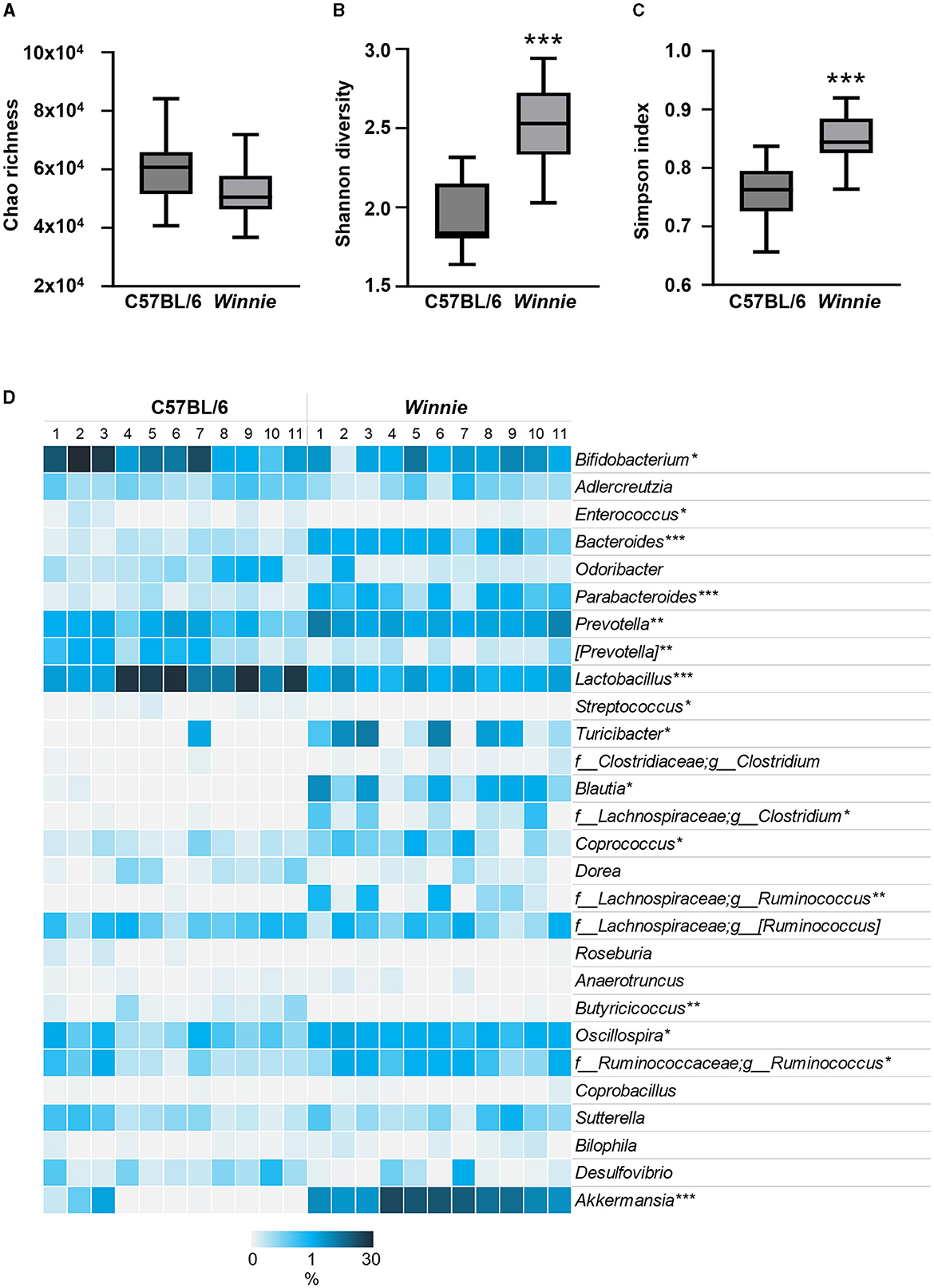
Figure 5. Changes to tryptophan and nicotinamide metabolism-associated microbiota at the genus level in fecal samples from Winnie mice. Microbiota communities were profiled from C57BL/6 and Winnie mice (n = 11/group). (A) Chao's richness estimate, (B) community diversity assessed by calculating the Shannon diversity, and (C) evenness by Simpson index based on abundance data. (D) Heat map representation of the percent relative abundance of tryptophan and nicotinamide metabolism-associated microbiota at the genus level in fecal samples from Winnie compared to C57BL/6 mice. Individual samples from C57BL/6 and Winnie mice are visualized in columns and genus level microbiota involved in tryptophan metabolism are represented by rows. Percent relative abundance was calculated by the number of ASVs by taxonomic classification at the genus level relative to the total number of ASVs per sample. Asterisks mark significant differences in the mean percent relative abundance between groups. *p<0.05, **p<0.01, ***p<0.001 when compared to C57BL/6 mice.
RNA-Seq was performed to determine changes in tryptophan and nicotinamide metabolism-associated gene expressions in the brains of Winnie mice (n = 6) vs. age-matched C57BL/6 littermates (n = 8). Visualization of the KEGG tryptophan and nicotinamide metabolism pathways showed changes to gene expressions in the Winnie mouse brain compared to brains from C57BL/6 mice (Figures 6, 7). Analysis of the transcriptome data in Winnie mouse brains revealed 11 DEGs associated with tryptophan metabolism compared to brains from C57BL/6 mice. Amine metabolic process was predisposed by the upregulation of Afmid, L-amino acid oxidase/monooxygenase (Laao), amine oxidase copper containing 1 (Aoc1), and cytochrome P450 family 1 subfamily A1 (Cyp1a1). Aromatic amino acid family metabolic and indole-containing compound processes were implicated by upregulated L-tryptophan hydroxylase (Tph1), Afmid, and Laao. The upregulated Afmid and Laao are involved in the tryptophan catabolic process, whereas upregulated Tph1, Afmid, dehydrogenase E1 and transketolase domain containing 1 (Dhtkd1), Laao, and Cyp1a1 affect carboxylic acid metabolic process in fatty acid pathways. The downregulation of Ido1 associates with the conversion of tryptophan to kynurenine. Both hydroxyacyl-CoA dehydrogenase (Hadh) and aldehyde dehydrogenase (NAD+) (Aldh2) are involved in the degradation pathways of fatty acids and branched-chain amino acids (valine, leucine, and isoleucine). Downregulation of Ido1, Hadh, and Aldh2 suggested a decrease in metabolic capacity and impacted oxidoreductase activity, affecting redox-sensitive signaling pathways. There were four DEGs associated with the nicotinamide metabolism pathway in the Winnie mouse brains when compared to brains from C57BL/6 mice. Aldehyde oxidase (Aox1) and Nmrk1 were upregulated, enhancing broader metabolic processes, while downregulated Pnp indicates impaired pyrimidine and purine metabolism.
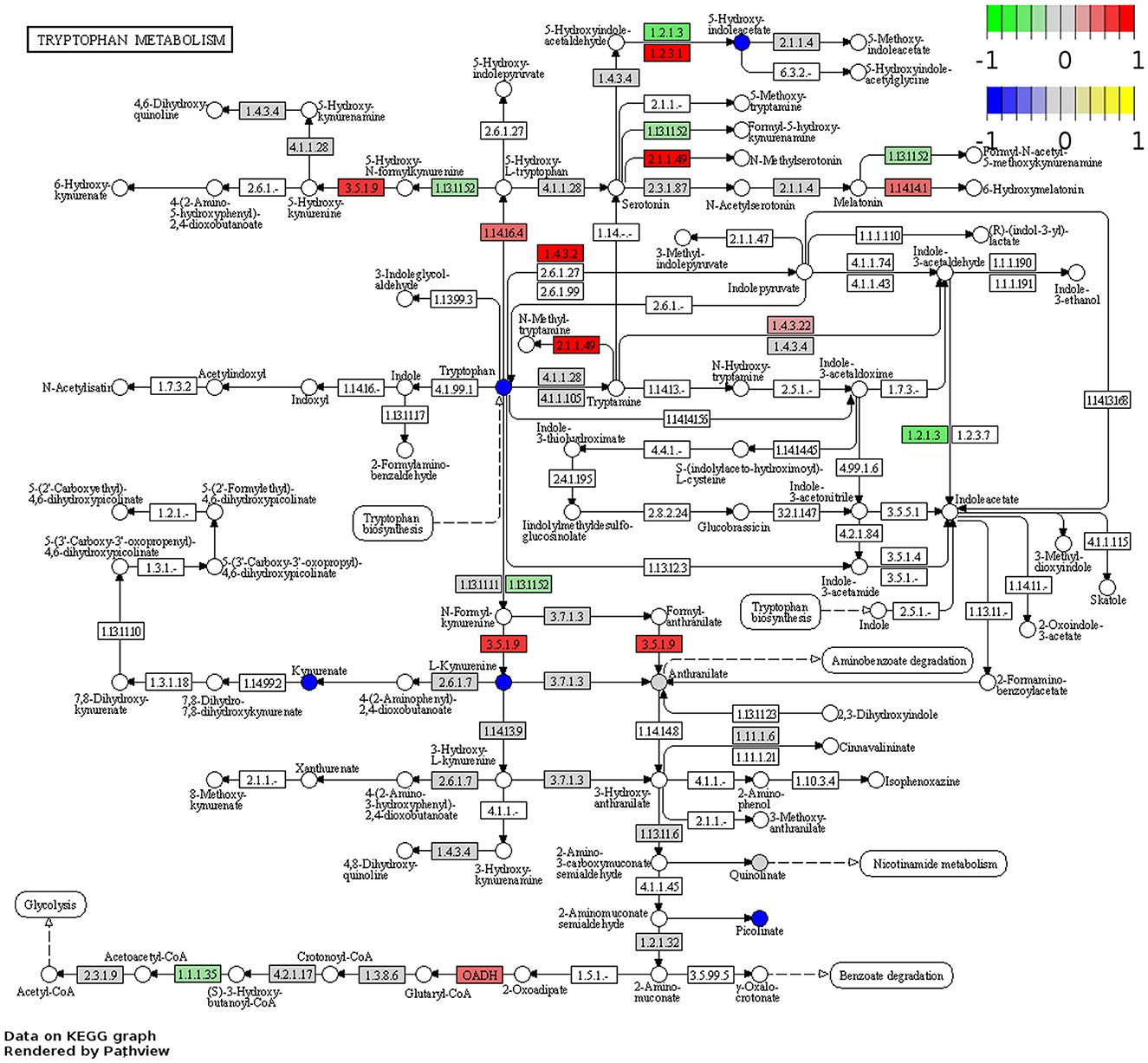
Figure 6. DEGs associated with tryptophan metabolism in the Winnie mouse brain. Pathview plot for the tryptophan metabolism KEGG pathway analysis of brain samples from Winnie mice (n = 6) vs. C57BL/6 littermates (n = 8). DEGs of the tryptophan metabolism KEGG pathways are colored according to their sign Log2FC × Log10P value changes. Rectangular nodes indicate gene expression data determined by RNA-seq. Positive values (red) indicate genes that are upregulated (tryptophan—EC:3.5.1.9, Afmid; EC:2.1.1.49, SAMe; EC:1.4.3.22, Aoc1; EC:1.14.16.4, TpH1; EC:1.4.3.2, Laao; EC:1.2.3.1, Aox; OADH, Dhtkd1; and EC:1.14.14.1, Cyp1a1, nicotinamide—EC:2.7.1.22 and EC:2.7.1.173, Nmrk1, and EC:1.2.3.1, Aox) and negative values (green) indicate genes that are downregulated (tryptophan—EC:1.13.11.52, Ido1, EC:1.2.1.3, Aldh2; EC:1.1.1.35, Hadh, nicotinamide—EC:2.4.2.1 Pnp; EC:3.2.2.6 and EC:249920, Cd38) in the Winnie mouse brain relative to brains from C57BL/6 mice. Circular nodes indicate the relative expression of tryptophan metabolism compounds.
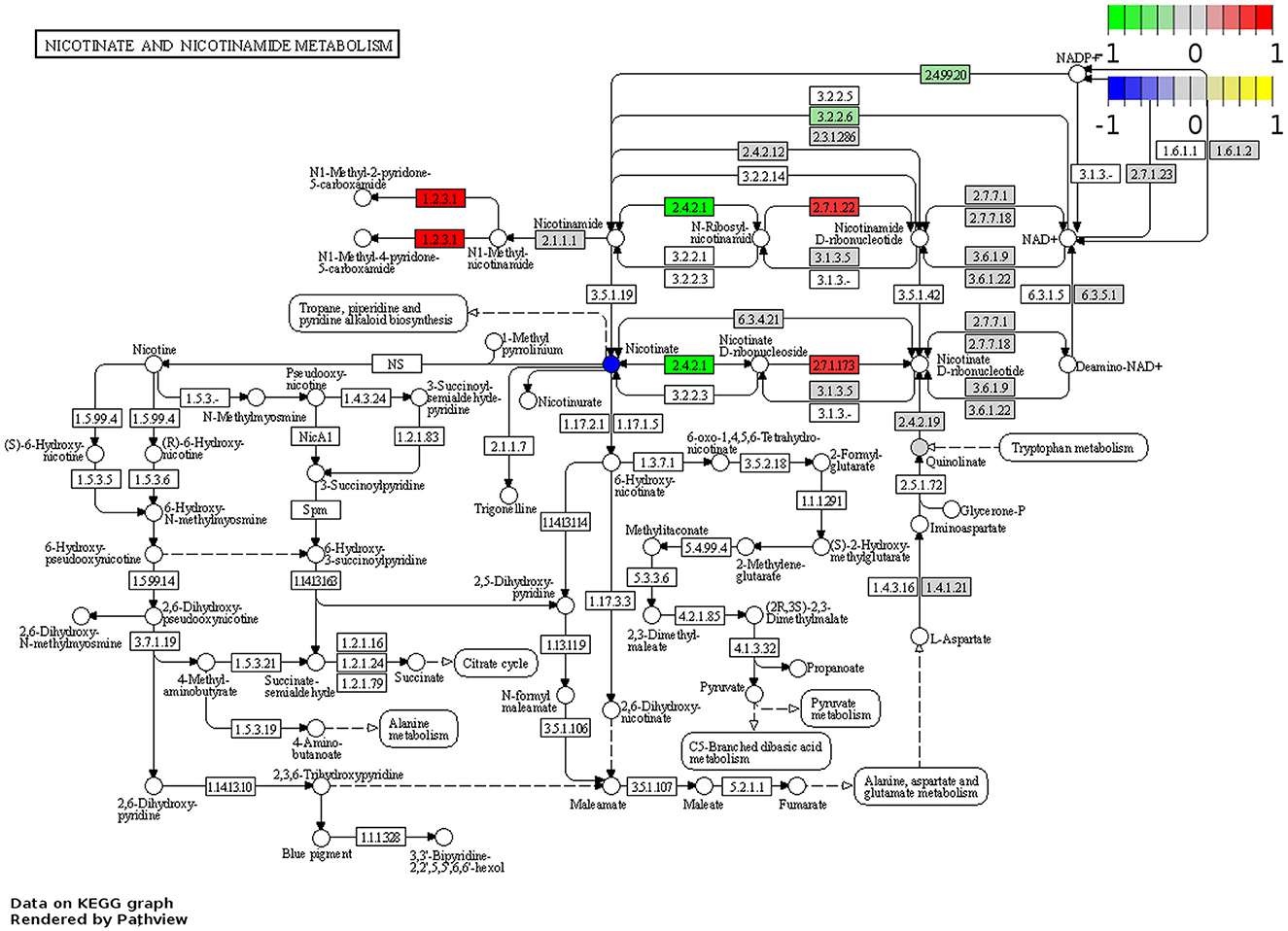
Figure 7. DEGs associated with nicotinamide metabolism in the Winnie mouse brain. Pathview plot for the nicotinate and nicotinamide metabolism KEGG pathway analysis of brain samples from Winnie mice (n = 6) vs. C57BL/6 littermates (n = 8). DEGs of the nicotinamide metabolism pathways are colored according to their sign Log2FC × Log10P value changes. Rectangular nodes indicate gene expression data determined by RNA-seq. Red represents upregulated gene expressions and green indicates genes that are downregulated in the Winnie mouse brain relative to brains from C57BL/6 mice. Circular nodes indicate the relative expression of nicotinamide metabolism compounds.
Metabolomics analysis was used to quantify tryptophan metabolite concentrations, including tryptophan, 5-HIAA, KYNA, kynurenine, QUIN, nicotinamide, and PIC, in the brains of Winnie mice and compared to brain samples from C57BL/6 littermates (n = 5–8 for both, Table 3, Figure 8). In the Winnie mouse brains, there were significantly lower concentrations of tryptophan (p < 0.001), 5-HIAA (p < 0.01), kynurenine (p < 0.05), KYNA (p < 0.01), PIC (p < 0.05), and nicotinamide (p < 0.05) when compared to brain samples from C57BL/6 mice. There was no difference in the concentration of QUIN between groups (Table 3, Figure 8). Changes in the concentration levels of tryptophan metabolites are reflected in changes in the expression of genes involved in these pathways.
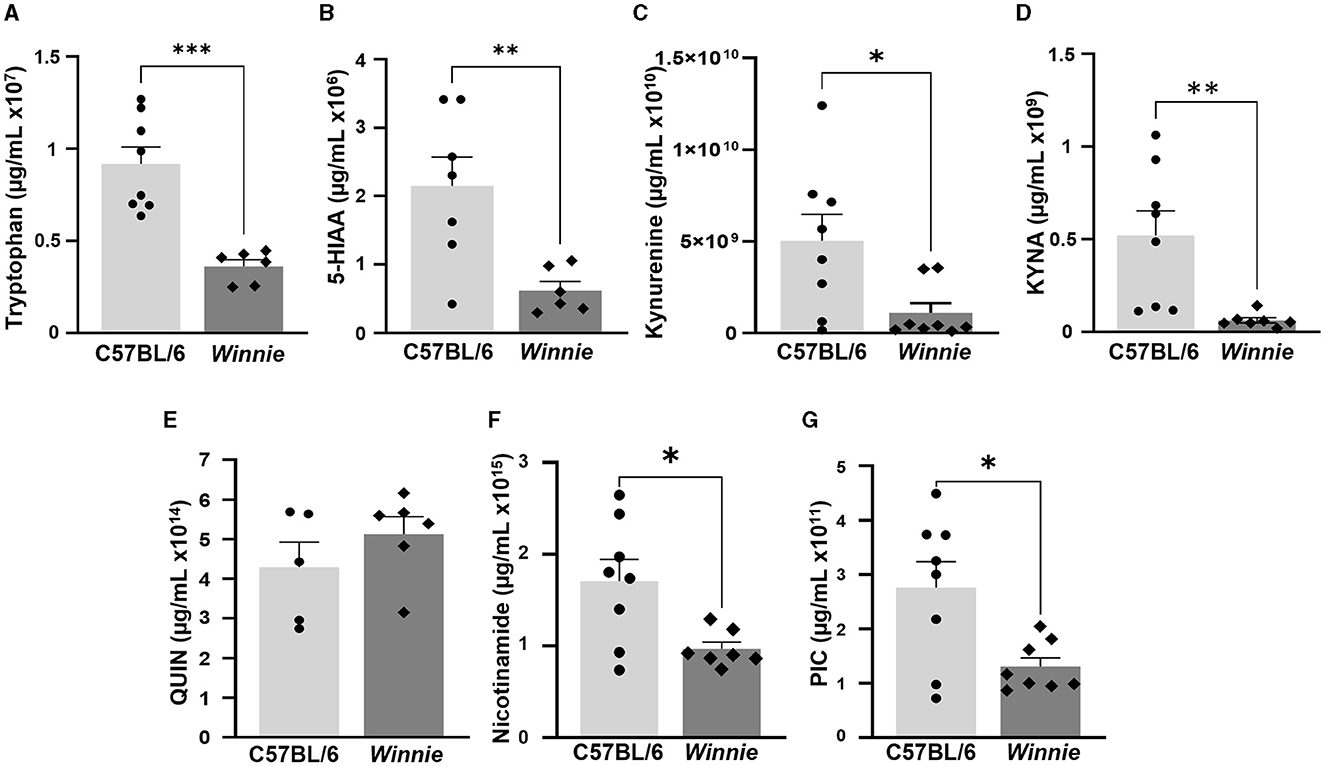
Figure 8. Changes in tryptophan metabolite concentrations in Winnie mouse brains. Quantification of tryptophan metabolites (μg/ml) measured by UFLC in Winnie mouse brains compared to brains from C57BL/6 mice (n = 5–8 for both). (A) Tryptophan, (B) 5-HIAA, (C) Kynurenine, (D) KYNA, (E) QUIN, (F) Nicotinamide, and (G) PIC. Data is expressed as mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001 when compared to C57BL/6 mice.
In the current study, we used Winnie mice, an experimental model of spontaneous chronic colitis established as highly representative of IBD, to identify changes in tryptophan and nicotinamide metabolism-associated genes and metabolites in the colon and brain, as well as explore alterations in the abundance of fecal microbiota involved in tryptophan metabolism. Our results demonstrated dysregulated tryptophan metabolism and de novo NAD+ synthesis corresponding to dysbiosis of the microbiota, indicating disruptions to the gut-brain axis associated with chronic intestinal inflammation.
Consistent with our findings of upregulated Ido1 expression in the Winnie mouse colon, IDO1 is increased in tissues from IBD patients and other animal models of colitis in response to inflammation (68–72). Upregulation of Ido1 corresponds to increased concentrations of tryptophan and kynurenine in colons from Winnie mice in this study, given that IDO1 is the first and rate-limiting step in tryptophan catabolism along the kynurenine pathway (20). Overexpression of IDO1 and subsequent increased kynurenine levels are implicated in inhibiting reactive effector T and natural killer (NK) cell responses, which are dysregulated in Winnie mice (37, 73). Previous studies have shown that species belonging to Lactobacillus and Bifidobacterium may shift the host tryptophan metabolism by suppressing the kynurenine pathway (24, 74). Specifically, Lactobacillus can reduce IDO1 activity in intestinal epithelial cells and decrease the production of kynurenine compounds (75, 76). Therefore, a reduction in the abundance of Lactobacillus and Bifidobacterium in fecal samples from Winnie mice in this study may be correlated to the upregulation of Ido1 and increased kynurenine in the distal colon. It is widely accepted that the kynurenine metabolism pathway of IBD patients is shifted toward QUIN compared to the healthy population (15). This suggests that the upregulation of Kynu observed in Winnie mice colons in this study promoted the conversion of kynurenine toward QUIN, a neurotoxic metabolite, which was correspondingly increased in the colons from Winnie mice. QUIN can increase the expression of N-methyl D-aspartate receptor 2B in the ENS and, in turn, leads to intestinal symptoms and mood disorders associated with gut inflammation (77). Furthermore, increased QUIN may be correlated with disease severity and a pro-inflammatory environment in the Winnie mouse colon, as previously demonstrated in mice with DSS-induced colitis (78). Although gut microbial phyla, such as Firmicutes and Bacteroidetes, contain organisms that encode for the Kynu gene (79), further meta-transcriptomic studies are required to determine any association between changes in microbial abundance and upregulation of Kynu in the Winnie mouse colon observed in this study.
Differential expression of several tryptophan metabolism-associated genes in the Winnie mouse colon, is also associated with oxidative stress, a pathophysiologic factor implicated in colonic inflammation in patients with IBD and Winnie mice (41, 80). For instance, Mao-b, involved in the production of hydrogen peroxide, was upregulated in the Winnie mouse, consistent with increased levels of the Mao substrate, tyramine, previously reported in fecal samples from Winnie mice (40). Elevated levels of tyramine and 5-HIAA are known to affect GI dysmotility (81, 82). In this study, the concentration of 5-HIAA was increased in the Winnie mouse colon, corresponding to upregulated Mao-b expression, as well as altered colonic function and impaired smooth muscle contractility previously shown in this model (35, 39). Additionally, our finding of downregulated Afmid in the Winnie mouse colon is supported by increased aldehyde, butanal, and propanol in Winnie mice feces (40). This aberration may be initiated by the upregulation of Mao-b and Aldh2 expression described in this study, thereby increasing the susceptibility of the colon to aldehyde-induced tissue damage (83). Correspondingly, our finding of downregulated Dhtkd1 may be associated with impaired mitochondrial function and increased reactive oxygen species (ROS) production, which have been previously described in the Winnie mouse colon (41, 84). Although levels of tryptophan are generally proportional to the expression of Dhtkd1 (85), the tryptophan concentration in the Winnie mouse colon was increased. Nicotinate interacts with the tryptophan pathway of de novo NAD+ biosynthesis from QUIN, which was also increased in Winnie mice colons in this study. Since this pathway is an alternative to the tryptophan degradation to 2-oxoadipate, reduced Dhtkd1 expression may increase the tryptophan flux to NAD+ synthesis (85).
In this study, the downregulation of S-adenosyl-L-methionine (SAMe) in the Winnie mouse colon may suggest an enhanced bacterial production of anionic sulfide, culminating in acute oxidative damage (86, 87). Alterations in sulfur metabolism and subsequent overproduction of hydrogen sulfide can be driven by Akkermansia, which were increased in fecal samples from Winnie mice in this study (88). Furthermore, reduced Bifidobacteria, [Prevotella], and Lactobacillus may correspond to the downregulation of SAMe in the Winnie mouse colon as decreased folate, butyrate, and acetate-producing microbiota can induce changes in host cell DNA methylation patterns via altered synthesis of epigenetically active metabolites necessary for generation of SAMe (89). Downregulation of L-tryptophan decarboxylase (Ddc) is reported to constrain the enzymatic activity of GI microbes, causing an imbalance in intestinal microbiome composition (62). Importantly, the synthesis of tryptamine is catalyzed by Ddc expressed by gut microbiota, such as members of the Lactobacillus, Clostridia, and Ruminococcus genera (63). In this study, downregulated Ddc may be associated with changes in the abundance of these bacteria in fecal samples from Winnie mice, reflecting disrupted tryptamine synthesis. However, further association analyses are required to confirm potential correlations. Consistent with our findings of downregulated Aoc1 in colons from Winnie mice, previous studies have reported decreased AOC1 in colon tissues from patients with UC (90, 91). Since decreased AOC1 diverts ornithine metabolism away from putrescine, our finding is substantiated by reduced putrescine levels previously reported in fecal samples from Winnie mice (40). Decreased diamine oxidase (DAO) activity is associated with changes to the abundance of Bifidobacterium, while certain strains of Lactobacillus are considered probiotic for DAO deficiency (92). Therefore, the downregulation of Aoc1 in the Winnie mouse colon may be correlated with the reduced abundance of Bifidobacterium and Lactobacillus observed in this study.
Our study revealed differential expression of genes involved in nicotinate and nicotinamide metabolism in the distal colon of Winnie mice. The upregulation of Nampt is supported by previous studies reporting elevated Nampt in colon tissues from IBD patients and DSS-induced colitis mice counteracting an increased cellular NAD turnover mediated by NAD-depleting enzymes (93–95). Nampt upregulation initiates activation of the nuclear factor kappa light chain enhancer of activated B cells (NF-kB) signaling pathway in the IL-6 and TNF-α feedback loop, leading to an escalation in IDO1 activity (20). This correlates to the upregulation of Ido1 and associated changes to microbial abundance observed in this study, thereby substantiating a mechanistic association between tryptophan catabolism and NAD+ synthesis in the Winnie mouse colon (96). Upregulation of Art2a amplifies the inflammatory/immune process (97). This contributes to depleted NAD+ availability, compromising mitochondrial function and triggering the release of ROS (98–100), consistent with the pathological conditions observed in Winnie mice (37). Furthermore, adenosine diphosphate (ADP)-ribosyltransferases are widely prevalent in the human gut microbiome and highly abundant in Bacteroides (101), thus the upregulation of ADP-ribosyltransferase 2a (Art2a) in the colon may be associated with the increased abundance of Bacteroides in fecal samples from Winnie mice in this study. Downregulation of Nmrk1 and Nmrk2 observed in the Winnie mouse colon may contribute to inflammation-associated ENS damage reported in a previous study (38), as in homeostatic conditions, Nmrk allosterically upregulates to safeguard neurons against axonopathy if NAD+ is depleted or under attack by NAD-consuming enzymes (102). Reduced expression of Naprt observed in our study can inhibit the production of NAD+ and prevent the protection of colon tissues from oxidative stress (103). The colitis in Winnie mice exhibits manifestations closely resembling IBD, such as tight junction damage, early intestinal cell death, and cellular stress responses (37, 39, 41, 104–109), which may be linked to glutamine deficiency. Nadsyn1, the gene encoding for a glutamate-dependent enzyme, was downregulated in Winnie mice, which suggests a response to glutamine deficiency (110). This notion is supported by a study in acute DSS-induced colitis mice that found glutamine sufficiency is required for the colonic epithelium to develop a cell-protective, antiapoptotic, and anti-inflammatory response to inflammatory damage (111).
Tryptophan metabolism is intricately linked with behavioral modulation, affecting mood, sleep, appetite, and cognitive function, therefore alterations in tryptophan metabolism-associated genes and tryptophan metabolites in the Winnie mouse brain demonstrate disruption of the gut-brain axis and may indicate behavioral changes associated with neurodegeneration and depression (25). Of significance, is the downregulation of Ido1 and Aldh2 in brains from Winnie mice, given IDO1's association with exploratory behavior, cognitive function, and depression (112–114), and Aldh2's role in detoxifying ROS and ethanol metabolism, processes relevant to neurodegeneration (115, 116). The reduction in Ido1 activity within the brain may serve as a protective mechanism to inhibit the over-degradation of tryptophan, an essential precursor to 5-HIAA (117). Since 5-HIAA was lower in Winnie mice brains, such regulation may ensure that serotonin levels remain sufficient to support standard brain functionality during systemic inflammation (112). In consistency with our findings, a reduced level of 5-HIAA in brains from mice with DSS-induced colitis is correlated with a decreased abundance of [Prevotella] and an increased abundance of Ruminococcus, indicating a close association between the gut microbiome and production of brain neurotransmitters (118). Additionally, Laao1 upregulation in Winnie mice brains may alter the ratio of tryptophan to large neutral amino acids (LNAA) influencing serotonin synthesis (119). This is compounded by reduced concentrations of both tryptophan and 5-HIAA in the brains of Winnie mice in this study. The upregulation of Tph1 in brains from Winnie mice is related to stress sensitivity and increased susceptibility to psychiatric pathologies (120), which are prevalent in IBD patients (121). Two of the dominant phyla, Firmicutes and Proteobacteria, both contain species with genes encoding TPH1 (79) and Lactobacillus has been reported to promote butyrate production, ultimately stimulating TPH1 activity by butyrate and regulating serotonin synthesis (61). However, further studies are required to establish any correlations between the upregulation of Tph1 and changes to the abundance of these microbiota in this study.
Differential expression of tryptophan metabolism-associated genes may indicate compensatory mechanisms with potential deleterious effects to preserve overall brain health in Winnie mice. Given that Aldh2, an enzyme involved in detoxification, is downregulated, the upregulation of Afmid, known to play a role in toxin elimination, could indicate action to restore some equilibrium to brain chemistry in Winnie mice (115, 122). Nonetheless, Aox was upregulated, indicating oxidative stress in the brains of Winnie mice. Plausibly, in an adaptive effort to optimize mitochondrial activity, Winnie mice allosterically upregulated Dhtkd1. However, variations in the Dhtkd1 gene have the potential to disrupt tryptophan metabolism and the equilibrium of neuroactive metabolites, contributing to the onset of depression or other neurological disorders (84). Additionally, upregulated SAMe in Winnie mice brains may be indicative of disturbed biological rhythms, by inhibiting methylation processes (123).
Several factors contributing to the dysregulation of de novo NAD+ synthesis were identified in the Winnie mouse brains, including reduced nicotinamide metabolites. While the overall activity of NAD+-consuming enzymes remained largely unaltered, downregulated Cd38 expression may be associated with lower levels of nicotinamide in Winnie in mice brains, potentially leading to behavioral changes previously observed in niacin deficient and Cd38−/− mice (124). Reduced nicotinamide has been associated with depression, anxiety, and psychological stress, as well as contributing to neurodegeneration and compromising cellular defense mechanisms against stress and injury (125–127). Despite downregulated Cd38, the underlying cause of the reduced nicotinamide levels in Winnie mice brains remains unclear. However, lower levels of nicotinamide aligned with the upregulation of Nmrk1, an enzyme known to increase its activity in response to NAD+ depletion (128), and Pnp downregulation, a major component in nicotinamide riboside metabolism (129). Further investigations are required to elucidate the synthesis of NAD+ and its interplay with nicotinamide in Winnie mice brains.
Lower levels of tryptophan and kynurenine in Winnie mice brains can indicate disruption in transport mechanisms of these metabolites across the blood-brain barrier or failure to sufficiently compete with other large amino acids sharing the LNAA transporter (130). These findings are significant as the metabolism of tryptophan, via the kynurenine pathway, serves as a critical link between peripheral inflammation and central nervous system alterations (25). In contrast to our results, elevated kynurenine levels have been observed in humans with major depressive disorder (131), as well as in acute DSS-induced colitis mice (132). Acute inflammation in DSS-induced colitis stimulates the tryptophan-kynurenine-KYNA pathway in the brain (132); therefore, our findings suggest that chronic colitis in Winnie mice has a diminished impact on this pathway, given that neuroprotective KYNA concentration was reduced.
To further elucidate the complex interactions of tryptophan metabolism and its systemic effects in IBD, subsequent investigations should encompass a multifaceted approach. A detailed comparison of tryptophan concentrations in colonic tissue, plasma, and cerebral structures would provide a more holistic understanding of the metabolic disturbances characteristic of chronic intestinal inflammation. While RNA-Seq and metabolomics have provided a foundational descriptive analysis, indicating changes to the regulation of tryptophan and nicotinamide-associated genes and metabolites, there is a necessity to extend these findings through various biochemical and molecular techniques. Systems biology approaches could synthesize data from genomics, proteomics, and metabolomics to model the biological networks that underpin tryptophan metabolism, providing predictive insights and a systemic understanding of disease pathology. Furthermore, the application of multi-omic analysis could unravel complex interactions and feedback loops, giving a more comprehensive understanding of the disease mechanisms at play. Western blotting and mass spectrometry would enable the quantification and confirmation of the proteins of interest, potentially unveiling post-translational modifications and the impact of gene regulation on protein expression. Additionally, immunohistochemistry would allow for the localization of these proteins within tissue contexts, offering insight into the cellular and tissue-specific distribution and the pathological significance of these findings.
This study demonstrates alterations in tryptophan and nicotinamide metabolism-associated genes, microbiota, and metabolites in Winnie mice with spontaneous chronic intestinal inflammation. Our findings signify the role of the gut-brain axis in gut inflammation and further establish the Winnie mouse as an experimental model highly representative of human IBD. We report changes in gene expression that contribute to the shift of tryptophan metabolism toward the kynurenine pathway, impacting NAD+ synthesis and revealed the dualistic influence of 5-HIAA in intestinal health. These findings connect these metabolic pathways to behavioral, cognitive, and emotional challenges experienced by IBD patients and suggest that tryptophan metabolism may exert a greater influence on the kynurenine and serotonin pathway than previously recognized. The insights gathered from this study, along with forthcoming data, can be utilized to enhance the identification and management of IBD through the development of targeted therapeutic interventions. Furthermore, these findings provide a compelling foundation for undertaking further detailed investigations into the association between intestinal inflammation, tryptophan metabolism, and behavior.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/, GSE244558 (colon data), GSE264317 (for brain data) and https://github.com/Nurgali-lab/C57-Win-APX-16SV3-4.git, C57-Win-APX-16SV3-4.
The animal study was approved by Victoria University Animal Experimentation Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
JD: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing. AR: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. RS: Formal analysis, Writing – review & editing. MD: Data curation, Formal analysis, Writing – review & editing. ND: Data curation, Formal analysis, Writing – review & editing. RE: Data curation, Formal analysis, Writing – review & editing. DK: Supervision, Writing – review & editing. VA: Supervision, Writing – review & editing. KN: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Crohn's & Colitis Foundation Senior Research Award (KN, Award number: 903433) and Victoria University postgraduate research scholarships to JD and MD.
The authors would like to thank Mr. Larruceo Bautista for his assistance with UFLC equipment and data collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1379335/full#supplementary-material
Supplementary Figure S1. Heat map representation of gene lists for tryptophan and nicotinamide-associated gene expressions. Gene lists for tryptophan and nicotinamide-associated gene expressions were generated by merging related gene lists from multiple databases accessed via the molecular signature database (MSigDB).
1. Powell N, Walker MM, Talley NJ. The mucosal immune system: master regulator of bidirectional gut-brain communications. Nat Rev Gastroenterol Hepatol. (2017) 14:143–59. doi: 10.1038/nrgastro.2016.191
2. Fakhoury M, Negrulj R, Mooranian A, Al-Salami H. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res. (2014) 7:113–20. doi: 10.2147/JIR.S65979
3. Subasinghe D, Nawarathne NM, Samarasekara D. Disease characteristics of inflammatory bowel disease (IBD). J Gastrointest Surg. (2022) 2011:1562–7. doi: 10.1007/s11605-011-1588-5
4. Tan Z, Zhu S, Liu C, Meng Y, Li J, Zhang J, et al. Causal link between inflammatory bowel disease and fistula: evidence from mendelian randomization study. J Clin Med. (2023) 12:2482. doi: 10.3390/jcm12072482
6. Kim ER, Chang DK. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol. (2014) 20:9872–81. doi: 10.3748/wjg.v20.i29.9872
7. Dubinsky MC, Dotan I, Rubin DT, Bernauer M, Patel D, Cheung R, et al. Burden of comorbid anxiety and depression in patients with inflammatory bowel disease: a systematic literature review. Expert Rev Gastroenterol Hepatol. (2021) 15:985–97. doi: 10.1080/17474124.2021.1911644
8. Collins SM, Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. (2009) 136:2003–14. doi: 10.1053/j.gastro.2009.01.075
9. Gracie DJ, Hamlin PJ, Ford AC. The influence of the brain-gut axis in inflammatory bowel disease and possible implications for treatment. Lancet Gastroenterol Hepatol. (2019) 4:632–42. doi: 10.1016/S2468-1253(19)30089-5
10. de Souza HSP, Fiocchi C, Iliopoulos D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat Rev Gastroenterol Hepatol. (2017) 14:739–49. doi: 10.1038/nrgastro.2017.110
11. Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. (2007) 132:397–414. doi: 10.1053/j.gastro.2006.11.002
12. Coates MD, Tekin I, Vrana KE, Mawe GM. Review article: the many potential roles of intestinal serotonin (5-hydroxytryptamine, 5-HT) signalling in inflammatory bowel disease. Aliment Pharm Ther. (2017) 46:569–80. doi: 10.1111/apt.14226
13. Wyler SC, Lord CC, Lee S, Elmquist JK, Liu C. Serotonergic control of metabolic homeostasis. Front Cell Neurosci. (2017) 11:277. doi: 10.3389/fncel.2017.00277
14. Martin AM, Young RL, Leong L, Rogers GB, Spencer NJ, Jessup CF, et al. The diverse metabolic roles of peripheral serotonin. Endocrinology. (2017) 158:1049–63. doi: 10.1210/en.2016-1839
15. Chen L-M, Bao C-H, Wu Y, Liang S-H, Wang D, Wu L-Y, et al. Tryptophan-kynurenine metabolism: a link between the gut and brain for depression in inflammatory bowel disease. J Neuroinflamm. (2021) 18:135. doi: 10.1186/s12974-021-02175-2
16. Hyland NP, Cavanaugh CR, Hornby PJ. Emerging effects of tryptophan pathway metabolites and intestinal microbiota on metabolism and intestinal function. Amino Acids. (2022) 54:57–70. doi: 10.1007/s00726-022-03123-x
17. Shajib MS, Chauhan U, Adeeb S, Chetty Y, Armstrong D, Halder SLS, et al. Characterization of serotonin signaling components in patients with inflammatory bowel disease. J Can Assoc Gastroenterol. (2019) 2:132–40. doi: 10.1093/jcag/gwy039
18. Abautret-Daly Á, Dempsey E, Parra-Blanco A, Medina C, Harkin A. Gut–brain actions underlying comorbid anxiety and depression associated with inflammatory bowel disease. Acta Neuropsychiatr. (2018) 30:275–96. doi: 10.1017/neu.2017.3
19. Gershon MD. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Curr Opin Endocrinol Diabetes Obes. (2013) 20:14–21. doi: 10.1097/MED.0b013e32835bc703
20. Ciorba MA. Indoleamine 2,3 dioxygenase in intestinal disease. Curr Opin Gastroenterol. (2013) 29:146–52. doi: 10.1097/MOG.0b013e32835c9cb3
21. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
22. Casén C, Vebø HC, Sekelja M, Hegge FT, Karlsson MK, Ciemniejewska E, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther. (2015) 42:71–83. doi: 10.1111/apt.13236
23. Bosi A, Banfi D, Bistoletti M, Giaroni C, Baj A. Tryptophan metabolites along the microbiota-gut-brain axis: an interkingdom communication system influencing the gut in health and disease. Int J Tryptophan Res. (2020) 13:1178646920928984. doi: 10.1177/1178646920928984
24. Gao K, Mu CL, Farzi A, Zhu WY. Tryptophan metabolism: a link between the gut microbiota and brain. Adv Nutr. (2020) 11:709–23. doi: 10.1093/advances/nmz127
25. Roth W, Zadeh K, Vekariya R, Ge Y, Mohamadzadeh M. Tryptophan metabolism and gut-brain homeostasis. Int J Mol Sci. (2021) 22:2973. doi: 10.3390/ijms22062973
26. Lamas B, Richard ML, Leducq V, Pham H-P, Michel M-L, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. (2016) 22:598. doi: 10.1038/nm.4102
27. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. (2017) 551:648–52. doi: 10.1038/nature24661
28. Sofia MA, Ciorba MA, Meckel K, Lim CK, Guillemin GJ, Weber CR, et al. Tryptophan metabolism through the kynurenine pathway is associated with endoscopic inflammation in ulcerative colitis. Inflamm Bowel Dis. (2018) 24:1471–80. doi: 10.1093/ibd/izy103
29. Johnson S, Imai S-I. NAD (+) biosynthesis, aging, and disease. F1000Res. (2018) 7:132. doi: 10.12688/f1000research.12120.1
30. Dobrovolsky VN, Bowyer JF, Pabarcus MK, Heflich RH, Williams LD, Doerge DR, et al. Effect of arylformamidase (kynurenine formamidase) gene inactivation in mice on enzymatic activity, kynurenine pathway metabolites and phenotype. Biochim Biophys Acta. (2005) 1724:163–72. doi: 10.1016/j.bbagen.2005.03.010
31. Zhang X, Liu X, Zhou W, Du Q, Yang M, Ding Y, et al. Blockade of IDO-Kynurenine-AhR axis ameliorated colitis-associated colon cancer via inhibiting immune tolerance. Cell Mol Gastroenterol Hepatol. (2021) 12:1179–99. doi: 10.1016/j.jcmgh.2021.05.018
32. Sun M, Ma N, He T, Johnston LJ, Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr. (2020) 60:1760–8. doi: 10.1080/10408398.2019.1598334
33. Wang G, Fan Y, Zhang G, Cai S, Ma Y, Yang L, et al. Microbiota-derived indoles alleviate intestinal inflammation and modulate microbiome by microbial cross-feeding. Microbiome. (2024) 12:59. doi: 10.1186/s40168-024-01750-y
34. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. (2018) 23:716–24. doi: 10.1016/j.chom.2018.05.003
35. Stavely R, Fraser S, Sharma S, Rahman AA, Stojanovska V, Sakkal S, et al. The onset and progression of chronic colitis parallels increased mucosal serotonin release via enterochromaffin cell hyperplasia and downregulation of the serotonin reuptake transporter. Inflamm Bowel Dis. (2018) 24:1021–34. doi: 10.1093/ibd/izy016
36. Heazlewood CK, Cook MC, Eri R, Price GR, Tauro SB, Taupin D, et al. Aberrant mucin assembly in mice causes endoplasmic reticulum stress and spontaneous inflammation resembling ulcerative colitis. PLoS Med. (2008) 5:e54. doi: 10.1371/journal.pmed.0050054
37. Eri RD, Adams RJ, Tran TV, Tong H, Das I, Roche DK, et al. An intestinal epithelial defect conferring ER stress results in inflammation involving both innate and adaptive immunity. Mucosal Immunol. (2011) 4:354–64. doi: 10.1038/mi.2010.74
38. Rahman AA, Robinson AM, Jovanovska V, Eri R, Nurgali K. Alterations in the distal colon innervation in Winnie mouse model of spontaneous chronic colitis. Cell Tissue Res. (2015) 362:497–512. doi: 10.1007/s00441-015-2251-3
39. Robinson AM, Rahman AA, Carbone SE, Randall-Demllo S, Filippone R, Bornstein JC, et al. Alterations of colonic function in the Winnie mouse model of spontaneous chronic colitis. Am J Physiol Gastrointest Liver Physiol. (2017) 312:G85–G102. doi: 10.1152/ajpgi.00210.2016
40. Robinson AM, Gondalia S, Karpe A, Eri R, Beale D, Morrison P, et al. Fecal microbiota and metabolome in a mouse model of spontaneous chronic colitis: relevance to human inflammatory bowel disease. Inflamm Bowel Dis. (2016) 22:2767–87. doi: 10.1097/MIB.0000000000000970
41. Sahakian L, Filippone RT, Stavely R, Robinson AM, Yan XS, Abalo R, et al. Inhibition of APE1/REF-1 redox signaling alleviates intestinal dysfunction and damage to myenteric neurons in a mouse model of spontaneous chronic colitis. Inflamma Bowel Dis. (2021) 27:388–406. doi: 10.1093/ibd/izaa161
42. Filippone R, Dargahi N, Eri R, Uranga-Ocio J, Bornstein J, Apostolopoulos V, et al. Potent CCR3 receptor antagonist, SB328437, suppresses colonic eosinophil chemotaxis and inflammation in the winnie murine model of spontaneous chronic colitis. Int J Mol Sci. (2022) 23:7780. doi: 10.3390/ijms23147780
43. Uhlig HH, Muise AM. Clinical genomics in inflammatory bowel disease. Trends Genet. (2017) 33:629–41. doi: 10.1016/j.tig.2017.06.008
44. Tsyganov K, Perry AJ, Archer SK, Powell D. RNAsik: a pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. JOSS. (2018) 3:583. doi: 10.21105/joss.00583
45. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
46. Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. (2019) 47:W199–205. doi: 10.1093/nar/gkz401
47. Powell D. Degust: Interactive RNA-seq Analysis. Drpowell/Degust 4.1.1 (4.1.1). Zenodo; Geneva (2019).
48. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
49. Kanehisa M, Goto S, KEGG. kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28:27–30. doi: 10.1093/nar/28.1.27
50. Luo W, Brouwer C. Pathview: an R/bioconductor package for pathway-based data integration and visualization. Bioinformatics. (2013) 29:1830–1. doi: 10.1093/bioinformatics/btt285
51. Stavely R, Robinson AM, Fraser S, Filippone RT, Stojanovska V, Eri R, et al. Bone marrow-derived mesenchymal stem cells mitigate chronic colitis and enteric neuropathy via anti-inflammatory and anti-oxidative mechanisms. Sci Rep. (2024) 14:6649. doi: 10.1038/s41598-024-57070-6
52. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. (2013) 41(Database issue):D991–5. doi: 10.1093/nar/gks1193
53. Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. (2002) 30:207–10. doi: 10.1093/nar/30.1.207
54. Peters LA, Perrigoue J, Mortha A, Iuga A, Song WM, Neiman EM, et al. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat Genet. (2017) 49:1437–49. doi: 10.1038/ng.3947
55. Borman P, Elder D. Q2(R1) Validation of analytical procedures. In: Teasdale A, Elder D, Nims RW, , editors. ICH Quality Guidelines. Hoboken, NJ: Wiley (2017), p. 127–66. doi: 10.1002/9781118971147.ch5
56. Guidance for Industry, Bioanalytical Method Validation. Center for Drug Evaluation and Research (CEDER). Silver Spring, MD: US Department of Health and Human Services, Food and Drug Administration (2001).
57. Veerasamy R, Sivadasan S, Karupiah S, Rajak H. Validation of analytical methods - strategies & importance. Int J Pharm Sci. (2010) 2:18–22.
58. Amjad M, Sarfraz S. A comprehensive review on HPLC method development, validation, optimization in pharmaceuticals. J Waste Manage Xenobio. (2023) 6:1–9. doi: 10.23880/oajwx-16000188
59. Cseh EK, Veres G, Szentirmai M, Nánási N, Szatmári I, Fülöp F, et al. HPLC method for the assessment of tryptophan metabolism utilizing separate internal standard for each detector. Anal Biochem. (2019) 574:7–14. doi: 10.1016/j.ab.2019.03.005
60. Li P, Bartlett MG. A review of sample preparation methods for quantitation of small-molecule analytes in brain tissue by liquid chromatography tandem mass spectrometry (LC-MS/MS). Anal Methods. (2014) 6:6183–207. doi: 10.1039/C4AY00915K
61. Hou Y, Li J, Ying S. Tryptophan metabolism and gut microbiota: a novel regulatory axis integrating the microbiome, immunity, and cancer. Metabolites. (2023) 13:1166. doi: 10.3390/metabo13111166
62. Williams Brianna B, Van Benschoten Andrew H, Cimermancic P, Donia Mohamed S, Zimmermann M, Taketani M, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. (2014) 16:495–503. doi: 10.1016/j.chom.2014.09.001
63. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:3294. doi: 10.1038/s41467-018-05470-4
64. Tran SM, Mohajeri MH. The role of gut bacterial metabolites in brain development, aging and disease. Nutrients. (2021) 13:732. doi: 10.3390/nu13030732
65. Kai L, Zheng W, Brandilyn AP, David BH, Tao W, Christopher CS, et al. Tryptophan metabolism, gut microbiota, and carotid artery plaque in women with and without HIV infection. AIDS. (2024) 38:223–33. doi: 10.1097/QAD.0000000000003596
66. Yin J, Song Y, Hu Y, Wang Y, Zhang B, Wang J, et al. Dose-dependent beneficial effects of tryptophan and its derived metabolites on akkermansia in vitro: a preliminary prospective study. Microorganisms. (2021) 9:1511. doi: 10.3390/microorganisms9071511
67. Yang F, DeLuca JAA, Menon R, Garcia-Vilarato E, Callaway E, Landrock KK, et al. Effect of diet and intestinal AhR expression on fecal microbiome and metabolomic profiles. Microb Cell Fact. (2020) 19:219. doi: 10.1186/s12934-020-01463-5
68. Gurtner GJ, Newberry RD, Schloemann SR, McDonald KG, Stenson WF. Inhibition of indoleamine 2,3-dioxygenase augments trinitrobenzene sulfonic acid colitis in mice. Gastroenterology. (2003) 125:1762–73. doi: 10.1053/j.gastro.2003.08.031
69. Wolf AM, Wolf D, Rumpold H, Moschen AR, Kaser A, Obrist P, et al. Overexpression of indoleamine 2,3-dioxygenase in human inflammatory bowel disease. Clin Immunol. (2004) 113:47–55. doi: 10.1016/j.clim.2004.05.004
70. Barceló-Batllori S, André M, Servis C, Lévy N, Takikawa O, Michetti P, et al. Proteomic analysis of cytokine induced proteins in human intestinal epithelial cells: implications for inflammatory bowel diseases. Proteomics. (2002) 2:551–60. doi: 10.1002/1615-9861(200205)2:5<551::AID-PROT551>3.0.CO;2-O
71. Ferdinande L, Demetter P, Perez-Novo C, Waeytens A, Taildeman J, Rottiers I, et al. Inflamed intestinal mucosa features a specific epithelial expression pattern of indoleamine 2,3-dioxygenase. Int J Immunopathol Pharmacol. (2008) 21:289–95. doi: 10.1177/039463200802100205
72. Proietti E, Pauwels RWM, de Vries AC, Orecchini E, Volpi C, Orabona C, et al. Modulation of indoleamine 2,3-dioxygenase 1 during inflammatory bowel disease activity in humans and mice. Int J Trypt Res. (2023) 16:11786469231153109. doi: 10.1177/11786469231153109
73. Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, et al. IDO1 and kynurenine pathway metabolites activate pi3k-akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Res. (2019) 79:1138–50. doi: 10.1158/0008-5472.CAN-18-0668
74. Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J Psychiatr Res. (2008) 43:164–74. doi: 10.1016/j.jpsychires.2008.03.009
75. Vujkovic-Cvijin I, Swainson LA, Chu SN, Ortiz AM, Santee CA, Petriello A, et al. Gut-resident lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. (2015) 13:1589–97. doi: 10.1016/j.celrep.2015.10.026
76. Valladares R, Bojilova L, Potts AH, Cameron E, Gardner C, Lorca G, et al. Lactobacillus johnsonii inhibits indoleamine 2,3-dioxygenase and alters tryptophan metabolite levels in BioBreeding rats. FASEB J. (2013) 27:1711–20. doi: 10.1096/fj.12-223339
77. Li P, Zheng J, Bai Y, Wang D, Cui Z, Li Y, et al. Characterization of kynurenine pathway in patients with diarrhea-predominant irritable bowel syndrome. Eur J Histochem. (2020) 64(s2). doi: 10.4081/ejh.2020.3132
78. Michaudel C, Danne C, Agus A, Magniez A, Aucouturier A, Spatz M, et al. Rewiring the altered tryptophan metabolism as a novel therapeutic strategy in inflammatory bowel diseases. Gut. (2023) 72:1296–307. doi: 10.1136/gutjnl-2022-327337
79. Shaw C, Hess M, Weimer BC. Microbial-derived tryptophan metabolites and their role in neurological disease: anthranilic acid and anthranilic acid derivatives. Microorganisms. (2023) 11:1825. doi: 10.3390/microorganisms11071825
80. Pereira C, Coelho R, Grácio D, Dias C, Silva M, Peixoto A, et al. DNA damage and oxidative DNA damage in inflammatory bowel disease. J Crohns Colitis. (2016) 10:1316–23. doi: 10.1093/ecco-jcc/jjw088
81. Hansen MB, Arif F, Gregersen H, Bruusgaard H, Wallin L. Effect of serotonin on small intestinal contractility in healthy volunteers. Physiol Res. (2008) 57:63–71. doi: 10.33549/physiolres.931177
82. Rafehi M, Faltraco F, Matthaei J, Prukop T, Jensen O, Grytzmann A, et al. Highly variable pharmacokinetics of tyramine in humans and polymorphisms in OCT1, CYP2d6, and MAO-A. Front Pharmacol. (2019) 10:1297. doi: 10.3389/fphar.2019.01297
83. Kim J, Chen C-H, Yang J, Mochly-Rosen D. Aldehyde dehydrogenase 2*2 knock-in mice show increased reactive oxygen species production in response to cisplatin treatment. J Biomed Sci. (2017) 24:33. doi: 10.1186/s12929-017-0338-8
84. Xu W, Zhu H, Gu M, Luo Q, Ding J, Yao Y, et al. DHTKD1 is essential for mitochondrial biogenesis and function maintenance. FEBS Lett. (2013) 587:3587–92. doi: 10.1016/j.febslet.2013.08.047
85. Artiukhov AV, Grabarska A, Gumbarewicz E, Aleshin VA, Kähne T, Obata T, et al. Synthetic analogues of 2-oxo acids discriminate metabolic contribution of the 2-oxoglutarate and 2-oxoadipate dehydrogenases in mammalian cells and tissues. Sci Rep. (2020) 10:1886. doi: 10.1038/s41598-020-58701-4
86. Schmedes A, Nielsen JN, Hey H, Brandslund I. Low S-adenosylmethionine concentrations found in patients with severe inflammatory bowel disease. Clin Chem Lab Med. (2004) 42:648–53. doi: 10.1515/CCLM.2004.111
87. Levitt MD, Furne J, Springfield J, Suarez F, DeMaster E. Detoxification of hydrogen sulfide and methanethiol in the cecal mucosa. J Clin Invest. (1999) 104:1107–14. doi: 10.1172/JCI7712
88. Hertel J, Harms AC, Heinken A, Baldini F, Thinnes CC, Glaab E, et al. Integrated analyses of microbiome and longitudinal metabolome data reveal microbial-host interactions on sulfur metabolism in Parkinson's disease. Cell Rep. (2019) 29:1767-77.e8. doi: 10.1016/j.celrep.2019.10.035
89. Paul B, Barnes S, Demark-Wahnefried W, Morrow C, Salvador C, Skibola C, et al. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenetics. (2015) 7:112. doi: 10.1186/s13148-015-0144-7
90. Schniers A, Goll R, Pasing Y, Sørbye SW, Florholmen J, Hansen T. Ulcerative colitis: functional analysis of the in-depth proteome. Clin Proteomics. (2019) 16:4. doi: 10.1186/s12014-019-9224-6
91. Mennigen R, Kusche J, Streffer C, Krakamp B. Diamine oxidase activities in the large bowel mucosa of ulcerative colitis patients. Agents Actions. (1990) 30:264–6. doi: 10.1007/BF01969056
92. Hrubisko M, Danis R, Huorka M, Wawruch M. Histamine intolerance-the more we know the less we know. A Review Nutrients. (2021) 13:2228. doi: 10.3390/nu13072228
93. Moschen AR, Kaser A, Enrich B, Mosheimer B, Theurl M, Niederegger H, et al. Visfatin, an adipocytokine with proinflammatory and immunomodulating properties. J Immunol. (2007) 178:1748–58. doi: 10.4049/jimmunol.178.3.1748
94. Gerner RR, Klepsch V, Macheiner S, Arnhard K, Adolph TE, Grander C, et al. NAD metabolism fuels human and mouse intestinal inflammation. Gut. (2018) 67:1813–23. doi: 10.1136/gutjnl-2017-314241
95. Colombo G, Clemente N, Zito A, Bracci C, Colombo FS, Sangaletti S, et al. Neutralization of extracellular NAMPT (nicotinamide phosphoribosyltransferase) ameliorates experimental murine colitis. J Mol Med. (2020) 98:595–612. doi: 10.1007/s00109-020-01892-0
96. Imai S. Nicotinamide phosphoribosyltransferase (NAMPT): a link between NAD biology, metabolism, and diseases. Curr Pharm Des. (2009) 15:20–8. doi: 10.2174/138161209787185814
97. Rosado MM, Bennici E, Novelli F, Pioli C. Beyond DNA repair, the immunological role of PARP-1 and its siblings. Immunology. (2013) 139:428–37. doi: 10.1111/imm.12099
98. Decker P, Muller S. Modulating poly (ADP-ribose) polymerase activity: potential for the prevention and therapy of pathogenic situations involving DNA damage and oxidative stress. Curr Pharm Biotechnol. (2002) 3:275–83. doi: 10.2174/1389201023378265
99. Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly (ADP-ribose) and PARP-1. Genes Dev. (2012) 26:417–32. doi: 10.1101/gad.183509.111
100. Ruszkiewicz JA, Bürkle A, Mangerich A. Fueling genome maintenance: on the versatile roles of NAD+ in preserving DNA integrity. J Biol Chem. (2022) 298:102037. doi: 10.1016/j.jbc.2022.102037
101. Brown EM, Arellano-Santoyo H, Temple ER, Costliow ZA, Pichaud M, Hall AB, et al. Gut microbiome ADP-ribosyltransferases are widespread phage-encoded fitness factors. Cell Host Microbe. (2021) 29:1351–65.e11. doi: 10.1016/j.chom.2021.07.011
102. Ratajczak J, Joffraud M, Trammell SAJ, Ras R, Canela N, Boutant M, et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat Commun. (2016) 7:13103. doi: 10.1038/ncomms13103
103. Colombo G, Gelardi ELM, Balestrero FC, Moro M, Travelli C, Genazzani AA. Insight into nicotinamide adenine dinucleotide homeostasis as a targetable metabolic pathway in colorectal cancer. Front pharmacol. (2021) 12:758320. doi: 10.3389/fphar.2021.758320
104. Kim M-H, Kim H. The roles of glutamine in the intestine and its implication in intestinal diseases. Int J Mol Sci. (2017) 18:1051. doi: 10.3390/ijms18051051
105. Stavely R, Sahakian L, Filippone RT, Stojanovska V, Bornstein JC, Sakkal S, et al. Oxidative stress-induced HMGB1 translocation in myenteric neurons contributes to neuropathy in colitis. Biomolecules. (2022) 12:1831. doi: 10.3390/biom12121831
106. Hou Y-C, Wu J-M, Wang M-Y, Wu M-H, Chen K-Y, Yeh S-L, et al. Glutamine supplementation attenuates expressions of adhesion molecules and chemokine receptors on t cells in a murine model of acute colitis. Mediators Inflamm. (2014) 2014:837107. doi: 10.1155/2014/837107
107. Fillmann H, Kretzmann NA, San-Miguel B, Llesuy S, Marroni N, González-Gallego J, et al. Glutamine inhibits over-expression of pro-inflammatory genes and down-regulates the nuclear factor kappaB pathway in an experimental model of colitis in the rat. Toxicology. (2007) 236:217–26. doi: 10.1016/j.tox.2007.04.012
108. Hu X, Deng J, Yu T, Chen S, Ge Y, Zhou Z, et al. Atf4 deficiency promotes intestinal inflammation in mice by reducing uptake of glutamine and expression of antimicrobial peptides. Gastroenterology. (2019) 156:1098–111. doi: 10.1053/j.gastro.2018.11.033
109. DeRoche TC, Xiao S-Y, Liu X. Histological evaluation in ulcerative colitis. Gastroenterology Rep. (2014) 2:178–92. doi: 10.1093/gastro/gou031
110. Lin J, Zhao L, Zhao S, Li S, Zhao Z, Chen Z, et al. Disruptive NADSYN1 variants implicated in congenital vertebral malformations. Genes. (2021) 12:1615. doi: 10.3390/genes12101615
111. Xue H, Sufit AJD, Wischmeyer PE. Glutamine therapy improves outcome of in vitro and in vivo experimental colitis models. J Parenter Enteral Nutr. (2011) 35:188–97. doi: 10.1177/0148607110381407
112. Too LK, Li KM, Suarna C, Maghzal GJ, Stocker R, McGregor IS, et al. Deletion of TDO2, IDO-1 and IDO-2 differentially affects mouse behavior and cognitive function. Behav Brain Res. (2016) 312:102–17. doi: 10.1016/j.bbr.2016.06.018
113. Fuertig RAD, Bergamini G, Cathomas F, Sigrist H, Seifritz E, Vavassori S, et al. Mouse chronic social stress increases blood and brain kynurenine pathway activity and fear behaviour: both effects are reversed by inhibition of indoleamine 2,3-dioxygenase. Brain Behav Immun. (2016) 54:59–72. doi: 10.1016/j.bbi.2015.12.020
114. Clark SM, Pocivavsek A, Nicholson JD, Notarangelo FM, Langenberg P, McMahon RP, et al. Reduced kynurenine pathway metabolism and cytokine expression in the prefrontal cortex of depressed individuals. J Psychiatry Neurosci. (2016) 41:386–94. doi: 10.1503/jpn.150226
115. Isse T, Matsuno K, Oyama T, Kitagawa K, Kawamoto T. Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol Clin Exp Res. (2005) 29:1959–64. doi: 10.1097/01.alc.0000187161.07820.21
116. Chen YC, Peng GS, Tsao TP, Wang MF, Lu RB, Yin SJ. Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics. (2009) 19:588–99. doi: 10.1097/FPC.0b013e32832ecf2e
117. Wichers MC, Maes M. The role of indoleamine 2,3-dioxygenase (IDO) in the pathophysiology of interferon-alpha-induced depression. J Psychiatry Neurosci. (2004) 29:11–7.
118. Chen X, Zhao H, Lu Y, Meng F, Lu Z, Lu Y. Surfactin mitigates dextran sodium sulfate-induced colitis and behavioral disorders in mice by mediating gut–brain-axis balance. J Agric Food Chem. (2023) 71:1577–92. doi: 10.1021/acs.jafc.2c07369
119. Usuda K, Kawase T, Shigeno Y, Fukuzawa S, Fujii K, Zhang H, et al. Hippocampal metabolism of amino acids by L-amino acid oxidase is involved in fear learning and memory. Sci Rep. (2018) 8:11073. doi: 10.1038/s41598-018-28885-x
120. Abumaria N, Ribic A, Anacker C, Fuchs E, Flügge G. Stress upregulates TPH1 but not TPH2 mRNA in the rat dorsal raphe nucleus: identification of two TPH2 mRNA splice variants. Cell Mol Neurobiol. (2008) 28:331–42. doi: 10.1007/s10571-007-9259-5
121. Colonnello V, Agostini A. Disease course, stress, attachment, and mentalization in patients with inflammatory bowel disease. Med Hypotheses. (2020) 140:109665. doi: 10.1016/j.mehy.2020.109665
122. Phillips RS, Iradukunda EC, Hughes T, Bowen JP. Modulation of enzyme activity in the kynurenine pathway by kynurenine monooxygenase inhibition. Front Mol Biosci. (2019) 6:3. doi: 10.3389/fmolb.2019.00003
123. Fukumoto K, Ito K, Saer B, Taylor G, Ye S, Yamano M, et al. Excess S-adenosylmethionine inhibits methylation via catabolism to adenine. Commun Biol. (2022) 5:313. doi: 10.1038/s42003-022-03280-5
124. Young GS, Kirkland JB. The role of dietary niacin intake and the adenosine-5′-diphosphate-ribosyl cyclase enzyme CD38 in spatial learning ability: is cyclic adenosine diphosphate ribose the link between diet and behaviour? Nutr Res Rev. (2008) 21:42–55. doi: 10.1017/S0954422408945182
125. Gasperi V, Sibilano M, Savini I, Catani MV. Niacin in the central nervous system: an update of biological aspects and clinical applications. Int J Mol Sci. (2019) 20:974. doi: 10.3390/ijms20040974
126. Liu B, Zhao G, Jin L, Shi J. Nicotinamide improves cognitive function in mice with chronic cerebral hypoperfusion. Front Neurol. (2021) 12:596641. doi: 10.3389/fneur.2021.596641
127. Klaidman LK, Mukherjee SK, Adams JD. Oxidative changes in brain pyridine nucleotides and neuroprotection using nicotinamide. Biochim Biophys Acta. (2001) 1525:136–48. doi: 10.1016/S0304-4165(00)00181-1
128. Diguet N, Trammell SAJ, Tannous C, Deloux R, Piquereau J, Mougenot N, et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation. (2018) 137:2256–73. doi: 10.1161/CIRCULATIONAHA.116.026099
129. Kropotov A, Kulikova V, Solovjeva L, Yakimov A, Nerinovski K, Svetlova M, et al. Purine nucleoside phosphorylase controls nicotinamide riboside metabolism in mammalian cells. J Biol Chem. (2022) 298:102615. doi: 10.1016/j.jbc.2022.102615
130. Feurté S, Gerozissis K, Regnault A, Paul FM. Plasma Trp/LNAA ratio increases during chronic ingestion of an alpha-lactalbumin diet in rats. Nutr Neurosci. (2001) 4:413–8. doi: 10.1080/1028415X.2001.11747377
131. Réus GZ, Jansen K, Titus S, Carvalho AF, Gabbay V, Quevedo J. Kynurenine pathway dysfunction in the pathophysiology and treatment of depression: evidences from animal and human studies. J Psychiatr Res. (2015) 68:316–28. doi: 10.1016/j.jpsychires.2015.05.007
Keywords: chronic intestinal inflammation, inflammatory bowel disease, colitis, gut-brain axis, tryptophan metabolism, nicotinamide metabolism, microbiota
Citation: Devereaux J, Robinson AM, Stavely R, Davidson M, Dargahi N, Ephraim R, Kiatos D, Apostolopoulos V and Nurgali K (2024) Alterations in tryptophan metabolism and de novo NAD+ biosynthesis within the microbiota-gut-brain axis in chronic intestinal inflammation. Front. Med. 11:1379335. doi: 10.3389/fmed.2024.1379335
Received: 31 January 2024; Accepted: 17 June 2024;
Published: 02 July 2024.
Edited by:
Shi Xue Dai, Guangdong Provincial People's Hospital, ChinaReviewed by:
Diego A. Esquivel-Hernandez, Metropolitan Autonomous University, MexicoCopyright © 2024 Devereaux, Robinson, Stavely, Davidson, Dargahi, Ephraim, Kiatos, Apostolopoulos and Nurgali. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kulmira Nurgali, a3VsbWlyYS5udXJnYWxpQHZ1LmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.