- 1Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Wollo University, Dessie, Ethiopia
- 2Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Jigjiga University, Jigjiga, Ethiopia
- 3Department of Medical Laboratory Science, Dessie Health Science College, Dessie, Ethiopia
- 4Department of Public Health, College of Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia
Introduction: Hepatocellular carcinoma (HCC) and liver cirrhosis (LC) stand as the primary causes of global mortality. Given their profound impact, the development of highly sensitive and specific circulating diagnostic markers becomes imperative to effectively identify and differentiate between cirrhosis and HCC. Accurate diagnosis is paramount in guiding appropriate therapeutic interventions. Hence, this study aimed to evaluate the potential of microRNAs (miRNAs) in discerning between HCC and LC.
Methods: This study followed the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines, with the protocol officially registered on PROSPERO under the reference number CRD42023417494. A thorough search across multiple databases like PubMed, Embase, Scopus, Wiley Online Library, and Science Direct was conducted to identify relevant studies published from January 1, 2018, to August 10, 2023. The included studies underwent methodological quality assessment using the Quality Assessment of Diagnostic Accuracy Studies 2 (QADAS-2) tool. The synthesis of pooled sensitivity, specificity, and other relevant diagnostic parameters employed a random-effects model and was conducted using Stata 14.0. Heterogeneity was assessed using I2 and Cochrane Q, with subsequent subgroup analysis and meta-regression performed to identify potential sources of observed heterogeneity. A sensitivity analysis was performed to assess the resilience of the findings. Furthermore, Deeks’ funnel plot was employed to evaluate publication bias.
Results: In this meta-analysis, we included fifteen publications, encompassing 787 HCC patients and 784 LC patients. The combined sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the curve (AUC) values of miRNAs in differentiating HCC from LC were 0.84 (95% CI: 0.78–0.88), 0.79 (95% CI: 0.73–0.84), 3.9 (95% CI: 3.0–5.2), 0.21 (95% CI: 0.14–0.29), 19.44 (95% CI: 11–34), and 0.88 (95% CI: 0.85–0.91), respectively. The results of the subgroup analysis revealed that upregulated miRNA levels and miRNA assessments specifically for individuals of European descent exhibited superior diagnostic performance.
Conclusion: The results of this study suggested that circulating miRNAs, especially those that are upregulated, have the potential to function as robust and promising biomarkers in the differentiation of HCC from LC.
Systematic review registration: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42023475954.
Introduction
Hepatocellular carcinoma (HCC) stands as the predominant form of liver cancer, commonly emerging as a consequence of liver cirrhosis (LC). Its association is notably linked to hepatitis virus infections and the presence of alcoholic or non-alcoholic fatty liver disease (1). According to the most recent global burden of disease statistics for 2020, liver cancer holds the sixth position among the most frequently diagnosed cancers and ranks as the third leading cause of cancer-related mortality worldwide. Approximately 905,700 individuals were diagnosed with liver cancer, resulting in 830,200 deaths (2).
Conversely, LC represents a late-stage scarring process wherein healthy liver tissue is replaced by nodules and scar tissue encircled by fibrous bands, stemming from prolonged liver injury and damage. The irreversible progression of cirrhosis may end in the development of HCC (3). It serves as the primary cause of liver-related deaths globally (4). In 2017, cirrhosis accounted for over 1.32 million deaths globally, with 440,000 in females and 883,000 in males, comprising 2.4% of the total global mortality for that year (5).
An association exists between liver cirrhosis and HCC, where cirrhosis may either precede the development of HCC or coexist with it (6). The diagnosis of HCC is frequently delayed as symptoms become noticeable only in the later stages or when the tumor is relatively larger. Consequently, both detection and treatment are often postponed, resulting in a diminished life expectancy for the patient (7). This delay is primarily attributed to the absence of an effective method for early diagnosis (8).
In general, the primary diagnostic pathways for HCC involve histopathological examination, blood biomarkers, and imaging techniques (9). Although serum alpha-fetoprotein (AFP) remains a widely used biomarker for HCC screening, early diagnosis, and therapeutic assessment (10), it has notable limitations, including low sensitivity and specificity. AFP can be elevated in some patients with cirrhosis or hepatic inflammation in the absence of a tumor, and it may not increase in 80% of small tumors (11, 12). Other methods, such as the gold standard liver biopsy and certain imaging platforms, face challenges like cost, invasiveness, limited availability in developing countries, and susceptibility to sampling errors and observer variations (13). Hence, developing sensitive and specific circulating diagnostic markers to identify and distinguish between cirrhosis and HCC is crucial, as the accurate diagnosis will play a pivotal role in determining the most suitable therapy.
The acknowledgment of microRNAs (miRNAs) has ushered in a new era of research focused on discovering novel non-invasive markers for cancer detection (14). Representing naturally occurring non-coding, single-stranded small RNA molecules ranging from 19 to 24 nucleotides in length (15), miRNAs play a crucial role in regulating posttranscriptional gene expression in the genome. They inhibit target genes by binding to the 3′ untranslated region (3’UTR) within messenger RNA (mRNA), leading to either mRNA degradation or the inhibition of protein translation (16, 17).
Altered miRNA expression has been observed in various cancer types, including lung, prostate, colon, breast, and liver tumors, influencing the activity of oncogenes and tumor suppressor genes and directly impacting carcinogenesis (18). In liver development, homeostasis, and pathophysiology, numerous miRNAs play essential roles (19). Due to their dysregulated expression, circulating miRNAs have been explored as potential biomarkers for cancer, including HCC, and can be identified in serum or plasma through non-invasive techniques (1). Several studies have demonstrated the potential of circulating miRNAs to differentiate between HCC and LC (20–22). Nevertheless, owing to inconsistencies observed in prior research, drawing reliable conclusions regarding the effectiveness of circulating miRNAs for distinguishing HCC from LC remains challenging. It is crucial to consider the need for aggregated data to provide a more comprehensive and conclusive assessment. Therefore, this study aimed to evaluate the discriminative potential of circulating miRNAs in distinguishing HCC from LC, utilizing recent data.
Methods
Study design and protocol registration
This study protocol was registered in PROSPERO (CRD42018104269) and was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplementary Table S1).
Search strategy
Electronic databases like PubMed, Embase, Scopus, Wiley Online Library, and ScienceDirect were searched to identify relevant articles reporting the diagnostic accuracy of circulating microRNA to discriminate HCC patients from LC patients from the time of inception to August 10, 2023. The following search terms were included: “circulating miRNAs” OR “circulating microRNAs” OR “circulating microRNA” OR “circulating miR*” OR “plasma miRNAs” OR “plasma microRNAs” OR plasma microRNA” OR “plasma miR*” OR “serum miRNAs” OR “serum microRNAs” OR “serum microRNA” OR “serum miR*” AND “diagnos*” AND “hepatocellular carcinoma” OR “HCC.” In addition, a manual search of relevant articles and references cited in these articles was conducted to identify all available studies. The detailed search is presented in Supplementary Table S2.
Eligibility criteria and quality assessment
The inclusion criteria encompassed the following criteria: (1) observational study; (2) human studies used miRNAs to discriminate HCC patients from LC patients; (3) false positive (FP), true positive (TP), false negative (FN), and true negative (TN) could be derived directly or calculated from the literature; and (4) studies published since January 1, 2018 G.C. The exclusion criteria were set as follows: (1) non-human studies; (2) full-text unavailability; (3) reviews, letters to editorials or conference proceedings; and (4) studies with inadequate information regarding diagnostic performance, sensitivity or specificity.
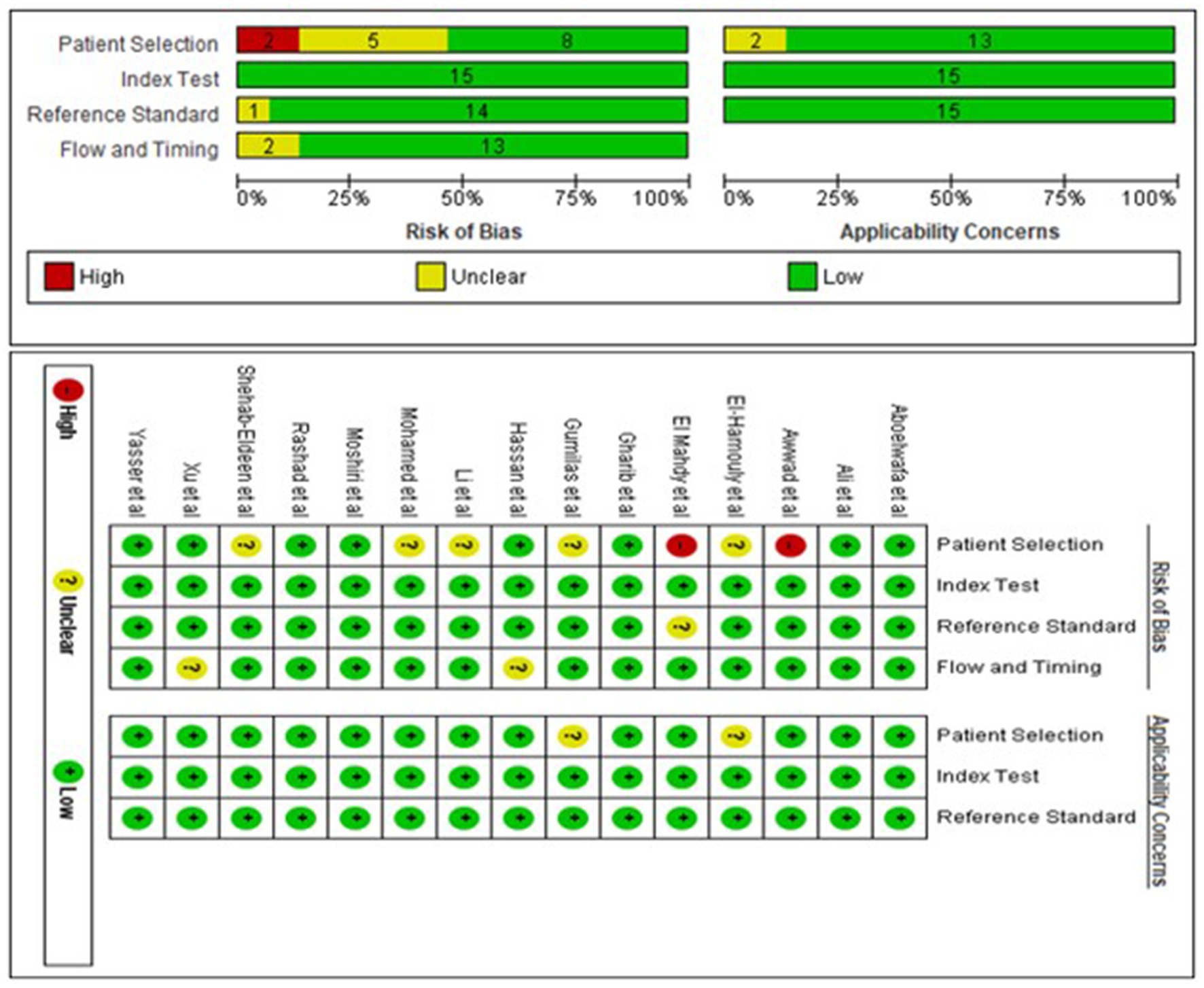
Two investigators (ZM and AG) used the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool (23) to independently assess the risk of bias and clinical applicability of included studies. The tool comprised four main components: (1) patient selection, (2) the index test, (3) the reference standard, and (4) flow and timing. Bias risk was graded as high (H), low (L), or unclear (U). Disagreements were resolved by agreement between the two investigators through negotiation or by involving a third reviewer (HE).
Study selection and data extraction
Two independent reviewers (MW and FG) systematically reviewed all studies with title and abstract for inclusion and the full text for the primary review. In cases of disagreements, the conclusion was finalized by a discussion with the third reviewer (MB). Then, data were extracted independently by two investigators (DA and EA) and from eligible studies, including first author, year of publication, country, miRNAs, type of samples, internal reference, cut-off values, sample size (HCC and LC patients), assay method, miRNAs expression, AUC with 95% confidence intervals (CIs), sensitivity, and specificity. The TP, FP, FN, and TN values were calculated using sensitivity, specificity and sample sizes.
Statistical analysis
Stata 14.0 software was used for the meta-analysis. Furthermore, Review Manager 5.4 was used to assess the quality of the included studies. Based on the random effect model, the pooled sensitivity, specificity, DOR, PLR, NLR, and corresponding 95% confidence interval (CI) of the included literature were determined using TP, FP, FN, and TN values (24). Summary receiver operating characteristic (SROC) curves were plotted to calculate the area under the curve (AUC) to test the pooled diagnostic value of miRNAs and to assess the presence of threshold effect. Cochrane Q test and I2 statistics were used to assess the heterogeneity between studies, with p-value less than 0.05 for Cochran-Q test and I2 > 50%, indicating significant heterogeneity between studies (25). The potential heterogeneity sources were analyzed through meta-regression and subgroup analyses. Additionally, sensitivity analysis was conducted to check the stability of the meta-analysis results. Deek’s quantitative funnel plot was used to assess the publication bias between studies. p < 0.05 denotes for the statistical significance. In addition, the clinical value of circulating miRNAs to discriminate hepatocellular carcinoma patients from liver cirrhosis patients was evaluated using Fagan’s plots.
Results
Search results, description, and quality assessment of the included studies
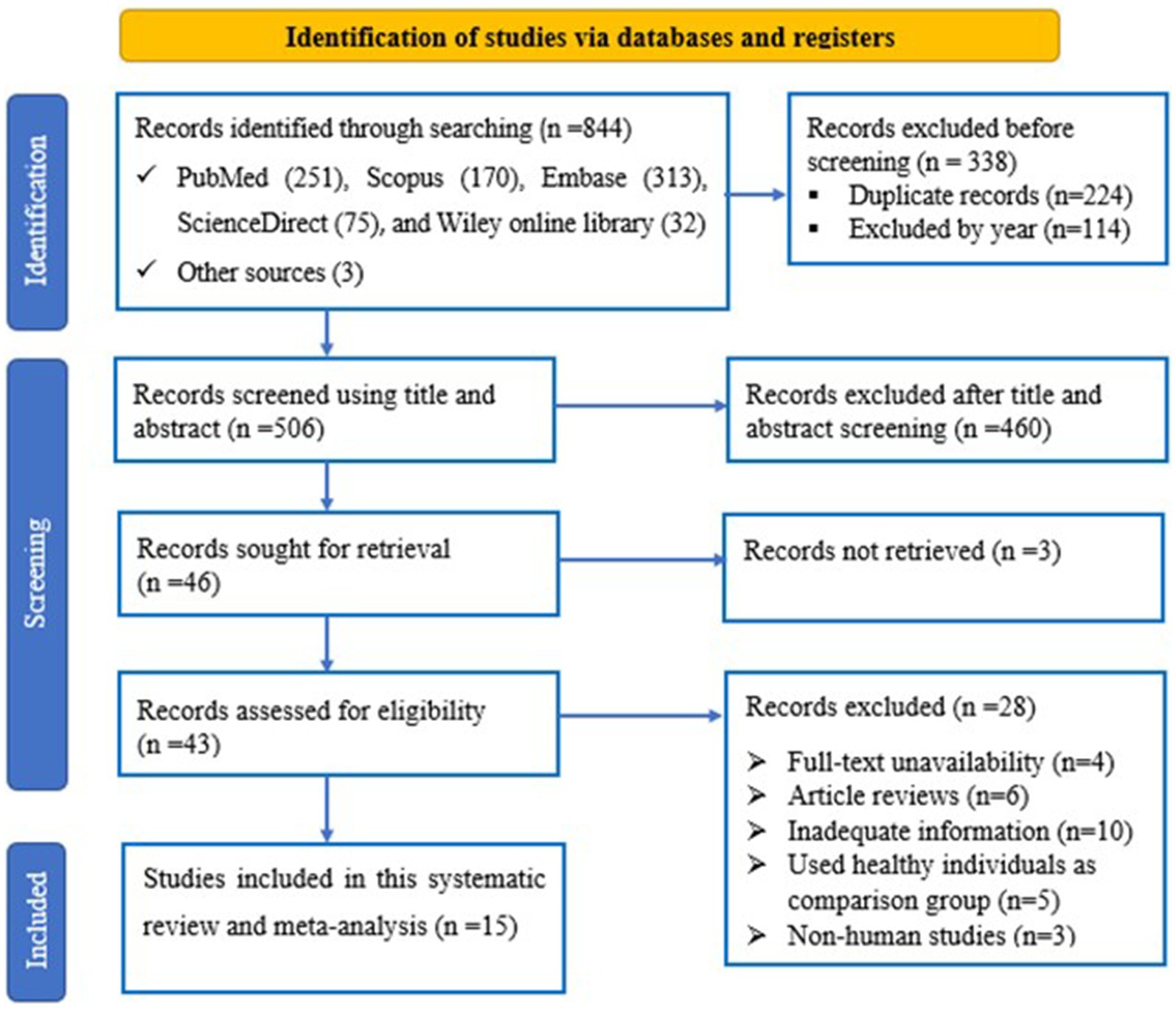
The flowchart represents the search and selection strategy for the study. The initial search resulted in a total of 844 studies, consisting of PubMed (n = 251), Scopus (n = 170), Embase (n = 313), ScienceDirect (n = 75) and Wiley online library (n = 32) articles, together with articles identified through relevant bibliography search (n = 3). 224 and 114 articles were excluded because of duplication and year, respectively. Furthermore, 460 articles were excluded because of title and abstract screening criteria. The full-text of the remaining 43 articles was reviewed. In addition, 28 articles were excluded from full-text review and finally 15 studies were considered for this meta-analysis (Figure 1).
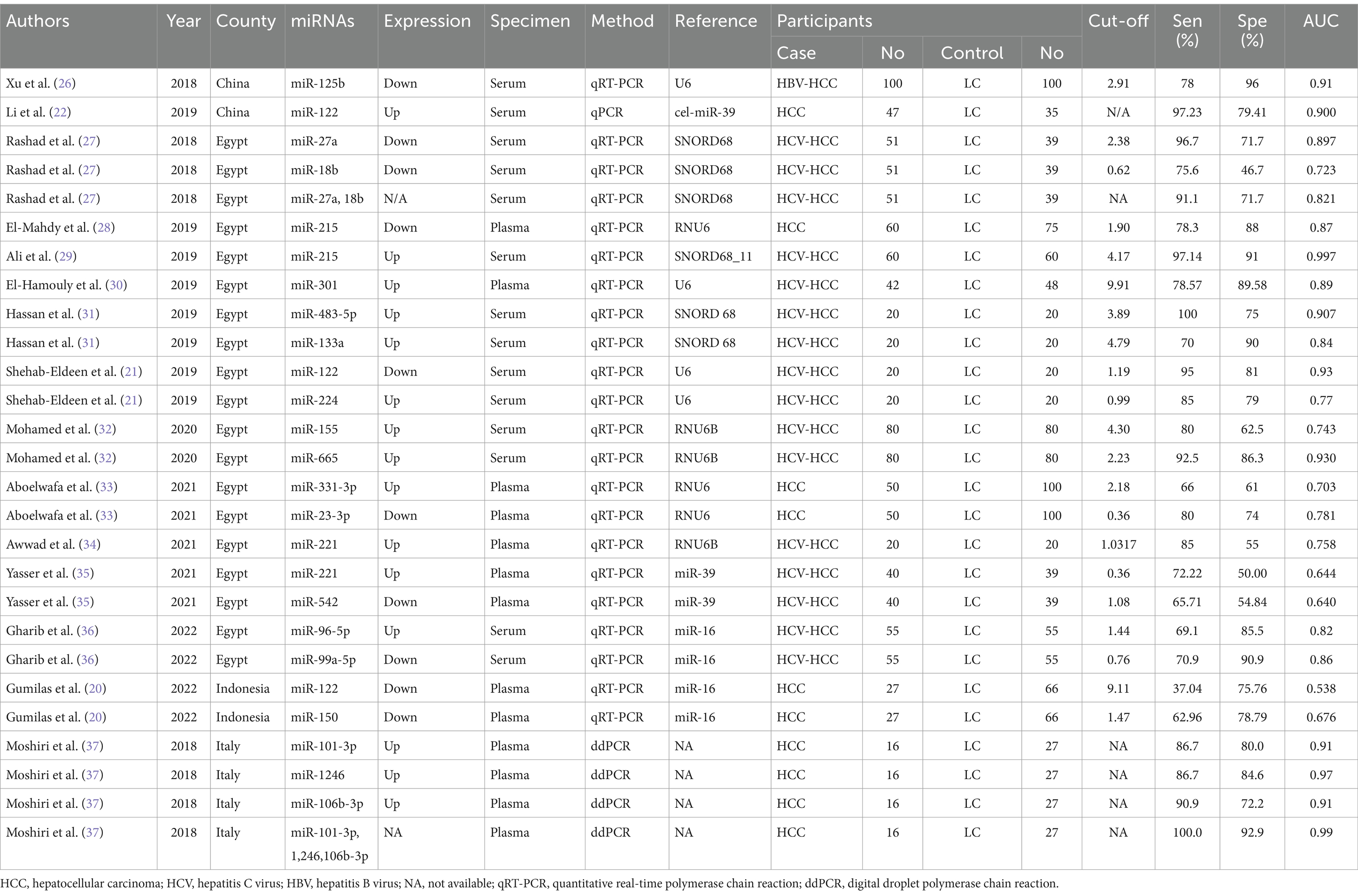
A total of 1,571 participants, including 787 HCC patients and 784 LC patients from 15 studies were included in the data analysis. The sample size ranged from 16 to 100 in the HCC cohort, and 20 to 100 in the LC cohort. Additionally, two studies were from China (22, 26), eleven studies were from Egypt (21, 27–36), one study was from Indonesia (20) and one study was from Italy (37). Seven studies used plasma samples, whereas four studies used serum samples for miRNA quantification. MiRNAs were measured using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Furthermore, fifteen miRNAs were upregulated and ten miRNAs were downregulated. The baseline characteristics of the included studies are summarized in Table 1. Based on the Quality Assessment of Diagnostic Accuracy Studies 2 evaluation tool, the RevMan 5.4 software was used to evaluate the quality of the included studies. The results are shown in Figure 2.
Diagnostic accuracy of miRNAs in distinguishing HCC patients from LC patients
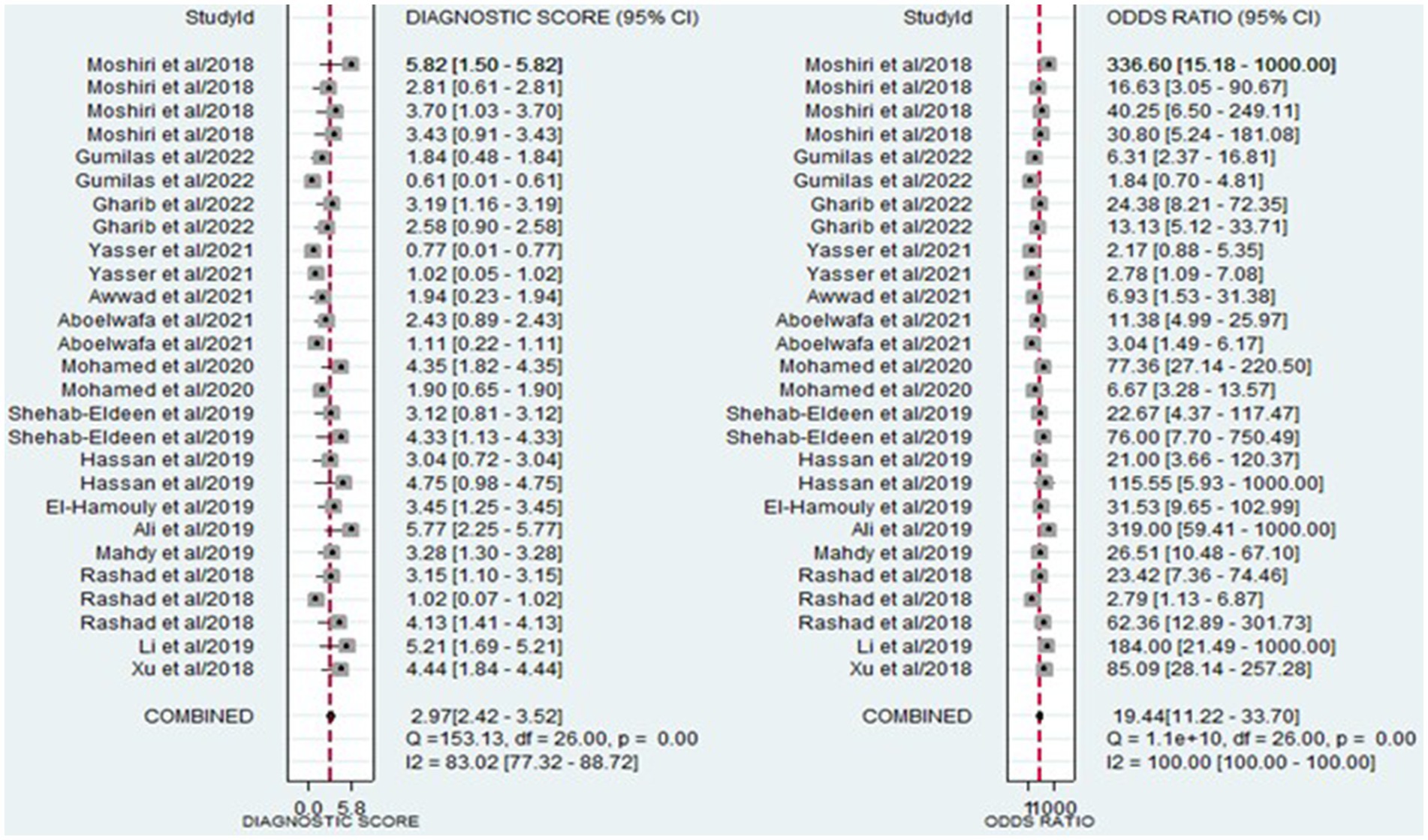
The analysis involved summarizing estimates of miRNA diagnostic accuracy in distinguishing between HCC and LC. The combined sensitivity and specificity for diagnosing HCC with miRNAs were 0.84 (95% CI: 0.78–0.88) and 0.79 (95% CI: 0.73–0.84), respectively (Figure 3). Notably, there was substantial heterogeneity observed among the studies, with an I2 value of 78.47% for sensitivity and 82.67% for specificity. Additionally, the pooled PLR, NLR, and DOR were 3.9 (95% CI: 3.0–5.2), 0.21 (95% CI: 0.14–0.29), and 19.44 (95% CI: 11–34), respectively (Figures 4, 5). The shape of the SROC curve did not exhibit the typical “shoulder-arm-like” pattern, suggesting that there is no evident threshold effect in this study. The AUC was 0.88 (95% CI: 0.85–0.91), indicating that overall, miRNAs demonstrate exceptional diagnostic accuracy (Figure 6A). Moreover, the Fagan nomogram was utilized to evaluate the efficacy of miRNA testing in confirming or ruling out the presence of HCC in patients. The results revealed that, when the pre-test probability was set at 20%, the post-test probabilities for the PLR and NLR were 50 and 5%, respectively. Consequently, miRNA testing assumes a crucial role in the initial screening of individuals with HCC (Figure 6B).
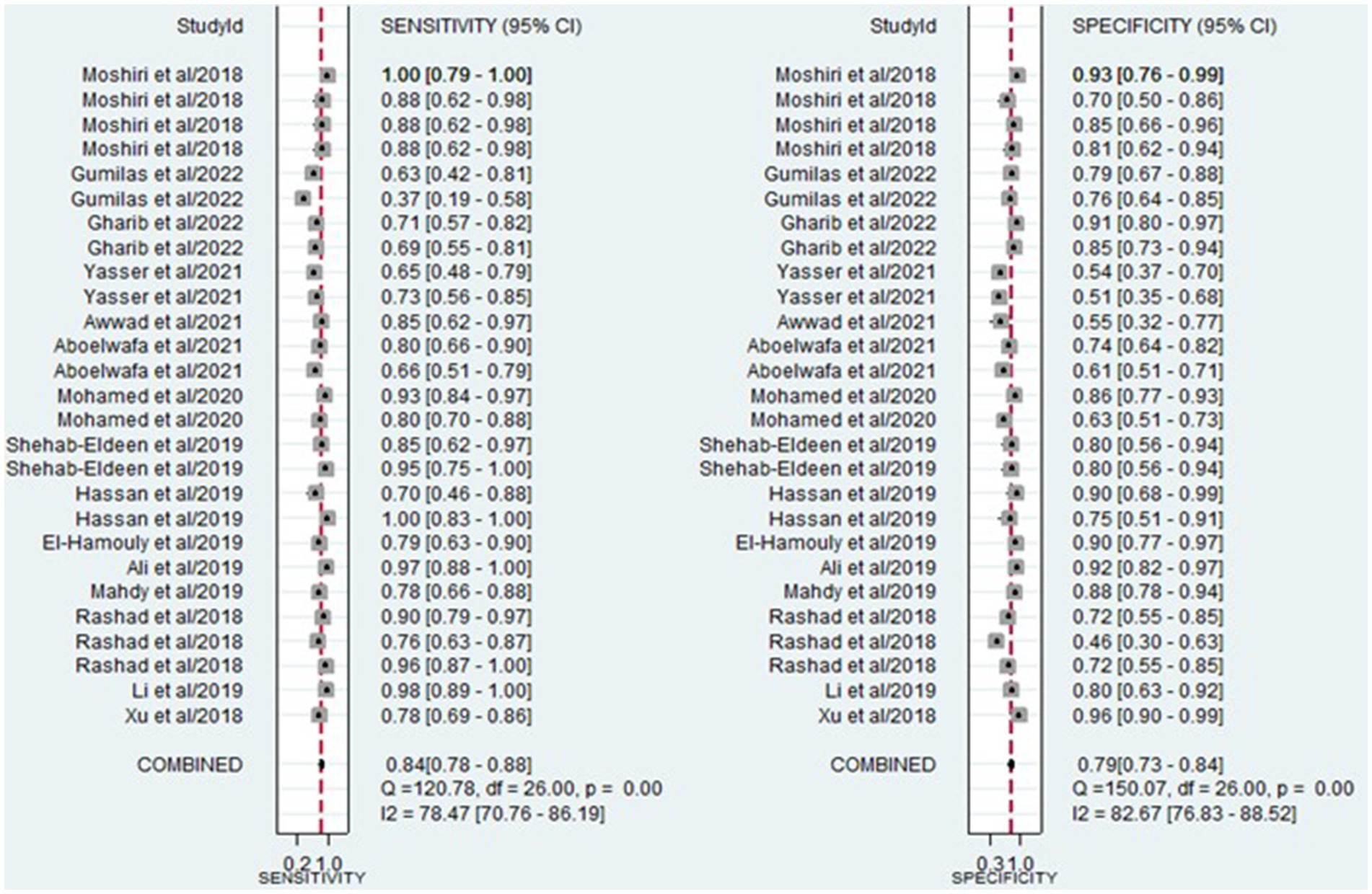
Figure 3. Forest plot illustrating the sensitivity and specificity of miRNAs in the discrimination of HCC and LC patients.
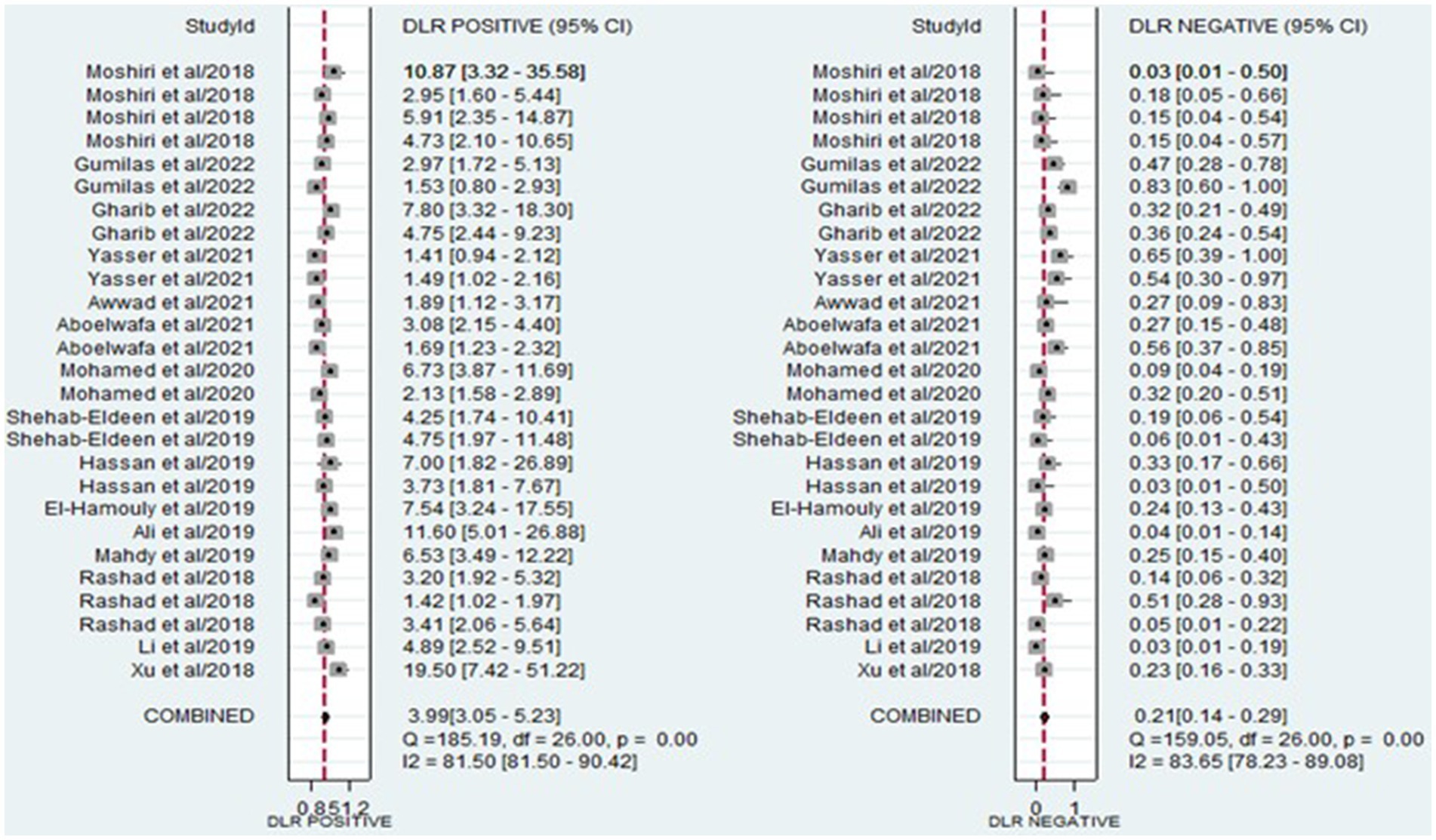
Figure 4. Forest plot illustrating the PLR and NLR of miRNAs in the discrimination of HCC and LC patients.
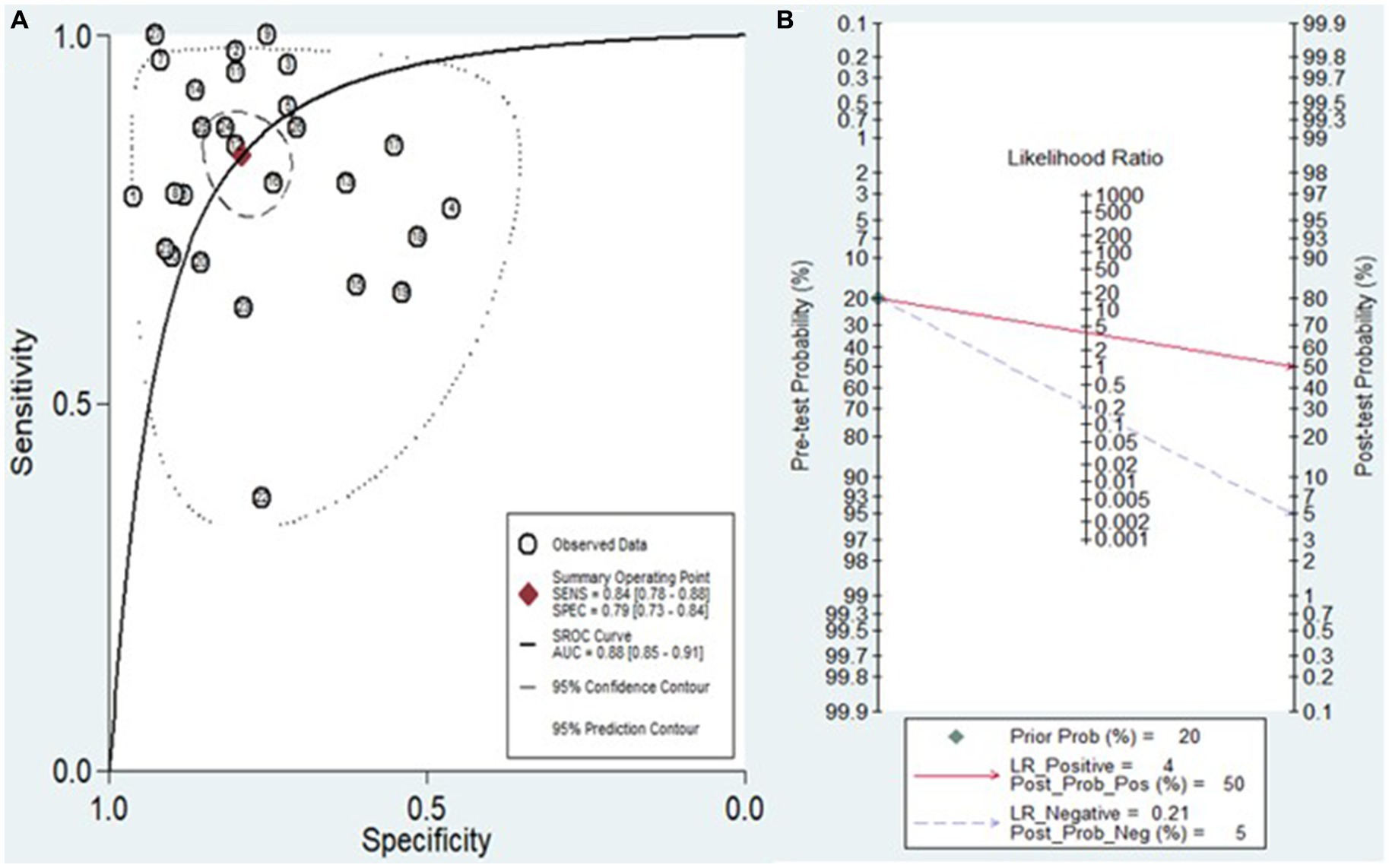
Figure 6. (A) Diagram of SROC curves illustrating the diagnostic performance of miRNAs in discriminating between HCC and LC patients. (B) The Fagan nomogram illustrates the capacity of miRNA testing to either confirm or exclude HCC in patients.
Subgroup analyses and meta-regression
In our pursuit to identify sources of heterogeneity among the studies, we conducted both subgroup analysis and meta-regression analysis. This involved categorizing studies based on ethnicity, sample source, regulation mode, miRNA profiling, sample size, cut-off value presence, and types of HCC.
During the subgroup analysis (Table 2), we observed that studies conducted on the European population exhibited higher overall diagnostic accuracy compared to those carried out on the Asian and African populations. The combined diagnostic values, along with their 95% confidence intervals, were reported as follows: sensitivity 0.91 (0.80–0.96), specificity 0.83 (0.72–0.90), PLR 5.3 (3.1–9.2), NLR 0.11 (0.04–0.27), DOR 49 (14–173), and an AUC of 0.94 (0.91–0.96). Additionally, individual miRNAs demonstrated the following aggregated diagnostic values for distinguishing between HCC and LC: sensitivity 0.83 (0.76–0.87), specificity 0.79 (0.73–0.84), PLR 3.9 (2.9–5.1), NLR 0.22 (0.16–0.31), DOR 17 (10–30), and an AUC of 0.87 (0.84–0.90). In contrast to downregulated miRNAs, upregulated miRNAs demonstrated superior overall diagnostic accuracy, with a sensitivity of 0.86 (0.79–0.91), specificity of 0.78 (0.71–0.84), PLR of 3.9 (2.8–5.5), NLR of 0.18 (0.12–0.28), DOR of 22 (11–44), and an AUC of 0.89 (0.86–0.91). Moreover, studies not reporting their cut-off values showed superior diagnostic accuracy compared to studies reporting their cut-off values, as indicated by sensitivity 0.93 (0.87–0.97), specificity 0.80 (0.72–0.86), PLR 4.7 (3.3–6.6), NLR 0.09 (0.04–0.17), DOR 54 (22–135), and an AUC of 0.93 (0.90–0.95).
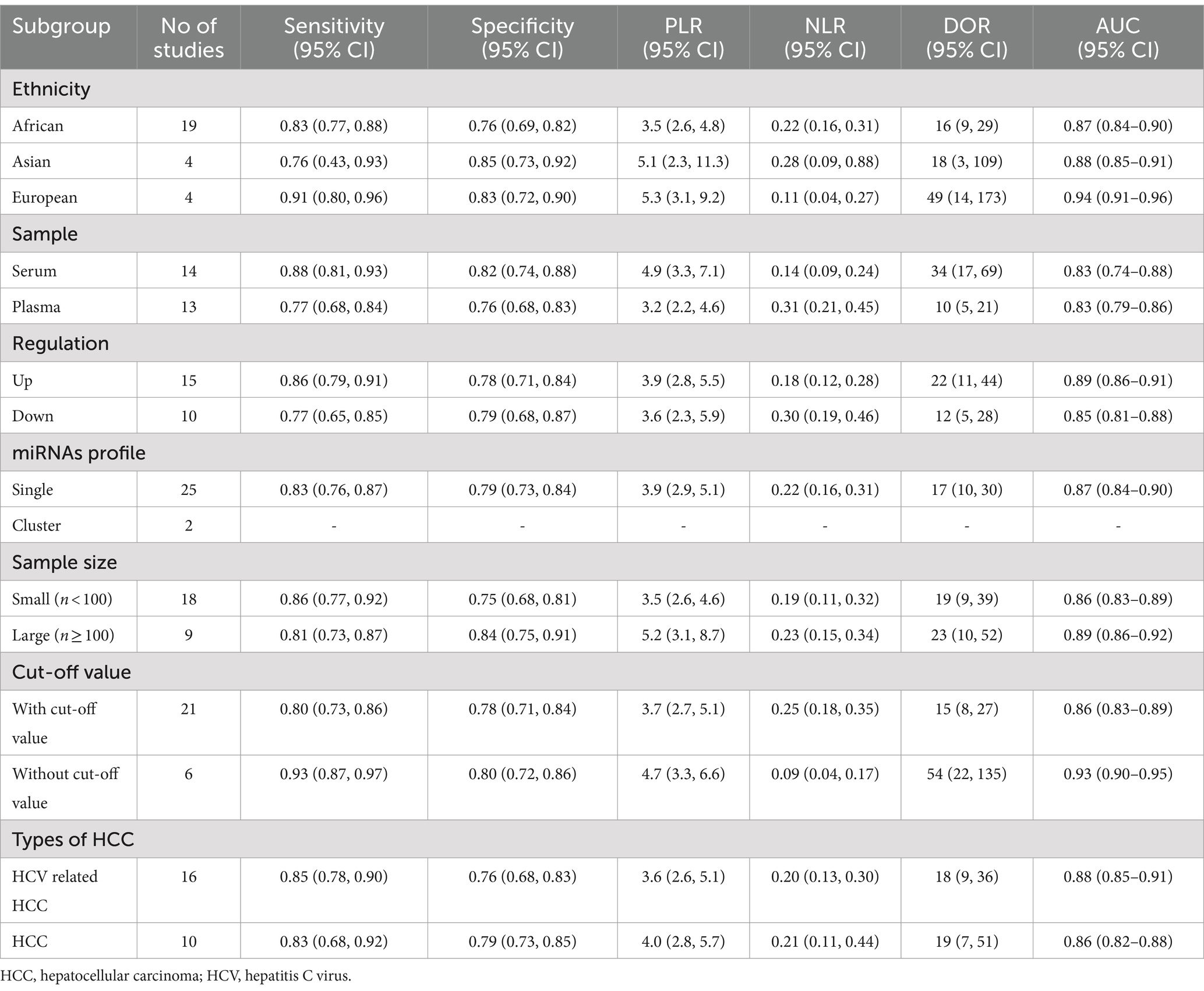
Table 2. Subgroup analysis of the diagnostic value of miRNAs in discriminating between HCC patients and LC patients.
Besides, the meta-regression analysis indicated that heterogeneity in sensitivity among the studies could be attributed to ethnicity and the setting of cut-off values. Conversely, factors such as ethnicity, sample source, regulation mode, sample size, cut-off value setting, and types of HCC were identified as reasons for heterogeneity in specificity among the studies (p < 0.05) (Figure 7).
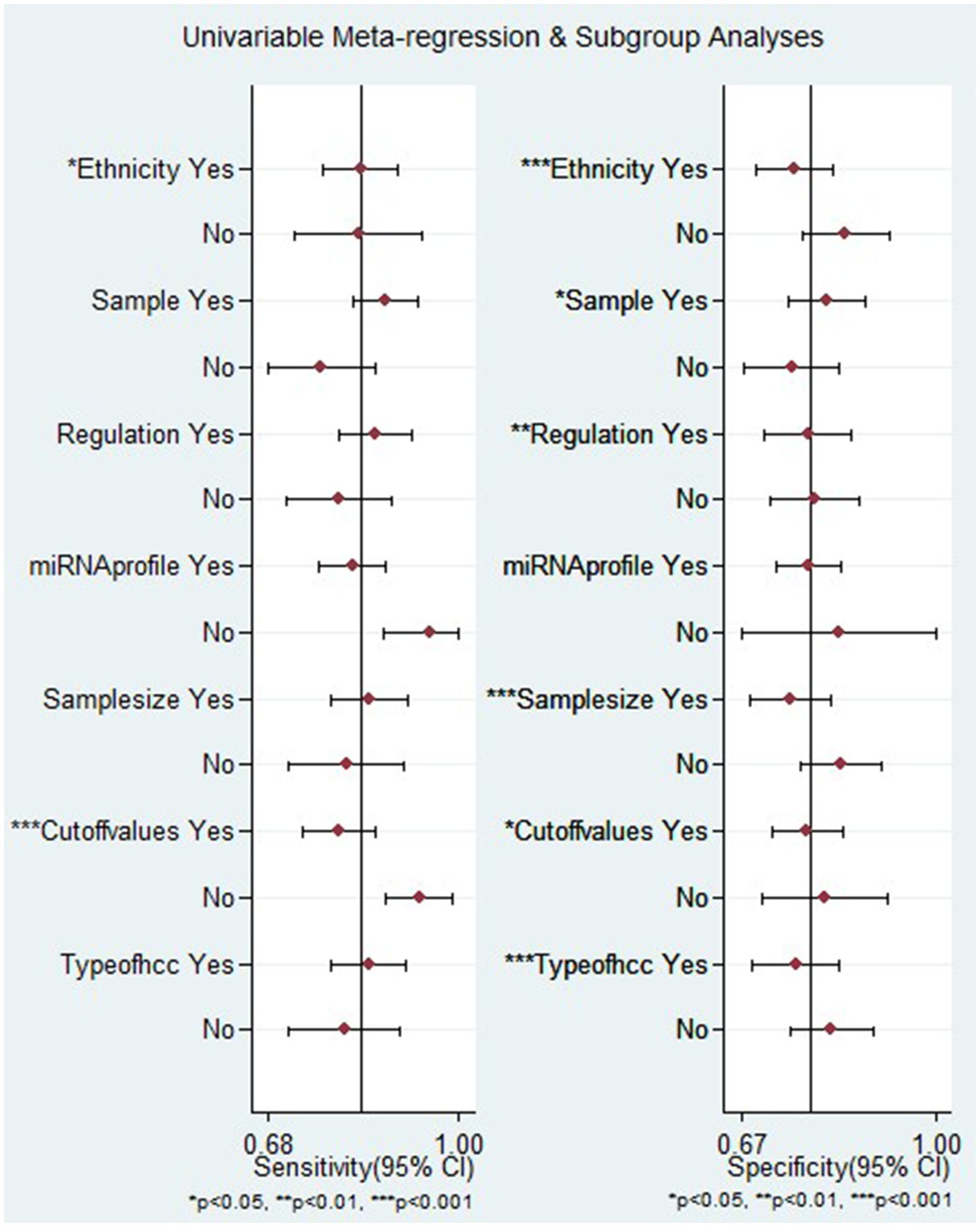
Figure 7. Meta-regression analysis examining the sensitivity and specificity of miRNAs in discriminating between HCC and LC patients.
Sensitivity analyses and publication bias
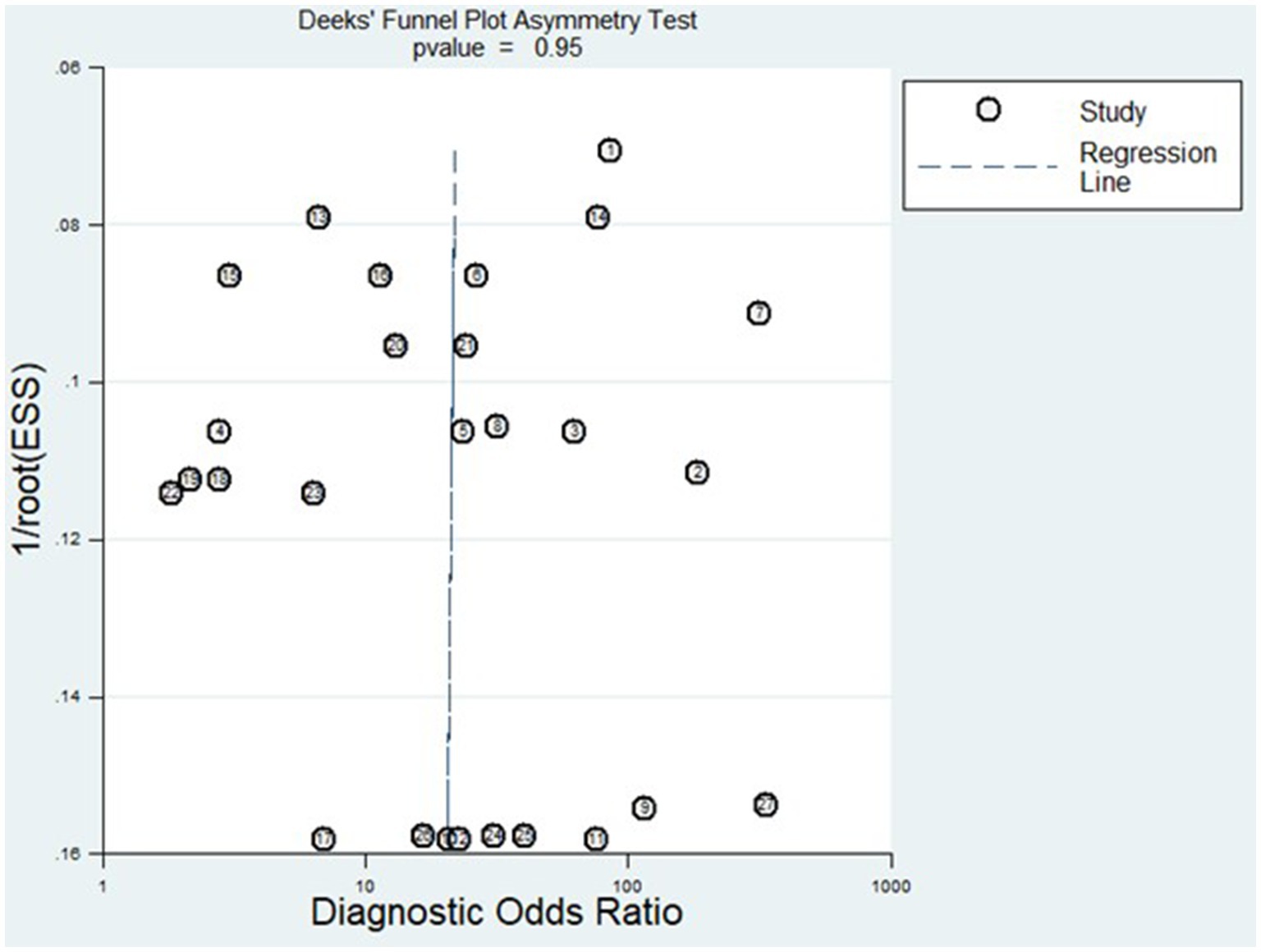
Figure 8 displays the outcomes of the sensitivity analysis. The goodness-of-fit and bivariate normal analysis, depicted in Figures 8A,B, affirm the robustness of the random effects model for meta-analysis. Furthermore, outlier detection points to two studies conducted by Xu et al. (miR-125b) (26) and Gumilas et al. (miR-122) (20) as potential sources of heterogeneity (Figure 8D). After removing these studies, the overall sensitivity remained unchanged at 0.85 (0.79–0.89), along with a specificity of 0.78 (0.72–0.83), PLR of 3.8 (3.0–5.0), NLR of 0.19 (0.14–0.27), DOR of 20 (11–34), and an AUC of 0.88 (0.85–0.91). This indicates that the sensitivity of the included studies was low, and the results became more robust and credible after excluding the identified outliers. Additionally, Figure 9 illustrates the Deeks’ funnel plot, serving as an assessment of publication bias. The p-value of 0.95 obtained from the analysis indicates the absence of significant publication bias in the included studies.
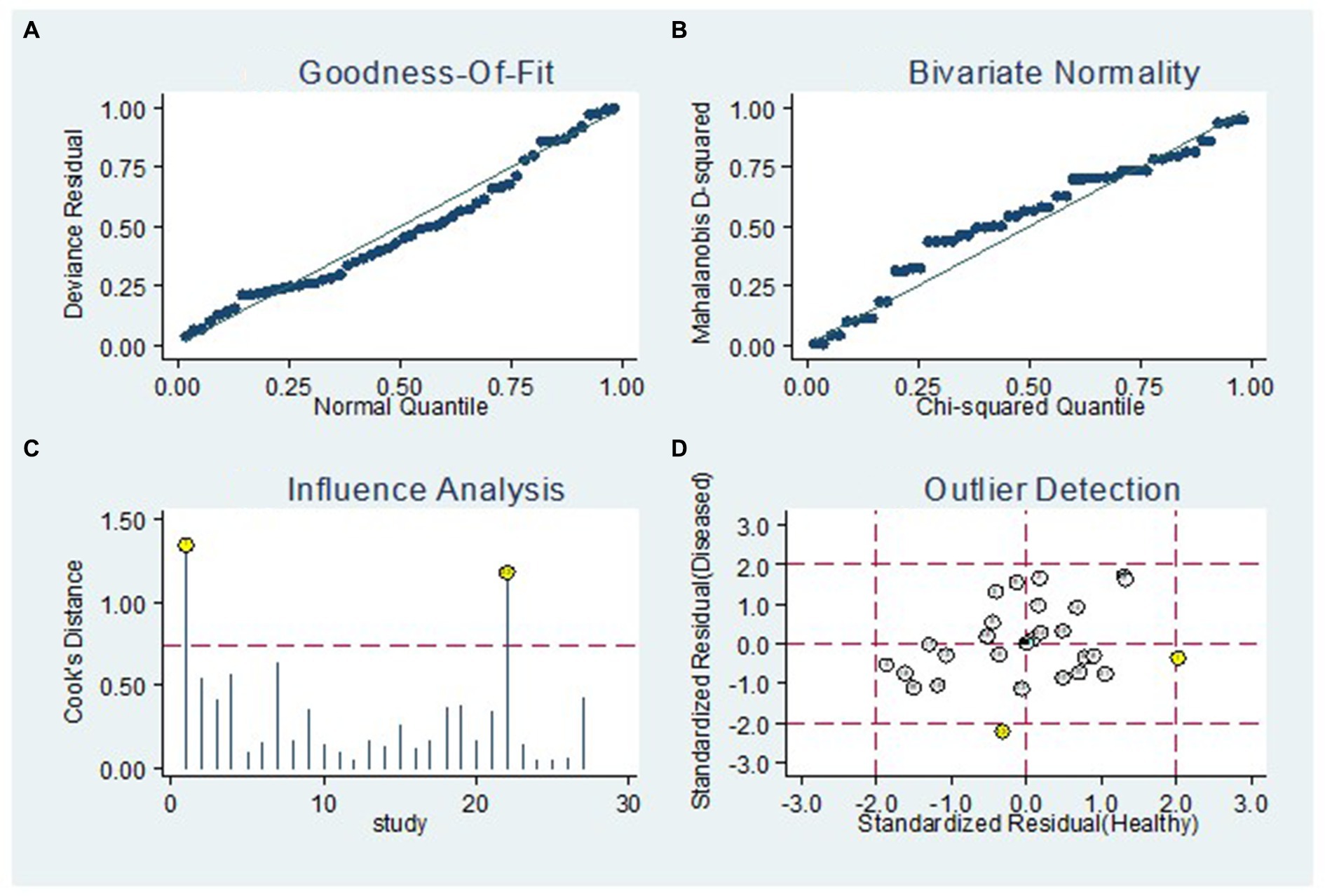
Figure 8. Sensitivity analysis. The diagram shows the (A) goodness-of-fit, (B) bivariate normality, (C) influence and (D) outlier detection analyses.
Discussion
Hepatocellular carcinoma ranks as the fourth major contributor to cancer-related deaths globally and stands as a prominent cause of mortality in individuals with cirrhosis (38). The definitive diagnosis of HCC is typically differentiated from cirrhosis through the use of advanced imaging techniques, including CT scans and MRI. This distinction relies on identifying enhancement patterns in the hepatic arterial phase (HAP) images (39). Liver biopsy is used for confirmation of diagnosis or exclusion of other lesions that may mimic HCC. Diagnosing HCC is often challenging as it tends to be identified after the onset of clinical deterioration. The silent and asymptomatic growth of HCC makes it difficult to detect in its early stages (40).
The identification of early-stage HCC is crucial for initiating aggressive intervention and improving overall survival rates (41). Hence, there is a critical need to identify a more precise and advanced non-invasive biomarker for the early diagnosis of HCC, enabling the differentiation of target groups from those with similar presentations with high sensitivity and specificity. Recent evidence has indicated that abnormal miRNA profiles are associated with the development, progression, and prognosis of various human cancers (42). Consequently, miRNA has garnered significant attention from experts in the diagnosis of HCC due to its notable specificity, repeatability, and accuracy (43). However, their stability as clinical biomarkers warrants careful consideration due to variations in sample processing conditions, methodology, and biological material sources (44, 45). While RNA molecules are generally unstable, prior research indicates that miRNAs exhibit exceptional stability in plasma and serum, demonstrating resistance to RNase activity, extreme pH conditions, and multiple freeze–thaw cycles (46, 47). Nonetheless, not all miRNAs maintain stability; they exhibit diverse stability profiles influenced by factors such as sequence, secondary structure, and associations with proteins or extracellular vesicles (48). This variability poses challenges in developing robust diagnostic assays, as unstable miRNAs can yield inconsistent results, impacting test reliability and reproducibility. Therefore, identifying stable miRNA biomarkers using longitudinal studies is crucial for enhancing diagnostic accuracy. Despite previous studies recommending miRNA for distinguishing HCC from liver cirrhosis, there is limited consistency among these studies, and the findings remain inconclusive. Therefore, we conducted a systematic review and meta-analysis to assess the potential of miRNAs in distinguishing between HCC and LC patients.
This study incorporated data from 15 research articles covering 27 studies, involving a total of 787 HCC patients and 784 LC patients. The overall summary estimate revealed that the pooled sensitivity and specificity of circulating miRNAs in distinguishing HCC from LC were 0.84 (95% CI: 0.78–0.88) and 0.79 (95% CI: 0.73–0.84), respectively. Additionally, the pooled PLR, NLR, DOR, and AUC from SROC were 3.9 (95% CI: 3.0–5.2), 0.21 (95% CI: 0.14–0.29), 19.44 (95% CI: 11–34), and 0.88 (95% CI: 0.85–0.91), respectively. The combined PLR of 3.9 suggests that a positive miRNA test is associated with a 3.9-fold increase in the likelihood of diagnosing HCC. Furthermore, the NLR of 0.21 indicates a 79% increase in the probability of diagnosing HCC with a negative miRNA test. Furthermore, the pooled DOR of 19.44 (greater than 1) and AUC value of 0.88 emphasize the robust diagnostic capacity of miRNAs for discriminating between HCC and LC patients. Considering all the diagnostic values collectively, these findings strongly suggest that miRNAs have the potential to function as diagnostic markers for distinguishing between HCC and LC patients. Similarly, other meta-analyses have explored the diagnostic biomarker potential of circulating miRNAs in blood for various conditions, including Leukemia (49), osteosarcoma (43), cervical intraepithelial neoplasia (50), bladder cancer (51), and ovarian cancer (52). This phenomenon may be attributed to the capacity of miRNAs to play a role in hematopoietic differentiation and modulate the expression of oncogenes or tumor suppressor genes (53).
Several meta-analyses have delved into the diagnostic utility of biomarkers like serum alpha-fetoprotein (AFP), protein induced by vitamin K absence or antagonist II (PIVKA II), and osteopontin (OPN) in distinguishing patients with and without HCC (54–56). Notably, Jang et al. found combined AUC values of 0.786 (95% CI 0.740–0.831) for AFP, 0.729 (95% CI 0.680–0.779) for PIVKA-II, and 0.660 (95% CI 0.606–0.713) for OPN in distinguishing patients with HCC from those with LC, signifying moderate diagnostic accuracy (56). In contrast, our investigation highlighted miRNAs as promising diagnostic markers, boasting an AUC of 0.88 (95% CI: 0.85–0.91), implying potentially superior discriminatory ability between HCC and LC patients. This significant disparity needs deeper exploration into the underlying biological mechanisms driving miRNA’s enhanced diagnostic potential compared to traditional biomarkers. Moreover, it emphasizes the critical need for stringent validation studies to ascertain the reliability and reproducibility of miRNA-based diagnostic methodologies. Such efforts are pivotal for advancing the clinical translation of miRNA-based diagnostics, potentially revolutionizing HCC diagnosis and patient care.
It is important to note that there was observed heterogeneity among the studies included in this study. Therefore, the impact of these confounding factors was investigated through meta-regression and subgroup analyses. The subgroup analysis by ethnicity indicated that studies conducted on the European population showed higher overall diagnostic accuracy compared to those carried out on the Asian and African populations. The pooled AUC was 0.94. The finding is supported by a meta-analysis conducted by Wu et al. (57). The discrepancy in overall diagnostic accuracy observed between studies may stem from a combination of genetic, environmental, and disease-related factors. Additionally, the subgroup analysis by miRNAs expression, upregulated miRNAs had favorable diagnostic efficacy compared to downregulated miRNAs in differentiating HCC from LC, with an AUC of 0.89. The result is in line with other meta-analyses (57, 58). This may be attributed to their heightened sensitivity as markers of abnormal cell growth and proliferation. Additionally, their frequent association with an oncogenic role reflects the activation of tumorigenic pathways, thereby enhancing their effectiveness in detecting cancerous cells.
Subsequently, the diagnostic efficacy of miRNAs based on sample type was explored, revealing that serum-derived miRNAs exhibited relatively similar diagnostic performance when compared to plasma-derived miRNAs, with AUCs of 0.83 (0.74–0.88) and 0.83 (0.79–0.86), respectively. Thus, the detection of miRNAs in the blood using both serum miRNA assays and plasma miRNA assays is equally deemed useful as noninvasive biological methods for the early diagnosis of HCC.
On the other hand, the subgroup analysis by cut-off value showed that studies not reporting their cut-off values demonstrated superior diagnostic accuracy compared to studies with established cut-off values, as evidenced by an AUC of 0.93. This may result from differences in the number of studies and the absence of standardized cut-off values, which could introduce heterogeneity and potentially impact the overall diagnostic accuracy.
Most studies incorporated in this meta-analysis employed qRT-PCR for the detection of circulating miRNA. This method emerges as the optimal choice for future applications due to its high sensitivity in identifying low copies of miRNAs in serum samples, a crucial factor for routine testing in clinical settings (59). On the other hand, the studies analyzed in this context employed various endogenous controls, potentially contributing to data heterogeneity. The absence of a universally accepted housekeeping control for miRNA, coupled with the ongoing controversy in selecting an appropriate reference (60), complicates the standardization process.
The meta-regression analysis indicated that heterogeneity in sensitivity among the studies could be attributed to ethnicity and the setting of cut-off values. Conversely, factors such as ethnicity, sample source, regulation mode, sample size, and types of HCC were identified as reasons for heterogeneity in specificity among the studies. The potential explanation might be that different ethnicities living in diverse environments and possessing varying genetic backgrounds, lifestyles, and dietary habits yield distinct miRNA expression profiles (51).
In addition, the Fagan nomogram outcomes suggest that with a pre-test probability set at 20%, a positive miRNA test result (PLR of 50%) substantially increases the likelihood of precise identification of HCC patients from LC patients. Conversely, a negative result (NLR of 5%) significantly diminishes the probability of misdiagnosing HCC patients from those with LC. These results emphasize the potential utility of miRNA testing as a valuable and precise tool for differentiation between HCC patients and individuals with LC. Future research efforts should focus on further validating and extending these findings within this specific context. If confirmed, the incorporation of miRNA testing into clinical protocols could represent a non-invasive and effective strategy for enhancing diagnostic accuracy in distinguishing between HCC and LC, facilitating more targeted and timely interventions in these two patient groups.
This meta-analysis offers numerous advantages. Firstly, it reveals that circulating miRNAs possess significant diagnostic potential in effectively distinguishing between HCC and LC patients. This discovery introduces a fresh outlook on the creation of biomarkers for the differentiation of HCC from LC. Secondly, the meta-analysis undertook a thorough assessment of miRNAs, incorporating subgroup analysis and regression analysis to examine influencing factors like ethnicity, sample source, regulation mode, miRNA profiling, sample size, presence of cut-off values, and various types of HCC. This comprehensive approach aimed to analyze and elucidate the origins of heterogeneity in the findings.
However, it is essential to recognize the limitations of this study. Firstly, variations in the cut-off parameters for miRNAs and internal reference controls across the included studies could serve as a potential source of heterogeneity. Secondly, the meta-analysis did not assess the distinctions in the diagnostic accuracy of miRNAs specifically in HCC cases with diverse clinicopathological features. Thirdly, the ethnicities of the participants were predominantly limited to Asian, European, and African populations. While the identified miRNAs demonstrated remarkable diagnostic value for distinguishing HCC from LC within these ethnic groups, their diagnostic performance may not be universally applicable to HCC patients worldwide. Fourthly, the lack of a substantial number of comparable miRNAs for pooling results hinders the identification of specific single miRNAs or a panel as the optimal diagnostic biomarkers for distinguishing HCC from LC. Therefore, it is crucial to interpret these findings with caution.
In conclusion, this meta-analysis offers evidence supporting the identification of circulating miRNAs as innovative and valuable biomarkers for distinguishing between HCC and LC patients. The detection of miRNAs, especially the upregulated ones, can be employed to distinguish HCC patients from LC. Nevertheless, it is imperative to conduct thorough functional assessments and additional prospective studies involving larger sample sizes and diverse ethnic groups to validate and expand upon these findings.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
EA: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. MB: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. MW: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. FG: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. ZM: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. MT: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. DA: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. DW: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. AG: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. HE: Writing – review & editing, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1359414/full#supplementary-material
References
1. Debes, JD, Romagnoli, PA, Prieto, J, Arrese, M, Mattos, AZ, Boonstra, A, et al. Serum biomarkers for the prediction of hepatocellular carcinoma. Cancers. (2021) 13:1681. doi: 10.3390/cancers13071681
2. Rumgay, H, Arnold, M, Ferlay, J, Lesi, O, Cabasag, CJ, Vignat, J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol. (2022) 77:1598–606. doi: 10.1016/j.jhep.2022.08.021
3. Bray, F, Ferlay, J, Soerjomataram, I, Siegel, RL, Torre, LA, and Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
4. Roth, GA, Abate, D, Abate, KH, Abay, SM, Abbafati, C, Abbasi, N, et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392:1736–88. doi: 10.1016/S0140-6736(18)32203-7
5. Sepanlou, SG, Safiri, S, Bisignano, C, Ikuta, KS, Merat, S, Saberifiroozi, M, et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. (2020) 5:245–66. doi: 10.1016/S2468-1253(19)30349-8
6. Dhanasekaran, R, Bandoh, S, and Roberts, LR. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. F1000Research. (2016):5. doi: 10.12688/f1000research.6946.1
7. Gani, RA. Hepatocellular carcinoma (HCC) surveillance–comprehensive management in liver cirrhosis patients. Ind J Gastroenterol Hepatol Digestive Endosc. (2020) 18:137–9. doi: 10.24871/1832017137-139
8. Wang, S, Yang, Y, Sun, L, Qiao, G, Song, Y, and Liu, B. Exosomal microRNAs as liquid biopsy biomarkers in hepatocellular carcinoma. Onco Targets Ther. (2020) 13:2021–30. doi: 10.2147/OTT.S232453
9. Zhang, N, Hu, Z, Qiang, Y, and Zhu, X. Circulating miR-130b-and miR-21-based diagnostic markers and therapeutic targets for hepatocellular carcinoma. Mol Genet Genomic Med. (2019) 7:e1012. doi: 10.1002/mgg3.1012
10. Wang, T, and Zhang, K-H. New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front Oncol. (2020) 10:1316. doi: 10.3389/fonc.2020.01316
11. Huang, JT, Liu, SM, Ma, H, Yang, Y, Zhang, X, Sun, H, et al. Systematic review and meta-analysis: circulating miRNAs for diagnosis of hepatocellular carcinoma. J Cell Physiol. (2016) 231:328–35. doi: 10.1002/jcp.25135
12. Berretta, M, Cavaliere, C, Alessandrini, L, Stanzione, B, Facchini, G, Balestreri, L, et al. Serum and tissue markers in hepatocellular carcinoma and cholangiocarcinoma: clinical and prognostic implications. Oncotarget. (2017) 8:14192–220. doi: 10.18632/oncotarget.13929
13. Schaffler-Schaden, D, Birsak, T, Zintl, R, Lorber, B, and Schaffler, G. Risk of needle tract seeding after coaxial ultrasound-guided percutaneous biopsy for primary and metastatic tumors of the liver: report of a single institution. Abdominal Radiol. (2020) 45:3301–6. doi: 10.1007/s00261-019-02120-1
14. Hauptman, N, and Glavac, D. MicroRNAs and long non-coding RNAs: prospects in diagnostics and therapy of cancer. Radiol Oncol. (2013) 47:311–8. doi: 10.2478/raon-2013-0062
15. Ye, Z, Li, Z-H, and He, S-Z. miRNA-1273g-3p involvement in development of diabetic retinopathy by modulating the autophagy-lysosome pathway. Med Sci Monitor. (2017) 23:5744–51. doi: 10.12659/MSM.905336
16. Suzuki, H, Katsura, A, Matsuyama, H, and Miyazono, K. MicroRNA regulons in tumor microenvironment. Oncogene. (2015) 34:3085–94. doi: 10.1038/onc.2014.254
17. Lu, TX, and Rothenberg, ME. MicroRNA. J Allergy Clin Immunol. (2018) 141:1202–7. doi: 10.1016/j.jaci.2017.08.034
18. Ye, J, Xu, M, Tian, X, Cai, S, and Zeng, S. Research advances in the detection of miRNA. J Pharmac Anal. (2019) 9:217–26. doi: 10.1016/j.jpha.2019.05.004
19. Niveditha, D, Jasoria, M, Narayan, J, Majumder, S, Mukherjee, S, Chowdhury, R, et al. Common and unique microRNAs in multiple carcinomas regulate similar network of pathways to mediate cancer progression. Sci Rep. (2020) 10:2331. doi: 10.1038/s41598-020-59142-9
20. Gumilas, NSA, Widodo, I, Ratnasari, N, and Heriyanto, DS. Potential relative quantities of miR-122 and miR-150 to differentiate hepatocellular carcinoma from liver cirrhosis. Non-coding RNA Research. (2022) 7:34–9. doi: 10.1016/j.ncrna.2022.01.004
21. Shehab-Eldeen, S, Nada, A, Abou-Elela, D, El-Naidany, S, Arafat, E, and Thoria, T. Diagnostic performance of microRNA-122 and microRNA-224 in hepatitis C virus-induced hepatocellular carcinoma (HCC). Asian Pacific journal of cancer prevention: APJCP. (2019) 20:2515–22. doi: 10.31557/APJCP.2019.20.8.2515
22. Li, J, Qiyu, S, Wang, T, Jin, B, and Li, N. Improving the detection of hepatocellular carcinoma using serum AFP expression in combination with GPC3 and micro-RNA miR-122 expression. Open Life Sci. (2019) 14:53–61. doi: 10.1515/biol-2019-0007
23. Freeman, K, Mistry, H, Tsertsvadze, A, Royle, P, McCarthy, N, Taylor-Phillips, S, et al. Multiplex tests to identify gastrointestinal bacteria, viruses and parasites in people with suspected infectious gastroenteritis: a systematic review and economic analysis. NIHR J Library. (2017) 21:1–188. doi: 10.3310/hta21230
24. Jackson, D, White, IR, and Thompson, SG. Extending DerSimonian and Laird’s methodology to perform multivariate random effects meta-analyses. Stat Med. (2010) 29:1282–97. doi: 10.1002/sim.3602
25. Higgins, JP, Thompson, SG, Deeks, JJ, and Altman, DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
26. Xu, L, Wei, B, Hui, H, and Liu, Y. Association of serum microRNA-125b and HBV-related hepatocellular carcinoma in Chinese Han patients. Int J Clin Exp Med. (2018) 11:3699–703.
27. Rashad, NM, El-Shal, AS, Shalaby, SM, and Mohamed, SY. Serum miRNA-27a and miRNA-18b as potential predictive biomarkers of hepatitis C virus-associated hepatocellular carcinoma. Mol Cell Biochem. (2018) 447:125–36. doi: 10.1007/s11010-018-3298-8
28. El Mahdy, HA, Abdelhamid, IA, Amen, AI, Abdelsameea, E, and Hassouna, MM. MicroRNA-215 as a diagnostic marker in Egyptian patients with hepatocellular carcinoma. Asian Pacific J Cancer Prevent. (2019) 20:2723–31. doi: 10.31557/APJCP.2019.20.9.2723
29. Ali, LH, Higazi, AM, Moness, HM, Farag, NM, Saad, ZM, Moukareb, HA, et al. Clinical significances and diagnostic utilities of both miR-215 and squamous cell carcinoma antigen-IgM versus alpha-fetoprotein in Egyptian patients with hepatitis C virus-induced hepatocellular carcinoma. Clin Exp Gastroenterol. (2019) 12:51–66. doi: 10.2147/CEG.S179832
30. El-Hamouly, MS, Azzam, AA, Ghanem, SE, El-Bassal, FI, Shebl, N, and Shehata, AM. Circulating microRNA-301 as a promising diagnostic biomarker of hepatitis C virus-related hepatocellular carcinoma. Mol Biol Rep. (2019) 46:5759–65. doi: 10.1007/s11033-019-05009-w
31. Hassan, AS, Elgendy, NA, Tawfik, NA, and Elnasser, AM. Serum miR-483-5p and miR-133a as biomarkers for diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. Egypt J Immunol. (2019) 26:31–40.
32. Mohamed, AA, Omar, AAA, El-Awady, RR, Hassan, SMA, Eitah, WMS, Ahmed, R, et al. MiR-155 and MiR-665 role as potential non-invasive biomarkers for hepatocellular carcinoma in Egyptian patients with chronic hepatitis C virus infection. J Transl Intern Med. (2020) 8:32–40. doi: 10.2478/jtim-2020-0006
33. Aboelwafa, RA, Ellakany, WI, Gamaleldin, MA, and Saad, MA. The expression of microRNA-331-3p and microRNA-23b3 in Egyptian patients with early-stage hepatocellular carcinoma in hepatitis C-related liver cirrhosis. Egypt Liver J. (2021) 11:49. doi: 10.1186/s43066-021-00122-7
34. Awwad, A, Salem, A, and Asser, S. Circulating microRNA-221 as a diagnostic biomarker for hepatitis C virus-related hepatocellular carcinoma. Microbes Infect Diseases. (2021) 2:68–76. doi: 10.21608/mid.2020.46774.1072
35. Yasser, MB, Abdellatif, M, Emad, E, Jafer, A, Ahmed, S, Nageb, L, et al. Circulatory miR-221 & miR-542 expression profiles as potential molecular biomarkers in hepatitis C virus mediated liver cirrhosis and hepatocellular carcinoma. Virus Res. (2021) 296:198341. doi: 10.1016/j.virusres.2021.198341
36. Gharib, AF, Eed, EM, Khalifa, AS, Raafat, N, Shehab-Eldeen, S, Alwakeel, HR, et al. Value of serum miRNA-96-5p and miRNA-99a-5p as diagnostic biomarkers for hepatocellular carcinoma. Int J General Med. (2022) 15:2427–36. doi: 10.2147/IJGM.S354842
37. Moshiri, F, Salvi, A, Gramantieri, L, Sangiovanni, A, Guerriero, P, De Petro, G, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. (2018) 9:15350–64. doi: 10.18632/oncotarget.24601
38. McGlynn, KA, Petrick, JL, and El-Serag, HB. Epidemiology of hepatocellular carcinoma. Hepatology. (2021) 73:4–13. doi: 10.1002/hep.31288
39. Cartier, V, and Aubé, C. Diagnosis of hepatocellular carcinoma. Diagn Interv Imaging. (2014) 95:709–19. doi: 10.1016/j.diii.2014.06.004
40. Bialecki, ES, and Di Bisceglie, AM. Diagnosis of hepatocellular carcinoma. HPB. (2005) 7:26–34. doi: 10.1080/13651820410024049
41. Ayuso, C, Rimola, J, Vilana, R, Burrel, M, Darnell, A, García-Criado, Á, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. (2018) 101:72–81. doi: 10.1016/j.ejrad.2018.01.025
42. Lieberman, J, Slack, F, Pandolfi, PP, Chinnaiyan, A, Agami, R, and Mendell, JT. Noncoding RNAs and cancer. Cell. (2013) 153:9–10. doi: 10.1016/j.cell.2013.03.019
43. Gao, S-S, Wang, Y-J, Zhang, G-X, and Zhang, W-T. Potential diagnostic value of miRNAs in peripheral blood for osteosarcoma: a meta-analysis. J Bone Oncol. (2020) 23:100307. doi: 10.1016/j.jbo.2020.100307
44. Rice, J, Roberts, H, Burton, J, Pan, J, States, V, Rai, SN, et al. Assay reproducibility in clinical studies of plasma miRNA. PLoS One. (2015) 10:e0121948. doi: 10.1371/journal.pone.0121948
45. Glinge, C, Clauss, S, Boddum, K, Jabbari, R, Jabbari, J, Risgaard, B, et al. Stability of circulating blood-based microRNAs–pre-analytic methodological considerations. PLoS One. (2017) 12:e0167969. doi: 10.1371/journal.pone.0167969
46. Mitchell, PS, Parkin, RK, Kroh, EM, Fritz, BR, Wyman, SK, Pogosova-Agadjanyan, EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. (2008) 105:10513–8. doi: 10.1073/pnas.0804549105
47. Chen, X, Ba, Y, Ma, L, Cai, X, Yin, Y, Wang, K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. (2008) 18:997–1006. doi: 10.1038/cr.2008.282
48. Coenen-Stass, AM, Pauwels, MJ, Hanson, B, Martin Perez, C, Conceição, M, Wood, MJ, et al. Extracellular microRNAs exhibit sequence-dependent stability and cellular release kinetics. RNA Biol. (2019) 16:696–706. doi: 10.1080/15476286.2019.1582956
49. Zhang, W-T, Zhang, G-X, and Gao, S-S. The potential diagnostic accuracy of circulating microRNAs for leukemia: a meta-analysis. Technol Cancer Res Treat. (2021) 20:15330338211011958.
50. Jiang, Y, Hu, Z, Zuo, Z, Li, Y, Pu, F, Wang, B, et al. Identification of circulating MicroRNAs as a promising diagnostic biomarker for cervical intraepithelial neoplasia and early cancer: a meta-analysis. Biomed Res Int. (2020) 2020:1–14. doi: 10.1155/2020/4947381
51. Shi, H-B, Yu, J-X, Yu, J-X, Feng, Z, Zhang, C, Li, G-Y, et al. Diagnostic significance of microRNAs as novel biomarkers for bladder cancer: a meta-analysis of ten articles. World J Surg Oncol. (2017) 15:1–10. doi: 10.1186/s12957-017-1201-9
52. Wang, X, Kong, D, Wang, C, Ding, X, Zhang, L, Zhao, M, et al. Circulating microRNAs as novel potential diagnostic biomarkers for ovarian cancer: a systematic review and updated meta-analysis. J Ovarian Res. (2019) 12:1–12. doi: 10.1186/s13048-019-0482-8
53. Schickel, R, Boyerinas, B, Park, S, and Peter, M. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. (2008) 27:5959–74. doi: 10.1038/onc.2008.274
54. Zhang, J, Chen, G, Zhang, P, Zhang, J, Li, X, Dn, G, et al. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. PLoS One. (2020) 15:e0228857. doi: 10.1371/journal.pone.0228857
55. Xing, H, Zheng, Y-J, Han, J, Zhang, H, Li, Z-L, Lau, W-Y, et al. Protein induced by vitamin K absence or antagonist-II versus alpha-fetoprotein in the diagnosis of hepatocellular carcinoma: a systematic review with meta-analysis. Hepatobiliary Pancreat Dis Int. (2018) 17:487–95. doi: 10.1016/j.hbpd.2018.09.009
56. Jang, ES, Jeong, S-H, Kim, J-W, Choi, YS, Leissner, P, and Brechot, C. Diagnostic performance of alpha-fetoprotein, protein induced by vitamin K absence, osteopontin, Dickkopf-1 and its combinations for hepatocellular carcinoma. PLoS One. (2016) 11:e0151069. doi: 10.1371/journal.pone.0151069
57. Wu, N, Sun, H, Sun, Q, Zhang, F, Ma, L, Hu, Y, et al. Circulating microRNAs as diagnostic biomarkers for melanoma: a systematic review and meta-analysis. BMC Cancer. (2023) 23:1–14. doi: 10.1186/s12885-023-10891-6
58. Ma, L, Wen, Y, Li, Z, Wu, N, and Wang, Q. Circulating microRNAs as potential diagnostic biomarkers for diabetic retinopathy: a meta-analysis. Front Endocrinol. (2022) 13:929924. doi: 10.3389/fendo.2022.929924
59. Mestdagh, P, Hartmann, N, Baeriswyl, L, Andreasen, D, Bernard, N, Chen, C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. (2014) 11:809–15. doi: 10.1038/nmeth.3014
Keywords: miRNAs, non-coding RNAs, diagnostic biomarkers, hepatocellular carcinoma, HCC, liver cancer, liver cirrhosis, meta-analysis
Citation: Alemayehu E, Belete MA, Walle M, Getu F, Mulatie Z, Teshome M, Anley DT, Weldehanna DG, Gedefie A and Ebrahim H (2024) Diagnostic accuracy of circulating miRNAs to discriminate hepatocellular carcinoma from liver cirrhosis: a systematic review and meta-analysis. Front. Med. 11:1359414. doi: 10.3389/fmed.2024.1359414
Edited by:
Gian Paolo Caviglia, University of Turin, ItalyReviewed by:
Rohini Mehta, Quest Diagnostics (United States), United StatesWei Li, Jilin University, China
Copyright © 2024 Alemayehu, Belete, Walle, Getu, Mulatie, Teshome, Anley, Weldehanna, Gedefie and Ebrahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ermiyas Alemayehu, ZXJtaXlhczAwMDlAZ21haWwuY29t
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Ermiyas Alemayehu
Ermiyas Alemayehu Melaku Ashagrie Belete
Melaku Ashagrie Belete Muluken Walle
Muluken Walle Fasil Getu
Fasil Getu Zewudu Mulatie1
Zewudu Mulatie1 Denekew Tenaw Anley
Denekew Tenaw Anley Daniel Gebretsadik Weldehanna
Daniel Gebretsadik Weldehanna Alemu Gedefie
Alemu Gedefie Hussen Ebrahim
Hussen Ebrahim