
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 07 November 2023
Sec. Precision Medicine
Volume 10 - 2023 | https://doi.org/10.3389/fmed.2023.1282820
 Uma Maheswara Rao Dindi1
Uma Maheswara Rao Dindi1 Suhadha Parveen Sadiq1
Suhadha Parveen Sadiq1 Sameer Al-Ghamdi2
Sameer Al-Ghamdi2 Naif Abdurhman Alrudian2
Naif Abdurhman Alrudian2 Salman Bin Dayel3
Salman Bin Dayel3 Abdulwahab Ali Abuderman4
Abdulwahab Ali Abuderman4 Mohammad Shahid4
Mohammad Shahid4 Thiyagarajan Ramesh4*
Thiyagarajan Ramesh4* Ravikumar Vilwanathan1*†
Ravikumar Vilwanathan1*†Introduction: Epigenetic enzymes can interact with a wide range of genes that actively participate in the progression or repression of a diseased condition, as they are involved in maintaining cellular homeostasis. Sirtuins are a family of Class III epigenetic modifying enzymes that regulate cellular processes by removing acetyl groups from proteins. They rely on NAD+ as a coenzyme in contrast to classical histone deacetylases (HDACs) (Class I, II, and IV) that depend on Zn+ for their activation, linking their function to cellular energy levels. There are seven mammalian sirtuin isoforms (Sirt1-7), each located in different subcellular compartments. Sirtuins have emerged as a promising target, given that inhibitors of natural and synthetic sources are highly warranted. Imidazole derivatives are often investigated as sirtuin regulators due to their ability to interact with the binding site and modulate their activity. Imidazole bestows many possible substitutions on its ring and neighboring atoms to design and synthesize derivatives with specific target selectivity and improved pharmacokinetic properties, optimizing drug development.
Materials and methods: Ligand preparation, protein preparation, molecular docking, molecular dynamics, density function theory (DFT) analysis, and absorption, distribution, metabolism, and excretion (ADME) analysis were performed to understand the interacting potential and effective stability of the ligand with the protein. RT-PCR and Western blot analyses were performed to understand the impact of ligands on the gene and protein expression of Class III HDAC enzymes.
Results and discussion: We evaluated the sirtuin inhibition activity of our in-house compound comprised of imidazole derivatives by docking the molecules with the protein data bank. ADME properties of all the compounds used in the study were evaluated, and it was found that all fall within the favorable range of being a potential drug. The molecule with the highest docking score was analyzed using DFT, and the specific compound was used to treat the non-small cell lung cancer (NSCLC) cell lines A549 and NCI-H460. The gene and protein expression data support the in-silico finding that the compound Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate has an inhibitory effect on nuclear sirtuins. In conclusion, targeting sirtuins is an emerging strategy to combat carcinogenesis. In this study, we establish that Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate possesses a strong inhibitory effect on nuclear sirtuins in NSCLC cell lines.
- Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate has a higher docking score, glide score, and glide energy compared to other compounds within the sirtuin family proteins.
- Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate has better stability when interacting with nuclear sirtuins.
- Imidazole derivative effectively reduces the viability of cancer cell lines A549 and NCI-H460 at lower concentrations compared to imidazole.
- Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate greatly affects sirtuins family members' gene expression and protein expression.
Imidazole derivatives have shown promising potential in combating various types of cancers. They can be designed and synthesized to specifically target molecular pathways and proteins that are crucial for the growth and survival of cancer cells (1, 2). This targeted approach minimizes damage to healthy cells and tissues, leading to fewer side effects. Imidazole derivatives have demonstrated the ability to inhibit cell proliferation and potentially induce apoptosis in breast cancer cells (3). They can inhibit angiogenesis, and by cutting off the blood supply to the tumor, these compounds can deprive the cancer cells of essential nutrients, thereby hindering their growth (4). Imidazole derivatives can sensitize cancer cells to the effects of chemotherapy and radiation therapy (5). They have demonstrated better antimicrobial, antiviral, anti-inflammatory, antifungal, antiparasitic, and anticancer properties compared to other synthetic compounds (6). The significance of imidazole derivatives lies in their diverse biological activities, structural versatility, and potential therapeutic applications. Using imidazole derivatives to study epigenetic enzymes to modulate their activity and restore normal gene expression patterns is an active area of research (7). Imidazole and its derivatives can either work as a friend or foe for epigenetic enzymes. Our previous research emphasized that heterocyclic imidazole derivatives intervene in classic histone deacetylase (HDAC) enzyme activity and inhibit their operations in lung cancer cell lines (8). However, class III-HDAC is the most active form in orchestrating the progression and suppression of various diseases (9). Furthermore, imidazole derivatives can function as activators or inhibitors of sirtuins. As activators, imidazole derivatives can typically bind to the protein and increase their enzymatic activity, leading to enhanced deacetylation or ADP-ribosylation of target proteins. On the other hand, as inhibitors, they can block sirtuin activity, leading to altered gene expression patterns and cellular functions. Sirtuins play a crucial role in metabolic pathways, including glucose and lipid metabolism (10). Activation of sirtuins by imidazole derivatives or other compounds may enhance metabolic efficiency and potentially offer therapeutic benefits in addressing metabolic disorders (11). Sirtuins have also been implicated in neuroprotection and cognitive function. Therefore, imidazole derivatives that modulate sirtuin activity may have potential applications in neurodegenerative diseases, such as Alzheimer's and Parkinson's, by promoting neuronal survival and enhancing cellular stress responses (12). Sirtuins can act as tumor suppressors by regulating cell cycle progression, DNA repair, and apoptosis (13). Sirtuin activators, including certain imidazole derivatives, may have anticancer properties by promoting tumor cell death and inhibiting tumor growth. Furthermore, based on the type of cancer tissues, the activity of sirtuins differs (14), making the development of effective and selective sirtuin regulators a complex task.
The development of sirtuin inhibitors as potential therapeutic agents for combating cancer, including lung cancer, is an active area of research (15). Inhibiting specific sirtuin isoforms may offer opportunities for novel cancer treatments (16). There are seven mammalian sirtuin isoforms (Sirt 1-7), each with unique functions and subcellular localizations (17). To develop effective inhibitors, it is crucial to understand the roles of individual sirtuin isoforms in lung cancer biology and identify those most relevant to disease progression. As sirtuins actively participate in cancer progression and regulation (18, 19), designing imidazole derivatives that can interfere with the activity of sirtuin has become imperative. Structure-based drug design and screening approaches can be used to identify small molecules with potential inhibitory activity against sirtuin isoforms. Imidazole derivatives and other chemical scaffolds can be modified and optimized to interact with the NAD+-binding site of the sirtuin isoform. Achieving isoform specificity is critical in the development of sirtuin regulators to avoid unwanted effects on non-target sirtuin isoforms, which may have distinct roles in normal cellular functions. Among the top 75% of the small-molecule medications containing heterocycles with nitrogen, imidazole ranks within the top 10, favoring a ring system that acts as essential building blocks for the development of novel drugs (20). In this context, multiple derivatives of imidazole were synthesized to study their characteristics with respect to cancer (21). Given that the action of imidazole derivatives on the modulation of sirtuins could have a beneficial effect in transformed cells, in the present study, we aimed to understand the inhibitory effect of our in-house imidazole derivatives on sirtuins.
An imidazole and six imidazole derivatives were generated in the ligand library. ChemDraw Pro 8.0 was used to draw the 3D structures of the ligands (Figure 1). The ligand structures were optimized using Maestro version 13.2.128, Release 2022-2, and the LigPrep module was performed with force field OPLS_2005.
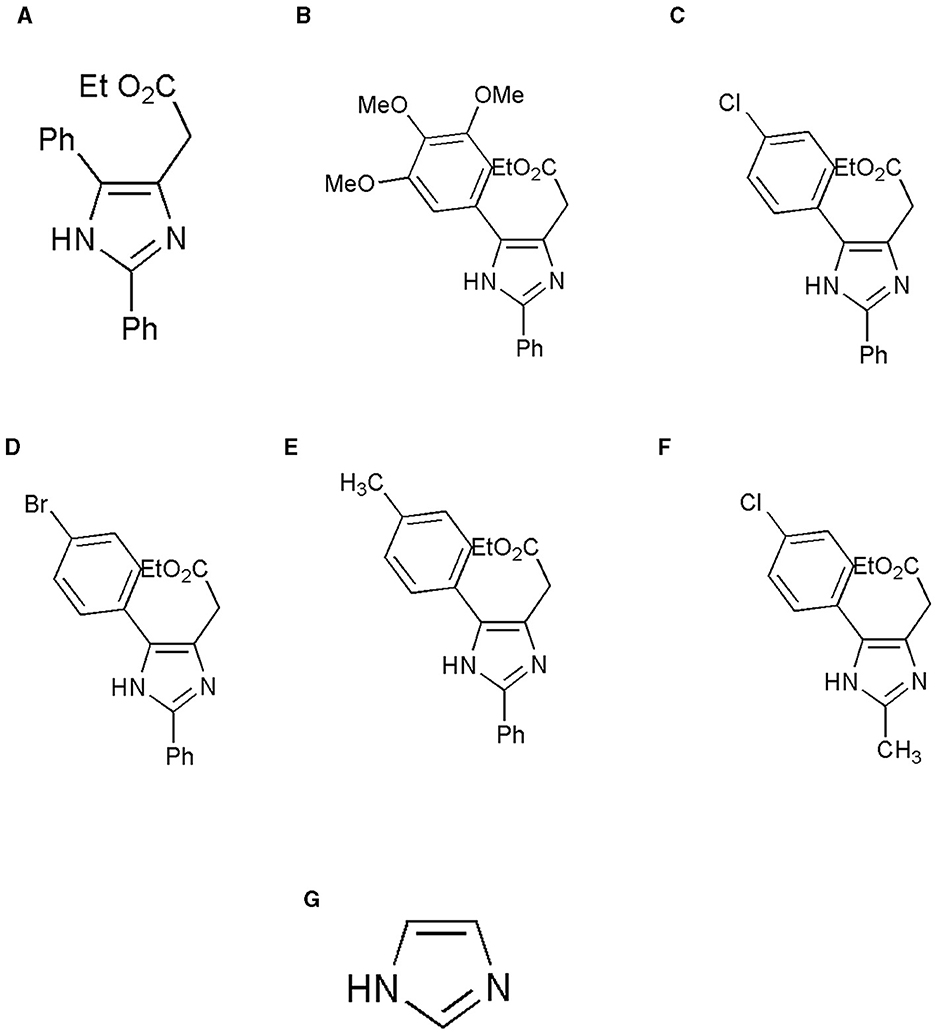
Figure 1. Structure of the ligands generated using ChemDraw is shown. (A) Ethyl 2-(2,5-diphenyl-1H-imidazole-4-yl) acetate, (B) Ethyl 2-[2-phenyl-5-(3,4,5-trimethoxyphenyl)-1H-imidazol-4-yl] acetate, (C) Ethyl 2-[5-(4-chlorophenyl)-2-phenyl-1H-imidazol-4-yl] acetate, (D) Ethyl 2-[5-(4-bromophenyl)-2-phenyl-1H-imidazol-4-yl] acetate, (E) Ethyl 2-{2-phenyl-5-[4-(trifluoromethyl) phenyl]-1H-imidazol-4-yl} acetate, (F) Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate, and (G) Imidazole. (A) Adapted from (8) with permission from Elsevier. Copyright ©2020 Elsevier. (F) Adapted with permission from (22). Copyright ©2014 American Chemical Society.
Barring Sirt4, the remaining six sirtuin isoforms were considered in the in-silico study. The protein data bank (PDB) structures of Sirt1, Sirt2, Sirt3, Sirt5, Sirt6, and Sirt7 were recovered from the Research Collaboratory for Structural Bioinformatics (RCSB) as 4I5I, 4RMH, 4JSR, 6LJK, 3K35, and 5IQZ, respectively. The proteins subjected to docking studies were pre-processed, optimized, removed water, and minimized using a protein preparation wizard. Water molecules, non-essential atoms, and attached ligands were annihilated. Further preparation was performed by adding missing atoms in the protein residues, fixing the alternate confirmations, and adding hydrogen. Grid generation surrounding the binding site was achieved as per the protocol followed in Maestro version 13.2.128, Release 2022-2.
Grid-based Ligand Docking with Energetics (GLIDE) Maestro version 13.2.128, Release 2022-2, was used to dock the desired imidazole derivatives with all six PDB structures. The ligand molecules were ranked based on their interactive score and energy in order to understand their interaction with protein molecules. Sirt 1-7 proteins, except Sirt4, were docked against the imidazole derivatives. Correspondence to the docking score, glide score, and glide energy of the ligand and the protein, poses were visualized using Maestro GUI.
Based on the docking score, the top ligand bound to all the sirtuins was subjected to molecular dynamic (MD) simulation using Desmond program version 7.0 (academic version). An orthorhombic periodic box of dimension 10 Å3 with solvent TIP3P at force field OPLS_2005 was set, and the neutralization of the system at stable pH was carried out by adding counter ions (Na+ and Cl–) to the system builder. The protein-ligand complex underwent minimization of energy and attained a pre-equilibrium state. Molecular dynamics simulations were conducted for a duration of 100 ns, utilizing a relaxation time of 1 ps and maintaining a consistent temperature of 300 kelvin. Throughout the simulation, a series of 1000 frames was generated using a time step of 20 ps, serving as the foundation for deriving average structures during the production phase. Additionally, the root mean square fluctuation (RMSF) and root mean square deviation (RMSD) were independently assessed for the protein-ligand complex structure and the protein structure. The purpose of this simulation was to investigate the dynamic stability of the complexes and was visualized by plotting RMSF and RMSD values against time.
The electronic structural characteristics play a pivotal role in elucidating the molecular interactions governing small molecule dynamics, molecular structure, and quantum properties. To delve into this, the Gaussian 09 software package was employed to conduct density functional theory (DFT) calculations on the primary ligand. Using the B3LYP-D3 hybrid functional and the 6-311G**++ (2d, 2p) basis set, the calculation encompassed the determination of the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) energy levels, collectively known as frontier orbitals. The HOMO represents the outermost electron of the ligand capable of donation upon binding to a protein, while the LUMO represents the region of the ligand with an affinity to accept electrons from the protein during complex formation. As an essential metric, the energy gap (HOMO-LUMO) was computed to clarify chemical potential, electron affinity, chemical stability, hardness, and the chemical potential of the ligand molecules.
The imidazole structure was constructed using Chemdraw, and various interactable R groups were added to the side chains. The structures were then exported into.sdf files and read in Schrodinger Maestro software version 13.2.128(2022-2). The structures were analyzed using the Qikprop module to understand their chemical properties. All the compounds underwent absorption, distribution, metabolism, and excretion (ADME) analysis where the molecular weight, number of hydrogen bonds accepted and donated, solvent accessible surface area (SASA), force-field overlapping scoring algorithm (FOSA), flexible inhibitor surface anchoring (FISA), percentage of human absorption, and the permeability to the gut and brain were calculated. This analysis indicated the ability of the domestic compounds to acquire the qualities of being a promiscuous drug.
The A549 (adenocarcinoma) and NCI-H460 (large cell carcinoma) human non-small-cell lung cancer cell lines used in this study were purchased from the National Center for Cell Science (NCCS) Pune, India. The reagents for cell culture, such as Dulbecco's modified eagle's medium (DMEM), fetal bovine serum (FBS), 10 × phosphate-buffered saline (pH 7.2), 10 × Trypsin-EDTA, and 100 × penicillin-streptomycin (pen strep, pH 7.2) antibiotic solution were from Hi-Media Laboratories, Mumbai, India. Cell culture plates were purchased from Wuxi NEST Biotechnology Co. Ltd., Jiangsu, China. The other plastic wares were purchased from Tarsons Pvt. Ltd.. The complete medium, which included DMEM, 10% FBS, and 1% pen strep solution, was used for culturing the cells. The cultured cell lines were maintained in a CO2 incubator at an atmospheric temperature of 37°C and supplied with 5% CO2.
The half maximal inhibitory concentration (IC50) is a quantitative measure of a substance (e.g., drug) to inhibit biological processes or biological components by 50%. The MTT assay is a well-known assay to determine IC50. The cell lines grown in monolayer were trypsinized and pelleted. The cell pellet was dissolved in a complete medium. The cells were then counted using the Trypan blue and hemacytometer. Approximately 1 × 104 cells were seeded in 96-well plates and incubated overnight in a CO2 incubator. After incubation, the old media were removed, and serum-free media were added to the wells. Imidazole and its derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate was synthesized as reported earlier (22) and were used to treat the cells seeded with different concentrations. Treated concentrations of imidazole ranged from 100 μM to 1000 μM, and its derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate concentration ranged from 50 μM to 500 μM and incubated for 24 h. After 24 h of treatment, 20 μL of 5 mg/mL concentration of MTT (3 (4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) was added to the wells, and incubated for 4 h. Later, the media were removed from the wells and 200 μL of dimethyl sulphoxide (DMSO) was added to dissolve the formazan crystals. After 30 min of incubation, the intensity of the purple color formed was measured by an ELISA plate reader (Bio-Rad Laboratories, Hercules, CA, USA) at 595 nm. The higher the intensity, the greater the number of viable cells, and the lower the intensity, the lesser the number of viable cells. MTT (RM1131) and DMSO (GRM5856) were purchased from Hi-Media Laboratories, Mumbai, India. The percentage of viable cells was calculated using the following formula:
% Cell viability = 100 – [(Mean OD of control cells – Mean OD of treated cells)/Mean OD of control cells × 100%]
The non-small cell lung cancer (NSCLC) cell lines A549 and NCI-H460 cells were seeded into culture plates using the complete medium. When the culture reached 80% confluency, the cells were treated with imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. The IC50 concentration of Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate in serum-free medium was used for treatment. The control was maintained in a serum-free medium without Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate was used as control. After the treatment period, total RNA was isolated from untreated (control) and Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate treated A549 and NCI-H460 cell lines using TRIzol reagent (RNA isoplus). The purity and quantity of the total RNA isolated was determined by BioPhotometer (Eppendorf, Hamburg, Germany). 1 μg of total RNA was taken for cDNA construction using the PrimeScript RT reagent kit following the manufacturer's instructions. Gene expression studies were performed in StepOnePlus real-time PCR system (Applied Biosystems, Thermo Fisher Scientific, MA, USA) using 2 × SYBR green master mix [TB Green Premix Ex Taq II (Tli RNase H Plus)]. The sample preparation was done according to the instructions in the datasheet provided by the manufacturer. Next, 0.5 μL of cDNA was used for each 10 μL reaction. The PCR condition was initiated by denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C – 60°C for 30 s (depending on the specific gene), and melt curve stage conditions set at 95°C for 15 s, 60°C for 60 s, and 95°C for 15 s. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. For normalization, the Ct value of the target gene was subtracted from the Ct value of GAPDH, which resulted in ΔCt values. The ΔCt values of the target were then subtracted from the ΔCt value of the control. This gave the ΔΔCt value, which is commonly used to find the mRNA expression. The expression is represented in fold change by 2(−ΔΔCt). The list of primers used in this study is provided in Table 1. Trizol for total RNA extraction (RNA isoplus, cat. # 9108), PrimeScript RT reagent kit (Perfect Real Time, Cat. # RR037A) for cDNA synthesis, and SYBR green master mix [TB Green® Premix Ex Taq™ II (Tli RNaseH Plus, Cat. #RR820A)] for gene expression studies were purchased from Takara Bio Inc. (Japan).
The A549 and NCI-H460 cell lines grown in monolayer were treated with Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate in serum-free medium, and untreated cell lines in serum-free medium were used as control samples. After the treatment period, the cells were washed with ice-cold PBS two times. Later, RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktails was added to the plates and incubated for 30 min in cold conditions. After incubation, the cell lysate was scraped using the cell scraper and collected into 1.5-mL tubes, which were then vortexed for 30 min at different intervals, maintaining cold conditions by keeping the tubes on ice. Later, the cell lysate was centrifugated at 12,000 g for 20 min at 4°C in the cooling centrifuge, and the supernatant containing the total protein was collected. The protein samples were quantified by Lowry's method. In total, 50 μg of protein from whole protein samples were used for the Western blot analysis. The 50 μg of whole protein was separated using SDS-PAGE (10% and 12% gels), and the separated proteins were transferred onto the nitrocellulose membrane by wet transfer. The membrane was blocked with 5% skimmed milk for 2 h, after which it was washed with 1 × TBST for 5 min. The washing was done three times. Following the TBST wash, the membrane was incubated overnight with specific primary antibodies at 4°C. Once the overnight incubation was over, the membrane was washed three times with 1 × TBST for 5 min each time. The alkaline phosphatase-conjugated secondary antibody specific to the primary antibody was added and incubated for 4 h at 4°C, after which the membrane was washed three times with 1 × TBST for 5 min each time. The blots were developed with BCIP/NBT chromogenic substrate, and the images were scanned. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to measure the intensity of the bands. The intensity of the measured target protein of the control sample was divided by the intensity of the measured βeta Actin of the control. Likewise, the intensity of the target protein of the treated sample was divided by the intensity of βeta-actin of the treated sample. The values obtained after nullifying with βeta-actin were used to represent protein expression in fold change by dividing the intensity of the target protein in the treated sample by that in the control sample. RIPA Lysis Buffer System (sc-24948) for protein isolation was purchased from Santa Cruz (CA, USA). Nitrocellulose Membrane (0.45μm, cat.log1620115) and Precision Plus Protein™ Kaleidoscope™ Prestained Protein Standards (#1610375) were purchased from Bio-Rad Laboratories (Hercules, California, United States). PageRuler Prestained Protein Ladder (cat.log 26616) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The bovine serum albumin, skimmed milk powder, and all the other fine chemicals used in the SDS and Western blotting were purchased from Sisco Research Laboratories Pvt. Ltd. (SRL, Maharashtra, India). The antibodies used in the study of Sirt 1 (9475), Sirt 2 (12650), Sirt 3 (5490), Sirt 5 (8782), Sirt 6 (12486), and Sirt 7 (5360) came from Cell Signaling Technology, USA. β-actin (MAB8929-SP) was purchased from Novus Biologicals (Briarwood, CO, USA). The secondary antibodies, such as goat anti-mouse IgG H&L (alkaline phosphatase) (ab97020) and goat anti-rabbit IgG H&L (alkaline phosphatase) (ab6722), were purchased from Abcam (Cambridge, UK). 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) substrate (B1911-100ML) was purchased from (Sigma-Aldrich Co, USA).
The graphical presentation of the data in this study was from biological replicates and presented as the mean ± SD done using ordinary one-way ANOVA with Tukey's multiple comparisons test by GraphPad Prism software (version 9.4.0, CA, USA). The significance was represented as *p < 0.05 and **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns (non-statically significant).
The structures of sirtuins retrieved from RCSB were scrutinized carefully. Multiple deposits of protein structure were found for the six sirtuin isoforms. The PDB structures 4I5I(Sirt1), 4RMH (Sirt2), 4JSR(Sirt3), 6LJK(Sirt5), 3K35(Sirt6), and 5IQZ(Sirt7) were selected because of better resolution and with zero mutation and superimposed to analyze the similarity and efficiency against all the deposited structures using Pymol software (PyMOL Molecular Graphics System, Version 2.5.5, Schrödinger, LLC). The binding pockets and the interactive amino acids present in the pockets were also visualized. While preparing the sirtuin protein, essential Zn+ metal and the NAD+ bound to the protein were allowed to be present within the structure, and the ligand of interest was docked against the binding site.
A molecular docking experiment was carried out using the Glide software in which the selected protein PDB was docked against the selected imidazole derivatives. Glide incorporates various scoring functions to assess the binding affinities of ligands within the active site (Figures 2, 3). These scoring functions consider factors such as van der Waals interactions, hydrogen bonding, electrostatic interactions, and hydrophobic interactions. Among all the compounds subjected to the docking, Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate showed higher docking score, glide score, and glide energy. Furthermore, the compound also had impressive interactions with all the sirtuins, with some concern for cellular localization (Table 2).
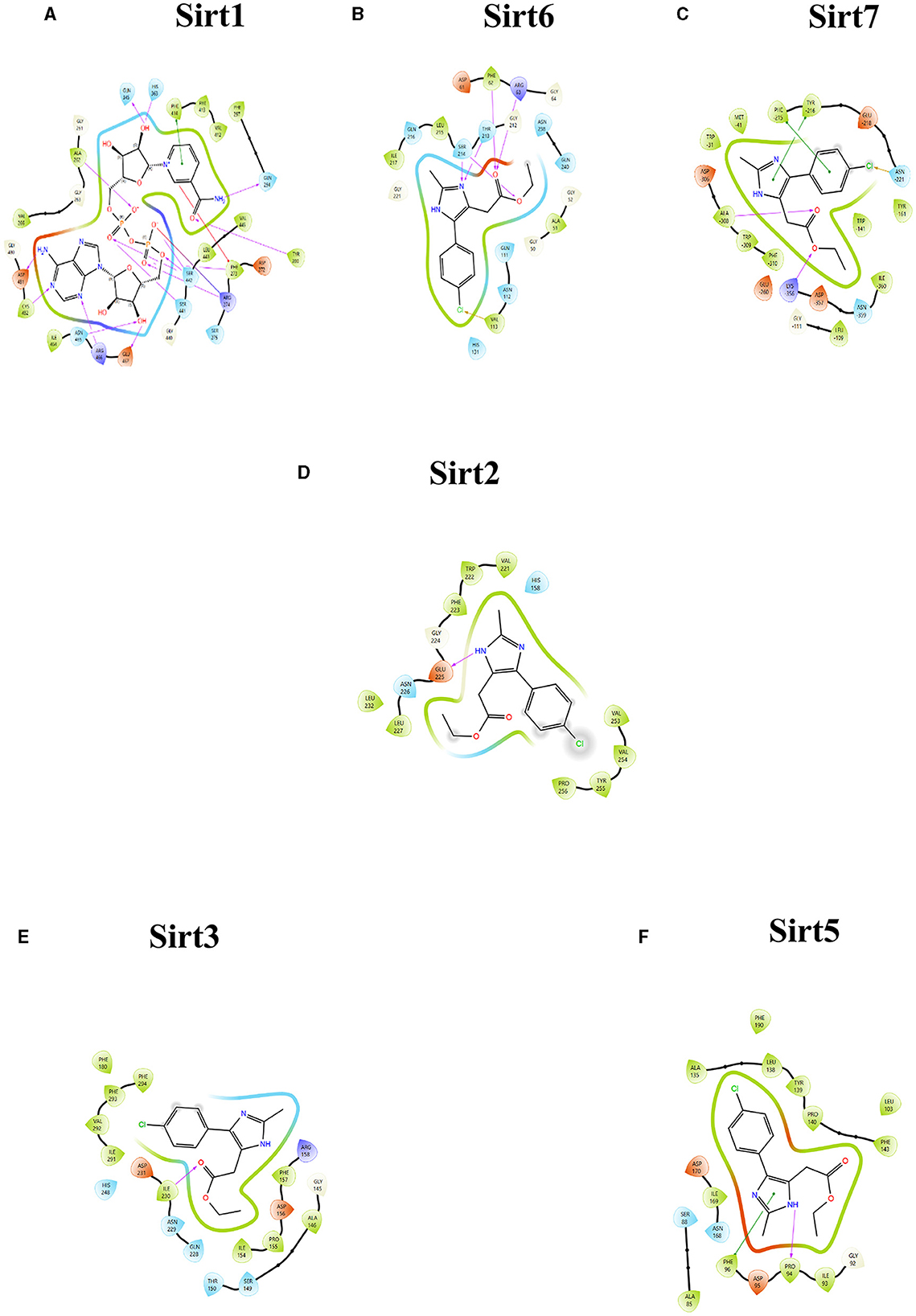
Figure 2. Ligand interaction between the sirtuins and the imidazole derivative. (A–C) Are the interactions of nuclear sirtuins Sirt1, Sirt6, and Sirt7, respectively. (D) Shows the ligand interaction with cytoplasmic sirtuin Sirt2. (E, F) Show the interaction of the mitochondrial sirtuins Sirt3 and Sirt5 with the ligand. The pink arrow indicates the hydrogen bonds and the green line depicts the pi-pi interaction formed between the ligand and the protein.
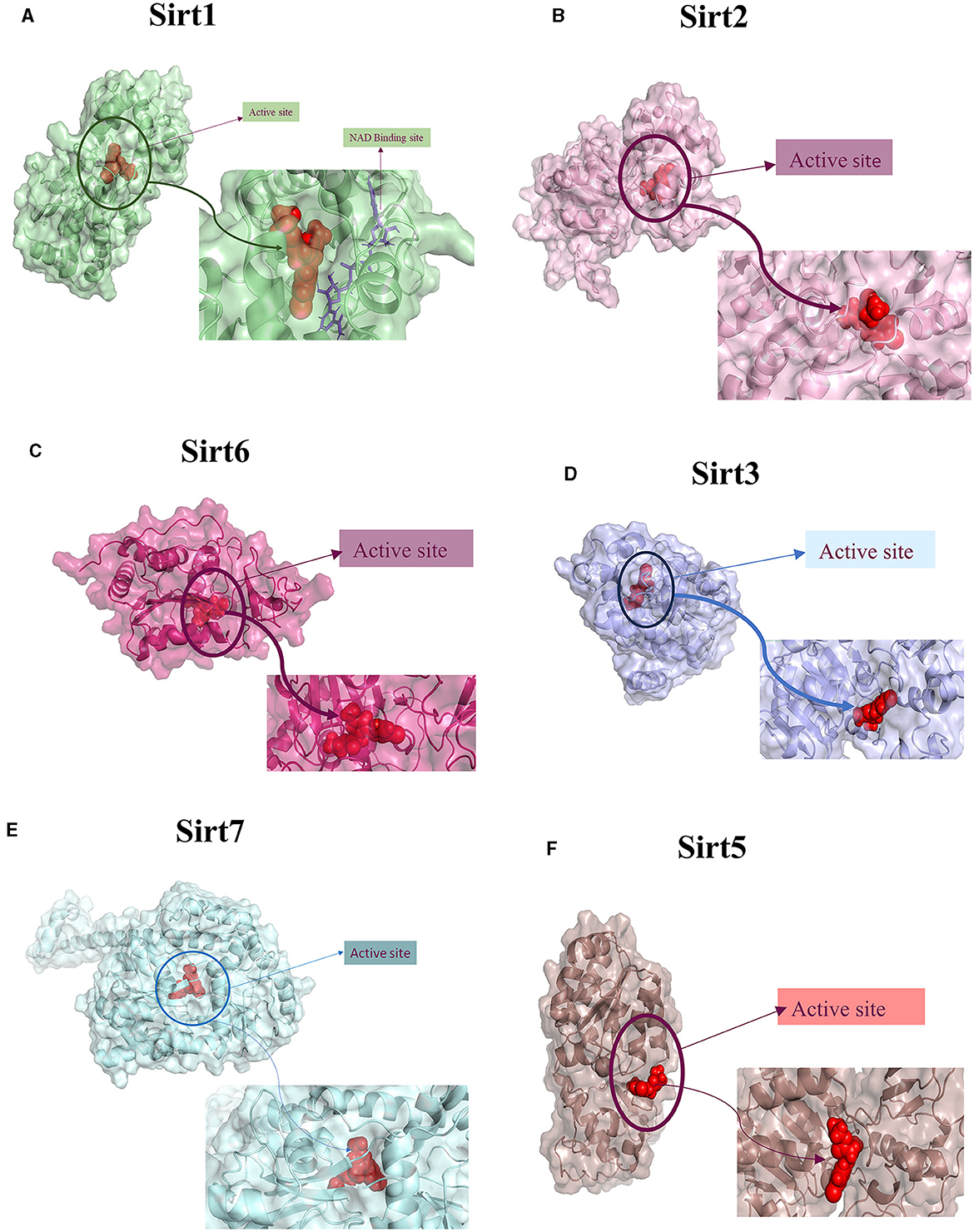
Figure 3. Protein-ligand interaction. 3D view of ligand fit into the binding pocket of the protein. The ligands bound to the active site of the protein are shown. (A) Sirt1, (B) Sirt2, (C) Sirt6, (D) Sirt3, (E) Sirt7, and (F) Sirt5.
MDS run with 100 ns simulation and 1000 frames generated a set of RMSD (Figure 4), RMSF (Figure 5), and protein-ligand interaction plots (Figures 6, 7). Sirt1, Sirt2, and Sirt6 were found to have stable simulation ranges with the increase of nanoseconds. Other sirtuins showed non-synchronous simulation stability.
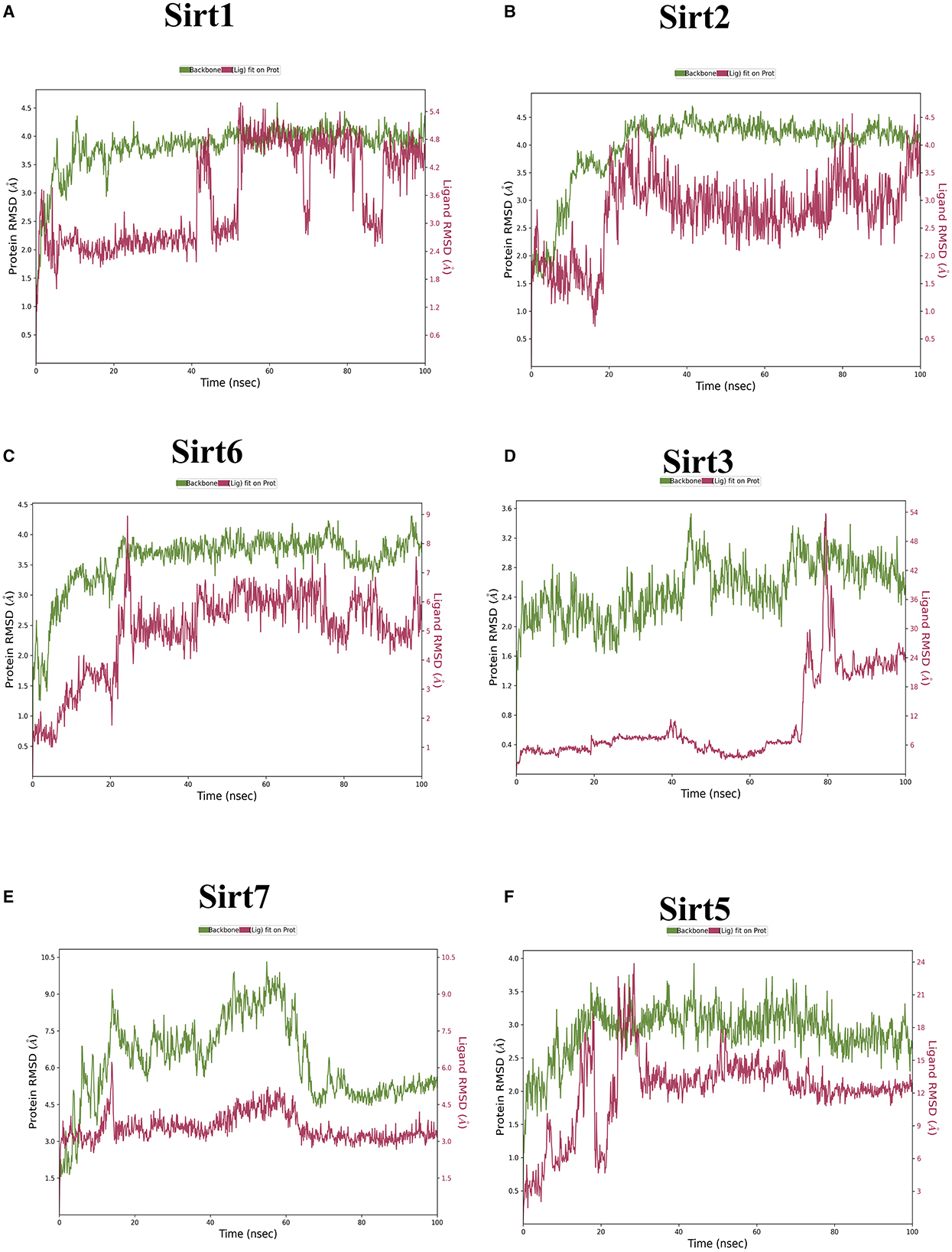
Figure 4. The root mean square deviation (RMSD). RMSD is a metric used to gauge the average change in atom displacement within a selected group of atoms in a specific frame in comparison to a reference frame. This calculation was performed for all frames in a given trajectory. The plot illustrates the progression of RMSD for the sirtuin protein (displayed on the left Y-axis in each graph plot). Meanwhile, ligand RMSD (depicted on the right Y-axis) provides insight into the stability of the ligand concerning the protein and its binding pocket. (A) Sirt1-ligand RMSD; (B) Sirt2-ligand RMSD; (C) Sirt6-ligand RMSD; (D) Sirt3-ligand RMSD; (E) Sirt7-ligand RMSD; and (F) Sirt5-ligand RMSD.
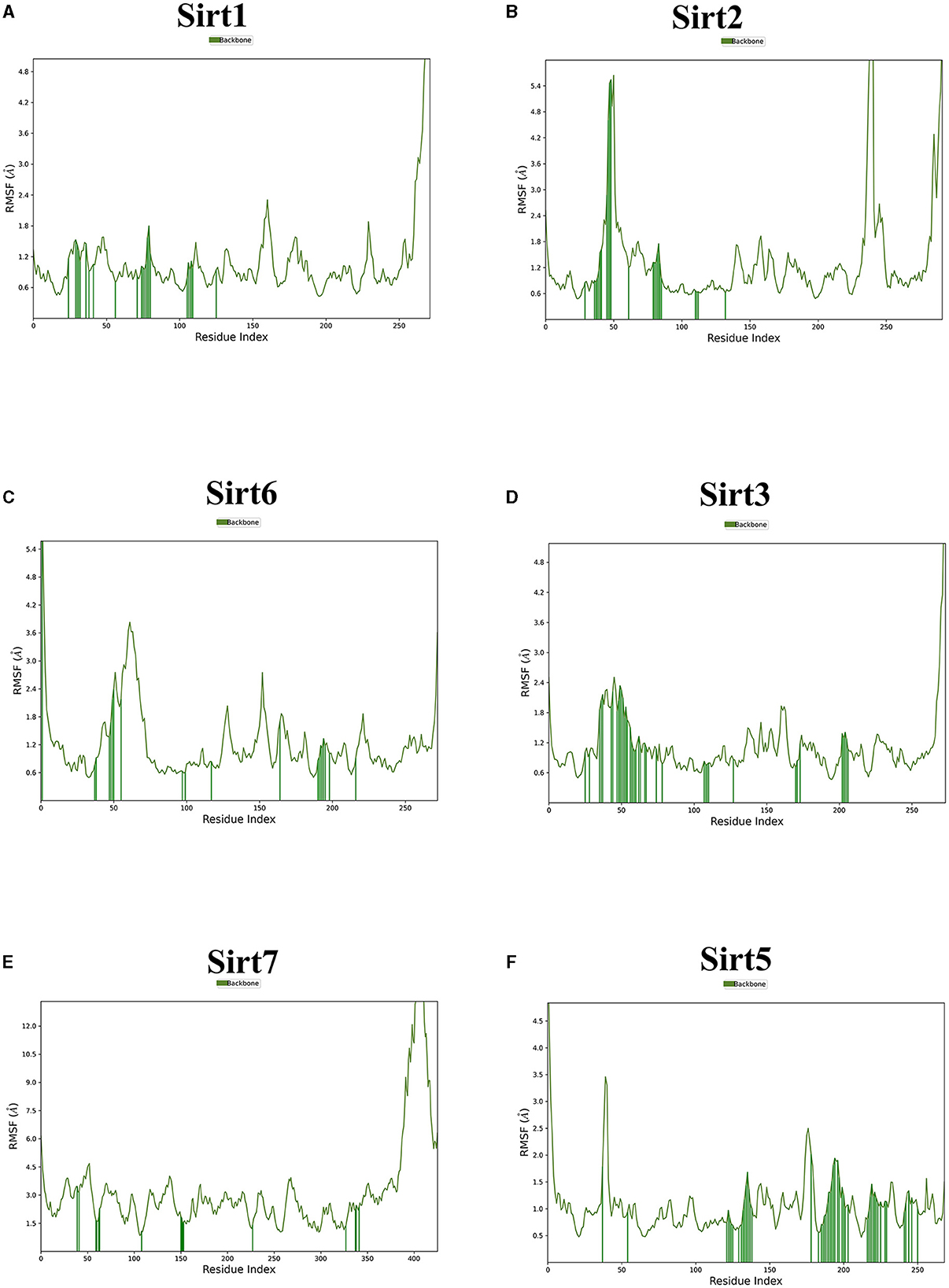
Figure 5. The root mean square fluctuation (RMSF). The parameter is useful for characterizing local changes along the protein chain. Sirtuin protein residues that interact with the ligand are marked with green-colored vertical bars. (A) Sirt1-ligand RMSF; (B) Sirt2-ligand RMSF; (C) Sirt6-ligand RMSF; (D) Sirt3-ligand RMSF; (E) Sirt7-ligand RMSF; and (F) Sirt5 -ligand RMSF.
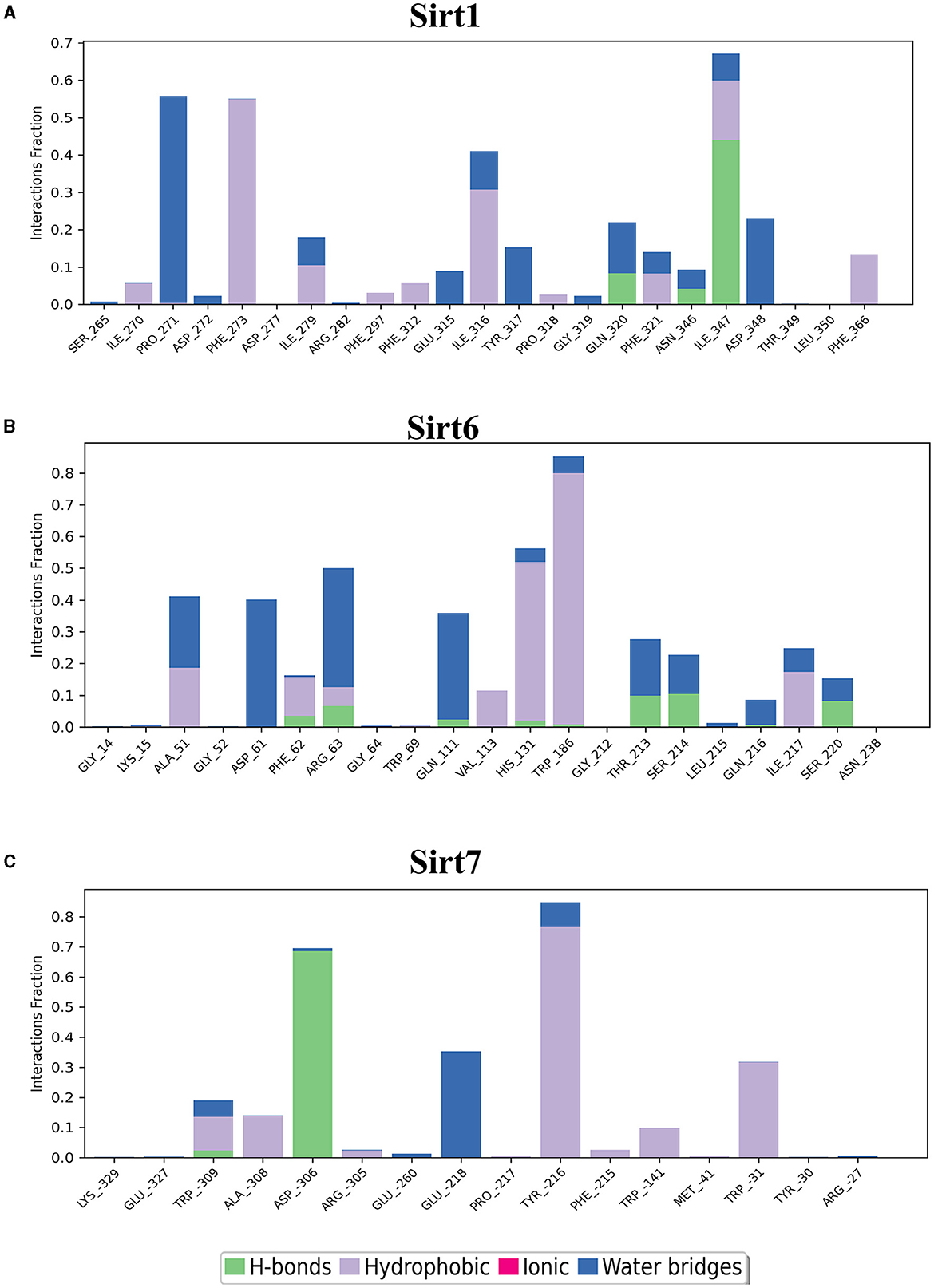
Figure 6. Nuclear sirtuin-ligand interaction. Protein interactions with the ligand can be monitored throughout simulations. These interactions can be categorized by type and summarized, as shown in the plot, which depicts hydrogen bonds and hydrophobic, ionic, and water bridges. (A) Sirt1, (B) Sirt6, and (C) Sirt7.
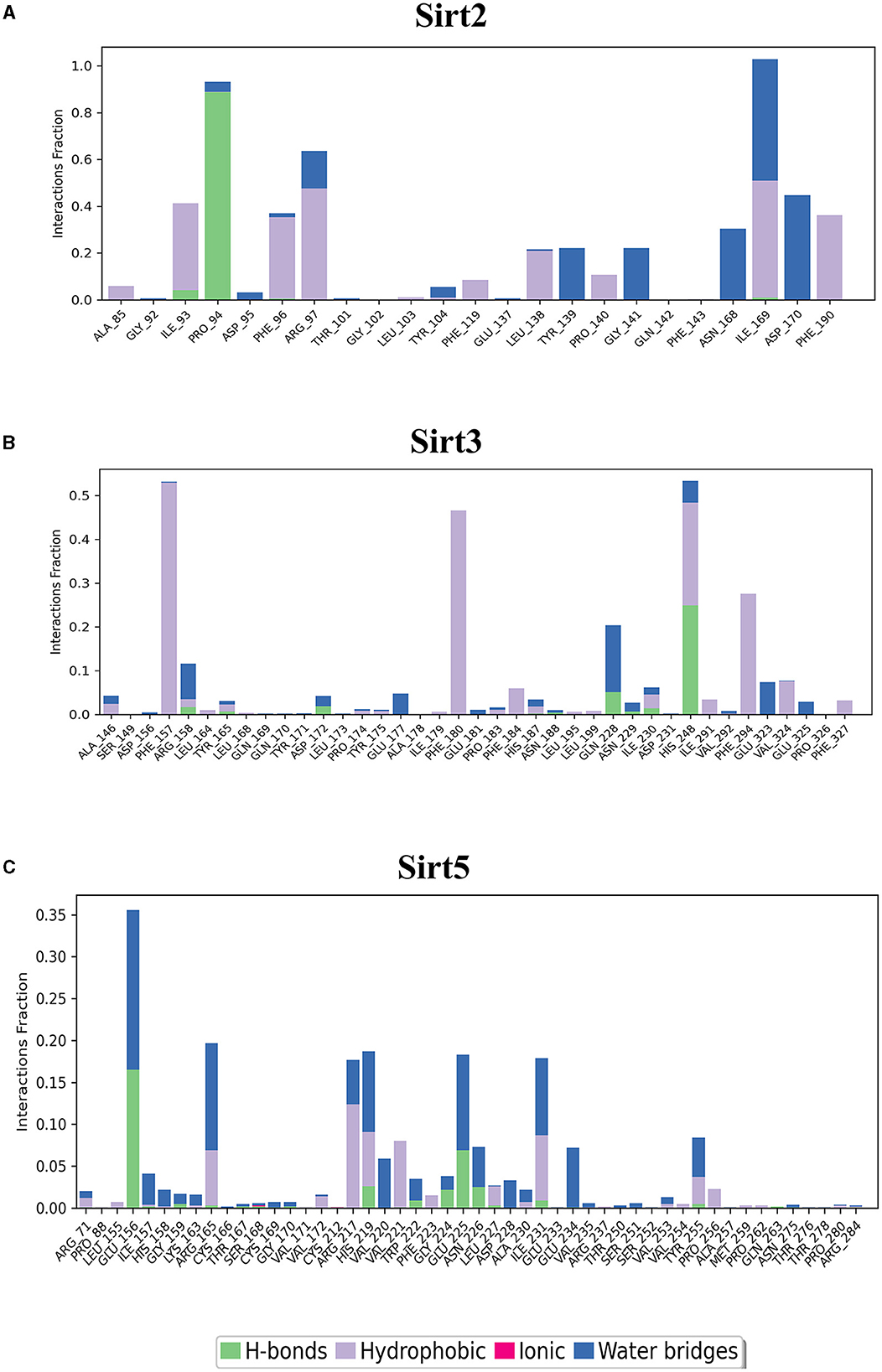
Figure 7. Cytoplasmic and mitochondrial sirtuin-ligand interaction. Protein interactions with the ligand can be monitored throughout simulations. These interactions can be categorized by type and summarized, as shown in the plot, which depicts hydrogen bonds and hydrophobic, ionic, and water bridges. (A) Sirt2; (B) Sirt3; (C) Sirt5.
DFT analysis is a promising method to investigate protein-ligand interactions from an atomistic-electronic perspective. It was carried out to understand the structure and relative energy of the ligand in the binding pocket as well as the electronic rearrangements that occur upon ligand binding. We also calculated the highest occupied molecular orbital of the ligand, which was approximately−0.27412, and the least unoccupied molecular orbital was−0.02618 (Figure 8).
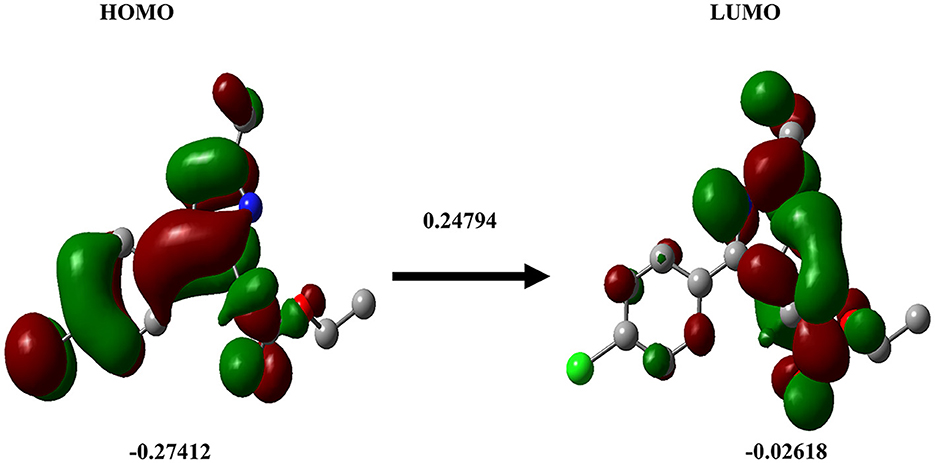
Figure 8. Density functional theory. The DFT calculation for the ligand Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate is shown. HOMO (left), LUMO (right).
ADME properties were analyzed for all the ligands that were investigated in this study. The results of the ADME analysis showed effective absorption, solubility, and the ability of the compound to dwell in the gut-blood barrier and blood-brain barrier (Table 3).
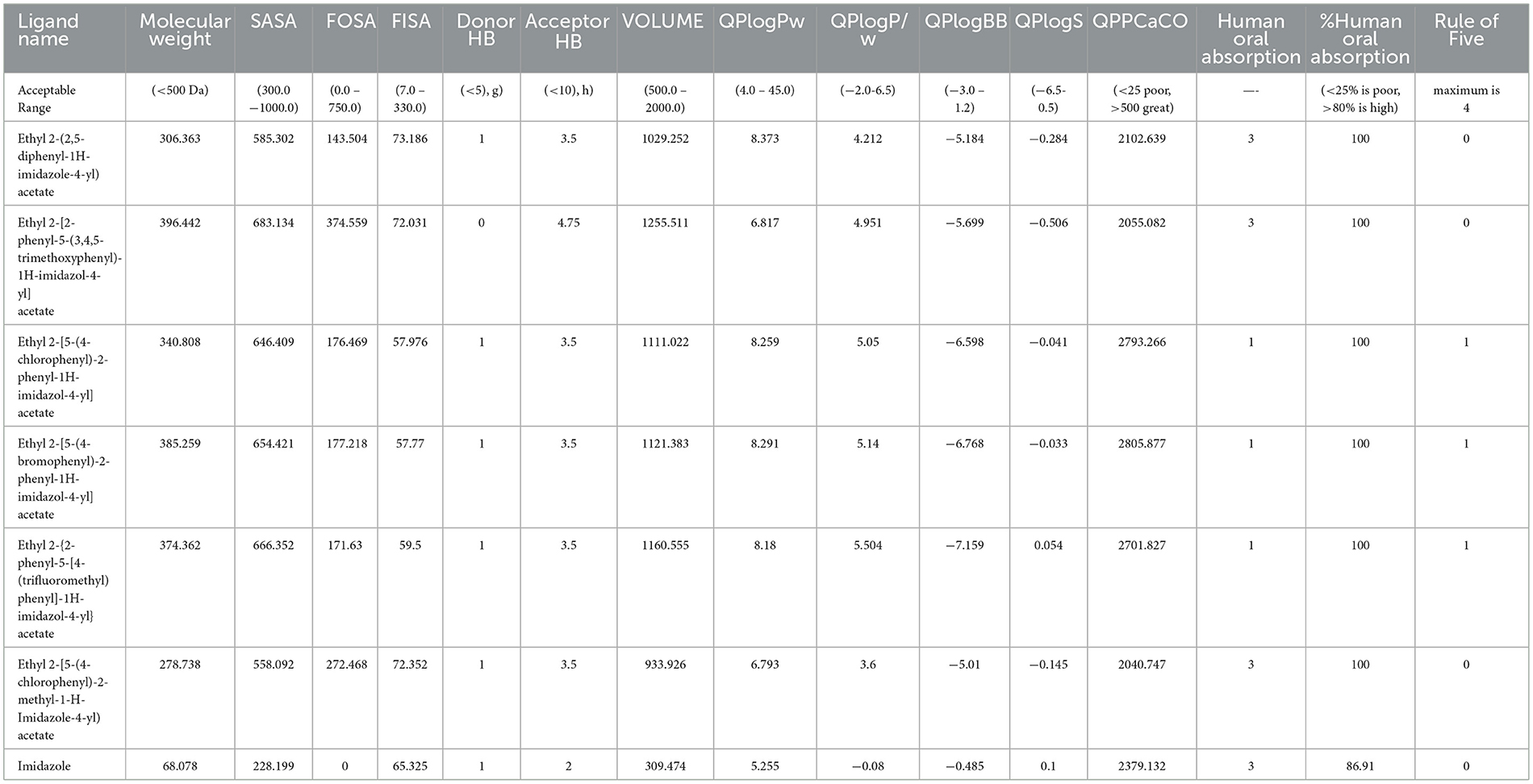
Table 3. ADMET analysis of the imidazole derivatives in comparison with imidazole parent structure emphasizing the efficiency of imidazole derivatives.
The cytotoxicity of imidazole and its derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate was determined in NSCLC cell lines A549 and NCI-H460 using MTT assay. The cells were treated with imidazole and imidazole derivatives of different concentrations for 24 h. The cell viability decreased gradually as concentration increased. The half maximal inhibitory concentration (IC50) of imidazole and its derivative was found to be 600 μM and 250 μM in the A549 cell line (Figures 9A, B) and 700 μM and 300 μM in the NCI-H460 cell line (Figures 9C, D). The imidazole derivative at lower concentrations reduced the cell viability compared to the parent compound imidazole in the NSCLC cell lines.
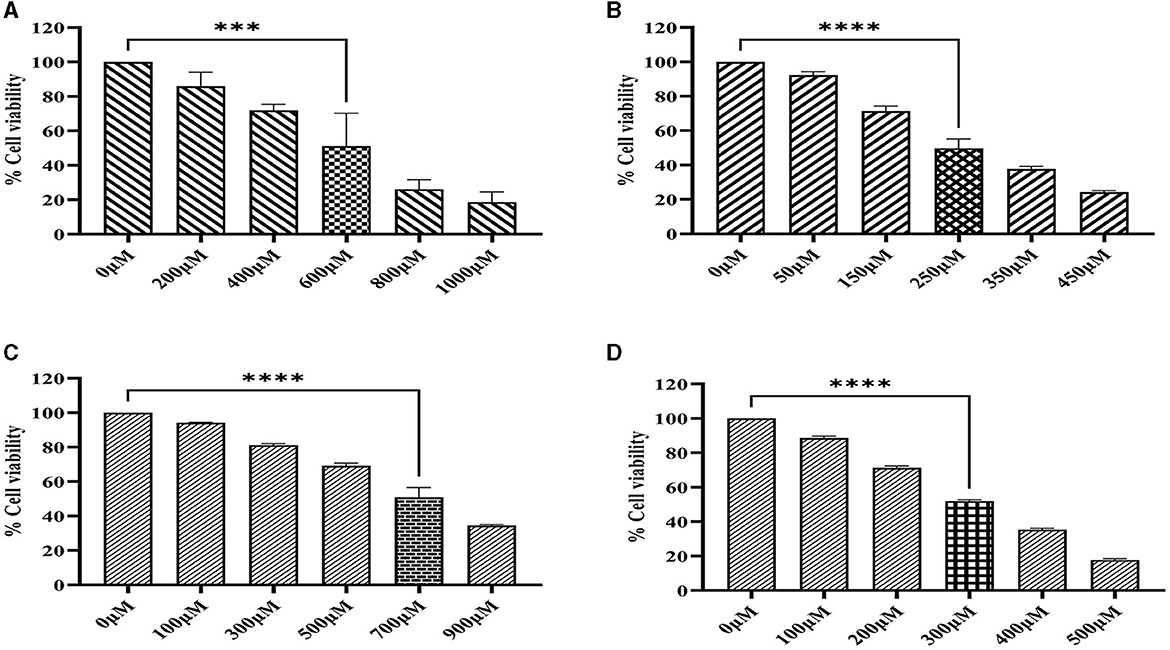
Figure 9. MTT assay. The cell viability activity of imidazole and imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate with different concertation for a treatment period of 24 h in NSCLC cell lines A549 and NCI-H460 cell lines. (A, B) Shows the cell viability activity of imidazole and imidazole derivatives in the A549 cell line. Whereas (C, D) show the cell viability activity of imidazole and imidazole derivatives in the NCI-H460 cell line. Data represent mean values ± SD. ***p < 0.001 and ****p < 0.0001.
Sirtuins are class III histone deacetylases with seven isoforms (Sirt 1-7). The role of sirtuins varies differently depending on the type of cancer. In this study, we explored the effect of imidazole derivative Ethyl 2-[5-(4-chlorophenSirtSirtyl)-2-methyl-1-H-Imidazole-4-yl] acetate on NSCLC cell line A549 and NCI-H460 cell lines. The gene and protein expression studies were carried out to understand the effect of the imidazole derivative. The results from the gene expression studies confirmed the downregulation of sirtuins on treatment with Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. Among all the sirtuins, Sirt1, Sirt2, and Sirt6 showed a prominent decrease in expression, followed by Sirt3, Sirt5, and Sirt4 in that order. Sirt7 was decreased non-significantly (Figure 10). Further Western blotting analysis was carried out to confirm the effect of Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate at the protein level. The expression of all the sirtuin members was decreased on treatment with Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. Among the different isoforms of sirtuins, Sirt1 and Sirt6 were greatly affected, followed by Sirt2, Sirt5, Sirt3, and Sirt7. Figure 11 shows the Western blot of all the isoforms of sirtuins. The decreased expression of Sirt1 was different in A549 and H460 cell lines, whereas Sirt6 was decreased to the same extent in A549 and NCI-H460 cell lines.
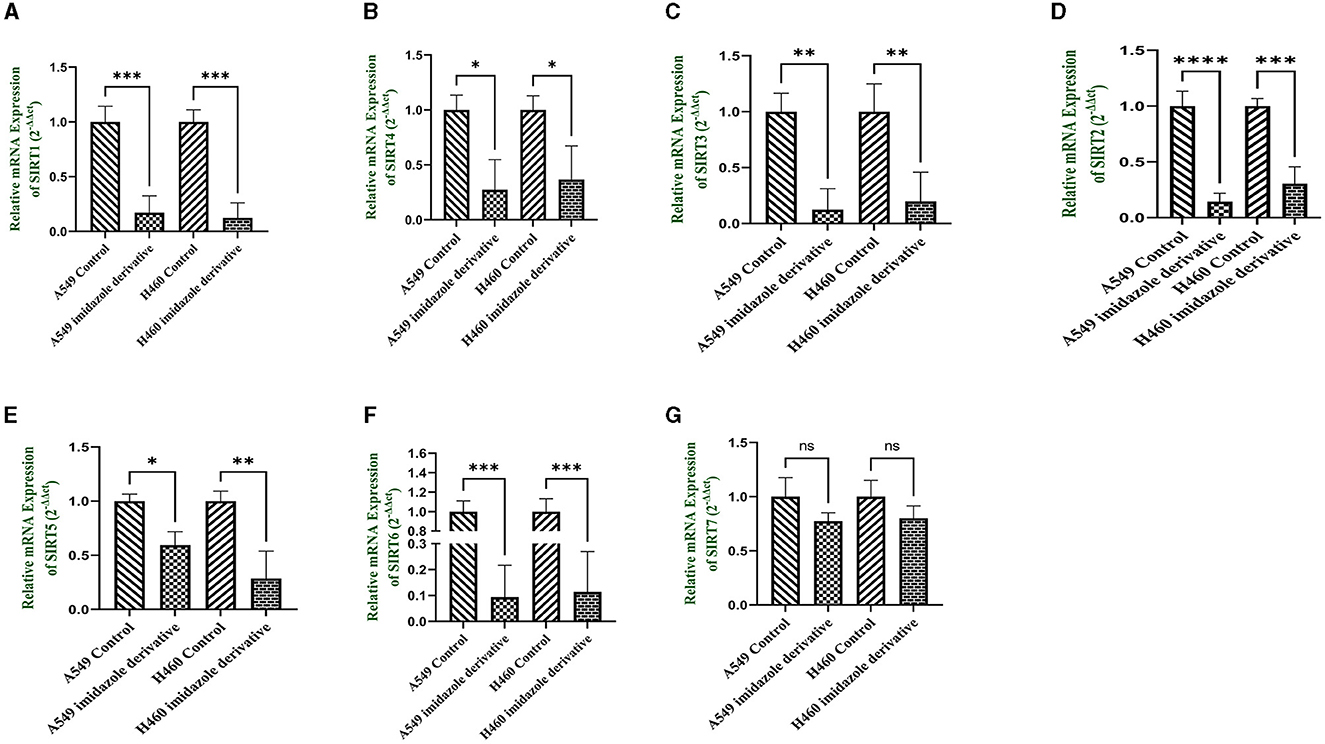
Figure 10. mRNA expression. Gene expression of the Class III histone deacetylase family (Sirt 1-7) was performed in quantitative Real-time PCR. The graphical representation shows expression of different sirtuins (A) Sirt1, (B) Sirt2, (C) Sirt3, (D) Sirt4, (E) Sirt5, (F) Sirt6, and (G) Sirt7 in NSCLC cell lines A549 and NCI-H460 (Data represents mean values ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, and ns, non-statistically significant).
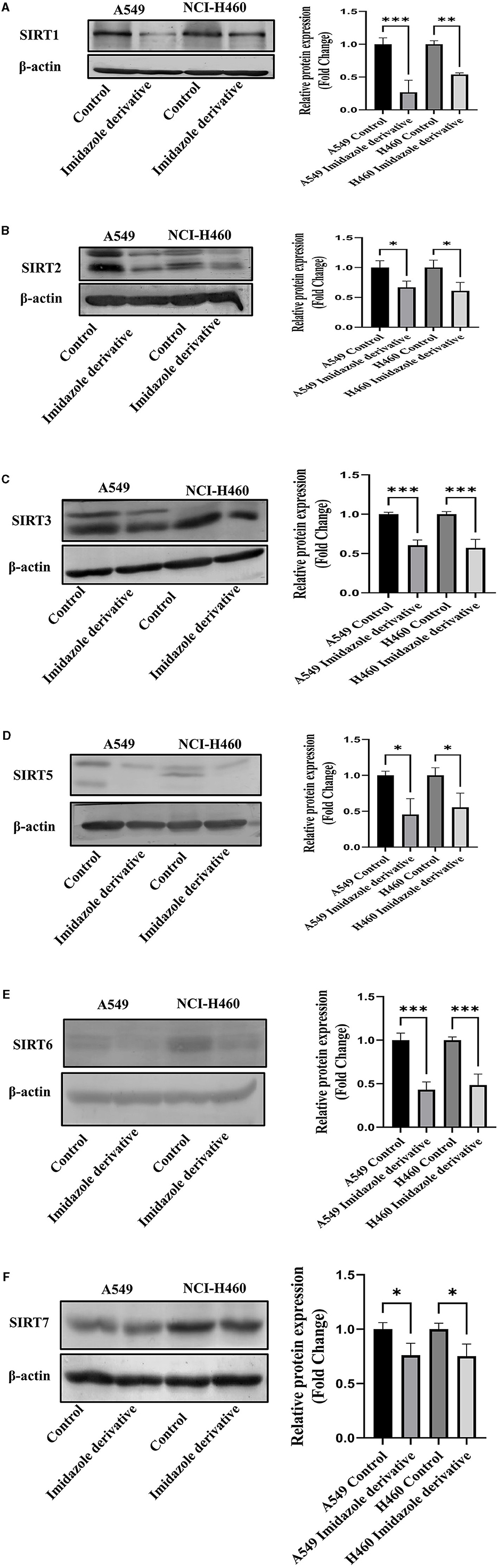
Figure 11. Protein expression. Western blotting was performed to understand the effect of Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate on protein expression of sirtuins. The results show the decrease in expression of (A) Sirt1, (B) Sirt2, (C) Sirt3, (D) Sirt5, (E) Sirt6, and (F) Sirt7 in Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate treated NSCLC cell lines A549 and NCI-H460 in comparison with control cells (untreated). β-actin was used as an endogenous control for normalization. Data represents mean values ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001.
The imidazole derivatives, designed and synthesized in-house, were subjected to Glide docking analysis. We found that in spite of the cellular localization of the sirtuins, all the compound structures interacted with the protein, especially compound Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate, which assured binding with all six sirtuin proteins. The glide gscore indicated that this compound had more affinity toward nuclear sirtuins Sirt1, Sirt6, and Sirt7 (−7.807, −6.544, and −5.616) and cytosolic sirtuin Sirt2 (−8.835).
The binding pockets of the protein comprised a set of amino acids (isoleucine, alanine, phenylalanine, tyrosine, tryptophan, glutamate, histidine, arginine, and aspartate) that form effective hydrogen bonds and pi-pi interaction with the ligand. Sirt1, Sirt6, Sirt7, and Sirt2 showed more hydrogen bond interactions, whereas the mitochondrial sirtuins Sirt3 and Sirt5 possessed the least interactive bonds.
Using the RMSD data, we determined the average distance between atoms in a simulated protein-ligand structure and a reference protein structure (23). This quantified the structural changes in the molecule during the simulation, commonly used to assess simulation stability and convergence. The RMSF values were also calculated to understand the average deviation of atom positions from their mean during the simulation, providing insights into the flexibility and dynamic behavior of various biomolecule regions (24).
The MD simulations investigated ligand interactions and, in particular, examined how a small molecule (ligand) interacted with a larger biomolecule (e.g., protein or nucleic acid). These ligand interactions encompassed hydrogen bonding, van der Waals forces, and electrostatic interactions, and understanding these interactions offered insights into binding strength, binding sites, and potential mechanisms of action. Using trajectory analysis, the complex RMSD for nuclear, mitochondrial, and cytoplasmic sirtuins was determined. The RMSD range for the sirtuin protein-ligand Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl) acetate complex was within 4.0 Å. All the RMSD graphs indicated that our compound had attained equilibrium and stayed at approximately 4 Å throughout the 100 ns simulation. This demonstrated the ability of our compound to effectively be stable-bound with all six sirtuins employed in the study.
Sirtuins are interconnected HDAC enzymes, and they take turns to achieve pathway activation and inhibition. For instance, multiple studies have reported that Sirt1 possesses power over other nuclear sirtuins. In the case of double-stranded break repair mechanisms, Sirt1 facilitates the mobilization of Sirt6 to double-strand breaks by deacetylation (25). One report suggested that Sirt1 and Sirt6 can bind to the N and C terminals of the Suv39h protein and regulate the repressor pathway of IkBα expression (26). In contrast, there are research reports that emphasize the importance of Sirt6 in regulating the random acetylation of Sirt1 with respect to lung cancer. Sirt1 can acetylate or deacetylate and regulate multiple pathways in lung cancer, like acetylation of AMP (27), AKT (28), PPAR γ (29), and more. Our gene expression and protein expression data suggest that the compound Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate cooperated well with nuclear sirtuins, which could favor the inhibition of nuclear sirtuins and activation of tumor suppression pathways. Inhibition of nuclear sirtuins can clearly influence the actions of other sirtuins.
The HOMO energy level represents the energy of the highest electron orbital that contains electrons in the molecule. It signifies the energy required to remove an electron from the highest occupied orbital. A lower (more negative) HOMO energy indicates that electrons are relatively tightly bound, which implies that the molecule is less likely to readily donate electrons. In terms of chemical reactivity, the HOMO energy level is associated with the molecule's ability to act as an electron donor in chemical reactions (30, 31). In our study, all the selected imidazole derivatives demonstrated values that surpassed the defined range. Based on a comprehensive analysis, it can be concluded that the compounds showed promising potential in terms of permeability. The ADME analysis revealed that all the compounds investigated in the study possessed favorable drug-likeness and did not violate Lipinski's rule of five. As a result, all the compounds were included in the docking study. From the ADME analysis and the docking score, it is evident that the imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate has greater interaction with all six sirtuins compared to imidazole and its other derivatives.
In the surface area analysis, several crucial components were assessed, including the total solvent accessible surface area (SASA), which accounts for the overall surface area available to solvents. Additionally, the hydrophobic component of SASA, known as FOSA, and its counterpart, the hydrophilic component called FISA, were examined (32). The evaluation of these components yielded values that largely fell within an acceptable range, as stipulated in the Qikprop manual provided by Schrödinger. In particular, the software's guidelines were referenced to ensure that the values obtained for SASA, FOSA, FISA, PISA, and volume align with established standards. Furthermore, the predictive permeability of the compounds was a focal point of investigation. This was achieved through the utilization of two key metrics: QPPCaco, a model designed for assessing gut-blood barrier permeability (33), and QPlogBB, which pertains to the brain-blood partition coefficient (34). QPPCaco values exceeding 500 are indicative of favorable permeability characteristics. Our investigation revealed that all the selected imidazole derivatives had values that surpassed the defined range. Based on a comprehensive analysis. It can be concluded that the compounds showed promising potential in terms of permeability. The ADME analysis and the docking score established that the imidazole derivative, Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate, had greater interaction with the sirtuins family compared to the imidazole.
Based on the Insilco result, we carried out in-vitro studies with Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate to explore its inhibitory effect on sirtuin family members (Sirt 1-7). Initially, we undertook an MTT assay to fix the IC50 concentration of the Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. An analysis of the respective MTT results for imidazole and imidazole derivative revealed that the imidazole derivative had a higher effect at lower concentrations in 24 h of treatment in A549 (250 μM) and NCI-H460 (300 μM) compared to the parent imidazole compound in A549 (600 μM) and NCI-H460 (700 μM) cell lines. Furthermore, gene and protein expression studies were carried out with IC50 concentration of imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. The gene expression results from qRT-PCR confirmed the decrease in the expression of sirtuins in NSCLC cell line A549 and NCI-H460.
Among the sirtuins, Sirt1, Sirt2, and Sirt6 were greatly reduced on treatment with Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate, comparted to Sirt3, Sirt5 and Sirt7. The results from the Western blot analysis also confirm the decreased expression of sirtuins, which correlated with the gene expression of sirtuins. Among the sirtuins, the protein expression of Sirt1 and Sirt6 was greatly reduced, which correlated with the gene expression of Sirt1 and Sirt6. A previous report from our lab crew explains that Sirt6 is highly expressed in NSCLC cell lines, and upon silencing Sirt6, cell cycle arrest and apoptosis are favored (35). Consequently, based on our current study, we believe that Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate potentially inhibits nuclear sirtuins, especially the expression of Sirt6 and regulates cancer progression.
Cancer is an indestructible target for biologists; every day, new drugs are out on the market. However, regulating compounds with potential are scarcely found. Our in-house compounds established potential interaction with epigenetic-modulating sirtuin enzymes that can inhibit cancer progression. Our in-silico data shows that the imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate perfects the Lipinski rule of five and possesses a high docking score, which confirms the interaction with all sirtuin isoforms. The molecular dynamics data further confirms the stability of the bound ligand with the protein. Similar to the in-silico study, the results from the in-vitro study confirm the inhibitory effect of imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate. Our study shows that the imidazole derivative is highly effective on nuclear sirtuins Sirt1 and Sirt6, as well as cytosolic sirtuin Sirt2. Furthermore, it has been found to be effective on other members of the sirtuins, namely Sirt3, Sirt5, and Sirt7, in both in-silico and in-vitro analysis. Therefore, based on the in-silico and in-vitro results, we claim that our in-house imidazole derivative Ethyl 2-[5-(4-chlorophenyl)-2-methyl-1-H-Imidazole-4-yl] acetate is a potential inhibitor of Class III HDACs, with particular reference to Sirt1, Sirt6, and Sirt2.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
UMRD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing—original draft, Writing—review and editing. SS: Methodology, Software, Validation, Writing—original draft. SA-G: Formal Analysis, Writing—review and editing. NA: Formal analysis, Writing—review and editing. SD: Formal analysis, Writing—review and editing. AA: Formal analysis, Writing—review and editing. MS: Formal analysis, Writing—review and editing. TR: Funding acquisition, Writing—review and editing. RV: Conceptualization, Funding acquisition, Project administration, Resources, Writing—original draft, Writing—review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
UMRD and RV acknowledge the support provided by Molecular Modelling Facility, RUSA-2.0 (Biological Sciences), Bharathidasan University. UMRD and RV acknowledge DST-PURSE Phase II, and DST-FIST, Government of India for their support in providing funds for the improvement of instrumentation facilities in our Institute and Department respectively. UMRD would like to acknowledge University Grants Commission (UGC) for Rajiv Gandhi National Fellowship (RGNF). RV would also like to acknowledge research grants from the Indian Council of Medical Research (ICMR), New Delhi, India. (Ref. No: 52/13/2020-BIO-BMS and Ref. No: VIR/ COVID-19/6/2021/ECD-I). The authors acknowledge Dr. K. Srinivasan, Professor, School of Chemistry, Bharathidasan University for providing the Imidazole derivative which was used in the present study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1282820/full#supplementary-material
HDAC, Histone Deacetylase; NSCLC, Non-Small Cell Lung Cancer; Sirt (1-7), Sirtuins (1-7); NAD, Nicotinamide Adenine Dinucleotide; PDB, Protein Data Bank; RCSB, Research Collaboratory for Structural Bioinformatics; OPLS, Optimized Potentials for Liquid Simulations; GLIDE, Grid-Based Ligand Docking with Energetics; MDS, Molecular Dynamics Simulation; DESMOND, Desmond Molecular Dynamics System; TIP3P, Transferable Intermolecular Potential with 3 Points; RMSD, Root Mean Square Deviation; RMSF, Root Mean Square Fluctuation; DFT, Density Functional Theory; ADME, Absorption, Distribution, Metabolism, Excretion; Ligprep, Ligand Preparation; B3LYP-D3, Becke 3-parameter hybrid functional combined with Lee-Yang-Parr correlation functional and D3 dispersion correction; HOMO, Highest Occupied Molecular Orbital; LUMO, Lowest Unoccupied Molecular Orbital; SASA, Solvent Accessible Surface Area; FISA, Flexible Inhibitor Surface Anchoring; FOSA, Force-Field Overlapping Scoring Algorithm; QPlog BB, Quantum Polarized Ligand-Based Binding Energy; QPlogS, Quantum Polarized Surface Area Solvation Energy; QplogPw, Quantum Polarized Water-Octanol Partitioning Energy; QPPCaCO, Quantum Polarized Partial Charge of Carbon in Carbonyl; HB, Hydrogen Bond.
1. Ali I, Lone MN, Aboul-Enein HY. Imidazoles as potential anticancer agents. Medchemcomm. (2017) 8:1742–73. doi: 10.1039/C7MD00067G
2. Sharma P, LaRosa C, Antwi J, Govindarajan R, Werbovetz KA. Imidazoles as potential anticancer agents: an update on recent studies. Molecules. (2021) 26:4213. doi: 10.3390/molecules26144213
3. Bryaskova R, Georgiev N, Philipova N, Bakov V, Anichina K, Argirova M, et al. Novel fluorescent benzimidazole-hydrazone-loaded micellar carriers for controlled release: impact on cell toxicity, nuclear and microtubule alterations in breast cancer cells. Pharmaceutics. (2023) 15:1753. doi: 10.3390/pharmaceutics15061753
4. Rajabi M, Mousa SA. The role of angiogenesis in cancer treatment. Biomedicines. (2017) 5:34. doi: 10.3390/biomedicines5020034
5. Azad A, Kong A. The therapeutic potential of imidazole or quinone-based compounds as radiosensitisers in combination with radiotherapy for the treatment of head and neck squamous cell carcinoma. Cancers (Basel). (2022) 14: 4694. doi: 10.3390/cancers14194694
6. Tolomeu HV, Fraga CAM. Imidazole: synthesis, functionalization and physicochemical properties of a privileged structure in medicinal chemistry. Molecules. (2023) 28:838. doi: 10.3390/molecules28020838
7. Sharma A, Kumar V, Kharb R, Kumar S, Sharma PC, Pathak DP. Imidazole derivatives as potential therapeutic agents. Curr Pharm Des. (2016) 22:3265–301. doi: 10.2174/1381612822666160226144333
8. Kandasamy S, Subramani P, Srinivasan K, Jayaraj J, Prasanth G, Muthusamy K, et al. Design and synthesis of imidazole based zinc binding groups as novel small molecule inhibitors targeting Histone deacetylase enzymes in lung cancer. J Mol Struct. (2020) 1214:128177. doi: 10.1016/j.molstruc.2020.128177
9. Dai H, Sinclair DA, Ellis JL, Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol Ther. (2018) 188:140–54. doi: 10.1016/j.pharmthera.2018.03.004
10. Zhu S, Dong Z, Ke X, Hou J, Zhao E, Zhang K, et al. The roles of sirtuins family in cell metabolism during tumor development. Semin Cancer Biol. (2019) 57:59–71. doi: 10.1016/j.semcancer.2018.11.003
11. Vu CB, Bemis JE, Disch JS, Ng PY, Nunes JJ, Milne JC, et al. Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem. (2009) 52:1275–83. doi: 10.1021/jm8012954
12. Morris BJ, Willcox BJ, Donlon TA. Genetic and epigenetic regulation of human aging and longevity. Biochim Biophys acta Mol basis Dis. (2019) 1865:1718–44. doi: 10.1016/j.bbadis.2018.08.039
13. Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. (2015) 15:608–24. doi: 10.1038/nrc3985
14. Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. (2011) 2:648–62. doi: 10.1177/1947601911417862
15. Villalba JM, Alcaín FJ. Sirtuin activators and inhibitors. Biofactors. (2012) 38:349–59. doi: 10.1002/biof.1032
16. Jiang Y, Liu J, Chen D, Yan L, Zheng W. Sirtuin inhibition: strategies, inhibitors, and therapeutic potential. Trends Pharmacol Sci. (2017) 38:459–72. doi: 10.1016/j.tips.2017.01.009
17. Hu J, Jing H, Lin H. Sirtuin inhibitors as anticancer agents. Future Med Chem. (2014) 6:945–66. doi: 10.4155/fmc.14.44
18. Wu Q-J, Zhang T-N, Chen H-H, Yu X-F, Lv J-L, Liu Y-Y, et al. The sirtuin family in health and disease. Signal Transduct Target Ther. (2022) 7:402. doi: 10.1038/s41392-022-01257-8
19. Sharma A, Mahur P, Muthukumaran J, Singh AK, Jain M. Shedding light on structure, function and regulation of human sirtuins: a comprehensive review. 3 Biotech. (2023) 13:29. doi: 10.1007/s13205-022-03455-1
20. Kerru N, Gummidi L, Maddila S, Gangu KK, Jonnalagadda SB. A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules. (2020) 25:1909. doi: 10.3390/molecules25081909
21. Malik MS, Alsantali RI, Jamal QMS, Seddigi ZS, Morad M, Alsharif MA, et al. New imidazole-based N-phenylbenzamide derivatives as potential anticancer agents: key computational insights. Front Chem. (2021) 9:808556. doi: 10.3389/fchem.2021.808556
22. Selvi T, Srinivasan K. Boron trifluoride mediated ring-opening reactions of trans-2-Aryl-3-nitro-cyclopropane-1,1-dicarboxylates. Synthesis of aroylmethylidene malonates as potential building blocks for heterocycles. J Org Chem. (2014) 79:3653–8. doi: 10.1021/jo402848v
23. Kirchmair J, Markt P, Distinto S, Wolber G, Langer T. Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection–what can we learn from earlier mistakes? J Comput Aided Mol Des. (2008) 22:213–28. doi: 10.1007/s10822-007-9163-6
24. Margreitter C, Oostenbrink C. MDplot: visualise molecular dynamics. R J. (2017) 9:164–86. doi: 10.32614/RJ-2017-007
25. Meng F, Qian M, Peng B, Peng L, Wang X, Zheng K, et al. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. Elife. (2020) 9:e55828. doi: 10.7554/eLife.55828
26. Santos-Barriopedro I, Bosch-Presegué L, Marazuela-Duque A, de la Torre C, Colomer C, Vazquez BN, et al. SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nat Commun. (2018) 9:101. doi: 10.1038/s41467-017-02586-x
27. Park S-J, Ahmad F, Philp A, Baar K, Williams T, Luo H, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. (2012) 148:421–33. doi: 10.1016/j.cell.2012.01.017
28. Pillai VB, Sundaresan NR, Gupta MP. Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ Res. (2014) 114:368–78. doi: 10.1161/CIRCRESAHA.113.300536
29. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. (2012) 150:620–32. doi: 10.1016/j.cell.2012.06.027
30. Prabhaharan M, Prabakaran AR, Gunasekaran S, Srinivasan S. DFT studies on vibrational spectra, HOMO-LUMO, NBO and thermodynamic function analysis of cyanuric fluoride. Spectrochim Acta A Mol Biomol Spectrosc. (2015) 136:494–503. doi: 10.1016/j.saa.2014.09.062
31. Mathammal R, Jayamani N, Geetha N. Molecular structure, NMR, HOMO, LUMO, and vibrational analysis of O-anisic acid and anisic acid based on DFT calculations. J Spectrosc. (2013) 2013:171735. doi: 10.1155/2013/171735
32. Fatima S, Gupta P, Sharma S, Sharma A, Agarwal SM. ADMET profiling of geographically diverse phytochemical using chemoinformatic tools. Future Med Chem. (2020) 12:69–87. doi: 10.4155/fmc-2019-0206
33. Telvekar VN, Bairwa VK, Satardekar K, Bellubi A. Novel 2-(2-(4-aryloxybenzylidene) hydrazinyl)benzothiazole derivatives as anti-tubercular agents. Bioorg Med Chem Lett. (2012) 22:649–52. doi: 10.1016/j.bmcl.2011.10.064
34. Jhala DD, Chettiar SS, Singh JK. Optimization and validation of an in vitro blood brain barrier permeability assay using artificial lipid membrane. J Bioequivalence Bioavailab. (2012) 2012:9. doi: 10.4172/jbb.S14-009
Keywords: imidazole, HDAC, inhibitors, epigenetics, sirtuins
Citation: Dindi UMR, Sadiq SP, Al-Ghamdi S, Alrudian NA, Dayel SB, Abuderman AA, Shahid M, Ramesh T and Vilwanathan R (2023) In-silico and in-vitro functional validation of imidazole derivatives as potential sirtuin inhibitor. Front. Med. 10:1282820. doi: 10.3389/fmed.2023.1282820
Received: 24 August 2023; Accepted: 21 September 2023;
Published: 07 November 2023.
Edited by:
Udhaya Kumar, Baylor College of Medicine, United StatesReviewed by:
Mahesh Gokara, Mayo Clinic Florida, United StatesCopyright © 2023 Dindi, Sadiq, Al-Ghamdi, Alrudian, Dayel, Abuderman, Shahid, Ramesh and Vilwanathan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravikumar Vilwanathan, cmF2aWt1bWFyYmR1QGdtYWlsLmNvbQ==; ZHJ2ckBiZHUuYWMuaW4=; Thiyagarajan Ramesh, ci50aGl5YWdhcmFqYW5AcHNhdS5lZHUuc2E=
†ORCID: Ravikumar Vilwanathan orcid.org/0000-0002-5134-6277
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.