- 1Digital Medical Research Center, School of Basic Medical Sciences, Fudan University, Shanghai, China
- 2Shanghai Key Lab of Medical Image Computing and Computer Assisted Intervention, Fudan University, Shanghai, China
- 3Department of Neurosurgery, Zhongshan Hospital, Fudan University, Shanghai, China
- 4Radiation Oncology Center, Huashan Hospital, Fudan University, Shanghai, China
Introduction: Precise delineation of glioblastoma in multi-parameter magnetic resonance images is pivotal for neurosurgery and subsequent treatment monitoring. Transformer models have shown promise in brain tumor segmentation, but their efficacy heavily depends on a substantial amount of annotated data. To address the scarcity of annotated data and improve model robustness, self-supervised learning methods using masked autoencoders have been devised. Nevertheless, these methods have not incorporated the anatomical priors of brain structures.
Methods: This study proposed an anatomical prior-informed masking strategy to enhance the pre-training of masked autoencoders, which combines data-driven reconstruction with anatomical knowledge. We investigate the likelihood of tumor presence in various brain structures, and this information is then utilized to guide the masking procedure.
Results: Compared with random masking, our method enables the pre-training to concentrate on regions that are more pertinent to downstream segmentation. Experiments conducted on the BraTS21 dataset demonstrate that our proposed method surpasses the performance of state-of-the-art self-supervised learning techniques. It enhances brain tumor segmentation in terms of both accuracy and data efficiency.
Discussion: Tailored mechanisms designed to extract valuable information from extensive data could enhance computational efficiency and performance, resulting in increased precision. It's still promising to integrate anatomical priors and vision approaches.
1. Introduction
Glioblastoma (GBM) is one of the most aggressive brain cancers among adults (1). Multi-parameter magnetic resonance imaging (MRI) provides valuable information for characterizing the size, invasiveness, and intrinsic heterogeneity of brain tumors (2, 3). Accurate delineation of GBM on multi-parameter MRI is crucial for clinical diagnosis and treatment, such as assisting surgical planning for maximum glioblastoma resection while preserving neurological function. However, the current clinical routine still relies on manual delineation, which is time-consuming and requires expert knowledge. There is a high demand for automatic brain tumor segmentation to enhance the efficiency of diagnostic procedures, facilitate surgical planning, and contribute to prognostic analyses (4).
In the last decade, there have been extensive studies on automatic brain tumor segmentation (5), and most of them are based on convolutional neural networks (CNNs) (6–8). However, due to limited receptive field, CNNs often struggle to capture long-range dependencies and global context (9, 10), potentially leading to inaccurate segmentation predictions. The recent success of transformer architecture in vision tasks (11, 12) has shown benefits in learning global contextual information. New network designs with vision transformers have emerged for medical image segmentation (13, 14) and achieved state-of-the-art (SOTA) performance in brain tumor segmentation (15–17). However, the supervised training of vision transformers typically requires a large amount of densely annotated images, otherwise there is a high risk of overfitting.
To combat the challenge of data scarcity in medical image segmentation, self-supervised learning (SSL) has proven to be a promising solution (18). In general, a pretext SSL task is designed to pre-train the network using unannotated data, and the learned encoder weights are further optimized in the downstream segmentation task. Since no manual annotation is needed for SSL, it can be applied to utilize large unannotated datasets. Recently, one of the most successful SSL frameworks is the masked language modeling (MLM), which has achieved great success in numerous natural language processing tasks with transformer-based architecture (19–21). Motivated by MLM, masked image modeling (MIM) was also proposed for pre-training vision transformers. In MIM, the model predicts masked image patches from unmasked patches. The prediction target can be either token features or raw pixel values of the masked patches. BEiT (22) utilizes a discrete variational autoencoder (dVAE) to transform all image patches into discrete tokens, which are then used to pre-train a vision transformer at the token level. However, tokenizing the image patches requires additional training of a dVAE. In contrast, He et al. (23) introduced the masked autoencoder (MAE), which randomly masks a subset of image patches and reconstructs the masked pixels from unmasked patches. The high masking ratio of MAE enables efficient pre-training of vision transformers with large annotated datasets. The success of MAE has motivated a series of variants in vision tasks (24–27) and applications in medical image analysis using MIM techniques. For instance, Tang et al. (28) utilized masked inpainting for the pre-training of a Swin UNETR (Shifted-window UNet transformer) in abdominal segmentation tasks. Chen et al. (29) compared multiple MIM approaches in abdominal segmentation. Zhou et al. (30) applied MAE pre-training with UNETR (UNet Transformer) and obtained performance gains in both abdominal and brain tumor segmentation.
Building a masked image is a crucial step in MIM pre-training. As shown in Figure 1, the smallest masking unit of MLM, such as BERT (19), is typically the vocabulary, which preserves contextual information. However, MIM employs random masking, which can disrupt the spatial context and regions with the same semantic meaning, given the absence of the concept of words commonly observed in MLM. This, in turn, makes it challenging for the representation learning process to obtain high-quality pretrained network, especially when the masking ratio reaches a high percentage. Recently, several studies demonstrated that the masking strategy has a substantial effect on model performance in downstream tasks (31, 32). Although random masking is widely used, recent advances have shown that appropriate masking strategies can achieve better performance, such as region-based masking (33), attention-based masking (34), and adaptive masking (AdaMAE) (31). These masking strategies take the patch context into account, leading to more effective and efficient pre-training.
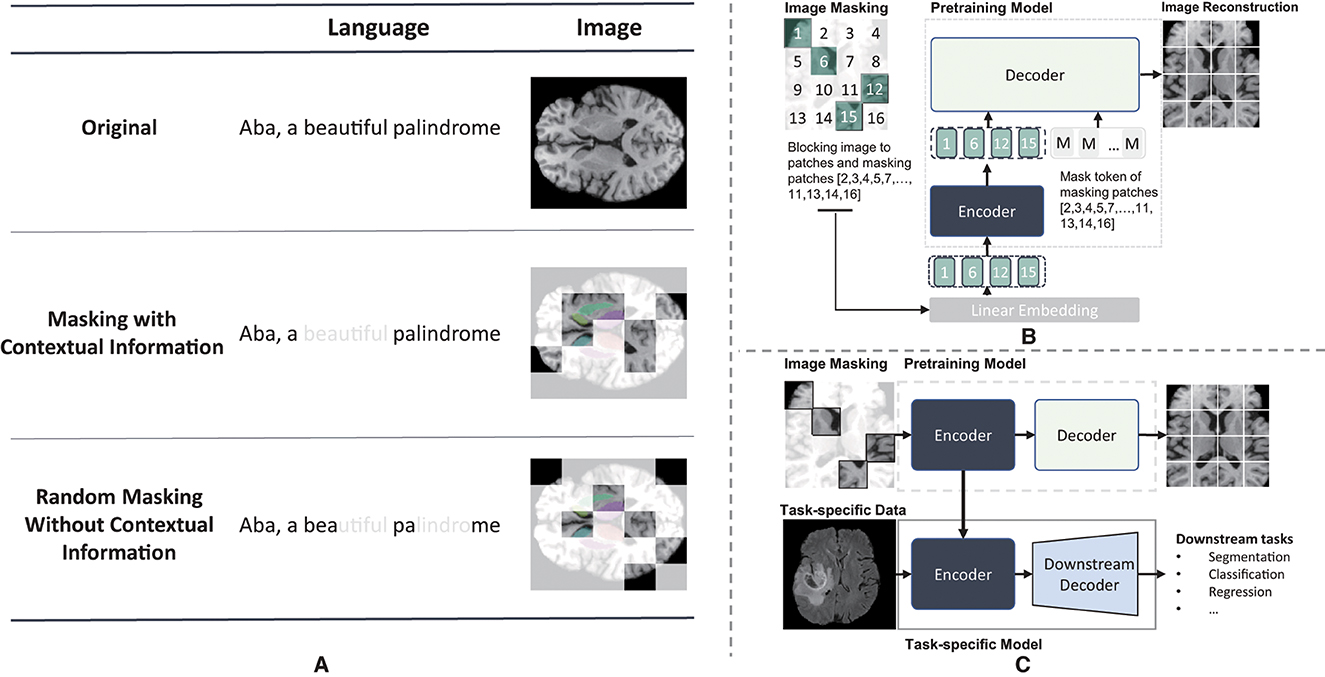
Figure 1. Illustration of maksed modeling pre-training and masked image modeling(MIM). (A) Different masking result of masked modeling pre-training. The light white regions represents the masked regions and the colored reigons of axial brain MR image represents different independent brain areas. (B) The pre-training illustraion of masked autoencoder. (C) the whole procedure of mim.
In the context of medical images, anatomical knowledge could help improve the pre-training. Huang et al. (35) incorporated the symmetry characteristics of brain structures into the pre-training by constructing symmetric positional encodings. However, few studies have integrated the more precise brain atlas (36) into the masking strategy. Inspired by the performance gains achieved by weighted masking strategies, we propose an anatomical prior-informed masking strategy for the MAE pre-training. We hypothesize that the tumor distribution among brain structures can guide the MAE pre-training, therefore improving the downstream brain tumor segmentation. To achieve this, we analyze the tumor occurrence in the SRI-24 space and establish an anatomical prior-informed probability map for image masking. This strategy allows us to select more informative patches for MAE pre-training. By combining the data-driven MAE with anatomical knowledge, we aim to improve the accuracy and data-efficiency of brain tumor segmentation.
In this study, our contributions are as follows:
(1) An anatomical prior-informed masking strategy is proposed to enhance the pre-training of masked autoencoder. This strategy is designed to preserve contextual information in 3D medical images and allows the pre-training process to concentrate on regions that are more relevant to the downstream segmentation task.
(2) By incorporating prior-informed weighted sampling, we construct an anatomical prior-informed masked autoencoder, referred to as API-MAE. This self-supervised pre-training approach utilizes 6,415 skull-stripped brain T1 MR images and combines data-driven reconstruction with anatomical priors.
(3) Inheriting the pretrained encoder weights, our method demonstrates superior performance in the downstream segmentation task on the BraTS21 dataset, outperforming several transformer models and surpassing state-of-the-art self-supervised learning methods. Subsequent experiments demonstrate that our method exhibits greater efficiency compared with a regular masked autoencoder and maintains a satisfactory trade-off between segmentation accuracy and computational consumption.
2. Methodology
2.1. Overview of proposed method
We propose a novel masking strategy for improved MAE pre-training and downstream brain tumor segmentation in MRI. As shown in Figure 2, our proposed method consists of two stages: (1) pre-training a masked autoencoder with anatomical prior-informed masking strategy on the unannotated dataset and (2) transferring the pre-trained weights of the encoder and fine-tuning the segmentation network on the annotated dataset.
2.2. Statistical analysis of tumor occurrence
2.2.1. Registration to standard brain template
To represent the anatomical priors, we first align all images with the standard brain template. The DICOM image data are transformed into Nifti format, and the brain is extracted using FSL tools (37). After that, we transform each image into the SRI-24 standard space (36) via affine registration. Using the optimized affine transformation matrix M*, all images are aligned in the SRI-24 space.
where Im represents the moving image, which corresponds to the MRI image of each sample. The fixed image, denoted as If, refers to the T1 template of the SRI-24 standard space. In this study, the operation C(Im, If) represents the cost function used to quantify disparities between the fixed image Im and the moving image during the registration optimization process, where a correction ratio is applied (38). The notation Affine(I; M) signifies the affine operation that maps the floating image I to the fixed image using the affine matrix M. Moreover, I represents the output registrated image.
2.2.2. Sampling weight map derived from brain tumor occurrence
We conduct a statistical analysis of enhanced tumor (ET) across BraTS21 dataset (39–41) and obatin a distribution map of ET occurrence in the SRI-24 standard space. To implement this analysis, we utilize a brain parcellation atlas building upon the parc116plus atlas (36). Some excessively small regions are merged into larger ones, resulting in 128 parcellation regions of the entire skull-stripped brain. To obtain the sampling probability of each voxel, the average sampling probability for each parcellation is defined as follows:
where Ri represents the i-th brain parcellation, PRi denotes the average sampling probability per volume of region Ri, fi, j is the occurrence frequency of the ET region in the j-th voxel within the i-th parcellation, VRi represents the volume of Ri, and NRi represents the number of voxel in Ri. Consequently, the sampling weight map W, depicted in Figure 3, can be generated by assigning voxels within the parcellation region Ri the identical probability value PRi.
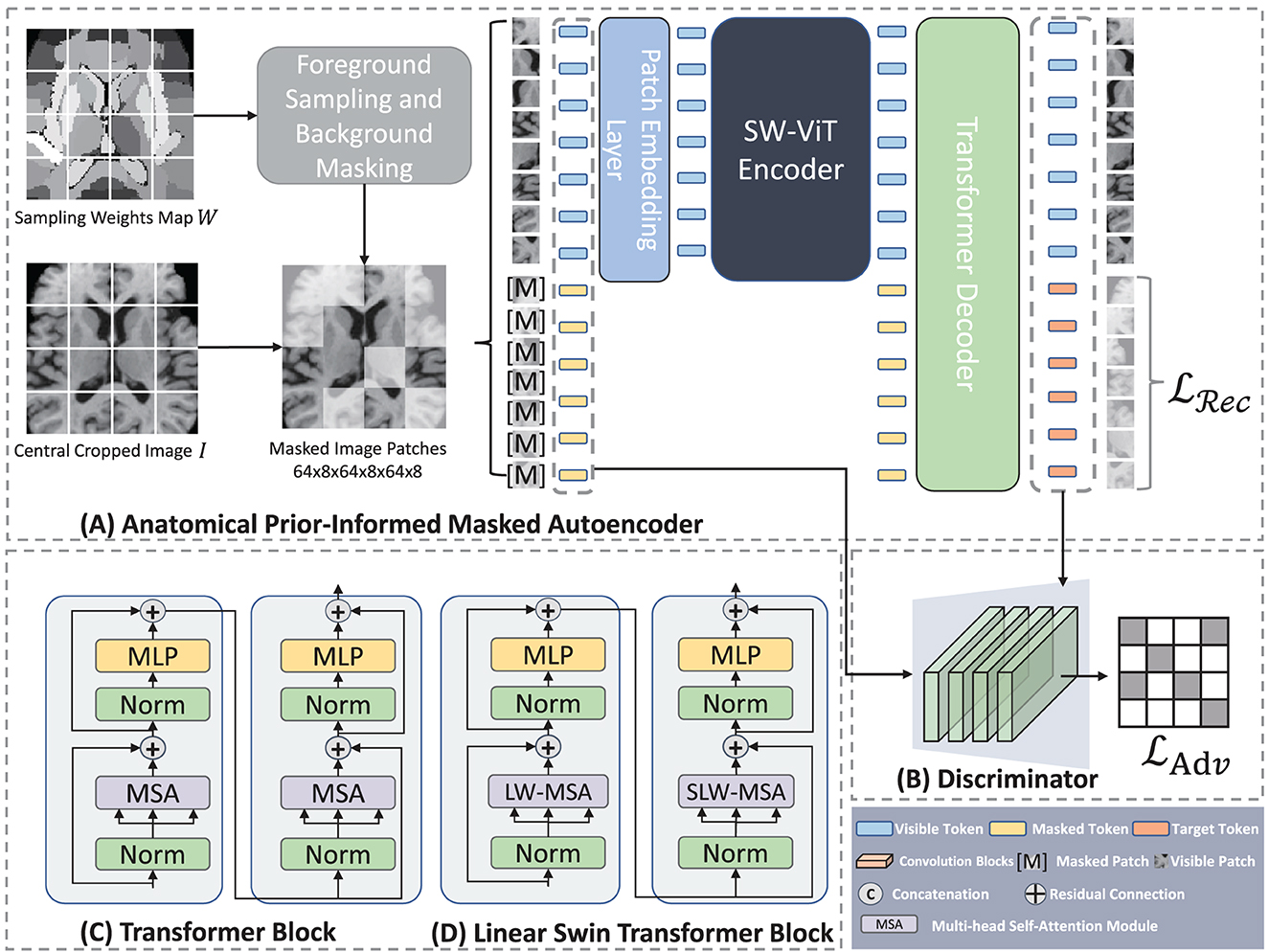
Figure 3. Architecture of Anatomical Prior-Informed Masked Autoencoder (API-MAE). (A) Is the architecture of Anatomical Prior-Informed Masked Autoencoder. (B) Is the Discriminator used for reconstruction. (C) If the Transformer block. (D) Is the linear Swin Transformer block.
2.3. Anatomical prior-informed masked auto-encoder
As shown in Figure 3, our proposed Anatomical Prior-Informed Masked AutoEncoder (API-MAE) consists of five components as follows: (1) Anatomical Prior-informed Masking, (2) Patch embedding, (3) Transformer Encoder, (4) Transformer Decoder, and (5) Discriminator.
2.3.1. Anatomical prior-informed masking strategy
Instead of the random masking strategy used in standard MAE pre-training, we propose a dedicated masking strategy to select informative patches based on the derived sampling weight map. The input image I and sampling weights map W are center-cropped with a size of 128, i.e., I ∈ ℝ128 × 128 × 128, W ∈ ℝ128 × 128 × 128. Subsequently, I and W are transformed into patches represented as and , respectively. Here, n signifies the quantity of patches, and the patch size is configured at 8, a choice consistent with previous studies (35). This configuration leads to n = 16 × 16 × 16, aligning with the concept of vision transformers (12) splitting the 2D image into 16 × 16 tokens. The sample probability of each patch is determined by the probability vector , where , and wi, j denotes the sampling weight of the j-th voxel within the i-th patch corresponding to the voxels xi, j of the image patch. Consequently, the visible patches that are fed into the encoder can be sampled as follows:
where represents visible patches sampled from the original image patches , and k = η·n represent the number of visible patches, η = 0.25 is the sampling ratio which aligned with the 75% masking ratio of MAE. The operation involves utilizing a multinomial probability distribution with the probability vector p to select tokens from for sampling, which then constitute the visible tokens. The sampling procedure is implemented using the multinomial API from PyTorch. As depicted in Figure 4, the prior-informed sampling maintains superior structural consistency compared to random masking, which is advantageous for the calculation of region-based sampling weights.
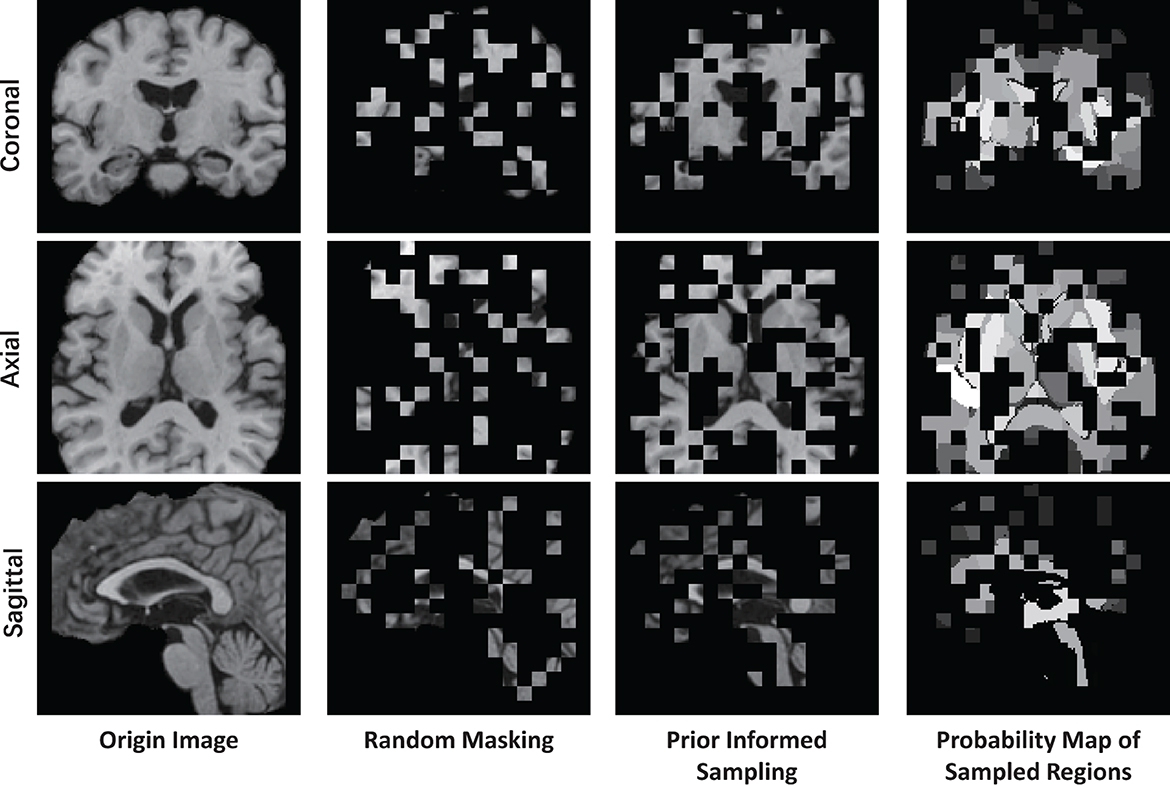
Figure 4. Example of visualizing a T1 MR image using different masking strategies with a masking ratio of 0.75. This image is center-cropped with a shape of 128 × 128 × 128, and each token has a patch size of 8 × 8 × 8.
2.3.2. Patch embedding
The input visible patches in are first flattened into one-dimensional vectors, then mapped to the feature dimension D via learnable patch tokenizer g(·). The input of the transformer encoder xenc is calculated as follows:
where , and PE is the sinusoidal positional encoding.
where pos = 1, 2, …, T represents the token position, i represents the i-th dimension.
2.3.3. Transformer encoder
We adopt a shifted window vision transformer, known as SW-ViT (35), as the transformer encoder in API-MAE. As shown in Figures 3C, D, the multi-head self-attention (MSA) in the original transformer block is replaced with linear window-based multi-head self-attention (LW-MSA) and shifted linear window-based multi-head self-attention (SLW-MSA) in the Swin transformer block. Both LW-MSA and SLW-MSA reduce parameters and computations among each head, which improves the network efficiency without significant accuracy loss. The transformer encoder serves as the feature extractor in API-MAE and the segmentation network. The output of the transformer encoder will undergo a linear projection to fit the higher feature dimension of the transformer decoder.
2.3.4. Transformer decoder
We use a shallow transformer decoder to reconstruct the original image in API-MAE. The inputs to the decoder consist of both visible tokens and masked tokens with positional encodings. The output of the decoder is the reconstructed image tokens for each input patch. The reconstruction loss function is the standard L2 loss:
where xi denotes the i-th image patch and m represents the number of masked tokens. It should be noted that only masked tokens are calculated for reconstructed loss.
2.3.5. Reconstruction Discriminator
Recent advancements in self-supervised learning, such as DiRA (42), have demonstrated that the collaborative learning of self-supervised and adversarial tasks can lead to a more generalizable representation, encompassing fine-grained semantic representation. Moreover, discriminators have been proven beneficial for the masked autoencoder (32, 43). In API-MAE, we introduced a reconstruction discriminator, envisioning its potential synergistic effect when integrated into MAE decoder. This combination aims to enhance the learning representation and improve visual quality of the reconstructed output. The discriminator is constructed as a shallower convolutional neural network, comprising five convolutional layers tasked with distinguishing between the reconstructed and real images. The adversarial loss employed for the discriminator is represented as an L2 loss as follows:
where xi is the i-th image patch, is the corresponding reconstructed patch, and n is the token number of the original image. Thus, the total loss of API-MAE is a combination of reconstruction loss and adversarial loss as follows:
2.4. Segmentation network
After the pre-training of API-MAE, we discard the transformer decoder and keep the transformer encoder for the brain tumor segmentation task. The architecture of the segmentation network is shown in Figure 5. The segmentation network contains three parts as follows: (1) encoder, which contains patch embedding and transformer blocks, (2) encoder propagation, and (3) decoder. The patch embedding layer maps the input multi-parameter MRI (i.e., T1, T1Gd, T2-FLAIR, and T2 image) patches to the embedding features. The transformer blocks share the same architecture and are initialized with the pre-training weight of the transformer encoder in API-MAE. The encoder propagation and decoder parts utilize features from the original image (i.e., z0) and specific transformer layers (2nd, 4th, 6th, 8th, and last layer, i.e., z2, z4, z6, z8, z12) to propagate features and segment the image into three target classes as follows: whole tumor (WT), tumor core (TC), and enhanced tumor (ET). To obtain better segmentation, the segmentation network adopts cross-entropy and Dice loss with deep supervision as the segmentation loss as follows:
where i represents the stage of deep supervision, denotes the prediction of stage i, and Si represents the ground truth resized to match the corresponding prediction.
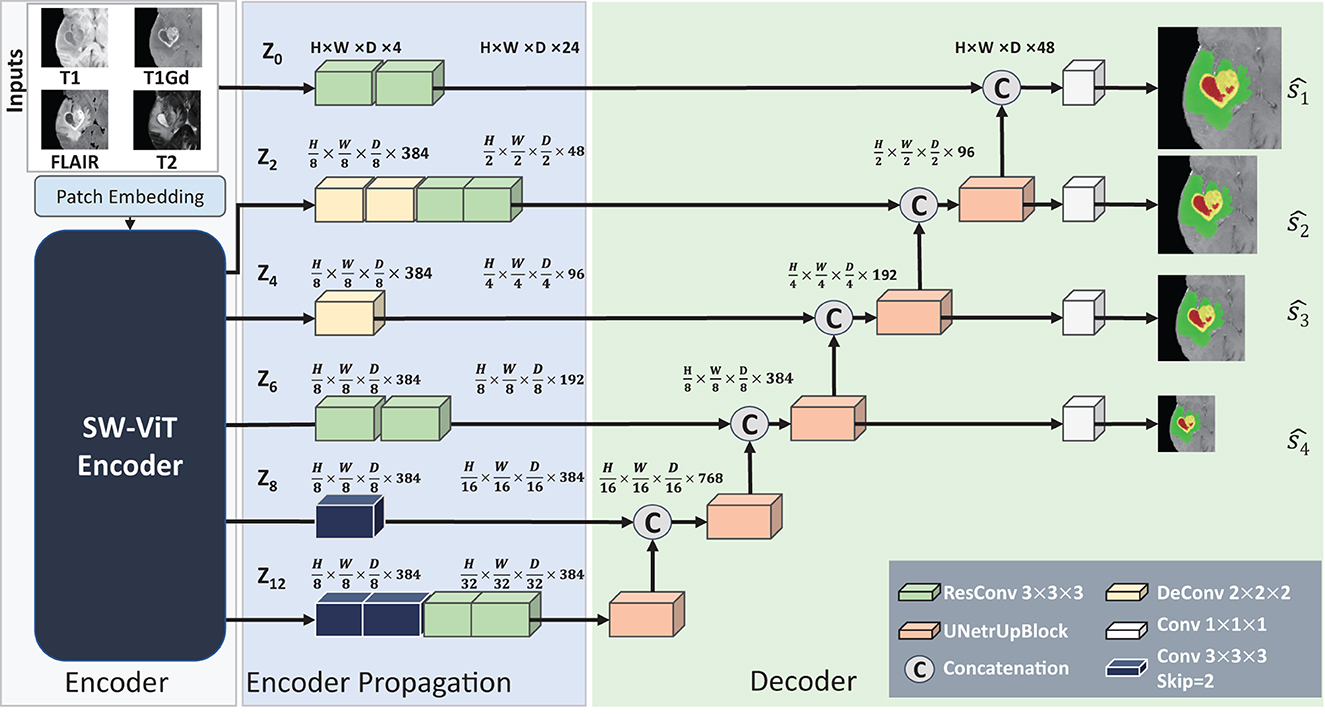
Figure 5. Architecture of the baseline segmentation network. This network is made up of three parts, i.e., the Encoder part for feature extraction, the Encoder Propagation part used for channel and spatial normalization and skip connection, and the Decoder parts used for upsampling and predicting the segmentation results. The convolution blocks with skip=2 in the Encoder Propagation part are used for downsampling, and the UNetrUpBlock used in the decoder part is used for upsampling and each block contains a deconvolution block and two residual convolution blocks.
3. Experiments
We pre-train the MAE model on an unannotated brain MRI dataset and evaluate the segmentation performance on an annotated brain tumor MRI dataset.
3.1. Datasets
3.1.1. ADNI dataset
Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset (44) is derived from a longitudinal multicenter study aimed at early detection and tracking of Alzheimer's disease (AD). In this study, we collected 7,945 skull-stripped T1 MR images and subsequently handpicked 6,415 images of superior visual quality for utilization in the pre-training dataset. This selection was made following a visual inspection of the registration results.
3.1.2. BraTS21 dataset
The BraTS21 dataset (39–41) consists of 1,251 multi-parameter MRI scans. Each case includes four different modalities as follows: a) native (T1), b) post-contrast T1-weighted (T1Gd), c) T2-weighted (T2), and d) T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) images, acquired from various protocols and scanners across multiple institutions. Each scan has been annotated by experienced radiologists with three different subregions as follows: enhancing tumor (ET), peritumoral edematous/invaded tissue (ED), and necrotic tumor core (NCR). In this study, we divide the 1,251 samples into training, validation, and testing sets at a ratio of 7:1:2, following previous studies (35).
3.2. Evaluation metrics
Both the volumetric metric dice similarity coefficient (DSC) and surface metric Hausdorff distance (HD) are used for performance evaluation. DSC quantifies the overlap between segmentation results and annotations in voxel space, while the 95th percentile of Hausdorff distance (HD95) measures the distances between the segmentation surface and ground-truth surface. The calculation of HD95 is performed by the MedPy package using the analysis framework from nnFormer (45).
3.3. Implementation details
Experimental settings: All the experiments are implemented using the PyTorch 1.2 framework. We use 4 NVIDIA A100 GPUs (40 GB VRAM) for MAE pre-training and NVIDIA RTX3090 GPU (24 GB VRAM) for segmentation training and inference.
Data preprocessing: In the preprocessing section, we employ affine registration to align individual images with the standard space. Here, the cost function during the image registration optimization is correlation ratio (38). To prevent the registration results from being flipped upside down, we defined the rotation search space for affine registration as follows: [−30°, 30°] for X-axis rotation, [−30°, 30°] for Y-axis rotation, and [−180°, 180°] for Z-axis rotation. This configuration is aimed to emphasize rotation in the X-Y plane and prevent upside-down flipping along the Z-axis. It performed effectively with our dataset of 6,415 pre-training samples. The registration optimization and transformation processing were executed using the FLIRT (46) toolbox from FSL. Trilinear interpolation was utilized to compute the intensity of new voxels during affine mapping. For the pre-training data, we employ the MONAI (47) library for data normalization and cropping. Additionally, we utilize the segmentation data preprocessing pipeline provided by nnUNet (7), to handle the multi-modality segmentation data.
Model architecture: In API-MAE, the transformer encoder contains 12 layers of linear swin transformer blocks with a feature dimension D = 384. The transformer decoder comprises 8 layers of vanilla transformer blocks with a feature dimension of 384. The discriminator consists of four convolution blocks with a kernel size of k = 3 and a convolution block with a kernel size of k = 1. In the segmentation network, the weights of encoder propagation and decoder parts are initialized with the He initialization (48).
Model training: For MAE training, the AdamW optimizer with a batch size of 12 is trained for 300 epochs. The initial learning rate is 1e-3. Weight decay of 5e-2 is also adopted for model regularization. For the segmentation procedure, we apply the (45) training framework and default parameter for 1,000 epochs.
4. Results
4.1. Pre-training results of anatomical prior-informed MAE
As presented in Figure 6, we note distinct differences in the spatial distribution of tumor occurrence within the SRI24 space. Specifically, gliomas are more frequently observed in the white matter regions of the middle and posterior sections of the brain, with comparatively lower frequencies in the brainstem and cerebellar regions. Table 1 shows the normalized probability of tumor occurrence among all 128 brain parcellations. Considering that ET is the most challenging region to segment, we employ the probability of the ET region for probabilistic masking.
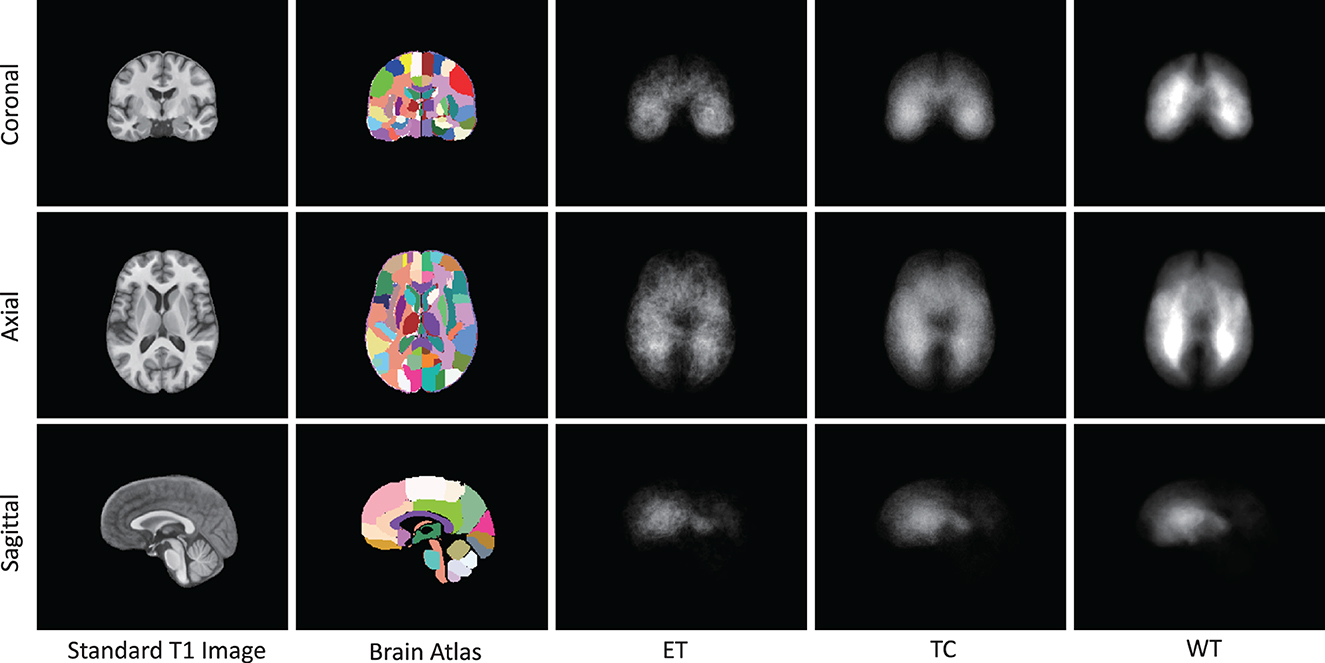
Figure 6. The occurrence frequency in SRI-24 standard space among 1,251 cases from BraTS21 dataset. The five columns represent the standard brain T1 MR image, brain atlas (enhanced parc116 plus) in SRI-24 Space, Enhanced Tumor (ET) occurrence, Tumor Core (TC) Tumor occurrence, and Whole Tumor (WT) occurrence, respectively.
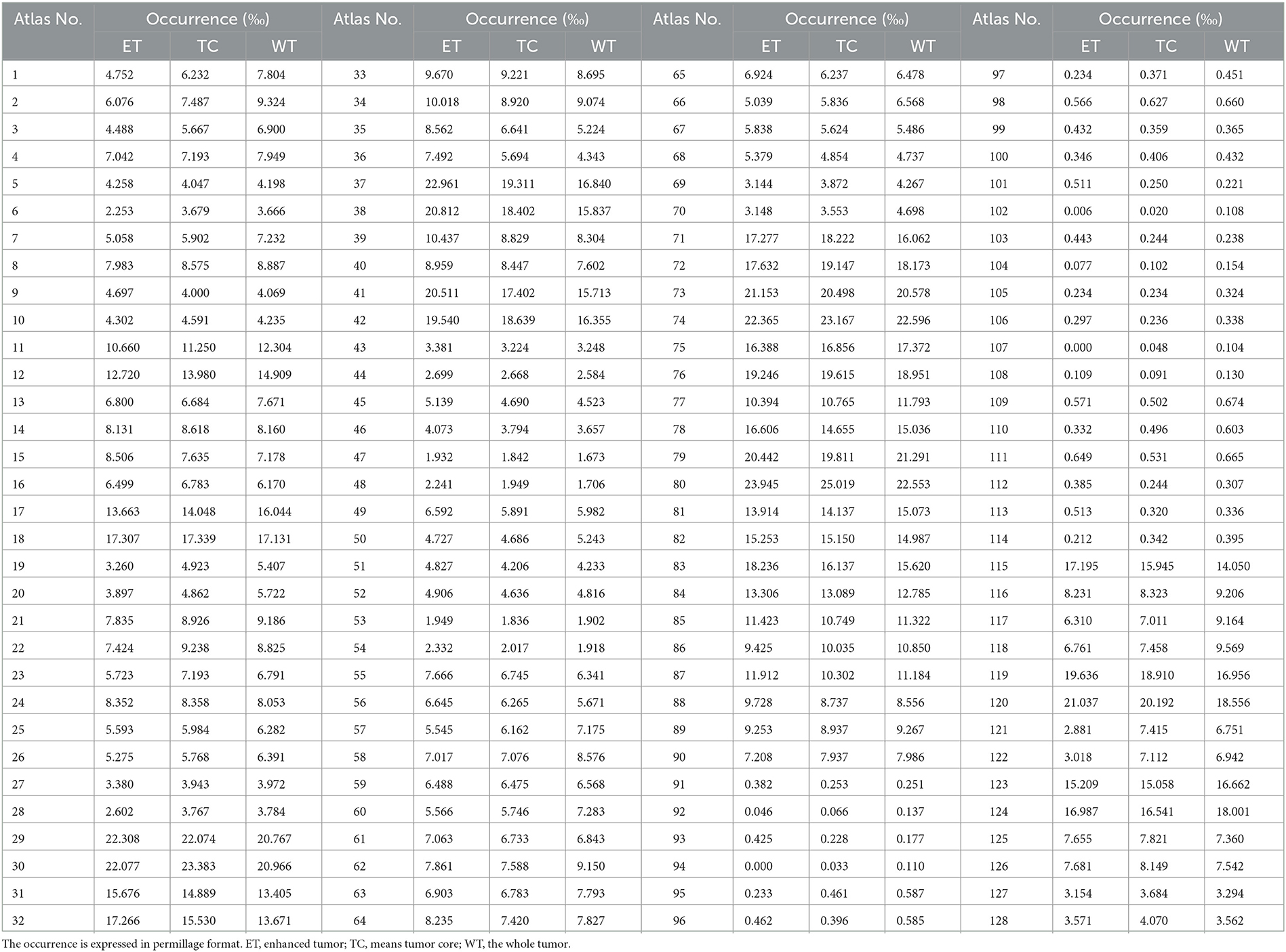
Table 1. The normalized occurrence of tumor regions within different brain parcellations in enhanced SRI-24 atlas analyzed from 1,251 training cases of the BraTS21 dataset.
The masking and reconstruction results are shown in Figure 7. It can be observed that random masking tends to distribute masked patches uniformly across the entire image, whereas our proposed weighted sampling strategy enables concentration on more valuable, concentrated, and relatively contiguous regions. The disruption of contextual information in random masking makes the reconstruction task challenging and results in a blurry reconstructed image. In contrast, the proposed weighted sampling method can maintain the integrity of semantic regions, allowing for better reconstruction results.
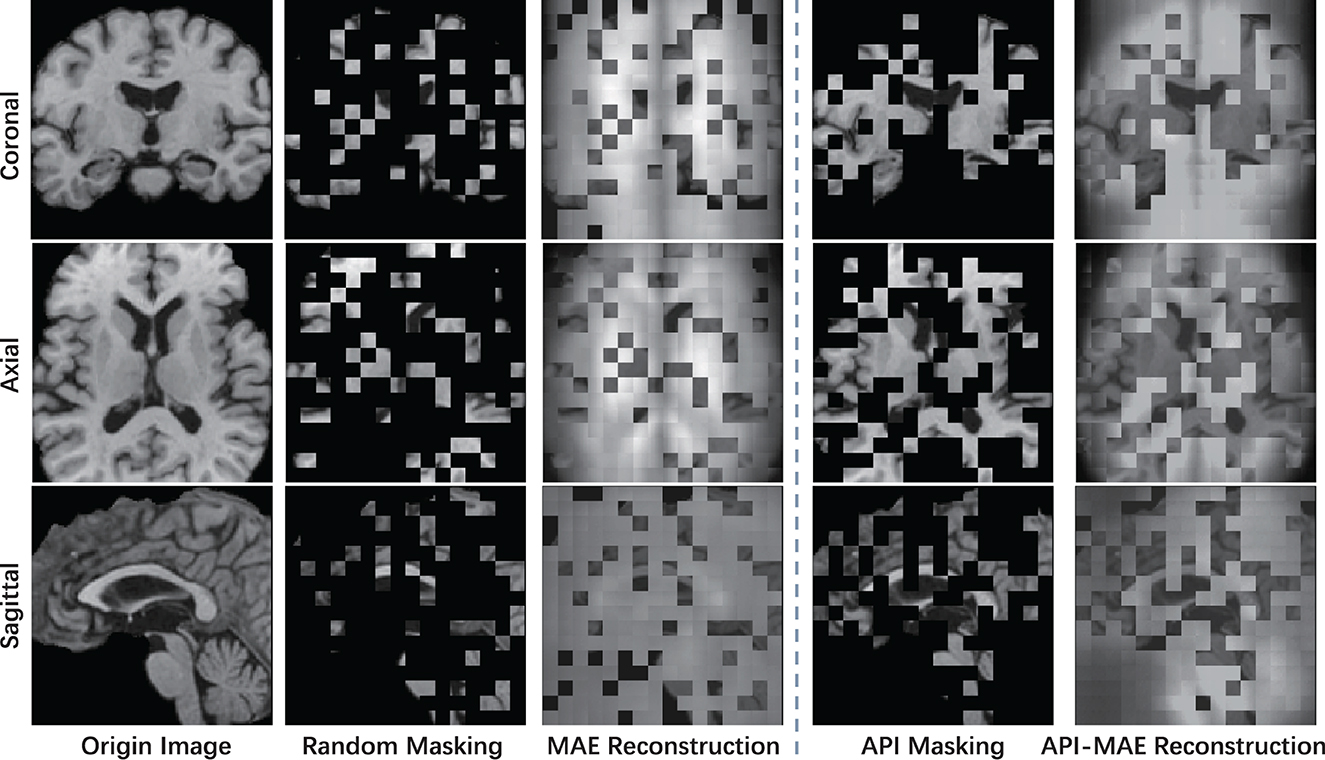
Figure 7. The visible example of masking images and the reconstruction results of MAE and API-MAE. The five columns represent the origin brain T1 MR image, the random masking strategy used in MAE, the masking image generated from API token sampling, and the reconstruction results of API-MAE, respectively.
4.2. Segmentation results on BraTS21 dataset
4.2.1. Segmentation performance on BraTS21 dataset
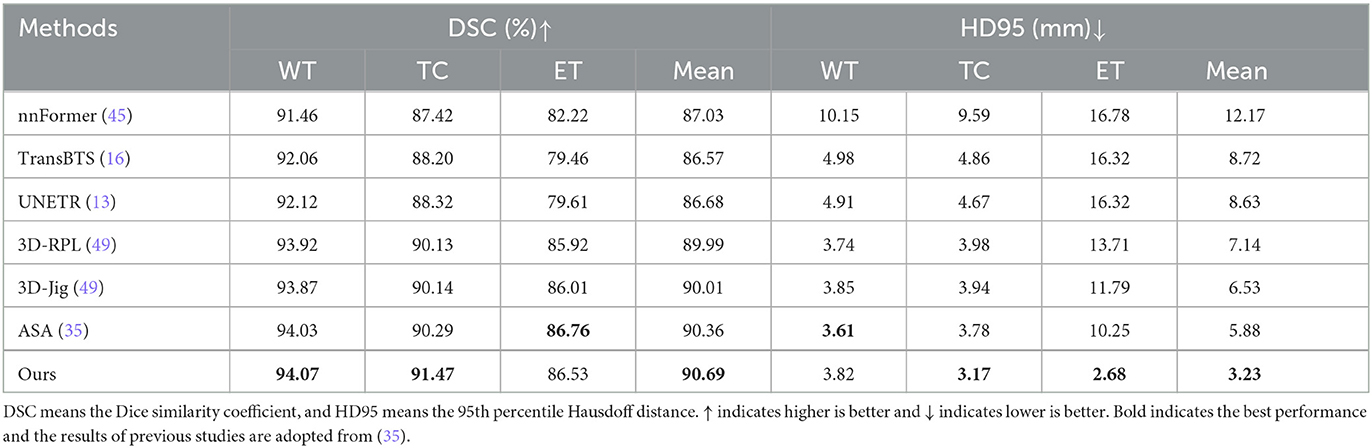
To validate the effectiveness of the proposed SSL pre-training approach in downstream segmentation task, we conducted validation experiments using the BraTS21 dataset. The downstream brain tumor segmentation network is initialized with the pre-trained API-MAE encoder weights and subsequently fine-tuned using the BraTS21 dataset. We conducted a comparison of our method against several transformer-based models, including nnFormer (45), TransBTS (16), and UNETR (13) without pre-training. Additionally, we compared against several SSL pre-training methods used in medical imaging, namely, 3D-RPL and 3D-Jig (49), as well as the current state-of-the-art ASA in brain tumor segmentation (35).
As shown in Table 2, we observed that the pre-trained models demonstrate better performance, and our proposed API-MAE achieved the best performance in terms of the Dice similarity coefficient (DSC) metrics for whole tumor (WT) and tumor core (TC) and the best average performance of all three regions.
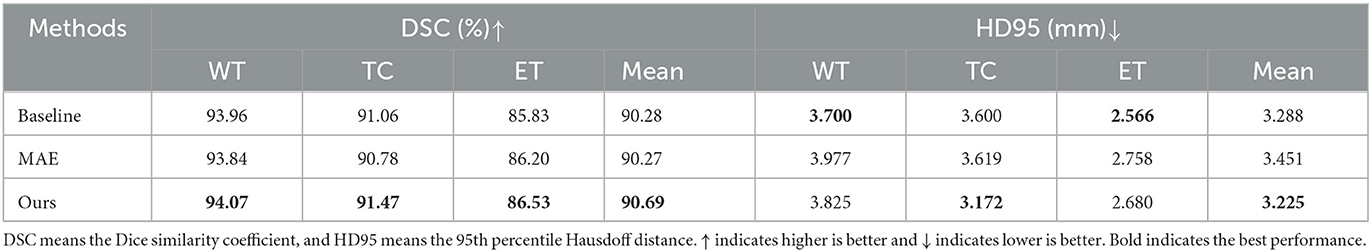
4.2.2. Ablation study on masking strategies
To evaluate the effectiveness of our proposed masking strategy, we conduct an ablation study on different MAE masking strategies. The comparison methods include the baseline without pre-training, MAE pre-trained with random masking, and our proposed API-MAE pre-trained with anatomical prior-informed masking strategy. Table 3 shows that our proposed API-MAE showed improved performance for all regions compared with vanilla MAE and baseline. This demonstrates the effectiveness of our anatomical prior-informed masking compared with the random masking strategy. However, the marginal improvement indicates that in the presence of enough annotated data (more than 1,000 cases in BraTS21), transformer-based models already achieve satisfactory performance, and the benefit of pre-training is not substantial.
4.2.3. Data-efficiency analysis
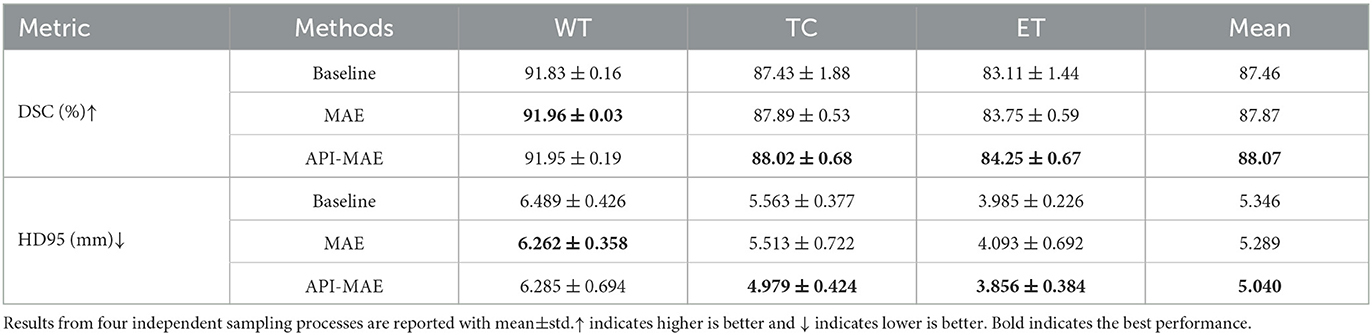
To validate the data efficiency of our pre-trained model, we further train the segmentation model on a small subset of the whole training dataset. We randomly sampled 100 cases from the original training cases, while the validation and testing sets were kept the same as the whole dataset. The compared methods include the baseline without pre-training, MAE pre-trained with random masking, and our proposed API-MAE pre-trained with anatomical prior-informed masking strategy. The sampling process is repeated four times to mitigate the selective bias.
The segmentation results on the small training set are shown in Table 4. It is observed that MAE pre-training benefits the segmentation performance and improves the model robustness in most scenarios. The improvement by pre-training is more prominent in this small-dataset setting compared with the whole dataset. The best segmentation performance for ET and TC regions is obtained by API-MAE, in terms of DSC metrics, which matches the purpose of using ET occurrence map for weighted sampling. As shown in Figure 8, training with the MAE paradigm tends to reduce the erroneous falsely predicted regions and reduce the prediction error of ET regions, particularly in difficult-to-segment regions.
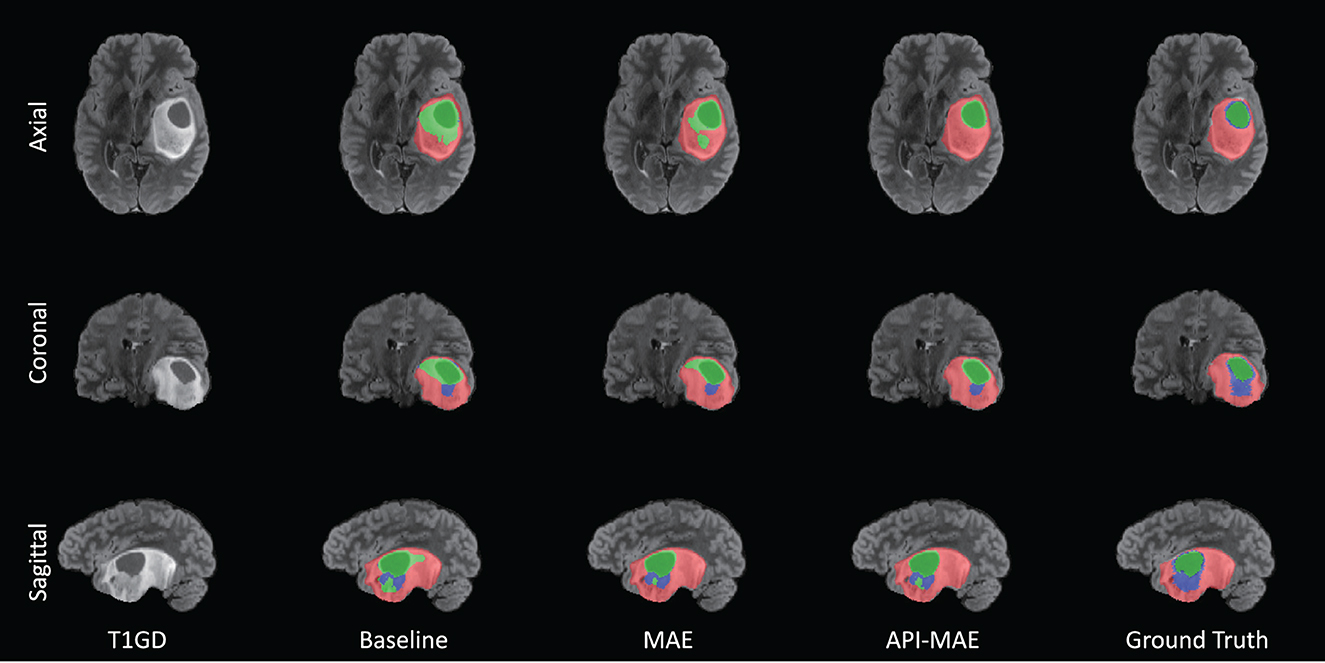
Figure 8. Example of tumor segmentation results from a testing image with 100 training cases. The three rows are from the axial, coronal, and sagittal views. The green region represents the necrotic tumor core (NCR), the blue region represents the Gd-enhancing tumor (ET), and the red region represents the peritumoral edematous/invaded tissue (ED).
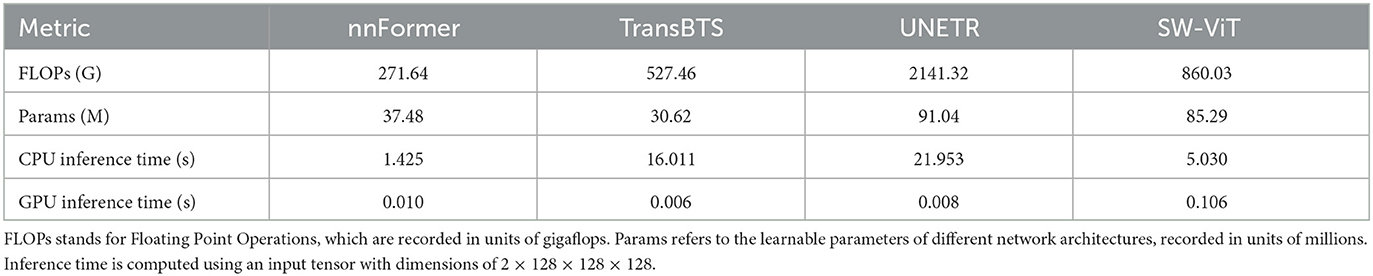
To further investigate the efficiency of proposed method, we conducted an efficiency analysis of the segmentation phase for the methods, as shown in Table 5. Since different SSL methods share the same segmentation network, specifically SW-ViT, the variations in performance arise from the encoder weights inherited from diverse SSL pre-training tasks. This comparison involves distinct network architectures, namely, nnFormer, TransBTS, UNETR, and SW-ViT. All the methods were reproduced using the original code on a local server equipped with an AMD Ryzen 9 5900X CPU (3.7 GHz), 128 GB RAM (DDR4 2400MT/s), and an NVIDIA RTX3090 GPU. For fair comparison, we modified UNETR by adjusting its input channels to 4 and configuring the patch size as 8 × 8 × 8, in alignment with SW-ViT. The computation consumption was calculated utilizing the thop package. This process entails inputting a tensor with dimensions of 2 × 128 × 128 × 128 into the network for computation and the standard segmentation procedure.
Combining the data from Tables 2, 4, we observe that nnFormer exhibits the best inference efficiency. This superiority can be attributed to the dimension of the embedding feature in the Transformer module of the network, which is [96, 192, 384, 768]. In contrast, other Transformer models often have embedding feature dimensions of 384 or 768. This relatively shallower transformer architecture contributes to its enhanced computational efficiency. However, it may result in slightly lower segmentation performance. Higher segmentation accuracy can be achieved in both WT and TC components in models with increased transformer layers. However, when using a high-layer transformer encoder such as UNETR, the number of floating point operations (FLOPs) and learnable parameters will increase rapidly. While the SW-ViT could reduce the FLOPs and parameters with the help of shifted window-based linear transformer modules. Enhanced with SSL pre-training tasks, particularly our proposed API-MAE, the methods using SW-ViT obtain the best segmentation performance while maintaining a favorable balance in terms of segmentation time consumption. Due to the presence of certain operations within the network architecture that do not parallelize efficiently during GPU computation, the proposed method does not achieve optimal computational efficiency on the GPU. However, the proposed method could attain decent CPU time consumption, which maintains a reasonable balance between accuracy and efficiency.
5. Discussion
Recently, transformer-based models have emerged as state-of-the-art methods for 3D medical image segmentation, owing to their superiority in modeling long-range dependencies and leveraging global contextual information over fully convolutional neural networks. However, such methods often rely on a vast of training data for network optimization. A major challenge in training such models is the limited availability of annotated data. In this study, we address this challenge by utilizing 6,415 unannotated T1-weighted MR images from the ADNI dataset for pre-training. Our approach consistently improved the segmentation accuracy in scenarios with both large and small training sets. Although only T1-weighted images are used for pre-training, the learned weights benefit the downstream brain tumor segmentation on multi-parameter MRI. This highlights the potential of pre-training for improved medical image segmentation.
The MAE used in computer vision typically employs random masking with a high masking ratio of 0.75 and utilizes 25% unmasked patches for encoder training. The high masking ratio can lead to the loss of contextual information in high-dimensional medical images, making image reconstruction challenging and potentially affecting the learning of generalizable features. Therefore, it is important to consider tailored sampling strategies that take into account the specific characteristics and requirements of the task at hand. In this study, we introduce an anatomical prior-informed masking strategy, where brain regions with higher tumor occurrence are more frequently sampled for pre-training. The experiments demonstrate that our proposed pre-training method enhances the performance of brain tumor segmentation, which outperforms other self-learning approaches. This indicates that incorporating anatomical priors into the pre-training stage leads to performance improvements in downstream tasks.
Additionally, our anatomical prior-informed sampling strategy can be considered as an attention mechanism in selecting valuable and task-related patches for MAE pre-training. In general, attention mechanisms usually help models filter out high-value information from large amount of data, thereby improving computational efficiency and performance and making computing more precise and efficient. Given a large number of image patches in the unannotated dataset, it is important to let the pre-training process attend the informative patches. By incorporating the tumor occurrence rate and brain template into the construction of an attentive sampling strategy, our approach integrates anatomical priors with masked image modeling pre-training. This enables efficient sampling and the most use of unannotated data.
There are some limitations of this study. Our proposed method requires the pre-registration of the sampling weighting map for each individual, a process typically executed on the CPU and incurring a time cost. In future study, this procedure can be expedited through the utilization of deep learning-based networks, enabling accurate and rapid registration. We showcase the advantage of integrating anatomical priors during the pre-training stage, leveraging only tumor occurrence information. In future, the exploration of more advanced anatomical priors, such as symmetric brain structure or active learning strategies (50), holds potential for further investigation.
6. Conclusion
In this study, we introduce a novel pre-training technique for brain tumor segmentation utilizing transformer networks. This technique involves the integration of an anatomical prior-informed masking strategy into the masked image modeling process. Informative image patches from brain parcellations with higher tumor occurrence are sampled more frequently, facilitating the mask autoencoder to focus on the regions of interest. The proposed approach demonstrates promising performance in the brain tumor segmentation task, surpassing compared self-learning methods.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
KW, HW, MW, and SW: main idea and study design. ZL and MP: organized the database, data inspection, and analysis. KW, ZL, and SW wrote the first draft of the manuscript. SL: built the initial code of the network training framework. MW, SW, and ZS: supervised, supported, and revised the manuscripts. All authors contributed to the manuscript revision, read, and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China under Grant 82072021 and SW was supported by Shanghai Sailing Programs of Shanghai Municipal Science and Technology Committee (22YF1409300).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Janjua TI, Rewatkar P, Ahmed-Cox A, Saeed I, Mansfeld FM, Kulshreshtha R, et al. Frontiers in the treatment of glioblastoma: past, present and emerging. Adv Drug Deliv Rev. (2021) 171:108–38. doi: 10.1016/j.addr.2021.01.012
2. Li C, Wang S, Yan JL, Piper RJ, Liu H, Torheim T, et al. Intratumoral heterogeneity of glioblastoma infiltration revealed by joint histogram analysis of diffusion tensor imaging. Neurosurgery. (2019) 85:524–34. doi: 10.1093/neuros/nyy388
3. Li C, Wang S, Liu P, Torheim T, Boonzaier NR, van Dijken BR, et al. Decoding the interdependence of multiparametric magnetic resonance imaging to reveal patient subgroups correlated with survivals. Neoplasia. (2019) 21:442–9. doi: 10.1016/j.neo.2019.03.005
4. Wang S, Dai C, Mo Y, Angelini E, Guo Y, Bai W. Automatic brain tumour segmentation and biophysics-guided survival prediction. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 5th International Workshop, BrainLes 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 17, 2019, Revised Selected Papers, Part II 5. Cham: Springer. (2020) p. 61–72.
5. Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y, et al. deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal. (2018) 43:98–111. doi: 10.1016/j.media.2017.10.002
6. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. Cham: Springer. (2015) p. 234–241.
7. Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. (2021) 18:203–11. doi: 10.1038/s41592-020-01008-z
8. Futrega M, Milesi A, Marcinkiewicz M, Ribalta P. Optimized U-Net for brain tumor segmentation. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, September 27, 2021, Revised Selected Papers, Part II. Cham: Springer. (2022) p. 15–29.
9. Azad R, Heidari M, Wu Y, Merhof D. Contextual attention network: transformer meets U-net. In: Lian C, Cao X, Rekik I, Xu X, Cui Z, editors. Machine Learning in Medical Imaging. Cham: Springer Nature Switzerland (2022). p. 377–86. doi: 10.1007/978-3-031-21014-3_39
10. Wu H, Pan J, Li Z, Wen Z, Qin J. Automated skin lesion segmentation via an adaptive dual attention module. IEEE Trans Med Imaging. (2021) 40:357–70. doi: 10.1109/TMI.2020.3027341
11. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Advances in Neural Information Processing Systems. (2017) p. 6000–10.
12. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: transformers for image recognition at scale. In: International Conference on Learning Representations. Ithaca, NY (2021). Available online at: https://openreview.net/forum?id=YicbFdNTTy
13. Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, et al. Unetr: Transformers for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. Waikoloa, HI: IEEE (2022) p. 574–584.
14. Lin A, Chen B, Xu J, Zhang Z, Lu G, Zhang D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans Instrum Meas. (2022) 71:1–15. doi: 10.1109/TIM.2022.3178991
15. Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, September 27, 2021, Revised Selected Papers, Part I. Cham: Springer. (2022) p. 272–284.
16. Wang W, Chen C, Ding M, Yu H, Zha S, Li J. Transbts: Multimodal brain tumor segmentation using transformer. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2021: 24th International Conference, Strasbourg, France, September 27-October 1, 2021, Proceedings, Part I 24. Cham: Springer. (2021) p. 109–119.
17. Dobko M, Kolinko DI, Viniavskyi O, Yelisieiev Y. Combining CNNs with transformer for multimodal 3D MRI brain tumor segmentation. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 7th International Workshop, BrainLes 2021, Held in Conjunction with MICCAI 2021, Virtual Event, September 27, 2021, Revised Selected Papers, Part II. Cham: Springer. (2022) p. 232–241. doi: 10.1007/978-3-031-09002-8_21
18. Liu X, Zhang F, Hou Z, Mian L, Wang Z, Zhang J, et al. Self-supervised learning: generative or contrastive. IEEE Trans Knowl Data Eng. (2021) 35:857–76. doi: 10.1109/TKDE.2021.3090866
19. Devlin J, Chang MW, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding. arXiv. (2018). doi: 10.48550/arXiv.1810.04805
20. Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al. Language models are few-shot learners. In: Proceedings of the Conference on Advances in Neural Information Processing Systems (NeuraIPS). MIT Press (2020). p. 1877–901.
21. Chung YA, Zhang Y, Han W, Chiu CC, Qin J, Pang R, et al. W2v-bert: Combining contrastive learning and masked language modeling for self-supervised speech pre-training. In: 2021 IEEE Automatic Speech Recognition and Understanding Workshop (ASRU). Cartagena: IEEE. (2021) p. 244-250.
22. Bao H, Dong L, Piao S, Wei F. Beit: Bert pre-training of image transformers. arXiv. (2021). doi: 10.48550/arXiv.2106.08254
23. He K, Chen X, Xie S, Li Y, Dollár P, Girshick R. Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. New Orleans, LA: IEEE (2022) p. 16000–16009.
24. Feichtenhofer C, Fan H, Li Y, He K. Masked autoencoders as spatiotemporal learners. In: Proceedings of the Conference on Advances in Neural Information Processing Systems (NeuraIPS). New Orleans, LA: MIT Press (2022).
25. Yu X, Tang L, Rao Y, Huang T, Zhou J, Lu J. Point-bert: Pre-training 3d point cloud transformers with masked point modeling. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. New Orleans, LA: IEEE (2022) p. 19313–22.
26. Chen X, Ding M, Wang X, Xin Y, Mo S, Wang Y, et al. Context autoencoder for self-supervised representation learning. arXiv. (2022). doi: 10.48550/arXiv.2202.03026
27. Tong Z, Song Y, Wang J, Wang L. Videomae: Masked autoencoders are data-efficient learners for self-supervised video pre-training. arXiv. (2022). doi: 10.48550/arXiv.2203.12602
28. Tang Y, Yang D, Li W, Roth HR, Landman B, Xu D, et al. Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). New Orleans, LA: IEEE (2022) p. 20730–40.
29. Chen Z, Agarwal D, Aggarwal K, Safta W, Balan MM, Brown K. Masked image modeling advances 3D medical image analysis. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV). Waikoloa, HI: IEEE (2023) p. 1970–80.
30. Zhou L, Liu H, Bae J, He J, Samaras D, Prasanna P. Self pre-training with masked autoencoders for medical image analysis. arXiv. (2022). doi: 10.48550/arXiv.2203.05573
31. Bandara WGC, Patel N, Gholami A, Nikkhah M, Agrawal M, Patel VM. AdaMAE: adaptive masking for efficient spatiotemporal learning with masked autoencoders. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Vancouver, BC: IEEE (2023). p. 14507–17.
32. Chen H, Zhang W, Wang Y, Yang X. Improving masked autoencoders by learning where to mask. arXiv. (2023). doi: 10.48550/arXiv.2303.06583
33. Qing Z, Zhang S, Huang Z, Wang X, Wang Y, Lv Y, et al. Mar: masked autoencoders for efficient action recognition. IEEE Trans Multimed. (2023) 1–16. doi: 10.1109/TMM.2023.3263288
34. Xu H, Ding S, Zhang X, Xiong H, Tian Q. Masked autoencoders are robust data augmentors. arXiv. (2022). doi: 10.48550/arXiv.2206.04846
35. Huang J, Li H, Li G, Wan X. Attentive symmetric autoencoder for brain MRI segmentation. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part V. Cham: Springer. (2022) p. 203–213.
36. Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. (2010) 31:798–819. doi: 10.1002/hbm.20906
37. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. (2012) 62:782–90. doi: 10.1016/j.neuroimage.2011.09.015
38. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. (2001) 5:143–56. doi: 10.1016/S1361-8415(01)00036-6
39. Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, et al. The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv. (2021). doi: 10.48550/arXiv.2107.02314
40. Bjoern HM, Andras J, Stefan B, Jayashree KC, Keyvan F, Justin K, et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging. (2015) 34:1993–2024. doi: 10.1109/TMI.2014.2377694
41. Lloyd CT, Sorichetta A, Tatem AJ. High resolution global gridded data for use in population studies. Scient Data. (2017) 4:1–17. doi: 10.1038/sdata.2017.1
42. Haghighi F, Taher MRH, Gotway MB, Liang J. DiRA: discriminative, restorative, and adversarial learning for self-supervised medical image analysis. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). New Orleans, LA, USA: IEEE. (2022) p. 20792–802.
43. Fei Z, Fan M, Zhu L, Huang J, Wei X, Wei X. Masked auto-encoders meet generative adversarial networks and beyond. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). Vancouver, BC: IEEE (2023) p. 24449–59.
44. Jack Jr CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J Magnet Reson. (2008) 27:685–91. doi: 10.1002/jmri.21049
45. Zhou HY, Guo J, Zhang Y, Han X, Yu L, Wang L, et al. nnFormer: volumetric medical image segmentation via a 3D transformer. IEEE Trans Image Proc. (2023) 32:4036–45. doi: 10.1109/TIP.2023.3293771
46. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. (2002) 17:825–41. doi: 10.1006/nimg.2002.1132
47. MONAI Consortium. MONAI: Medical Open Network for AI. (2020). Available online at: https://github.com/Project-MONAI/MONAI (accessed April 24, 2023).
48. He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE International Conference on Computer Vision. Santiago: IEEE (2015) p. 1026–1034.
49. Taleb A, Loetzsch W, Danz N, Severin J, Gaertner T, Bergner B, et al. 3d self-supervised methods for medical imaging. In: Proceedings of the Conference on Advances in neural information processing systems (NeuraIPS). MIT Press (2020). p. 1815872.
Keywords: masked autoencoder, anatomical priors, transformer, brain tumor segmentation, magnetic resonance image, self-supervised learning
Citation: Wang K, Li Z, Wang H, Liu S, Pan M, Wang M, Wang S and Song Z (2023) Improving brain tumor segmentation with anatomical prior-informed pre-training. Front. Med. 10:1211800. doi: 10.3389/fmed.2023.1211800
Received: 25 April 2023; Accepted: 21 August 2023;
Published: 13 September 2023.
Edited by:
Diwei Zhou, Loughborough University, United KingdomReviewed by:
Zeju Li, University of Oxford, United KingdomSafa Elsheikh, Loughborough University, United Kingdom
Copyright © 2023 Wang, Li, Wang, Liu, Pan, Wang, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo Wang, c2h1b3dhbmdAZnVkYW4uZWR1LmNu; Zhijian Song, empzb25nQGZ1ZGFuLmVkdS5jbg==
†These authors have contributed equally to this work
 Kang Wang
Kang Wang Zeyang Li
Zeyang Li Haoran Wang
Haoran Wang Siyu Liu
Siyu Liu Mingyuan Pan
Mingyuan Pan Manning Wang
Manning Wang Shuo Wang
Shuo Wang Zhijian Song
Zhijian Song