- 1Laboratory Sciences and Services Division, International Centre for Diarrhoeal Disease Research, Bangladesh (icddr, b), Dhaka, Bangladesh
- 2Department of Biochemistry and Microbiology, North South University, Dhaka, Bangladesh
- 3Department of Infection Biology, London School of Hygiene and Tropical Medicine, London, United Kingdom
- 4Department of Infectious Disease Epidemiology, London School of Hygiene and Tropical Medicine, London, United Kingdom
Klebsiella aerogenes, a nosocomial pathogen, is increasingly associated with extensive drug resistance and virulence profiles. It is responsible for high morbidity and mortality. This report describes the first successfully treated case of community-acquired urinary tract infection (UTI) caused by Klebsiella aerogenes in an elderly housewife with Type-2 diabetes (T2D) from Dhaka, Bangladesh. The patient was empirically treated with intravenous ceftriaxone (500 mg/8 h). However, she did not respond to the treatment. The urine culture and sensitivity tests, coupled with bacterial whole-genome sequencing (WGS) and analysis, revealed the bacteria to be K. aerogenes which was extensively drug-resistant but was susceptible to carbapenems and polymyxins. Based on these findings, meropenem (500 mg/8 h) was administered to the patient, who then responded to the treatment and recovered successfully without having a relapse. This case raises awareness of the importance of diagnosis of not-so-common etiological agents, correct identification of the pathogens, and targeted antibiotic therapy. In conclusion, correctly identifying etiological agents of UTI using WGS approaches that are otherwise difficult to diagnose could help improve the identification of infectious agents and improve the management of infectious diseases.
1. Introduction
Community and hospital-acquired urinary tract infections (UTIs) are predicted to afflict approximately 405 million individuals around the globe. It has caused over 0.23 million deaths in 2019, leading to 5.2 million disability-adjusted life years (1). The treatment of UTIs usually consists of broad-spectrum antibiotics. Globally, the incidence of UTIs caused by multidrug-resistant pathogens is rising alarmingly. These infections can also be life-threatening in some cases (2). In 2022, a study of community-acquired urinary tract infections in Bangladesh revealed that Escherichia coli was the most prevalent bacterial pathogen, followed by Streptococcus Spp., Klebsiella Spp., Enterococcus Spp., Pseudomonas Spp., Staphylococcus Spp., Enterobacter Spp., Proteus Spp., Acinetobacter Spp., Staphylococcus saprophyticus, Staphylococcus aureus, Corynebacterium, and Serratia (3). A wide variety of bacterial infections may cause UTIs, reinforcing the need for the early identification of the etiological agent to properly manage UTIs (3–5).
There are abundant reports on the prevalence of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in UTIs. However, reports on Klebsiella aerogenes are limited (6–8). K. aerogenes is a gram-negative, facultative anaerobe, motile, rod-shaped, non-spore-forming member of the Enterobacteriaceae family. It was formerly Enterobacter aerogenes but was renamed Klebsiella aerogenes as it was more closely related to Klebsiella species than Enterobacter species (9, 10). K. aerogenes is a part of the ESKAPE group of pathogens (11) that significantly impact public health. K. aerogenes is ubiquitous, a habitant of the human gastrointestinal tract, and a prominent opportunistic pathogen causing nosocomial infections than community-acquired infections. It can cause UTIs, skin and soft tissue and respiratory infections, and bloodstream infections in immune-compromised or patients with damaged intestinal mucosa (12, 13). K. aerogenes has been associated with high mortality rates in patients from intensive care units (14).
Community-acquired urinary tract infections account for a large proportion of infectious diseases in the female population worldwide (15). The likelihood of UTIs in women increases with age; ~13% of women aged between 60 and 70 years old experience bacteriuria at least once in their lifetime (16). Urine glucose also promotes bacterial growth and colonization. Consequently, infections in diabetic patients may lead to extended hospital stays, recurrence, renal complications, bloodstream infection and septic shock (17). According to rough estimates, UTIs are fifteen times more likely to occur in diabetic patients, emphasizing the necessity for prompt identification and management of UTIs (18).
Herein, we describe the first successfully treated case of community-acquired UTI due to extensively drug-resistant (XDR) K. aerogenes in an elderly housewife suffering from Type-2 diabetes (T2D) in Bangladesh.
2. Case presentation
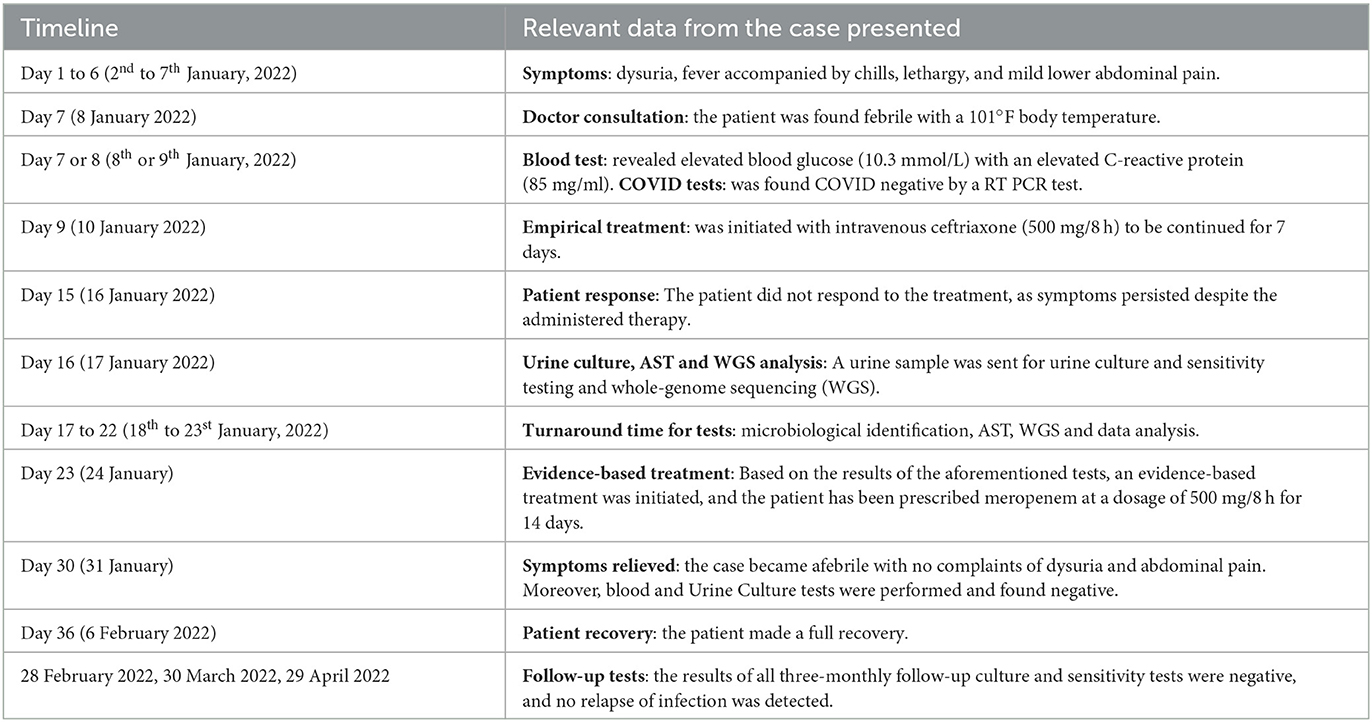
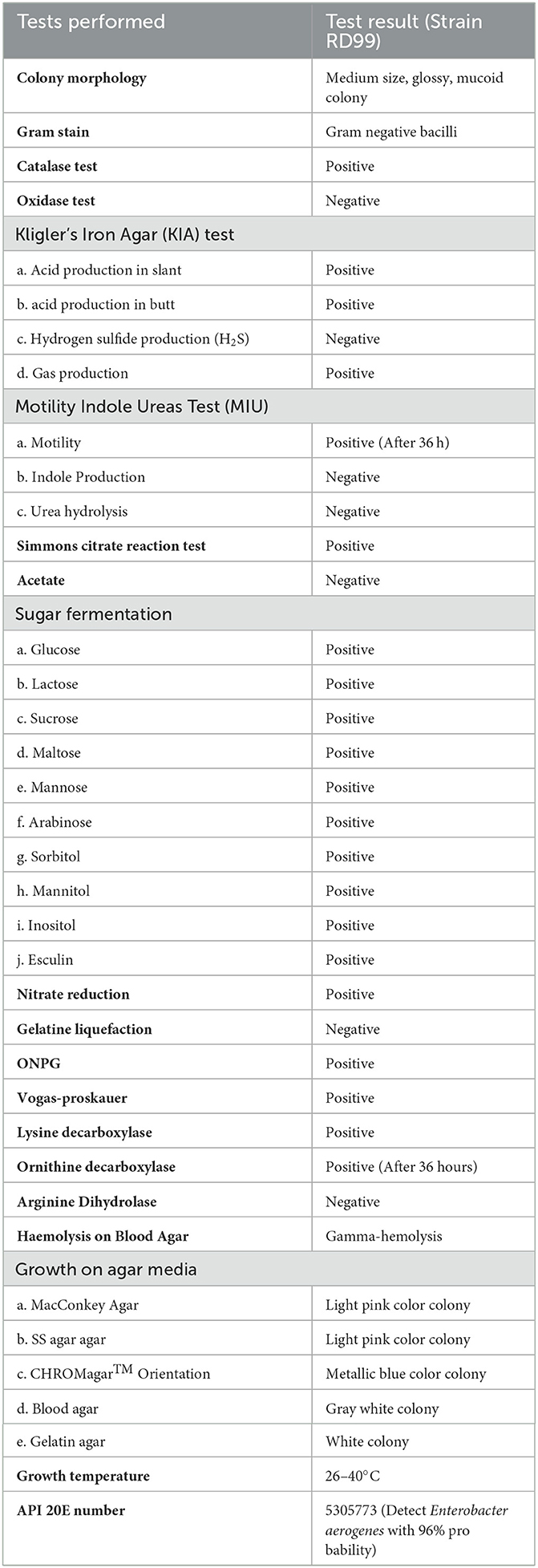
A 63-year-old female housewife from Narayanganj district of Dhaka division in Bangladesh attended the outpatient department of a local private hospital on January 2022. Her main complaints were dysuria, fever with chills, lethargy and mild lower abdominal pain for 6 days. She had a history of hypertension and T2D for 4 years. She was on medication for hypertension [Bisoprolol Hemifumarate (5 mg)] and diabetes [Linagliptin plus Metformin Hydrochloride (2.5 mg plus 500 mg)]. On consultation, the patient was found febrile with a 101°F body temperature, and her pulse, respiratory rate, blood pressure and oxygen saturation were detected to be 109 pulses/min, 20 breaths/min, 128/84 mmHg, and 97% on room air, respectively. She did not have a recent history of a hospital visit, travel, contact with the pet, or ingesting raw food. The patient provided a blood sample for a laboratory investigation. An initial hematological assessment of the blood specimen demonstrated that the patient's hemoglobin was 10.3 g/dl and the erythrocyte sedimentation rate was 50 mm/h. In particular, the number of leukocytes was 13,000 cells/mm3, with 81% neutrophils, 26% lymphocytes, 3% eosinophils, and 2% monocytes. The platelet counts, as well as other blood cell variables, were normal. Blood biochemistry analysis showed random blood glucose raised at 10.3 mmol/L and C-reactive protein elevated at 85 mg/ml with normal hepatorenal function parameters. A COVID-19 screening test was done per the hospital requirements and a negative result was obtained (19, 20). Preliminarily, it was suspected to be a case of UTI. Therefore, the patient was empirically treated with intravenous ceftriaxone (500 mg/8 h) for 7 days. However, she did not respond to the treatment and symptoms like increased frequency of painful and burning micturition, fever, and abdominal pain continued while she was admitted to the hospital (Table 1). Therefore, a clean catch midstream urine sample was obtained and sent to the laboratory for routine microscopic examination and microbiological investigation. The urine sample was turbid and revealed the presence of 20 to 30 pus cells per high-power field. The urine sample was inoculated onto 5% sheep blood agar, MacConkey's agar, and CHROMagar Orientation agar plates and incubated the plates at 37°C in an aerobic environment (21). After overnight incubation, a significant (>105 CFU/ml) growth of single-type colonies was observed on all culture plates. The bacteria produced 2–3mm, circular, raised, mucoid, pale pink color colonies on MacConkey agar, gray-white colonies on a blood agar plate with gamma hemolysis and metallic blue pigmented colonies on a CHROMagar Orientation plate. Based on the colony morphology on MacConkey and CHROMagar Orientation agar plates, we suspected it would be either Klebsiella or Enterobacter species. Gram staining revealed gram-negative bacilli. Further, biochemical characterization was carried out as described previously (22). The bacterium was finally identified as K. aerogenes based on its gram-negative morphology, motility, utilization of citrate, decarboxylation of Ornithine, reduction of nitrate and fermentation of glucose and lactose with the production of acid and gas (Table 2) (23). The strain was further tested with the API 20E test (BioMérieux, France), and it confirmed the strain as Enterobacter aerogenes with 96% certainty. Antibiotic susceptibility testing followed the Kirby-Bauer disc diffusion method using 21 antibiotic disks representing 11 classes (24–27). The K. aerogenes (RD99) strain was resistant to most antibiotic classes tested, including aminoglycosides, β-Lactams (penicillin), β lactams (cephalosporins), amphenicol, fluoroquinolones, folate drugs, tetracyclines, phosphonic acid, and glycycline. However, the strain was susceptible to carbapenems and polymyxins (Table 3).
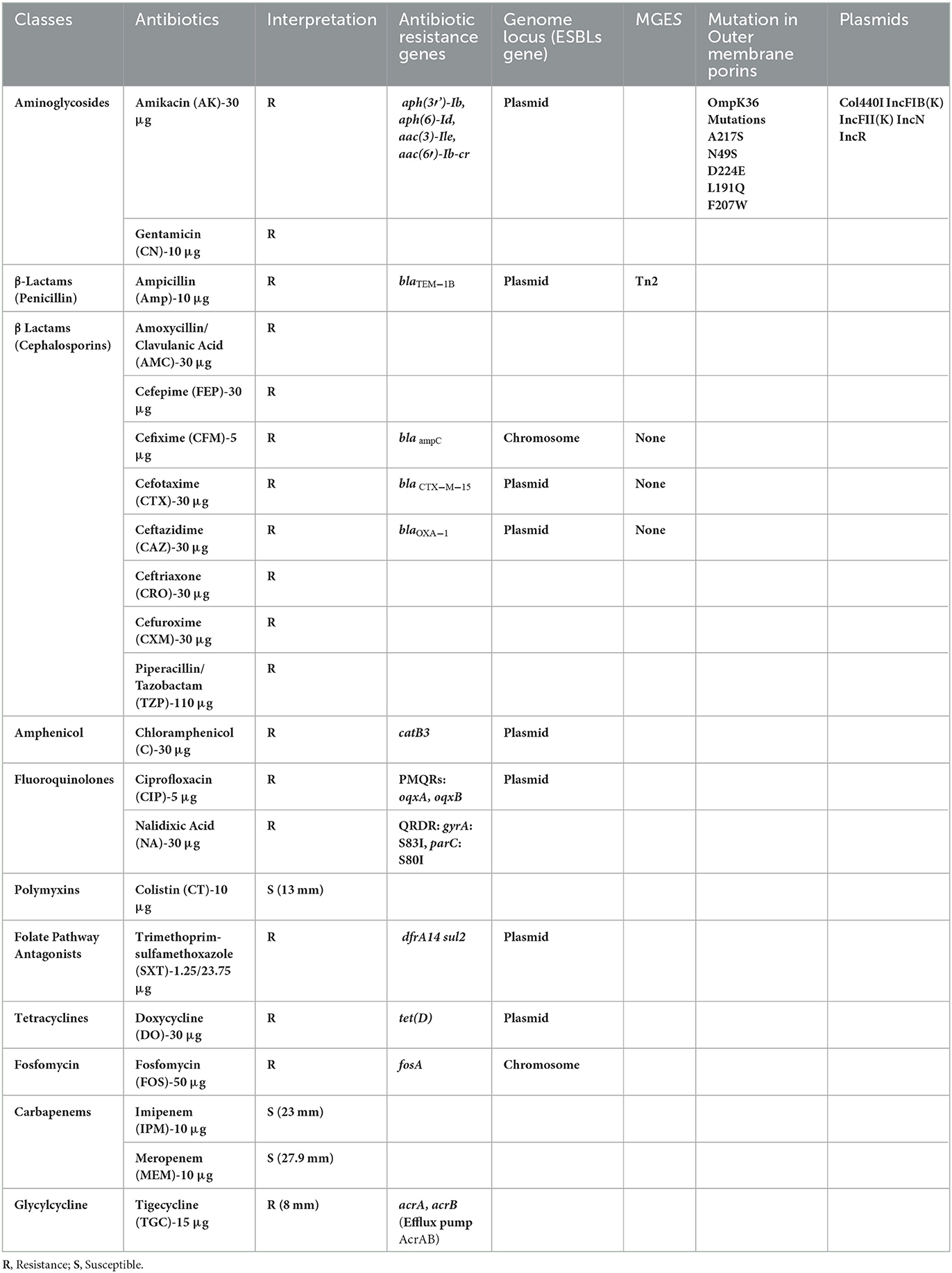
Table 3. Antibiotic susceptibility, resistome profile, mobilome and plasmid replicons in Klebseilla aerogenes (RD99) isolated from Dhaka, Bangladesh.
The K. aerogenes (RD99) strain was further subjected to bacterial whole-genome sequencing (WGS) using an Illumina NextSeq 500 platform at icddr,b using an established protocol (28, 29). The raw genome sequence data is publicly available at GenBank with accession number: JAQQRH000000000. The quality control, assembly of sequenced data into draft genomes and annotation were performed using established bioinformatic pipelines (28–30). The FastANI tool was used for taxonomy identification, and K. aerogenes was confirmed with a 98.81 Average Nucleotide Identity (ANI) value (31).
The in-silico analysis predicted sequence types (ST) and clonal complex using the PubMLST database (https://pubmlst.org/kaerogenes/), capsule polysaccharides using the Kaptive database (32), and plasmid replicons using Plasmid Finder 2.1 (33). The virulence genes were identified using Virulence Factor Database (VFDB) (34). The resistance genes were identified by ABRicate tool v1.0.1 (https://github.com/tseemann/abricate) (35) using Resfinder (36), and NCBI AMR Finder (37) database. Mobile genetic elements using Mobile Element Finder v1.0.3 (38), point mutations using PointFinder software (39), prophage using PHASTER (40), and integrons using IntegronFinder 2.0 (41). Default parameters were applied for all software unless otherwise mentioned. We aligned six K. aerogenes isolates (including RD99), 15 publicly available Klebsiella species, and 5 Enterobacter species genomes with K. pneumoniae ATCC 25955 as reference genome using Snippy v4.4.0 (42) and Gubbins v3.2 5 (43). A phylogenetic tree was constructed using RaxML v8.2.12, employing the Generalized Time Reversible substitution model and the GAMMA distribution for rate heterogeneity (44). The tree was visualized using iTOL (45).
The K. aerogenes strain (RD99) genome sequence is 5,490,860 bp with a GC content of 54.8%. It comprises 73 contigs, having 5,348 genes, of which 5,224 coding DNA sequences (CDSs) and 124 RNAs. In silico profiling revealed that RD99 is a K. aerogenes strain and belonged to ST4 and the CC1 clonal complex group. More than 60% (108/175) of the reference virulence genes were detected in the RD99 strain. It harbored the fimA-K operon, involved in adherence to human mucosal or epithelial surfaces, iutA, basG, ent, fep, and iro, which encodes aerobactin, acinetobactin, enterobactin and salmochelin siderophores, rcsAB, regulators of mucoid phenotype A. We also found SenB, an intrinsic gene encoding toxin (Table 4).
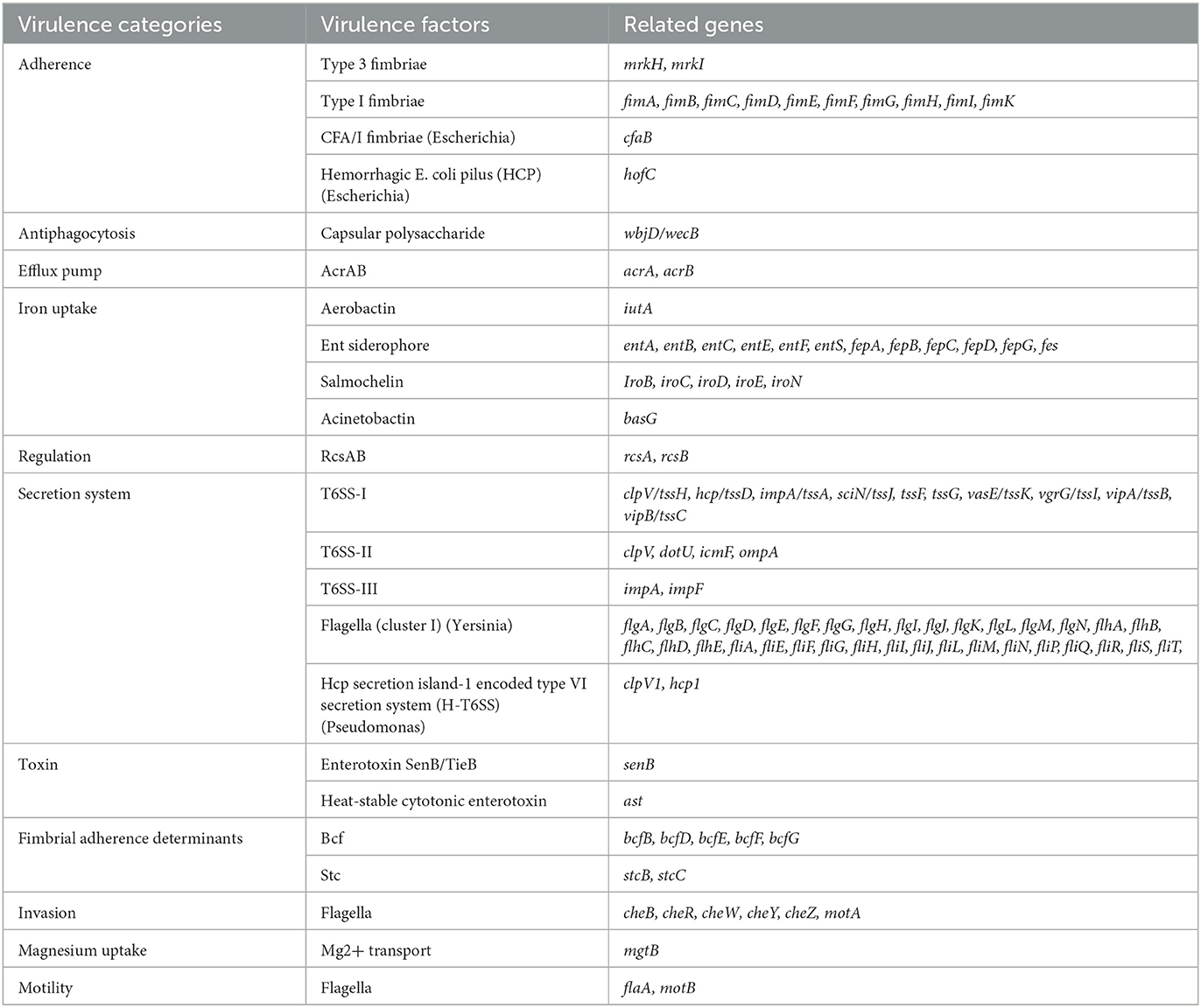
Table 4. Genome based virulence gene screening of Klebsiella aerogenes (strain RD99) isolated from Dhaka, Bangladesh.
A total of 10 prophage elements were detected, including 3 intact, 5 incomplete and 2 questionable phages and a Class 1 integron was identified. The strain RD99 harbored 17 antibiotic resistance genes that included aminoglycosides resistance genes aph(3′)-Ib, aph(6)-Id, aac(3)-Ile, aac(6′)-Ib-cr, β-Lactams resistance genes, blaTEM − 1B, blaCTX − M−15, blaOXA − 1, blaampC, folate pathway antagonists resistance genes, dfrA14, sul2, plasmid-mediated fluoroquinolones resistance genes, oqxA, oqxB, amphenicol resistance gene catB3, fosfomycin resistance gene fosA, and tetracyclines resistance gene tet(D). The efflux pumps AcrAB-associated genes acrA and acrB were also detected in this strain that would mediate tigecycline resistance. The genes blaampC and fosA were located on the chromosome. Other resistance genes were located on plasmids, including blaTEM − 1B, blaCTX − M−15, and blaOXA − 1. We also identified the TN2 transposon element linked with the blaTEM − 1B gene. In addition, we identified amino acid substitutions at codon positions S83I in gyrA and S80I in parC. We also found mutations in ompK36 gene. No mutations were detected in ompK35 and ompK37 genes. In agreement with the phenotypic results, we did not detect any genes conferring resistance to polymyxins and carbapenem antibiotics. We identified five plasmid replicons in RD99 genome, which were Col440I, IncFIB(K), IncFII(K), IncN, IncR (Table 2). The strain RD99 demonstrated OL102 and KL186 antigens.
We conveyed the results of the above analysis, particularly its phenotypic and genotypic features, to the physician. Based on this, the therapy course was changed to meropenem (500 mg/8 h) for 14 days. After 8 days of effective antibiotic treatment and appropriate management of diabetes, the patient became afebrile with no complaints of dysuria and abdominal pain. She was discharged from the hospital and advised to complete the course of antibiotics to avoid the recurrence of the infection. After 14th day the patient recovered completely and demonstrated a negative urine and blood culture. A monthly urine culture was advised for 3 months (one urine CS/month) to assess the efficacy of treatment and the recurrence of infection. All monthly follow-ups (for 3 months) of urine culture and sensitivity tests were negative, and the patient did not experience infection recurrence.
3. Discussion
K. aerogenes, previously known as Enterobacter aerogenes, is phylogenetically closer to Klebsiella species than Enterobacter species (10). K. aerogenes is often misidentified as K. pneumoniae due to delayed expression of certain phenotypic characteristics (46). This report identified a K. aerogenes strain RD99, with delayed motility and ornithine decarboxylase-positivity. Misidentification is also a concern while utilizing automated technologies like Vitek2 and Phoenix (46). Therefore, we carried out bacterial WGS for accurate taxonomic identification. The resulting data revealed that RD99 is 98% similar to the K. aerogenes KCTC 2190 type strain. In a phylogenetic analysis, all K. aerogenes genomes analyzed belonged to the same clade. Our analysis confirmed that the Enterobacter aerogenes genomes were more closely related to Klebsiella species than Enterobacter species (Figure 1). Based on this study, we propose that the WGS and 16S rRNA gene sequencing techniques (21) may be utilized as an alternative to phenotypic techniques for identifying K. aerogenes if confusion arises while employing sophisticated bacteriological identification methods such as API 20E, Vitek2, and Phoenix.
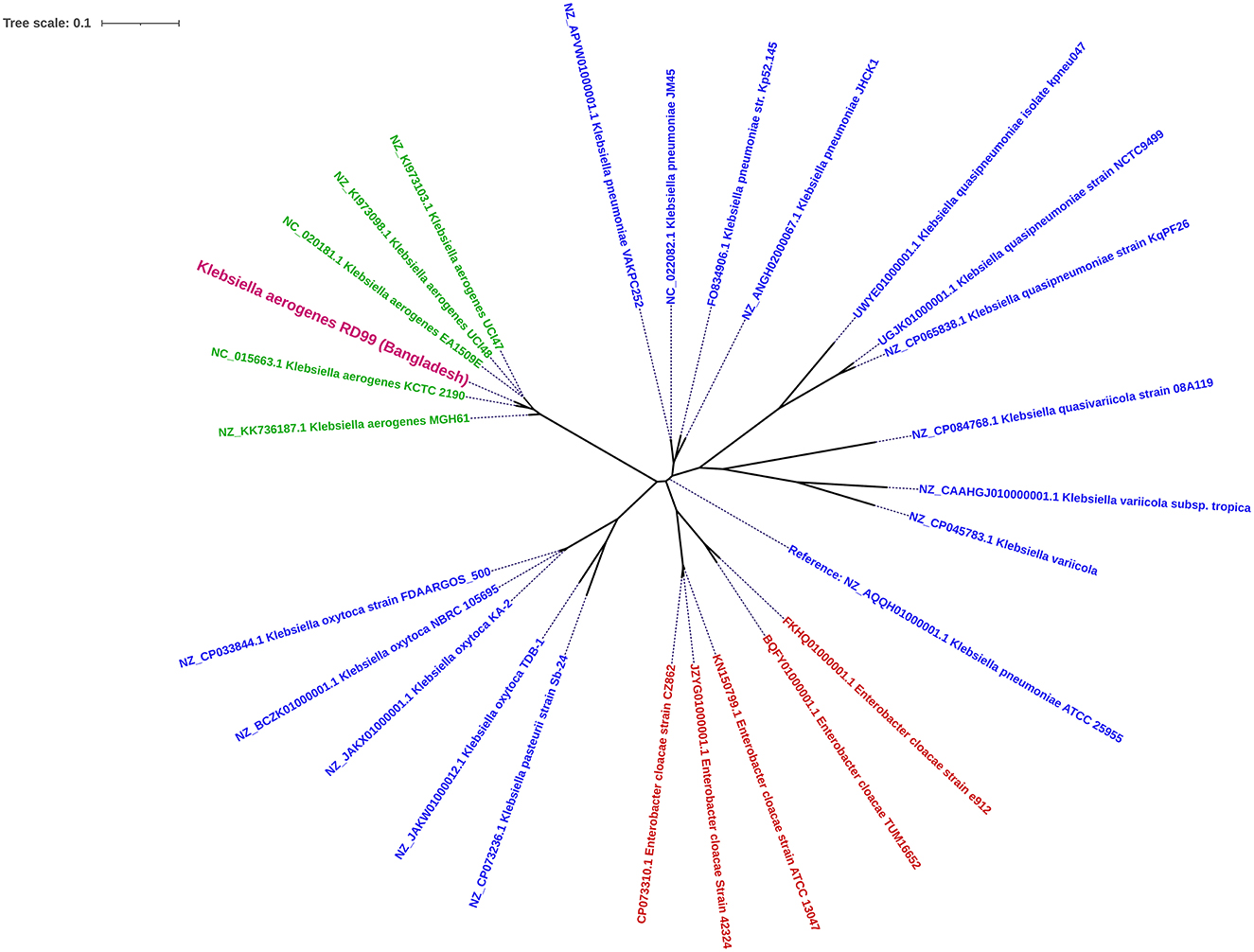
Figure 1. Core-genome SNP-based maximum likelihood phylogenetic tree illustrating the homology between the genome of Klebsiella aerogenes RD99 (Violet Color) and other reported genome sequences of Klebsiella aerogenes retrieved from NCBI. klebsiella aerogenes (Green color) and Enterobacter spp (Red color).
During the consultation, the subject denied any recent history of hospital stays; therefore, she most likely had a community-acquired UTI. However, community-acquired Klebsiella aerogenes infection cases are rare, and most reported cases were associated with hospital-acquired infections (12). The patient professed that she was involved with household chores and may have come into contact with environmental sources like contaminated water. K. aerogenes is ubiquitous in the environment and human gastrointestinal tract (47). Moreover, a study on the bacteriological quality of drinking water samples across Bangladesh identified K. aerogenes from tube wells, a common drinking water source in Bangladesh (48). In addition, poor sanitation practices, such as open defecation, might contribute to spreading germs, resulting in UTIs. Diabetes mellitus has been recognized as a predisposing factor for UTIs. Particularly, those women suffering from T2D are more susceptible to UTIs (49–51). In the case presented here, T2D could be a reason for the serious complications and long recovery from UTI.
The WGS analysis of strain RD99 revealed a similar virulence pattern as reported for earlier K. aerogenes (52). The fim gene cluster was identified in our strain, which produces Type 1 fimbriae and the adhesive subunit FimH. These are key virulence factors responsible for urinary tract infection and biofilm formation (53). The identified Type 3 fimbriae would be involved in adhesion to human tissue structures and biofilm formation, particularly in immunocompromised catheterised patients (54). Siderophores such as aerobactin, acinetobactin, enterobactin, and salmochelin were identified as critical for bacterial virulence (52). Capsular and mucopolysaccharides also play a decisive role in the pathogenesis of K. aerogenes, promoting successful establishment of infection by resisting phagocytosis (55). This study also identified the T6SS gene cluster that produces a functioning system that manipulates host cells during pathogenesis and kills competing bacteria (56). The three intact bacteriophages identified in our strain are Klebsiella phage ST16-OXA48phi5.4, Enterobacteria phage mEp237 and Escherichia phage phiV10. Since most Escherichia phage phiV10 is commonly found in E. coli O157:H7 strain (57), its presence in K. aerogenes may indicate a horizontal gene transfer (HGT) event.
It is difficult for clinicians to select the appropriate antimicrobial therapy to treat infections in older patients with co-morbidities like T2D, and the empirical therapy might not be fully effective (16). Klebsiella infections develop rapidly, causing multiple organ failures and death. The selection of effective antibiotics with the right dose and course is critical for disease management in older patients to minimize the adverse effects of drugs. Thus, we have evaluated the phenotypic and genotypic antimicrobial resistance features of strain RD99 to initiate antimicrobial therapy. The strain RD99 was classified as XDR using established criteria (58). We observed that the strain RD99 was resistant to most antibiotics with corresponding antibiotic resistance genes, including aminoglycosides, β lactams, amphenicol, fluoroquinolones, and folate pathway antagonists, tetracyclines, phosphonic and glycylcycline antibiotics. However, it was sensitive to carbapenems and polymyxin with no corresponding resistance genes.
Moreover, an inducible chromosomal blaAmpC ß-lactamase gene was detected in the RD99 isolate. The high levels of resistance to cephalosporins, even in the presence of a ß-lactamase inhibitor (59), could be due to the blaAmpC ß-lactamase gene. Most antibiotic resistance genes found in strain RD99, including blaTEM − 1B, blaCTX − M−15, blaOXA − 1 were found on plasmids, except blaampC and fosA genes located on the chromosome. The gene acrAB encoding multidrug efflux pump is associated with reduced susceptibility to tigecycline (60). The antibiotic sensitivity tests and antibiotic resistance determinants suggested that the strain RD99 was sensitive to carbapenems.
Studies have shown that the presence of the ompK36 mutation in Klebsiella aerogenes is associated with increased resistance to carbapenems, a critical class of antibiotics used to treat serious infections caused by gram-negative bacteria (52). However, in the present study, drug sensitivity test revealed that K. aerogenes isolates were all sensitive to carbapenem antibiotics. We did not choose imipenem for an anti-infection treatment because the size of its inhibition zone was just close to threshold, this could be due to ompK36 mutation observed. However, meropenem produced a larger inhibition zone (Table 2). Therefore, meropenem was selected for the management of UTI of the case presented.
In conclusion, the identification of K. aerogenes RD99 strain, which had a delayed motility activity and ornithine decarboxylase-positivity, was confirmed through WGS, revealing that this technique may be used as an alternative to phenotypic methods. The patient presented with a community-acquired urinary tract infection, possibly due to poor sanitation practices, contaminated water, and medical conditions such as T2D. Clinicians should initiate an appropriate antimicrobial therapy to treat such infections, as such pathogens are associated with extensive AMR and virulence profiles. Moreover, the strain RD99 was classified as XDR and was resistant to almost all classes of antibiotics. To our knowledge, this is the first in-depth whole-genome-based characterization of K. aerogenes isolated from a clinical urine sample in Bangladesh.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, JAQQRH000000000.
Ethics statement
The patient provided written informed consent for the publishing of this case report.
Author contributions
RM conceptualized the study, performed all in vitro tests, carried out whole genome sequencing and drafted the manuscript. AH contributed to discussions and manuscript writing. BB was responsible for obtaining the patient's consent and case history. BB and RM performed bioinformatics analysis, interpreted the data, and generated tables and figure. JP, SC, TC, and DM participated in the discussions and reviewed the manuscript. DM supervised the research. The final paper has been reviewed and approved by all authors.
Funding
The work is funded through a Royal Society International Collaboration Award (ref. ICA\R1\191309).
Acknowledgments
icddr,b is grateful to the governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support for its operations and research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zeng Z, Zhan J, Zhang K, Chen H, Cheng S. Global, regional, and national burden of urinary tract infections from 1990 to 2019: an analysis of the global burden of disease study 2019. World J Urol. (2022) 40:755–63. doi: 10.1007/s00345-021-03913-0
2. Mazzariol A, Bazaj A, Cornaglia G. Multi-drug-resistant gram-negative bacteria causing urinary tract infections: a review. J Chemother. (2017) 29:2–9. doi: 10.1080/1120009X.2017.1380395
3. Islam MA, Islam MR, Khan R, Amin MB, Rahman M, Hossain MI, et al. Prevalence, etiology and antibiotic resistance patterns of community-acquired urinary tract infections in Dhaka, Bangladesh. PLoS ONE. (2022) 17:e0274423. doi: 10.1371/journal.pone.0274423
4. Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis. (2004) 38:1150–8. doi: 10.1086/383029
5. Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. (2010) 7:653–60. doi: 10.1038/nrurol.2010.190
6. Pulcrano G, Pignanelli S, Vollaro A, Esposito M, Iula VD, Roscetto E, et al. Isolation of Enterobacter aerogenes carrying blaTEM-1 and blaKPC-3 genes recovered from a hospital intensive care unit. Apmis. (2016) 124:516–21. doi: 10.1111/apm.12528
7. Santo Pereira R, Dias VC, Ferreira-Machado AB, Resende JA, Bastos AN, Bastos LQA, et al. Physiological and molecular characteristics of carbapenem resistance in Klebsiella pneumoniae and Enterobacter aerogenes. J Infect Dev Ctries. (2016) 10:592–9. doi: 10.3855/jidc.6821
8. Gu H, Cai Q, Dai X, Wang H, Xu W, Cao X, et al. A case report of Klebsiella aerogenes-caused lumbar spine infection identified by metagenome next-generation sequencing. BMC Infect Dis. (2022) 22:616. doi: 10.1186/s12879-022-07583-0
9. Tindall B, Sutton G, Garrity G. Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int J Syst Evol. (2017) 67:502–4. doi: 10.1099/ijsem.0.001572
10. Wesevich A, Sutton G, Ruffin F, Park LP, Fouts DE, Fowler Jr VG, et al. Newly named Klebsiella aerogenes (formerly Enterobacter aerogenes) is associated with poor clinical outcomes relative to other Enterobacter species in patients with bloodstream infection. J Clin Microbiol. (2020) 58:e00582–20. doi: 10.1128/JCM.00582-20
11. Mann A, Malik S, Rana J, Nehra K. Whole genome sequencing data of Klebsiella aerogenes isolated from agricultural soil of Haryana, India. Data in Brief. (2021) 38:107311. doi: 10.1016/j.dib.2021.107311
12. Jha P, Kim C-M, Kim D-M, Chung J-H, Yoon N-R, Jha B, et al. Transmission of Enterobacter aerogenes septicemia in healthcare workers. Springerplus. (2016) 5:1–4. doi: 10.1186/s40064-016-3011-x
13. Li E, Wei X, Ma Y, Yin Z, Li H, Lin W, et al. Isolation and characterisation of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci Rep. (2016) 6:1–11. doi: 10.1038/srep28338
14. Davin-Regli A, Pagès J-M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. (2015) 6:392. doi: 10.3389/fmicb.2015.00392
15. Søraas A, Sundsfjord A, Sandven I, Brunborg C, Jenum PA. Risk factors for community-acquired urinary tract infections caused by ESBL-producing enterobacteriaceae–a case–control study in a low prevalence country. PLoS ONE. (2013) 8:e69581. doi: 10.1371/journal.pone.0069581
16. Mody L, Juthani-Mehta M. Urinary tract infections in older women: a clinical review. JAMA. (2014) 311:844–54. doi: 10.1001/jama.2014.303
17. Nitzan O, Elias M, Chazan B, Saliba W. Urinary tract infections in patients with type 2 diabetes mellitus: review of prevalence, diagnosis, and management. Diabetes Metab Syndr Obes. (2015) 8:129–36. doi: 10.2147/DMSO.S51792
18. Jung C, Brubaker L. The etiology and management of recurrent urinary tract infections in postmenopausal women. Climacteric. (2019) 22:242–9. doi: 10.1080/13697137.2018.1551871
19. Das P, Satter SM, Ross AG, Abdullah Z, Nazneen A, Sultana R, et al. A case series describing the recurrence of COVID-19 in patients who recovered from initial illness in Bangladesh. Trop Med Infect Dis. (2021) 6:41. doi: 10.3390/tropicalmed6020041
20. Mazumder R, Abdullah A, Hossain ME, Rahman MM, Bin Manjur OH, Rahman M, et al. Genome sequencing identified a SARS-CoV-2 lineage B. 1.1. 7 strain with a high number of mutations from dhaka. Bangladesh Microbiology Resource Announcements. (2021) 10:e00345–00321. doi: 10.1128/MRA.00345-21
21. Mazumder R, Sadique T, Sen D, Mozumder P, Rahman T, Chowdhury A, et al. Agricultural injury-associated Chromobacterium violaceum infection in a Bangladeshi Farmer. Am J Trop Med Hyg. (2020) 103:1039–42. doi: 10.4269/ajtmh.20-0312
22. Mazumder R, Hussain A, Phelan JE, Campino S, Haider S, Mahmud A, et al. Non-lactose fermenting Escherichia coli: following in the footsteps of lactose fermenting E. coli high-risk clones. Front Microbiol. (2022) 13:1027494. doi: 10.3389/fmicb.2022.1027494
23. Davin-Regli A, Lavigne J-P, Pagès J-M. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. (2019) 32:e00002–19. doi: 10.1128/CMR.00002-19
24. Weinstein MP. Performance Standards For Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute (2019).
25. Jones RN, Ferraro MJ, Reller LB, Schreckenberger PC, Swenson JM, Sader HS. Multicenter studies of tigecycline disk diffusion susceptibility results for Acinetobacter spp. J Clin Microbiol. (2007) 45:227–30. doi: 10.1128/JCM.01588-06
26. Galani I, Kontopidou F, Souli M, Rekatsina P-D, Koratzanis E, Deliolanis J, et al. Colistin susceptibility testing by Etest and disk diffusion methods. Int J Antimicrob Agents. (2008) 31:434–9. doi: 10.1016/j.ijantimicag.2008.01.011
27. Babaei S, Haeili M. Evaluating the performance characteristics of different antimicrobial susceptibility testing methodologies for testing susceptibility of gram-negative bacteria to tigecycline. BMC Infect Dis. (2021) 21:1–8. doi: 10.1186/s12879-021-06338-7
28. Mazumder R, Hussain A, Abdullah A, Islam MN, Sadique MT, Muniruzzaman S, et al. International high-risk clones among extended-spectrum β-lactamase–producing Escherichia coli in Dhaka, Bangladesh. Front Microbiol. (2021) 12:736464. doi: 10.3389/fmicb.2021.736464
29. Mazumder R, Abdullah A, Hussain A, Ahmed D, Mondal D. Draft genome sequence of Chromobacterium violaceum RDN09, isolated from a patient with a wound infection in Bangladesh. Microbiol Resour Announc. (2020) 9:e00957–e00920. doi: 10.1128/MRA.00957-20
30. Baddam R, Sarker N, Ahmed D, Mazumder R, Abdullah A, Morshed R, et al. Genome dynamics of Vibrio cholerae isolates linked to seasonal outbreaks of cholera in Dhaka, Bangladesh. MBio. (2020) 11:e03339–e03319. doi: 10.1128/mBio.03339-19
31. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. (2018) 9:5114. doi: 10.1038/s41467-018-07641-9
32. Wick RR, Heinz E, Holt KE, Wyres KL. Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J Clin Microbiol. (2018) 56:e00197–e00118. doi: 10.1128/JCM.00197-18
33. Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
34. Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. (2005) 33:D325–8. doi: 10.1093/nar/gki008
35. Seemann T. ABRicate: mass screening of contigs for antimicrobial virulence genes. Department of Microbiology Immunology, The University of Melbourne, Melbourne, Australia (2018). Available online at: https://github.com/tseemann/abricate (accessed 28 February, 2019).
36. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
37. Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother. (2019) 63:e00483–e00419. doi: 10.1128/AAC.00483-19
38. Johansson MH, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, Petersen TN. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J Antimicrob Chemoth. (2021) 76:101–9. doi: 10.1093/jac/dkaa390
39. Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemoth. (2017) 72:2764–8. doi: 10.1093/jac/dkx217
40. Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. (2016) 44:W16–21. doi: 10.1093/nar/gkw387
41. Cury J, Jové T, Touchon M, Néron B, Rocha EP. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. (2016) 44:4539–50. doi: 10.1093/nar/gkw319
43. Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. (2015) 43:e15. doi: 10.1093/nar/gku1196
44. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
45. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. (2016) 44:W242–5. doi: 10.1093/nar/gkw290
46. Claeys G, De Baere T, Wauters G, Vandecandelaere P, Verschraegen G, Muylaert A, et al. Extended-Spectrum beta-lactamase (ESBL) producing Enterobacter aerogenes phenotypically misidentified as Klebsiella pneumoniae or K. terrigena. BMC Microbiol. (2004) 4:49. doi: 10.1186/1471-2180-4-49
47. Pan F, Xu Q, Zhang H. Emergence of NDM-5 producing carbapenem-resistant Klebsiella aerogenes in a pediatric hospital in Shanghai, China. Front Public Health. (2021) 9:621527. doi: 10.3389/fpubh.2021.621527
48. Parvez A, Liza S, Marzan M, Ahmed A, Rahman MS. Bacteriological quality of drinking water samples across Bangladesh. Arch Clin Microbiol. (2016) 7:9.
49. Soh PN, Vidal F, Huyghe E, Gourdy P, Halimi J, Bouhanick B. Urinary and genital infections in patients with diabetes: how to diagnose and how to treat. Diabetes Metab. (2016) 42:16–24. doi: 10.1016/j.diabet.2015.07.002
50. Khalid F, Bushra R, Ghayas S, Mushtaque M. Case report: urinary tract infection in a diabetic postmenopausal woman with multiple episodes of recurrence: an antimicrobial susceptibility dispute. Clin Med Insights Case Rep. (2022) 15:11795476221112819. doi: 10.1177/11795476221112819
51. Worby CJ, Schreiber Iv HL, Straub TJ, Van Dijk LR, Bronson RA, Olson BS, et al. Longitudinal multi-omics analyses link gut microbiome dysbiosis with recurrent urinary tract infections in women. Nat Microbiol. (2022) 7:630–9. doi: 10.1038/s41564-022-01107-x
52. Passarelli-Araujo H, Palmeiro JK, Moharana KC, Pedrosa-Silva F, Dalla-Costa LM, Venancio TM. Genomic analysis unveils important aspects of population structure, virulence, and antimicrobial resistance in Klebsiella aerogenes. FEBS J. (2019) 286:3797–810. doi: 10.1111/febs.15005
53. Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. (2009) 77:5016–24. doi: 10.1128/IAI.00585-09
54. El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathologie Biologie. (2013) 61:209–16. doi: 10.1016/j.patbio.2012.10.004
55. Wyres K, Wick R, Gorrie C, Jenney A, Follador R, Thomson N, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. (2016) 2:e000102. doi: 10.1099/mgen.0.000102
56. Enany S, Zakeer S, Diab AA, Bakry U, Sayed AA. Whole genome sequencing of Klebsiella pneumoniae clinical isolates sequence type 627 isolated from Egyptian patients. PLoS ONE. (2022) 17:e0265884. doi: 10.1371/journal.pone.0265884
57. Zhang D, Coronel-Aguilera CP, Romero PL, Perry L, Minocha U, Rosenfield C, et al. The use of a novel NanoLuc-based reporter phage for the detection of Escherichia coli O157: H7. Sci Rep. (2016) 6:33235. doi: 10.1038/srep33235
58. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas M, Giske C, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
Keywords: Klebsiella aerogenes, community-acquired urinary tract infection, extensive drug resistance (XDR), ESBLs (extended spectrum β-lactamases), type 2 diabetes mellitus, Bangladesh
Citation: Mazumder R, Hussain A, Bhadra B, Phelan J, Campino S, Clark TG and Mondal D (2023) Case report: A successfully treated case of community-acquired urinary tract infection due to Klebsiella aerogenes in Bangladesh. Front. Med. 10:1206756. doi: 10.3389/fmed.2023.1206756
Received: 16 April 2023; Accepted: 05 June 2023;
Published: 26 June 2023.
Edited by:
Mohsin Khurshid, Government College University, Faisalabad, PakistanReviewed by:
Junping Peng, Institute of Pathogen Biology (CAMS), ChinaTimothy Kudinha, Charles Sturt University, Australia
Copyright © 2023 Mazumder, Hussain, Bhadra, Phelan, Campino, Clark and Mondal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dinesh Mondal, ZGluNjNkQGljZGRyYi5vcmc=
 Razib Mazumder
Razib Mazumder Arif Hussain
Arif Hussain Bithika Bhadra
Bithika Bhadra Jody Phelan
Jody Phelan Susana Campino
Susana Campino Taane G. Clark
Taane G. Clark Dinesh Mondal
Dinesh Mondal