- 1Department of Hematology, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, China
- 2Shanghai Yuanqi Biomedical Technology Co., Ltd., Shanghai, China
- 3National Clinical Research Center for Hematologic Diseases, The First Affiliated Hospital of Soochow University, Suzhou, China
Objective: Diagnosis classification and risk stratification are crucial in the prognosis prediction and treatment selection of acute myeloid leukemia (AML). Here, we used a database of 536 AML patients to compare the 4th and 5th WHO classifications and the 2017 and 2022 versions of ELN guidance.
Methods: AML patients were classified according to the 4th and 5th WHO classifications, as well as the 2017 and 2022 versions of the European LeukemiaNet (ELN) guidance. Kaplan–Meier curves with log-rank tests were used for survival analysis.
Results: The biggest change was that 25 (5.2%), 8 (1.6%), and 1 (0.2%) patients in the AML, not otherwise specified (NOS) group according to the 4th WHO classification, were re-classified into the AML-MR (myelodysplasia-related), KMT2A rearrangement, and NUP98 rearrangement subgroups based on the 5th WHO classification. Referring to the ELN guidance, 16 patients in the favorable group, six patients in the adverse group, and 13 patients in the intermediate group based on the 2017 ELN guidance were re-classified to the intermediate and adverse groups based on the 2022 ELN guidance. Regrettably, the Kaplan–Meier curves showed that the survival of intermediate and adverse groups could not be distinguished well according to either the 2017 or 2022 ELN guidance. To this end, we constructed a risk model for Chinese AML patients, in which the clinical information (age and gender), gene mutations (NPM1, RUNX1, SH2B3, and TP53), and fusions (CBFB::MYH11 and RUNX1::RUNX1T1) were included, and our model could help divide the patients into favorable, intermediate, and adverse groups.
Conclusion: These results affirmed the clinical value of both WHO and ELN, but a more suitable prognosis model should be established in Chinese cohorts, such as the models we proposed.
Introduction
Acute myeloid leukemia (AML) represents the most common type of acute leukemia in adults worldwide (1). Accumulated evidence has revealed that genetic abnormalities, such as gene mutations and fusions play crucial roles in the pathogenesis of AML, causing hyperproliferation and maturational arrest of myeloid precursor cells (2, 3). The 4th revision of the World Health Organization (WHO) classification of hematologic malignancies divides AML into 11 subgroups based on genetic abnormalities (4). Recently, the 5th revision of the WHO classification was published (5), and several alterations were made based on AML, such as the subgroups of KMT2A/MECOM/NUP98 rearrangement and CEBPA mutation, as well as the AML, myelodysplasia-related (MR) subgroup.
In the present study, we used a dataset of 536 consecutive subjects with AML initially diagnosed using the 2016 WHO criteria to compare how these subjects would be classified using the 2022 WHO criteria. In addition, we compared the prognostic classification of AML according to the 2017 and 2022 versions of the European Leukemia Net (ELN) guidelines (6, 7) and established a new prognostic model including clinical information, gene mutations, and fusions for Chinese AML.
Patients and methods
Patients
A total of 536 patients with primary AML were included in this study between September 2013 and February 2021 from the First Affiliated Hospital of Xi'an Jiaotong University. The patients were diagnosed with AML according to the 4th or 5th version of the WHO guidance (4, 5) and followed up until July 2022. General clinical characteristics [age, sex, the proportion of blasts in bone marrow (BM) samples, karyotype, white blood cell (WBC) counts, red blood cell (RBC) counts, hemoglobin, blood platelet counts (PLT), activated partial thromboplastin time (APTT), prothrombin time, thrombin time, fibrinogen, fibrinogen degradation product, D-dimer, and survival time], structural variations [5/5q deletion (-5/-5q),−7/-7q, inv (involvement) (16)(p13q22), t (translocation) (16;16)(p13;q22), t(8;21)(q22;q22), t(9;22)(q34.1;q11.2), inv(3)(p21p26), and t(3;3)(q21;q26)], and fusion genes (CBFB-MYH11, BCR-ABL1, KMT2A-PARTNER, TLS-ERG, PML-RARA, RUNX1-RUNXT1, NUP98-PARTNER, DEK-NUP214, FIPIL1-PDGFRα, AML1-ETO, NPM1-RARα, PLZF-RARα, and SET-NUP214) were collected and shown as Table 1. After manual evaluation of the karyotype, fusion, and mutation, patients were assigned to respective 4th/5th WHO and ELN 2017/2022 risk groups. This study was approved by the First Affiliated Hospital of Xi'an Jiaotong University. Informed consent forms were signed by each patient.
Targeted sequencing
Genomic DNA (gDNA) was extracted from the formalin-fixed paraffin-embedded (FFPE) or fresh BM samples, followed by the targeted sequencing of 38 genes (ASXL1, BCOR, BCORL1, CBL, CEBPA, CSF3R, DNMT3A, ETV6, EV11, MECOM, EZH2, FLT3, FLT3-ITD, FLT3-TKD, GATA2, HOX11, IDH1, IDH2, JAK2, KIT, KMT2A, KMT2A-PTD, KRAS, MPL, MYC, NF1, NPM1, NRAS, NTRK3, RUNX1, SF3B1, SH2B3, SRSF2, TET2, TP53, TPMT, U2AF1, and ZRSR2) on the Novaseq (Illumina, USA) sequencing platform. The original sequencing was aligned with the human reference genome GRCh37. Single nucleotide variations (SNVs) and insertion and deletion (Indels) were screened by Shanghai Rightongene Biotechnology Co., Ltd. (Shanghai, China) based on the filtering conditions: (1) SNVs or Indels with a mutation allele frequency (MAF) of ≥0.001 in databases of 1,000 genomes project, 1,000 genome East Asian, ExAC all, or ExAC East Asian were removed; (2) SNVs or Indels with a variant allele frequency (VAF) of ≥ 1% were retained; (3) dbSNP (v147) sites present in COSMIC database were retained; and (4) SNPs or Indels including stopgain, stoploss, frameshift, non-frameshift, and splicing sites were retained.
Establishment of a prognostic risk scoring system for Chinese AML
To establish the training and test cohorts, 536 samples were randomly divided into two groups, the training/validation set (70%) and the test set (30%), respectively. The training set was subjected to 10-fold cross-validation to account for variability and provide risk estimates. The mutated genes (ASXL1, BCOR, BCORL1, CBL, CEBPA, CSF3R, DNMT3A, ETV6, EV11, MECOM, EZH2, FLT3, FLT3-ITD, FLT3-TKD, GATA2, HOX11, IDH1, IDH2, JAK2, KIT, KMT2A, KMT2A-PTD, KRAS, MPL, MYC, NF1, NPM1, NRAS, NTRK3, RUNX1, SF3B1, SH2B3, SRSF2, TET2, TP53, TPMT, U2AF1, and ZRSR2), rearrangements (AML1-ETO, BCR-ABL1, CBFB-MYH11, DEK-NUP214, KMT2A rearrangement, MECOM rearrangement, NPM1-RARα, NUP98 rearrangement, PLZF-RARα, RUNX1-RUNXT1, and SET-NUP214), and clinicopathologic features (age, sex, proportion of BM blasts, hemoglobin, WBC, and PLT) were included in the models. Conventional Cox regression was used to train the models for assessing survival with the selected variables by Lasso using the “Glmnet” package. The optimal cutoff values for risk score were determined using the X-tile software (Version 3.6.1, Yale University, USA) (8); thereafter, the patients were divided into favorable, intermediate, and adverse groups.
Statistical analysis
The statistical analysis was performed using GraphPad Prism software v.6 (GraphPad, San Diego, CA, United States). Kaplan–Meier curves with log-rank tests were used to analyze the OS and PFS of AML patients in different groups. A P-value of < 0.05 was considered significant.
Results
AML classification according to the 4th and 5th WHO classification
The 5th WHO classification made some changes in the diagnostic criteria of AML, including (1) persons with KMT2A, MECOM, and NUP98 rearrangements and NPM1 mutation were diagnosed with AML regardless of the percentage of blasts; (2) the definition of AML with CEBPA mutation was changed to include both biallelic (biCEBPA) and single mutation of CEBPA located in the basic leucine zipper (bZIP) region (smbZIP-CEBPA); (3) the previous classification of AML with mutated RUNX1 was abolished; (4) the classification of AML with myelodysplasia-related changes (AML-MRC) has been changed to AML-MR, and the mutations in ASXL1, BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 genes were newly defined as the defining cytogenetic abnormalities; (5) the classification of AML with other defined genetic alterations was newly added; and (6) the classification of AML, not otherwise specified (NOS) was replaced with AML, defined by differentiation.
A total of 485 and 487 of the 536 patients were classified according to the 4th and 5th WHO classifications, respectively, as the mutation location of CEBPA or the VAF of FLT3-ITD is not available. Six subgroups, AML with RUNX1::RUNX1T1 (n = 70), AML with CBFB::MYH11 (n = 29), AML with DEK::NUP214 (n = 5), AML with BCR::ABL1 (n = 4), AML with GATA2::MECOM (n = 2), and AML with NPM1 (n = 89), remained unchanged according to the 5th WHO, although the subgroup of AML with GATA2::MECOM was changed to AML with MECOM rearrangement. The group that changed the most was the AML, NOS (not otherwise specified) subgroup (n = 212); in detail, 8 patients were divided into the AML with KMT2A-rearrangement subgroup, 25 patients were divided into the AML-MR subgroup, and one case was divided into the AML with NUP98-rearrangement subgroup based on the 5th WHO classification (Figure 1A, Table 2).
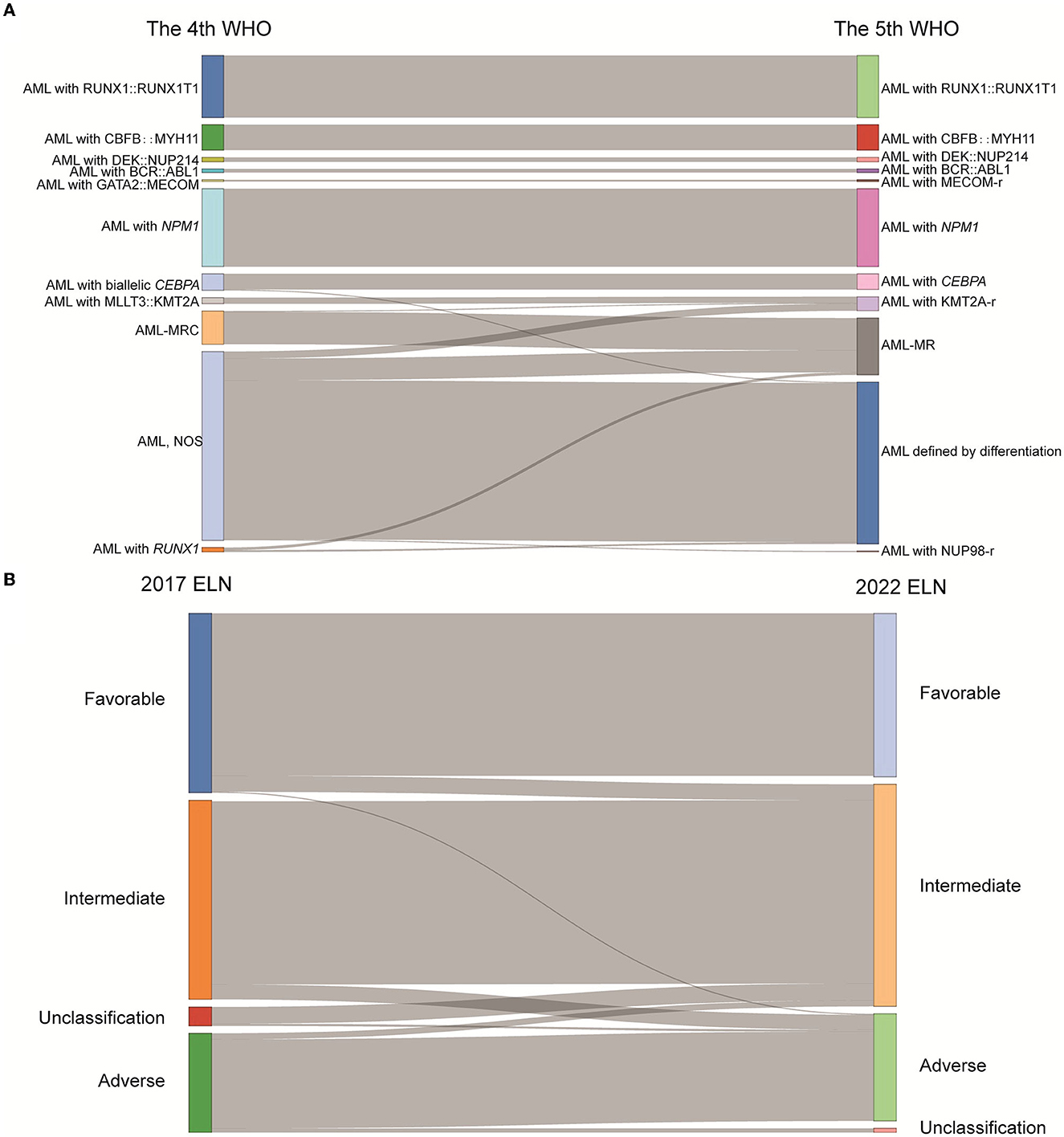
Figure 1. The classification of AML patients based on the WHO and ELN guidance. (A) Sankey diagram demonstrated the relationship between AML patients' subtypes defined in the 4th and 5th WHO classifications. (B) Sankey diagram demonstrated the relationship between AML patients' subtypes defined in the 2017 ELN and 2022 ELN guidance (WHO, World Health Organization; ELN, European Leukemia Network; AML, acute myeloid leukemia; MECOM-r, MECOM rearrangement; KMT2A-r, KMT2A rearrangement; NUP98-r, NUP98 rearrangement; AML-MRC, AML with myelodysplasia-related changes; AML-MR, AML, myelodysplasia-related; NOS, not otherwise specified).
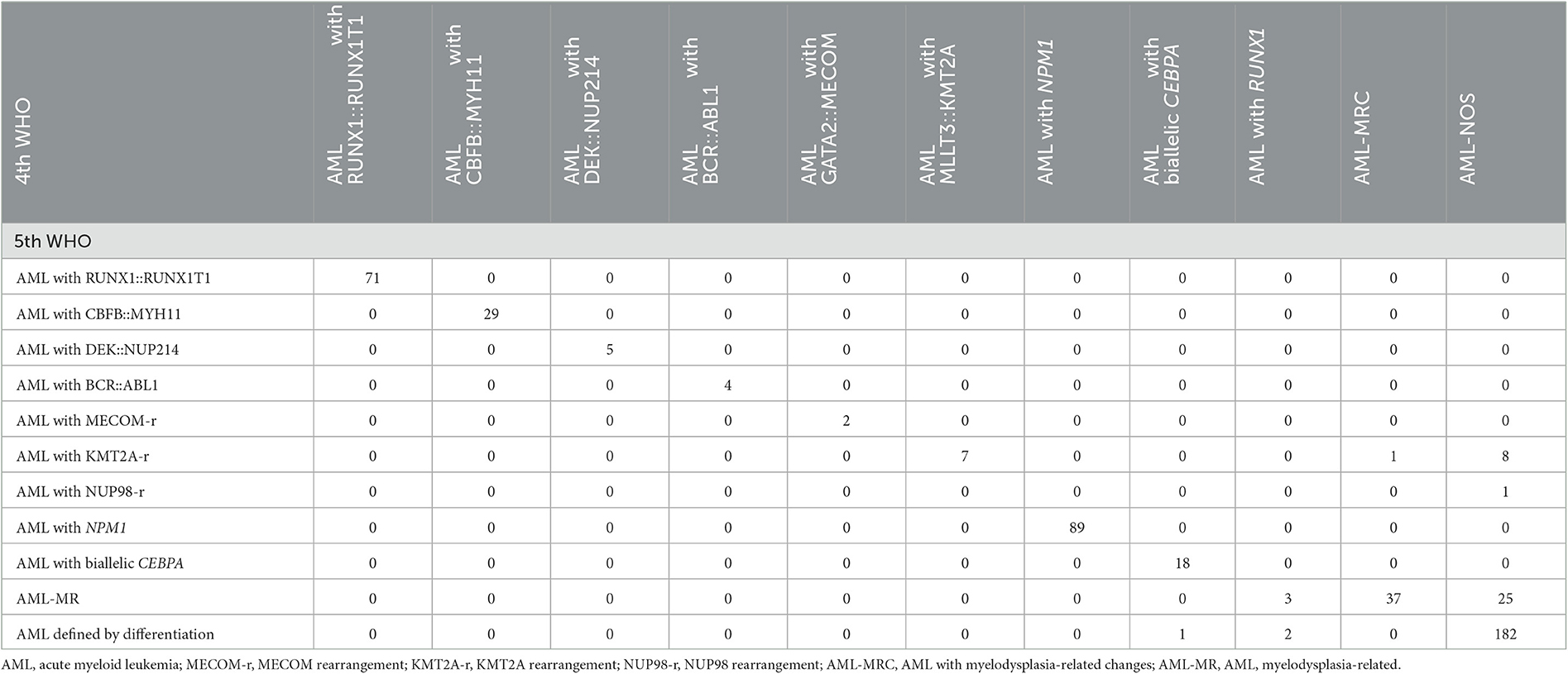
Table 2. Re-stratification matrix of the number of AML patients classified in each of the 4th WHO 2016 classification and each of the 5th WHO classification (n = 485).
Risk stratification of AML according to the 2017 and 2022 ELN guidelines
Diagnostic criteria were largely unchanged in the new proposal, except for the following: (1) persons with FLT3-ITD were classified as intermediate regardless of the VAF; (2) the previously defined favorable risk of biCEBPA was changed to bZIP in-frame mutated CEBPA; (3) KAT6A::CREBBP and mutated ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, or ZRSR2 were added as adverse events; and (4) VAF ≥ 10% was defined as an additional condition for the adverse event of TP53 mutation.
A total of 483 and 498 patients were submitted to risk stratification based on the 2017 and 2022 ELN guidance, respectively. The stratification for most patients (442/501) according to the 2022 ELN guidance was consistent with the 2017 ELN. In contrast, 16 patients in the favorable group based on the 2017 ELN guidance were regrouped into the intermediate group based on the 2022 ELN guidance due to the low VAF value of FLT3-ITD and the lack of the bZIP in-frame mutated CEBPA. In addition, 13 patients in the intermediate group based on the 2017 ELN guidance were regrouped into the adverse group based on the 2022 ELN guidance due to the mutations of BCOR, SRSF2, U2AF1, and ZRSR2. Seventeen unclassified patients (the detailed information of CEBPA and FLT3-ITD unknown) based on the 2017 ELN guidance were regrouped into the intermediate group (Figure 1B, Table 3).
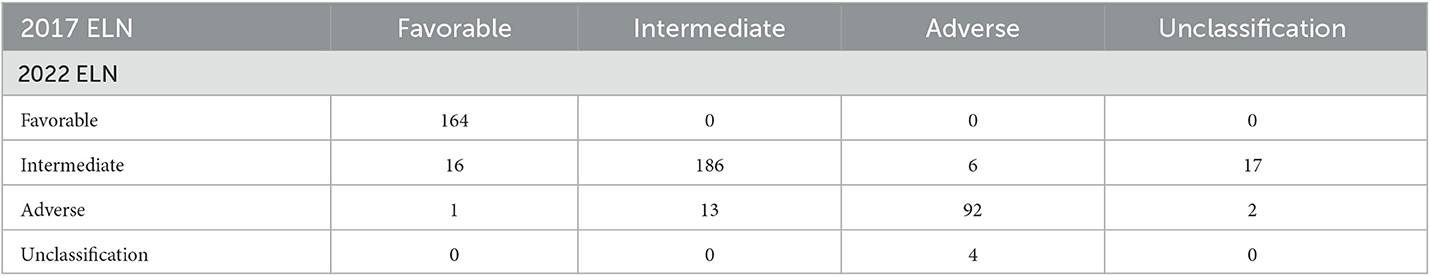
Table 3. Re-stratification matrix of the number of AML patients classified in each of the 2017 ELN classification and each of the 2022 ELN classification (n = 501).
Prognosis analysis according to the WHO classification and ELN guidance
Moreover, we evaluated the PFS and OS of subgroups based on the WHO classification and ELN guidance. Both PFS and OS rates were higher in AML patients with RUNX1::RUNX1T1, CBFB::MYH11, and biCEBPA, while the PFS and OS rates were lower for patients with MLLT3::KMT2A and GATA2::MECOM according to the 4th WHO classification (Figure 2A). Similarly, the PFS and OS rates for patients with RUNX1::RUNX1T1, CBFB::MYH11, and biCEBPA were higher, and the prognosis for patients of the AML-MR and AML defined by differentiation subgroups was worse according to the 5th WHO classification (Figure 2B). From the ELN subgroups, both PFS and OS curves of patients of the intermediate and adverse groups were not very distinguishable according to the 2017 (Figure 3A) and 2022 ELN guidance (Figure 3B), although the OS and PFS curves for the favorable group and other groups could be distinguished well. Also, we assessed whether the therapeutic means affected the prognosis of AML patients. Hematopoietic stem cell transplantation (HCT) significantly improved the PFS and OS of patients in favorable, intermediate, and adverse groups as compared with the non-HCT patients. However, the PFS and OS curves for the non-HCT patients of the intermediate and adverse groups remained not very distinguishable (Figure 3C). These results affirmed the clinical value of both WHO and ELN, but a better prognosis model should be established in Chinese cohorts.
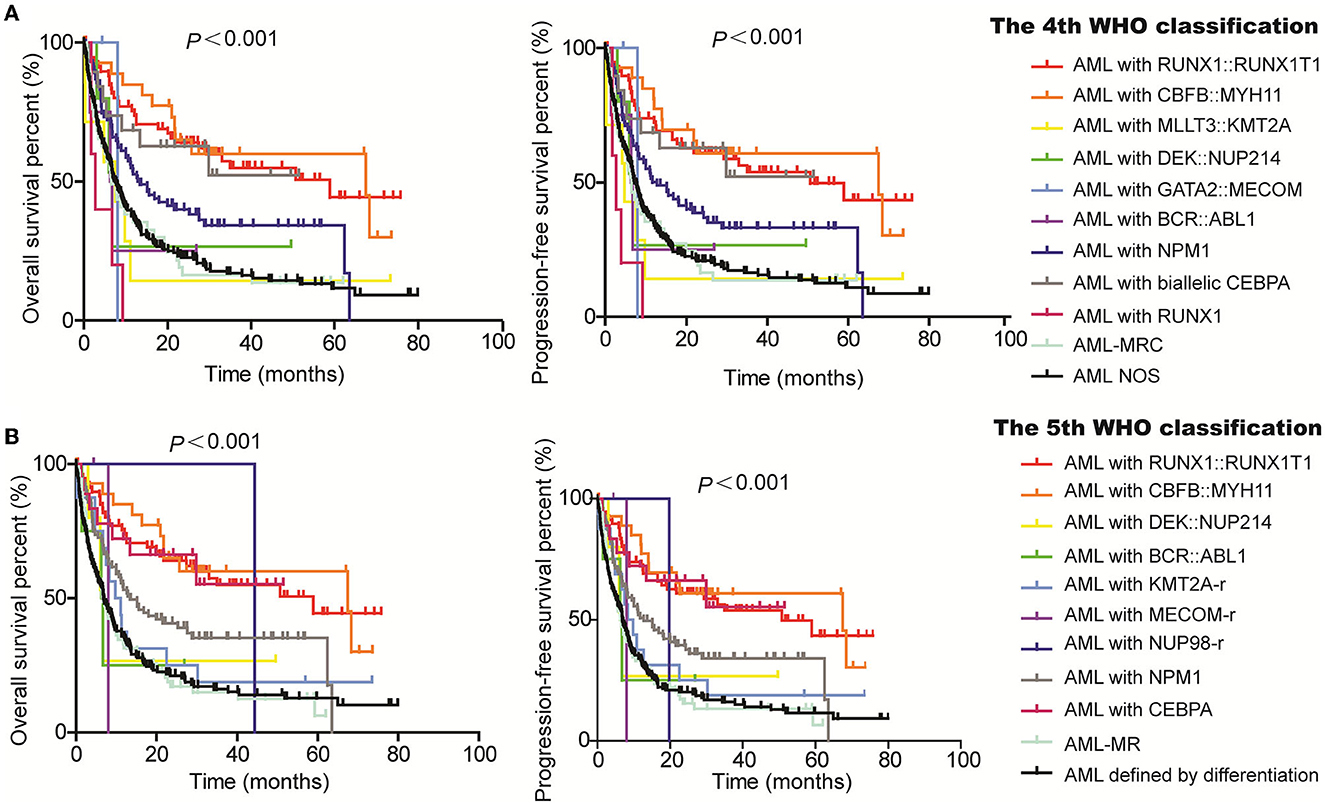
Figure 2. Assessment of the value of 4th and 5th WHO classifications on the PFS and OS of AML patients. Kaplan–Meier curves were used to assess the OS and PFS of patients of the favorable, intermediate, and adverse groups according to the 4th (A) and 5th WHO classifications [(B); WHO, World Health Organization; AML, acute myeloid leukemia; MECOM-r, MECOM rearrangement; KMT2A-r, KMT2A rearrangement; NUP98-r, NUP98 rearrangement; AML-MRC, AML with myelodysplasia-related changes; AML-MR, AML, myelodysplasia-related; NOS, not otherwise specified].
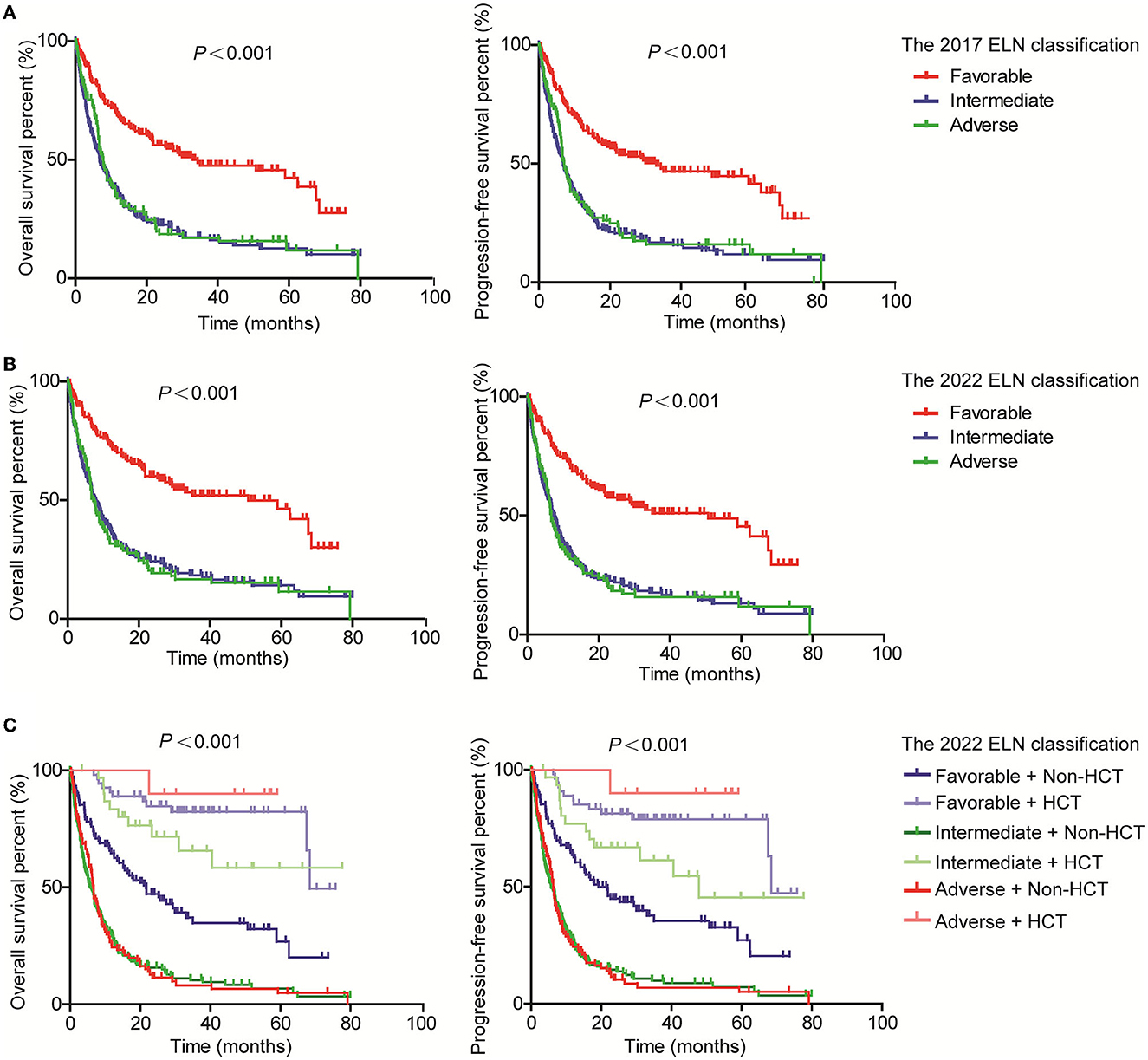
Figure 3. Assessment of the value of 2017 and 2022 ELN guidance on the PFS and OS of AML patients. Kaplan–Meier curves were used to assess the OS and PFS of patients of the favorable, intermediate, and adverse groups according to the 2017 (A) and 2022 (B) ELN guidance, together with transplantation [(C); ELN, European Leukemia Network; HCT, hematopoietic stem cell transplantation].
Establishment of the prognosis models for Chinese AML patients
To establish a better prognosis model for Chinese patients with newly diagnosed AML, mutation, rearrangements, and clinicopathologic features were considered. For the PFS model, risk score was first calculated according to the following formula, risk score = 0.324134*sex (male = 1, female = 2) + 0.025315*age- 0.88687*CBFB:: MYH11-0.801312*NPM1+0.89387*RUNX1-0.757403*RUNX1::RUNX1T1+1.244844*SH2B3+1.120662*TP53, in which positive was defined as “1” and negative was defined as “0” in terms of gene mutation and fusion. Then, the patients were divided into three groups, favorable (risk score ≤ 1.80), intermediate (risk score < 1.8 < 2.37), and adverse (risk score ≥ 2.37). As shown in Figure 4A, the PFS time of the favorable group was obviously longer than the intermediate and adverse groups, as well as the intermediate group vs. the adverse group in both training and test sets. Furthermore, the nomogram demonstrated the contributions of the selected factors to the 1-, 3-, and 5-year PFS probability (Figure 4B).
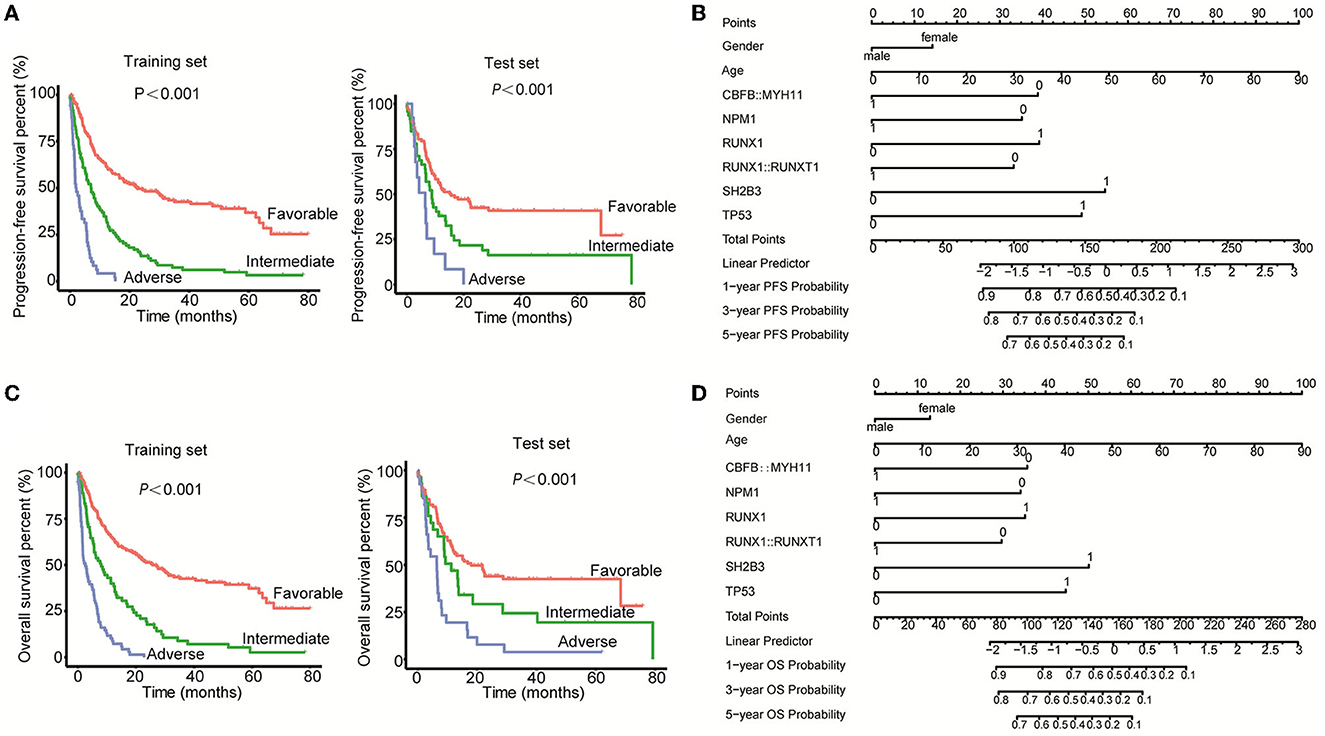
Figure 4. Establishment of the prognosis models for Chinese AML patients. (A) Kaplan–Meier curves were used to assess the PFS of patients of the favorable, intermediate, and adverse groups in both the training set and test set. (B) The nomogram model was applied to demonstrate the contributions of the selected factors to the 1-, 3-, and 5-year PFS probability. (C) Kaplan–Meier curves were used to assess the OS of patients of the favorable, intermediate, and adverse groups in both the training set and the test set. (D) The nomogram model was applied to demonstrate the contributions of the selected factors to the 1-, 3-, and 5-year OS probability.
For the OS model, the risk score was first calculated using the formula = 0.319677*sex (male = 1, female = 2) + 0.027509*age- 0.883355*CBFB::MYH11-0.84456*NPM1+0.870257*RUNX1-0.73479*RUNX1::RUNX1T1+1.240181*SH2B3+1.106195*TP53. After calculating the risk score of each patient, the patients were divided into three groups, favorable (≤ 1.94), intermediate (< 1.94 < 2.33), and adverse (≥2.33). The PFS time of the favorable group was obviously higher as compared with the intermediate and adverse groups, as well as the intermediate group vs. the adverse group in both the training set and test set (Figure 4C). In addition, the nomogram demonstrated the contributions of the selected factors to the 1, 3, and 5-year OS probability (Figure 4D). Collectively, we constructed a prognosis model for Chinese AML patients through the integration of the mutations, fusions, and clinical information.
Discussion
Recently, the 5th WHO guidance was issued, which has made some modifications to the classification of hematologic malignancies including AML. Herein, we compared the 4th and 5th WHO in the Chinese AML classification. The biggest change was that 25 (5.2%), 8 (1.6%), and 1 (0.2%) patients in the AML, NOS group according to the 4th WHO, were re-classified into the AML-MR, KMT2A rearrangement, and NUP98 rearrangement subgroups, respectively. Still, 38% (185/486) of patients were divided into the AML defined by differentiation subgroup according to the 5th WHO classification due to the limitation of sequencing technology earlier in the years. However, it was decreased compared to the proportion of AML, NOS (44.5%, 216/485). With the advances in gene detection technologies, patients are divided into more precise categories with clearer treatment strategies and prognoses (9–11). For instance, the mutations of ASXL1, BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 genes are added as the defining cytogenetic abnormalities, and patients carrying one of which are considered a member of the AML-MR subgroup, while the subgroup defined by RUNX1 mutation is abolished. It has been reported that some somatic mutations, including ASXL1, BCOR, EZH2, SF3B1, SRSF2, STAG2, U2AF1, and ZRSR2 mutations are associated with an adverse prognosis of AML (12–14). RUNX1 mutations were previously reported to be linked to unfavorable outcomes in AML patients (15). However, increasing evidence has demonstrated that normal RUNX1 is also implicated in leukemogenesis. Sood et al. (16) indicated that leukemic cells of core-binding factor AML and certain types of leukemia with KMT2A rearrangements require normal RUNX1 to survive. Wesely et al. (17) showed that RUNX1 was of importance in maintaining leukemia stem cells across various genetic subgroups in AML. Thus, either mutations or normal RUNX1 is essential in AML development.
In addition, we compared the risk stratification of AML patients according to the 2017 and 2022 ELN guidance. In total, 16 (3.3%) patients in the favorable group and 13 (2.7%) patients in the intermediate group were re-classified to the intermediate and adverse groups based on the 2022 ELN guidance, while 6 (1.2%) patients in the adverse group were re-grouped into the intermediate group based on the 2022 ELN guidance. Regrettably, both the PFS and OS curves of the intermediate and adverse groups were not very distinguishable in our cohort even after excluding the patients who received HCT, which significantly improved the prognosis of AML, as previously reported (18, 19). This result further highlights the high heterogeneity of AML, but it also cannot exclude the reasons for inadequate detection means as early as 2013 and the small sample size.
Based on the above results, we constructed the prognosis model for Chinese AML patients through the integration of mutations, rearrangements, and clinicopathologic features. Finally, the mutations of NPM1, RUNX1, SH2B3, and TP53 genes, fusions of CBFB::MYH11 and RUNX1::RUNX1T1, and the clinical factors of sex and age were selected as the important influencing factors of both PFS and OS. Among them, CBFB::MYH11, RUNX1::RUNX1T1, and NPM1 mutations were the protective factors, while the mutations of RUNX1, SH2B3, and TP53 genes were adverse factors, which were consistent with the ELN guidance (6, 7). In addition, we found that older adults and female patients were also two adverse factors of PFS and OS for Chinese AML patients. Stabellini et al. (20) recently studied the effect of sex on the survival of adults with AML and found that male patients had a lower risk of death than female patients (aHR = 0.41) in a total of 1,020 AML patients (57.4% male patients). This was consistent with our study, which demonstrated that being male was a protective factor for both PFS and OS in Chinese AML patients. In addition, Bin et al. (21) showed that age served as an independent prognostic factor to predict the 1-, 3-, and 5-year survival of AML patients, which was consistent with our study.
Collectively, this study compared the 4th and 5th WHO, as well as the 2017 and 2022 ELN guidance in Chinese AML patients. Although the classification and risk stratification were improved and defined by the 5th WHO classification and 2022 ELN guidance, the risk models were not very suitable. Based on this, we established a risk model for Chinese AML patients, which included age, sex, mutations (NPM1, RUNX1, SH2B3, and TP53), and fusions (CBFB::MYH11 and RUNX1::RUNX1T1). This model could easily help divide the patients into favorable, intermediate, and adverse groups, which may be suitable for Chinese AML patients.
Data availability statement
The data presented in the study are deposited in the Genome Sequence Archive for Human (GSA-Human) repository, accession number “HRA004529”, https://ngdc.cncb.ac.cn/gsa-human/browse/HRA004529.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
XW and JieW wrote the paper. SW, JuZ, JiZ, DW, ML, SZ, YC, HL, HZ, JinW, and WW provided the data. BX and GL analyzed the data. HW, HX, and PH reviewed the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Key Research and Development of Shaanxi Province (2022SF-13) and the Translational Research Grant of NCRCH (2021WWC01).
Acknowledgments
We thank Shanghai Rightongene Biomedical Technology Co. Ltd (Shanghai, China) for help with sequencing and data analysis.
Conflict of interest
BX, GL, and HX were employed by Shanghai Yuanqi Biomedical Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nagel G, Weber D, Fromm E, Erhardt S, Lubbert M, Fiedler W, et al. Epidemiological, genetic, and clinical characterization by age of newly diagnosed acute myeloid leukemia based on an academic population-based registry study (Amlsg Bio). Ann Hematol. (2017) 96:1993–2003. doi: 10.1007/s00277-017-3150-3
2. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. (2015) 373:1136–52. doi: 10.1056/NEJMra1406184
3. Song MK, Park BB, Uhm JE. Clinical efficacies of FLT3 inhibitors in patients with acute myeloid leukemia. Int J Mol Sci. (2022) 23:2012708. doi: 10.3390/ijms232012708
4. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
5. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia. (2022) 36:1703–19. doi: 10.1038/s41375-022-01613-1
6. Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. (2017) 129:424–47. doi: 10.1182/blood-2016-08-733196
7. Dohner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. (2022) 140:1345–77. doi: 10.1182/blood.2022016867
8. Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. (2004) 10:7252–9. doi: 10.1158/1078-0432.CCR-04-0713
9. Meena JP, Pathak N, Gupta AK, Bakhshi S, Gupta R, Makkar H, et al. Molecular evaluation of gene mutation profiles and copy number variations in pediatric acute myeloid leukemia. Leuk Res. (2022) 122:106954. doi: 10.1016/j.leukres.2022.106954
10. Mrozek K. Molecular cytogenetics in acute myeloid leukemia in adult patients: Practical implications. Pol Arch Intern Med. (2022) 132:7–8. doi: 10.20452/pamw.16300
11. Castano-Diez S, Lopez-Guerra M, Bosch-Castaneda C, Bataller A, Charry P, Esteban D, et al. Real-world data on chronic myelomonocytic leukemia: Clinical and molecular characteristics, treatment, emerging drugs, and patient outcomes. Cancers. (2022) 14:174107. doi: 10.3390/cancers14174107
12. Montalban-Bravo G, Kanagal-Shamanna R, Class CA, Sasaki K, Ravandi F, Cortes JE, et al. Outcomes of acute myeloid leukemia with myelodysplasia related changes depend on diagnostic criteria and therapy. Am J Hematol. (2020) 95:612–22. doi: 10.1002/ajh.25769
13. Lindsley RC, Mar BG, Mazzola E, Grauman PV, Shareef S, Allen SL, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. (2015) 125:1367–76. doi: 10.1182/blood-2014-11-610543
14. Yokoyama K, Shimizu E, Yokoyama N, Nakamura S, Kasajima R, Ogawa M, et al. Cell-lineage level-targeted sequencing to identify acute myeloid leukemia with myelodysplasia-related changes. Blood Adv. (2018) 2:2513–21. doi: 10.1182/bloodadvances.2017010744
15. Rungjirajittranon T, Siriwannangkul T, Kungwankiattichai S, Leelakanok N, Rotchanapanya W, Vittayawacharin P, et al. Clinical outcomes of acute myeloid leukemia patients harboring the RUNX1 mutation: Is it still an unfavorable prognosis? A cohort study and meta-analysis. Cancers. (2022) 14:215239. doi: 10.3390/cancers14215239
16. Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. (2017) 129:2070–82. doi: 10.1182/blood-2016-10-687830
17. Wesely J, Kotini AG, Izzo F, Luo H, Yuan H, Sun J, et al. Acute myeloid leukemia IPSCS reveal a role for RUNX1 in the maintenance of human leukemia stem cells. Cell Rep. (2020) 31:107688. doi: 10.1016/j.celrep.2020.107688
18. Heini AD, Porret N, Zenhaeusern R, Winkler A, Bacher U, Pabst T. Clonal hematopoiesis after autologous stem cell transplantation does not confer adverse prognosis in patients with AML. Cancers. (2021) 13:133190. doi: 10.3390/cancers13133190
19. Zhang YY, Mo XD, Zhang XH, Xu LP, Wang Y, Yan CH, et al. FLT3 internal tandem duplication does not impact prognosis after haploidentical allogeneic hematopoietic stem cell transplantation in AML patients. Bone Marrow Transplant. (2019) 54:1462–70. doi: 10.1038/s41409-019-0456-x
20. Stabellini N, Tomlinson B, Cullen J, Shanahan J, Waite K, Montero AJ, et al. Sex differences in adults with acute myeloid leukemia and the impact of sex on overall survival. Cancer Med. (2022) 12:6711–21. doi: 10.1002/cam4.5461
Keywords: acute myeloid leukemia, World Health Organization, European Leukemia Net, risk model, prognosis
Citation: Wang X, Wang J, Wei S, Zhao J, Xin B, Li G, Zhao J, Wu D, Luo M, Zhao S, Chen Y, Liu H, Zhang H, Wang J, Wang W, Wang H, Xiong H and He P (2023) The latest edition of WHO and ELN guidance and a new risk model for Chinese acute myeloid leukemia patients. Front. Med. 10:1165445. doi: 10.3389/fmed.2023.1165445
Received: 14 February 2023; Accepted: 30 May 2023;
Published: 23 June 2023.
Edited by:
Ahmet Emre Eskazan, Istanbul University-Cerrahpasa, TürkiyeReviewed by:
Sinan Mersin, Dr. Ersin Arslan Education and Research Hospital, TürkiyeSelin Küçükyurt, Istanbul University Cerrahpasa, Türkiye
Copyright © 2023 Wang, Wang, Wei, Zhao, Xin, Li, Zhao, Wu, Luo, Zhao, Chen, Liu, Zhang, Wang, Wang, Wang, Xiong and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaiyu Wang, d2h5bWVkQDEyNi5jb20=; Hui Xiong, eGlvbmdodWlAcmlnaHRvbmdlbmUuY29t; Pengcheng He, aGVwY0AxNjMuY29t
†These authors share first authorship
 Xiaoning Wang
Xiaoning Wang Jie Wang
Jie Wang Suhua Wei
Suhua Wei Juan Zhao
Juan Zhao Beibei Xin2
Beibei Xin2 Haibo Liu
Haibo Liu