- 1Malaria Branch, Division of Parasitic Diseases and Malaria, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 2Centre for Health Informatics, Computing, and Statistics, Lancaster University, Lancaster, United Kingdom
- 3Department of Infection Biology, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom
- 4Department of Mathematics & Information Science, Warsaw University of Technology, Warsaw, Poland
- 5Centro de Estatística e Aplicações da Universidade de Lisboa (CEAUL), Lisbon, Portugal
Editorial on the Research Topic
Current research on serological analyses of infectious diseases
Introduction
Serology based on antibody detection or quantification is a key research tool in the analysis of human infectious diseases. In Public Health and Epidemiology, it allows the estimation of the disease burden beyond the classical measures based on the presence or frequency of active infections in the population (1, 2). It also allows the prediction of when individuals were previously infected for tailoring novel disease control strategies (3, 4). In Medicine, it can assist in diagnosis (5), in the inference of disease etiology and pathology (6–8), and in the stratification of patients for better disease management and treatment (9). All these research opportunities motivated a discussion about the creation of a World Serum bank for infectious diseases (10–12).
Until recently, the enzyme-linked immunoassay (ELISA) and other related tests were at the core of the research made in infectious diseases. These tests typically detect or quantify antibodies against a single antigen. Nowadays high-throughput serological technologies, such as microarrays and multiplex bead assays, are becoming competing rivals of these standard tests due to the possibility of measuring multiple antibodies in the same biological sample at a reasonable cost. As such, these new technologies are giving rise to multiple system serology analyses (13–16).
In this Research Topic, we took the pulse of current research of infectious diseases based on serology with a special focus on applications in Medicine, Epidemiology, and Public Health. Among more than 20 submissions received, we were able to collect 9 original Research Topics (Figure 1A) and one systematic review (Figure 1B). These papers featured diverse infectious diseases, including neglected, re-emerging, tropical, established and novel. We were pleased to have at least one study investigating populations from different continents. This suggests that the findings of this Research Topic are likely to benefit multiple human populations globally. Below we provide a brief context of the published studies.
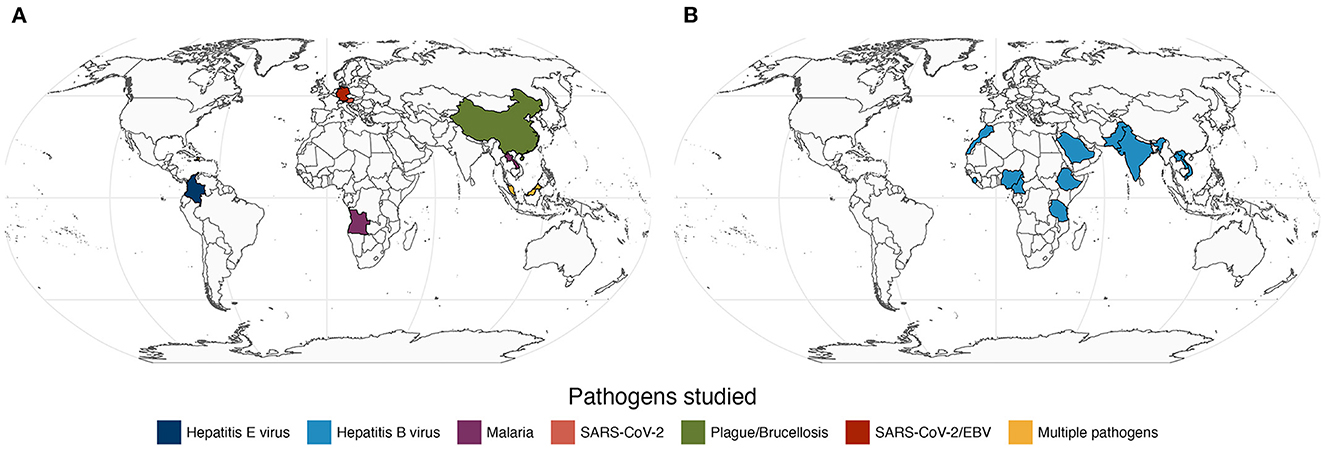
Figure 1. Map of the study populations and the respective pathogens featured in 9 original research papers (A) and in one systematic review (B) related to seroprevalence of the Hepatitis B virus among health workers (Maamor et al.). Note that Germany contributed with two independent papers (Girl et al. and Sepúlveda et al.) in plot A.
Serological studies using standard antibody assays
Notwithstanding the technological advances in serology, standard antibody assays seem the only viable option for investigating many diseases or populations from low- and middle-income countries (LMIC). This idea is illustrated in three independent studies included in the Research Topic. In one study, Qin et al. used serum agglutination tests to estimate the seroprevalence of animal plague and brucellosis in the neglected populations of Qinghai-Tibet plateau; these two neglected diseases are re-emerging in the area and, therefore, the reported findings have the potential to support more effective disease control interventions in the affected populations. In another study, Fernández Villaslobo et al. estimated the seroprevalence of Hepatitis E virus in children and adolescents in Colombia's capital city of Bogotá using commercial ELISA. Several study limitations were identified by the authors (e.g., massive school closure due to COVID-19 pandemic), but the findings suggested a low seroprevalence of IgG antibodies (~1%) and a single case of active infection (detected by IgM reactivity) among the study participants. It would be interesting to know whether these findings hold true if antibodies against multiple antigens were measured in the study. In a third study, Maamor et al. summarized data of 25 reports on the seroprevalence of Hepatitis B virus among health workers. These reports were restricted to African and Asian LMIC (Figure 1B) and, therefore, it is no surprise that the respective data were based on ELISA or rapid antibody tests whose performance can be affected by transport and storage conditions among other factors.
In the emergency of containing a disease outbreak, it is desirable to conduct a large-scale epidemiological study in real time. In this scenario, classical serological assays are invaluable tools in the field due to their low cost, the use of relatively simple technology, and the easiness of protocol's standardization across participating labs. The valuable use of these assays was illustrated in the peak of the COVID-19 pandemic with the fast execution of studies using ELISA protocols or rapid immunochromatography tests (17–19). In this scenario, it is fundamental to know the performance of available assays or tests in the field, as done in the United Kingdom and Denmark (20, 21). With a similar purpose of these benchmark studies, Girl et al. evaluated the performance of 2 lateral flow assays and 2 surrogate ELISA tests to detect SARS-CoV-2-neutralizing antibodies using data from more than 300 German individuals. Notwithstanding the current dampening of the COVID-19 threat in many parts of the world, this and similar studies provide a solid basis of evidence for facing future resurgences of SARS-CoV-2 in the respective populations.
Serological studies using high-throughput antibody assays
High-throughput antibody technologies are becoming more popular among the scientific community. Our Research Topic captures somehow this increasing popularity given that more than half of the published studies featured the use of these avant-garde assays in different epidemiological contexts. Similar to a nationwide seroprevalence study from Spain (22), Willeit et al. illustrated the benefits of using a commercial high-throughput and automated immunoassay to screen rapidly antibodies against the spike protein of SARS-CoV-2 B1.351 variant in almost 2,500 individuals from an Austrian district. In Sepúlveda et al., more than 3,000 different antibody responses to the common Epstein-Barr virus were screened in German patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) and healthy controls using a seroarray. The rationale for conducting a massive antibody screening lies on the fact that this complex and neglected disease remains without a diagnostic biomarker. This study highlighted two candidate antibody targets (EBNA4_0529 and EBNA6_0070) that could not only help in diagnosing a large subset of ME/CFS patients, but also further support the autoimmune hypothesis for the pathogenesis of this disease (23). In the realm of tropical diseases, Rogier et al. conducted a detailed analysis of multiple antibody responses to the malaria-causing Plasmodium falciparum in Angolan children using a multiplex bead assay. The respective findings provide additional evidence for the current understanding of malaria immunity. These and other findings from the literature set the foundation to innovate on malaria vaccine development and immune-related treatments. In another study of Malaria, Byrne et al. reported data from more than 5,000 individuals from the Lao People's Democratic Republic using a multiplex bead assay. This study illustrates how multiplex data can be used to construct risk maps that could detect important foci of infection for close surveillance and future interventions.
The most impressive applications of multiplex serological assays in this Research Topic are related to two studies where data allowed to investigate multiple diseases at the same time; other examples of studies based on similar ideas can be found elsewhere (24–26). In Chan et al., the study was performed in the Malaysian province of Sabah and contemplated the sampling of more than 10,000 individuals. The data comprised antibodies against twelve antigens related to 6 neglected tropical diseases, including lymphatic filariasis, yaws, and trachoma. In another study, Chan et al. extended the number of measured antibodies to investigate the epidemiology of 11 pathogens in Haiti. This study integrates a series of research efforts to support malaria elimination in the country (27–31). This study showed the benefit of using multiplex data to screen secondary diseases in a single timepoint, thus, avoiding the negative effects (e.g., increasing cost and participation fatigue) of conducting multiple sampling in the same population.
Conclusion
This Research Topic provides an interesting contrast between studies using standard serological tools and those using more advanced technology. In our perspective, this contrast is particularly important to judge the pros and cons of using one or another technology across different research contexts. Above all, the availability of more advanced serological technology is a great opportunity to foster collaboration among researchers and to enhance capacity building (e.g., lab automation and data analysis skills) where it is needed the most.
In summary, this Research Topic shows the increasing popularity of high-throughput serological data and how these can be useful in the support of public health and epidemiological interventions (e.g., creating of multiple risk maps based on different antibodies to identify the key foci of infection). It also shows how the same type of data can expand our current knowledge on the pathogenesis and diagnosis not only of infectious disease but also of diseases with unknown etiology, as illustrated for ME/CFS.
Author contributions
NS drafted the editorial. All authors revised and approved the final version of the editorial.
Funding
NS received funding from the Fundação para a Ciência e Tecnologia, Portugal (ref. UIDB/00006/2020), and the Polish National Agency for Academic Exchange, Poland (ref. PPN/ULM/2020/1/00069/U/00001) during the editorial work related to this Research Topic.
Acknowledgments
We would like to thank all the authors, reviewers, and study participants who contributed one way or another to this Research Topic. We also thank João Malato for helping with the design of Figure 1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wiegand RE, Deng Y, Deng X, Lee A, Meyer WA, Letovsky S, et al. Estimated SARS-CoV-2 antibody seroprevalence trends and relationship to reported case prevalence from a repeated, cross-sectional study in the 50 states and the District of Columbia, United States-October 25, 2020-February 26, 2022. Lancet Reg Heal Am. (2023) 18:104. doi: 10.1016/j.lana.2022.100403
2. Corran P, Coleman P, Riley E, Drakeley C. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol. (2007) 23:575–82. doi: 10.1016/j.pt.2007.08.023
3. Longley RJ, White MT, Takashima E, Brewster J, Morita M, Harbers M, et al. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat Med. (2020) 26:741–9. doi: 10.1038/s41591-020-0841-4
4. Helb DA, Tetteh KKA, Felgner PL, Skinner J, Hubbard A, Arinaitwe E, et al. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc Natl Acad Sci U S A. (2015) 112:E4438–47. doi: 10.1073/pnas.1501705112
5. Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. (2020) 92:1518–24. doi: 10.1002/jmv.25727
6. Bjornevik K, Cortese M, Healy BC, Kuhle J, Mina MJ, Leng Y, et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. (2022) 375:296–301. doi: 10.1126/science.abj8222
7. Loebel M, Eckey M, Sotzny F, Hahn E, Bauer S, Grabowski P, et al. Serological profiling of the EBV immune response in Chronic Fatigue Syndrome using a peptide microarray. PLoS ONE. (2017) 12:e0179124. doi: 10.1371/journal.pone.0179124
8. Ruprecht K, Wunderlich B, Gieß R, Meyer P, Loebel M, Lenz K, et al. Multiple sclerosis: the elevated antibody response to Epstein-Barr virus primarily targets, but is not confined to, the glycine-alanine repeat of Epstein-Barr nuclear antigen-1. J Neuroimmunol. (2014) 272:56–61. doi: 10.1016/j.jneuroim.2014.04.005
9. Domingues TD, Grabowska AD, Lee JS, Ameijeiras-Alonso J, Westermeier F, Scheibenbogen C, et al. Herpesviruses Serology Distinguishes Different Subgroups of Patients From the United Kingdom Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Biobank. Front Med. (2021) 8:686736. doi: 10.3389/fmed.2021.686736
10. Metcalf CJE, Farrar J, Cutts FT, Basta NE, Graham AL, Lessler J, et al. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet. (2016) 388:728–30. doi: 10.1016/S0140-6736(16)30164-7
11. de Lusignan S, Correa A. Opportunities and challenges of a World Serum Bank. Lancet. (2017) 389:250–1. doi: 10.1016/S0140-6736(17)30046-6
12. Coates KM. Opportunities and challenges of a World Serum Bank. Lancet. (2017) 389:251–2. doi: 10.1016/S0140-6736(17)30052-1
13. Wine Y, Horton AP, Ippolito GC, Georgiou G. Serology in the 21st Century: The Molecular-Level Analysis of the Serum Antibody Repertoire. Curr Opin Immunol. (2015) 35:89. doi: 10.1016/j.coi.2015.06.009
14. Pittala S, Morrison KS, Ackerman ME. Systems Serology for Decoding Infection and Vaccine-Induced Antibody Responses to HIV-1. Curr Opin HIV AIDS. (2019) 14:253. doi: 10.1097/COH.0000000000000558
15. Selva KJ, van de Sandt CE, Lemke MM, Lee CY, Shoffner SK, Chua BY, et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat Commun. (2021) 12:1–14. doi: 10.1038/s41467-021-22236-7
16. Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. (2021) 595:283–8. doi: 10.1101/2020.12.10.20247205
17. Gonçalves J, Sousa RL, Jacinto MJ, Silva DA, Paula F, Sousa R, et al. Evaluating SARS-CoV-2 Seroconversion Following Relieve of Confinement Measures. Front Med. (2020) 7:971. doi: 10.3389/fmed.2020.603996
18. Stringhini S, Wisniak A, Piumatti G, Azman AS, Lauer SA, Baysson H, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): a population-based study. Lancet. (2020) 396:313–9. doi: 10.1016/S0140-6736(20)31304-0
19. Khalagi K, Gharibzadeh S, Khalili D, Mansournia MA, Mirab Samiee S, Aghamohamadi S, et al. Prevalence of COVID-19 in Iran: results of the first survey of the Iranian COVID-19 Serological Surveillance programme. Clin Microbiol Infect. (2021) 27:1666–71. doi: 10.1016/j.cmi.2021.06.002
20. Ainsworth M, Andersson M, Auckland K, Baillie JK, Barnes E, Beer S, et al. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. (2020) 20:1390–400. doi: 10.1016/S1473-3099(20)30634-4-4
21. Harritshøj LH, Gybel-Brask M, Afzal S, Kamstrup PR, Jørgensen CS, Thomsen MK, et al. Comparison of 16 serological SARS-CoV-2 immunoassays in 16 clinical laboratories. J Clin Microbiol. (2021) 59:2596–616. doi: 10.1128/JCM.02596-20
22. Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. (2020) 396:535–44. doi: 10.1016/S0140-6736(20)32266-2
23. Sotzny F, Blanco J, Capelli E, Castro-Marrero J, Steiner S, Murovska M, et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome – Evidence for an autoimmune disease. Autoimmun Rev. (2018) 17:601–9. doi: 10.1016/j.autrev.2018.01.009
24. O'Neal AJ, Glass KA, Emig CJ, Vitug AA, Henry SJ, Shungu DC, et al. Survey of Anti-Pathogen Antibody Levels in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Proteomes. (2022) 10:21. doi: 10.3390/proteomes10020021
25. Brenner N, Mentzer AJ, Butt J, Braband KL, Michel A, Jeffery K, et al. Validation of Multiplex Serology for human hepatitis viruses B and C, human T-lymphotropic virus 1 and Toxoplasma gondii. PLoS ONE. (2019) 14:407. doi: 10.1371/journal.pone.0210407
26. Mentzer AJ, Brenner N, Allen N, Littlejohns TJ, Chong AY, Cortes A, et al. Identification of host-pathogen-disease relationships using a scalable multiplex serology platform in UK Biobank. Nat Commun. (2022) 13:1818. doi: 10.1038/s41467-022-29307-3
27. van den Hoogen LL, Présumé J, Romilus I, Mondélus G, Elismé T, Sepúlveda N, et al. Quality control of multiplex antibody detection in samples from large-scale surveys: the example of malaria in Haiti. Sci Rep. (2020) 10:1–10. doi: 10.1038/s41598-020-57876-0
28. Rogier E, Van Den Hoogen L, Herman C, Gurrala K, Joseph V, Stresman G, et al. High-throughput malaria serosurveillance using a one-step multiplex bead assay. Malar J. (2019) 18:3027. doi: 10.1186/s12936-019-3027-0
29. van den Hoogen LL, Stresman G, Présumé J, Romilus I, Mondélus G, Elismé T, et al. Selection of Antibody Responses Associated With Plasmodium falciparum Infections in the Context of Malaria Elimination. Front Immunol. (2020) 11:928. doi: 10.3389/fimmu.2020.00928
30. Druetz T, Stresman G, Ashton RA, Joseph V, Van Den Hoogen L, Worges M, et al. The Immediate Effects of a Combined Mass Drug Administration and Indoor Residual Spraying Campaign to Accelerate Progress Toward Malaria Elimination in Grande-Anse, Haiti. J Infect Dis. (2022) 225:1611–20. doi: 10.1093/infdis/jiab259
Keywords: microarray, immunochromatography test, enzyme-linked immunosorbent assay (ELISA), multiplex bead array, neglected diseases, tropical diseases, SARS-CoV-2, disease diagnosis and analysis
Citation: Rogier EW, Giorgi E, Tetteh K and Sepúlveda N (2023) Editorial: Current research on serological analyses of infectious diseases. Front. Med. 10:1154584. doi: 10.3389/fmed.2023.1154584
Received: 30 January 2023; Accepted: 07 February 2023;
Published: 17 February 2023.
Edited and reviewed by: Shisan Bao, The University of Sydney, Australia
Copyright © 2023 Rogier, Giorgi, Tetteh and Sepúlveda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuno Sepúlveda,  Ti5TZXB1bHZlZGFAbWluaS5wdy5lZHUucGw=
Ti5TZXB1bHZlZGFAbWluaS5wdy5lZHUucGw=
 Eric William Rogier
Eric William Rogier Emanuele Giorgi
Emanuele Giorgi Kevin Tetteh
Kevin Tetteh Nuno Sepúlveda
Nuno Sepúlveda