- 1School of Biomedical Engineering, Science and Health Systems, Drexel University, Philadelphia, PA, United States
- 2Biomedical Engineering, Bucknell University, Lewisburg, PA, United States
- 3Division of Molecular Biology - Research Services, PrimBio Research Institute, Exton, PA, United States
- 4Department of Nursing, College of Nursing and Health Professions, Drexel University, Philadelphia, PA, United States
- 5Department of Surgery, College of Medicine, Drexel University, Philadelphia, PA, United States
Introduction: Low-frequency, low-intensity ultrasound has been previously shown to promote healing of chronic wounds in humans, but mechanisms behind these effects are poorly understood. The purpose of this study was to evaluate gene expression differences in debrided human venous ulcer tissue from patients treated with low-frequency (20 kHz), low-intensity (100 mW/cm2) ultrasound compared to a sham treatment in an effort to better understand the potential biological mechanisms.
Methods: Debrided venous ulcer tissue was collected from 32 subjects one week after sham treatment or low-frequency, low-intensity ultrasound treatment. Of these samples, 7 samples (3 ultrasound treated and 4 sham treated) yielded sufficient quality total RNA for analysis by ultra-high multiplexed PCR (Ampliseq) and expression of more than 24,000 genes was analyzed. 477 genes were found to be significantly differentially expressed between the ultrasound and sham groups using cut-off values of p < 0.05 and fold change of 2.
Results and Discussion: The top differentially expressed genes included those involved in regulation of cell metabolism, proliferation, and immune cell signaling. Gene set enrichment analysis identified 20 significantly enriched gene sets from upregulated genes and 4 significantly enriched gene sets from downregulated genes. Most of the enriched gene sets from upregulated genes were related to cell-cell signaling pathways. The most significantly enriched gene set from downregulated genes was the inflammatory response gene set. These findings show that therapeutic ultrasound influences cellular behavior in chronic wounds as early as 1 week after application. Considering the well-known role of chronic inflammation in impairing wound healing in chronic wounds, these results suggest that a downregulation of inflammatory genes is a possible biological mechanism of ultrasound-mediated venous chronic wound healing. Such increased understanding may ultimately lead to the enhancement of ultrasound devices to accelerate chronic wound healing and increase patient quality of life.
Introduction
Chronic venous leg ulcers (VLUs), defined as wounds that do not heal in 4–6 weeks, are a significant health care burden and affect 1–2% of the worldwide adult population with increased incidence in women and older adults (1–3). Because of their slow healing time, VLUs cause substantial socioeconomic impact, costing up to $2 billion per year in the United States alone (4, 5). The current standard of care for chronic VLUs includes weekly wound dressing changes, the use of compression bandages, and sharp tissue debridement (5). Although 93% of VLUs will heal in 12 months (6), the recurrence rate within 3 months is 70% (6). There is a need for more effective chronic wound therapies.
One promising therapeutic approach is the application of low-frequency, low-intensity ultrasound (LFLI US), which has been shown in clinical trials to significantly accelerate wound healing of chronic VLUs (7–10). In a double-blind study, patients with VLUs were treated with a non-operational sham device or with LFLI US operating at 20 kHz and 100 mW/cm2 once a week for 15 min or 45 min for 8 weeks (8). Patients in the 15-min treatment group experienced full wound closure after 4 weeks (n = 5), while patients in the sham group saw an increase in wound size (n = 5), and patients in the 45-min treatment group did not see full wound closure in the duration of the study (n = 5) (8). Despite these promising clinical results, the optimal operating parameters of therapeutic ultrasound are still unknown, and the mechanisms are poorly understood. Furthermore, these mechanisms depend on ultrasound exposure parameters, such as frequency, spatial distribution of the pressure amplitude or intensity, and time duration. A better understanding of the mechanisms of action by which therapeutic ultrasound promotes wound closure could lead to optimization of ultrasound device parameters and increase the rate of wound healing.
Some studies have explored the potential biological mechanism of US in animal models (9, 11). For example, Lyu et al. demonstrated that US stimulation (operating at 2 MHz) increased wound healing in a diabetic rat model, and suggested that the effect might be via the Rac1 mechanotransduction pathway, which was also increased (9). Moreover, Maan et al. showed that the delivery of low frequency (40 kHz) US through a saline mist increased wound healing in diabetic mice, and suggested that stimulation of angiogenesis might be a potential mechanism (11). These potential mechanisms also have been put forth as mediators of US-stimulated bone fracture healing, an indication in which LFLI US has been more thoroughly evaluated compared to chronic wounds (12, 13). However, wound healing in animals, particularly rodents, is fundamentally different than wound healing in humans as it primarily occurs via contraction (14, 15) and because of differences in the inflammatory response to injury (16). Therefore, there is a need to investigate mechanisms of wound healing in humans.
The goal of this study was to explore the possible hypotheses for the biological mechanisms of therapeutic ultrasound-assisted wound healing in human patients by analyzing gene expression in tissue collected from VLUs. Expression of a panel of inflammation-related genes has been previously linked to healing in ultrasound-treated human diabetic foot ulcers using tissue collected during routine wound debridement (17). Therefore, in this study, we similarly analyzed debrided wound tissue and assessed the whole transcriptome to identify possible mechanisms. We collected debrided human chronic wound tissue 1 week after treatment as part of a double-blinded study in which patients were treated with ultrasound operating at 20 kHz and 100 mW/cm2 (SPTP) or a nonoperational sham device. This tissue was processed using an ultra-high multiplexed PCR (Ampliseq), which was selected because of its favorable profile in detecting transcriptional changes in even low-input samples (18).
Materials and methods
Study design
As part of an ongoing double-blind human clinical study investigating the efficacy of therapeutic ultrasound for chronic wound healing (ClinicalTrials.gov Identifier: NCT03041844), 48 patients with chronic VLUs were recruited from Drexel University Comprehensive Wound Healing Program. The study protocol was reviewed and approved by the Drexel University Institutional Review Board (IRB).
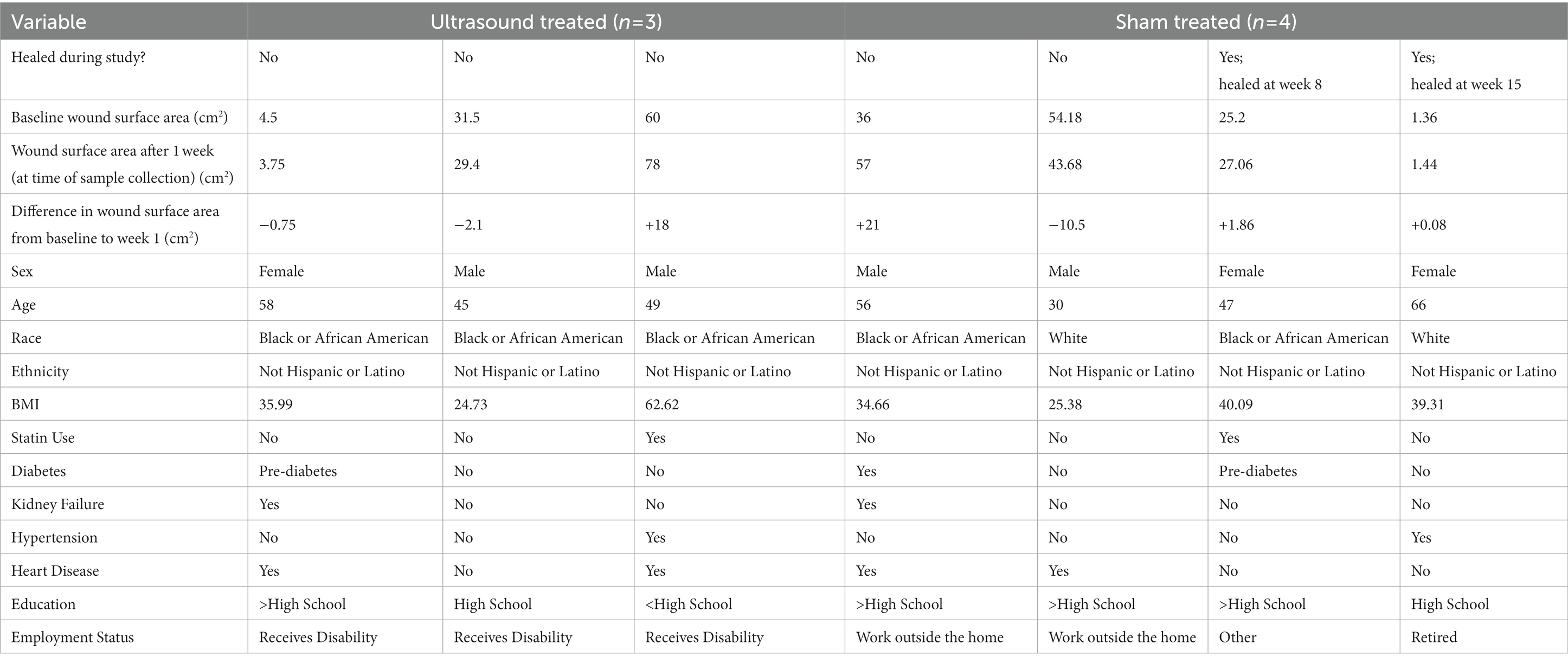
Inclusion criteria for participants to enroll into the study were as follows: have a VLU that has been documented for at least 8 weeks without complete re-epithelialization, a VLU size of larger than 0.75 cm2, VLU must be present on the lower extremities non-weight-bearing areas. The exclusion criteria included: VLU is secondary to any connective tissue disorder or blood dyscrasias, severe vascular insufficiency (ankle-brachial index lower than 0.75 or toe-brachial index below 0.5), active and untreated infection, acute deep venous thrombosis, cutaneous malignancy present on involved extremity, active (or past 6 months) cancer treatment, presence of both a diabetic ulcer and venous ulcer on the same extremity, known allergy to Tegaderm (a polyurethane dressing), pregnancy, individuals younger than 18 years of age, prisoners, individuals not able to read or speak English, Spanish, or Mandarin, and adults unable to consent. Finally, subjects with concomitant arterial disease were excluded by the presence of a palpable pedal pulse in the extremity on physical examination or a toe/bracial index of 0.6 or greater. Subjects were asked for a list of their comorbidities or the subject’s list of comorbidities were extracted from the physician’s medical record. Subject demographics are shown in Table 1.
Weekly study visits took place in an outpatient wound clinic. Subjects randomized to the therapeutic ultrasound group were topically treated with 20 kHz, 100 mW/cm2 for 15 min while patients in the sham group were treated with a non-operating device for 15 min. The ultrasound devices used in this study have been previously described in detail in Ngo et al. (10). After completion of the ultrasound or sham treatment, the vascular surgeon performed tissue debridement using a surgical scalpel and participants received standard wound care. All tissue samples analyzed in this study were collected 1 week following ultrasound or sham treatment. Subjects who did not return to the clinic 1 week following ultrasound or sham treatment were considered lost to follow up.
Of the 48 subjects enrolled in the study, 16 subjects were lost to follow up resulting in 32 tissue samples processed (Figure 1). Tissue samples were processed using the methods described in subsequent sections. Of the 32 processed samples, 16 did not yield RNA of sufficient quantity, and of those, 9 did not yield RNA of sufficient quality for library preparation. Thus, 7 tissue samples were ultimately analyzed, and these comprised 3 from the ultrasound treatment group and 4 from the sham group. Healing outcome was determined by considering complete wound closure at week 16 or earlier as healed and incomplete wound closure at that time point as non-healed.
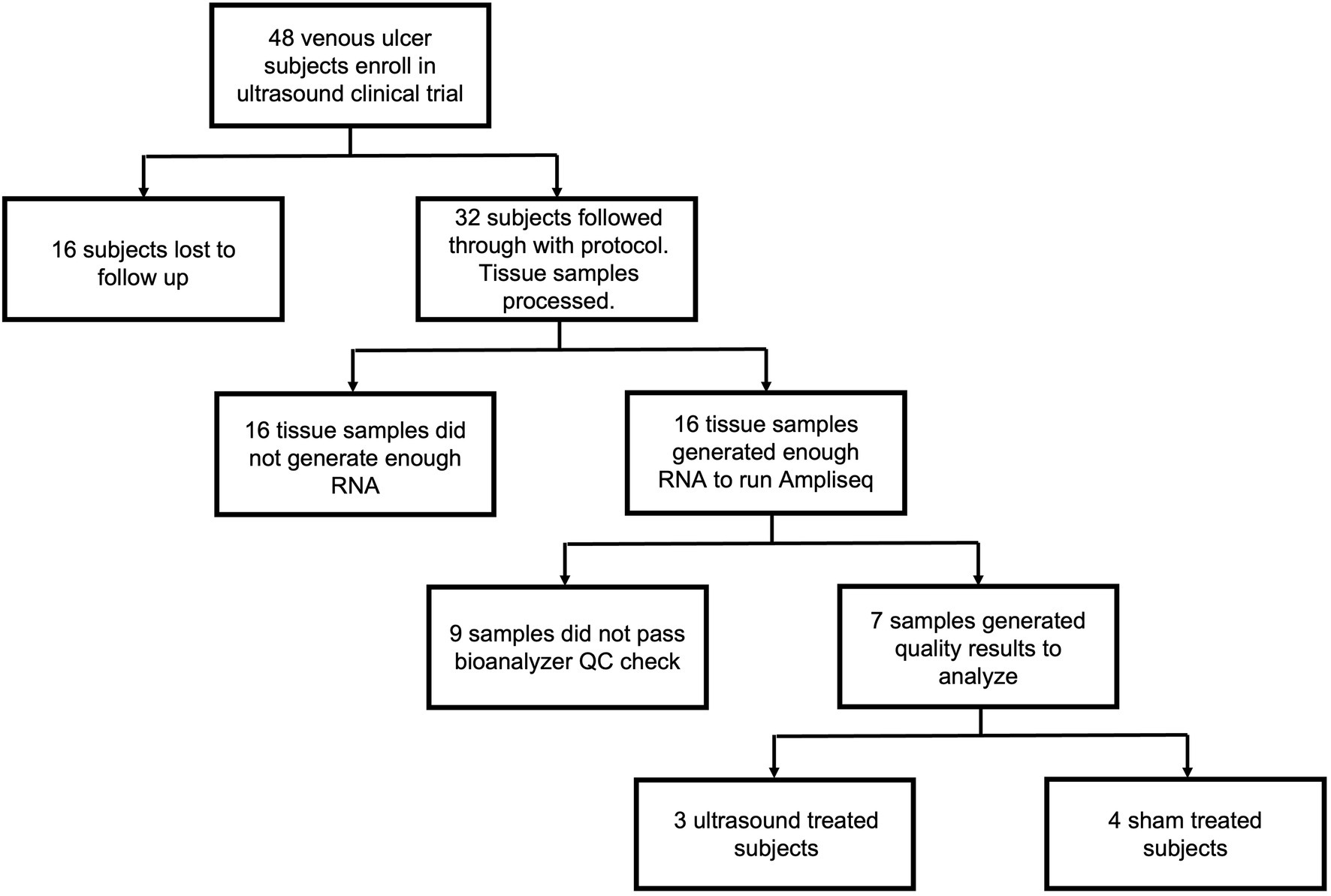
Figure 1. Flow chart outlining subject enrollment, sample collection, quality control, and final gene expression analysis for ultrasound-treated subjects (n = 3) and sham-treated subjects (n = 4).
Debrided tissue collection
All participants underwent VLU debridement by a vascular surgeon as part of standard wound care regimen during each visit. Sharp debridement was conducted using a scalpel or curette after removing the overlying biofilm and necrotic tissue. The entirety of the debrided tissue was collected, as opposed to only wound bed or edge, because separate debridement of different wound regions is not routinely conducted in clinical practice, although it is noted that there are biological differences between the wound regions (19, 20). For similar reasons, intact skin tissue was not collected. Finally, wound histology was not conducted so that sample collection methods would not deviate from the normal standard of care debridement procedures. In addition, all of the debrided tissue was required to isolate sufficient quantity and quality of RNA for gene expression profiling. Debrided tissue was immediately stored in RNAlater® Solution (Ambion, Austin, TX, United States) at 4°C overnight before long term storage at −80°C prior to RNA extraction and gene expression analysis.
RNA extraction
The collected tissue samples were thawed at room temperature and total RNA was extracted using chloroform and Trizol method followed by purification with the Qiagen RNeasy kit (Qiagen, Inc., CA, United States) according to the manufacturer’s instructions. DNA was inactivated using DNAse I Amplification Grade (Invitrogen, Carsbad, CA, United States). Tissue samples that yielded less than 10 ng of total RNA were omitted from further analysis because of insufficient RNA quantity, and this included 16 out of the 32 tissue samples processed.
Library preparation
cDNA libraries were constructed using Ion Ampliseq Transcriptome Human Gene Expression Kit from Thermo Fisher (MA, United States, Cat# A26325) according to manufacturer’s recommended protocol. Briefly, 10 ng of total RNA was reverse transcribed at 42°C for 30 min. After reverse transcription the cDNA was amplified by PCR using Ion Ampliseq Transcriptome Human Gene Expression Core Panel primers that amplified the specific targets (step 1: 99°C for 2 min; step 2: 99°C for 15 s, 60°C for 16 min for 16 cycles; step 3: Hold at 10°C). Next, the primers were partially digested as directed. Following the partial digestion of primers, adapters and bar codes were ligated to the cDNA. The cDNA was then purified using AMPure XP reagent (Beckman Coulter IN, United States, Cat#A63881) and the recommended protocol. The purified cDNA libraries were then amplified by PCR using 1X Library Amp Mix and 2 uL of 25X Library Amp Primers with the conditions as follows: Step 1: 98°C for 2 min; Step 2: 98°C for 15 s, 64°C for 1 min; steps 2–3 for 5 cycles and then hold at 10°C. The amplified cDNA libraries were purified using Nucleic Acid binding beads, binding buffers and processed on Agilent 2,100 Bioanalyzer (Agilent CA, United States) to determine the yield and size distribution of each library.
Bioanalyzer library quality control
The quality of each final library was assessed using the Agilent® dsDNA High Sensitivity Kit (Agilent CA, United States, CAT#5067–4,626). All samples had very low RIN numbers (range: 1–3), but they did not differ between the ultrasound and sham groups, so we further investigated sample quality by examining the DNA library directly. A typical Ampliseq Trancriptome library is shown in Supplementary Figure S1. The molar concentration is determined and lower percentages of DNA (50–160 bp range) indicate a higher quality library. Those with less than 50% of the library in this region were deemed acceptable for templating and sequencing. 13 out of the 16 samples were deemed acceptable after the bioanalyzer quality control and were templated and sequenced.
Templating, enrichment, and sequencing
Approximately 50 pM of pooled barcoded libraries were used for templating using Thermo Fisher Ion 550 Kit-Chef (ThermoFisher, MA, United States, Cat.# A34541) and the manufacturer’s recommended protocol. Briefly, 50 pM of pooled libraries were combined and 25 ul of each sample was loaded onto the Ion Chef. Next, all reagents for the Ion 550 Chef Kit were loaded onto the Ion Chef (ThermoFisher, MA, United States) and the run was performed. The Ion Chef templated, enriched, and loaded the sample onto a 550 chip. After 15 h the Chef paused, and QC was performed on the unenriched samples. The beads were then isolated, and quality assessment was performed on the Qubit instrument (ThermoFisher, MA, United States) to determine the percentage of beads that were polyclonal. All samples at this point passed QC. After polyclonal assessment the Ion Chef resumed running and loaded the samples onto a 550 chip. The loaded chip was then placed into an Ion S5 sequencer, and the run was started using an Ion torrent Ampliseq transcriptome run plan that was configured based on type of library, species, number of run flows required, type of plug-in required, adapter-trimming as well as other parameters specific to the Ampliseq transcriptome run.
Alignment and data analysis
After completion of the run, the raw sequence files (fastq) were aligned to the human transcriptome (hg19) reference sequences by the StrandNGS (CA, United States) software using the default parameters. The genes and transcript annotations that were used were retrieved from the Ensembl data base. Aligned BAM files were used for further analysis. Quality control was assessed by the Strand NGS program, which determined the pre-and post-alignment quality of the reads for each sample. The aligned reads were then filtered based on alignment score (≥90), match count (≤1), number of N’s allowed in reads (≤0). Aligned reads that failed these vendor quality control guidelines were removed. Only 7 of the 13 samples sequenced showed aligned sequence greater than 60% (vendor quality control guideline). After this filtering, the aligned reads were normalized and quantified using the DEseq algorithm by the StrandNGS program. Next statistical analysis was performed using the Benjamini Hochberg false discovery rate and Moderated T-test to determine significant differentially expressed genes (p < 0.05) between ultrasound-treated and sham-treated groups. After significant DEGs were identified, significant fold change between ultrasound treated subjects and sham treated subjects was determined and significant genes with a fold change >4 were clustered. Additionally, genes that had a significant fold change of 2 or higher were listed. Gene Set Enrichment Analysis (GSEA) was performed using the StrandNGS software and Broad Institute databases.
Data availability
Raw and processed data have been deposited in GEO (accession number GSE222503).
Results
Global gene expression analysis
A total of 7 human venous leg ulcer samples were analyzed (n = 3 ultrasound; n = 4 sham) for significantly different gene expression when comparing ultrasound-treated patients to sham-treated subjects. 477 significantly differentially expressed genes (DEGs) were identified with a fold change greater than 2 and p < 0.05 (Figure 2, Supplementary Tables S1, S2). Hierarchical clustering of DEGs showed that the ultrasound-and sham-treated subjects clustered separately. We have also displayed the top 20 upregulated and downregulated protein encoding DEGs (Table 2). Many of these genes are involved in regulation of metabolism (PLA2G2A, MT3, GGT3P, RTBDN, TM4SF5, PGM3, AWAT1, CYP21A2, SLC8B1, HGD, ACSM3, AK8, PISD, PLB1), cell proliferation (CA9, TM4SF5, RORA, USP51, CGREF1, PHACTR3, CDC25A, UBE2V1), and immune cell signaling (GGT3P, IFIT1B, GPA33, RUFY4, P2RY12, CLEC1B).
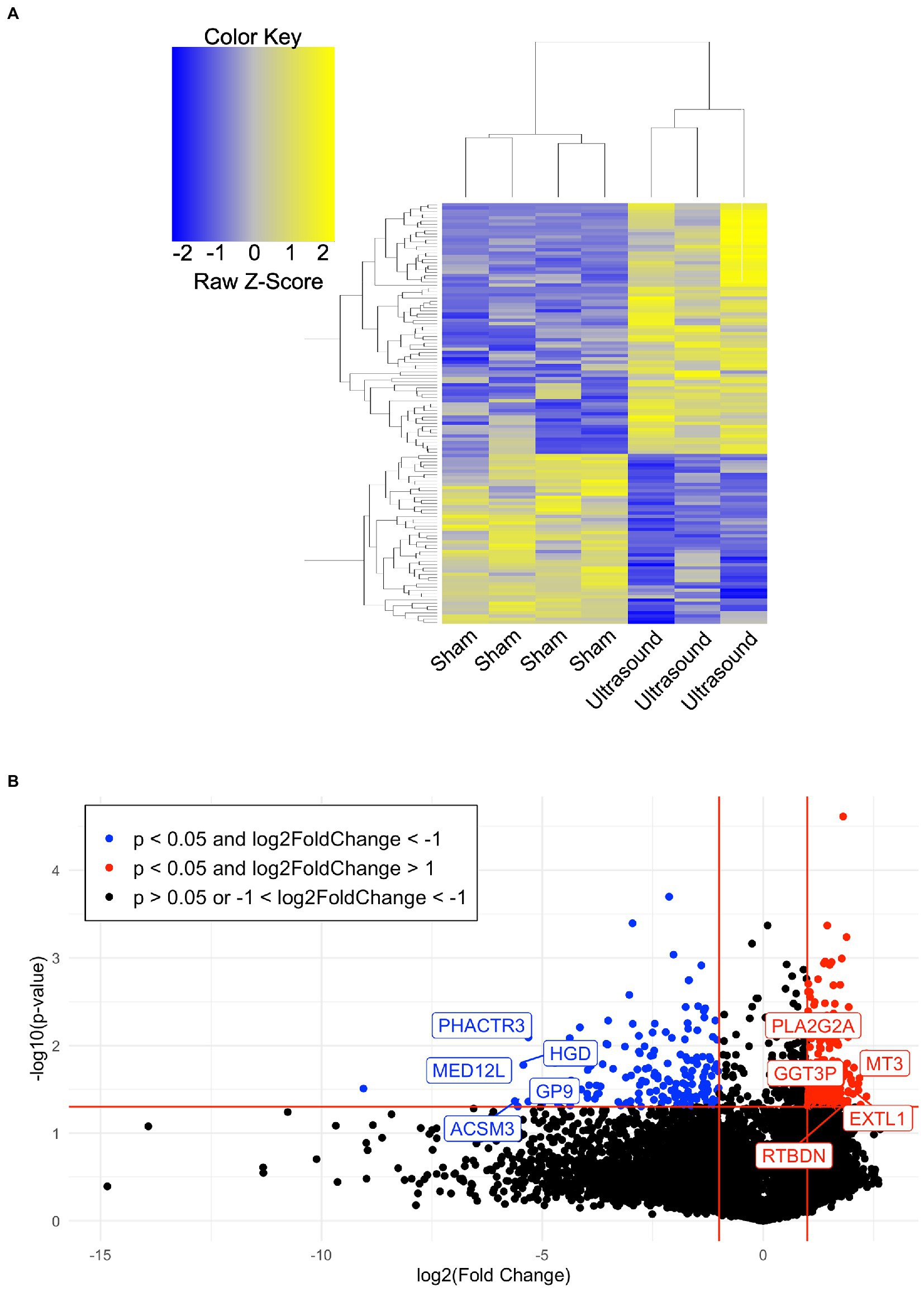
Figure 2. (A) Hierarchical clustering of DEGs (p < 0.05, absolute value of fold change >4) when comparing the ultrasound to sham group. (B) Volcano plot of value of p vs. fold change (FC) of gene expression in ultrasound compared to sham group. Lines indicate FC greater than 2 and p < 0.05 using moderated T-test with Benjamini Hochberg FDR multiple correction test.
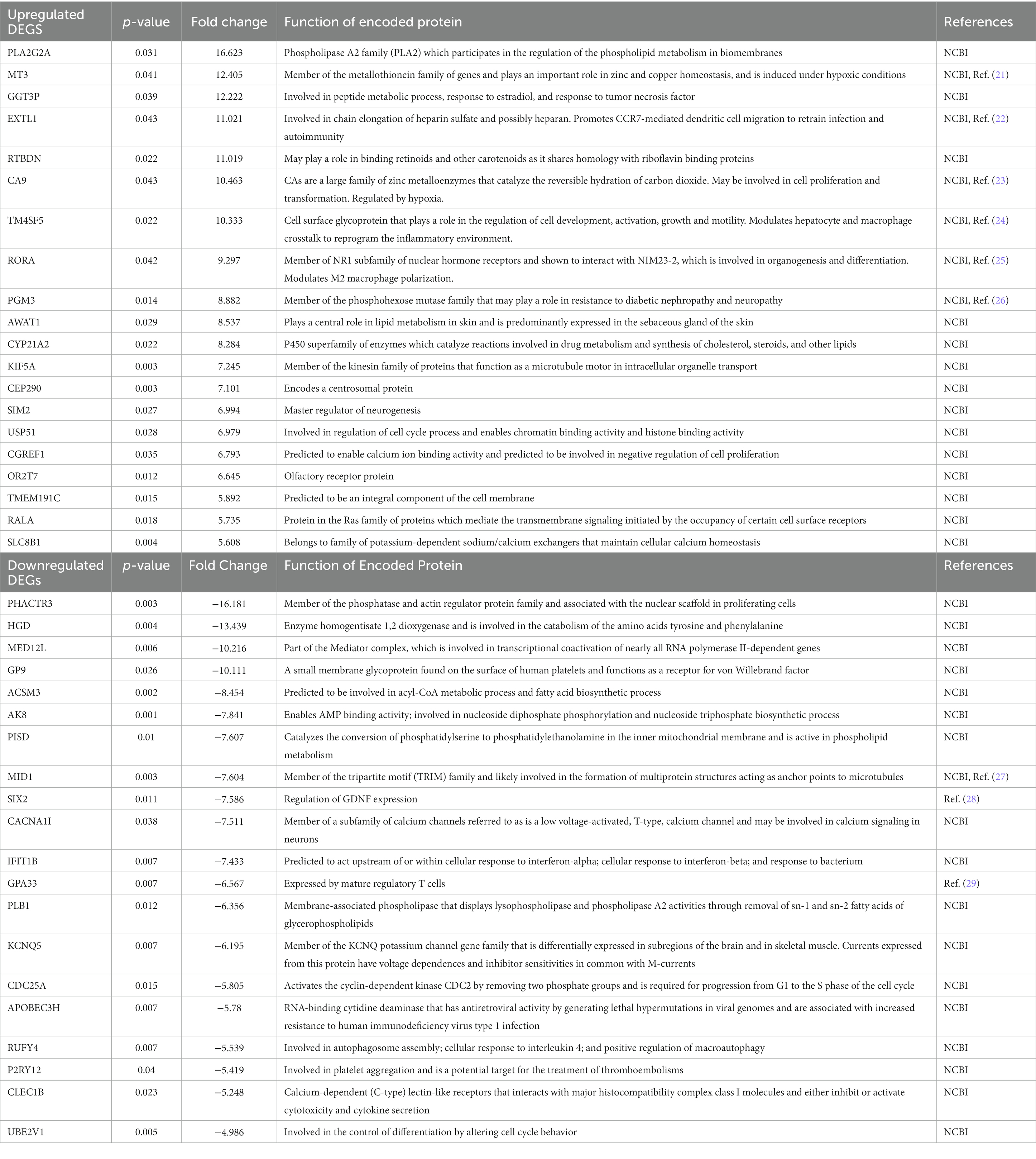
Table 2. Top 20 upregulated and downregulated protein encoding DEGs (p < 0.05) in ultrasound-treated vs. sham-treated wounds.
We employed gene set enrichment analysis to determine any significantly enriched sets of genes with similarly grouped functions, using gene sets listed on the Broad Institute database. 20 significantly enriched gene sets from upregulated DEGs were identified (Figure 3A). Ten DEGs that most contributed to the enriched gene sets are shown in Figure 3B. Four significantly enriched gene sets from downregulated DEGs were identified (Figure 4A) with 10 DEGs that most contributed to the enriched gene sets (Figure 4B). Of note, the most enriched gene set from downregulated DEGs was the inflammatory response gene set, which includes 761 hallmark genes.
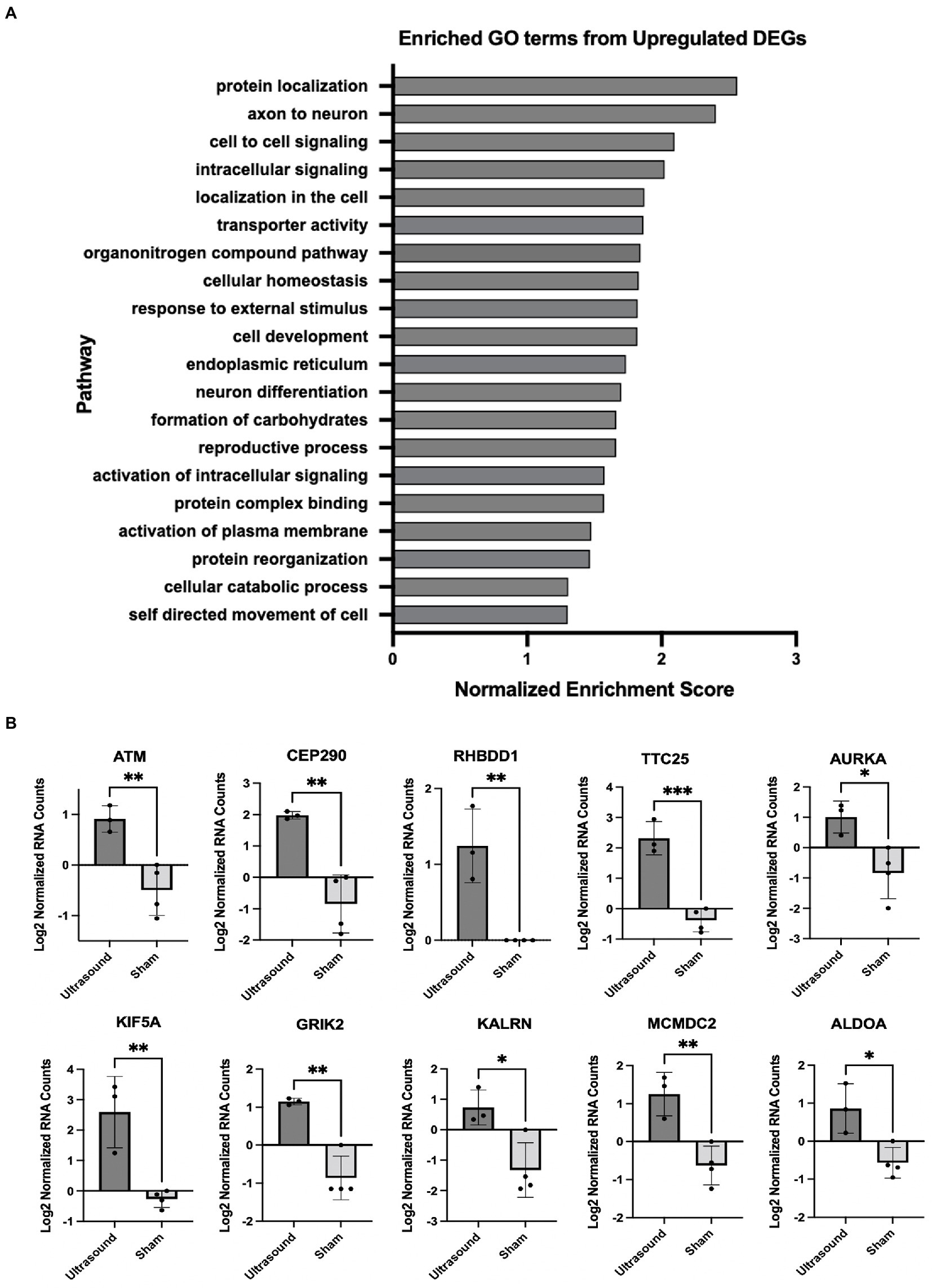
Figure 3. (A) Enriched GO terms identified from significantly upregulated DEGs (p < 0.05). (B) DEGs that contributed the most to the enriched gene sets; *p < 0.05; **p < 0.01, ***p < 0.001.
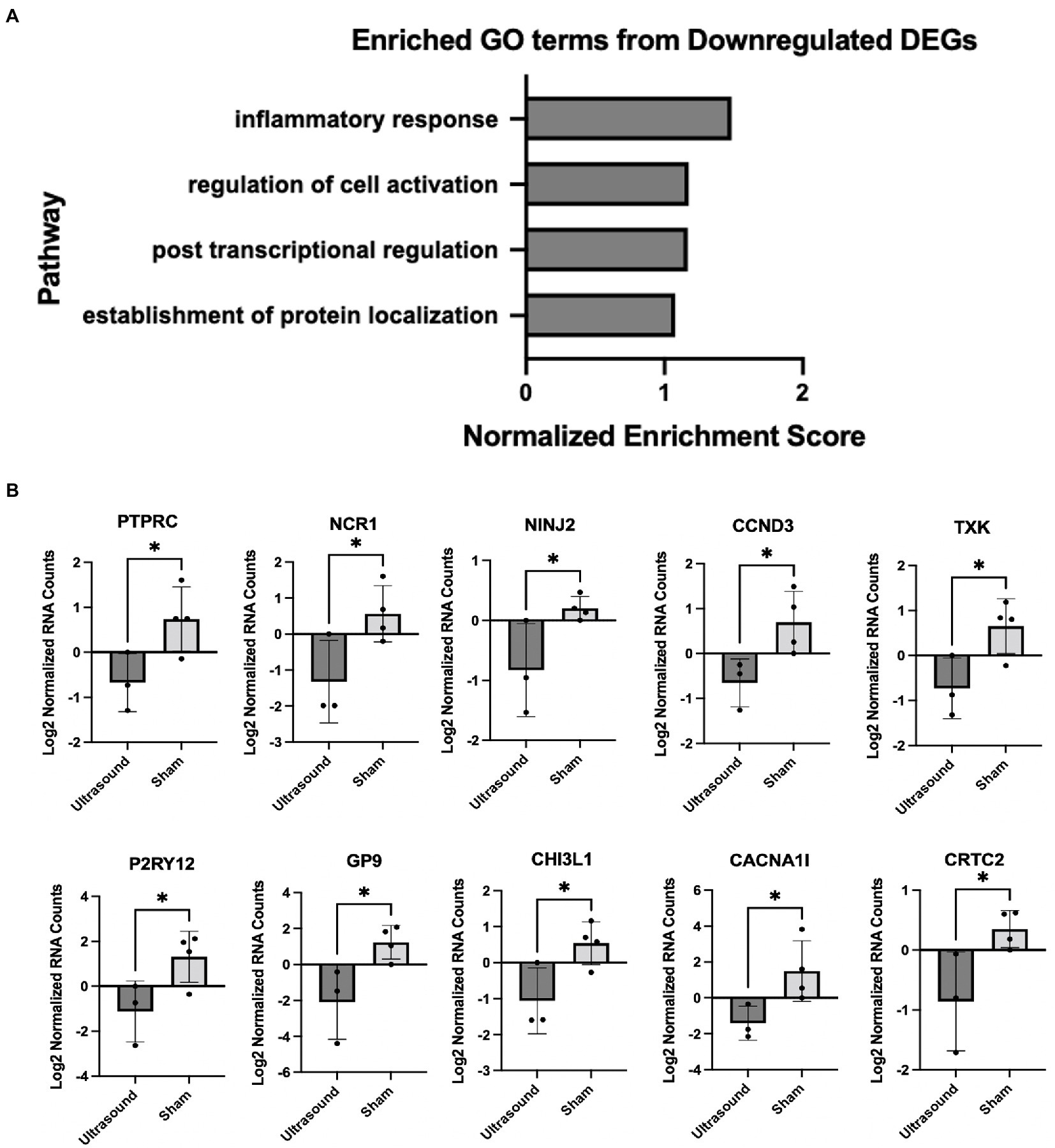
Figure 4. (A) Enriched gene sets identified from significantly downregulated DEGs (p < 0.05). (B) DEGs that contributed the most to the enriched gene sets; *p < 0.05.
Discussion
Gene set enrichment analyses of human chronic venous leg wound tissue showed significant differences in participants treated with LFLI US compared to participants treated with a sham device. These results are important because they provide the foundation for future studies to investigate possible mechanisms of action behind the beneficial effects of ultrasound. In particular, the inflammatory response gene set was significantly downregulated in the ultrasound-treated patients compared to sham-treated patients as early as 1 week after treatment. These results suggest that a decrease in inflammation is a possible biological mechanism of ultrasound-mediated chronic venous leg ulcer wound healing, although this hypothesis will need to be rigorously tested in follow-up studies. These data contribute to the understanding of how therapeutic ultrasound modulates cells in human chronic wounds and are particularly essential in the absence of adequate animal models for human VLUs. With additional understanding of the biological mechanism of therapeutic ultrasound on cells, ultrasound parameters can be tailored to maximize chronic wound healing, ultimately leading to an increase in the quality of life for patients and a decrease in wound care associated costs.
Our findings that LFLI US significantly downregulates the inflammatory response gene set corroborates the findings of Yang et al., who found low-intensity ultrasound to globally activate anti-inflammatory genes revealed through Ingenuity Pathway Analysis (IPA) on in-vitro experimental data found in a widely available microRNA database (30). Furthermore, Hundt et al. reported a significant downregulation in genes associated with inflammation after high-intensity (total acoustical power of 120 W) ultrasound treatment applied directly to muscle tissue in mice (31). The downregulation of inflammation would be expected to be beneficial for chronic wound healing as sustained inflammation has been shown to be a major impediment of healing in chronic wounds (32, 33) and decreasing inflammation has been associated with improved healing outcomes in diabetic foot ulcers (17, 34). Modulation of inflammation has been suggested to be a primary mediator of ultrasound-stimulated bone fracture healing (12). Additionally, the top protein encoding genes (Table 2) can be used to motivate future studies in understanding pathway specific mechanisms. For example, many of the upregulated genes in ultrasound-treated wounds were related to metabolism, which has been recently linked to regulation of wound healing in animal models of diabetic wounds (35). Some of these genes are also related to inflammation via metabolic regulation. For example, the top upregulated gene, PLA2G2A, is a member of the PLA2 phosolipase family of proteins that regulate synthesis of bioactive lipids that regulate inflammation in wound healing (36). The second most upregulated gene, MT3, encodes metallothionein 3, which is critical for inhibiting the pro-inflammatory phenotype and promoting the “pro-healing” phenotype of macrophages (21). In considering the literature and our findings, we suggest that the possible biological mechanisms of ultrasound-assisted healing may include regulation of inflammation. Additional studies will need to be conducted to determine specific pathways responsible for these effects.
Although our findings identified several statistically significant genes and pathways that were affected by LFLI US, this study should be considered preliminary. While the study started with the recruitment of 48 venous ulcer patients, due to patient loss to follow up and poor tissue quality, the final analysis included samples from only 3 ultrasound-treated and 4 sham-treated subjects, thus limiting the statistical power of the study. Moreover, 0 of the 3 ultrasound-treated subjects and 2 of the 4 sham-treated subjects healed during the study, so it is possible that healing outcome could have influenced the results. However, this risk may be partially reduced by the fact that the samples were collected at 1 week after initial treatment and several weeks away from healing or the end of the study. Additionally, differences in subject demographics could have influenced the results, and the low number of replicates prevented statistical analysis of each factor between groups. Much larger studies are required to definitively conclude that the observed effects result from ultrasound treatment and not some other factor including healing outcome. Technical limitations include that this study analyzed whole tissue, which limits the understanding of individual cell types that contribute to the observed differences, and that the selected platform (Ampliseq) only analyzes protein-encoding genes as opposed to the whole transcriptome. Hence, future work exploring cell specific mechanisms should utilize single cell analysis instead of whole tissue and using whole transcriptomic sequencing if possible. Finally, we only analyzed tissue collected at a single time point (1 week after initial treatment), and other changes would be expected to occur over time. These limitations notwithstanding, these results show that low-frequency, low-intensity therapeutic ultrasound significantly affects human chronic wounds on the cellular level and these results can be used to further develop therapeutic ultrasound and maximize the potential for healing.
Conclusion
Ultrasound treatment influenced cellular processes in human chronic VLUs as early as 1 week after treatment. Wound tissue samples from subjects treated with ultrasound exhibited increased cell–cell signaling and decreased inflammatory response gene sets. These results provide a foundation for future studies examining these potential mechanisms in depth.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving human participants were reviewed and approved by Drexel University Institutional Review Board. The patients/participants provided their written informed consent to participate in this study.
Author contributions
OB, KS, PL, MW, RD-G, and MN designed the study. OB and ZA conducted the experiments and analyzed the data. OB, ZA, and KS wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by NIH F31 AR074847 and NIH R01 NR015995. The content of this study is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Acknowledgments
The authors would like to thank clinical physicians, nurses, and staff for their technical assistance in collecting the human wound tissue and the patients for their participation in this study.
Conflict of interest
PL declares pending patent applications related to the ultrasound technology described in this work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1144182/full#supplementary-material
References
1. Fonder, MA, Lazarus, GS, Cowan, DA, Aronson-Cook, B, Kohli, AR, and Mamelak, AJ. Treating the chronic wound: a practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol. (2008) 58:185–206. doi: 10.1016/j.jaad.2007.08.048
2. Ligi, D, Croce, L, and Mannello, F. Chronic venous disorders: the dangerous, the good, and the diverse. Int J Mol Sci. (2018) 19:2544. doi: 10.3390/ijms19092544
3. Zhao, R, Liang, H, Clarke, E, Jackson, C, and Xue, M. Inflammation in chronic wounds. Int J Mol Sci. (2016) 17:2085. doi: 10.3390/ijms17122085
4. Nicolaides, AN. The Most severe stage of chronic venous disease: an update on the Management of Patients with venous leg ulcers. Adv Ther. (2020) 37:19–24. doi: 10.1007/s12325-020-01219-y
5. Valencia, IC, Falabella, A, Kirsner, RS, and Eaglstein, WH. Chronic venous insufficiency and venous leg ulceration. J Am Acad Dermatol. (2001) 44:401–24. doi: 10.1067/mjd.2001.111633
6. Franks, P, B, J, Collier, M, Gethin, G, Haesler, E, Jawien, A, et al. Management of Patients with Venous leg Ulcers: challenges and current best practices. J Wound Care. (2016) 25:S1–S67.
7. Weichenthal, M, Mohr, P, Stegmann, W, and Breitbart, EW. Low-frequency ultrasound treatment of chronic venous ulcers. Wound Repair Regen. (1997) 5:18–22. doi: 10.1046/j.1524-475X.1997.50107.x
8. Samuels, JA, Weingarten, MS, Margolis, DJ, Zubkov, L, Sunny, Y, Bawiec, CR, et al. Low-frequency (<100 kHz), low-intensity (<100 mW/cm(2)) ultrasound to treat venous ulcers: a human study and in vitro experiments. J Acoust Soc Am. (2013) 134:1541–7. doi: 10.1121/1.4812875
9. Lyu, W, Ma, Y, Chen, S, Li, H, Wang, P, Chen, Y, et al. Flexible ultrasonic patch for accelerating chronic wound healing. Adv Healthc Mater. (2021) 10:e2100785. doi: 10.1002/adhm.202100785
10. Ngo, O, N, E, Gunasekaran, V, Sankar, P, Putterman, M, Lafontant, A, et al. Development of low frequency (20-100 kHz) clinically viable ultrasound applicator for chronic wound treatment. IEEE transactions ultrasound ferroelectric frequency. Control. (2019) 66:572–80. doi: 10.1109/TUFFC.2018.2836311
11. Maan, ZN, Januszyk, M, Rennert, RC, Duscher, D, Rodrigues, M, Fujiwara, T, et al. Noncontact, low-frequency ultrasound therapy enhances neovascularization and wound healing in diabetic mice. Plast Reconstr Surg. (2014) 134:402e–11e. doi: 10.1097/PRS.0000000000000467
12. Harrison, A, Lin, S, Pounder, N, and Mikuni-Takagaki, Y. Mode & mechanism of low intensity pulsed ultrasound (LIPUS) in fracture repair. Ultrasonics. (2016) 70:45–52. doi: 10.1016/j.ultras.2016.03.016
13. Pounder, NM, and Harrison, AJ. Low intensity pulsed ultrasound for fracture healing: a review of the clinical evidence and the associated biological mechanism of action. Ultrasonics. (2008) 48:330–8. doi: 10.1016/j.ultras.2008.02.005
14. Dunn, L, Prosser, HC, Tan, JT, Vanags, LZ, Ng, MK, and Bursill, CA. Murine model of wound healing. J Vis Exp. (2013) 75:e50265. doi: 10.3791/50265
15. Grada, A, Mervis, J, and Falanga, V. Research techniques made simple: animal models of wound healing. J Invest Dermatol. (2018) 138:2095–2105.e1. doi: 10.1016/j.jid.2018.08.005
16. Takao, K, and Miyakawa, T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. (2015) 112:1167–72. doi: 10.1073/pnas.1401965111
17. Bajpai, A, Nadkarni, S, Neidrauer, M, Weingarten, MS, Lewin, PA, and Spiller, KL. Effects of non-thermal, non-cavitational ultrasound exposure on human diabetic ulcer healing and inflammatory gene expression in a pilot study. Ultrasound Med Biol. (2018) 44:2043–9. doi: 10.1016/j.ultrasmedbio.2018.05.011
18. Wang, J, Rieder, SA, Wu, J, Hayes, S, Halpin, RA, de los Reyes, M, et al. Evaluation of ultra-low input RNA sequencing for the study of human T cell transcriptome. Sci Rep. (2019) 9:8445. doi: 10.1038/s41598-019-44902-z
19. Charles, CA, Tomic-Canic, M, Vincek, V, Nassiri, M, Stojadinovic, O, Eaglstein, WH, et al. A gene signature of nonhealing venous ulcers: potential diagnostic markers. J Am Acad Dermatol. (2008) 59:758–71. doi: 10.1016/j.jaad.2008.07.018
20. Theocharidis, G, Thomas, BE, Sarkar, D, Mumme, HL, Pilcher, WJR, Dwivedi, B, et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat Commun. (2022) 13:181. doi: 10.1038/s41467-021-27801-8
21. Chowdhury, D, Alrefai, H, Landero Figueroa, JA, Candor, K, Porollo, A, Fecher, R, et al. Metallothionein 3 controls the phenotype and metabolic programming of alternatively activated macrophages. Cell Rep. (2019) 27:3873–3886.e7. doi: 10.1016/j.celrep.2019.05.093
22. Liu, J, Cheng, Y, Zhang, X, Chen, Y, Zhu, H, Chen, K, et al. Glycosyltransferase Extl1 promotes CCR7-mediated dendritic cell migration to restrain infection and autoimmunity. Cell Rep. (2023) 42:111991. doi: 10.1016/j.celrep.2023.111991
23. M Said, H, T Supuran, C, Hageman, C, Staab, A, Polat, B, Katzer, A, et al. Modulation of carbonic anhydrase 9 (CA9) in human brain cancer. Curr Pharm Des. (2010) 16:3288–99. doi: 10.2174/138161210793429788
24. Kim, E, Um, H, Park, J, Jung, JW, Kim, JE, Lee, H, et al. TM4SF5-dependent crosstalk between hepatocytes and macrophages to reprogram the inflammatory environment. Cell Rep. (2021) 37:110018. doi: 10.1016/j.celrep.2021.110018
25. Wang, ZG, Deng, MS, Su, JQ, Liu, DB, and Zhou, Y. Exosomal miR-181a-5p derived from SAOS-2 cells promotes macrophages M2 polarization by targeting RORA. Kaohsiung J Med Sci. (2022):124–33. doi: 10.1002/kjm2.12623.
26. Lee, H, Cai, F, Kelekar, N, Velupally, NK, and Kim, J. Targeting PGM3 as a novel therapeutic strategy in KRAS/LKB1 co-mutant lung cancer. Cells. (2022) 11:176. doi: 10.3390/cells11010176
27. Baldini, R, Mascaro, M, and Meroni, G. The MID1 gene product in physiology and disease. Gene. (2020) 747:144655. doi: 10.1016/j.gene.2020.144655
28. Brodbeck, S, Besenbeck, B, and Englert, C. The transcription factor Six2 activates expression of the Gdnf gene as well as its own promoter. Mech Dev. (2004) 121:1211–22. doi: 10.1016/j.mod.2004.05.019
29. Opstelten, R, de Kivit, S, Slot, MC, van den Biggelaar, M, Iwaszkiewicz-Grześ, D, Gliwiński, M, et al. GPA33: a marker to identify stable human regulatory T cells. J Immunol. (2020) 204:3139–48. doi: 10.4049/jimmunol.1901250
30. Yang, Q, Nanayakkara, GK, Drummer, C, Sun, Y, Johnson, C, Cueto, R, et al. Low-intensity ultrasound-induced anti-inflammatory effects are mediated by several new mechanisms including gene induction, Immunosuppressor cell promotion, and enhancement of exosome biogenesis and docking. Front Physiol. (2017) 8:818. doi: 10.3389/fphys.2017.00818
31. Hundt, W, Y, E, Steinbach, S, Bednarski,, and Guccione, S. Comparison of continuous vs. pulsed focused ultrasound in treated muscle tissue as evaluated by magnetic resonance imaging, histological analysis, and microarray analysis. Mol Imaging. (2008) 18:993–1104. doi: 10.1007/s00330-007-0848-y
32. Sindrilaru, A, Peters, T, Wieschalka, S, Baican, C, Baican, A, Peter, H, et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest. (2011) 121:985–97. doi: 10.1172/JCI44490
33. Liu, Z, Zhang, L, Toma, MA, Li, D, Bian, X, Pastar, I, et al. Integrative small and long RNA omics analysis of human healing and nonhealing wounds discovers cooperating microRNAs as therapeutic targets. elife. (2022) 11:e80322. doi: 10.7554/eLife.80322
34. Nassiri, S, Zakeri, I, Weingarten, MS, and Spiller, KL. Relative expression of Proinflammatory and Antiinflammatory genes reveals differences between healing and nonhealing human chronic diabetic foot ulcers. J Invest Dermatol. (2015) 135:1700–3. doi: 10.1038/jid.2015.30
35. Willenborg, S, Sanin, DE, Jais, A, Ding, X, Ulas, T, Nüchel, J, et al. Mitochondrial metabolism coordinates stage-specific repair processes in macrophages during wound healing. Cell Metab. (2021) 33:2398–2414.e9. doi: 10.1016/j.cmet.2021.10.004
Keywords: venous leg ulcer, ultrasound, gene expression, ampliseq, translational medicine
Citation: Boerman O, Abedin Z, DiMaria-Ghalili RA, Weingarten MS, Neidrauer M, Lewin PA and Spiller KL (2023) Gene expression changes in therapeutic ultrasound-treated venous leg ulcers. Front. Med. 10:1144182. doi: 10.3389/fmed.2023.1144182
Edited by:
Piul Rabbani, Grossman School of Medicine, New York University, United StatesReviewed by:
Catherine Anders, United States Department of Veterans Affairs, United StatesGeorgios Theocharidis, Beth Israel Deaconess Medical Center and Harvard Medical School, United States
Copyright © 2023 Boerman, Abedin, DiMaria-Ghalili, Weingarten, Neidrauer, Lewin and Spiller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter A. Lewin, bGV3aW5wYUBkcmV4ZWwuZWR1; Kara L. Spiller, c3BpbGxlckBkcmV4ZWwuZWR1
 Olivia Boerman
Olivia Boerman Zahidur Abedin
Zahidur Abedin Rose Ann DiMaria-Ghalili
Rose Ann DiMaria-Ghalili Michael S. Weingarten5
Michael S. Weingarten5 Peter A. Lewin
Peter A. Lewin Kara L. Spiller
Kara L. Spiller