- 1Harvey Cushing Neurooncology Research Laboratories, Department of Neurosurgery, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
- 2Department of Molecular Biology, Massachusetts General Hospital, Boston, MA, United States
- 3Molecular Biology Core Facilities, Dana-Farber Cancer Institute, Boston, MA, United States
- 4Candel Therapeutics, Needham, MA, United States
- 5Department of Neurology and Interdisciplinary Neuro-Oncology, University Hospital Tübingen, Hertie Institute for Clinical Brain Research, Eberhard Karls University Tübingen, Tübingen, Germany
- 6Department of Pathology and Laboratory Medicine, Legorreta Cancer Center, Brown University, Providence, RI, United States
Rationale: CAN-2409 is a locally delivered oncolytic therapy, which results in vaccination against the injected tumor. CAN-2409 consists of a non-replicating adenovirus armed with the Herpes virus thymidine kinase, which metabolizes ganciclovir into a phosphorylated nucleotide that is incorporated into the tumor cell’s genome, thereby inflicting immunogenic cancer cell death. While CAN-2409’s immunological impact has been well characterized, its effects on the tumor cells transcriptome remains unknown. We compared the transcriptomic landscape after treatment of glioblastoma models with CAN-2409 in vitro and in vivo to assess how the interplay with the tumor microenvironment influences CAN-2409-mediated transcriptome alterations.
Methods: We performed RNA-Seq with CAN-2409 treated patient-derived glioma stem-like cells and tumors of C57/BL6 mice and compared KEGG pathway usage and differential gene expression focusing on immune cell and cytokine profiles. T-cell -killing assays were performed to assess candidate effectors.
Results: PCA analysis showed distinct clustering of control and CAN-2409 samples under both conditions. KEGG pathway analysis revealed significant enrichment for p53 signaling and cell cycle pathway, with similar dynamics for key regulators of both pathways in vitro and in vivo, including MYC, CCNB1, PLK1 and CDC20. Selected alterations (PLK1 and CCNB1) were validated at the protein level. Cytokine expression analysis revealed upregulation of pro-inflammatory IL12a under both conditions; immune cell gene profiling showed reduction of myeloid associated genes. T-cell-killing assays showed increased killing in the presence of IL-12.
Conclusion: CAN-2409 significantly alters the transcriptome both in vitro and in vivo. Comparison of pathway enrichment revealed mutual and differential utilization of pathways under both conditions, suggesting a modulating influence on the cell cycle in tumor cells, and of the tumor microenvironment on the transcriptome in vivo. IL-12 synthesis likely depends on interactions with the tumor microenvironment, and it facilitates CAN-2409 cell killing. This dataset provides potential to understand resistance mechanisms and identify potential biomarkers for future studies.
1. Introduction
Glioblastoma is the most frequent malignant brain tumor with an incidence of 3.19/100,000 in the United States (1). Despite extensive research activity and clinical trials in the recent years, mass debulking surgery followed by concomitant radiochemotherapy with temozolomide (2) remains standard of care. CAN-2409 is a non-replicating adenovirus expressing the Herpes simplex virus-thymidine kinase (tk). When administered together with a prodrug (acyclovir, ganciclovir or valacyclovir), CAN-2409 catalyzes its phosphorylation into “false” nucleotides that become incorporated into tumor cell DNA during repair or mitosis, eventually causing immunogenic tumor cell death associated with an influx of CD3+ T-cells (3). CAN-2409 has been shown to significantly prolong median overall survival compared to SoC in a phase II study for primary glioblastoma (4). A recent clinical trial of CAN-2409 combined with nivolumab demonstrated the induction of a systemic immune response after CAN-2409 administration to the resection bed (NCT03576612,) (5). CAN-2409 inflicts changes upon the transcriptome through incorporation of a toxic nucleotide, but these have not yet been evaluated for assessment of further therapeutic implications. The aim of this work was to characterize the impact of CAN-2409 on the transcriptome using in vitro and in vivo models of glioblastoma both to identify therapeutically relevant alterations with a focus on how the interplay between CAN-2409 and the tumor microenvironment further contributes to transcriptomic modifications.
2. Materials and methods
2.1. Cell culture
In vitro experiments were performed with the glioblastoma stem cell like cell (GSC) line G9_pCDH (6). The cell line was either cultured with Neurobasal Medium (Life technologies), supplemented with B27 (Invitrogen), 1% Glutamax (Invitrogen), 20 ng/mL EGF (Peprotech), 20 ng/mL FGF (Peprotech), Primocin (Invivogen) and Plasmocin (Invivogen) or Dulbecco’s modified Eagle’s Medium (DMEM) (Life technologies), supplemented with 10% heat-inactivated fetal bovine serum, Plasmocin and Primocin (both Invivogen). For in vivo experiments GL261fluc cells were utilized and cultured with DMEM as mentioned above. All cell lines were incubated at 37°C and 5% CO2 and tested for mycoplasma infection on a regular basis.
2.2. Reagents
For in vitro experiments, cells were treated with CAN-2409 (MOI 100) and 10 μg/mL ganciclovir, for in vivo experiments, 3 μL of AdV-tk with 2 × 108 vector particles [vp]/μL were injected intracranially and animals treated with 20 mg/kg bodyweight ganciclovir intraperitoneally 2x/day for 7 days. Both AdV-tk and ganciclovir were kindly provided by Candel Therapeutics Inc. (Needham, MA, United States).
2.3. Animal experiments
For generation of in vivo samples, 100,000 GL261fluc cells were injected intracranially in 2 μL HBSS 2 mm right lateral, 1 mm frontal to the bregma, and 3 mm deep in the brains of albino C57/BL6 mice. Tumor growth was monitored by bioluminescence imaging with the Perkin-Elmer IVIS Lumina 3 after intraperitoneal of injection of D-Luciferin (#LUCK-1G, Gold Biotechnology).
14 days post-tumor implantation, CAN-2409 (treatment group) or HBSS (control) was stereotactically injected intratumorally by, followed by 7 days treatment with ganciclovir in the treatment group as described above. All animal experiments were performed according to BWH Center for Comparative Medicine IACUC guidelines. Animals were euthanized on the third day after end of therapy.
2.4. RNA isolation
For generation of in vitro samples, 500,000 G9_pCDH cells were seeded in triplicates and treated as mentioned before. For generation of in vivo samples, tumors, respectively, normal brain of the corresponding brain region were excised. RNA of both in vitro and in vivo samples was isolated with the RNeasy mini kit (#74104, Qiagen). RNA concentration was measured with NanoDrop and quality verified using an Agilent Bioanalyzer. 3 in vivo RNA samples per group were used for further analyses.
2.5. RNA Seq
Libraries were prepared using Roche Kapa mRNA HyperPrep sample preparation kits from 100 ng of purified total RNA according to the manufacturer’s protocol. The finished dsDNA libraries were quantified by Qubit fluorometer, Agilent TapeStation 2,200, and RT-qPCR using the Kapa Biosystems library quantification kit according to manufacturer’s protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 runs with single-end 75 bp reads by the Dana-Farber Cancer Institute Molecular Biology Core Facilities. Sequenced reads were aligned to the UCSC hg19 reference genome assembly and gene counts were quantified using STAR (v2.5.1b) (7). Differential gene expression testing was performed by DESeq2 (v1.10.1) (8) and normalized read counts (FPKM) were calculated using cufflinks (v2.2.1) (9). RNAseq analysis was performed using the VIPER snakemake pipeline (10).
2.6. Immunoblots
Lysates of G9pCDH cells and G98 cells were generated with 8 M urea buffer (8 M urea, 1% Triton 100, 50 mM Tris-base ph = 7.5, 100 mM NaCl, 1 mM EGTA, 1 mM RDTA, 50 mM beta-glycerophosphate), supplemented with protease inhibitors (Millipore-Sigma #539134) and phosphatase inhibitors (Millipore-Sigma/Roche #4906845001) after treatment with CAN-2409, AdVtk (MOI 500) or mock for 48 h. Protein concentration was determined with Bradford assay. Primary antibodies used were anti-CCNB1 (Cell Signaling Technologies #12231), anti-PLK1 (Cell Signaling Technologies, #4513), anti-c-MYC (Cell Signaling Technologies #5605) and Alexa Fluor 488-labeled anti-GAPDH (#sc-365,062, Santa Cruz), anti-rabbit (Cell Signaling Technologies #7074) was used a as secondary antibody. Band signal intensity quantification was carried out with Bio-Rad Image Lab software in triple measurements.
2.7. T-cell killing assays ± recombinant human IL-12
Human CD8+-T-cells were isolated from healthy blood donors’ PBMCs with Ficoll-Paque PLUS (GE Healthcare) and human CD8+-T-Cell Isolation Kit (Miltenyi Biotec) and subsequently activated with Dynabeads™ Human T-Activator CD3/CD28 for T-Cell Expansion and Activation (Gibco) (ratio 1:1) and human IL-2 (Peprotech) (10 ng/mL). G9_pCDH cells were seeded in a density of 750 cell/well in ultra-low-attachment 96-well plates (Corning). CAN-2409 and/or CD8+-T-cells together with human recombinant IL-12 (R&D Systems) were added 48 h later. Spheres were incubated for 6 days further with daily acquisition of microscope images (Nikon TI, magnification 4x). Sphere area was assessed with ImageJ.
2.8. Statistics
Further statistical analysis of RNA Seq data was performed using R. Analysis of T-cell killing assays was performed with unpaired t-tests in Graph Pad Prism (version 9).
2.9. Graphics
Illustrations were generated with BioRender (publication agreements numbers OS24UGAKQ0, MS24UGAGGH, TI24UGABY0).
3. Results
3.1. CAN-2409 is associated with downregulation of transcription associated pathways in vitro and in vivo
To assess the effect of CAN-2409 on the glioblastoma cell transcriptome, we isolated total RNA after treatment in vitro and in vivo and performed RNA-Seq and compared with mock treated controls (Figure 1A). Initial PCA analysis observed an outlier in the in vivo sample batch, that was excluded for further analyses (Supplementary Figure S1A). Subsequent PCA analysis showed a similar pattern with a clear separation of treatment and control samples in both conditions (Figures 1B,C) with CAN-2409 treatment explaining 87.8 and 89.9% of the sample clustering in vitro and in vivo, respectively.
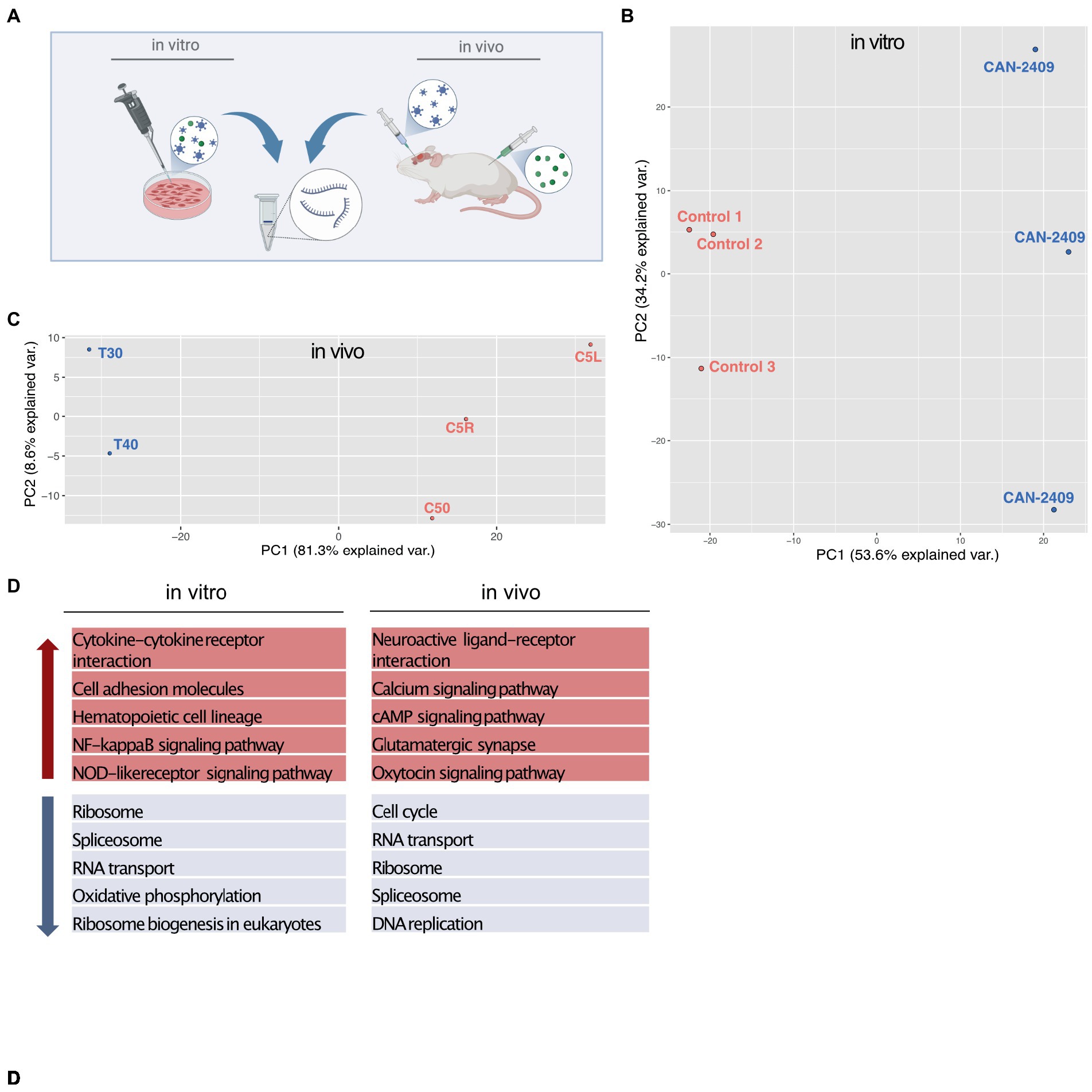
Figure 1. (A) Experimental setup In vitro samples were isolated from G9_pCDH glioma cells after 72 h incubation time with CAN-2409. In vivo samples were generated from brains of albino C57/BL6 mice 3 days after treatment with CAN-2409. (B,C) PCA analysis in vitro and in vivo both sample sets display spatial segregation depending on the treatment, explaining 87.8% of sample clustering in vitro and 89.9% in vivo. D TOP 5 KEGG pathways in vitro and in vivo Up (red arrow) and down (blue arrow) regulated pathways in vitro and in vivo.
KEGG pathway analysis for genes with fold changes >0 (positive) and < 0 (negative) revealed transcription related pathways among the top 10 downregulated pathways (Figure 1D), while only one pathway –cytokine-cytokine receptor interaction appeared to be upregulated in vitro (data not shown). In vivo, we also observed a downregulation of transcription associated pathways, with an additional and highly significant downregulation of the KEGG pathway “cell cycle.” Among the top 10 upregulated pathways in vivo were cell signaling and cell–cell communication pathways (Figure 1D).
3.2. Significant downregulation of p53 pathway after CAN-2409 treatment
To narrow down the extensive CAN-2409 associated effects on the transcriptome, both datasets were further filtered. For any subsequent analyses, only DEGs with an adjusted value of p <0.05 were considered and then divided into upregulated (log2 fold change >1) and downregulated (log2 fold change <1). In vitro, we detected a total of 135 downregulated and 553 upregulated genes; in vivo, 1,284 downregulated and 2,125 upregulated genes were observed (Figure 2A).
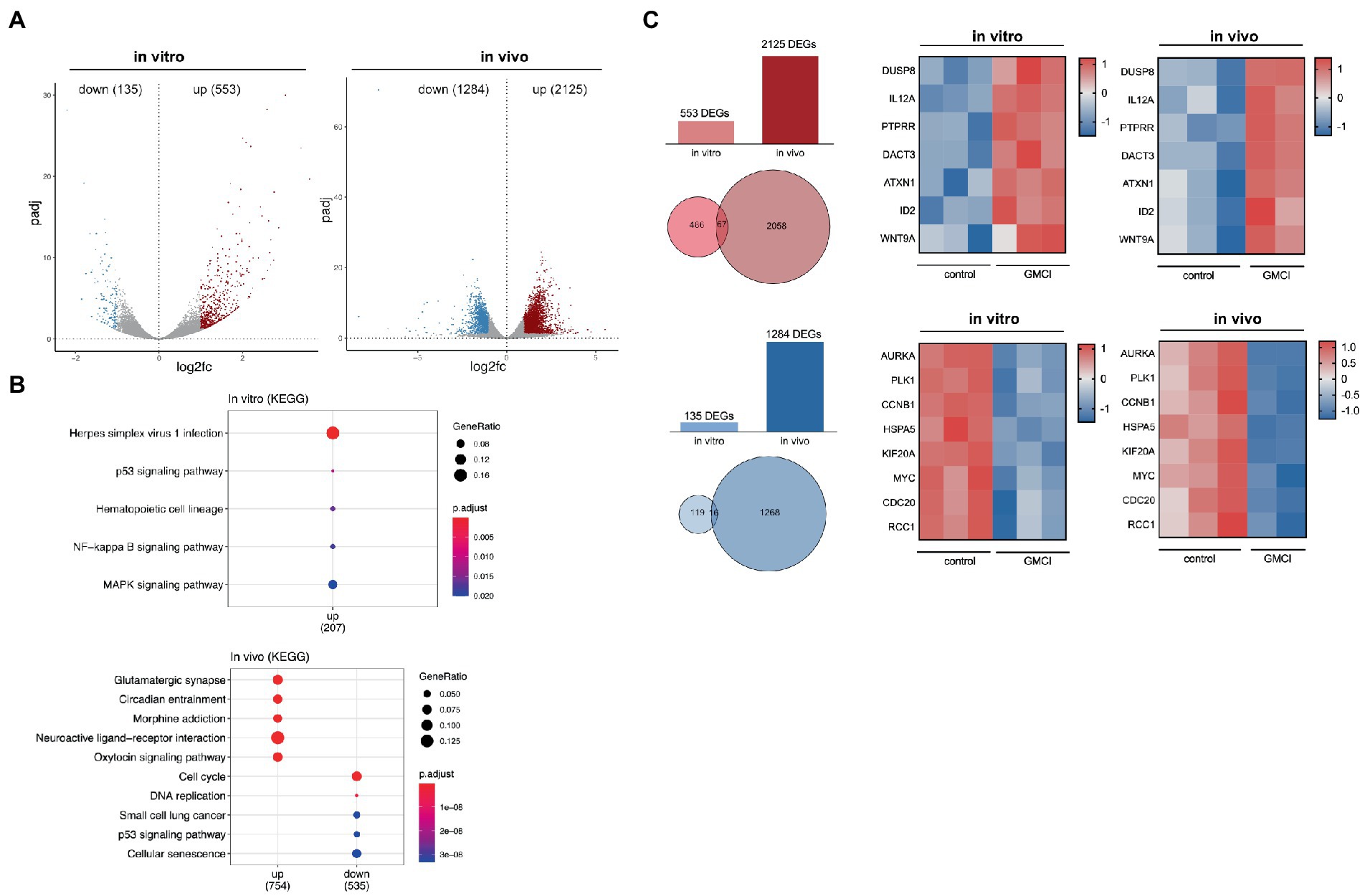
Figure 2. (A) Distribution of DEGs in vitro and in vivo After applying filter criteria (p < 0.05, log2FC > 1/< −1) we detected 135 downregulated and 553 upregulated DEGs in vitro and 1,284 downregulated and 2,125 upregulated DEGs in vivo. (B) TOP 5 KEGG pathways of significant DEGs in vitro and in vivo No downregulated KEGG pathways were detected in vitro, while an enrichment, i.e., for “p53 signaling” and “MAPK signaling” was observed. In vivo, among others downregulation of “Cell cycle,” “DNA replication” and “p53 signaling.” (C) Comparison of significant DEGs in vitro and in vivo 67 common upregulated and 16 downregulated genes were detected, a relevant selection is displayed in adjacent heatmaps.
To track differential pathway enrichment, we applied a second KEGG pathway analysis only with the significant DEGs as mentioned above. In vitro, we observed an upregulation of the KEGG pathways “Herpes simplex virus infection,” “p53 signaling,” “NFkB signaling” and “MAPK signaling,” while no downregulated pathways were detected. In vivo, KEGG pathways “cell cycle,” “DNA replication,” “small cell lung cancer,” “p53 signaling” and “cellular senescence” scored highest among the downregulated, while the upregulated pathways were dominated by “Glutamatergic synapse,” “circadian entrainment,” “morphine addiction,” “neuroactive ligand-receptor interaction” and “oxytocin signaling” (Figure 2B).
Although KEGG pathway enrichment analysis revealed converse regulation for the p53 pathway in vitro and in vivo, strategically important genes that participate in p53 signaling, but also regulate cell cycle progression show similar dynamics: among the 16 commonly downregulated genes were AURKA, MYK, CCNB1, PLK1, HSPA5, RCC1 and CDC20. Within the 67 commonly upregulated genes, we detected DUSP8, PTPRR, DACT3, ATXN1, ID2, WNT9a and the proinflammatory cytokine IL12a (Figure 2C).
3.3. CAN-2409 halts mitotic progression and induces apoptosis
As outlined, CAN-2409 is associated with significant alterations of key molecules of the p53 signaling pathway in vitro, including CDKN1a (triggering G1), GADD45 (blocking progression into S-phase), FAS and CASP10 (inducing apoptosis) (Figure 3A).
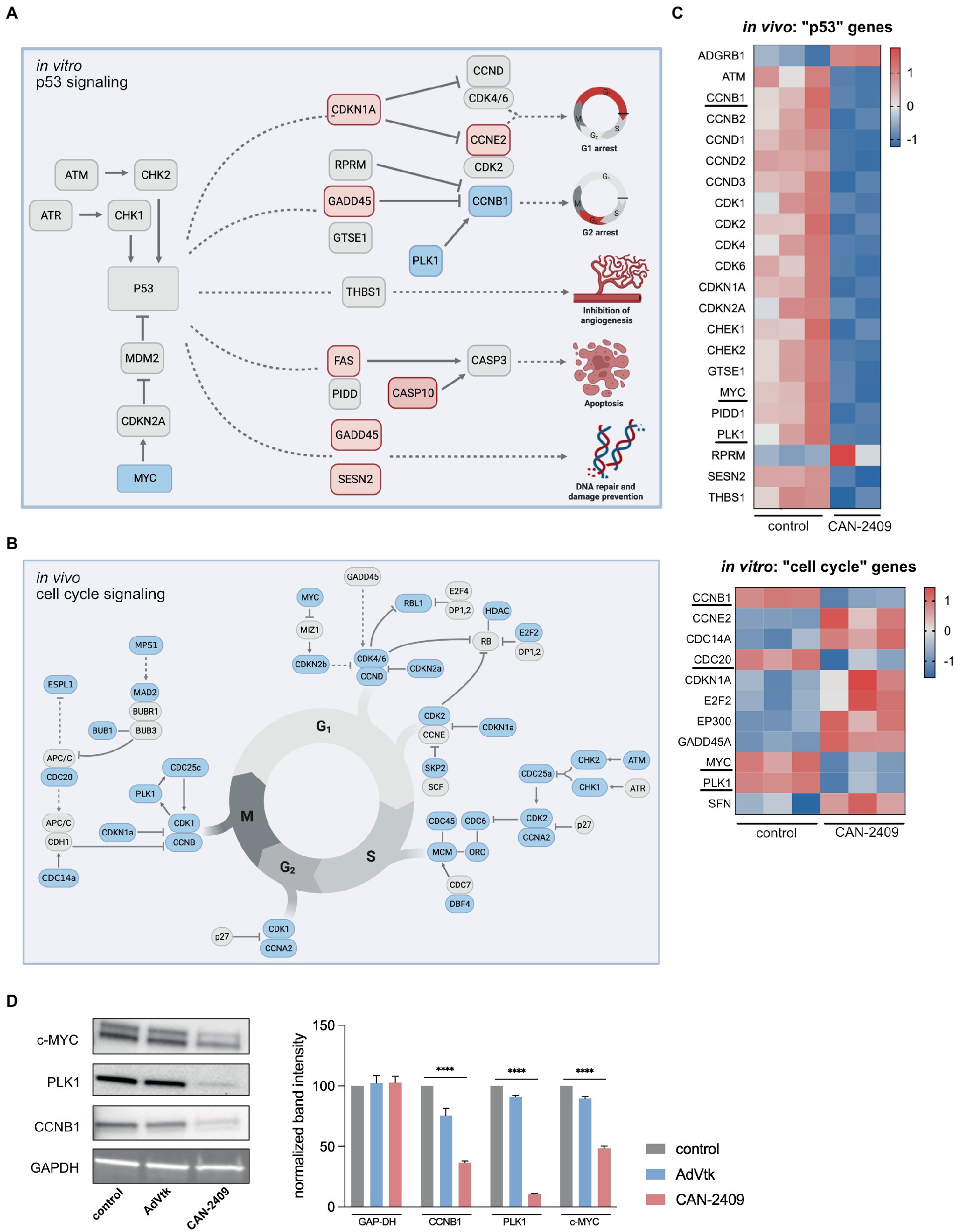
Figure 3. (A) Enrichment of p53 signaling pathway in vitro Key players of p53 signaling are upregulated including CDKN1A, GADD45 and FAS, while MYC is downregulated, culminating in G1/2 arrest, increased apoptosis and DNA repair. Modified after KEGG pathway “p53 signaling.” (B) Reduction of cell signaling pathway in vivo We observed a global downregulation of cell cycle related genes in vivo, including checkpoint modulators of G1/S-phase (CDK4/6-CCND), S/G2-phase (CDK2-CCNA) and G2/M-phase (CDK1/CCNB), pointing towards reduced mitotic activity. (C) Heatmaps of p53 signaling related genes in vivo and cell cycle related genes in vitro Compared to in vitro, the majority of p53 signaling related genes is downregulated after treatment with CAN-2409. A similar dynamic under both conditions is displayed for PLK1, CCNB1 and MYC which are part of both p53 and cell cycle signaling. Commonly downregulated genes are highlighted. (D) Immunoblots of relevant cell cycle targets. Immunoblots of G9_pCDH after 48 h incubation with CAN-2409 show reduced protein abundance of CCNB1, PLK1, and c-MYC compared to control. Multiple unpaired t-tests of normalized band intensities showed significance compared to control (CCNB1, PLK1, and c-MYC p < 0.000001). N = 1, triple measurement of band signal intensity.
In vivo, KEGG pathway “Cell cycle” was significantly enriched, with a broad downregulation of genes participating in this pathway noted, including checkpoint modulators at the transition from G1 to S-phase (CDK4/6-CCND), S to G2-phase (CDK2-CCNA) and G2 to M-phase (CDK1/CCNB), suggesting a general decrease in mitotic activity (Figure 3B). While cyclin-dependent kinases are not differentially expressed, a reduced expression of central cell cycle regulators like CCNB1, CDC20, PLK1, and MYC was observed in vitro, as those genes are also part of p53 signaling (Figure 3C); moreover, we also observed upregulation of CDKN1A and GADD45 -both inhibitors of cell cycle progression which contribute to cell cycle arrest in G1 and S-phase. Although CAN-2409 is associated with different nuances of pathway enrichment under both conditions, common strategic genes are similarly altered – namely CCNB1, PLK1 and MYC (Figure 3C), pointing towards similar transcriptome modifications in vitro and in vivo, which was confirmed on the protein level as well (Figure 3D), clearly demonstrating reduced expression of CCNB1, PLK1 and MYC after treatment with CAN-2409 compared to control or AdVtk alone. These findings were confirmed in another GSC cell line (G98) with reduced expression of PLK1 and CCNB1 after CAN-2409 compared to control, while MYC was not altered (Supplementary Figure S2).
3.4. CAN-2409 leads to significantly increased expression of IL-12A in vitro and in vivo
Glioblastoma exhibits a distinct cytokine profile with certain pro-tumorigenic cytokines contributing to its malignant features such as IL-6, IL-10 and TGF-β, while others are expressed at lower levels and may have tumor suppressing properties, e.g., IL-4 and IL-12 (11).
In vitro, we observed an upregulation of IL12A, IL6, and VEGFC, while no clear pattern for TGFß, HIF1a and VEGFA/B could be defined; significant differential expression could only be noted for IL12A and IL6 (Figure 4A). As the in vitro samples are lacking the influence of immune cells, we further analyzed the in vivo datasets for a more accurate depiction of cytokine pattern changes, as these include not only tumor mass, but also infiltrating immune cells. Here, we observed -in animals treated with CAN-2409 -an upregulation of the anti-tumorigenic cytokine related genes IL12A (significant: log2FC 3.279160537, value of p 0.00054542, adj value of p 0.002450255) and IL12B (not significant), but also of pro-tumorigenic related genes for IL6, CSF2 and VEGF. However, the majority of pro-tumorigenic associated genes including TGFß, IL10, VEGFA and HIF1A were downregulated (Figure 4B). Similar to the in vitro dataset, only IL12A appeared to be significantly overexpressed.
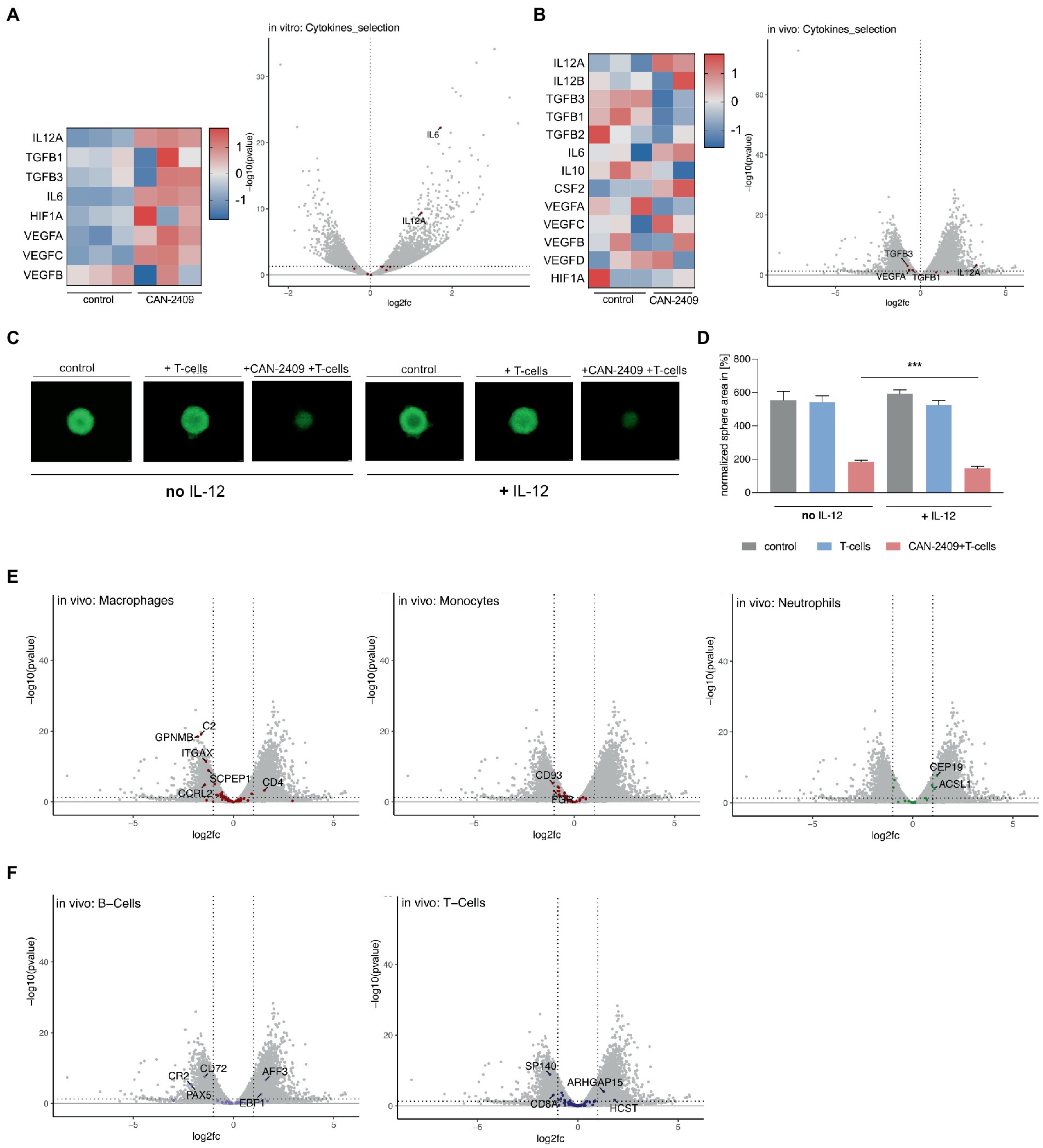
Figure 4. (A) Cytokine gene expression in vitro Increased expression of IL12A, TGFB, IL6, HIF1A and VEGF after treatment with CAN-2409 compared to control (heatmap). Adjacent Volcano plot demonstrating significant increased expression for only IL12A and IL6 (cut off: p < 0.05, log2(fc) > 1/<−1). (B) Cytokine expression in vivo Increased expression of IL12A, partly IL12B, IL6, CSF2 and partly of VEGF as well as decreased expression of TGFB, IL10 and HIF1A after treatment with CAN-2409 compared to control (heatmap). Adjacent Volcano plot showing significant increased expression for IL12A and decreased expression for VEGF and TGFB1/3 (cut off: p < 0.05, log2(fc) > 1/<−1). (C,D) Efficiency of T-cell killing by CAN-2409 is enhanced by concomitant IL-12 T-cell killing assays with G9_pCDH (green) showed significant reductions of the tumor sphere area under the combination treatment with IL-12 and CAN-2409 compared to the same treatment without IL-12 (p = 0.0008 – p = 0.0728) scale bar = 500 μm. N = 3, exemplary images and graph. (E) Diminished macrophage/monocyte signature after CAN-2409 treatment in vivo Volcano plots of macrophage, monocyte and neutrophil associated genes showing significant decreased expression after treatment with CAN-2409; for neutrophils an enhanced expression of neutrophil associated genes CEP19 and ACSL1 in accordance with Nirmal et al. (12). (F) Differential regulation of B-and T-cell associated genes in vivo For B-cells a diminished expression for CR2, CD72 and PAX5 with concomitant heightened expression of AFF3 and EBF1 was noticed. Interestingly, in T-cell signature analysis CD8a together with SP140 were downregulated, whereas ARHGAP15 and HCST were upregulated.
CAN-2409 has been shown to induce a Th1-like immune response in vivo, including increased production of IL-12 (13). While we observed differential expression of cytokine regulating genes under both conditions, IL12A, encoding for a subunit of IL-12 (14), was stably overexpressed in vitro and in vivo. In vivo, we also observed an (although not significantly) increased expression of IL12B; for successful translation of the cytokine IL-12 on protein level, both IL12A and IL12B mRNA transcripts are necessary, therefore one can assume that in contrast to the in vitro data, above outlined in vivo expression patterns are biologically relevant, i.e., leading to increased IL-12 production on the protein level. Moreover, the concordant expression dynamics of IL12A under both conditions point towards IL12A being a central molecule for CAN-2409’s mode of action. To confirm the importance of this cytokine for CAN-2409’s therapeutic efficiency, we validated the effect of IL-12 and CAN-2409 in T-cell killing assays in the presence and absence of recombinant human IL-12 (Figure 4C). Statistical analysis on day 6 of co-culture with CD8+ T cells showed a significant reduction of the tumor sphere area under the combination treatment with IL-12 compared to the same treatment without IL-12 in all cell lines (G9_pCDH, p = 0.0008**** –0.0728 ns, n = 3) under co-culture with IL-12 (Figure 4D). This suggests that indeed the efficacy of CAN-2409 is enhanced by IL-12.
3.5. Decreased expression of macrophage and monocyte gene signatures after CAN-2409 in vivo
The characteristics of glioblastoma are to a certain extent defined by the interaction of tumor cells and the surrounding tumor microenvironment; especially macrophages and monocytes – which account for up to 50% of the tumor mass (15) and contribute to the malignant features of glioblastoma. To investigate CAN-2409’s effect on the tumor microenvironment’s transcriptome, we analyzed our in vivo data for expression of immune cell signatures according to (12) (detailed tables in Supplementary material S2). We observed significantly reduced expression of myeloid signature genes, whereas we saw the opposite for neutrophils and no distinct dynamics for B-cell or T-cell cell related genes.
The comparison with previously described monocyte and macrophage gene sets (12) revealed overexpression of CD4, whereas the majority of genes detected were either not altered or significantly downregulated, like inflammation associated C2 (complement protein), GPNMB (involved in inflammation (16), associated with promoting CSC features (17)), ITGAX (encoding CD11c surface protein (18)), SCPEP1 and CCRL2 (altering M1/2 polarization (19), promoting T-cell mediated anti-tumor immunity (20))(macrophages) and CD93 (general myeloid marker (21)) and FGR (protooncogene, normal myeloid marker (22))(monocytes). For neutrophils, upregulation of CEP19 and ASCL1 was observed (Figure 4E).
In our previous studies of tumor cellular composition by mass cytometry, B-cells were shown to be increased upon treatment with CAN-2409 (23). Compared to above mentioned datasets, a decrease of B-cell related genes CD72, PAX5 and CR2 (24–26) was noticed, while AFF3 and EBP1 were upregulated. CAN-2409’s immune cell related effects are mediated via CD8+ T-cells, but also an increase in Tregs was reported previously (27); here we observed a downregulation of T-cell related genes SP140 and CD8a and an increase of ARHGAP15 and HCST (Figure 4F). Thus our in vivo studies show molecular transcriptomic alterations indicative of changes in the tumor microenvironment in response to CAN-2409 treatment.
4. Discussion
CAN-2409 has been shown to induce immunogenic cell death in glioblastoma and transform the immunologically “cold” into a “hot” tumor microenvironment. Although this is achieved by interfering with tumor cell DNA through incorporation of an antimetabolite -the HSV-tk metabolized prodrug ganciclovir, acyclovir or valacyclovir. The resulting alterations of the tumor transcriptome have not been assessed. CAN-2409 has been thoroughly characterized in vitro and in vivo, especially regarding it immunological implications in previous publications (13, 27, 28). We conducted an RNA-Seq analysis of both in vitro and in vivo samples to characterize the CAN-2409 mediated transcriptional modifications and further alterations caused by interaction with the tumor microenvironment.
In the current study, we showed that CAN-2409 is associated with a downregulation of transcriptionally relevant pathways under both conditions. More detailed analysis revealed a specific in vitro downregulation of p53 signaling and in vivo downregulation of cell cycle signaling, with reduced expression of MYC, CCNB1, CDC20 and PLK1 in both conditions. Analyses for cytokine expression profiles indicated upregulation of IL12a in vitro and in vivo, with IL12b showing increased expression in vivo, although not being significantly overexpressed. Immune cell signature profiling with the in vivo data pointed towards diminished expression of macrophage and monocyte relevant genes.
Ganciclovir, which together with CAN-2409 (Ad.HSV-tk) constitutes the CAN-2409 therapeutic regimen, inflicts DNA damage, culminating in activation of ATR and ATM (29). The latter are involved in DNA damage control and hence mitotic regulation (30). ATR inhibition further increased CAN-2409’s tumor cell killing abilities (23).
The HSV-tk/ganciclovir gene suicide system was shown to be associated with cell cycle arrest in the G1/S-phase: Zeng et al. reported that after treatment of a breast cancer cell line with a Tet-on-HSV-tk/GCV system, an increment of the S-phase status was noticed along with increased expression of CDKN1a and decreased expression of CDK1, CCNB and PCNA, suggesting that HSV-tk/GCV’s modus operandi includes cell cycle inhibition (31). These reports are complemented with our findings: in vitro, an overexpression of CDKN1a as well as decreased expression of CCNB1 (which was also the case for the in vivo data) was observed. Whereas CDK1 was not significantly altered in vitro, its expression was diminished in vivo. While we noticed some discrepancies in expression between in vitro and in vivo, cell cycle and p53 relevant genes, i.e., CCNB1, PLK1, MYC, CDC20 were similarly downregulated in both conditions, supporting the hypothesis that indeed tumor cell killing features of this gene suicide system are induced via disruption of the cell cycle, which also translated onto the protein level.PLK1, which phosphorylates CCNB1 (32), is overexpressed in several cancers (33). PLK1 depletion as well as CCNB1 silencing have been reported to mediate apoptosis and G2/M arrest (34–36). Inhibition of PLK1 combined with radiotherapy (37) or temozolomide chemotherapy (38) increased the efficiency of those treatments in glioblastoma models.
As previously shown, CAN-2409 induces a Th1-like immune response, i.e., increased expression of IL-12, IL-2, IFN-γ, TNF-α, and GM-CSF (13). IL-12 specifically is considered as an anti-tumorigenic cytokine in various malignant diseases (39). Kerkar et al. reported that the tumor microenvironment was considerably altered towards inflammation by in vitro activated IL-12 producing CD8+ T-cells (40). Current strategies to further enhance previously established immunotherapies for glioma include intratumoral delivery or increase of IL-12 (41–43). Here, we show an increase of IL12a in vitro and IL12a and IL12b in vivo. IL-12 consists out of two subunits, light chain p35 (encoded by IL12a) and heavy chain p40 (encoded by IL12b) (14). For a physiologically functioning IL-12, both subunits are necessary. While we found upregulation of IL12a in vitro, we observed upregulation for both IL12a and IL12b in vivo, i.e., the latter is presumptively associated with increased biologically active cytokine expression of IL-12 which has been previously reported by Vile et al. (13). We also showed that upregulation of IL-12 is an integral part of CAN-2409’s mode of action (Figures 4A–D). Indeed treatment with CAN-2409 under addition of recombinant IL-12 increased tumor cell killing abilities in presence of activated T-cells significantly. IL-12 therefore might serve as a potential biomarker in patients treated with CAN-2409.
The glioblastoma microenvironment is characterized by a large proportion of myeloid cells, such as macrophages. Tumor associated macrophages (TAMs) contribute to the malignant features of glioblastoma by secreting pro-tumorigenic factors, e.g., IL-10 and TGF-β (44). Treatment with CAN-2409 has been shown to reduce the number of TAMs, which can be reversed by concomitant treatment with dexamethasone (28) or enhanced by parallel inhibition of the ATR-CHK1 axis (23). RNA Seq analyses for immune cell profiles confirm previous observations on a transcriptomic level, as we demonstrated a reduction of macrophage/monocyte related genes after treatment with CAN-2409 (Figure 4E).
Taken together our findings show that CAN-2409 broadly changes the tumor cell transcriptome -which is further modulated by the interplay with the tumor microenvironment. Our observations suggest that the CAN-2409 induced transcriptomic alterations make a major contribution to the treatment’s tumor cell killing abilities and complements its immunogenic impact.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found under GEO accession number GSE230661 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE230661).
Ethics statement
The animal study was reviewed and approved by IACUC Brigham and Women's Hospital Boston, United States.
Author contributions
SL, MK, and EC: study conceptualization, design, and project administration. MK, MN, MZ, ZH, and MH: methodology. MK and SL: investigation and writing–original draft. MK, ZH, and MH: data analysis. SL, EC, and GT: funding acquisition. SL and ZH: supervision. MK, SL, EC, MZ, MN, EC, FB, PT, MH, ZH, and GT: writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by NCI P01CA069246. This research was partially funded by NCI R50 CA243706-02 (MN) and Else Kröner Fresenius Forschungskolleg (2015_Kolleg_14) 9 (GT).
Acknowledgments
The authors thank Candel Therapeutics (Needham, MA, United States) for providing CAN-2409 and ganciclovir used in this study.
Conflict of interest
FB and PT are employees of Candel Therapeutics.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1140352/full#supplementary-material
References
1. Tamimi, AF, and Juweid, M. Epidemiology and outcome of glioblastoma. Glioblastoma. (2017):143–53. doi: 10.15586/CODON.GLIOBLASTOMA.2017.CH8
2. Stupp, R, Mason, WP, van den Bent, MJ, Weller, M, Fisher, B, Taphoorn, MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. (2005) 352:987–96. doi: 10.1056/NEJMoa043330
3. Antonio Chiocca, E, Aguilar, LK, Bell, SD, Kaur, B, Hardcastle, J, Cavaliere, R, et al. Phase IB study of gene-mediated cytotoxic immunotherapy adjuvant to up-front surgery and intensive timing radiation for malignant glioma. J Clin Oncol. (2011) 29:3611–9. doi: 10.1200/JCO.2011.35.5222
4. Wheeler, LA, Manzanera, AG, Bell, SD, Cavaliere, R, McGregor, JM, Grecula, JC, et al. Phase II multicenter study of gene-mediated cytotoxic immunotherapy as adjuvant to surgical resection for newly diagnosed malignant glioma. Neuro-Oncology. (2016) 18:1137–45. doi: 10.1093/neuonc/now002
5. Wen, P, Duault, C, Gonzalez-Kozlova, E, Brennan, K, Holmes, T, Kim-Schulze, S, et al. 1477 first efficacy and multi-omic analysis data from phase 1 clinical trial of oncolytic viral immunotherapy with CAN-2409 + valacyclovir in combination with nivolumab and standard of care in newly diagnosed high-grade glioma. J Immunother Cancer. (2022) 10:A1533–3. doi: 10.1136/JITC-2022-SITC2022.1477
6. Williams, SP, Nowicki, MO, Liu, F, Press, R, Godlewski, J, Abdel-Rasoul, M, et al. Indirubins decrease glioma invasion by blocking migratory phenotypes in both the tumor and stromal endothelial cell compartments. Cancer Res. (2011) 71:5374–80. doi: 10.1158/0008-5472.CAN-10-3026
7. Dobin, A, Davis, CA, Schlesinger, F, Drenkow, J, Zaleski, C, Jha, S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
8. Love, MI, Huber, W, and Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/S13059-014-0550-8
9. Trapnell, C, Williams, BA, Pertea, G, Mortazavi, A, Kwan, G, van Baren, MJ, et al. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms HHS public access Author manuscript. Nat Biotechnol. (2010) 28:511–5. doi: 10.1038/nbt.1621
10. Cornwell, MI, Vangala, M, Taing, L, Herbert, Z, Köster, J, Li, B, et al. VIPER: visualization pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics. (2018) 19:135. doi: 10.1186/S12859-018-2139-9
11. Albulescu, R, Codrici, E, Popescu, ID, Mihai, S, Necula, LG, Petrescu, D, et al. Cytokine patterns in brain tumour progression. Mediat Inflamm. (2013) 1:7. doi: 10.1155/2013/979748
12. Nirmal, AJ, Regan, T, Shih, BB, Hume, DA, Sims, AH, and Freeman, TC. Immune cell gene signatures for profiling the microenvironment of solid tumors. Cancer Immunol Res. (2018) 6:1388–400. doi: 10.1158/2326-6066.CIR-18-0342
13. Vile, RG, Castleden, S, Marshall, J, Camplejohn, R, Uptov, C, and Chong, H. Generation of an anti-tumour immune response in a non-immunogenic tumour: HSVtk killing in vivo stimulates a mononuclear cell infiltrate and a Th1-like profile of intratumoural cytokine expression. Int J Cancer. (1997) 71:267–74. doi: 10.1002/(SICI)1097-0215(19970410)71:2<267::AID-IJC23>3.0.CO;2-D
14. Yuzhalin, AE, and Kutikhin, AG. Interleukin-12: clinical usage and molecular markers of cancer susceptibility. Growth Factors. (2012) 30:176–91. doi: 10.3109/08977194.2012.678843
15. Ma, J, Chen, CC, and Li, M. Molecular sciences macrophages/microglia in the glioblastoma tumor microenvironment. Int J Mol Sci. (2021) 22:5775. doi: 10.3390/ijms22115775
16. Saade, M, Araujo de Souza, G, Scavone, C, and Kinoshita, PF. The role of GPNMB in inflammation. Front Immunol. (2021) 12:674739. doi: 10.3389/FIMMU.2021.674739
17. Allavena, P, Digifico, E, and Belgiovine, C. Macrophages and cancer stem cells: a malevolent alliance. Mol Med. (2021) 121:27. doi: 10.1186/s10020-021-00383-3
18. Shen, X, Qiu, Y, Wight, AE, Kim, H-J, Cantor, H, Colonna, M, et al. Definition of a mouse microglial subset that regulates neuronal development and proinflammatory responses in the brain. Cancer Immunol Res. (2018) 119:e2116241119. doi: 10.1073/pnas.2116241119/-/DCSupplemental
19. Mazzon, C, Zanotti, L, Wang, L, del Prete, A, Fontana, E, Salvi, V, et al. CCRL2 regulates M1/M2 polarization during EAE recovery phase. J Leukoc Biol. (2016) 99:1027–33. doi: 10.1189/JLB.3MA0915-444RR
20. Yin, W, Li, Y, Song, Y, Zhang, J, Wu, C, Chen, Y, et al. CCRL2 promotes antitumor T-cell immunity via amplifying TLR4-mediated immunostimulatory macrophage activation. Proc Natl Acad Sci U S A. (2021) 118:e2024171118. doi: 10.1073/pnas.2024171118/-/DCSupplemental
21. Tenner, AJ, Bohlson, SS, Silva, R, and Fonseca, MI. Is detected in human plasma human myeloid cells and the soluble form CD93 is rapidly shed from the surface of. J Immunol. (2005) 175:1239–47. doi: 10.4049/jimmunol.175.2.1239
22. Notario, V, Gutkind, JS, Imaizumi, M, Katamine, S, and Robbins, KC. Expression of thefgr Protooncogene product as a function of Myelomonocytic cell maturation. J Cell Biol. (1989) 109:3129–36. doi: 10.1083/jcb.109.6.3129
23. Koch, MS, Zdioruk, M, Nowicki, MO, Griffith, AM, Aguilar-Cordova, E, Aguilar, LK, et al. Perturbing DDR signaling enhances cytotoxic effects of local oncolytic virotherapy and modulates the immune environment in glioma. Mol Ther Oncolytics. (2022) 26:275–88. doi: 10.1016/J.OMTO.2022.07.009
24. Bondada, S, Sander Brown Building, A, and Wu Subbarao Bondada, H-J. Positive and negative roles of CD72 in B cell function. Immunol Res. (2002) 25:155–66. doi: 10.1385/IR:25:2:155
25. Cobaleda, C, Schebesta, A, Delogu, A, and Busslinger, M. Pax5: the guardian of B cell identity and function. Nat Immunol. (2007) 8:463–70. doi: 10.1038/ni1454
26. Erdei, A, Kovács, KG, Nagy-Baló, Z, Lukácsi, S, Mácsik-Valent, B, Kurucz, I, et al. New aspects in the regulation of human B cell functions by complement receptors CR1, CR2, CR3 and CR4. Immunol Lett. (2021) 237:42–57. doi: 10.1016/J.IMLET.2021.06.006
27. Speranza, M, Passaro, C, Ricklefs, F, Kasai, K, Klein, SR, Kaufmann, JK, et al. Preclinical investigation of combined gene-mediated cytotoxic immunotherapy and immune checkpoint blockade in glioblastoma. Neuro Oncol. (2018) 20:225–35. doi: 10.1093/neuonc/nox139
28. Koch, MS, Zdioruk, M, Nowicki, MO, Griffith, AM, Aguilar, E, Aguilar, LK, et al. Systemic high-dose dexamethasone treatment may modulate the efficacy of intratumoral viral oncolytic immunotherapy in glioblastoma models. J Immunother Cancer. (2022) 10:e003368. doi: 10.1136/jitc-2021-003368
29. Ladd, B, O’konek, JJ, Ostruszka, LJ, and Shewach, DS. Unrepairable DNA double Strand breaks initiate cytotoxicity with HSV-TK/ganciclovir. Cancer Gene Ther. (2017) 18:751–9. doi: 10.1038/cgt.2011.51
30. Abraham, RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. (2001) 15:2177–96. doi: 10.1101/gad.914401
31. Zeng, ZJ, Xiang, SG, Xue, WW, Li, Hde, Ma, N, Ren, ZJ, et al. The cell death and DNA damages caused by the Tet-on regulating HSV-tk/GCV suicide gene system in MCF-7 cells. Biomed Pharmacother (2014) 68:887–892. doi: 10.1016/j.biopha.2014.07.018
32. Toyoshima-Morimoto, F, Taniguchi, E, Shinya, N, Iwamatsu, A, and Nishida, E. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature. (2001) 410:215–20. doi: 10.1038/35065617
33. Elizabeth, R, Gutteridge, A, Ndiaye, MA, Liu, X, and Ahmad, N. Plk1 inhibitors in cancer therapy: from laboratory to clinics. Mol Cancer Ther. (2016) 15:1427–35. doi: 10.1158/1535-7163.MCT-15-0897
34. Zhang, H, Zhang, X, Li, X, Meng, WB, Bai, ZT, Rui, SZ, et al. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J Cell Physiol. (2018) 234:619–31. doi: 10.1002/jcp.26816
35. Liu, X, and Erikson, RL. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci U S A. (2003) 13:5789–94.
36. Jung, Y, Kraikivski, P, Shafiekhani, S, Terhune, SS, and Dash, RK. ARTICLE crosstalk between Plk1, p53, cell cycle, and G2/M DNA damage checkpoint regulation in cancer: computational modeling and analysis. NPJ Syst Biol Appl. (2021) 7:46. doi: 10.1038/s41540-021-00203-8
37. Pezuk, JA, Brassesco, MS, Morales, AG, Carvalho De Oliveira, J, Francisco De Oliveira, H, Scrideli, CA, et al. Inhibition of polo-like kinase 1 induces cell cycle arrest and sensitizes glioblastoma cells to ionizing radiation. Cancer Biother Radiopharm. (2013) 28:516–22. doi: 10.1089/cbr.2012.1415
38. Pandey, A, Tripathi, SC, Mai, J, Hanash, SM, Shen, H, Mitra, S, et al. Combinatorial effect of PLK1 inhibition with Temozolomide and radiation in glioblastoma. Cancers (Basel). (2021) 13:5114. doi: 10.3390/cancers13205114
39. Tugues, S, Burkhard, SH, Ohs, I, Vrohlings, M, Nussbaum, K, Vom Berg, J, et al. New insights into IL-12-mediated tumor suppression. Cell Death Differ. (2015) 22:237–46. doi: 10.1038/cdd.2014.134
40. Kerkar, SP, Goldszmid, RS, Muranski, P, Chinnasamy, D, Yu, Z, Reger, RN, et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J Clin Invest. (2011) 121:4746–57. doi: 10.1172/JCI58814DS1
41. Agliardi, G, Rita Liuzzi, A, Hotblack, A, de Feo, D, Núñez, N, Stowe, CL, et al. Intratumoral IL-12 delivery empowers CAR-T cell immunotherapy in a pre-clinical model of glioblastoma. Nat Commun. (2021) 12:444. doi: 10.1038/s41467-020-20599-x
42. Chiocca, EA, Yu, JS, Lukas, RV, Solomon, IH, Ligon, KL, Nakashima, H, et al. Regulatable interleukin-12 gene therapy in patients with recurrent high-grade glioma: results of a phase 1 trial. Sci Transl Med. (2019) 11:eaaw5680. doi: 10.1126/SCITRANSLMED.AAW5680
43. Chiocca, EA, Gelb, AB, Chen, CC, Rao, G, Reardon, DA, Wen, PY, et al. Combined immunotherapy with controlled interleukin-12 gene therapy and immune checkpoint blockade in recurrent glioblastoma: an open-label, multi-institutional phase I trial. Neuro-Oncology. (2022) 24:951–63. doi: 10.1093/NEUONC/NOAB271
Keywords: transcriptomics, CAN-2409, oncolytic virotherapy, glioma, oncoimmunology
Citation: Koch MS, Zdioruk M, Nowicki MO, Hoetker MS, Herbert ZT, Barone F, Tak PP, Chiocca EA, Tabatabai G and Lawler SE (2023) Uncovering transcriptomic landscape alterations of CAN-2409 in in vitro and in vivo glioma models. Front. Med. 10:1140352. doi: 10.3389/fmed.2023.1140352
Edited by:
Silvia Di Angelantonio, Sapienza University of Rome, ItalyReviewed by:
Maria Rosito, Sapienza University of Rome, ItalyMaria Graciela Castro, Department of Neurosurgery, School of Medicine, Michigan Medicine, University of Michigan, United States
Copyright © 2023 Koch, Zdioruk, Nowicki, Hoetker, Herbert, Barone, Tak, Chiocca, Tabatabai and Lawler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean E. Lawler, c2Vhbl9sYXdsZXJAYnJvd24uZWR1
 Marilin S. Koch
Marilin S. Koch Mykola Zdioruk1
Mykola Zdioruk1 Michael S. Hoetker
Michael S. Hoetker Zachary T. Herbert
Zachary T. Herbert Ghazaleh Tabatabai
Ghazaleh Tabatabai