- 1Department of Gastroenterology, Guang' anmen Hospital, China Academy of Traditional Chinese Medical Sciences, Beijing, China
- 2Office of Good Clinical Practice, Guang' anmen Hospital, China Academy of Traditional Chinese Medical Sciences, Beijing, China
Background: Omics has emerged as a promising biological science to shed light on the etiology, pathogenesis, and treatment of ulcerative colitis (UC). At present, although research on the omics of UC has drawn global attention, there is still a lack of bibliometric analysis in this field. This study aimed to access the trends and hotspots of omics in UC research.
Method: Publications related to omics in UC from 1 January 2000 to 15 October 2022 were retrieved from the Web of Science Core Collection database. VOSviewer, CiteSpace, and the online bibliometric analysis platform “Bibliometrix” were adopted to extract and visualize information.
Results: A total of 385 publications were finally included and the annual number of publications fluctuated. The trend in publications increased rapidly after 2019. The United States showed its dominant position in several publications, total citations, and international collaborations. The top five research organizations for publications on the research of omics in UC were Harvard Medical School, the Icahn School of Medicine at Mount Sinai, Karolinska Institutet, the Brigham and Women's Hospital, and the Massachusetts General Hospital. Ashwin Ananthakrishnan from the Massachusetts General Hospital was the most productive author, and Séverine Vermeire from the Catholic University of Leuven was co-cited most often. Inflammatory bowel disease was the most popular and co-cited journal in this field. The reference with citation bursts and trend topics showed that “ulcerative colitis,” “inflammatory bowel disease,” “microbiome,” “transcriptomics,” “genomics,” “metabolomics,” “proteomics,” “dysbiosis,” “biomarkers,” “loci,” and “therapy” are currently research hotspots.
Conclusion: Our study presents several important insights into the research trends and developments in the field of omics in UC, which will provide key information for further research.
Introduction
Omics is a rapidly evolving, comprehensive, and emerging field of study in biological science that encompasses genomics, transcriptomics, proteomics, metabolomics, and microbiomics (1). More specifically, genomics is the study of the structure, function, and inheritance of an organism's entire genome (2). Transcriptomics examines all messenger RNA molecules qualitatively or quantitatively in one cell, tissue, or organism (3). In contrast, proteomics evolved from genomics and focuses on quantifying proteins/peptides, modification, and interaction in multiple sample types by MS-based methods or high-throughput analyses (4, 5). Metabolomics is the large-scale study of multiple small molecules, such as amino acids, fatty acids, organic acids, and ketones, which are the end products of complex biochemical processes (6). More than 1,000 species of microbial cells constitute human gut microbiota, and the amount of genes present in the microbial community is 100 times greater than the human genome (7). Microbiomics, driven by the development of genomic sequencing technology, is the science of characterizing the microbial community (8). Overall, each type of omics science is typically used to identify, characterize, and quantify all biological molecules, which are associated with diseases. Omics can explore markers of disease and advance our understanding of biological pathways or processes.
Ulcerative colitis (UC) is a chronic disease where the colon and rectum become inflamed and develop ulcers or sores (9). UC occurs worldwide, with increasing incidence and prevalence (10). The prevalence rates of UC in the United States range from 214 to 286 cases per 100,000 from 2000 to 2011 (11, 12). The characteristics of UC in remission are without symptoms. However, in the period of relapsing, typical gastrointestinal presentation of UC includes bloody diarrhea, rectal urgency, mucus in the stool, and variable degrees of abdominal pain. Until now, the pathogenesis of UC is still poorly understood, which may be related to abnormal reactions of the immune system, environmental factors, and genetics (13–15).
The omics is a good way to gain insights into the etiology, pathogenesis, and treatment of UC, especially with a large number of research articles published per year. The bibliometric analysis first appeared in the late 19th and early 20th centuries (16). Nowadays, bibliometric methods are frequently adopted to decipher and map the cumulative scientific knowledge, the impact of a set of researchers, and obtain hot topics of certain research (17). The benefit of bibliometric analysis is to enable and empower scholars to gain an overview of the hotspot, co-authorship, co-citation, and the development of the specific field, which would build firm foundations for advancing a field. However, the research of omics in UC has not been assessed through bibliometric analysis. Therefore, in this study, a bibliometric analysis of publications on omics in UC was carried out. The aim of this study was to assess global trends and hotspots of omics in UC research.
Methods
Data source and search strategy
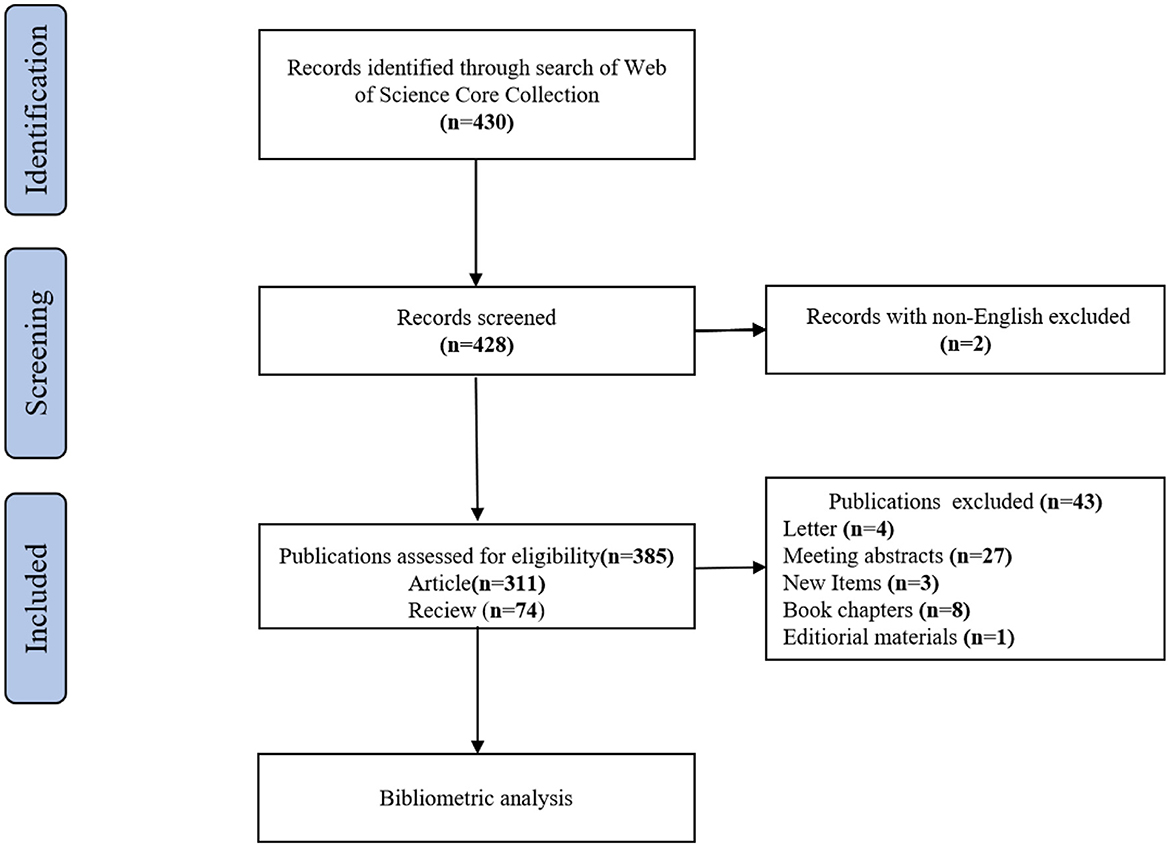
A search was conducted on the Web of Science Core Collection (WoSCC) database (https://www.webofscience.com/wos/woscc/basic-search) from 1 January 2000 to 15 October 2022 (18). The search formula was ((((((TS = (Genomics)) OR TS = (Metabolomic)) OR TS = (Transcriptomics)) OR TS = (Proteomics)) OR TS = (Microbiomics)) AND TS = (ulcerative colitis)) AND LA = (English). Only articles and review articles were included in this study. To avoid bias due to daily database updates, all searches were performed on the same day. The literature screening procedure is depicted in Figure 1.
Data analysis
In the present study, VOSviewer (version 1.6.18), CiteSpace (version 6.1.R3), and Bibliometrix (version 3.2.1) (https://www.bibliometrix.org) were applied to analyze the data from the literature (19). The literature's authors, organizations, titles, abstracts, keywords, journals, and cited references were downloaded in plain text. VOSviewer, developed at Leiden University Centre for Science and Technology Studies, is an open-source software tool for mapping and visualizing bibliometric networks (20, 21). In our study, VOSviewer was used to analyze the co-authorship (authors, organizations, and countries), co-occurrence (author keywords), bibliographic coupling (sources), and co-citation (cited references, cited sources, and cited authors). In the networks constructed by VOSviewer, items such as author, country, organization, and keywords were represented by nodes, and the links were called edges reflecting the degree of collaboration or co-citation of each item. In order to optimize the figures display clearly, a minimum threshold was set and no more than 200 nodes were displayed in each network map. For example, 37 countries, 99 organizations, 49 most productive authors, 52 author keywords, 62 most frequently used journals, 40 co-cited references, 193 co-cited journals, and 127 co-authors were shown in the visualization maps. CiteSpace, developed by Professor Chaomei Chen of Drexel University, is a freely available application for visualizing and analyzing the literature of a scientific domain (22, 23). In this study, the dual-map overlay of journals and citation bursts was built based on CiteSpace, which helped to identify emerging trends and the distribution of academic journals in real time. The CiteSpace settings were as follows: time span (2000–2022), years per slice (1); link strength (Cosine), link scope (within slices); and selection criteria (a modified g-index in each slice). R package “Bibliometrix” was adopted to perform a global distribution of publications and the thematic evolution analysis (24). Let us install the R package and start biblioshiny digiting: library (Bibliometrix) biblioshiny. Information on scientific studies extracted from the WoSCC database. The figures were created using “biblioshiny,” a shiny application that performed a web interface for the Bibliometrix. In addition, Microsoft Office Excel 2019 was used to analyze the annual publications. The 2022 impact factor (IF) and JCR division of journals were obtained from the Web of Science group (InCites, Journal Citation Reports).
Results
Quantitative analysis of publication
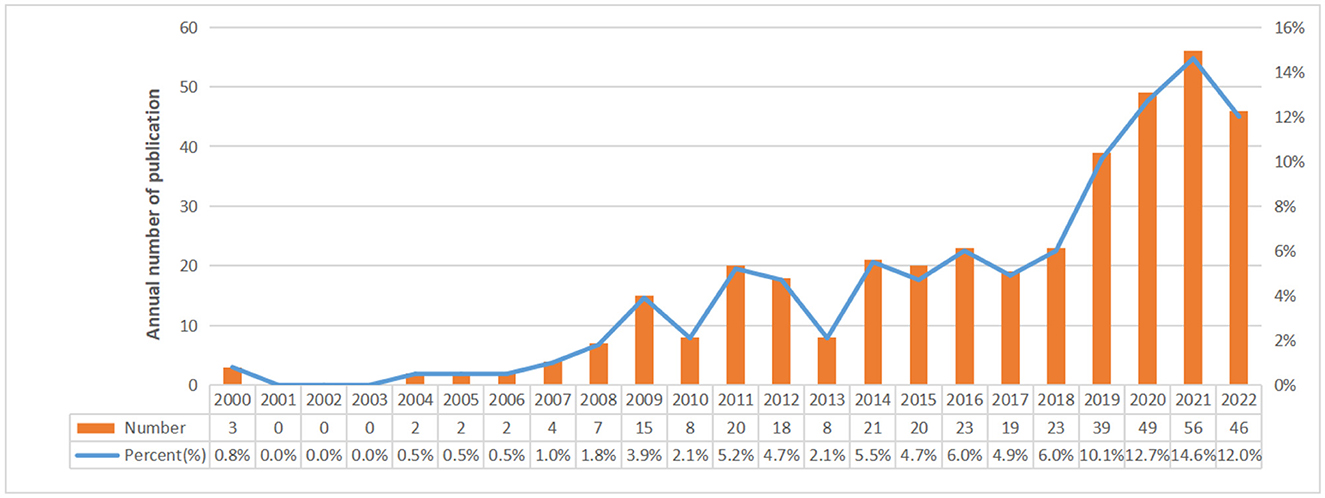
Finally, there were a total of 385 studies included, comprising 311 articles and 74 reviews on the research of omics in UC. The annual number of publications from 2000 to 2022 is shown in Figure 2. From 2000 to 2007, it was the initial stage where the annual publication number was relatively small. The number of publications from 2008 to 2018 showed continued instability, with an average annual publication number of 16.5. An outbreak of omics in UC research was witnessed from 2019 to 2022. The statistic showed the number of publications in the field of omics in UC peaked in 2021.
Countries and institutions analysis
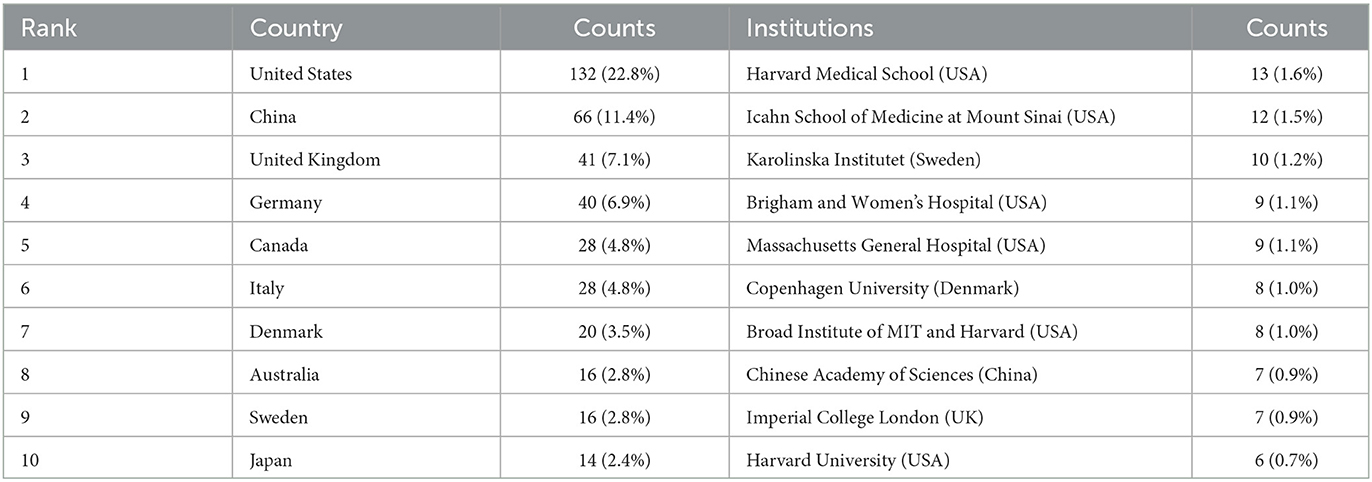
In total, 49 countries and 809 institutions are involved in the research of omics in UC. The top 10 leading countries were distributed in North America, Asia, and Europe, as shown in Table 1. The United States (USA) (132, 22.8%) had the largest number of publications. China was in second place with 66 publications, followed by the United Kingdom (41, 7.1%) and Germany (40, 6.9%). Figure 3A presents the network of countries on the research of omics in UC, with the minimum threshold of two documents of a country. Among the 49 countries, 37 met the threshold. The network offered a clear image of seven clusters in the countries, with the highest link strength. The figure suggested the frequent coupling among the United States, United Kingdom, Germany, Italy, and Canada.
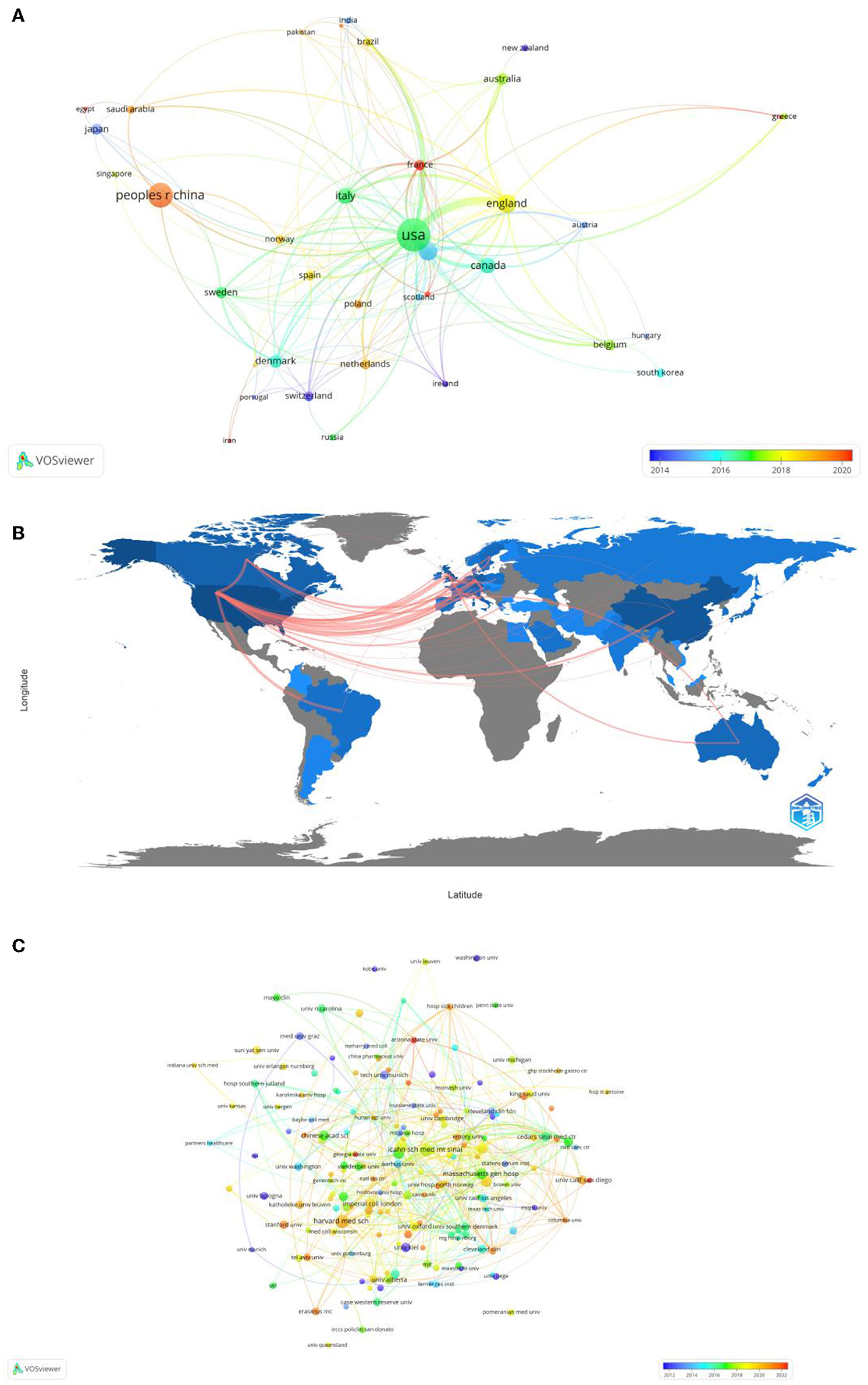
Figure 3. (A) Map of visualization of countries on research of omics in UC. (B) Country collaboration map. (C) Map of visualization of institutions on research of omics in UC.
Subsequently, a collaborative network was constructed according to the number and relationship of publications in each country (Figure 3B). The world collaboration map also highlighted collaboration and networking among countries. The blue color on the map referred to the collaboration between different countries. In addition, the pink lines between the countries reflected the degree of collaboration between the authors. The United States had the highest number of collaborations with the United Kingdom researchers (16), followed by Germany (13), Canada (8), and Brazil (7).
The 10 institutions with the largest number of publications are also presented in Table 1. Among the institutions, the top five research organizations for publications on the research of omics in UC were Harvard Medical School (13, 1.6%), the Icahn School of Medicine at Mount Sinai (12, 1.5%), Karolinska Institutet (10, 1.2%), the Brigham and Women's Hospital (9, 1.1%), and the Massachusetts General Hospital (9, 1.1%).
In Figure 3C, the institutions that met the thresholds of more than or equal to three minimum number of documents of the organization were included. The network displayed the organizations that had collaborated on the scientific documents. The size of the node referred to the total number of publications and the size of the link reflected the number of collaborations. Harvard Medical School had the highest number of scientific collaborations with international organizations, followed by the Icahn School of Medicine at Mount Sinai, the Massachusetts General Hospital, and Harvard University.
Authors and co-cited authors
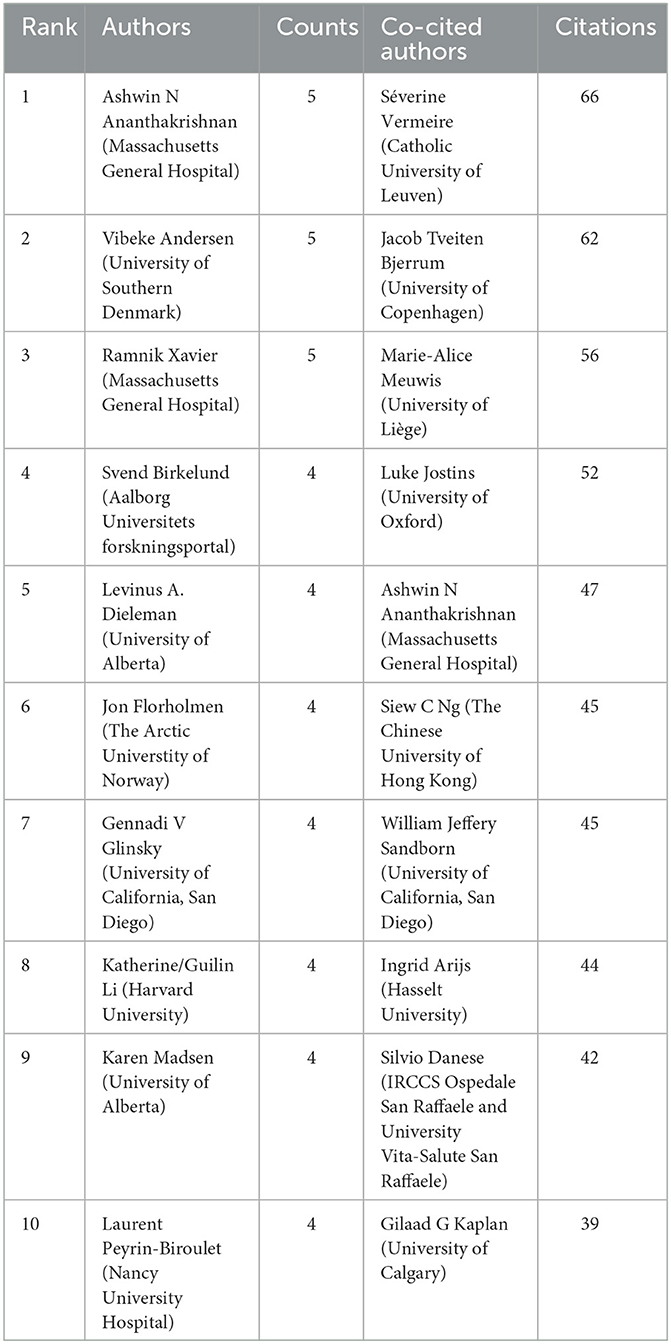
A total of 2,607 authors have participated in the research of omics in UC. Detailed information of the top 10 writers based on publications is presented in Table 2. Three of them, including Ashwin Ananthakrishnan (Massachusetts General Hospital), Vibeke Andersen (University of Southern Denmark), and Ramnik Xavier (Massachusetts General Hospital) had published five articles. The author's collaborative network is presented in Figure 4A. Ashwin Ananthakrishnan, Katherine Li, Carrie Brodmerkel, Shannon E Telesco, and Carmen Argmann had the largest nodes due to the most related publications. A close collaboration existed among multiple authors. Ashwin Ananthakrishnan and Ramnik Xavier were working together. Katherine Li had close cooperation with Carrie Brodmerkel.
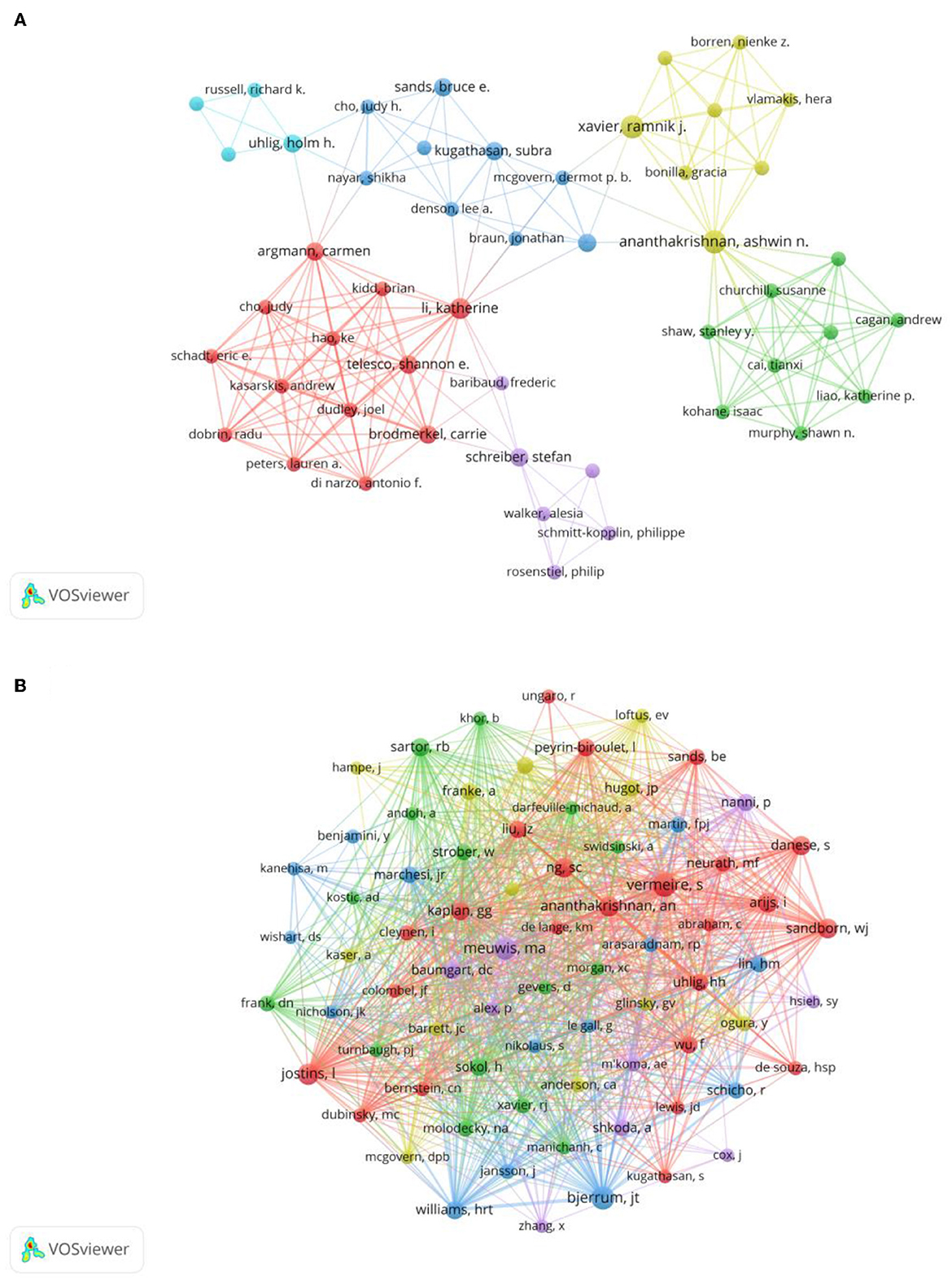
Figure 4. (A) Network of authors on research of omics in UC. (B) Network of co-cited authors on research of omics in UC.
The top 10 co-authors are presented in Table 2. The most co-cited author was Séverine Vermeire (n = 66), followed by Jacob Tveiten Bjerrum (n = 62), Marie-Alice Meuwis (n = 56), and Luke Jostins (n = 52). Authors with minimum co-citations of equal to 15 were filtered to map co-author network graphs (Figure 4B). As shown in Figure 4B, the authors were presented as the nodes in the network. The size of the author node represented citation counts. There were also close collaborations among different co-cited authors, such as Séverine Vermeire, William Jeffery Sandborn, Marie-Alice Meuwis, and Jacob Tveiten Bjerrum.
Journals and co-cited journals
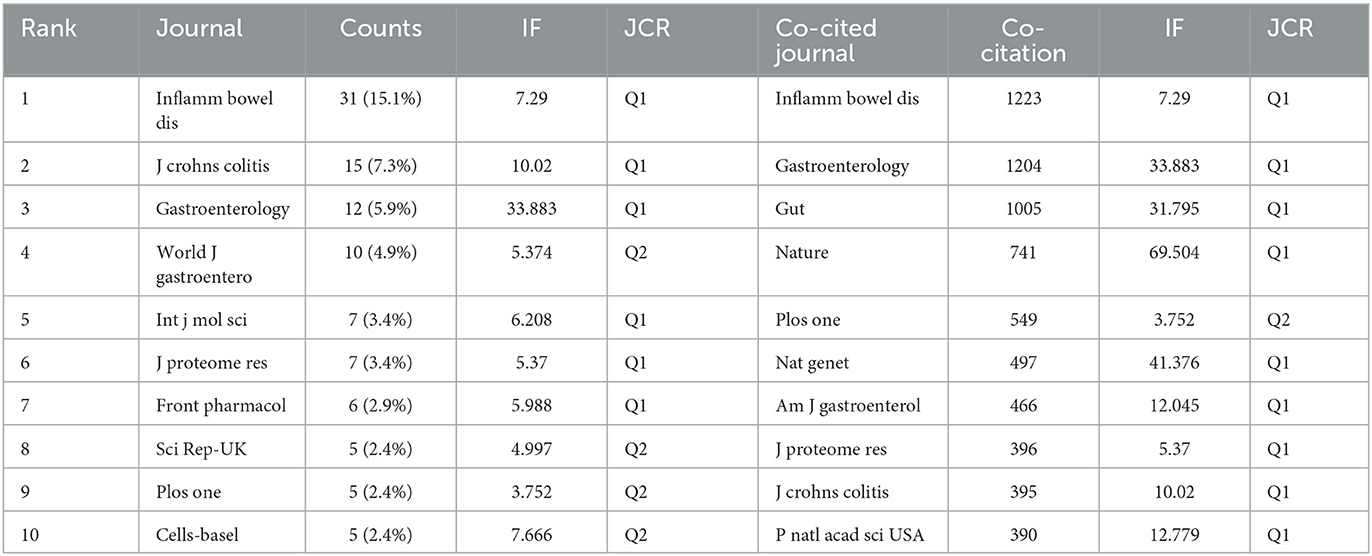
In total, publications related to omics in UC were published in 205 journals. The top 10 journals are presented in Table 3. Inflammatory bowel disease had published the most articles (n = 31, 15.1%), followed by Journals of Crohn's and Colitis (n = 12, 5.9%), Gastroenterology (n = 12, 5.9%), and World Journal of Gastroenterology (n = 10, 4.9%). The top 10 productive journals had an impact factor (IF) ranging from 3.752 to 33.883. Furthermore, the active journals appeared in the Journal Citation Reports (JCR) Q1 or Q2. The journal network is shown in Figure 5A.
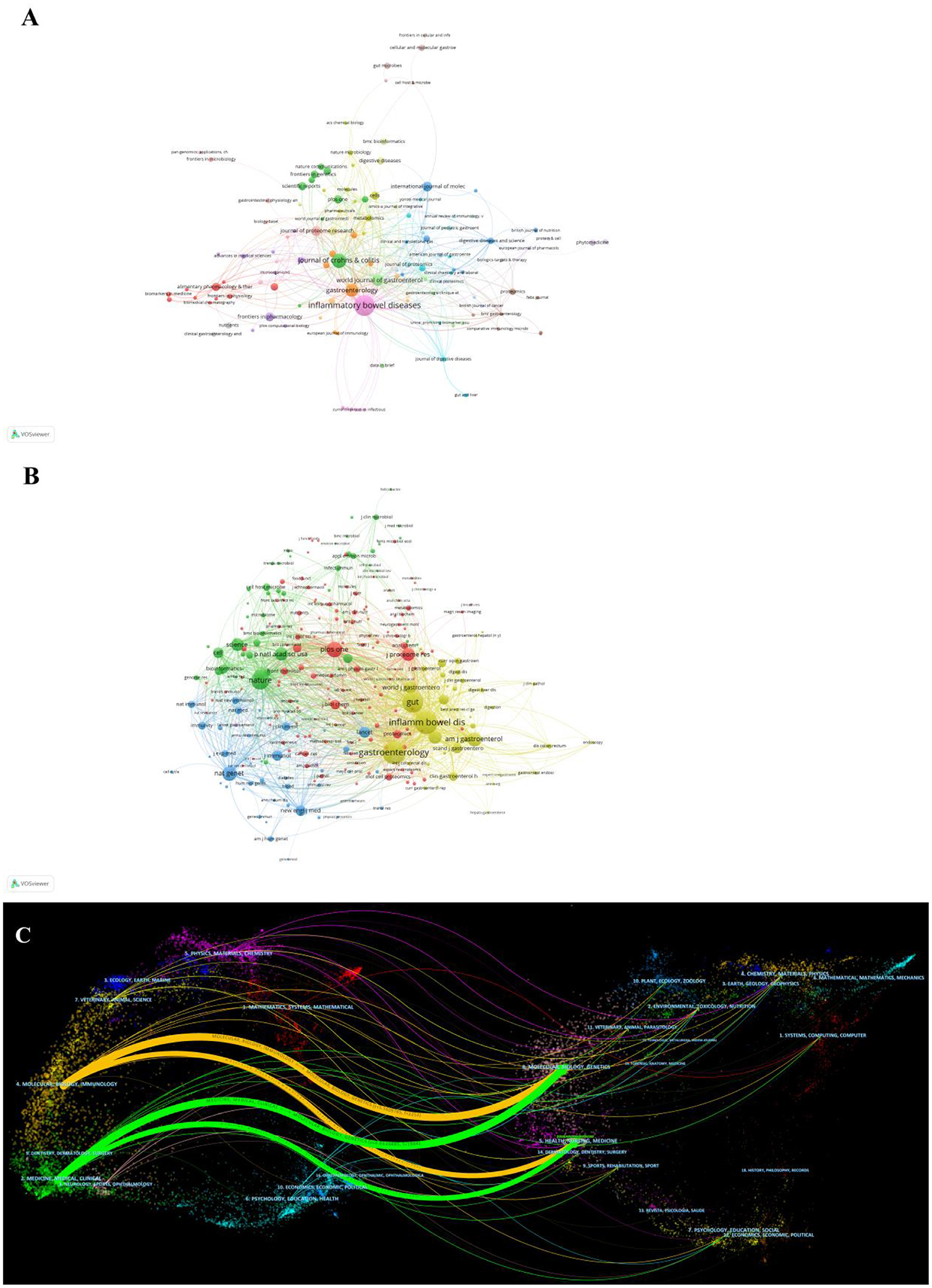
Figure 5. (A) Network of journals on research of omics in UC. (B) Network of co-cited journals on research of omics in UC. (C) The dual-map overlay of journals on research of omics in UC.
Co-citation was called as the frequency with which two documents were cited together. The co-citation analysis of journals was considered the assessment of the influence within a particular field. As shown in Table 3, among the 2,913 co-cited journals, inflammatory bowel disease had the most co-citations (1,223), followed by Gastroenterology (1,204) and gut (1,005). Moreover, Nature (IF = 69.504) has the highest impact factor, followed by Nature Genetics (41.376). Figure 5B presents the co-citation network, which is filtered by journals' minimum co-citation equal to 20.
The dual-map overlay map is the discipline co-occurrence network, which simultaneously displays both the journals and co-cited journals. As shown in Figure 5C, the dual-map overlay of journals showed two main citation paths. The orange path, articles published in Molecular Biology/ Immunology area, and the cited publications were mostly published in journals in the fields of Molecular Biology/Genetics. The green path represented the research published in Medicine/Medical/Clinical and was mainly cited by the literature in Molecular Biology/Genetics/Health/Nurse/ Medicine.
Co-cited references
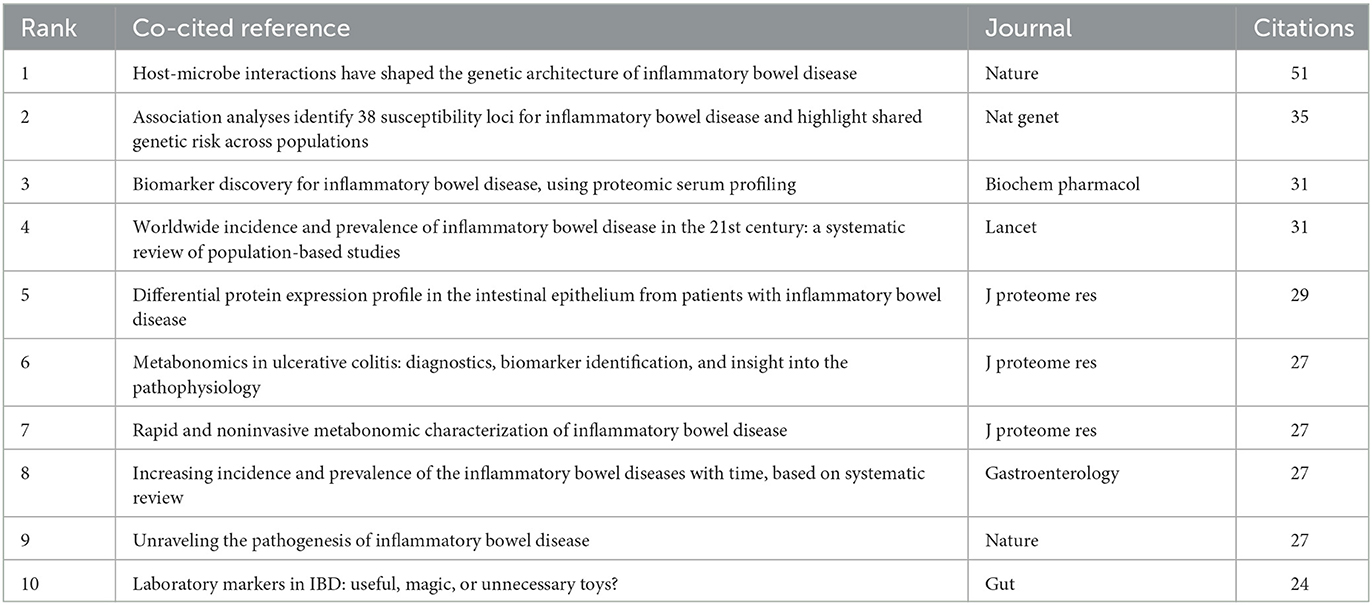
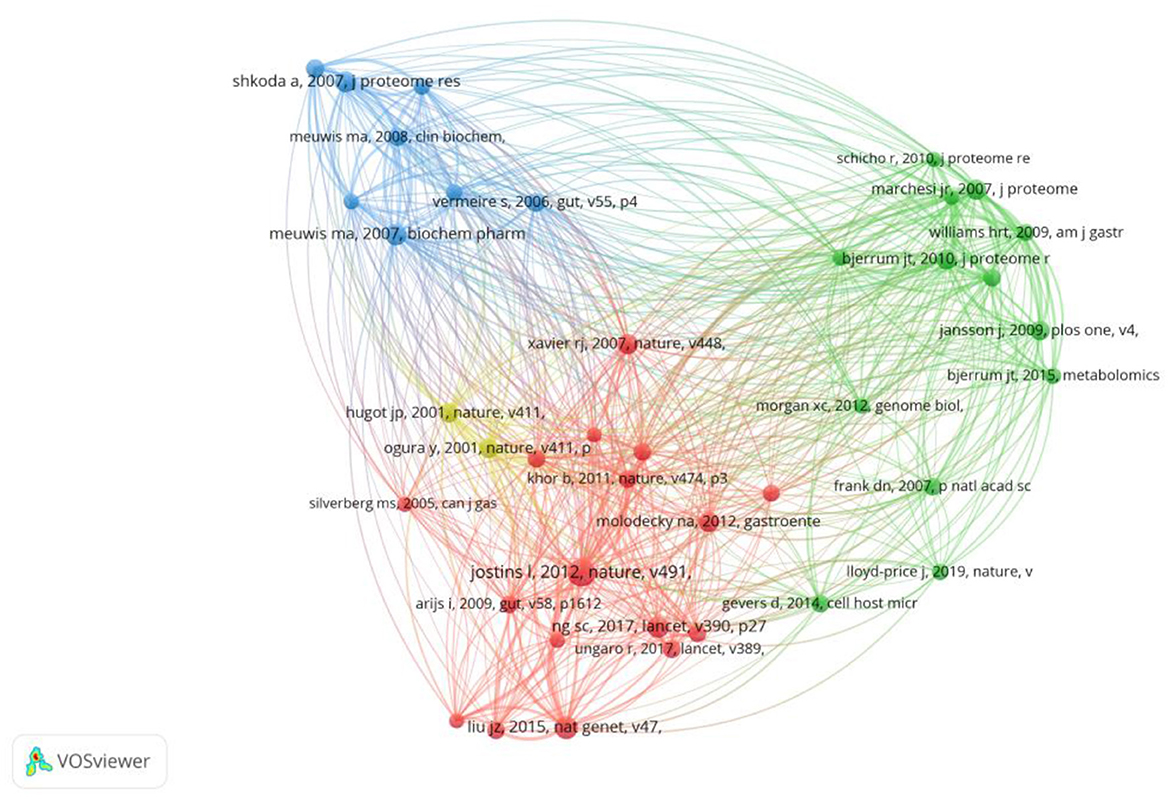
Co-cited references are considered as two documents cited together. From 2000 to 2022, there were 19,478 co-cited references on the topic of omics and ulcerative colitis. Of those, 26 studies had more than 20 citations. Table 4 presents the top 10 co-cited references with the most citations. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease (15) published in Nature, was the most frequently co-cited (n = 51), followed by association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations (25), published in Nature Genetics, and biomarker discovery for inflammatory bowel disease, using proteomic serum profiling (26) published in Biochemical Pharmacology. The co-cited reference network map of references with a co-citation of more than or equal to 15 was constructed (Figure 6).
Reference with citation bursts
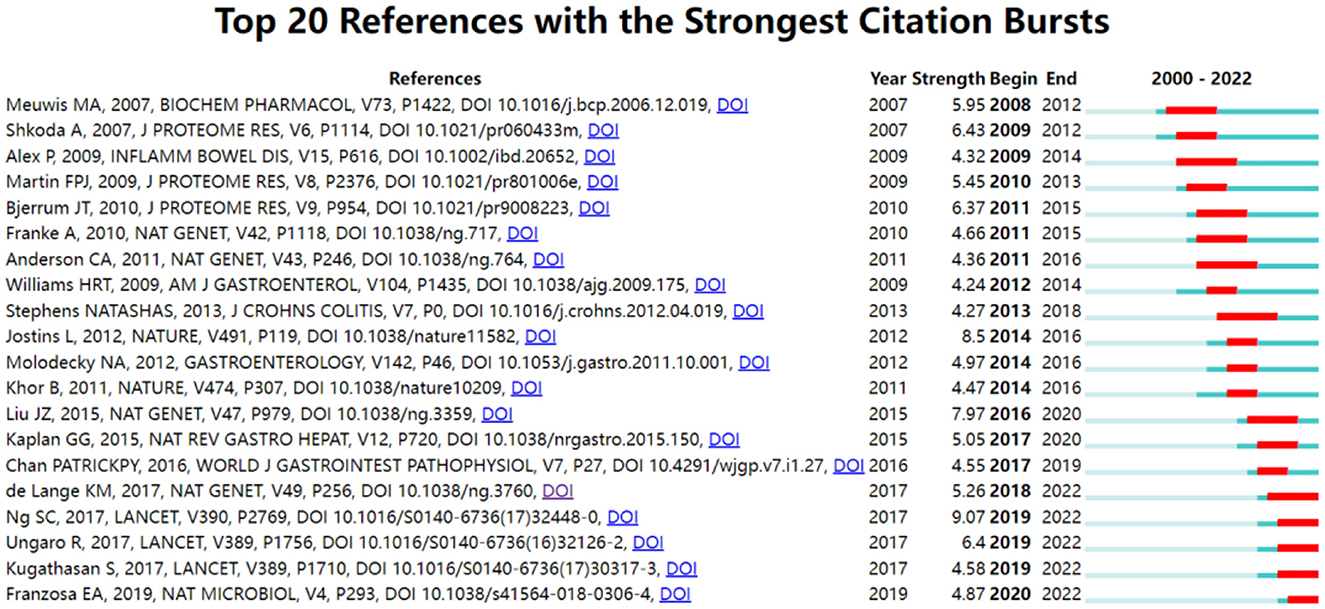
Citation bursts refer to a frequency surge of publications over a period of time, which can reflect the dynamics of a certain field in part. In our study, Figure 7 shows the top 20 references with the strongest citation bursts, which are identified by CiteSpace. As shown in Figure 7, the blue line represented the time interval, and the red line depicted the year of the beginning and end of each citation burst. Citation bursts appeared as early as 2007, the strength of these 20 references ranged from 4.24 to 9.07. The reference with the strongest citation burst of 9.07 entitled “Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies” published in The Lancet, with citation burst from 2019 to 2022, was written by Ng et al. (27). The second strongest citation burst (strength = 8.5) was entitled “Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease” with citation burst from 2016 to 2020, which was published in Nature (15).
Analysis of keywords and frontiers
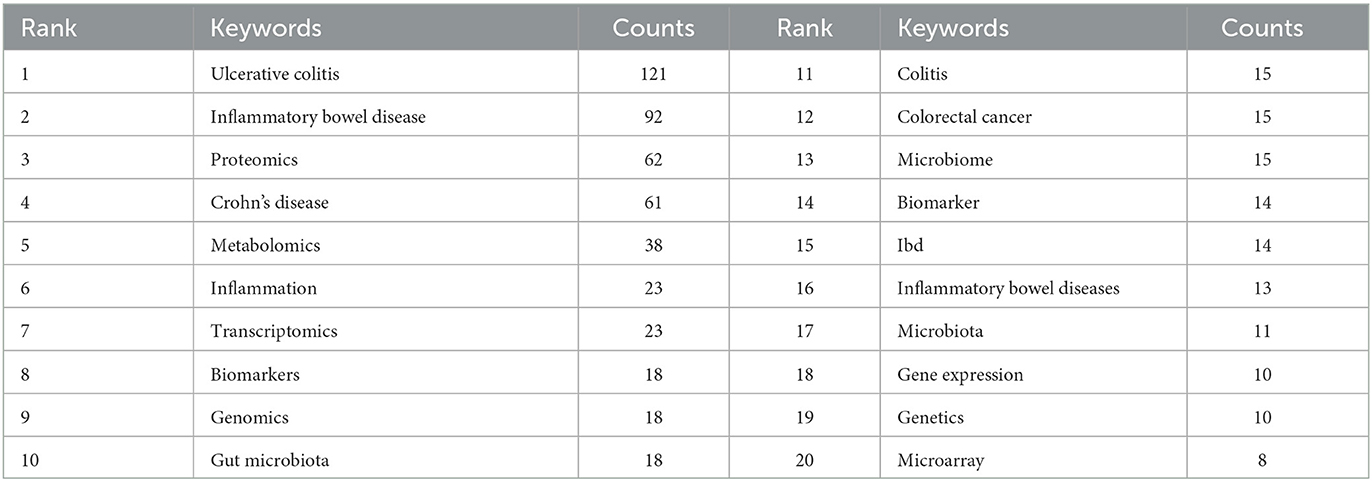
A co-occurrence keywords map has been created based on VOSviewer to quickly capture research hotspots, as shown in Figures 8A, B. In total, 924 author keywords were found, and 53 keywords met the threshold of a minimum number of occurrences more than or equal to four. As shown in Figure 8A, six clusters were obtained in total, which was indicating six research directions. The five closest keywords in those five main clusters (red, green, blue, purple, and yellow) were as follows: (1) colorectal cancer, innate immunity, gene expression, rheumatoid arthritis, and microarray; (2) transcriptomics, inflammatory bowel disease, microbiome, bioinformatics, and systems biology; (3) ulcerative colitis, Crohn's disease, proteomics, biomarkers, and mass spectrometry; (4) inflammatory bowel disease, metabolomics, gut microbiota, microbiota, and metabolome; (5) inflammation, genomics, genetics, colitis, and extracellular matrix. Table 5 highlights the 20 most occurring keywords. In addition, the trend topic analysis of the keywords was conducted in order to gain further insights into the trends in the field of omics in UC (Figure 8C). From 2008 to 2011, the main keywords were systemic lupus erythematosus, necrosis factor alpha, and antineutrophil cytoplasmic antibodies. High-throughput sequencing has greatly assisted in research on proteomics, metabolomics, transcriptomics, genomics, and gut microbiota since 2012. Notably, these keywords of dysbiosis, gut microbiota, loci, expression, and inflammation have remarkably represented the current research hotspots of omics in UC in the past 3 years (2020–2022).
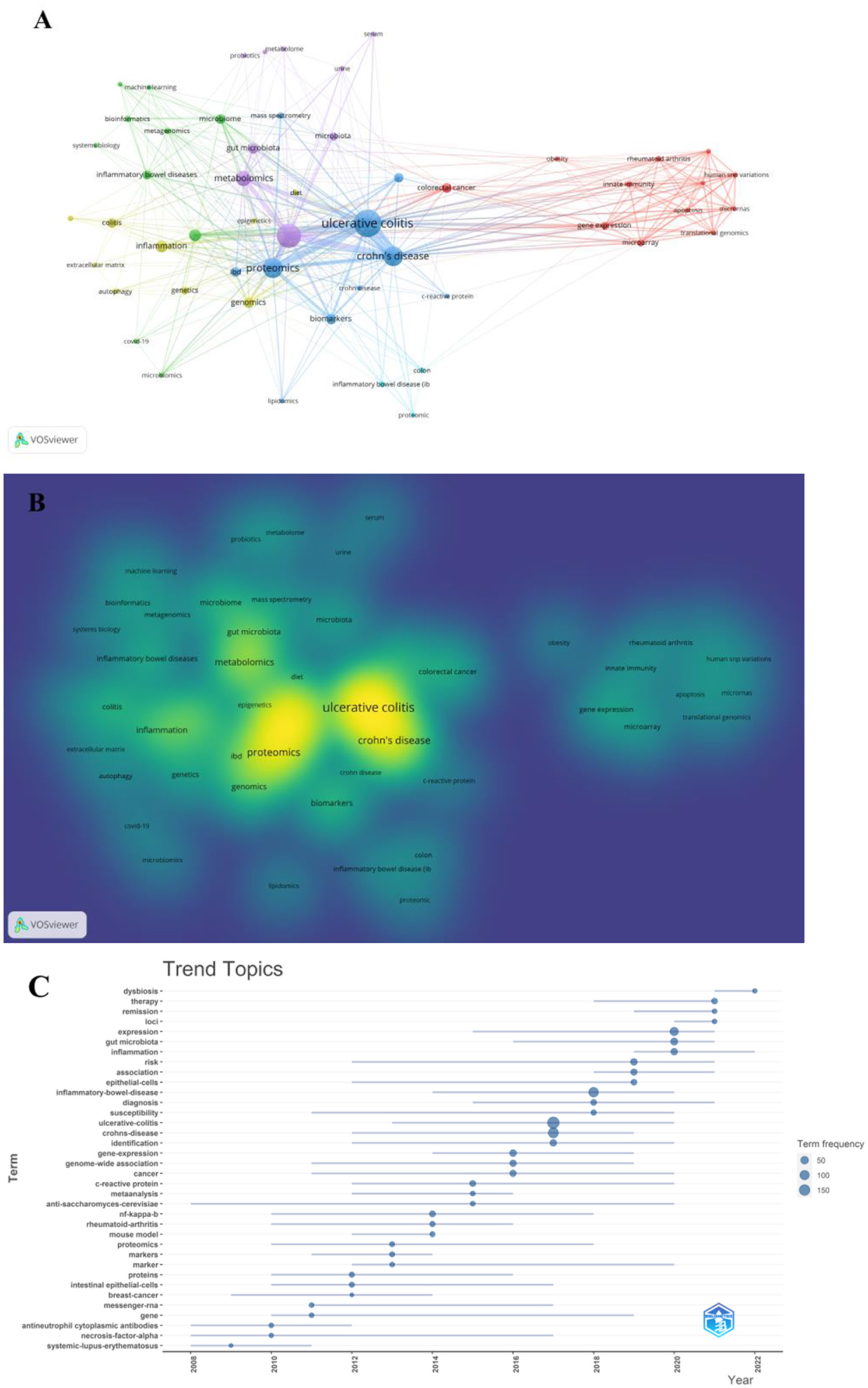
Figure 8. (A) Network of keyword on research of omics in UC. (B) Density map of keyword on research of omics in UC. (C) Trend topics.
Discussion
This study adopted VOSviewer, CiteSpace, and Bibliometrix to conduct a bibliometric analysis of 385 articles from the Web of Science core database in order to identify the research hotspots and new research trends in the field of omics in UC. The results showed that the annual publications from 2000 to 2007 were extremely rare, indicating that the research foundation of omics in UC was lacking. From 2008 to 2018, the trend in publications in this field fluctuated, with an average annual publication of 16.5 articles. According to the analysis of the citation number of articles and H-index, we found the publications with high citations were located in 2012 and 2017. Therefore, during the period of 2008 to 2018, research of omics in UC was in explosive period. The number of related publications increased rapidly after 2019, indicating that research of omics in UC had entered the maturity stage of development and drew more and more researchers' attention.
The publications show a dynamic trend and change varying with years as well as the difference among the different countries. The United States, Europe, and Asia were the main regions and countries conducting research on omics in UC, especially the United States. The United States was globally dominant in publication outputs, cited authors, international collaborations, and total citations. When it came to publication numbers, the United States, China, United Kingdom, and Germany ranked favorably. Furthermore, we noticed that the United States had active cooperation with the United Kingdom, Germany, Canada, and Brazil. Although China ranked second with a large number of publications, and international collaborations and citation publications were relatively low, which may indicate that Chinese researchers should carry out extensive cooperation with foreign research institutions and promote the development of omics in UC with high-quality publications. Among the top 10 leading institutions, six institutions were located in the USA, explaining the reasons for and rapid development of omics in UC in the USA. Harvard Medical School, the Icahn School of Medicine at Mount Sinai, the Massachusetts General Hospital, and the Brigham and Women's Hospital have extensive experience in this research field. The top 10 leading institutions are quite well research platforms for collaboration and further learning.
To deeply understand the research of omics in UC, the top 10 prolific authors are summarized. Ashwin Ananthakrishnan and Ramnik Xavier are both working at the Massachusetts General Hospital. They focus on research of epidemiology and outcomes of inflammatory bowel diseases and therapy personalized medicine. Their publications of “Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases”(28), “Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease”(29), and “Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease”(15) are associated with multi-omics profiles to facilitate therapeutics for patients and serve as targets for new therapies. Vibeke Andersen, from the University of Southern Denmark, has paid much attention to differential genetic architecture and new therapeutic targets in inflammatory bowel disease (15, 30, 31). The three authors have a close collaboration. The publication with the most citations “Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease” was co-authored by them.
From the perspective of the co-author, Séverine Vermeireis was the most frequently cited author, followed by Jacob Tveiten Bjerrum and Jacob Tveiten Bjerrum. Séverine Vermeire coming from the Catholic University of Leuven led a research team that focused on the genetics and pharmacogenetics of inflammatory bowel disease (32). Jacob Tveiten Bjerrum is interested in the research of characterizing ulcerative colitis by metabonomics. For example, one of his publications is “Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals” (33). Marie-Alice Meuwis has paid attention to proteomics studies of inflammatory bowel diseases (34, 35). Thus, the achievements of the top 10 leading co-authors laid the foundation for the research of omics in UC.
Meanwhile, most of the research on omics in UC was published in inflammatory bowel diseases (IF = 7.29, Q1), followed by Journals of Crohn's and Colitis (IF = 10.02, Q1) and Gastroenterology (IF = 33.883, Q1). The journals and co-cited journals can be divided into four groups: gastrointestinal-related journals, omics professional journals, biology and molecular journals, and comprehensive journals. Compared with journals, the citation of the journals is more high-quality international journals. From the category of journals, it demonstrates that gut, genome, and proteome are new important research areas.
Hotspots and frontiers
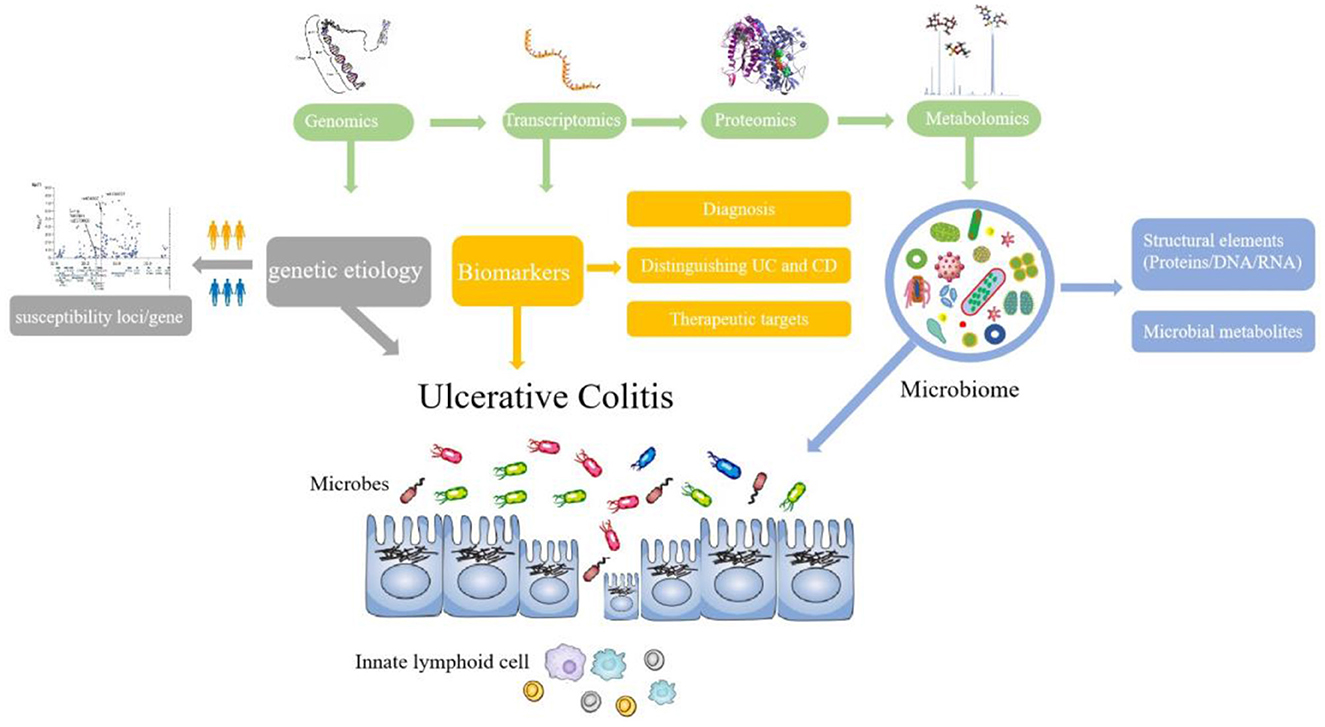
References with citation bursts and keywords represent frontiers and hotspots within a particular field (36). The reference with strong citation bursts “Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies” pointed out that IBD was a global disease with increasing incidence and prevalence in different regions (27). The publication “Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease” was undertaken a meta-analysis of Crohn's disease and ulcerative colitis genome-wide association scans to emphasize the relationship and pathways shared between host mucosal immune system and microbes (15). In addition, keywords can also quickly shed light on the distribution and evolution of hotspots in the research field of omics in UC. Table 5 mainly illustrated the following keywords: ulcerative colitis, inflammatory bowel disease, proteomics, metabolomics, inflammation, transcriptomics, biomarkers, genomics, and gut microbiota. Based on keyword clusters and trend topic analysis, we summarized that the research of omics in UC mainly focused on the following aspects (Figure 9):
1. To monitor the changes in UC microbial community composition and function,
Evidence has suggested that dysbiosis of intestinal microbiota resulting in excessive intestinal inflammation contributes to the complex pathogenesis of UC (37). In patients with UC, the microbial community composition, functional diversity, and stability are compromised (38). For example, some specific bacteria associated with Firmicutes are decreased greatly while Bacteroidetes bacteria and facultative anaerobes display certain increments (39). In addition, an increase in Desulfovibrio and pathogenic bacteria such as Fusobacterium varium were also found in patients with UC (40). Omics is a good way to explore and map the intestinal microbiota change in patients with UC. Genomics, such as metagenomic analysis and microbiome genome-wide association study (mGWAS), has provided new insights into host–microbiota interactions. Several recent efforts have been made to construct genomes collection of gut microbes associated with healthy humans using metagenome sequencing, as a reference database for gut microbiota studies (41). Microbiome genome-wide association study (mGWAS) could not only identify some heritable bacterial taxa but also characterize the composition and gene contents of the intestinal microbiome associated with human health or disease (42). The interactions of intestinal microbiota and host cells occur through metabolite production, such as short-chain fatty acids (SCFAs) which have a close relationship with inflammation in the host digestive tract. SCFAs exert beneficial effects on the intestinal immune system, including regulating the recognition of intestinal epithelial cells and secretion of repair cytokine (43). Previous studies revealed that dysbiotic microbiota in patients with IBD was mainly linked to decreased abundance of SCFA species, which may affect other important metabolic pathways (44). Metabolomics focuses on the functional status of host–microbial relationships in order to identify key drivers of metabolites and metabolic pathways. The co-cited reference “Metabonomics in Ulcerative Colitis: Diagnostics, Biomtherapyntification, And Insight into the Pathophysiology” pointed out that patients with active UC showed antioxidants and a range of amino acids increased, while lipid, glycerophosphocholine (GPC), myo-inositol, and betaine decreased (45). In addition, the gut microbiota appears to be a promising target in the treatment of UC. Evidence was shown that stemming from a multi-omics approach, the overabundance of proteases originating from the bacterium Bacteroides vulgatus contributed to UC disease activity, which gain an understanding of functional microbiota alterations that drive UC and provided a strategy to treat UC (46).
2. To explore the genetic etiology of UC
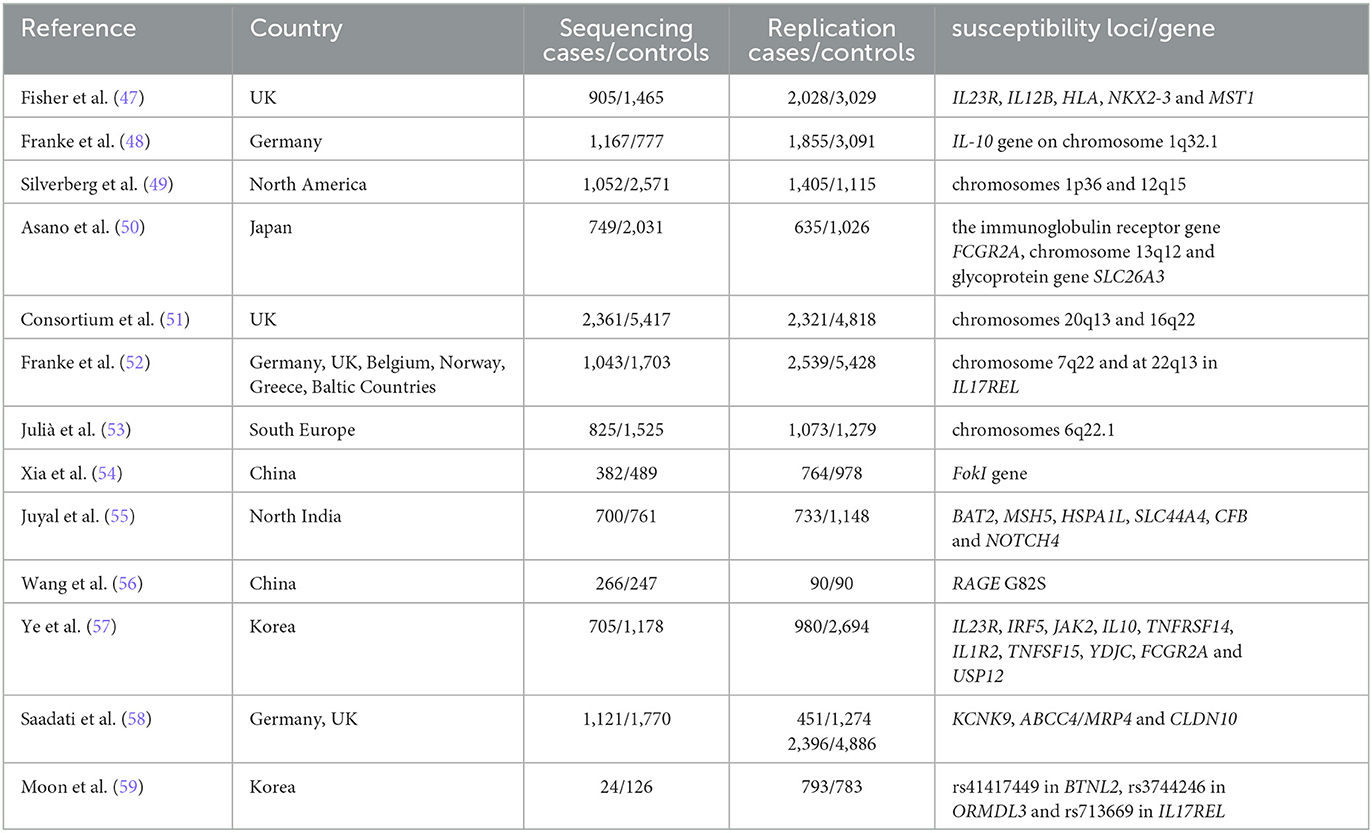
Genetic susceptibility increased the chance of developing a certain disease through genetic variation. The genetic epidemiological data clearly show that genetic susceptibility to the etiology of UC is inherited. Owing to the development of genome-wide association studies (GWAS), many potential culprit genes in UC have been successfully identified, as shown in Table 6.
3. To diagnose and distinguish UC and Crohn's disease
Until now, the diagnosis of UC is based on clinical implications, laboratory analysis, histopathological investigation, and imaging examination. Currently, studies have demonstrated that omics can be a diagnostic tool in active and quiescent UC and also provide a differential diagnosis of Crohn's disease (CD) and UC. The most common approach to explore UC biomarkers, especially proteomics, metabonomics, and metagenomics, is to assess relative differences in proteins, metabolites, and genes between patients with UC and controls (healthy or patients with CD). Han et al. (60) compared the protein profiles of colonic mucosa in three individuals (UC, CD, and healthy controls). Twenty-seven potential biomarkers for UC, 37 biomarkers for CD and bone marrow proteoglycan (PRG2), L-plastin (LCP1), and proteasome activator subunit 1 (PSME1) for active CD were identified. In addition, UC has been associated with dramatic changes in the gut microbiota changes in the gut metabolome and gene. Thus, it is a good way to explore biomarkers associated with diagnosis and distinguishing UC through a combination of 16S rRNA gene sequencing, shotgun metagenomics, and metabolomics (61, 62). Research on candidate biomarkers for UC could not only provide insights into UC pathogenesis but also future therapeutic targets.
Furthermore, with the progress in different omics fields, it is being recognized that multi-omics variously combined with two or more omics data during analysis could provide more valuable assistance in the diagnosis, biological processes, and treatment of UC. Multi-omics profiles could demonstrate dynamic changes in gut microbiota, as well as molecular disruptions in microbial transcription, metabolite pools, and levels of antibodies in host serum during UC activity (28). Moreover, a biomarker could develop to predict disease evolution and guide stratified therapeutic approaches. Multi-omics profiles such as fecal metagenomics, serum metabolomics, and proteomics markers serve as targets for newer therapies of UC (29, 63).
Currently, there are multiple challenges in UC management due to the course of the disease and its outcome. Therefore, there is a growing need for personalized approaches to enable timely therapy and avoid a one-size-fits-all standard of treatment and care. Omics, a systems biology approach, will help to identify and validate potential biomarkers that promote personalized treatment for UC (64). With the increase of research in UC through omics technology, network biology could become a useful tool for analyzing patient data generated from various omics platforms to improve risk assessment, disease monitoring, and personalized treatment for patients with UC.
Limitations
This study also has several limitations inherent in bibliometrics. First, a few relevant studies not included in the WoSCC database are ignored. However, WoSCC is a multidisciplinary, core journal citation index database covering approximately 34,000 journals worldwide (65). The search results can be exported from the WoSCC database and then imported into other software tools for further analysis. The WoSCC database is thought to be the most commonly used and appropriate database for bibliometric analyses. Second, only studies published in English were included, which may mean non-English publications were underestimated. Moreover, bibliometric analysis has its own weaknesses and bias. The publications with high frequency and citations may be cited for both negative as well as positive reasons. The recent publications are underrepresented due to time constraints. Nevertheless, our study still provides researchers with great objective information and insights.
Conclusion
For this study, we used bibliometrics to analyze and evaluate the related publications on omics and UC. Generally, the number of publications in this field fluctuated. The publications increased rapidly after 2019, indicating that the research of omics in UC had attracted global attention. In total, 49 countries, 809 institutions, 2,607 authors, and 205 journals were represented in all 385 articles. The United States and China ranked favorably. The USA had active cooperation with the United Kingdom, Germany, Canada, and Brazil. Harvard Medical School topped the list of institutions with the most publications. Ashwin Ananthakrishnan from the Massachusetts General Hospital was the most productive author and Séverine Vermeire from the Catholic University of Leuven was the co-cited author most often. Moreover, inflammatory bowel disease is the most popular and co-cited journal in this field. Through the analysis of references with citation bursts and trend topics, we find “ulcerative colitis,” “inflammatory bowel disease,” “microbiome,” “transcriptomics,” “genomics,” “metabolomics,” “proteomics,” “dysbiosis,” “biomarkers,” “loci,” and “therapy” are currently research hotspots. Further studies on gut microbiota and the pathological mechanism of omics will promote understanding and targeted therapies for UC.
Author contributions
HZ and SL design this study. HZ and YN collected and analyzed the data. HZ, HJ, and HL participated in writing the original draft. SL reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Aardema MJ, MacGregor JT. Toxicology and genetic toxicology in the new era of “toxicogenomics”: impact of “-omics” technologies. Mutat Res. (2002) 499:13–25. doi: 10.1016/s0027-5107(01)00292-5
2. Horgan RP, Kenny LC. ‘Omic' technologies: genomics, transcriptomics, proteomics and metabolomics. Obstet Gynaecol. (2011) 13:189–95. doi: 10.1576/toag.13.3.189.27672
3. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489:57–74. doi: 10.1038/nature11247
4. Chen WG, White FM. Proteomic analysis of cellular signaling. Expert Rev Proteomics. (2004) 1:343–54. doi: 10.1586/14789450.1.3.343
5. Selevsek N, Chang CY, Gillet LC, Navarro P, Bernhardt OM, Reiter L, et al. Reproducible and consistent quantification of the Saccharomyces cerevisiae proteome by SWATH-mass spectrometry. Mol Cell Proteomics. (2015) 14:739–49. doi: 10.1074/mcp.M113.035550
6. Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, Siuzdak G. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol. (2018) 36:316–20. doi: 10.1038/nbt.4101
7. Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. (2008) 6:431–40. doi: 10.1038/nrmicro1872
8. Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. (2015) 3:31. doi: 10.1186/s40168-015-0094-5
9. Silverberg MS, Satsangi J, Ahmad T, Arnott IDR, Bernstein CN, Brant SR, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a working party of the 2005 Montreal world congress of gastroenterology. Can J Gastroenterol. (2005) 19:5. doi: 10.1155/2005/269076
10. Conrad K, Roggenbuck D, Laass MW. Diagnosis and classification of ulcerative colitis. Autoimmun Rev. (2014) 13:463–6. doi: 10.1016/j.autrev.2014.01.028
11. Shivashankar R, Tremaine WJ, Harmsen WS, Loftus Jr EV. Incidence and prevalence of Crohn's disease and ulcerative colitis in Olmsted County, Minnesota from 1970 through 2010. Clin Gastroenterol Hepatol. (2017) 15:857–63. doi: 10.1016/j.cgh.2016.10.039
12. Loftus CG, Loftus Jr EV, Harmsen SW, Zinsmeister AR, Tremaine WJ, Melton LJ 3rd, et al. Update on the incidence and prevalence of Crohn's disease and ulcerative colitis in Olmsted County, Minnesota, 1940–2000. Inflamm Bowel Dis. (2007) 13:254–61. doi: 10.1002/ibd.20029
13. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. (2011) 474:307–17. doi: 10.1038/nature10209
14. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
15. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. (2012) 491:119–24. doi: 10.1038/nature11582
16. De Bellis N. Bibliometrics and Citation Analysis: From the Science Citation Index to Cybermetrics. Scarecrow Press (2009). p. 23–49. ISBN 978-0-8108-6714-7.
17. Ma D, Yang B, Guan B, Song L, Liu Q, Fan Y, et al. A bibliometric analysis of pyroptosis from 2001 to 2021. Front Immunol. (2021) 12:731933. doi: 10.3389/fimmu.2021.731933
18. Mulet-Forteza C, Genovart-Balaguer J, Mauleon-Mendez E, Merigo JM. A bibliometric research in the tourism, leisure and hospitality fields. J Business Res. (2019) 101:819–27. doi: 10.1016/j.jbusres.2018.12.002
19. Qin Y, Zhang Q, Liu Y. Analysis of knowledge bases and research focuses of cerebral ischemia-reperfusion from the perspective of mapping knowledge domain. Brain Res Bull. (2020) 156 15–24. doi: 10.1016/j.brainresbull.2019.12.004
20. Van Eck NJ, Waltman L. Software Survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. (2010) 84:523–38. doi: 10.1007/s11192-009-0146-3
21. Van Eck NJ, Waltman L. VOSviewer Manual: Manual for VOSviewer version 16 15 Leiden: Centre for Science and Technology Studies (CWTS) of Leiden University. (2020).
22. Chen C, Song M. Visualizing a field of research: a methodology of systematic scientometric reviews. PLoS ONE. (2019) 14:e0223994. doi: 10.1371/journal.pone.0223994
23. Chen C. Science mapping: a systematic review of the literature. J Data Inform Sci. (2017) 2:1–40. doi: 10.1515/jdis-2017-0006
24. Zhang X-L, Zheng Y, Xia M-L, Wu Y-N, Liu X-J, Xie S-K, et al. Knowledge domain and emerging trends in vinegar research: a bibliometric review of the literature from WoSCC. Foods. (2020) 9:166. doi: 10.3390/foods9020166
25. Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. (2015) 47:979–86. doi: 10.1038/ng.3359
26. Meuwis M-A, Fillet M, Geurts P, de Seny D, Lutteri L, Chapelle J-P, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. (2007) 73:1422–33. doi: 10.1016/j.bcp.2006.12.019
27. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. (2017) 390:2769–78. doi: 10.1016/S0140-6736(17)32448-0
28. Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. (2019) 569:655–62. doi: 10.1038/s41586-019-1237-9
29. Lee JWJ, Plichta D, Hogstrom L, Borren NZ, Lau H, Gregory SM, et al. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe. (2021) 29:1294–304. doi: 10.1016/j.chom.2021.06.019
30. Somineni HK, Nagpal S, Venkateswaran S, Cutler DJ, Okou DT, Haritunians T, et al. Whole-genome sequencing of African Americans implicates differential genetic architecture in inflammatory bowel disease. Am J Hum Genet. (2021) 108:431–45. doi: 10.1016/j.ajhg.2021.02.001
31. Chen E, Chuang L-S, Giri M, Villaverde N, Hsu N-Y, Sabic K, et al. Inflamed ulcerative colitis regions associated with MRGPRX2-mediated mast cell degranulation and cell activation modules, defining a new therapeutic target. Gastroenterology. (2021) 160:1709–24. doi: 10.1053/j.gastro.2020.12.076
32. Cleynen I, Jüni P, Bekkering GE, Nüesch E, Mendes CT, Schmied S, et al. Genetic evidence supporting the association of protease and protease inhibitor genes with inflammatory bowel disease: a systematic review. PLoS ONE. (2011) 6:e24106. doi: 10.1371/journal.pone.0024106
33. Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Günther U, et al. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. (2015) 11:122–33. doi: 10.1007/s11306-014-0677-3
34. Merli A-M, Vieujean S, Massot C, Blétard N, Calvo FQ, Baiwir D, et al. Solute carrier family 12 member 2 as a proteomic and histological biomarker of dysplasia and neoplasia in ulcerative colitis. J Crohns Colitis. (2021) 15:287–98. doi: 10.1093/ecco-jcc/jjaa168
35. Pierre N, Salée C, Massot C, Blétard N, Mazzucchelli G, Smargiasso N, et al. Proteomics highlights common and distinct pathophysiological processes associated with ileal and colonic ulcers in Crohn's disease. J Crohns Colitis. (2020) 14:205–15. doi: 10.1093/ecco-jcc/jjz130
36. Miao Y, Zhang Y, Yin L. Trends in hepatocellular carcinoma research from 2008 to 2017: a bibliometric analysis. PeerJ. (2018) 6:e5477. doi: 10.7717/peerj.5477
37. Shen Z-H, Zhu C-X, Quan Y-S, Yang Z-Y, Wu S, Luo W-W, et al. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J Gastroenterol. (2018) 24:5–14. doi: 10.3748/wjg.v24.i1.5
38. Hansen J, Gulati A, Sartor RB. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol. (2010) 26:564–71. doi: 10.1097/MOG.0b013e32833f1195
39. Gryaznova MV, Solodskikh SA, Panevina AV, Syromyatnikov MY, Dvoretskaya YD, Sviridova TN, et al. Study of microbiome changes in patients with ulcerative colitis in the Central European part of Russia. Heliyon. (2021) 7:e06432. doi: 10.1016/j.heliyon.2021.e06432
40. Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. (2017) 51:413. doi: 10.1146/annurev-genet-110711-155532
41. Hiseni P, Rudi K, Wilson RC, Hegge FT, Snipen L. HumGut: a comprehensive human gut prokaryotic genomes collection filtered by metagenome data. Microbiome. (2021) 9:1–12. doi: 10.1186/s40168-021-01114-w
42. Awany D, Allali I, Dalvie S, Hemmings S, Mwaikono KS, Thomford NE, et al. Host and microbiome genome-wide association studies: current state and challenges. Front Genet. (2019) 9:637. doi: 10.3389/fgene.2018.00637
43. Zhang Z, Zhang H, Chen T, Shi L, Wang D, Tang D. Regulatory role of short-chain fatty acids in inflammatory bowel disease. Cell Commun Signal. (2022) 20:1–10. doi: 10.1186/s12964-021-00806-y
44. Sasso GL, Khachatryan L, Kondylis A, Battey JND, Sierro N, Danilova NA, et al. Inflammatory bowel disease–associated changes in the gut: focus on Kazan patients. Inflamm Bowel Dis. (2021) 27:418–33. doi: 10.1093/ibd/izaa188
45. Bjerrum JT, Nielsen OH, Hao F, Tang H, Nicholson JK, Wang Y, et al. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J Proteome Res. (2010) 9:954–62. doi: 10.1021/pr9008223
46. Mills RH, Dulai PS, Vázquez-Baeza Y, Sauceda C, Daniel N, Gerner RR, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. (2022) 7:262–76. doi: 10.1038/s41564-021-01050-3
47. Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. (2008) 40:710–2. doi: 10.1038/ng.145
48. Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. (2008) 40:1319–23. doi: 10.1038/ng.221
49. Silverberg MS, Cho JH, Rioux JD, McGovern DPB, Wu J, Annese V, et al. Ulcerative colitis–risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. (2009) 41:216–20. doi: 10.1038/ng.275
50. Asano K, Matsushita T, Umeno J, Hosono N, Takahashi A, Kawaguchi T, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. (2009) 41:1325–9. doi: 10.1038/ng.482
51. Consortium UIG, Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. (2009) 41:1330–4. doi: 10.1038/ng.483
52. Franke A, Balschun T, Sina C, Ellinghaus D, Häsler R, Mayr G, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat Genet. (2010) 42:292–4. doi: 10.1038/ng.553
53. Julià A, Domènech E, Chaparro M, García-Sánchez V, Gomollón F, Panés J, et al. A genome-wide association study identifies a novel locus at 6q22. 1 associated with ulcerative colitis. Hum Mol Genet. (2014) 23:6927–34. doi: 10.1093/hmg/ddu398
54. Xia S-L, Yu L-Q, Chen H, Hu D-Y, Shao X-X, Guo M-D, et al. Association of vitamin D receptor gene polymorphisms with the susceptibility to ulcerative colitis in patients from Southeast China. J Recept Signal Transduct. (2015) 35:530–5. doi: 10.3109/10799893.2014.975248
55. Juyal G, Negi S, Sood A, Gupta A, Prasad P, Senapati S, et al. Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut. (2015) 64:571–9. doi: 10.1136/gutjnl-2013-306625
56. Wang J, Zeng J, Wang H, Ye S, Bi Y, Zhou Y, et al. Genetic polymorphisms of RAGE and risk of ulcerative colitis in a Chinese population. Immunol Lett. (2016) 170:88–94. doi: 10.1016/j.imlet.2015.09.003
57. Ye BD, Choi H, Hong M, Yun WJ, Low H-Q, Haritunians T, et al. Identification of ten additional susceptibility loci for ulcerative colitis through immunochip analysis in Koreans. Inflamm Bowel Dis. (2016) 22:13–9. doi: 10.1097/MIB.0000000000000584
58. Saadati HR, Wittig M, Helbig I, Häsler R, Anderson CA, Mathew CG, et al. Genome-wide rare copy number variation screening in ulcerative colitis identifies potential susceptibility loci. BMC Med Genet. (2016) 17:1–10. doi: 10.1186/s12881-016-0289-z
59. Moon CM, Kim SW, Ahn JB, Ma HW, Che X, Kim TI, et al. Deep of ulcerative colitis-associated genes identifies novel variants in candidate genes in the korean population. Inflamm Bowel Dis. (2018) 24:1706–17. doi: 10.1093/ibd/izy122
60. Han N-Y, Choi W, Park J-M, Hee Kim EH, Lee H, Hahm K-B. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. J Dig Dis. (2013) 14:166–74. doi: 10.1111/1751-2980.12035
61. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol.. (2019) 4:293–305. doi: 10.1038/s41564-018-0306-4
62. Galipeau HJ, Caminero A, Turpin W, Bermudez-Brito M, Santiago A, Libertucci J, et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology. (2021) 160:1532–45. doi: 10.1053/j.gastro.2020.12.004
63. Lacroix V, Cassard A, Mas E, Barreau F. Multi-omics analysis of gut microbiota in inflammatory bowel diseases: what benefits for diagnostic, prognostic and therapeutic tools? Int J Mol Sci. (2021) 22:11255. doi: 10.3390/ijms222011255
64. Lamb CA, Saifuddin A, Powell N, Rieder F. The future of precision medicine to predict outcomes and control tissue remodeling in inflammatory bowel diseas. Gastroenterology. (2022) 5:1525–42. doi: 10.1053/j.gastro.2021.09.077
Keywords: bibliometrics, omics, ulcerative colitis, inflammatory bowel disease, CiteSpace, VOSviewer, Bibliometrix
Citation: Zhang H, Ni Y, Ji H, Liu H and Liu S (2023) Research trends of omics in ulcerative colitis: A bibliometric analysis. Front. Med. 10:1115240. doi: 10.3389/fmed.2023.1115240
Received: 03 December 2022; Accepted: 20 February 2023;
Published: 27 March 2023.
Edited by:
Giorgia Bodini, San Martino Hospital (IRCCS), ItalyReviewed by:
Chuanxing Xiao, Xiamen University, ChinaPaulina Krawiec, Medical University of Lublin, Poland
Copyright © 2023 Zhang, Ni, Ji, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoneng Liu, bGl1c2hhb25lbmdAMTI2LmNvbQ==
 He Zhang
He Zhang Yuanyuan Ni1
Yuanyuan Ni1