
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 08 August 2022
Sec. Ophthalmology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.906554
This article is part of the Research Topic Clinical Applications of Artificial Intelligence in Retinal and Optic Nerve Disease View all 6 articles
 Jimmy S. Chen1,2
Jimmy S. Chen1,2 Sally L. Baxter1,2*
Sally L. Baxter1,2*Advances in technology, including novel ophthalmic imaging devices and adoption of the electronic health record (EHR), have resulted in significantly increased data available for both clinical use and research in ophthalmology. While artificial intelligence (AI) algorithms have the potential to utilize these data to transform clinical care, current applications of AI in ophthalmology have focused mostly on image-based deep learning. Unstructured free-text in the EHR represents a tremendous amount of underutilized data in big data analyses and predictive AI. Natural language processing (NLP) is a type of AI involved in processing human language that can be used to develop automated algorithms using these vast quantities of available text data. The purpose of this review was to introduce ophthalmologists to NLP by (1) reviewing current applications of NLP in ophthalmology and (2) exploring potential applications of NLP. We reviewed current literature published in Pubmed and Google Scholar for articles related to NLP and ophthalmology, and used ancestor search to expand our references. Overall, we found 19 published studies of NLP in ophthalmology. The majority of these publications (16) focused on extracting specific text such as visual acuity from free-text notes for the purposes of quantitative analysis. Other applications included: domain embedding, predictive modeling, and topic modeling. Future ophthalmic applications of NLP may also focus on developing search engines for data within free-text notes, cleaning notes, automated question-answering, and translating ophthalmology notes for other specialties or for patients, especially with a growing interest in open notes. As medicine becomes more data-oriented, NLP offers increasing opportunities to augment our ability to harness free-text data and drive innovations in healthcare delivery and treatment of ophthalmic conditions.
Adoption of electronic health records (EHRs) and advances in ocular imaging technology have revolutionized healthcare delivery in ophthalmology and resulted in significantly increased data available for clinical care and research (1). Moreover, the breadth of available data has resulted in large, multimodal datasets that have enabled the revolution in “big data” analytics (1, 2). The American Academy of Ophthalmology (AAO) and National Institutes of Health (NIH) have supported this movement with the development of large, processed EHR-based datasets such as the Intelligent Research in Sight (IRIS) Registry (3, 4) and the All of Us research program (5). Research efforts using these datasets have largely focused on retrospective association analysis and trends in care (6–14). Large datasets have also been used to develop predictive artificial intelligence (AI) models. The majority of these applications within ophthalmology have focused on image-based AI including diagnosis of diabetic retinopathy (15, 16), age-related macular degeneration (17, 18), retinopathy of prematurity (19, 20), and glaucoma (21–23), among others. Though structured datasets (such as extracted tabular data from EHRs) and large image datasets have been studied extensively in ophthalmic big data applications, far fewer AI studies in ophthalmology have utilized unstructured, or free-text, data such as EHR clinical notes from office visits (24–27). Because clinical notes represent the majority of provider documentation regarding each office visit, there remains a large amount of untapped free-text data (up to 80% of data in the EHR) that may be useful in predictive AI or analytics (28).
Natural language processing (NLP) is a subfield of AI focused on extracting and processing text data, including written and spoken words. While NLP as a linguistic concept originated in the early 1900s, it did not gain widespread interest until the last few decades with the proliferation of computer-based and AI algorithms. Within medicine, NLP has primarily been used for information retrieval (IR, otherwise known as search) (29, 30), text extraction for analytic studies, and AI algorithm development, though recent studies have focused on more complex tasks such as question-answering and summarization. Furthermore, there is a dearth of studies exploring the use of NLP in ophthalmology. Because ophthalmology is a high-volume medical and surgical subspecialty, there are significant opportunities to take advantage of the wealth of available data to develop text-based technologies with the potential to improve patient care and enhance future research.
The purpose of this study was to introduce ophthalmologists and researchers to natural language processing by (1) reviewing current ophthalmic applications of NLP, and (2) discussing future opportunities for NLP in ophthalmology.
In simple terms, the goal of NLP is to learn meaning from a set of words. However, the distinction between NLP, AI, and machine learning (ML) is often unclear. Broadly, AI is the branch of computer science that deals with teaching computers to perform tasks ordinarily performed by humans (31, 32). ML is a branch of AI that deals with developing models for automated prediction of a given task (33–35). Within ML, modeling can be performed using neural networks, which have the ability to learn from large amounts of data without explicitly defined features, an area known as deep learning (DL) (33, 36). While several NLP techniques that do not utilize modeling such as ML or DL exist, some NLP can be used to perform modeling with ML or DL, using free-text (raw or processed) as input rather than images or pre-defined features (i.e., tabular data, or data in tables) (27, 37). This intersection of ML, DL, and NLP is shown in Figure 1.
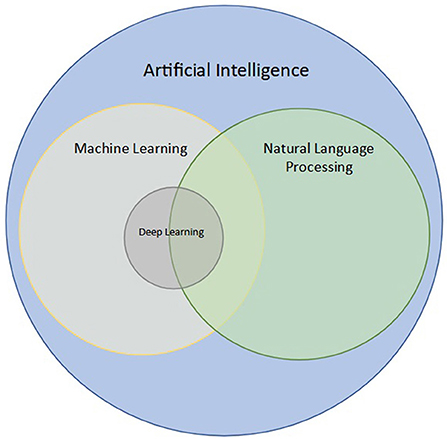
Figure 1. Intersection of natural language processing (NLP) with artificial intelligence (AI), machine learning (ML), and deep learning (DL). NLP is a branch of AI concerned with processing and analyzing text data. ML is a subfield of AI aimed at modeling data, and DL is a subfield of ML that uses neural networks to analyze large datasets. NLP techniques may utilize ML and DL when used for classification of words, sentences, or even paragraphs.
Before the advent of computational NLP techniques, search methods originally focused on simple keyword extraction. At the most basic level, this was analogous to the “find” function in a word processor, where a body of text was searched for all instances of a specific word or phrase, often described as a regular expression, or regex, in computer science. Relatively more advanced search could be performed using rule-based search, or conditional searching, such as extracting a word if it was in the sentence with another word. In fact, this concept of search, otherwise known as information retrieval (IR), is an important cornerstone of NLP (38, 39). However, the primitive methods described above are limited by the need for manual search input and a prior understanding of the text involved.
More sophisticated methods of text extraction require understanding the context of each input word in a body of text. This is most commonly done by labeling specific words as entities, which can include person, location, etc., a technique known as named entity recognition (NER). This is often done in conjunction with relation extraction (RE), which focuses on how phrases relate to others (i.e., patient underwent “4 cycles” of chemotherapy, where 4 cycles defines duration). However, these techniques often require pre-processing words within a given text to their simplest form. This usually begins with tokenization, or splitting a body of text into its individual words, and transforming all words to lowercase. Further text-preprocessing includes stemming (reducing a word down to its base form often with misspellings; i.e., “changes,” “changing” becomes “change”), lemmatization (simplifying a word down to its simplest form - i.e., “changes” to “change”, or “different” to “differ”), as well as stop-word removal (i.e., removing common words to simplify data analysis; most commonly articles like “a” and “the” are removed). Once a text has been pre-processed, NER techniques can be used to perform tasks such as de-identification, automated search, or annotating specific words (i.e., medications in progress notes). De-identification in particular has been a recent focus of research in NLP (40–42), and typically involves using text negation, or censoring out specific words of interest such as patient health information (PHI). NER can also be augmented by tagging each word's part of speech (43). In medicine, existing NLP models for NER such as MedEx (44) and MedLEE (45), which identify medications and diagnostic entities for billing, respectively, have been previously developed without ML. Off-the-shelf NER models for medical information extraction have also been provided by Amazon Comprehend Medical and require no prior programming knowledge, which has implications for increasing the accessibility for NLP engagement to the general public.
Recently, NLP techniques have utilized ML and DL to perform more intelligent and complex textual tasks. For example, several state-of-the-art algorithms have utilized ML and DL to create more robust and efficient NER algorithms, including open-source software libraries such as spaCy (46). However, these algorithms are unable to recognize similarities and differences between words (i.e., “happy” is similar to “joy” but different from “sad”). A simple method to capture word similarity is a bag-of-words approach, commonly implemented as term frequency-inverse document frequency (TF-IDF). In this approach, a numerical value is essentially assigned to each unique word, though this approach is limited by its ability to recognize synonyms and more complex relationships between words. To address this gap, word embedding was developed. Simply put, word embeddings, such as word2Vec (47), are developed as a result of DL algorithms that learn to assign a numerical distance to 2 words, and are trained to do so on many combinations of words based on the corpora of text used for training. These algorithms have previously been fine-tuned on several datasets including Google News and a combination of EHR and biomedical corpora (48). Current state-of-the-art word embedding algorithms have utilized more complex neural networks, known as transformers, to automate complex analysis of contexts between words. These algorithms, the most common of which is known as Bidirectional Encoder Representations from Transformers (BERT), introduce the idea of attention, of the ability to focus on specific words and their complex relationships, and have transformed our ability to perform text processing (49). BERT models have also been trained on biomedical text and include: clinicalBERT trained on EHR notes (50) and bioBERT trained on biomedical publications (51). Common applications of word embedding algorithms include tasks such as: question-answering, summarization (52–57), topic modeling (58–61), creating recommendation systems (62–64), chatbots (65–68), voice recognition (i.e., speech-to-text) (69, 70), text translation (71, 72), ranking texts for relevance based on a search query (73–75), and sentiment (emotion) analysis (76–78). A summary of these aforementioned described techniques and applications is shown in Figure 2.
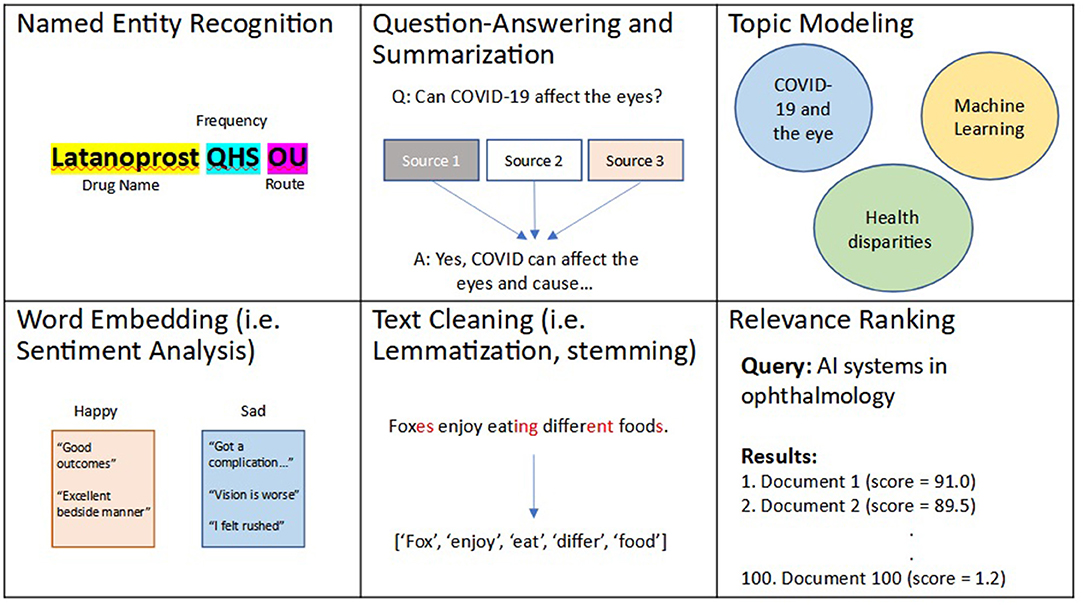
Figure 2. Examples of natural language processing (NLP) techniques and applications. Natural language processing, or NLP, is an area of artificial intelligence (AI) that deals with processing and analyzing textual data. Several NLP techniques include: relevance ranking, named entity recognition (NER), text cleaning, word embedding, which has applications in question-answering, summarization, topic modeling, among several other use cases.
To conduct this narrative review, a keyword-based and medical subject headings (MeSH)-based search of Pubmed and Google Scholars was performed in March 2022 using a combination of the following terms: “ophthalmology”, “optometry”, “eye”, “natural language processing”, and “NLP” to identify studies that used NLP in an ophthalmic context. These terms were combined in several different combinations and permutations in both search engines to yield an initial yield of 22 studies. Studies were included if they described original research using NLP in ophthalmology. All studies designed as a literature review or prototype description were excluded. Ancestor search was performed on included studies to further broaden our references. Both authors (JSC and SLB) manually reviewed each study's title, abstract, and manuscript text to validate the relevance of the studies to both ophthalmology and NLP. Data extracted from each study included: the authors and year of publication, study aim, NLP techniques used, performance, and study conclusions. Disagreements were resolved by discussion. This methodology is summarized in Figure 3.
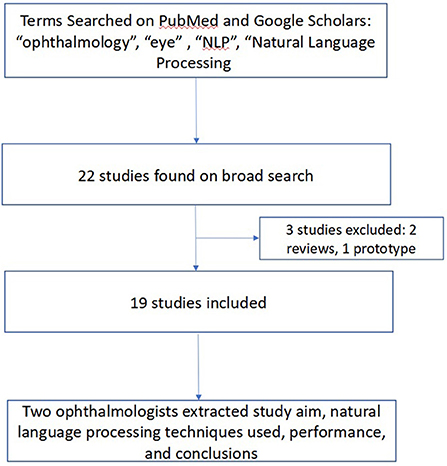
Figure 3. Methodology for Review of Ophthalmic Studies Utilizing Natural Language Processing (NLP). We searched PubMed and Google Scholars, augmented by ancestor search for studies related to use of NLP in ophthalmology applications.
Overall, 19 studies using NLP in ophthalmology were identified in the literature. These studies were published between 2000 and 2022, of which the majority (n = 11, 58%) were published within the last 3 years (2019–2022). Initial NLP studies did not use ML and focused mostly on algorithmic text extraction of relevant text from clinical notes using rule-based search and keyword extraction for parameters such as visual acuity (VA) (79–81), demographic data (i.e., age, sex) as well as clinical data (i.e., intraocular pressure, visual acuity) related to glaucoma (82) and cataract identification (83). Subsequent studies focused on using similar algorithmic rule-based search retrieving text relevant to the diagnosis and identification of several diseases such as herpes zoster ophthalmicus (84), pseudoexfoliation syndrome (85), microbial keratitis (25), and fungal endophthalmitis (24). While most published work has focused on extracting information from clinical visit notes (24, 84, 86), Stein et al. extracted a combination of unstructured data, problem lists, clinical notes, and billing code documentation for multi-modal extraction of pseudoexfoliation syndrome (85). Other use cases for text extraction using search included identifying antibiotics used for and post-operative complications of cataract surgery (87), extracting eye laterality and medications of patients who underwent cataract surgery (88), as well as for triaging ophthalmology referrals (89).
In the last 3 years, more recent studies have begun using ML for more sophisticated applications of NLP. For example, Wang et al. created the first word embeddings specific to ophthalmology using ophthalmology publications and EHR notes and found that DL models trained on ophthalmology-specific word embeddings outperformed those trained on previous word embeddings trained on general vocabulary for predicting prognosis of low-vision (90). These embeddings were also later used in combination with structural, tabular data from the EHR to refine models predicting low-vision prognosis (91). This idea of combining structured and unstructured data from the EHR was also applied to predicting glaucoma progression using similar methods described earlier (92). Additionally, Lin et al. recently applied an existing DL framework for NER to accurately extract entities relevant to ophthalmic medications (F1 score = 0.95) for glaucoma patients and simulated successful medication reconciliation as an application of this NLP model (93). The F score has become an increasingly popular metric to evaluate model performance in NLP, and measures both the precision (positive predictive value) and recall (sensitivity). These F scores can be weighted (with weights appended to the score name - i.e., F1, F2 scores) to increase the importance of maximizing either precision or recall. Other recent studies utilizing ML/DL with NLP included topic modeling to define groups of topics pertaining to ophthalmology publications during the COVID-19 pandemic (94), as well as sentiment analysis of user emotions from an ophthalmology forum (95). Topic modeling uses unsupervised learning, or machine learning without explicitly labeled data, to cluster documents by topic. In work performed by Hallak et al., the authors used a statistical model called Latent Dirichlet Allocation (LDA) to identify ocular manifestations of COVID-19, viral transmission, patient care, and practice management during the COVID-19 pandemic as relevant topics in ophthalmology over 2020–2021 (94). Additionally, in work by Nguyen et al., a cloud-based NLP program called Watson was utilized to associate emotions with extracted keywords from ophthalmology forums and demonstrated that NLP can be used to understand patient perspectives on care. A summary of these studies is shown in Table 1.
Ophthalmology is a surgical subspecialty that could significantly benefit from applications of NLP, though there is a relative scarcity of published studies compared to those exploring NLP in other areas of medicine. Future avenues of exploration within ophthalmology include: (1) more complex use cases for text extraction, (2) translating notes both in terms of language, as well as (3) applications to assist with patient interaction.
While most studies within ophthalmology have focused on searching for specific keywords or entities, text extraction can be more broadly used for other use cases. For example, cohort selection, particularly for rare diseases, is a necessary prerequisite for clinical trial recruitment, and has been facilitated in the past by NLP algorithms reviewing EHR notes. In the 2018 National NLP Clinical Challenge for cohort identification, the highest performing model achieved an F-score of 0.9 for identifying cohorts using various criteria (96). Within inherited retinal diseases, cohort identification has been recognized internationally as an important goal in research with rapid advances in gene therapy; (97) however, a previously published current cohort identification study within this space focused on simple keyword search without use of more sophisticated NLP techniques (98). Additionally, drug repurposing has long been of interest to the medical community (99–101), and has been employed in mouse models for inherited retinal diseases (102, 103) as well as hypothesis testing for ocular protection against COVID-19 (104). While ophthalmology stands to greatly benefit from drug repurposing (105), the majority of applications using NLP have been published exploring novel drug use in cancer (106, 107) and COVID-19 (108, 109). However, within ophthalmology (110), one study by Brilliant et al. retrospectively demonstrated that L-DOPA could have protective effects against development AMD. Although drugs were quickly repurposed owing to the urgency of the COVID-19 pandemic, there remains a need for further exploration and prospective validation of potential drug candidates for repurposing both within ophthalmology and other specialties. As our techniques and capabilities for big data collection and analytics rapidly advances, more research is needed in both cohort identification and drug repurposing using NLP techniques and may have important implications in accelerating new innovations in ophthalmology.
NLP is also positioned to address challenges in interpreting documentation in the EHR by facilitating improved communication and understanding of clinician notes. NLP techniques centered around word embeddings have recently been utilized to develop question-answering (111–114), as well as summarizing large bodies of text such as clinical notes (52–54), and scientific publications (55, 115). With the advent of the Open Notes movement, a movement supporting transparent documentation among patients, families, and clinicians (116–118), and the 21st Century Cures Act of 2021 (119), which mandated patient accessibility to their clinical notes, there has been an increasing emphasis on patient involvement and advocacy in their own care. However, previous work in ophthalmology exploring clinician attitudes toward Open Notes revealed concerns that patients would have a difficult time understanding their records (120). In fact, the terminology used in ophthalmology notes have been anecdotally difficult to understand even among clinicians in other specialties, reflected by the creation of tools used by non-ophthalmologists to help “translate” ophthalmology notes by replacing common abbreviations used in ophthalmology (121). Summarization techniques may be useful to translate notes into patient-friendly language or even other languages (71) and may improve patient engagement in their healthcare, especially in underrepresented populations (72). However, in a systematic review by Mishra et al., the authors found that current work in NLP-based summarization focused largely on summarizing biomedical literature (97% of published work) as opposed to clinical data from the EHR (3% of published work), reflecting a need for work in NLP-based summarization in the clinical domain (56). Because ophthalmologists utilize specialized knowledge that is not commonly known to clinicians in other specialties, ophthalmology, as well as primary care specialties, stand to benefit significantly from tools that could summarize ophthalmic notes using NLP. Additionally, question-answering may have a role in extracting key data that would be most useful to facilitate management plans by primary care providers. While these technologies have numerous potential benefits, iterative testing and stakeholder participation will be needed to ensure that these NLP applications are useful and trustworthy by their users.
Patient interaction and patient-physician relationships remain the hallmark of medicine, but in areas with limited resources, NLP may be able to augment knowledge dissemination and assist clinician workflows. For example, chatbots using NLP have been previously developed to help patients with triaging concerns related to inflammatory bowel disease (65), recommending medical specialties based on symptoms (66), and other uses cases including depression symptom monitoring (67). Similar NLP-based chatbots may potentially be developed to assist with ophthalmic treatment monitoring and medication adherence as well as triaging ophthalmic symptoms for evaluation, particularly in areas where ophthalmology services may not be readily available. Additionally, NLP has been explored in the context of digital scribes, which have the potential to reduce physician burnout and increase patient satisfaction (122, 123). The burden of EHR-based documentation in ophthalmology has been well-described previously (124–127). A growing number of companies including Microsoft, Google, Amazon, IBM, Mozilla, DeepScribe, Suki, and Robin Healthcare have developed NLP-based scribes with speech recognition and smart medical assistants (122, 128–130). Because the majority of these NLP-based scribes are still in development, performance data to date is limited, though recent data from DeepScribe suggested that the model had an error rate of 18%, which is significantly lower than error rates from existing models by IBM and Mozilla (38–65%) (122). Development of these scribes have been complicated by technical challenges (i.e., audio quality, audio-to-text transcription) as well as conversational challenges (i.e., meaningful summarization, extracting topics from often fragmented conversations) (123). A recent study by Dusek et al. showed that scribe use in ophthalmology was associated with increased documentation efficiency (131). Automated scribes may potentially further increase documentation efficiency, and may be able to provide additional value if integrated with automated text extraction for providing relevant clinical information. Augmedix is another company attempting to integrate both remote scribing while providing data via Google Glass, though no NLP methods are currently used (132). Future research may focus on integrating NLP into these technologies to fully develop a “computer-based assistant” to assist with documentation, which may allow clinicians to focus on their relationship with the patient. Specifically in ophthalmology, counseling patients on preventing blindness, which remains the leading feared condition among American patients (133), requires significant investment in patient-physician relationships, and automated documentation could improve the quality of these relationships. These tools could also have additional value from a clinical workflow standpoint as ophthalmology is a high-volume specialty that requires processing of several data points and imaging modalities. While both chatbots and digital scribes are promising for optimizing the patient-physician relationship, significant refinement and iterative development of these systems is required before clinical deployment is feasible.
Though the future of NLP is exciting across all medical specialties including ophthalmology, there are important limitations that existing and future applications must address before use in the clinical setting. First, natural text is highly variable and error prone. Previous studies have shown that both dictated (7% error rate) (134) and written clinical notes (135) frequently contain errors such as documented actions that were not performed during the visit, findings from the visit that were not charted, as well as grammatical and typographical errors. Further, text often contains uses of words that can be used in different contexts, including colloquialisms, irony, sarcasm, and synonyms. These linguistic nuances often are difficult for NLP algorithms to distinguish. Second, word embeddings in NLP are often trained in specific domains [i.e., scientific publications (29), documents from web search (136)]. This importantly impacts how word embeddings interpret relationships between words, as different words may have different meanings in other contexts. Third, NLP models trained using DL or ML require huge datasets. These datasets are often difficult to acquire, and often need to be collected for a variety of settings (i.e., multi-institutional) to train a robust, generalizable model. Transformers, the current state-of-the-art DL method for NLP, require large datasets, with prior studies demonstrating worse performance on more limited datasets (137). Fourth, the majority of NLP applications have currently been developed in the English language (138). To promote equity in care and reduce healthcare disparities, more research is needed in developing NLP applications in non-English languages (71, 138–140), which has the potential to benefit populations with limited access to healthcare resources.
NLP within ophthalmology is in its nascent stages of development and has already demonstrated potential in augmenting our ability to analyze free-text data from the EHR and improve predictive modeling with AI. As data from the EHR continues to grow, there remains significant opportunities to use NLP to improve our quality of research, “big data” analytics, and ultimately patient outcomes. However, there remain significant limitations of NLP that future work will need to address. More research and ongoing interdisciplinary collaborations will be needed to eventually translate NLP innovations into deployable solutions in the clinic.
JC drafted the manuscript. SB provided overall supervision and guidance. All authors conceived the study, analyzed the data, interpreted the data, reviewed the manuscript, and revised for important intellectual content. All authors contributed to the article and approved the submitted version.
This work was supported by NIH Grant DP5OD029610 (Bethesda, MD, USA) and an unrestricted departmental grant from Research to Prevent Blindness (New York, NY).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lee CS, Brandt JD, Lee AY. Big data and artificial intelligence in ophthalmology: where are we now? Ophthalmol Sci. (2021) 1:1–3. doi: 10.1016/j.xops.2021.100036
2. Cheng CY, Soh ZD, Majithia S, Thakur S, Rim TH, Tham YC, et al. Big data in ophthalmology. Asia Pac J Ophthalmol. (2020) 9:304. doi: 10.1097/APO.0000000000000304
3. Chiang MF, Sommer A, Rich WL, Lum F, Parke DW II. The 2016 American Academy of Ophthalmology IRIS® Registry (intelligent research in sight) database: characteristics and methods. Ophthalmology. (2018) 125:1143–8. doi: 10.1016/j.ophtha.2017.12.001
4. Parke DW, Rich WL, Sommer A, Lum F. The American Academy of Ophthalmology's IRIS® Registry (intelligent research in sight clinical data): a look back and a look to the future. Ophthalmology. (2017) 124:1572–4. doi: 10.1016/j.ophtha.2017.08.035
5. All All of Us Research Program Investigators, Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, et al. The “All of Us” research program. N Engl J Med. (2019) 381:668–76. doi: 10.1056/NEJMsr1809937
6. Chang TC, Parrish RK, Fujino D, Kelly SP, Vanner EA. Factors associated with favorable laser trabeculoplasty response: IRIS registry analysis. Am J Ophthalmol. (2021) 223:149–58. doi: 10.1016/j.ajo.2020.10.004
7. Leng T, Gallivan MD, Kras A, Lum F, Roe MT, Li C, et al. Ophthalmology and COVID-19: the impact of the pandemic on patient care and outcomes: an IRIS® Registry Study. Ophthalmology. (2021) 128:1782–4. doi: 10.1016/j.ophtha.2021.06.011
8. Rao P, Lum F, Wood K, Salman C, Burugapalli B, Hall R, et al. Real-world vision in age-related macular degeneration patients treated with single anti–VEGF drug type for 1 year in the IRIS registry. Ophthalmology. (2018) 125:522–8. doi: 10.1016/j.ophtha.2017.10.010
9. Pershing S, Lum F, Hsu S, Kelly S, Chiang MF, Rich WL III, et al. Endophthalmitis after Cataract Surgery in the United States: A Report from the Intelligent Research in Sight Registry, 2013–2017. Ophthalmology. (2020) 127:151–8. doi: 10.1016/j.ophtha.2019.08.026
10. Baxter SL, Saseendrakumar BR, Paul P, Kim J, Bonomi L, Kuo TT, et al. Predictive analytics for glaucoma using data from the all of US research program. Am J Ophthalmol. (2021) 227:74–86. doi: 10.1016/j.ajo.2021.01.008
11. Chan AX, Radha Saseendrakumar B, Ozzello DJ, Ting M, Yoon JS, Liu CY, et al. Social determinants associated with loss of an eye in the United States using the All of Us nationwide database. Orbit. (2021) 1–6. doi: 10.1080/01676830.2021.2012205
12. Lee EB, Hu W, Singh K, Wang SY. The association among blood pressure, blood pressure medications, and glaucoma in a nationwide electronic health records database. Ophthalmology. (2022) 129:276–84. doi: 10.1016/j.ophtha.2021.10.018
13. Delavar A, Radha Saseendrakumar B, Weinreb RN, Baxter SL. Racial and ethnic disparities in cost-related barriers to medication adherence among patients with glaucoma enrolled in the National Institutes of Health All of Us Research Program. JAMA Ophthalmol. (2022) 140:354–61. doi: 10.1001/jamaophthalmol.2022.0055
14. McDermott JJ IV, Lee TC, Chan AX, Ye GY, Shahrvini B, Saseendrakumar BR, et al. Novel association between opioid use and increased risk of retinal vein occlusion using the National Institutes of Health All of Us Research Program. Ophthalmol Sci. (2022) 2:1–8. doi: 10.1016/j.xops.2021.100099
15. Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. Npj Digit Med. (2018) 1:39. doi: 10.1038/s41746-018-0040-6
16. Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. (2016) 316:2402–10. doi: 10.1001/jama.2016.17216
17. Burlina PM, Joshi N, Pacheco KD, Freund DE, Kong J, Bressler NM. Use of deep learning for detailed severity characterization and estimation of 5-year risk among patients with age-related macular degeneration. JAMA Ophthalmol. (2018) 136:1359–66. doi: 10.1001/jamaophthalmol.2018.4118
18. Lee CS, Baughman DM, Lee AY. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. (2017) 1:322–7. doi: 10.1016/j.oret.2016.12.009
19. Brown JM, Campbell JP, Beers A, Chang K, Ostmo S, Chan RVP, et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. (2018) 136:803–10. doi: 10.1001/jamaophthalmol.2018.1934
20. Chen JS, Coyner AS, Ostmo S, Sonmez K, Bajimaya S, Pradhan E, et al. Deep learning for the diagnosis of stage in retinopathy of prematurity: accuracy and generalizability across populations and cameras. Ophthalmol Retina. 5:1027–35.
21. Medeiros FA, Jammal AA, Thompson AC. From machine to machine: an OCT-trained deep learning algorithm for objective quantification of glaucomatous damage in fundus photographs. Ophthalmology. (2019) 126:513–21. doi: 10.1016/j.ophtha.2018.12.033
22. Christopher M, Bowd C, Belghith A, Goldbaum MH, Weinreb RN, Fazio MA, et al. Deep learning approaches predict glaucomatous visual field damage from OCT Optic nerve head en face images and retinal nerve fiber layer thickness maps. Ophthalmology. (2020) 127:346–56. doi: 10.1016/j.ophtha.2019.09.036
23. Christopher M, Bowd C, Proudfoot JA, Belghith A, Goldbaum MH, Rezapour J, et al. Deep learning estimation of 10-2 and 24-2 visual field metrics based on thickness maps from macula optical coherence tomography. Ophthalmology. 128:1534–48.
24. Baxter SL, Klie AR, Radha Saseendrakumar B, Ye GY, Hogarth M. Text processing for detection of fungal ocular involvement in critical care patients: cross-sectional study. J Med Int Res. (2020) 22:e18855. doi: 10.2196/18855
25. Maganti N, Tan H, Niziol LM, Amin S, Hou A, Singh K, et al. Natural language processing to quantify microbial keratitis measurements. Ophthalmology. (2019) 126:1722–4. doi: 10.1016/j.ophtha.2019.06.003
26. Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc. (2020) 27:457–70. doi: 10.1093/jamia/ocz200
27. Yang LWY, Ng WY, Foo LL, Liu Y, Yan M, Lei X, et al. Deep learning-based natural language processing in ophthalmology: applications, challenges and future directions. Curr Opin Ophthalmol. (2021) 32:397–405. doi: 10.1097/ICU.0000000000000789
28. Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. (2013) 309:1351–2. doi: 10.1001/jama.2013.393
29. Roberts K, Alam T, Bedrick S, Demner-Fushman D, Lo K, Soboroff I, et al. Searching for scientific evidence in a pandemic: an overview of TREC-COVID. J Biomed Inform. (2021) 121:103865. doi: 10.1016/j.jbi.2021.103865
30. Gundlapalli AV, Carter ME, Palmer M, Ginter T, Redd A, Pickard S, et al. Using natural language processing on the free text of clinical documents to screen for evidence of homelessness among US veterans. AMIA Annu Symp Proc AMIA Symp. (2013) 2013:537–46.
31. Amisha, Malik P, Pathania M, Rathaur VK. Overview of artificial intelligence in medicine. J Fam Med Prim Care. (2019) 8:2328–31. doi: 10.4103/jfmpc.jfmpc_440_19
32. Ramesh AN, Kambhampati C, Monson JRT, Drew PJ. Artificial intelligence in medicine. Ann R Coll Surg Engl. (2004) 86:334–8. doi: 10.1308/147870804290
33. Choi RY, Coyner AS, Kalpathy-Cramer J, Chiang MF, Campbell JP. Introduction to machine learning, neural networks, and deep learning. Transl Vis Sci Technol. (2020) 9:14. doi: 10.1167/tvst.9.2.14
34. Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, et al. Artificial intelligence and machine learning in clinical development: a translational perspective. NPJ Digit Med. (2019) 2:69. doi: 10.1038/s41746-019-0148-3
35. Helm JM, Swiergosz AM, Haeberle HS, Karnuta JM, Schaffer JL, Krebs VE, et al. Machine learning and artificial intelligence: definitions, applications, and future directions. Curr Rev Musculoskelet Med. (2020) 13:69–76. doi: 10.1007/s12178-020-09600-8
37. Lin WC, Chen JS, Chiang MF, Hribar MR. Applications of artificial intelligence to electronic health record data in ophthalmology. Transl Vis Sci Technol. (2020) 9:13. doi: 10.1167/tvst.9.2.13
38. Hersh WR, Greenes RA. Information retrieval in medicine: state of the art. Comput Comput Med Pract. (1990) 7:302–11.
39. Hersh W. Information Retrieval: A Biomedical and Health Perspective. 4th ed. New York, NY: Springer (2020).
40. Gupta A, Lai A, Mozersky J, Ma X, Walsh H, DuBois JM. Enabling qualitative research data sharing using a natural language processing pipeline for deidentification: moving beyond HIPAA Safe Harbor identifiers. JAMIA Open. (2021) 4:ooab069. doi: 10.1093/jamiaopen/ooab069
41. Norgeot B, Muenzen K, Peterson TA, Fan X, Glicksberg BS, Schenk G, et al. Protected Health Information filter (Philter): accurately and securely de-identifying free-text clinical notes. NPJ Digit Med. (2020) 3:57. doi: 10.1038/s41746-020-0258-y
42. Yang X, Lyu T, Li Q, Lee CY, Bian J, Hogan WR, et al. A study of deep learning methods for de-identification of clinical notes in cross-institute settings. BMC Med Inform Decis Mak. (2019) 19:232. doi: 10.1186/s12911-019-0935-4
43. Suzuki M, Komiya K, Sasaki M, Shinnou H. Fine-tuning for named entity recognition using part-of-speech tagging. In: Proceedings of the 32nd Pacific Asia Conference on Language, Information and Computation. Association for Computational Linguistics (2018). Available online at: https://aclanthology.org/Y18-1072 (accessed March 25, 2022).
44. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. (2010) 17:19–24. doi: 10.1197/jamia.M3378
45. Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. (2004) 11:392–402. doi: 10.1197/jamia.M1552
46. Honnibal M, Montani I. spaCy 2: natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. (2017). doi: 10.5281/zenodo.3358113
47. Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. (2013). Available online at: http://arxiv.org/abs/1301.3781 (accessed March 23, 2022).
48. Wang Y, Liu S, Afzal N, Rastegar-Mojarad M, Wang L, Shen F, et al. A comparison of word embeddings for the biomedical natural language processing. J Biomed Inform. (2018) 87:12–20. doi: 10.1016/j.jbi.2018.09.008
49. Devlin J, Chang MW, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. (2019). Available online at: http://arxiv.org/abs/1810.04805 (accessed October 14, 2020).
50. Alsentzer E, Murphy J, Boag W, Weng WH, Jindi D, Naumann T, et al. Publicly available clinical BERT embeddings. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop. Minneapolis, MN: Association for Computational Linguistics (2019). p. 72–8.
51. Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. (2019) 36:1234–40. doi: 10.1093/bioinformatics/btz682
52. Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. J Am Med Inform Assoc. (2015) 22:938–47. doi: 10.1093/jamia/ocv032
53. Liang J, Tsou CH, Poddar A. A novel system for extractive clinical note summarization using EHR data. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop. Minneapolis, MN: Association for Computational Linguistics (2019). p. 46–54.
54. Guo Y, Qiu W, Wang Y, Cohen T. Automated Lay Language Summarization of Biomedical Scientific Reviews. (2022). Available online at: http://arxiv.org/abs/2012.12573 (accessed March 16, 2022).
55. Gupta V, Bharti P, Nokhiz P, Karnick H. SumPubMed: summarization dataset of PubMed scientific articles. In: Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing: Student Research Workshop. Bangkok: Association for Computational Linguistics (2021). p. 292–303.
56. Mishra R, Bian J, Fiszman M, Weir CR, Jonnalagadda S, Mostafa J, et al. Text summarization in the biomedical domain: a systematic review of recent research. J Biomed Inform. (2014) 52:457–67. doi: 10.1016/j.jbi.2014.06.009
57. Bui DDA, Del Fiol G, Hurdle JF, Jonnalagadda S. Extractive text summarization system to aid data extraction from full text in systematic review development. J Biomed Inform. (2016) 64:265–72. doi: 10.1016/j.jbi.2016.10.014
58. Tighe PJ, Sannapaneni B, Fillingim RB, Doyle C, Kent M, Shickel B, et al. Forty-two million ways to describe pain: topic modeling of 200,000 pubmed pain-related abstracts using natural language processing and deep learning–based text generation. Pain Med. (2020) 21:3133–60. doi: 10.1093/pm/pnaa061
59. Liu L, Tang L, Dong W, Yao S, Zhou W. An overview of topic modeling and its current applications in bioinformatics. SpringerPlus. (2016) 5:1608. doi: 10.1186/s40064-016-3252-8
60. Muchene L, Safari W. Two-stage topic modelling of scientific publications: a case study of University of Nairobi, Kenya. PLoS ONE. (2021) 16:e0243208. doi: 10.1371/journal.pone.0243208
61. Harpaz R, Callahan A, Tamang S, Low Y, Odgers D, Finlayson S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf. (2014) 37:777–90. doi: 10.1007/s40264-014-0218-z
62. Ochoa JGD, Csiszár O, Schimper T. Medical recommender systems based on continuous-valued logic and multi-criteria decision operators, using interpretable neural networks. BMC Med Inform Decis Mak. (2021) 21:186. doi: 10.1186/s12911-021-01553-3
63. De Croon R, Van Houdt L, Htun NN, Štiglic G, Vanden Abeele V, Verbert K. Health recommender systems: systematic review. J Med Internet Res. (2021) 23:e18035. doi: 10.2196/18035
64. Feng X, Zhang H, Ren Y, Shang P, Zhu Y, Liang Y, et al. The deep learning-based recommender system “Pubmender” for choosing a biomedical publication venue: development and validation study. J Med Internet Res. (2019) 21:e12957. doi: 10.2196/12957
65. Zand A, Sharma A, Stokes Z, Reynolds C, Montilla A, Sauk J, et al. An exploration into the use of a chatbot for patients with inflammatory bowel diseases: retrospective cohort study. J Med Internet Res. (2020) 22:e15589. doi: 10.2196/15589
66. Lee H, Kang J, Yeo J. Medical specialty recommendations by an artificial intelligence chatbot on a smartphone: development and deployment. J Med Internet Res. (2021) 23:e27460. doi: 10.2196/27460
67. Sennaar K. Chatbots for Healthcare - Comparing 5 Current Applications. Emerj Artificial Intelligence Research. Available online at: https://emerj.com/ai-application-comparisons/chatbots-for-healthcare-comparison/ (accessed March 16, 2022).
68. Comendador BEV, Francisco BMB, Medenilla JS, SharleenMae T, Nacion Serac TBE. Pharmabot : a pediatric generic medicine consultant chatbot. J Autom Control Eng. (2014) 3:137–40. doi: 10.12720/joace.3.2.137-140
69. Blackley SV, Huynh J, Wang L, Korach Z, Zhou L. Speech recognition for clinical documentation from 1990 to 2018: a systematic review. J Am Med Inform Assoc. (2019) 26:324–38. doi: 10.1093/jamia/ocy179
70. Goss FR, Blackley SV, Ortega CA, Kowalski LT, Landman AB, Lin CT, et al. A clinician survey of using speech recognition for clinical documentation in the electronic health record. Int J Med Inf. (2019) 130:103938. doi: 10.1016/j.ijmedinf.2019.07.017
71. Soto X, Perez-de-Viñaspre O, Labaka G, Oronoz M. Neural machine translation of clinical texts between long distance languages. J Am Med Inform Assoc. (2019) 26:1478–87. doi: 10.1093/jamia/ocz110
72. Dew KN, Turner AM, Choi YK, Bosold A, Kirchhoff K. Development of machine translation technology for assisting health communication: a systematic review. J Biomed Inform. (2018) 85:56–67. doi: 10.1016/j.jbi.2018.07.018
73. Pang L, Lan Y, Guo J, Xu J, Xu J, Cheng X. DeepRank: a new deep architecture for relevance ranking in information retrieval. In: Proc 2017 ACM Conf Inf Knowl Manag. Singapore City (2017). p. 257−66.
74. Fiorini N, Canese K, Starchenko G, Kireev E, Kim W, Miller V, et al. Best match: new relevance search for PubMed. PLoS Biol. (2018) 16:e2005343. doi: 10.1371/journal.pbio.2005343
75. Chen JS, Hersh WR. A comparative analysis of system features used in the TREC-COVID information retrieval challenge. J Biomed Inform. (2021) 117:103745. doi: 10.1016/j.jbi.2021.103745
76. Acosta MJ, Castillo-Sánchez G, Garcia-Zapirain B, de la Torre Díez I, Franco-Martín M. Sentiment analysis techniques applied to raw-text data from a Csq-8 questionnaire about mindfulness in times of COVID-19 to improve strategy generation. Int J Environ Res Public Health. (2021) 18:6408. doi: 10.3390/ijerph18126408
77. Petersen CL, Halter R, Kotz D, Loeb L, Cook S, Pidgeon D, et al. Using natural language processing and sentiment analysis to augment traditional user-centered design: development and usability study. JMIR MHealth UHealth. (2020) 8:e16862. doi: 10.2196/16862
78. Zunic A, Corcoran P, Spasic I. Sentiment analysis in health and well-being: systematic review. JMIR Med Inform. (2020) 8:e16023. doi: 10.2196/16023
79. Smith DH, Johnson ES, Russell A, Hazlehurst B, Muraki C, Nichols GA, et al. Lower visual acuity predicts worse utility values among patients with type 2 diabetes. Qual Life Res. (2008) 17:1277–84. doi: 10.1007/s11136-008-9399-1
80. Mbagwu M, French DD, Gill M, Mitchell C, Jackson K, Kho A, et al. Creation of an accurate algorithm to detect snellen best documented visual acuity from ophthalmology electronic health record notes. JMIR Med Inform. (2016) 4:e14. doi: 10.2196/medinform.4732
81. Gaskin GL, Pershing S, Cole TS, Shah NH. Predictive modeling of risk factors and complications of cataract surgery. Eur J Ophthalmol. (2016) 26:328–37. doi: 10.5301/ejo.5000706
82. Barrows Jr RC, Busuioc M, Friedman C. Limited parsing of notational text visit notes: ad-hoc vs. NLP approaches. In: Proc AMIA Symp. Los Angeles, CA (2000). p. 51–5.
83. Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. (2012) 19:225–34. doi: 10.1136/amiajnl-2011-000456
84. Zheng C, Luo Y, Mercado C, Sy L, Jacobsen SJ, Ackerson B, et al. Using natural language processing for identification of herpes zoster ophthalmicus cases to support population-based study. Clin Experiment Ophthalmol. (2019) 47:7–14. doi: 10.1111/ceo.13340
85. Stein JD, Rahman M, Andrews C, Ehrlich JR, Kamat S, Shah M, et al. Evaluation of an algorithm for identifying ocular conditions in electronic health record data. JAMA Ophthalmol. (2019) 137:491–7. doi: 10.1001/jamaophthalmol.2018.7051
86. Ashfaq HA, Lester CA, Ballouz D, Errickson J, Woodward MA. Medication accuracy in electronic health records for microbial keratitis. JAMA Ophthalmol. (2019) 137:929–31. doi: 10.1001/jamaophthalmol.2019.1444
87. Liu L, Shorstein NH, Amsden LB, Herrinton LJ. Natural language processing to ascertain two key variables from operative reports in ophthalmology. Pharmacoepidemiol Drug Saf. (2017) 26:378–85. doi: 10.1002/pds.4149
88. Wang SY, Pershing S, Tran E, Hernandez-Boussard T. Automated extraction of ophthalmic surgery outcomes from the electronic health record. Int J Med Inf. (2020) 133:104007. doi: 10.1016/j.ijmedinf.2019.104007
89. Tan Y, Bacchi S, Casson RJ, Selva D, Chan W. Triaging ophthalmology outpatient referrals with machine learning: a pilot study. Clin Exp Ophthalmol. (2020) 48:169–73. doi: 10.1111/ceo.13666
90. Wang S, Tseng B, Hernandez-Boussard T. Development and evaluation of novel ophthalmology domain-specific neural word embeddings to predict visual prognosis. Int J Med Inf. (2021) 150:104464. doi: 10.1016/j.ijmedinf.2021.104464
91. Gui H, Tseng B, Hu W, Wang SY. Looking for low vision: predicting visual prognosis by fusing structured and free-text data from electronic health records. Int J Med Inf. (2022) 159:104678. doi: 10.1016/j.ijmedinf.2021.104678
92. Wang S, Tseng B, Hernandez-Boussard T. Deep learning approaches for predicting glaucoma progression using electronic health records and natural language processing. Ophthalmol Sci. 2:1–9.
93. Lin WC, Chen JS, Kaluzny J, Chen A, Chiang MF, Hribar MR. Extraction of active medications and adherence using natural language processing for glaucoma patients. AMIA Annu Symp Proc. (2022) 2021:773–82.
94. Hallak JA, Scanzera AC, Azar DT, Chan RVP. Artificial intelligence in ophthalmology during COVID-19 and in the post COVID-19 era. Curr Opin Ophthalmol. (2020) 31:447–53. doi: 10.1097/ICU.0000000000000685
95. Nguyen AXL, Trinh XV, Wang SY, Wu AY. Determination of patient sentiment and emotion in ophthalmology: infoveillance tutorial on web-based health forum discussions. J Med Internet Res. (2021) 23:e20803. doi: 10.2196/20803
96. Chen L, Gu Y, Ji X, Lou C, Sun Z, Li H, et al. Clinical trial cohort selection based on multi-level rule-based natural language processing system. J Am Med Inform Assoc. (2019) 26:1218–26. doi: 10.1093/jamia/ocz109
97. Thompson DA, Iannaccone A, Ali RR, Arshavsky VY, Audo I, Bainbridge JWB, et al. Advancing clinical trials for inherited retinal diseases: recommendations from the Second Monaciano Symposium. Transl Vis Sci Technol. (2020) 9:2. doi: 10.1167/tvst.9.7.2
98. Bremond-Gignac D, Lewandowski E, Copin H. Contribution of electronic medical records to the management of rare diseases. BioMed Res Int. (2015) 2015:954283. doi: 10.1155/2015/954283
99. Rodriguez S, Hug C, Todorov P, Moret N, Boswell SA, Evans K, et al. Machine learning identifies candidates for drug repurposing in Alzheimer's disease. Nat Commun. (2021) 12:1033. doi: 10.1038/s41467-021-21330-0
100. Subramanian S, Baldini I, Ravichandran S, Katz-Rogozhnikov DA, Natesan Ramamurthy K, Sattigeri P, et al. A natural language processing system for extracting evidence of drug repurposing from scientific publications. Proc AAAI Conf Artif Intell. (2020) 34:13369–81. doi: 10.1609/aaai.v34i08.7052
101. Jarada TN, Rokne JG, Alhajj R. A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J Cheminformatics. (2020) 12:46. doi: 10.1186/s13321-020-00450-7
102. Rabiee Behnam, Anwar Khandaker N, Xiang Shen, Ilham Putra, Mingna Liu, Rebecca Jung, et al. Gene dosage manipulation alleviates manifestations of hereditary PAX6 haploinsufficiency in mice. Sci Transl Med. (2020) 12:eaaz4894. doi: 10.1126/scitranslmed.aaz4894
103. Leinonen HO, Zhang J, Gao F, Choi EH, Einstein EE, Einstein DE, et al. A disease-modifying therapy for retinal degenerations by drug repurposing. Invest Ophthalmol Vis Sci. (2021) 62:3157.
104. Napoli PE, Mangoni L, Gentile P, Braghiroli M, Fossarello M. A panel of broad-spectrum antivirals in topical ophthalmic medications from the drug repurposing approach during and after the coronavirus disease 2019 era. J Clin Med. (2020) 9:1–16. doi: 10.3390/jcm9082441
105. Novack GD. Repurposing medications. Ocul Surf. (2021) 19:336–40. doi: 10.1016/j.jtos.2020.11.012
106. Xu H, Aldrich MC, Chen Q, Liu H, Peterson NB, Dai Q, et al. Validating drug repurposing signals using electronic health records: a case study of metformin associated with reduced cancer mortality. J Am Med Inform Assoc. (2015) 22:179–91. doi: 10.1136/amiajnl-2014-002649
107. Subramanian S, Baldini I, Ravichandran S, Katz-Rogozhnikov DA, Ramamurthy KN, Sattigeri P, et al. Drug Repurposing for Cancer: An NLP Approach to Identify Low-Cost Therapies. (2019). Available online at: http://arxiv.org/abs/1911.07819 (accessed March 16, 2022).
108. Jang WD, Jeon S, Kim S, Lee SY. Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc Natl Acad Sci USA. (2021) 118:e2024302118. doi: 10.1073/pnas.2024302118
109. Venkatesan P. Repurposing drugs for treatment of COVID-19. Lancet Respir Med. (2021) 9:e63. doi: 10.1016/S2213-2600(21)00270-8
110. Brilliant MH, Vaziri K, Connor TB Jr, Schwartz SG, Carroll JJ, McCarty CA, et al. Mining retrospective data for virtual prospective drug repurposing: L-DOPA and age-related macular degeneration. Am J Med. (2016) 129:292–8. doi: 10.1016/j.amjmed.2015.10.015
111. Cairns BL, Nielsen RD, Masanz JJ, Martin JH, Palmer MS, Ward WH, et al. The MiPACQ clinical question answering system. Annu Symp Proc. (2011) 2011:171–80.
112. Sarrouti M, Ouatik El Alaoui S. SemBioNLQA: a semantic biomedical question answering system for retrieving exact and ideal answers to natural language questions. Artif Intell Med. (2020) 102:101767. doi: 10.1016/j.artmed.2019.101767
113. Wen A, Elwazir MY, Moon S, Fan J. Adapting and evaluating a deep learning language model for clinical why-question answering. JAMIA Open. (2020) 3:16–20. doi: 10.1093/jamiaopen/ooz072
114. Tang R, Nogueira R, Zhang E, Gupta N, Cam P, Cho K, et al. Rapidly Bootstrapping a Question Answering Dataset for COVID-19. (2020). Available online at: http://arxiv.org/abs/2004.11339 (accessed May 4, 2020).
115. Chen YP, Chen YY, Lin JJ, Huang CH, Lai F. Modified bidirectional encoder representations from transformers extractive summarization model for hospital information systems based on character-level tokens (AlphaBERT): development and performance evaluation. JMIR Med Inform. (2020) 8:e17787. doi: 10.2196/17787
116. Walker J, Leveille S, Bell S, Chimowitz H, Dong Z, Elmore JG, et al. OpenNotes after 7 years: patient experiences with ongoing access to their clinicians' outpatient visit notes. J Med Internet Res. (2019) 21:e13876. doi: 10.2196/13876
117. DesRoches CM, Bell SK, Dong Z, Elmore J, Fernandez L, Fitzgerald P, et al. Patients managing medications and reading their visit notes: a survey of OpenNotes participants. Ann Intern Med. (2019) 171:69–71. doi: 10.7326/M18-3197
118. Esch T, Mejilla R, Anselmo M, Podtschaske B, Delbanco T, Walker J. Engaging patients through open notes: an evaluation using mixed methods. BMJ Open. (2016) 6:e010034. doi: 10.1136/bmjopen-2015-010034
119. Commissioner of the 21st Century Cures Act. FDA (2020). Available online at: https://www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act (accessed March 23, 2022).
120. Radell JE, Tatum JN, Lin CT, Davidson RS, Pell J, Sieja A, et al. Risks and rewards of increasing patient access to medical records in clinical ophthalmology using OpenNotes. Eye. (2021) 1–8. doi: 10.1038/s41433-021-01775-9
121. Ophthalmology Abbreviations List and Note Translator. EyeGuru. Available online at: https://eyeguru.org/translator/ (accessed March 23, 2022).
122. van Buchem MM, Boosman H, Bauer MP, Kant IMJ, Cammel SA, Steyerberg EW. The digital scribe in clinical practice: a scoping review and research agenda. Npj Digit Med. (2021) 4:57. doi: 10.1038/s41746-021-00432-5
123. Quiroz JC, Laranjo L, Kocaballi AB, Berkovsky S, Rezazadegan D, Coiera E. Challenges of developing a digital scribe to reduce clinical documentation burden. Npj Digit Med. (2019) 2:114. doi: 10.1038/s41746-019-0190-1
124. Read-Brown S, Hribar MR, Reznick LG, Lombardi LH, Parikh M, Chamberlain WD, et al. Time requirements for electronic health record use in an academic ophthalmology center. JAMA Ophthalmol. (2017) 135:1250–7. doi: 10.1001/jamaophthalmol.2017.4187
125. Baxter SL, Gali HE, Mehta MC, Rudkin SE, Bartlett J, Brandt JD, et al. Multicenter analysis of electronic health record use among ophthalmologists. Ophthalmology. (2021) 128:165–6. doi: 10.1016/j.ophtha.2020.06.007
126. Baxter SL, Gali HE, Chiang MF, Hribar MR, Ohno-Machado L, El-Kareh R, et al. Promoting quality face-to-face communication during ophthalmology encounters in the electronic health record era. Appl Clin Inform. (2020) 11:130–41. doi: 10.1055/s-0040-1701255
127. Lim MC, Boland MV, McCannel CA, Saini A, Chiang MF, David Epley K. Adoption of electronic health records and perceptions of financial and clinical outcomes among ophthalmologists in the united states. JAMA Ophthalmol. (2018) 136:164–70. doi: 10.1001/jamaophthalmol.2017.5978
128. Sil A, Lin XV, , eds. Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies: Demonstrations. Association for Computational Linguistics (2021). Available online at: https://aclanthology.org/2021.naacl-demos.0 (accessed March 25, 2022).
129. Ferracane E, Konam S. Towards Fairness in Classifying Medical Conversations into SOAP Sections. (2020). Available online at: http://arxiv.org/abs/2012.07749 (accessed March 16, 2022).
130. Patel D, Konam S, Selvaraj SP. Weakly Supervised Medication Regimen Extraction from Medical Conversations. (2020). Available online at: http://arxiv.org/abs/2010.05317 (accessed March 16, 2022).
131. Dusek HL, Goldstein IH, Rule A, Chiang MF, Hribar MR. Clinical documentation during scribed and nonscribed ophthalmology office visits. Ophthalmol Sci. (2021) 1:1–8. doi: 10.1016/j.xops.2021.100088
132. Augmedix. Automated Medical Documentation and Data Services. Available online at: https://augmedix.com/ (accessed March 17, 2022).
133. Scott AW, Bressler NM, Ffolkes S, Wittenborn JS, Jorkasky J. Public attitudes about eye and vision health. JAMA Ophthalmol. (2016) 134:1111–8. doi: 10.1001/jamaophthalmol.2016.2627
134. Zhou L, Blackley SV, Kowalski L, Doan R, Acker WW, Landman AB, et al. Analysis of errors in dictated clinical documents assisted by speech recognition software and professional transcriptionists. JAMA Netw Open. (2018) 1:e180530. doi: 10.1001/jamanetworkopen.2018.0530
135. Weiner SJ, Wang S, Kelly B, Sharma G, Schwartz A. How accurate is the medical record? A comparison of the physician's note with a concealed audio recording in unannounced standardized patient encounters. J Am Med Inform Assoc. (2020) 27:770–5. doi: 10.1093/jamia/ocaa027
136. Bajaj P, Campos D, Craswell N, Deng L, Gao J, Liu X, et al. MS MARCO: A Human Generated MAchine Reading COmprehension Dataset. (2018). Available online at: http://arxiv.org/abs/1611.09268 (accessed October 11, 2020).
137. Ganesan AV, Matero M, Ravula AR, Vu H, Schwartz HA. Empirical evaluation of pre-trained transformers for human-level NLP: the role of sample size and dimensionality. Proc Conf Assoc Comput Linguist North Am Chapter Meet. (2021) 2021:4515–32. doi: 10.18653/v1/2021.naacl-main.357
138. Névéol A, Dalianis H, Velupillai S, Savova G, Zweigenbaum P. Clinical natural language processing in languages other than English: opportunities and challenges. J Biomed Semant. (2018) 9:12. doi: 10.1186/s13326-018-0179-8
139. Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G, et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med. (2019) 25:433–8. doi: 10.1038/s41591-018-0335-9
140. Prunotto A, Schulz S, Boeker M. Automatic generation of german translation candidates for SNOMED CT textual descriptions. Stud Health Technol Inform. (2021) 281:178–82. doi: 10.3233/SHTI210144
Keywords: natural language processing, ophthalmology, artificial intelligence, machine learning, big data, informatics, data science
Citation: Chen JS and Baxter SL (2022) Applications of natural language processing in ophthalmology: present and future. Front. Med. 9:906554. doi: 10.3389/fmed.2022.906554
Received: 28 March 2022; Accepted: 31 May 2022;
Published: 08 August 2022.
Edited by:
Tiarnan Keenan, National Eye Institute (NIH), United StatesReviewed by:
Meghna Jani, The University of Manchester, United KingdomCopyright © 2022 Chen and Baxter. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sally L. Baxter, UzFiYXh0ZXJAaGVhbHRoLnVjc2QuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.