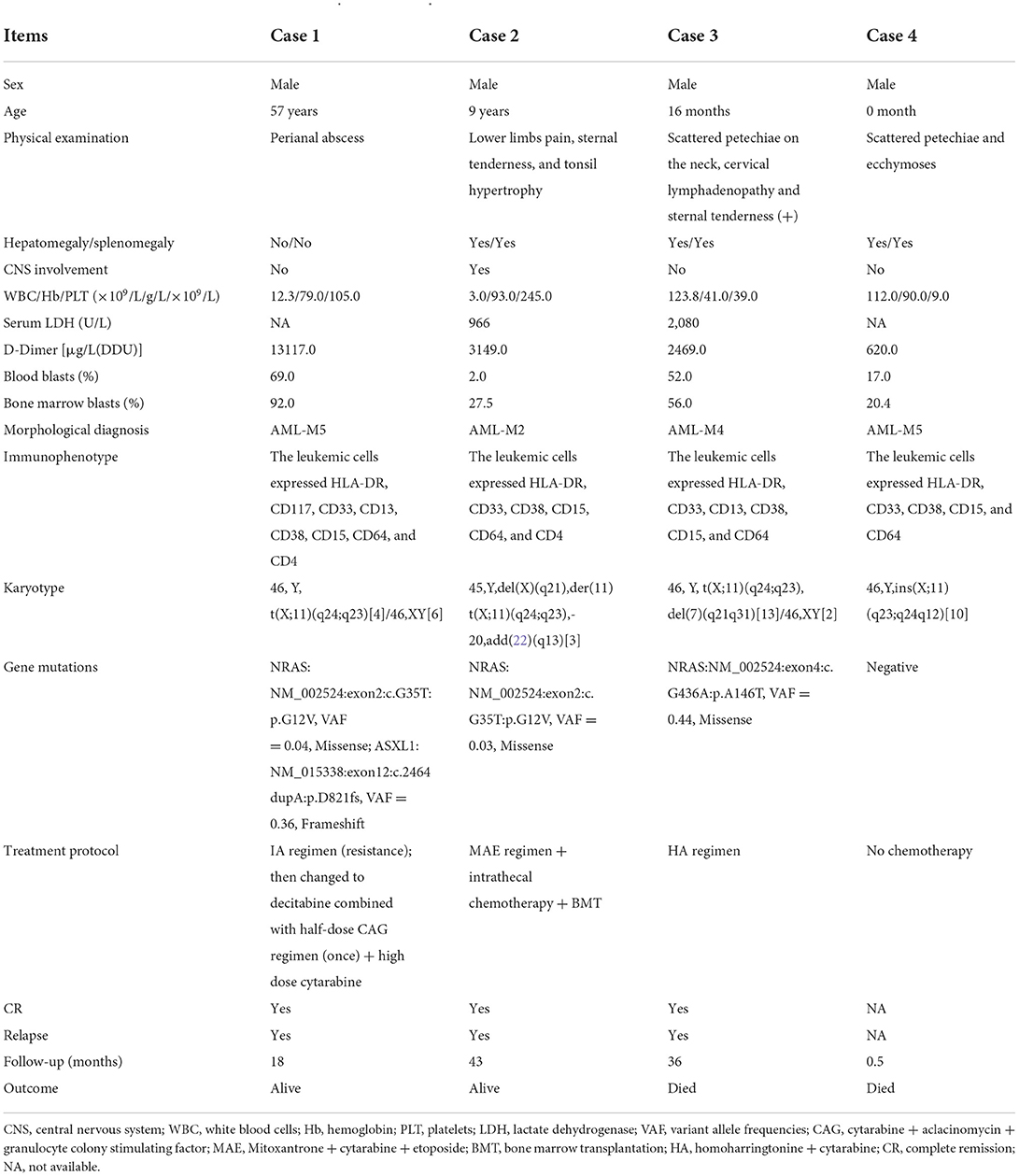
- 1Department of Hematology Laboratory, Shengjing Hospital of China Medical University, Shenyang, China
- 2Division of Hematology, Department of Medicine, Shengjing Hospital of China Medical University, Shenyang, China
Background: The KMT2A-SEPT6 fusion gene is a relatively rare genetic event in leukemia. Its clinical characteristics and prognosis, especially the profile of co-occurring gene mutations remain unclear.
Methods: We retrospectively analyzed the characteristics of four cases carrying KMT2A-SEPT6 in our hospital, and provided a literature review.
Results: All the four patients were diagnosed with acute myeloid leukemia (AML) and harbored X chromosome and 11 chromosome rearrangements, they all manifested high levels of D-dimer. Three of four patients had NRAS mutations while one patient with congenital AML did not. Of the four cases, one developed drug resistance, one suffered relapse after bone marrow transplantation (BMT) and two died. Combined with other cases reported in the literature, we found that of all patients diagnosed with AML, 90.9% were children (≤9 years old). Patients with white blood cells ≥20.0 × 109/L or diagnosed with M4 had a shorter overall survival (P < 0.05). Age, whether to receive BMT, and the chromosome rearrangement patterns had no significant effect on overall survival (P > 0.05).
Conclusions: KMT2A-SEPT6 was more commonly observed in pediatric AML patients, some of which may co-occur with NRAS mutations. The prognosis was related to the white blood cell levels and the leukemia subtype, but was not related to age or BMT. More cases need to be accumulated to better understand the profile in KMT2A-SEPT6-positive AML.
Introduction
Acute myeloid leukemia (AML) is a malignant tumor that originates from myeloid blood cells and is characterized by abnormally increased leukemic cells in bone marrow or peripheral blood (1). Cytogenetic and molecular abnormalities commonly occur in AML patients. Based on genetic mutations and specific chromosomal rearrangements, the World Health Organization (WHO) divides AML with recurrent genetic abnormalities into different subgroups (2). Translocations involving the lysine (K)-specific methyltransferase 2A gene (KMT2A), previously known as mixed lineage leukemia (MLL), are common chromosomal abnormalities in AML (3, 4). The KMT2A gene at 11q23 has many partner genes, of which over 80 have been identified (5).
Among them, SEPT6 located at Xq24 which is involved in the formation of the KMT2A arrangement t(X;11)(q22-24;q23), is extremely rare in AML (6). SEPT6 is a member of Septins family, an evolutionarily conserved family of GTP-binding proteins that associate with cell polarity, cytokinesis and oncogenesis (7, 8). Five septin genes (SEPT2, SEPT5, SEPT6, SEPT9, and SEPT11) were identified as KMT2A fusion partner (9). SEPT6, which plays a role in actin and microtubule cytoskeletons, is involved in infectious diseases, Down's syndrome, schizophrenia, bipolar disorder and cancers, including leukemia and lymphoma (10, 11). To our knowledge, only a limited number of KMT2A-SEPT6-positive cases have been documented in the literature. Most cases are children while only one adult case has been reported (6, 12–21). The exact role of KMT2A-SEPT6 in hematopoietic cells and its effect on leukemogenesis are still unknown. There is little information on the clinical features, treatment strategies and prognosis of such patients, and accompanying gene mutations carried by such patients have not been described.
We retrospectively analyzed data from four acute leukemia patients carrying the KMT2A-SEPT6 fusion gene who were treated in our hospital, especially gene mutation information. Additionally, we reviewed cases in the literature together with our cases to provide evidence for potential therapeutic strategies.
Materials and methods
Case selection
We collected four patients harboring the KMT2A-SEPT6 gene from a pool of 1,656 leukemia patients within our hematological disease database in the past 4 years, which were referred to as cases 1–4. The diagnostic criteria were according to the WHO classification of tumors of hematopoietic and lymphoid tissues (22). We conducted a retrospective analysis and systematic summary with information on morphology, flow cytometric analysis, cytogenetics, molecular biology, and other related laboratory test results.
Literature review
We conducted a literature search on PubMed with the keywords “KMT2A-SEPT6,” “MLL-SEPT6,” “KMT2A-SEPTIN6,” “MLL-SEPTIN6” or “t(X;11)” to gather related case reports.
Statistical analysis
Kaplan–Meier method was used for survival evaluation. The log-rank test was used to assess the difference between groups. P < 0.05 was considered statistically significant. All data were analyzed with SPSS Statistics, version 21 (StatSoft).
Results
Clinical presentation
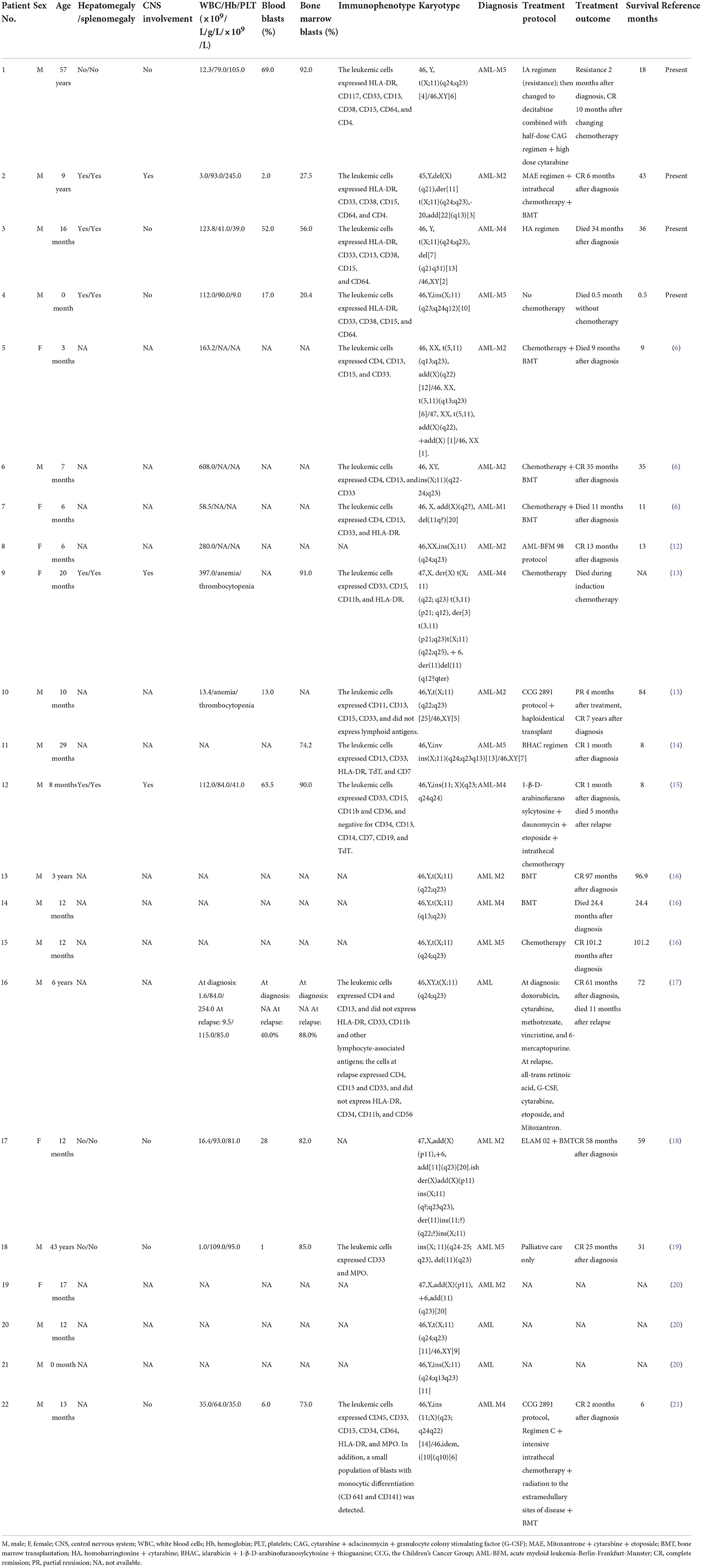
All KMT2A-SEPT6-positive cases (cases 1–4) were male with ages ranging from 0 to 57 years, and their detailed information is summarized in Table 1. All cases had manifestations of fever and pale complexion. The three pediatric cases (cases 2, 3, and 4) were accompanied by hepatosplenomegaly, and two cases (cases 3 and 4) had scattered petechiae and ecchymoses. Case 1 was an elderly male patient with perianal abscess and diabetes in addition to the above symptoms. Case 2 was accompanied by pain in both lower limbs, tenderness of the sternum and hypertrophy of the tonsils. In case 3, multiple lymph nodes were palpable on the bilateral neck, he was positive for sternal tenderness, and he also had hyperuricemia and acute bronchial pneumonia. Case 4 was a newborn delivered by cesarean section due to a “decreased fetal heart rate.” The birth weight was 3,100 g. The infant had no spontaneous breath at birth and was generally cyanotic. He restored spontaneous respiration under assisted ventilation, and he developed persistent pulmonary arterial hypertension and neonatal pneumonia. His parents were healthy and had no history of genetic disease.
Laboratory results showed that three cases (cases 1, 3, and 4) had high white blood cell (WBC) counts, anemia, and low platelet counts. One patient (case 2) had low hemoglobin levels and WBC counts but normal platelet counts, and two patients (cases 2 and 3) had increased serum lactate dehydrogenase levels. The D-dimer levels of all four patients were increased.
Morphological evaluation
All cases exhibited morphological characteristics of AML (Figure 1). Leukemia cells have the morphological characteristics of blast cells, including medium to large nuclei with round and often eccentric nuclei, loose chromatin, abundant basophilic cytoplasm, and Auer bodies can be seen in some cells. The French American British (FAB) morphological classification of each case was M5 (cases 1 and 4), M2 (case 2) or M4 (case 3). Three cases (cases 1, 3, and 4) showed hypercellular bone marrow, and the other case (case 2) had severe hypocellular bone marrow. Case 1 and case 4 were evaluated as M5 and revealed 92.0 and 20.4% blasts in marrow aspirate and 69.0 and 17.0% blasts in peripheral blood, respectively (Table 1; Figures 1A,D). Case 2 was evaluated as M2; the marrow aspirate revealed 27.5% myeloblasts and the peripheral blood exhibited 2.0% blasts (Table 1; Figure 1B). Case 3 was evaluated as M4; the marrow aspirate showed 56.0% blasts and the peripheral blood exhibited 52.0% blasts (Table 1; Figure 1C).
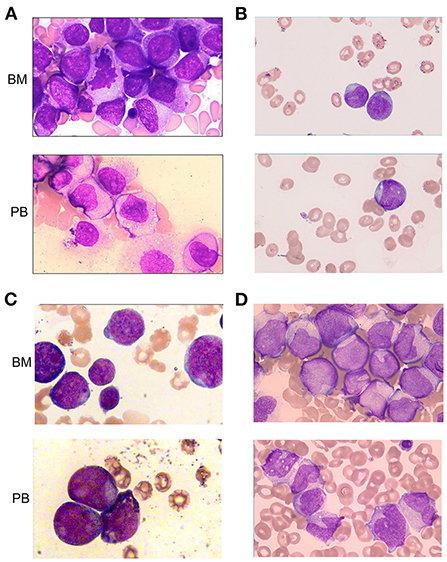
Figure 1. Morphologic evaluation of leukemic cells at diagnosis (Wright–Giemsa stain, × 1,000). (A–D) represent cases 1, 2, 3, and 4, respectively. BM, bone marrow; PB, peripheral blood.
Flow cytometric analysis
Flow cytometric analysis revealed the presence of myeloid blasts in bone marrow samples from all four patients (Table 1; Figure 2). The percentage of blasts was highest at 92% in case 1 and lowest at 4.2% in case 2. All cases were positive for CD33, CD15, and CD64, indicating myeloid lineage, and CD13 was positive only in case 1 and case 3. All the cases were positive for CD38 and HLA-DR. CD117 was positive only in case 1, and CD34 was negative in all four cases.
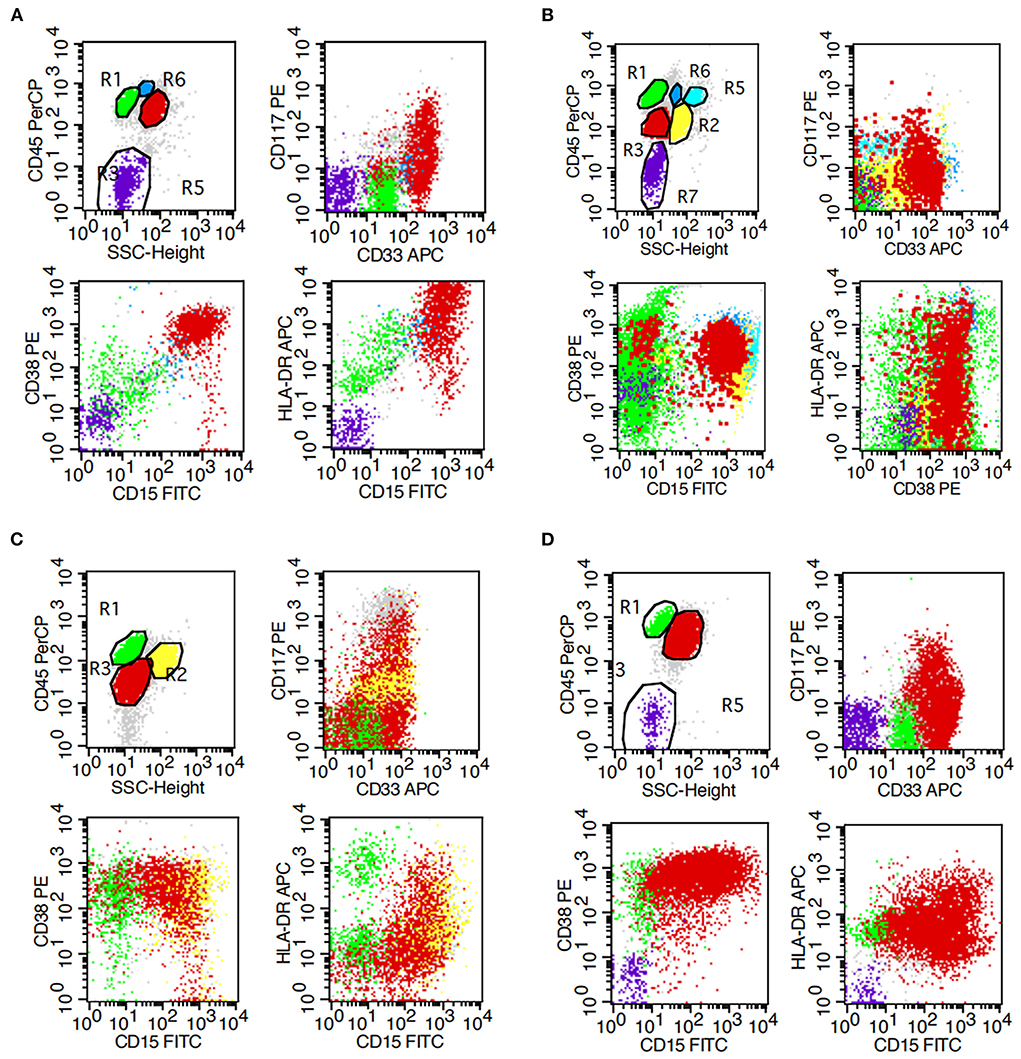
Figure 2. Flow cytometry results of bone marrow. (A–D) represent cases 1, 2, 3, and 4, respectively.
Cytogenetic analysis
The results showed that all four cases had clonal abnormalities of the X chromosome and chromosome 11 or complex karyotype abnormalities (Table 1; Figure 3). In case 1, the metaphase cells exhibited abnormalities of t(X;11)(q24;q23) (Figure 3A). The metaphase cells collected in case 2 showed 45, Y, del(X)(q21), der(11)t(X;11)(q24;q23), −20, add (22)(q13) (Figure 3B). In case 3, in addition to the abnormal karyotype of t(X;11)(q24;q23), thirteen metaphase cells had del(7)(q21q31) (Figure 3C). The metaphase cells in case 4 showed an abnormal karyotype of 46, Y, ins(X;11)(q23;q24q12) (Figure 3D).

Figure 3. Karyotype analysis results of bone marrow. (A–D) represent cases 1, 2, 3, and 4, respectively.
Molecular analysis
We performed molecular biology tests, including screening of fusion genes and next-generation sequencing (NGS) analysis, on all cases (Table 1). The KMT2A-SEPT6 fusion gene was detected in all four cases. The NGS panel included a total of 20 frequently mutated genes in AML: ASXL1, CEBPA, DNMT3A, EZH2, FLT3, IDH1, IDH2, KIT, NPM1, PHF6, RUNX1, TET2, TP53, BCOR, GATA2, KMT2A, KRAS, NRAS, PDGFRA, and WT1. The sequencing depth was 2,000 ×. The results showed that three cases (cases 1–3) harbored NRAS (NM_002524) mutations, and the mutation sites of cases 1 and 2 were both NRAS G12V, with variant allele frequencies (VAFs) of 0.04 and 0.03, respectively. In addition, case 1 was also accompanied by an insertion mutation of ASXL1 D821 with a VAF of 0.36. The reexamination results at the third month showed that the KMT2A-SEPT6 gene remained positive but its expression level dropped from 100 to 24.4%. The ASXL1 D821 VAF dropped to 0.01, and the NRAS G12V gene mutation was not detected. Four months later, the patient achieved complete remission (CR), the ASXL1 D821 VAF dropped to 0.003, and both the NRAS mutation and KMT2A-SEPT6 fusion gene were negative. The NRAS A146T mutation in case 3 occurred in exon 4 with a VAF of 0.44. KMT2A-SEPT6 gene and NRAS gene mutations both turned negative on the ninth month after diagnosis. The patient relapsed on the twenty-seventh month with positive KMT2A-SEPT6 and NRAS A146T mutation (VAF = 0.27). In case 4, no pathogenic gene mutations were detected.
Clinical course
The treatment and follow-up information of all cases (cases 1–4) is shown in Table 1. Three patients (cases 1–3) received chemotherapy, patient 2 subsequently received a bone marrow transplantation (BMT), and patient 4 was a newborn and did not receive any chemotherapy. The clinical follow-up period ranged from 0.5 to 43 months, with a median of 27 months. Case 1 received induction chemotherapy of IA (idarubicin + standard cytarabine) regimen, and revealed non-remission with blasts from 92.0 to 40.5% in bone marrow. Then, he received D-CAG (decitabine combined with low-dose cytarabine + aclacinomycin + granulocyte colony stimulating factor) and achieved CR. The KMT2A-SEPT6 gene expression reduced from 100 to 24.4%, and the NRAS G12V gene mutation was negative after the second chemotherapy. Subsequently, he received high-dose cytarabine consolidation therapy and an intrathecal injection (cytarabine + dexamethasone + methotrexate) for central nervous system infiltration prevention. The patient revealed a normal karyotype and negative molecular results of KMT2A-SEPT6 rearrangement and NRAS mutation, and sustained CR for 10 months. However, he relapsed 3 months later. From this time point the patients had a stable blast count of 5.0%-10.5% to the last follow-up. Case 2 received MAE (mitoxantrone + standard cytarabine + etoposide) regimen and reached CR. Then he received high-dose cytarabine consolidation therapy. After that, the patient received allogeneic hematopoietic stem cell transplantation (allo-HSCT). The conditioning regime was modified BU/CY + ATG (busulfan/cyclophosphamide + antithymocyte globulin, peking regime). The donor was his father, HLA 5/10, A+ to B+. We used CsA + MTX + MMF (cyclosporine + methotrexate + mycophenolate mofetil) for Graft-Versus-Host Disease (GVHD) prophylactic. The process of transplantation was well-off, and bone marrow assessment results were negative continuously. However, 19 months after transplantation, he relapsed with central nervous system leukemia. Following five courses of intrathecal injection, the child achieved CR again. Case 3 received HA (homoharringtonine + standard cytarabine) regimen and intrathecal injection (cytarabine + dexamethasone + methotrexate). He reached CR 4 weeks after diagnosis. This patient relapsed 27 months later and died 36 months after diagnosis. Case 4 had dyspnea at birth, and he was on assisted ventilation and given blood infusion to improve anemia and thrombocytopenia. The newborn's condition did not improve during the treatment, and the child died 2 weeks later.
Literature review
A total of 22 KMT2A-SEPT6-positive cases were included in this literature review, including four cases in our report and eighteen cases from the literature (6, 12–21). Tables 2, 3 list the detailed clinical information of all patients and clinical features of evaluable patients.
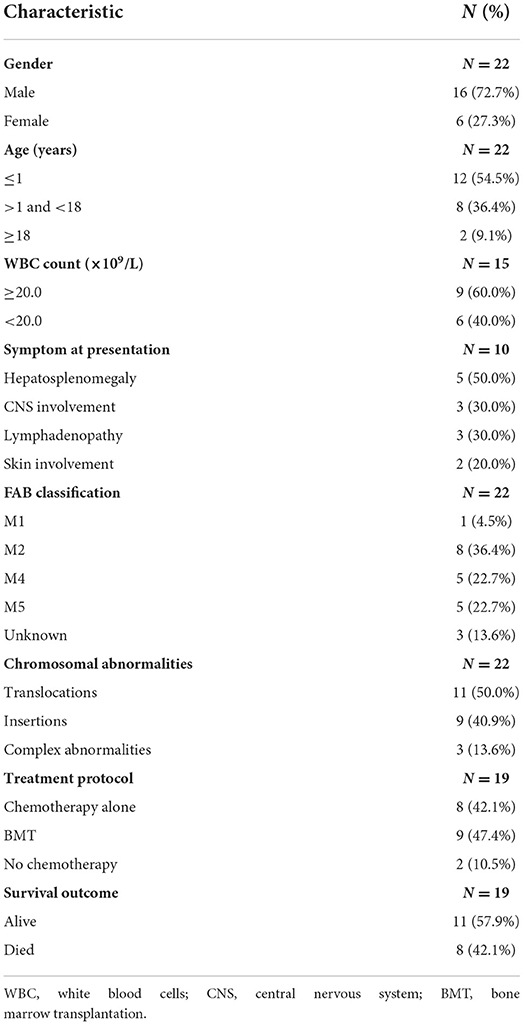
The age of the patients ranged from 0 to 57 years (median = 1 year), with a male–female ratio of nearly 3:1 (16 males vs. 6 females). Twenty patients (90.9%) were children (≤9 years old), including twelve (54.5%) infants (≤1 year old). The majority of the patients manifested leukocytosis (range 1–608 × 109/L), anemia (range 41–109 g/L) and low platelet counts (range 9–254 × 109/L). According to the high WBC index (23), nine (60.0%) of the fifteen cases with WBC count information were defined as high WBC levels. Twelve cases were not provided with a description of clinical features. Of the remaining 10 cases, five children (50.0%) had splenomegaly and hepatomegaly, and three patients (30.0%) had lymphadenopathy. Central nervous system involvement was observed in three children (30.0%), and skin involvement was observed in two cases (20.0%). All patients were diagnosed with AML (twenty children and two adults) according to the former FAB classification: five patients (three children and two adults, 22.7%) with M5, five children (22.7%) with M4, eight children (36.4%) with M2, one child (4.5%) with M1, and three children (13.6%) unknown. All the cases had available cytogenetic information, and chromosomal translocations (eleven cases) were the most common chromosomal rearrangements, followed by chromosomal insertions (nine cases). Among them, Xq24 (nine cases) and 11q23 (fourteen cases) were the most frequently involved chromosomal bands. Three cases (13.6%) demonstrated complex abnormalities. Eight patients (42.1%) received chemotherapy alone. Nine patients (47.4%) received BMT. Eight patients (42.1%) died.
Of all 22 cases, 18 cases had clinical follow-up with a median period of 27.7 months (0.5–101.5 months). Kaplan-Meier survival analysis was performed on eighteen cases with complete follow-up information (Figure 4A). As of the final follow-up, the median survival time was 72 months.
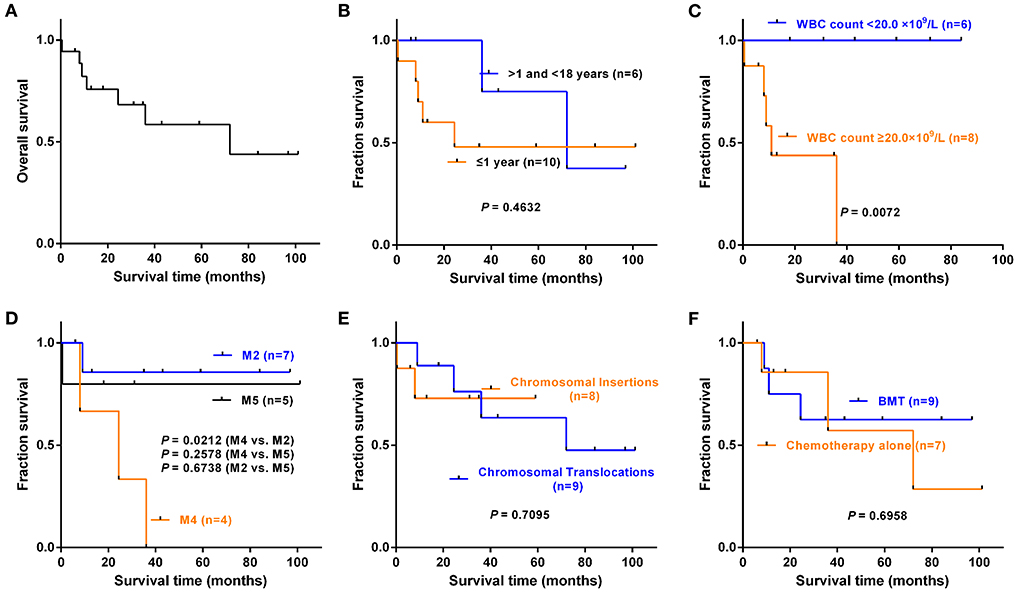
Figure 4. Kaplan–Meier survival analysis of eighteen cases with complete follow-up information. These included fourteen reported cases with clinical follow-up and four cases in our series. (A) Overall survival of eighteen cases. (B) Infant group (≤1 year) vs. pediatric group (>1 and < 18 years old), P = 0.4632. (C) WBC count < 20.0 × 109/L vs. ≥20.0 × 109/L, P = 0.0072. (D) FAB subtype. M4 vs. M2, P = 0.0212; M4 vs. M5, P = 0.2578; M2 vs. M5, P = 0.6738. (E) Chromosomal insertion vs. chromosomal translocation, P = 0.7095. (F) Chemotherapy alone vs. BMT, P = 0.6958.
To understand the impact of different clinical features on overall survival (OS), we grouped the patients according to the characteristics in Table 3, and groups with fewer than three patients and cases with incomplete follow-up information were not included in the statistics. The results showed that there was no statistical significance in OS between the infant group (≤1 year old) and pediatric group (>1 and < 18 years old) (P = 0.4632, Figure 4B). The OS of patients with WBC levels ≥20.0 × 109/L was much shorter than that of patients with WBC levels < 20.0 × 109/L (P = 0.0072, Figure 4C). The patients with M4 had a shorter OS than those with M2 (P = 0.0212, Figure 4D). However, there were no significant differences in OS between the chromosomal translocation group and the chromosomal insertion group (P = 0.7095, Figure 4E) or between the patients who received chemotherapy alone and those who received BMT (P = 0.6958, Figure 4F).
Discussion
The KMT2A gene is a frequent target of rearrangement in human leukemia, especially in infant and pediatric leukemia (14, 24, 25). These rearrangements include fusions with many partner genes but rarely involve the gene SEPT6 on the X chromosome. The KMT2A gene and SEPT6 gene are vulnerable to damage to form translocations associated with infant AML.
In this study, we described four cases of AML with the KMT2A-SEPT6 fusion gene. The FAB subtypes were mainly M2, M4, and M5, which was consistent with the literature. To the best of our knowledge, only two adult patients have been reported, including one case in our series. Most of the cases that have been reported were children, and 54.5% were infant patients (≤1 year old). Among these cases, 60.0% had a high level of WBC, and 30% manifested central nervous system involvement, which were similar to the clinical features of KMT2A-rearranged AML patients. The findings of Balgobind et al. (26) showed that KMT2A-rearranged AML patients usually exhibit a high tumor burden, including organomegaly, a high median WBC count and central nervous system involvement. The present study included the largest number of KMT2A-SEPT6 cases to date. The patients' NGS test results were not provided except ours, and we also tracked the patients' molecular biological examination results. Three of four cases in our series had NRAS mutations, while one case with congenital AML did not.
NRAS G12V is required in the leukemia self-renewal process, independent of its effects on growth and survival (27). Compared with other subtypes of leukemia, acute leukemia with KMT2A translocations (such as KMT2A-AF4, and KMT2A-AF9) harbored the fewest number of mutations, in which NRAS mutations commonly co-occur (27, 28). In our series, we identified NRAS mutations in KMT2A-SEPT6-positive AML patients for the first time, and most of the mutation sites appeared at codons 12 and 145. The former site is a hotspot mutant of NRAS and the latter site has also been reported (29, 30). The VAF of NARS mutation decreased as the patient's condition improved, indicating that there was no underlying residual clonal hematopoiesis after chemotherapy. When the patients achieved CR, they also turned negative. The underlying connection between NRAS mutations and KMT2A-SEPT6 and whether non-congenital KMT2A-SEPT6-positive AML patients all have NRAS mutations remain to be further studied in a larger cohort in the future.
KMT2A gene rearrangement in AML usually indicates poor prognosis (5, 31). The prognostic significance of NRAS mutations in AML patients remains unclear (32–35). Of the four cases in our series, one developed drug resistance at first, one suffered relapse after BMT and two died, showing unsatisfactory therapeutic effect. However, whether the outcomes of patients with KMT2A-SEPT6 were aggravated by the concurrence of NRAS mutations needs a follow-up study. Kaplan–Meier curves demonstrated that the pediatric group (>1 and < 18 years old) did not show better OS than the infant group (≤1 year old). Age may not be an independent prognostic factor for survival. Most of the patients received chemotherapy, nine of them received BMT, but three of them eventually died. The OS of patients between the chemotherapy alone group and the BMT group did not show a significant difference, which suggested that BMT may not improve the survival time of such patients. This was consistent with several studies and meta-analyses that suggested that BMT does not improve survival in patients with KMT2A rearrangement (36, 37). In addition, chromosome rearrangement patterns had no significant effect on the OS of patients. However, we found that a higher white blood cell count at the initial diagnosis was associated with a shorter OS. Moreover, FAB classification also has an impact on the prognosis of patients. A trend for worse OS was observed in M4 patients.
Our study has some limitations. First, this was a retrospective study, coupled with a limited number of reported cases and incomplete clinical information, resulting in small sample sizes in some subgroups, which may lead to false negative results. Second, our NGS detection only covers the twenty most frequently mutated genes in AML, and the prognostic effects of some critical genes may be neglected. Third, gene mutation information in the reported cases was not available and cases in our series were detailed but limited by sample size. Therefore, our findings need to be combined with more cases for further analysis in the future.
In conclusion, the KMT2A-SEPT6 fusion gene was more commonly observed in pediatric patients diagnosed with AML. NRAS mutations were observed in these patients, most frequently of the NRAS G12V hotspot mutation. Whether NRAS mutations are related to the occurrence of KMT2A-SEPT6-positive AML is currently unclear. The prognosis was related to the subtype of leukemia and WBC levels, but may not be related to age. BMT may not improve survival in these patients. More cases should be accumulated and summarized to better understand the profile in KMT2A-SEPT6-positive AML.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Shengjing Hospital of China Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
SF and FC performed the study concept and design. FC developed the methodology and wrote the paper. FC and YY acquired, analyzed and interpreted the data, and performed the statistical analysis. SF reviewed and revised the paper. All authors read and approved the final paper.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) [grant number: 82070165] and 345 Talent Project of Shengjing Hospital [grant number: M0957].
Acknowledgments
We thank Yue Zhao for his help in manuscript editing, and Yu Fu, Xuan Liu, and Minyu Zhang for their expert technical assistance. All individuals referenced here have no publication-related funding source, industry relations, or conflicts of interest.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Culp-Hill R, D'Alessandro A, Pietras EM. Extinguishing the embers: targeting AML metabolism. Trends Mol Med. (2021) 27:332–44. doi: 10.1016/j.molmed.2020.10.001
2. Hasserjian RP. Controversies in the recent (2016) World Health Organization classification of acute myeloid leukemia. Best Pract Res Clin Haematol. (2021) 34:101249. doi: 10.1016/j.beha.2021.101249
3. Matsuo H, Yoshida K, Fukumura K, Nakatani K, Noguchi Y, Takasaki S, et al. Recurrent CCND3 mutations in MLL-rearranged acute myeloid leukemia. Blood Adv. (2018) 2:2879–89. doi: 10.1182/bloodadvances.2018019398
4. Wander P, Arentsen-Peters STCJM, Pinhan?os SS, Koopmans B, Dolman MEM, Ariese R, et al. High-throughput drug screening reveals Pyrvinium pamoate as effective candidate against pediatric MLL-rearranged acute myeloid leukemia. Transl Oncol. (2021) 14:101048. doi: 10.1016/j.tranon.2021.101048
5. Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. (2018) 32:273–84. doi: 10.1038/leu.2017.213
6. Ono R, Taki T, Taketani T, Kawaguchi H, Taniwaki M, Okamura T, et al. SEPTIN6, a human homolog to mouse Septin6, is fused to MLL in infant acute myeloid leukemia with complex chromosomal abnormalities involving 11q23 and Xq24. Cancer Res. (2002) 62:333–7.
7. Ivanov AI, Le HT, Naydenov NG, Rieder F. Novel functions of the septin cytoskeleton: shaping up tissue inflammation and fibrosis. Am J Pathol. (2021) 191:40–51. doi: 10.1016/j.ajpath.2020.09.007
8. Macara IG, Baldarelli R, Field CM, Glotzer M, Hayashi Y, Hsu SC, et al. Mammalian septins nomenclature. Mol Biol Cell. (2002) 13:4111–3. doi: 10.1091/mbc.e02-07-0438
9. Cerveira N, Bizarro S, Teixeira MR. MLL-SEPTIN gene fusions in hematological malignancies. Biol Chem. (2011) 392:713–24. doi: 10.1515/BC.2011.072
10. Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu Z, et al. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. (2014) 4:7546. doi: 10.1038/srep07546
11. Ono R, Ihara M, Nakajima H, Ozaki K, Kataoka-Fujiwara Y, Taki T, et al. Disruption of Sept6, a fusion partner gene of MLL, does not affect ontogeny, leukemogenesis induced by MLL-SEPT6, or phenotype induced by the loss of Sept4. Mol Cell Biol. (2005) 25:10965–78. doi: 10.1128/MCB.25.24.10965-10978.2005
12. Borkhardt A, Teigler-Schlegel A, Fuchs U, Keller C, König M, Harbott J, et al. An ins(X;11)(q24;q23) fuses the MLL and the Septin 6/KIAA0128 gene in an infant with AML-M2. Genes Chromosomes Cancer. (2001) 32:82–8. doi: 10.1002/gcc.1169
13. Slater DJ, Hilgenfeld E, Rappaport EF, Shah N, Meek RG, Williams WR, et al. MLL-SEPTIN6 fusion recurs in novel translocation of chromosomes 3, X, and 11 in infant acute myelomonocytic leukemia and in t(X;11) in infant acute myeloid leukemia, and MLL genomic breakpoint in complex MLL-SEPTIN6 rearrangement is a DNA topoisomerase II cleavage site. Oncogene. (2002) 21:4706–14. doi: 10.1038/sj.onc.1205572
14. Kim HJ, Ki CS, Park Q, Koo HH, Yoo KH, Kim EJ, et al. MLL/SEPTIN6 chimeric transcript from inv ins(X;11)(q24;q23q13) in acute monocytic leukemia: report of a case and review of the literature. Genes Chromosomes Cancer. (2003) 38:8–12. doi: 10.1002/gcc.10235
15. Fu JF, Liang DC, Yang CP, Hsu JJ, Shih LY. Molecular analysis of t(X;11)(q24;q23) in an infant with AML-M4. Genes Chromosomes Cancer. (2003) 38:253–9. doi: 10.1002/gcc.10272
16. Harrison CJ, Cuneo A, Clark R, Johansson B, Lafage-Pochitaloff M, Mugneret F, et al. Ten novel 11q23 chromosomal partner sites. European 11q23 workshop participants. Leukemia. (1998) 12:811–22. doi: 10.1038/sj.leu.2401017
17. Nakata Y, Mori T, Yamazaki T, Suzuki T, Okazaki T, Kurosawa Y, et al. Acute myeloid leukemia with hypergranular cytoplasm accompanied by t(X;11)(q24;q23) and rearrangement of the MLL gene. Leuk Res. (1999) 23:85–8. doi: 10.1016/S0145-2126(98)00131-3
18. Cerveira N, Lisboa S, Correia C, Bizarro S, Santos J, Torres L, et al. Genetic and clinical characterization of 45 acute leukemia patients with MLL gene rearrangements from a single institution. Mol Oncol. (2012) 6:553–64. doi: 10.1016/j.molonc.2012.06.004
19. De Braekeleer E, Meyer C, Douet-Guilbert N, Basinko A, Le Bris MJ, Morel F, et al. Identification of MLL partner genes in 27 patients with acute leukemia from a single cytogenetic laboratory. Mol Oncol. (2011) 5:555–63. doi: 10.1016/j.molonc.2011.08.003
20. Cerveira N, Micci F, Santos J, Pinheiro M, Correia C, Lisboa S, et al. Molecular characterization of the MLL-SEPT6 fusion gene in acute myeloid leukemia: identification of novel fusion transcripts and cloning of genomic breakpoint junctions. Haematologica. (2008) 93:1076–80. doi: 10.3324/haematol.12594
21. Kadkol SS, Bruno A, Oh S, Schmidt ML, Lindgren V. MLL-SEPT6 fusion transcript with a novel sequence in an infant with acute myeloid leukemia. Cancer Genet Cytogenet. (2006) 68:162–7. doi: 10.1016/j.cancergencyto.2006.02.020
22. Arber DA, Brunning RD, Le Beau MM, et al. Acute myeloid leukemia and related precursor neoplasms. In: Swerdlow SH, Campo E, Harris NL, et al. editors. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Revised 4th ed. France: IARC Press (2017). p. 130–71.
23. Nguyen S, Leblanc T, Fenaux P, Witz F, Blaise D, Pigneux A, et al. A white blood cell index as the main prognostic factor in t(8;21) acute myeloid leukemia (AML): a survey of 161 cases from the French AML Intergroup. Blood. (2002) 99:3517–23. doi: 10.1182/blood.V99.10.3517
24. Antunes ETB, Ottersbach K. The MLL/SET family and haematopoiesis. Biochim Biophys Acta Gene Regul Mech. (2020) 1863:194579. doi: 10.1016/j.bbagrm.2020.194579
25. Rice S, Roy A. MLL-rearranged infant leukemia: a 'thorn in the side' of a remarkable success story. Biochim Biophys Acta Gene Regul Mech. (2020) 1863:194564. doi: 10.1016/j.bbagrm.2020.194564
26. Balgobind BV, Zwaan CM, Pieters R, Van den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. (2011) 25:1239–48. doi: 10.1038/leu.2011.90
27. Sachs Z, LaRue RS, Nguyen HT, Sachs K, Noble KE, Mohd Hassan NA, et al. NRASG12V oncogene facilitates self-renewal in a murine model of acute myelogenous leukemia. Blood. (2014) 124:3274–83. doi: 10.1182/blood-2013-08-521708
28. Trentin L, Bresolin S, Giarin E, Bardini M, Serafin V, Accordi B, et al. Deciphering KRAS and NRAS mutated clone dynamics in MLL-AF4 pediatric leukemia by ultra deep sequencing analysis. Sci Rep. (2016) 6:34449. doi: 10.1038/srep34449
29. Bacher U, Haferlach T, Schoch C, Kern W, Schnittger S. Implications of NRAS mutations in AML: a study of 2502 patients. Blood. (2006) 107:3847–53. doi: 10.1182/blood-2005-08-3522
30. Wang S, Wu Z, Li T, Li Y, Wang W, Hao Q, et al. Mutational spectrum and prognosis in NRAS-mutated acute myeloid leukemia. Sci Rep. (2020) 10:12152. doi: 10.1038/s41598-020-69194-6
31. Wong NM, So CWE. Novel therapeutic strategies for MLL-rearranged leukemias. Biochim Biophys Acta Gene Regul Mech. (2020) 1863:194584. doi: 10.1016/j.bbagrm.2020.194584
32. Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. (2012) 150:264–78. doi: 10.1016/j.cell.2012.06.023
33. Liu X, Ye Q, Zhao XP, Zhang PB, Li S, Li RQ, et al. RAS mutations in acute myeloid leukemia patients: a review and meta-analysis. Clin Chim Acta. (2019) 489:254–60. doi: 10.1016/j.cca.2018.08.040
34. Damm F, Heuser M, Morgan M, Wagner K, Görlich K, Grosshennig A, et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. (2011) 117:4561–8. doi: 10.1182/blood-2010-08-303479
35. Li TT, Li J, Geng YH, Zhang F, Liu L, Yang YL. NRAS gene expression and its clinical significance in patients with acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. (2020) 28:76–81. doi: 10.19746/j.cnki.issn.1009-2137.2020.01.013
36. Winters AC, Bernt KM. MLL-rearranged leukemias-an update on science and clinical approaches. Front Pediatr. (2017) 5:4. doi: 10.3389/fped.2017.00004
Keywords: acute myeloid leukemia, gene rearrangement, KMT2A-SEPT6, mutations, NRAS
Citation: Chen F, Yang Y and Fu S (2022) Clinical profile in KMT2A-SEPT6-positive acute myeloid leukemia: Does it often co-occur with NRAS mutations? Front. Med. 9:890959. doi: 10.3389/fmed.2022.890959
Received: 15 March 2022; Accepted: 30 August 2022;
Published: 21 September 2022.
Edited by:
Håkon Reikvam, University of Bergen, NorwayReviewed by:
Nirmalya Saha, University of Michigan, United StatesTor Henrik Anderson Tvedt, Haukeland University Hospital, Norway
Copyright © 2022 Chen, Yang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuang Fu, Y29vbGVyZnNwcmV0dHlAMTI2LmNvbQ==
 Fang Chen
Fang Chen Ying Yang2
Ying Yang2 Shuang Fu
Shuang Fu