
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 04 March 2022
Sec. Translational Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.845094
This article is part of the Research Topic microRNAs as predictive Bio-Markers in Metabolic Signalling Pathways in Tumour Progression View all 4 articles
Genetic and epigenetic characteristics are core factors of cancer. MicroRNAs (miRNAs) are small non-coding RNAs which regulate gene expression at the post-transcriptional level via binding to corresponding mRNAs. Recently, increasing evidence has proven that miRNAs regulate the occurrence and development of human cancer. Here, we mainly review the abnormal expression of miR-625 in a variety of cancers. In summarizing the role and potential molecular mechanisms of miR-625 in various tumors in detail, we reveal that miR-625 is involved in a variety of biological processes, such as cell proliferation, invasion, migration, apoptosis, cell cycle regulation, and drug resistance. In addition, we discuss the lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks and briefly explain the specific mechanisms of competing endogenous RNAs. In conclusion, we reveal the potential value of miR-625 in cancer diagnosis, treatment, and prognosis and hope to provide new ideas for the clinical application of miR-625.
Cancer is a severe health problem worldwide given its increasing incidence and high mortality (1). According to reports, in 2018, there were 9.6 million cancer-related deaths and 18.1 million new cancer cases all over the world (2). Isabelle Soerjomataram and Freddie Bray recently pointed out that the number of cancer patients worldwide is predicted to increase in the next 50 years. It is predicted that 34 million new cancer cases will be diagnosed by 2070, double the number in 2018 (3). Although the understanding of cancer biology is increasing, and efforts are being made to develop more effective and targeted diagnosis, treatment, and prevention strategies, cancer is still the main cause of severe social pressure and a substantial economic burden (4). Current research focuses on finding new diagnostic and prognostic biomarkers and potential molecular targets.
In recent years, evidence has shown that non-coding RNAs, as epigenetic factors, are important in the occurrence and development of cancer, including microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). Among them, miRNAs are the most studied (5–7). miRNAs, with about 22 nucleotides in length, control numerous biological processes, including proliferation, differentiation, and apoptosis (8–11). As a kind of non-coding RNA, miRNAs mainly interact with complementary sequences in the 3′-untranslated regions (3′-UTRs) of corresponding mRNAs through their seed regions and play a role in inhibiting gene expression at the post-transcriptional level (12, 13). Since a single miRNA can interact with hundreds of mRNAs simultaneously, aberrant miRNA expression is involved in the occurrence and development of numerous diseases, especially cancer (14). The expression of most miRNAs is decreased in cancers; for example, miRNA-4317 is low in gastric cancer (GC) and exerts a tumor suppressor effect through inhibiting cell proliferation by targeting ZNF322 (15). The expression of miR-186 is reduced in hepatocellular carcinoma (HCC), and it can inhibit the occurrence and development of HCC by targeting yes-associated protein 1 (YAP1) (16). miR-186 is also expressed at low levels in breast cancer (BC), and low expression of miR-186 is closely associated with the poor prognosis of BC (17). Some miRNAs are increased in cancers; for example, miR-106b-5p is highly expressed in glioma, non-small-cell lung cancer (NSCLC), HCC, and other tumors (18–21). miR-140-5p is upregulated in renal cell carcinoma and promotes tumor cell progression through the miR-140-5p/KLF9/KCNQ1 axis (22).
miR-625 is a recently discovered miRNA that is widely involved in countless human diseases, including cancer. miRNAs are a group of non-coding RNA with a length of ~22 nucleotides (23–25). They guide the RNA-induced silencing complex (RISC) to degrade mRNA by base pairing with target gene mRNAs, thus inhibiting the expression of the target genes (26, 27). Single-stranded RNA monomers (pre-miRNAs) of ~70–100 bases in size and have a hairpin structure (28). These pre-miRNAs are processed by Dicer enzyme digestion to form mature miRNAs (28–30). For some miRNA precursors, the two separate arms target different sites and comprise the functional mature miRNA; these arms are represented by the notation−5p and−3p, as processed from the 5′ end arm and the 3′ end arm, respectively (e.g., miR-625-5p or miR-625-3p) (28, 31).
In this review, we comprehensively summarize the expression and functions of miR-625 in a variety of cancer types and its underlying mechanisms. Importantly, we will discuss the lncRNA-miRNA-mRNA and circRNA-miRNA-mRNA networks and briefly discuss the specific mechanism of competing endogenous RNAs (ceRNAs). Overall, this article reveals the powerful potential of miR-625 as a cancer biomarker and therapeutic target and aims to provide some suggestions for further basic research or clinical applications.
Recently, many studies have shown that miR-625 expression is aberrant in tumor tissues compared to nontumor tissues. Researchers often use quantitative real-time RT-PCR (qRT-PCR) to detect the expression of miRNAs in the tissues and plasma of cancer patients (32, 33). Furthermore, they also perform further verification at the cellular level. We found that miR-625 expression was decreased in most cancers and increased in a small number of cancers. In malignant pleural mesothelioma (MPM) (34, 35) and thyroid cancer (TC) (36, 37), miR-625 was found to be upregulated. It is downregulated in bladder cancer (38), nasopharyngeal carcinoma (NPC) (39), lung cancer (40–46), HCC (47), cervical cancer (CC) (48, 49), osteosarcoma (50), melanoma (51–54), laryngeal squamous cell carcinoma (LSCC) (55), acute myeloid leukemia (AML) (56), BC (57–59), glioma (60, 61), esophageal cancer (EC) (62–64), clear cell renal cell carcinoma (65), GC (66–70), and pancreatic ductal adenocarcinoma (PDAC) (71). In colorectal cancer (CRC), the expression of miR-625 seems to vary, being either high or low. Lou et al. found that miR-625 expression was low in cancer tissues compared with adjacent normal tissues (72). Shang et al. also revealed that miR-625-5p was downregulated in CRC (73). In addition, other researchers found that miR-625-3p expression was increased in CRC (74–76). Furthermore, Rasmussen reported that miR-625-3p was not dysregulated between cancer and non-cancer samples (77). These conflicting findings may be attributed to the different action sites of miR-625 and differences in sample size and source. The expression status of and other parameters related to miR-625 in different cancers are depicted in Table 1.
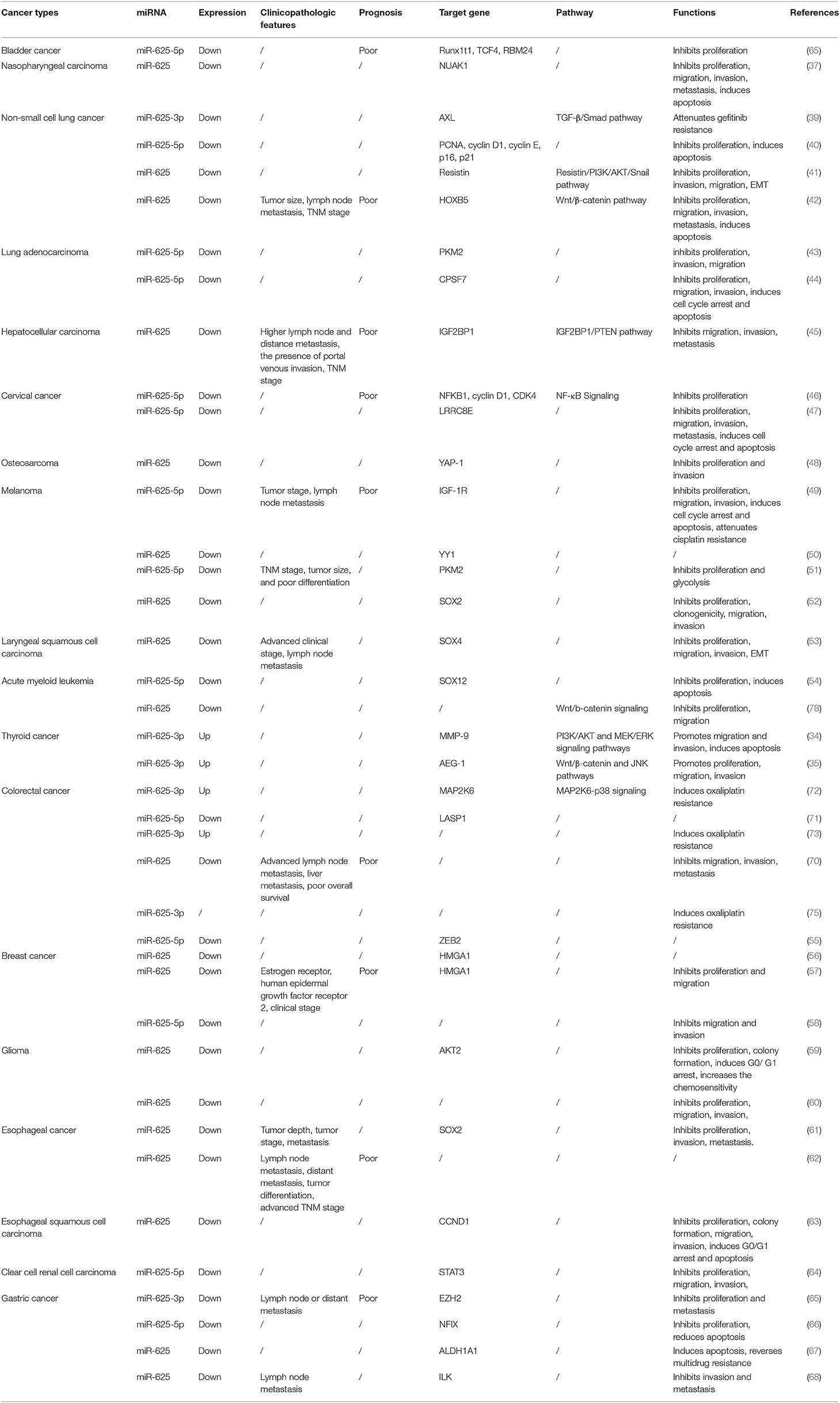
Table 1. The expression, clinical significance, function, and mechanism of miR-625 in different cancers.
Lung cancer is one of the most malignant tumors in the world, and it is also a leading cause of cancer-associated deaths among males and females (79, 80). Lung cancers are categorized into two major histological types: NSCLC and small-cell lung cancer (81). NSCLC accounts for approximately 85% of all lung cancers (82, 83). An increasing number of studies have revealed the anti-cancer effect of miR-625 in NSCLC. Xiaoxia Tan found that low expression of miR-625 was related to advanced clinical characteristics and poor overall survival (OS) of patients with NSCLC (44). In addition, in vitro and in vivo studies have proven that miR-625 suppresses cell proliferation, migration and invasion and induces apoptosis in NSCLC. Further experiments proved that miR-625 inactivated the Wnt/β-catenin pathway by targeting Homeobox B5 (HOXB5), thereby exerting a tumor suppressor effect in NSCLC. Homeobox (HOX) genes are a family of transcription factors (84), and HOXB5 belongs to the HOX gene family (85, 86). Zhang et al. previously reported that knockdown of HOXB5 inhibited β-catenin expression and its downstream targets cyclin D1 and c-Myc in A549 cells (87). HOXB5 significantly promoted NSCLC cell growth, invasion, metastasis, and epithelial-mesenchymal transition (EMT), partly through the Wnt/β-catenin signaling pathway. Another study revealed that miR-625 suppresses NSCLC cell metastasis by obstructing the resistin/PI3K/AKT/Snail pathway and by decreasing EMT (43). Lung adenocarcinoma (LAC) is the most common histological subtype of NSCLC (88). Xue and Yang et al. reported that miR-625 was involved in the process by which lncRNAs promoted the progression of LAC (45, 46). Interestingly, miR-625 shows obvious anti-inflammatory effect in both lung injury model and asthma model, through completely different molecular mechanisms (89, 90). And there is an inseparable relationship between inflammation and tumorigenesis. This also explains anti-tumor effect of miR-625 in the deeper mechanism.
In addition to participating in the regulation of a series of biological processes, miR-625 is also closely related to drug resistance and disease diagnosis. Du et al. found that miR-625-3p overexpression reversed gefitinib resistance (41). At present, gefitinib is the preferred treatment NSCLC patients with for epidermal growth factor receptor (EGFR) mutation (91, 92). Mechanistically, miR-625-3p overexpression was found to inhibit the EMT induced by TGF-β1 and enhance gefitinib sensitivity by targeting AXL (41). Roth et al. conducted a blood-based microRNA expression profile analysis, and the results showed that miR-625* was lower in NSCLC patients than in healthy controls or those with benign disease (40). In addition, the levels of miR-625* were noticeably lower in patients who smoked and large-cell lung cancer patients than in nonsmoking patients and adenocarcinoma patients, respectively. Therefore, miR-625* expression can be used as a blood-based marker for disease diagnosis. Figure 1 summarizes the regulatory mechanism of miR-625 in lung cancer. These results reveal that miR-625 is a novel diagnostic and prognostic biomarker and treatment target for lung cancer.
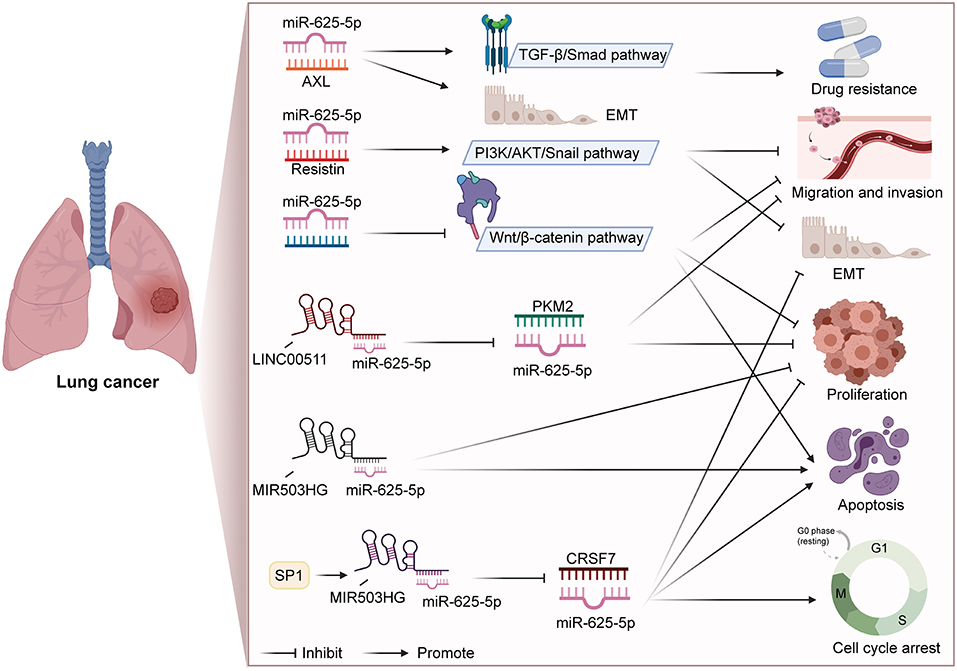
Figure 1. Summary diagram of the roles and mechanisms of miR-625 in lung cancer (Created with BioRender.com).
Cervical cancer is the fourth most common malignant tumor in women and is the main cause of gynecological tumor-related death in the world (93, 94). The most common predisposing cause of CC is high-risk human papillomavirus (HPV) infection (95, 96). Persistent HPV infection can lead to chronic inflammation, thereby causing cervical intraepithelial neoplasia (CIN) as well as cervical cancer (97). Although great progress has been made in the prevention, diagnosis, and treatment of CC, the OS rate of patients is still unsatisfactory, partly due to its late detection and late recurrence (98, 99). In recent years, there have been many studies on miRNAs in CC. Researchers have tried to find new tumor markers for the early diagnosis of CC to improve the disease detection rate and reduce the mortality rate. Li et al. found that miR-625-5p expression was significantly low in CC tissues and cell lines (48). The downregulation of miR-625-5p is linked to unfavorable clinical prognosis of CC patients. Overexpression of miR-625-5p suppresses cell proliferation in cervical carcinoma. Another study confirmed that the LINC00958/miR-625-5p/LRRC8E axis participated in CC cell proliferation and metastasis (49).
Melanoma arises from the deterioration of melanocytes located in the basement of the epidermis (100). It is the most aggressive form of skin cancer (101, 102). Pyruvate kinase (PK) participates in the transformation of phosphoenolpyruvate to pyruvate and is a rate-limiting enzyme in the last step of the glycolysis process (103, 104). Pyruvate kinase m2 (PKM2) is an alternative splice variant of PK (105). Evidence from clinical in vitro and in vivo studies shows that PKM2 is an important molecule in processes related to progression of cancer, such as glucose metabolism and apoptosis (78, 106, 107). Zhang et al. found a negative correlation between the expression of miR-625-5p and PKM2 in clinical melanoma specimens (53). They further revealed that miR-625-5p inhibited the proliferation, lactic acid production, ATP production, and glucose consumption of melanoma cells by targeting PKM2. Another study also revealed a tumor suppressor role of miR-625 in melanoma. miR-625 can inhibit melanoma cell proliferation, wound healing, migration, and tumorigenicity. Mechanistically, miR-625 plays a role at least in part by inhibiting SOX2 (54). In addition, a relationship between the expression of miR-625 and clinical characteristics has been noted. The results show that miR-625 is associated with tumor size, lymph node metastasis, TNM stage, and differentiation (51, 53).
AML is a malignant hematopoietic system disease with high morbidity and mortality (108, 109). Gain-of-function assays suggested that after transfection with miR-625-5p, the proliferation of U937 and HL60 cells was significantly reduced, whereas the miR-625-5p + SOX12 group had the opposite pattern. Therefore, miR-625-5p can regulate AML cell growth by targeting SOX12 (56). Ma et al. also pointed out that miR-625 participated in the occurrence and development of AML through Wnt/β-catenin signaling (110).
Although miR-625 is down-regulated in most human diseases, its expression in TC is increased (36, 37). Fang et al. investigated the function, molecular mechanism, and signaling pathways of miR-625 in TC (37). The researchers found that miR-625-3p overexpression promotes cancer cell proliferation, migration, and invasion by upregulating the expression of astrocyte elevated gene 1 (AEG-1). Moreover, overexpression of AEG-1 promotes the activation of Wnt/β-catenin and JNK pathways (37). Another study showed that icariin (ICA) exerted tumor inhibitory effect by blocking TC cell proliferation, and metastasis by suppressing miR-625-3p. In addition, ICA can also inactivate the PI3K/AKT and MEK/ERK signaling pathways by regulating miR-625-3p in CC cells (36).
An increasing number of researchers are exploring the link between miRNAs and CRC and possible mechanistic targets. miR-625-3p was found to play an oncogenic role in CRC: it could promote cell migration and invasion and induce oxaliplatin resistance (74–76). The SCAI/E-cadherin/MMP-9 pathways (76) and MAP2K6-p38 signaling are involved in this process (74). Lou et al. revealed the tumor suppressor effect of miR-625 in CRC (72). The researchers found that miR-625 was obviously downregulated in CRC tissues and cell lines. The expression of miR-625 had an inverse relationship with the lymph node metastasis and liver metastasis status. The univariate and multivariate analysis results both showed that miR-625 could be used as an independent prognostic factor for CRC. In vivo and in vitro experiments also revealed that miR-625 inhibited invasion and migration. Similarly, another study reported that miR-625-5p was involved in the inhibition of CRC development (73).
BC is the most frequently diagnosed malignancy in females worldwide (80, 111). At present, the main treatment strategies include surgical resection, radiotherapy and chemotherapy, hormone therapy, and targeted biological therapy (112). Although the prognosis of BC patients has improved, BC is still the main cause of cancer-related deaths in women (113, 114). Zhou and his colleagues conducted clinical studies and in vitro cell function experiments, revealing the clinical value of miR-625 in BC. They found that miR-625 expression was decreased in BC and related to poor outcomes. The decreased expression level of miR-625 was closely associated with estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) expression and clinical stage (59). In addition, miR-625 inhibits cell proliferation and migration by regulating HMGA1, its downstream target. Mechanistically, HMGA1 transfers YAP to the nucleus by regulating the activity of cyclin E2, thereby promoting cell migration and invasiveness (115). Wu and Qi et al. also reported that miR-625 exerted a tumor suppressor effect in BC (57, 58).
Glioma accounts for ~80% of brain malignancies and is the most common intracranial tumor (116–118). It is characterized by high malignancy, strong invasiveness, and poor prognosis (119, 120). Studies have found that circDENND2A promotes hypoxia-induced migration and invasion of U87MG and A172 cells. However, miR-625-5p inhibited these effects and had a tumor suppressor effect in gliomas (60). In addition, Zhang and his colleagues conducted a series of studies to explore the role of miR-625 in glioma. They used BALB/c nude mice to conduct in vivo experiments, and the results showed that miR-625 inhibited tumor growth and angiogenesis. In vitro functional experiments revealed that miR-625 suppressed glioma cell proliferation and colony formation and induced G0/G1 arrest, thereby influencing cell cycle progression (61). Moreover, miR-625 enhanced temozolomide (TMZ) chemosensitivity by targeting AKT2. Drug resistance has always been a difficult problem in cancer treatment (121). Therefore, miR-625 can be used as a treatment target for glioma.
miR-625 has been reported to be downregulated in EC. Low miR-625 expression was significantly correlated with tumor stage, tumor depth, and metastasis. Functionally, miR-625 inhibits tumor cell proliferation, migration, and metastasis but does not affect apoptosis. Mechanistically, miR-625 works by directly binding to the 3′-UTR of SOX2 (63). Chuan Li evaluated the correlation of miR-625 expression with clinicopathological features in 169 pairs of ESCC tissues and adjacent non-tumor tissues (64). Low miR-625 expression is closely linked to lymph node and distant metastasis, poor tumor differentiation, and advanced TNM stage. Moreover, the 5-year OS rate in the low expression group is 38.1%, compared with 68.8% in the high expression group. All these results suggest that down-regulation of miR-625 may serve as a novel biomarker to predict tumor progression and poor prognosis in EC patients.
Accumulating evidence has shown that miR-625 is involved in many processes in the development and progression of GC. Wang et al. reported the tumor suppressor effect of miR-625 in GC (70). In clinical application studies, it was found that low expression of miR-625 was related to lymph node metastasis. In mechanistic studies, the results showed that miR-625 was an important regulator of the migration and invasion potential of GC cells. miR-625 inhibits the migration and invasion of cells by regulating ILK. Furthermore, miR-625 is also involved in influencing multidrug resistance (MDR). According to a study report, miR-625 reverses MDR in GC cells by inhibiting ALDH1A1 (69). Consistent with previous studies, Li and colleagues also found that low miR-625-3p had a close relationship with lymph node and distant metastasis. A Kaplan-Meier survival curve analysis indicated that low miR-625-3p expression was remarkably associated with poor prognosis in GC patients. In addition, miR-625-3p was found to regulate the proliferation and migration of GC cells through the inhibition of EZH2 expression (67).
In bladder cancer (38), NPC (39), HCC (47), osteosarcoma (50), LSCC (55), ccRCC (65), and PDAC (71), miR-625 plays a protective role. Up-regulation of miR-625 can affect tumor progression through different mechanisms of action. Zhou et al. found that a decrease in miR-625 was obviously associated with lymph node and distant metastasis, the presence of portal venous invasion, advanced TNM stage, and unfavorable OS (47). Further investigation revealed that the miR-625/IGF2BP1/PTEN axis participated in the occurrence and development of HCC. Moreover, we know that liver cirrhosis is an important risk factor for HCC. The latest research shows that the expression level of miR-625 is related to the levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST), and the results of receiver operating characteristic (ROC) analysis show that miR-625 has ideal sensitivity and specificity in the diagnosis of liver cirrhosis (122). Therefore, we have reason to believe that miR-625 will contribute to the early diagnosis of HCC. EMT is a complicated trans-differentiation process that is a hallmark of cancer (123, 124). Cancer cells gain the ability to migrate and invade through this process (125, 126). Li et al. point out that up-regulated miR-625 promoted E-cadherin and inhibited N-cadherin and vimentin, suppressing EMT of LSCC cells (55). Interestingly, through a systematic review and a qualitative meta-analysis, a series of miRNAs have been found which have the potential to diagnose malignant mesothelioma, including miR-625. Although this result was found several years ago and had limitation, it still can confirm the biomarker potential of miR-625 again. In addition, researchers found that miR-625 worked by targeting Sex-determining region Y-box 4 (SOX4). Researchers have found that miR-625-3p could be used as a marker to assess the prognosis of cancer (34, 35).
Salmena et al. first proposed the ceRNA hypothesis, which concerns how RNA interacts through microRNA response elements (MREs) (127). As mentioned above, miRNAs mainly inhibit the expression of target genes at the post-transcriptional level (12, 13). ceRNAs can function as natural miRNA sponges and competitively bind to and deactivate miRNAs through MREs, hence influencing the mRNA level of target genes (128–130). Theoretically, any RNA molecule may become an active ceRNA if it shares miRNA binding sites with other RNAs (131). Currently, the most studied ceRNAs are lncRNAs, circRNAs, pseudogenes, and protein-coding transcripts (131–135). Cancer is usually related to abnormal gene expression at the transcriptional and post-transcriptional levels (124). Gene expression is a key determinant of cell phenotype (136). In recent years, studies have found that ceRNAs play an important role in the pathogenesis and development of cancer by affecting the expression of carcinogenic and tumor suppressor genes. Figure 2 shows the ceRNA network of circRNAs/lncRNAs-miR-625-mRNAs clearly and comprehensively.
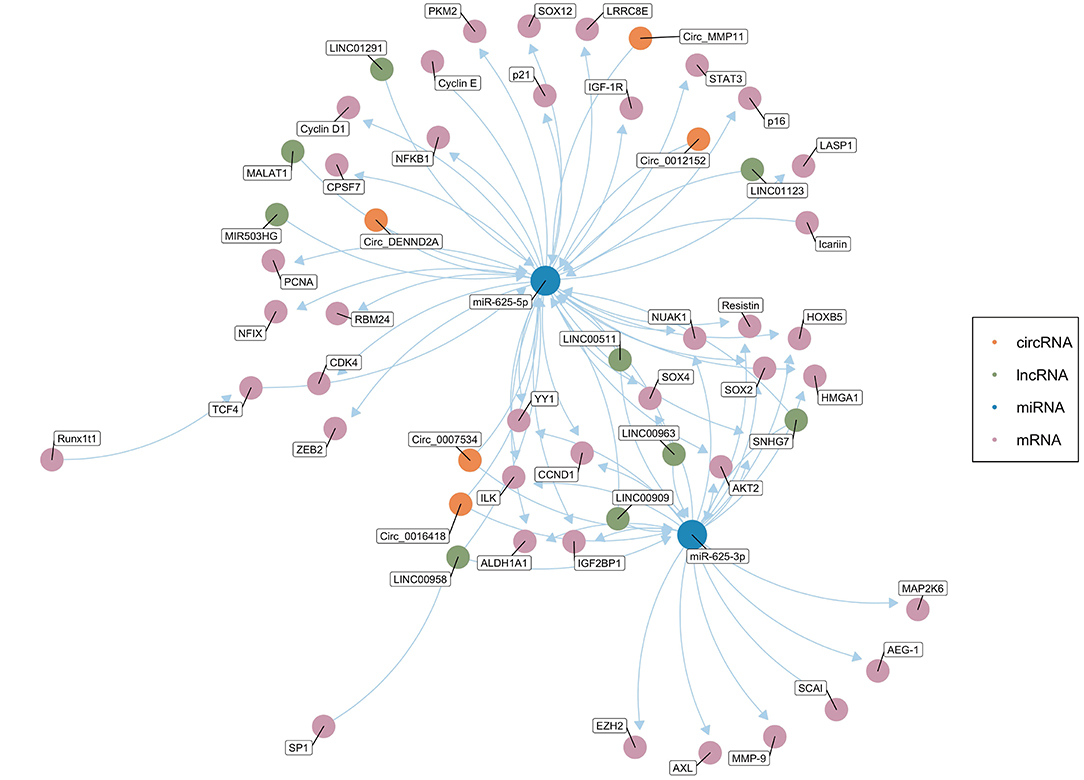
Figure 2. The competing endogenous RNA (ceRNA) network of circRNAs/lncRNAs-miR-625-mRNAs. The green dots represent lncRNAs, and the orange dots represent circRNAs. The blue dots represent miR-625-5p or miR-625-3p. The pink dots represent mRNAs (Created with BioRender.com).
Table 2 summarizes known circRNA-miR-625-mRNA networks. Y Zou evaluated the expression of circ0016418 in skin melanoma (52). The qRT-PCR results showed that compared with that in adjacent normal tissues, the circ0016418 in melanoma tissues was remarkably higher. Further research revealed that circ0016418 exerted a carcinogenic effect by regulating the miR-625/YY1 axis. Shang et al. found that down-regulation of circ0012152 suppressed proliferation and induced apoptosis of AML cells via the miR-625-5p/SOX12 axis (56). In addition, the circMMP11/miR-625-5p/ZEB2 axis is involved in the BC cell proliferation, migration and apoptosis (57). Moreover, circDENND2A promotes the migration and invasion of glioma cells by sponging miR-625-5p (60). Hao et al. found that circ0007534 was significantly related to PDAC stage and lymph node infiltration (71). The results of survival analysis shows that an increase in circ0007534 indicates a poor prognosis. Mechanistically, the carcinogenic function of circ0007534 partly depends on the regulation of miR-625 and miR-892b.
Table 3 summarizes known lncRNA-miR-625-mRNA networks. In LAC, ccRCC, and GC, LINC00511 exerts carcinogenic effects by sponging miR-625. Members of the LINC00511/miR-625-5p/PKM2 axis may be helpful therapeutic targets for LAC (45). Similarly, Huanghao Deng pointed out that the LINC00511/miR-625/CCND1 axis participated in ccRCC progression and was a potential therapeutic target (65). LINC00511 promotes GC progression through the miR-625-5p/STAT3 axis and miR-625-5p/NFIX axis (66, 68). In NPC (39), LAC (46), and CC (49), LINC00958 functions as a ceRNA that can promote the growth, migration, and invasion of tumor cells by sponging miR-625. The study found that an increase in circulating miR-625-3p and a decrease in lncRNA GAS5 were significantly related to MPM progression. Reduced GAS5 is significantly associated with shorter OS and progression-free survival. The study also revealed the potential value of GAS5 in patients treated with platinum-adjuvant chemotherapy (34). Dao et al. reported that knockdown of the lncRNA MIR503HG inhibited proliferation and induced apoptosis of NSCLC cells by regulating miR-625-5p and miR-489-3p (42). Ma and his colleagues explored the clinical significance of LINC00909 expression in AML patients. The results showed that LINC00909 was an independent prognostic indicator of OS for AML patients. Further research found that LINC00909 promoted disease progression by regulating the miR-625/b-catenin axis (110). Wang et al. reported that expression of the lncRNA SNHG7 was enhanced in ESCC, promoting ESCC cell proliferation, and metastasis by regulating miR-625 (62). In addition, the lncRNA MALAT1/miR-625-5p/NF-κB, LINC01291/miR-625-5p/IGF-1R, LINC01123/miR-625-5P/LASP1 and LINC00963/miR-625/HMGA1 pathways play important roles in CC, melanoma, CRC, and BC, respectively (48, 51, 58, 73).
In recent years, increasing evidence has shown that miRNAs participate in the occurrence and development of cancers. miRNAs are abnormally expressed in cancers and are widely involved in a variety of biological processes, including proliferation, migration, invasion, cell cycle regulation, apoptosis, and intracellular metabolism. Some abnormally expressed miRNAs are closely related to clinical features and can also be used as independent markers of disease prognosis. Further understanding the miRNA biogenesis plays a vital role in the follow-up miRNA drug research and exploring the functions of miRNA in the occurrence and development of cancers. Therefore, we summarized the latest miRNA biogenesis process in Figure 3. In addition, miRNAs also play an important role in tumor treatment, especially in drug resistance. For example, Wang et al. found that FER1L4 could suppress miR-106a-5p/miR-372-5p expression, activating the E2F1-mediated NF-κB pathway and thus leading to drug resistance in liver cancer (137). Luo et al. also reported that FOXO3a-miRNA feedback could lead to Herceptin resistance in BC (138). In CRC, it was reported that the HSF1/miR-135b-5p axis could promote oxaliplatin resistance via the MUL1/ULK1 pathway (139). Interestingly, chronic accumulation of senescent cells and the concomitant senescence-associated secretory phenotype contribute to tumor microenvironment remodeling. In this process, the levels of a large number of related miRNAs have changed greatly. After measurement the level of miRNAs one by one, it is found that the level of miR-625 continues to decline with cell senescence. The trend of miR-625 in cell senescence is consistent with the trend of miR-625 in tumors (140). So, we can guess that the level of miR-625 may be a part of the tumor microenvironment affecting the occurrence and development of cancer.
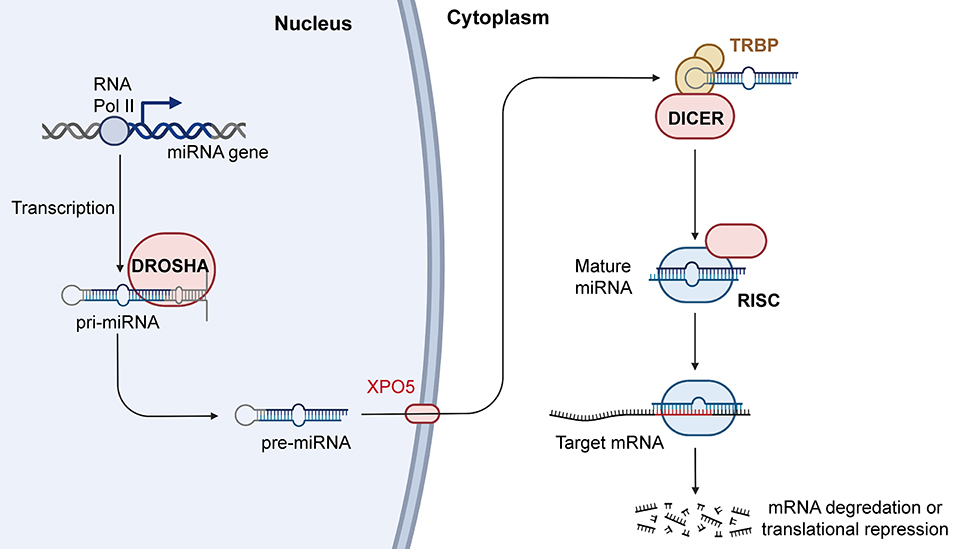
Figure 3. Biogenesis and mechanism of miRNA. RNAPII transcribes miRNA genes into pri-miRNAs, and the pri-miRNAs are cleaved by Drosha into the pre-miRNAs. Pre-miRNAs are transferred from nucleus to cytoplasm with the help of XPO5, and Dicer can process the pre-miRNAs into mature miRNAs. Mature miRNAs then combine with AGO2 to form RISCs, which further play an important role in regulating gene expression. RNAPII, RNA polymerase II; XPO5, exportin-5; AGO2, argonaute RISC catalytic component 2; RISC, RNA induced silencing complex (Created with BioRender.com).
There has been increasing research on miR-625. miR-625 can function as a tumor suppressor or a tumor-promoting factor. The abnormal expression of miR-625 in whole blood, plasma, urine, and other samples of tumor patients can be assessed to diagnose cancer with non-invasive methods (34, 35). In fact, according to the existing research progress, we believe that the clinical diagnostic biomarker value of miR-625 will be expected to play in the clinic in the near future. The future research direction should focus on the statistical work before clinical application and clinical application verification. LncRNAs and circRNAs participate in the occurrence and progression of various cancers by regulating miR-625. In addition to ncRNAs, icariin also promotes the development of cancer through the regulation of miR-625 (36). In addition, we believe that miR-625 is an extraordinarily valuable target for cancer treatment, especially in the treatment of drug resistance. Wu et al. found that inhibiting miR-625-5p or up-regulating IGF-1R could offset the regulatory effect of siLINC01291 on the sensitivity of melanoma cells to cisplatin chemotherapy (51). In NSCLC (41), CRC (75), and GC (69), miR-625 has also been reported to affect drug resistance.
In this review, the expression and function of miR-625 in cancer were reviewed, and the related molecular mechanisms were briefly discussed. However, due to the limited number of existing studies and the dual role of miR-625 in the development of some tumors, a large number of basic experiments, animal models, and clinical studies are needed to further reveal and verify its function and clinical significance. Overall, miR-625 is expected to be a promising new target for cancer treatment.
WG, YH, and SZ designed and guided the review. MZ wrote the manuscript. YH and FX drew the mechanism diagrams. WG and YH edited and reviewed the manuscript. All authors read and approved the final manuscript.
This work was supported by Leading Talents of Zhongyuan Science and Technology Innovation (214200510027), Henan Provincial Medical Science and Technology Research Plan (SBGJ2018002 and SBGJ202102117), Youth Talent Lifting Project of Henan Province (2021HYTP059), Key Scientific Research Project of Henan Higher Education Institutions of China (21A320026), Henan Medical Science and Technology Joint Building Program (LHGJ20210324), Science and Technology Innovation Talents in Henan Universities (19HASTIT003), Outstanding Foreign Scientist Studio in Henan Province (GZS2020004), and the Gandan Xiangzhao Research Fund (GDXZ2022002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The figure is created with BioRender.com. We sincerely appreciate all the participators in our work.
1. Perez-Amado CJ, Bazan-Cordoba A, Hidalgo-Miranda A, Jimenez-Morales S. Mitochondrial heteroplasmy shifting as a potential biomarker of cancer progression. Int J Mol Sci. (2021) 22:7369. doi: 10.3390/ijms22147369
2. Robertson AG, Rendina LM. Gadolinium theranostics for the diagnosis and treatment of cancer. Chem Soc Rev. (2021) 50:4231–44. doi: 10.1039/D0CS01075H
3. Soerjomataram I, Bray F. Planning for tomorrow: global cancer incidence and the role of prevention 2020-2070. Nat Rev Clin Oncol. (2021) 18:663–72. doi: 10.1038/s41571-021-00514-z
4. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
5. Pajares MJ, Alemany-Cosme E, Goni S, Bandres E, Palanca-Ballester C, Sandoval J. Epigenetic Regulation of microRNAs in Cancer: Shortening the Distance from Bench to Bedside. Int J Mol Sci. (2021) 22:7350. doi: 10.3390/ijms22147350
6. Leite DJ, Ninova M, Hilbrant M, Arif S, Griffiths-Jones S, Ronshaugen M, et al. Pervasive microRNA duplication in chelicerates: insights from the embryonic microRNA repertoire of the spider parasteatoda tepidariorum. Genome Biol Evol. (2016) 8:2133–44. doi: 10.1093/gbe/evw143
7. Malayaperumal S, Sriramulu S, Banerjee A, Pathak S. Over-expression of MicroRNA-122 inhibits proliferation and induces apoptosis in colon cancer cells. Microrna. (2020) 9:354–62. doi: 10.2174/2211536609666201209152228
8. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci. (2020) 21:1723. doi: 10.3390/ijms21051723
9. Suzuki H, Maruyama R, Yamamoto E, Kai M. Epigenetic alteration and microRNA dysregulation in cancer. Front Genet. (2013) 4:258. doi: 10.3389/fgene.2013.00258
10. Mirzaei S, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Ranjbar A, et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. (2021) 509:63–80. doi: 10.1016/j.canlet.2021.03.025
11. Mirzaei S, Zarrabi A, Asnaf SE, Hashemi F, Zabolian A, Hushmandi K, et al. The role of microRNA-338-3p in cancer: growth, invasion, chemoresistance, and mediators. Life Sci. (2021) 268:119005. doi: 10.1016/j.lfs.2020.119005
12. Ji L, Lin Z, Wan Z, Xia S, Jiang S, Cen D, et al. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. (2020) 11:250. doi: 10.1038/s41419-020-2413-4
13. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. (2015) 15:321–33. doi: 10.1038/nrc3932
14. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. (2011) 8:467–77. doi: 10.1038/nrclinonc.2011.76
15. Hu X, Zhang M, Miao J, Wang X, Huang C. miRNA-4317 suppresses human gastric cancer cell proliferation by targeting ZNF322. Cell Biol Int. (2018) 42:923–30. doi: 10.1002/cbin.10870
16. Ruan T, He X, Yu J, Hang Z. MicroRNA-186 targets Yes-associated protein 1 to inhibit Hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett. (2016) 11:2941–5. doi: 10.3892/ol.2016.4312
17. Sun WJ, Zhang YN, Xue P. miR-186 inhibits proliferation, migration, and epithelial-mesenchymal transition in breast cancer cells by targeting Twist1. J Cell Biochem. (2019) 120:10001–9. doi: 10.1002/jcb.28283
18. Liu F, Gong J, Huang W, Wang Z, Wang M, Yang J, et al. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. (2014) 33:4813–22. doi: 10.1038/onc.2013.428
19. Wei K, Pan C, Yao G, Liu B, Ma T, Xia Y, et al. MiR-106b-5p Promotes Proliferation and Inhibits Apoptosis by Regulating BTG3 in Non-Small Cell Lung Cancer. Cell Physiol Biochem. (2017) 44:1545–58. doi: 10.1159/000485650
20. Lee J, Kim HE, Song YS, Cho EY, Lee A. miR-106b-5p and miR-17-5p could predict recurrence and progression in breast ductal carcinoma in situ based on the transforming growth factor-beta pathway. Breast Cancer Res Treat. (2019) 176:119–30. doi: 10.1007/s10549-019-05192-1
21. Zhu HR, Huang RZ Yu XN, Shi X, Bilegsaikhan E, Guo HY, et al. Microarray Expression Profiling of microRNAs Reveals Potential Biomarkers for Hepatocellular Carcinoma. Tohoku J Exp Med. (2018) 245:89–98. doi: 10.1620/tjem.245.89
22. Huang C, Li J, Zhang X, Xiong T, Ye J, Yu J, et al. The miR-140-5p/KLF9/KCNQ1 axis promotes the progression of renal cell carcinoma. FASEB J. (2020) 34:10623–39. doi: 10.1096/fj.202000088RR
23. Yang X, Chen Y, Chen L. The Versatile Role of microRNA-30a in Human Cancer. Cell Physiol Biochem. (2017) 41:1616–32. doi: 10.1159/000471111
24. Li X, Zhou S, Fan T, Feng X. lncRNA DGCR 5/miR27a3p/BNIP3 promotes cell apoptosis in pancreatic cancer by regulating the p38 MAPK pathway. Int J Mol Med. (2020) 46:729–39. doi: 10.3892/ijmm.2020.4632
25. Min J, Han TS, Sohn Y, Shimizu T, Choi B, Bae SW, et al. microRNA-30a arbitrates intestinal-type early gastric carcinogenesis by directly targeting ITGA2. Gastric Cancer. (2020) 23:600–13. doi: 10.1007/s10120-020-01052-w
26. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. (2019) 20:5–20. doi: 10.1038/s41580-018-0059-1
27. Kim YJ, Maizel A, Chen X. Traffic into silence: endomembranes and post-transcriptional RNA silencing. EMBO J. (2014) 33:968–80. doi: 10.1002/embj.201387262
28. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. (2012) 13:358–69. doi: 10.1038/nrg3198
29. Costa C, Teodoro M, Rugolo CA, Alibrando C, Giambo F, Briguglio G, et al. MicroRNAs alteration as early biomarkers for cancer and neurodegenerative diseases: New challenges in pesticides exposure. Toxicol Rep. (2020) 7:759–67. doi: 10.1016/j.toxrep.2020.05.003
30. Wulff BE, Sakurai M, Nishikura K. Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat Rev Genet. (2011) 12:81–5. doi: 10.1038/nrg2915
31. Ansel KM. RNA regulation of the immune system. Immunol Rev. (2013) 253:5–11. doi: 10.1111/imr.12062
32. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
33. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. (2011) 25:1915–27. doi: 10.1101/gad.17446611
34. Kresoja-Rakic J, Szpechcinski A, Kirschner MB, Ronner M, Minatel B, Martinez VD, et al. miR-625-3p and lncRNA GAS5 in Liquid Biopsies for Predicting the Outcome of Malignant Pleural Mesothelioma Patients Treated with Neo-Adjuvant Chemotherapy and Surgery. Noncoding RNA. (2019) 5:41. doi: 10.3390/ncrna5020041
35. Michaela B, Kirschner YYC, Badrian B. Increased circulating miR-625-3p: a potential biomarker for patients with malignant pleural mesothelioma. J Thorac Oncol. (2012) 7:1184–91. doi: 10.1097/JTO.0b013e3182572e83
36. Fang L, Xu W, Kong D. Icariin inhibits cell proliferation, migration and invasion by down-regulation of microRNA-625-3p in thyroid cancer cells. Biomed Pharmacother. (2019) 109:2456–63. doi: 10.1016/j.biopha.2018.04.012
37. Fang L, Kong D, Xu W. MicroRNA-625-3p promotes the proliferation, migration and invasion of thyroid cancer cells by up-regulating astrocyte elevated gene 1. Biomed Pharmacother. (2018) 102:203–11. doi: 10.1016/j.biopha.2018.03.043
38. Yin YW, Liu KL, Lu BS Li W, Niu YL, Zhao CM, et al. RBM24 exacerbates bladder cancer progression by forming a Runx1t1/TCF4/miR-625-5p feedback loop. Exp Mol Med. (2021) 53:933–46. doi: 10.1038/s12276-021-00623-w
39. Chen M, Xu Z, Zhang Y, Zhang X. LINC00958 promotes the malignancy of nasopharyngeal carcinoma by sponging microrna-625 and thus upregulating NUAK1. Onco Targets Ther. (2019) 12:9277–90. doi: 10.2147/OTT.S216342
40. Roth C, Stuckrath I, Pantel K, Izbicki JR, Tachezy M, Schwarzenbach H. Low levels of cell-free circulating miR-361-3p and miR-625* as blood-based markers for discriminating malignant from benign lung tumors. PLoS ONE. (2012) 7:e38248. doi: 10.1371/journal.pone.0038248
41. Du W, Sun L, Liu T, Zhu J, Zeng Y, Zhang Y, et al. The miR6253p/AXL axis induces nonT790M acquired resistance to EGFRTKI via activation of the TGFbeta/Smad pathway and EMT in EGFRmutant nonsmall cell lung cancer. Oncol Rep. (2020) 44:185–95. doi: 10.3892/or.2020.7579
42. Dao R, Wudu M, Hui L, Jiang J, Xu Y, Ren H, et al. Knockdown of lncRNA MIR503HG suppresses proliferation and promotes apoptosis of non-small cell lung cancer cells by regulating miR-489-3p and miR-625-5p. Pathol Res Pract. (2020) 216:152823. doi: 10.1016/j.prp.2020.152823
43. Zhao Y, Zheng R, Ning D, Xie F. MiR-625 inhibits tumor cell invasion, migration and EMT by negatively regulating the expression of resistin in non-small cell lung. Cancer Manag Res. (2020) 12:4171–80. doi: 10.2147/CMAR.S248251
44. Tan X, Jiang L, Wu X, Feng W, Lin Q. MicroRNA-625 inhibits the progression of non-small cell lung cancer by directly targeting HOXB5 and deactivating the Wnt/β-catenin pathway. Int J Mol Med. (2019) 44:346–56. doi: 10.3892/ijmm.2019.4203
45. Xue J, Zhang F. LncRNA LINC00511 plays an oncogenic role in lung adenocarcinoma by regulating PKM2 expression via sponging miR-625-5p. Thorac Cancer. (2020) 11:2570–9. doi: 10.1111/1759-7714.13576
46. Yang L, Li L, Zhou Z, Liu Y, Sun J, Zhang X, et al. SP1 induced long non-coding RNA LINC00958 overexpression facilitate cell proliferation, migration and invasion in lung adenocarcinoma via mediating miR-625-5p/CPSF7 axis. Cancer Cell Int. (2020) 20:24. doi: 10.1186/s12935-020-1099-0
47. Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ, Liu LL, et al. miR-625 suppresses tumour migration and invasion by targeting IGF2BP1 in hepatocellular carcinoma. Oncogene. (2015) 34:965–77. doi: 10.1038/onc.2014.35
48. Li Y, Ding Y, Ding N, Zhang H, Lu M, Cui X, et al. MicroRNA-625-5p sponges lncRNA MALAT1 to inhibit cervical carcinoma cell growth by suppressing NF-kappa B signaling. Cell Biochem Biophys. (2020) 78:217–25. doi: 10.1007/s12013-020-00904-7
49. Wang L, Zhong Y, Yang B, Zhu Y, Zhu X, Xia Z, et al. LINC00958 facilitates cervical cancer cell proliferation and metastasis by sponging miR-625-5p to upregulate LRRC8E expression. J Cell Biochem. (2020) 121:2500–9. doi: 10.1002/jcb.29472
50. Luo Z, Wu G, Zhang D, Liu J, Ran R. microRNA625 targets Yesassociated protein 1 to suppress cell proliferation and invasion of osteosarcoma. Mol Med Rep. (2018) 17:2005–11. doi: 10.3892/mmr.2017.8079
51. Wu L, Li K, Lin W, Liu J, Qi Q, Shen G, et al. Long noncoding RNA LINC01291 promotes the aggressive properties of melanoma by functioning as a competing endogenous RNA for microRNA-625-5p and subsequently increasing IGF-1R expression. Cancer Gene Ther. (2021). doi: 10.1038/s41417-021-00313-9
52. Zou Y, Wang S-S, Wang J. CircRNA_0016418 expedites the progression of human skin melanoma via miR-625/YY1 axis. Eur Rev Med Pharmacol Sci. (2019) 23:10918–30. doi: 10.26355/eurrev_201912_19795
53. Zhang H, Feng C, Zhang M, Zeng A, Si L, Yu N, et al. miR-625-5p/PKM2 negatively regulates melanoma glycolysis state. J Cell Biochem. (2019) 120:2964–72. doi: 10.1002/jcb.26917
54. Fan Y, Fa Z, Xu J, Yu H, Li P, Fang W, et al. microRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget. (2017) 8:13253–63. doi: 10.18632/oncotarget.14710
55. Li Y, Tao C, Dai L, Cui C, Chen C, Wu H, et al. MicroRNA-625 inhibits cell invasion and epithelial-mesenchymal transition by targeting SOX4 in laryngeal squamous cell carcinoma. Biosci Rep. (2019) 39:BSR20181882. doi: 10.1042/BSR20181882
56. Shang Z, Ming X, Wu J, Xiao Y. Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematol Oncol. (2021) 39:539–48. doi: 10.1002/hon.2895
57. Qi L, Sun B, Yang B, Lu S. CircMMP11 regulates proliferation, migration, invasion, and apoptosis of breast cancer cells through miR-625-5p/ZEB2 axis. Cancer Cell Int. (2021) 21:133. doi: 10.1186/s12935-021-01816-z
58. Wu Z, Wang W, Wang Y, Wang X, Sun S, Yao Y, et al. Long noncoding RNA LINC00963 promotes breast cancer progression by functioning as a molecular sponge for microRNA-625 and thereby upregulating HMGA1. Cell Cycle. (2020) 19:610–24. doi: 10.1080/15384101.2020.1728024
59. Zhou WB, Zhong CN, Luo XP, Zhang YY, Zhang GY, Zhou DX, et al. miR-625 suppresses cell proliferation and migration by targeting HMGA1 in breast cancer. Biochem Biophys Res Commun. (2016) 470:838–44. doi: 10.1016/j.bbrc.2016.01.122
60. Su H, Zou D, Sun Y, Dai Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol Biol Lett. (2019) 24:24. doi: 10.1186/s11658-019-0149-x
61. Zhang J, Zhang J, Zhang J, Qiu W, Xu S, Yu Q, et al. MicroRNA-625 inhibits the proliferation and increases the chemosensitivity of glioma by directly targeting AKT2. Am J Cancer Res. (2017) 7:1835–49.
62. Wang Y, Bao D, Wan L, Zhang C, Hui S, Guo H. Long non-coding RNA small nucleolar RNA host gene 7 facilitates the proliferation, migration, and invasion of esophageal cancer cells by regulating microRNA-625. J Gastrointest Oncol. (2021) 12:423–32. doi: 10.21037/jgo-21-147
63. Wang Z, Qiao Q, Chen M, Li X, Wang Z, Liu C, et al. miR-625 down-regulation promotes proliferation and invasion in esophageal cancer by targeting Sox2. FEBS Lett. (2014) 588:915–21. doi: 10.1016/j.febslet.2014.01.035
64. Li C, Li DC, Che SS, Ma K, Wang YJ, Xia LH, et al. The decreased expression of miR-625 predicts poor prognosis of esophageal squamous cell carcinoma. Int J Clin Exp Med. (2015) 8:9560–4.
65. Deng H, Huang C, Wang Y, Jiang H, Peng S, Zhao X, et al. LINC00511 promotes the malignant phenotype of clear cell renal cell carcinoma by sponging microRNA-625 and thereby increasing cyclin D1 expression. Aging. (2019) 11:5975–91. doi: 10.18632/aging.102156
66. Cui N, Sun Q, Liu H, Li L, Guo X, Shi Y, et al. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered. (2021) 12:2915–27. doi: 10.1080/21655979.2021.1940611
67. Li Y, Zhou HC, Zhang Y, Huang H. MicroRNA-625-3p inhibits gastric cancer metastasis through modulating EZH2. Eur Rev Med Pharmacol Sci. (2020) 24:1177–85. doi: 10.26355/eurrev_202002_20169
68. Chen Z, Wu H, Zhang Z, Li G, Liu B. LINC00511 accelerated the process of gastric cancer by targeting miR-625-5p/NFIX axis. Cancer Cell Int. (2019) 19:351. doi: 10.1186/s12935-019-1070-0
69. Gong X, Xu B, Zi L, Chen X. miR-625 reverses multidrug resistance in gastric cancer cells by directly targeting ALDH1A1. Cancer Manag Res. (2019) 11:6615–24. doi: 10.2147/CMAR.S208708
70. Wang M, Li C, Nie H, Lv X, Qu Y, Yu B, et al. Down-regulated miR-625 suppresses invasion and metastasis of gastric cancer by targeting ILK. FEBS Lett. (2012) 586:2382–8. doi: 10.1016/j.febslet.2012.05.050
71. Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y, et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. (2019) 120:3780–9. doi: 10.1002/jcb.27658
72. Lou X, Qi X, Zhang Y, Long H, Yang J. Decreased expression of microRNA-625 is associated with tumor metastasis and poor prognosis in patients with colorectal cancer. J Surg Oncol. (2013) 108:230–5. doi: 10.1002/jso.23380
73. Shang T, Zhou X, Chen W. LINC01123 promotes the progression of colorectal cancer via miR-625-5p/LASP1 axis. Cancer Biother Radiopharm. (2021) 36:765–73. doi: 10.1089/cbr.2020.3740
74. Rasmussen MH, Lyskjaer I, Jersie-Christensen RR, Tarpgaard LS, Primdal-Bengtson B, Nielsen MM, et al. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat Commun. (2016) 7:12436. doi: 10.1038/ncomms12436
75. Iben Lyskjær MHR, Claus L. Andersen. Putting a brake on stress signaling: miR-625-3p as a biomarker for choice of therapy in colorectal cancer Epigenomics. (2016) 8:1449–52. doi: 10.2217/epi-2016-0128
76. Zheng H, Ma R, Wang Q, Zhang P, Li D, Wang Q, et al. MiR-625-3p promotes cell migration and invasion via inhibition of SCAI in colorectal carcinoma cells. Oncotarget. (2015) 6:27805–15. doi: 10.18632/oncotarget.4738
77. Rasmussen MH, Jensen NF, Tarpgaard LS, Qvortrup C, Romer MU, Stenvang J, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. (2013) 7:637–46. doi: 10.1016/j.molonc.2013.02.016
78. Yang W, Lu Z. Regulation and function of pyruvate kinase M2 in cancer. Cancer Lett. (2013) 339:153–8. doi: 10.1016/j.canlet.2013.06.008
79. Zhang P, Shao G, Lin X, Liu Y, Yang Z. MiR-338-3p inhibits the growth and invasion of non-small cell lung cancer cells by targeting IRS2. Am J Cancer Res. (2017) 7:53–63.
80. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
81. Fiteni F, Anota A, Westeel V, Bonnetain F. Methodology of health-related quality of life analysis in phase III advanced non-small-cell lung cancer clinical trials: a critical review. BMC Cancer. (2016) 16:122. doi: 10.1186/s12885-016-2152-1
82. Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. (2015) 10:1243–60. doi: 10.1097/JTO.0000000000000630
83. Ettinger DS, Akerley W, Bepler G. Non-small cell lung cancer. J Natl Compr Canc Netw. (2010) 8:740–801. doi: 10.6004/jnccn.2010.0056
84. Samuel S, Naora H. Homeobox gene expression in cancer: insights from developmental regulation and deregulation. Eur J Cancer. (2005) 41:2428–37. doi: 10.1016/j.ejca.2005.08.014
85. Kam MK, Lui VC. Roles of Hoxb5 in the development of vagal and trunk neural crest cells. Dev Growth Differ. (2015) 57:158–68. doi: 10.1111/dgd.12199
86. Kam MK, Cheung M, Zhu JJ, Cheng WW, Sat EW, Tam PK, et al. Homeobox b5 (Hoxb5) regulates the expression of Forkhead box D3 gene (Foxd3) in neural crest. Int J Biochem Cell Biol. (2014) 55:144–52. doi: 10.1016/j.biocel.2014.09.002
87. Zhang B, Li N, Zhang H. Knockdown of Homeobox B5 (HOXB5) Inhibits Cell Proliferation, Migration, and Invasion in Non-Small Cell Lung Cancer Cells Through Inactivation of the Wnt/beta-Catenin Pathway. Oncol Res. (2018) 26:37–44. doi: 10.3727/096504017X14900530835262
88. Pisapia P, Pepe F, Troncone G, Malapelle U. Predictive biomarkers for molecular pathology in lung cancer. Biomark Med. (2020) 14:253–7. doi: 10.2217/bmm-2019-0490
89. Tang B, Xu Q, Xuan L. Circ 0001434 RNA reduces inflammation in acute lung injury model through Wnt/β-catenin and NF-κB by miR-625-5p. Int J Clin Exp Pathol. (2019) 12:3290–300.
90. Qian FH, Deng X, Zhuang QX, Wei B, Zheng DD. miR6255p suppresses inflammatory responses by targeting AKT2 in human bronchial epithelial cells. Mol Med Rep. (2019) 19:1951–7. doi: 10.3892/mmr.2019.9817
91. Sanford M, Scott LJ. Gefitinib: a review of its use in the treatment of locally advanced/metastatic non-small cell lung cancer. Drugs. (2009) 69:2303–28. doi: 10.2165/10489100-000000000-00000
92. Watanabe H, Tamura T, Kagohashi K, Takayashiki N, Kurishima K, Satoh H, et al. Successful treatment of EGFR-mutated non-small cell lung cancer with reduced-dose gefitinib: a case report. Exp Ther Med. (2015) 10:386–8. doi: 10.3892/etm.2015.2499
93. Sakuragi N, Murakami G, Konno Y, Kaneuchi M, Watari H. Nerve-sparing radical hysterectomy in the precision surgery for cervical cancer. J Gynecol Oncol. (2020) 31:e49. doi: 10.3802/jgo.2020.31.e49
94. Zhou P, Liu W, Cheng Y, Qian D. Nanoparticle-based applications for cervical cancer treatment in drug delivery, gene editing, and therapeutic cancer vaccines. Wiley Interdiscip Rev Nanomed Nanobiotechnol. (2021) 13:e1718. doi: 10.1002/wnan.1718
95. Bava SV, Thulasidasan AK, Sreekanth CN, Anto RJ. Cervical cancer: A comprehensive approach towards extermination. Ann Med. (2016) 48:149–61. doi: 10.3109/07853890.2016.1145796
96. Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. (2003) 16:1–17. doi: 10.1128/CMR.16.1.1-17.2003
97. Tewari KS, Monk BJ. New strategies in advanced cervical cancer: from angiogenesis blockade to immunotherapy. Clin Cancer Res. (2014) 20:5349–58. doi: 10.1158/1078-0432.CCR-14-1099
98. van Dyk S, Narayan K, Bernshaw D, Kondalsamy-Chennakesavan S, Khaw P, Lin MY, et al. Clinical outcomes from an innovative protocol using serial ultrasound imaging and a single MR image to guide brachytherapy for locally advanced cervix cancer. Brachytherapy. (2016) 15:817–24. doi: 10.1016/j.brachy.2016.07.008
99. Yadav N, Parveen S, Banerjee M. Potential of nano-phytochemicals in cervical cancer therapy. Clin Chim Acta. (2020) 505:60–72. doi: 10.1016/j.cca.2020.01.035
101. Fattore L, Costantini S, Malpicci D, Ruggiero CF, Ascierto PA, Croce CM, et al. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget. (2017) 8:22262–78. doi: 10.18632/oncotarget.14763
102. Sandru F, Draghici CC, Predescu T, Magdalena Constantin M, Petca RC, Constantin T, et al. Regressive melanoma in a female patient: a case report. Exp Ther Med. (2020) 20:87–90. doi: 10.3892/etm.2020.8675
103. Sun Y, Chen Y, Xu M, Liu C, Shang H, Wang C. Shenmai injection supresses glycolysis and enhances cisplatin cytotoxicity in cisplatin-resistant A549/DDP cells via the AKT-mTOR-c-Myc signaling pathway. Biomed Res Int. (2020) 2020:9243681. doi: 10.1155/2020/9243681
104. Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. (2008) 452:230–3. doi: 10.1038/nature06734
105. Li Z, Sun X. Non-Coding RNAs Operate in the Crosstalk Between Cancer Metabolic Reprogramming and Metastasis. Front Oncol. (2020) 10:810. doi: 10.3389/fonc.2020.00810
106. Li Z, Yang P, Li Z. The multifaceted regulation and functions of PKM2 in tumor progression. Biochim Biophys Acta. (2014) 1846:285–96. doi: 10.1016/j.bbcan.2014.07.008
107. Kwon OH, Kang TW, Kim JH, Kim M, Noh SM, Song KS, et al. Pyruvate kinase M2 promotes the growth of gastric cancer cells via regulation of Bcl-xL expression at transcriptional level. Biochem Biophys Res Commun. (2012) 423:38–44. doi: 10.1016/j.bbrc.2012.05.063
108. Kreitz J, Schonfeld C, Seibert M, Stolp V, Alshamleh I, Oellerich T, et al. Metabolic plasticity of acute myeloid leukemia. Cells. (2019) 8:805. doi: 10.3390/cells8080805
109. Prada-Arismendy J, Arroyave JC, Rothlisberger S. Molecular biomarkers in acute myeloid leukemia. Blood Rev. (2017) 31:63–76. doi: 10.1016/j.blre.2016.08.005
110. Ma L, Wang YY, Jiang P. LncRNA LINC00909 promotes cell proliferation and metastasis in pediatric acute myeloid leukemia via miR-625-mediated modulation of Wnt/beta-catenin signaling. Biochem Biophys Res Commun. (2020) 527:654–61. doi: 10.1016/j.bbrc.2020.05.001
111. Ghoncheh M, Pournamdar Z, Salehiniya H. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev. (2016) 17:43–6. doi: 10.7314/APJCP.2016.17.S3.43
112. Liu H, Ye H. Screening of the prognostic targets for breast cancer based co-expression modules analysis. Mol Med Rep. (2017) 16:4038–44. doi: 10.3892/mmr.2017.7063
113. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. (2010) 60:277–300. doi: 10.3322/caac.20073
114. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. (2013) 49:1374–403. doi: 10.1016/j.ejca.2012.12.027
115. Pegoraro S, Ros G, Ciani Y, Sgarra R, Piazza S, Manfioletti G. A novel HMGA1-CCNE2-YAP axis regulates breast cancer aggressiveness. Oncotarget. (2015) 6:19087–101. doi: 10.18632/oncotarget.4236
116. Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. (2012) 205:613–21. doi: 10.1016/j.cancergen.2012.10.009
117. Kim YW, Liu TJ, Koul D, Tiao N, Feroze AH, Wang J, et al. Identification of novel synergistic targets for rational drug combinations with PI3 kinase inhibitors using siRNA synthetic lethality screening against GBM. Neuro Oncol. (2011) 13:367–75. doi: 10.1093/neuonc/nor012
118. Wu G, Song X, Liu J, Li S, Gao W, Qiu M, et al. Expression of CD44 and the survival in glioma: a meta-analysis. Biosci Rep. (2020) 40:BSR20200520. doi: 10.1042/BSR20200520
119. Yang C, Zheng J, Liu X, Xue Y, He Q, Dong Y, et al. Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via staufen1-mediated mRNA decay. Mol Ther Nucleic Acids. (2020) 20:866–78. doi: 10.1016/j.omtn.2020.05.004
120. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. (2010) 60:166–93. doi: 10.3322/caac.20069
121. Hamasy A, Wang Q, Blomberg KE, Mohammad DK Yu L, Vihinen M, et al. Substitution scanning identifies a novel, catalytically active ibrutinib-resistant BTK cysteine 481 to threonine (C481T) variant. Leukemia. (2017) 31:177–85. doi: 10.1038/leu.2016.153
122. Ghaempoor M, Arefian E, Mohebbi SR, Shanaki M, Kazerouni F, Hatami B. Circulating miR-625 as an Emerging Biomarker for Liver Cirrhosis. Clin Lab. (2019) 65:453–9. doi: 10.7754/Clin.Lab.2018.180704
123. Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. (2017) 11:28–39. doi: 10.1002/1878-0261.12017
124. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
125. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. (2009) 9:265–73. doi: 10.1038/nrc2620
126. Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. (2009) 9:274–84. doi: 10.1038/nrc2622
127. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
128. Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. (2010) 20:R858–61. doi: 10.1016/j.cub.2010.08.052
129. Gerard C, Gonze D, Lemaigre F, Novak B. A model for the epigenetic switch linking inflammation to cell transformation: deterministic and stochastic approaches. PLoS Comput Biol. (2014) 10:e1003455. doi: 10.1371/journal.pcbi.1003455
130. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. (2014) 505:344–52. doi: 10.1038/nature12986
131. Wang Y, Hou J, He D, Sun M, Zhang P, Yu Y, et al. The emerging function and mechanism of ceRNAs in cancer. Trends Genet. (2016) 32:211–24. doi: 10.1016/j.tig.2016.02.001
132. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. (2011) 147:358–69. doi: 10.1016/j.cell.2011.09.028
133. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
134. Marques AC, Tan J, Lee S, Kong L, Heger A, Ponting CP. Evidence for conserved post-transcriptional roles of unitary pseudogenes and for frequent bifunctionality of mRNAs. Genome Biol. (2012) 13:R102. doi: 10.1186/gb-2012-13-11-r102
135. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP, et al. coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. (2010) 465:1033–8. doi: 10.1038/nature09144
136. Zou C, Wan Y, He L, Zheng JH, Mei Y, Shi J, et al. RBM38 in cancer: role and mechanism. Cell Mol Life Sci. (2021) 78:117–28. doi: 10.1007/s00018-020-03593-w
137. Wang X, Chen Y, Dong K, Ma Y, Jin Q, Yin S, et al. Effects of FER1L4-miR-106a-5p/miR-372-5p-E2F1 regulatory axis on drug resistance in liver cancer chemotherapy. Mol Ther Nucleic Acids. (2021) 24:449–61. doi: 10.1016/j.omtn.2021.02.006
138. Luo L, Zhang Z, Qiu N, Ling L, Jia X, Song Y, et al. Disruption of FOXO3a-miRNA feedback inhibition of IGF2/IGF-1R/IRS1 signaling confers Herceptin resistance in HER2-positive breast cancer. Nat Commun. (2021) 12:2699. doi: 10.1038/s41467-021-23052-9
139. Wang H, Wang X, Zhang H, Deng T, Liu R, Liu Y, et al. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene. (2021) 40:4695–708. doi: 10.1038/s41388-021-01898-z
Keywords: miR-625, cancer, proliferation, therapeutic target, ceRNA
Citation: Zhang M, Xiong F, Zhang S, Guo W and He Y (2022) Crucial Roles of miR-625 in Human Cancer. Front. Med. 9:845094. doi: 10.3389/fmed.2022.845094
Received: 29 December 2021; Accepted: 09 February 2022;
Published: 04 March 2022.
Edited by:
Victoria Bunik, Lomonosov Moscow State University, RussiaReviewed by:
Milad Ashrafizadeh, Sabanci University, TurkeyCopyright © 2022 Zhang, Xiong, Zhang, Guo and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhi Guo, ZmNjZ3Vvd3pAenp1LmVkdS5jbg==; Yuting He, ZmNjaGV5dDFAenp1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.