
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CASE REPORT article
Front. Med. , 04 March 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.839389
This article is part of the Research Topic Emerging SARS-COV-2 Variants: Genomic Variations, Transmission, Pathogenesis, Clinical Impact and Interventions View all 65 articles
 Zoraida Fernandez1*†
Zoraida Fernandez1*† Gislene G. C. Lichs2†
Gislene G. C. Lichs2† Claudia S. Zubieta1,3†
Claudia S. Zubieta1,3† Ana B. Machado4†
Ana B. Machado4† Mia A. Ferreira4†
Mia A. Ferreira4† Natalia Valente4†
Natalia Valente4† Thayssa Keren4†
Thayssa Keren4† Ighor Arantes4†
Ighor Arantes4† Valeria Nacife4†
Valeria Nacife4† Elisa Cavalcante Pereira4
Elisa Cavalcante Pereira4 Luciana Appolinario4†
Luciana Appolinario4† Thays E. J. Lacerda4†
Thays E. J. Lacerda4† Marilda M. Siqueira4†
Marilda M. Siqueira4† Ana O. P. Esposito2†
Ana O. P. Esposito2† Luiz H. F. Demarchi2†
Luiz H. F. Demarchi2† Marina C. S. U. Zardin2†
Marina C. S. U. Zardin2† Crhistinne C. M. Goncalves5†
Crhistinne C. M. Goncalves5† Livia M. A. Maziero6†
Livia M. A. Maziero6† Luciana A. F. Miziara7†
Luciana A. F. Miziara7† Felipe G. Naveca8†
Felipe G. Naveca8† Alex Pauvolid-Corrêa4,9†
Alex Pauvolid-Corrêa4,9† Paola C. Resende4†
Paola C. Resende4† Alexsandra R. M. Favacho1†
Alexsandra R. M. Favacho1†The present study investigated a SARS-CoV-2 infection in placenta and fetal samples from an early pregnancy miscarriage in Midwest Brazil. The Gamma variant was isolated and fully sequenced from the placenta sample, but not from fetal samples. Our findings highlight potential adverse perinatal outcomes caused by SARS-CoV-2 Gamma infection during pregnancy.
The emergence of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in December of 2019 in Wuhan, China, and its rapid worldwide spread resulted in a pandemic of coronavirus disease (COVID-19) with severe consequences for global public health (1). COVID-19 may vary from mild to severe and life-threatening conditions, especially in the elderly or patients with comorbidities (2).
Most COVID-19 patients have mild clinical signs and symptoms such as fever, cough, dyspnea, lymphopenia, and systemic effects (3). Pregnant women have an increased risk of developing pneumonia due to inherent physiology and immune response variations, which may lead to adverse obstetric and neonatal outcomes (4). COVID-19 during pregnancy may cause hypertensive disorders with placental inflammation distinct from typical preeclampsia (5). The placenta is usually an effective barrier that prevents fetuses' infection from spreading viruses. However, there is a theoretical risk of vertical transmission in SARS-CoV-2 since the ACE2 receptor is widely expressed in the organ, as reported for SARS (6). The use of antibody tests has provided new evidence that vertical transmission of SARS-CoV-2 can occur, and some studies have demonstrated an increase in perinatal death, mainly related to prematurity. Placenta samples taken from COVID-19 pregnant patients at mid-trimester and swabs and biopsies after spontaneous fetal loss at 19 weeks gestation have also tested positive for SARS-CoV-2 by molecular and immunohistochemical methods (5, 7). Fetal complications include miscarriage (2%), intrauterine growth restriction (10%), and premature birth (39%), and stillbirth, which is likely due to damage to the placenta (3).
Reports of SARS-CoV-2 infection during pregnancy remain limited, and the consequences of fetal infections caused by Variants of Concern (VOC) have been even less assessed. The present study describes virus isolation and whole-genome sequencing of SARS-CoV-2 Gamma variant from a placenta sample of an early pregnancy miscarriage.
On February 22, 2021, a 31-week pregnant woman tested positive for SARS-CoV-2 by real-time RT-PCR performed in a private laboratory of Campo Grande, State of Mato Grosso do Sul (MS), Midwest Brazil. On February 23, she felt unwell and sought medical care and gestational follow-up. During clinical evaluation, the patient presented fever around 38°C, benign vital signs, headache, mild cough, eupneic, blood pressure of 110/70, heart rate of 78, and resting pulse oximetry of 99%. The fetal heart rate was 132 beats per minute, active and reactive. On February 25, a decreased fetal movement was reported during medical ultrasound examination, followed by fetal death. At the time of the miscarriage, the pregnant woman had not yet been vaccinated against SARS-CoV-2.
Fetal nasopharyngeal swabs and placenta samples were sent to the Central Public Health Laboratory of MS (LACEN-MS) for further investigation. The RNA, of both samples, was extracted using Loccus and Extracta Fast Kit (MVXA-P096) and tested by a multiplex real time RT-PCR using the AllplexTM 2019-nCoV Assay (Seegene) and further confirmed by the multiplex EDx kit (Biomanguinhos/Fiocruz) (8). Subsequently, positive samples were submitted to a screening by real-time RT-PCR for detection targeting a deletion in the ORF1b gene (NSP6: S106del, G107del, F108del) found in the VOCs P.1 (Gamma); B.1.1.7 (Alpha) and B.1.351 (Beta) (9). For confirmation and complementary analysis, placenta, and fetuses' samples were sent to the Laboratory of Respiratory Viruses and Measles (LVRS) of Fiocruz, Brazil's National Reference Laboratory, and WHO Reference Laboratory for Coronavirus. At LVRS, positive samples were retested by real-time RT-PCR using the EDx kit (Biomanguinhos), and the CDC protocol (10). Positive samples were submitted to whole-genome sequencing and virus isolation in cell culture.
The viral transport media (VTM) of the nasopharyngeal swab and a small fragment of the placenta were assayed in a biosafety laboratory level 3 (NB3), for viral cytopathic effect (CPE) on Vero E6 cell cultures as previously described (11). The placenta fragment was macerated, resuspended in Dulbecco's Modified Eagle Medium (DMEM), and clarified by centrifugation for inoculation. Samples were inoculated in Vero cells seeded in 25 cm2 flasks and maintained at 37°C with 5% CO2 for 1 h to optimize virus adsorption. Then, maintenance media supplemented by 2% FBS was added, and cell cultures were inspected daily under an inverted microscope for CPE in a total of two 4-day blind passages.
Sequencing was conducted using Illumina protocols previously established and used by the Fiocruz COVID-19 Genomic Surveillance Network to acquire high-quality genomes (P.C. Resende, unpub. data, https://doi.org/10.1101/2020.04.30.069039). The FASTQ reads obtained were imported into the CLC Genomics Workbench version 20.0.4 (QIAGEN), trimmed, and mapped against the reference sequence EPI_ISL_402124 (hCoV-19/Wuhan/WIV04/2019) available in EpiCoV database in the GISAID (https://www.gisaid.org/). The contig was refined using the InDels and Structural Variants module then, the Local Realignment module and the final consensus were obtained. The SARS-CoV-2 lineage characterization was performed by Pango Lineage (12). To characterize phylogenetically both sequences, a maximum likelihood tree was inferred with a dataset composed of various SARS-CoV-2 lineages circulating in Brazil (n = 189). The tree was constructed with IQ-TREE v. 2.1.3 (13), and statistical support of its topology estimated with the approximate likelihood-ratio test (aLRT) (14). The nucleotide and amino acid features were observed by the online tools NextClade (https://clades.nextstrain.org/) and CoVSurver (https://mendel.bii.a-star.edu.sg/METHODS/corona/beta/).
The fetal nasopharyngeal swabs were negative for SARS-CoV-2 by real time RT-PCR, but the placenta sample was SARS-CoV-2 positive by real time RT-PCR (Table 1) and the virus was isolated. CPE caused by SARS-CoV-2 was confirmed by real-time RT-PCR of culture supernatant. In cases where no CPE was observed, real-time PCR was performed four days after inoculation to confirm the absence of virus replication (Figure 1). Whole-genome sequencing confirmed infection by SARS-CoV-2 Gamma lineage confirming the deletions at the ORF1a gene (9). The obtained nucleotide sequences of the placenta fragment (EPI_ISL_2274091) and the cell culture isolate (EPI_ISL_2863796) (Figure 1) were subsequently used alongside a panel of Brazilian sequences of SARS-CoV-2 lineages in the inference of a maximum likelihood tree (Figure 2). Both samples branched among highly supported (aLRT = 100) Gamma lineage branches, corroborating their Pango Lineage classification (12).

Table 1. Real time RT-PCR for SARS-CoV-2 in samples from an early pregnancy miscarriage in Midwest Brazil.
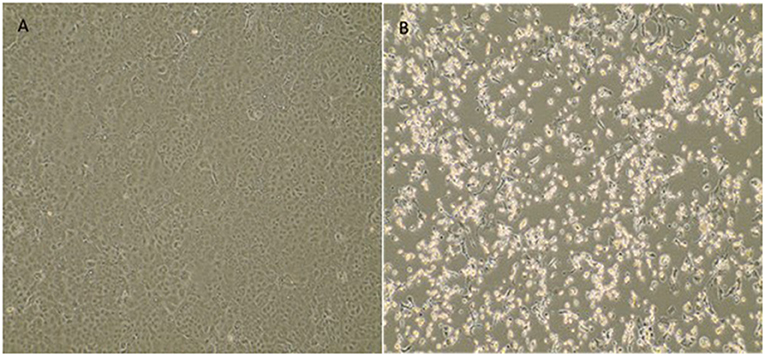
Figure 1. Cytopathic effect of SARS-CoV-2 variant of concern Gamma (P.1) from a placenta sample in Vero E6 cell culture. (A) Vero E6 cell cultures (negative control). (B) Cytopathic effects consisting of rounding and detachment of cells in VERO E6 cultures 3 days after the third passage. EVOS™ XL Core Imaging System—Thermo Fisher Scientific (10×).
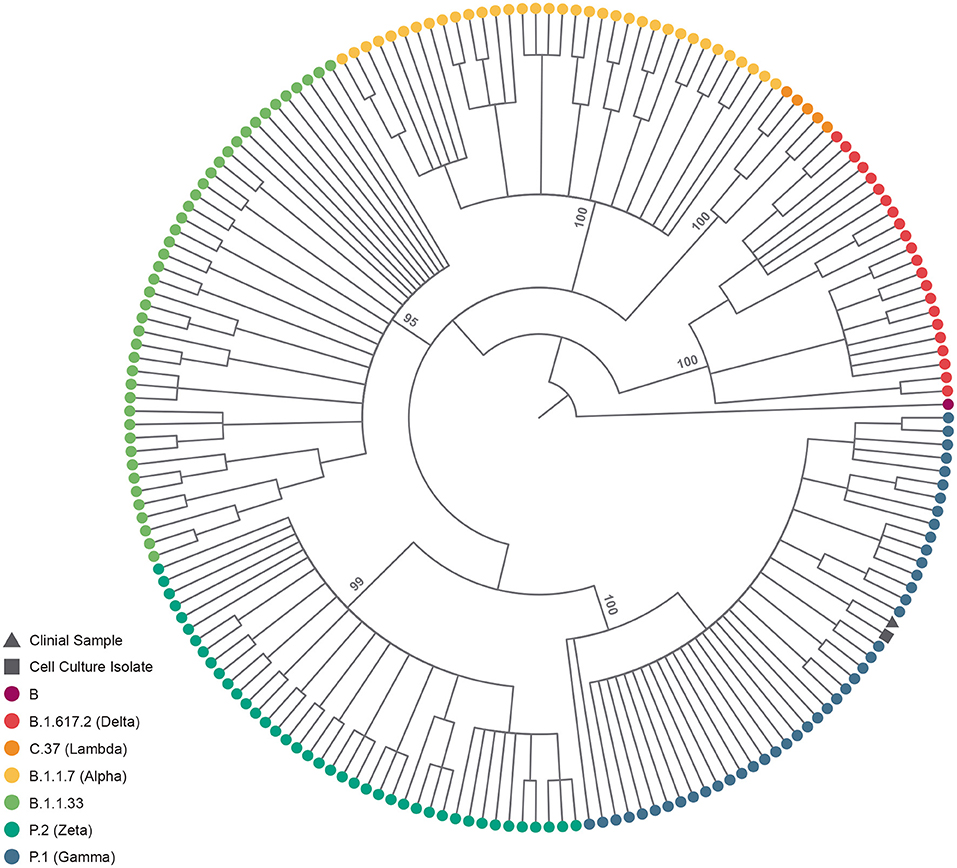
Figure 2. SARS-CoV-2 lineage assignment of placenta sample and its cell culture isolate. Cladogram-transformed maximum likelihood tree (n = 191) of SARS-CoV-2 complete genomes (n = 29,412). The tree encopasses Brazillian sequences of PangoLineage designated B.1.617.2 (n = 22) VOC Delta, C.37 (n = 5) VOI Labmda, B.1.1.7 (n = 35) VOC Aplha, B.1.1.33 (n = 43), P.2 (n = 40) VOI Zeta and P.1 (n = 45) VOC Gamma, identified by their tip colors. The clinical sample (EPI_ISL_2274091) and cell culture isolate (EPI_ISL_2863796) are identified by their tip shapes. Statistical support (approximate likelihood-ratio test, aLRT) of lineage branches is annotated in the tree.
An analysis of a subset of viral sequences from the consortium Fiocruz COVID-19 Genomics Surveillance Network of the Brazilian Ministry of Health (http://www.genomahcov.fiocruz.br/) and the Federal University of state of Mato Grosso do Sul revealed that between March and April 2021, 68 (83%) out of 82 SARS-CoV-2 sequences from MS were VOC Gamma. Additionally, an assessment of the molecular composition of both sequences by the NextClade algorithm revealed that as they bare all synapomorphic signatures of the VOC Gamma, two additional amino-acid substitutions are shared, both in ORF1a, P885L and T2121I.
Here, we report a case of COVID-19 during an early pregnancy miscarriage confirmed by virus isolation and the SARS-CoV-2 whole-genome sequencing of placenta samples. The placenta is an effective maternal-neonatal barrier against the virus even in the presence of a severe infection. However, the placental damage induced by the SARS-CoV-2 may have detrimental effects for the neonate independently of vertical transmission (15).
Despite the increasing number of published studies on COVID-19 infection during pregnancy, other studies are needed to better understand the effect of different variants of the virus on pregnant women and the complications they may develop. Data from clinical studies and systematic review (16–19) shown the effect of SARS-CoV-2 infection in terms of maternal and fetal outcomes. The data have shown that SARS-CoV-2 infection in pregnancy, when compared with non-pregnant women, is asso-ciated with a small increase in risk to the mother, and the most common clinical features are fever, cough, myalgia, and shortness of breath (16, 19). However, there are reports of cases of women who developed severe disease and needed to be admitted to ICU (16). The recommendations for pregnant women with COVID-19 are based on previous experiences with SARS-CoV and MERS-CoV infections. Some of principles of management include early isolation, oxygen therapy, avoidance of fluid overload, antibiotics for secondary bacterial infection risk, fetal and uterine contraction monitoring, and early mechanical ventilation for progressive respiratory failure. To prevent the infection by the virus it is advisable avoid crowed areas, postpone those medical consultations that are not essential and if it is possible consult the doctor virtually. The vaccination against SARS-CoV-2 is of great importance to reduce risks of serious illness, premature birth, and abortions (20).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
This study was approved by the FIOCRUZ-IOC Ethics Committee (68118417.6.0000.5248 and CAAE 32333120.4.0000.5190) and the Ministry of Health of Brazil SISGEN (A1767C3). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
CG was employed by Secretaria de Estado de Saúde (SES).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank all the health care workers and scientists who have worked hard to deal with this pandemic threat, the GISAID team, and all the EpiCoV database's submitters. We also appreciate the support of the Fiocruz COVID-19 Genomic Surveillance Network (http://www.genomahcov.fiocruz.br/) members, the Respiratory Viruses Genomic Surveillance. General Coordination of the Laboratory Network (CGLab), Brazilian Ministry of Health (MoH), Brazilian States Central Laboratories (LACENs) for the partnership in viral surveillance in Brazil and the DECIT/MS.
1. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. (2020) 382:1199–207. doi: 10.1056/nejmoa2001316
2. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Published erratum appears in Lancet;395:1038. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
3. Arthurs AL, Jankovic-Karasoulos T, Roberts CT. COVID-19 in pregnancy: What we know from the first year of the pandemic. Biochim Biophys Acta Mol Basis Dis. (2021) 2021:166248. doi: 10.1016/j.bbadis.2021.166248
4. Jamieson DJ, Honein MA, Rasmussen SA, Williams JL, Swerdlow DL, Biggerstaff MS, et al. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet. (2009) 374:451–8. doi: 10.1016/S0140-6736(09)61304-0
5. Hosier H, Hosier H, Farhadian SF, Morotti RA, Deshmukh U, Lu-Culligan A, et al. SARS-CoV-2 infection of the placenta. J Clin Invest. (2020) 130:4947–53. doi: 10.1172/JCI139569
6. Dashraath P, Wong JLJ, Lim MXK, Lim LM, Li S, Biswas A, et al. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am J Obstet Gynecol. (2020) 222:521–31. doi: 10.1016/j.ajog..03.021
7. Whittaker E, Bamford A, Kenny J, Kaforou M, Jones CE, Shah P, et al. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. (2020) 324:259–69. doi: 10.1001/jama.2020.10369
8. Vogels CBF, Breban MI, Ott IM, Alpert T, Petrone ME, Watkins AE, et al. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. (2021) 19:e3001236. doi: 10.1371/journal.pbio.3001236
9. Naveca FG, Nascimento V, de Souza VC, Corado AL, Nascimento F, Silva G, et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat Med. (2021) 27:1230–8. doi: 10.1038/s41591-021-01378-7
10. Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. (2020) 26:1654–65. doi: 10.3201/eid2608.201246
11. Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Severe acute respiratory syndrome coronavirus 2 from a patient with coronavirus disease, United States. Emerg Infec Dis. (2020) 26:1266–73. doi: 10.3201/eid2606.200516
12. Rambaut A, Holmes EC, O'toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. (2020) 5:1403–7. doi: 10.1038/s41564-020-0770-5
13. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Haeseler AV, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. (2020) 37:1530–4. doi: 10.1093/molbev/msaa015
14. Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. (2006) 55:539–52. doi: 10.1080/10635150600755453
15. Cribiù FM, Erra R, Pugni L, Rubio-Perez C, Alonso L, Simonetti S, et al. Severe SARS-CoV-2 placenta infection can impact neonatal outcome in the absence of vertical transmission. J Clin Invest. (2021) 131:e145427. doi: 10.1172/JCI145427
16. Bellos I, Pandita A, Panza R. Maternal and perinatal outcomes in pregnant women infected by SARS-CoV-2: a meta-analysis. Eur J Obstet Gynecol Reprod Biol. (2020) 256:194–204. doi: 10.1016/j.ejogrb.2020.11.038
17. Juan J, Gil MM, Rong Z, Zhang Y, Yang H, Poon LC. Effect of coronavirus disease 2019 (COVID-19) on maternal, perinatal and neonatal outcomes: a systematic review. Ultrasound Obstet Gynecol. (2020) 56:15–27. doi: 10.1002/uog.22088
18. Do Amaral WN, de Moraes CL, Rodrigues APS, Noll M, Arruda JT, Mendonça CR. Maternal coronavírus infections and neonates born to mothers with SARS-CoV-2: a systematic review. Healthcare. (2020) 8:511. doi: 10.3390/healthcare8040511
19. Parums DV. Editorial: Maternal SARS-CoV-2 infection and pregnancy outcomes from current global study data. Med Sci Monit. (2021) 27:e933831. doi: 10.12659/MSM.933831
Keywords: SARS-CoV-2, gamma variant, viral isolation, Brazil, placenta, fetal death
Citation: Fernandez Z, Lichs GGC, Zubieta CS, Machado AB, Ferreira MA, Valente N, Keren T, Arantes I, Nacife V, Pereira EC, Appolinario L, Lacerda TEJ, Siqueira MM, Esposito AOP, Demarchi LHF, Zardin MCSU, Goncalves CCM, Maziero LMA, Miziara LAF, Naveca FG, Pauvolid-Corrêa A, Resende PC and Favacho ARM (2022) Case Report: SARS-CoV-2 Gamma Isolation From Placenta of a Miscarriage in Midwest, Brazil. Front. Med. 9:839389. doi: 10.3389/fmed.2022.839389
Received: 19 December 2021; Accepted: 20 January 2022;
Published: 04 March 2022.
Edited by:
Pragya Dhruv Yadav, National Institute of Virology (ICMR), IndiaReviewed by:
Kamran Zaman, Indian Council of Medical Research (ICMR), IndiaCopyright © 2022 Fernandez, Lichs, Zubieta, Machado, Ferreira, Valente, Keren, Arantes, Nacife, Pereira, Appolinario, Lacerda, Siqueira, Esposito, Demarchi, Zardin, Goncalves, Maziero, Miziara, Naveca, Pauvolid-Corrêa, Resende and Favacho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zoraida Fernandez, emZlcm5hbmRlekBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.