- 1Division of Nephrology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Division of Nutrition, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Gut dysbiosis is defined as disorders of gut microbiota and loss of barrier integrity, which are ubiquitous on pathological conditions and associated with the development of various diseases. Kidney diseases are accompanied with gut dysbiosis and metabolic disorders, which in turn contribute to the pathogenesis and progression of kidney diseases. Microbial alterations trigger production of harmful metabolites such as uremic toxins and a decrease in the number of beneficial ones such as SCFAs, which is the major mechanism of gut dysbiosis on kidney diseases according to current studies. In addition, the activation of immune responses and mitochondrial dysfunction by gut dysbiosis, also lead to the development of kidney diseases. Based on the molecular mechanisms, modification of gut dysbiosis via probiotics, prebiotics and synbiotics is a potential approach to slow kidney disease progression. Fecal microbiota transplantation (FMT) and genetic manipulation of the gut microbiota are also promising choices. However, the clinical use of probiotics in kidney disease is not supported by the current clinical evidence. Further studies are necessary to explore the causal relationships of gut dysbiosis and kidney diseases, the efficiency and safety of therapeutic strategies targeting gut-kidney axis.
Introduction
Kidney disease is a major public health concern worldwide and is associated with the high morbidity and mortality, with the various pathogenic mechanisms related to immune responses, oxidative stress, inflammation, metabolic disturbance and so on (1–5). However, lack of effective strategies for the prevention and management of renal disease is still an important issue urgently needed to be addressed. For example, how to slow the progression of chronic kidney disease (CKD) is tough due to the current therapeutic strategies just aiming at the decrease on proteinuria and the control of elevated blood pressure (6). Thus, novel targets and treatments become the hot spot in mitigating the growing burden of kidney diseases.
Human gastrointestinal (GI) tract contains more than 100 trillion microorganisms, which forms a microbial community with abundance and diversity (7). Bacteria are one of microorganisms and classified on the level of phyla, classes, orders, families, genera, and species. The dominant bacteria on phylum level are Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia. Among these, Firmicutes and Bacteroidetes predominate and represent 90% of the bacteria in gut (8). Gut microbiota composition is highly variable and maintain dynamic balance in the physiologic condition. However, gut microbiota variations tend to be one of the important implications in intestinal and extra-intestinal disorders. These variations are defined as gut dysbiosis, which includes disorders of gut microbiota composition and loss of intestinal barrier integrity (9–11). For examples, pathogenic bacteria were increased, and uremia toxin levels were elevated under the condition of CKD. The disruption of intestinal epithelium integrity enhances harmful gut-derived metabolites into circulation, in turn, aggravating kidney injury (9). Gut dysbiosis have dual identities as not only the contributing factors to kidney disease progression, but also increasing the risk for various kidney diseases especially in individuals with genetic predisposition (12–16). Therefore, understanding the complex relationships between gut dysbiosis and kidney diseases may provide us with potential therapeutic strategies.
Recent technological advances in omics have arisen our knowledge of gut microbiota. The next generation sequencing technologies, including 16S ribosomal RNA (rRNA) or DNA and metagenomics sequencing analysis identified a large number of gut microorganisms and differential species between healthy subjects and diseased individuals (11). In addition, metabolomics sequencing has become the most common technique to explore the alterations of gut metabolites, while single-cell sequencing is widely used to dig out the specific molecular mechanisms (17, 18).
In this review, we aimed to describe the manifestation of gut dysbiosis in kidney diseases according to numerous clinical studies and animal experiments. Furthermore, we discussed the molecular mechanisms of gut dysbiosis contributing to the pathogenesis and progression of kidney diseases. Finally, we focused on the potential therapeutic strategies targeting gut dysbiosis on kidney disease treatments.
Gut Dysbiosis in Kidney Diseases
16S rRNA or DNA sequencing is the common method to evaluate microbial diversity and identify differential microbes in patients compared with healthy control (HC) subjects. Numerous observational clinical studies demonstrated that patients with CKD, in general, exhibited a prominent reduction in abundance and diversity of fecal microbiota (19–23). At the phylum level, the abundance of Actinobacteria and Firmicutes was reduced in the CKD groups compared with the HC group, while that of Verrucomicrobia Fusobacteria and Proteobacteria was increased (19, 24). Moreover, there were some differences among the altered gut microbiota at the genus and species level in individuals with CKD (23). The individual differences caused by genetic and environmental factors might account for the variety and inconsistence of results. Beyond that, the severity and etiology of CKD in clinical stages and pathological classifications also led to the different manifestations of gut dysbiosis in the enrolled-in patients. For patients with CKD of 3–5 stages, Lactobacillus, Clostridium IV, Paraprevotella, Clostridium sensu stricto, Desulfovibrio, and Alloprevotella were abundant in the fecal samples, while Akkermansia and Parasutterella were enriched in those of HC subjects (19, 23). Another study indicated that fecal microbiota had significantly higher abundance of Citrobacter, Coprobacillus and lower abundance of Prevotella spp., Faecalibacterium prausnitzii, Roseburia spp. in patients with CKD of 3b-4 stages compared to HC subjects (20). A group of microbial species were enriched in patients with end-stage renal disease (ESRD), including Eggerthella lenta, Flavonifractor spp, Alistipes spp, Ruminococcus spp and Fusobacterium spp, while several species were depleted, including Prevotella spp, Clostridium spp, Roseburia spp, Faecalibacterium prausnitzii and Eubacterium rectale (21). CKD patients with dietary protein restriction showed an increase in intestinal Escherichia, Shigella, and Klebsiella and a decrease in Blautia (25). When accompanied with idiopathic nephrotic syndrome (INS), patients with CKD held decreased Megamonas, Megasphaera, Akkermansia, Lachnospira, Roseburia and Fusobacterium compared with CKD patients without INS (24). The significant gut microbiota disorders also varied among kidney diseases with distinct pathological patterns. For examples, Escherichia-Shigella and unclassified Defluviitaleaceae were increased in IgA nephropathy (IgAN) compared with HC, whereas Roseburia, unclassified Lachnospiraceae, sensu stricto Clostridium, and Fusobacterium were decreased. Escherichia-Shigella, Peptostreptococcaceae, Streptococcus, and unclassified Enterobacteriaceae were increased in MN, whereas Lachnospira, unclassified Lachnospiraceae, sensu stricto Clostridium, and Veillonella were decreased. When comparing IgAN with MN, Megasphaera and Bilophila were increased, while Megamonas, Veillonella, Klebsiella, and Streptococcus were decreased in IgAN (26). These results described the landscape of intestinal flora in kidney diseases. Identification of differential bacteria is the first step to dig out the potential gut-associated target on treatments of kidney diseases.
Whether the altered gut microbiota is correlated with clinical characteristics of kidney diseases is of much importance, which determines the significance of gut dysbiosis in the diagnosis and treatments of renal diseases. The correlation analysis demonstrated that the butyrate-producing bacteria, such as Faecalibacterium and Prevotella, were negatively related to the serum level of C-reactive protein (CRP) and Cystatin C (CysC) in ESRD patients (27). In addition, Akkermansia was negatively correlated with inflammatory indicators like interleukin-10 production in CKD patients, suggesting Akkermansia might be a novel biomarker of CKD (19, 22). In diabetic nephropathy (DN), the genus Escherichia-Shigella and the genus Prevotella 9 levels in feces might be used to distinguish DN from diabetes mellitus, which may contribute to the early diagnosis of DN (28). Further, Anaerosporobacter might be a harmful factor and Blautia might be a protective factor in DN due to the positive and negative correlation with 24-h urinary protein content, respectively (29). In individuals with IgAN, Prevotella was positively correlated with serum albumin level, while Klebsiella, Citrobacter, and Fusobacterium were negatively correlated. Furthermore, Bilophila was identified positively correlating with crescents in the Oxford classification of IgAN. Similarly, the negative correlation was existed between Escherichia-Shigella and proteinuria in individuals with MN. Bacteroides and Klebsiella exhibited positive correlation with different MN stages. Thus, the alterations of gut microbiota might also serve as implications of clinical and pathological severity of IgAN and MN (26). These observations indicate that gut microbiota can be used as a novel kind of biomarker for kidney diseases in diagnosis and prognosis. However, large-scale prospective clinical studies are needed to further confirm the reliability of setting the microbial biomarkers.
Dialysis, as one of the effective therapeutic strategies for patients with ESRD, has been showed to improve gut dysbiosis. In patients treated with hemodialysis (HD), the numbers of fecal Bifidobacterial and Lactobacillus acidophilus were higher, while Escherichia coli and Enterococcus faecalis numbers were lower than those of patients without HD treatment. Compared with ordinary hemodialysis, combination treatment with hemoperfusion dialysis could further improve gut microbial disorders with higher number of fecal Lactobacillus acidophilus and lower number of Escherichia coli (30).
In addition, individuals with peritoneal dialysis (PD) presented with different changes of gut microbiota compared with patients with HD. Fecal Proteobacteria was decreased in HD patients, while increased in PD patients (31). It also showed that Bacteroidetes were significantly deceased in HD patients compared with pre-dialysis patients. HD was observed reversing the dysbiosis in pre-dialysis patients (32). Further analysis demonstrated that Bacteroides and Phascolarctobacterium were related to cardiovascular mortality in patients with dialysis (32). The number of fecal Dorea, Clostridium, and SMB53 were associated with peritonitis in PD patients, suggesting gut dysbiosis predicts prognosis of patient with PD (32). These findings imply that monitoring gut dysbiosis in individuals with CKD is necessary for providing specific treatment suggestions.
To determine the exact causations between gut dysbiosis and kidney diseases, renal-injured germ-free or antibiotic-treated rodents were transplanted with microbiota from CKD patients or healthy controls. The results showed that CKD patient-derived microbiota induced higher levels of serum uremic toxins and aggravated renal fibrosis as well as oxidative stress more than that from healthy controls (21). The pathogenic role of gut microbiota from CKD patients was confirmed by fecal microbiota transplantation. Moreover, microbiome changes induced by CKD might be exacerbated in the process of renal transplantation with immunosuppression (33). A study on renal transplantation patients indicated that spousal pairs with similar microbial composition had better 6-month allograft function and lower morbidity of post-transplantation infection. Correlation analysis showed that the pre-transplantation microbial similarity in donors and recipients hold a fantastic accuracy in the prediction of the estimated glomerular filtration rate (eGFR) at 6-months post transplantation (21, 34). These data suggest that gut dysbiosis is not only an intestinal manifestation in kidney diseases but also an important factor that leads to disease progression. Thus, it is necessary to explore the molecular mechanisms of gut dysbiosis on aggravating renal diseases and treat patients with kidney diseases by modifying gut microbiota.
Molecular Mechanisms of Gut Dysbiosis on Kidney Diseases
It is obvious that kidney diseases are accompanied by gut dysbiosis, which is a promotive factor involving in the progression of diseases. However, as the manifestations appeared in distal organs, how gut dysbiosis impacts on kidney disease progression becomes an urgent demand. The development of metabolomics sequencing helps us to explore the link underlining molecular basis of gut-kidney crosstalk. Multi-omics combined analysis showed that altered gut microbial species linked to intestinal, circulating and renal metabolites, including uremic toxins, short-chain fatty acids and trimethylamine (TMA) (21, 35–39).
Uremic Toxins
Excessive production of uremic toxins is a consequence of gut microbiota alteration, including indoxyl sulfate (IS) and p-cresyl sulfate (PCS) (11). Mice with depleted gut microbiota were transplanted with microbiota from CKD patients, finally detected higher levels of serum uremic toxins, suggesting that altered gut microbiota might aggravate CKD progression by increasing the production of uremic toxins. Moreover, two of the species, Eggerthella lenta and Fusobacterium nucleatum, were identified to increase uremic toxins productions and deteriorate CKD progression (21). Protein post-translational modifications in bacteria have been detected and characterized associated with nitrogen metabolism (40, 41). Based on the fact, Lior Lobel et al. found that microbial tryptophanase activity was posttranslationally modified by a high sulfur amino acid-containing diet, which reduced the activity of uremic toxin production and ameliorated CKD progression in mice (35). However, a study showed that serum uremic toxins levels were upregulated in patients with different stages of CKD, while remained the same in fecal and urine samples (36). Therefore, an increase in these toxins cannot be entirely explained by increase in bacterial generation in gut. The decrease of fractional clearance due to the renal function decline in CKD patients appeared to increase uremic toxins levels in serum. Meanwhile, the disruption of intestinal epithelial barriers was also one of the important causes to the influx of uremic toxins into circulation (36). Nevertheless, daily medications for CKD patients also interferes the metabolism of these uremic toxins. For examples, two-week canagliflozin treatment for adenine-induced renal failure in mice did not prevent the impaired renal function. However, it significantly reduced the plasma levels of IS and PCS, meanwhile, reversed gut dysbiosis (42). These data suggest that increased production of uremic toxins caused by gut dysbiosis might contribute to CKD progression despite the causes for the increase of these toxins are various.
Short Chain Fatty Acids
SCFAs are generated from indigestible dietary carbohydrate fibers by the intestinal microbiota, mainly including acetate, propionate and butyrate (43). Depletion of gut microbiota using antibiotics in diabetic rats markedly reduced serum acetate levels. As expected, fecal microbiota transplantation from the healthy donor also effectively decreased serum acetate levels. These animal experiments indicate tight correlations between gut dysbiosis and SCFA level alterations. Furthermore, SCFAs were showed to play a critical role in slowing CKD progression. Butyrate was almost three times lower in serum of CKD patients than that of HC. A negative correlation was observed between butyrate level and renal function, which was improved by supplementation with extra butyrate (44). Our previous study confirmed that oral administration of the probiotics Lactobacillus casei Zhang (Lac.z) increased SCFA levels in serum and kidney, which prevented acute kidney injury (AKI) and delayed renal fibrosis progression (45). Thus, regulation of gut dysbiosis, manifested as inducing an increase in SCFA-producing bacteria, may lead to higher levels of SCFAs in circulation and kidney, further reducing tubular cell injury and tubulointerstitial fibrosis.
TMA and TMAO
Nutrient precursors, abundant in red meat and a Western diet, such as phospatidylcholine choline and carnitine, were used by gut microbiota as a carbon fuel source to generate TMA as a waste product, which was subsequently absorbed into circulation and converted within the liver to trimethylamine N-oxide (TMAO) (46, 47). Dietary supplementation of TMAO significantly aggravated the impairment of renal function and the progression of renal fibrosis, reflecting as the decline of eGFR, and the increase of CysC, albumin/creatinine and renal tubulointerstitial fibrosis score (37). Transplantation microbiota from CKD patients to rats were confirmed to accelerate CKD progression by increasing TMAO production (44). The specific medication like iodomethylcholine, a gut microbiota-dependent choline TMA-lyase mechanism-based inhibitor, reversed choline and adenine diet-induced gut microbial community composition related to TMA and markedly suppressed TMA generation, subsequently TMAO level, thus alleviating renal functional impairment (37, 48). In addition, ranitidine and finasteride also inhibited the synthesis and metabolism of TMAO to exhibit potential protective effects on CKD and cardiovascular complications (49). These data show that TMA and TMAO production is dependent on gut microbiota and become a potential target to attenuate renal disease progression.
A rigorous study was performed that patients with CKD were highly selected with strict inclusion criteria to eliminate the influence of the confounding factors on the microbial composition changes. The results showed that gut Lactobacillaceae family displayed a stepwise change in relative richness responding to renal insufficiency from mild, moderate to severe group. In addition, the uremic toxin pathway was associated with microbial changes in CKD patients (50). Despite that, the question remains to be answered whether these metabolite alterations are the results of gut dysbiosis and trigger kidney disease development or they just come with the occurrence and progression of diseases. More designed studies, larger-size samples and multidimensional analysis are needed to clarify the causal or concomitant relationships among gut dysbiosis, metabolite changes and kidney disease progression.
Gut is abundant in what called gut-associated lymphoid tissue, suggesting gut dysbiosis and disease-related metabolite alterations may impact on kidney diseases by activating immune responses. TLR7-dependent translocation of Lactobacillus reuteri was confirmed to induce the increase in plasmacytoid dendritic cells (pDCs) and interferon signaling, which worsened autoimmune responses in systemic lupus erythematosus (SLE). While dietary resistant starch suppressed the abundance and translocation of Lactobacillus reuteri, resulting in a decrease in pDCs, which were benefit to patients with SLE (51). It also mitigated inflammation via expansion of Tregs to slow CKD progression (52). However, another study showed that treatment with a mixture of five Lactobacillus strains skewed the Treg-Th17 balance toward a Treg-dominant phenotype in the kidney of MRL/lpr mice, suggesting that gut microbiota protected against lupus nephritis by expanding Tregs (53). Although the percentage of Tregs was not different between CKD and control mice, pro-inflammatory/resident macrophages increased in the colon of CKD mice. Moreover, probiotics could restore the high percentage of pathogenic macrophages and increased regulatory dendritic cells in the colon. These immune-related changes suppressed systemic inflammation and kidney fibrosis (54). Depletion of gut microbiota with broad-spectrum antibiotics in mice induced lower levels of F4/80 and chemokine receptors CX3CR1 and CCR2 in renal resident macrophages and bone marrow monocytes compared with control mice. In addition, the migratory capacity of monocytes was decreased in gut bacteria-depleted mice. These inflammatory changes with gut microbiota depletion protected mice against renal ischemia reperfusion injury (55). Our previous study also confirmed that the reno-protective role of the probiotics Lac.z was dependent on the decrease in renal macrophages, including inflammatory subset, intermediate-stage subset, and proliferating subset. Further, we used CCR2-KO mice to confirm that the protection was partially dependent on inhibition of CCR2+ macrophages. In addition, Lac.z-induced increase in SCFA levels contributed to the reno-protective role by interaction with the classical SCFA-related receptors, GPR43 and GPR109a expressed on neutrophils, and macrophages, which resulted in the anti-inflammatory effect (45). In mice with crescentic glomerulonephritis, gut-derived Th17 cells were confirmed to migrate into kidney via S1PR1-dependent CCL20/CCR6 axis and trigger autoimmune responses in kidney. Depletion of intestinal Th17 cells in germ-free and antibiotic-treated mice ameliorated autoimmune-mediated renal disease, whereas expansion of these cells on condition of Citrobacter rodentium infection exacerbated pathology (56, 57). Therefore, gut dysbiosis may deteriorate kidney diseases by interacting with immune cells.
The metabolic disturbances in CKD patients, such as oxidative stress and inflammation, promote vascular damage, which is potentially linked to the mitochondrial dysfunction in the enterocytes. Many factors have been reported to control mitochondrial function such as peroxisome proliferator activated receptor gamma coactivator 1 alpha (PGC-1α), which increases the mitochondrial electron transport chain and DNA copy numbers (58). The disturbances in the biogenesis, bioenergetics, morphology and degradation of mitochondria were reported along with PGC-1α expression reduction in CKD (59, 60). Bacterial infections were developed in parallel with mitochondrial dysfunction, which hints that the relationship of gut dysbiosis and mitochondrial dysfunction (61, 62). Gut dysbiosis-induced bacterial metabolites like hydrogen sulfide, uremic toxins, bile acids and lipopolysaccharide interfered with mitochondrial dysfunction by increasing ROS production. Thus, the gut microbiota-mitochondria axis is also important for CKD treatment (63). However, the profound mechanisms are needed to be further explored. In summary, gut dysbiosis affects kidney diseases in the manner of metabolite alterations, immune response activation and mitochondrial dysfunction, which is shown in Figure 1.
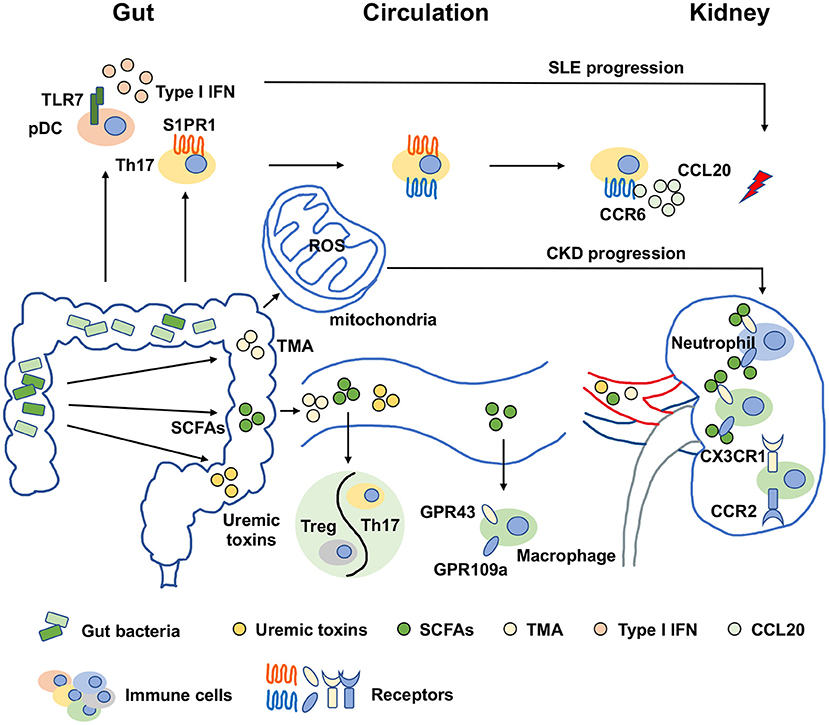
Figure 1. Molecular mechanisms of gut dysbiosis on kidney diseases. The molecular mechanisms of gut dysbiosis on kidney diseases were major focused on two aspects. One was that gut dysbiosis-induced metabolic disorders, manifested as an increase in harmful metabolites such as TMA and uremic toxins and a decrease in beneficial metabolites such as SCFAs, which might directly promote the pathogenesis and progression of kidney diseases. The other was related to immune response activation. Gut dysbiosis and metabolic disorders could expand or activate immune cells by binding specific receptors. For examples, SCFAs regulated macrophages in kidney in a GPR43 and GPR109a-dependent manners, and the activation of pDCs were dependent on TLR7 to induce IFN production on the condition of gut dysbiosis. In addition, another novel perspective has arisen recently that gut dysbiosis-induced the increase of ROS production resulted in mitochondrial dysfunction, which was also important for CKD progression. SCFAs, short chain fatty acids; TMA, trimethylamine; pDC, plasmacytoid dendritic cell; Treg, regulatory T cell; Th17, type 17 of T helper cell; IFN, interferon; ROS, reactive oxygen species; TLR7, toll-like receptor 7; S1PR1, sphingosine-1-phosphate receptor 1; CCR6, chemokine receptor 6; CCL20, C-C chemokine ligand 20; CX3CR1, CX3C chemokine receptor 1; CCR2, C-C chemokine receptor 2; GPR43, G-protein coupled receptor 43; GPR109a, G-protein coupled receptor 109a; SLE, systemic lupus erythematosus; CKD, chronic kidney disease.
Therapeutic Strategies Targeting Gut Dysbiosis on Kidney Diseases
Based on the data from 16S rRNA or DNA sequencing and metagenomic sequencing, microbial dysbiosis can be corrected by exogenous probiotics supplementation to prevent kidney diseases. Among that, Lactobacillales and Bifidobacteri are the most common probiotics. In patients with stable CKD stage 3a, compared to placebo group, probiotics treatment increased fecal Lactobacillales and Bifidobacteria concentrations, induced significant improvements of CRP, iron status, intact parathyroid hormone (iPTH) and β2-microglobulin with possible beneficial effects on cardiovascular outcomes, particularly when the early treatment was performed (64). We found that Lac.z intervention on individuals with CKD in stage 3–5 resulted in a lower serum CysC. Supplementation with Lactobacillus restored the abundance of SCFA-producing bacteria, leading to increased SCFA levels in gut, circulation, and kidney (45, 65). Another group gavaged the CKD rats with Bifidobacterium animalis A6 and found that the abundances of two toxin-driving species, E. lenta and Fusobacterium spp were significantly decreased even though the overall microbial composition was not significantly altered. Meanwhile, serum levels uraemic toxins, creatinine and urea were decreased and renal fibrosis and glomerulosclerosis were reduced (21). In hyperuricemic animals, probiotics containing uricolytic bacteria lowed serum uric acid with benefit on blood pressure and renal disease (66). In obese-induced kidney injury, Lactobacillus paracasei HII01 supplementation alleviated kidney inflammation, endoplasmic reticulum stress, and apoptosis, resulting in improved kidney function. In addition, the probiotics also induced the attenuation of hyperlipidemia, systemic inflammation, and insulin resistance, along with gut dysbiosis improvement (67). Moreover, a mixture of five Lactobacillus strains (Lactobacillus oris, Lactobacillus rhamnosus, Lactobacillus reuteri, Lactobacillus johnsonii, and Lactobacillus gasseri) contributed to decreased IL-6 and increased IL-10 production with an anti-inflammatory environment in the gut and circulation. Moreover, Lactobacillus treatment increased IL-10 and decreased immune deposit in the kidney of MRL/lpr mice (53). However, the safety and dosage of probiotics need to be strictly controlled due to the liveness of probiotics. Despite that the efficiency of probiotics administration on various renal diseases was confirmed by numerous animal experiments, the placebo-controlled, two-blind, and randomized clinical trials are in serious lack, and current studies show few direct efficiencies such as preventing eGFR decline or serum creatinine increase and urea, which are the stumbling block of applications of probiotics (68). The studies on the effects of probiotics on CKD were listed at Table 1.
Prebiotics is the organic matter that promote the metabolism and proliferation of beneficial bacteria in host. Synbiotics is the combination of probiotics and prebiotics. Prebiotics diets, such as butyrate-releasing high-amylose maize starch diet, were proved to be highly effective for protection against kidney injury (74). Beneficial polyphenols and fructooligosaccharide, as the essential prebiotics in our regular diet, inhibited pathogenic bacteria, and improved inflammation, thus preventing CKD progression (75, 76). In a mouse kidney ischemia/reperfusion model, D-serine, one of the prebiotics, was identified to mitigate AKI via suppressing hypoxia-induced tubular damage and promoting posthypoxic tubular cell proliferation. Meanwhile, D-serine levels in circulation was significantly correlated with renal function decrease in AKI patients (77). Butyrate prevented proteinuria by preserving podocytes of adriamycin nephropathy model in a GPR109a-dependent manner (74). In addition, dietary fiber protected against diabetic nephropathy by SCFAs-mediated activation of GPR43 and GPR109a (78). Synbiotics diets containing prebiotics (glutamine, dietary fiber, and oligosaccharide) and probiotics (Bifidobacterium longum strain) preserved renal function decline and lowed serum IS. Moreover, this kind of synbiotics was proved without serious adverse effects (79). Bupleurum polysaccharides ameliorated diabetic nephropathy induced by STZ (80). Prebiotic–gum acacia (GA) treatment successfully reversed CKD-induced gut dysbiosis and increased butyrate production (81, 82).
In summary, probiotics, prebiotics and synbiotics can be widely applicated to treat kidney diseases by improving gut dysbiosis and metabolic disorders. The species-related selection of these treatments should be dependent on variety of diseases and individuals. Large numbers of rigorous clinical trials are needed to further confirm the effects and safety and optimize the methods and durations of treatment. In addition, bacteria are essentially living organisms and regulated by genes. An interesting research was reported to introduce the targeted genetic manipulation of Bacteroides species in the human gut, which provides a brand-new perspective for gut microbiota interventions to prevent kidney diseases (83).
Different from probiotics treatment to supply several specific bacteria, FMT consists of fecal infusion from healthy donor. So far, the recurrent Clostridium difficile infection has been the only condition on which FMT is accurately efficient with more than 80% of efficiency rate (84). However, no clear evidence supporting a FMT approach in CKD. In CKD mice, FMT decreased PCS accumulation in circulation and improved glucose tolerance, but there was no alteration in renal function (85). FMT and probiotic treatments both targeting on microbiome reconstitution, partly. Suez J et al. found that post antibiotic perturbation, FMT arose a rapid and near-complete recovery within days after administration, while probiotics induced a delayed and persistently incomplete one due to the imperfection of colonization (86). The term and accompanied treatments, especially antibio-therapies, impact on the FMT application. Further studies, especially double-blind, placebo-controlled, are needed to explore the FMT efficiency and adverse events, donor selection and feces management, the timing course and feces amount to ensure that FMT can be a promising option in CKD treatment.
Conclusions
In this review, we described that gut dysbiosis existed in various kidney diseases, generally manifesting as imbalance between beneficial bacteria and pathogenic bacteria. In many conditions, gut dysbiosis was also accompanied by metabolic disorders, which were the important mediators in gut-kidney axis. Metabolite alterations not only directly impact on kidney disease progression but also indirectly regulate immune responses. Gut-derived immune cells might also migrate to kidney where they served as pathogenic factors. The regulation of gut dysbiosis by probiotics, prebiotics, synbiotics is confirmed to be beneficial to kidney diseases. While the value of FMT remained to be verified.
Nevertheless, there are still several questions to be solved. Firstly, the exact causal relationship between gut dysbiosis and kidney diseases are needed to be confirmed to consider the specific dysbiosis as the target to treat diseases. Secondly, the deeper understanding of the mechanisms of the crosstalk between gut and kidney is needed to explore. Finally, the further clinical trials are needed to confirm the efficiency and safety of probiotics, prebiotics, synbiotics and FMT.
Author Contributions
CC wrote the manuscript. YY supervised the work and provided financial support. RZ designed the work and provided financial support. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grants 81770684, 81974087, 81770681, and 81974086).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Luyckx VA, Cherney DZI, Bello AK. Preventing CKD in developed countries. Kidney Int Rep. (2020) 5:263–77. doi: 10.1016/j.ekir.2019.12.003
2. Tecklenborg J, Clayton D, Siebert S, Coley SM. The role of the immune system in kidney disease. Clin Exp Immunol. (2018) 192:142–50. doi: 10.1111/cei.13119
3. Hickey FB, Martin F. Role of the immune system in diabetic kidney disease. Curr Diab Rep. (2018) 18:20. doi: 10.1007/s11892-018-0984-6
4. Tomas-Simo P, D'Marco L, Romero-Parra M, Tormos-Muñoz MC, Sáez G, Torregrosa I, et al. Oxidative stress in non-dialysis-dependent chronic kidney disease patients. Int J Environ Res Public Health. (2021) 18:7806. doi: 10.3390/ijerph18157806
5. Nigam SK, Bush KT. Uraemic syndrome of chronic kidney disease: altered remote sensing and signalling. Nat Rev Nephrol. (2019) 15:301–16. doi: 10.1038/s41581-019-0111-1
6. Knauf F, Brewer JR, Flavell RA. Immunity, microbiota and kidney disease. Nat Rev Nephrol. (2019) 15:263–74. doi: 10.1038/s41581-019-0118-7
7. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
8. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7:14. doi: 10.3390/microorganisms7010014
9. Ikee R, Sasaki N, Yasuda T, Fukazawa S. Chronic kidney disease, gut dysbiosis, and constipation: a burdensome triplet. Microorganisms. (2020) 8:1862. doi: 10.3390/microorganisms8121862
10. Richards EM, Pepine CJ, Raizada MK, Kim S. The gut, its microbiome, and hypertension. Curr Hypertens Rep. (2017) 19:36. doi: 10.1007/s11906-017-0734-1
11. Chen YY, Chen DQ, Chen L, Liu JR, Vaziri ND, Guo Y, et al. Microbiome-metabolome reveals the contribution of gut-kidney axis on kidney disease. J Transl Med. (2019) 17:5. doi: 10.1186/s12967-018-1756-4
12. Chaves LD, McSkimming DI, Bryniarski MA, Honan AM, Abyad S, Thomas SA, et al. Chronic kidney disease, uremic milieu, and its effects on gut bacterial microbiota dysbiosis. Am J Physiol Renal Physiol. (2018) 315:F487–502. doi: 10.1152/ajprenal.00092.2018
13. Nishiyama K, Aono K, Fujimoto Y, Kuwamura M, Okada T, Tokumoto H, et al. Chronic kidney disease after 5/6 nephrectomy disturbs the intestinal microbiota and alters intestinal motility. J Cell Physiol. (2019) 234:6667–78. doi: 10.1002/jcp.27408
14. Kim SM, Song IH. The clinical impact of gut microbiota in chronic kidney disease. Korean J Intern Med. (2020) 35:1305–16. doi: 10.3904/kjim.2020.411
15. Anders HJ. 2019 Update in basic kidney research: microbiota in chronic kidney disease, controlling autoimmunity, kidney inflammation and modelling the glomerular filtration barrier. Nephrol Dial Transplant. (2020) 35:4–9. doi: 10.1093/ndt/gfz219
16. Coppo R. The gut-kidney axis in IgA nephropathy: role of microbiota and diet on genetic predisposition. Pediatr Nephrol. (2018) 33:53–61. doi: 10.1007/s00467-017-3652-1
17. Cresci GA, Bawden E. Gut microbiome: what we do and don't know. Nutr Clin Pract. (2015) 30:734–46. doi: 10.1177/0884533615609899
18. Kanemitsu Y, Mishima E, Maekawa M, Matsumoto Y, Saigusa D, Yamaguchi H, et al. Comprehensive and semi-quantitative analysis of carboxyl-containing metabolites related to gut microbiota on chronic kidney disease using 2-picolylamine isotopic labeling LC-MS/MS. Sci Rep. (2019) 9:19075. doi: 10.1038/s41598-019-55600-1
19. Li FX, Wang MH, Wang JP Li RS, Zhang YQ. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front Cell Infect Microbiol. (2019) 9:206. doi: 10.3389/fcimb.2019.00206
20. Wu IW, Gao SS, Chou HC, Yang HY, Chang LC, Kuo YL, et al. Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics. (2020) 10:5398–411. doi: 10.7150/thno.41725
21. Wang X, Yang S, Li S, Zhao L, Hao YL, Qin JJ, et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut. (2020) 69:2131–42. doi: 10.1136/gutjnl-2019-319766
22. Margiotta E, Miragoli F, Callegari ML, Vettoretti S, Caldiroli L, Meneghini M, et al. Gut microbiota composition and frailty in elderly patients with chronic kidney disease. PLoS ONE. (2020) 15:e0228530. doi: 10.1371/journal.pone.0228530
23. Lun H, Yang W, Zhao S, Jiang M, Xu M, Liu F, et al. Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen. (2019) 8:e00678. doi: 10.1002/mbo3.678
24. Zhang J, Luo D, Lin Z, Zhou WY, Rao JL Li YQ, et al. Dysbiosis of gut microbiota in adult idiopathic membranous nephropathy with nephrotic syndrome. Microb Pathog. (2020) 147:104359. doi: 10.1016/j.micpath.2020.104359
25. Jiang S, Wang B, Sha T, Li X. Changes in the intestinal microbiota in patients with stage 5 chronic kidney disease on a low-protein diet and the effects of human to rat fecal microbiota transplantation. Med Sci Monit. (2020) 26:e921557. doi: 10.12659/MSM.921557
26. Dong R, Bai M, Zhao J, Wang D, Ning X, Sun S, et al. Comparative study of the gut microbiota associated with immunoglobulin a nephropathy and membranous nephropathy. Front Cell Infect Microbiol. (2020) 10:557368. doi: 10.3389/fcimb.2020.557368
27. Jiang S, Xie S, Lv D, Wang P, He H, Zhang T, et al. Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci Rep. (2017) 7:2870. doi: 10.1038/s41598-017-02989-2
28. Tao S, Li L, Li L, Liu Y, Ren Q, Shi M, et al. Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition. Acta Diabetol. (2019) 56:581–92. doi: 10.1007/s00592-019-01316-7
29. Li Y, Su X, Gao Y, Lv CX, Gao ZW, Liu YP, et al. The potential role of the gut microbiota in modulating renal function in experimental diabetic nephropathy murine models established in same environment. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165764. doi: 10.1016/j.bbadis.2020.165764
30. He H, Xie Y. Effect of different hemodialysis methods on microbiota in uremic patients. Biomed Res Int. (2020) 2020:6739762. doi: 10.1155/2020/6739762
31. Simoes-Silva L, Araujo R, Pestana M, Soares-Silva I, Sampaio-Maia B. The microbiome in chronic kidney disease patients undergoing hemodialysis and peritoneal dialysis. Pharmacol Res. (2018) 130:143–51. doi: 10.1016/j.phrs.2018.02.011
32. Luo D, Zhao W, Lin Z, Wu JH, Lin HC Li YJ, et al. The effects of hemodialysis and peritoneal dialysis on the gut microbiota of end-stage renal disease patients, and the relationship between gut microbiota and patient prognoses. Front Cell Infect Microbiol. (2021) 11:579386. doi: 10.3389/fcimb.2021.579386
33. Campbell PM, Humphreys GJ, Summers AM, Konkel JE, Knight CG, Augustine T, et al. Does the microbiome affect the outcome of renal transplantation? Front Cell Infect Microbiol. (2020) 10:558644. doi: 10.3389/fcimb.2020.558644
34. Kim JE, Kim HE, Cho H, Park JI, Kwak MJ, Kim BY, et al. Effect of the similarity of gut microbiota composition between donor and recipient on graft function after living donor kidney transplantation. Sci Rep. (2020) 10:18881. doi: 10.1038/s41598-020-76072-8
35. Lobel L, Cao YG, Fenn K, Glickman JN, Garrett WS. Diet posttranslationally modifies the mouse gut microbial proteome to modulate renal function. Science. (2020) 369:1518–24. doi: 10.1126/science.abb3763
36. Gryp T, De Paepe K, Vanholder R, Kerckhof FM, Biesen WV, Wiele TV, et al. Gut microbiota generation of protein-bound uremic toxins and related metabolites is not altered at different stages of chronic kidney disease. Kidney Int. (2020) 97:1230–42. doi: 10.1016/j.kint.2020.01.028
37. Gupta N, Buffa JA, Roberts AB, Sangwan N, Skye SM Li L, et al. Targeted inhibition of gut microbial trimethylamine n-oxide production reduces renal tubulointerstitial fibrosis and functional impairment in a murine model of chronic kidney disease. Arterioscler Thromb Vasc Biol. (2020) 40:1239–55. doi: 10.1161/ATVBAHA.120.314139
38. Sun CY, Lin CJ, Pan HC, Lee CC, Lu SC, Hsieh YT, et al. Clinical association between the metabolite of healthy gut microbiota, 3-indolepropionic acid and chronic kidney disease. Clin Nutr. (2019) 38:2945–8. doi: 10.1016/j.clnu.2018.11.029
39. Hu X, Xie Y, Xiao Y, Zeng W, Gong Z, Du J. Longitudinal analysis of fecal microbiome and metabolome during renal fibrotic progression in a unilateral ureteral obstruction animal model. Eur J Pharmacol. (2020) 886:173555. doi: 10.1016/j.ejphar.2020.173555
40. Macek B, Forchhammer K, Hardouin J, Weber-Ban E, Grangeasse C, Mijakovic I. Protein post-translational modifications in bacteria. Nat Rev Microbiol. (2019) 17:651–64. doi: 10.1038/s41579-019-0243-0
41. Pisithkul T, Patel NM, Amador-Noguez D. Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol. (2015) 24:29–37. doi: 10.1016/j.mib.2014.12.006
42. Mishima E, Fukuda S, Kanemitsu Y, Saigusa D, Mukawa C, Asaji K, et al. Canagliflozin reduces plasma uremic toxins and alters the intestinal microbiota composition in a chronic kidney disease mouse model. Am J Physiol Renal Physiol. (2018) 315:F824–33. doi: 10.1152/ajprenal.00314.2017
43. Pluznick JL. Gut microbiota in renal physiology: focus on short-chain fatty acids and their receptors. Kidney Int. (2016) 90:1191–8. doi: 10.1016/j.kint.2016.06.033
44. Wang S, Lv D, Jiang S, Jiang JP, Liang M, Hou FF, et al. Quantitative reduction in short-chain fatty acids, especially butyrate, contributes to the progression of chronic kidney disease. Clin Sci. (2019) 133:1857–70. doi: 10.1042/CS20190171
45. Zhu H, Cao C, Wu Z, Zhang H, Sun Z, Wang M, et al. The probiotic L. casei zhang slows the progression of acute and chronic kidney disease. Cell Metab. (2021) 33:2091–3. doi: 10.1016/j.cmet.2021.08.015
46. Tang WH, Wang Z, Kennedy DJ, Wu YP, Buffa JA, Agatisa-Boyle B, et al. Gut microbiota-dependent Trimethylamine N-Oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. (2015) 116:448–55. doi: 10.1161/CIRCRESAHA.116.305360
47. Kim RB, Morse BL, Djurdjev O, Tang M, Muirhead N, Barrett B, et al. Advanced chronic kidney disease populations have elevated Trimethylamine N-Oxide levels associated with increased cardiovascular events. Kidney Int. (2016) 89:1144–52. doi: 10.1016/j.kint.2016.01.014
48. Zhang W, Miikeda A, Zuckerman J, Jia X, Charugundla S, Zhou Z, et al. Inhibition of microbiota-dependent TMAO production attenuates chronic kidney disease in mice. Sci Rep. (2021) 11:518. doi: 10.1038/s41598-020-80063-0
49. Liu J, Lai L, Lin J, Zheng JJ, Nie XL, Zhu XY, et al. Ranitidine and finasteride inhibit the synthesis and release of Trimethylamine N-Oxide and mitigates its cardiovascular and renal damage through modulating gut microbiota. Int J Biol Sci. (2020) 16:790–802. doi: 10.7150/ijbs.40934
50. Yacoub R, Nadkarni GN, McSkimming DI, Chaves LD, Abyad S, Bryniarski MA, et al. Fecal Microbiota Analysis of Polycystic Kidney Disease Patients According to Renal Function: A Pilot Study. Exp Biol Med. (2019) 244:505–13. doi: 10.1177/1535370218818175
51. Zegarra-Ruiz DF, El Beidaq A, Iniguez AJ, Ricco MLD, Vieira SM, Ruff WE, et al. A diet-sensitive commensal lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. (2019) 25:113–27 e6. doi: 10.1016/j.chom.2018.11.009
52. Snelson M, Kellow NJ, Coughlan MT. Modulation of the gut microbiota by resistant starch as a treatment of chronic kidney diseases: evidence of efficacy and mechanistic insights. Adv Nutr. (2019) 10:303–20. doi: 10.1093/advances/nmy068
53. Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. (2017) 5:73. doi: 10.1186/s40168-017-0300-8
54. Yang J, Lim SY, Ko YS, Lee HY, Oh SW, Kim MG, et al. Intestinal barrier disruption and dysregulated mucosal immunity contribute to kidney fibrosis in chronic kidney disease. Nephrol Dial Transplant. (2019) 34:419–28. doi: 10.1093/ndt/gfy172
55. Emal D, Rampanelli E, Stroo I, Butter LM, Teske GJ, Claessen N, et al. Depletion of gut microbiota protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. (2017) 28:1450–61. doi: 10.1681/ASN.2016030255
56. Krebs CF, Paust HJ, Krohn S, Koyro T, Brix SR, Riedel JH, et al. Autoimmune renal disease is exacerbated by S1P-receptor-1-dependent intestinal Th17 cell migration to the kidney. Immunity. (2016) 45:1078–92. doi: 10.1016/j.immuni.2016.10.020
57. Rahbar Saadat Y, Hejazian M, Bastami M, Hosseinian Khatibi SM, Ardalan M, Zununi Vahed S. The role of microbiota in the pathogenesis of lupus: does it impact lupus nephritis? Pharmacol Res. (2019) 139:191–8. doi: 10.1016/j.phrs.2018.11.023
58. Clark A, Mach N. The crosstalk between the gut microbiota and mitochondria during exercise. Front Physiol. (2017) 8:319. doi: 10.3389/fphys.2017.00319
59. Su Z, Klein JD, Du J, Franch HA, Zhang L, Hassounah F, et al. Chronic kidney disease induces autophagy leading to dysfunction of mitochondria in skeletal muscle. Am J Physiol Renal Physiol. (2017) 312:F1128–40. doi: 10.1152/ajprenal.00600.2016
60. Eirin A, Lerman A, Lerman LO. The emerging role of mitochondrial targeting in kidney disease. Handb Exp Pharmacol. (2017) 240:229–50. doi: 10.1007/164_2016_6
61. Zorov DB, Plotnikov EY, Silachev DN, Zorova LD, Pevzner IB, Zorov SD, et al. Microbiota and mitobiota. Putting an equal sign between mitochondria and bacteria. Biochemistry. (2014) 79:1017–31. doi: 10.1134/S0006297914100046
62. Lobet E, Letesson JJ, Arnould T. Mitochondria: a target for bacteria. Biochem Pharmacol. (2015) 94:173–85. doi: 10.1016/j.bcp.2015.02.007
63. Mafra D, Borges NA, Lindholm B, Stenvinkel P. Mitochondrial dysfunction and gut microbiota imbalance: an intriguing relationship in chronic kidney disease. Mitochondrion. (2019) 47:206–9. doi: 10.1016/j.mito.2018.11.006
64. Simeoni M, Citraro ML, Cerantonio A, Deodato F, Provenzano M, Cianfrone P, et al. An open-label, randomized, placebo-controlled study on the effectiveness of a novel probiotics administration protocol (ProbiotiCKD) in patients with mild renal insufficiency (stage 3a of CKD). Eur J Nutr. (2019) 58:2145–56. doi: 10.1007/s00394-018-1785-z
65. Huang H, Li K, Lee Y., Chen M. Preventive effects of lactobacillus mixture against chronic kidney disease progression through enhancement of beneficial bacteria and downregulation of gut-derived uremic toxins. J Agric Food Chem. (2021) 69:7353–66. doi: 10.1021/acs.jafc.1c01547
66. Garcia-Arroyo FE, Gonzaga G, Munoz-Jimenez I, Blas-Marron MG, Silverio O, Tapia E, et al. Probiotic supplements prevented oxonic acid-induced hyperuricemia and renal damage. PLoS ONE. (2018) 13:e0202901. doi: 10.1371/journal.pone.0202901
67. Wanchai K, Yasom S, Tunapong W, Chunchai T, Eaimworawuthikul S, Thiennimitr P, et al. Probiotic Lactobacillus paracasei HII01 protects rats against obese-insulin resistance-induced kidney injury and impaired renal organic anion transporter 3 function. Clin Sci. (2018) 132:1545–63. doi: 10.1042/CS20180148
68. McFarlane C, Ramos CI, Johnson DW, Campbell KL. Prebiotic, probiotic, and synbiotic supplementation in chronic kidney disease: a systematic review and meta-analysis. J Ren Nutr. (2019) 29:209–20. doi: 10.1053/j.jrn.2018.08.008
69. Andrade-Oliveira V, Amano MT, Correa-Costa M, Castoldi A, Felizardo RJF, Almeida DC. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol. (2015) 26:1877–88. doi: 10.1681/ASN.2014030288
70. Wang IK, Wu YY, Yang YF, Ting IW, Lin CC, Yen TH, et al. The effect of probiotics on serum levels of cytokine and endotoxin in peritoneal dialysis patients: a randomised, double-blind, placebo-controlled trial. Benef Microbes. (2015) 6:423–30. doi: 10.3920/BM2014.0088
71. Rossi M, Johnson DW, Morrison M, Pascoe EM, Coombes JS, Forbes JM, et al. Synbiotics easing renal failure by improving gut microbiology (SYNERGY): a randomized trial. Clin J Am Soc Nephrol. (2016) 11:223–31. doi: 10.2215/CJN.05240515
72. Takayama F, Taki K, Niwa T. Bifidobacterium in gastro-resistant seamless capsule reduces serum levels of indoxyl sulfate in patients on hemodialysis. Am J Kidney Dis. (2003) 41(Suppl. 1):S142–5. doi: 10.1053/ajkd.2003.50104
73. Nakabayashi I, Nakamura M, Kawakami K, Ohta T, Kato I, Uchida K, et al. Effects of synbiotic treatment on serum level of p-cresol in haemodialysis patients: a preliminary study. Nephrol Dial Transplant. (2011) 26:1094–8. doi: 10.1093/ndt/gfq624
74. Felizardo RJF, de Almeida DC, Pereira RL, Watanabe IKM, Doimo NTS, Ribeiro WR, et al. Gut microbial metabolite butyrate protects against proteinuric kidney disease through epigenetic- and GPR109a-mediated mechanisms. FASEB J. (2019) 33:11894–908. doi: 10.1096/fj.201901080R
75. Bao N, Chen F, Dai D. The regulation of host intestinal microbiota by polyphenols in the development and prevention of chronic kidney disease. Front Immunol. (2019) 10:2981. doi: 10.3389/fimmu.2019.02981
76. Ramos CI, Armani RG, Canziani MEF, Dalboni MA, Dolenga CJR, Nakao LS, et al. Effect of prebiotic (fructooligosaccharide) on uremic toxins of chronic kidney disease patients: a randomized controlled trial. Nephrol Dial Transplant. (2019) 34:1876–84. doi: 10.1093/ndt/gfy171
77. Nakade Y, Iwata Y, Furuichi K, Mita M, Hamase K, Konno R, et al. Gut microbiota-derived D-serine protects against acute kidney injury. JCI Insight. (2018) 3:e97957. doi: 10.1172/jci.insight.97957
78. Li YJ, Chen X, Kwan TK, Loh YW, Singer J, Liu YZ, et al. Dietary fiber protects against diabetic nephropathy through short-chain fatty acid-mediated activation of G Protein-Coupled Receptors GPR43 and GPR109A. J Am Soc Nephrol. (2020) 31:1267–81. doi: 10.1681/ASN.2019101029
79. Iwashita Y, Ohya M, Yashiro M, Sonou T, Kawakami K, Nakashima Y, et al. Dietary changes involving Bifidobacterium longum and other nutrients delays chronic kidney disease progression. Am J Nephrol. (2018) 47:325–32. doi: 10.1159/000488947
80. Feng Y., Weng H, Ling L, Zeng T, Zhang YY, Chen DF, et al. Modulating the gut microbiota and inflammation is involved in the effect of bupleurum polysaccharides against diabetic nephropathy in mice. Int J Biol Macromol. (2019) 132:1001–11. doi: 10.1016/j.ijbiomac.2019.03.242
81. Lakshmanan AP, Al Za'abi M, Ali BH, Terranegra A. The influence of the prebiotic gum acacia on the intestinal microbiome composition in rats with experimental chronic kidney disease. Biomed Pharmacother. (2021) 133:110992. doi: 10.1016/j.biopha.2020.110992
82. Hung TV, Suzuki T. Dietary fermentable fibers attenuate chronic kidney disease in mice by protecting the intestinal barrier. J Nutr. (2018) 148:552–61. doi: 10.1093/jn/nxy008
83. Bencivenga-Barry NA, Lim B, Herrera CM, Trent MS, Goodman AL. Genetic manipulation of wild human gut bacteroides. J Bacteriol. (2020) 202:e00544–19. doi: 10.1128/JB.00544-19
84. Caggiano G, Cosola C, Di Leo V, Gesualdo M, Gesualdo L. Microbiome modulation to correct uremic toxins and to preserve kidney functions. Curr Opin Nephrol Hypertens. (2020) 29:49–56. doi: 10.1097/MNH.0000000000000565
85. Barba C, Soulage CO, Caggiano G, Glorieux G, Fouque D, Koppe L. Effects of fecal microbiota transplantation on composition in mice with CKD. Toxins. (2020) 12:741. doi: 10.3390/toxins12120741
Keywords: gut microbiota, chronic kidney disease, metabolic disorders, probiotics, FMT
Citation: Cao C, Zhu H, Yao Y and Zeng R (2022) Gut Dysbiosis and Kidney Diseases. Front. Med. 9:829349. doi: 10.3389/fmed.2022.829349
Received: 10 December 2021; Accepted: 09 February 2022;
Published: 03 March 2022.
Edited by:
Chenyang Wang, Nanjing University, ChinaReviewed by:
Laetitia Koppe, Université Claude Bernard Lyon 1, FranceMoshe Levi, Georgetown University, United States
Copyright © 2022 Cao, Zhu, Yao and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Yao, eWFveWluZ2trQDEyNi5jb20=; Rui Zeng, emVuZ3IxMjZAMTI2LmNvbQ==
 Chujin Cao
Chujin Cao Han Zhu
Han Zhu Ying Yao
Ying Yao Rui Zeng
Rui Zeng