
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 29 April 2022
Sec. Gastroenterology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.827297
This article is part of the Research Topic The Emerging Role of Liquid Biopsy in Gastrointestinal, Pancreatic and Liver Cancers View all 6 articles
 Razvan Iacob1,2,3†
Razvan Iacob1,2,3† Matei Mandea1†
Matei Mandea1† Speranta Iacob1,2,3
Speranta Iacob1,2,3 Catalina Pietrosanu1,4
Catalina Pietrosanu1,4 Doru Paul5
Doru Paul5 Razvan Hainarosie1,4*
Razvan Hainarosie1,4* Cristian Gheorghe1,2,3
Cristian Gheorghe1,2,3Squamous cell carcinomas of the esophagus (ESCC) and of the head and neck (HNSCC) are two neoplasms that share common risk factors and have the same embryological origin, but a very different prognosis, the 5-year survival of HNSCC being almost double (40–50%) compared to the 5-year survival of ESCC (20%). Current guidelines emphasize the importance of screening for ESCC in patients diagnosed with head and neck cancers. A liquid biopsy is a novel tool for diagnosis, prognostic stratification, and personalized therapy. Liquid biopsy biomarkers for these two malignancies could help both their early detection, facilitate residual disease identification, and provide prognosis information. The present systematic review of the literature was aimed at describing the liquid biopsy biomarkers present in these two malignancies, with an emphasis on potential clinical applications.
Squamous cell carcinoma of the esophagus (ESCC) and of the head and neck are common neoplastic pathologies being responsible of over 500,000 deaths per year, with an increased incidence in the past few years. The head and neck squamous cell carcinoma (HNSCC) can arise from tissues of the oral cavity, oropharynx, hypopharynx, larynx, and nasopharynx. According to the GLOBOCAN 2020, the incidence of HNSCC has increased worldwide in the past decade, with more than 800,000 new cases every year, with cancer of the oral cavity and lips being the most frequent (1). The most important risk factors are smoking, human papillomavirus (HPV) infection, and alcohol abuse, with an increased number of cases in males (2–4). The diagnosis is often made in a late stage due to suboptimal conventional biomarkers for early-stage disease. 40% of patients are diagnosed when lymph nodes metastasis is present and the outcome is poor (2, 3). Currently, the diagnosis consists of imaging methods associated with tissue biopsy (3). Despite recent advances in locoregional therapies, up to 60% of HNSCC tumors will have a locoregional recurrence and additional 20% will develop distant metastasis, leading to treatment failure (5). The tumor HPV assessed by p16 expression is currently the most widely used biomarker in HNSCC (6). Overall, HNSCC has a 40–50% 5-year survival.
The ESCC is often diagnosed in a late stage, with a <20% survival at 5 years from diagnosis. The incidence has decreased in the past decade due to smoking cessation, but there are still over 600,000 new cases diagnosed every year (1). ESCC ranks as the sixth cause of cancer-related mortality worldwide, with an age-standardized rate/100,000 (ASR) of 5.6. Currently, screening for ESCC is recommended also for patients with a personal or familial history of head and neck cancers and is based on upper gastrointestinal endoscopy. The use of carbohydrate antigen 19-9 (CA 19-9) and carcinoembryonic antigen (CEA) do not have good sensitivity in detecting early cases of ESCC and are not recommended as routine screening tests in clinical practice (7–9).
Although HNSCC and ESCC share common risk factors, such as tobacco usage, alcohol abuse, male sex, and other risk factors are specific for each type, such as HPV infection and poor oral hygiene for HNSCC and hot food and beverages and changes in the oral microbiome for ESCC, respectively (4, 6, 8, 10).
Several studies have documented that the presence of HNSCC and ESCC in the same patient leads to decreased longevity and increased morbidity, especially when surgical resection is conducted for both the malignancies (11, 12). Currently, high-resolution chromoendoscopy or virtual chromoendoscopy using NBI or I-SCAN techniques is the gold standard for early diagnosis of ESCC and endoscopic resection of early ESCC is the best therapeutic option (13). As radiation therapy conducted in HNSCC might induce radiation esophagitis with or without strictures, alternative methods of molecular diagnosis that could indicate the potential coexistence of ESCC are clearly useful. Therefore, the identification of markers for early diagnosis and posttreatment disease monitoring are of paramount importance for both the malignancies, as early diagnosis is the cornerstone of curative therapies. Considering that the two anatomical sites share a common embryological origin and common risk factors, one could expect to find common molecular markers that could be used for the early detection and prognosis of both the malignancies. At the same time, tumor-specific molecular markers would further refine our ability to stratify patients both for the diagnostic and prognostic purposes and for treatment that is different in these two conditions.
Liquid biopsy has recently emerged as a new non-invasive technique of detecting blood circulating biomarkers. There are multiple applications of liquid biopsy currently investigated both for ESCC and HNSCC (3, 14, 15), for example, detection of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and exosomes-based biomarkers (16, 17).
Circulating tumor cells represent cancer cells derived from the primary site of neoplasia that are shed in the blood stream or in the lymphatics and are responsible for metastatic progression of the disease (16, 17). They can be used as diagnostic or prognosis markers, being useful for estimating the tumor dynamics. The detection of CTCs is based on markers expression on the surface and in the cytoplasm of the CTCs (17).
Cell-free DNA is released in the bloodstream during necrosis and apoptosis of cells. The tumoral source of cfDNA is represented by ctDNA and it is present along cfDNA originating from normal cells, as a fraction of the total circulating free DNA. Valuable tumor information like somatic mutations or methylation profiles could be detected through specific methods such as next-generation sequencing (NGS) and are important for early disease detection, detection of residual disease following therapy, personalized therapeutic tailoring, and prognostic estimation (15, 16, 18).
Exosomes are stable extracellular vesicles of small size (40–150 nm), released by cells in various body fluids, containing proteins, microRNAs (miRNA), messenger RNA (mRNA), DNA, and long noncoding RNA (lncRNA) (16). miRNAs and lncRNAs are included in the category of regulatory noncoding RNAs. miRNA consists of 20–24 nucleotides, with important roles in cell differentiation, proliferation, apoptosis as well as in tumor suppression or promotion, by regulating the expression of multiple target genes (16, 19, 20). LncRNAs are RNAs consisting of over 200 nucleotides that do not code proteins and are released by tumoral cells also via exosomes (20).
The scope of the present review was to systematically perform a literature search to identify biomarkers of liquid biopsy significant for diagnosis and prognosis of ESCC and HNSCC, respectively, with a special focus on biomarkers shared by the two cancers. Pubmed and CrossRef were used to search for original articles that were published between 2001 and 2021 using HNSCC and ESCC medical subject heading (MeSH) terms combined with terms liquid biopsy, CTCs, cfDNA, circulating-tumoral DNA, exosomes, and miRNA. The systematic literature search was conducted in March to August 2021. The literature was initially screened for types of liquid biopsy used in both the HNSCC and ESCC, further investigating whether markers are common or different depending on the type of cancer investigated.
Two excellent recent publications, a systematic review and a meta-analysis have addressed the utility of CTCs in diagnostic and prognosis of HNSCC (17, 21). Most representative papers on this subject consist of nine prospective cohort studies and one retrospective cohort study (5, 22–30). The studies are heterogeneous in terms of disease staging, including either patients diagnosed in stages I–IV or only advanced disease cases (stages III–IV). Similarly, significant heterogeneity was noted about the timing of the study, before, during, or after treatment, or about the types of treatment (surgery, radiotherapy, or chemotherapy).
Several techniques were used for CTC isolation, such as negative enrichment immunofluorescence in-situ hybridization (Ne imFISH), the use of CellSearch or Clearcell FX platforms, by staining with immunofluorescent antibodies to specific cytokeratins, or by detection of Epidermal Growth Factor Receptor (EGFR) transcripts (23, 25, 26, 29, 31). The most frequently used definition of positive CTCs in HNSCC was by positivity for epithelial cell adhesion molecule (EpCAM) that is absent in normal blood mononuclear cells and negative CD45 staining that is specific to blood mononuclear cells (24, 32). The CellSearch system (Menarini Silicon Biosystems, Bologna, Italy) is, to date, the single Food and Drug Administration (FDA)-approved CTC detection and quantification platform. It performs immunomagnetic enrichment using ferrofluids with EpCAM antibodies coupled to an analyzer that captures images of isolated cells after staining with specific fluorescent antibodies. After immunomagnetic enrichment, recovered cells are permeabilized and stained with a nuclear stain (DAPI), and a fluorescent antibody conjugate against CD45 (leukocyte marker) and cytokeratins 8, 18, and 19 (epithelial markers). To be considered a CTC, a cell must be EpCAM positive, stain positive for nuclear DAPI, have a positive cytoplasmic appearance for cytokeratins and a negative leucocytic CD45 staining, having a diameter larger than 5 μm. The detection of more than 3–5 positive cells/7.5 ml of blood defines a positive sample and the detection limit is of 1 CTC/7.5 ml Ethylenediaminetetraacetic acid (EDTA)-blood (22, 31–34). The use of leucocytic marker CD45 for negative selection combined with immunofluorescence staining methods that use antibodies against N-cadherin, CD133, and multi-cytokeratin was also employed for the detection of CTCs in HNSCC (30).
We have shown in Table 1 the most significant studies published on CTCs in HNSCC and ESCC, using the CellSearch platform. In both the malignancies, the level of CTCs detected correlated with poor prognosis, shorter disease-free survival, or reduced response to treatment, the data being more robust for ESCC (35, 37, 39, 41).
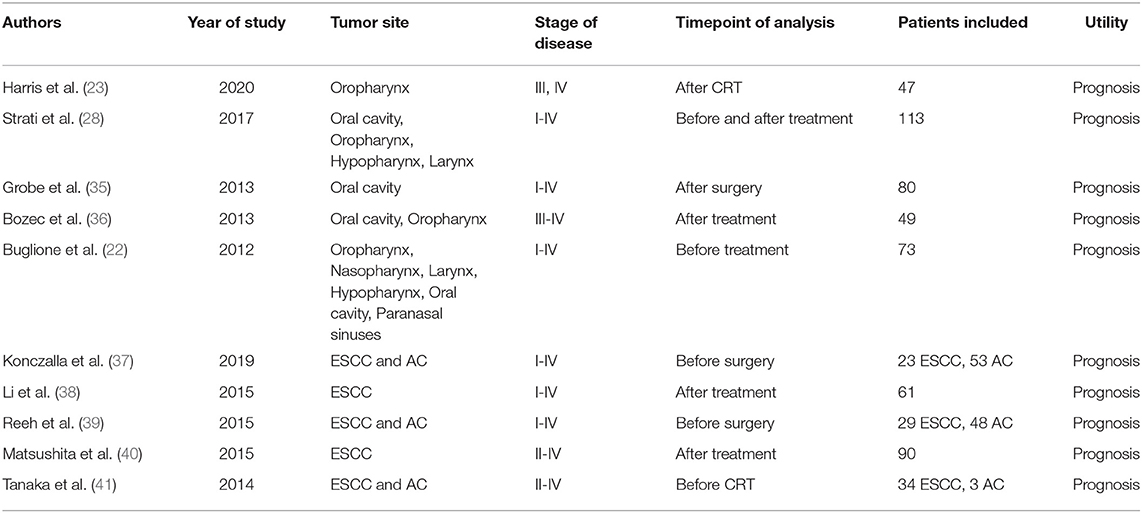
Table 1. Studies on circulating tumor cells in head and neck squamous cell carcinoma (HNSCC) and esophagus squamous cell carcinoma (ESCC) using CellSearch platform.
In several studies, expression of the programmed death ligand 1 (PD-L1) immunotherapy target in CTCs was investigated, in order to identify possible responders to pembrolizumab therapy and prognostic estimation (26, 28). A study from Strati et al. has quantified PD-L1 expression on CTCs in peripheral blood samples of patients with HNSCC, using direct imaging through the CellSearch system, showing that the positivity to PD-L1 is associated with poor prognosis (28).
The microfluidic platform system (CTC-Chip) and the isolation by size of epithelial tumor cells filtration methods (Isolation by Size of Tumor cells - ISET) use the size of the CTC specific to each tumor type. The use of dimension detecting techniques can help identify both the circulating tumor micro-emboli and clusters, which may express the CD45 marker in some of the cells, therefore escaping methods that use CD45 antibodies negative selection (21, 28).
The utility of detecting CTCs was, in most of the studies, for prognostic purpose or personalized therapy, with no current proven benefit for early diagnosis of HNSCC. The most important roles of detecting CTCs are for identifying future targets of treatment, to evaluate treatment response and disease recurrence (21, 32, 34). Further translational applications consist in establishing in vitro disease models based on circulating cancer stem cells that could be used for drug development and personalized therapy. Proposed head and neck cancer stem cell transcription factors and biomarkers are CD44, CD133, ALDH, cMET, Oct-4, Nanog, Sox2 (42, 43). The identification of CTC subpopulations in patients with recurrent HNSCC may have future applications in developing novel antitumoral targeted therapies.
The current approach to therapy in ESCC is dependent on accurate estimation of the tumor staging and is conducted in a multidisciplinary way, involving endoscopist, radiologist, oncologist, and surgeon. An accurate pretherapeutic staging providing a clear picture of the disease spread is critical for optimal therapeutic choice, between endoscopic resection, surgery, radiochemotherapy, or systemic chemotherapy. As extraesophageal tumor recurrence is found in only approximately 50% of cases, one could imagine that micrometastases are frequently missed by current imaging modalities and responsible for frequent distant disease relapse (44). The detection of CTCs can improve our prognostic ability and enhance staging systems in ESCC. Our systematic literature search has identified several prospective cohort studies, published between 2007 and 2019 about the clinical utility of CTCs in ESCC. The studies included patients diagnosed in all stages of the disease, between I and IV as well as at different time points during the disease course, before or after treatment, consisting in surgery, radiotherapy, or chemotherapy (45, 46).
For detection and isolation of CTCs, there were used techniques selecting CTCs based on their antigenic surface profile and immune magnetic procedures or methods that use physical properties of the cells, such as cell size, specific density (gradient centrifugation enrichment), cellular electric charge properties (dielectrophoretic procedures), and the deformation ability (chip-based microfluidic CTC enrichment methods). Dedicated CTC isolation platforms such as CellSearch system and CanPatrol have also been used (40, 46–48). Usually, for detection of CTCs similar markers were used as in HNSCC, consisting in CD45 negativity, and positivity for DAPI, CEP8, and cytokeratins (CK18, CK19, and CK8) and EpCAM (38, 40, 45, 46). The use of CanPatrol system by Chen et al. further classified CTCs in epithelial, mesenchymal, and hybrid, using expression of markers such as EpCAM, CK, vimentin, and Twist by fluorescent probes. The authors found that mesenchymal CTC count is directly correlated with clinical stage and response to chemotherapy (47).
Several studies have investigated the expression of survivin in CTCs. Survivin is a protein in the family of the inhibitors of apoptosis and is frequently expressed in numerous carcinomas and only rarely within normal cells. It can be detected in many types of samples such as blood, urine, and sputum. Survivin expressing CTCs are associated with invasiveness, high nodal status, and shorter overall survival (48, 49), however, no study concerning survivin expression in HNSCC was identified, so far. Similarly to HNSCC, the majority of the studies indicated that higher detection of CTCs count directly correlates with metastasis, recurrence, shorter disease-free survival, worse response to chemoradiotherapy (CRT), regardless of the expressing marker used for CTC detection (18, 38, 40, 46, 48–50).
The presence of CTCs in the blood is found in both the HNSCC and ESCC and, according to current knowledge, CTCs detection cannot discriminate between the two conditions.
Cell-free DNA from the tumoral origin is called ctDNA and is represented by 140–170 bp DNA fragments that originate from necrotic or apoptotic tumoral cells. It can be used to detect tumor-specific genomic alterations such as somatic mutations and methylation profiles. It has been shown that the detection of specific genomic alterations of driver oncogenes, such as TP53, EGFR, and KRAS in ctDNA, has clinical utility for cancer screening, early diagnosis, and targeted treatment approach, identifying in patients with lung cancer, colorectal cancer, or pancreatic cancer subgroups that would benefit of specific chemotherapy regimens. According to the Catalogue of Somatic Mutations in Cancer (COSMIC) database, the most frequently mutated genes in HNSCC are TP53 (44%), LRP1B (21%), NOTCH1 (20%), FAT1 (19%), and KMT2D (16%). More than 550 somatic variants in TP53 have been described so far in HNSCC, the most frequently reported being R175H, R248Q, R248W, and R273H (51). One of the most important prognostic roles of identifying genomic alterations in cfDNA resides in the prediction of minimal residual disease, which is known as microscopic tumoral cells that are still present after a potentially curative treatment and that are associated with disease recurrence (52, 53).
Several PCR and NGS sequencing methods could be used to detect somatic mutations in cfDNA, such as digital droplet PCR (DDPCR), Competitive allele-specific TaqMan PCR (CAST-PCR), and NGS techniques with novel modifications and adaptations, such as safe-sequencing systems (Safe-SeqS) or Plasma sequencing (Plasma-Seq) (54–56). The various techniques have different limits of detection and could be used to investigate specific genomic alteration or a panel of target sequences. As shown in Table 2, the NGS and specified PCR techniques have been recently used to screen for and validate hotspot mutations in a targeted panel of genes, in HNSCC, by several study groups, including Braig et al. (61) and Wang et al. (63). Wang et al. have investigated genomic alterations in TP53, CDKN2A, HRAS, NRAS, PIK3CA, and HPV16 DNA in plasma or saliva from 93 patients with HNSCC, documenting a potential role for diagnosis of invasive HNSCC for this gene panel (63). Braig et al. identified by NGS that mutations in the RAS family genes are found in a subgroup of patients with HNSCC and correlate with disease progression, also after cetuximab treatment (61).
Mazurek et al. and Perdomo et al. have shown that somatic alterations of either KRAS and EGFR or TP53, NOTCH1, CDKN2A, CAP8, and PTEN could be used for the diagnostic purpose by liquid biopsy in HNSCC (59, 62). Perdomo et al. have proposed that TP53 genomic alterations could be a suitable biomarker for early detection of HNSCC in patients with HPV negative (59).
Aberrant methylation profiles are genomic alteration marking the early events of carcinogenesis and quantitative measure of methylation in specific gene loci has emerged as a novel tool for early diagnosis, prognosis, and disease monitoring (60). Methylation can be determined using different technologies, such as quantitative PCR (qPCR) and methylation sensitive high-resolution melting PCR (MS-HRM PCR) (53, 60). In HNSCC, the most frequently investigated methylated targets were SEPT9, SHOX2, DAPK1, RASSF1A, and CDKN2A genes (53, 58, 60). Gene methylations of SEPT9 and SHOX2 were identified in patients with different types of HNSCC, regardless of tumor stage, and hypermethylation could be used as a valuable biomarker for diagnosis and prognosis. For diagnostic purpose, studies have shown a good specificity, over 90%, but a rather low sensitivity (about 50%) (58, 60). By investigating methylation profiles in cfDNA in patients with nasopharyngeal-squamous cell carcinoma, Tian et al. and Yang et al. have proposed that investigating a panel of markers leads to better results that a single methylation target (65, 66). Methylation of CDKN2A had a sensitivity of 22.5% and a specificity of 97%, methylation of DAPK1, a sensitivity of 51.4%, and a specificity of 90%, whereas the panel consisting of CDKN2A, DLEC1, DAPK1, and UCHL1 was found to have the highest specificity and sensitivity for HNSCC detection (66). In another study, the use of a methylation panel consisting of RASSF1, WIF1, DAPK1, and RARB2 had a sensitivity of 95.8% and a detection rate of early-stage disease of 90%, showing great promise for clinical application. The highest specificity was identified for methylations of RASSF1A gene (65).
Tumor-specific gene alterations detection in ESCC are currently explored as a novel type of biomarker useful for diagnosis and prognosis. In the COSMIC database, there are to date 2,236 samples of ESCC genomic profiles reported, indicating that the most frequently mutated genes are TP53 (58%), KMT2D (18%), NOTCH1 (16%), LRP1B (14%), and FAT1 (14%), similar to HNSCC top 5 genes. The most frequently identified TP53 somatic mutations are R175H, R282W, Y220C, R213*, R248Q, R248W, R273H, and R342* (51). Of these, R175H, R248Q, R248W, and R273H somatic alterations are also the most frequently encountered TP53 alterations in HNSCC (18, 67). A recent systematic review has presented the clinical utility of circulating cfDNA as a novel biomarker for esophageal cancer (67). In total, 59 abstracts and 20 full papers have been reviewed including ESCC and esophageal adenocarcinoma cases. Seven studies have included patients with ESCC.
The most significant studies reporting the utility of cfDNA in ESCC are depicted in Table 3. In a cohort of 137 patients with ESCC, Tian et al. have shown that the use of 5-hydroxymethylcytosine (5hmc)-based biomarkers has clinical utility for diagnosis of ESCC, achieving a sensitivity of 93.75% and a specificity of 85.71% (Area Under Curve = 0.972). It has been shown that 5hmC, the oxidative product of 5-methylcytosine (5 mC), reflects the epigenetic features in patients with cancer, as it is a relatively stable intermediate of active DNA demethylation, being currently regarded as a novel epigenetic hallmark of cancer (73).
In a pilot cohort of 13 patients with ESCC patients prior of resection and neoadjuvant therapy, Ueda et al. have shown that NGS using a panel of 53 cancer-related genes is an effective tool for detecting somatic alterations in plasma cfDNA. The assay was able to detect variant frequencies of 0.12–7.2% and the use of cfDNA analysis in clinical assessments of the tumor burden could help predict tumor recurrence in ESCC (74). The best diagnostic utility was identified for the presence of mutations in the following 4 genes TP53, FAT3, MLL3, and AJUBA. Using these somatic alterations, the authors achieved 78.9% sensitivity, 100% specificity, and 92.3% diagnostic accuracy. TP53 is also a frequently mutated target in HNSCC, but FAT1 and FAT4 rather than FAT3 are more frequently mutated in HNSCC, according to COSMIC.
In the study of Hagi et al., molecular barcode sequencing of the TP53 gene enabled comprehensive and highly sensitive detection of ctDNA in patients with ESCC, prior to resection and neoadjuvant therapy as well as after therapy (68).
Luo et al. have designed a sequencing panel with improved targeting, identifying significantly mutated genes by meta-analysis of 532 ESCC genomes. The proposed gene panel consists of 90 genes, achieving 94 and 75% of sensitivity, respectively when detecting at least 1 or 2 mutant genes in patients with ESCC. Thus, the gene panel proposed by Luo et al. could be useful in ESCC diagnosis and disease monitoring following surgery (75). A comparison of pre- and postoperative plasma ctDNA using deep NGS sequencing has showed that some ctDNA mutations had a lower occurrence frequency or even disappeared postoperatively, providing useful cfDNA targets for disease recurrence.
Meng et al. have shown that cfDNA could potentially be used to monitor disease load in patients with ESCC, as somatic mutations could be detected in cfDNA of patients with stages IIA to IIIB presurgery, but at a lower frequency in cfDNA of the same patients postsurgery. Using an NGS panel of 483 genes they have shown that somatic alteration could be detected in cfDNA in genes involved in traditional cancer-related pathways: PI3K-Akt/mammalian target of rapamycin (mTOR) signaling, genes encoding proteoglycans or focal adhesion molecules, genes involved in FoxO signaling, chemokine signaling pathways, or Janus kinase - signal transducer and activator of transcription protein (JAK-STAT) signaling.
With the primary aim to determine whether detection of ctDNA after CRT is associated with risk of tumor progression Azad et al. have analyzed by NGS 802 hot spots in 607 genes for Single nucleotide polymorphisms (SNPs) previously associated with esophageal adenocarcinoma or squamous cell carcinoma, including hot spots in LRP1B and TP53 frequently identified in HNSCC. The study group comprised of 10 ESCC cases out of 45 esophageal cancers. The authors have shown that the median proportion of tumor-derived DNA out of the total cell-free DNA before treatment was only 0.07%, further documenting the fact that ultrasensitive assays are needed for the assessment of ctDNA from localized esophageal tumors. The detection of ctDNA was associated with tumor progression, metastasis, and disease-specific survival (76).
Due to the lack of studies including patients with both conditions, currently, it is difficult to discriminate between HNSCC and ESCC based only on ctDNA mutation analysis.
MicroRNAs are currently among the most promising biomarkers studied in many types of cancers, including ESCC and HNSCC, with diagnostic and prognostic values. The miRNAs are regulatory molecules found in tumoral cells and in the blood stream, carried by exosomes and function as translation repressors. MiRNAs can have multiple roles, being functional regulators of cell proliferation, interacting with many signaling pathways either as inhibitors or stimulators. Therefore, they can be upregulated or downregulated in various types of tumors (3, 77). MiRNAs can be quantified from formalin-fixed paraffin-embedded (FFPE) tumoral samples as well as from exosome or serum liquid biopsies, by qRT-PCR (78–80). Exosomes could be isolated using ultracentrifugation, filtration, chromatography, magnetic associated cell sorting, fluorescence, colorimetric ELISA assays, and other methods and are excellent miRNA carriers (3). Total exosomal RNA extraction followed by RT-PCR miRNA quantification is currently one of the best liquid biopsy approach for diagnosis and prognosis estimation in patients with cancer (81).
Our systematic literature search has identified 20 miRNAs and one lncRNA (HOTAIR) relevant for HNSCC and ESCC liquid biopsies. These miRNAs have multiple gene targets, in both ESCC and HNSCC.
miR-10 is a promising biomarker for ESCC and HNSCC, targeting T-cell lymphoma invasion and metastasis-inducing protein1 (TIAM1). In ESCC mir-10 down-regulates TIAM1, reducing its expression. Increased TIAM1 expression is currently considered a negative prognostic factor in ESCC, as it is associated with increased histology grade, advanced clinical stages, and lymph node metastasis (82). Increased miR-10 is a good prognostic indicator, being associated with the suppression of tumor growth and metastasis in ESCC via TIAM1 repression. In HNSCC, miR-10 was identified in patients with oral squamous cell carcinoma, having the same target as in ESCC, TIAM1, but also HOXD10, RhoA/RhoC, and KLF4. miR-10 has also diagnostic utility as it achieved 81.2% sensitivity and 80% specificity in the diagnosis of ESCC and 94.4% sensitivity and 80% specificity for HNSCC detection (83, 84). The main miRNA targets with diagnostic utility in HNSCC and ESCC and proposed signaling pathways are presented in Table 4.
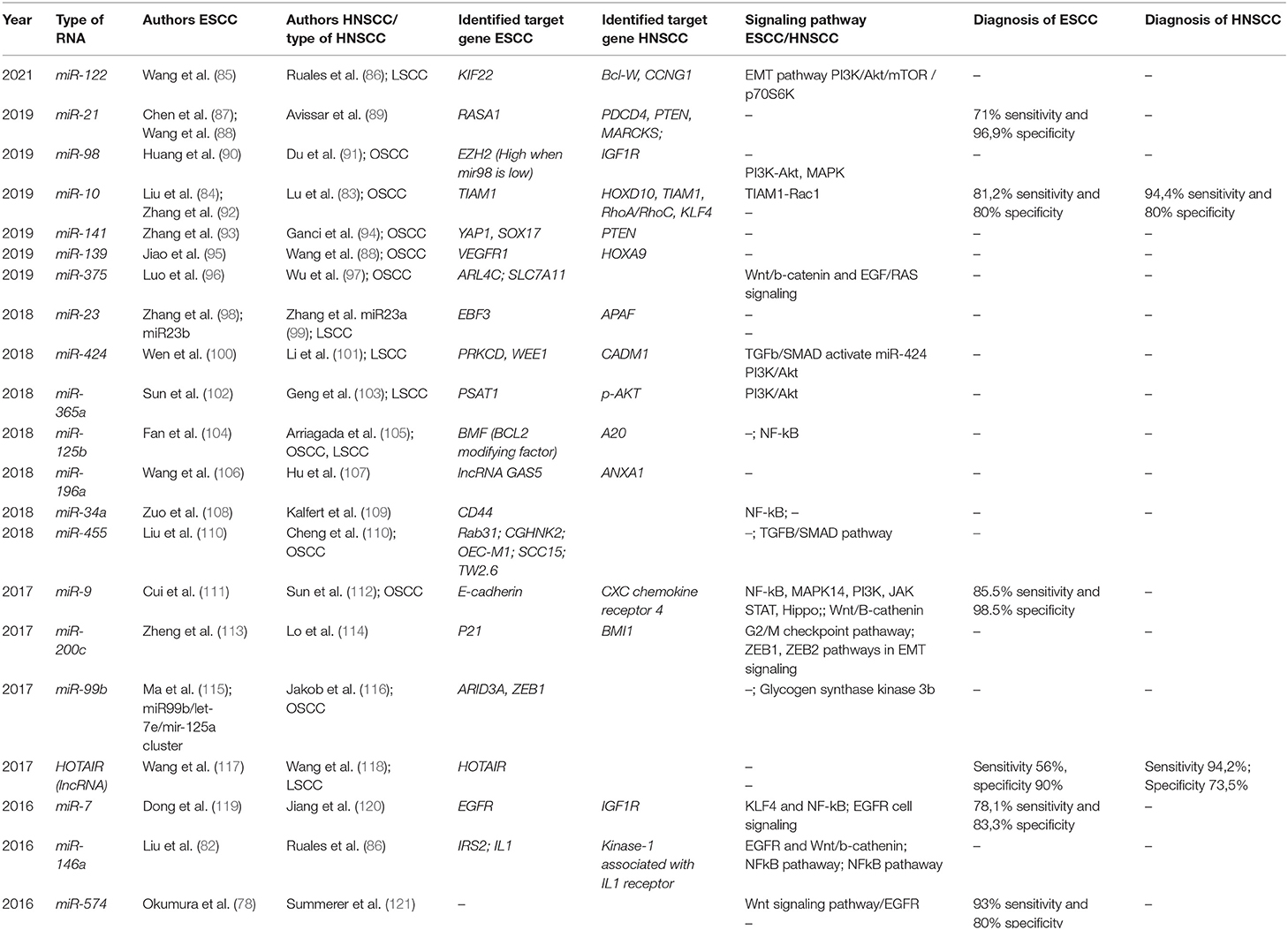
Table 4. MicroRNA and lncRNA liquid biopsy studies in ESCC/HNSCC—targets and signaling pathways, and diagnostic utility.
A significant signaling pathway for ESCC and HNSCC pathogenesis is controlled by the transcription factor NF-kB and was found to be regulated by miR-7, miR-9, miR-146a, and miR-34a in ESCC and by miR-125b and miR146a in HNSCC, respectively. Thus, miR-146a is involved in both malignancies via NF-kB pathway regulation, but it seems to have an opposite behavior, as it is downregulated in ESCC and up-regulated in HNSCC (82, 86, 105, 108, 111, 119).
Furthermore, among the evaluated miRNA targets, miR-9, miR-21, miR-23, miR-141, miR-424, and miR-196a were upregulated in both the cancers, whereas miR-7, miR-98, miR-122, miR-139, mir-200c, and miR375 were downregulated in both the ESCC and HNSCC. These miRNA targets were associated with metastasis, invasion, tumoral cell growth, and poor prognosis in both the tumor types (85–87, 89, 90, 93–95, 98–101, 106, 107, 111–114, 122). Other miRNA targets could have utility for diagnosis or prognostic stratification as they were up-regulated in HNSCC and down-regulated in ESCC: miR-10, miR-365a, miR-125b, miR-146a, miR-34a, miR-455, and miR574. On the other hand, miR-99b was upregulated in ESCC and downregulated in HNSCC (78, 82–84, 86, 102–105, 108–110, 121, 123).
The potential roles of miRNAs in diagnosis, prognosis, and as possible therapeutic targets are given in Table 5. Of particular interest as a possible target for treatment is miR-200c since it was associated in both types of SCC with increased radiosensitivity. It controls G2/M checkpoint in DNA replication via p21 and BMI1 as well as ZEB1 and ZEB2 signaling pathways. miR-200c is downregulated in aggressive SCCs and its increased expression is associated with a better prognosis and a better response to radiotherapy (113, 114).
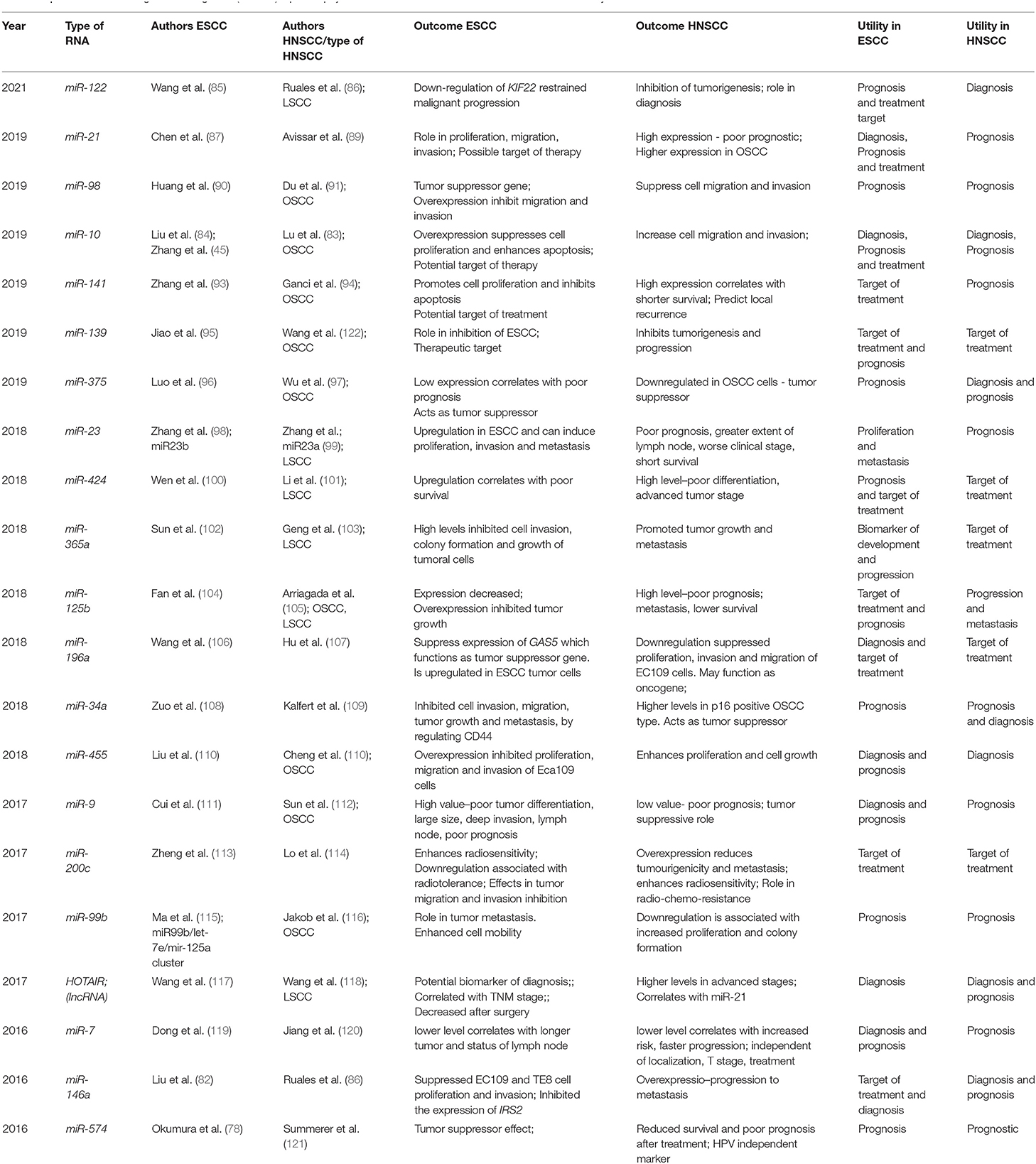
Table 5. MicroRNA and long non-coding RNA (lncRNA) liquid biopsy studies in ESCC/HNSCC—outcomes and clinical utility.
Homebox - HOX transcript antisense RNA (HOTAIR), a lncRNA was identified as a valuable biomarker for both HNSCC and ESCC, having regulatory effects in tumoral cells, by controlling proliferation, invasion, and differentiation. HOTAIR expression was directly correlated to the TNM stage. Its serum expression in patients at risk could be used also as a diagnostic tool, with a sensitivity of 56 and 94.2% and specificity of 90 and 73.5% for ESCC and for HNSCC, respectively (15, 117, 118).
In the future, circulating non-coding RNA expression could be one of the best methods to discriminate between ESCC and HNSCC and robust research studies including patients with both conditions and cohorts in which the two malignancies occur synchronous or metachronous are mandatory.
Liquid biopsy as a field of research could be an important tool in the battle of discovering new biomarkers for early diagnosis, since both the HNSCC and ESCC are currently diagnosed in a late stage of disease, with advanced tumor locoregional invasion, associated with complications, metastases, and poor survival. Currently, in the approved clinical guidelines, there are no biomarkers proposed for early diagnosis in patients that are at risk. There are excellent reviews focused on liquid biopsy applications for individual malignancies. Recently, Mishra et al. have reviewed clinical applications of different liquid biopsy multifunctional biomarkers in head and neck cancers such as: somatic DNA mutations-based markers, methylation profiles, mitochondrial DNA-based biomarkers, exosomes and EVs, transcriptomic, nongenomic based biomarkers as well as CTCs, and viral-based biomarkers (124). Our extensive literature search, however, has identified no study, so far, addressing the topic of common or specific molecular biomarkers validated in patient cohorts with synchronous or metachronous HNSCC and ESCC, or at least studies including patients with both malignancies. This is still an unmet need, although one could imagine significant clinical applications in terms of early diagnosis and prognostic stratification. The present review is the first that has aimed to analyze current knowledge regarding the utility of liquid biopsy in both conditions and to provide potential targets for further research. The review was focused on common markers, as these could be identified based on published literature, in the lack of studies systematically addressing both malignancies with the same panel of molecular markers.
The current applications, strengths, and limitations of liquid biopsies have been recently reviewed by Martins et al. (125). The main strength of liquid biopsies resides in the non-invasive character of the procedures that could be applied repeatedly to capture the molecular dynamics of the tumors. This is a major point with clinical impact considering the changing molecular characteristics of the tumors subjected to multimodal therapies. In comparison to traditional biopsies, liquid biopsies have the potential to better capture the intratumor molecular variability at different time points during follow-up. However, liquid biopsies has not yet fully entered daily practice as they require expensive equipment and trained personnel to conduct tests and interpret the results. There are also technology-specific limitations. CTCs are rarely found in the circulation and their isolation is difficult and costly. For cfDNA pre-analytical protocols lack standardization whereas the cfDNA template is found in specific body fluids at very low concentrations, leading to the low sensitivity of detection. Techniques to identify and isolate RNA species are still under development and one of the main challenges is the instability of the RNA molecule leading to difficulties in the identification of low abundance RNA species. The isolation and characterization of extracellular vesicles show great promise in the field of liquid biopsy. EVs could derive specifically from the tumor cells and are excellent carriers of tumor-related molecular information, but the lack of standardization of preanalytical protocols is still challenging.
The detection and quantification of CTCs have a minor role as a diagnostic tool, since they are usually discovered in the late stages of the disease and are related to extensive tumor spread and metastasis, having mainly a prognostic utility. The use of the FDA-approved Cellsearch platform has a good applicability for both ESCC and HNSCC.
Circulating tumor DNA analysis shows promising perspectives for early diagnosis as studies have already indicated both common and specific somatic alterations but identified somatic variants should be further validated in independent series comprising both malignancies.
The investigation of methylation profiles seems to have the best utility for early diagnosis of both pathological conditions. Two years ago, using a ctDNA plasma methylation platform, the feasibility of cancer detection up to four years before the clinical cancer diagnostic in five common malignancies including esophageal cancer has been demonstrated (126). Almost at the same time, the results of an article that is a potential game changer have been published in the Annals of Oncology (127). The study included 6689 participants (2482 with more than 50 types of cancer and 4207 without cancer. The study used methylation patterns in cfDNA. The specificity of cancer detection was close to 100% and the sensitivity was 67.3% in 12 types of cancer including ESCC and HNSCC. The study did not specify whether any patients with both ESCC and HNSCC were included. Aside from methylation, TP53 somatic alterations could be proposed for screening purposes along with other somatic variants in frequently mutated genes such as KMT2D, NOTCH1, LRP1B, and FAT1. Specific polymorphisms could be integrated in screening panels with applicability for early diagnosis, detection of residual disease following surgery, or treatment response after radio-chemotherapy.
Given the relationship of some head and neck cancers with viral infections, (Epstein-Barr virus (EBV) for nasopharynx cancers and human papilloma virus (HPV) for SCC of the oropharynx), it is likely that in the near future biomarkers related to these infections will be used routinely in the diagnostic and management of viral related cancers. Five years ago, a landmark article published in New England Journal of Medicine demonstrated the feasibility and usefulness of screening plasma EBV ctDNA in a population at high risk for nasopharynx cancer (128). Although less than 1/1000 HPV carriers develop cancer, at least in theory, a similar technique may be used also for early detection of HPV ctDNA in a population at risk, for example HPV carrier men (129).
Possibly the best field of research is translational regulation by noncoding RNAs, with applications for early diagnosis and prognostic stratification of both the patients with HNSCC and ESCC. There are currently multiple miRNA targets identified that behave similarly in both cancers as well as targets with opposite effects, some of them showing promising utility even as therapeutic targets.
However, none of the biomarkers described in this review can be used individually to robustly discriminate between ESCC and HNSCC. Further research is needed to integrate currently available molecular targets in biomarker panels, with clinical applicability for screening purposes and early diagnosis, detection of residual disease, and prognostic estimation. Given the different prognostic of these conditions, the discovery of specific molecular markers for each of these two malignancies remains an unmet need of paramount importance.
RI and MM have conducted the systematic literature search and generated the first draft of the manuscript. SI has updated CTC section of the manuscript and provided clinical background for the ESCC sections. CP has updated the cfDNA section. RH has conceived the general plan of the article, updated the miRNA section, and provided clinical background for the HNSCC sections. DP and CG have supervised the study and provided critical comments and review of the manuscript. All authors edited the manuscript and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. (2020).
2. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. (2016) 91:386–96. doi: 10.1016/j.mayocp.2015.12.017
3. Yang W-Y, Feng L-F, Meng X, Chen R, Xu W-H, Hou J, et al. Liquid biopsy in head and neck squamous cell carcinoma: circulating tumor cells, circulating tumor DNA, and exosomes. Expert Rev Mol Diagn. (2020) 20:1213–27. doi: 10.1080/14737159.2020.1855977
4. McDermott JD, Bowles DW. Epidemiology of head and neck squamous cell carcinomas: impact on staging and prevention strategies. Curr Treat Options Oncol. (2019) 20:43. doi: 10.1007/s11864-019-0650-5
5. Kulasinghe A, Kenny L, Perry C, Thiery J-P, Jovanovic L, Vela I, et al. Impact of label-free technologies in head and neck cancer circulating tumour cells. Oncotarget. (2016) 7:71223–34. doi: 10.18632/oncotarget.12086
6. McMullen C, Chung CH, Hernandez-Prera JC. Evolving role of human papillomavirus as a clinically significant biomarker in head and neck squamous cell carcinoma. Expert Rev Mol Diagn. (2019) 19:63–70. doi: 10.1080/14737159.2019.1559056
7. Ibuki Y, Nishiyama Y, Tsutani Y, Emi M, Hamai Y, Okada M, et al. Circulating microRNA/isomiRs as novel biomarkers of esophageal squamous cell carcinoma. PLoS ONE. (2020) 15:e0231116. doi: 10.1371/journal.pone.0231116
8. Abnet CC, Arnold M, Wei W-Q. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. (2018) 154:360–73. doi: 10.1053/j.gastro.2017.08.023
9. Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. (2015) 149:1700–15. doi: 10.1053/j.gastro.2015.08.054
10. Swiecicki PL, Brennan JR, Mierzwa M, Spector ME, Brenner JC. Head and neck squamous cell carcinoma detection and surveillance: advances of liquid biomarkers. Laryngoscope. (2019) 129:1836–43. doi: 10.1002/lary.27725
11. Lim H, Kim DH, Jung HY, Gong EJ, Na HK, Ahn JY, et al. Clinical significance of early detection of esophageal cancer in patients with head and neck cancer. Gut Liver. (2015) 9:159–65. doi: 10.5009/gnl13401
12. Katada C, Muto M, Nakayama M, Tanabe S, Higuchi K, Sasaki T, et al. Risk of superficial squamous cell carcinoma developing in the head and neck region in patients with esophageal squamous cell carcinoma. Laryngoscope. (2012) 122:1291–6. doi: 10.1002/lary.23249
13. Lecleire S, Antonietti M, Iwanicki-Caron I, Duclos A, Lemoine F, Pessot FL, et al. Lugol chromo-endoscopy versus narrow band imaging for endoscopic screening of esophageal squamous-cell carcinoma in patients with a history of cured esophageal cancer: a feasibility study. Dis esophagus Off J Int Soc Dis Esophagus. (2011) 24:418–22. doi: 10.1111/j.1442-2050.2010.01164.x
14. Wang H-C, Yeh T-J, Chan L-P, Hsu C-M, Cho S-F. Exploration of feasible immune biomarkers for immune checkpoint inhibitors in head and neck squamous cell carcinoma treatment in real world clinical practice. Int J Mol Sci. (2020) 21:7621. doi: 10.3390/ijms21207621
15. Chu L-Y, Peng Y-H, Weng X-F, Xie J-J, Xu Y-W. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J Gastroenterol. (2020) 26:1708–25. doi: 10.3748/wjg.v26.i15.1708
16. Kong L, Birkeland AC. Liquid biopsies in head and neck cancer: current state and future challenges. Cancers. (2021) 13:1874. (2021). doi: 10.3390/cancers13081874
17. Payne K, Pugh M, Brooks J, Batis N, Taylor G, Nankivell P, et al. Circulating tumour cell expression of immune markers as prognostic and therapeutic biomarkers in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Int J Mol Sci. (2020) 21:8229. doi: 10.3390/ijms21218229
18. Su P-J, Wu M-H, Wang H-M, Lee C-L, Huang W-K, Wu C-E, et al. Circulating tumour cells as an independent prognostic factor in patients with advanced oesophageal squamous cell carcinoma undergoing chemoradiotherapy. Sci Rep. (2016) 6:31423. doi: 10.1038/srep31423
19. Feng Q, Zhang H, Yao D, Chen W-D, Wang Y-D. Emerging role of non-coding rnas in esophageal squamous cell carcinoma. Int J Molecular Sci. (2019) 21:258. doi: 10.3390/ijms21010258
20. Ghafouri-Fard S, Gholipour M, Taheri M, Shirvani Farsani Z. MicroRNA profile in the squamous cell carcinoma: prognostic and diagnostic roles. Heliyon. (2020) 6:e05436. doi: 10.1016/j.heliyon.2020.e05436
21. Perumal V, Corica T, Dharmarajan AM, Sun Z, Dhaliwal SS, Dass CR, et al. Circulating tumour cells (CTC), head and neck cancer and radiotherapy; future perspectives. Cancers. (2019) 11:367. doi: 10.3390/cancers11030367
22. Buglione M, Grisanti S, Almici C, Mangoni M, Polli C, Consoli F, et al. Circulating tumour cells in locally advanced head and neck cancer: preliminary report about their possible role in predicting response to non-surgical treatment and survival. Eur J Cancer. (2012) 48:3019–26. doi: 10.1016/j.ejca.2012.05.007
23. Harris EJ, Huang J, Carroll E, Lowe AC, Chau NG, Rabinowits G, et al. Circulating tumor cell analysis in locally advanced and metastatic squamous cell carcinoma of the head and neck. Laryngoscope Investig Otolaryngol. (2020) 5:1063–9. doi: 10.1002/lio2.448
24. Inhestern J, Oertel K, Stemmann V, Schmalenberg H, Dietz A, Rotter N, et al. Prognostic role of circulating tumor cells during induction chemotherapy followed by curative surgery combined with postoperative radiotherapy in patients with locally advanced oral and oropharyngeal squamous cell cancer. PLoS ONE. (2015) 10:e0132901. doi: 10.1371/journal.pone.0132901
25. Jatana KR, Balasubramanian P, Lang JC, Yang L, Jatana CA, White E, et al. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Arch Otolaryngol Head Neck Surg. (2010) 136:1274–9. doi: 10.1001/archoto.2010.223
26. Kulasinghe A, Kapeleris J, Kimberley R, Mattarollo SR, Thompson EW, Thiery J-P, et al. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med. (2018) 7:5910–9. doi: 10.1002/cam4.1832
27. Kulasinghe A, Zhou J, Kenny L, Papautsky I, Punyadeera C. Capture of circulating tumour cell clusters using straight microfluidic chips. Cancers (Basel). (2019) 11:89. doi: 10.3390/cancers11010089
28. Strati A, Koutsodontis G, Papaxoinis G, Angelidis I, Zavridou M, Economopoulou P, et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann Oncol. (2017) 28:1923–33. doi: 10.1093/annonc/mdx206
29. Tinhofer I, Konschak R, Stromberger C, Raguse J-D, Dreyer JH, Jöhrens K, et al. Detection of circulating tumor cells for prediction of recurrence after adjuvant chemoradiation in locally advanced squamous cell carcinoma of the head and neck. Ann Oncol Off J Eur Soc Med Oncol. (2014) 25:2042–7. doi: 10.1093/annonc/mdu271
30. Weller P, Nel I, Hassenkamp P, Gauler T, Schlueter A, Lang S, et al. Detection of circulating tumor cell subpopulations in patients with head and neck squamous cell carcinoma (HNSCC). PLoS ONE. (2014) 9:e113706. doi: 10.1371/journal.pone.0113706
31. Zhou S, Wang L, Zhang W, Liu F, Zhang Y, Jiang B, et al. Circulating tumor cells correlate with prognosis in head and neck squamous cell carcinoma. Technol Cancer Res Treat. (2021) 20:1533033821990037. doi: 10.1177/1533033821990037
32. Wu X-L, Tu Q, Faure G, Gallet P, Kohler C, Bittencourt MDC. Diagnostic and prognostic value of circulating tumor cells in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Sci Rep. (2016) 6:20210. doi: 10.1038/srep20210
33. Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol. (2008) 26:3213–21. doi: 10.1200/JCO.2007.15.8923
34. Economopoulou P, Kotsantis I, Kyrodimos E, Lianidou ES, Psyrri A. Liquid biopsy: an emerging prognostic and predictive tool in Head and Neck Squamous Cell Carcinoma (HNSCC). Focus on Circulating Tumor Cells (CTCs) Oral Oncol. (2017) 74:83–9. doi: 10.1016/j.oraloncology.2017.09.012
35. Gröbe A, Blessmann M, Hanken H, Friedrich RE, Schön G, Wikner J, et al. Prognostic Relevance of Circulating Tumor Cells in Blood and Disseminated Tumor Cells in Bone Marrow of Patients with Squamous Cell Carcinoma of the Oral Cavity. Clin Cancer Res. (2014) 20:425 LP−433. doi: 10.1158/1078-0432.CCR-13-1101
36. Bozec A, Ilie M, Dassonville O, Long E, Poissonnet G, Santini J, et al. Significance of circulating tumor cell detection using the CellSearch system in patients with locally advanced head and neck squamous cell carcinoma. Eur Arch Oto-Rhino-Laryngology. (2013) 270:2745–9. doi: 10.1007/s00405-013-2399-y
37. Konczalla L, Ghadban T, Effenberger KE, Wöstemeier A, Riethdorf S, Uzunoglu FG, et al. Prospective comparison of the prognostic relevance of circulating tumor cells in blood and disseminated tumor cells in bone marrow of a single patient's cohort with esophageal cancer. Ann Surg. (2021) 273:299–305. doi: 10.1097/SLA.0000000000003406
38. Li H, Song P, Zou B, Liu M, Cui K, Zhou P, et al. Circulating tumor cell analyses in patients with esophageal squamous cell carcinoma using epithelial marker-dependent and -independent approaches. Medicine (Baltimore). (2015) 94:e1565. doi: 10.1097/MD.0000000000001565
39. Reeh M, Effenberger KE, Koenig AM, Riethdorf S, Eichstädt D, Vettorazzi E, et al. Circulating tumor cells as a biomarker for preoperative prognostic staging in patients with esophageal cancer. Ann Surg. (2015) 261:1124–30. doi: 10.1097/SLA.0000000000001130
40. Matsushita D, Uenosono Y, Arigami T, Yanagita S, Nishizono Y, Hagihara T, et al. Clinical significance of circulating tumor cells in peripheral blood of patients with esophageal squamous cell carcinoma. Ann Surg Oncol. (2015) 22:3674–80. doi: 10.1245/s10434-015-4392-8
41. Tanaka M, Takeuchi H, Osaki Y, Hiraiwa K, Nakamura R, Oyama T, et al. Prognostic significance of circulating tumor cells in patients with advanced esophageal cancer. Esophagus. (2015) 12:352–9. doi: 10.1007/s10388-014-0482-0
42. Patel SS, Shah KA, Shah MJ, Kothari KC, Rawal RM. Cancer stem cells and stemness markers in oral squamous cell carcinomas. Asian Pac J Cancer Prev. (2014) 15:8549–56. doi: 10.7314/APJCP.2014.15.20.8549
43. Major AG, Pitty LP, Farah CS. Cancer stem cell markers in head and neck squamous cell carcinoma. Stem Cells Int. (2013) 2013:319489. doi: 10.1155/2013/319489
44. Makowiec F, Baier P, Kulemann B, Marjanovic G, Bronsert P, Zirlik K, et al. Improved long-term survival after esophagectomy for esophageal cancer: influence of epidemiologic shift and neoadjuvant therapy. J Gastrointest Surg Off J Soc Surg Aliment Tract. (2013) 17:1193–201. doi: 10.1007/s11605-013-2212-7
45. Zhang Y, Li J, Wang L, Meng P, Zhao J, Han P, et al. Clinical significance of detecting circulating tumor cells in patients with esophageal squamous cell carcinoma by EpCAM-independent enrichment and immunostaining-fluorescence in situ hybridization. Mol Med Rep. (2019) 20:1551–60. doi: 10.3892/mmr.2019.10420
46. Qiao Y, Li J, Shi C, Wang W, Qu X, Xiong M, et al. Prognostic value of circulating tumor cells in the peripheral blood of patients with esophageal squamous cell carcinoma. Onco Targets Ther. (2017) 10:1363–73. doi: 10.2147/OTT.S129004
47. Chen W, Li Y, Yuan D, Peng Y, Qin J. Practical value of identifying circulating tumor cells to evaluate esophageal squamous cell carcinoma staging and treatment efficacy. Thorac Cancer. (2018) 9:956–66. doi: 10.1111/1759-7714.12771
48. Yin X-D, Yuan X, Xue J-J, Wang R, Zhang Z-R, Tong J-D. Clinical significance of carcinoembryonic antigen-, cytokeratin 19-, or survivin-positive circulating tumor cells in the peripheral blood of esophageal squamous cell carcinoma patients treated with radiotherapy. Dis esophagus Off J Int Soc Dis Esophagus. (2012) 25:750–6. doi: 10.1111/j.1442-2050.2012.01326.x
49. Cao M, Yie S-M, Wu S-M, Chen S, Lou B, He X, et al. Detection of survivin-expressing circulating cancer cells in the peripheral blood of patients with esophageal squamous cell carcinoma and its clinical significance. Clin Exp Metastasis. (2009) 26:751–8. doi: 10.1007/s10585-009-9274-7
50. Liu Z, Jiang M, Zhao J, Ju H. Circulating tumor cells in perioperative esophageal cancer patients: quantitative assay system and potential clinical utility. Clin cancer Res an Off J Am Assoc Cancer Res. (2007) 13:2992–7. doi: 10.1158/1078-0432.CCR-06-2072
51. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. (2019) 47:D941–7. doi: 10.1093/nar/gky1015
52. Galot R, Machiels J-PH. Current applications and challenges of circulating tumor DNA (ctDNA) in squamous cell carcinoma of the head and neck (SCCHN). Cancer Treat Rev. (2020) 85:101992. doi: 10.1016/j.ctrv.2020.101992
53. Pall AH, Jakobsen KK, Grønhøj C, von Buchwald C. Circulating tumour DNA alterations as biomarkers for head and neck cancer: a systematic review. Acta Oncol. (2020) 59:845–50. doi: 10.1080/0284186X.2020.1742930
54. Quan P-L, Sauzade M, Brouzes E. dPCR: a technology review. Sensors (Basel). (2018) 18:1271. doi: 10.3390/s18041271
55. Slatko BE, Gardner AF, Ausubel FM. Overview of next-generation sequencing technologies. Curr Protoc Mol Biol. (2018) 122:e59. doi: 10.1002/cpmb.59
56. Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. (2011) 108:9530–5. doi: 10.1073/pnas.1105422108
57. Schmidt H, Kulasinghe A, Allcock RJN, Tan LY, Mokany E, Kenny L, et al. A Pilot study to non-invasively track PIK3CA mutation in head and neck cancer. Diagnostics. (2018) 8:79. doi: 10.3390/diagnostics8040079
58. de Vos L, Gevensleben H, Schröck A, Franzen A, Kristiansen G, Bootz F, et al. Comparison of quantification algorithms for circulating cell-free DNA methylation biomarkers in blood plasma from cancer patients. Clin Epigenetics. (2017) 9:125. doi: 10.1186/s13148-017-0425-4
59. Perdomo S, Avogbe PH, Foll M, Abedi-Ardekani B, Lescher Facciolla V, Anantharaman D, et al. Circulating tumor DNA detection in head and neck cancer: evaluation of two different detection approaches. Oncotarget. (2017) 8:72621–32. doi: 10.18632/oncotarget.20004
60. Schröck A, Leisse A, de Vos L, Gevensleben H, Dröge F, Franzen A, et al. Free-circulating methylated DNA in blood for diagnosis, staging, prognosis, and monitoring of head and neck squamous cell carcinoma patients: an observational prospective cohort study. Clin Chem. (2017) 63:1288–96. doi: 10.1373/clinchem.2016.270207
61. Braig F, Voigtlaender M, Schieferdecker A, Busch C-J, Laban S, Grob T, et al. Liquid biopsy monitoring uncovers acquired RAS-mediated resistance to cetuximab in a substantial proportion of patients with head and neck squamous cell carcinoma. Oncotarget. (2016) 7:42988–95. doi: 10.18632/oncotarget.8943
62. Mazurek AM, Rutkowski T, Fiszer-Kierzkowska A, Małusecka E, Składowski K. Assessment of the total cfDNA and HPV16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. (2016) 54:36–41. doi: 10.1016/j.oraloncology.2015.12.002
63. Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. (2015) 7:293ra104. doi: 10.1126/scitranslmed.aaa8507
64. Mydlarz WK, Hennessey PT, Wang H, Carvalho AL, Califano JA. Serum biomarkers for detection of head and neck squamous cell carcinoma. Head Neck. (2016) 38:9–14. doi: 10.1002/hed.23842
65. Yang X, Dai W, Kwong DL, Szeto CYY, Wong EH, Ng WT, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J cancer. (2015) 136:E127–35. doi: 10.1002/ijc.29192
66. Tian F, Yip SP, Kwong DLW, Lin Z, Yang Z, Wu VWC. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol. (2013) 37:708–13. doi: 10.1016/j.canep.2013.05.012
67. Yuan Z, Wang X, Geng X, Li Y, Mu J, Tan F, et al. Liquid biopsy for esophageal cancer: Is detection of circulating cell-free DNA as a biomarker feasible? Cancer Commun. (2021) 41:3–15. doi: 10.1002/cac2.12118
68. Hagi T, Kurokawa Y, Takahashi T, Saito T, Yamashita K, Tanaka K, et al. Molecular barcode sequencing for highly sensitive detection of circulating tumor DNA in patients with esophageal squamous cell carcinoma. Oncology. (2020) 98:222–9. doi: 10.1159/000504808
69. Tian X, Sun B, Chen C, Gao C, Zhang J, Lu X, et al. Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res. (2018) 28:597–600. doi: 10.1038/s41422-018-0014-x
70. Li X, Zhou F, Jiang C, Wang Y, Lu Y, Yang F, et al. Identification of a DNA methylome profile of esophageal squamous cell carcinoma and potential plasma epigenetic biomarkers for early diagnosis. PLoS ONE. (2014) 9:e103162. doi: 10.1371/journal.pone.0103162
71. Liu J-B, Qiang F-L, Dong J, Cai J, Zhou S-H, Shi M-X, et al. Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol. (2011) 17:4917–21. doi: 10.3748/wjg.v17.i44.4917
72. Hibi K, Taguchi M, Nakayama H, Takase T, Kasai Y, Ito K, et al. Molecular Detection of strong. promoter methylation in the serum of patients with esophageal squamous cell carcinoma. Clin Cancer Res. (2001) 7:3135–8.
73. Vasanthakumar A, Godley LA. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. (2015) 208:167–77. doi: 10.1016/j.cancergen.2015.02.009
74. Ueda M, Iguchi T, Masuda T, Nakahara Y, Hirata H, Uchi R, et al. Somatic mutations in plasma cell-free DNA are diagnostic markers for esophageal squamous cell carcinoma recurrence. Oncotarget. (2016) 7:62280–91. doi: 10.18632/oncotarget.11409
75. Luo H, Li H, Hu Z, Wu H, Liu C, Li Y, et al. Noninvasive diagnosis and monitoring of mutations by deep sequencing of circulating tumor DNA in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. (2016) 471:596–602. doi: 10.1016/j.bbrc.2016.02.011
76. Azad TD, Chaudhuri AA, Fang P, Qiao Y, Esfahani MS, Chabon JJ, et al. Circulating tumor DNA analysis for detection of minimal residual disease after chemoradiotherapy for localized esophageal cancer. Gastroenterology. (2020) 158:494–505.e6. doi: 10.1053/j.gastro.2019.10.039
77. de Carvalho AC, Scapulatempo-Neto C, Maia DCC, Evangelista AF, Morini MA, Carvalho AL, et al. Accuracy of microRNAs as markers for the detection of neck lymph node metastases in patients with head and neck squamous cell carcinoma. BMC Med. (2015) 13:108. doi: 10.1186/s12916-015-0350-3
78. Okumura T, Kojima H, Miwa T, Sekine S, Hashimoto I, Hojo S, et al. The expression of microRNA 574-3p as a predictor of postoperative outcome in patients with esophageal squamous cell carcinoma. World J Surg Oncol. (2016) 14:228. doi: 10.1186/s12957-016-0985-3
79. Hsu PJ, Yan K, Shi H, Izumchenko E, Agrawal N. Molecular biology of oral cavity squamous cell carcinoma. Oral Oncol. (2020) 102:104552. doi: 10.1016/j.oraloncology.2019.104552
80. Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS ONE. (2014) 9:e92292. doi: 10.1371/journal.pone.0092292
81. Zhang H, Li T, Zheng L, Huang X. Biomarker MicroRNAs for diagnosis of oral squamous cell carcinoma identified based on gene expression data and microrna-mrna network analysis. Comput Math Methods Med. (2017) 2017:9803018. doi: 10.1155/2017/9803018
82. Liu H, Ren G, Zhu L, Liu X, He X. The upregulation of miRNA-146a inhibited biological behaviors of ESCC through inhibition of IRS2. Tumour Biol J Int Soc Oncodevelopmental Biol Med. (2016) 37:4641–7. doi: 10.1007/s13277-015-4274-5
83. Lu Y-C, Chen Y-J, Wang H-M, Tsai C-Y, Chen W-H, Huang Y-C, et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev Res. (2012) 5:665 LP−674. doi: 10.1158/1940-6207.CAPR-11-0358
84. Liu Y, Wang X, Jiang X, Yan P, Zhan L, Zhu H, et al. Tumor-suppressive microRNA-10a inhibits cell proliferation and metastasis by targeting Tiam1 in esophageal squamous cell carcinoma. J Cell Biochem. (2019) 120:7845–57. doi: 10.1002/jcb.28059
85. Wang J, Yu P-Y, Yu J-P, Luo J-D, Sun Z-Q, Sun F, et al. KIF22 promotes progress of esophageal squamous cell carcinoma cells and is negatively regulated by miR-122. Am J Transl Res. (2021) 13:4152–66.
86. Salazar-Ruales C, Arguello J-V, López-Cortés A, Cabrera-Andrade A, García-Cárdenas JM, Guevara-Ramírez P, et al. Salivary microRNAs for early detection of head and neck squamous cell carcinoma: a case-control study in the high altitude mestizo ecuadorian population. Biomed Res Int. (2018) 2018:9792730. doi: 10.1155/2018/9792730
87. Chen X, Cai S, Li B, Zhang X, Li W, Liang H, et al. MicroRNA-21 regulates the biological behavior of esophageal squamous cell carcinoma by targeting RASA1. Oncol Rep. (2019) 41:1627–37. doi: 10.3892/or.2018.6944
88. Wang K, Chen D, Meng Y, Xu J, Zhang Q. Clinical evaluation of 4 types of microRNA in serum as biomarkers of esophageal squamous cell carcinoma. Oncol Lett. (2018) 16:1196–204. doi: 10.3892/ol.2018.8720
89. Avissar M, McClean MD, Kelsey KT, Marsit CJ. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. (2009) 30:2059–63. doi: 10.1093/carcin/bgp277
90. Huang S-D, Yuan Y, Zhuang C-W, Li B-L, Gong D-J, Wang S-G, et al. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol Cancer. (2012) 11:51. doi: 10.1186/1476-4598-11-51
91. Du Y, Li Y, Lv H, Zhou S, Sun Z, Wang M. miR-98 suppresses tumor cell growth and metastasis by targeting IGF1R in oral squamous cell carcinoma. Int J Clin Exp Pathol. (2015) 8:12252–9.
92. Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. (2010) 56:1871–9. doi: 10.1373/clinchem.2010.147553
93. Zhang J-H, Xia H-B. Lentiviral-mediated overexpression of microRNA-141 promotes cell proliferation and inhibits apoptosis in human esophageal squamous cell carcinoma. Recent Pat Anticancer Drug Discov. (2019) 14:170–6. doi: 10.2174/1574892814666181231142136
94. Ganci F, Sacconi A, Manciocco V, Sperduti I, Battaglia P, Covello R, et al. MicroRNA expression as predictor of local recurrence risk in oral squamous cell carcinoma. Head Neck. (2016) 38 Suppl 1:E189–97. doi: 10.1002/hed.23969
95. Jiao W, Zhang J, Wei Y, Feng J, Ma M, Zhao H, et al. MiR-139-5p regulates VEGFR and downstream signaling pathways to inhibit the development of esophageal cancer. Dig liver Dis Off J Ital Soc Gastroenterol Ital Assoc Study Liver. (2019) 51:149–56. doi: 10.1016/j.dld.2018.07.017
96. Luo H-S, Wu D-H. Identification of miR-375 as a potential prognostic biomarker for esophageal squamous cell cancer: A bioinformatics analysis based on TCGA and meta-analysis. Pathol Res Pract. (2019) 215:512–8. doi: 10.1016/j.prp.2019.01.009
97. Wu Y, Sun X, Song B, Qiu X, Zhao J. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med. (2017) 6:1686–97. doi: 10.1002/cam4.1110
98. Zhang J, Zhang Y, Tan X, Zhang Q, Liu C, Zhang Y. MiR-23b-3p induces the proliferation and metastasis of esophageal squamous cell carcinomas cells through the inhibition of EBF3. Acta Biochim Biophys Sin (Shanghai). (2018) 50:605–14. doi: 10.1093/abbs/gmy049
99. Zhang X-W, Liu N, Chen S, Wang Y, Zhang Z-X, Sun Y-Y, et al. High microRNA-23a expression in laryngeal squamous cell carcinoma is associated with poor patient prognosis. Diagn Pathol. (2015) 10:22. doi: 10.1186/s13000-015-0256-6
100. Wen J, Hu Y, Liu Q, Ling Y, Zhang S, Luo K, et al. miR-424 coordinates multilayered regulation of cell cycle progression to promote esophageal squamous cell carcinoma cell proliferation. EBioMedicine. (2018) 37:110–24. doi: 10.1016/j.ebiom.2018.10.043
101. Li Y, Liu J, Hu W, Zhang Y, Sang J, Li H, et al. miR-424-5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. (2019) 12:10441–53. doi: 10.2147/OTT.S224325
102. Sun C, Zhang X, Chen Y, Jia Q, Yang J, Shu Y. MicroRNA-365 suppresses cell growth and invasion in esophageal squamous cell carcinoma by modulating phosphoserine aminotransferase 1. Cancer Manag Res. (2018) 10:4581–90. doi: 10.2147/CMAR.S157858
103. Geng J, Liu Y, Jin Y, Tai J, Zhang J, Xiao X, et al. MicroRNA-365a-3p promotes tumor growth and metastasis in laryngeal squamous cell carcinoma. Oncol Rep. (2016) 35:2017–26. doi: 10.3892/or.2016.4617
104. Fan Y-X, Bian X-H, Qian P-D, Chen Z-Z, Wen J, Luo Y-H, et al. MicroRNA-125b inhibits cell proliferation and induces cell apoptosis in esophageal squamous cell carcinoma by targeting BMF. Oncol Rep. (2018) 40:61–72. doi: 10.3892/or.2018.6413
105. González-Arriagada WA, Olivero P, Rodríguez B, Lozano-Burgos C, de Oliveira CE, Coletta RD. Clinicopathological significance of miR-26, miR-107, miR-125b, and miR-203 in head and neck carcinomas. Oral Dis. (2018) 24:930–9. doi: 10.1111/odi.12872
106. Wang K, Li J, Xiong G, He G, Guan X, Yang K, et al. Negative regulation of lncRNA GAS5 by miR-196a inhibits esophageal squamous cell carcinoma growth. Biochem Biophys Res Commun. (2018) 495:1151–7. doi: 10.1016/j.bbrc.2017.11.119
107. Hu C, Peng J, Lv L, Wang X, Zhou Y, Huo J, et al. miR-196a regulates the proliferation, invasion and migration of esophageal squamous carcinoma cells by targeting ANXA1. Oncol Lett. (2019) 17:5201–9. doi: 10.3892/ol.2019.10186
108. Zuo J, Zhu K, Wang Y, Yu Z. MicroRNA-34a suppresses invasion and metastatic in esophageal squamous cell carcinoma by regulating CD44. Mol Cell Biochem. (2018) 443:139–49. doi: 10.1007/s11010-017-3218-3
109. Kalfert D, Pesta M, Kulda V, Topolcan O, Ryska A, Celakovsky P, et al. MicroRNA profile in site-specific head and neck squamous cell cancer. Anticancer Res. (2015) 35:2455–63.
110. Liu Y, Tang Y, Li P. Inhibitory effect of microRNA-455-5p on biological functions of esophageal squamous cell carcinoma Eca109 cells via Rab31. Exp Ther Med. (2018) 16:4959–66. doi: 10.3892/etm.2018.6820
111. Cui Y, Xue Y, Dong S, Zhang P. Plasma microRNA-9 as a diagnostic and prognostic biomarker in patients with esophageal squamous cell carcinoma. J Int Med Res. (2017) 45:1310–7. doi: 10.1177/0300060517709370
112. Sun L, Liu L, Fu H, Wang Q, Shi Y. Association of decreased expression of serum miR-9 with poor prognosis of oral squamous cell carcinoma patients. Med Sci Monit. (2016) 22:289–94. doi: 10.12659/MSM.895683
113. Zheng R, Liu Y, Zhang X, Zhao P, Deng Q. miRNA-200c enhances radiosensitivity of esophageal cancer by cell cycle arrest and targeting P21. Biomed Pharmacother. (2017) 90:517–23. doi: 10.1016/j.biopha.2017.04.006
114. Lo W-L, Yu C-C, Chiou G-Y, Chen Y-W, Huang P-I, Chien C-S, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol. (2011) 223:482–95. doi: 10.1002/path.2826
115. Ma J, Zhan Y, Xu Z, Li Y, Luo A, Ding F, et al. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. (2017) 398:37–45. doi: 10.1016/j.canlet.2017.04.006
116. Jakob M, Mattes LM, Küffer S, Unger K, Hess J, Bertlich M, et al. MicroRNA expression patterns in oral squamous cell carcinoma: hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck. (2019) 41:3499–515. doi: 10.1002/hed.25866
117. Wang W, He X, Zheng Z, Ma X, Hu X, Wu D, et al. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol Cancer. (2017) 16:75. doi: 10.1186/s12943-017-0643-6
118. Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, et al. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. (2014) 31:148. doi: 10.1007/s12032-014-0148-8
119. Dong W, Li B, Wang J, Song Y, Zhang Z, Fu C, et al. Diagnostic and predictive significance of serum microRNA-7 in esophageal squamous cell carcinoma. Oncol Rep. (2016) 35:1449–56. doi: 10.3892/or.2015.4499
120. Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. (2010) 432:199–205. doi: 10.1042/BJ20100859
121. Summerer I, Unger K, Braselmann H, Schuettrumpf L, Maihoefer C, Baumeister P, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer. (2015) 113:76–82. doi: 10.1038/bjc.2015.111
122. Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J Cell Mol Med. (2017) 21:3730–40. doi: 10.1111/jcmm.13282
123. Cheng C-M, Shiah S-G, Huang C-C, Hsiao J-R, Chang J-Y. Up-regulation of miR-455-5p by the TGF-β-SMAD signalling axis promotes the proliferation of oral squamous cancer cells by targeting UBE2B. J Pathol. (2016) 240:38–49. doi: 10.1002/path.4752
124. Mishra V, Singh A, Chen X, Rosenberg AJ, Pearson AT, Zhavoronkov A, et al. Application of liquid biopsy as multi-functional biomarkers in head and neck cancer. Br J Cancer. (2022) 126:361–70. doi: 10.1038/s41416-021-01626-0
125. Martins I, Ribeiro IP, Jorge J, Gonçalves AC, Sarmento-Ribeiro AB, Melo JB, et al. Liquid biopsies: Applications for cancer diagnosis and monitoring. Genes (Basel). (2021) 12:1–20. doi: 10.3390/genes12030349
126. Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol Off J Eur Soc Med Oncol. (2020) 31:745–59. doi: 10.1016/j.annonc.2020.02.011
127. Chen X, Gole J, Gore A, He Q, Lu M, Min J, et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. (2020) 11:3475. doi: 10.1038/s41467-020-17316-z
128. Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of plasma epstein-barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. (2017) 377:513–22. doi: 10.1056/NEJMoa1701717
Keywords: liquid biopsy, esophageal squamous cell carcinoma, head and neck squamous cell carcinoma, cell free DNA, circulating tumor cells, exosomes
Citation: Iacob R, Mandea M, Iacob S, Pietrosanu C, Paul D, Hainarosie R and Gheorghe C (2022) Liquid Biopsy in Squamous Cell Carcinoma of the Esophagus and of the Head and Neck. Front. Med. 9:827297. doi: 10.3389/fmed.2022.827297
Received: 01 December 2021; Accepted: 15 February 2022;
Published: 29 April 2022.
Edited by:
Weiren Luo, The Second Affiliated Hospital of Southern University of Science and Technology, ChinaReviewed by:
Mercedes Camacho, Institut de Recerca de l'Hospital de la Santa Creu i Sant Pau, SpainCopyright © 2022 Iacob, Mandea, Iacob, Pietrosanu, Paul, Hainarosie and Gheorghe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Razvan Hainarosie, cmF6dmFuLmhhaW5hcm9zaWVAdW1mY2Qucm8=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.