
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 19 May 2022
Sec. Hematology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.810612
This article is part of the Research Topic Immune Thrombocytopenia (ITP) - Diagnosis and Treatment View all 8 articles
 Xiaomin Yu1†
Xiaomin Yu1† Qingyun Zheng2†
Qingyun Zheng2† Yun He2
Yun He2 Dandan Yu1
Dandan Yu1 Guolin Chang1
Guolin Chang1 Cheng Chen3
Cheng Chen3 Laixi Bi3
Laixi Bi3 Jia Lv4
Jia Lv4 Misheng Zhao5*
Misheng Zhao5* Xiangyang Lin1*
Xiangyang Lin1* Liqing Zhu1*
Liqing Zhu1*Objective: To determine whether gut microbiota, fatty metabolism and cytokines were associated with immune thrombocytopenia (ITP).
Methods: In total, 29 preliminarily diagnosed ITP patients and 33 healthy volunteers were enrolled. Fecal bacterial were analyzed based on 16S rRNA sequencing. Plasma cytokines and motabolites were analyzed using flow cytometry and liquid chromatography-mass spectrometry (LC-MS), respectively.
Results: Bacteroides, Phascolarctobacterium, and Lactobacillus were enriched at the genus level in ITP patients, while Ruminococcaceae UCG-002, Eubacterium coprostanoligeues, Megamonas, and Lachnospiraceae NC2004 were depleted. At the phylum level, the relative abundance of Proteobacteria and Chloroflexi increased in ITP patients, while Firmicutes, Actinobacteria, and the Firmicutes/Bacteroidetes ratio decreased. Plasma levels of 5-hydroxyeicosatetraenoic acid (5-HETE), 6-trans-12-epi-leukotriene B4 (6t,12e-LTB4), and resolvin D2 (RvD2) were upregulated, and stachydrine, dowicide A, dodecanoylcarnitine were downregulated in ITP patients. Furthermore, RvD2 is positively correlated with order Bacteroidetes VC2.1 Bac22, 5-HETE is positively correlated with genus Azospirillum, and 6t,12e-LTB4 is positively correlated with genus Cupriavidus. In addition, stachydrine is positively correlated with family Planococcaceae, dowicide A is positively correlated with class MVP-15, and dodecanoylcarnitine is positively correlated with order WCHB1-41. Plasma levels of interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) were upregulated in ITP patients.
Conclusion: Our study revealed a relationship between microbiota and fatty metabolism in ITP. Gut microbiota may participate in the pathogenesis of ITP through affecting cytokine secretion, interfering with fatty metabolism.
Immune thrombocytopenia (ITP) is characterized by low peripheral blood platelet count. It is an acquired autoimmune disorder which can lead to easy and excessive bruising and bleeding. The increased platelet clearance arises through diverse mechanisms, such as antiplatelet autoantibodies production, and T lymphocyte dysfunction. It was reported that receptors for the Fc fragment of IgG result in the removal of platelets coated with immunoglobulin (1) and the activation of phagocytosis in the reticuloendothelial system (2). In addition to B cell dependent autoimmune mechanisms, it is well established that cellular immune pathophysiology of ITP is associated with several T-cell abnormalities, including uncontrolled activation of T helper cells, deficiency of T regulatory cells, and increase of CD8 + T cell targeting megakaryocytes, etc. (2, 3). Another mechanism that contributes to the platelet clearance is platelet activation (4). In two pilot studies, it was reported that platelet activation markers, such as P-selectin, CD62 and CD11b expression, were significantly higher in ITP patients (5, 6). In the ITP mouse model, Fc-independent platelet activation was observed, which led to platelet clearance in the liver (7). This partially explained the fact that the risk of venous thromboembolism is twice higher in ITP patients than in healthy population (8). More recently, COVID-19 vaccines have been linked with serious vaccine-induced ITP (9), which is believed to be mediated by activated platelets (10, 11). However, the initial trigger of platelets activation has not yet to be determined.
Emerging data has demonstrated a strong association among platelet function, metabolites of gut microbiota, and lipid metabolism (12). Two gut microbiota derived-metabolites, trimethylamine N-oxide (13) and phenylacetylglutamine (14) were shown to directly contribute to platelet activation related phenotypes. It is worth noticing that the metabolite phenylacetylglutamine was discovered through untargeted metabolomics, a powerful and unbiased platform for discovery of pathways linked to diseases. Using the similar strategy, analysis of a cohort of 534 healthy adult Dutch volunteers provided evidence for the importance of intestinal microbiota in regulating platelet function (15). More than 20% metabolites were significantly associated with platelet activation markers and the majority involved lipids. In a more recent study, clinical findings with experimental data suggested that gut microbiota dysbiosis may be an important mechanism for the high platelet reactivity in patients treated with ticagrelor, a first-line drug for the treatment of acute ST elevation myocardial infarction (16). For example, the abundance of gut microbiota species Methylbacillus, Sphingomonas, and Staphylococcus was negatively correlated with serum salicylate, which was previously found to induce ITP and inhibit platelet production in megakaryocytes (17).
Platelet metabolism and reactivity critically depend on lipid composition (18). For example, arachidonic acid (AA), a polyunsaturated fatty acid constituting an important part of phospholipid-content in cell membranes, is associated with the enzyme-derived oxidized metabolites (19). It enters the cell, activates the platelet and exerts in platelet function and fate. Up to 25% of phospholipid fatty acids are AA in resting platelets, the concentration of which may influence normal cellular functions and development of platelet diseases (20). Other metabolites like prostacyclin (PGI2) and prostaglandin D2 (PGD2) also inhibit platelet activation (21, 22) while 12(S)-HETE produced by 12-LOX was found to impact the platelet function (23). Besides, it was observed that high-density lipoprotein (HDL) reduced platelet aggregation in response to a variety of agonists, while oxidized HDL exerted the opposite effect binding to a similar receptor (24). In general, specific lipid species play important roles in enhancing stability of the domains and stabilizing signaling events in platelets, which permit the feasibility of a systematic assessment on platelet-lipid analysis (18).
To date, the analysis of gut microbes and lipid metabolism in modulation of ITP-associated platelet function has not yet been reported. The present study should provide new insights in the pathogenesis mechanisms associated with gut microbiota, cytokine, fatty metabolism in ITP, and therefore could improve differential diagnosis and support further treatment decision, such as traditional Chinese medicine, for ITP patients.
Fecal and plasma samples were collected from patients with newly diagnosed ITP before systemic treatment at the First Affiliated Hospital of Wenzhou Medical University. None of the recruited patients and healthy volunteers presented any other autoimmune diseases or intestinal and metabolic diseases that could affect the gut microbiota. The protocol was approved by the Ethics Committee of Wenzhou Medical University (KY2021-048), and informed consent was obtained from all participants. ITP was diagnosed according to the criteria established by the American Society of Hematology (ASH), which identifies as an autoimmune disorder characterized by an isolated platelet count < 100×109/L, with or without bleeding manifestations, in the absence of other causes or disorders that may be associated with thrombocytopenia (25). In total, 29 patients were enrolled in the present study. In addition, 33 volunteers were recruited after a routine physical examination. All healthy controls presented no gut problems such as chronic diarrhea, constipation, gas, heartburn, bloating (26), and none had taken any antibiotics or probiotics within 3 months of this study. Moreover, there were no significant differences between the two groups in terms of sex, age, smoking history, and alcohol or dietary intake.
For cytokine analysis, venous blood (2 mL) was collected and centrifuged to obtain 200 μL of serum. Cytokines, including interleukin (IL)-2, IL-4, IL-6, IL-10, TNF-α, and interferon (IFN)-γ, were detected by employing a cytometric bead array human inflammation kit according to the manufacturer’s instructions (BD Parmingen, San Diego, CA, United States) and operated using AimPlex™ bead-based multiparametric flow cytometry (EPICS-Elite, Beckman-Coulter, United States). For determining the protein concentration, sample fluorescence signals were compared with those of a standard curve generated from serial dilutions of a known concentration of the analyte.
Genomic DNA was extracted based on the V4 hypervariable region of 16S rDNA using a fecal genomic DNA extraction kit (QIAGEN), with 0.8% agarose gel electrophoresis performed to separate DNA. Then, to amplify the bacterial 16S rRNA from diluted DNA extracts, we used template-specific sequences 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′GGACTACHVGGGTWTCTAAT-3′). Polymerase chain reaction (PCR) amplification was performed using a 25 μL reaction volume with the following composition: 1 × PCR buffer, 1.5 mM MgCl2, 0.4 μM dNTPs, 1.0 μL forward primer, 1.0 μL reverse primer, 0.5 U KOD-Plus-Neo enzyme (TOYOBO), and 10 ng template. The reaction was initiated at 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 54°C for 30 s, and elongation at 72°C for 30 s, with a final extension step at 72°C for 5 min. Each sample was assessed in triplicate. PCR products were mixed with 1/6 volume 6 × loading buffer and detected by 2% agarose gel electrophoresis. These amplicons were recycled using QIAquick Gel Extraction Kit (QIAGEN), quantified using Qubit@ 2.0 Fluorometer (Thermo Scientific), and finally mixed equally. The library was prepared using TruSeq DNA PCR-Free Sample Prep Kit (FC-1213001/3003) (Illumina Inc., San Diego, CA). After quantification and detection, the sequencing reaction was conducted using the PE250 pattern of Hiseq Rapid SBS Kit v2 (FC-402-4023 500 Cycle) (Illumina Inc., San Diego, CA).
After sequencing, high-quality sequences were filtered for subsequent analysis and assembled from raw data. Alpha diversity [Chao1, phylogenetic diversity (PD), Simpson, Shannon] and beta diversity (PCoA) were analyzed using Usearch and QIIME. Statistics and plotting were completed using R6, Python, and Java. Next, to determine the species composition and diversity, high-quality sequences were clustered with a cut-off of 97% similarity, resulting in a total of 6,327 operational taxonomic units (OTUs). For each OTU, the sequence that occurred most frequently was selected as the representative OTU sequence. Annotation analysis was performed using UCLUST and SILVA databases. Multiple sequence alignment was performed using PyNAST, with the phylogenetic tree constructed using FastTree. Samples were homogenized and resampled with the standard of the least amount.
After plasma samples were thawed on ice, 120 μL of precooled 50% methanol buffer was added to 20 μL of samples. Then, samples were vortexed for 1 min, incubated for 10 min at room temperature and stored overnight at −20°C. The mixture of metabolites was centrifuged at 4,000 × g for 20 min to obtain the supernatant. Subsequently, supernatants were transferred into new 96-well plates and stored at −80°C for subsequent liquid chromatography-mass spectrometry (LC-MS) analysis. For pooled quality control (QC) samples, 10 μL of each extraction mixture were combined and mixed.
All samples were analyzed using the TripleTOF 5600 Plus high-resolution tandem mass spectrometer (SCIEX, Warrington, United Kingdom), utilizing both positive and negative ion modes. Chromatographic separation was performed in an ultra-performance liquid chromatography (UPLC) system (SCIEX, United Kingdom) using an ACQUITY UPLC T3 column (100 mm × 2.1 mm, 1.8 μm; Waters, United Kingdom) for reversed-phase separation, with a constant column temperature of 35°C. The mobile phase consisted of solvent A (water, 0.1% formic acid) and solvent B (acetonitrile, 0.1% formic acid). The UPLC elution conditions were optimized and set as follows: flow rate, 0.4 mL/min; 0–0.5 min, 5% solvent B; 0.5–7 min, 5–100% solvent B; 7–8 min, 100% solvent B; 8–8.1 min, 100–5% solvent B; 8.1–10 min, 5% solvent. The TripleTOF 5600 Plus System was used to detect eluted metabolites. The curtain gas was set at 30 PSI, with ion source gases 1 and 2 at 60 PSI. The temperature of the interface heater was 650°C. The ion spray floating voltage was set at 5 kV for the positive ion mode and −4.5 kV for the negative ion mode.
MS data were acquired in Information Dependent Acquisition mode. The time-of-flight mass ranged between 60 and 1,200 Da. Survey scans were acquired over 0.15 s, and up to 12 product ion scans were collected if the 100 counts/s threshold was exceeded, with a 1 + charge state. The total cycle time was fixed at 0.56 s. Four-time bins were summed for each scan at an impulsator frequency of 11 kHz by monitoring the 40 GHz multichannel TDC detector with four-anode/channel detectors. Dynamic exclusion was performed for 4 s. During the entire acquisition period, the mass accuracy was calibrated for every 20 samples. In addition, a QC sample was analyzed every 10 samples to evaluate the LC-MS stability.
The acquired LC-MS data was pretreated using XCMS software (SCIEX). Raw data was converted into mzXML format and processed using the XCMS, CAMERA, and metaX toolboxes included in R software. Each ion was identified using comprehensive information regarding the retention time and m/z. The intensity of each peak was recorded, and a three-dimensional matrix containing arbitrarily assigned peak indices (retention time –m/z pairs), sample names (observations), and ion intensity information (variables) were generated. Then, the information was matched to in-house and public databases. The open-access databases, KEGG (Kyoto Encyclopedia of Genes and Genomes)1,2 and HMDB (Human Metabolome Database)3, were employed to annotate metabolites by matching the exact molecular mass data (m/z) to those from the database, within a threshold of 10 ppm. Peak intensity data was further pre-processed using metaX software. Features detected in 30% of samples were removed. Group datasets were normalized before analysis. Data normalization was performed for all samples using a probabilistic quotient normalization algorithm. Then, QC-robust spline batch correction was performed using QC samples. The P-value was analyzed using the Student’s t-test, then adjusted for multiple tests using a false discovery rate (Benjamini–Hochberg) to select different metabolites. Supervised partial least-squares discriminant analysis was performed using metaX to identify variables that differed between groups. The variable importance in projection was calculated, and a cut-off value of 1.0 was set to select important features.
The results were statistically analyzed using SPSS version 22.0 (IBM Corp., Armonk, NY, United States). Data are expressed as mean ± standard error of the mean. Significant differences in clinical characteristics and cytokine levels between healthy controls and ITP patients were analyzed by independent-sample t-test. A P-value of <0.05 was considered statistically significant. We used Spearman correlation analysis to filter differential metabolites and gut microbiota.
The present study recruited 62 subjects, including 29 ITP patients (13 men, 16 women) and 33 healthy controls (17 men, 16 women) (Table 1). No statistically significant difference was observed between the patient group and control group in terms of gender (male: 45% vs. male: 52%, P = 0.599) and age (39.50 ± 15.98 vs. 39.60 ± 15.87, P = 0.989) (Table 1). Serum levels of total protein (TP), alanine aminotransferase (ALT), alkaline phosphatase (ALP), gamma-glutamyl transpeptidase (GGT), glucose (GLU), urea, total cholesterol (TC), total triglyceride (TG), and low-density lipoprotein (LDL) were similar between patients and healthy controls. Levels of total bilirubin (TB, 9.93 ± 4.17 vs. 13.11 ± 4.33, P = 0.007), indirect bilirubin (IB, 5.86 ± 2.66 vs. 10.37 ± 3.67, P<0.001), albumin (ALB, 40.78 ± 4.94 vs. 45.21 ± 2.62, P<0.001), creatinine (Crea, 58.62 ± 16.92 vs. 69.93 ± 15.70, P = 0.012), uric acid (UA, 287.24 ± 87.61 vs. 345.37 ± 91.31, P = 0.018), and high-density lipoprotein (HDL, 1.11 ± 0.35 vs. 1.34 ± 0.27, P = 0.011) were lower in ITP patients compared to healthy controls (Table 1). In addition, levels of direct bilirubin (DB, 4.07 ± 1.85 vs. 2.74 ± 0.90, P = 0.001), globulin (GLB, 32.67 ± 6.85 vs. 29.23 ± 2.48, P = 0.016), and aspartate aminotransferase (AST, 21.57 ± 5.89 vs. 18.33 ± 3.28, P = 0.015) were higher in ITP patients than healthy controls (Table 1).
Alpha and beta diversities were calculated to evaluate the structural differences of gut microbial community between ITP patients and healthy controls. Chao 1 is a measure of total richness and is particularly useful owing to a valid variance, which can be employed to calculate confidence intervals. PD whole tree reflects the sum of all branch-lengths on the constructed phylogenetic tree from all taxa. Simpson considers both species abundance (Richness) and uniformity (Evenness). Shannon reflects species numbers and evenness of species abundance. As shown in Figure 1A, PD whole tree, Simpson, and Shannon had a significant difference between two groups, which represent the richness and diversity of gut microbiome community. The beta diversity in Figure 1B was also showed differences according to the principal coordinates analysis (PCoA). A total of 2,149 OTUs were found to be shared between ITP patients and healthy controls, accounting for a similarity level of more than 93% (Figure 1C).
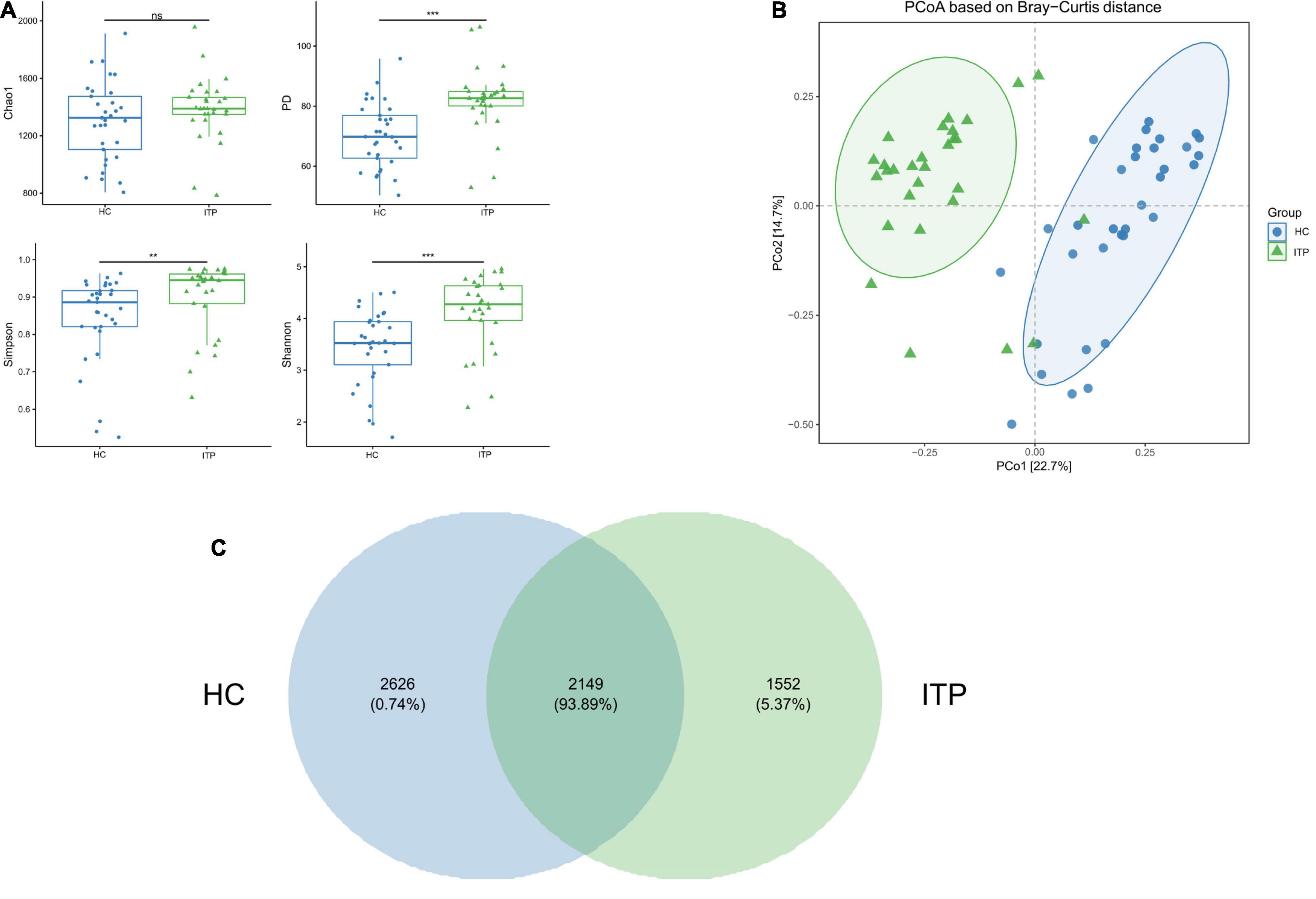
Figure 1. Significant differences of gut microbial community between ITP patients and healthy controls. (A) The alpha diversity indexes include Chao 1, PD whole tree, Simpson, and Shannon. There are significant differences in PD whole tree, Simpson, and Shannon, except for Chao1. (B) PCoA is a convenient method to observe the degree of difference and the rule of variation among samples. Each dot represents a sample. The more similar the microbial community composition, the closer the dots. The plot shows a distinct difference between ITP patients and healthy controls. (C) A Venn plot is used to demonstrate the bacterial microbiota structure, assessing which species are common and unique among groups or samples. **p < 0.01, ***p < 0.001.
The top 10 ITP-associated bacterium at phylum level were shown in Figure 2A. Proteobacteria (P = 0.002), Chloroflexi (P < 0.001) were significantly high in ITP patients, while Firmicutes (P < 0.001), Actinobacteria (P < 0.001) were lower compared to healthy controls. Correspondingly, the Firmicutes/Bacteroidetes ratio was decreased. Next, we explored ITP-associated microbiome changes at genus level. As shown in Figure 2B, Bacteroides (P = 0.020), Phascolarctobacterium (P = 0.008), and Lactobacillus (P < 0.001) were significant high in ITP patients. While the Ruminococcaceae UCG-002 (P < 0.001), Eubacterium coprostanoligeues (P = 0.011), Megamonas (P < 0.001), and Lachnospiraceae NC2004 (P = 0.002) were significantly decreased.
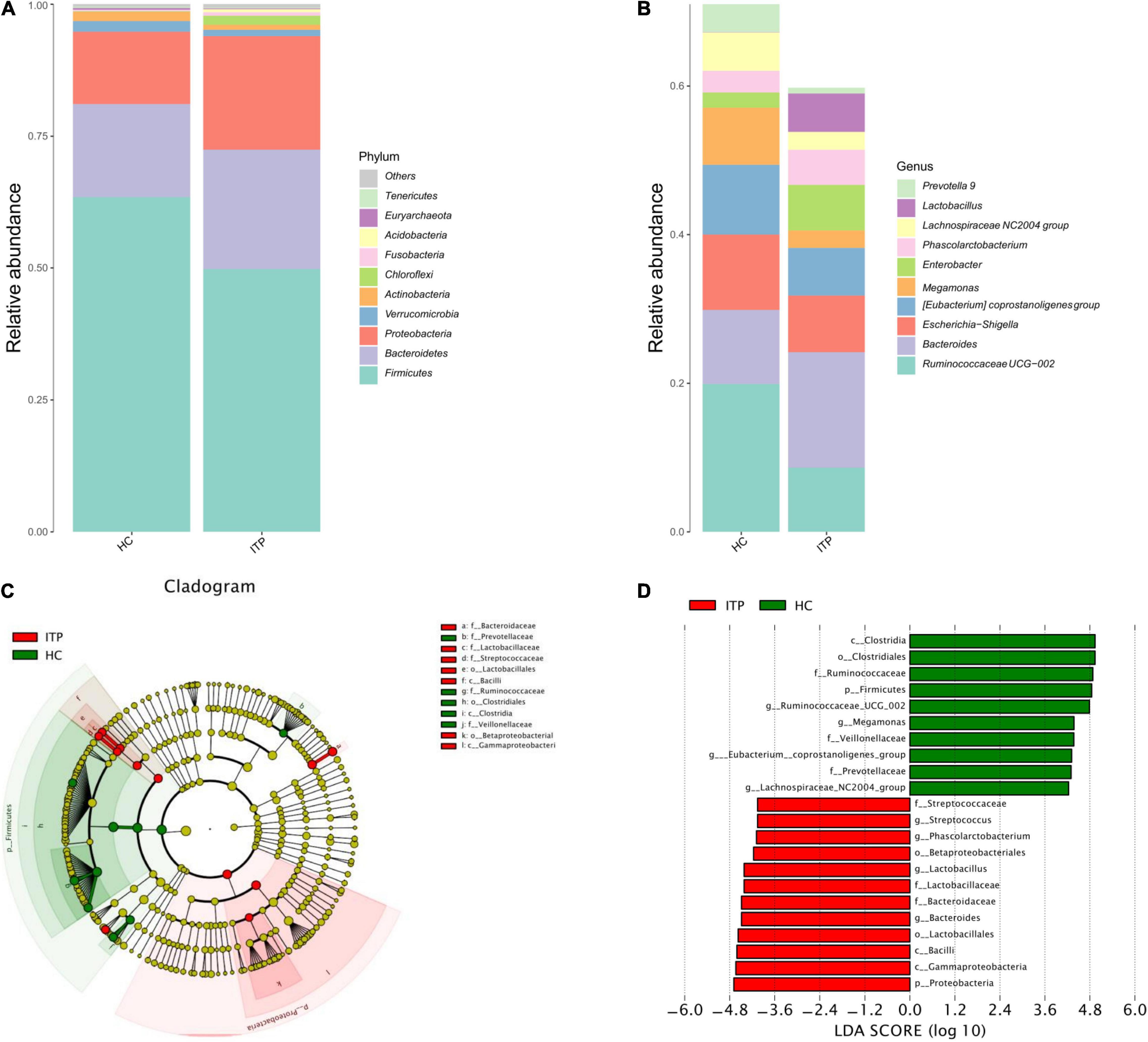
Figure 2. Differences of gut microbiota between ITP patients and healthy controls at phylum and genus level, respectively. (A) Relative abundance of bacterial microbiota in two groups at phylum level. (B) Relative abundance of bacterial microbiota in two groups at genus level. (C) The cladogram is constructed according to the hierarchical structure of classified data. (D) A linear discriminant analysis effect size (LEfSe) between ITP patients (ITP, green) and healthy controls (HC, red) revealed taxonomic abundances between groups (LDA > 4.0).
The cladogram was used for discovering the high-dimensional biomarkers that substantially contributed to the differences between groups. The main taxonomic groups of ITP patients and healthy controls were highlighted, respectively. Accordingly, Lactobacillaceae (family) and Streptococcaceae (family) belong to Lactobacillales (order), which belong to Bacilli (class) is highlighted (Figure 2C).
Next, we compared taxonomic abundances between ITP patients and healthy controls by employing linear discriminant analysis effect size (LEfSe), which was used to quantitatively analyze the important species that significantly differed among groups. The linear discriminant analysis (LDA) score correlates taxa abundancy which have the potential to be a biomarker. We listed those taxa which were meeting a significant LDA threshold more than 4 (LDA score > 4.0, P < 0.05). In this study, LEfSe analysis revealed more abundant Proteobacteria in ITP patients at phylum level, and more abundant Bacteroides, Lactobacillus, Phascolarctobacterium, and Streptococcus at genus level (Figure 2D).
Next, we explored the alteration of metabolites from gut microbiota in ITP. Specific diversity of metabolites was found between ITP patients and healthy controls. The metabolite expression levels were identified by volcano plot. As shown in Figure 3A, totally 423 metabolites which had a fold change more than 2.0 (P < 0.05) were identified from 4,786 metabolites. Among them, 337 were upregulated and 86 were downregulated (Figure 3B).
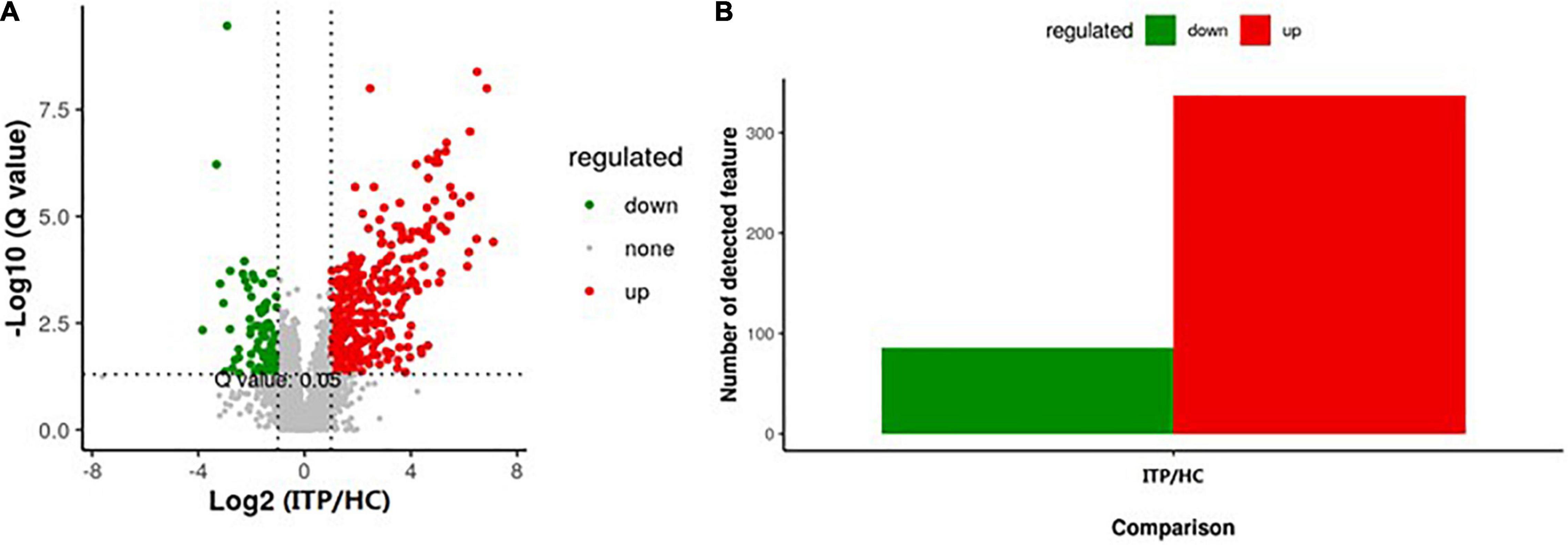
Figure 3. Alterations of metabolites of gut microbiota in ITP patients. (A) The vertical lines corresponded to a 2.0-fold change (up and down). The horizontal line represented a P-value of less than 0.05. The red and green dots represented upregulation and downregulation of microbiota, respectively. (B) Overall, 337 metabolites were significantly upregulated, while 86 were significantly downregulated.
Furthermore, serum IL-2, IL-6, IL-4, IL-10, TNF-α, and IFN-γ were detected to reflect the immune response of ITP patients which were shown in Figure 4. IL-6 and TNF-α were significant higher in ITP patients compared to the healthy controls (P < 0.001, P < 0.001, respectively). While IL-2, IL-4, IL-10, and IFN-γ had no significant difference between two groups. However, a trend of down regulation of IL-4 and IFN-γ were observed in ITP patients, even they failed to reach a statistical significance.
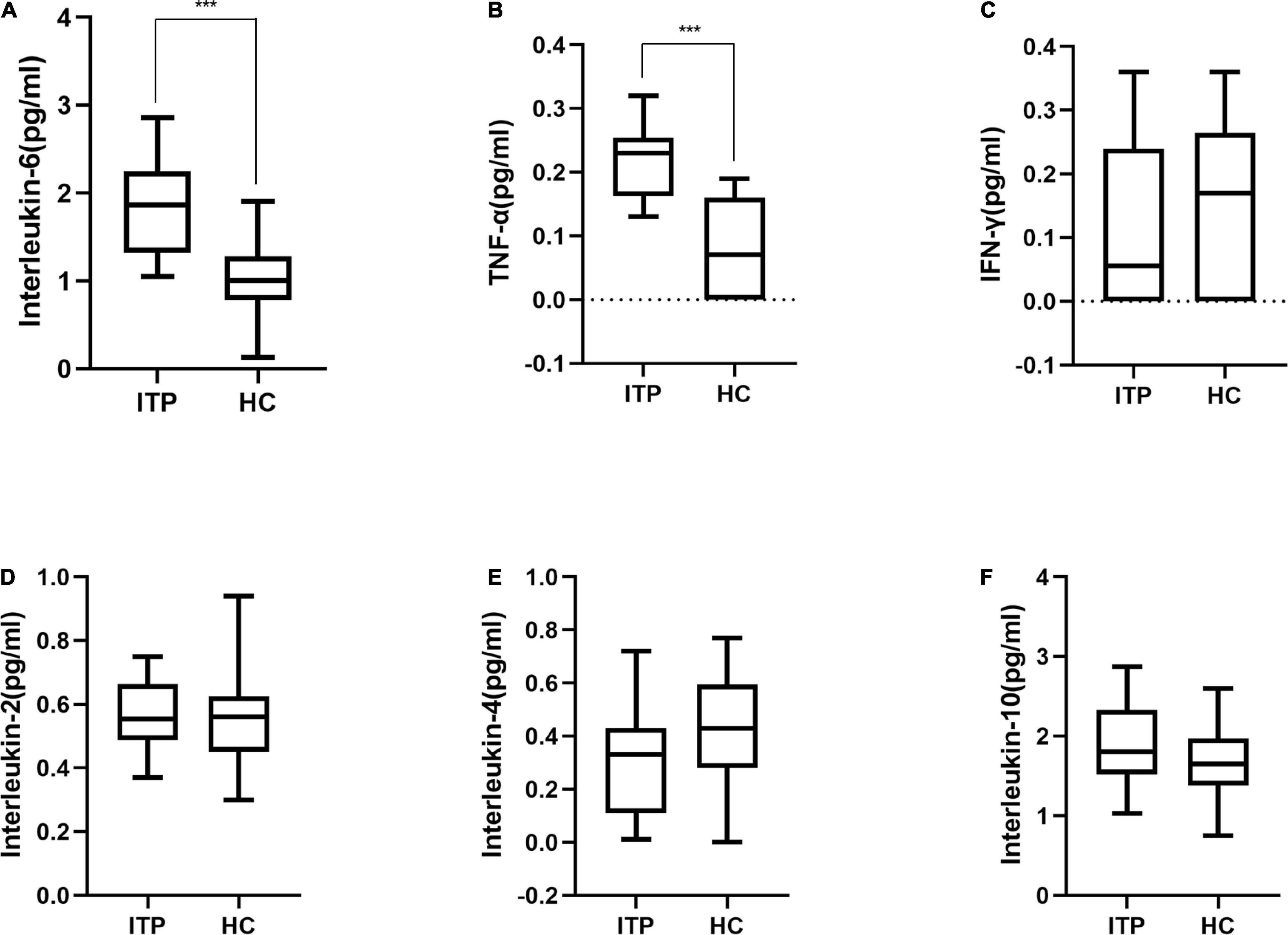
Figure 4. Box plot of cytokines in ITP patients and healthy controls. (A) Interleukin-6, (B) TNF-α, (C) IFN-γ, (D) Interleukin-2, (E) Interleukin-4, (F) Interleukin-10. These cytokines were all detected by flow cytometry. ***p < 0.001.
We then investigated whether the metabolic pathway could help predict the clinical prognosis of ITP. The PCA plot showed metabolites of ITP patients are different from healthy controls. Univariate analysis, fold change (Ratio ≥ 2 or ≤ 1/2), t-test (P value<0.05), and Variable Important for the Projection (VIP ≥ 1) were obtained by multivariate analysis, which were employed to filter the representative metabolic parameters. Compared with healthy controls, fatty acyls compounds including 5-HETE, RvD2, and 6t,12e-LTB4 were significantly upregulated. While stachydrine, dowicide and dodecanoylcarnitine were significantly downregulated. Hence, we speculated that lipid metabolism dysfunction might be occurred in the pathogenesis of ITP (Table 2).
In present study, interactions between gut microbiota and metabolites were investigated through metabolic and 16S sequencing analyses. Spearman correlation was performed to filter differential metabolites and gut microbiota. The Spearman’s rank correlation coefficient was showed as heatmaps of Figure 5. It was showed that RvD2, 5-HETE, 6t,12e-LTB4, stachydrine, dowicide A and dodecanoylcarnitineis were highly correlated with order Bacteroidetes VC2.1 Bac22 (r = 0.91, P < 0.05), genus Azospirillum (r = 0.91, P < 0.05), genus Cupriavidus (r = 0.90, P < 0.05), family Planococcaceae (r = 0.88, P < 0.05), class MVP-15 (r = 0.90, P < 0.05), order WCHB1-41 (r = 0.85, P < 0.05), respectively (Table 3).
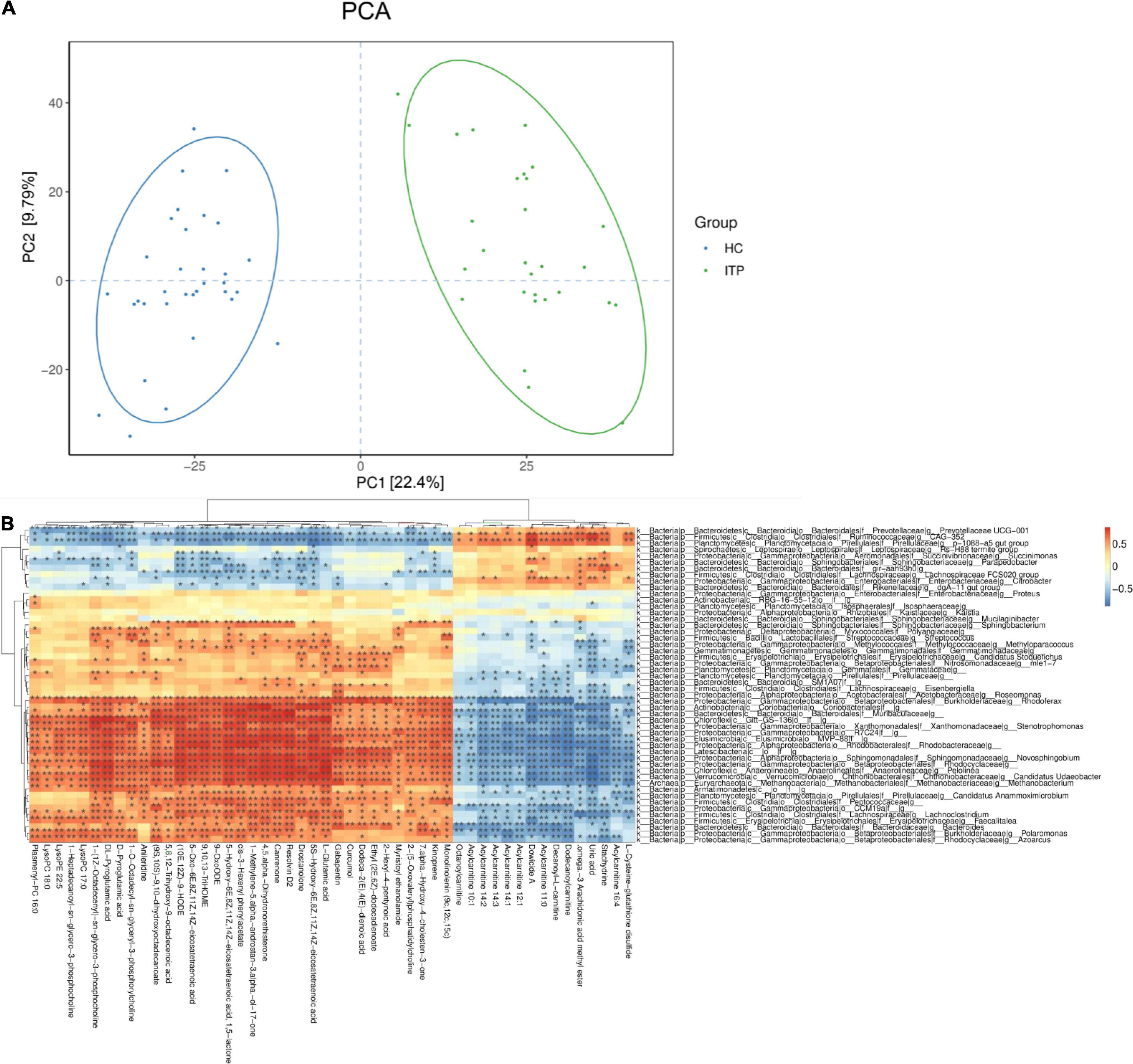
Figure 5. Relationship between serum metabolites and gut microbiota in ITP. (A) The PCA of ITP patients and healthy controls. (B) Heatmap of gut microbiota and metabolites. This heatmap shows a data matrix, where colored areas provided an overview of numeric differences.
Herein, we investigated the relationship among the gut microbiome, serum metabolism, cytokines in both the healthy people (n = 33) and ITP patients (n = 29), an autoimmune disorder. In addition to genetic and environmental triggers, accumulating evidence has suggested gut microbiota as another important causative element influencing the development of autoimmunity (27) as well as hematologic diseases (28). For example, there was evidence showing that gut bacterium contribute to type 1 diabetes development probably through translocation into pancreatic lymph nodes (28). For another example, inflammatory bowel disease with intestinal inflammation was reported to result in thrombocytopenia (29) and treatment led to recovery of the platelet number (30) indicating the link between hematology and gut microbiome. Indeed, Malnick et al. reported a case, in which the patient underwent fecal microbial transplantation twice from the same donor (31). Each time the patient developed transient ITP, with the platelet level decreasing and antiplatelet antibodies increasing rapidly after transplantation. These results, especially the recurrence of ITP after the second transplantation, strongly indicated that the gut microbiota is associated with the pathogenesis of ITP. However, the causality has not yet been established, with some arguments that the change of gut microbiome is only an outcome of disease progression rather than an initiator or exacerbator of disease.
Our results revealed that gut microbiota (diversity, richness, evenness, and PCA plot) and metabolites of fatty acids in ITP patients significantly differed from those in normal controls. Specifically, Bacteroides, Phascolarctobacterium and Lactobacillus were increased at the genus level, while Ruminococcaceae UCG-002, Eubacterium coprostanoligeues, Megamonas and Lachnospiraceae NC2004 were decreased. At the phylum level, the relative abundance of Proteobacteria and Chloroflexi was increased in ITP patients, while Firmicutes, Actinobacteria and the Firmicutes/Bacteroidetes ratio were decreased. It was reported that Firmicutes (32) and Firmicutes/Bacteroidetes ratio (33) were associated with Th17-Tregs trans-differentiation, which is regulated by IL-6 (34). Interestingly, we found that IL-6 levels were significantly elevated in ITP patients when compared with healthy controls, which is consistent to the other reports showing that IL-6 upregulation is related to autoimmune diseases, such as rheumatoid arthritis, multiple sclerosis, and asthma (35). TNF-α is another cytokine that correlates with Treg frequencies in chronic ITP (36) and its secretion is stimulated by Bacteroides (37). It was evident that fecal microbiota transplantation reduced TNF-α signaling (38). Our results also showed that the Bacteroides composition and TNF-α level were elevated in ITP patients. For the fatty metabolites, those that were upregulated in ITP patients included RvD2, eicosatetraenoic acids, Monolinolein, and phosphatidylcholine, all of which can be regulated by gut microbiota (39). Interestingly, RvD2 was recently shown to reduce platelet activation and preserve platelet function (40). Eicosatetraenoic acid, also named arachidonic acid, is considered to regulate platelet homeostasis (41). In addition, eicosatetraenoic acids (41), Monolinolein (42), phosphatidylcholine (43), are all proposed to be the key compounds contributing to platelet activation. Nevertheless, at this moment, our understanding is still at its relative infancy regarding the exact roles of gut microbiota and fatty metabolism in the development of ITP. Apparently, this requires additional and longer-term studies in the future.
In summary, we attempt to obtain a comprehensive insight of altered gut microbiota and fatty metabolism in ITP patients. Our study may provide information for the treatment of ITP. Indeed, ITP is considered as purpura disease and differentiated as spleen deficiency in the TCMs. Nowadays in China, Many TCM drugs are routinely used to treat ITP via tonifying the spleen and qi, which may reduce the frequency and severity of bleeding (44) (Table 4). Meanwhile, TCM treatments were proven to be associated with changes in gut microbiota (61), and serum metabolic (62). Our data along with other literatures suggested that changes of gut microbiota in ITP patients may increase the toxicity of specific TCM drugs. For example, Bacteroides metabolizes geniposide to genipin with stronger hepatic toxic effect (45) and thus, geniposide-containing TCM formulations should be cautiously used to treat ITP. Further studies are needed with larger number of patients and animal experiments to establish the causality among gut microbiota, fatty metabolism and ITP.
Over the years, several researchers have explored the relationship between gut microbiota, metabolites, traditional Chinese medicine and autoimmune diseases. Advances in sequencing technology have made it possible to further explore the role of gut microbiota in autoimmune diseases. Our research revealed that gut microbiota and metabolites are associated with the pathogenesis of ITP and the gut microbiota might influence the metabolism of TCMs in ITP patients. Further experiments are required to determine the mechanisms of microbial species, the pathway of metabolome, the interaction between the gut microbiota and TCMs, and the pathogenesis of ITP.
The data presented in the study are deposited in NCBI Sequence Read Archive, accession number PRJNA816310.
The studies involving human participants were reviewed and approved by the Ethics Committee in Clinical Research (ECCR) of the First Affiliated Hospital of Wenzhou Medical University. The patients/participants provided their written informed consent to participate in this study.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This work was sponsored by grants from the Natural Science Foundation of Zhejiang Province (LY22H200004), Medical Health Science and Technology Project of Zhejiang Province (2022KY205), Traditional Chinese Medicine Science and Technology Program Project of Zhejiang Province (2021ZB181), and Wenzhou Science and Technology Project (Y2020570).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Yu X, Menard M, Prechl J, Bhakta V, Sheffield WP, Lazarus AH. Monovalent Fc receptor blockade by an anti-Fcγ receptor/albumin fusion protein ameliorates murine ITP with abrogated toxicity. Blood. (2016) 127:132–8. doi: 10.1182/blood-2015-08-664656
2. Chow L, Aslam R, Speck ER, Kim M, Cridland N, Webster ML, et al. A murine model of severe immune thrombocytopenia is induced by antibody- and CD8+ T cell-mediated responses that are differentially sensitive to therapy. Blood. (2010) 115:1247–53. doi: 10.1182/blood-2009-09-244772
3. Swinkels M, Rijkers M, Voorberg J, Vidarsson G, Leebeek FWG, Jansen AJG. Emerging concepts in immune thrombocytopenia. Front Immunol. (2018) 9:880. doi: 10.3389/fimmu.2018.00880
4. Sun S, Urbanus RT, Ten Cate H, de Groot PG, de Laat B, Heemskerk JWM, et al. Platelet activation mechanisms and consequences of immune thrombocytopenia. Cells. (2021) 10:3386. doi: 10.3390/cells10123386
5. Bonnard G, Babuty A, Collot R, Costes D, Drillaud N, Eveillard M, et al. Platelet features allow to differentiate immune thrombocytopenia from inherited thrombocytopenia. Ann Hematol. (2021) 100:2677–82. doi: 10.1007/s00277-021-04651-4
6. Lozano ML, Garabet L, Fernandez-Perez MP, De Los Reyes-García AM, Diaz-Lozano P, Garcia-Barbera N, et al. Platelet activation and neutrophil extracellular trap (NET) formation in immune thrombocytopenia: is there an association? Platelets. (2020) 31:906–12. doi: 10.1080/09537104.2019.1696456
7. Li J, van der Wal DE, Zhu G, Xu M, Yougbare I, Ma L, et al. Desialylation is a mechanism of Fc-independent platelet clearance and a therapeutic target in immune thrombocytopenia. Nat Commun. (2015) 6:7737. doi: 10.1038/ncomms8737
8. Rodeghiero F. Is ITP a thrombophilic disorder? Am J Hematol. (2016) 91:39–45. doi: 10.1002/ajh.24234
9. Greinacher A, Selleng K, Palankar R, Wesche J, Handtke S, Wolff M, et al. Insights in ChAdOx1 nCoV-19 vaccine-induced immune thrombotic thrombocytopenia. Blood. (2021) 138:2256–68. doi: 10.1182/blood.2021013231
10. Ostrowski SR, Søgaard OS, Tolstrup M, Stærke NB, Lundgren J, Østergaard L, et al. Inflammation and platelet activation after COVID-19 vaccines - possible mechanisms behind vaccine-induced immune thrombocytopenia and thrombosis. Front Immunol. (2021) 12:779453. doi: 10.3389/fimmu.2021.779453
11. Uzun G, Pelzl L, Singh A, Bakchoul T. Immune-mediated platelet activation in COVID-19 and vaccine-induced immune thrombotic thrombocytopenia. Front Immunol. (2022) 13:837629. doi: 10.3389/fimmu.2022.837629
12. Duttaroy AK. Role of gut microbiota and their metabolites on atherosclerosis, hypertension and human blood platelet function: a review. Nutrients. (2021) 13:144. doi: 10.3390/nu13010144
13. Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. (2016) 165:111–24. doi: 10.1016/j.cell.2016.02.011
14. Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM, et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell. (2020) 180:862–77.e22. doi: 10.1016/j.cell.2020.02.016
15. Vadaq N, Schirmer M, Tunjungputri RN, Vlamakis H, Chiriac C, Ardiansyah E, et al. Untargeted plasma metabolomics and gut microbiome profiling provide novel insights into the regulation of platelet reactivity in healthy individuals. Thromb Haemost. (2021). [Epub ahead of print]. doi: 10.1055/a-1541-3706
16. Zhang X, Zhang X, Tong F, Cai Y, Zhang Y, Song H, et al. Gut microbiota induces high platelet response in patients with ST segment elevation myocardial infarction after ticagrelor treatment. Elife. (2022) 11:e70240. doi: 10.7554/eLife.70240
17. Kazama I, Baba A, Endo Y, Toyama H, Ejima Y, Matsubara M, et al. Salicylate inhibits thrombopoiesis in rat megakaryocytes by changing the membrane micro-architecture. Cell Physiol Biochem. (2015) 35:2371–82. doi: 10.1159/000374039
18. Peng B, Geue S, Coman C, Münzer P, Kopczynski D, Has C, et al. Identification of key lipids critical for platelet activation by comprehensive analysis of the platelet lipidome. Blood. (2018) 132:e1–12. doi: 10.1182/blood-2017-12-822890
19. Ramström S. Arachidonic acid causes lysis of blood cells and ADP-dependent platelet activation responses in platelet function tests. Platelets. (2019) 30:1001–7. doi: 10.1080/09537104.2018.1557614
20. Paes AMA, Gaspar RS, Fuentes E, Wehinger S, Palomo I, Trostchansky A. Lipid metabolism and signaling in platelet function. Adv Exp Med Biol. (2019) 1127:97–115. doi: 10.1007/978-3-030-11488-6_7
21. Midgett C, Stitham J, Martin K, Hwa J. Prostacyclin receptor regulation–from transcription to trafficking. Curr Mol Med. (2011) 11:517–28. doi: 10.2174/156652411800615144
22. Stitham J, Midgett C, Martin KA, Hwa J. Prostacyclin: an inflammatory paradox. Front Pharmacol. (2011) 2:24. doi: 10.3389/fphar.2011.00024
23. Van Doren L, Nguyen N, Garzia C, Fletcher EK, Stevenson R, Jaramillo D, et al. Lipid receptor GPR31 (G-Protein-Coupled Receptor 31) regulates platelet reactivity and thrombosis without affecting hemostasis. Arterioscler Thromb Vasc Biol. (2021) 41:e33–45. doi: 10.1161/ATVBAHA.120.315154
24. Valiyaveettil M, Kar N, Ashraf MZ, Byzova TV, Febbraio M, Podrez EA. Oxidized high-density lipoprotein inhibits platelet activation and aggregation via scavenger receptor BI. Blood. (2008) 111:1962–71. doi: 10.1182/blood-2007-08-107813
25. Rodeghiero F, Ruggeri M. ITP and international guidelines: what do we know, what do we need? Presse Med. (2014) 43:e61–7. doi: 10.1016/j.lpm.2014.02.004
26. De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. (2013) 8:e76993. doi: 10.1371/journal.pone.0076993
27. Mu Q, Kirby J, Reilly CM, Luo XM. Leaky gut as a danger signal for autoimmune diseases. Front Immunol. (2017) 8:598. doi: 10.3389/fimmu.2017.00598
28. Manzo VE, Bhatt AS. The human microbiome in hematopoiesis and hematologic disorders. Blood. (2015) 126:311–8. doi: 10.1182/blood-2015-04-574392
29. Millstead J, Kamat A, Duffner U, Abdel-Mageed A, Freswick P, Dickens D, et al. WD repeat domain 1 (WDR1) deficiency presenting as a cause of infantile inflammatory bowel disease. J Pediatr Gastroenterol Nutr. (2020) 71:e113–7. doi: 10.1097/MPG.0000000000002826
30. Uzzan M, Galicier L, Gornet JM, Oksenhendler E, Fieschi C, Allez M, et al. Autoimmune cytopenias associated with inflammatory bowel diseases: insights from a multicenter retrospective cohort. Dig Liver Dis. (2017) 49:397–404. doi: 10.1016/j.dld.2016.12.006
31. Malnick SD, Oppenheim A, Melzer E. Immune thrombocytopenia caused by fecal microbial transplantation in a patient with severe recurrent Clostridium difficile infection. J Clin Gastroenterol. (2015) 49:888–9. doi: 10.1097/MCG.0000000000000404
32. Johnson BM, Gaudreau MC, Al-Gadban MM, Gudi R, Vasu C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol. (2015) 181:323–37. doi: 10.1111/cei.12609
33. Ruiz L, López P, Suárez A, Sánchez B, Margolles A. The role of gut microbiota in lupus: what we know in 2018? Expert Rev Clin Immunol. (2018) 14:787–92. doi: 10.1080/1744666X.2018.1519395
34. Zheng X, Wang D. The adenosine A2A receptor agonist accelerates bone healing and adjusts Treg/Th17 cell balance through interleukin 6. Biomed Res Int. (2020) 2020:2603873. doi: 10.1155/2020/2603873
35. Zhu XM, Shi YZ, Cheng M, Wang DF, Fan JF. Serum IL-6, IL-23 profile and Treg/Th17 peripheral cell populations in pediatric patients with inflammatory bowel disease. Pharmazie. (2017) 72:283–7. doi: 10.1691/ph.2017.6957
36. Zhong H, Bussel J, Yazdanbakhsh K. In vitro TNF blockade enhances ex vivo expansion of regulatory T cells in patients with immune thrombocytopenia. Br J Haematol. (2015) 168:274–83. doi: 10.1111/bjh.13126
37. Lin CH, Chen CC, Chiang HL, Liou JM, Chang CM, Lu TP, et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J Neuroinflammation. (2019) 16:129. doi: 10.1186/s12974-019-1528-y
38. Sun MF, Zhu YL, Zhou ZL, Jia XB, Xu YD, Yang Q, et al. Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: gut microbiota, glial reaction and TLR4/TNF-α signaling pathway. Brain Behav Immun. (2018) 70:48–60. doi: 10.1016/j.bbi.2018.02.005
39. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7. doi: 10.1126/science.1223813
40. Reddoch-Cardenas KM, Sharma U, Salgado CL, Cantu C, Darlington DN, Pidcoke HF, et al. Use of specialized pro-resolving mediators to alleviate cold platelet storage lesion. Transfusion. (2020) 60:S112–8. doi: 10.1111/trf.15750
41. Djuricic I, Calder PC. Beneficial outcomes of omega-6 and omega-3 polyunsaturated fatty acids on human health: an update for 2021. Nutrients. (2021) 13:2421. doi: 10.3390/nu13072421
42. Trostchansky A, Moore-Carrasco R, Fuentes E. Oxidative pathways of arachidonic acid as targets for regulation of platelet activation. Prostaglandins Other Lipid Mediat. (2019) 145:106382. doi: 10.1016/j.prostaglandins.2019.106382
43. Mao G, Songdej N, Voora D, Goldfinger LE, Del Carpio-Cano FE, Myers RA, et al. Transcription Factor RUNX1 regulates platelet PCTP (Phosphatidylcholine Transfer Protein): implications for cardiovascular events: differential effects of RUNX1 variants. Circulation. (2017) 136:927–39. doi: 10.1161/CIRCULATIONAHA.116.023711
44. Zhang Y, Wang J, Gao J, Guo M, Chen H, Liu J, et al. Jianpi Yiqi Shexue ameliorates immune thrombocytopenia related fatigue by regulating mitochondrial function. Ann Palliat Med. (2021) 10:156–68. doi: 10.21037/apm-20-2153
45. Kang MJ, Khanal T, Kim HG, Lee DH, Yeo HK, Lee YS, et al. Role of metabolism by human intestinal microflora in geniposide-induced toxicity in HepG2 cells. Arch Pharm Res. (2012) 35:733–8. doi: 10.1007/s12272-012-0418-y
46. Abdel-Hafez AA, Nakamura N, Hattori M. Biotransformation of phorbol by human intestinal bacteria. Chem Pharm Bull. (2002) 50:160–4. doi: 10.1248/cpb.50.160
47. Mills SW, Montgomery SH, Morck DW. Evaluation of the effects of short-chain fatty acids and extracellular pH on bovine neutrophil function in vitro. Am J Vet Res. (2006) 67:1901–7. doi: 10.2460/ajvr.67.11.1901
48. Bae EA, Shin JE, Kim DH. Metabolism of ginsenoside Re by human intestinal microflora and its estrogenic effect. Biol Pharm Bull. (2005) 28:1903–8. doi: 10.1248/bpb.28.1903
49. Cyong J, Matsumoto T, Arakawa K, Kiyohara H, Yamada H, Otsuka Y. Anti-Bacteroides fragilis substance from rhubarb. J Ethnopharmacol. (1987) 19:279–83. doi: 10.1016/0378-8741(87)90005-5
50. Zhao Q, Kretschmer N, Bauer R, Efferth T. Shikonin and its derivatives inhibit the epidermal growth factor receptor signaling and synergistically kill glioblastoma cells in combination with erlotinib. Int J Cancer. (2015) 137:1446–56. doi: 10.1002/ijc.29483
51. Zhou SS, Xu J, Zhu H, Wu J, Xu JD, Yan R, et al. Gut microbiota-involved mechanisms in enhancing systemic exposure of ginsenosides by coexisting polysaccharides in ginseng decoction. Sci Rep. (2016) 6:22474. doi: 10.1038/srep22474
52. Gao Y, Wang G, Wang T, Li G, Lin J, Sun L, et al. A 26-week 20(S)-ginsenoside Rg3 oral toxicity study in Beagle dogs. Regul Toxicol Pharmacol. (2020) 110:104522. doi: 10.1016/j.yrtph.2019.104522
53. Jin Y, Jung SY, Kim YJ, Lee DY, Min JW, Wang C, et al. Microbial ketonization of ginsenosides F1 and C-K by Lactobacillus brevis. Antonie Van Leeuwenhoek. (2014) 106:1215–21. doi: 10.1007/s10482-014-0291-4
54. Zhang C, Shao H, Li D, Xiao N, Tan Z. Role of tryptophan-metabolizing microbiota in mice diarrhea caused by Folium sennae extracts. BMC Microbiol. (2020) 20:185. doi: 10.1186/s12866-020-01864-x
55. Sun Z, Li J, Dai Y, Wang W, Shi R, Wang Z, et al. Indigo naturalis alleviates dextran sulfate sodium-induced colitis in rats via altering gut microbiota. Front Microbiol. (2020) 11:731. doi: 10.3389/fmicb.2020.00731
56. Qi Y, Chen L, Gao K, Shao Z, Huo X, Hua M, et al. Effects of Schisandra chinensis polysaccharides on rats with antibiotic-associated diarrhea. Int J Biol Macromol. (2019) 124:627–34. doi: 10.1016/j.ijbiomac.2018.11.250
57. Shao J, Liu Y, Wang H, Luo Y, Chen L. An integrated fecal microbiome and metabolomics in T2DM rats reveal antidiabetes effects from host-microbial metabolic axis of EtOAc extract from Sophora flavescens. Oxid Med Cell Longev. (2020) 2020:1805418. doi: 10.1155/2020/1805418
58. Liu CS, Liang X, Wei XH, Jin Z, Chen FL, Tang QF, et al. Gegen Qinlian decoction treats diarrhea in piglets by modulating gut microbiota and short-chain fatty acids. Front Microbiol. (2019) 10:825. doi: 10.3389/fmicb.2019.00825
59. Xiao S, Liu C, Chen M, Zou J, Zhang Z, Cui X, et al. Scutellariae radix and coptidis rhizoma ameliorate glycolipid metabolism of type 2 diabetic rats by modulating gut microbiota and its metabolites. Appl Microbiol Biotechnol. (2020) 104:303–17. doi: 10.1007/s00253-019-10174-w
60. Wei X, Tao J, Xiao S, Jiang S, Shang E, Zhu Z, et al. Xiexin Tang improves the symptom of type 2 diabetic rats by modulation of the gut microbiota. Sci Rep. (2018) 8:3685. doi: 10.1038/s41598-018-22094-2
61. Zhu LQ, Zhang L, Zhang J, Chang GL, Liu G, Yu DD, et al. Evodiamine inhibits high-fat diet-induced colitis-associated cancer in mice through regulating the gut microbiota. J Integr Med. (2021) 19:56–5. doi: 10.1016/j.joim.2020.11.001
Keywords: immune thrombocytopenia, gut microbiota, fatty metabolism, 16S rRNA, cytokines
Citation: Yu X, Zheng Q, He Y, Yu D, Chang G, Chen C, Bi L, Lv J, Zhao M, Lin X and Zhu L (2022) Associations of Gut Microbiota and Fatty Metabolism With Immune Thrombocytopenia. Front. Med. 9:810612. doi: 10.3389/fmed.2022.810612
Received: 07 November 2021; Accepted: 27 April 2022;
Published: 19 May 2022.
Edited by:
Nora V. Butta, University Hospital La Paz Research Institute (IdiPAZ), SpainReviewed by:
Chyntia Olivia Maurine Jasirwan, Universitas Indonesia, IndonesiaCopyright © 2022 Yu, Zheng, He, Yu, Chang, Chen, Bi, Lv, Zhao, Lin and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Misheng Zhao, emhhb21pc2hlbmdAMTI2LmNvbQ==; Xiangyang Lin, bGlueHkxOTY4QDEyNi5jb20=; Liqing Zhu, emh1bGlxaW5nQHd6aG9zcGl0YWwuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.