- 1Department of Dermatology, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China
- 2Department of Dermatology, Shishi Municipal General Hospital, Quanzhou, China
- 3Central Laboratory, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China
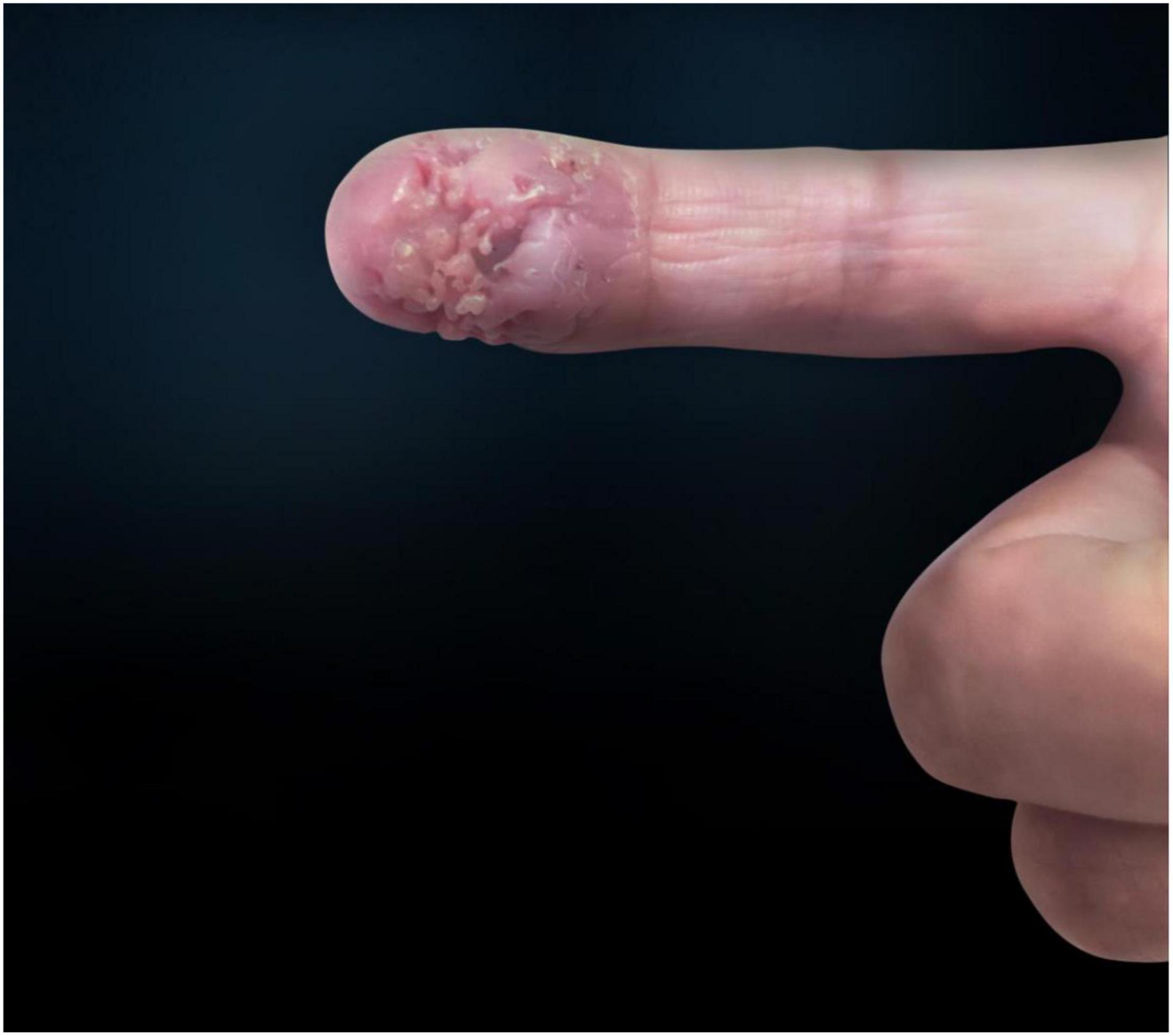
Proteus mirabilis, the most widespread species of all Proteus spp. bacteria, is proven to be one of the most universal pathogens in chronic wounds. In this case, a woman in her 40s consulted a physician about an asymptomatic ulceration with a stalactite appearance at the distal end of the index finger after she was exposed to a needle when vaccinating chickens. The patient did not response to ceftazidime. Physical examination revealed a well-demarcated violescent ulceration with a stalactite appearance at the distal end of the index finger. A biopsy of the lesion showed dense infiltration of multinucleated giant cells, histiocytes, and lymphocytes in the dermis. The result of metagenomics next-generation sequencing (NGS) showed 306 unique sequence reads of P. mirabilis, covering 33.49% of the nucleotide sequences. The pathogen was identified as P. mirabilis, which was resistant to ceftazidime. The patient was treated with ciprofloxacin hydrochloride and improved considerably. This case reported a distinctive cutaneous lesion of P. mirabilis on human infection and showed a successful use of NGS in P. mirabilis.
Introduction
Proteus mirabilis is the most widespread species of Proteus spp. bacteria (1). Culture, biochemical tests, and multiplex PCR can be used for diagnosis of P. mirabilis (2–4). Metagenomics next-generation sequencing (NGS) has the potential to identify and quantify new or unexpected pathogens in challenging situations such as culture- and PCR-negative results (5–8).
Case Report
A 47-year-old woman presented with an asymptomatic ulceration with a stalactite appearance at the distal of end of the index finger for 30 days. She reported no dyspnea, fever, arthralgias, or other systemic symptoms. Prior to admission to our department, the patient received ceftazidime 2 mg twice daily, but her lesion worsened. One month earlier, the patient had a needle-stick injury while vaccinating chickens. However, she did not report any exposure to the vaccine. Physical examination revealed a well-demarcated violescent ulceration with a stalactite appearance at the distal end of the index finger (Figure 1). Biopsy of the lesion showed a dense infiltration of multinucleated giant cells, histiocytes, and lymphocytes in the dermis (Figure 2). A culture performed on the cutaneous lesion was negative in this case. Metagenomics NGS of the specimen was performed (Supplementary Methods). Four negative and positive control samples were subjected to the same procedures. NGS result showed that P. mirabilis had 306 unique sequence reads, covering 33.49% of the nucleotide sequences (Figure 3). The pathogen was identified as P. mirabilis, which was resistant to ceftazidime. The patient was treated with ciprofloxacin hydrochloride (at a dose of 1.5 g three times daily for 20 days), and her lesion showed substantial improvement (Supplementary Figure 1). After 2 weeks, dermatology life quality index (DLQI) decreased rapidly from 15 to 2.
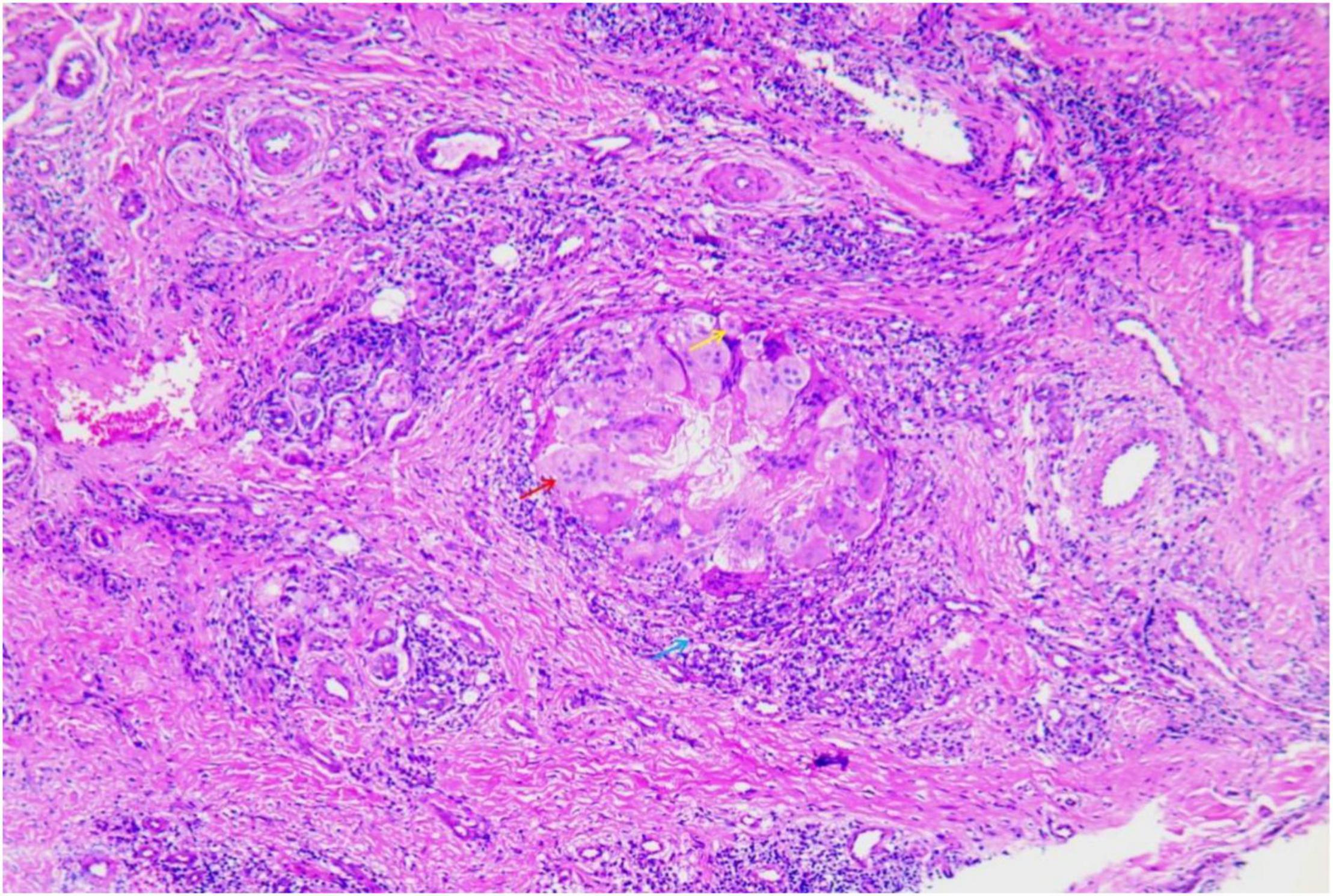
Figure 2. Biopsy of the lesion showed dense infiltration of multinuclear giant cells (red arrow), histiocytes (yellow arrow), and lymphocytes (blue arrow) in the dermis (hematoxylin and eosin stain, ×400).
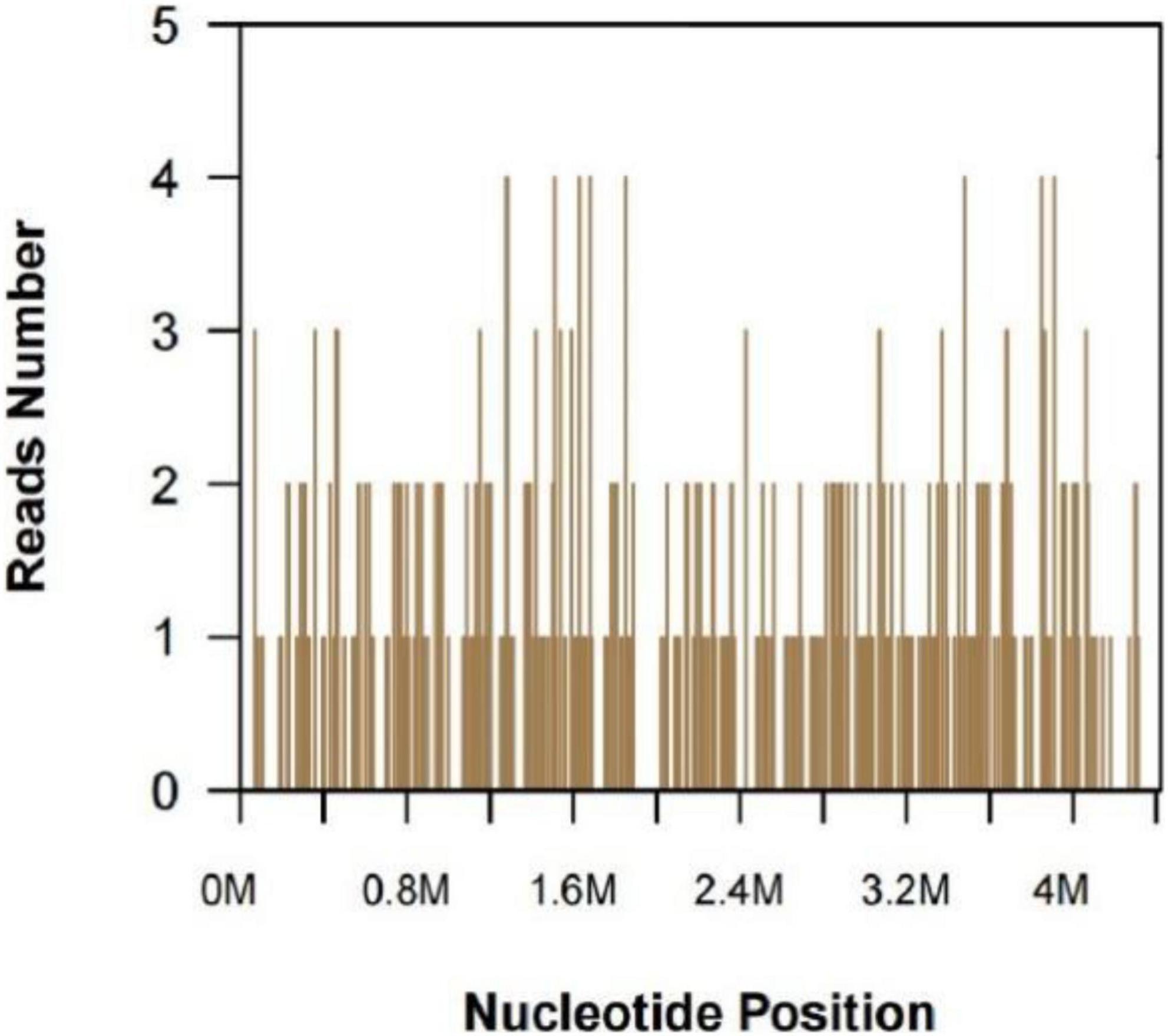
Figure 3. Next-generation sequencing and metagenomics analysis. Proteus mirabilis specific reads and its nucleotide position along the Proteus mirabilis genome.
Discussion
Proteus mirabilis is one of the most universal pathogens in chronic trauma, accounting for 8.0% (9). Humans were infected with P. mirabilis from consumption and slaughter of wild and domestic animals (mammals, birds, reptiles, amphibians, insects, and seafood) (10). The phenotypic and virulence gene characteristics in human and chicken isolates of P. mirabilis are similar (11). Cellulitis has been reported in broiler chickens (12). However, features of skin lesions in human P. mirabilis infection are rarely described. Culture is an important diagnostic tool for P. mirabilis, which has a characteristic swarming motility and distinct fishy smell (2). There are also both traditional and kit-based biochemical tests in addition to molecular-based multiplex PCR, which can be used for diagnosis of P. mirabilis (3).
In this case, the patient had a history of a needle-stick injury and a specific exanthem of asymptomatic ulceration with stalactite appearance. The antibiotic (ceftazidime) was ineffective, and the culture was negative. P. mirabilis may be in a viable but non-culturable (VBNC) state. It was a state of bacteria where bacteria under certain conditions of starvation or stress could enter a state of no division and, therefore, become non-culturable but in the same time keep their viability and sometimes their ability to cause infection (13). Thus, VBNC of P. mirabilis was a possible reason for non-culturability of the bacteria and may also explain the failure of ceftazidime treatment, which required actively dividing bacteria to performed their antimicrobial effect (14, 15).
We chose to perform mNGS to blindly detect the causative agent, as PCR required multiple tests. Besides, NGS has the potential to predict antibiotic resistance. Many resistant genes coding beta-lactamases were detected in P. mirabilis, including TEM genes, VEB-1 genes, OXA genes, CTX-M genes and CIT genes, with the TEM genes being the most common resistant gene (16). In conclusion, this case reported a characteristic cutaneous lesion of P. mirabilis after human infection and showed the successful use of NGS in P. mirabilis. Further studies need to enlarge the sample size to support the clinical utility of NGS metagenomic testing.
Data Availability Statement
The datasets presented in this study can be found in the online repository at: https://www.ncbi.nlm.nih.gov/guide under accession number PRJNA813626.
Ethics Statement
The studies involving human participants were reviewed and approved by the Medical Technology Clinical Application Ethics Committee of The First Affiliated Hospital of Fujian Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
LZ, YD, CJ, and DC conceptualized and designed the study. LZ and YD wrote the manuscript. TG, BC, CJ, and DC revised the manuscript critically for important intellectual content. ML, QX, and TL collected clinical pictures and analyzed the data. All authors have read and approved the final manuscript.
Funding
This work was supported by grants from the Natural Science Foundation of Fujian Province (No. 2020J02053).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all study participants involved in facilitating and running this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.801086/full#supplementary-material
Supplementary Figure 1 | Data of therapeutic interventions.
References
1. Lin MF, Liou ML, Kuo CH, Lin YY, Chen JY, Kuo HY. Antimicrobial susceptibility and molecular epidemiology of Proteus mirabilis isolates from three hospitals in Northern Taiwan. Microb Drug Resist. (2019) 25:1338–46. doi: 10.1089/mdr.2019.0066
2. Jamil RT, Foris LA, Snowden J. Proteus Mirabilis Infections. StatPearls. Treasure Island, FL: StatPearls Publishing. Copyright © 2022, StatPearls Publishing LLC (2022).
3. Yang R, Xu G, Wang X, Qing Z, Fu L. Establishment and application of a dual TaqMan Real-Time PCR method for Proteus mirabilis and proteus vulgaris. Pol J Microbiol. (2020) 69:293–300. doi: 10.33073/pjm-2020-032
4. Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, et al. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother. (2017) 61:e1697–1616. doi: 10.1128/aac.01697-16
5. Michel C, Raimo M, Lazarevic V, Gaïa N, Leduc N, Knoop C, et al. Case report: about a case of hyperammonemia syndrome following lung transplantation: could metagenomic next-generation sequencing improve the clinical management? Front Med (Lausanne). (2021) 8:684040. doi: 10.3389/fmed.2021.684040
6. Kolb M, Lazarevic V, Emonet S, Calmy A, Girard M, Gaïa N, et al. Next-generation sequencing for the diagnosis of challenging culture-negative endocarditis. Front Med (Lausanne). (2019) 6:203. doi: 10.3389/fmed.2019.00203
7. Imai A, Gotoh K, Asano Y, Yamada N, Motooka D, Fukushima M, et al. Comprehensive metagenomic approach for detecting causative microorganisms in culture-negative infective endocarditis. Int J Cardiol. (2014) 172:e288–9. doi: 10.1016/j.ijcard.2013.12.197
8. Fukui Y, Aoki K, Okuma S, Sato T, Ishii Y, Tateda K. Metagenomic analysis for detecting pathogens in culture-negative infective endocarditis. J Infect Chemother. (2015) 21:882–4. doi: 10.1016/j.jiac.2015.08.007
9. Guan H, Dong W, Lu Y, Jiang M, Zhang D, Aobuliaximu Y, et al. Distribution and antibiotic resistance patterns of pathogenic bacteria in patients with chronic cutaneous wounds in China. Front Med (Lausanne). (2021) 8:609584. doi: 10.3389/fmed.2021.609584
10. Drzewiecka D. Significance and roles of proteus spp. Bacteria in natural environments. Microb Ecol. (2016) 72:741–58. doi: 10.1007/s00248-015-0720-6
11. Barbour EK, Hajj ZG, Hamadeh S, Shaib HA, Farran MT, Araj G, et al. Comparison of phenotypic and virulence genes characteristics in human and chicken isolates of Proteus mirabilis. Pathog Glob Health. (2012) 106:352–7. doi: 10.1179/2047773212y.0000000042
12. Sanches MS, Baptista AAS, de Souza M, Menck-Costa MF, Justino L, Nishio EK, et al. Proteus mirabilis causing cellulitis in broiler chickens. Braz J Microbiol. (2020) 51:1353–62. doi: 10.1007/s42770-020-00240-1
13. Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. (2014) 5:258. doi: 10.3389/fmicb.2014.00258
14. Wilks SA, Koerfer VV, Prieto JA, Fader M, Keevil CW. Biofilm development on urinary catheters promotes the appearance of viable but nonculturable bacteria. mBio. (2021) 12:e3584–3520. doi: 10.1128/mBio.03584-20
15. Wasfi R, Abdellatif GR, Elshishtawy HM, Ashour HM. First-time characterization of viable but non-culturable Proteus mirabilis: induction and resuscitation. J Cell Mol Med. (2020) 24:2791–801. doi: 10.1111/jcmm.15031
Keywords: Proteus mirabilis, cutaneous exanthem, next generation sequencing, Proteus spp. bacteria, human infection
Citation: Zhang L, Dai Y, Lin M, Xu Q, Lin T, Gong T, Cheng B, Ji C and Cai D (2022) Case Report: An Ulceration With a Stalactite Appearance on the Index Finger. Front. Med. 9:801086. doi: 10.3389/fmed.2022.801086
Received: 24 October 2021; Accepted: 09 March 2022;
Published: 18 April 2022.
Edited by:
Li Ang, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Ammar Husami, Cincinnati Children’s Hospital Medical Center, United StatesReham Wasfi, October University for Modern Sciences and Arts, Egypt
Copyright © 2022 Zhang, Dai, Lin, Xu, Lin, Gong, Cheng, Ji and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Ji, amljaGFvZnlAZmptdS5lZHUuY24=; Donghua Cai, Y2FpZGhkb2N0b3JAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Liangliang Zhang1†
Liangliang Zhang1† Qiuyun Xu
Qiuyun Xu Chao Ji
Chao Ji