- 1Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education, Fuzhou, China
- 2Fujian Key Laboratory of Tumor Microbiology, Department of Medical Microbiology, Fujian Medical University, Fuzhou, China
- 3Fujian Medical University Union Hospital, Fuzhou, China
Helicobacter pylori causes gastric infections in more than half of the world's population. The bacterium's survival in the stomach is mediated by the abundant production of urease to enable acid acclimation. In this study, our transcriptomic analysis demonstrated that the expression of urease structural proteins, UreA and UreB, is induced by the autoinducer AI-2 in H. pylori. We also found that the orphan response regulator HP1021 is downregulated by AI-2, resulting in the induction of urease expression. HP1021 represses the expression of urease by directly binding to the promoter region of ureAB, ranging from −47 to +3 with respect to the transcriptional start site. The study findings suggest that quorum sensing via AI-2 enhances acid acclimation when bacterial density increases, and might enable bacterial dispersal to other sites when entering gastric acid.
Introduction
Helicobacter pylori is a microaerobic, gram-negative, gastric pathogen that colonizes more than half of the world's population (1, 2). Infection with H. pylori is closely related to the onset of many gastrointestinal diseases, including chronic atrophic gastritis, peptic ulcer, gastric cancer, and gastric mucosa-associated lymphoma (3). Many virulence factors are involved in the pathogenesis of H. pylori. For example, flagella enable the bacteria to penetrate the gastric mucosa and migrate to gastric epithelial cells (4, 5). Additionally, several outer membrane proteins, such as BabA, SabA, AlpA, AlpB, HopZ, HopQ, and OipA, are involved in bacterium-host interactions that facilitate bacterial adhesion to gastric epithelial cells (6–11). Further, H. pylori synthesizes and delivers cytotoxin-associated gene A (CagA) protein to gastric epithelial cells through a type IV secretion system (T4SS), which activates the signal transduction pathways involved in the development of gastric cancer (12). T4SS is critical to activating the proinflammatory response via delivery of ADP-heptose, an intermediate metabolite of lipopolysaccharide, to the host cell (13). Finally, H. pylori secretes another major toxin, vacuolating cytotoxin A (VacA), which enhances colonization by inducing apoptosis, autophagy, and cell death, as well as disruption of the cytoskeletal architecture in human gastric adenocarcinoma (AGS) cells (14–16).
Survival of H. pylori under harsh acidic conditions in the stomach is critical, H. pylori has developed a unique acid acclimation mechanism through the production of urease and regulation of its activity. Urease is a 550 kDa, multimeric protein that can hydrolyze urea to ammonia and carbonic acid (17). The urease gene cluster comprises two operons: ureAB and ureIEFGH. The ureAB operon encodes the two structural subunits of urease: UreA (26.5 kDa) and UreB (60.3 kDa). Encoded by the ureIEFGH operon, UreE, UreF, UreG, and UreH are four accessory proteins involved in facilitating nickel transfer and insertion into apo-urease (18), while UreI is an inner membrane protein that forms a proton-gated urea channel to increase the influx of urea in acidic conditions (19). After urea enters the cytoplasm, it is hydrolyzed by urease to generate ammonia, maintaining cytoplasmic pH homeostasis (20, 21).
Urease is abundant in H. pylori, accounting for ~8% of total protein, and is critical for the survival and growth of H. pylori in acidic gastric environments (22). Indeed, urease-deficient H. pylori are unable to achieve successful colonization in gastric epithelial cells (23–25). Acidic pH induces the expression of urease through the two-component system, ArsRS, which directly activates the expression of urease (26). A cis-encoded antisense sRNA, 5' ureB-sRNA downregulates ureAB transcription by enhancing transcriptional termination of UreAB mRNA (27, 28). Besides, NikR directly activates the transcription of ureAB by binding to the promoter region of ureAB in the presence of nickel (29). However, the regulation of urease expression by other factors remains to be elucidated.
Quorum sensing is a cell-to-cell communication mechanism utilized by bacteria to coordinate group behaviors, including bioluminescence production, biofilm formation, bacterial competence, toxin production, and motility (30–35). Quorum sensing involves the secretion and accumulation of extracellular chemical signals (named autoinducers) as bacterial density increases, recognition of the autoinducers, signal transduction, and regulation of bacterial behavior. Gram-negative bacteria secrete and recognize autoinducers such as N-acylated homoserine lactone (AHL), which is the most common class of autoinducer (36, 37). Another autoinducer present in most gram-negative bacteria, AI-2, is derived from 4,5-dihydroxy-2,3-pentanedione (DPD), which is synthesized by LuxS and has been proposed to be involved in interspecies communication. LuxS is reportedly responsible for the synthesis of AI-2 in H. pylori (38–40). Studies have shown that AI-2 serves as a chemical repellent and also participates in the regulation of H. pylori chemotaxis and motility (41–43). Further, AI-2 inhibits biofilm formation and promotes bacterial dispersal in H. pylori (44, 45). We recently reported that AI-2 inhibits CagA expression and bacterial adhesion, thus attenuating the H. pylori-induced inflammatory response in gastric epithelial cells (46). Taken together, these results suggest that AI-2 plays an important role in the regulation of H. pylori pathogenesis.
Therefore, the goal of the current study was to investigate the role of AI-2 in H. pylori gene regulation, thus determining the underlying mechanism that regulates urease expression and further elucidating the factors responsible for H. pylori virulence.
Materials and Methods
Bacterial Strains and Growth Conditions
H. pylori strain 26695 and its isogenic mutants Δhp1021, Δfur, Δhp0564, ΔhrcA, ΔrpoN, and ΔluxSΔhp1021 were routinely maintained on Columbia blood agar plates supplemented with 5% sheep blood at 37 °C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2). For liquid culture, H. pylori strains were cultured in Brucella broth (BD Biosciences, San Jose, CA, USA) supplemented with 10% fetal bovine serum (PAN Seratech, Aidenbach, Germany) (BB+FBS) with gentle agitation (120 rpm). Kanamycin (5 μg/mL) or chloramphenicol (4 μg/mL) were added to H. pylori isogenic or complemented mutant strains when necessary. The Escherichia coli strain BL21 (DE3) (Novagen, Darmstadt, Germany) was cultured in Luria broth (LB) under aerobic conditions at 37°C with agitation (230 rpm).
Construction of H. pylori Strain 26695 Isogenic Mutants Δhp1021, Δfur, Δhp0564, ΔhrcA, ΔrpoN, and ΔluxSΔhp1021
Construction of H. pylori strain 26695 isogenic mutants was performed as previously described (46). Briefly, to construct the Δhp1021 mutant, primers hp1021-up-F and hp1021-up-R were used to amplify the upstream sequence of hp1021 using the genomic DNA of H. pylori strain 26695 as the template; primers hp1021-down-F and hp1021-down-R were used to amplify the downstream sequence of hp1021; and primers hp1021-aphA3-F and hp1021-aphA3-R were used to amplify the kanamycin resistance gene cassette (aphA-3). AphA-3 was amplified from pHel3 purchased from Addgene (Addgene plasmid 102961) (47). Then, pBluescript II SK (–) (Stratagene, La Jolla, CA, USA) was linearized by digestion with KpnI and HindIII (New England Biolabs, Ipswich, MA, USA). The upstream and downstream fragments and the kanamycin resistance gene cassette were ligated to lineariz pBluescript II SK(-) using the ClonExpress Ultra One Step Cloning Kit (Vazyme Biotech, Nanjing, China), generating pBluescript-hp1021KO, which was subsequently transfected into E. coli DH5α. Colonies were selected on LB agar plates containing kanamycin (25 μg/mL). After confirmation by colony PCR and sequencing, pBluescript-hp1021KO was extracted and transformed to H. pylori strain 26695 by electroporation. The hp1021 knockout mutant (Δhp1021) was selected on Columbia agar plates supplemented with kanamycin (5 μg/mL), and deletion of hp1021 was confirmed by DNA sequencing.
Construction of Δfur, Δhp0564, ΔhrcA, and ΔrpoN were performed as described above, with the following changes: primers fur-up-F, fur-up-R, fur-down-F, fur-down-R, fur-aphA3-F, and fur-aphA3-R were used to construct Δfur; primers hp0564-up-F, hp0564-up-R, hp0564-down-F, hp0564-down-R, hp0564-aphA3-F, and hp0564-aphA3-R were used to construct Δhp0564; primers hrcA-up-F, hrcA-up-R, hrcA-down-F, hrcA-down-R, hrcA-aphA3-F, and hrcA-aphA3-R were used to construct ΔhrcA; and primers rpoN-up-F, rpoN-up-R, rpoN-down-F, rpoN-down-R, rpoN-aphA3-F, and rpoN-aphA3-R were used to construct ΔrpoN.
Construction of ΔluxSΔhp1021 was performed by introducing upstream and downstream fragments of the hp1021 coding region flanked by a non-polar chloramphenicol acetyltransferase gene (CAT) amplified from pHel2 into ΔluxS. The plasmid pHel2 purchased from Addgene (Addgene plasmid 102960). Primers hp1021-up-F1 and hp1021-up-R1, and hp1021-down-F1 and hp1021-down-R1 were used to amplify the hp1021 upstream and downstream sequences, respectively. Primers hp1021-CAT-F and hp1021-CAT-R were used to amplify CAT. Allelic exchange of hp1021 was performed as described above, and Columbia agar plates with chloramphenicol (4 μg/mL) were used to select ΔluxSΔhp1021 mutant colonies.
Chromosomal Complementation of Hp1021 in Δhp1021 (Δhp1021/Hp1021C)
For complementation of HP1021 in Δhp1021, the hp1021 coding sequence ligated downstream of the ureAB promoter region was introduced to the chromosome in the intergenic region between hp0204 and hp0203, as previously reported (48, 49). Primers hp1021C-up-F, and hp1021C-up-R, and hp1021C-down-F, and hp1021C-down-R, were used to amplify the sequences upstream and downstream of the insertion locus, respectively. Primers hp1021C-PureA-F and hp1021C-PureA-R were used to amplify the ureA promoter; primers hp1021C-F and hp1021C-R were used to amplify the hp1021 coding sequence; and primers hp1021C-CAT-F and hp1021C-CAT-R were used to amplify CAT. Allelic exchange was performed as described above. Insertion of hp1021-CAT was selected with chloramphenicol (4 μg/mL) and confirmed by DNA sequencing. HP1021 expression was confirmed by evaluation of the HP1021 mRNA level.
RNA Isolation
H. pylori strains were cultured on Columbia agar plates for 3 days, followed by collection of cells, resuspension in Brucella broth with an initial OD600 of ~0.2, and culturing for another 8 h until the cells reached the exponential growth phase. To prepare the ΔluxS+AI-2 mutant, H. pylori ΔluxS was resuspended in Brucella broth and cultured for 4 h, then 80 μM AI-2 was added, followed by culturing for another 4 h. Bacterial cells were subsequently collected for total RNA isolation using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. RNA quality was confirmed by gel electrophoresis, and the RNA concentration was quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Reverse Transcription and Quantitative PCR (QPCR) Analysis
cDNA synthesis was performed using the HiScript Q RT SuperMix for qPCR (Vazyme Biotech) according to the manufacturer's instructions. Briefly, 1 μg isolated total RNA was treated with 4 × gDNA Wiper Mix to eliminate genomic DNA contamination. Reverse transcription was performed by adding 5 × HiScript qRT SuperMix II. To quantify the UreA, UreB, and HP1021 mRNA levels, qPCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech) with the Mx3000P QPCR system (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. PCR amplification consisted of an initial denaturation step for 60 s at 95°C, followed by a 40-cycle reaction: 95°C for 5 s, 60°C for 34 s. Gene expression levels were calculated using the 2−ΔΔCT method, while 16S rRNA was used as an endogenous control gene. qPCR was conducted in accordance with the guidelines (50). Relative RNA levels were presented as gene expression levels of the target genes normalized to that of the wild type. The primers used for each target gene are listed in Supplementary Table 1.
Transcriptomic Analysis
Overnight cultures of H. pylori wild type or ΔluxS mutant were resuspended in Brucella broth with an initial OD600 of ~0.2 and cultured for 8 h until the log phase of growth. RNA samples were prepared using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Four samples containing two biological replicates were included in the transcriptomic analysis. RNA quality was verified using the 2100 Bioanalyzer (Agilent Technologies). All RNA sequencing (RNAseq) and alignment procedures were performed by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The reads were mapped against the reference genome of H. pylori strain 26695 (51). The relative RNA level was measured in reads per kilobase per million mapped reads (RPKM). Genes were considered LuxS-regulated genes when |log2fold change| > 1.0, while the P-value < 0.05.
SDS-PAGE and Western Blotting
For SDS-PAGE, H. pylori strains were cultured as described above, cells were collected, and bacterial lysates were prepared by ultrasonication. Cellular debris was removed by centrifugation, and the protein concentration was quantified using the BCA Protein Assay Kit (Beyotime Biotech, Shanghai, China). After boiling, 10 μg cell lysate for each sample plus a 180-kDa pre-stained protein marker (Vazyme) was resolved on a 10% SDS-PAGE gel and stained with Coomassie Blue Super Fast Staining Solution (Beyotime Biotech). Gels were analyzed using a GS-900 calibrated densitometer (Bio-Rad Laboratories, Hercules, CA, USA) to determine UreA protein levels.
For western blotting, proteins were transferred to a PVDF membrane (Millipore, Billerica, MA, USA) and blocked for 1 h with TBS-T buffer (50 mM Tris, 150 mM NaCl, 0.05% Tween-20) containing 5% BSA (Medix, Shanghai, China). Anti-UreB primary antibody (1:50,000; Abcam, Shanghai, China) was added and incubated overnight at 4°C. After washing with TBS-T buffer three times, the membranes were incubated with anti-rabbit IgG, HRP-linked secondary antibody (1:2,000; Cell Signaling, Danvers, MA, USA) for 1 h at room temperature. The membranes were washed three times with TBS-T, and UreB protein signals were detected using a WesternBright ECL Kit (Advansta Inc., San Jose, CA, USA) and visualized using the ImageQuant LAS 4000 mini system (GE Life Sciences, Piscataway, NJ, USA).
Urease Activity Assay
Urease activity of bacterial lysates were determined through measurement of ammonia production using phenol-hypochlorite methods. Briefly, bacteria were cultured in BB+FBS until logarithmic growth phase as described above, and were subsequently collected and resuspended in 300 μl HEPES buffer (pH 7.5). Cell lysates were obtained by sonication while the concentration was quantified using BCA Protein Assay Kit (Beyotime Biotech). For the determination of urease activity, 20 μl cell lysate was first incubated with the urea solution for 1 h at 37°C using urease activity detection kit (Solarbio, Beijing, China) according to the manufacturer's instructions. The phenol-hypochlorite reaction was then performed for 20 min at room temperature to detect the released ammonia. The absorbance at 630 nm was measured using an EnSightTM Multimode Plate Reader (PerkinElmer, Waltham, MA, USA). The urease activity (U/mg protein) are expressed as μg NH3/min/mg protein.
Expression and Purification of Strep-HP1021
For the expression of Strep-HP1021, the DNA fragment containing the coding region of hp1021 was amplified with primers hp1021CDS-F and hp1021CDS-R, and cloned into pASK-IBA7plus (IBA Lifesciences, Göttingen, Germany). The resulting expression vector, pASK-IBA7-HP1021, was subsequently transfected into the E. coli strain BL21 (DE3). Colonies were selected on LB agar plates containing ampicillin, and the presence of the expression vector was confirmed by colony PCR and DNA sequencing. For induction and purification of Strep-HP1021, overnight cultures of E. coli strain BL21 (DE3) carrying the expression vector were washed and resuspended in 250 mL fresh LB medium. Then, the bacteria were cultured at 37°C with agitation (250 rpm) until the exponential growth phase with an OD600 of ~0.6. Tetracycline (0.2 μg/mL) was added to induce protein expression. After 3 h, cells were harvested by centrifugation at 6,000 rpm for 10 min at 4 °C. Bacterial cell lysates were obtained by ultrasonication, and Strep-HP1021 was purified using Strep-Tactin Sepharose resin (IBA Lifesciences) according to the manufacturer's instructions. The purified Strep-HP1021 was dialyzed using Tube-O-DIALYZER (G-Biosciences, St. Louis, MO, USA) with washing buffer (100 mM Tris-Cl, 150 mM NaCl, 10 mM EDTA). The purity of Strep-HP1021 was assessed by SDS-PAGE followed by Coomassie blue staining, and the Strep-HP1021 concentration was measured using the BCA Protein Assay Kit (Beyotime Biotech).
Electrophoretic Mobility Shift Assay (EMSA)
The promoter region of ureAB was determined according to the transcriptional start site reported by Sharma et al. (52). A 252-bp DNA fragment containing the promoter region of ureA (ranging from −252 to −1 with respect to the start codon) was amplified by PureA-F and PureA-AF700-R, which was labeled at the 5'-end with Alexa Fluor 700 (AF700) (Thermo Fisher Scientific). EMSA was performed by incubating 12 fmol AF700-labeled DNA probe with increasing concentrations of Strep-HP1021 (0, 25, 50, 100, 200, 300 nM) in 20 μL reaction mixture (50 mM Tris-Cl, 250 mM KCl, and 5 mM DTT, pH 7.0) at 37°C for 30 min. For each sample, 100 ng/μL sheared salmon sperm DNA (Solarbio) were supplied. DNA probe without labeling was amplified with PureA-F and PureA-R and was used in excess (25-, 50-, and 100-fold concentrations) as a cold probe. The binding reaction was terminated by adding loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, and 40% dextrose) and the sample mixtures were resolved on a 4% native polyacrylamide gel. DNA bands were visualized using the Odyssey CLx imaging system (LI-COR Biosciences, Lincoln, NE, USA) with an excitation wavelength of 700 nm, and the binding between HP1021 and the AF700-labeled ureA promoter region was analyzed using Image Studio version 5.2 software (LI-COR Biosciences).
DNase I Footprinting Assay
To identify the specific binding site for HP1021, a DNase I footprinting assay was performed. The promoter region of ureA was first amplified using PureA-F and PureA-R, and the obtained PCR product was then ligated to the pLB-Simple Vector using the Lethal Based Simple Fast Cloning Kit (Tiangen Biotech, Beijing, China) to construct pLB-PureA. The promoter region of ureA was amplified using primers pLB-PureA-F and pLB-PureA-FAM-R, which was labeled at its 5'-end with 6-carboxyfluorescein (FAM) (Invitrogen). The FAM-labeled PCR product was then purified, and its concentration was quantified using a NanoDrop One spectrophotometer (Thermo Fisher Scientific). The DNase I footprinting assay was performed as previously described (53). Briefly, 50 ng probe was incubated with or without Strep-HP1021 (5 μg) in a total volume of 40 μL for 30 min at 37°C. For each sample, 100 ng/μL sheared salmon sperm DNA were supplied. Then, 10 μL solution containing 0.015 units DNase I (Promega, Madison, WI, USA) and 100 nmol CaCl2 was added, followed by incubation at 37°C for 1 min. The digestion was terminated by the addition of 140 μL stop solution (200 mM sodium acetate, 30 mM EDTA, and 0.15% SDS). Digested DNA was extracted with phenol/chloroform, precipitated with 70% ethanol, and finally dissolved in 10 μL HiDi formamide (Applied Biosystems, Foster City, CA, USA) with 1 μL GeneScan-LIZ 500 standard (Applied Biosystems). The samples were then analyzed by capillary electrophoresis using a 3730 DNA Analyzer (Applied Biosystems), while the HP1021 binding site was analyzed using Peak Scanner version 1.0 software (Applied Biosystems).
Site-Directed Mutagenesis
The HP1021 binding site on ureA promoter were mutated using Mut Express MultiS Fast Mutagenesis Kit V2 (Vazyme Biotech) according to the manufacturer's instructions. The plasmid pLB-PureA containing the ureA promoter region was used as template while primers pLB-PureAmt-F and pLB-PureAmt-R (listed in Supplementary Table 1) carrying the substituted nucleotides were used. The resulting plasmid pLB-PureAmt were confirmed by DNA sequencing. AF700-labled PureAmt was obtained by PCR amplification using pLB-PureAmt as template while PureA-F and PureA-AF700-R as primers.
Statistical Analysis
All results were analyzed using GraphPad Prism version 7.0 software (GraphPad Software, La Jolla, CA, USA). Student's t-test was used to evaluate the statistical significance of differences between transcript levels of target genes in two groups. A p-value < 0.05 was considered statistically significant.
Results
Identification of LuxS-Regulated Genes Through Transcriptomic Profiling
To elucidate the regulatory role of LuxS in H. pylori, transcriptomic analysis was performed on wild type and ΔluxS mutant strains cultured in Brucella broth for 8 h with an initial OD600 of 0.2. The raw RNA-seq data were deposited in the NCBI SRA database (BioProject: PRJNA603208). Comparative analysis revealed 187 differentially expressed genes (DEGs) between the wild type and ΔluxS mutant strains (Figure 1A), suggesting that LuxS affects the expression of numerous genes. Among them, 122 DEGs were upregulated in the wild type strain, while 65 DEGs were downregulated (listed in Table 1). Gene ontology (GO) analysis indicated that DEGs upregulated by LuxS were mainly involved in establishment of localization, aromatic compound biosynthetic process, peptide transport, cilium or flagellum-dependent cell motility, and amine metabolic process (Figure 1B). LuxS-downregulated genes were mainly involved in oxidation reduction activity, drug metabolic process, amino acid metabolic process, and ATP metabolic process (Figure 1C). We have also performed qPCR to confirmed the expression of these genes, same conclusion is obtained (listed in Supplementary Table 2).
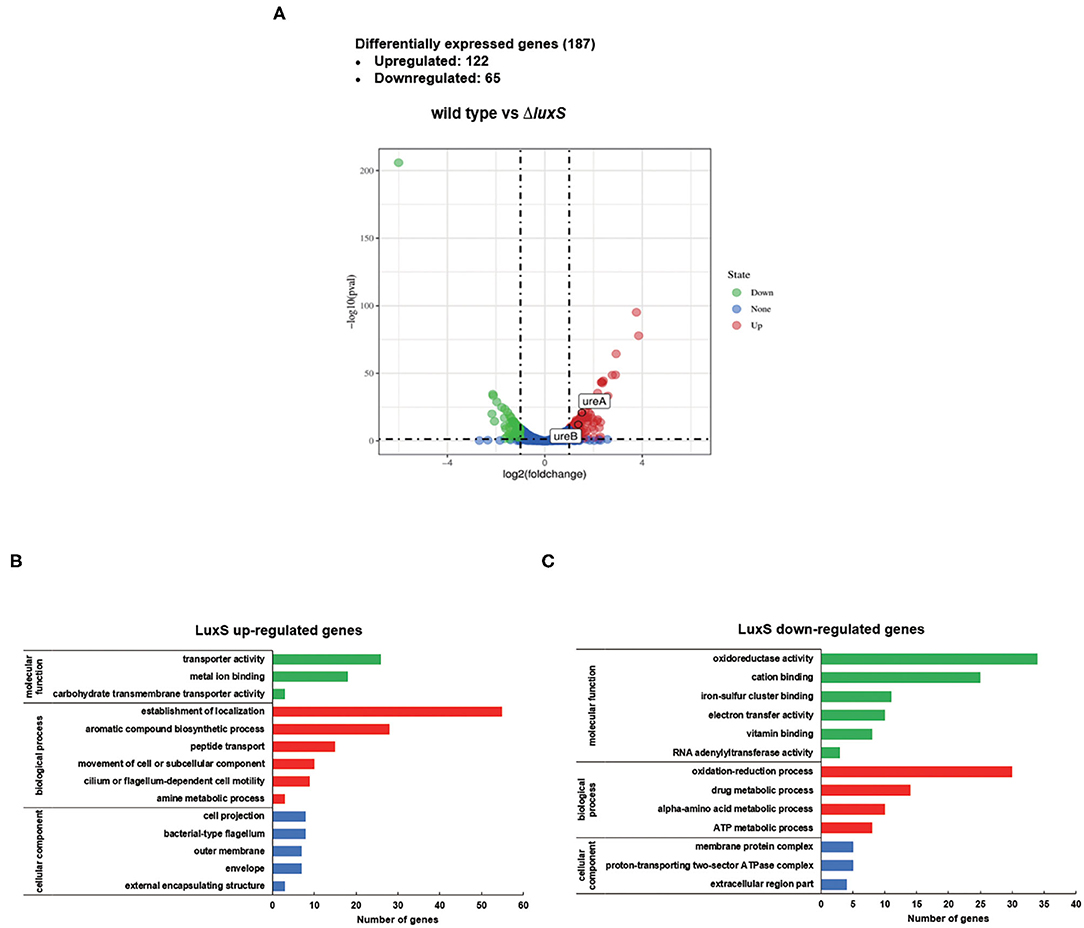
Figure 1. Differentially expressed genes (DEGs) regulated by LuxS identified by RNA sequencing. (A) Volcano plot comparing gene transcripts from wild type and ΔluxS mutant strains of H. pylori. The dashed transverse line indicates where P-value = 0.05 and the two vertical dotted lines represent |log2FoldChange| = 1. Genes of |log2FoldChange| > 1 and P-value < 0.05 are highlighted in red (significantly upregulated) and green (significantly downregulated). ureA and ureB genes are mapped in upregulated regions. Functional classification of (B) LuxS-upregulated DEGs and (C) LuxS-downregulated DEGs based on transcriptomic profiling. The green, red, and blue bars represent DEGs classified as molecular function, biological process, and cellular component categories, respectively.
AI-2 Induces UreAB Expression and Bacterial Urease Activity in H. pylori
From the transcriptomic analysis, we determined that UreA and UreB expression was lower in the ΔluxS mutant than in the wild type strain (Table 1), suggesting that quorum sensing might be important for acid acclimation in H. pylori. To confirm this hypothesis, we first investigated the expression of UreA and UreB in the wild type and ΔluxS mutant strains by qPCR (Figure 2A). The ΔluxS mutant strain displayed only half the mRNA expression levels of UreA and UreB as in the wild type strain, which concurred with the results of the transcriptomic analysis. We then complemented LuxS by introducing a luxS coding sequence downstream of the ureAB promoter region and verified UreA and UreB expression. The results demonstrated that LuxS complementation restored UreA and UreB expression levels in the ΔluxS mutant to those in the wild type strain (Supplementary Figure 1). Since LuxS is involved in both quorum sensing as an AI-2 synthase and in non-quorum sensing as part of the activated methyl cycle, we hypothesized that AI-2 might be responsible for activation of urease expression. To examine this possibility, chemical complementation of LuxS was performed by supplementing the ΔluxS mutant with AI-2 (ΔluxS+AI-2). The results revealed that supplementation with AI-2 restored UreA and UreB expression levels in the ΔluxS mutant to those in the wild type strain (Figure 2A), suggesting that LuxS induced urease expression through AI-2. We also performed western blotting to investigate the UreB expression regulated by AI-2, we found that protein level of UreB in ΔluxS was approximately half of the levels in wild type and ΔluxS+AI-2 (Figures 2B,C). To ensure that the decreased UreAB expression might lead to a lower bacterial urease activity, we harvested the cell lysates, and measured bacterial urease activity. The result showed that the urease activity was lower in ΔluxS compared to wild type and ΔluxS+AI-2 (Figure 2D). These results suggested that AI-2 induced the expression of UreAB, leading to an upregulated urease activity in H. pylori.
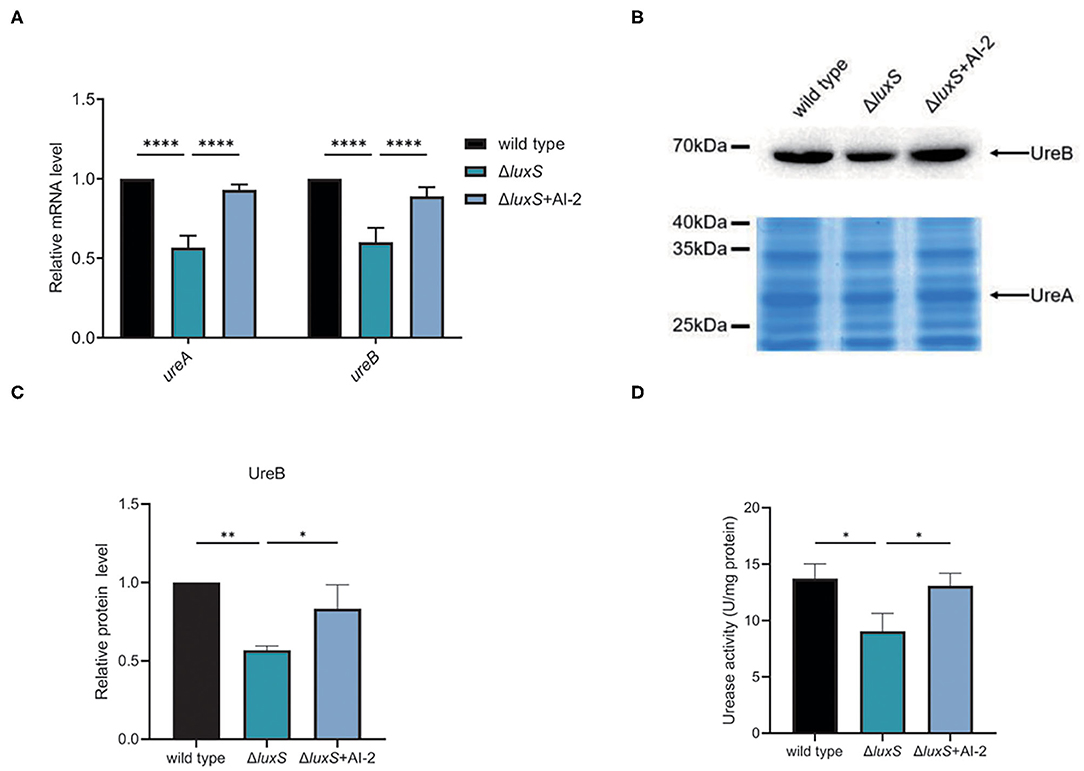
Figure 2. AI-2 regulated the expression of urease. (A) Transcriptional levels of UreA and UreB regulated by AI-2, quantified by qPCR. Relative mRNA levels represent mRNA levels of UreA and UreB in the mutant strains normalized to those in the wild type strain. (B) Protein levels of UreA and UreB regulated by AI-2. Expression of UreB was determined by western blotting, with the arrow indicating protein bands of UreB. Expression of UreA were detected by SDS-PAGE followed by staining with Coomassie brilliant blue, with the arrow indicating protein bands of UreA. (C) Densitometric analysis of UreB protein bands. Total protein bands stained with Coomassie brilliant blue were used as loading control. (D) Determination of urease activity in cell lysates. Values are shown as averages of three biological independent experiments and bars represent standard deviations. ****P < 0.0001, **P < 0.01, *P < 0.05.
AI-2 Downregulates Expression of Transcriptional Regulator HP1021, Which Depresses Urease Expression
Since AI-2 promoted the transcription of ureAB, we suspected that AI-2 might induce the expression of urease by regulating the expression of transcriptional regulators. H. pylori strain 26695 harbors 16 transcriptional regulators (54), which we examined in our transcriptomic analysis (Supplementary Table 3) and confirmed by qPCR (Figure 3A). The expression levels of HrcA (HP0111) and RpoN (HP0714) were lower in the ΔluxS mutant strain than those in the wild type strain, whereas those of HP1021, Fur (HP1027), and HP0564 were higher. We then determined which transcriptional regulator might be involved in the regulation of urease expression. We constructed Δhp1021, Δfur, ΔrpoN, ΔhrcA, and Δhp0564 mutant strains, and investigated the expression of UreA and UreB in the mutant and wild type strains (Figures 3B–D). Compared to the wild type strain, the expression of UreA and UreB was similar at the RNA and protein levels in the Δfur, ΔrpoN, ΔhrcA, and Δhp0564 mutant strains, suggesting that Fur, RpoN, HrcA, and HP0564 were not involved in the regulation of urease expression. However, the mRNA expression levels of UreA and UreB were 3.5 and 2.5 times higher while the protein level of UreB was 2.4 times higher in the Δhp1021 mutant strain than in the wild type strain, respectively. We also performed chromosomal complementation of HP1021 in the Δhp1021 mutant strain (Δhp1021/hp1021C), revealing that complementation with HP1021 restored the expression levels of UreA and UreB to those in the wild type strain (Figures 3E–G). The urease activity was confirmed, and were consistent with the higher urease activity in the Δhp1021 mutant strain compared to wild type strain and Δhp1021/hp1021C (Figure 3H). These results demonstrated that HP1021 represses the expression of urease, leading to decreased bacterial urease activity.
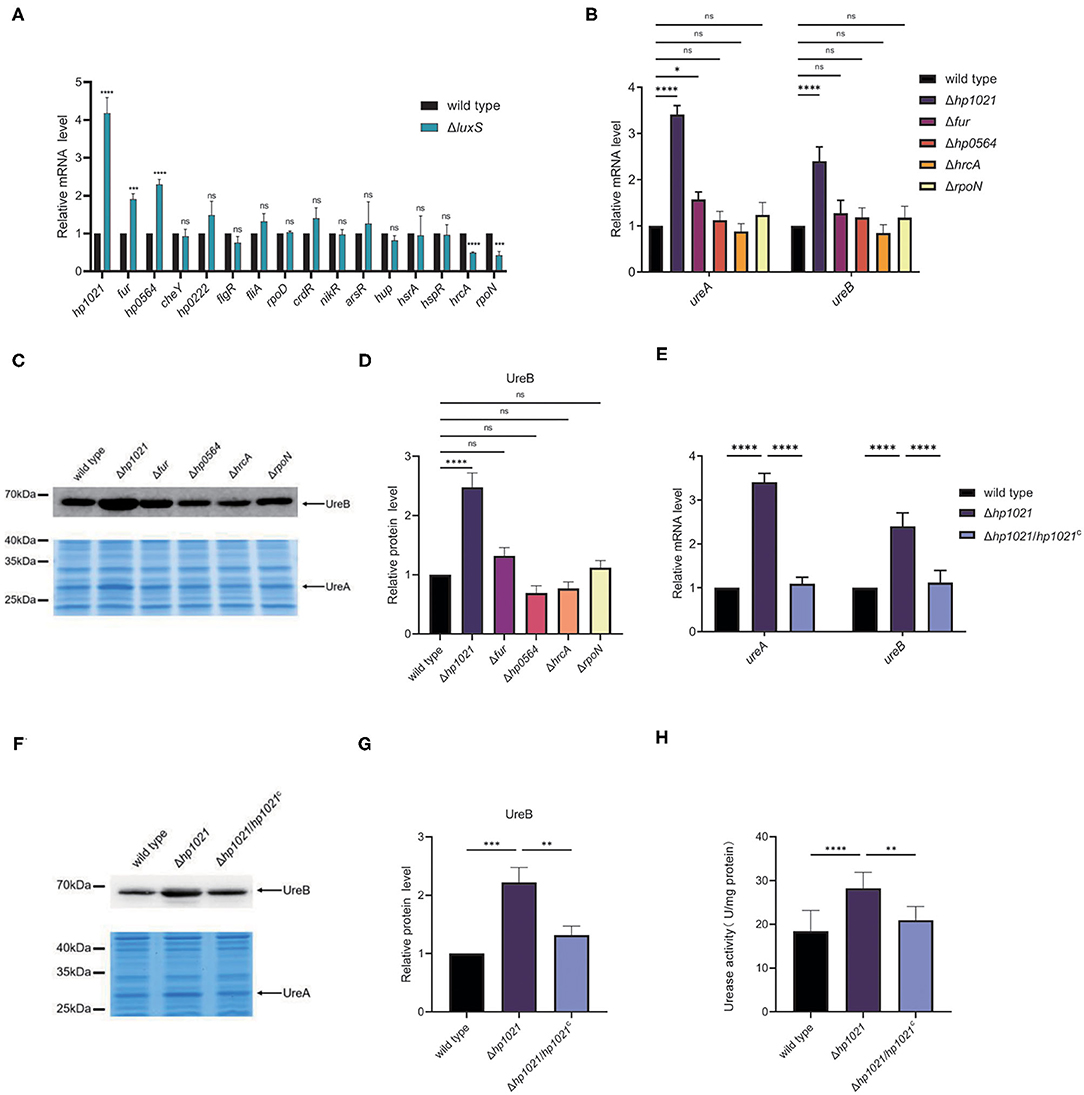
Figure 3. HP1021 inhibited urease expression while AI-2 downregulated HP1021 expression. (A) The mRNA expression levels of transcriptional regulators determined by qPCR. (B) The mRNA expression levels of UreA and UreB regulated by transcriptional regulators, as determined by qPCR. Relative RNA levels represent the mRNA levels in the mutant strains normalized to those in the wild type strain. Values represent averages of three biological independent experiments and bars represent standard deviations. (C) Expression of UreA and UreB proteins regulated by transcriptional regulators. UreB expression was detected by Western blotting, with the arrow indicating UreB protein bands. Expression of UreA was detected by SDS-PAGE followed by staining with Coomassie brilliant blue. The arrow indicates UreA protein bands. (D) Densitometric analysis of UreB protein bands. Total protein bands stained with Coomassie brilliant blue were used as loading control. (E) The mRNA expression of UreA and UreB regulated by HP1021. (F) Expression of UreA and UreB proteins regulated by HP1021. (G) Densitometric analysis of UreB protein bands. Total protein bands stained with Coomassie brilliant blue were used as loading control. (H) Determination of urease activity in cell lysates. Values represent averages of three biological independent experiments and bars represent standard deviations. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns, non-significant.
AI-2 Induced Urease Expression Through HP1021
Next, we investigated whether HP1021 was responsible for the induction of urease expression by AI-2. We first confirmed the expression of HP1021 in the wild type and ΔluxS mutant strains supplemented with or without AI-2. AI-2 supplementation resulted in decreased HP1021 expression, suggesting that HP1021 expression was downregulated by AI-2 (Figure 4A). Complementation of LuxS also restored the HP1021 expression in ΔluxS to the level similar with that in wild type strain (Supplementary Figure 1). Since HP1021 was the only transcriptional regulator regulated by AI-2 while manipulating the expression of urease, we investigated whether HP1021 was responsible for the regulation of urease expression by AI-2. For this purpose, we constructed the ΔluxSΔhp1021 mutant strain and compared the expression of UreA and UreB in the wild type and ΔluxS, Δhp1021, and ΔluxSΔhp1021 mutant strains (Figure 4B). The mRNA expression levels of UreA and UreB were similar in the Δhp1021 and ΔluxSΔhp1021 mutant strains, but higher than in the ΔluxS mutant strain, suggesting that HP1021 lies downstream of LuxS and plays an indispensable role in the regulation of urease expression by AI-2. We also confirmed these results at the protein level through western blotting and SDS-PAGE, and measured bacterial urease activity, supporting the conclusion that AI-2 induced urease expression and urease activity through HP1021 (Figures 4C–E).
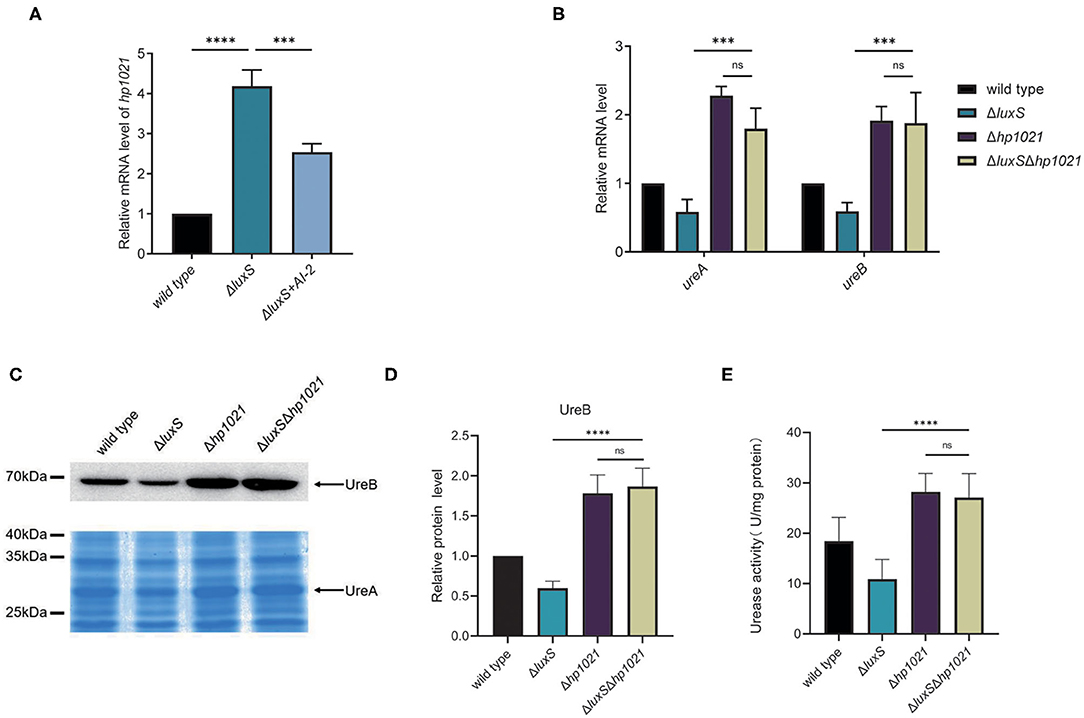
Figure 4. LuxS induced the expression of urease through HP1021. (A) mRNA expression levels of HP1021 regulated by LuxS and supplemented with AI-2, measured by qPCR. Relative mRNA levels represent mRNA levels of HP1021 in the mutant strain and mutant strain supplemented with AI-2 normalized to those in the wild type strain. (B) mRNA expression of UreA and UreB regulated by LuxS and HP1021. Relative mRNA levels represent mRNA levels of UreA and UreB in the mutant strains normalized to those in the wild type strain. (C) Expression of UreA and UreB proteins regulated by LuxS and HP1021. UreB expression was detected by western blotting, and protein bands are indicated with the arrow. UreA expression was measured by SDS-PAGE gel followed by staining with Coomassie brilliant blue. The arrow indicates UreA protein bands. (D) Densitometric analysis of UreB protein bands. Total protein bands stained with Coomassie brilliant blue were used as loading control. (E) Determination of urease activity in cell lysates. Values are the averages from three biological independent experiments and bars represent standard deviations. ****P < 0.0001, ***P < 0.001, ns, non-significant.
HP1021 Directly Binds to the Promoter Region of ureAB
HP1021 is known as an atypical orphan response regulator involved in the initiation of DNA replication in H. pylori by directly binding to the promoter region of oriC (55, 56). Therefore, we investigated whether HP1021 repressed urease expression by directly binding to the promoter region of ureAB. The expression of Strep-tagged HP1021 was induced through a TetR induction system and HP1021 was purified by Strep-tag affinity chromatography (Figure 5A). Promoter fragments of ureAB with 5'-labeled AF700 were then incubated with increasing amounts of purified HP1021. The results demonstrated that the DNA probes were fully bound by HP1021 when the concentration of HP1021 was increased to 200 nM (Figure 5B). To verify the specific affinity between HP1021 and the ureAB promoter fragments, excess non-labeled probe was used as a cold probe, with the results indicating that the cold probe relieved the AF700-labeled PureA bound by HP1021 (Figure 5B). These results suggested that HP1021 had a high binding affinity to the promoter region of ureAB, resulting in the repression of ureAB transcription.
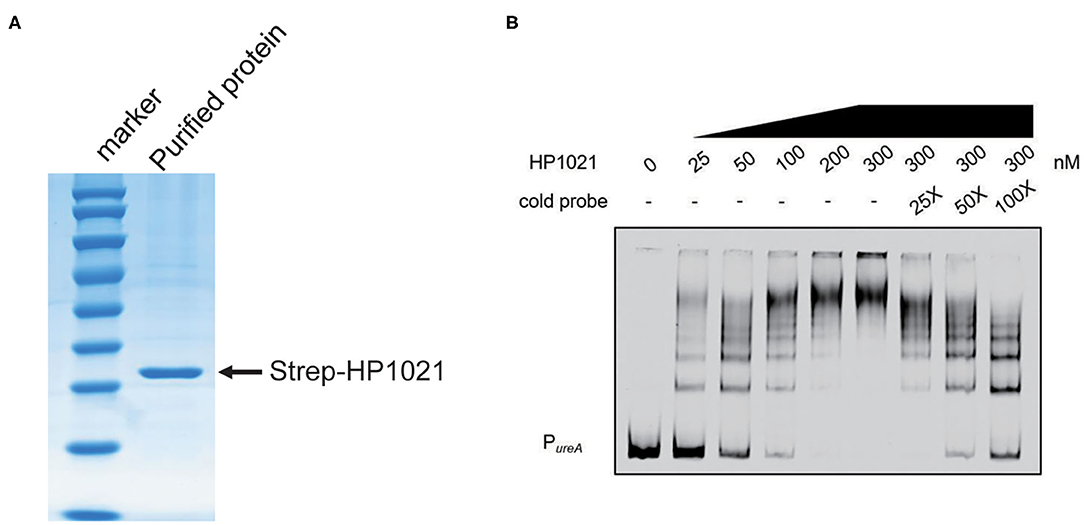
Figure 5. HP1021 bound directly to the ureA promoter region. (A) Coomassie-stained gel of purified Strep-tagged HP1021 protein, indicated by the arrow. (B) Electrophoretic mobility shift assay (EMSA) results of the interaction between HP1021 and the ureAB promoter. Fragments of the 300-bp ureAB promoter region labeled at the 5'-end with AF700 were incubated with increasing concentrations of Strep-HP1021 in the presence or absence of unlabeled ureAB promoter (cold probe). The arrow indicates bands representing 5' AF700-labeled ureAB probe bound by HP1021.
Identification of HP1021 Binding Site on ureAB Promoter Region
To elucidate the molecular mechanism by which HP1021 repressed urease expression, we performed a DNase I footprinting assay using dye primer sequencing to identify the binding site of HP1021 on the promoter region of ureAB (Figure 6A). The results demonstrated that HP1021 protected the promoter region of ureAB (5'-CGCTTCTGTTAATCTTAGTAAATCAAAACATTGCTACAATTACATCCAAC-3'), ranging from −47 to +3 with respect to the transcriptional start site, as previously reported (52), overlapping the −10 and −35 consensus sequences (Figure 6B). The HP1021 protected DNA sequence revealed three HP1021 binding boxes harboring five putative reported consensus HP1021 binding sequence in sense and antisense strands (57). These results suggested that multiple HP1021 proteins bound to the promoter region of ureAB, strongly inhibiting ureAB transcription by preventing RNA polymerase binding. We then prepared a mutant probe (PureAmt) through site-directed mutagenesis to replace nucleotides on the PureA (Figure 6C), and performed EMSA to investigate the interaction between HP1021 and PureAmt (Figure 6D). The results indicated that HP1021 did not interact with PureAmt, suggesting that the putative HP1021 binding motifs were critical to the interaction between HP1021 and the promoter region of ureAB.
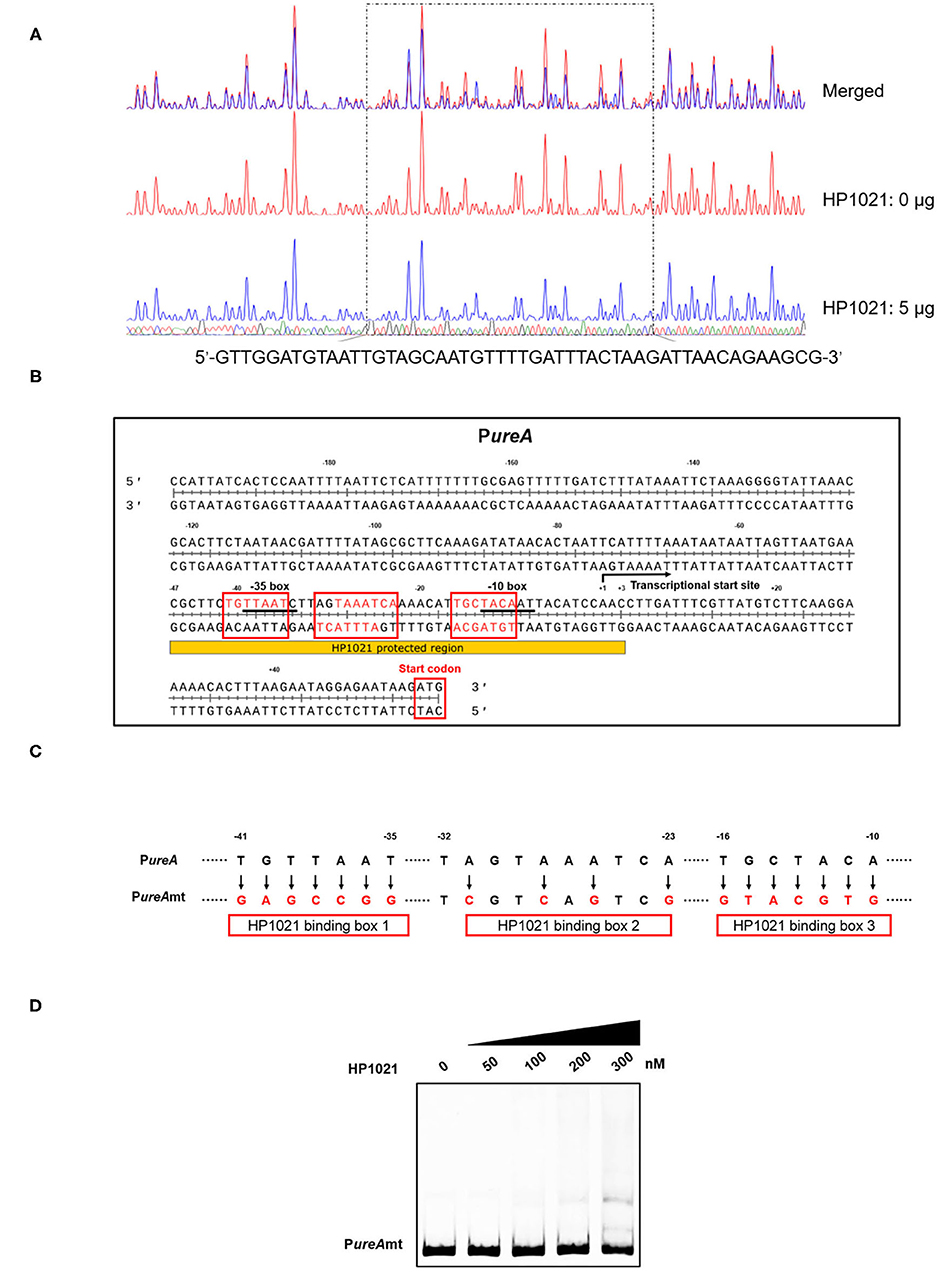
Figure 6. Determination of the binding site of HP1021 on the ureAB promoter region. (A) Dye primer-based DNase I footprinting assay of HP1021 and the ureAB promoter region. Protected fluorograms were obtained by incubating fragments of the ureAB promoter region with or without HP1021. The nucleotide sequence of the protected region is shown below. (B) Analysis of the HP1021 binding site and consensus HP1021 binding boxes. Sequence shown is PureA with numbers represent the position related to the transcriptional start site. The consensus −10 and −35 box are underlined, while the HP1021 protected sequence is indicated. The transcriptional start site was indicated with arrow; three HP1021 binding boxes were indicated with red box, while the putative consensus HP1021 binding sequences within were shown with red. (C) Mutagenesis of the HP1021 binding site. The replaced nucleotides are indicated with arrows and the mutation probe (PureAmt) is shown. (D) Electrophoretic mobility shift assay (EMSA) results of the interaction between HP1021 and PureAmt probe. PureAmt labeled at the 5'-end with AF700 was incubated with increasing concentrations of Strep-HP1021 protein. The arrow indicates PureAmt probe bands.
Discussion
H. pylori is a human pathogen that colonizes the stomach and can cause chronic infection, leading to the occurrence and development of gastric diseases such as gastric cancer (1, 2). More than 50% of the world's population is infected with H. pylori, and people tend to carry this pathogen without intervention. Indeed, ~80–90% of infected individuals exhibit no significant clinical symptoms (58). These findings suggest that H. pylori has successfully adapted to the gastric environment and is capable of avoiding the host immune defense system. However, the disease outcomes of H. pylori infection are strongly associated with factors such as host genetics, the environment, and diet; therefore, understanding the pathogenesis of H. pylori necessitates elucidating the mechanisms that regulate its virulence.
Urease plays a key role in acid acclimation for H. pylori, as it hydrolyzes intracellular urea to ammonia and carbon dioxide, buffering the cytoplasm under the acidic environment (59). Urease expression is abundant in H. pylori, but it varies in different environments. Urease expression can be directly or indirectly activated through the ArsRS two-component system in response to external acidic pH (26–28), and also by intracellular nickel concentrations through NikR (60). The results of these studies suggest that higher expression of urease is required under low pH and high nickel conditions. In the current study, we determined that the expression of urease was induced by the quorum-sensing signal molecule AI-2 (Figure 2). AI-2 reportedly activates the expression of flagella to increase the motility of H. pylori, thus enabling cells to disperse to other sites when bacterial density increases (41, 42, 61–64). Dispersal of bacteria is important for the survival of H. pylori since nutrients and space become limited when bacterial density increases. Previous studies reported that AI-2 also inhibits biofilm formation and promotes bacterial dispersal (44, 65). Therefore, we suspect that when AI-2 activates bacterial movement at high cell density, it also upregulates urease expression to combat acidic gastric juice.
In this study, we found that Fur, HP0564, HrcA, and RpoN were not involved in the regulation of urease expression (Figures 3B,C). Fur is a transcriptional regulator involved in iron homeostasis, suggesting that iron is not involved in the regulation of urease expression in H. pylori, which concurs with previously reported results that iron has no effect on urease expression (66). HP0564 is a transcriptional regulator with an unknown regulon, HrcA is involved in heat shock response, and RpoN is a sigma factor that is responsive to nutrient stress and regulates flagellar expression (67–69). However, other transcriptional regulators involved in urease expression warrant further investigation, as other environmental conditions might alter urease expression. Urease regulation has been studied in other bacteria, including by carbon storage regulator (CsrA) in Yersinia pseudotuberculosis and through Fur in Helicobacter hepaticus (70, 71). These results suggest that nutrient conditions are closely related to urease expression. Studies have shown that urease expression is not only involved in initial colonization, but is also critical for maintaining chronic infection (72). Therefore, the expression of urease is likely critical for the pathogenesis of H. pylori, although the environmental factors involved in its regulation need to be further explored.
Quorum sensing plays an important role in the regulation of bacterial behavior, such as the expression of virulence factors (73). The transcriptomic study indicated that LuxS represses the expression hydrogenase (HP0631-0632, HP0635) and iron-dependent superoxide dismutase (SodB), suggesting that LuxS is involved in iron related cellular activity. Expression of flagellar related genes including FlgL, MotA, FliS, MotB, FliD, FlaG and FlgK are also activated by LuxS (Supplementary Table 2), suggesting that LuxS not only activated the flagellar formation, but also controls movement of H. pylori. We also noticed that multidrug efflux pump components HP0607 and HP1082 are also repressed in ΔluxS, suggesting that quorum sensing might be also associated with drug resistance in H. pylori. These results suggest that LuxS plays important roles in regulating the growth and pathogenesis of H. pylori.
The molecular mechanism of quorum sensing has been studied in various pathogens, including Vibrio cholerae and Pseudomonas aeruginosa. In general, quorum sensing systems consist of autoinducer synthases, autoinducer signals, and response regulators that alter gene expression (74). In V. cholerae, the transcriptional regulator LuxO activates autoinducer signaling through the transcription of four quorum-sensing regulatory sRNAs, which regulates the quorum-sensing master regulator HapR (75, 76). In H. pylori, we found that HP1021 is a transcriptional regulator that responds to AI-2, and to the best of our knowledge, HP1021 is the first transcriptional regulator identified in H. pylori that plays an important role in quorum sensing regulation. HP1021 reportedly regulates the expression of numerous genes, including genes involved in Fe-S assembly, acetone metabolism, DNA replication initiation, and virulence gene cagA (56, 57). Quorum sensing in H. pylori might regulate these processes through HP1021 repressed by AI-2. In this study, we found that AI-2 cannot restore the expression of HP1021 in ΔluxS to the level similar to wild type (Figure 4), this might attribute to the concentration of exogenous supplementation of AI-2 decreased followed by bacterial growth, resulted in decreased AI-2 function as discussed in our previous work (46). However, according to our results, AI-2 is sufficient to restore urease expression in ΔluxS through HP1021 (Figure 2). In this study, we focused on the molecular mechanism by which AI-2 regulates urease expression, but the roles of other transcriptional regulators, such as Fur, RpoN, HrcA, and HP0564, remain to be elucidated.
Our study findings indicated that HP1021 is a negative regulator of urease expression by directly binding to the promoter region of ureAB. To our knowledge, HP1021 is the first transcriptional regulator that can directly exert a repressive effect on urease structural genes' expression in H. pylori. The EMSA results demonstrated that the probes were fully bound in the presence of 200 nM Strep-HP1021 (Figure 5B), suggesting high binding affinity between HP1021 and the ureA promoter. We also observed multiple band shifts, indicating that HP1021 bound to multiple sites on the ureA promoter. Studies have reported multiple band shifts for the binding of HP1021 to the promoter regions of oriC, hyuA, and katA, with 2–3 consensus HP1021 binding boxes identified on the probe fragments, while single band shifts were observed for gluP, as only one HP1021 binding site was found on its promoter region (56, 57). The results of the DNase I footprinting assay indicated that HP1021 protected DNA fragments ranging from −47 to +3 with respect to the transcriptional start site, overlapping the −10 and −35 consensus sequences. This suggests that binding of HP1021 inhibits ureAB transcription by preventing RNA polymerase binding. DNA analysis revealed the presence of three HP1021 binding boxes, suggesting that multiple HP1021 molecules bind simultaneously, thus repressing urease expression.
In conclusion, our study findings indicate that AI-2 upregulates urease expression at the mRNA and protein levels through repression of HP1021 in H. pylori. Further, HP1021 is the first transcriptional regulator identified in H. pylori that plays a role in quorum sensing circuits. HP1021 directly binds to the promoter region of ureAB, resulting in repression of urease expression. Our data suggest that AI-2 might play an important role in facilitating bacterial dispersal to a new niche by upregulation of bacterial urease expression and activity to neutralize intracellular pH when encountering gastric juice.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA603208.
Author Contributions
YW and FS designed the study. HY, XH, and XiaocZ performed the experiments. XX and XiaoyZ analyzed the data. YW and HY wrote the manuscript. All authors have read and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81701980 and 82072316), the Natural Science Foundation of Fujian Province, China (Grant Nos. 2019J01295 and 2018J01837), Key Projects of the Natural Science Foundation of Fujian Province, China (Grant No. 2020J02019), and Fujian Medical University Talent Startup Fund (XRCZX2017027, XJ2021011202, and 2017XQ1008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Editage (https://www.editage.cn/) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.790994/full#supplementary-material
References
1. Sugano K. Screening of gastric cancer in Asia. Best Pract Res Clin Gastroenterol. (2015) 29:895–905. doi: 10.1016/j.bpg.2015.09.013
2. Camilo V, Sugiyama T, Touati E. Pathogenesis of Helicobacter pylori infection. Helicobacter. (2017) 22:e12405. doi: 10.1111/hel.12405
3. Muhammad JS, Sugiyama T, Zaidi SF. Gastric pathophysiological ins and outs of Helicobacter pylori: a review. J Pak Med Assoc. (2013) 63:1528–33.
4. Basso D, Plebani M, Kusters JG. Pathogenesis of Helicobacter pylori infection. Helicobacter. (2010) 15(Suppl. 1):14–20. doi: 10.1111/j.1523-5378.2010.00781.x
5. Gu H. Role of flagella in the pathogenesis of Helicobacter pylori. Curr Microbiol. (2017) 74:863–9. doi: 10.1007/s00284-017-1256-4
6. Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, et al. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. (1998) 279:373–7. doi: 10.1126/science.279.5349.373
7. Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. (1999) 27:3325–33. doi: 10.1093/nar/27.16.3325
8. de Jonge R, Durrani Z, Rijpkema SG, Kuipers EJ, van Vliet AHM, Kusters JG. Role of the Helicobacter pylori outer-membrane proteins AlpA and AlpB in colonization of the guinea pig stomach. J Med Microbiol. (2004) 53(Pt. 5):375–9. doi: 10.1099/jmm.0.45551-0
9. Aspholm M, Olfat FO, Nordén J, Sondén B, Lundberg C, Sjöström R, et al. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. (2006) 2:e110. doi: 10.1371/journal.ppat.0020110
10. Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, et al. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun. (2011) 79:3106–16. doi: 10.1128/IAI.01275-10
11. Koniger V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, et al. Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol. (2016) 2:16188. doi: 10.1038/nmicrobiol.2016.233
12. Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. (2004) 4:688–94. doi: 10.1038/nrc1433
13. Pfannkuch L, Hurwitz R, Traulsen J, Sigulla J, Poeschke M, Matzner L, et al. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J. (2019) 33:9087–99. doi: 10.1096/fj.201802555R
14. Pai R, Cover TL, Tarnawski AS. Helicobacter pylori vacuolating cytotoxin (VacA) disorganizes the cytoskeletal architecture of gastric epithelial cells. Biochem Biophys Res Commun. (1999) 262:245–50. doi: 10.1006/bbrc.1999.1194
15. Cho SJ, Kang NS, Park SY, Kim BO, Rhee DK, Pyo S. Induction of apoptosis and expression of apoptosis related genes in human epithelial carcinoma cells by Helicobacter pylori VacA toxin. Toxicon. (2003) 42:601–11. doi: 10.1016/j.toxicon.2003.08.003
16. Chauhan N, Tay ACY, Marshall BJ, Jain U. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: an overview. Helicobacter. (2019) 24:e12544. doi: 10.1111/hel.12544
17. Dunn BE, Campbell GP, Perez-Perez GI, Blaser MJ. Purification and characterization of urease from Helicobacter pylori. J Biol Chem. (1990) 265:9464–9. doi: 10.1016/S0021-9258(19)38872-6
18. Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed J. (2016) 39:14–23. doi: 10.1016/j.bj.2015.06.002
19. Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. (2000) 287:482–5. doi: 10.1126/science.287.5452.482
20. Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. (2002) 123:187–95. doi: 10.1053/gast.2002.34218
21. Stingl K, De Reuse H. Staying alive overdosed: how does Helicobacter pylori control urease activity? Int J Med Microbiol. (2005) 295:307–15. doi: 10.1016/j.ijmm.2005.06.006
22. Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. (1991) 59:2470–5. doi: 10.1128/iai.59.7.2470-2475.1991
23. Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. (1994) 62:3586–9. doi: 10.1128/iai.62.8.3586-3589.1994
24. Schoep TD, Fulurija A, Good F, Lu W, Himbeck RP, Schwan C, et al. Surface properties of Helicobacter pylori urease complex are essential for persistence. PLoS ONE. (2010) 5:e15042. doi: 10.1371/journal.pone.0015042
25. Uberti AF, Olivera-Severo D, Wassermann GE, Scopel-Guerra A, Moraes JA, Barcellos-de-Souza P, et al. Pro-inflammatory properties and neutrophil activation by Helicobacter pylori urease. Toxicon. (2013) 69:240–49. doi: 10.1016/j.toxicon.2013.02.009
26. Pflock M, Kennard S, Delany I, Scarlato V, Beier D. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect Immun. (2005) 73:6437–45. doi: 10.1128/IAI.73.10.6437-6445.2005
27. Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J Bacteriol. (2011) 193:40–51. doi: 10.1128/JB.00800-10
28. Wen Y, Feng J, Sachs G. Helicobacter pylori 5'ureB-sRNA, a cis-encoded antisense small RNA negatively regulates ureAB expression by transcription termination. J Bacteriol. (2013) 195:444–52. doi: 10.1128/JB.01022-12
29. Jones MD, Li Y, Zamble DB. Acid-responsive activity of the Helicobacter pylori metalloregulator NikR. Proc Natl Acad Sci USA. (2018) 115:8966–71. doi: 10.1073/pnas.1808393115
30. Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. (1994) 13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x
31. Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. (1994) 77:207–16. doi: 10.1016/0092-8674(94)90313-1
32. Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. (2003) 50:101–4. doi: 10.1046/j.1365-2958.2003.03688.x
33. He Y, Frye JG, Strobaugh TP, Chen CY. Analysis of AI-2/LuxS-dependent transcription in Campylobacter jejuni strain 81-176. Foodborne Pathog Dis. (2008) 5:399–415. doi: 10.1089/fpd.2008.0106
34. Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. (2011) 65:389–410. doi: 10.1146/annurev-micro-090110-102908
35. Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, et al. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol. (2016) 70:299–316. doi: 10.1146/annurev-micro-102215-095708
36. Henke JM, Bassler BL. Bacterial social engagements. Trends Cell Biol. (2004) 14:648–56. doi: 10.1016/j.tcb.2004.09.012
37. Deryabin D, Galadzhieva A, Kosyan D, Duskaev G. Plant-derived inhibitors of AHL-mediated quorum sensing in bacteria: modes of action. Int J Mol Sci. (2019) 20:5588. doi: 10.3390/ijms20225588
38. Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. (1999) 96:1639–44. doi: 10.1073/pnas.96.4.1639
39. Forsyth MH, Cover TL. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect Immun. (2000) 68:3193–9. doi: 10.1128/IAI.68.6.3193-3199.2000
40. Joyce EA, Bassler BL, Wright A. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J Bacteriol. (2000) 182:3638–43. doi: 10.1128/JB.182.13.3638-3643.2000
41. Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. (2007) 189:6109–17. doi: 10.1128/JB.00246-07
42. Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology. (2011) 157(Pt. 9):2445–55. doi: 10.1099/mic.0.049353-0
43. Tsang J, Hoover TR. Themes and variations: regulation of rpon-dependent flagellar genes across diverse bacterial species. Scientifica. (2014) 2014:681754. doi: 10.1155/2014/681754
44. Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, et al. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio. (2015) 6:e00379. doi: 10.1128/mBio.00379-15
45. Sweeney EG, Nishida A, Weston A, Bañuelos MS, Potter K, Conery J, et al. Agent-based modeling demonstrates how local chemotactic behavior can shape biofilm architecture. mSphere. (2019) 4:e00285-19. doi: 10.1128/mSphere.00285-19
46. Wen Y, Huang H, Tang T, Yang H, Wang X, Huang X, et al. AI-2 represses CagA expression and bacterial adhesion, attenuating the Helicobacter pylori-induced inflammatory response of gastric epithelial cells. Helicobacter. (2021) 26:e12778. doi: 10.1111/hel.12778
47. Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol Gen Genet. (1998) 257:519–28. doi: 10.1007/s004380050677
48. Langford ML, Zabaleta J, Ochoa AC, Testerman TL, McGee DJ. In vitro and in vivo complementation of the Helicobacter pylori arginase mutant using an intergenic chromosomal site. Helicobacter. (2006) 11:477–93. doi: 10.1111/j.1523-5378.2006.00441.x
49. Kim J, Kim SW, Jang S, Merrell DS, Cha JH. Complementation system for Helicobacter pylori. J Microbiol. (2011) 49:481–6. doi: 10.1007/s12275-011-1196-9
50. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
51. Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. (1997) 388:539. doi: 10.1038/41483
52. Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. (2010) 464:250–55. doi: 10.1038/nature08756
53. Wang Y, Cen XF, Zhao GP, Wang J. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol. (2012) 194:5237–44. doi: 10.1128/JB.00989-12
54. De la Cruz MA, Ares MA, von Bargen K, Panunzi LG, Martinez-Cruz J, Valdez-Salazar HA, et al. Gene expression profiling of transcription factors of Helicobacter pylori under different environmental conditions. Front Microbiol. (2017) 8:615. doi: 10.3389/fmicb.2017.00615
55. Pflock M, Bathon M, Schar J, Muller S, Mollenkopf H, Meyer TF, et al. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J Bacteriol. (2007) 189:2339–49. doi: 10.1128/JB.01827-06
56. Donczew R, Makowski Ł, Jaworski P, Bezulska M, Nowaczyk M, Zakrzewska-Czerwińska J, et al. The atypical response regulator HP1021 controls formation of the Helicobacter pylori replication initiation complex. Mol Microbiol. (2015) 95:297–312. doi: 10.1111/mmi.12866
57. Szczepanowski P, Noszka M, Zyła-Uklejewicz D, Pikuła F, Nowaczyk-Cieszewska M, Krezel A, et al. HP1021 is a redox switch protein identified in Helicobacter pylori. Nucleic Acids Res. (2021) 49:6863–79. doi: 10.1093/nar/gkab440
58. Fischbach W, Malfertheiner P. Helicobacter pylori Infection. Dtsch Arztebl Int. (2018) 115:429–36. doi: 10.3238/arztebl.2018.0429
59. Khan S, Karim A, Iqbal S. Helicobacter urease: niche construction at the single molecule level. J Biosci. (2009) 34:503–11. doi: 10.1007/s12038-009-0069-4
60. Zambelli B, Danielli A, Romagnoli S, Neyroz P, Ciurli S, Scarlato V. High-affinity Ni2+ binding selectively promotes binding of Helicobacter pylori NikR to its target urease promoter. J Mol Biol. (2008) 383:1129–43. doi: 10.1016/j.jmb.2008.08.066
61. Loh JT, Forsyth MH, Cover TL. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect Immun. (2004) 72:5506–10. doi: 10.1128/IAI.72.9.5506-5510.2004
62. Lee WK, Ogura K, Loh JT, Cover TL, Berg DE. Quantitative effect of luxS gene inactivation on the fitness of Helicobacter pylori. Appl Environ Microbiol. (2006) 72:6615–22. doi: 10.1128/AEM.01291-06
63. Osaki T, Hanawa T, Manzoku T, Fukuda M, Kawakami H, Suzuki H, et al. Mutation of luxS affects motility and infectivity of Helicobacter pylori in gastric mucosa of a Mongolian gerbil model. J Med Microbiol. (2006) 55(Pt. 11):1477–85. doi: 10.1099/jmm.0.46660-0
64. Shen F, Hobley L, Doherty N, Loh JT, Cover TL, Sockett RE, et al. In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiol. (2010) 10:210. doi: 10.1186/1471-2180-10-210
65. Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. (2004) 186:3124–32. doi: 10.1128/JB.186.10.3124-3132.2004
66. Szczebara F, Dhaenens L, Armand S, Husson MO. Regulation of the transcription of genes encoding different virulence factors in Helicobacter pylori by free iron. FEMS Microbiol Lett. (1999) 175:165–70. doi: 10.1111/j.1574-6968.1999.tb13615.x
67. Borin BN, Krezel AM. Structure of HP0564 from Helicobacter pylori identifies it as a new transcriptional regulator. Proteins. (2008) 73:265–8. doi: 10.1002/prot.22159
68. Sun Y, Liu S, Li W, Shan Y, Li X, Lu X, et al. Proteomic analysis of the function of sigma factor σ54 in Helicobacter pylori survival with nutrition deficiency stress in vitro. PloS ONE. (2013) 8:e72920. doi: 10.1371/journal.pone.0072920
69. Roncarati D, Scarlato V. The interplay between two transcriptional repressors and chaperones orchestrates helicobacter pylori heat-shock response. Int J Mol Sci. (2018) 19:1702. doi: 10.3390/ijms19061702
70. Belzer C, van Schendel BA, Kuipers EJ, Kusters JG, van Vliet AH. Iron-responsive repression of urease expression in Helicobacter hepaticus is mediated by the transcriptional regulator Fur. Infect Immun. (2007) 75:745–52. doi: 10.1128/IAI.01163-06
71. Dai Q, Xu L, Xiao L, Zhu K, Song Y, Li C, et al. RovM and CsrA negatively regulate urease expression in Yersinia pseudotuberculosis. Front Microbiol. (2018) 9:348. doi: 10.3389/fmicb.2018.00348
72. Debowski AW, Walton SM, Chua EG, Tay AC, Liao T, Lamichhane B, et al. Helicobacter pylori gene silencing in vivo demonstrates urease is essential for chronic infection. PLoS Pathog. (2017) 13:e1006464. doi: 10.1371/journal.ppat.1006464
73. Azimi S, Klementiev AD, Whiteley M, Diggle SP. Bacterial quorum sensing during infection. Annu Rev Microbiol. (2020) 74:201–19. doi: 10.1146/annurev-micro-032020-093845
74. Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. (2009) 43:197–222. doi: 10.1146/annurev-genet-102108-134304
75. Ball AS, Chaparian RR, van Kessel. J. C. Quorum sensing gene regulation by LuxR/HapR master regulators in vibrios. J Bacteriol. (2017) 199:e00105-17. doi: 10.1128/JB.00105-17
Keywords: Helicobacter pylori, quorum sensing, LuxS, AI-2, urease, HP1021
Citation: Yang H, Huang X, Zhang X, Zhang X, Xu X, She F and Wen Y (2022) AI-2 Induces Urease Expression Through Downregulation of Orphan Response Regulator HP1021 in Helicobacter pylori. Front. Med. 9:790994. doi: 10.3389/fmed.2022.790994
Received: 13 November 2021; Accepted: 07 March 2022;
Published: 01 April 2022.
Edited by:
Anna Magdalena Zawilak-Pawlik, Hirszfeld Institute of Immunology and Experimental Therapy (PAN), PolandReviewed by:
Guangqiang Wang, University of Shanghai for Science and Technology, ChinaDavide Roncarati, University of Bologna, Italy
Copyright © 2022 Yang, Huang, Zhang, Zhang, Xu, She and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feifei She, c2hlZmVpZmVpQHllYWgubmV0; Yancheng Wen, aGl0d3ljQHFxLmNvbQ==
†These authors have contributed equally to this work
 Huang Yang
Huang Yang Xiaoxing Huang1,2†
Xiaoxing Huang1,2† Xiaochuan Zhang
Xiaochuan Zhang Yancheng Wen
Yancheng Wen