- 1Laboratory of Experimental Oncology, Research Institute of Molecular and Cellular Medicine, People’s Friendship University of Russia (RUDN University), Moscow, Russia
- 2Laboratory of Combined Treatment, N.N. Blokhin National Medical Research Center of Oncology of Ministry of Health of Russian Federation, Moscow, Russia
- 3Department of Biotechnology, Sirius University of Science and Technology, Sochi, Russia
- 4Engelhardt Institute of Molecular Biology of the Russian Academy of Sciences, Moscow, Russia
- 5A.N. Belozersky Institute of Physicochemical Biology, M.V. Lomonosov Moscow State University, Moscow, Russia
- 6Faculty of Bioengineering and Bioinformatics, M.V. Lomonosov Moscow State University, Moscow, Russia
- 7Department of Biological Chemistry, Sechenov First Moscow State Medical University, Moscow, Russia
Amino acid deprivation therapy (AADT) is a promising strategy for developing novel anticancer treatments, based on variations in metabolism of healthy and malignant cells. L-asparaginase was the first amino acid-degrading enzyme that received FDA approval for the treatment of acute lymphoblastic leukemia (ALL). Arginase and arginine deiminase were effective in clinical trials for the treatment of metastatic melanomas and hepatocellular carcinomas. Essential dependence of certain cancer cells on methionine explains the anticancer efficacy of methionine-g-lyase. Along with significant progress in identification of metabolic vulnerabilities of cancer cells, new amino acid-cleaving enzymes appear as promising agents for cancer treatment: lysine oxidase, tyrosine phenol-lyase, cysteinase, and phenylalanine ammonia-lyase. However, sensitivity of specific cancer cell types to these enzymes differs. Hence, search for prognostic and predictive markers for AADT and introduction of the markers into clinical practice are of great importance for translational medicine. As specific metabolic pathways in cancer cells are determined by the enzyme expression, some of these enzymes may define the sensitivity to AADT. This review considers the known predictors for efficiency of AADT, emphasizing the importance of knowledge on cancer-specific amino acid significance for such predictions.
Introduction
More than 50 years of research on cancer cell metabolism has concluded that a deficiency of certain amino acids inhibits the growth and proliferation of tumor cells much more than that of normal cells (1). Hence, àmino-acid degrading enzymes have been studied for their potential to treat cancer since 1960s, starting with a report of Broome et al. on antilymphoma effects of L-asparaginase (2, 3). Metabolic reprogramming commonly occurs in tumor cells to sustain the high nutritional requirements for carcinogenesis and growth (4). Many cancer cells develop an auxotrophic response to certain amino acids, such as methionine, arginine, and asparagine (5–7). For the treatment of acute lymphoblastic leukemia (ALL), bacterial L-asparaginase has been approved since 1970s (8). Arginine-depleting enzymes are suggested to treat metastatic melanoma (9). Arginine deiminase and recombinant human arginase 1 that deplete serum arginine, have recently been evaluated in phase II clinical trials (10, 11). Summarizing available information on the molecular mechanisms of the cancer-specific action of these amino-acid-degrading enzymes, our review draws attention to the amino-acid-replenishing counterparts among potential predictors of the efficacy of anticancer action of the amino acid-degrading enzymes.
L-asparagine depletion therapy
In normal cells, L-asparagine is produced by asparagine synthetase (EC 6.3.5.4), which catalyzes the synthesis of asparagine from aspartate, using glutamine as a nitrogen source (Figure 1A). Leukemic and certain other types of cancer cells are dependent on exogenous sources of asparagine due to low expression of the asparagine-synthetase-encoding gene ASNS (12, 13).
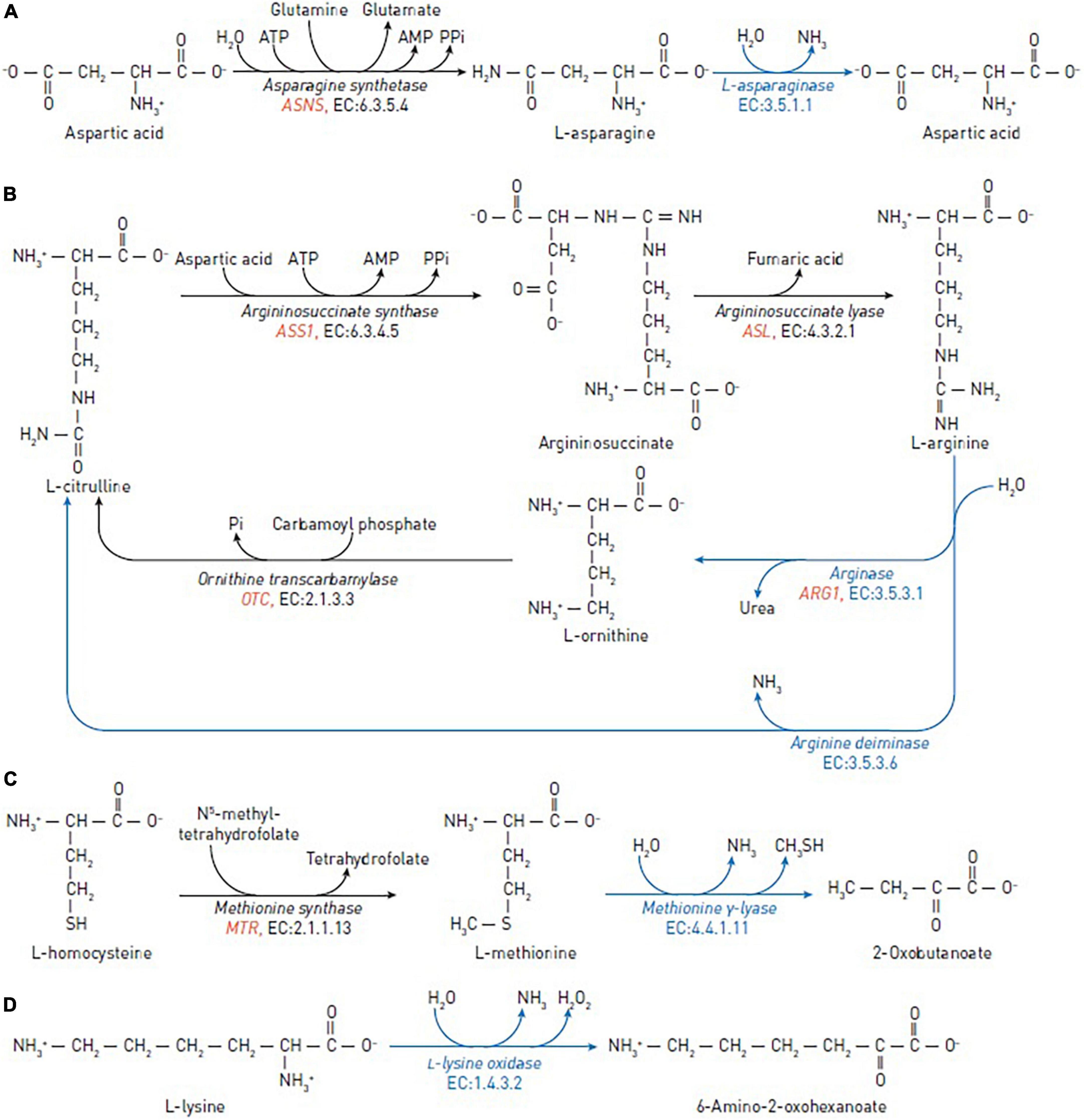
Figure 1. Reactions catalyzed by the amino-acid degrading enzymes used in cancer therapy (blue arrows) and reactions catalyzed by human enzymes that may affect the efficacy of the anticancer enzymes (black arrows). (A) The reactions of asparagine synthetase and asparaginase in asparagine depletion therapy. (B) The reactions of argininosuccinate synthase, argininosuccinate lyase, ornithine transcarbamylase, arginine deiminase and arginase in arginine depletion therapy. (C) The reactions of methionine synthase and methionine γ-lyase in methionine depletion therapy. (D) The reaction of L-lysine oxidase in lysine depletion therapy. The human genes encoding enzymes discussed as biomarkers are in red italic.
During the last 50 years, bacterial asparaginase have been widely used for acute lymphoblastic leukemia treatment (8). This enzyme has recently received attention in the treatment of advanced extra-nodal NK/T-cell lymphoma (14, 15). In vitro, it is shown that asparaginase-resistant cells become more sensitive to asparagine depletion when ASNS expression is downregulated, as observed in lymphoblasts (K562) and non-Hodgkin’s lymphoma (Karpas299) cells (16, 17). Immunohistochemistry of ASNS protein is available to assess the enzyme expression (18), in addition to estimation of mRNA levels. Tissue microarrays have been utilized to identify asparagine-synthetase-low cancer cells within a number of solid cancer subtypes in non-hematological malignancies (19). Based on this study, asparagine synthetase level in the tumor cells is proposed as a predictive biomarker for their sensitivity to asparaginase therapy (19). In 2006, FDA approved the first-line treatment of patients with ALL by chemically modified form of E. coli asparaginase (Oncaspar®). The modification by polyethylene glycol (i.e., pegylation) extends half-life of asparaginase in vivo (20). Erythrocytes-encapsulated asparaginase (GRASPA®, Erytech) represents another innovative formulation, that has been investigated for treatment of solid tumors, including pancreatic cancer (21). Conjugation with heparin-binding peptides or directed mutagenesis are also employed for the therapeutic usage of asparaginase isolated from bacterial sources (22–25).
A wide range of ASNS expression in different tissues is reported. Particularly high levels of expression are detected in the brain, testes, thyroid, and normal exocrine pancreatic cells. Acute lymphoblastic leukemia and hepatocytes typically have low ASNS expression (26, 27), and more than 50% of pancreatic ductal adenocarcinomas have very low ASNS expression (28). Thus, asparaginase may be suggested as effective drug for the treatment of pancreatic ductal adenocarcinomas lacking ASNS expression. Limited asparaginase efficiency in fighting many solid cancers, such as prostate and ovarian cancer, is believed to be due to the medium/high expression of ASNS in these cancer types (29, 30). Moreover, it has been shown that ASNS hypermethylation results in low ASNS protein expression in liver and gastric cancer cells, making them more susceptible to asparaginase therapy in vitro and in vivo (31). However, many studies have demonstrated that acute lymphoblastic leukemia can still be inhibited by asparaginase even when ASNS is expressed (32, 33). Additionally, no association was found between the asparaginase sensitivity and various levels of asparagine synthetase in acute myeloid leukemia subgroups in human cancer cells, probably explained by post-translational control of asparagine synthetase (34, 35). Asparagine auxotrophy not only has the apparent implication in heightened sensitivity to asparagine depletion and, therefore, to asparaginase therapy, but also implies the cancer cells’ tight reliance on external supplies of the amino acid even under normal growth conditions (26).
In summary, ASNS expression is suggested as a marker for clinical prediction of asparaginase resistance (19), supported by majority of the studies revealing a strong negative correlations of the asparaginase efficacy with the ASNS gene expression. High ASNS expression may contribute to asparaginase resistance of tumor cells. Low ASNS expression in tumor cells, including certain solid cancers, such as pancreatic ductal adenocarcinomas, could be used to suggest patients the treatment with asparaginase-added chemotherapy.
L-arginine depletion therapy
L-arginine is a crucial semi-essential amino acid involved in a variety of physiological functions, including cellular proliferation, through the arginine-dependent signaling pathways. These pathways involve the arginine-dependent generation of nitric oxide and polyamines, as well as activation of mTOR, a nutrient-sensing kinase strongly implicated in tumorigenesis. Arginine is synthesized from citrulline in two steps: (1) Argininosuccinate synthase converts L-citrulline and aspartic acid to argininosuccinate; (2) argininosuccinate lyase converts argininosuccinate to arginine and fumaric acid (Figure 1B).
Arginine deiminase (EC 3.5.3.6) and arginase (EC 3.5.3.1) degrade arginine. Arginine deiminase is widely distributed in bacterial organisms and certain anaerobic eukaryotes, and is isolated from a variety of sources, such as Pseudomonas putida, Giardia intestinalis, Streptococcus pyogenes, Mycoplasma spp. (36–39) and others. Arginine deiminase catalyzes the irreversible conversion of arginine to L-citrulline and ammonia (40). This process produces anti-tumor effect in a wide range of human cancers, including hepatoma, malignant melanoma, malignant fibrosarcoma, squamous cell carcinoma, nasopharyngeal carcinoma, and lung carcinoma in vitro and in vivo (41, 42). Arginine deiminase has been effective in phase II clinical trials for metastatic melanoma, hepatocellular carcinoma, and malignant mesothelioma (42–46). Therapeutic usage of pegylated arginine deiminase, possessing antiproliferative action against human leukemia cells (47).
Arginase is a manganese-dependent enzyme catalyzing the arginine conversion to ornithine and urea. Recombinant human arginase 1 has previously been shown in vitro to suppress non-Hodgkin’s lymphoma cells (48), prostate cancer cells (LNCaP, DU-145, and PC-3) (49), melanoma cells, laryngeal squamous cell carcinoma (50), leukemia cells (51), non-small cell lung cancer (NSCLC) (52), and ovarian cancer cells (53).
Arginine-depriving enzymes, such as arginase and arginine deiminase, may be useful to fight cancer cells (54) which lack significant levels of the arginine-replenishing enzymes: the ASS-encoded argininosuccinate synthase and ASL-encoded argininosuccinate lyase. On the other hand, overexpression of arginase in cells significantly increases the concentration of L-ornithine, that is recycled to arginine by ornithine transcarbamylase/argininosuccinate synthase (Table 1). These arginine re-synthesizing reactions provide resistance to the arginase treatment in several malignancies (5).
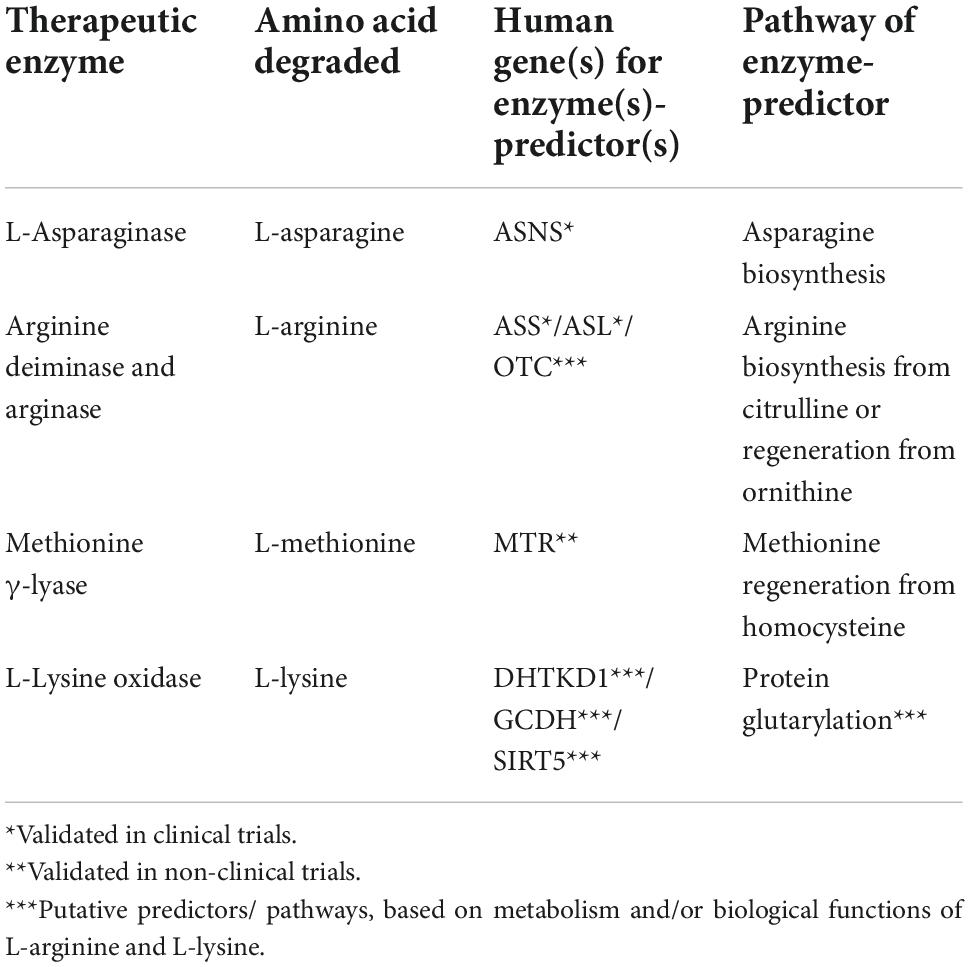
Table 1. Enzyme expression as predictive markers for treatment cancer with amino-acid degrading enzymes.
Deficiencies of argininosuccinate synthase and/or OTC-encoded ornithine transcarbamylase are regarded to be prognostic biomarkers and predictors of sensitivity to the arginine deprivation (55). Human melanoma (56), hepatocellular carcinoma (57), colon cancer (HT29) (58) and prostate carcinoma (59) have been demonstrated to be sensitive to arginine depletion by arginine deiminase due to low or negligible expression of ASS/ASL genes in vitro and in vivo (60–62). In contrast, arginine deiminase is ineffective in the treatment of cancers with high or medium expression of ASS, like ovarian (63), and colon cancer in vitro and in vivo (SW480 and HCT116) (58).
More than a 75% reduction of argininosuccinate synthase activity in cancer cells, compared to their healthy counterparts, is a positive prognostic marker for arginine deiminase efficacy (46). Expression of ASL and OTC genes may also provide valuable information to predict the efficacy of arginine-depletion therapy (52, 64).
L-methionine depletion therapy
L-methionine is an essential amino acid that contains a sulfur atom and participates in such a crucial function as DNA methylation. In normal cells, methionine can be recycled by re-methylation of homocysteine, catalyzed by the cobalamin-dependent enzyme methionine synthase (Figure 1C) or by betaine-homocysteine methyltransferase in the liver (65). Experiments show that many cancer cells, including leukemia (L1210 and J111) (66), breast (MDA-MB231, MCF7, SKBR3, and T47D) (67), lung (A2182 and SK-LU), kidney (A498), CNS (SK-N-SH), prostate (PC-3), and colon (SK-CO-1 and loVo) (68) cancer cells, cannot proliferate when methionine in growth medium is replaced with homocysteine in vitro.
Methionine γ-lyase (EC 4.4.1.11) is a bacterial pyridoxal-5’-phosphate-dependent enzyme which catalyzes γ-elimination of L-methionine to generate α-ketobutyric acid, methyl mercaptan and ammonia (69). The enzyme has been isolated from Pseudomonas putida (70), Trichomonas vaginalis (71), Clostridium sporogenes (72), Entamoeba histolytica (73), Citrobacter freundii (74), Clostridium tetani (75), and others. Methionine γ-lyase from Pseudomonas putida inhibited the growth of neuroblastoma (LAN-1 and NMB-7) (76), Yoshida sarcoma and lung cancer (H460) (77), advanced breast cancer (78, 79), renal cancer and lymphoma (78), human colon cancer xenografts (HCT15, HT29, COLO205, and SW620) (80) and glioblastoma (81). Pegylation of methionine γ-lyase was used to increase serum half-life and reduce immunological reactions in vivo (82). No clinical toxicity was found after treatment with methionine γ-lyase in a pilot phase I trial on human cancer patients (83).
Compared to normal tissues, cancer cells have a higher requirement for methionine synthase activity and may thus be more sensitive to methionine synthase inhibition (84–87). Large number of tumor cell lines, including melanoma, glioblastoma, colon, lung, breast, bladder, and kidney tumors, lack the normal pathway of methionine re-synthesis (88, 89). Methionine synthase is encoded by MTR gene whose polymorphism may affect DNA methylation and thus contribute to cancer development (90). Furthermore, the A2756G (rs1805087) substitution in the MTR gene plays a role in the progression of breast and prostate cancer via the pathway of the methyl group transfer, which is involved in both DNA methylation and DNA synthesis (91, 92). Ile22Met mutation (A66G) of the MTRR-encoded methionine synthase reductase is linked to folate, vitamin B6, or vitamin B12 levels in colorectal cancer. However, no statistically significant correlation between the Ile22Met mutant and risk of pancreatic cancer is reported (93). Instead, pancreatic cancer risk is influenced by His595Tyr mutation of methionine synthase reductase (94). MTRR gene suppression may be effective in the treatment of pancreatic ductal adenocarcinomas (94).
Thus, analysis of available data suggests that expression of methionine synthase could predict sensitivity of cancer cells to methionine-cleaving enzymes. Cells with low expression of methionine synthase are expected to be more sensitive to treatment with methionine γ-lyase. However, further studies are needed to establish the diagnostic significance.
Lysine depletion therapy
L-lysine is an essential and abundant amino acid in humans. In addition to proteinogenesis (95), L-lysine may be used for ketogenesis. An important energy source under starvation (96, 97), ketogenesis is also involved in responses of cancer cells to therapeutic agents (98). Besides, L-lysine catabolism through the DHTKD1-encoded 2-oxoadipate dehydrogenase produces glutaryl-CoA for protein glutarylation (97, 99, 100). Particularly glutarylation of histones, associated with gene activation (99), and regulation of pyruvate dehydrogenase by glutarylation (100) may be involved in metabolic transformation of cancer cells, causing their specific sensitivity to L-lysine depletion. L-lysine α-oxidases (EC 1.4.3.14) catalyze the oxidative deamination of L-lysine, resulting in the production of α-keto-ε-aminocaproate, ammonia, and H2O2 (Figure 1D). Over the last 40 years, several biological effects of L-lysine α-oxidases have been described, including antiviral, antimicrobial, anti-protozoa, anti-metastatic, and antitumor (101–105).
L-Lysine α-oxidase from Trichoderma cf. aureoviride Rifai has significant cytotoxicity against the following human cancer cell lines: K562, LS174T, HT29, SCOV3, PC3, and MCF7 in vitro (102) and PC12 (106). Human colon cancer xenografts HCT116 and LS174T, as well as breast adenocarcinoma T47D, demonstrated high sensitivity to L-lysine α-oxidase (102). Ophiophagus hannah venom-derived L-lysine α-oxidase inhibited the growth and proliferation of PC-3 prostate cancer xenografts (107). Depending on the dosage, the enzyme from Agkistrodon acutus suppressed the development of hepatoma 22, sarcoma 180, and Ehrlich carcinoma (108). In many species, L-lysine α-oxidase from Trichoderma harzianum Rifai and Tr. viride Y244-2 reduced malignant properties of solid tumors (105, 109). The most susceptible murine transplantable tumors were melanoma B16, breast adenocarcinoma Ca755, ascitic hepatoma 22, cervical cancer RSHM5, and colon carcinoma AKATOL (110). It may be hypothesized that the susceptibility is linked to the L-lysine-dependent induction of specific metabolism of these cancer cells by protein glutarylation. In this case, expression of the enzymes determining the levels of glutaryl-CoA, i.e., the proteins encoded by the DHTKD1 and GCDH genes, and protein deglutarylation, i.e., SIRT5 protein, may comprise the markers of efficiency of the L-lysine-depleting therapies.
Conclusion
Recent discoveries of the molecular pathways of amino acid metabolism and their regulation in tumor cells highlight specific features of tumor metabolism that may be used for prediction of efficacies of therapeutic strategies based on depletion of amino acids. An increased need for amino acids caused by rapid proliferation of cancer cells, contributes to metabolic abnormalities of these cells. In comparison to traditional anticancer treatments, those involving the amino-acid-degrading enzymes offer several advantages: (1) potent effects on specific amino acids indispensable for cancer cells; (2) low toxicity; (3) usage for combinatorial therapies; (4) existence of biochemical markers to predict the treatment responses. The predictors are expression of genes, such as ASNS for asparagine depletion therapy; ASL, ASS, and OTC for arginine depletion therapy; MTR for methionine depletion therapy. More clinical research is necessary to extend the list of such biomarkers and assess their prognostic value demonstrated in pilot studies.
Author contributions
VP, LQ, and EM wrote the manuscript. VB reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement on subsidies No. 075-15-2021-1060 of 28 September 2021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dhankhar R, Gupta V, Kumar S, Kapoor RK, Gulati P. Microbial enzymes for deprivation of amino acid metabolism in malignant cells: biological strategy for cancer treatment. Appl Microbiol Biotechnol. (2020) 104:2857–69. doi: 10.1007/s00253-020-10432-2
2. Broome JD. Evidence that the L-asparaginase of guinea pig serum is responsible for its antilymphoma effects. I. Properties of the L-asparaginase of guinea pig serum in relation to those of the antilymphoma substance. J Exp Med. (1963) 118:99–120. doi: 10.1084/jem.118.1.99
3. Old LJ, Boyse EA, Campbell HA, Daria GM. Leukaemia-inhibiting properties and L-asparaginase activity of sera from certain South American rodents. Nature. (1963) 198:801. doi: 10.1038/198801a0
4. Macintyre AN, Rathmell JC. Activated lymphocytes as a metabolic model for carcinogenesis. Cancer Metab. (2013) 1:5. doi: 10.1186/2049-3002-1-5
5. Cheng P, Lam TL, Lam WM, Tsui SM, Cheng AW, Lo WH, et al. Pegylated recombinant human arginase (rhArg-PEG 5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Cancer Res. (2007) 67:309–17. doi: 10.1158/0008-5472.CAN-06-1945
6. Rytting M. Peg-asparaginase for acute lymphoblastic leukemia. Expert Opin Biol Ther. (2010) 10:833–9. doi: 10.1517/14712591003769808
7. Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature. (2019) 572:397–401. doi: 10.1038/s41586-019-1437-3
8. Whitecar JP Jr., Bodey GP, Harris JE, Freireich EJ. L-Asparaginase. N Engl J Med. (1970) 282:732–4. doi: 10.1056/NEJM197003262821307
9. Yoon J-K, Frankel AE, Feun LG, Ekmekcioglu S, Kim KB. Arginine deprivation therapy for malignant melanoma. Clin Pharmacol. (2013) 5:11–9. doi: 10.2147/CPAA.S37350
10. Synakiewicz A, Stachowicz-Stencel T, Adamkiewicz-Drozynska E. The role of arginine and the modified arginine deiminase enzyme ADI-PEG 20 in cancer therapy with special emphasis on Phase I/II clinical trials. Expert Opin Invest Drugs. (2014) 23:1517–29. doi: 10.1517/13543784.2014.934808
11. Chan SL, Cheng PNM, Liu AM, Chan LL, Li L, Chu CM, et al. A phase II clinical study on the efficacy and predictive biomarker of pegylated recombinant arginase on hepatocellular carcinoma. Invest New Drugs. (2021) 39:1375–82. doi: 10.1007/s10637-021-01111-8
12. Haskell CM, Canellos GP. l-asparaginase resistance in human leukemia–asparagine synthetase. Biochem Pharmacol. (1969) 18:2578–80. doi: 10.1016/0006-2952(69)90375-x
13. Prager MD, Bachynsky N. Asparagine synthetase in normal and malignant tissues: correlation with tumor sensitivity to asparaginase. Arch Biochem Biophys. (1968) 127:645–54. doi: 10.1016/0003-9861(68)90273-7
14. Pokrovsky VS, Vinnikov D. L-Asparaginase for newly diagnosed extra-nodal NK/T-cell lymphoma: systematic review and meta-analysis. Expert Rev Anticancer Ther. (2017) 17:759–68. doi: 10.1080/14737140.2017.1344100
15. Yamaguchi M, Suzuki R, Oguchi M. Advances in the treatment of extranodal NK/T-cell lymphoma, nasal type. Blood. (2018) 131:2528–40. doi: 10.1182/blood-2017-12-791418
16. Song P, Ye L, Fan J, Li Y, Zeng X, Wang Z, et al. Asparaginase induces apoptosis and cytoprotective autophagy in chronic myeloid leukemia cells. Oncotarget. (2015) 6:3861–73. doi: 10.18632/oncotarget.2869
17. Hermanova I, Zaliova M, Trka J, Starkova J. Low expression of asparagine synthetase in lymphoid blasts precludes its role in sensitivity to L-asparaginase. Exp Hematol. (2012) 40:657–65. doi: 10.1016/j.exphem.2012.04.005
18. Fu Y, Ding L, Yang X, Ding Z, Huang X, Zhang L, et al. Asparagine synthetase-mediated L-asparagine metabolism disorder promotes the perineural invasion of oral squamous cell carcinoma. Front Oncol. (2021) 11:637226. doi: 10.3389/fonc.2021.637226
19. Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. (2000) 24:236–44. doi: 10.1038/73439
20. Jarrar M, Gaynon PS, Periclou AP, Fu C, Harris RE, Stram D, et al. Asparagine depletion after pegylated E. coli asparaginase treatment and induction outcome in children with acute lymphoblastic leukemia in first bone marrow relapse: a children’s oncology group study (CCG-1941). Pediatr Blood Cancer. (2016) 47:141–6. doi: 10.1002/pbc.20713
21. Thomas X, Le Jeune C. Erythrocyte encapsulated l-asparaginase (GRASPA) in acute leukemia. Int J Hematol Oncol. (2016) 5:11–25. doi: 10.2217/ijh-2016-0002
22. Zhdanov DD, Pokrovsky VS, Pokrovskaya MV, Alexandrova SS, Eldarov MA, Grishin DV, et al. Rhodospirillum rubruml-asparaginase targets tumor growth by a dual mechanism involving telomerase inhibition. Biochem Biophys Res Commun. (2017) 492:282–8. doi: 10.1016/j.bbrc.2017.08.078
23. Sannikova EP, Bulushova NV, Cheperegin SE, Gubaydullin II, Chestukhina GG, Ryabichenko VV, et al. The modified heparin-binding L-asparaginase of wolinella succinogenes. Mol Biotechnol. (2016) 58:528–39. doi: 10.1007/s12033-016-9950-1
24. Sidoruk KV, Pokrovsky VS, Borisova AA, Omeljanuk NM, Aleksandrova SS, Pokrovskaya MV, et al. Creation of a producent, optimization of expression, and purification of recombinant Yersinia pseudotuberculosis L-asparaginase. Bull Exp Biol Med. (2011) 152:219–23. doi: 10.1007/s10517-011-1493-7
25. Pokrovskaya MV, Pokrovsky VS, Aleksandrova SS, Sokolov NN, Zhdanov DD. Molecular analysis of L-asparaginases for clarification of the mechanism of action and optimization of pharmacological functions. Pharmaceutics. (2022) 14:599. doi: 10.3390/pharmaceutics14030599
26. Chiu M, Taurino G, Bianchi MG, Kilberg MS, Bussolati O. Asparagine synthetase in cancer: beyond acute lymphoblastic leukemia. Front Oncol. (2020) 9:1480. doi: 10.3389/fonc.2019.01480
27. Milman HA, Cooney DA. The distribution of l-asparagine synthetase in the principal organs of several mammalian and avian species. Biochem J. (1974) 142:27–35. doi: 10.1042/bj1420027
28. Dufour E, Gay F, Aguera K, Scoazec JY, Horand F, Lorenzi PL, et al. Pancreatic Tumor Sensitivity to Plasma L-Asparagine Starvation. Pancreas. (2012) 41:940–8. doi: 10.1097/MPA.0b013e318247d903
29. Sircar K, Huang H, Hu L, Cogdell D, Dhillon J, Tzelepi V, et al. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. Am J Pathol. (2012) 180:895–903. doi: 10.1016/j.ajpath.2011.11.030
30. Lorenzi PL, Llamas J, Gunsior M, Ozbun L, Reinhold WC, Varma S, et al. Asparagine synthetase is a predictive biomarker of L-asparaginase activity in ovarian cancer cell lines. Mol Cancer Ther. (2008) 7:3123–8. doi: 10.1158/1535-7163.MCT-08-0589
31. Li H, Ning S, Ghandi M, Kryukov GV, Gopal S, Deik A, et al. The landscape of cancer cell line metabolism. Nat Med. (2019) 25:850–60. doi: 10.1038/s41591-019-0404-8
32. Stams WAG, den Boer ML, Beverloo HB, Meijerink JP, Stigter RL, van Wering ER, et al. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. (2003) 101:2743–7. doi: 10.1182/blood-2002-08-2446
33. Krall AS, Xu S, Graeber TG, Braas D, Christofk HR. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun. (2016) 7:11457. doi: 10.1038/ncomms11457
34. Dübbers A, Würthwein G, Müller HJ, Schulze-Westhoff P, Winkelhorst M, Kurzknabe E, et al. Asparagine synthetase activity in paediatric acute leukaemias: AML-M5 subtype shows lowest activity. Br J Haematol. (2000) 109:427–9. doi: 10.1046/j.1365-2141.2000.02015.x
35. Okada S, Hongo T, Yamada S, Watanabe C, Fujii Y, Ohzeki T, et al. In vitro efficacy of L-asparaginase in childhood acute myeloid leukaemia. Br J Haematol. (2003) 123:802–9. doi: 10.1046/j.1365-2141.2003.04703.x
36. Wolf C, Brüss M, Hänisch B, Göthert M, von Kügelgen I, Molderings GJ, et al. Molecular basis for the antiproliferative effect of agmatine in tumor cells of colonic, hepatic, and neuronal origin. Mol Pharmacol. (2007) 71:276–83. doi: 10.1124/mol.106.028449
37. Ni Y, Li Z, Sun Z, Zheng P, Liu Y, Zhu L, et al. Expression of arginine deiminase from Pseudomonas plecoglossicida CGMCC2039 in Escherichia coli and its anti-tumor activity. Curr Microbiol. (2009) 58:593–8. doi: 10.1007/s00284-009-9376-0
38. Knodler LA, Schofield PJ, Edwards MR. L-Arginine transport and metabolism in Giardia intestinalis support its position as a transition between the prokaryotic and eukaryotic kingdoms. Microbiology. (1995) 141:2063–70. doi: 10.1099/13500872-141-9-2063
39. Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, et al. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun. (2000) 68:2441–8. doi: 10.1128/IAI.68.5.2441-2448.2000
40. Galkin A, Kulakova L, Sarikaya E, Lim K, Howard A, Herzberg O. Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target. J Biol Chem. (2004) 279:14001–8. doi: 10.1074/jbc.M313410200
41. Takaku H, Takase M, Abe S, Hayashi H, Miyazaki K. In vivo anti-tumor activity of arginine deiminase purified from Mycoplasma arginini. Int J Cancer. (1992) 51:244–9. doi: 10.1002/ijc.2910510213
42. Sugimura K, Ohno T, Kusuyama T, Azuma I. High sensitivity of human melanoma cell lines to the growth inhibitory activity of mycoplasmal arginine deiminase in vitro. Melanoma Res. (1992) 2:191–6. doi: 10.1097/00008390-199209000-00007
43. Ott PA, Carvajal RD, Pandit-Taskar N, Jungbluth AA, Hoffman EW, Wu BW, et al. Phase I/II study of pegylated arginine deiminase (ADI-PEG 20) in patients with advanced melanoma. Invest New Drugs. (2013) 31:425–34. doi: 10.1007/s10637-012-9862-2
44. Feun LG, Savaraj N, Marini A, Wu C, Robles C, Herrera C, et al. Phase II study of pegylated arginine deiminase (ADI-PEG20), a novel targeted therapy for melanoma. J Clin Oncol. (2006) 24:8045. doi: 10.1200/jco.2006.24.18_suppl.8045
45. Delman KA, Brown TD, Thomas M, Ensor CM, Holtsberg FW, Bomalaski JS, et al. Phase I/II trial of pegylated arginine deiminase (ADI-PEG20) in unresectable hepatocellular carcinoma. J Clin Oncol. (2005) 23:4139. doi: 10.1200/jco.2005.23.16_suppl.4139
46. Szlosarek PW, Steele JP, Nolan L, Gilligan D, Taylor P, Spicer J, et al. Arginine deprivation with pegylated arginine deiminase in patients with argininosuccinate synthetase 1-deficient malignant pleural mesothelioma: a randomized clinical trial. JAMA Oncol. (2017) 3:58–66. doi: 10.1001/jamaoncol.2016.3049
47. Shen LJ, Beloussow K, Shen WC. Modulation of arginine metabolic pathways as the potential anti-tumor mechanism of recombinant arginine deiminase. Cancer Lett. (2006) 231:30–5. doi: 10.1016/j.canlet.2005.01.007
48. Zeng X, Li Y, Fan J, Zhao H, Xian Z, Sun Y, et al. Recombinant human arginase induced caspase-dependent apoptosis and autophagy in non-Hodgkin’s lymphoma cells. Cell Death Dis. (2013) 4:e840. doi: 10.1038/cddis.2013.359
49. Matos A, Carvalho M, Bicho M, Ribeiro R. Arginine and arginases modulate metabolism, tumor microenvironment and prostate cancer progression. Nutrients. (2021) 13:4503. doi: 10.3390/nu13124503
50. Lin C, Wang Z, Li L, He Y, Fan J, Liu Z, et al. The role of autophagy in the cytotoxicity induced by recombinant human arginase in laryngeal squamous cell carcinoma. Appl Microbiol Biotechnol. (2015) 99:8487–94. doi: 10.1007/s00253-015-6565-6
51. Li Y, Zeng X, Wang S, Fan J, Wang Z, Song P, et al. Blocking autophagy enhanced leukemia cell death induced by recombinant human arginase. Tumor Biol. (2016) 37:6627–35. doi: 10.1007/s13277-015-4253-x
52. Shen W, Zhang X, Fu X, Fan J, Luan J, Cao Z, et al. A novel and promising therapeutic approach for NSCLC: recombinant human arginase alone or combined with autophagy inhibitor. Cell Death Dis. (2017) 8:e2720. doi: 10.1038/cddis.2017.137
53. Nasreddine G, El-Sibai M, Abi-Habib RJ. Cytotoxicity of [HuArgI (co)-PEG5000]-induced arginine deprivation to ovarian cancer cells is autophagy dependent. Invest New Drugs. (2020) 38:10–9. doi: 10.1007/s10637-019-00756-w
54. Bach SJ, Lasnitzki I. Some aspects of the role of arginine and arginase in mouse carcinoma 63. Enzymologia. (1947) 12:198–205.
55. Qiu F, Huang J, Sui M. Targeting arginine metabolism pathway to treat arginine-dependent cancers. Cancer Lett. (2015) 364:1–7. doi: 10.1016/j.canlet.2015.04.020
56. Savaraj N, Wu C, Kuo MT, You M, Wangpaichitr M, Robles C, et al. The relationship of arginine deprivation, argininosuccinate synthetase and cell death in melanoma. Drug Target Insights. (2007) 2:119–28.
57. Curley SA, Bomalaski JS, Ensor CM, Holtsberg FW, Clark MA. Regression of hepatocellular cancer in a patient treated with arginine deiminase. Hepatogastroenterology. (2003) 50:1214–6.
58. Alexandrou C, Al-Aqbi SS, Higgins JA, Boyle W, Karmokar A, Andreadi C, et al. Sensitivity of colorectal cancer to arginine deprivation therapy is shaped by differential expression of urea cycle enzymes. Sci Rep. (2018) 8:12096. doi: 10.1038/s41598-018-30591-7
59. Kim RH, Coates JM, Bowles TL, McNerney GP, Sutcliffe J, Jung JU, et al. Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Cancer Res. (2009) 69:700–8. doi: 10.1158/0008-5472.CAN-08-3157
60. Yoon C-Y, Shim YJ, Kim EH, Lee JH, Won NH, Kim JH, et al. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. Int J Cancer. (2007) 120:897–905. doi: 10.1002/ijc.22322
61. Park I-S, Kang SW, Shin YJ, Chae KY, Park MO, Kim MY, et al. Arginine deiminase: a potential inhibitor of angiogenesis and tumour growth. Br J Cancer. (2003) 89:907–14. doi: 10.1038/sj.bjc.6601181
62. Dillon BJ, Prieto VG, Curley SA, Ensor CM, Holtsberg FW, Bomalaski JS, et al. Incidence and distribution of argininosuccinate synthetase deficiency in human cancers: a method for identifying cancers sensitive to arginine deprivation. Cancer. (2004) 100:826–33. doi: 10.1002/cncr.20057
63. Szlosarek PW, Grimshaw MJ, Wilbanks GD, Hagemann T, Wilson JL, Burke F, et al. Aberrant regulation of argininosuccinate synthetase by TNF-alpha in human epithelial ovarian cancer. Int J Cancer. (2007) 121:6–11. doi: 10.1002/ijc.22666
64. Wang Z, Shi X, Li Y, Fan J, Zeng X, Xian Z, et al. Blocking autophagy enhanced cytotoxicity induced by recombinant human arginase in triple-negative breast cancer cells. Cell Death Dis. (2014) 5:e1563. doi: 10.1038/cddis.2014.503
65. Pajares MA, Pérez-Sala D. Betaine homocysteine S-methyltransferase: just a regulator of homocysteine metabolism? Cell Mol Life Sci. (2006) 63:2792–803. doi: 10.1007/s00018-006-6249-6
66. Halpern BC, Clark BR, Hardy DN, Halpern RM, Smith RA. The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc Natl Acad Sci USA. (1974) 71:1133–6. doi: 10.1073/pnas.71.4.1133
67. Lien EC, Ghisolfi L, Geck RC, Asara JM, Toker A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci Signal. (2017) 10:eaao6604. doi: 10.1126/scisignal.aao6604
68. Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun. (1983) 117:429–34. doi: 10.1016/0006-291X(83)91218-4
69. Tanaka H, Esaki N, Soda K. Properties of L-methionine gamma-lyase from Pseudomonas ovalis. Biochemistry. (1977) 16:100–6. doi: 10.1021/bi00620a016
70. Kudou D, Misaki S, Yamashita M, Tamura T, Takakura T, Yoshioka T, et al. Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution. J Biochem. (2007) 141:535–44. doi: 10.1093/jb/mvm055
71. Lockwood BC, Coombs GH. Purification and characterization of methionine y-lyase from Trichomonas vaginalis. Biochem J. (1991) 279:675–82.
72. Revtovich S, Anufrieva N, Morozova E, Kulikova V, Nikulin A, Demidkina T. Structure of methionine γ-lyase from Clostridium sporogenes. Acta Crystallogr :F Struct Biol Commun. (2016) 72:65–71. doi: 10.1107/S2053230X15023869
73. Sato D, Karaki T, Shimizu A, Kamei K, Harada S, Nozaki T. Crystallization and preliminary X-ray analysis of L-methionine gamma-lyase 1 from Entamoeba histolytica. Acta Crystallogr Sect F Struct Biol Cryst Commun. (2008) 64:697–9. doi: 10.1107/S1744309108018691
74. Mamaeva DV, Morozova EA, Nikulin AD, Revtovich SV, Nikonov SV, Garber MB, et al. Structure of Citrobacter freundii L-methionine gamma-lyase. Acta Crystallogr F Struct Biol Cryst Commun. (2005) 61:546–9. doi: 10.1107/S1744309105015447
75. Morozova EA, Kulikova VV, Yashin DV, Anufrieva NV, Anisimova NY, Revtovich SV, et al. Kinetic parameters and cytotoxic activity of recombinant methionine γ-Lyase from Clostridium tetani, Clostridium sporogenes, Porphyromonas gingivalis and Citrobacter freundii. Acta Nat. (2013) 5:92–8.
76. Hu J, Cheung N-KV. Methionine depletion with recombinant methioninase: in vitro and in vivo efficacy against neuroblastoma and its synergism with chemotherapeutic drugs. Int J Cancer. (2009) 124:1700–6. doi: 10.1002/ijc.24104
77. Tan Y, Xu M, Guo H, Sun X, Kubota T, Hoffman RM. Anticancer efficacy of methioninase in vivo. Anticancer Res. (1996) 16:3931–6.
78. Tan Y, Zavala J Sr., Han Q, Xu M, Sun X, Tan X, et al. Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. (1997) 17:3857–60.
79. Tan Y, Zavala J Sr., Xu M, Zavala J Jr., Hoffman RM. Serum methionine depletion without side effects by methioninase in metastatic breast cancer patients. Anticancer Res. (1996) 16:3937–42.
80. Tan Y, Sun X, Xu M, Tan X, Sasson A, Rashidi B, et al. Efficacy of recombinant methioninase in combination with cisplatin on human colon tumors in nude mice. Clin Cancer Res. (1999) 5:2157–63.
81. Kokkinakis DM, Schold SC Jr., Hori H, Nobori T. Effect of long-term depletion of plasma methionine on the growth and survival of human brain tumor xenografts in athymic mice. Nutr Cancer. (1997) 29:195–204. doi: 10.1080/01635589709514624
82. Tan Y, Sun X, Xu M, An Z, Tan X, Han Q, et al. Polyethylene glycol conjugation of recombinant methioninase for cancer therapy. Protein Expr Purif. (1998) 12:45–52. doi: 10.1006/prep.1997.0805
83. Hoffman RM, Tan Y, Li S, Han Q, Zavala JSr., Zavala JJr. Pilot phase I clinical trial of methioninase on high-stage cancer patients: rapid depletion of circulating methionine. Methods Mol Biol. (2019) 1866:231–42. doi: 10.1007/978-1-4939-8796-2_17
84. Baldwin CM, Perry CM. Pemetrexed: a review of its use in the management of advanced non-squamous non-small cell lung cancer. Drugs. (2009) 69:2279–302. doi: 10.2165/11202640-000000000-00000
85. Gonen N, Assaraf YG. Antifolates in cancer therapy: structure, activity and mechanisms of drug resistance. Drug Resist Updat. (2012) 15:183–210. doi: 10.1016/j.drup.2012.07.002
86. Visentin M, Zhao R, Goldman ID. The antifolates. Hematol Oncol Clin North Am. (2012) 26:629–48. doi: 10.1016/j.hoc.2012.02.002
87. Sakura T, Hayakawa F, Sugiura I, Murayama T, Imai K, Usui N, et al. High-dose methotrexate therapy significantly improved survival of adult acute lymphoblastic leukemia: a phase III study by JALSG. Leukemia. (2018) 32:626–32. doi: 10.1038/leu.2017.283
88. Breillout F, Antoine E, Poupon MF. Methionine dependency of malignant tumors: a possible approach for therapy. J Natl. Cancer Inst. (1990) 82:1628–32. doi: 10.1093/jnci/82.20.1628
89. Poirson-Bichat F, Gonçalves RA, Miccoli L, Dutrillaux B, Poupon MF. Methionine depletion enhances the antitumoral efficacy of cytotoxic agents in drug-resistant human tumor xenografts. Clin Cancer Res. (2000) 6: 643–53.
90. Harmon DL, Shields DC, Woodside JV, McMaster D, Yarnell JW, Young IS, et al. Methionine synthase D919G polymorphism is a significant but modest determinant of circulating homocysteine concentrations. Genet Epidemiol. (1999) 17:298–309. doi: 10.1002/(SICI)1098-2272(199911)17:43.0.CO;2-V
91. Yu K, Zhang J, Zhang J, Dou C, Gu S, Xie Y, et al. Methionine synthase A2756G polymorphism and cancer risk: a meta-analysis. Eur J Hum Genet. (2010) 18:370–8. doi: 10.1038/ejhg.2009.131
92. Hosseini M. Role of polymorphism of methyltetrahydrofolate-homocysteine methyltransferase (MTR) A2756G and breast cancer risk. Pol J Pathol. (2013) 64:191–5. doi: 10.5114/pjp.2013.38138
93. Gaughan DJ, Kluijtmans LA, Barbaux S, McMaster D, Young IS, Yarnell JW, et al. The methionine synthase reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. (2001) 157:451–6. doi: 10.1016/s0021-9150(00)00739-5
94. Ohnami S, Sato Y, Yoshimura K, Ohnami S, Sakamoto H, Aoki K, et al. His595Tyr Polymorphism in the methionine synthase reductase (MTRR) gene is associated with pancreatic cancer risk. Gastroenterology. (2008) 135:477–88. doi: 10.1053/j.gastro.2008.04.016
95. Smriga M, Ando T, Akutsu M, Furukawa Y, Miwa K, Morinaga Y. Oral treatment with L-lysine and L-arginine reduces anxiety and basal cortisol levels in healthy humans. Biomed Res. (2007) 28:85–90. doi: 10.2220/biomedres.28.85
96. Noda C, Ichihara A. Control of ketogenesis from amino acids. IV. Tissue specificity in oxidation of leucine, tyrosine, and lysine. J Biochem. (1976) 80:1159–64. doi: 10.1093/oxfordjournals.jbchem.a131371
97. Bhatt DP, Mills CA, Anderson KA, Henriques BJ, Lucas TG, Francisco S, et al. Deglutarylation of glutaryl-CoA dehydrogenase by deacylating enzyme SIRT5 promotes lysine oxidation in mice. J Biol Chem. (2022) 298:101723. doi: 10.1016/j.jbc.2022.101723
98. Palma FR, Ratti BA, Paviani V, Coelho DR, Miguel R, Danes JM, et al. AMPK-deficiency forces metformin-challenged cancer cells to switch from carbohydrate metabolism to ketogenesis to support energy metabolism. Oncogene. (2021) 40:5455–67. doi: 10.1038/s41388-021-01943-x
99. Xu Y, Shi Z, Bao L. An expanding repertoire of protein acylations. Mol Cell Proteomics. (2022) 21:100193. doi: 10.1016/j.mcpro.2022.100193
100. Boyko AI, Karlina IS, Zavileyskiy LG, Aleshin VA, Artiukhov AV, Kaehne T, et al. Delayed impact of 2-oxoadipate dehydrogenase inhibition on the rat brain metabolism is linked to protein glutarylation. Front Med. (2022) 9:896263. doi: 10.3389/fmed.2022.896263
101. Chen WM, Lin CY, Sheu SY. Investigating antimicrobial activity in Rheinheimera sp. due to hydrogen peroxide generated by l-lysine oxidase activity. Enzyme Microb Technol. (2010) 46:487–93. doi: 10.1016/j.enzmictec.2010.01.006
102. Pokrovsky VS, Treshalina HM, Lukasheva EV, Sedakova LA, Medentzev AG, Arinbasarova AY, et al. Enzymatic properties and anticancer activity of L-lysine α-oxidase from Trichoderma cf. aureoviride Rifai BKMF-4268D. Anti Cancer Drugs. (2013) 24:846–51. doi: 10.1097/CAD.0b013e328362fbe2
103. Selishcheva AA, Alekseev SB, Smirnova IP, Podboronov VM. Antiherpetic activity of l-lysine-alpha-oxidase in different dosage forms. Antibiot Chemoter. (2003) 48:9–12.
104. Umanskii VY, Khaduev SKH, Zaletok SP, Balitskii KP, Berdinskikh NK, Berezov TT. Antimetastic effect of L-lyasine alpha-oxidase. Biull Eksp Biol Med. (1990) 109:458–9.
105. Lukasheva EV, Babayeva G, Karshieva SS, Zhdanov DD, Pokrovsky VS. L-Lysine α-Oxidase: enzyme with anticancer properties. Pharmaceuticals. (2021) 14:1070. doi: 10.3390/ph14111070
106. Lukasheva EV, Ribakova YS, Fedorova TN, Makletsova MG, Arinbasarova AY, Medentzev AG, et al. The effect of L-Lysine alpha-oxidase from Trichoderma cf. aureoviride Rifai VKM F-4268D on the rat pheochromocytoma PC12 cell line. Biomed Chem. (2014) 8:130–3. doi: 10.1134/S1990750814020061
107. Lee ML, Fung SY, Chung I, Pailoor J, Cheah SH, Tan NH. King cobra (Ophiophagus hannah) venom L-amino acid oxidase induces apoptosis in PC-3 cells and suppresses PC-3 solid tumor growth in a tumor xenograft mouse model. Int J Med Sci. (2014) 11:593–601. doi: 10.7150/ijms.8096
108. Zhang L, Wu WT. Isolation and characterization of ACTX-6: a cytotoxic L-amino acid oxidase from agkistrodon acutus snake venom. Nat Prod Res. (2008) 22:554–63. doi: 10.1080/14786410701592679
109. El-fakharany EM, Afifi M. Purification, characterization and antitumor activity of L-lysine alpha- oxidase from 2 Trichoderma harzianum Rifai AUMC No 848. Sci Eng Res. (2018) 5:350–67.
Keywords: amino acid deprivation therapy, L-asparaginase, arginine deiminase, arginase, methionine γ-lyase, L-lysine oxidase, asparagine synthetase, methionine synthase
Citation: Pokrovsky VS, Abo Qoura L, Morozova E and Bunik VI (2022) Predictive markers for efficiency of the amino-acid deprivation therapies in cancer. Front. Med. 9:1035356. doi: 10.3389/fmed.2022.1035356
Received: 02 September 2022; Accepted: 18 October 2022;
Published: 03 November 2022.
Edited by:
Jing He, Guangzhou Medical University, ChinaReviewed by:
Stefan Broer, Australian National University, AustraliaCopyright © 2022 Pokrovsky, Abo Qoura, Morozova and Bunik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vadim S. Pokrovsky, di5wb2tyb3Zza3lAcm9uYy5ydQ==
 Vadim S. Pokrovsky
Vadim S. Pokrovsky Louay Abo Qoura
Louay Abo Qoura Elena Morozova
Elena Morozova Victoria I. Bunik
Victoria I. Bunik