
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 24 October 2022
Sec. Pathology
Volume 9 - 2022 | https://doi.org/10.3389/fmed.2022.1034594
This article is part of the Research TopicPathological Reactions of Cytotoxic Lymphoid Cells as Universal Therapeutic Targets in Cancer and Autoimmune DiseaseView all 11 articles
 Eric S. Geanes1
Eric S. Geanes1 Stacey A. Krepel1
Stacey A. Krepel1 Rebecca McLennan1
Rebecca McLennan1 Stephen Pierce1,2
Stephen Pierce1,2 Santosh Khanal1
Santosh Khanal1 Todd Bradley1,2,3,4*
Todd Bradley1,2,3,4*Diffuse large B-cell lymphoma (DLBCL), the most common form of lymphoma, is typically treated with chemotherapy combined with the immunotherapy rituximab, an antibody targeting the B cell receptor, CD20. Despite the success of this treatment regimen, approximately a third of DLBCL patients experience either relapse or have refractory disease that is resistant to rituximab, indicating the need for alternative therapeutic strategies. Here, we identified that CD74 and IL4R are expressed on the cell surface of both CD20 positive and CD20 negative B cell populations. Moreover, genes encoding CD74 and IL4R are expressed in lymphoma biopsies isolated from all stages of disease. We engineered bispecific antibodies targeting CD74 or IL4R in combination with rituximab anti-CD20 (anti-CD74/anti-CD20 and anti-IL4R/anti-CD20). Bispecific antibody function was evaluated by measuring direct induction of apoptosis, antibody-dependent cellular phagocytosis (ADCP), and antibody-dependent cellular cytotoxicity in both rituximab-sensitive and rituximab-resistant DLBCL cell lines. Both anti-CD74/anti-CD20 and anti-IL4R/anti-CD20 were able to mediate ADCC and ADCP, but CD74-targeting therapeutic antibodies could also mediate direct cytotoxicity. Overall, this study strongly indicates that development of bispecific antibodies that target multiple B cell receptors expressed by lymphoma could provide improved defense against relapse and rituximab resistance.
About 4% of all cancer diagnoses in the United States each year are classified as Non-Hodgkin’s lymphomas (NHL) (1). The most common type of NHL is diffuse large B cell lymphoma (DLBCL), which represents 40% of all newly diagnosed lymphomas annually (2). There are many subtypes of DLBCL, which are based on gene expression profiling and location of initiation (3–5). Despite being genetically and phenotypically diverse, most patients with DLBCL are treated with common therapy regardless of the subtypes involved. However, with the advances in immunotherapy, more targeted therapeutic approaches are being implemented.
One such therapeutic is the monoclonal antibody rituximab, which targets the B cell marker CD20 (6). Rituximab was the first monoclonal antibody approved by the United States Food and Drug Administration (FDA) for use in cancer treatment in the late 1990’s, and in the early 2000’s, it was added to the standard DLBCL chemotherapeutic regimen, CHOP, which includes cyclophosphamide, vincristine, doxorubicin, and prednisone (7–13). With the addition of rituximab, R-CHOP demonstrated a 10-year disease-free survival of approximately 64%, an improvement compared to the 42.5% disease-free survival with CHOP treatment alone (8). Despite this success, 30–40% of DLCBL patients experience either relapse or refractory disease with R-CHOP treatment (14–18). There have been various mechanisms proposed for the development of rituximab resistance, including a decrease in complement-dependent cytotoxicity, resistance to killing via antibody-dependent cell-mediated cytotoxicity, and resistance to apoptosis (19). One of the most strongly supported hypotheses attributes resistance to the loss of CD20 expression on B cells following initial rituximab treatment (19–21). Importantly, rituximab targets only select subpopulations of B cells and is not a pan-B cell therapeutic (22). For example, plasma B cells do not express CD20 and are not targeted by rituximab. This has led to the development of immunotherapies targeting other B cells markers to have broader clinical application. Due to the phenotypically diverse nature of DLCBL subtypes, it has been suggested that targeting multiple B cell specific pathways or receptors would be a more effective treatment (3). Additionally, development of resistance may prove to be more difficult if there are multiple therapeutic targets, as these targets would need to be simultaneously mutated or expression level reduced to effectively escape treatment (3).
Bispecific antibodies are a new immunotherapy that allows for expression of an antibody that targets two cell surface receptors, simultaneously. There are multiple mechanisms associated with bispecific antibody efficacy, including complement activation; recruitment of macrophages for antibody-dependent cellular phagocytosis (ADCP); recruitment of natural killer (NK) cells or T-cells for antibody-dependent cell-mediated cytotoxicity (ADCC); apoptosis activation; and priming for cross-presentation by antigen-presenting cells (19, 23, 24). One advantage of bispecific antibodies over more conventional treatments is the decreased risk of resistance against two different targets.
In this study, using a bispecific antibody approach, we sought to broaden the therapeutic benefit of rituximab by developing novel lymphoma-targeted bispecific antibodies. We identified CD74 and IL4R as surface receptors that are expressed on B cell populations that have CD20 or do not have CD20 expression, respectively. Moreover, we found that these markers are expressed in lymphomas and could serve as potential therapeutic targets on B cells. We engineered bispecific antibodies against these targets in combination with anti-CD20. We then evaluated these antibodies for functional targeting of lymphoma cell lines.
Single cell RNA sequencing (scRNA-seq) data was acquired from a previously published dataset of scRNA-seq performed on peripheral blood mononuclear cells (PBMCs) from 12 healthy and HIV-infected subjects that were deidentified from Duke University (25). Original sample collection was reviewed and approved by the Duke Medicine Institutional Review Board. The data is publicly available at SRA BioProject ID: PRJNA681021. PBMCs were thawed, washed and placed in single-cell suspensions with PBS + 0.04% bovine serum albumin (BSA). Cellular suspensions were loaded on a GemCode Single-Cell instrument (10X Genomics, Pleasanton, CA, USA) to generate single-cell beads in emulsion. Single-cell RNA-seq libraries were then prepared using a GemCode Single Cell 3′ Gel bead and library kit version 2 (10X Genomics). Single-cell barcoded cDNA libraries were quantified by quantitative PCR (Kappa Biosystems, Potters Bar, UK) and sequenced on an Illumina (San Diego, CA, USA) NextSeq 500 (26–29). Read lengths were 26 bp for read 1, 8 bp i7 index, and 98 bp read 2. Cells were sequenced to greater than 50,000 reads per cell.
After sequencing, the Cell Ranger Single Cell Software Suite (version 2.1.1) was used to generate sequencing FASTQ files and to perform sample de-multiplexing, barcode processing, reference alignment and single-cell 3′ gene counting (30). Reads were aligned to the human genome (GRCH38). Samples were aggregated using the CellRanger Aggr function to create a single matrix of cell barcodes and gene counts for the groups. During the process each library was normalized for mapped sequencing depth. In order to control for variation in the number of reads per sample (sequencing depth), reads were subsampled from higher-depth libraries until they all had an equal number of reads per cell that were confidently mapped to the transcriptome. Finally, in order to control for technical variation and correct for any batch effects we used the Seurat analysis pipeline Multi CCA method to regress out cell-cell variation in gene expression. The union of variable genes across all individual samples were utilized to renormalize the data.
Matrices of cell barcodes and gene counts generated by Cell Ranger were loaded into Seurat R package (v3.2.3) for graph-based cell clustering, dimensionality reduction and data visualization (31–33). We filtered low quality cells that had lower than 200 expressed transcripts and percentage of mitochondrial genes expressed greater than 20% and for the primary cell model we reduced this threshold to greater than 10% mitochondrial genes. We included up to 45 PCA dimensions for the PBMCs and 48 PCA for the primary cell model for downstream graph-based clustering and UMAP visualization. All other parameters we followed the default Seurat recommendations. We then selected cells that were B cells using CD79A transcript expression and utilized this subset of cells for further analysis. Genes that correlated with MS4A1 or CXCR4 transcript expression were calculated by Pearson correlation and corrected for multiple comparisons using Bonferroni. Graphs and plots were generated using the Seurat and ggplot2 (v3.3.3) R packages and Graphpad Prism version 8 (Graphpad Software, San Diego, CA, USA). The single-cell RNA seq unprocessed reads have been deposited in the NCBI SRA database under the BioProject ID: PRJNA681021. Code and other processed file formats are available from corresponding author/s upon reasonable request.
Lymphoma cDNA Array I and Array II were purchased from OriGene Technologies, Inc. (Rockville, MD, USA). The 2 panels consisted of cDNA from the following tumor samples: 12 normal, 15 stage IE, 41 stage I, 1 stage IIB, 12 stage IIE, 6 stage II, 1 stage III, 7 stage IV, and 1 N/R. Plates with lyophilized cDNA were warmed to room temperature, centrifuged for 30 s at 1,000 revolutions per minute, and pellets from each well suspended in 30 μl of qPCR master mix. For a single 30 μl reaction, the qPCR master mix consisted of 15 μl of 2x Taqman Fast Advanced Master Mix (Applied Biosystems, Waltham, MA, USA), 1.5 μl of 20x Taqman probe (Thermo Fisher Scientific, Waltham, MA, USA), and 13.5 μl of PCR-grade water (Invitrogen, Waltham, MA, USA). Probes utilized in the assay targeted IGSF9 (Hs00325279_m1), CD209 (Hs01588349_m1), MS4A1 (Hs00544819_m1), IL4R (Hs00965056_m1), and CD74 (Hs00269961_m1). An 18S probe (Hs03003631_g1) was used as a housekeeping control. Only 1 probe was used per master mix. Following resuspension, plates were vortexed, centrifuged and placed on ice. Plates were loaded into the QuantStudio 12 Flex (Applied Biosystems). After loading, the reactions underwent an initial 50°C for 2 min activation followed by a 95°C for 10 min pre-soak for 1 cycle. The remaining 42 cycles were performed in two steps: denaturing at 95°C for 15 s followed by annealing at 60°C for 1 min. Data was analyzed by subtracting the Ct values of 18S from the Ct value of the target gene. This difference (x) was transformed via the function 2^(-x) and plotted on a logarithmic scale to illustrate target gene expression relative to 18S for each sample.
SU-DHL-4 (CRL-2957), NU-DUL-1 (CRL-2969), and SU-DHL-8 (CRL-2961) cell lines were purchased from the Non-Hodgkin’s Lymphoma Cell Line Panel at ATCC (Manassas, VA, USA) and maintained in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). All cell lines were cultured at 37°C under 5% CO2. Media was refreshed every 48 h.
RNA extraction was performed according to manufacturer protocol using RNeasy Plus Mini Kit with the additional use of Qiashredder from Qiagen (Hilden, Germany). cDNA synthesis was performed using SuperScript™ VILO™ cDNA Synthesis kit and High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). RT-PCR was performed using TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific) and the following Advanced Biosystems Taqman Probes: 18S TaqMan Probe (FAM-MGB) Assay ID: Hs99999901_s1, MS4A1 TaqMan Probe (FAM-MGB) Assay ID: Hs00544819_m1, CD74 TaqMan Probe (FAM-MGB) Assay ID: Hs00269961_m1, IL4R TaqMan Probe (FAM-MGB) Assay ID: Hs00965056_m1.
Rituximab was purchased from Creative Biolabs (Shirley, NY, USA) and Invivogen (San Diego, CA, USA). Milatuzumab (TAB-763), Dupilumab (TAB-021ML) were purchased from Creative Biolabs. Bispecific antibodies were produced in collaboration with Creative Biolabs. Expression vectors encoding the antibody heavy and light chain gene sequences were transiently transfected and expressed in HEK293F cells. Secreted antibody was purified by Protein A affinity chromatography, ultrafiltration and then subjected to 0.2-micron sterile filtration. Purified antibodies were stored at PBS, pH 7.4. For sequences used for each heavy and light chain in each of the anti-CD20/anti-CD74 bispecific antibody and anti-CD20/anti-IL4R bispecific antibodies (see Supplementary Data 1). Quality of bispecific antibodies was measured by Creative Biolabs by reducing SDS-PAGE and size exclusion chromatography-HPLC (Supplementary Figure 1). AffiniPure Goat Anti-Human IgG, Fcγ fragment specific (109-005-098) cross-linking antibody was purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA).
Enzyme-linked immunosorbent assays (ELISAs) were performed using the following antigens and antibodies: recombinant CD20 full length protein (Acro Biosystems, Newark, DE, USA), recombinant CD74 protein (R&D Systems, Minneapolis, MN, USA), recombinant IL4R protein (R&D Systems), human Anti-CD20 antibody (Invivogen), Milatuzumab/Anti-CD74 Antibody (Creative Biolabs), Dupilumab/IL4R monoclonal antibody (Creative Biolabs), bispecific anti-CD20/anti-CD74 antibody (Creative Biolabs), bispecific anti-CD20/anti-IL4R antibody (Creative Biolabs). Antigens were all diluted to 2 μg/mL in 0.1 M sodium bicarbonate and incubated on high-binding plates (Corning Inc., Corning, NY, USA) overnight at 4 degrees. Antibodies were diluted to 33.3 μg/mL in superblock buffer with sodium azide followed by subsequent 1:3 dilutions until a final dilution of 0.565 ng/mL. Secondary Goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, Inc.) dilutions were done in superblock buffer without sodium azide within range of manufacturer’s recommendations at 1:50,000 dilution. SureBlue Reserve Microwell Substrate (VWR, Radnor, PA, USA) was added and incubated in the dark for 15 min. Immediately after, 0.33 N HCl Acid Stop solution was added to the plate and absorbance was measured at 450 nm.
Cells were seeded at 50,000 cells per well in 24 well dishes. 10 μg/mL of respective antibody and AffiniPure Goat Anti-Human IgG, Fcγ fragment specific cross-linking antibody (Jackson ImmunoResearch Laboratories, Inc.) was added to wells and incubated for 8 h. Cells were collected and incubated in the Muse Annexin V reagent according to the manufacture’s protocol. Total cell death was measured and recorded on the Muse Cell Analyzer (Luminex Corporation, Austin, TX, USA) in reference to crosslinking antibody control wells with the addition of only crosslinking antibody.
ADCC Bioassay Core Kit (Promega, Madison, WI, USA) was performed using the manufacturer protocol with the addition of SU-DHL-4 (CRL-2957), NU-DUL-1 (CRL-2969), and SU-DHL-8 (CRL-2961) cell lines as target cells purchased from ATCC. ADCC Bioassay Complete Kit (Promega) was performed using the manufacturer protocol with the provided Raji target cells. Antibodies were treated at 1 μg/mL and subsequently diluted at a 1:3 ratio until a final concentration of 0.152 ng/mL. Respective target cells were incubated with antibody and modified Jurkat NFAT-luc FcγRIIIa effector cells for 6 h. Bio-Glo luciferase assay reagent was added to each well and luminescence was measured.
ADCP Bioassay Complete Kit (Promega) was performed using the manufacturer protocol with the provided Raji target cells and modified Jurkat NFAT-luc FcγRIIa-H effector cells. Antibodies were treated at 1 μg/mL and subsequently diluted at a 1:3 ratio until a final concentration of 0.152 ng/mL. Raji target cells were incubated with antibody and modified Jurkat NFAT-luc FcγRIIa-H effector cells for 6 h. Bio-Glo luciferase assay reagent was added to each well and luminescence was measured.
To identify putative B cell specific targets for the engineering of bispecific antibodies against DLBCL, we performed expression analysis of B cells isolated from healthy individuals using single cell RNA sequencing from a prior published study (SRA BioProject ID: PRJNA681021). CD20, the target of rituximab, is encoded by the MS4A1 gene. Initial evaluation of the MS4A1 levels showed heterogeneous expression within the B cell subpopulations (Figure 1A). CD20 and the chemokine receptor CXCR4 are critical for B cell trafficking and are molecular targets for cancer immunotherapies (34–37). Moreover, a prior study demonstrated that cells with high CXCR4 expression were less responsive to rituximab treatment suggesting an inverse expression pattern (38, 39). Therefore, we examined the expression CXCR4 and confirmed an inverse relationship between CXCR4 and CD20, with a low Pearson correlation score of 0.008 (Figure 1A and Supplementary Table 1). CXCR4 is known to be expressed by many other immune cell types such as neutrophils and T cells, making it less specific therapeutic targeting of B cells (40–44). Thus, we focused on putative targets that correlated highly with CXCR4 expression, were expressed on the B cell surface, and had FDA-approved monoclonal antibodies available. CD74 and IL4R were identified as candidates for further investigation, with expression that overlapped with CXCR4 and Pearson correlation scores of 0.325 and 0.307, respectively (Figures 1B,C and Supplementary Table 1). Additionally, CD74 was broadly expressed and correlated highly with MS4A1 expression while IL4R did not, with correlation scores of 0.523 and −0.064, respectively (Supplementary Table 1). Thus, both CD74 and IL4R could target B cells, even when CD20 is low or not present.
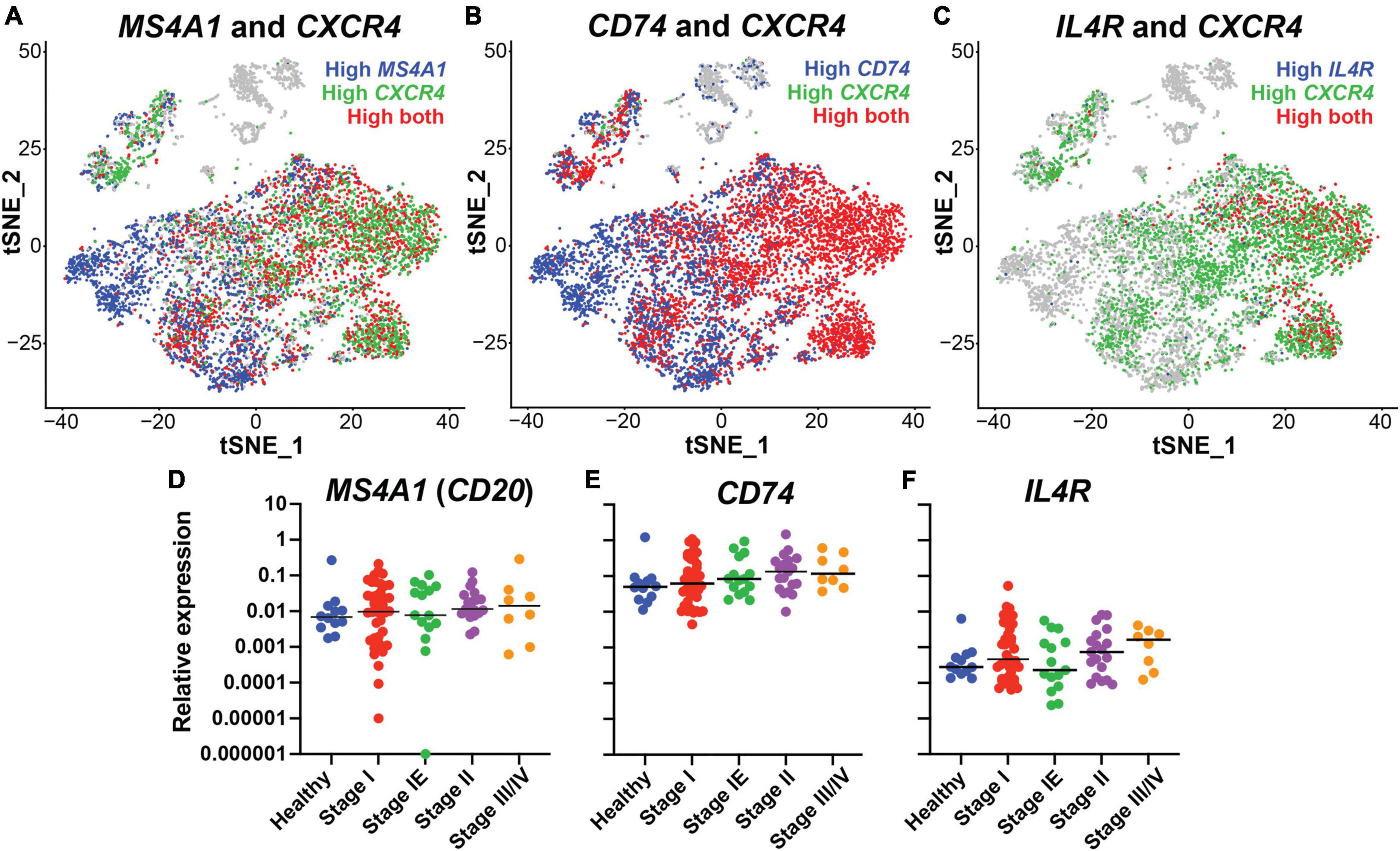
Figure 1. CD74 and IL4R identified as good candidates for bispecific antibody development, in conjunction with CD20, by single cell RNA sequencing. (A) tSNE plot of MS4A1 (Blue) and CXCR4 (Green). (B) tSNE plot of CD74 (Blue) and CXCR4 (Green). (C) tSNE plot of IL4R (Blue) and CXCR4 (Green). (D) Relative expression of MS4A1 in samples collected from healthy donors and donors with lymphoma at different stages of disease. Each dot represents one individual. (E) Relative expression of CD74 in samples collected from healthy donors and donors with lymphoma at different stages of disease. (F) Relative expression of IL4R in samples collected from healthy donors and donors with lymphoma at different stages of disease. n = 12 healthy, 41 stage I, 15 stage IE, 19 stage II, 8 stage III/IV.
Next, we determined MS4A1 (CD20), CD74 and IL4R gene expression levels in 84 lymphoma samples across different tumor stages (stage I-III/IV) and compared to expression in 12 healthy tissue controls using quantitative PCR of tissue biopsies. We found that all three genes were expressed in lymphoma samples of all stages, with no significant difference compared to healthy control samples. CD74 had the highest relative expression level followed by MS4A1 and IL4R. There were no significant correlations with gene expression and tumor stage, although CD74 expression did trend higher in later tumor stages (Figures 1D–F). These results demonstrated that with CD20, genes encoding CD74 and IL4R are highly expressed in lymphoma tissues, in addition to being broad B cell markers, and could represent candidate therapeutic targets for lymphoma.
There are therapeutic antibodies that target CD74 (milatuzumab), IL4R (dupilumab), and CD20 (rituximab) that are approved for clinical use. We utilized these antibody combinations to engineer bispecific antibodies to combine anti-CD74 with anti-CD20 and anti-IL4R with anti-CD20 (Figure 2A). After bispecific antibody expression and purification, we determined antibody binding specificity against CD20, CD74, and IL4R recombinant protein antigens using ELISA. Using area under the curve (AUC) to compare binding response, anti-CD20 (rituximab) (AUC 78.1), bispecific anti-CD20/anti-CD74 (AUC 70.1) and bispecific anti-CD20/anti-IL4R (AUC 79.8) all bound to the recombinant CD20 with no appreciable non-specific binding from either anti-CD74 (milatuzumab) (AUC 1.6) or anti-IL4R (dupilumab) (AUC 1.5) antibodies (Figure 2B). Anti-CD74 (AUC 131.9) and bispecific anti-CD20/anti-CD74 (AUC 132.5) bound exclusively to CD74 with comparable levels of binding (Figure 2C). Likewise, anti-IL4R (AUC 132.6) and bispecific anti-CD20/anti-IL4R (AUC 133) bound exclusively and comparably to IL4R (Figure 2D). These data confirmed antibody specificity and similar binding levels between the monoclonal, single target antibodies and the bispecific antibodies.
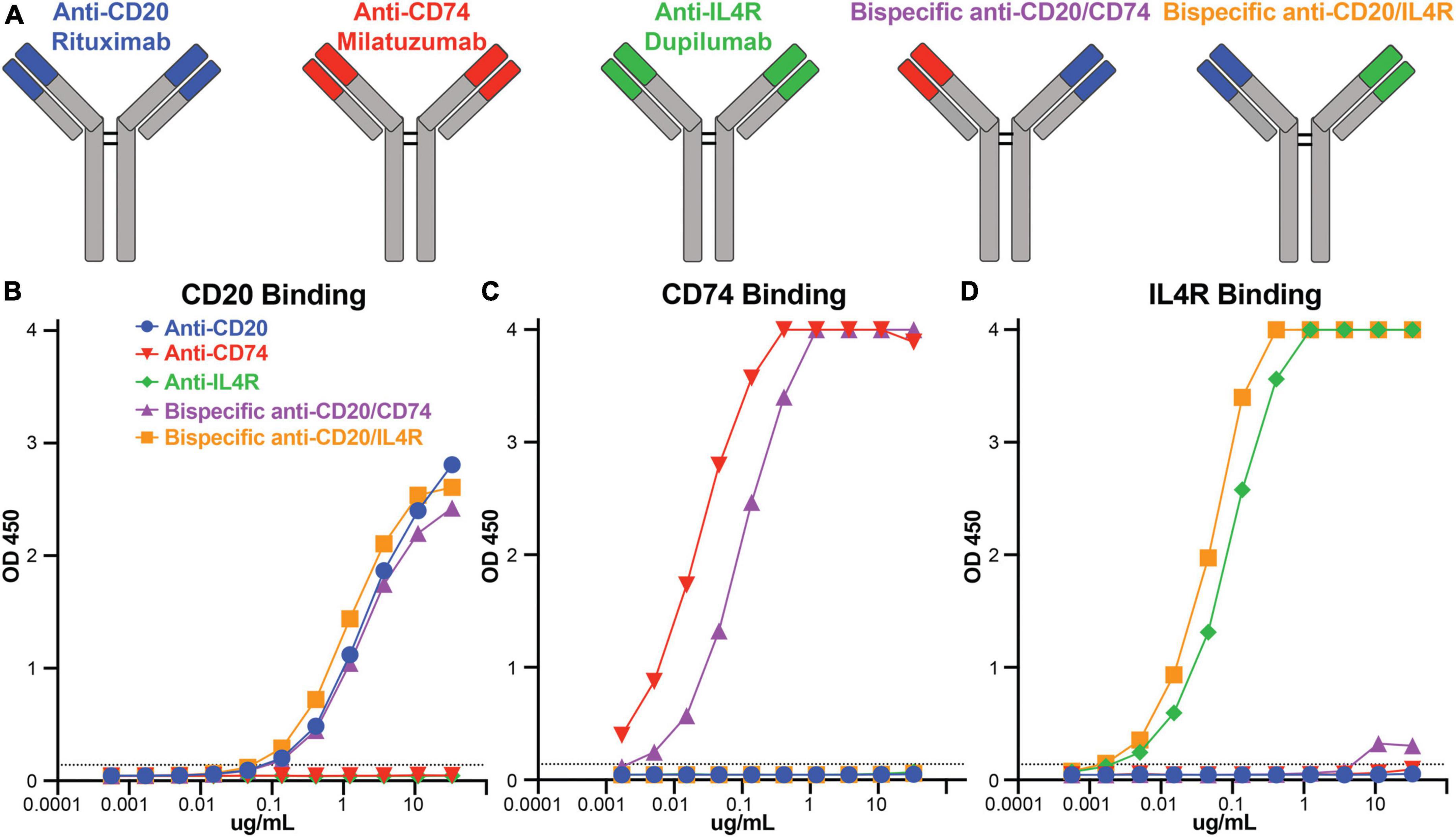
Figure 2. Bispecific antibody construction and antibody binding responses. (A) Schematic representation of rituximab, milatuzumab, dupilumab, and the bispecific antibodies constructed with anti-CD20/anti-CD74 heavy chains and anti-CD20/anti-IL4R heavy chains. (B) Line graph of OD450 absorbance obtained by ELISA measuring antibody binding to CD20 at serial dilutions. (C) Line graph of OD450 absorbance obtained by ELISA measuring antibody binding to CD74 at serial dilutions. (D) Line graph of OD450 absorbance obtained by ELISA measuring antibody binding to IL4R at serial dilutions. Background absorbance, represented by the dashed line, was considered 3X blank well OD450 measurement.
We determined the ability of the antibodies to mediate direct cytotoxicity of three lymphoma cell lines by measuring apoptosis using annexin V in the presence of a cross-linking antibody. We utilized a rituximab-sensitive (SU-DHL-4), rituximab-intermediate (NH-DUL-1) and rituximab-resistant (SU-DHL-8) cell lines (38). First, we determined the expression of MS4A1, CD74, and IL4R in each cell line using qPCR (Supplementary Figure 2). MS4A1 (CD20) gene expression was detected in all three cell lines but was reduced in the rituximab-intermediate and resistant cell lines (Supplementary Figure 2). CD74 expression was detectable in all three cell lines, with no significant difference in expression level between the different cell lines. Similarly, IL4R was detectable in all three cell lines, and like MS4A1, was reduced in the rituximab-intermediate and resistant cell lines (Supplementary Figure 2). This data suggested that reduced gene expression of CD20 and IL4R in the NH-DUL-1 and SU-DHL-8 cell lines could contribute to the observed resistance to antibody therapy. Next, we measured antibody-mediated direct cytotoxicity of the cell lines. We found that the anti-CD20 antibody induced 25.5% apoptosis of the rituximab-sensitive SU-DHL-4 cell line (Figure 3A), 18.3 and 2.3% apoptosis of the intermediate (NH-DUL-1) and resistant (SU-DHL-8) cell lines, respectively (Figures 3B,C). Anti-CD74 antibody also induced direct cytotoxicity of SU-DHL-4 cell line at 8.6% and had higher percent cytotoxicity compared to rituximab (Figure 3A), of the NU-DUL-1 and SU-DHL-8 cell lines with 22.1 and 16.2% induced apoptosis, respectively (Figures 3B,C). Anti-IL4R antibody did not directly induce apoptosis in any of the tested cell lines with 2.7, 0.4, and 0.4% for the SU-DHL-4, NU-DUL-1 and SU-DHL-8 cell lines, respectively (Figures 3A–C). This data indicated that antibodies that are cross-linked to CD20 or CD74 could induce direct cellular cytotoxicity of lymphoma cells, whereas antibodies targeting IL4R could not. Moreover, reduced expression of CD20 on lymphoma cells reduced this cytotoxicity by anti-CD20 antibody.
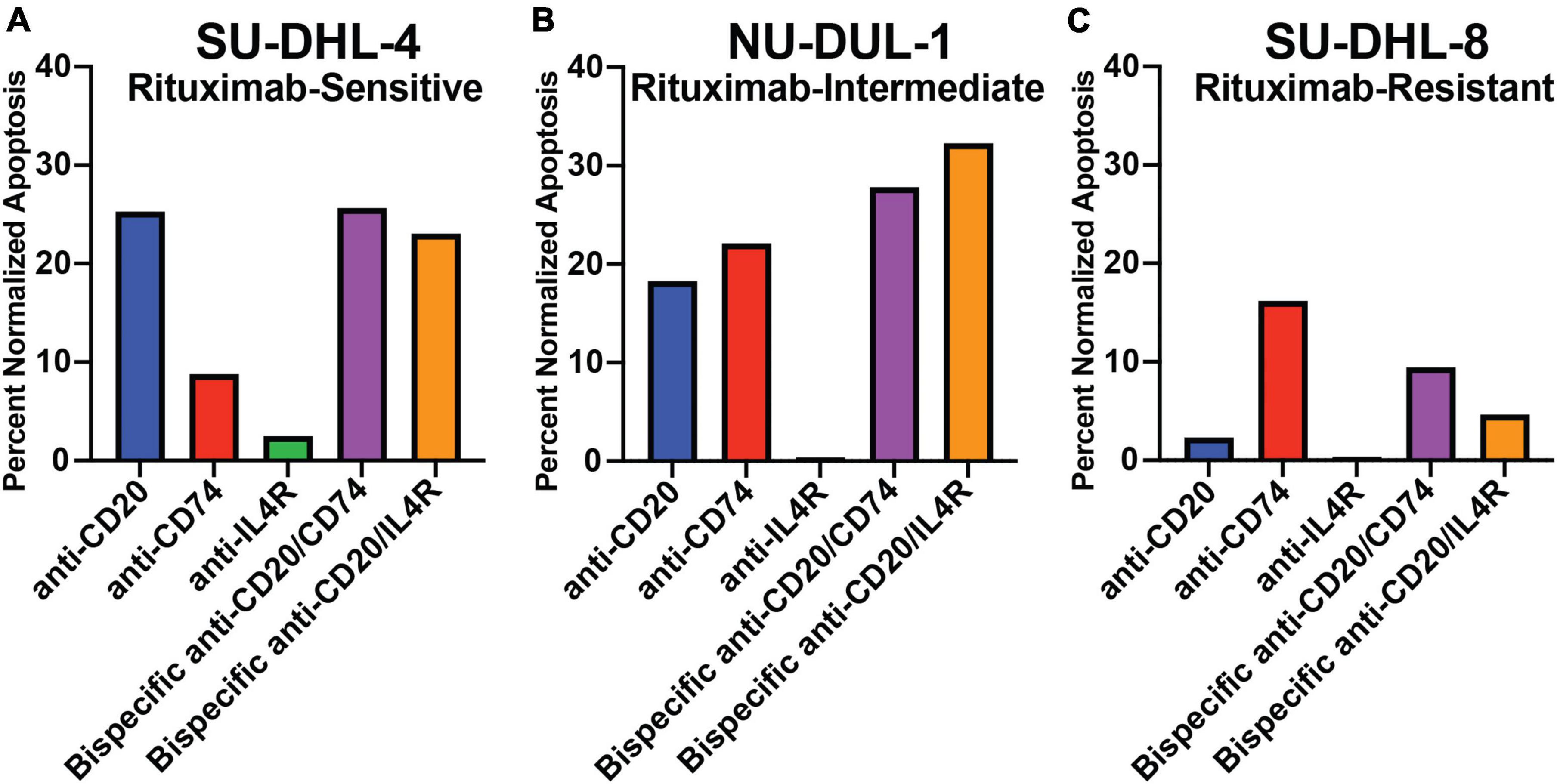
Figure 3. Apoptotic response to crosslinking antibodies within varying rituximab-resistant lymphoma cell lines, measured by annexin V staining. (A) Bar graph of the percent of total apoptosis within rituximab-sensitive, high CD20 expressing, SU-DHL-4 cells after 8 h of exposure to antibodies and crosslinking antibodies. (B) Bar graph of the percent of total apoptosis within rituximab-intermediate, intermediate CD20 expressing, NU-DUL-1 cells after 8 h of exposure to antibodies and crosslinking antibodies. (C) Bar graph of the percent of total apoptosis within rituximab-resistant, low CD20 expressing, SU-DHL-8 cells after 8 h of exposure to antibodies and crosslinking antibodies. All bar graphs normalized to respective cell line crosslinking antibody control.
The bispecific antibodies anti-CD20/anti-CD74 or anti-CD20/anti-IL4R could also mediate direct toxicity of the SU-DHL-4 cell line with 23.6 and 21.9% apoptosis, respectively, albeit not higher than anti-CD20 alone (25.5% apoptosis) (Figure 3A). However, the two bispecific antibodies had higher percent apoptosis of the rituximab-resistant cell lines NU-DUL-1 and SU-DHL-8 compared to anti-CD20 alone (Figures 3B,C). These results demonstrated that bispecific antibodies targeting CD20 and either CD74 or IL4R could mediate direct cytotoxicity of lymphoma cells, but when using a rituximab resistant lymphoma cell line (SU-DHL-8), bispecific anti-CD20/anti-CD74 or anti-CD74 alone induced the highest levels of apoptosis.
In addition to direct cytotoxicity of lymphoma cells by engaging cellular receptors, therapeutic antibodies can engage and recruit effector cells through Fc-Fc receptor interactions to mediate antibody-dependent cellular cytotoxicity (ADCC) or ADCP. We utilized a cell-based assay employing a reporter gene that generates luciferase downstream of the Fc receptor pathway (Promega). For ADCC, FcγRIIIa is engaged and for ADCP, FcγRIIa-H is primarily engaged (Figure 4A). We utilized these cell lines as proxies for ADCC and ADCP activities. We found that anti-CD20 alone could mediate ADCC with increasing concentrations of antibody using Raji (AUC 50,712), SU-DHL-4 (11,125), NU-DUL-1 (AUC 6,535), and SU-DHL-8 (AUC 22,929) lymphoma targets (Figure 4B). The two bispecific antibodies (anti-CD20/anti-CD74 and anti-CD20/anti-IL4R) could also induce ADCC, but at lower levels than anti-CD20 alone. Anti-CD20/anti-CD74 bispecific ADCC AUC for Raji, SU-DHL-4, NU-DUL-1 and SU-DHL-8 cell targets were: 10,512, 1,351, 1,798, and 9,815, respectively (Figure 4B). Anti-CD20/anti-IL4R bispecific ADCC AUC for Raji, SU-DHL-4, NU-DUL-1 and SU-DHL-8 cell targets were: 10,001, 2,123, 1,623, and 11,610, respectively (Figure 4B). Similarly, anti-CD20 and the bispecific antibodies could induce ADCP of Raji cell targets using our cell reporter system, with anti-CD20 having a higher magnitude of response (Figure 4C). The AUC for anti-CD20, bispecific anti-CD20/anti-CD74, and bispecific anti-CD20/anti-IL4R being 1,679, 482, and 510, respectively. These data showed that the anti-CD20 monoclonal antibody and both bispecific antibodies containing anti-CD20 could induce ADCC and ADCP immune responses, although the bispecific antibodies had lower magnitude of ADCC and ADCP at the same antibody concentrations.
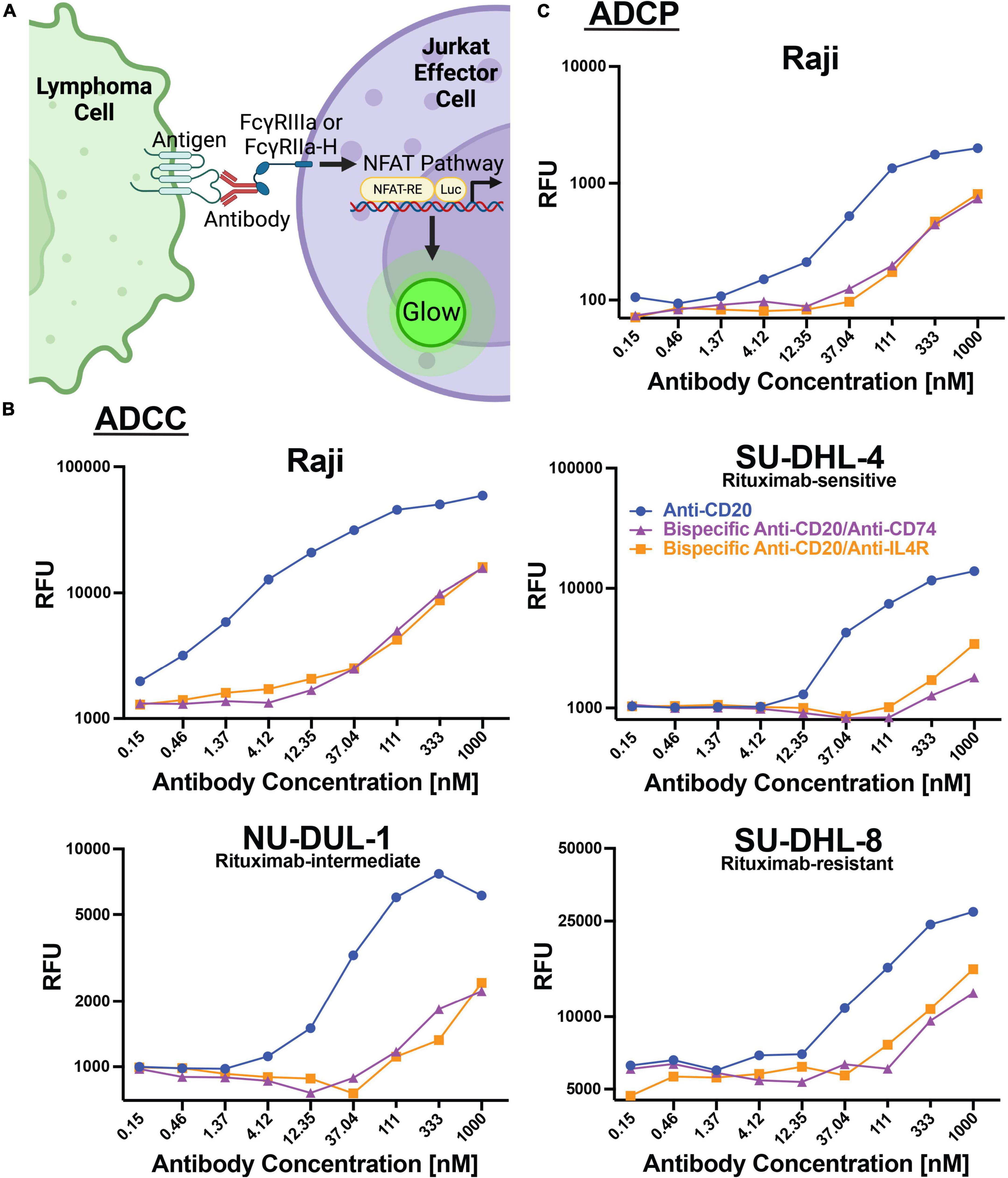
Figure 4. Antibodies induce ADCC and ADCP responses within lymphoma cell lines. (A) Schematic representation of the assays used to measure ADCC and ADCP response within modified Jurkat effector cells. Created with BioRender.com. (B) Line graphs of the relative fluorescent units (RFU) of luciferase produced by ADCC activation of FcγRIIIa modified Jurkat cells when incubated with Raji, SU-DHL-4, NU-DUL-1, or SU-DHL-8 DLBCL cell lines and serial diluted anti-CD20 (Blue), bispecific anti-CD20/anti-CD74 (Purple), or bispecific anti-CD20/anti-IL4R (Orange) antibodies. (C) Line graph of the relative fluorescent units (RFU) of luciferase produced by ADCC activation of FcγRIIa-H modified Jurkat cells when incubated with Burkitt’s lymphoma Raji cells and serial diluted anti-CD20 (Blue), bispecific anti-CD20/anti-CD74 (Purple), or bispecific anti-CD20/anti-IL4R (Orange) antibodies.
Although therapeutic applications of bispecific antibodies are relatively new, the dual nature of the antibodies could be predicted to decrease the ability of lymphomas to escape treatment or develop resistance as two cellular targets would need to be escaped. Besides tagging cells for destruction, bispecific antibodies may also be designed to deliver nanoparticles or drugs to target cells (45). Additionally, some treatments such as CAR-T cells require significant modifications specific to each patient, a costly and time-consuming series of steps that may be avoided with the use of bispecific antibodies (46, 47). Here, we investigated the ability of anti-CD20/anti-CD74 and anti-CD20/anti-IL4R bispecific antibodies to bind their designated targets on lymphoma cells and to kill their targeted cells via different mechanisms.
Using B cells isolated from healthy patients, we explored other potential B cell markers that previously have FDA-approved antibody therapies available and focused on CD74 and IL4R. CD74, a transmembrane glycoprotein that functions as a survival receptor, is highly expressed in malignant B cells (48, 49). Stein et al. demonstrated survival of immune-deficient mice with lymphoma was significantly improved when treated with anti-CD74 antibody, especially when administered in conjunction with rituximab (50). Milatuzumab, the first anti-CD74 monoclonal antibody approved by the FDA for clinical practice, is effective at treating aggressive B cell malignancies such as multiple myeloma, especially in combination with rituximab (49–51). When PBMC samples from patients were treated with milatuzumab, naïve and memory B cells were bound by milatuzumab 98.3 and 97% of the time, respectively (52). Although IL4R expression was limited to a smaller population of total B cells, it provided an opportunity to target cells otherwise capable of escaping rituximab treatment due to their low CD20 expression. Cells treated with the IL4 antagonist, APG201, were more susceptible to chemotherapeutics, suggesting a role for IL4 pathway signaling in chemotherapy resistance (53). Furthermore, anti-IL4R treatments reduced inflammatory cell recruitment, improved measurable lung function and decreased overall asthma symptoms in both humans and monkeys (54, 55). Consequently, both targets would help expand the range of targeted cells compared to current rituximab treatment.
The bispecific antibodies, anti-CD74/anti-CD20 and anti-IL4R/anti-CD20, caused significantly more apoptosis than anti-CD20 alone in both the rituximab intermediate (NU-DUL-1) and rituximab resistant (SU-DHL-8) cell, while the rituximab-sensitive (SU-DHL-4) cell line showed comparable levels of apoptosis between the bispecific antibodies and anti-CD20. These data demonstrate the potential benefit of using bispecific antibodies in conditions of rituximab resistance without losing significant apoptosis induction in conditions of continued rituximab sensitivity. It is interesting to note that the relative patterns of apoptosis induction by anti-CD74 across the cell lines appear to be negatively correlated with CD20 expression: the rituximab-resistant cell line exhibited higher killing by anti-CD74 compared to anti-CD20, while the rituximab-sensitive cell line exhibited lower apoptosis by anti-CD74. Milatuzumab, which has previously been shown to reduce cell growth and proliferation of B cells (52), mediated direct apoptosis of the lymphoma cells in this study. The anti-IL4R drug, dupilumab, an antagonist for the IL4 signaling pathway (56, 57), did not robustly elicit cytotoxicity here. This highlights both CD20 and CD74 as targets to facilitate direct killing of lymphoma cells without the need for effector cells. The significant utility of bispecific antibodies is demonstrated here as anti-IL4R treatment only was quite ineffective, while using an anti-CD20/anti-IL4R bispecific antibody caused apoptosis significantly higher than—or at least comparable to—the anti-CD20 treatment in all cell lines. Gupta et al. presented similar results in other bispecific antibodies, with very little apoptosis after treatment with monoclonal antibodies for CD20 or CD74, but anti-CD20/anti-CD74 bispecific antibodies displayed 3–4 times higher apoptosis than either monoclonal antibody alone (58).
ADCC experiments revealed high activity with anti-CD20 treatment, though the bispecific antibodies showed decreased activity in all cell lines. The decrease in ADCC of the bispecific antibodies could be due to the having only a single Fab arm of the antibody for each target resulting in reduced antibody affinity or other binding kinetic attributes. The decreased effectiveness of the bispecific antibody containing anti-CD74 may also be due to the rapid internalization causing difficulty for the effector cells to detect cell surface antibodies, therefore making cell recruitment unlikely (59–61). Stein et al. explicitly demonstrated that anti-CD74 antibody treatment did not produce significant ADCC in Raji cells when cocultured with purified human leukocyte populations from peripheral blood samples (50). Here, ADCC experiments utilized only T-lymphocytes, however, it is possible that ADCC via NK cells would occur with anti-CD74 treatment (62, 63). ADCP activity was high in Raji cells when treated with anti-CD20, with both anti-CD20/anti-CD74 and anti-CD20/anti-IL4R bispecific antibodies having lower than anti-CD20 but still elevated ADCP response. These bispecific antibodies have shown to be effective in all methods of cell killing evaluated, similar to those results of anti-CD20 alone.
Bispecific antibodies are already being tested in clinical trials and are demonstrating their utility in a broad range of diseases (64–68). The bispecific anti-CD20/anti-CD74 antibody has been tested in mantle cell lymphoma, an NHL sub-type and has significantly improved survival of mice with lymphoma (58). Furthermore, an anti-IL4Rα/anti-IL5 bispecific antibody decreased the number of recruited lymphocytes and eosinophils during asthmatic reactions in mice more effectively than when either antibody was administered alone or concurrently, demonstrating the ability of IL4R bispecific antibodies to be effective in alleviating symptoms of asthma (54). Finally, an anti-CD20/anti-CD3 bispecific antibody targeted lymphoma and cytotoxic T-cells, showed high killing capacity both in vivo and in vitro, even with very low cell surface expression of CD20; anti-CD20 alone was unable to cause significant cell death (46). Phase I clinical trials of this anti-CD20/anti-CD3 antibody, Glofitamab, showed that at high doses, nearly 50% of previously treated patients with aggressive NHL had complete recovery after treatment, and 81% of those remained disease free past 2 years following treatment (47). Previous reports in mantle cell lymphoma cell lines show distinct cytoskeletal dynamics, ROS generation, and disruptions of NF-κB pathways after treatment with rituximab and milatuzumab (51). Future studies would benefit from the use of single-cell RNA sequencing to identify unique downstream pathways of cell death induced by these bispecific anti-CD20/anti-CD74 and anti-CD20/anti-IL4R antibodies.
There were several key limitations of our study that should be addressed in future work. One major limitation is that we evaluated the function of the antibodies in in vitro model systems. While these models could identify promising therapeutic targets and antibodies, in vivo evaluation of the antibodies in animal models will confirm the therapeutic efficacy of these bispecific antibodies. Moreover, in vivo studies will be needed to determine any off-target, and other toxicities, that could be caused by these antibodies. Studies in vivo will also be required to determine the pharmacokinetics of these antibodies in order to determine the clinical feasibility of using them for treatment. Lastly, we evaluated cytotoxicity of several lymphoma cell lines that may not represent primary tumors of broad types. Further study with primary tumors or other types of B-cell derived tumors will be required.
In summary, bispecific antibodies targeting CD74 or IL4R could mediate ADCC and ADCP, while anti-CD74 targeting antibodies could mediate direct cellular cytotoxicity similar to anti-CD20. This observation, coupled with higher expression of CD74 on lymphoma cells, leads to anti-CD74 and anti-CD20 immunotherapies as better therapeutic targets. To further develop these bispecific antibodies as a potential future therapeutic, it would be beneficial to evaluate the level of acquired resistance that develops with the use of prolonged bispecific antibodies both in vitro and in vivo. These data demonstrate that the dual specificity of engineered bispecific antibodies is an effective future prospect of cancer immunotherapy.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author. The single-cell RNA Sequencing Data SRA BioProject ID: PRJNA681021.
TB and EG: conceptualization. EG, SAK, and SP: laboratory experimentation. EG and SK: data analysis. TB, RM, SAK, and EG: writing. All authors contributed to the article and approved the submitted version.
This work was supported through internal institutional funds from the Children’s Mercy Research Institute and Children’s Mercy Kansas City.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.1034594/full#supplementary-material
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. (2022) 72:7–33.
3. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. (2000) 403:503–11.
4. Khanal S, Bradley T. A prognostic gene signature for predicting survival outcome in diffuse large B-cell lymphoma. Cancer Genet. (2021) 252-253:87–95. doi: 10.1016/j.cancergen.2021.01.001
6. Sacchi S, Federico M, Dastoli G, Fiorani C, Vinci G, Clo V, et al. Treatment of B-cell non-hodgkin’s lymphoma with anti CD 20 monoclonal antibody rituximab. Crit Rev Oncol Hematol. (2001) 37:13–25. doi: 10.1016/s1040-8428(00)00069-x
7. Coleman M, Lammers PE, Ciceri F, Jacobs IA. Role of rituximab and rituximab biosimilars in diffuse large B-cell lymphoma. Clin Lymphoma Myeloma Leuk. (2016) 16:175–81. doi: 10.1016/j.clml.2016.01.004
8. Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the groupe d’etudes des lymphomes de l’adulte. Blood. (2010) 116:2040–5. doi: 10.1182/blood-2010-03-276246
9. Griffin MM, Morley N. Rituximab in the treatment of non-hodgkin’s lymphoma–a critical evaluation of randomized controlled trials. Expert Opin Biol Ther. (2013) 13:803–11. doi: 10.1517/14712598.2013.786698
10. Lee L, Crump M, Khor S, Hoch JS, Luo J, Bremner K, et al. Impact of rituximab on treatment outcomes of patients with diffuse large b-cell lymphoma: a population-based analysis. Br J Haematol. (2012) 158:481–8. doi: 10.1111/j.1365-2141.2012.09177.x
11. Lindenmeyer LP, Hegele V, Caregnato JP, Wust D, Grazziotin L, Stoll P. Follow-up of patients receiving rituximab for diffuse large B cell lymphoma: an overview of systematic reviews. Ann Hematol. (2013) 92:1451–9. doi: 10.1007/s00277-013-1811-4
12. Press OW, Leonard JP, Coiffier B, Levy R, Timmerman J. Immunotherapy of non-hodgkin’s lymphomas. Hematology Am Soc Hematol Educ Program. (2001) 2001:221–40.
13. Sehn LH, Donaldson J, Chhanabhai M, Fitzgerald C, Gill K, Klasa R, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. (2005) 23:5027–33. doi: 10.1200/JCO.2005.09.137
14. Cai Q, Westin J, Fu K, Desai M, Zhang L, Huang H, et al. Accelerated therapeutic progress in diffuse large B cell lymphoma. Ann Hematol. (2014) 93:541–56.
15. Cultrera JL, Dalia SM. Diffuse large B-cell lymphoma: current strategies and future directions. Cancer Control. (2012) 19:204–13.
16. Davis TA, Grillo-Lopez AJ, White CA, McLaughlin P, Czuczman MS, Link BK, et al. Rituximab anti-CD20 monoclonal antibody therapy in non-hodgkin’s lymphoma: safety and efficacy of re-treatment. J Clin Oncol. (2000) 18:3135–43. doi: 10.1200/JCO.2000.18.17.3135
17. Falgas A, Pallares V, Unzueta U, Cespedes MV, Arroyo-Solera I, Moreno MJ, et al. A CXCR4-targeted nanocarrier achieves highly selective tumor uptake in diffuse large B-cell lymphoma mouse models. Haematologica. (2020) 105:741–53. doi: 10.3324/haematol.2018.211490
18. Feugier P, Van Hoof A, Sebban C, Solal-Celigny P, Bouabdallah R, Ferme C, et al. Long-term results of the R-CHOP study in the treatment of elderly patients with diffuse large B-cell lymphoma: a study by the groupe d’etude des lymphomes de l’adulte. J Clin Oncol. (2005) 23:4117–26. doi: 10.1200/JCO.2005.09.131
20. Beers SA, French RR, Chan HT, Lim SH, Jarrett TC, Vidal RM, et al. Antigenic modulation limits the efficacy of anti-CD20 antibodies: implications for antibody selection. Blood. (2010) 115:5191–201. doi: 10.1182/blood-2010-01-263533
21. Davis TA, Czerwinski DK, Levy R. Therapy of B-cell lymphoma with anti-CD20 antibodies can result in the loss of CD20 antigen expression. Clin Cancer Res. (1999) 5:611–5.
22. Leandro MJ. B-cell subpopulations in humans and their differential susceptibility to depletion with anti-CD20 monoclonal antibodies. Arthritis Res Ther. (2013) 15 Suppl 1:S3. doi: 10.1186/ar3908
23. Cruz JW, Damko E, Modi B, Tu N, Meagher K, Voronina V, et al. A novel bispecific antibody platform to direct complement activity for efficient lysis of target cells. Sci Rep. (2019) 9:12031. doi: 10.1038/s41598-019-48461-1
24. Del Bano J, Chames P, Baty D, Kerfelec B. Taking up cancer immunotherapy challenges: bispecific antibodies, the path forward? Antibodies (Basel). (2015) 5:1. doi: 10.3390/antib5010001
25. Pollara J, Khanal S, Edwards RW, Hora B, Ferrari G, Haynes BF, et al. Single-cell analysis of immune cell transcriptome during HIV-1 infection and therapy. BMC Immunol (2022) 23:48. doi: 10.1186/s12865-022-00523-2
26. Bradley T, Ferrari G, Haynes BF, Margolis DM, Browne EP. Single-cell analysis of quiescent hiv infection reveals host transcriptional profiles that regulate proviral latency. Cell Rep. (2018) 25:107–17.e3. doi: 10.1016/j.celrep.2018.09.020
27. Bradley T, Kuraoka M, Yeh CH, Tian M, Chen H, Cain DW, et al. Immune checkpoint modulation enhances HIV-1 antibody induction. Nat Commun. (2020) 11:948.
28. Bradley T, Peppa D, Pedroza-Pacheco I, Li D, Cain DW, Henao R, et al. RAB11FIP5 expression and altered natural killer cell function are associated with induction of HIV broadly neutralizing antibody responses. Cell. (2018) 175:387–99.e17. doi: 10.1016/j.cell.2018.08.064
29. Han Q, Bradley T, Williams WB, Cain DW, Montefiori DC, Saunders KO, et al. Neonatal Rhesus macaques have distinct immune cell transcriptional profiles following HIV envelope immunization. Cell Rep. (2020) 30:1553–69.e6. doi: 10.1016/j.celrep.2019.12.091
30. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. (2017) 8:14049.
31. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. (2015) 161:1202–14. doi: 10.1016/j.cell.2015.05.002
32. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. (2015) 33:495–502.
33. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single-cell data. Cell. (2019) 177:1888–1902.e21.
34. Beider K, Ribakovsky E, Abraham M, Wald H, Weiss L, Rosenberg E, et al. Targeting the CD20 and CXCR4 pathways in non-hodgkin lymphoma with rituximab and high-affinity CXCR4 antagonist BKT140. Clin Cancer Res. (2013) 19:3495–507. doi: 10.1158/1078-0432.CCR-12-3015
35. Nie Y, Waite J, Brewer F, Sunshine MJ, Littman DR, Zou YR. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J Exp Med. (2004) 200:1145–56. doi: 10.1084/jem.20041185
36. Reinholdt L, Laursen MB, Schmitz A, Bodker JS, Jakobsen LH, Bogsted M, et al. The CXCR4 antagonist plerixafor enhances the effect of rituximab in diffuse large B-cell lymphoma cell lines. Biomark Res. (2016) 4:12. doi: 10.1186/s40364-016-0067-2
37. Tedder TF, Streuli M, Schlossman SF, Saito H. Isolation and structure of a cDNA encoding the B1 (CD20) cell-surface antigen of human B lymphocytes. Proc Natl Acad Sci USA. (1988) 85:208–12. doi: 10.1073/pnas.85.1.208
38. Laursen MB, Reinholdt L, Schonherz AA, Due H, Jespersen DS, Grubach L, et al. High CXCR4 expression impairs rituximab response and the prognosis of R-CHOP-treated diffuse large B-cell lymphoma patients. Oncotarget. (2019) 10:717–31. doi: 10.18632/oncotarget.26588
39. Pavlasova G, Borsky M, Seda V, Cerna K, Osickova J, Doubek M, et al. Ibrutinib inhibits CD20 upregulation on CLL B cells mediated by the CXCR4/SDF-1 axis. Blood. (2016) 128:1609–13. doi: 10.1182/blood-2016-04-709519
40. Eash KJ, Means JM, White DW, Link DC. CXCR4 is a key regulator of neutrophil release from the bone marrow under basal and stress granulopoiesis conditions. Blood. (2009) 113:4711–9. doi: 10.1182/blood-2008-09-177287
41. Arieta Kuksin C, Gonzalez-Perez G, Minter LM. CXCR4 expression on pathogenic T cells facilitates their bone marrow infiltration in a mouse model of aplastic anemia. Blood. (2015) 125:2087–94. doi: 10.1182/blood-2014-08-594796
42. Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carre C, et al. RNA-Seq signatures normalized by MRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. (2019) 26:1627–40.e7. doi: 10.1016/j.celrep.2019.01.041
43. Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. (2018) 175:1701–15.e16.
44. Susek KH, Karvouni M, Alici E, Lundqvist A. The role of CXC chemokine receptors 1-4 on immune cells in the tumor microenvironment. Front Immunol. (2018) 9:2159. doi: 10.3389/fimmu.2018.02159
45. Tekewe A, Saleh M, Kassaye M. Proteins and peptides as targeting carriers in anticancer drug delivery: a review. Int J Pharm Sci Res. (2013) 4:1–18.
46. Sun LL, Ellerman D, Mathieu M, Hristopoulos M, Chen X, Li Y, et al. Anti-CD20/CD3 T cell-dependent bispecific antibody for the treatment of B cell malignancies. Sci Transl Med. (2015) 7:287ra70. doi: 10.1126/scitranslmed.aaa4802
47. Hutchings M, Morschhauser F, Iacoboni G, Carlo-Stella C, Offner FC, Sureda A, et al. Glofitamab, a novel, bivalent CD20-targeting T-cell-engaging bispecific antibody, induces durable complete remissions in relapsed or refractory B-cell lymphoma: a phase I trial. J Clin Oncol. (2021) 39:1959–70. doi: 10.1200/JCO.20.03175
48. Starlets D, Gore Y, Binsky I, Haran M, Harpaz N, Shvidel L, et al. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. (2006) 107:4807–16. doi: 10.1182/blood-2005-11-4334
49. Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J, et al. CD74: a new candidate target for the immunotherapy of B-cell neoplasms. Clin Cancer Res. (2007) 13:5556s–63s. doi: 10.1158/1078-0432.CCR-07-1167
50. Stein R, Qu Z, Cardillo TM, Chen S, Rosario A, Horak ID, et al. Antiproliferative activity of a humanized anti-CD74 monoclonal antibody, hLL1, on B-cell malignancies. Blood. (2004) 104:3705–11. doi: 10.1182/blood-2004-03-0890
51. Alinari L, Yu B, Christian BA, Yan F, Shin J, Lapalombella R, et al. Combination anti-CD74 (milatuzumab) and anti-CD20 (rituximab) monoclonal antibody therapy has in vitro and in vivo activity in mantle cell lymphoma. Blood. (2011) 117:4530–41. doi: 10.1182/blood-2010-08-303354
52. Frolich D, Blassfeld D, Reiter K, Giesecke C, Daridon C, Mei HE, et al. The anti-CD74 humanized monoclonal antibody, milatuzumab, which targets the invariant Chain of MHC II complexes, alters B-cell proliferation, migration, and adhesion molecule expression. Arthritis Res Ther. (2012) 14:R54. doi: 10.1186/ar3767
53. Natoli A, Lupertz R, Merz C, Muller WW, Kohler R, Krammer PH, et al. Targeting the IL-4/IL-13 signaling pathway sensitizes Hodgkin lymphoma cells to chemotherapeutic drugs. Int J Cancer. (2013) 133:1945–54. doi: 10.1002/ijc.28189
54. Godar M, Deswarte K, Vergote K, Saunders M, de Haard H, Hammad H, et al. A bispecific antibody strategy to target multiple type 2 cytokines in asthma. J Allergy Clin Immunol. (2018) 142:1185–193.e4. doi: 10.1016/j.jaci.2018.06.002
56. Le Floc’h A, Allinne J, Nagashima K, Scott G, Birchard D, Asrat S, et al. Dual blockade of IL-4 and IL-13 with dupilumab, an IL-4Ralpha antibody, is required to broadly inhibit type 2 inflammation. Allergy. (2020) 75:1188–204. doi: 10.1111/all.14151
57. Kim JE, Jung K, Kim JA, Kim SH, Park HS, Kim YS. Engineering of anti-human interleukin-4 receptor alpha antibodies with potent antagonistic activity. Sci Rep. (2019) 9:7772. doi: 10.1038/s41598-019-44253-9
58. Gupta P, Goldenberg DM, Rossi EA, Cardillo TM, Byrd JC, Muthusamy N, et al. Dual-targeting immunotherapy of lymphoma: potent cytotoxicity of anti-CD20/CD74 bispecific antibodies in mantle cell and other lymphomas. Blood. (2012) 119:3767–78. doi: 10.1182/blood-2011-09-381988
59. Roche PA, Teletski CL, Stang E, Bakke O, Long EO. Cell surface HLA-DR-invariant chain complexes are targeted to endosomes by rapid internalization. Proc Natl Acad Sci USA. (1993) 90:8581–5. doi: 10.1073/pnas.90.18.8581
60. Ong GL, Goldenberg DM, Hansen HJ, Mattes MJ. Cell surface expression and metabolism of major histocompatibility complex class II invariant chain (CD74) by diverse cell lines. Immunology. (1999) 98:296–302.
61. Binsky I, Haran M, Starlets D, Gore Y, Lantner F, Harpaz N, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci USA. (2007) 104:13408–13. doi: 10.1073/pnas.0701553104
62. Karakikes I, Morrison IE, O’Toole P, Metodieva G, Navarrete CV, Gomez J, et al. Interaction of HLA-DR and CD74 at the cell surface of antigen-presenting cells by single particle image analysis. FASEB J. (2012) 26:4886–96. doi: 10.1096/fj.12-211466
63. Jiang YZ, Couriel D, Mavroudis DA, Lewalle P, Malkovska V, Hensel NF, et al. Interaction of natural killer cells with MHC class II: reversal of HLA-DR1-mediated protection of K562 transfectant from natural killer cell-mediated cytolysis by brefeldin-A. Immunology. (1996) 87:481–6. doi: 10.1046/j.1365-2567.1996.483556.x
64. Heiss MM, Strohlein MA, Jager M, Kimmig R, Burges A, Schoberth A, et al. Immunotherapy of malignant ascites with trifunctional antibodies. Int J Cancer. (2005) 117:435–43.
66. Heiss MM, Murawa P, Koralewski P, Kutarska E, Kolesnik OO, Ivanchenko VV, et al. The trifunctional antibody catumaxomab for the treatment of malignant ascites due to epithelial cancer: results of a prospective randomized phase II/III trial. Int J Cancer. (2010) 127:2209–21. doi: 10.1002/ijc.25423
67. Acheampong DO. Bispecific antibody (bsAb) construct formats and their application in cancer therapy. Protein Pept Lett. (2019) 26:479–93.
Keywords: bispecific antibody, lymphoma, diffuse large B cell lymphoma, rituximab, cancer therapy, CD20, CD74, IL4R
Citation: Geanes ES, Krepel SA, McLennan R, Pierce S, Khanal S and Bradley T (2022) Development of combinatorial antibody therapies for diffuse large B cell lymphoma. Front. Med. 9:1034594. doi: 10.3389/fmed.2022.1034594
Received: 01 September 2022; Accepted: 05 October 2022;
Published: 24 October 2022.
Edited by:
Bernhard F. Gibbs, University of Oldenburg, GermanyReviewed by:
Cheng-Rong Yu, National Eye Institute (NIH), United StatesCopyright © 2022 Geanes, Krepel, McLennan, Pierce, Khanal and Bradley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Todd Bradley, dGNicmFkbGV5QGNtaC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.