- 1Center for Clinical Laboratory, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2School of Gusu, The Affiliated Suzhou Hospital of Nanjing Medical University, Suzhou Municipal Hospital, Nanjing Medical University, Suzhou, China
- 3Department of Laboratory Medicine, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Objective: To assess the association between lipid metabolism and fetal fraction, which is a critical factor in ensuring a highly accurate non-invasive prenatal testing (NIPT), and on the rate of screen failures or “no calls” in NIPT.
Methods: A total of 4,514 pregnant women at 12–26 weeks of gestation underwent NIPT sequencing and serum lipid measurements. Univariate analysis and multivariate regression models were used to evaluate the associations of serum lipid concentrations with the fetal fraction and the rate of screen failures.
Results: The fetal fraction decreased with increased low-density lipoprotein cholesterol and triglyceride (TG) levels, which were significant factors (standardized coefficient: −0.11). Conversely, high-density lipoprotein cholesterol and the interval between the two tests were positively correlated with the fetal fraction. The median fetal fraction was 10.88% (interquartile range, 8.28–13.89%) and this decreased with TG from 11.56% at ≤1.10 mmol/L to 9.51% at >2.30 mmol/L. Meanwhile, multivariate logistic regression analysis revealed that increased TG levels were independently associated with the risk of screen failures. The rate of screen failures showed an increase with TG levels from 1.20% at ≤1.70 mmol/L to 2.41% at >2.30 mmol/L.
Conclusions: The fetal fraction and the rate of screen failures in NIPT are affected by TG levels. Meanwhile, in pregnant women with high TG levels, delaying the time between NIPT blood collections can significantly increase the fetal fraction.
Introduction
Massively parallel sequencing of cell-free DNA (cfDNA) in maternal blood is a non-invasive prenatal testing (NIPT) method that has been applied in screening for trisomy 21, 18, and 13 (1, 2). In trisomic pregnancies, cfDNA derived from the extra fetal chromosome leads to a higher proportion of fetal DNA than in disomic pregnancies (3). Indeed, the proportional increase caused by the trisomy chromosome is equal to half the fetal fraction (4). Thus, the accurate identification of fetal aneuploidy depends largely on the detection of the fetal fraction using the NIPT method (5, 6).
The American College of Medical Genetics and Genomics defines low fetal fractions as less than 4% (<4%) (7), which would result in test failures and false-negative results (8). This is assuming that NIPT screening has a 100% trisomy 21 (T21) detection rate at different failure rates. When the failure rate is 1, 5, or 10%, the corresponding detection rate of the screening population declines to 99, 95, and 90%, respectively, assuming that the incidence of T21 among failures is the same as that reported in the literature (9). Indeed, recent evidence suggests that a 99.7% detection rate for T21 (357 of 358) was observed in a study of 58,048 pregnant women with whole-genome sequencing-based NIPT. There were 13 T21 cases with a fetal fraction of <4%. The sensitivities for T21 decreased from 99.7 to 96.4% when considering screening failure cases (10, 11). Therefore, screening failure will negatively influence NIPT performance, and it is recommended to find a variety of other strategies to reduce this rate.
Researchers have mainly focused on two aspects to reduce the rate of screen failures. First, Lo et al. performed plasma DNA size analysis and found that the most significant difference between fetal and maternal cfDNA was a decrease in the 166-bp peak relative to the 143-bp peak (12, 13). From this, they have developed a new NIPT method to enrich shorter cfDNA fragments (<140 bp) to significantly improve the fetal fraction (by 2.3-fold) and analytical performance of NIPT (11, 14–17). However, this new method has not been applied in clinical practice. Second, other researchers have studied the effects of maternal and fetal characteristics and experimental factors on fetal fraction with the aim of adjusting these factors to obtain higher numbers of fetal-derived cfDNA. Fetal fraction can be influenced by maternal body mass index (BMI), gestational age (GA), anticoagulation therapy, blood collection, and fetal aneuploidy (14, 18, 19). Based on prior research, one potential way to reduce the incidence of screening failure is to repeat test failures by sequencing a second sample. The second sample can either be collected in conjunction with the first sample or from a redraw after initial failure due to a low fetal fraction (20). However, recent studies have shown that only 56–66% of repeated NIPT after a second blood draw are successful, suggesting that other factors may also affect the fetal fraction. Serum lipid levels, the main source of cfDNA in the plasma of pregnant women, are extensively associated with peripheral leukocyte counts, and this may affect the fetal fraction.
The aims of this study on 4,514 pregnant women undergoing NIPT at 12–26 weeks of gestation were, first, to examine the possible effects of lipid metabolism on the fetal fraction and second, to estimate the proportion of pregnancies at a high risk for screen failure.
Materials and Methods
Study Population
This was a single-center, retrospective study investigating pregnant women who underwent NIPT to screen for trisomy 21, 18, 13 and serum lipid measurement at 12–26 weeks of gestation. Data on 4,514 pregnancies with male fetuses were collected between January 2016 and December 2020 at the Suzhou Municipal Hospital. The study was approved by the Reproductive Medicine Ethics Committee of the Suzhou Municipal Hospital (ID: K-2021-032-H01).
Procedure and Data Collection
Serum samples were collected after overnight fasting and analyzed for total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), very low-density lipoprotein cholesterol (VLDL-C), and triglyceride (TG) levels using Beckman-Coulter AU5800 (Beckman Coulter, Brea, USA) according to the manufacturer's instructions.
All patients received pretest counseling and were given informed consent by genetic counselors before testing for NIPT. Genetic counseling primarily included topics on elderly pregnancy, previous pregnancy affected by fetal aneuploidy, and ultrasonic detection for aneuploidy or fetal abnormalities. The pregnant women screened using NIPT underwent ultrasound before 14 weeks of gestation to determine the number of fetuses, chorionicity, GA, and maternal and fetal characteristics. Medical history including the maternal age, weight, BMI, number of pregnancies, singleton or twin pregnancies, GA, and the method of conception were also recorded.
Blood samples (10 mL) were collected on-site from pregnant women who gave their consent following pretest counseling provided by genetic counselors. cfDNA was extracted using the QIAamp DSP DNA Blood Mini Kit (Qiagen) according to the manufacturer's instructions. The library was constructed using polymerase chain reaction. cfDNA and library concentrations were measured using the QubitTM dsDNA HS Kit (Invitrogen, Carlsbad, CA, USA). Sequencing libraries were then sequenced using the Ion Proton system or the BGISEQ-500 (MGI, China) system. The fetal fraction was evaluated by calculating the proportion of chromosome Y reads. The fetal fraction was reported to be >4%. In samples with a fetal fraction <4%, the laboratory did not generate a risk assessment. All samples had a fetal karyotype (pregnancy with a positive NIPT result) or clinical follow-up results.
Statistical Analysis
Descriptive data are presented as median and interquartile range for non-normally continuous variables and as absolute values and percentages for categorical variables. The square root (√) of the measured fetal fraction was obtained to make the distribution a Gaussian normality to be assessed using a probability plot. Univariate analysis and multivariate linear regression models were used to examine the associations of the fetal fraction with TG, TC, HDL-C, LDL-C, VLDL-C, maternal weight (kg), GA (weeks), and interval between two tests (weeks). Variance inflation factor (VIF) was used to evaluate multicollinearity; VIF values ≥ 4 require further investigation, whereas VIF values > 10 indicate severe multicollinearity and need to be corrected. Maternal weight and TG levels were highly correlated, and the relationship between GA and the time interval between the two tests was also the same; weight and gestational week were not included in the multiple linear regression model analysis. TG levels were categorized as ≤ 1.10, 1.11–1.70, 1.71–2.30, and >2.30 mmol/L. We also computed estimates and 95% confidence intervals (CIs) for the mean differences in fetal fraction for each category of TG levels. A total of three models were used. Model 1 was a univariate linear regression of the relationship between the TG level and fetal fraction. Model 2 was adjusted for HDL-C and LDL-C levels. In model 3, the time interval between the two tests (weeks) was added on the basis of model 2.
Multiple logistic regression was used to determine the associations of TG, HDL-C, LDL-C and the time interval between the two tests (weeks) with the test failure rate. SPSS version 26.0 (IBM Corp, Armonk, NY, USA) was used for the data analysis. All P-values were two-sided, and statistical significance was set at P < 0.05.
Results
Sample Characteristics
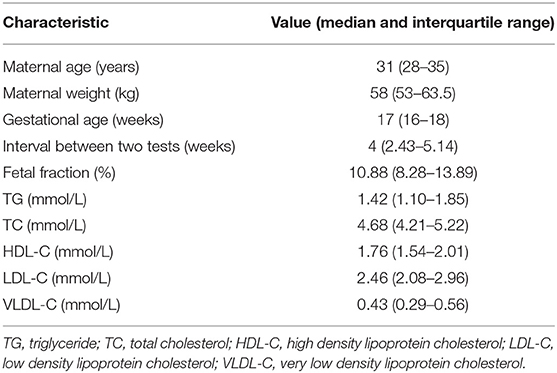
The maternal characteristics of the study population are presented in Table 1, while the frequency distribution of the fetal fraction is shown in Figure 1. The results from 4,514 pregnancies with male fetuses showed that the mean maternal age, weight, NIPT sampling GA, time interval between two tests (weeks), TG, TC, HDL-C, LDL-C, and VLDL-C levels, and fetal DNA concentration were 31 years (28–35), 58 kg (53–63.5), 17 weeks (16–18), 4 weeks (2.43–5.14), 1.42 mmol/L (1.10–1.85), 4.68 mmol/L (4.21–5.22), 1.76 mmol/L (1.54–2.01), 2.46 mmol/L (2.08–2.96), 0.43 mmol/L (0.29–0.56), and 10.88% (8.28–13.89), respectively.
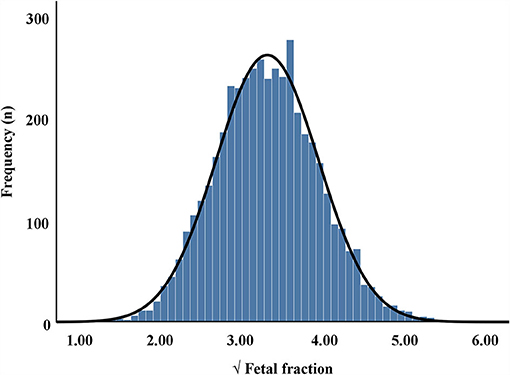
Figure 1. Frequency distribution of square root (√) of fetal fraction in maternal plasma cell-free DNA.
Relationship Between Lipid Metabolism and Fetal Fraction
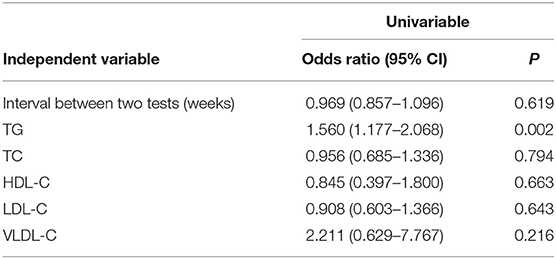
Univariate regression analysis demonstrated that the fetal fraction was negatively correlated with maternal weight (regression coefficient = −0.024, P < 0.001), TG (regression coefficient = −0.135, P < 0.001), TC (regression coefficient = −0.032, P = 0.008), LDL-C (regression coefficient = −0.065, P < 0.001), and VLDL-C (regression coefficient = −0.195, P < 0.001) levels and positively correlated with GA (regression coefficient = 0.034, P < 0.001), time interval between the two tests (regression coefficient = 0.021, P < 0.001), and HDL-C (regression coefficient = 0.129, P < 0.001; Table 2) level. Since the maternal weight was related to the TG level (r = 0.221, P < 0.001), it was not included in the multiple linear regression analysis. NIPT sampling GA and the interval between the two tests were highly correlated (r = 0.697, P < 0.001). It is still under consideration whether lengthening the interval will change the effect of lipid metabolism on NIPT. Therefore, only the time interval between the two tests was included in the multiple linear regression study.
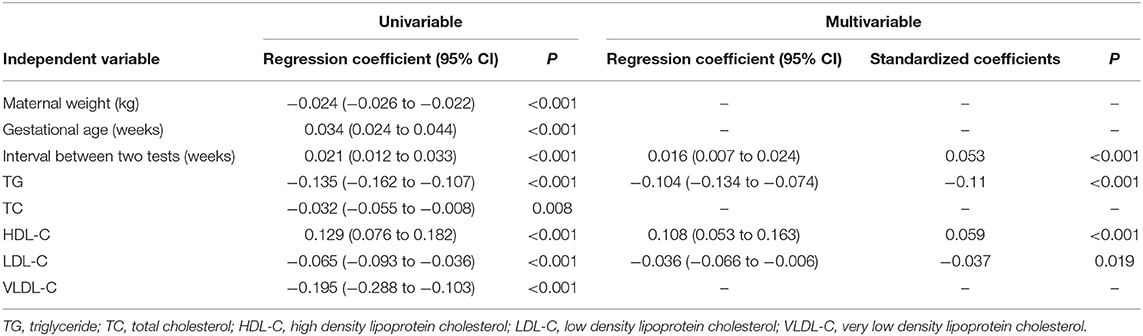
Table 2. Regression analysis of factors from maternal characteristics and lipid metabolism for predicting √fetal fraction in 4,514 pregnancies with male fetuses.
Moreover, multivariable regression analysis demonstrated that the fetal fraction was negatively correlated with the TG (standardized coefficients = −0.11, P < 0.001) and LDL-C (standardized coefficients = −0.037, P = 0.019) levels and positively correlated with the time interval between the two tests (standardized coefficients = 0.053, P < 0.001) and the HDL-C (standardized coefficients = 0.059, P < 0.001; Table 2) level.
Association Between TG and Fetal Fraction
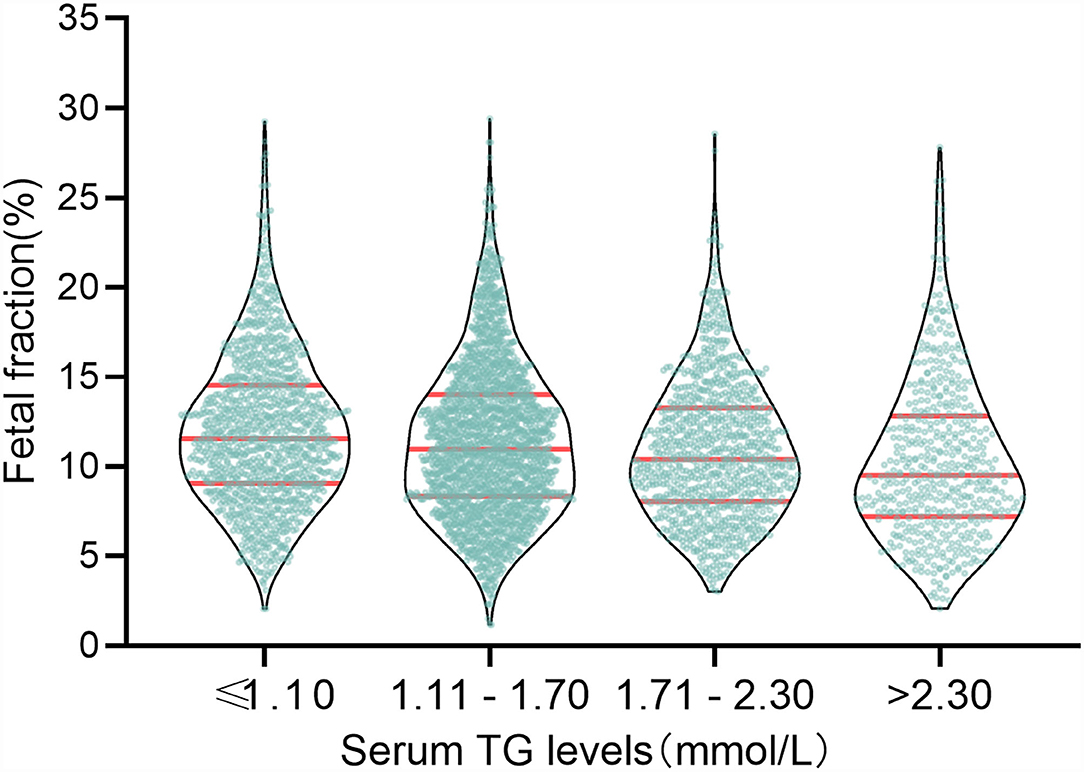
The above results verify that the TG level was negatively correlated with fetal fraction in NIPT (Figure 2). TG levels were classified into four categories: ≤ 1.10, 1.11–1.70, 1.71–2.30, and >2.30 mmol/L. The numbers of pregnant women with TG levels in these four categories were 1163, 1923, 930, and 498, respectively. The mean fetal fractions across the TG categories were 11.56% (9.06–14.53), 10.96% (6.33–14.02), 10.41% (8.05–13.27), and 9.51% (7.22– 12.81), respectively (Figure 3).
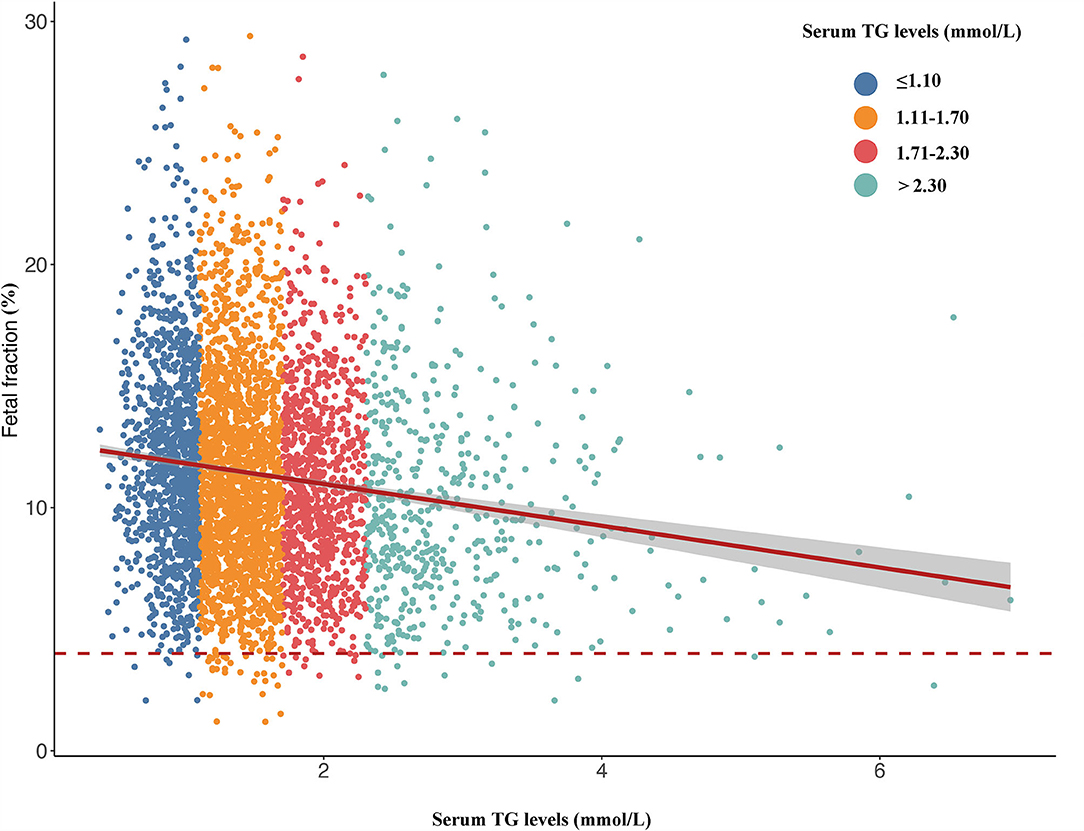
Figure 2. Scatterplots of fetal fraction according to different groups of TG levels with lines of best fit from the linear regression.
The multivariable-adjusted (model 2) mean √fetal fraction differences across the categories of TG levels were −0.075 (95% CI: −0.121 to −0.028) for TG levels of 1.11–1.70 mmol/L, −0.125 (95% CI: −0.183 to −0.067) for TG levels of 1.71–2.30 mmol/L, and −0.228 (95% CI: −0.298 to −0.158) for TG levels > 2.30 mmol/L compared with TG levels ≤ 1.10 mmol/L (Ptrend < 0.0001). Nevertheless, the mean √fetal fraction differences were −0.067 (95% CI: −0.114 to −0.020) for TG levels of 1.11–1.70 mmol/L, −0.108 (95% CI: −0.167 to −0.050) for TG levels of 1.71–2.30 mmol/L, and −0.206 (95% CI: −0.277 to −0.135) for TG levels > 2.30 mmol/L compared with TG levels ≤ 1.10 mmol/L (Ptrend < 0.0001) after adjusting for confounding factors plus the time interval between two tests (weeks) (model 3, Figure 4), suggesting that delaying the time of NIPT blood collection significantly decreases the mean fetal fraction differences between the different TG groups, especially in pregnant women with elevated TG levels by about 10%.
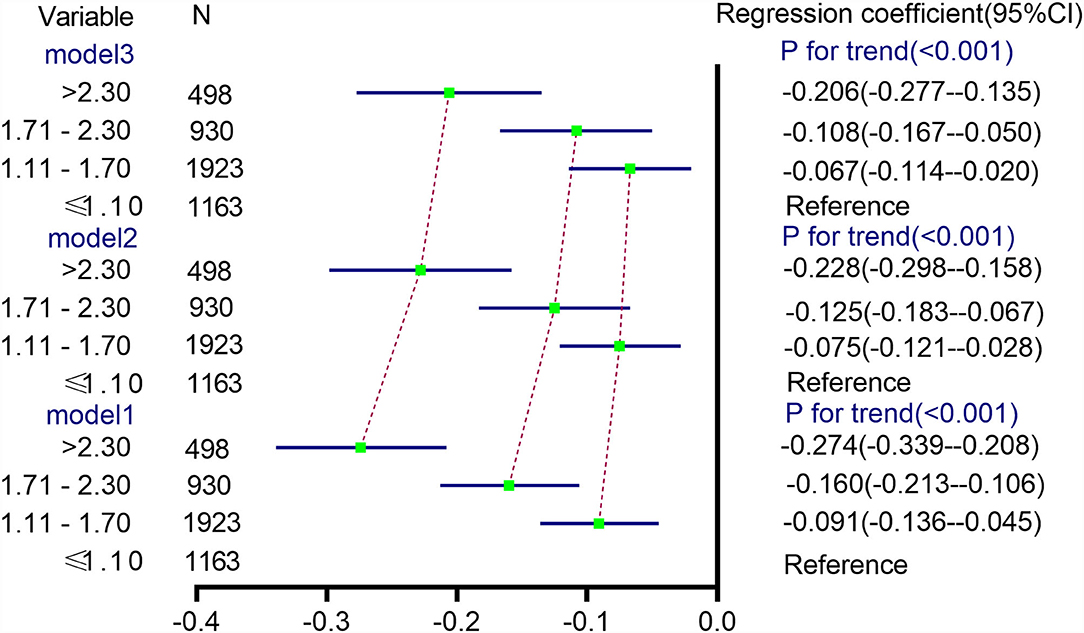
Figure 4. Differences in √fetal fraction according to TG categories. Model 1 was a univariate linear regression of the relationship between the TG level and √fetal fraction. Model 2 was adjusted for HDL-C and LDL-C levels. In model 3, the time interval between two tests (weeks) was added on the basis of model 2.
Association Between TG and Test Failure Rate of NIPT
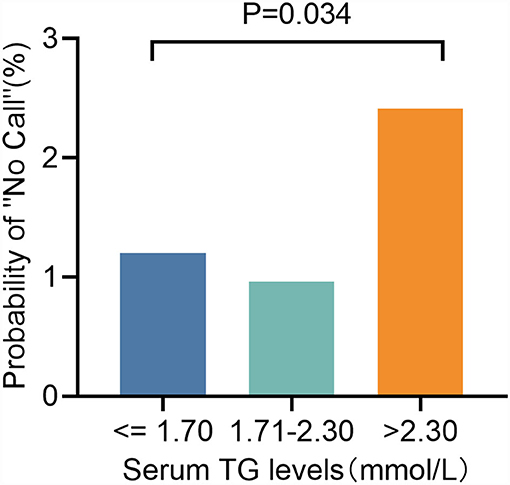
In the logistic regression analyses, augmented TG [odds ratio (OR): 1.560, 95% CI: 1.177–2.068, P = 0.002; Table 3] levels were significant predictors of test failure. We then classified TG levels into the following three categories: ≤1. 70, 1.71–2.30, and >2.30 mmol/L. Although they are still low, the rate of screen failures increased with TG levels from 1.20% at ≤1.70 mmol/L to 2.41% (P = 0.034) at >2.30 mmol/L (2.41%, P = 0.034) (Figure 5).
Discussion
Accurate identification of fetal aneuploidy depends on sufficient fetal fraction using the NIPT method. To date, several strategies have been used to increase the relative abundance of fetal cfDNA and reduce the failure rate of NIPT (14, 20–22). Welker et al. (22) performed a new NIPT method that sequenced shorter cfDNA fragments (<140 bp) to significantly improve the fetal fraction, with an average fetal fraction increase of 2.3-fold. Moreover, the zero sample had a fetal fraction < 4% when screened with fetal fraction amplification, whereas 3.7% of the same patients had fetal fraction < 4% without fetal fraction amplification. However, the original fetal fraction information may be lost when using this new technology, and further studies are needed to evaluate this. Studies on factors affecting fetal fraction aim to increase the relative abundance of fetal cfDNA by regulating these factors, which may reduce the test failure rate. To the best of our knowledge, this is the largest and most extensive study to determine the associations of lipid metabolism with the fetal fraction and test failure rate in the NIPT method. Among the 4,514 pregnancies with male fetuses, TG, which was a significant factor (standardized coefficient: −0.11), and LDL-C levels were negatively correlated with fetal fraction. Conversely, the HDL-C level and the time interval between the two tests were positively correlated with fetal fraction. Hence, laboratories should be aware that excessive TG concentrations will reduce fetal-derived cfDNA and affect the accuracy of NIPT in screening aneuploidy.
Our results suggested that the fetal fraction was negatively associated with TG and LDL-C levels and positively correlated with the HDL-C level. In the logistic regression analyses, we found that an augmented TG level (OR: 1.560, 95% CI: 1.177–2.068, P = 0.002; Table 3) was a significant predictor of test failure. After classifying the TG levels, we found that the rate of screen failures increased with an increase in TG. Additionally, complex changes occurred in the lipid profiles during pregnancy. Brizzi et al. (23) investigated the changes in lipoproteins and lipids in women during normal pregnancy and compared their results with those obtained in nulliparous women with similar age. Pregnant women had increased TG levels vs. nulliparous and primiparous women (24). Interestingly, in the MESA (25), NHANES (26) and UK Biobank studies (27) TG demonstrated the strongest positive association with leukocyte count. Hu et al. (28) also found that peripheral leukocyte counts are extensively associated with serum lipid levels, with patterns differing by leukocyte subsets. In view of the close relationship between TG and leukocyte counts, the data indicate that TG may be directly involved in leukogenesis. cfDNA in the peripheral blood sample of a pregnant woman is derived from three tissues: the placenta, maternal bone marrow, and fetus. Moreover, cfDNA derived from the white-cell lineage typically contributes to more than 70%, sometimes even to up to 90% (29, 30). As such, we speculate that lipids during pregnancy, especially TG levels, affect the fetal fraction by participating in leukogenesis. At present, there are limited reports on the relationship between TG and low fetal fraction. Our study provides a starting point for more in-depth exploration of the impact of lipid metabolism on NIPT in the future.
The positive association between the time interval between the two tests and fetal fraction is compatible with a previous study (31). Although the fetal fraction changes slowly with GA, the study indicates that by 8–9 days after the initial draw, there is sufficient cfDNA to expect a 55–60% chance of success. In this study, our results indicated that delaying the time of NIPT blood collection significantly decreased (about 10%) the mean fetal fraction differences between the TG groups, especially in pregnant women with elevated TG (>2.30 mmol/L) levels. It is more likely that apoptotic placental cells (trophoblasts) are a major source of fetal-derived cfDNA in maternal plasma (32), and the number of apoptotic placental cells is proportional to the mass. Therefore, as the interval increases, the weight of the placenta increases and its apoptotic release increases. However, the test failure differences between the TG groups were not influenced by the time interval between the two tests. This may be because there are fewer failed samples which can affect the statistical performance. In addition, due to limitations within our clinical data, the following considerations should be noted: First, the relationship between lipid metabolism and fetal fraction was studied according to data obtained at from a single center. Larger samples are recommended for further evaluation. Second, although we adjusted for many contributing variables, such as biochemical placental markers (free β-hCG, PAPP-A, and PlGF) and in vitro fertilization conception (33), other relevant variables may have been overlooked.
Conclusion
In summary, our results show that TG and LDL-C levels were negatively correlated with fetal fraction, while HDL-C level and the time interval between the two tests were positively correlated with fetal fraction. Lower TG level and delay in the time between NIPT blood collections may be more conducive to obtaining sufficient fetal fractions, thereby providing a way to effectively attenuate test failures and further improve test analytical performance.
Data Availability Statement
The datasets for this article are not publicly available to assure patient confidentiality and participant privacy. Requests to access the datasets should be directed to Ting Wang, Ymlvd3RAbmptdS5lZHU=.cn.
Ethics Statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
JC, LQ, JJ, and SZ: conception, design, collection, and assembly of data. PC: administrative support. HT, ZY, and JS: provision of study materials or patients. TW and YL: data analysis and interpretation. All authors: manuscript writing and final approval of manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 81901632 and 82001576); Suzhou Science and Technology Support Program (SYS2019095, SYS2019098, BE2019683, and SS2019066); Jiangsu Science and Technology Support Program (SBE2019740167); Suzhou Clinical Medical Expert Team (SZYJTD201708); Jiangsu Provincial Medical Innovation Team (CXTDB2017013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Allyse MA, Wick MJ. Noninvasive prenatal genetic screening using cell-free DNA. JAMA. (2018) 320:591–2. doi: 10.1001/jama.2018.9418
2. Hoskovec JM, Swigert AS. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med. (2018) 379:2282. doi: 10.1056/NEJMc1812266
3. Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks' gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. (2013) 41:26–32. doi: 10.1002/uog.12331
4. Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. (2013) 33:667–74. doi: 10.1002/pd.4126
5. van Beek DM, Straver R, Weiss MM, Boon EMJ, Huijsdens-van Amsterdam K, Oudejans CBM, et al. Comparing methods for fetal fraction determination and quality control of NIPT samples. Prenat Diagn. (2017) 37:769–73. doi: 10.1002/pd.5079
6. La Verde M, De Falco L, Torella A, Savarese G, Savarese P, Ruggiero R, et al. Performance of cell-free DNA sequencing-based non-invasive prenatal testing: experience on 36,456 singleton and multiple pregnancies. BMC Med Genomics. (2021) 14:93. doi: 10.1186/s12920-021-00941-y
7. Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med. (2016) 18:1056–65. doi: 10.1038/gim.2016.97
8. Khalil A, Archer R, Hutchinson V, Mousa HA, Johnstone ED, Cameron MJ, et al. Noninvasive prenatal screening in twin pregnancies with cell-free DNA using the IONA test: a prospective multicenter study. Am J Obstet Gynecol. (2021) 225:79.e1–79.e13. doi: 10.1016/j.ajog.2021.01.005
9. Karlsson F, Ahola T, Dahlberg J, Prensky L, Moilanen H, Spalding H. Evaluation of repeat testing of a non-sequencing based NIPT test on a Finnish general-risk population. Acta Obstet Gynecol Scand. (2021) 100:1497–1500. doi: 10.1111/aogs.14125
10. Hancock S, Ben-Shachar R, Adusei C, Oyolu CB, Evans EA, Kang HP, et al. Clinical experience across the fetal-fraction spectrum of a non-invasive prenatal screening approach with low test-failure rate. Ultrasound Obstet Gynecol. (2020) 56:422–30. doi: 10.1002/uog.21904
11. Jelsema R, Demko ZP, Billings PR. Re: Clinical experience across the fetal-fraction spectrum of a non-invasive prenatal screening approach with low test-failure rate. Ultrasound Obstet Gynecol. (2020) 56:471–2. doi: 10.1002/uog.22104
12. Jiang P, Lo YMD. The long and short of circulating cell-free DNA and the ins and outs of molecular diagnostics. Trends Genet. (2016) 32:360–71. doi: 10.1016/j.tig.2016.03.009
13. Sun K, Jiang P, Wong AIC, Cheng YKY, Cheng SH, Zhang H, et al. Size-tagged preferred ends in maternal plasma DNA shed light on the production mechanism and show utility in noninvasive prenatal testing. Proc Natl Acad Sci USA. (2018) 115:E5106–e14. doi: 10.1073/pnas.1804134115
14. Hu P, Liang D, Chen Y, Lin Y, Qiao F, Li H, et al. An enrichment method to increase cell-free fetal DNA fraction and significantly reduce false negatives and test failures for non-invasive prenatal screening: a feasibility study. J Transl Med. (2019) 17:124. doi: 10.1186/s12967-019-1871-x
15. Qiao L, Zhang Q, Liang Y, Gao A, Ding Y, Zhao N, et al. Sequencing of short cfDNA fragments in NIPT improves fetal fraction with higher maternal BMI and early gestational age. Am J Transl Res. (2019) 11:4450–9.
16. Qiao L, Mao J, Liu M, Liu Y, Song X, Tang H, et al. Experimental factors are associated with fetal fraction in size selection noninvasive prenatal testing. Am J Transl Res. (2019) 11:6370–81.
17. Qiao L, Yu B, Liang Y, Zhang C, Wu X, Xue Y, et al. Sequencing shorter cfDNA fragments improves the fetal DNA fraction in noninvasive prenatal testing. Am J Obstet Gynecol. (2019) 221:345.e1–345.e11. doi: 10.1016/j.ajog.2019.05.023
18. Stokowski R, White K, Hacker C, Doshi J, Schmid M. Hemolysis and fetal fraction in cell-free DNA blood collection tubes for noninvasive prenatal testing. Mol Diagn Ther. (2020) 24:185–90. doi: 10.1007/s40291-020-00446-x
19. Lopes JL, Lopes GS, Enninga EAL, Kearney HM, Hoppman NL, Rowsey RA. Most noninvasive prenatal screens failing due to inadequate fetal cell free DNA are negative for trisomy when repeated. Prenat Diagn. (2020) 40:831–7. doi: 10.1002/pd.5693
20. Hopkins MK, Koelper N, Caldwell S, Dyr B, Dugoff L. Obesity and no call results: optimal timing of cell-free DNA testing and redraw. Am J Obstet Gynecol. (2021) 225:417.e1–10. doi: 10.1016/j.ajog.2021.05.027
21. Galeva S, Gil MM, Konstantinidou L, Akolekar R, Nicolaides KH. First-trimester screening for trisomies by cfDNA testing of maternal blood in singleton and twin pregnancies: factors affecting test failure. Ultrasound Obstet Gynecol. (2019) 53:804–9. doi: 10.1002/uog.20290
22. Welker NC, Lee AK, Kjolby RAS, Wan HY, Theilmann MR, Jeon D, et al. High-throughput fetal fraction amplification increases analytical performance of noninvasive prenatal screening. Genet Med. (2021) 23:443–50. doi: 10.1038/s41436-020-01009-5
23. Brizzi P, Tonolo G, Esposito F, Puddu L, Dessole S, Maioli M, et al. Lipoprotein metabolism during normal pregnancy. Am J Obstet Gynecol. (1999) 181:430–4. doi: 10.1016/S0002-9378(99)70574-0
24. Ghio A, Bertolotto A, Resi V, Volpe L, Di Cianni G. Triglyceride metabolism in pregnancy. Adv Clin Chem. (2011) 55:133–53. doi: 10.1016/B978-0-12-387042-1.00007-1
25. Lai YC, Woollard KJ, McClelland RL, Allison MA, Rye KA, Ong KL, et al. The association of plasma lipids with white blood cell counts: results from the multi-ethnic study of atherosclerosis. J Clin Lipidol. (2019) 13:812–20. doi: 10.1016/j.jacl.2019.07.003
26. Andersen CJ, Vance TM. Gender dictates the relationship between serum lipids and leukocyte counts in the national health and nutrition examination survey 1999-2004. J Clin Med. (2019) 8:365. doi: 10.3390/jcm8030365
27. Tucker B, Sawant S, McDonald H, Rye KA, Patel S, Ong KL, et al. The association of serum lipid and lipoprotein levels with total and differential leukocyte counts: results of a cross-sectional and longitudinal analysis of the UK Biobank. Atherosclerosis. (2021) 319:1–9. doi: 10.1016/j.atherosclerosis.2020.12.016
28. Hu W, Zhang P, Su Q, Li D, Hang Y, Ye X, et al. Peripheral leukocyte counts vary with lipid levels, age and sex in subjects from the healthy population. Atherosclerosis. (2020) 308:15–21. doi: 10.1016/j.atherosclerosis.2020.07.009
29. Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. (2015) 112:E5503–12. doi: 10.1073/pnas.1508736112
30. Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY, Jiang P, et al. DNA of erythroid origin is present in human plasma and informs the types of anemia. Clin Chem. (2017) 63:1614–23. doi: 10.1373/clinchem.2017.272401
31. Benn P, Valenti E, Shah S, Martin K, Demko Z. Factors associated with informative redraw after an initial no result in noninvasive prenatal testing. Obstet Gynecol. (2018) 132:428–35. doi: 10.1097/AOG.0000000000002728
32. Lim JH, Lee BY, Kim JW, Han YJ, Chung JH, Kim MH, et al. Evaluation of extraction methods for methylated cell-free fetal DNA from maternal plasma. J Assist Reprod Genet. (2018) 35:637–41. doi: 10.1007/s10815-018-1114-8
Keywords: non-invasive prenatal testing, fetal fraction, cell-free DNA, triglyceride, multivariable linear regression models
Citation: Cao J, Qiao L, Jin J, Zhang S, Chen P, Tang H, Yu Z, Shi J, Wang T and Liang Y (2022) Lipid Metabolism Affects Fetal Fraction and Screen Failures in Non-invasive Prenatal Testing. Front. Med. 8:811385. doi: 10.3389/fmed.2021.811385
Received: 10 November 2021; Accepted: 17 December 2021;
Published: 12 January 2022.
Edited by:
Marco La Verde, Università degli Studi della Campania “Luigi Vanvitelli”, ItalyCopyright © 2022 Cao, Qiao, Jin, Zhang, Chen, Tang, Yu, Shi, Wang and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuting Liang, bGlhbmd5dXRpbmc2NjZAMTI2LmNvbQ==; Ting Wang, Ymlvd3RAbmptdS5lZHUuY24=
†These authors have contributed equally to this work
 Jun Cao1†
Jun Cao1† Longwei Qiao
Longwei Qiao Ting Wang
Ting Wang Yuting Liang
Yuting Liang