- Department of Dermatology, Faculty of Medicine and Graduate School of Medicine, Hokkaido University, Sapporo, Japan
Tripartite motif (TRIM) proteins play important roles in a wide range of cell physiological processes, such as signal transduction, transcriptional regulation, innate immunity, and programmed cell death. TRIM29 protein, encoded by the ATDC gene, belongs to the RING-less group of TRIM protein family members. It consists of four zinc finger motifs in a B-box domain and a coiled-coil domain, and makes use of the B-box domain as E3 ubiquitin ligase in place of the RING. TRIM29 was found to be involved in the formation of homodimers and heterodimers in relation to DNA binding; additional studies have also demonstrated its role in carcinogenesis, DNA damage signaling, and the suppression of radiosensitivity. Recently, we reported that TRIM29 interacts with keratins and FAM83H to regulate keratin distribution. Further, in cutaneous SCC, the expression of TRIM29 is silenced by DNA methylation, leading to the loss of TRIM29 and promotion of keratinocyte migration. This paper reviews the role of TRIM family proteins in malignant tumors, especially the role of TRIM29 in cutaneous SCC.
Introduction
Tripartite motif (TRIM) proteins play important roles in signal transduction, transcriptional regulation, innate immunity, programmed cell death, and other cell physiological processes (1). TRIM protein dysfunction contributes to various morbid states, including viral disease, malignancy, cardiovascular disease, and neuropsychiatric disorders (2–4). More than 80 TRIM protein genes have been reported in humans. The majority of the N-terminal alignment in TRIM proteins is comprised of a RING-finger domain, one or two B boxes (the zinc-binding motif), and a coiled-coil region (5–7). The TRIM family proteins are composed of E3 ubiquitin ligase activity among the RING-finger domain. With that said, not all TRIM proteins present with a RING-finger domain; some of them are RING-less. TRIM family proteins can be subclassified into groups I to XI due to the high variability in their C-terminal functional domains, whereas the SPRY domain is predominantly conserved (6, 8). Newly discovered evidence has revealed that TRIM family proteins play a fundamental role in oncogenesis (9–11). Intriguingly, TRIM family proteins can promote either oncogenesis or tumor-suppression depending on the cancer-specific TRIM proteins. This article provides a basic overview of TRIM family proteins and discusses about their role in malignancies.
Trim Family Proteins and TRIM29
A growing number of reports have revealed that the up- or down-regulation of TRIM proteins can promote or hinder tumorigenesis (7, 12–14). Depending on the target of the specific TRIM protein, increased expression of certain TRIM family members (e.g., TRIM25, TRIM27, and TRIM29) in tumor tissue promotes oncogenesis, and this has been correlated with poor clinical outcomes. In contrast, other TRIM proteins (e.g., TRIM3, TRIM29, and TRIM40) were found to have a tumor-suppressive effect. This dichotomic phenomenon is simultaneously observed for specific TRIM proteins (e.g., TRIM29) in different cancers.
Ataxia telangiectasia is an uncommon autosomal recessive inherited disorder that affects the nervous system, the immune response, DNA repair, and the risk of cancer (15). The ataxia-telangiectasia group D complementing (ATDC) gene resides on human chromosome 11q23 and is involved in ataxia telangiectasia. TRIM29 protein, the product of the ATDC gene, belongs to the RING-less group of TRIM family proteins. It consists of four zinc finger motifs in a B-box domain and a coiled-coil domain (Figure 1A), and it makes use of the B-box domain as E3 ubiquitin ligase in place of the RING (16, 17). TRIM29 was found to be involved in the formation of homodimers or heterodimers in relation to DNA binding; additional studies have also demonstrated its role in carcinogenesis, DNA damage signaling, and the suppression of radiosensitivity (18).
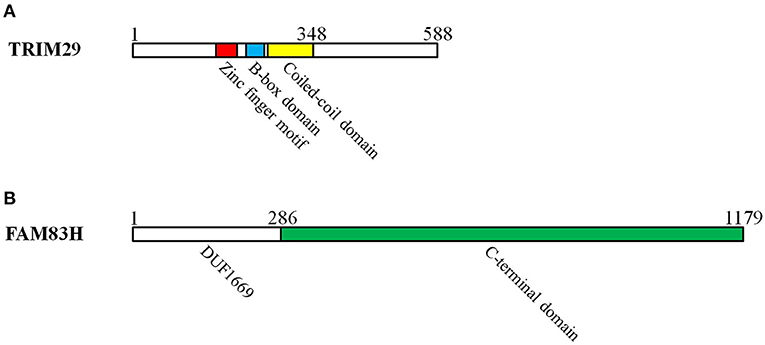
Figure 1. The domains of TRIM29 and FAM83H interact with keratin in cutaneous squamous cell carcinoma. (A) TRIM29 is pertinent to RING-less group of TRIM family proteins, which consist of four zinc finger motifs in the B-box domain and a coiled-coil domain. The zinc finger, B-box, coiled-coil, and C-terminal domains of TRIM29 are all essential for the construction of the TRIM29–keratin–FAM83H complex. (B) FAM83 members have a common N-terminal domain (domain unknown function 1669: DUF1669) and a C-terminal domain. The C-terminal domain of FAM83H is required for the formation of the TRIM29–keratin–FAM83H complex.
TRIM29 Protein and Innate Immunity
Innate immunity is crucial in protecting the human respiratory tract from infection. TRIM29 downregulates macrophage activation against the invasion of bacteria and viruses in the respiratory system. TRIM29 directly interacts with NEMO/IKBKG in lysosomes and induces its subsequent ubiquitination and proteolytic degradation. The expressions of downstream IFN-1 and proinflammatory cytokines are then suppressed (19). Similarly, persistent infection by a DNA virus (e.g., EBV and HSV-1) may be caused by TRIM29-induced STING1 degradation in the cGAS/STING/TBK1/IRF3 signaling pathway (20, 21). TRIM29 binds to the mitochondrial antiviral signaling protein (MAVS) and provokes its ubiquitination and degradation, leading to inhibition of the host innate defense mechanism in RNA viruses (22). In CMV-induced NK cells, TRIM29 promotes the decay of TGF-β activated kinase 1 binding protein 2 (TAB2) to decrease IFN-γ production by NK cells (23). Taken together, these studies indicate that TRIM29 is targeted by bacteria and viruses for chronic infection and immune evasion.
TRIM29 Negatively Modulates p53
TRIM29 plays an important role in cell cycle regulation, mitosis, DNA repair, and radioresistance. Three main pathways have been identified where TRIM29 contributes to cancer formation. First, histone deacetylase 9 (HDAC9) helps to remove the acetyl group from TRIM29, decreasing the ability of TRIM29 to bind to p53, which results in the increased expression of p53-regulated genes and finally the repression of cell proliferation (24). Once TRIM29 interacts with p53, the cytoplasmic sequestration of p53 occurs and its translocation to the nucleus decreases (25). Second, TRIM29 binds to Tat-interactive protein 60 (TIP60), enhancing its breakdown, and prevents the acetylation of p53 by TIP60. The reduction of acetylated p53 causes cell growth and transformation. Lastly, the overexpression of TRIM29 in HCT116 cell lines suppresses apoptosis elicited by ultraviolet exposure (26). TRIM29 serves as a scaffold protein for recruiting DNA repair proteins into chromatin for DNA damage repair in response to ionizing radiation (27). Therefore, TRIM29 is thought to be a tumorigenic factor that negatively modulates p53, induces cell transformation, increases radiation resistance, and mounts an anti-apoptotic response.
TRIM29 Protein in Malignancies
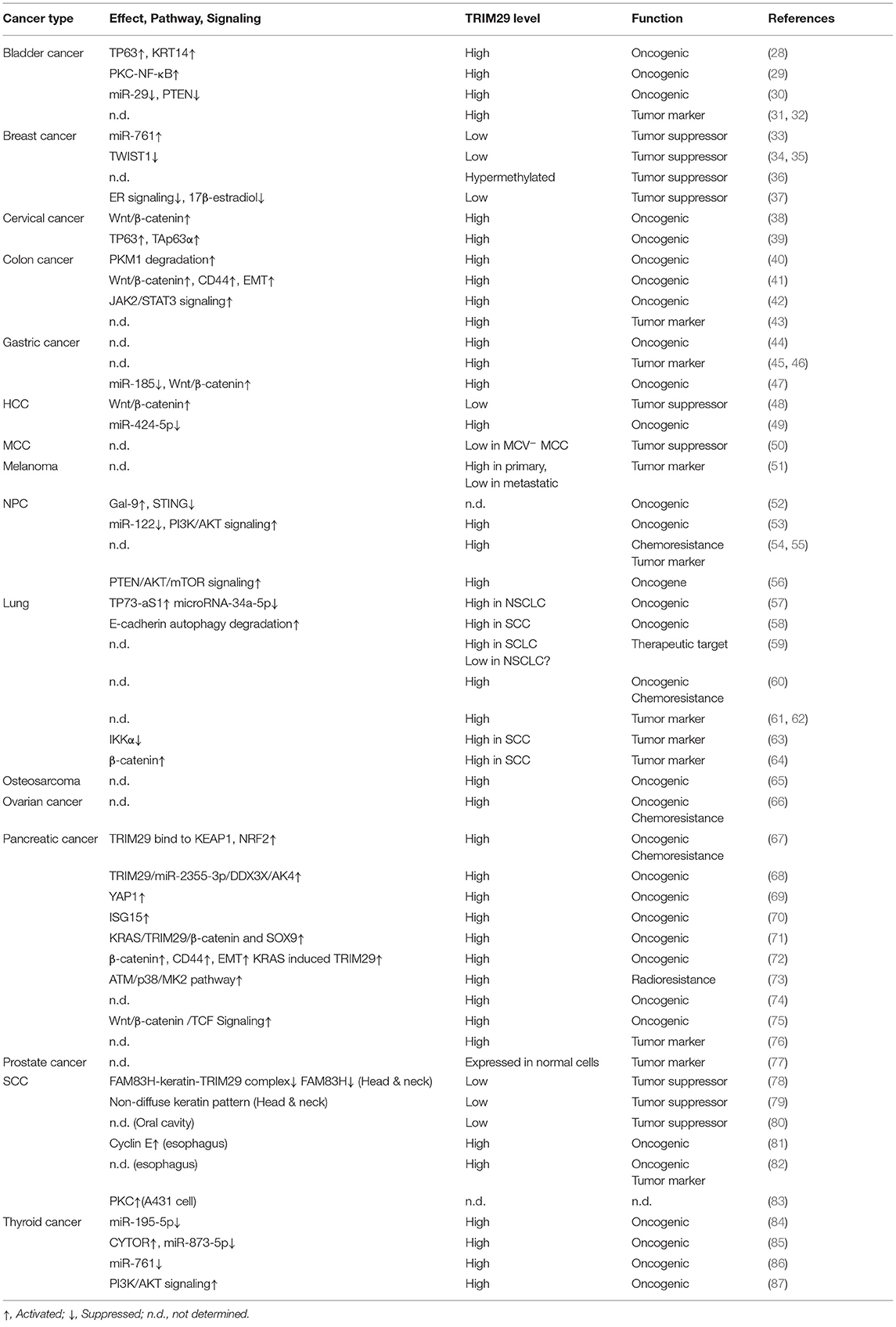
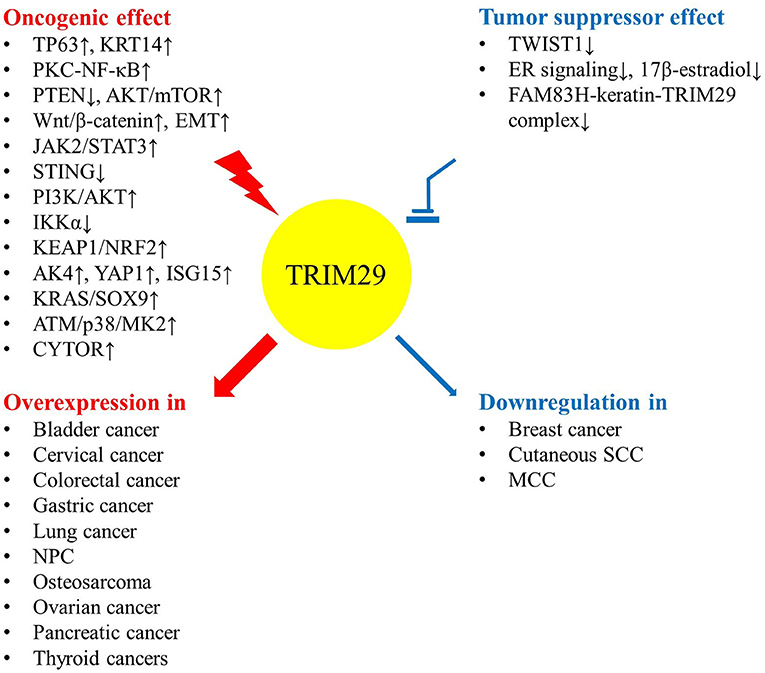
The expression level and function of TRIM29 contribute to human malignancies in various ways (Table 1). Even though the dysregulation of TRIM29 protein in cancer has not been fully elucidated, there are some common conditions that provide evidence for the multifaceted roles of TRIM protein (10). Elevated TRIM29 expression is accompanied by more invasive phenotypes of malignancies, including bladder, cervical, colorectal, gastric, lung, ovarian, pancreatic, and thyroid cancers; osteosarcoma; and nasopharyngeal carcinoma (NPC). Conversely, TRIM29 suppresses cell expansion, migration, and invasion in breast cancer and Merkel cell carcinoma (MCC). The low expression of this gene is associated with poor prognosis. This evidence potentially supports the dual effect of TRIM29, which can act as either an oncogene or a tumor suppressor gene, depending on the tumor type (Figure 2).
Oncogenic Effect of TRIM29
Some studies have indicated that upregulation of TRIM29 is associated with an advanced stage and poor outcomes in many malignancies. In bladder cancer, the overexpression of TRIM29 suppresses miR-29 and subsequently activates DNA methyltransferase 3A (DNMT3A), resulting in DNA methylation and silencing of tumor suppressor PTEN (30–32). TRIM29 upregulates cyclin D1/E and Bcl family proteins by the PKC-NF-κB signaling pathway to assist cell proliferation as well as inhibit apoptosis (29). The invasive phenotype of bladder cancer occurs via the binding of TP63 to TRIM29 and keratin14 (KRT14), along with the enhancement of their expression (28). In cervical cancer cells, TRIM29 mediates the p63 pathway and activates Wnt/β-catenin signaling to promote cell proliferation, colony formation, migration, and invasion (38, 39). The expression of TRIM29 in colorectal cancer (CRC) tissue is significantly higher and correlates with tumor progression and poor prognosis (43). TRIM29 enhances the degradation of pyruvate kinase M1 (PKM1) and the reduction of PKM1/PKM2 ratio, which leads to stronger malignant behavior, enhanced PKM2-mediated aerobic glycolysis (the Warburg effect), and weakened oxidative phosphorylation (40). TRIM29 also plays a pro-tumorigenic role through JAK2/STAT3 signaling, induced epithelial to mesenchymal transition (EMT) via the Wnt/β-catenin signaling pathway, and CD44 expression in CRC (41, 42). In gastric cancer, TRIM29 functions as an oncogene to activate Wnt/β-catenin signaling, which can be silenced by miR-185 (44–47). The expression level of TRIM29 protein is particularly related to advanced malignant tumor stage and cancer drug resistance in NPC (54, 55). miR-335-5p and miR-15b-5p suppress the metastasis of NPC by targeting TRIM29 and inhibit the activity of the PI3K/AKT/mTOR signaling pathway (56); miR-122 inhibits tumor progression by downregulating TRIM29 and repressing the activity of PI3K/AKT signaling (53). Enrichment of galectin-9 (Gal-9) in the microenvironment and serum triggers the TRIM29-mediated K48-linked ubiquitination of STING and is associated with a more aggressive phenotype of NPC (52). In lung cancer, the upregulation of TRIM29 can cause oncogenic activity and chemoresistance (59–62). TRIM29 may also enhance tumor progression and metastasis by interaction with β-catenin and modifying E-cadherin autophagy degradation (58, 64). IKKα suppresses TRIM29 and p63 in an epigenetic manner to prevent lung squamous cell carcinoma (SCC) development, but TP73-AS1 serves as a molecular sponge for miR-34a-5p to prevent TRIM29 suppression (57, 63). In osteosarcoma and ovarian cancer, TRIM29 induces EMT and chemoresistance, respectively (65, 66).
TRIM29 is thought to be a molecular marker and oncogene for pancreatic ductal adenocarcinoma (74, 76). It binds to Disheveled-2 (Dvl-2) and components of the β-Catenin destruction complex to stabilize β-catenin, resulting in the activation of Wnt/β-catenin/TCF signaling pathway (75). The activation of β-catenin signaling further upregulates CD44, which induces EMT characterized by the expression of Zeb1 and Snail1 (72). TRIM29 can also directly bind to Yes-associated protein 1 (YAP1) to reduce its ubiquitination and degradation, and thus promote the proliferation of pancreatic cancer cells (69). In the presence of oncogenic KRAS, TRIM29 subsequently upregulates SRY-Box transcription factor 9 (SOX9), which accelerates the formation of pancreatic intraepithelial neoplasia and progression to invasive carcinoma (71). The DNA damage sensor ataxia telangiectasia mutated (ATM) is activated by exposure to ionizing radiation, and it causes MAPKAP kinase 2 (MK2) kinase phosphorylation in a p38 kinase-dependent process. The phosphorylated MK2 induces TRIM29 phosphorylation, resulting in a radioresistant response by pancreatic cancer cells (73). TRIM29 interacts with Kelch-like ECH-associated protein 1 (KEAP1) to prevent the degradation of nuclear factor erythroid 2-related factor 2 (NRF2), which promotes antioxidant response element and enhanced chemoresistance (67). The cancer stem cell-like features of pancreatic ductal adenocarcinoma are maintained by the autocrine stimulation of extracellular interferon-stimulated gene 15 (ISG15), which is modulated by TRIM29 via Calpain-3 (CAPN3)-mediated posttranslational degradation (70). In addition, TRIM29 controls the transcripts of adenylate kinase 4 (AK4) via posttranscriptional regulation to alter the bioenergetics in pancreatic cancer (68). In thyroid carcinoma, high expression of TRIM29 activates PI3K/AKT signaling, which circumvents cell cycle arrest at the G0/G1 phase and enhances cancer cell resistance to chemotherapy (87). LncRNA HOXA11-AS has been identified as a molecular sponge that competes with endogenous miR-761, which downregulates TRIM29 (86). Similarly, TRIM29 inhibits miR-873 biogenesis by LncRNA CYTOR sponging to upregulate FN1 and promote the progression of papillary thyroid cancer cells (85). In papillary thyroid carcinoma, LncRNA 00324 is characterized as a microRNA sponge to miR-195-5p to maintain the oncogenic effect of TRIM29 (84).
Tumor-Suppressive Effect of TRIM29
TRIM29 can antagonize 17β-estradiol-induced cell proliferation, interrupt the induction of estrogen-responsive genes, and block the upregulation of the TWIST1 oncogene in response to hypoxic stress (34, 35, 37). However, TRIM29 is underexpressed in primary breast tumors. It is silenced by gene hypermethylation or by a specific microRNA, miR-761, leading to the aggressive behavior of breast cancer (33, 36). In Merkel cell polyomavirus-uninfected (MCV−) MCC, more aggressive disease may be linked to the loss of TRIM29 expression (50). More studies are warranted to verify these early findings.
Equivocal or Unknown Status of TRIM29 in Specific Malignancy
The function of TRIM29 in hepatocellular carcinoma (HCC) is ambiguous, and it has been reported that the silencing of TRIM29 is associated with cell proliferation, tumor formation, cell migration, and cell invasion. TRIM29 possesses a tumor-suppressor feature via inhibition of the Wnt/β-catenin signaling pathway (48). However, another study indicated that miR-424-5p, a tumor-suppressor miRNA, targets the TRIM29 gene and reverses its oncogenic effects (49). Therefore, more evidence is required to elucidate the exact role of TRIM29 in HCC. Interestingly, TRIM29 expression was found to be lower in metastatic melanoma but higher in primary melanoma. The status of TRIM29 in melanoma also needs further confirmation (51). In prostate cancer, TRIM29 is expressed in normal cells but not in cancer cells (77).
TRIM29 Protein and SCC
Cutaneous squamous cell carcinoma (cSCC), the second most common non-melanoma skin malignancy, contributes to more than 20% of skin cancers in Caucasian populations (88). cSCC derives from the malignant transformation of epithelial keratinocytes, and cumulative UV damage in the sun-exposed epithelium is the major cause of cSCC (89, 90). Other risks inclined toward morbidity include male gender, elderly age, Fitzpatrick skin type I to III, immunocompromise, hematologic cancer, HPV infection, chronic wounds, scars, several hereditary syndromes (e.g., epidermolysis bullosa, oculocutaneous albinism, xeroderma pigmentosum), and environmental exposure to arsenic or coal tar. Regarding the genetic changes, cSCC is the second most mutated skin cancer after basal cell carcinoma. The most frequent mutation in cSCC is the tumor suppressor gene p53. UV-induced p53 mutation prevents keratinocytes from apoptosis and promotes the precancerous clonal expansion of abnormal cells. The two other common tumor suppressor genes (CDKN2A and NOTCH), as well as the oncogenic RAS gene, all participate in the initial event of oncogenesis. Epidermal growth factor receptor (EGFR) gene amplification along with protein overexpression is highly correlated with aggressive phenotypes and poor outcomes (91). As a consequence of the accumulation of these altered alleles, the skin in the affected area evolves from hyperplasia and dysplasia to invasive carcinoma.
An early study showed that the quantity of TRIM29 is markedly higher in epidermoid cells (cSCC cell line A431) than in fibroblast cell lines. TRIM29 is the substrate of serine/threonine protein kinase C (PKC), and it may regulate the cell response to ionizing radiation via the PKC signaling pathway, but the role of TRIM29 in tumorigenesis has not been determined (83). Furthermore, elevated TRIM29 expression is associated with tumor growth and proliferation via the regulation of cyclin E in esophageal SCC patients. TRIM29 might be a useful marker for evaluating prognosis (81, 82). The oncogenic effect of TRIM29 is also found in lung SCC. Conversely, new evidence points to the tumor-suppressive effect of TRIM29 in cSCC (78–80). This may be due to tissue-specific gene regulation differences between internal organs and keratinocytes.
Expression of TRIM29 and FAM83H in CSCC
The expression of TRIM29 is decreased in cutaneous head and neck SCC but is normal in healthy skin and benign tumors. Unusual hypermethylation in the CpG island of the TRIM29 promoter suppresses the expression of TRIM29. Decreased TRIM29 is related to more aggressive clinical behavior (79). Such results strongly suggest that TRIM29 is a useful diagnostic and prognostic biomarker for cSCC. Furthermore, the subcellular localization of TRIM29 is mainly found in the cytoplasm, consistent with TRIM29 acting as a tumor suppressor in breast and prostate cancers. The level of TRIM29 in the cytosol alters the distribution pattern of keratin in cSCC, with low expression showing a perinuclear pattern and high expression presenting a diffuse pattern. Another experiment confirmed that keratin-interacting protein FAM83H (Family With Sequence Similarity 83 Member H) could be a new interactor with TRIM29 (Figure 1B) (78). FAM83H expression is also lower in cSCC than in normal skin, and the level shows an association with different cancer stages. FAM83H knockdown increase the solubility of keratin 5 and 14 in the soluble fraction of cancer cell lysates. The change in solubility of keratin could assist in the redistribution of keratins during cell division (92). This would mean that the loss of FAM83H induces the reorganization of keratin and promotes SCC cell migration, invasion, and metastasis in vivo. Additionally, FAM83H knockdown enhances the level of integrin β4 at the wound edge, which reinforce keratinocytes migration close to that edge (93). This means that FAM83H may also be an essential molecule in the cell motility and oncogenesis of cSCC.
The TRIM29–Keratin–FAM83H Complex Regulates Keratin Distribution
Keratin filaments are of fundamental importance in protecting epithelial cells from damage or stress. Some current studies provide supporting evidence that keratin plays a role in the regulation of cell polarization, migration, and signal transduction (94). The distribution of intracellular keratin is involved in cancer invasion, metastasis, and treatment response. In cSCC, TRIM29 has been proven to colocalize with keratin 5, keratin 14, and FAM83H in the cytosol, and to form the TRIM29–keratin–FAM83H complex. The zinc finger, B-box, coiled-coil, and C-terminal domains of TRIM29 and the C-terminal domain of FAM83H are all essential for the construction of the TRIM29–keratin–FAM83H complex (Figure 1), and FAM83H or TRIM29 binds to keratins separately (78, 79). FAM83H and TRIM29 can be characterized as sharing similar features in the regulation of keratin distribution and cell migration. Consequently, the TRIM29–keratin–FAM83H complex is thought to regulate keratin distribution in cSCC (Figure 3).
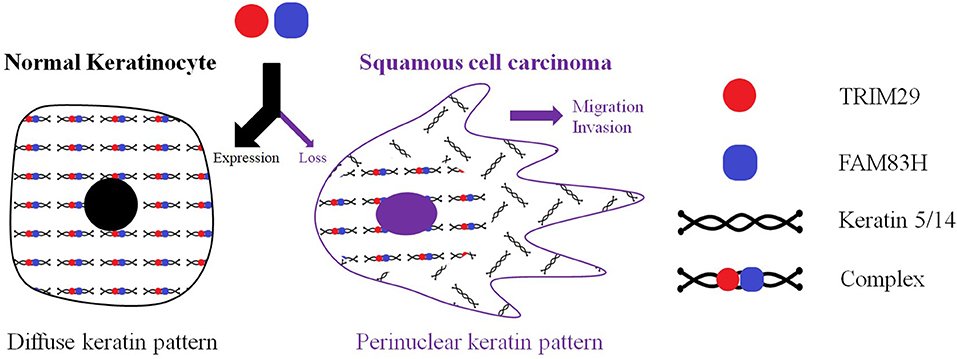
Figure 3. TRIM29 was proven to colocalize with keratin 5, keratin 14, and FAM83H in the cytosol, and to form the TRIM29–keratin–FAM83H complex. FAM83H and TRIM29 can be characterized as sharing similar features in regulating keratin distribution and cell migration. The TRIM29–keratin–FAM83H complex is thought to regulate the keratin distribution in cutaneous SCC.
Conclusion
We have summarized recent advances in our understanding of the multiple roles of TRIM29 in innate immunity and oncogenesis. In the past few years, more studies have confirmed that TRIM29 has “two-faced” effects in various malignancies. With reference to the finding that low TRIM29 expression is closely associated with poor prognosis in cSCC, it stands for promising therapeutic targets and modern tumor markers for early identification and risk evaluation. The modulation of the TRIM29–keratin–FAM83H complex could be an interventional strategy, via DNA and RNA, as well as epigenetic therapy. Also, injury to the epidermis provokes the keratin filaments to redistribute from a diffuse cytoplasmic pattern to a perinuclear distribution and prompts the upregulation of integrin β4, which cause keratinocytes to migrate to the wound. The TRIM29–keratin–FAM83H complex seems to be involved in wound healing. This raises the novel idea that developing a TRIM29 inhibitor could contribute to wound healing by disturbing the formation of this complex. We expect these concepts to be gradually put to practical use.
Author Contributions
C-YH: designed this project, acquired the data and references, and wrote the manuscript. TY: designed this project and wrote the manuscript. HU: supervised this project, analyzed the data, and wrote the manuscript. All authors contributed sufficiently and met the criteria for authorship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Hatakeyama S TRIM. Family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. (2017) 42:297–311. doi: 10.1016/j.tibs.2017.01.002
2. Watanabe M, Hatakeyama S. TRIM proteins and diseases. J Biochem. (2016) 161:135–44. doi: 10.1093/jb/mvw087
3. Zhang JR, Li XX, Hu WN, Li CY. Emerging role of TRIM family proteins in cardiovascular disease. Cardiology. (2020) 145:390–400. doi: 10.1159/000506150
4. Wan T, Li X, Li Y. The role of TRIM family proteins in autophagy, pyroptosis, and diabetes mellitus. Cell Biol Int. (2021) 45:913–26. doi: 10.1002/cbin.11550
5. Ikeda K, Inoue S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv Exp Med Biol. (2012) 770:27–37. doi: 10.1007/978-1-4614-5398-7_3
6. Esposito D, Koliopoulos MG, Rittinger K. Structural determinants of TRIM protein function. Biochem Soc Trans. (2017) 45:183–91. doi: 10.1042/BST20160325
8. Di Rienzo M, Romagnoli A, Antonioli M, Piacentini M, Fimia GM. TRIM proteins in autophagy: selective sensors in cell damage and innate immune responses. Cell Death Differ. (2020) 27:887–902. doi: 10.1038/s41418-020-0495-2
10. Liang C, Dong H, Miao C, Zhu J, Wang J, Li P, et al. TRIM29 as a prognostic predictor for multiple human malignant neoplasms: a systematic review and meta-analysis. Oncotarget. (2018) 9:12323–32. doi: 10.18632/oncotarget.23617
11. Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells. (2020) 38:165–73. doi: 10.1002/stem.3109
12. Venuto S, Merla G. E3 ubiquitin ligase TRIM proteins, cell cycle and mitosis. Cells. (2019) 8:510. doi: 10.3390/cells8050510
13. Eberhardt W, Haeussler K, Nasrullah U, Pfeilschifter J. Multifaceted roles of TRIM proteins in colorectal carcinoma. Int J Mol Sci. (2020) 21:7532. doi: 10.3390/ijms21207532
14. Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, et al. TRIM40 promotes neddylation of IKKγ and is downregulated in gastrointestinal cancers. Carcinogenesis. (2011) 32:995–1004. doi: 10.1093/carcin/bgr068
15. Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. (1995) 268:1749–53. doi: 10.1126/science.7792600
16. Leonhardt EA, Kapp LN, Young BR, Murnane JP. Nucleotide sequence analysis of a candidate gene for ataxia-telangiectasia group D (ATDC). Genomics. (1994) 19:130–6. doi: 10.1006/geno.1994.1022
17. Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem Cell Biol. (1998) 76:351–8. doi: 10.1139/o98-021
18. Yang H, Palmbos PL, Wang L, Kim EH, Ney GM, Liu C, et al. ATDC (ataxia telangiectasia group D complementing) promotes radioresistance through an interaction with the RNF8 Ubiquitin ligase. J Biol Chem. (2015) 290:27146–57. doi: 10.1074/jbc.M115.665489
19. Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin R, et al. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat Immunol. (2016) 17:1373–80. doi: 10.1038/ni.3580
20. Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat Commun. (2017) 8:945. doi: 10.1038/s41467-017-00101-w
21. Li Q, Lin L, Tong Y, Liu Y, Mou J, Wang X, et al. TRIM29 negatively controls antiviral immune response through targeting STING for degradation. Cell Discov. (2018) 4:13. doi: 10.1038/s41421-018-0010-9
22. Xing J, Zhang A, Minze LJ, Li XC, Zhang Z. TRIM29 negatively regulates the type I IFN production in response to RNA virus. J Immunol. (2018) 201:183–92. doi: 10.4049/jimmunol.1701569
23. Dou Y, Xing J, Kong G, Wang G, Lou X, Xiao X, et al. Identification of the E3 ligase TRIM29 as a critical checkpoint regulator of NK cell functions. J Immunol. (2019) 203:873–80. doi: 10.4049/jimmunol.1900171
24. Yuan Z, Peng L, Radhakrishnan R, Seto E. Histone deacetylase 9 (HDAC9) regulates the functions of the ATDC (TRIM29) protein. J Biol Chem. (2010) 285:39329–38. doi: 10.1074/jbc.M110.179333
25. Yuan Z, Villagra A, Peng L, Coppola D, Glozak M, Sotomayor EM, et al. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. (2010) 30:3004–15. doi: 10.1128/MCB.01023-09
26. Sho T, Tsukiyama T, Sato T, Kondo T, Cheng J, Saku T, et al. TRIM29 negatively regulates p53 via inhibition of Tip60. Biochim Biophys Acta. (2011) 1813:1245–53. doi: 10.1016/j.bbamcr.2011.03.018
27. Masuda Y, Takahashi H, Sato S, Tomomori-Sato C, Saraf A, Washburn MP, et al. TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin. Nat Commun. (2015) 6:7299. doi: 10.1038/ncomms8299
28. Palmbos PL, Wang Y, Bankhead Iii A, Kelleher AJ, Wang L, Yang H, et al. ATDC mediates a TP63-regulated basal cancer invasive program. Oncogene. (2019) 38:3340–54. doi: 10.1038/s41388-018-0646-9
29. Tan ST, Liu SY, Wu B. TRIM29 overexpression promotes proliferation and survival of bladder cancer cells through NF-κB signaling. Cancer Res Treat. (2016) 48:1302–12. doi: 10.4143/crt.2015.381
30. Palmbos PL, Wang L, Yang H, Wang Y, Leflein J, Ahmet ML, et al. ATDC/TRIM29 drives invasive bladder cancer formation through miRNA-mediated and epigenetic mechanisms. Cancer Res. (2015) 75:5155–66. doi: 10.1158/0008-5472.CAN-15-0603
31. Huang W, Williamson SR, Rao Q, Lopez-Beltran A, Montironi R, Eble JN, et al. Novel markers of squamous differentiation in the urinary bladder. Hum Pathol. (2013) 44:1989–97. doi: 10.1016/j.humpath.2013.04.005
32. Fristrup N, Birkenkamp-Demtröder K, Reinert T, Sanchez-Carbayo M, Segersten U, Malmström PU, et al. Multicenter validation of cyclin D1, MCM7, TRIM29, and UBE2C as prognostic protein markers in non-muscle-invasive bladder cancer. Am J Pathol. (2013) 182:339–49. doi: 10.1016/j.ajpath.2012.10.017
33. Guo GC, Wang JX, Han ML, Zhang LP, Li L. microRNA-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression. Cell Oncol (Dordr). (2017) 40:157–66. doi: 10.1007/s13402-016-0312-6
34. Dükel M, Streitfeld WS, Tang TC, Backman LR, Ai L, May WS, et al. The breast cancer tumor suppressor TRIM29 is expressed via ATM-dependent signaling in response to hypoxia. J Biol Chem. (2016) 291:21541–52. doi: 10.1074/jbc.M116.730960
35. Ai L, Kim WJ, Alpay M, Tang M, Pardo CE, Hatakeyama S, et al. TRIM29 suppresses TWIST1 and invasive breast cancer behavior. Cancer Res. (2014) 74:4875–87. doi: 10.1158/0008-5472.CAN-13-3579
36. Avraham A, Cho SS, Uhlmann R, Polak ML, Sandbank J, Karni T, et al. Tissue specific DNA methylation in normal human breast epithelium and in breast cancer. PLoS ONE. (2014) 9:e91805. doi: 10.1371/journal.pone.0091805
37. Liu J, Welm B, Boucher KM, Ebbert MT, Bernard PS. TRIM29 functions as a tumor suppressor in nontumorigenic breast cells and invasive ER+ breast cancer. Am J Pathol. (2012) 180:839–47. doi: 10.1016/j.ajpath.2011.10.020
38. Xu R, Hu J, Zhang T, Jiang C, Wang HY. TRIM29 overexpression is associated with poor prognosis and promotes tumor progression by activating Wnt/β-catenin pathway in cervical cancer. Oncotarget. (2016) 7:28579–91. doi: 10.18632/oncotarget.8686
39. Masuda Y, Takahashi H, Hatakeyama S. TRIM29 regulates the p63-mediated pathway in cervical cancer cells. Biochim Biophys Acta. (2015) 1853:2296–305. doi: 10.1016/j.bbamcr.2015.05.035
40. Han J, Zhao Z, Zhang N, Yang Y, Ma L, Feng L, et al. Transcriptional dysregulation of TRIM29 promotes colorectal cancer carcinogenesis via pyruvate kinase-mediated glucose metabolism. Aging. (2021) 13:5034–54. doi: 10.18632/aging.202414
41. Sun J, Zhang T, Cheng M, Hong L, Zhang C, Xie M, et al. TRIM29 facilitates the epithelial-to-mesenchymal transition and the progression of colorectal cancer via the activation of the Wnt/β-catenin signaling pathway. J Exp Clin Cancer Res. (2019) 38:104. doi: 10.1186/s13046-019-1098-y
42. Xu W, Xu B, Yao Y, Yu X, Cao H, Zhang J, et al. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol Rep. (2016) 36:1411–8. doi: 10.3892/or.2016.4941
43. Jiang T, Tang HM, Lu S, Yan DW, Yang YX, Peng ZH. Up-regulation of tripartite motif-containing 29 promotes cancer cell proliferation and predicts poor survival in colorectal cancer. Med Oncol. (2013) 30:715. doi: 10.1007/s12032-013-0715-4
44. Wang C, Zhou Y, Chen B, Yuan W, Huang J. Prognostic value of tripartite motif containing 29 expression in patients with gastric cancer following surgical resection. Oncol Lett. (2018) 15:5792–8. doi: 10.3892/ol.2018.8059
45. Zhao MX, Ding SG, Liu LN, Wang Y, Zhang J, Zhang HJ, et al. [Predicative value of expression of TrkB and TRIM29 in biopsy tissues from preoperative gastroscopy in lymph node metastasis of gastric cancer]. Zhonghua Yi Xue Za Zhi. (2012) 92:376–9.
46. Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, et al. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. (2007) 14:2543–9. doi: 10.1245/s10434-007-9461-1
47. Qiu F, Xiong JP, Deng J, Xiang XJ. TRIM29 functions as an oncogene in gastric cancer and is regulated by miR-185. Int J Clin Exp Pathol. (2015) 8:5053–61.
48. Xu M, Hu J, Zhou B, Zhong Y, Lin N, Xu R. TRIM29 prevents hepatocellular carcinoma progression by inhibiting Wnt/β-catenin signaling pathway. Acta Biochim Biophys Sin. (2019) 51:68–77. doi: 10.1093/abbs/gmy151
49. Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C, et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. (2019) 224:1–11. doi: 10.1016/j.lfs.2019.03.028
50. Toptan T, Cantrell PS, Zeng X, Liu Y, Sun M, Yates NA, et al. Proteomic approach to discover human cancer viruses from formalin-fixed tissues. JCI Insight. (2020) 5:e143003. doi: 10.1172/jci.insight.143003
51. Xia Y, Zhao J, Yang C. Identification of key genes and pathways for melanoma in the TRIM family. Cancer Med. (2020) 9:8989–9005. doi: 10.1002/cam4.3545
52. Zhang CX, Huang DJ, Baloche V, Zhang L, Xu JX, Li BW, et al. Galectin-9 promotes a suppressive microenvironment in human cancer by enhancing STING degradation. Oncogenesis. (2020) 9:65. doi: 10.1038/s41389-020-00248-0
53. Yang Y, Li Q, Guo L. MicroRNA-122 acts as tumor suppressor by targeting TRIM29 and blocking the activity of PI3K/AKT signaling in nasopharyngeal carcinoma in vitro. Mol Med Rep. (2018) 17:8244–52. doi: 10.3892/mmr.2018.8894
54. Wang KMsd, Chen ZMsd, Long LMsd, Tao YMsd, Wu QMsd, Xiang MMsd, et al. iTRAQ-based quantitative proteomic analysis of differentially expressed proteins in chemoresistant nasopharyngeal carcinoma. Cancer Biol Ther. (2018) 19:809–24. doi: 10.1080/15384047.2018.1472192
55. Chen Z, Long L, Wang K, Cui F, Zhu L, Tao Y, et al. Identification of nasopharyngeal carcinoma metastasis-related biomarkers by iTRAQ combined with 2D-LC-MS/MS. Oncotarget. (2016) 7:34022–37. doi: 10.18632/oncotarget.9067
56. Zhou XM, Sun R, Luo DH, Sun J, Zhang MY, Wang MH, et al. Upregulated TRIM29 promotes proliferation and metastasis of nasopharyngeal carcinoma via PTEN/AKT/mTOR signal pathway. Oncotarget. (2016) 7:13634–50. doi: 10.18632/oncotarget.7215
57. Luo S, Shen M, Chen H, Li W, Chen C. Long non-coding RNA TP73-AS1 accelerates the progression and cisplatin resistance of non-small cell lung cancer by upregulating the expression of TRIM29 via competitively targeting microRNA-34a-5p. Mol Med Rep. (2020) 22:3822–32. doi: 10.3892/mmr.2020.11473
58. Xu W, Chen B, Ke D, Chen X. TRIM29 mediates lung squamous cell carcinoma cell metastasis by regulating autophagic degradation of E-cadherin. Aging. (2020) 12:13488–501. doi: 10.18632/aging.103451
59. Shriwash N, Singh P, Arora S, Ali SM, Ali S, Dohare R. Identification of differentially expressed genes in small and non-small cell lung cancer based on meta-analysis of mRNA. Heliyon. (2019) 5:e01707. doi: 10.1016/j.heliyon.2019.e01707
60. Liu C, Huang X, Hou S, Hu B, Li H. Silencing of tripartite motif (TRIM) 29 inhibits proliferation and invasion and increases chemosensitivity to cisplatin in human lung squamous cancer NCI-H520 cells. Thorac Cancer. (2015) 6:31–7. doi: 10.1111/1759-7714.12130
61. Song X, Fu C, Yang X, Sun D, Zhang X, Zhang J. Tripartite motif-containing 29 as a novel biomarker in non-small cell lung cancer. Oncol Lett. (2015) 10:2283–8. doi: 10.3892/ol.2015.3623
62. Ring BZ, Seitz RS, Beck RA, Shasteen WJ, Soltermann A, Arbogast S, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol. (2009) 22:1032–43. doi: 10.1038/modpathol.2009.60
63. Xiao Z, Jiang Q, Willette-Brown J, Xi S, Zhu F, Burkett S, et al. The pivotal role of IKKα in the development of spontaneous lung squamous cell carcinomas. Cancer Cell. (2013) 23:527–40. doi: 10.1016/j.ccr.2013.03.009
64. Zhou ZY, Yang GY, Zhou J, Yu MH. Significance of TRIM29 and β-catenin expression in non-small-cell lung cancer. J Chin Med Assoc. (2012) 75:269–74. doi: 10.1016/j.jcma.2012.04.015
65. Zeng SX, Cai QC, Guo CH, Zhi LQ, Dai X, Zhang DF, et al. High expression of TRIM29 (ATDC) contributes to poor prognosis and tumor metastasis by inducing epithelial-mesenchymal transition in osteosarcoma. Oncol Rep. (2017) 38:1645–54. doi: 10.3892/or.2017.5842
66. Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. (2021) 1868:118878. doi: 10.1016/j.bbamcr.2020.118878
67. Purohit V, Wang L, Yang H, Li J, Ney GM, Gumkowski ER, et al. ATDC binds to KEAP1 to drive NRF2-mediated tumorigenesis and chemoresistance in pancreatic cancer. Genes Dev. (2021) 35:218–33. doi: 10.1101/gad.344184.120
68. Hao L, Zhang Q, Qiao HY, Zhao FY, Jiang JY, Huyan LY, et al. TRIM29 alters bioenergetics of pancreatic cancer cells via cooperation of miR-2355-3p and DDX3X recruitment to AK4 transcript. Mol Ther Nucleic Acids. (2021) 24:579–90. doi: 10.1016/j.omtn.2021.01.027
69. Deng X, Fu X, Teng H, Fang L, Liang B, Zeng R, et al. E3 ubiquitin ligase TRIM29 promotes pancreatic cancer growth and progression via stabilizing Yes-associated protein 1. J Transl Med. (2021) 19:332. doi: 10.1186/s12967-021-03007-w
70. Sun J, Yan J, Qiao HY, Zhao FY, Li C, Jiang JY, et al. Loss of TRIM29 suppresses cancer stem cell-like characteristics of PDACs via accelerating ISG15 degradation. Oncogene. (2020) 39:546–59. doi: 10.1038/s41388-019-0992-2
71. Wang L, Yang H, Zamperone A, Diolaiti D, Palmbos PL, Abel EV, et al. ATDC is required for the initiation of KRAS-induced pancreatic tumorigenesis. Genes Dev. (2019) 33:641–55. doi: 10.1101/gad.323303.118
72. Wang L, Yang H, Abel EV, Ney GM, Palmbos PL, Bednar F, et al. ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. (2015) 29:171–83. doi: 10.1101/gad.253591.114
73. Wang L, Yang H, Palmbos PL, Ney G, Detzler TA, Coleman D, et al. ATDC/TRIM29 phosphorylation by ATM/MAPKAP kinase 2 mediates radioresistance in pancreatic cancer cells. Cancer Res. (2014) 74:1778–88. doi: 10.1158/0008-5472.CAN-13-2289
74. Sun H, Dai X, Han B. TRIM29 as a novel biomarker in pancreatic adenocarcinoma. Dis Markers. (2014) 2014:317817. doi: 10.1155/2014/317817
75. Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. (2009) 15:207–19. doi: 10.1016/j.ccr.2009.01.018
76. Chen Y, Zheng B, Robbins DH, Lewin DN, Mikhitarian K, Graham A, et al. Accurate discrimination of pancreatic ductal adenocarcinoma and chronic pancreatitis using multimarker expression data and samples obtained by minimally invasive fine needle aspiration. Int J Cancer. (2007) 120:1511–7. doi: 10.1002/ijc.22487
77. Kanno Y, Watanabe M, Kimura T, Nonomura K, Tanaka S, Hatakeyama S. TRIM29 as a novel prostate basal cell marker for diagnosis of prostate cancer. Acta Histochem. (2014) 116:708–12. doi: 10.1016/j.acthis.2013.12.009
78. Tokuchi K, Kitamura S, Maeda T, Watanabe M, Hatakeyama S, Kano S, et al. Loss of FAM83H promotes cell migration and invasion in cutaneous squamous cell carcinoma via impaired keratin distribution. J Dermatol Sci. (2021) 104:112–21. doi: 10.1016/j.jdermsci.2021.09.007
79. Yanagi T, Watanabe M, Hata H, Kitamura S, Imafuku K, Yanagi H, et al. Loss of TRIM29 alters keratin distribution to promote cell invasion in squamous cell carcinoma. Cancer Res. (2018) 78:6795–806. doi: 10.1158/0008-5472.CAN-18-1495
80. Harris TM, Du P, Kawachi N, Belbin TJ, Wang Y, Schlecht NF, et al. Proteomic analysis of oral cavity squamous cell carcinoma specimens identifies patient outcome-associated proteins. Arch Pathol Lab Med. (2015) 139:494–507. doi: 10.5858/arpa.2014-0131-OA
81. Lai W, Zheng X, Huang Q, Wu X, Yang M. Down-regulating ATDC inhibits the proliferation of esophageal carcinoma cells. Eur Rev Med Pharmacol Sci. (2014) 18:3511–6.
82. Lai W, Zhao J, Zhang C, Cui D, Lin J, He Y, et al. Upregulated ataxia-telangiectasia group D complementing gene correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Dis Esophagus. (2013) 26:817–22. doi: 10.1111/j.1442-2050.2012.01400.x
83. Laderoute KR, Knapp AM, Green CJ, Sutherland RM, Kapp LN. Expression of the ATDC (ataxia telangiectasia group D-complementing) gene in A431 human squamous carcinoma cells. Int J Cancer. (1996) 66:772–8. doi: 10.1002/(SICI)1097-0215(19960611)66:6<772::AID-IJC11>3.0.CO
84. Xu J, Li Z, Su Q, Zhao J, Ma J. Suppression of long noncoding RNA LINC00324 restricts cell proliferation and invasion of papillary thyroid carcinoma through downregulation of TRIM29 via upregulating microRNA-195-5p. Aging. (2020) 12:26000–11. doi: 10.18632/aging.202219
85. Wu T, Zhang DL, Wang JM, Jiang JY, Du X, Zeng XY, et al. TRIM29 inhibits miR-873-5P biogenesis via CYTOR to upregulate fibronectin 1 and promotes invasion of papillary thyroid cancer cells. Cell Death Dis. (2020) 11:813. doi: 10.1038/s41419-020-03018-3
86. Yin X, Zhang J, Li C, Zhang Z, Jin T, Song L, et al. LncRNA HOXA11-AS accumulation-induced microRNA-761 downregulation regulates cell growth by targeting TRIM29 in papillary thyroid cancer. Am J Transl Res. (2019) 11:6826–37.
87. Xu J, Li Z, Su Q, Zhao J, Ma J. TRIM29 promotes progression of thyroid carcinoma via activating P13K/AKT signaling pathway. Oncol Rep. (2017) 37:1555–64. doi: 10.3892/or.2017.5364
88. Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the U.S. population, 2012. JAMA Dermatol. (2015) 151:1081–6. doi: 10.1001/jamadermatol.2015.1187
89. Gutzmer R, Wiegand S, Kölbl O, Wermker K, Heppt M, Berking C. Actinic keratosis and cutaneous squamous cell carcinoma. Dtsch Arztebl Int. (2019) 116:616–26. doi: 10.3238/arztebl.2019.0616
90. Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. (2018) 78:237–47. doi: 10.1016/j.jaad.2017.08.059
91. Corchado-Cobos R, García-Sancha N, González-Sarmiento R, Pérez-Losada J, Cañueto J. Cutaneous squamous cell carcinoma: from biology to therapy. Int J Mol Sci. (2020) 21:2956. doi: 10.3390/ijms21082956
92. Skalli O, Goldman RD. Recent insights into the assembly, dynamics, and function of intermediate filament networks. Cell Motil Cytoskeleton. (1991) 19:67–79. doi: 10.1002/cm.970190202
93. Elaimy AL, Wang M, Sheel A, Brown CW, Walker MR, Amante JJ, et al. Real-time imaging of integrin β4 dynamics using a reporter cell line generated by Crispr/Cas9 genome editing. J Cell Sci. (2019) 132:jcs231241. doi: 10.1242/jcs.231241
Keywords: TRIM29, ATDC, squamous cell carcinoma, cell migration, keratin, FAM83H
Citation: Hsu C-Y, Yanagi T and Ujiie H (2021) TRIM29 in Cutaneous Squamous Cell Carcinoma. Front. Med. 8:804166. doi: 10.3389/fmed.2021.804166
Received: 28 October 2021; Accepted: 02 December 2021;
Published: 20 December 2021.
Edited by:
Salvador Gonzalez, University of Alcalá, SpainReviewed by:
Philippe Lefrançois, McGill University, CanadaChangmin Peng, George Washington University, United States
Copyright © 2021 Hsu, Yanagi and Ujiie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teruki Yanagi, eWFuYWdpQG1lZC5ob2t1ZGFpLmFjLmpw
 Che-Yuan Hsu
Che-Yuan Hsu Teruki Yanagi
Teruki Yanagi Hideyuki Ujiie
Hideyuki Ujiie