
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 07 January 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.786096
This article is part of the Research TopicBurden of Diarrhoeal DiseasesView all 11 articles
 Jing Ai1,2†
Jing Ai1,2† Yuanzhao Zhu3†
Yuanzhao Zhu3† Jianguang Fu1,2†
Jianguang Fu1,2† Xiaoqing Cheng1,2†
Xiaoqing Cheng1,2† Xuefeng Zhang1,2
Xuefeng Zhang1,2 Hong Ji1,2
Hong Ji1,2 Wendong Liu1,2
Wendong Liu1,2 Jia Rui3
Jia Rui3 Jingwen Xu3
Jingwen Xu3 Tianlong Yang3
Tianlong Yang3 Yao Wang3
Yao Wang3 Xingchun Liu3
Xingchun Liu3 Meng Yang3
Meng Yang3 Shengnan Lin3
Shengnan Lin3 Xiaohao Guo3
Xiaohao Guo3 Changjun Bao1,2*
Changjun Bao1,2* Qun Li4*
Qun Li4* Tianmu Chen3*
Tianmu Chen3*Objective: To describe the epidemiological characteristics of norovirus outbreaks in Jiangsu Province, utilize the total attack rate (TAR) and transmissibility (Runc) as the measurement indicators of the outbreak, and a statistical difference in risk factors associated with TAR and transmissibility was compared. Ultimately, this study aimed to provide scientific suggestions to develop the most appropriate prevention and control measures.
Method: We collected epidemiological data from investigation reports of all norovirus outbreaks in Jiangsu Province from 2012 to 2018 and performed epidemiological descriptions, sequenced the genes of the positive specimens collected that were eligible for sequencing, created a database and calculated the TAR, constructed SEIAR and SEIARW transmission dynamic models to calculate Runc, and performed statistical analyses of risk factors associated with the TAR and Runc.
Results: We collected a total of 206 reported outbreaks, of which 145 could be used to calculate transmissibility. The mean TAR in was 2.6% and the mean Runc was 12.2. The epidemiological characteristics of norovirus outbreaks showed an overall increasing trend in the number of norovirus outbreaks from 2012 to 2018; more outbreaks in southern Jiangsu than northern Jiangsu; more outbreaks in urban areas than in rural areas; outbreaks occurred mostly in autumn and winter. Most of the sites where outbreaks occurred were schools, especially primary schools. Interpersonal transmission accounted for the majority. Analysis of the genotypes of noroviruses revealed that the major genotypes of the viruses changed every 3 years, with the GII.2 [P16] type of norovirus dominating from 2016 to 2018. Statistical analysis of TAR associated with risk factors found statistical differences in all risk factors, including time (year, month, season), location (geographic location, type of settlement, type of premises), population (total number of susceptible people at the outbreak site), transmission route, and genotype (P < 0.05). Statistical analysis of transmissibility associated with risk factors revealed that only transmissibility was statistically different between sites.
Conclusions: The number of norovirus outbreaks in Jiangsu Province continues to increase during the follow-up period. Our findings highlight the impact of different factors on norovirus outbreaks and identify the key points of prevention and control in Jiangsu Province.
Norovirus, initially known as winter vomiting disease, was identified as the cause of the outbreak in 1972 when researchers observed 27-nm-virus particles by immune electron microscopy in the infected fecal filtrate from an acute outbreak of gastroenteritis in Norwalk, Ohio, in 1968, and named “Norwalk virus” (1, 2). Norovirus is a single-stranded and unenveloped RNA virus, which can be classified into 10 genogroups (GI-GX) and 49 genotypes with most of the infections in humans caused by GI and GII genogroups, which could be further divided to 9 and 27 genotypes, respectively (3). Currently, no drugs effectively treat norovirus.
The disease burden of norovirus is becoming increasingly serious worldwide and in China, causing ~684 million cases and 212,000 deaths, annually (4). Compared with other countries, China has a higher prevalence of norovirus (19.8–21.0%) (5). The annual report of the National Statutory Infectious Diseases Reporting System, found a significant decrease in the incidence of infectious diarrhea caused by cholera, dysentery and typhoid fever between 2006 and 2016; on the other hand, the number of cases caused by viral pathogens increased from 741,809 to 1,017,962 (6). Nowadays, norovirus is replacing rotavirus as the main pathogen of viral acute gastroenteritis, which indicates that norovirus outbreaks remain an important public health problem in China. Understanding the characteristics of the disease is key to the effective implementation of disease prevention and control. Existing studies have found significant differences in the epidemiological characteristics of norovirus outbreaks between China and other countries. First, whole populations are susceptible to norovirus infection (7); however, the outbreak sites are different. In developed countries, norovirus outbreaks occur mainly in medical institutions, such as long-term care facilities and hospitals (8, 9), whereas in China, norovirus outbreaks mainly occur in school environments and nurseries (10–13). Second, different age groups face different risks of infection; particularly, one study found that the highest incidence was observed in children aged <5 years (14). Other studies have found that children younger than 12 years are more susceptible and infection is common in children under 2 years old in developing countries (15, 16). Additionally, differences were observed between the genotypes. In the past two decades, the GII.4 virus has caused norovirus outbreaks in most age groups worldwide (17). However, norovirus genotypes are constantly changing in China with the emergence of new genotypes and differences in genotypes observed in different regions.
In recent years, an increase has been observed in the incidence rate of infectious diarrhea in Jiangsu Province. In a study on norovirus and meteorological factors in Jiangsu Province, bacterial culture and viral nucleic acid testing were performed on 6,640 stool specimens collected, and 1,193 positive specimens were obtained, of which the positive rate for viruses was greater than that for bacteria; and among the positive specimens for viruses, the positive rate for norovirus was higher than that for rotavirus (18). Norovirus has a heavy disease burden in Jiangsu Province. Currently, researchers in Jiangsu Province have made significant progress in studying the molecular epidemiological characteristics of norovirus outbreaks (19–22). However, epidemiological characterization of large sample sizes is still limited for a large number of reported norovirus outbreaks in Jiangsu Province. Previous epidemiological studies on norovirus outbreaks usually calculated the total attack rate (TAR) or duration of the outbreak as the dependent variable and collected risk factors associated with the infection as independent variables for statistical analysis, and finally found the epidemiological characteristics of norovirus outbreaks and factors influencing the severity of outbreaks in a certain area during a certain period of time, providing a scientific basis for controlling norovirus outbreaks (23–25). However, we believe that studies on the TAR do not completely reflect the transmissibility of the virus, and it is necessary to construct mathematical models to quantify the transmissibility of norovirus in outbreaks. Current mathematical models used to calculate the transmissibility of infectious diseases include agent-based models and ordinary differential equation models. In our previous study, the transmissibility of norovirus in interpersonal transmission and water transmission modes was described by constructing ordinary differential equation models (26). Therefore, in order to identify as many risk factors as possible and determine the epidemiological characteristics of norovirus outbreaks in Jiangsu province, we collected all outbreak investigation reports from 2012 to 2018 in Jiangsu Province, sequenced all positive specimens eligible for sequencing to determine the genotype, determined the TAR of each outbreak, and constructed transmission dynamics models for different transmission routes to calculate the transmissibility, and performed statistical analyses of risk factors associated with the TAR and transmissibility.
Our study is an observational study that combines fieldwork and transmissibility. This study consists of three parts, the logical relationship of which is shown in Figure 1. According to the investigation report, we established a database, numbered each outbreak, and transformed the description of the outbreak into tabular data. We could find out epidemiological data, demographic data and influencing factors from investigation reports. In order to identify genotypes, we also collected and detected samples. From the report, we could know exactly the transmission route of each outbreak, and select different transmission models according to different transmission routes. The independent variable was the data extracted from each investigation report; the dependent variables were the TAR and reproduction number (R0). The TAR was calculated according to the outbreak data, and R0 was calculated using the transmission dynamics model. In this study, prevalence was used for model fitting. The transmission dynamics model aims to refine the transmission process of infectious diseases in the whole population, so as to explain the transmission mechanism of infectious diseases. According to the influencing factors in the report, we explored whether each influencing factor would affect the TAR and R0. Chi-square analysis was used to compare the rates of multiple samples, and analysis of variance was used to compare the mean of multiple samples.
The outbreak database (Supplementary Table 1) contained the city where the outbreak occurred, the number of people affected by the outbreak (susceptible people), the number of cases, the number of deaths, the location type, the onset date of the initial case, the transmission route and genotype. The data source of the outbreak database was mainly based on investigation reports. Contents of the report mainly included the investigation process and contents of the aggregated epidemic and outbreak, mainly including the basic situation investigation of the epidemic occurrence institution, on-site epidemiological investigation, hygienic investigation and epidemic termination evaluation, as well as the sample collection and laboratory test results involved in the investigation. All contents of the investigation report were specified in the “Guidelines on outbreak investigation, prevention and control of norovirus infection” issued by China Center for Disease Control and Prevention (27).
Case definition:
(1) Suspected case: That is, acute gastroenteritis cases, defined as those who defecate ≥ 3 times within 24 h and have character changes (dilute watery stool), and/or vomit ≥ 2 times within 24 h. (2) Clinical diagnostic cases: In the cluster epidemic or outbreak caused by norovirus infection, the cases that meet the definition of suspected cases and are epidemiologically related to laboratory diagnosed cases. (3) Laboratory diagnosed cases: In suspected cases or clinical diagnostic cases, stool, anal swab or vomit samples are positive for norovirus nucleic acid or ELISA antigen.
Judgment criteria of cluster epidemic and outbreak:
(2) Cluster epidemic: Within 3 days, 5 or more epidemiologically linked cases of norovirus infection occur in the same school, childcare institution, medical institution, nursing home, factory, construction site, cruise ship, community or village, and other crowded places or sites, of which at least 2 are laboratory diagnosed cases. (2) Outbreak: 20 or more epidemiologically linked cases of norovirus infection in the same school, childcare institution, medical institution, nursing home, factory, construction site, cruise ship, community or village, and other crowded places or sites within 7 days, of which at least 2 are laboratory diagnosed cases. (3) If no laboratory testing capability for norovirus is available or in the early stage of outbreak detection, a suspected outbreak or outbreak of norovirus infection can be determined if the following four characteristics of Kaplan Criteria are met: (1) more than half of the patients present vomiting symptoms; (2) average incubation period of 24–48 h; (3) average duration of illness 12–60 h; (4) exclude bacterial, parasitic and other pathogenic infections. The sensitivity of the Kaplan Criteria for identifying norovirus outbreaks is 68% and the specificity is 99%.
All stool specimens collected from outbreaks would be tested for norovirus. The stool suspensions were diluted 10-fold with saline solution. Viral nucleic acids were extracted and tested using the MagMAXTM-96 Viral RNA Isolation Kit (Applied Biosystems, Foster City, CA) and the Qiagen Probe reverse-transcriptase polymerase chain reaction (RT-PCR) Kit (Qiagen, Hilden, Germany) on a 7500 real-time PCR platform (Applied Biosystems, Foster City, CA) with primers as described (28).
In our study, norovirus nucleic acid was detected in samples collected from each outbreak. All norovirus-positive samples were detected with a region of 543 bp (GI) or 557 bp (GII) targeted at the ORF1/ORF2 junction of the viral genome by one-step RT-PCR for norovirus genotyping (29). If sequencing failed, the VP1 gene in ORF2 was amplified using the previously described primers G1SKF/G1SKR(GI) or G2SKF/G2SKR(GII) (30). However, the CT values of some samples were >33, the viral load was low, and sufficient amplification products could not be obtained, meaning that sequencing conditions were not met and sequencing failed. The genotypes were determined using the norovirus automated genotyping tool (http://www.rivm.nl/mpf/norovirus/typing tool) and the human calicivirus typing tool (http://norovirus.ng.philab.cdc.gov). We selected 25 representative gene sequences for gene polymorphism demonstration, which covered all genotypes of norovirus causing norovirus outbreaks in Jiangsu Province from 2012 to 2018. Phylogenetic trees were constructed using the maximum likelihood algorithm with 1,000 bootstrap replicates and a Kimura2-parameter model in MEGA 7.0 (31); the norovirus reference sequences were obtained from the GenBank database. Nucleotide sequences obtained from this study were deposited in GenBank under the accession numbers MZ373204-MZ373210 and MZ373215-MZ373233.
Similar to a previous study (32), a susceptible-exposed-symptomatic-asymptomatic-recovered (SEIAR) and a susceptible-exposed-symptomatic-asymptomatic-recovered-transmission media (SEIARW) model was employed for the simulation. The SEIAR model of interpersonal transmission and SEIARW model of water or food transmission were used to assess the transmissibility of norovirus in each outbreak. In this study, Berkeley Madonna 8.3.18 (developed by Robert Macey and George Oster of the University of California at Berkeley) was used for curve fitting and simulation, and SPSS 21.0 (IBM Company, U.S.A.) was used to calculate the coefficient of determination (R2) and P-value.
In the SEIAR model, the population was divided into five groups: susceptible (S), exposed (E), symptomatic (I), asymptomatic (A), and recovered (R)
The assumptions of the model are as follows:
A susceptible person becomes the exposed person after coming into contact with the symptomatic and asymptomatic infected person at the speed of βSI and βSκA. Where β represents the probability of transmission per contact, and κ represents the transmissibility of asymptomatic cases compared to symptomatic cases.
After the incubation period (1/ω) and latent period (1/ω′), the exposed persons become symptomatic or asymptomatic; where ρ represents the proportion of asymptomatic infections.
Symptomatic and asymptomatic cases recovered after a period of 1/γ and 1/γ′.
The flowchart of the model is shown in Figure 2A.
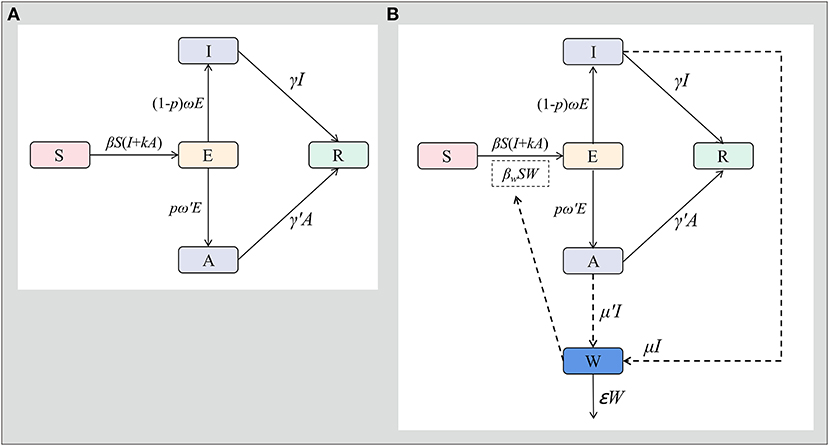
Figure 2. SEIAR model and SEIARW model for norovirus transmission in Jiangsu Province. [A: susceptible-exposed-symptomatic-asymptomatic-recovered model (SEIAR model), B: susceptible-exposed-symptomatic-asymptomatic-recovered-transmission media model (SEIARW model)].
The equations of the model are as follow:
In the SEIARW model, the water or food transmission medium (W) is added.
The assumptions of the model are as follows:
Both symptomatic and asymptomatic cases contaminate water or food by shedding pathogens into compartment W at shedding rates of μI and μ'A. μ and μ' represent shedding coefficients.
When exposed to contaminated water or food, the susceptible people will become exposed people at the rate of βwSW, which represents the coefficient of water or food transmission.
The flowchart of the model is shown in Figure 2B.
The equations of the model are as follow:
Model 1 included seven parameters: β, k, ω, ω′, ρ, γ and γ′ (Table 1). β was adjusted using the actual outbreak data and model. Previous researches have shown that the incubation period of norovirus was 12–48 h (33, 34), therefore, our model chose 1 d as the incubation period (ω = 1). A study showed that norovirus started to shed within an average of 36 h (range: 18–110) after infection, and the virus could be discharged for approximately 26 d (range: 11–54) (35), thus, the latent and infectious period was 1 d and 26 d, so ω′=1, γ′ = 0.03846. The course of norovirus infectious diarrhea is generally 1–5 d (36, 37), with some cases reaching 4–6 d (38, 39). According to the Kaplan principle of norovirus infectious diarrhea diagnosis commonly used in the United States (40), the average course of the disease is 1–3 d. In this study, 3 d were used for the modeling study, namely γ = 0.3333. The asymptomatic infection ratio of this disease potentially reaches 30% (35, 41, 42); therefore, ρ = 0.3 was used.
Model 2 included 14 parameters. The normalized 10 parameters were the parameters that required specific values, which were b, bw, κ, ω, ω′, ρ, γ, γ′, c, and ε. Among them, b and bwwere fitted using the actual outbreak data and model.
The shedding rate of the asymptomatic compared to the infectious was c, and μ′ = c/μ (43); subsequently, the specific value of the parameter was calculated by fitting the actual epidemic data. The survival time of norovirus in the external environment was approximately about 7–12 d (44–46), and the longest was 21–28 d (47–52). In this study, 10 d were used for the study; therefore, ε = 0.1 was used.
In this study, R0 was used to evaluate the transmissibility of norovirus. In Model 1, the R0 formula is expressed as follows:
In Model 2, R0 formula is expressed as follows:
R0 was divided into two parts (Runc and Rcon) where Runc and Rcon represent the uncontrolled and controlled R0, respectively. The epidemic curve was divided into two stages based on the increase and decrease. The increasing stage of the epidemic curve indicates that the intervention measures are yet to exert their effects; therefore, it is a better representation of the real transmissibility of the disease. Therefore, we used the data for epidemiological analysis and recorded the data as Runc.
TAR was used to evaluate the effects of various preventive and control measures. where n is the cumulative number of cases and N is the number of susceptible people in an outbreak.
The formula of TAR is:
All statistical analyses were performed using SPSS 21.0 (IBM Company, U.S.A.). For TAR, the chi-square test was used for comparison between the groups. Groups with significant differences were analyzed using the Bonferroni method for further pairwise comparisons. Statistical significance was set at P < 0.05 for both between group and pairwise comparisons. For R0, a homogeneity test of variance was first performed. If the variance was homogeneous, analysis of variance was used for inter-group comparison. Simultaneously, the LSD method was used for further pairwise comparison. If the homogeneity of variance was not satisfied, the Kruskal Wallis rank-sum test was used for inter-group comparison.
From 2012 to 2018, data from 206 norovirus outbreaks were collected in Jiangsu Province; all were able to calculate for the TAR. The epidemic curves of 206 outbreaks are showed in Figure 3. However, only 145 met the conditions for calculating transmissibility, and the basic reproduction number was calculated. The fitting effects of 145 outbreaks are shown in Supplementary Figure 1 which found the mean R2 was 0.76 and all P-values were significant.
For risk factors for TAR and transmissibility, we have the following findings (Tables 2, 3).
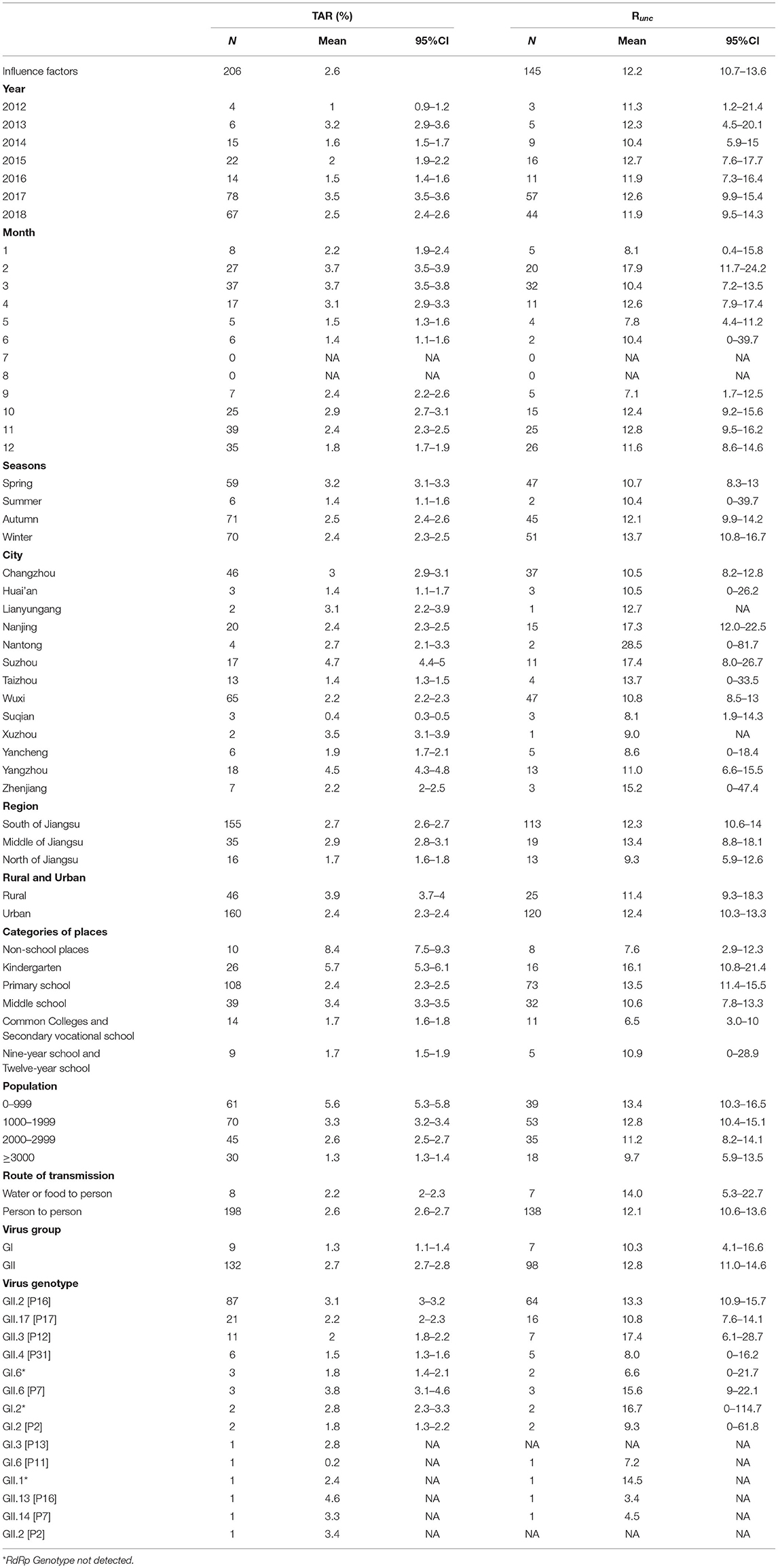
Table 2. TAR and transmissibility and influence factors in 206 outbreaks of norovirus in Jiangsu Province.
The mean TAR of 206 outbreaks was 2.6% and the mean Runc of 145 outbreaks was 12.2 (95% confidence interval: 10.7–13.6) from 2012 to 2018.
The map showed significantly more outbreaks in southern Jiangsu compared to northern Jiangsu; furthermore, the TAR and Runc showed the highest trend in southern Jiangsu (Figure 4). Wuxi City reported the most outbreaks (65 outbreaks), and Xuzhou City reported the least outbreaks (two outbreaks). There was a significant difference in mean TAR between cities (χ2 = 1,458.876, P < 0.05). The highest and lowest mean TAR were found in Suzhou City (4.7%) and Suqian City (0.4%), respectively. There was no significant difference in transmissibility between cities. Regarding location, the TAR between different cities was statistically different (χ2 = 193.783, P < 0.05), the highest and lowest mean TAR were observed in central and northern Jiangsu, respectively.
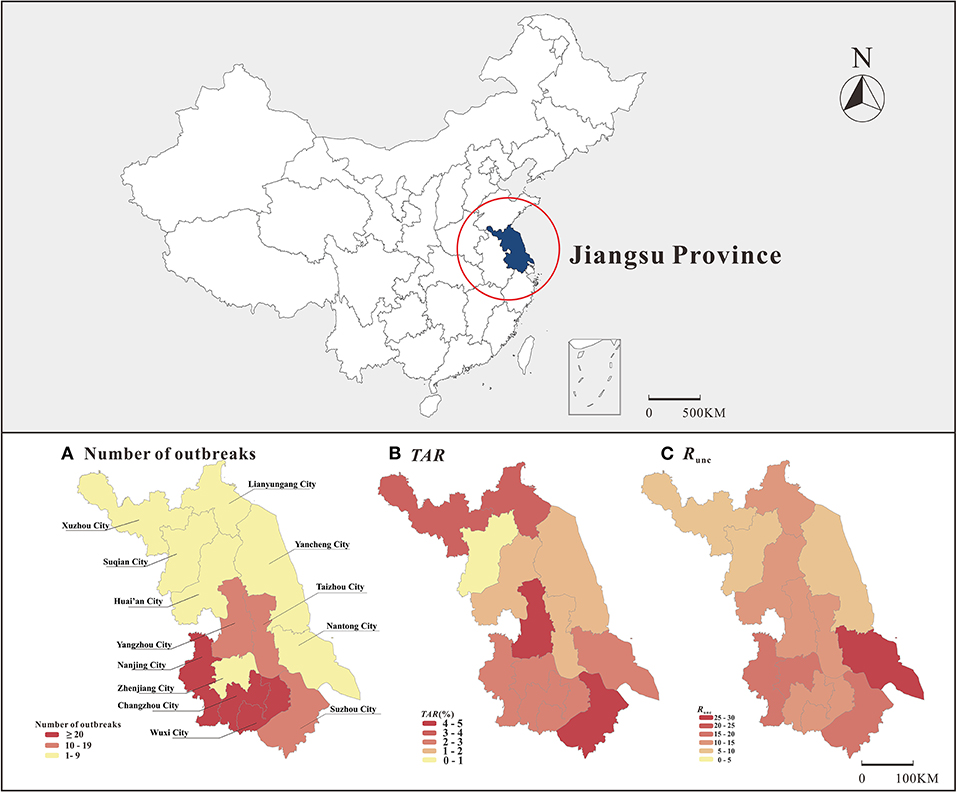
Figure 4. Distribution of number of outbreaks, TAR and transmissibility on map of Jiangsu Province (A: number of outbreaks, B: TAR, C: Runc).
The number of outbreaks showed a significant upward trend from 2012 to 2018. A big turning point was observed in 2016–2017, with 14 and 78 outbreaks reported, respectively. There was a significant difference in mean TAR between the years analyzed (χ2 = 998.138, P < 0.05). Chi-square test pairwise comparison results showed differences between all years except 2013 and 2017, and 2014 and 2016. The mean TAR and Runc tortuously changed over time, with a peak every 3 years.
In the four seasons, the number of outbreaks in summer was the lowest (six outbreaks); more outbreaks were observed in autumn and winter, 71 and 70, respectively. There was a significant difference in the mean TAR between seasons (χ2 = 237.62, P < 0.05). The summer had the lowest mean TAR. The mean TAR was found to be high from February to March and October (3.7, 3.7, 2.9%, respectively). The season significantly affected the TAR in all seasons except between autumn and winter. There was no significant difference in transmissibility between seasons. Outbreaks with high Runc were concentrated in February, November, and April (17.9, 12.8, and 12.6, respectively).
Most outbreaks occurred in urban areas (160 outbreaks) compared to rural areas (46 outbreaks). The mean TAR between rural and urban areas was significantly different (χ2 = 502.826, P < 0.05). The mean TAR in rural areas was higher compared to urban areas (3.9 and 2.4%, respectively). The mean Runc in rural areas and urban areas were 11.4 and 12.4, respectively.
Among 206 outbreaks, 196 occurred in schools, and ten occurred in places other than schools. There was a significant difference in the mean TAR between the categories of places analyzed (χ2 = 1,565.869, P < 0.05). Regarding the type of location, no significant difference was observed between common colleges and secondary vocational schools and 9-year school and 12-year school; however, other places significantly affected the mean TAR. The mean TAR in non-school places was 8.4%. Among the schools, kindergartens had the highest mean TAR (5.7%). There was no significant difference in transmissibility between the categories of places analyzed. Among the outbreak sites, the mean Runc for kindergartens was 16.1, the mean Runc observed in common colleges and secondary vocational schools was 6.5. Furthermore, the mean Runc of non-schools was only 7.6.
Most of the outbreaks occurred in places with a small number of susceptible people, 61 and 70 outbreaks occurred in places with a number of <1,000 and 1,000–2,000 susceptible people, respectively, and the least outbreaks occurred in places with a small number of susceptible people, only 30. A comparison of the different numbers of susceptible people regarding the mean TAR showed statistically significant differences (χ2 = 22.19, P < 0.05). The highest and lowest TAR means were found in places with <1,000 people (5.6%) and places with more than 3,000 people (1.3%), respectively. Further, there was no significant difference in transmissibility between places with differing numbers of susceptible people, the mean Runc of places with <1,000 people was 13.4, and the mean Runc of places with more than 3,000 people was 9.7.
Regarding the transmission routes, 198 outbreaks were interpersonal transmission and eight were waterborne or foodborne transmission. The mean TAR was significantly different between different transmission routes (χ2 = 2,296.419, P < 0.05). The mean TAR of waterborne or foodborne transmission was lower compared to interpersonal transmission (2.2 and 2.6%, respectively). There was no significant difference in transmissibility between transmission routes. The mean Runc of water or food transmission route and human transmission route was 14.0 and 12.1.
Genotype sequencing was completed in 141 outbreaks; among which, the GII genotype caused the highest number of outbreaks (132/141). Among the GII genotypes, the GII.2 [P16] genotype caused the most outbreaks (87 outbreaks). As shown in Figure 5, the outbreaks in 2012–2014 were mainly caused by the GII.4 [P31] and GII.P17 genotypes, and the majority of outbreaks in 2015–2016 were caused by the GII.17 [P17] genotype, while a change occurred after 2016 when the GII.2 [P16] genotype became dominant. In 2017 and 2018, outbreaks caused by the GII.2 [P16] genotype accounted for 91.0% (50/55) and 73.5% (36/49) of all outbreaks in that year, respectively. The GII.13 [P16] genotype had the highest mean TAR (4.6%), while GI.6 [P11] had the lowest (0.2%). Furthermore, the mean Runc of GII.3 [P12] was 17.4, and the mean Runc of GII.13 [P16] was 3.4.
To show the genetic diversity of norovirus in Jiangsu Province, 25 norovirus sequences with representative genotypes were analyzed using MEGA 7.0 software. Phylogenetic trees were constructed based on the partial RdRp gene (230 bp) and VP1 gene (280 bp) using the maximum likelihood method. As shown in Supplementary Figure 2, eight genotypes had discordant capsid and polymerase genotypes and were considered intergenotype recombinant strains (GI.6 [P11], GII.2 [P16], GII.3 [P12], GII.4 Sydney [P31], GII.6 [P7], GII.13 [P16], GI.3 [P13], and GII.14 [P7]), and three genotypes had accordant capsid and polymerase genotypes (GI.2 [P2], GII.2 [P2], and GII.17 [P17]).
In this study, we used TAR and a transmission dynamics model to explore which factors would influence the norovirus outbreaks in Jiangsu Province.
Regarding time, the number of reported cases increased sharply in 2017 and 2018. Jiangsu Province began to establish a norovirus surveillance system in 2012, using a unified case definition, outbreak determination principles, and laboratory testing methods for surveillance, so we consider our description that the number of norovirus outbreaks is increasing is consistent with the real epidemiologic characteristics. Meanwhile, we observed that the GII.2 [P16] genotype caused more than half of the norovirus outbreaks in Jiangsu province in 2017 and 2018. Previously, a study on norovirus outbreak surveillance in China had already found that the number of norovirus outbreaks increased substantially at the end of 2016, greatly exceeding the number reported in the same month in the previous 4 years, and the majority of these outbreaks were associated with GII.2 [P16] (53). The researchers analyzed this finding and concluded that the GII.2 [P16] genotype evolved extremely fast, which most likely led to a sudden increase of this virus (54). We, therefore, considered that the prevalence of the GII.2 [P16] genotype is responsible for the surge in the number of norovirus outbreaks in Jiangsu Province between 2017 and 2018.
Regarding the season, in our study, TAR was significantly lower during summer, and norovirus outbreaks in Jiangsu Province mainly occurred in the colder weather in spring, autumn, and winter, which is consistent with the results of previous studies (55, 56). Regarding the regional distribution, the increased outbreaks in southern Jiangsu despite the higher TAR in central Jiangsu may be attributed to the increased sensitivity to disease surveillance and more timely detection and reporting of the outbreaks in southern Jiangsu. Regarding rural and urban sites, the mean TAR in rural areas was higher compared to urban areas, which may be related to poor sanitation in rural areas and the untimely adoption of control measures.
Consistent with previous studies (8–13), the outbreaks we monitored occurred more often in schools than in hospitals and long-term care facilities, this phenomenon is largely due to the fact that: (1) In China (56), schools have a more standard outbreak screening system in which the government requires schools to check and screen students for symptoms of fever, vomiting, or diarrhea every morning. In contrast, hospitals or long-term care facilities do not see similar mechanisms for disease detection and reporting. (2) Long-term care facilities are important health care settings outside China. In the United States (57), long-term care facilities serve more than 2 million Americans each year, and local health units often provide infection control courses to facility staff. Ontario requires by law that each health unit must provide in-service training to long-term care facilities at least once a year. In France (58), epidemiological surveillance of acute gastroenteritis outbreaks in various types of nursing homes has been conducted since November 2010. Well-established facilities and systematic services make such settings capable of detecting and reporting norovirus outbreaks.
Most outbreaks occurred in primary schools, which is consistent with a previous study on norovirus outbreaks in China in 2014–2017 (13). Kindergarten has a higher TAR and transmissibility than other types of schools for the following reasons: (1) Studies have found that students in kindergartens and primary schools are more likely to vomit when infected with norovirus (59), and some children in kindergartens may become infected with the virus by going around when vomiting occurs due to poor hygiene awareness. (2) Kindergartens are small and therefore have a high population density and higher exposure among younger children. We found that the mean TAR of norovirus outbreaks was highest in outbreaks with <1,000 susceptible people, and of the 61 outbreaks reported in Jiangsu Province, each with <1,000 susceptible people, the highest proportion of outbreaks occurred in kindergartens, so it can be assumed that small crowded places such as kindergartens are more likely to cause the spread of the outbreak. (3) Different types of schools are composed of students of different ages. Since children under 5 years of age are most susceptible (60), and most of the students in kindergartens in China are children aged 3–6 years old, norovirus outbreaks are more likely to occur in kindergartens.
Jiangsu Provincial Center for Disease Control and Prevention has undertaken a national foodborne and waterborne norovirus outbreak surveillance program. Once an outbreak of norovirus occurs, the corresponding outbreak investigation will be started, the cases, water and food will be sampled, and the prevention and control measures will be implemented. However, due to the high economic level and satisfactory sanitary conditions in Jiangsu Province, norovirus outbreaks caused by water or food are rare, accounting for only eight (3.9%) of all reported outbreaks. Among them, water source outbreaks are caused by barreled water contamination rather than pipe network contamination, and foodborne outbreaks are caused by a small number of kitchen workers infecting and polluting the food, and not from the food itself. These conditions led to a small significant difference in TAR among outbreaks of different transmission routes.
The genetic diversity of noroviruses can be reflected by differences in epidemiology (61). Surveillance of norovirus genotypes in our study showed that the major genotypes of noroviruses in Jiangsu province change every 2–3 years. The following characteristics of the changes in the genotypes causing norovirus outbreaks have been noted: since 2002, GII.4 has been the predominant genotype in norovirus outbreaks in many countries. In the winter of 2014, the predominant norovirus strain in China changed to GII.17. Subsequently, in the winter of 2016 (2016–17), GII.2 [P16] became the predominant genotype causing norovirus outbreaks (56). Norovirus outbreaks caused by the GII.2 [P16] genotype were also found in other regions and countries outside mainland China, such as Hong Kong, Taiwan, Germany, Japan, France, the United States, and Australia. This genotype has also become the major genotype causing norovirus outbreaks in Hong Kong, Taiwan, and Germany (54). The changes in genotypes we observed in Jiangsu province are temporally consistent with the changes in genotypes throughout China. Epidemiological surveillance and vaccines are essential to control human pathogens (62). However, it has been found that different genotypes show different evolutionary patterns, and the production of new variants of GII.4 genotype every 2–3 years will always lead to a population that is always susceptible to GII.4 genotype of norovirus. Once an immune barrier is established, individuals will no longer be susceptible to the GII.2 virus, which is classified as a static virus (63, 64). Based on this view, our future studies intend to observe this through continuous monitoring of genotypes and changes in population incidence levels in combination with other factors.
The mean Runc of norovirus outbreaks in Jiangsu Province was 12.2, which was close to the data from a previous study on outbreaks (2 to >14) (65). In our previous study, we used the model fitting method to adjust the value of κ (interpersonal transmission coefficient) to be closer to 0. Finally, we obtained transmissibility of 1.94 for norovirus outbreaks in the community, 3.44 for transmissibility in schools and 4.91 for transmissibility by water (26); these results were all smaller compared to the values obtained in the current study. This difference can be explained by another study in which researchers found that the transmissibility of norovirus was largely influenced by the structure of the model and the weight of asymptomatic infections (66). We, therefore, considered that differences in parameter values in the model and differences in sample size could be responsible for the higher transmissibility of norovirus outbreaks in Jiangsu Province.
According to our research, further improvements are required in the sensitivity monitoring in northern Jiangsu. According to the epidemiological characteristics of norovirus in Jiangsu Province, the surveillance of norovirus outbreaks should be strengthened in all seasons of the year. Furthermore, health education and health promotion should be accomplished before autumn and winter where the incidence of norovirus is high, especially in rural areas. For schools, kindergartens have the highest transmissibility, followed by primary schools. This phenomenon mainly considers the immunological differences between age groups and health habits. Older students who are not in kindergartens and primary schools, as well as teachers, should be more aware of norovirus and need to know how to handle vomit properly to prevent the spread of aerosols. Also, students who have gathered should be evacuated promptly and kept away from the vomit. Furthermore, suspected cases of norovirus infection should be isolated, especially kitchen staff. Finding out changes of dominant genotype has occurred is important for exploring the epidemiological characteristics of norovirus outbreaks in Jiangsu Province, however, not every positive specimen collected in an outbreak met the criteria for genetic sequencing and could be used to determine the genotype of the norovirus causing the outbreak. Therefore, tracking changes in the major genotypes causing outbreaks requires further surveillance.
The number of norovirus outbreaks in Jiangsu Province has generally been on the rise during the follow-up period, so prevention and control remain an important issue. Currently, factors such as year, month, season, region, urban and rural type, place type, the population of outbreak sites, route of transmission, and virus genotype significantly impact the TAR of norovirus outbreaks; notably, different types of schools influence transmissibility. The dominant genotype of the virus will change every 3 years and surveillance of norovirus genotypes should be further strengthened to explore the transmissibility of different genotypes. Our study explored how these factors affect the TAR and transmissibility of norovirus outbreaks in Jiangsu Province, suggesting current priorities for the prevention and control of norovirus outbreaks in Jiangsu Province and future issues to be addressed in the surveillance of outbreaks.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
CB, QL, TC, JA, YZ, JF, and XC contributed to conception and design of the study. JA, JF, XC, XZ, HJ, and WL collected data. TC, QL, YZ, JA, JR, TY, YW, JX, XL, MY, SL, and XG analyzed the data. CB, TC, JA, YZ, JF, and XC wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This study was partly supported by the Key Medical Discipline of Epidemiology (No. ZDXK A2016008), the National Major S&T Projects (No. 2018ZX10714-002), the Jiangsu Provincial Medical Youth Talent (No. QNRC2016542), the Project of Jiangsu Commission of health (Z2019006), the NHC Key Laboratory of Echinococcosis Prevention and Control (No. 2020WZK2001), and project S202110384382 supported by XMU Training Program of Innovation and Entrepreneurship for Undergraduates.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank all study participants for providing the data and the field investigators for collecting the data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.786096/full#supplementary-material
1. Kapikian AZ, Wyatt RG, Dolin R, Thornhill TS, Kalica AR, Chanock RM. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J Virol. (1972) 10:1075–81. doi: 10.1128/jvi.10.5.1075-1081.1972
2. Kapikian AZ. The discovery of the 27-nm Norwalk virus: an historic perspective. J Infect Dis. (2000) 181:S295–302. doi: 10.1086/315584
3. Chhabra P, de Graaf M, Parra GI, Chan MC, Green K, Martella V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. (2019) 100:1393–406. doi: 10.1099/jgv.0.001318
4. Mattison CP, Cardemil CV, Hall AJ. Progress on norovirus vaccine research: public health considerations and future directions. Expert Rev Vaccines. (2018) 17:773–84. doi: 10.1080/14760584.2018.1510327
5. Saupe AA, Rounds J, Sorenson A, Hedeen N, Bagstad E, Reinberg R, et al. Outbreak of norovirus gastroenteritis associated with ice cream contaminated by frozen raspberries from China; Minnesota, USA, 2016. Clin Infect Dis. (2021) 73:e3701–7. doi: 10.1093/cid/ciaa821
6. Zhou HL, Zhen SS, Wang JX, Zhang CJ, Qiu C, Wang SM, et al. Burden of acute gastroenteritis caused by norovirus in China: A systematic review. J Infect. (2017) 75:216–24. doi: 10.1016/j.jinf.2017.06.004
7. de Rougemont A, Ambert-Balay K, Belliot G, Pothier P. [Norovirus infections: an overview]. Med Sci. (2010) 26:73–8. doi: 10.1051/medsci/201026173
8. Barclay L, Park GW, Vega E, Hall A, Parashar U, Vinjé J, et al. Infection control for norovirus. Clin Microbiol Infect. (2014) 20:731–40. doi: 10.1111/1469-0691.12674
9. Lindsay L, Wolter J, De Coster I, Van Damme P, Verstraeten T. A decade of norovirus disease risk among older adults in upper-middle and high income countries: a systematic review. BMC Infect Dis. (2015) 15:425. doi: 10.1186/s12879-015-1168-5
10. Li B, Xiao D, Li Y, Wu X, Qi L W., Tang, et al. Epidemiological analysis of norovirus infectious diarrhea outbreaks in Chongqing, China, from 2011 to 2016. J Infect Public Health. (2020) 13:46–50. doi: 10.1016/j.jiph.2019.06.019
11. Gao Z, Liu B, Yan H, Li W, Jia L, Tian Y, et al. Norovirus outbreaks in Beijing, China, from 2014 to 2017. J Infect. (2019) 79:159–66. doi: 10.1016/j.jinf.2019.05.019
12. Li J, Gao X, Ye YL, Wan T, Zang H, Mo PH, et al. An acute gastroenteritis outbreak associated with person-to-person transmission in a primary school in Shanghai: first report of a GI5 norovirus outbreak in China. BMC Infect Dis. (2018) 18:316. doi: 10.1186/s12879-018-3224-4
13. Lian Y, Wu S, Luo L, Lv B, Liao Q, Li Z, et al. Epidemiology of norovirus outbreaks reported to the public health emergency event surveillance system, China, 2014–2017. Viruses. (2019) 11:342. doi: 10.3390/v11040342
14. Allen DJ, Harris JP. Methods for ascertaining norovirus disease burdens. Hum Vaccin Immunother. (2017) 13:2630–6. doi: 10.1080/21645515.2017.1360455
15. Santos VS, Gurgel RQ, Cavalcante SM, Kirby A, Café LP, Souto MJ, et al. Acute norovirus gastroenteritis in children in a highly rotavirus-vaccinated population in Northeast Brazil. J Clin Virol. (2017) 88:33–8. doi: 10.1016/j.jcv.2016.10.015
16. Mans J, Armah GE, Steele AD, Taylor MB. Norovirus Epidemiology in Africa: a review. PLoS ONE. (2016) 11:e0146280. doi: 10.1371/journal.pone.0146280. [Epub ahead of print].
17. Pringle K, Lopman B, Vega E, Vinje J, Parashar UD, Hall AJ. Noroviruses: epidemiology, immunity and prospects for prevention. Future Microbiol. (2015) 10:53–67. doi: 10.2217/fmb.14.102
18. Fang X, Ai J, Liu W, Ji H, Zhang X, Peng Z, et al. Epidemiology of infectious diarrhoea and the relationship with etiological and meteorological factors in Jiangsu Province, China. Sci Rep. (2019) 9:19571. doi: 10.1038/s41598-019-56207-2
19. Fu JG, Shi C, Xu C, Lin Q, Zhang J, Yi QH, et al. Outbreaks of acute gastroenteritis associated with a re-emerging GII.P16-GII.2 norovirus in the spring of 2017 in Jiangsu, China. PLoS ONE. (2017) 12:e0186090. doi: 10.1371/journal.pone.0186090
20. Shi C, Feng WH, Shi P, Ai J, Guan HX, Sha D, et al. An acute gastroenteritis outbreak caused by GII17 norovirus in Jiangsu Province, China. Int J Infect Dis. (2016) 49:30–2. doi: 10.1016/j.ijid.2016.05.004
21. Fu J, Bao C, Huo X, Hu J, Shi C, Lin Q, et al. Increasing recombinant strains emerged in norovirus outbreaks in Jiangsu, China: 2015-2018. Sci Rep. (2019) 9:20012. doi: 10.1038/s41598-019-56544-2
22. Fu JG, Shi C, Sha D, Shi P, Bao CJ Ai J. [Genetic characterization of norovirus isolated in an outbreak of gastroenteritis in Jiangsu province]. Zhonghua Liu Xing Bing Xue Za Zhi. (2018) 39:72–4. doi: 10.3760/cma.j.issn.0254-6450.2018.01.015
23. Petrignani M, van Beek J, Borsboom G, Richardus JH, Koopmans M. Norovirus introduction routes into nursing homes and risk factors for spread: a systematic review and meta-analysis of observational studies. J Hosp Infect. (2015) 89:163–78. doi: 10.1016/j.jhin.2014.11.015
24. Geng Q, Shen Y, Shi P, Gao YM, Feng WH, Xiao Y, et al. Epidemiologic features and influencing factors of norovirus outbreaks in the City of Wuxi, China from 2014 to 2018. Am J Trop Med Hyg. (2021). doi: 10.4269/ajtmh.20-1371
25. Wu CY, Chi H, Liu CC, Huang YC, Huang YC, Lin HC, et al. Clinical characteristics and risk factors for children with norovirus gastroenteritis in Taiwan. J Microbiol Immunol Infect. (2021) 54:909–17. doi: 10.1016/j.jmii.2020.07.013
26. Chen T, Gu H, Leung RK, Liu R, Chen Q, Wu Y, et al. Evidence-based interventions of norovirus outbreaks in China. BMC Public Health. (2016) 16:1072. doi: 10.1186/s12889-016-3716-3
27. Liao Q, Ran L, Jin M, Cui S, Yuan J, Ma H, et al. [Guidelines on outbreak investigation, prevention and control of norovirus infection (2015)]. Zhonghua Yu Fang Yi Xue Za Zhi. (2016) 50:7–16. doi: 10.3760/cma.j.issn.0253-9624.2016.01.003
28. Fu J, Ai J, Jin M, Jiang C, Zhang J, Shi C, et al. Emergence of a new GII.17 norovirus variant in patients with acute gastroenteritis in Jiangsu, China, September 2014 to March 2015. Euro Surveill. (2015) 20:21157. doi: 10.2807/1560-7917.ES2015.20.24.21157
29. Cannon JL, Barclay L, Collins NR, Wikswo ME, Castro CJ, Magaña LC, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol. (2017) 55:2208–21. doi: 10.1128/JCM.00455-17
30. Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. (2002) 100:107–14. doi: 10.1016/S0166-0934(01)00404-9
31. Kumar S, Stecher G. Tamura K. MEGA7: molecular evolutionary genetics analysis version 70 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
32. Tsang TK, Chen TM, Longini IM Jr, Halloran ME, Wu Y, Yang Y. Transmissibility of norovirus in urban versus rural households in a large community outbreak in China. Epidemiology. (2018) 29:675–83. doi: 10.1097/EDE.0000000000000855
33. Caul EO. Small round structured viruses: airborne transmission and hospital control. Lancet. (1994) 343:1240–2. doi: 10.1016/S0140-6736(94)92146-6
34. Hutson AM, Atmar RL, Estes MK. Norovirus disease: changing epidemiology and host susceptibility factors. Trends Microbiol. (2004) 12:279–87. doi: 10.1016/j.tim.2004.04.005
35. Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, et al. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. (2008) 14:1553–7. doi: 10.3201/eid1410.080117
36. Atmar RL, Estes MK. The epidemiologic and clinical importance of norovirus infection. Gastroenterol Clin North Am. (2006) 35:275–90, viii. doi: 10.1016/j.gtc.2006.03.001
37. Kaplan JE, Schonberger LB, Varano G, Jackman N, Bied J, Gary GW. An outbreak of acute nonbacterial gastroenteritis in a nursing home. Demonstration of person-to-person transmission by temporal clustering of cases. Am J Epidemiol. (1982) 116:940–8. doi: 10.1093/oxfordjournals.aje.a113496
38. Rockx B, De Wit M, Vennema H, Vinjé J, De Bruin E, Van Duynhoven Y, et al. Natural history of human calicivirus infection: a prospective cohort study. Clin Infect Dis. (2002) 35:246–53. doi: 10.1086/341408
39. Lopman BA, Reacher MH, Vipond IB, Sarangi J, Brown DW. Clinical manifestation of norovirus gastroenteritis in health care settings. Clin Infect Dis. (2004) 39:318–24. doi: 10.1086/421948
40. Kaplan JE, Feldman R, Campbell DS, Lookabaugh C, Gary GW. The frequency of a Norwalk-like pattern of illness in outbreaks of acute gastroenteritis. Am J Public Health. (1982) 72:1329–32. doi: 10.2105/AJPH.72.12.1329
41. Graham DY, Jiang X, Tanaka T, Opekun AR, Madore HP, Estes MK. Norwalk virus infection of volunteers: new insights based on improved assays. J Infect Dis. (1994) 170:34–43. doi: 10.1093/infdis/170.1.34
42. Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis. (2009) 9:63. doi: 10.1186/1471-2334-9-63
43. Chen T, Leung RK, Zhou Z, Liu R, Zhang X, Zhang L. Investigation of key interventions for shigellosis outbreak control in China. PLoS ONE. (2014) 9:e95006. doi: 10.1371/journal.pone.0095006
44. Mattison K, Karthikeyan K, Abebe M, Malik N, Sattar SA, Farber JM, et al. Survival of calicivirus in foods and on surfaces: experiments with feline calicivirus as a surrogate for norovirus. J Food Prot. (2007) 70:500–3. doi: 10.4315/0362-028X-70.2.500
45. DallingJ. A review of environmental contamination during outbreaks of Norwalk-like virus. Br J Infect Control. (2004) 5:9–13. doi: 10.1177/14690446040050020301
46. D'Souza DH, Sair A, Williams K, Papafragkou E, Jean J, Moore C, et al. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int J Food Microbiol. (2006) 108:84–91. doi: 10.1016/j.ijfoodmicro.2005.10.024
47. Wu HM, Fornek M, Schwab KJ, Chapin AR, Gibson K, Schwab E, et al. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect Control Hosp Epidemiol. (2005) 26:802–10. doi: 10.1086/502497
48. Jones EL, Kramer A, Gaither M, Gerba CP. Role of fomite contamination during an outbreak of norovirus on houseboats. Int J Environ Health Res. (2007) 17:123–31. doi: 10.1080/09603120701219394
49. Gallimore CI, Taylor C, Gennery AR, Cant AJ, Galloway A, Iturriza-Gomara M, et al. Environmental monitoring for gastroenteric viruses in a pediatric primary immunodeficiency unit. J Clin Microbiol. (2006) 44:395–9. doi: 10.1128/JCM.44.2.395-399.2006
50. Kuusi M, Nuorti JP, Maunula L, Minh Tran NN, Ratia M, Karlsson J, et al. A prolonged outbreak of Norwalk-like calicivirus (NLV) gastroenteritis in a rehabilitation centre due to environmental contamination. Epidemiol Infect. (2002) 129:133–8. doi: 10.1017/S0950268802007276
51. Cheesbrough JS, Green J, Gallimore CI, Wright PA, Brown DW. Widespread environmental contamination with Norwalk-like viruses (NLV) detected in a prolonged hotel outbreak of gastroenteritis. Epidemiol Infect. (2000) 125:93–8. doi: 10.1017/S095026889900432X
52. Green J, Wright PA, Gallimore CI, Mitchell O, Morgan-Capner P, Brown DW. The role of environmental contamination with small round structured viruses in a hospital outbreak investigated by reverse-transcriptase polymerase chain reaction assay. J Hosp Infect. (1998) 39:39–45. doi: 10.1016/S0195-6701(98)90241-9
53. Ao Y, Wang J, Ling H, He Y, Dong X, Wang X, et al. Norovirus GII.P16/GII.2-associated gastroenteritis, China, 2016. Emerg Infect Dis. (2017) 23:1172–5. doi: 10.3201/eid2307.170034
54. Ao Y, Cong X, Jin M, Sun X, Wei X, Wang J, et al. Genetic analysis of reemerging GII.P16-GII.2 noroviruses in 2016-2017 in China. J Infect Dis. (2018) 218:133–43. doi: 10.1093/infdis/jiy182
55. Harris JP. Norovirus surveillance: an epidemiological perspective. J Infect Dis. (2016) 213(Suppl. 1):S8–11. doi: 10.1093/infdis/jiv452
56. Jin M, Wu S, Kong X, Xie H, Fu J, He Y, et al. Norovirus outbreak surveillance, China, 2016-2018. Emerg Infect Dis. (2020) 26:437–45. doi: 10.3201/eid2603.191183
57. Greig JD, Lee MB. Enteric outbreaks in long-term care facilities and recommendations for prevention: a review. Epidemiol Infect. (2009) 137:145–55. doi: 10.1017/S0950268808000757
58. Nicolay N, Boulet L, Le Bourhis-Zaimi M, Badjadj-Kab L, Henry L, Erouart S, et al. The role of dependency in a norovirus outbreak in a nursing home. Eur Geriatr Med. (2018) 9:837–44. doi: 10.1007/s41999-018-0120-9
59. Wang Y, Hao L, Pan L, Xue C, Liu Q, Zhao X, et al. Age, primary symptoms, and genotype characteristics of norovirus outbreaks in Shanghai schools in 2017. Sci Rep. (2018) 8:15238. doi: 10.1038/s41598-018-33724-0
60. Lopman BA, Adak GK, Reacher MH, Brown DW. Two epidemiologic patterns of norovirus outbreaks: surveillance in England and wales, 1992-2000. Emerg Infect Dis. (2003) 9:71–7. doi: 10.3201/eid0901.020175
61. van Beek J, de Graaf M, Al-Hello H, Allen DJ, Ambert-Balay K, Botteldoorn N, et al. Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis. (2018) 18:545–53. doi: 10.1016/S1473-3099(18)30059-8
62. Green KY. Norovirus surveillance comes of age: the impact of NoroNet. Lancet Infect Dis. (2018) 18:482–3. doi: 10.1016/S1473-3099(18)30062-8
63. Parra GI, Squires RB, Karangwa CK, Johnson JA, Lepore CJ, Sosnovtsev SV, et al. Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog. (2017) 13:e1006136. doi: 10.1371/journal.ppat.1006136
64. Zhou HL, Chen LN, Wang SM, Tan M, Qiu C, Qiu TY, et al. Prevalence and evolution of noroviruses between 1966 and 2019, implications for vaccinedesign. Pathogens. (2021) 10:1012. doi: 10.3390/pathogens10081012
65. Heijne JC, Teunis P, Morroy G, Wijkmans C, Oostveen S, Duizer E, et al. Enhanced hygiene measures and norovirus transmission during an outbreak. Emerg Infect Dis. (2009) 15:24–30. doi: 10.3201/eid1501.080299
Keywords: infectious diseases, norovirus, total attack rate, transmission dynamics, risk factors
Citation: Ai J, Zhu Y, Fu J, Cheng X, Zhang X, Ji H, Liu W, Rui J, Xu J, Yang T, Wang Y, Liu X, Yang M, Lin S, Guo X, Bao C, Li Q and Chen T (2022) Study of Risk Factors for Total Attack Rate and Transmission Dynamics of Norovirus Outbreaks, Jiangsu Province, China, From 2012 to 2018. Front. Med. 8:786096. doi: 10.3389/fmed.2021.786096
Received: 29 September 2021; Accepted: 01 December 2021;
Published: 07 January 2022.
Edited by:
Amir Abdoli, Jahrom University of Medical Sciences, IranReviewed by:
Leena Maunula, University of Helsinki, FinlandCopyright © 2022 Ai, Zhu, Fu, Cheng, Zhang, Ji, Liu, Rui, Xu, Yang, Wang, Liu, Yang, Lin, Guo, Bao, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjun Bao, YmFvMjAwMF9jbkAxNjMuY29t; Qun Li, bGlxdW5AY2hpbmFjZGMuY24=; Tianmu Chen, MTM2OTg2NjVAcXEuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.