
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Med. , 03 December 2021
Sec. Gastroenterology
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.754959
This article is part of the Research Topic Manipulation of Gut Microbiota as a Key Target to Intervene on the Onset and Progression of Digestive System Diseases, Volume II View all 5 articles
 Yingyun Yang1†
Yingyun Yang1† Ruoyu Ji1†
Ruoyu Ji1† Xinyu Zhao2
Xinyu Zhao2 Xinyuan Cao1
Xinyuan Cao1 Qiang Wang1
Qiang Wang1 Qingwei Jiang1
Qingwei Jiang1 Yizhen Zhang1
Yizhen Zhang1 Weiyang Zheng1
Weiyang Zheng1 Xi Wu1*
Xi Wu1* Aiming Yang1*
Aiming Yang1*Background: The gastric microbiota profile alters during gastric carcinogenesis. We aimed to identify the alterations in the alpha diversity and relative abundance of bacterial phyla and genera of gastric microbiota in the development of gastric cancer (GC).
Methods: The systematic review was performed based on a published protocol with the registration number CRD42020206973. We searched through PubMed, EMBASE and Cochrane databases, as well as conference proceedings and references of review articles (May 2021) for observational studies reporting either the relative abundance of bacterial phyla or genera, or alpha diversity indexes in both GC and non-cancer groups. Selection of studies and data extraction were performed independently by two researchers, with disagreements resolved through discussion. Risk of bias was assessed using the self-modified Newcastle-Ottawa Scale. Results of random-effects meta-analyses were presented as mean differences (MD).
Results: Our systematic review included 751 GC patients and 792 non-cancer patients from 14 case-control studies. Gastric cancer group had fewer operational taxonomic units (OTUs) (MD = −68.52, 95%CI: −126.65 to −10.39) and a lower Simpson index (MD = −0.13, 95%CI: −0.20 to −0.07) compared with non-cancer group. At the phylum level, gastric cancer group had a higher abundance of Firmicutes (MD = 7.11, 95%CI: 1.76 to 12.46). At the genus level, Streptococcus (MD = 3.03, 95%CI: 0.07 to 6.00) and Lactobacillus (MD = 5.15, 95%CI: 1.27 to 9.04) were found to be enriched in GCgroup. The relative abundance of the rest bacterial phyla or genera analyzed in our study did not significantly differ between two groups. Subgroup analyses indicated that the source of samples was the major source of interstudy heterogeneity.
Conclusion: This systematic review suggested that gastric microbiota dysbiosis occurred in gastric carcinogenesis, with alpha diversity declined and microbiota composition altered.
Increasing evidence has illustrated that the diversity and composition of gastric mucosal microbiota alter in gastric carcinogenesis. However, the changing pattern remains poorly understood as the findings differed across studies. This systematic review was performed based on a peer-reviewed and published protocol with the aim of evaluating the differences in the alpha diversity and relative abundance of bacterial phyla and genera between GC tissues and non-cancer tissues. Based on 14 studies with a total of 1,543 patients, our results indicated that alpha diversity declined in GC tissues and the microbiota composition altered at both phylum and genus levels. Our findings provided new insights into the involvements of gastric microbiota in gastric carcinogenesis as well as the prevention, diagnosis and treatment strategies for GC at the microbiological level.
The human gastrointestinal tract is a complicated ecosystem harboring numerous microorganisms. Gut microbes play essential roles in diverse physiological processes and are also involved in disease occurrence and development (1). The stomach had long been considered as a sterile organ until the discovery of Helicobacter pylori (H. pylori). With the advancement of high-throughput sequencing technology, a unique and complex gastric microbiota composition has been gradually uncovered (2).
Gastric cancer (GC) is the fifth most prevalent and the third most lethal malignancy worldwide (3). As postulated by Correa's model, normal gastric mucosa goes through the progressive stages from non-atrophic gastritis, atrophic gastritis, intestinal metaplasia, intraepithelial neoplasia and eventually to GC (4). Numerous studies have implicated H. pylori infection in this multistep process (5). However, only a minor fraction of patients with H. pylori infection ultimately develop GC (6), and the eradication of H. pylori does not completely prevent carcinogenesis (7, 8). Therefore, a growing number of studies have focused on the contribution of gastric microbiota dysbiosis to GC development (9, 10).
To date, the knowledge on the role of gastric microbiota dysbiosis in gastric tumorigenesis is still insufficient. Studies have noticed consecutive shifts in gastric microbiota profile during cancer development, with microbial diversity and composition changed (9, 11). However, the gastric microbiota is diverse and dynamic, which could be affected by multiple factors and differs geographically and ethnically (12, 13). Discrepancies across studies and limited sample sizes have compromised a clear understanding of this issue. Moreover, identifying the changes in gastric microbiota profile may help in prevention, diagnosis and treatment of GC. This underscores the need to perform a systematic review and meta-analysis for quantitative evaluation of changes in the diversity and composition of gastric mucosal microbiota in gastric carcinogenesis.
We performed the systematic review based on a peer-reviewed and published protocol (14) with the registration number CRD42020206973 and complied with the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) statement (15). Reporting items were detailed in the PRISMA checklist (Supplementary Material).
The purpose of this review was to evaluate the differences in the diversity of gastric microbiota and relative abundance of bacterial phyla and genera between GC tissues and non-cancer tissues.
We searched through PubMed, EMBASE and Cochrane databases. The search strategy in PubMed was: ((“microbiome” OR “microbial” OR “microbiota” [MeSH Terms]) OR “microflora” OR “bacterial” OR “dysbiosis”) AND (“gastric” [MeSH Terms] OR “stomach” OR “upper digestive tract” OR “upper gastrointestinal tract”) AND ((“lesion” OR “cancer” [MeSH Terms] OR “neoplasia” OR “neoplasms” OR “malignancy” OR “tumor” OR “carcinoma” OR “adenocarcinoma” OR “premalignancy” OR “premalignant” OR “tumorigenesis” OR “carcinogenesis”) OR “intestinal metaplasia” OR “gastritis”). The search strategy was adapted for EMBASE and Cochrane databases. We also searched conference proceedings and the references of review articles for relevant studies. The last search update was May 2021.
We included observational human studies. Eligible studies must contain both GC tissues and non-cancer tissues that were confirmed by clinical and histological evaluations. Histological diagnoses of non-cancer tissues complied with the updated Sydney system (16) and the revised Vienna classification system (17). The source of samples was limited to gastric biopsy samples (surgical or endoscopic). The sequencing technology was limited to high-throughput sequencing methods. Regarding the phenomenon of interest, studies must report either the relative abundance of bacteria at the phylum or genus level, or at least one of the alpha diversity indexes (the number of operational taxonomic units (OTUs), Shannon index, Chao 1, Simpson index, etc.) in both groups. Detailed inclusion and exclusion criteria were described in the published study protocol (14). Study selection was conducted by two researchers (YYY and RYJ) independently, with disagreements resolved through discussion.
We extracted study information including publication (authors, year, journal title), study design (patient inclusion and exclusion criteria, source of samples, grouping and the sample size of each, sequencing technology) and bias control. We extracted patient characteristics including demographics (age, sex, country or region, race/ethnicity, comorbidities), lesion location, clinical and histological diagnosis and H. pylori infection status (determined by 13C urea breath test or histological assessment). We also extracted outcome data including relative abundance of bacterial phyla or genera and alpha diversity indexes.
We made full use of available materials for data extraction. If required information was not clearly or completely recorded, we contacted the corresponding author and co-authors via e-mail. Data extraction was conducted by two researchers (YYY and RYJ) independently, with disagreements resolved through discussion.
We assessed the risk of bias using a self-modified Newcastle-Ottawa Scale (NOS) (14) which was adapted with the intention of best evaluating our phenomenon of interest (Supplementary Material). The risk of bias was evaluated from three domains: selection, comparability and exposure (or outcome), and each study was awarded with a maximum of 11 scores. Risk of bias assessment was conducted by two researchers (YYY and RYJ) independently, with disagreements resolved through discussion.
Basic characteristics and the phenomenon of interest of included studies were firstly tabulated. The mean differences [MD] with 95% confidence intervals [CI] were calculated as our effect measurements. If data were reported as the median with interquartile range, we converted them into the mean with standard deviation through a recommended formula (18). Considering large interstudy heterogeneity, we utilized the random-effects model. We evaluated heterogeneity across studies using the Cochrane chi-square (χ2) and quantified with the I2 statistics (19). I2 values of 25, 50 and 75% represented low, moderate and high heterogeneity, respectively (20). Publication bias was statistically examined by Egger's test (21). We conducted the following subgroup analyses to explore potential sources of heterogeneity: mean age, H. pylori infection status, study population, source of samples, sample size and study quality. Univariate meta-regression analyses were furthermore conducted to identify heterogeneity sources across studies. Multivariate meta-regression analyses were not performed due to limited number of included studies. All analyses except Egger's test and meta-regression analyses were performed using Review Manager 5.3.3 (Nordic Cochrane Centre, Copenhagen, Denmark) and STATA version 16 (StataCorp, College Station, TX) was used for Egger's test and meta-regression analyses. P < 0.05 was considered statistically significant.
The electronic search yielded a total of 7,568 potentially relevant studies, and 5 additional studies were identified through reference lists (Figure 1). All records were imported into Endnote with 1,024 duplicates removed. After evaluating the eligibility of the studies by reading the titles and abstracts, 6,520 studies were eliminated. Studies evaluating fecal samples (8, 22, 23), oral samples (24–26), blood samples (27) or gastric wash samples (28) were further excluded. Among the remaining 21 studies, 14 reported sufficient data available for meta-analysis (29–42), while seven did not (43–49). We have so far contacted the corresponding authors and other co-authors for additional information three times, unfortunately we received no reply. Therefore, 14 studies met our inclusion criteria and were ultimately included.
All included studies were case-control studies with a median NOS score of 8 (Range: 5–9) (Table 1). Altogether, 1,543 patients, with 751 GC patients and 792 non-GC patients were included. Only one study enrolled a mixture of Chinese and Mexican population (37), while patients of the remaining studies were composed of pure Asian populations. None of the included studies enrolled patients who had recently received antibiotic treatments or chemotherapy prior to recruiments. The samples in the reviewed studies were obtained either as endoscopic biopsies (29–34, 36, 39–42) or surgical biopsies (35, 37, 38). High-throughput sequencing methods were applied by all studies.
Regarding the phenomenon of interest (Table 2), 10 studies (32–40, 42) reported at least one of the alpha diversity indexes and nine studies (29–31, 34, 37, 39–42) reported the relative abundace of bacteria at the phylum or genus level. Four alpha diversity indexes (OTUs, Chao 1, Shannon index, Simpon index), five bacterial phyla (Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria) and eight bacterial genera (Helicobacter, Streptococcus, Lactobacillus, Veillonella, Prevotella, Sphingomonas, Fusobacterium, Neisseria) were further analyzed in the quantitative analysis.
We performed random-effects meta-analyses based on nine studies (32–40) evaluating Shannon index, six studies (32, 34–36, 38, 40) evaluating Chao 1, seven studies (32–35, 37, 40, 42) evaluating OTUs and four studies (32, 34, 35, 40) evaluating Simpson index (Figure 2). The microbiome of GC group had similar Shannon index compared with non-cancer group (MD = 0.00, 95%CI: −0.48 to 0.48, P > 0.99, I2 = 91%). GC group had siginificantly fewer OTUs compared with non-cancer group (MD = −68.52, 95%CI: −126.65 to −10.39, P = 0.02; I2 = 95%). The decrease of Chao 1 in GC group was not siginificant (MD = −130.46, 95%CI: −270.82 to 9.91, P = 0.07; I2 = 96%). Simpson index siginificantly declined in the GC group (MD = −0.13, 95%CI: −0.20 to −0.07, P < 0.001), with no evidence of between study heterogeneity (I2 = 0%).
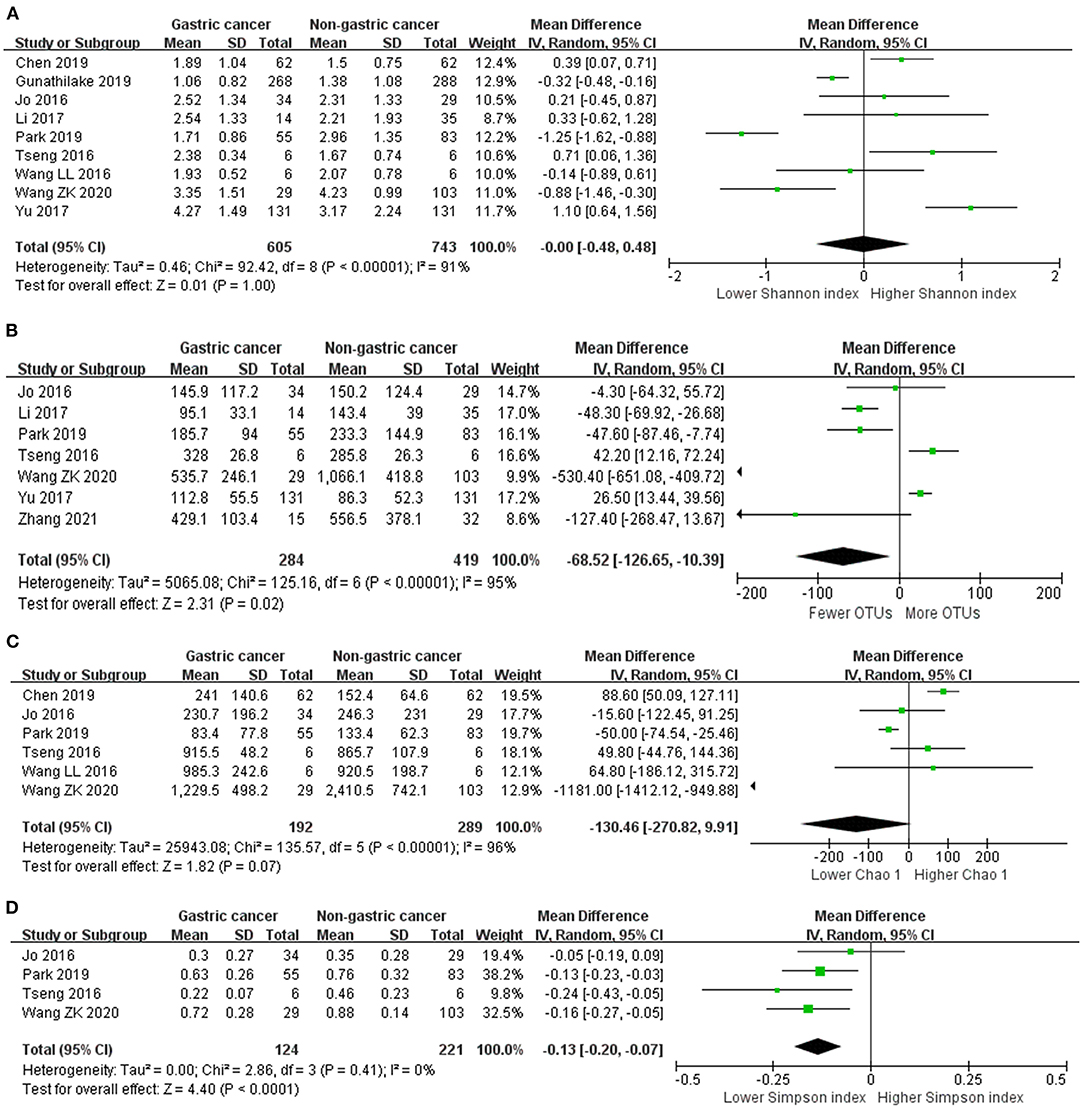
Figure 2. Forest plot for changes in alpha diversity indexes including Shannon index (A), OTUs (B), Chao 1 (C), and Simpson index (D) between gastric cancer and non-gastric cancer groups.
Egger's test suggested no significant publication bias for Shannon index (P = 0.499), Chao 1 (P = 0.857), OTUs (P = 0.459) and Simpson index (P = 0.560).
We performed random-effects meta-analyses based on five studies (29, 34, 37, 40, 42) evaluating the relative abundance of Proteobacteria, Firmicutes, Bacteroidetes and Fusobacteria, and four (29, 34, 40, 42) studies evaluating the relative abundance of Actinobacteria (Figure 3). The microbiome of GC group was composed of less Proteobacteria compared with non-cancer group (MD = −12.02, 95%CI: −27.37 to 3.33, P = 0.12; I2 = 83%), however, the difference was not significant. Conversely, Firmicutes was significantly enriched in GC group (MD = 7.11, 95%CI: 1.76 to 12.46, P = 0.009; I2 = 70%). Non-siginificant difference was found in the relative abundance of Bacteroidetes (MD = 1.86, 95%CI: −2.49 to 6.21, P = 0.40; I2 = 75%), Fusobacteria (MD = 0.82, 95%CI: −0.31 to 1.95, P = 0.15; I2 = 83%) and Actinobacteria (MD = 0.65, 95%CI: −1.09 to 2.40, P = 0.46; I2 = 74%) between two groups.
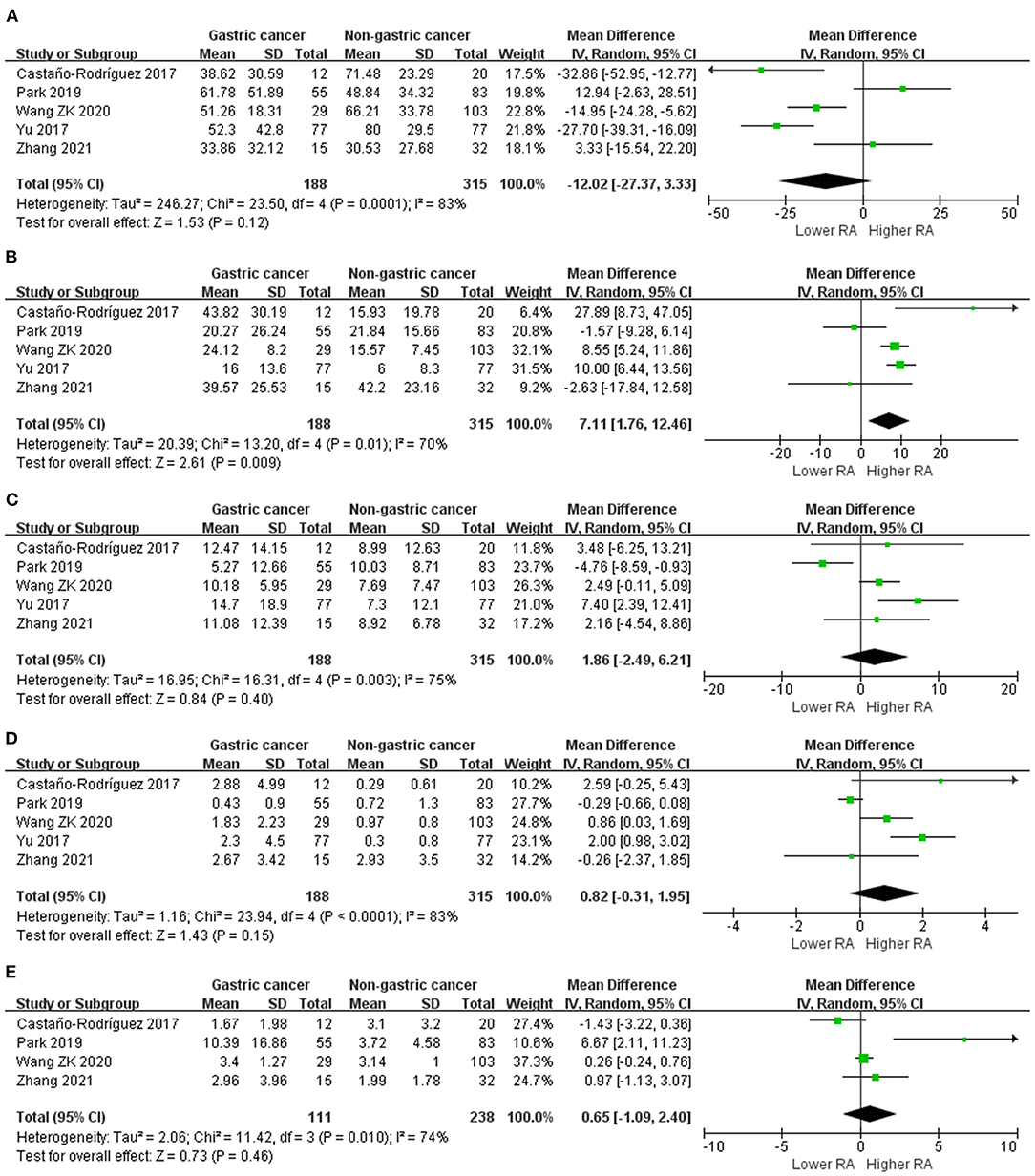
Figure 3. Forest plot for changes in relative abundance of bacterial phyla including Proteobacteria (A), Firmicutes (B), Bacteroidetes (C), Fusobacteria (D), and Actinobacteria (E) between gastric cancer and non-gastric cancer groups.
Egger's test suggested no significant publication bias for the relative abundance of Proteobacteria (P = 0.742), Firmicutes (P = 0.853), Bacteroidetes (P = 0.835), Fusobacteria (P = 0.823) and Actinobacteria (P = 0.197).
Random-effects meta-analysis based on seven studies (29–31, 34, 37, 39, 42) revealed a non-significantly lower abundance of Helicobacter in GC group (MD = −13.40, 95%CI: −28.24 to 1.45, P = 0.08, I2 = 89%). Regarding other bacterial genera, Streptococcus (MD = 3.03, 95%CI: 0.07 to 6.00, P = 0.04; I2 = 66%) and Lactobacillus (MD = 5.15, 95%CI: 1.27 to 9.04, P = 0.009; I2 = 40%) were found to be enriched in GC group. While for Veillonella, Prevotella, Sphingomonas, Fusobacterium and Neisseria, no significant differences were found between two groups (Supplementary Material).
Results of subgroup analyses and univariate meta-regression analyses indicated that mean age, sources of samples, study population, sample size, study quality and H. pylori infection status all contributed to heterogeneities across studies to varying degrees (Table 3; Supplementary Material).
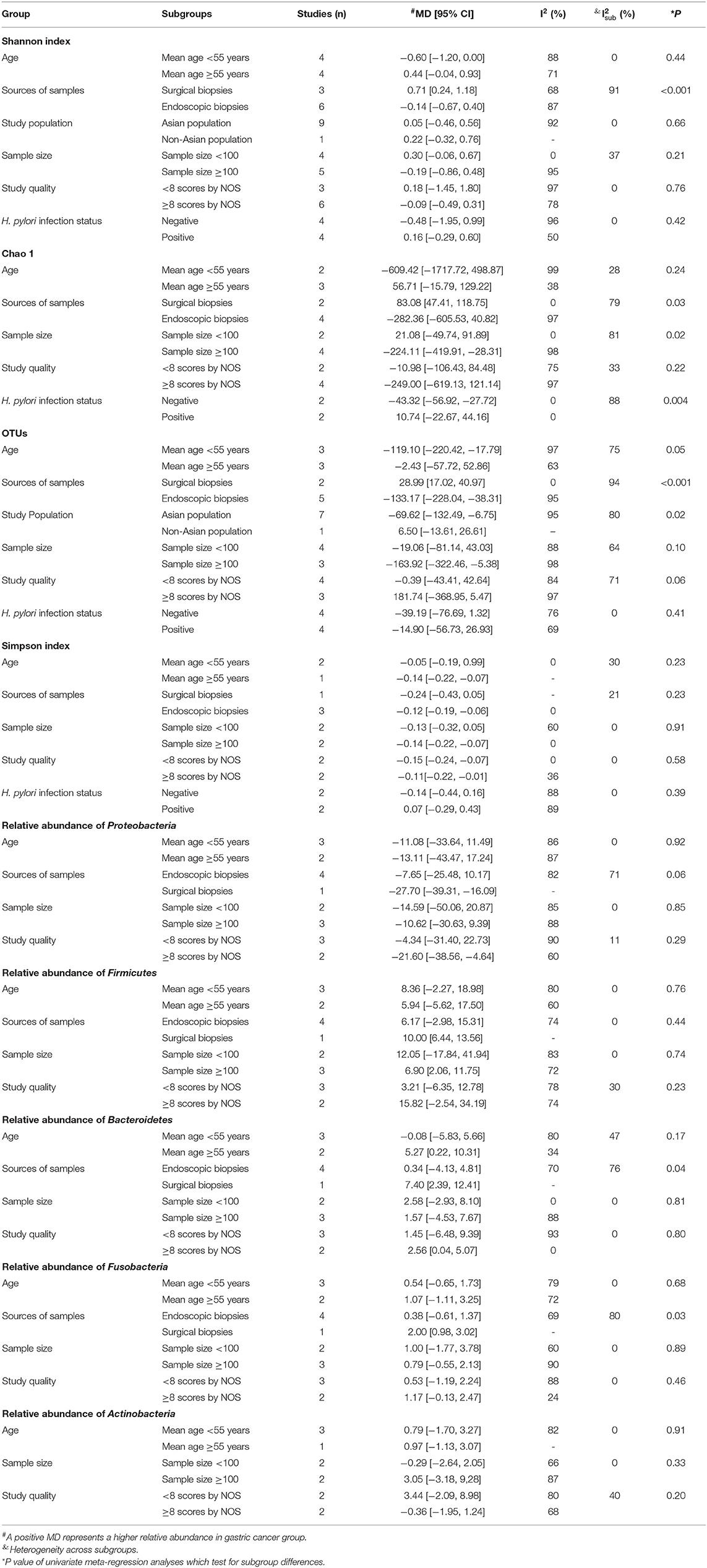
Table 3. Subgroup analyses and univariate meta-regression analyses of changes in alpha diversity indexes and relative abundance of bacterial phyla.
Sources of samples were the major source of heterogeneity for Shannon index and OTUs. In the surgical subgroup, GC tissues had higher Shannon index (MD = 0.71, 95%CI: 0.24 to 1.18) and more OTUs (MD = 28.99, 95%CI: 17.02 to 40.97) compared with non-cancer tissues. However, in the endoscopic subgroup, GC tissues had similar Shannon index (MD = −0.14, 95%CI: −0.67 to 0.40) and less OTUs (MD = −133.17, 95%CI: −228.04 to −38.31) compared with non-cancer tissues. The study population was also a source of interstudy heterogeneity for OTUs. In the Asian population, GC tissues had fewer OTUs (MD = −69.62, 95%CI: −132.49 to −6.75) compared with non-cancer tissues, which was not observed in the non-Asian population (MD = 6.50, 95%CI: −13.61 to 26.61).
Sources of samples, sample size and H. pylori infection status contributed to heterogeneity for Chao 1. In the surgical subgroup, GC tissues had higher Chao 1 (MD = 83.08, 95%CI: 47.41 to 118.75), but no significant difference was observed in the endoscopic subgroup (MD = −282.36, 95%CI: −605.53 to 40.82). Lower Chao 1 (MD = −224.11, 95%CI: −419.91 to −28.31) in GC tissues was observed in the large sample size subgroup, but not in the small sample size subgroup (MD = 21.08, 95%CI: −49.74 to 91.89). In the H. pylori negative subgroup, Chao 1 (MD = −43.32, 95%CI: −56.92 to −27.72) significantly declined in GC tissues, while no significant difference was observed in the H. pylori positive subgroup (MD = 10.74, 95%CI: −22.67 to 44.16).
Sources of samples was also the major source of heterogeneity for the relative abundance of Bacteroidetes and Fusobacteria. In the surgical subgroup, Bacteroidetes (MD = 7.40, 95%CI: −2.39 to 12.41) and Fusobacteria (MD = 2.00, 95%CI: 0.98 to 3.02) was found to be enriched in GC tissues, while the differences in the abundance of Bacteroidetes (MD = 0.34, 95%CI: −4.13 to 4.81) and Fusobacteria (MD = 0.38, 95%CI: −0.61 to 1.37) was not significant in the endoscopic subgroup.
Consecutive alternations in the diversity and composition of gastric microbiota have been observed during GC development (50). In this meta-analysis, we identified the changes in the diversity and composition of gastric microbiome during gastric carcinogenesis based on 14 case-control studies with a total of 1,543 patients. The results demonstrated that alpha diversity declined during gastric carcinogenesis. Compared with non-cancer group, GC group had a higher abundance of Firmicutes at the phylum level, and enrichments of Streptococcus and Lactobacillus at the genus level. The relative abundance of the rest bacterial phyla or genera analyzed in our study did not significantly differ between groups.
Decline in the microbial diversity has been reported in a range of gastrointestinal diseases including inflammatory bowel disease (51, 52) and colorectal cancer (53), which could be a sign of microbial dysbiosis. We evaluted the the differences in four alpha diversity indexes between cancer and non-cancer groups. OTUs and Simpson index significantly declined in cancer group, and we also observed a non-significant downward trend in Chao 1. During gastric carcinogenesis, the disruption of gastric homeostasis leads to decreased gastric acidity and dysregulated metabolic functions (44, 54), which makes the microhabitats of the tumor site no longer suitable for the colonization of bacteria.
Previous studies have reported controversial results on differences of gastric microbiota composition between GC and non-cancer patients (9). Discrepancies across studies were comprehensively and quantitavely analyzed in the present meta-analysis. Proteobacteria and Firmicutes are two dominant bacterial phylum of the gastric microbiota in patients with or without GC (28, 38, 49). Helicobacter is the major component of Proteobacteria, and its abundance was found to be inversely correlated with the abundance of Firmicutes (47). The current view posits that gastric carcinogenesis is accompanied by a gradual loss of Helicobacter (especially H. pylori) colonization (10), which may explain the depletion of Proteobacteria and the enrichment of Firmicutes in the GC group as demonstrated by our analyses.
At the genus level, the GC tissue was characterized by significant enrichments of Streptococcus and Lactobacillus. Streptococcus is the first inhabitant of the human oral cavity (55). The abundance of Veillonella, which is another dominant genus of the oral microbiota (56), was higher in the cancer group, although it did not achieve statistical significance. Overabundance of oral bacteria was found to be correlated with a spectrum of malignancies including but not limited to colorectal cancer (57), pancreatic cancer (58, 59) and lung cancer (60). Though the relationship between oral microbiome and GC has not yet been clarified, oral microbiome has been considered as a potential biomarker for non-invasive diagnosis of GC (44). Streptococcus is also categorized as lactic acid bacteria (LAB) together with Lactobacillus. The enrichments of LAB increase microbially-derived lactate, which is not only an important energy source for cancer cells, but is also involved in multiple steps of carcinogenesis by promoting inflammation, angiogenesis, metastasis and immune evasion (61). Additionally, LAB may result in DNA damage by increasing the level of reactive oxygen species (ROS) (62).
Due to the diverse and dynamic nature of gastric microbiota, we conducted several subgroup analyses as well as meta-regression analyses to investigate potential sources of heterogeneity. Our analyses indicated that the source of samples (surgical vs. endoscopic) was the major source of heterogeneity for both alpha diversity indexes and abundance of bacterial phyla. Notably, peritumor non-cancer tissues in the surgical subgroup had even lower Shannon index, Chao 1 and fewer OTUs compared with tumor tissues, which is contrary to the overall findings as well as the results of the endoscopic subgroup. Considering that in studies based on surgical samples, the comparison of diversity was carried out between cancer tissues and peritumor non-cancer tissues, the above findings might be explained by the overgrowth of certain bacteria (for example, oral bacteria or LAB as identified by our analyses) at the tumor site in the context of gastric bacterial dysbiosis.
Our systematic review and meta-analysis quantitavely assesses the alterationsin the diversity and composition of gastric microbiota during gastric carcinogenesis. Hence, the present study has several clinical implications. Firstly, to clarify the changing regularity of gastric microbiota composition during carcinogenesis, and to identify specific microorganisms involved in this process, which may provide hints for the pathogenesis of GC and the exploration of potential microbial therapy targets. Secondly, the detection of changes in gastric microbiota, especially the overgrowth of certain bacteria (eg, Streptococcus, Lactobacillus, Veillonella), might assist the diagnosis of GC. The present study has several limitations that should be noted. Despite our best efforts, several studies were not included in the quantitative analysis due to lack of sufficient data, which might lead to bias to our results. Also, there is substantial heterogeneity across studies, even though the source of heterogeneity was partly identified by subgroup analyses. Moreover, gastric microbiota, especially non-H. pylori bacteria is a relatively young field, thus the number of studies and available data included are limited, adding to the difficulty to perform stable meta-analyses, subgroup analyses and meta-regression analyses, as well as to evaluate the correlation between gastric microbiota and the prognosis of GC which is an important but open clinical issue. Finally, we only enrolled observational studies, limiting the establishment of a cause-and-effect relationship.Therefore, with continuous publication of articles, the update of the meta-analysis is warranted.
In summary, our review found that the diversity and composition of gastric microbiota differed between GC and non-GC tissues. Dysbiosis of gastric microbiota occurred in GC tissues, this was reflected by decreased alpha diversity and enrichments or depletions of certain bacteria. The update of meta-analysis is warranted with more articles published.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YY: study concept and design, data extraction and interpretation, and drafting the article and critical revision. RJ: data extraction and statistical analysis and interpretation of the data and drafting the article. XZ: study design, statistical analysis, and revision of the article for important intellectual content. XC: data extraction and statistical analysis. QW and WZ: study design, and revision of the article for important intellectual content. QJ: revision of the article for important intellectual content. YZ: data extraction and interpretation of the data. XW and AY: study concept, design, and interpretation of the data revision of the article for important intellectual content. All the authors contributed to the review and revision of the manuscript and approved the submission.
This work was supported by the National Natural Science Foundation of China (General Program, Grant Number 82073184), Peking Union Medical College Hospital Youth Program (Grant Number pumch201911356) and Beijing Science and Technology Program (Grant Number Z181100001618013). The sponsors have not been involved in study design, data collection, data analysis and result interpretation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.754959/full#supplementary-material
1. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
2. Ianiro G, Molina-Infante J, Gasbarrini A. Gastric microbiota. Helicobacter. 2015 20(Suppl. 1):68–71. doi: 10.1111/hel.12260
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
4. Correa P. Human gastric carcinogenesis: a multistep and multifactorial process–first American Cancer Society Award Lecture on cancer epidemiology and prevention. Cancer Res. (1992) 52:6735–40.
5. Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, et al. Management of Helicobacter pylori infection-the Maastricht V/florence consensus report. Gut. (2017) 66:6–30. doi: 10.1136/gutjnl-2016-312288
6. Shah MA. Gastric cancer: The gastric microbiota - bacterial diversity and implications. Nat Rev Gastroenterol Hepatol. (2017) 14:692–3. doi: 10.1038/nrgastro.2017.140
7. Ma JL, Zhang L, Brown LM Li JY, Shen L, Pan KF, et al. Fifteen-year effects of Helicobacter pylori, garlic, and vitamin treatments on gastric cancer incidence and mortality. J Natl Cancer Inst. (2012) 104:488–92. doi: 10.1093/jnci/djs003
8. Gao JJ, Zhang Y, Gerhard M, Mejias-Luque R, Zhang L, Vieth M, et al. Association between gut microbiota and helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol. (2018) 8:202. doi: 10.3389/fcimb.2018.00202
9. Dias-Jácome E, Libânio D, Borges-Canha M, Galaghar A, Pimentel-Nunes P. Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria - a systematic review. Rev Esp Enferm Dig. (2016) 108:530–40. doi: 10.17235/reed.2016.4261/2016
10. Li J, Perez Perez GI. Is there a role for the non-helicobacter pylori bacteria in the risk of developing gastric cancer? Int J Mol Sci. (2018) 19:1353. doi: 10.3390/ijms19051353
11. Zhang S, Shi D, Li M, Li Y, Wang X, Li W. The relationship between gastric microbiota and gastric disease. Scand J Gastroenterol. (2019) 54:391–6. doi: 10.1080/00365521.2019.1591499
12. Nardone G, Compare D, Rocco A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. (2017) 2:298–312. doi: 10.1016/S2468-1253(16)30108-X
13. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473:174–80. doi: 10.1038/nature09944
14. Ji R, Zhao X, Cao X, Zhang Y, Yang Y. Changes in gastric mucosal microbiota in gastric carcinogenesis: a systematic review protocol. BMJ Open. (2021) 11:e045810. doi: 10.1136/bmjopen-2020-045810
15. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. (2009) 339:b2535. doi: 10.1136/bmj.b2535
16. Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol. (1996) 20:1161–81. doi: 10.1097/00000478-199610000-00001
17. Dixon MF. Gastrointestinal epithelial neoplasia: Vienna revisited. Gut. (2002) 51:130–1. doi: 10.1136/gut.51.1.130
18. Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. (2014) 14:135. doi: 10.1186/1471-2288-14-135
19. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. (2002) 21:1539–58. doi: 10.1002/sim.1186
20. Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
21. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
22. Liang W, Yang Y, Wang H, Wang H, Yu X, Lu Y, et al. Gut microbiota shifts in patients with gastric cancer in perioperative period. Medicine (Baltimore). (2019) 98:e16626. doi: 10.1097/MD.0000000000016626
23. Qi YF, Sun JN, Ren LF, Cao XL, Dong JH, Tao K, et al. Intestinal microbiota is altered in patients with gastric cancer from Shanxi Province, China. Dig Dis Sci. (2019) 64:1193–203. doi: 10.1007/s10620-018-5411-y
24. Cui J, Cui H, Yang M, Du S, Li J, Li Y, et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell. (2019) 10:496–509. doi: 10.1007/s13238-018-0596-6
25. Hu J, Han S, Chen Y, Ji Z. Variations of tongue coating microbiota in patients with gastric cancer. Biomed Res Int. (2015) 2015:173729. doi: 10.1155/2015/173729
26. Sun JH Li XL, Yin J, Li YH, Hou BX, Zhang Z. A screening method for gastric cancer by oral microbiome detection. Oncol Rep. (2018) 39:2217–24. doi: 10.3892/or.2018.6286
27. Dong Z, Chen B, Pan H, Wang D, Liu M, Yang Y, et al. Detection of microbial 16S rRNA gene in the serum of patients with gastric cancer. Front Oncol. (2019) 9:608. doi: 10.3389/fonc.2019.00608
28. Hu YL, Pang W, Huang Y, Zhang Y, Zhang CJ. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun metagenomics. Front Cell Infect Microbiol. (2018) 8:433. doi: 10.3389/fcimb.2018.00433
29. Castaño-Rodríguez N, Goh KL, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. (2017) 7:15957. doi: 10.1038/s41598-017-16289-2
30. Gantuya B, El Serag HB, Matsumoto T, Ajami NJ, Uchida T, Oyuntsetseg K, et al. Gastric mucosal microbiota in a Mongolian population with gastric cancer and precursor conditions. Aliment Pharmacol Ther. (2020) 51:770–80. doi: 10.1111/apt.15675
31. Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, et al. Increased abundance of clostridium and fusobacterium in gastric microbiota of patients with gastric cancer in Taiwan. Sci Rep. (2018) 8:158. doi: 10.1038/s41598-017-18596-0
32. Jo HJ, Kim J, Kim N, Park JH, Nam RH, Seok YJ, et al. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than helicobacter pylori in the gastric carcinogenesis. Helicobacter. (2016) 21:364–74. doi: 10.1111/hel.12293
33. Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK. Alterations in gastric microbiota after H. Pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep. (2017) 7:44935. doi: 10.1038/srep44935
34. Park CH, Lee AR, Lee YR, Eun CS, Lee SK, Han DS. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter. (2019) 24:e12547. doi: 10.1111/hel.12547
35. Tseng CH, Lin JT, Ho HJ, Lai ZL, Wang CB, Tang SL, et al. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci Rep. (2016) 6:20701. doi: 10.1038/srep20701
36. Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. (2016) 28:261–6. doi: 10.1097/MEG.0000000000000542
37. Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, et al. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Microbiol. (2017) 7:302. doi: 10.3389/fcimb.2017.00302
38. Chen XH, Wang A, Chu AN, Gong YH, Yuan Y. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol. (2019) 10:1261. doi: 10.3389/fmicb.2019.01261
39. Gunathilake MN, Lee J, Choi IJ, Kim YI, Ahn Y, Park C, et al. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci Rep. (2019) 9:13589. doi: 10.1038/s41598-019-50054-x
40. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the gastric mucosal microbiome associated with histological stages of gastric carcinogenesis. Front Microbiol. (2020) 11:997. doi: 10.3389/fmicb.2020.00997
41. Wang L, Xin Y, Zhou J, Tian Z, Liu C, Yu X, et al. Gastric mucosa-associated microbial signatures of early gastric cancer. Front Microbiol. (2020) 11:1548. doi: 10.3389/fmicb.2020.01548
42. Zhang X, Li C, Cao W, Zhang Z. Alterations of gastric microbiota in gastric cancer and precancerous stages. Front Cell Infect Microbiol. (2021) 11:559148. doi: 10.3389/fcimb.2021.559148
43. Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. (2014) 4:4202. doi: 10.1038/srep04202
44. Coker OO Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. (2018) 67:1024–32. doi: 10.1136/gutjnl-2017-314281
45. Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. (2009) 58(Pt 4):509–16. doi: 10.1099/jmm.0.007302-0
46. Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. (2014) 19:407–16. doi: 10.1111/hel.12145
47. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. (2018) 67:226–36. doi: 10.1136/gutjnl-2017-314205
48. Kim J, Kim N, Jo HJ, Park JH, Nam RH, Seok YJ, et al. An Appropriate cutoff value for determining the colonization of Helicobacter pylori by the pyrosequencing method: comparison with conventional methods. Helicobacter. (2015) 20:370–80. doi: 10.1111/hel.12214
49. Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. (2019) 40:336–48. doi: 10.1016/j.ebiom.2018.12.034
50. Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, et al. Systematic review: gastric microbiota in health and disease. Aliment Pharmacol Ther. (2020) 51:582–602. doi: 10.1111/apt.15650
51. Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
52. Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. (2011) 141:227–36. doi: 10.1053/j.gastro.2011.04.011
53. Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. (2013) 105:1907–11. doi: 10.1093/jnci/djt300
54. Engstrand L, Graham DY. Microbiome and gastric cancer. Dig Dis Sci. (2020) 65:865–73. doi: 10.1007/s10620-020-06101-z
55. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC Yu WH, et al. The human oral microbiome. J Bacteriol. (2010) 192:5002–17. doi: 10.1128/JB.00542-10
56. Mashima I, Kamaguchi A, Nakazawa F. The distribution and frequency of oral veillonella spp. in the tongue biofilm of healthy young adults. Curr Microbiol. (2011) 63:403–7. doi: 10.1007/s00284-011-9993-2
57. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. (2015) 6:8727. doi: 10.1038/ncomms9727
58. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. (2018) 67:120–7. doi: 10.1136/gutjnl-2016-312580
59. Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. (2013) 34:2193–7. doi: 10.1093/carcin/bgt249
60. Hosgood HD, Cai Q, Hua X, Long J, Shi J, Wan Y, et al. Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax. (2020) 76:256–63. doi: 10.1136/thoraxjnl-2020-215542
61. Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. (2019) 1872:188309. doi: 10.1016/j.bbcan.2019.07.004
Keywords: gastric microbiota, gastric cancer, dysbiosis, stomach microhabitat, meta-analysis
Citation: Yang Y, Ji R, Zhao X, Cao X, Wang Q, Jiang Q, Zhang Y, Zheng W, Wu X and Yang A (2021) Alterations in Gastric Mucosal Microbiota in Gastric Carcinogenesis: A Systematic Review and Meta-Analysis. Front. Med. 8:754959. doi: 10.3389/fmed.2021.754959
Received: 28 August 2021; Accepted: 15 November 2021;
Published: 03 December 2021.
Edited by:
Avtar Singh Meena, Centre for Cellular & Molecular Biology (CCMB), IndiaReviewed by:
Aftab Alam, University at Buffalo, United StatesCopyright © 2021 Yang, Ji, Zhao, Cao, Wang, Jiang, Zhang, Zheng, Wu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi Wu, eGl3YmpAYWxpeXVuLmNvbQ==; Aiming Yang, eWFuZ2FtMjAyMEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.