- 1Graduate Institute of Physiology, College of Medicine, National Taiwan University, Taipei, Taiwan
- 2Department of Anesthesiology, Hsinchu MacKay Memorial Hospital, Hsinchu, Taiwan
- 3Department of Electrical Engineering, Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan
- 4Graduate Institute of Biotechnology, College of Bioresources and Agriculture, National Taiwan University, Taipei, Taiwan
- 5Bioinformatics and Biostatistics Core, Center of Genomic and Precision Medicine, National Taiwan University, Taipei, Taiwan
- 6Graduate Institute of Biomedical Electronics and Bioinformatics, College of Electrical Engineering and Computer Science, National Taiwan University, Taipei, Taiwan
- 7College of Biomedical Engineering, China Medical University, Taichung, Taiwan
- 8Graduate Institute of Epidemiology and Preventive Medicine, College of Public Health, National Taiwan University, Taipei, Taiwan
Background: Breast cancer is the most common malignancy and a leading cause of death among women. The majority of patients require surgery, and retrospective studies have revealed an association between anaesthetic techniques during surgery and clinical outcomes. Local anaesthetics (LAs) influence carcinogenesis by interacting with non-coding RNAs (ncRNAs). However, the detailed mechanisms underlying the association between LAs and ncRNAs remain unclear.
Methods: In this study, the effects of two commonly used LAs, lidocaine and bupivacaine, on the malignancy of MCF-7 breast cancer cells were investigated. The expression profiles of the microRNAs (miRNAs) that responded to treatment with LAs were determined through next-generation sequencing.
Results: Data from the functional assay revealed that the LAs suppressed the proliferation of MCF-7 cells. The result of next-generation sequencing revealed that 131 miRNAs were upregulated, following treatment with the LAs. Validation using polymerase chain reaction (PCR) identified miR-187-5p as a potential biomarker, and it was selected for further analyses. Prediction with bioinformatics tools and luciferase reporter assays revealed that MYB is a direct target gene of miR-187-5p. Based on the hypothesis that lncRNAs acts as miRNA sponges, the target lncRNA, DANCR, of miR-187-5p was predicted using DIANA-LncBase v2 and validated using luciferase reporter assays. In addition, the reciprocal suppressive effect between DANCR and miR-187-5p was determined.
Conclusions: This study suggests that one of the anti-tumour mechanisms of lidocaine and bupivacaine is mediated through the DANCR-miR-187-5p-MYB axis. This may provide a novel molecular mechanism of tumour suppression in breast cancer.
Introduction
Breast cancer is one of the most common malignancies and a leading cause of death among women (https://gco.iarc.fr/). Most patients require surgery; however, residual disease from scattered micro-metastases and tumour cells related to surgery is usually inevitable (1). Experimental and clinical retrospective studies indicate an association between the types of anaesthetic techniques used during the cancer resection surgery and outcomes (1, 2). There are conflicting results on the administration of volatile anaesthetics and opioids and cancer development, metastasis, and recurrence (3, 4). Regional anaesthesia is hypothesised to attenuate immunosuppression and surgical stress by minimising the requirement for opioids and volatile anaesthetics, therefore improving the long-term postoperative outcomes (2). The overall survival for different local anaesthetics (LAs) varies among the clinical studies; however, laboratory studies suggest that LAs have direct inhibitory effects on tumour cells (5, 6).
Lidocaine and bupivacaine are commonly used amide-type LAs for regional anaesthesia and peripheral nerve blockade, providing excellent perioperative pain relief, especially in breast cancer surgery (7). The mechanisms underlying the inhibitory effect of LAs on cancer cell proliferation, migration, and metastasis, are elucidated (5, 8, 9). However, the precise mechanism remains unclear; it could involve sodium channel blockade, DNA demethylation (10, 11), or interactions between LAs and non-coding RNAs (ncRNAs). Lidocaine inhibits the growth and invasion of gastric carcinoma cells by upregulating miR-145 (12). It inhibits proliferation and induces apoptosis in colorectal cancer cells by upregulating miR-520a-3p and targeting EGFR (13).
Dysregulated gene expression is a major hallmark of cancer. ncRNAs play a critical role in tumourigenesis, growth, and progression (14–16). NcRNAs are categorised as small ncRNAs (sncRNAs) and long ncRNAs (lncRNAs) with a cut-off length of 200 bp; they are directly or indirectly involved in diverse biological processes through epigenetic, transcriptional, and post-transcriptional mechanisms (17). MicroRNAs (miRNAs) are a type of short ncRNAs that regulate cellular proliferation, differentiation, apoptosis, adhesion, epithelial-mesenchymal transition (EMT), and metastasis in various cancers (18). The competing endogenous RNA (ceRNA) theory (19) states that miRNAs recognise miRNA response elements (MREs) in different RNA molecules, and induce target repression through miRNA-RNA-induced silencing complex (RISC)-mediated degradation. Therefore, the transcriptional regulation of diverse RNAs, including mRNAs and ncRNAs, can regulate biological processes through a novel MRE-mediated mechanism (19).
The tumour-suppressive effects of lidocaine and bupivacaine were studied in MCF-7 cells. The effects of these LAs on the function of miR-187-5p, Differentiation Antagonising Non-Protein Coding RNA (DANCR), and MYB were investigated for elucidating the underlying mechanism. Our findings provide novel insights into the process of tumour suppression for future breast cancer therapy.
Methods
Cell Culture
MCF-7 cells were purchased from Bioresource Collection and Research Center (Hsinchu, Taiwan), and HEK-293T cells were provided by Dr. Shau-Ping Lin (Institute of Biotechnology, National Taiwan University, Taiwan). All cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, Thermo Fisher, CA, USA) containing 1% penicillin-streptomycin (Gibco) and 10% foetal bovine serum (FBS) (HyClone, Logan, UT, USA). The culture plates were maintained at 37°C in a humidified atmosphere of 5% CO2 and were routinely tested for Mycoplasma sp. using Mycoplasma polymerase chain reaction (PCR) Detection Kit (ABM Inc., Vancouver, BC, Canada).
Cell Viability Assay
MCF-7 cells were seeded at 5 ×103 cell into 96-well plates 1 day before lidocaine or bupivacaine (Sigma, St. Louis, MO, USA) treatment at the indicated concentrations. LAs and the controls were diluted in 200 μL DMEM for each well. For the thiazolyl blue tetrazolium bromide (MTT) colorimetric assay, 5 mg/mL of MTT (Sigma) were added to each well and incubated at 37°C for 4 h. The supernatants were removed and replaced with dimethyl sulfoxide (DMSO) (Sigma). The absorbance was measured using a microtiter plate reader (BioTek, Winooski, VT, USA) at 570 nm.
Wound Healing Assays
MCF-7 cells were seeded at a density of 2.5 ×104 cells/well in a medium containing 10% FBS and incubated overnight. The culture inserts were removed and an image of the gap area at 0 h was captured. The cells were further incubated at 37°C in a 5% CO2 incubator, and the images were captured at 12, 24, and 36 h, respectively. The cell migration ability in the gap area was quantified using ImageJ v1.8.0 (National Institutes of Health, USA).
Cell Migration
Migration assays were performed using SPLInsertTM Hanging plate (SPL Life Sciences, Pocheon, South Korea). The upper chamber of the transwell unit was seeded at 4 ×104 cells/well in 0.2 mL serum-free DMEM. The lower chambers were loaded with 0.6 mL DMEM containing 10% FBS as chemo-attractant. The MCF-7 cells were incubated at 37°C for 36 h. A methanol: acetic acid (3:1) mixture was added to the lower chamber for fixing the cells by incubating at 25°C for 10 min. The non-migrating cells on the inner transwell membrane were removed carefully, and the membranes were stained with 1 mL of 0.5% crystal violet. The stained cells were solubilized with 10% acetic acid, imaged, and analysed using ImageJ v1.8.0.
Western Blotting
The proteins were extracted from the cells lysed with radioimmunoprecipitation assay buffer (RIPA) Lysis Buffer (EMD Millipore, Billerica, MA, USA) and the concentration was determined using Coomassie Protein Assay Reagent (Thermo Fisher, Waltham, MA, USA). The extracted protein samples (20 μg) were separated using 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (GE healthcare, Chicago, IL, USA). The membranes were incubated with primary rabbit polyclonal antibodies against c-MYB (Proteintech, Rosemont, IL, USA) and β-actin (GeneTex, Irvine, CA, USA), at 4°C overnight; the samples were hybridised with horseradish peroxidase (HRP) conjugated anti-rabbit IgG (GeneTex) at 25°C for 1 h. The blotted proteins were detected using an enhanced chemiluminescence (ECL) system (Millipore, Billerica, MA, USA) equipped with a BioSpectrum Imaging System (UVP, Upland, CA, USA). The images were analysed using ImageJ v1.8.0.
RNA Extraction, Reverse Transcription, and Quantitative Real Time Polymerase Chain Reaction
The RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA, USA) and reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). The RNA quality was detected by a spectrophotometer (NanoDrop; Thermo Scientific) with the A260/A280 ratio between 1.8~2.0. Samples were kept in RNase free water buffer at neutral pH. All RNA samples were stored at −80°C. One μg of total RNA was reverse-transcribed by High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). SuperScript IV Reverse Transcriptase (Invitrogen) was used for reverse transcription of the miRNAs in accordance with the manufacturer's instructions. Two point five percent of each reaction was used as template for quantitative PCR with OmicsGreen qPCR MasterMix (OmicsBio, New Taipei City, Taiwan) and the reactions were performed on StepOnePlus Real-Time PCR System (Thermo Fisher, Waltham, MA, USA). The primers were checked for only generating a single melting curve peak and detailed information are provided in Supplementary Table 1. The relative expression was normalised to that of 18S rRNA or U6 snRNA, using the 2−ΔΔCt method.
Library Preparation and Sequencing
The total RNA was extracted using TRIZOL reagent (Invitrogen). The small RNA library was constructed from total RNA (2 μg) using the TruSeq Small RNA Library Prep Kit (Illumina Inc., San Diego, CA, USA), according to the manufacturer's instructions. A total of 75 single-read nucleotides were obtained from each miRNA library using NextSeq500 (Illumina Inc., CIC bioGUNE, Bilbao, Spain). The sequencing data were submitted to the Gene Expression Omnibus (GEO) (accession number: GSE171282).
Sequencing Data Analyses
The raw Illumina FASTQ sequencing reads were mapped to the miRBase mature miRNA reference (http://www.mirbase.org/) using Partek Flow™ v5.0 (Partek Inc., St. Louis, MO, USA) with Bowtie alignment algorithm. The Bowtie alignment method was generally thought to be optimal for sequences less than 50 bp. The adapter sequences were first removed from each FASTQ reads, and the remaining bases were trimmed from the 3′ end with a minimum Phred quality score of 20. A minimum read-length philtre of 15-bases in length was used in this work. A minimum seed length of 10 that was consistent with the standard setting provided by the Partek Flow pipeline was chosen for the Bowtie aligner. The remaining reads were aligned to the human genome reference (RefSeq Hg19). Raw miRNA expression reads were normalised by scale normalisation and then processed by log2 transformation. Then, miRNAs with extremely low expression (log2 <5 in all samples) were removed from further analysis. Then, differentially expressed miRNA profiles between groups were identified by log2 fold-change value ≧2X or < -1X and P-value <0.05 using a two-sided Student's t-test. Principal component analysis using the expression values of total miRNAs was used to visualise the similarity of different groups.
Transfection
MCF-7 cells were cultured in an antibiotic-free medium to 70–80% confluence in 6-well plates (2.5 ×105 cells/well) and transfected with the 0.025 nmol of miR-187-5p mimic (GenePharma, Shanghai, China) or negative control (GenePharma), using Lipofectamine 2000 as the transfection reagent (Invitrogen), following the manufacturer's instructions.
The MCF-7 cells were transfected with 0.5 nmol of pcDNA-DANCR or the empty vector (Dharmacon, New Taipei city, Taiwan) using jetPRIME (Polyplus-transfection, New York, NY, USA) reagent, following the manufacturer's instructions. After 4 h, the medium was replaced with fresh medium containing serum. The cellular RNA expression after 48 h was determined using qRT-PCR.
Plasmid Construct and Site-Directed Mutagenesis
To determine promoter activity by luciferase assay, the luciferase expression plasmid pMIR-REPORT-c-MYB 3′UTR and pMIR-REPORT-DANCR was purchased from the BioMed Resource Core of the 1st Core Facility Lab, National Taiwan University College of Medicine (Taipei, Taiwan). Briefly, c-MYB 3′UTR region encompassing 2,493–3,684 bp relative to the transcription start site was amplified from the human genomic DNA by PCR. The c-MYB 3′UTR was subcloned into pMIR-REPORT vector, and the final construct was called pMIR-REPORT-c-MYB 3′UTR. DANCR encompassing 1–915 bp relative to the transcription start site was amplified from the human genomic DNA by PCR and the final construct was called pMIR-REPORT-DANCR.
miR-187-5p binding sites of c-MYB 3′UTR were located at 2,710–2,734 and 3,634–3,656 bp. Besides, miR-187-5p binding sites of DANCR were located at 134–157 and 380–408 bp. To determine binding activity of miR-187-5p by luciferase assay, the luciferase expression plasmid pMIR-REPORT-c-MYB 3′UTR S1, pMIR-REPORT-c-MYB 3′UTR S2, pMIR-REPORT-c-MYB 3′UTR S12, pMIR-REPORT-DANCR S1, pMIR-REPORT-DANCR S2, and pMIR-REPORT-DANCR S12 was purchased from the BioMed Resource Core of the 1st Core Facility Lab, NTU-CM (Taipei, Taiwan). In addition, the mutated sequences were validated by sequencing.
Luciferase Reporter Assay
Luciferase assays were performed with HEK-293T cells using the Dual-Glo® luciferase reporter assay system (Promega, Fitchburg, WI, USA). Cells (5 ×104/well) were transfected with 100 ng reporter constructs containing the wild-type or mutant-type of MYB, the wild-type or mutant-type of DANCR and 2 ng Ranilla luciferase plasmid (BioMed Resource Core of the 1st Core Facility Lab, National Taiwan University College of Medicine, Taiwan), using the jetPRIME (Polyplus-transfection) reagent. The cells were additionally transfected with 0.025 nmol of miR-187-5p (GenePharma). After 24 h, the cells were lysed and the Renilla luciferase signals were used for normalisation, according to manufacturer's protocol.
Statistical Analyses
The experimental data are presented as the mean ± standard deviation (SD) of at least three independent experiments. The genes with statistically significant expression patterns in the different samples were determined by a two-tailed Student's t-test using GraphPad Prism v5 (GraphPad Software, Inc., La Jolla, CA, USA). p-values < 0.05 were considered statistically significant.
Results
LAs Significantly Suppressed Cell Viability and Migration
To determine the effects of lidocaine and bupivacaine on MCF-7 cells, the cell viability following treatment with different concentrations of lidocaine or bupivacaine for 6 h was determined. Lidocaine and bupivacaine inhibited the growth of MCF-7 cells in a dose-dependent manner (Figure 1A). The median lethal dose (LD50) of lidocaine (4 mM) and bupivacaine (1 mM) were used as the concentrations for the following experiments, respectively. Wound healing and transwell invasion assays were performed using MCF-7 cells treated with lidocaine or bupivacaine for 12, 24, and 36 h. Both the LAs significantly reduced the migration and invasive ability of MCF-7 cells (Figures 1B,C), suggesting suppression of malignancy in the MCF-7 cells.
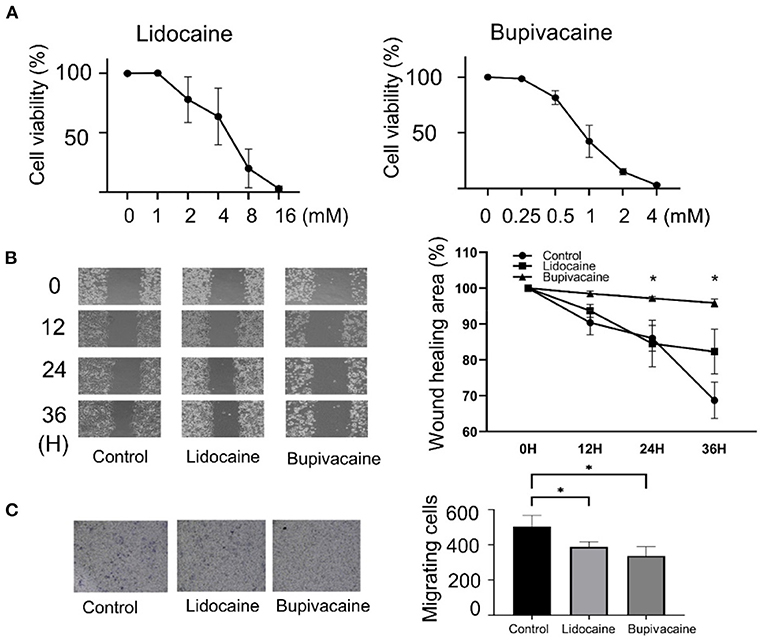
Figure 1. Lidocaine and bupivacaine inhibited the proliferation and migration of MCF-7 cells. (A) Cell proliferation determined using the MTT assay. The growth of MCF-7 cells measured after 6 h of treatment with serially diluted concentrations of lidocaine or bupivacaine. The proliferation rate was normalised to that at 0 h. (B) Wound healing assay. Left: Images after 0, 12, 24, and 36 h of treatment with lidocaine (4 mM) or bupivacaine (1 mM). Right: The migratory ability quantified using the reduction in wound size over time by ImageJ software v1.8.0. (C) Transwell migration assay. Right: The cells were seeded after 24 h of treatment with lidocaine (4 mM) or bupivacaine (1 mM). Left: Cell migration measured after 36 h of seeding. All data are presented as mean ± SD (n = 3). *p < 0.05.
Differential miRNA Expression Profiles Following LA Treatment
To identify the miRNAs that mediated the effects of the LAs on MCF-7 cells, next-generation sequencing was used for identifying the differentially expressed miRNAs, following lidocaine or bupivacaine treatment. Principal component analysis (PCA) was performed for the samples (lidocaine, bupivacaine, and control groups, nine dots in total) using the data for 2,522 differentially expressed miRNAs in the three groups (Figure 2A). The miRNAs in the same group showed similar expression profiles, while the expression profiles in the lidocaine- and bupivacaine-treated groups were distinct from those of the control.
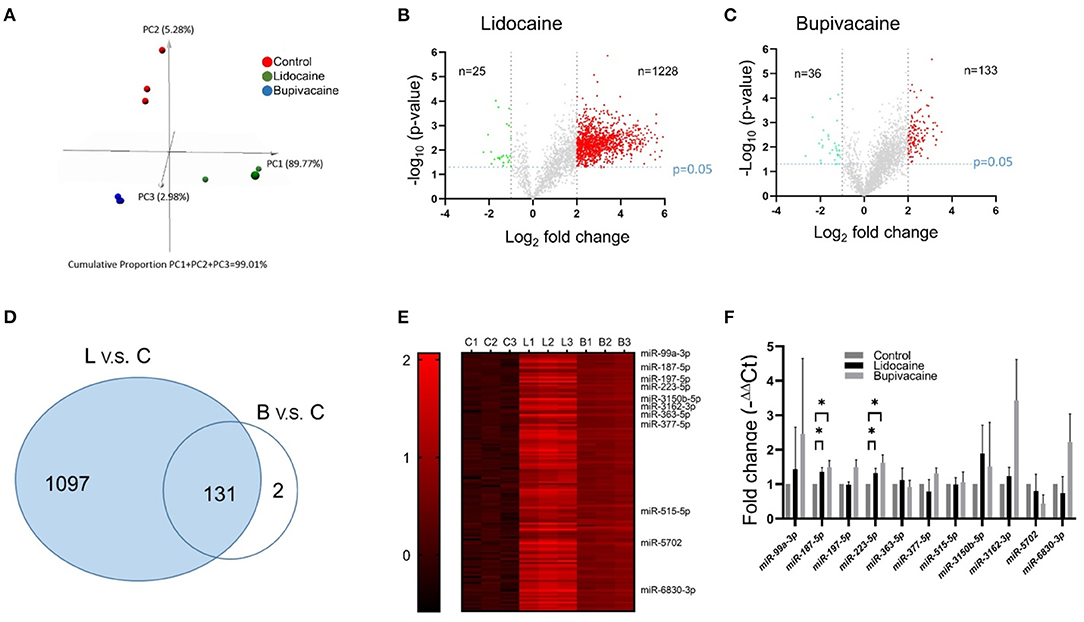
Figure 2. Identification of miRNAs in MCF-7 cells that responded to treatment with lidocaine or bupivacaine. (A) PCA of differentially expressed miRNAs in response to treatment with lidocaine (4 mM) or bupivacaine (1 mM). (B,C) Volcano plot of the differentially expressed miRNAs in response to treatment with lidocaine (B) or bupivacaine (C) determined by next-generation sequencing. (D) Venn diagram of the differentially expressed miRNAs. The expression of 131 miRNAs was upregulated following treatment with lidocaine or bupivacaine. (E) Heat map of the upregulated miRNAs (n = 131) that responded to treatment with both lidocaine and bupivacaine. (F) PCR validation of the selective differentially expressed miRNAs in MCF-7 cells, following treatment with lidocaine and bupivacaine. The expression levels measured using qRT-PCR and normalised to that of U6 snRNA. L, lidocaine; B, bupivacaine; C, control. All data are presented as mean ± SD (n = 3). *p < 0.05.
The miRNAs that were differentially expressed in the lidocaine- and bupivacaine-treated groups were filtered using the criteria: fold change ≥ 2.0X and p-value ≤ 0.05. The expression of 1,228 and 133 miRNAs was significantly upregulated in the lidocaine (Figure 2B) and bupivacaine groups (Figure 2C), respectively (red dots). The expression of 25 and 36 miRNAs was significantly downregulated (fold change < -1X and p-value ≤ 0.05) in the lidocaine (Figure 2B) and bupivacaine groups (Figure 2C), respectively (green dots). The genes that were not differentially expressed are depicted in grey. Among the upregulated miRNAs, 131 were common to both the lidocaine and bupivacaine groups (Figure 2D). The number of downregulated miRNAs common to both the groups was low; therefore, only the upregulated miRNAs were selected for subsequent studies. The expression of the selected miRNAs is depicted using a heatmap (Figure 2E).
The data from next-generation sequencing was validated by selecting the miRNAs whose expression was significantly higher and for which experimentally validated data were available. The expression of only miR-187-5p and miR-223-5p was significantly (p < 0.05) higher than that of the control. The expression of the other miRNAs examined using qRT-PCR was not statistically different between the lidocaine and bupivacaine groups (Figure 2F).
LAs Downregulated MYB Expression via miR-187-5p
To identify the genes regulated by miR-187-5p and ascertain their functional relevance, we used the TargetScan tool (http://www.targetscan.org/vert_72/) for identifying the downstream target genes. The MYB transcription factor, a proto-oncogene, was a predicted target gene of miR-187-5p. In vitro and in vivo studies indicated that MYB plays an important role in different cancers. MYB overexpression and dysregulation is observed in almost all breast tumours, and is associated with poor prognosis (20, 21). MYB also regulates cell proliferation, differentiation, and angiogenesis (22), and therefore, was selected for further studies.
qRT-PCR and western blotting were used to investigate the mechanism underlying the influence of miR-187-5p on cellular function. The mRNA and protein levels of MYB were significantly downregulated in the LA-treated groups (Figures 3A,B). The levels of MYB mRNA were significantly reduced when miR-187-5p was overexpressed (Figure 3C). Using bioinformatics analysis, we validated MYB as a target gene of miR-187-5p; two binding sites (2,710–2,732 nt and 3,634–3,656 nt) were predicted in the 3′-UTR of MYB, which were complementary to the seed sequence of miR-187-5p (Figure 3D). The direct interactions between miR-187-5p and MYB were examined using luciferase reporter plasmids containing the wild-type MYB (MYB-wt) and binding site mutants (MYB-site1-mut, MYB-site2-mut, and MYB-site1+2-mut). The luciferase activity of MYB-wt decreased significantly, with the transfection of miR-187-5p mimics; however, they had no influence on MYB-site1, MYB-site2, and MYB-site1+2 (Figure 3E), indicating that MYB is a direct target gene of miR-187-5p. The LAs, lidocaine and bupivacaine, upregulated the expression of miR-187-5p, directly downregulating the expression of MYB.
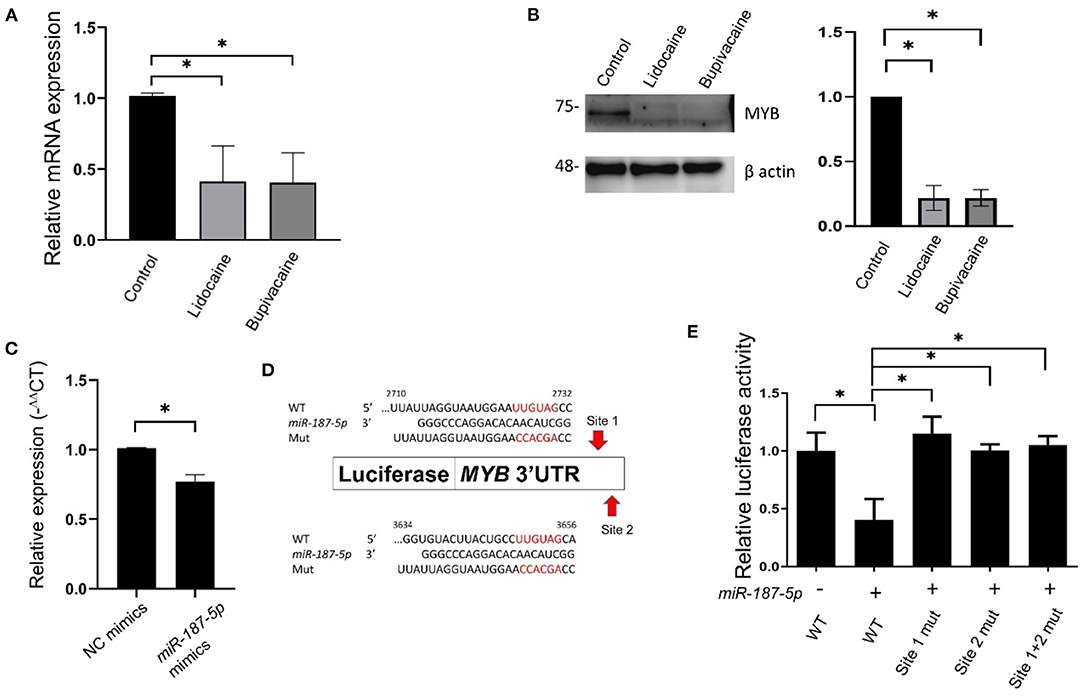
Figure 3. Downregulation of MYB in MCF-7 cells following lidocaine and bupivacaine treatment. (A) The relative mRNA levels of MYB in MCF-7 cells following treatment with lidocaine (4 mM) or bupivacaine (1 mM). The expression levels of MYB were measured using qRT-PCR and normalised to that of 18S rRNA. (B) Western blots of MYB in MCF-7 cells following treatment with the LAs. Right: graphical representation of the results of western blotting. Left: quantification of the western blots. β actin was used as the loading control. (C) Relative expression levels of MYB in MCF-7 cells overexpressing miR-187-5p. The cells were transfected with 2 μg of miR-187-5p or control mimics. The expression levels were detected using qRT-PCR and normalised to that of 18S rRNA. (D) Schematic diagram of the putative binding site of miR-187-5p in the 3′-UTR region (2,493 ~ 3,684 bp) of MYB. (E) Luciferase reporter assays of MYB 3′-UTR in HEK-293T cells overexpressing miR-187-5p. The HEK-293T cells were transfected with miR-187-5p mimics, firefly luciferase plasmids, and Renilla luciferase vectors. The relative activity of firefly luciferase was measured and normalised to that of Renilla luciferase. The data are presented as mean ± SD (n = 3). *p < 0.05.
DANCR Decoys miR-187-5p by Reciprocal Suppression
LncRNAs act as ceRNAs and influence mRNA levels by sequestering miRNAs that target both lncRNA and mRNAs. To identify the ceRNAs of miR-187-5p, we used DIANA-LncBase v2 (23) for predicting the lncRNAs that bind to miR-187-5p. Five possible lncRNAs were predicted and validated using qRT-PCR (Figure 4A). The expression of DANCR was downregulated by the LAs, while that of the HIF1A-AS2 and SNHG1 lncRNAs was upregulated; however, these lncRNAs did not function as ceRNAs of miR-187-5p. In addition, the expression of DANCR significantly decreased when the expression of miR-187-5p increased (Figure 4B).
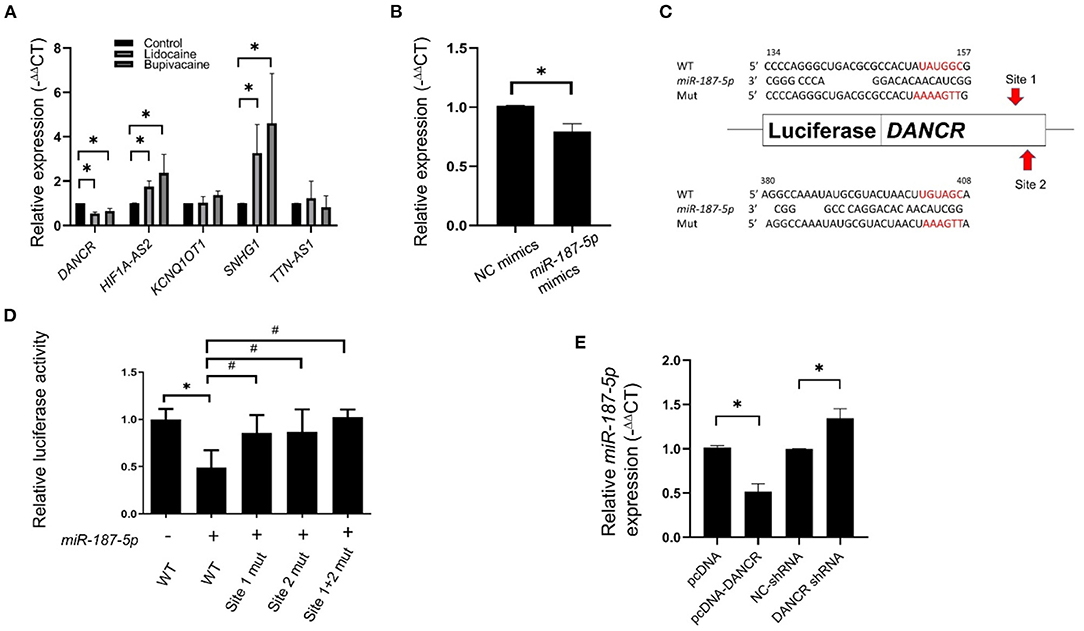
Figure 4. Expression of DANCR lncRNA was downregulated in MCF-7 cells following treatment with lidocaine or bupivacaine. (A) The relative expression levels of selective lncRNAs in MCF-7 cells following treatment with lidocaine (4 mM) or bupivacaine (1 mM). The expression levels were measured using qRT-PCR and normalised to that of 18S rRNA. (B) Relative expression levels of DANCR in MCF-7 cells overexpressing miR-187-5p. The cells were transfected with 2 μg of miR-187-5p or control mimics. The expression levels were detected using qRT-PCR and normalised to that of 18S rRNA. (C) Schematic diagram of the putative binding site of miR-187-5p in the binding region of DANCR. (D) Luciferase reporter assays of DANCR in HEK-293T cells overexpressing miR-187-5p. The HEK-293T cells were transfected with miR-187-5p mimics, firefly luciferase plasmids, and Renilla luciferase vectors. The relative activity of firefly luciferase was measured and normalised to that of Renilla luciferase. (E) The relative expression levels of miR-187-5p in MCF-7 cells overexpressing DANCR or shRNAs against DANCR. The expression levels were detected using qRT-PCR and normalised to that of U6 snRNA. The data are presented as mean ± SD (n = 3). *p < 0.05.
Luciferase reporter assays were performed for validating the association between DANCR and miR-187-5p. Prediction revealed two potential binding sites of miR-187-5p at 134–157 nt and 380–408 nt of DANCR, which were individually positioned upstream of the luciferase reporter genes (Figure 4C). The binding site mutants (sites 1-mut, 2-mut, and 1+2-mut) were constructed and transfected into HEK-293T cells. The luciferase activity of DANCR-wt was significantly reduced by the miR-187-5p mimics; however, they had no effect on sites 1-mut, 2-mut, and 1+2-mut (Figure 4D), indicating that the DANCR lncRNA sponges miR-187-5p by direct binding.
The overexpression of DANCR suppressed miR-187-5p expression, while the downregulation of DANCR increased miR-187-5p expression (Figure 4E). These results demonstrated a reciprocal inhibitory relationship between DANCR and miR-187-5p, and that miR-187-5p could mediate the inhibitory effect of the LAs on MCF-7 cell viability and migration.
Discussion
The LAs, lidocaine and bupivacaine, inhibited MCF-7 cell proliferation and migration. Genomic studies and validation experiments revealed that the LAs promoted miR-187-5p expression, which downregulated MYB expression. Additionally, the LAs reduced the expression of DANCR lncRNA, which could be a ceRNA of miR-187-5p. Our study demonstrated a novel mechanism underlying the inhibitory effect of LAs on cancer cell proliferation, migration, and invasion, mediated via the DANCR-miR-187-5p-MYB regulatory axis.
LAs inhibit cancer cell proliferation, migration, and invasion by regulating miRNA expression (12, 13). Lidocaine inhibits proliferation and induces apoptosis in retinoblastoma cells by modulating the miR-520a-3p/EGFR axis (24). Bupivacaine inhibits gastric cancer progression by regulating the circ_0000376/miR-145-5p axis (25). Using NGS and qRT-PCR, we demonstrated that lidocaine and bupivacaine upregulated miR-187-5p expression. MiR-187-5p regulates cellular proliferation and apoptosis in lung cancer (26), hepatocellular carcinoma (27), bladder cancer (28), and acute lymphoblastic leukaemia (29), and is a possible indicator of drug sensitivity in breast cancer (30). This study is the first to identify that miR-187-5p regulates the inhibitory effect of LAs on the proliferation, migration, and invasion of MCF-7 cells.
Bioinformatics-based prediction revealed that MYB is a target gene of miR-187-5p, consistent with the reports that MYB functions as an oncogene in different tumours, including breast cancer (21, 31–33). MYB could be inhibited in MCF-7 cells through the upregulation of miR-187-5p by lidocaine or bupivacaine. These LAs reduced the mRNA and protein levels of MYB, and the miR-187-5p mimics significantly reduced MYB expression in MCF-7 cells. The luciferase reporter assay confirmed that miR-187-5p directly targets the 3′-UTR of MYB.
LncRNAs interact with various molecules and have key roles in regulating signalling processes at the epigenetic, transcriptional, and post-transcriptional levels (34, 35). A novel regulatory mechanism elucidates that all types of transcripts, including mRNAs, lncRNAs, and circular RNAs, can act as ceRNAs, forming a complex RNA interactome involving different RNA species, for regulating gene expression (35). The lncRNA-miRNA-mRNA crosstalk could have a prominent role in the anti-tumour effects of LAs. Among the lncRNAs, DANCR, encoded by a gene located in human chromosome 4q12, plays an important role in different cancers (36). The DANCR-SOCS3-EZH2 axis regulates the inflammatory phenotype and breast cancer cell metastasis (37). DANCR promotes progressive osteosarcoma by functioning as a ceRNA and sponging miR-335-5p and miR-1972, regulating ROCK1 expression (38). In this study, the expression of DANCR was inhibited by the LAs, and a reciprocal suppressive effect was observed between DANCR and miR-187-5p. The luciferase reporter assay confirmed that DANCR was the target of miR-187-5p. Therefore, the upregulation of miR-187-5p and downregulation of DANCR could mediate the inhibitory effect of LAs on the proliferation, migration, and invasion of MCF-7 cells. Together, we concluded that the DANCR-miR-187-5p-MYB axis is activated by lidocaine and bupivacaine; the proposed schematic diagram is presented in Figure 5.
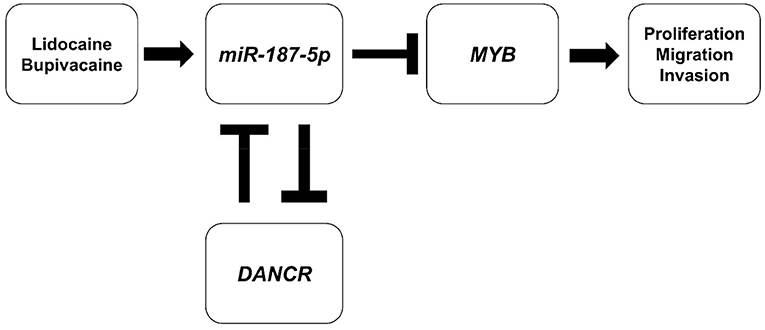
Figure 5. Schematic diagram of the proposed mechanism underlying the inhibitory effect of LAs. Lidocaine and bupivacaine suppress the malignancy of MCF-7 breast cancer cells by upregulating miR-187-5p, which in turn downregulates the expression of MYB (MYB proto-oncogene, transcription factor) proteins and the DANCR (Differentiation Antagonising Non-Protein Coding RNA) lncRNA.
This study has certain limitations. First, LAs are chemically categorised into amide-ester and amide-amide groups, and the new generation LAs, including ropivacaine and levobupivacaine, are used clinically. Only the older generation LAs, lidocaine and bupivacaine, were considered. LAs have variable anti-tumour effects and mechanisms of action in different cancers (39). Further studies are necessary for confirming whether the DANCR-miR-187-5p-MYB axis mediates the inhibitory effects of other LAs. Second, there are several breast cancer types; and therefore, further studies are necessary for confirming the effects of lidocaine and bupivacaine in other breast cancer cell lines and in an animal model, which may improve upon the present findings. Third, although luciferase studies proved the interaction between DANCR-miR-187-5p-MYB axis, whether the anti-tumour properties of LAs can be reversed requires more experiments to prove, such as using antagomir to inhibit miR-187-5p or overexpressing MYB in MCF-7 cells treated with LAs.
This study demonstrated that lidocaine and bupivacaine inhibited the growth and metastasis of breast cancer cells. The anti-tumour properties of these LAs were partially attributed to the upregulation of miR-187-5p, which inhibited MYB signalling by directly binding to MYB. Screening for the possible targets of miR-187-5p using a dual-luciferase reporter assay revealed that the DANCR lncRNA directly targets miR-187-5p, suggesting that DANCR could act as a miR-187-5p sponge. In summary, we report that the DANCR-miR-187-5p-MYB axis may be activated by lidocaine and bupivacaine.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov/geo/, GSE171282.
Author Contributions
C-YL and L-CL: conception and design of experiments and manuscript preparation. C-YL and W-TT: performed experiments. C-YL and Y-YC: data analyses. L-CL, M-HT, and EC: contributed reagents, materials, and analytical tools. All the authors reviewed and approved the final version of the manuscript.
Funding
This work was supported by a grant from the Ministry of Science and Technology [MOST 109-2320-B-002-016-MY3]. These funding sources had no role in the design of this study and will not have any role during its execution, analyses, interpretation of the data, or decision to submit results.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the technical assistance provided by the Biomedical Resource Core at the 1st Core Facility Lab, NTU College of Medicine.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.732817/full#supplementary-material
Abbreviations
LA, local anaesthetic; ncRNA, non-coding RNA; miRNA, microRNA; PCR, polymerase chain reaction; sncRNA, small ncRNA; lncRNA, long ncRNA; EMT, epithelial-mesenchymal transition; ceRNA, competing endogenous RNA; MRE, miRNA response element; RISC, RNA-induced silencing complex; MYB, MYB proto-oncogene, transcription factor; DANCR, Differentiation Antagonising Non-Protein Coding RNA; qRT-PCR, quantitative real time polymerase chain reaction; PCA, principal component analysis.
References
1. Sessler DI, Pei L, Huang Y, Fleischmann E, Marhofer P, Kurz A, et al. Recurrence of breast cancer after regional or general anaesthesia: a randomised controlled trial. Lancet. (2019) 394:1807–15. doi: 10.1016/S0140-6736(19)32313-X
2. Wall T, Sherwin A, Ma D, Buggy DJ. Influence of perioperative anaesthetic and analgesic interventions on oncological outcomes: a narrative review. Br J Anaesth. (2019) 123:135–50. doi: 10.1016/j.bja.2019.04.062
3. Connolly C, Buggy DJ. Opioids and tumour metastasis: does the choice of the anesthetic-analgesic technique influence outcome after cancer surgery? Curr Opin Anaesthesiol. (2016) 29:468–74. doi: 10.1097/ACO.0000000000000360
4. Dockrell L, Buggy DJ. The role of regional anaesthesia in the emerging subspecialty of onco-anaesthesia: a state-of-the-art review. Anaesthesia. (2021) 76(Suppl. 1):148–59. doi: 10.1111/anae.15243
5. Li R, Xiao C, Liu H, Huang Y, Dilger JP, Lin J. Effects of local anesthetics on breast cancer cell viability and migration. BMC Cancer. (2018) 18:666. doi: 10.1186/s12885-018-4576-2
6. Xuan W, Zhao H, Hankin J, Chen L, Yao S, Ma D. Local anesthetic bupivacaine induced ovarian and prostate cancer apoptotic cell death and underlying mechanisms in vitro. Sci Rep. (2016) 6:26277. doi: 10.1038/srep26277
7. FitzGerald S, Odor PM, Barron A, Pawa A. Breast surgery and regional anaesthesia. Best Pract Res Clin Anaesthesiol. (2019) 33:95–110. doi: 10.1016/j.bpa.2019.03.003
8. D'Agostino G, Saporito A, Cecchinato V, Silvestri Y, Borgeat A, Anselmi L„ et al. Lidocaine inhibits cytoskeletal remodelling and human breast cancer cell migration. Br J Anaesth. (2018) 121:962–8. doi: 10.1016/j.bja.2018.07.015
9. Lucchinetti E, Awad AE, Rahman M, Feng J, Lou PH, Zhang L, et al. Antiproliferative effects of local anesthetics on mesenchymal stem cells: potential implications for tumor spreading and wound healing. Anesthesiology. (2012) 116:841–56. doi: 10.1097/ALN.0b013e31824babfe
10. Chamaraux-Tran TN, Piegeler T. The amide local anesthetic lidocaine in cancer surgery-potential antimetastatic effects and preservation of immune cell function? A narrative review. Front Med. (2017) 4:235. doi: 10.3389/fmed.2017.00235
11. Lirk P, Hollmann MW, Fleischer M, Weber NC, Fiegl H. Lidocaine and ropivacaine, but not bupivacaine, demethylate deoxyribonucleic acid in breast cancer cells in vitro. Br J Anaesth. (2014) 113(Suppl. 1):i32–8. doi: 10.1093/bja/aeu201
12. Sui H, Lou A, Li Z, Yang J. Lidocaine inhibits growth, migration and invasion of gastric carcinoma cells by up-regulation of miR-145. BMC Cancer. (2019) 19:233. doi: 10.1186/s12885-019-5431-9
13. Qu X, Yang L, Shi Q, Wang X, Wang D, Wu G. Lidocaine inhibits proliferation and induces apoptosis in colorectal cancer cells by upregulating mir-520a-3p and targeting EGFR. Pathol Res Pract. (2018) 214:1974–9. doi: 10.1016/j.prp.2018.09.012
14. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
15. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. (2020) 20:303–22. doi: 10.1038/s41568-020-0253-2
16. Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. (2016) 17:719–32. doi: 10.1038/nrg.2016.134
17. Han Li C, Chen Y. Small and long non-coding RNAs: Novel targets in perspective cancer therapy. Curr Genomics. (2015) 16:319–26. doi: 10.2174/1389202916666150707155851
18. Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci. (2015) 22:9. doi: 10.1186/s12929-015-0113-7
19. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
20. Li Y, Jin K, van Pelt GW, van Dam H, Yu X, Mesker WE, et al. c-Myb enhances breast cancer invasion and metastasis through the Wnt/beta-catenin/axin2 pathway. Cancer Res. (2016) 76:3364–75. doi: 10.1158/0008-5472.CAN-15-2302
21. Mitra P. Transcription regulation of MYB: a potential and novel therapeutic target in cancer. Ann Transl Med. (2018) 6:443. doi: 10.21037/atm.2018.09.62
22. Liu X, Xu Y, Han L, Yi Y. Reassessing the potential of Myb-targeted anti-cancer therapy. J Cancer. (2018) 9:1259–66. doi: 10.7150/jca.23992
23. Paraskevopoulou MD, Vlachos IS, Karagkouni D, Georgakilas G, Kanellos I, Vergoulis T, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. (2016) 44:D231–8. doi: 10.1093/nar/gkv1270
24. Xia W, Wang L, Yu D, Mu X, Zhou X. Lidocaine inhibits the progression of retinoblastoma in vitro and in vivo by modulating the miR-520a-3p/EGFR axis. Mol Med Rep. (2019) 20:1333–42. doi: 10.3892/mmr.2019.10363
25. Ju C, Zhou J, Miao H, Chen X, Zhang Q. Bupivacaine suppresses the progression of gastric cancer through regulating circ_0000376/miR-145-5p axis. BMC Anesthesiol. (2020) 20:275. doi: 10.1186/s12871-020-01179-4
26. Mao M, Wu Z, Chen J. MicroRNA-187-5p suppresses cancer cell progression in non-small cell lung cancer (NSCLC) through down-regulation of CYP1B1. Biochem Biophys Res Commun. (2016) 478:649–55. doi: 10.1016/j.bbrc.2016.08.001
27. Gong J, Mou T, Wu H, Wu Z. Brg1 regulates murine liver regeneration by targeting miR-187-5p dependent on Hippo signalling pathway. J Cell Mol Med. (2020) 24:11592–602. doi: 10.1111/jcmm.15776
28. Li Z, Lin C, Zhao L, Zhou L, Pan X, Quan J, et al. Oncogene miR-187-5p is associated with cellular proliferation, migration, invasion, apoptosis and an increased risk of recurrence in bladder cancer. Biomed Pharmacother. (2018) 105:461–9. doi: 10.1016/j.biopha.2018.05.122
29. Lou Y, Liu L, Zhan L, Wang X, Fan H. miR-187-5p regulates cell growth and apoptosis in acute lymphoblastic leukemia via DKK2. Oncol Res. (2016) 24:89–97. doi: 10.3727/096504016X14597766487753
30. Uhr K, Prager-van der Smissen WJC, Heine AAJ, Ozturk B, van Jaarsveld MTM, Boersma AWM, et al. MicroRNAs as possible indicators of drug sensitivity in breast cancer cell lines. PLoS ONE. (2019) 14:e0216400. doi: 10.1371/journal.pone.0216400
31. Jia H, Wu D, Zhang Z, Li S. Regulatory effect of the MAFG-AS1/miR-150-5p/MYB axis on the proliferation and migration of breast cancer cells. Int J Oncol. (2021) 58:33–44. doi: 10.3892/ijo.2020.5150
32. Miao RY, Drabsch Y, Cross RS, Cheasley D, Carpinteri S, Pereira L, et al. MYB is essential for mammary tumorigenesis. Cancer Res. (2011) 71:7029–37. doi: 10.1158/0008-5472.CAN-11-1015
33. Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. (2008) 8:523–34. doi: 10.1038/nrc2439
34. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. (2017) 1008:47–74. doi: 10.1007/978-981-10-5203-3_2
35. Volovat SR, Volovat C, Hordila I, Hordila DA, Mirestean CC, Miron OT, et al. MiRNA and lncRNA as potential biomarkers in triple-negative breast cancer: a review. Front Oncol. (2020) 10:526850. doi: 10.3389/fonc.2020.526850
36. Yan Y, Shi Q, Yuan X, Xue C, Shen S, He Y, et al. An emerging therapeutic target for cancer. Am J Transl Res. (2020) 12:4031–42.
37. Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. (2020) 14:309–28. doi: 10.1002/1878-0261.12622
38. Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. (2018) 17:89. doi: 10.1186/s12943-018-0837-6
Keywords: miR-187-5p, MYB, DANCR, bupivacaine, lidocaine
Citation: Lin C-Y, Tseng W-T, Chang Y-Y, Tsai M-H, Chuang EY, Lu T-P and Lai L-C (2022) Lidocaine and Bupivacaine Downregulate MYB and DANCR lncRNA by Upregulating miR-187-5p in MCF-7 Cells. Front. Med. 8:732817. doi: 10.3389/fmed.2021.732817
Received: 29 June 2021; Accepted: 20 December 2021;
Published: 13 January 2022.
Edited by:
Ata Murat Kaynar, University of Pittsburgh, United StatesReviewed by:
Johanna Catharina Duvigneau, University of Veterinary Medicine Vienna, AustriaBetina Orman, Universidad de Buenos Aires, Argentina
Copyright © 2022 Lin, Tseng, Chang, Tsai, Chuang, Lu and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang-Chuan Lai, bGxhaUBudHUuZWR1LnR3
 Chiao-Yi Lin
Chiao-Yi Lin Wen-Ting Tseng
Wen-Ting Tseng Yao-Yin Chang3
Yao-Yin Chang3 Eric Y. Chuang
Eric Y. Chuang Tzu-Pin Lu
Tzu-Pin Lu Liang-Chuan Lai
Liang-Chuan Lai