
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 30 July 2021
Sec. Precision Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.699672
This article is part of the Research Topic Computational Genomics and Structural Bioinformatics in Personalized Medicines View all 18 articles
 Nourah H. Al Qahtani1
Nourah H. Al Qahtani1 Sayed AbdulAzeez2
Sayed AbdulAzeez2 Noor B. Almandil3
Noor B. Almandil3 Norah Fahad Alhur2
Norah Fahad Alhur2 Hind Saleh Alsuwat2
Hind Saleh Alsuwat2 Hatoon Ahmed Al Taifi1
Hatoon Ahmed Al Taifi1 Ahlam A. Al-Ghamdi1
Ahlam A. Al-Ghamdi1 B. Rabindran Jermy4
B. Rabindran Jermy4 Mohamed Abouelhoda5,6
Mohamed Abouelhoda5,6 Shazia Subhani5,6
Shazia Subhani5,6 Lubna Al Asoom7
Lubna Al Asoom7 J. Francis Borgio2,8*
J. Francis Borgio2,8*Family trio next-generation sequencing-based variant analysis was done to identify the genomic reason on unexplained recurrent pregnancy loss (RPL). A family (dead fetus and parents) from Saudi Arabia with an earlier history of three unexplained RPLs at the ninth week of pregnancy was included in the study. Whole-genome sequencing (WGS) of a dead fetus and the parents was done to identify the pathogenic variation and confirmed through Sanger sequencing. WGS of dead fetus identifies a novel homozygous exonic variation (NM_017419.3:c.680G>T) in ASIC5 (acid-sensing ion channel subunit family member 5) gene; the parents are heterozygous. Newly designed ARMS PCR followed by direct sequencing confirms the presence of heterozygous in one subject and absence of homozygous novel mutation among randomly selected healthy Saudis. The second family with heterozygous was confirmed with three unexplained RPLs. Pathogenicity analysis of R227I amino acid substitution in ASIC5 protein through molecular docking and interaction analysis revealed that the mutations are highly pathogenic, decrease the stability of the protein, and prevent binding of amiloride, which is an activator to open the acid-sensing ion channel of ASIC5. The identified rare and novel autosomal recessive mutation, c.680G>T:p.R227I (ASIC5Saudi), in two families confirm the ASIC5 gene association with RPL and can be fatal to the fetus.
Recurrent pregnancy loss (RPL), or recurrent miscarriage (RM) is described as three or more sequential unpremeditated abortions before 20 weeks of gestation (1), a condition termed “habitual abortion” or “repeated spontaneous abortions” (2). RPL affects couples at propagative age around the world. The etiologies of RPL in Saudis or Arabs and other populations tend to be multifactorial. Factors including genetic abnormalities (3–10), placental anomalies (11–13), psychological trauma and stressful life events (14), and certain coagulation and immunoregulatory protein defects (15–18) were reported to be associated with RPL among women in the Gulf region. In some populations, other factors have been studied, such as anatomical, endocrine, hormonal problems, infection, smoking and alcohol consumption, and exposure to environmental factors, and these factors could increase the risk of RPL (19). Several studies have reported the relationship between various causes of recurrent miscarriage among Saudis and the rest of the population; however, 30–50% of RPLs were unexplained (5, 19). More studies on RPL only can reveal the cause. The objective of the study is to analyze the genetic basis of a family from Saudi Arabia with an earlier history of recurrent pregnancy loss at the ninth week of pregnancy using next-generation sequencing [whole-genome sequencing (WGS)] by complete analysis of whole genome of the fetus and parents followed by rigorous bioinformatics and confirmatory analyses (20–36). The study reports a novel homozygous exonic variation in the ASIC5 gene in a dead fetus, while the parents are heterozygous.
The study was approved by the Institutional Review Boards Committee of the Imam Abdulrahman Bin Faisal University (IRB-2017-13-137).
A family with a past history of three miscarriages has been included in the study with a written consent from the father and mother. During the fourth pregnancy, the mother experienced a similar type of miscarriage at the ninth week of pregnancy. Tissue (separated cautiously from maternal tissue to avoid contamination) samples and blood samples were collected from the fetus (proband) and parents, respectively. Miscarriage sample was collected in an RNAprotect Cell Reagent (Qiagen, Hilden, Germany). The DNAs of the samples were isolated, and the most prevalent genetic disease, hemoglobinopathies, were screened using the Sanger sequencing. Genes (functional variants and deletions in HBB, HBA1, HBA2, ATRX, and HBD) related to the most prevalent mutations have been found to be normal. Hence, the WGS was done for the miscarriage tissue, mother, and father genomes.
The trio analysis has been carried out using the best practice GATK pipeline (20). The program Fastx (http://hannonlab.cshl.edu/fastx_toolkit) was used to filter low-quality reads. Then the reads were aligned to the reference human genome (hg19) using the program BWA (21). The GATK haplotype caller was used to call the variants. The resulting variants were then annotated using in-house developed workflow including the following three sets of data sources:
1. Public databases: These were collected from the Annovar packages, and they include the basic positional information about genes and related proteins. They also include information from the dbSNP database, the 1000 Genome database, ExAC, and gnomAD databases. Annovar also includes predictions of the functional effect of the variants from the tools Polyphan, Sift, CADD, and MetaSVM. In addition to Annovar, we used the clinvar and OMIM databases to annotate the variants and genes with up-to-data medical information.
2. In-house databases: We annotated the variants using the Saudi Human Genome Program variant DB to check for variant frequency in the Saudi population (22– 24).
3. Commercial databases: We used the HGMD database to annotate the variant with clinical information.
After variant annotation, we ran filters according to the ACMG (American College of Medical Genetics and Genomics) guidelines. We excluded variants that are intergenic, synonymous, appearing more than 5% in population databases, or not damaging (as predicted by CADD, Polyphen, SIFT, and MetaSVM). We also ran extra trio analysis to filter the variants according to the autosomal recessive, de novo, compound heterozygous, and x-linked. After applying these filters, the remaining variants were examined manually to match the annotated clinical information to the fetus phenotype.
Whole-genome result was confirmed using Sanger sequencing. The presence of the homozygous NM_017419.3:c.680G>T in the proband and heterozygous in the parents were confirmed using Sanger sequencing. Highly specific primers (ASIC5F: 5′-CAGATAAAAACATGTTTCCATACATCTTCAG-3′ and ASIC5R: 5′- TTGTGGCATGAACATTCCCTGGA-3′) were designed, and the selected region of the gene was amplified [PCR recipe: MOLEQULE-ON absolute master mix 12.5 μl, ASIC5F 1 μl (10 nM), ASIC5R 1 μl (10 nM), DNA Template 25 ng, and Dis H2O to 25 μl; temperature profile: 95°C for 10 min; 35 cycles of 95°C/60 s, 60°C/60 s, 72°C/60 s; and 72°C for 5 min] and sequenced using BigDye Terminator Cycle Sequencing Kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Amplified PCR product (691 bp) of the ASIC5 gene region was purified and sequenced using Genetic Analyzer 3500 (Thermo Fisher Scientific, Inc.) at the Department of Genetic Research, Institute for Research and Medical Consultations, Imam Abdulrahman Bin Faisal University (Dammam, Saudi Arabia). Sequences were analyzed using mutation surveyor software (Softgenetics, US) and DNA sequencing analysis software v.5.3 (Applied Biosystem; Thermo Fisher Scientific, Inc.).
The amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) was designed (primers will be available on request) to screen the presence of NM_017419.3:c.680G>T among healthy Saudis (n = 200). The subjects positive for the presence of NM_017419.3:c.680G>T was confirmed through Sanger sequencing using primers (ASIC5F and ASIC5R). This is also to confirm the absence of the homozygous NM_017419.3:c.680G>T in the healthy Saudi subjects randomly selected.
The homology modeling of wild (p.R227) and mutant (p.R227I) ASIC5 protein was performed using Swiss Model server (25), validated using PROCHECK (26). The structural functional annotations were completed using SAS-sequence server (27), ProFunc (28), and PDBsum (29). Mutant structures were generated using Swiss-PDB Viewer and PyMol (30). Energy minimization for the wild and mutants was estimated using GROMACS (31). Evolutionary conservation and functional aspect analysis of the R227 residue in the wild-type protein was performed using the ConSurf (32). PROVEAN and I-Mutant were used for analyzing the impact on the biological function of a protein due to an amino acid substitution R227I (33, 34). AutoDock Vina was used for molecular docking of the ligand with wild type and mutant ASIC5 protein (35), and the molecular visualization was done in PyMol and LigPlot (36).
The family with a history of three unexplained miscarriages was included in the study. The couple is consanguineous but not first-degree relatives. There was no history of genetic and chronic diseases in the couple. The family was identified with a similar type of unknown spontaneous abortion at the ninth week of pregnancy. The mother was 30 years at the time of the fourth unexplained spontaneous miscarriage; the father was 34. The previous three unexplained miscarriages and the fourth were also of similar gestation. At this gestation, the gender of the proband cannot be determined even after miscarriage. The mother is devoid of uterine or cervical abnormalities. In order to identify the cause of the recurrent spontaneous abortion, WGS was done for the mother, father, and proband. The WGS of the trio (proband and parents) samples has revealed an inheritance of NM_017419.3:c.680G>T mutation in the ASIC5 gene from the parents (Figure 1A and Supplementary Table 1). Various heterozygous mutations observed in the proband are listed in the Supplementary Material, which were inherited either from the mother or father (Supplementary Table 2). The WGS result of NM_017419.3:c.680G>T variation in exon 4 of the ASIC5 gene has been confirmed through the Sanger sequencing (Figure 1B). The father and the mother were found to be carriers (heterozygous) of the c.680G>T:p.R227I at the ASIC5 gene, while the proband was homozygous to c.680G>T:p.R227I (GenBank: MN251164; ClinVar: SCV000930628; SNP ID: rs1248841709) (Figure 1). The name of the novel variant was validated using Mutalyzer 2.0.32.
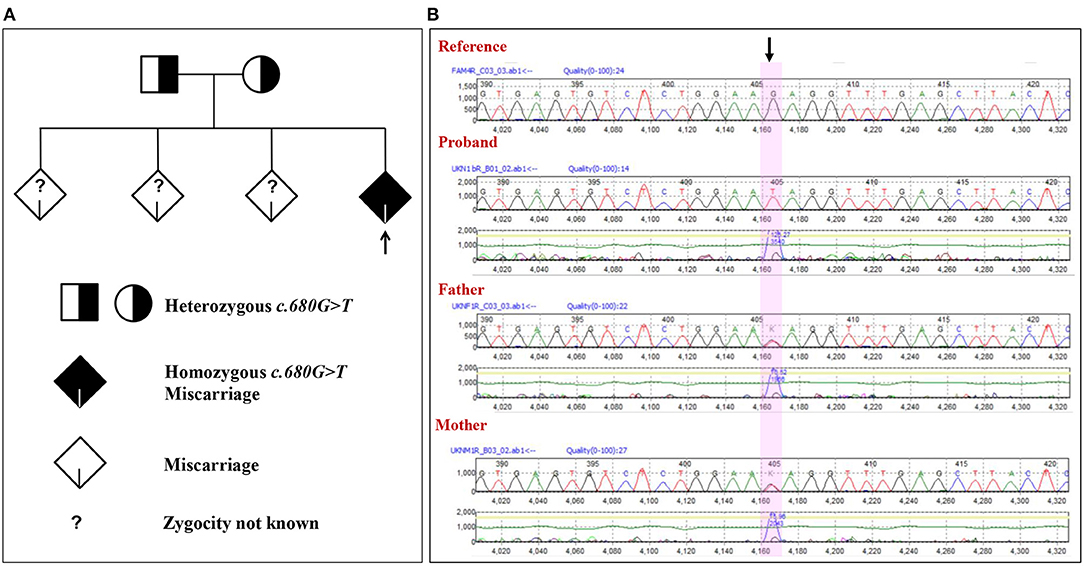
Figure 1. Novel mutation in the ASIC5 gene (NM_017419.3:c.680G>T) in the family. (A) Phylogenic analysis of the family with the NM_017419.3:c.680G>T mutation in the ASIC5 gene. (B) Electropherogram of the sequence c.664 to c.695 of exon 4 at the ASIC5 gene of the proband and the parents. The highlighted nucleotide with arrow indicates the position of the NM_017419.3:c.680G>T. The proband is homozygous for the NM_017419.3:c.680G>T. The mother and father are heterozygous for the NM_017419.3:c.680G>T.
In order to confirm the absence of the homozygous NM_017419.3:c.680G>T among the living population, a total of 200 healthy Saudis were selected randomly and checked for the mutation at the c.680 position in the ASIC5 gene using ARMS-PCR followed by Sanger sequencing. The results of the ARMS-PCR and direct sequencing of 200 healthy Saudis in the c.680 position in the ASIC5 gene revealed the absence of homozygous NM_017419.3:c.680G>T. Furthermore, this mutation is novel to the SHGP (Saudi Human Genome Program) database (about 9,500 cases). This suggests that the discovered mutation NM_017419.3:c.680G>T is rare, and their absence of a homozygous state in the healthy Saudis is validated. Furthermore, a female subject was observed with a heterozygous NM_017419.3:c.680G>T in the ASIC5 gene. The female subject with heterozygous mutations is a single daughter, and her mother experienced the unexplained RPL similar with the earlier family in the ninth week of pregnancy consecutively three times.
The predicted structure of the wild ASIC5 on the Ramachandran plot showed ϕ/Ψ angles of 83.1% residues in the most favored regions, 15.4% in the additional allowed regions, 1.1% in the generously allowed regions, and 0.3% in the disallowed regions (Figure 2B). The total residue span of the secondary structure consist of 23.0% residues involved in the formation of the strands, 23.0% residues in alpha helices, 2.6% residues in 3–10 helices, and 51.5% residues in other structural moieties. Analysis of secondary structure in ProFunc showed the presence of 3 β-sheets, 4 β-hairpins, 1 psi loop, 3 β-bulges, 14 strands, 14 helices, 5 helix–helix interactions, 34 β-turns, and 9 γ-turns. Homology modeling of the mutant structure (R227I) of the ASIC5 showed deviations from the wild type; the mutant structure on the Ramachandran plot showed ϕ/Ψ angles of 84.3% residues in the most favored regions, 14.3% in the additional allowed regions, 1.1% in the generously allowed regions, and 0.3% in the disallowed regions. The total residue span of the secondary mutant structure consisting of 22.7% residues involving the formation of the strands, 23.7% residues in alpha helices, 1.8% residues in 3–10 helices, and 51.8% residues in other structural moieties. Analysis of the secondary structure of the mutant in ProFunc showed the presence of 3 β-sheets, 4 β-hairpins, 1 psi loop, 2 β-bulges, 14 strands, 13 helices, 5 helix–helix interactions, 42 β-turns, and 8 γ-turns.
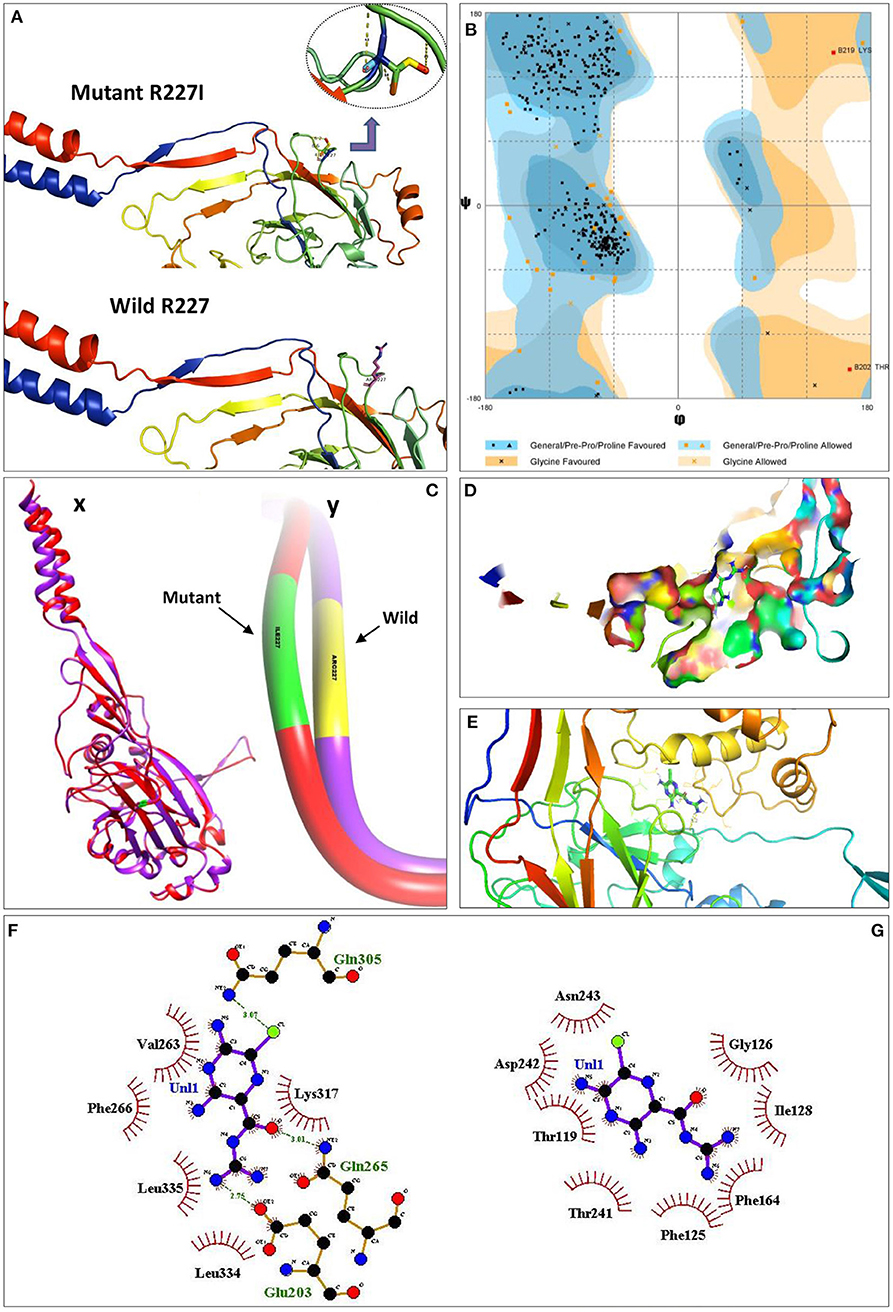
Figure 2. Pathogenicity analysis of R227I mutation in the ASIC5 protein through molecular docking and interaction analysis. (A) Structural models of the wild (R227) and mutated (R227I) ASIC5 proteins. (B) Ramachandran plot for the predicted structure of the ASIC5 protein. Eighty-three porterage residues of the ASIC5 protein are in the most favored regions. Cx, superimposed structures of the wild (R227) and mutated (R227I) ASIC5 proteins; Cy, deviated region of R227I from R227 on superimposed wild and mutant ASIC5. (D) Amiloride with ASIC5 at the active binding site. (E) 3D amiloride with surrounding amino acids of ASIC5 protein. (F,G) Protein–ligand interaction. (F) Wild ASIC5 (R227) protein with ligand, amiloride. (G) Mutant ASIC5 (R227I) protein with ligand, amiloride.
ConSurf analysis revealed that R227I is a functional residue, which is highly conserved and exposed. A total of 97 HMMER hits were considered for this analysis, while 91 of them were unique, including the query. PROVEAN analysis showed that R227I is a deleterious amino acid substitution as evident from PROVEAN score −3.830. I-Mutant analysis predicted that the free energy change value (DDG) between wild type and mutant type was less than zero (DDG < 0), which declares the decrease in protein stability. Wild (RMSD = 0.045 Å) and mutant (RMSD = 0.088 Å) proteins were superimposed, quantitative measure of similarity analysis revealed an increase of 95.56% root-mean-square deviation of atomic positions in the mutant (Figure 2C).
Molecular docking studies of wild (p.R227) and mutant (p.R227I) ASIC5 protein with amiloride, a potent inhibitor of acid-sensing ion channel proteins, were performed, and it was observed that the binding behavior of amiloride with the mutant model compared with the wild-type model was completely different (Figure 2). R227 residue is not directly involved in binding with the ligand, but it assists atomic interactions through binding of the ligand with protein molecules at specific sites (Figure 2F). In particular, a halogen bonding occurs between the chlorine atom (colored green) of amiloride with the amino group (NH2) of Gln305 (colored blue). The oxygen atom of the carbonyl group (colored red) of amiloride interacts with the hydrogen of the amino group (NH2) of Gln265 through N–H···O hydrogen bonding. In a similar fashion, the hydrogen of amiloride interacts with the oxygen group of Glu203. However, the R227I prevents the binding of ligand with the ASIC5 molecule at a specific site (Figure 2G). In this mutant model, an alteration in the protein coordination site occurs (Gly126 and Asn243) and, therefore, fails to coordinate with amiloride functional groups.
Studies on tissues of miscarriage specimens from women with RPL observed the chromosomal aberrations from 29 to 46% of miscarriage tissues, while majority of the RPL may be due to alternative mechanisms or other than chromosomal aberrations (37–39). The present observation suggests that coding variants in ASIC5 gene can be one among the alternative mechanisms for RPL. The role of the acid-sensing ion channel subunit family member 5 (ASIC5) or ACCN5 or bile acid-sensitive ion channel (BASIC) gene in humans, in general, and the development of the fetus, in particular, is scanty (40–42). Very limited studies are available on the gene ASIC5 and related expression. This gene, ASIC5, was reported to be expressed in the amniotic fluid (43), fetal gut, brain, liver, heart, ovary, and testis (44). ASIC5 is overexpressed in the fetal gut (41.0) and plasma (27.5). ASIC5 was observed to a key player in the physiology of unipolar brush cells of the vestibulocerebellum (42, 45, 46). The complete functions of the ASIC5 gene and its product are yet to be identified (40–42). Animal studies on the autosomal recessive mouse mutant of the gene encoding the L-type calcium channel revealed that the homozygous mutant animals die at birth; however, the heterozygous for the mutant is not distinguishable from that of wild animals (47). The study resembles the present observation of the heterozygous mutant of the healthy parents, while death of the fetus with a homozygous mutant in the gene belongs to the amiloride-sensitive Na+ channel. The R227I prevents the binding of amiloride with ASIC5 protein. However, more confirmatory studies are mandatory to prove the failure in amiloride-R227I (ASIC5) binding in wet lab, which is mandatory for an activator to open its own channel (41, 48). Acid-sensing ion channel subunit channels play an important role in the fetal developmental pathology due to acidosis; furthermore, prolonged acidosis is significantly associated with mortality of the fetus (49, 50). Increased apoptosis was observed in the retina due to the mutant ASIC2 gene compared with the wild type (51). Mammalian degenerin (MDEG) or ASIC2 (acid-sensing ion channel subunit 2) gene mutant study on the development of Xenopus reported that the Xenopus oocytes with ASIC2 mutation start to maturate and die (52). This indicates the pathophysiology of the mutation in the acid-sensing ion channel subunit genes.
Earlier reports reported that in 39% of the Saudi females who had RPL, the origin of the patient in the study was unexplained or had no identifiable cause (5). Various reasons including genetic factors were stated for recurrent pregnancy loss among Saudi women (3, 4, 6, 14, 18). Consanguineous marriages are also considerably (p = 0.046) impacting (3). Genome-wide association study (GAWS) revealed the association of ASIC5 (p = 0.0029; Supplementary Table 3) and level of manganese (53, 54). Furthermore, the level of manganese in the placental tissue of Saudi women with recurrent pregnancy loss was significantly (p < 0.0001) decreased (11). This suggests that the identified mutation in ASIC5 might have played a role in the level of manganese in the present women. A recent study on the prognosis markers of glioblastoma revealed the expression of ASIC5 as associated prognosis markers (55). ASIC5 was found to be activated in the ethanol-(100 mM)-exposed neonatal rat cardiomyocytes along with other six molecules (CYP2A6, PRL, CHRNA4, CNR1, CRH, and SLC40A1) (56). Low (in 50%) ASIC5 protein expression in melanoma were observed with <4% mutation rates (57).
Preparing the mutated animal model to study the impact of the mutant on the fetal development is not available in our laboratory, which is a limitation of the study. Hence, the region with the mutation, c.680G>T in the ASIC5 gene, was screened using ARMS-PCR followed by sequencing using designed primers to identify the presence of c.680G>T in randomly selected Saudis in the study region, which confirms the absence of the homozygous NM_017419.3:c.680G>T and the presence of heterozygous NM_017419.3:c.680G>T in a female subject and her mother with RPL. The study confirms the influence of the association of the novel exonic mutation with RPL. However, nationwide studies are mandatory to identify the prevalence of this rare mutation and mutations in this gene among unexplained miscarriages cases and their impact on the recurrent pregnancy loss and fetal development. This can reveal the role of ASIC5. The protein–protein interaction analysis of ASIC5 protein, with the protein observed with the mutation in the proband using STRING, revealed lack of interaction (Supplementary Table 2). However, the analysis using STRING cannot reveal any specific impact of mutated protein–protein interactions due to specific amino acid changes (58).
Based on the earlier reports on the member of the DEG/ENaC (degenerin/epithelial sodium channel) protein family and the current observations, it may be concluded that the R227I amino acid substitution in the ASIC5 is highly deleterious; the mutant ASIC5 showed decreased stability and of the protein and prevents the binding of amiloride, a potent inhibitor of acid-sensing ion channel proteins (59).
The observed novel ASIC5 gene-coding variant (ASIC5Saudi) in two families confirm the ASIC5 association with the results of RPL. Hence, this mutation is pathogenic, which may cause serious illness to the fetus and cause fetal mortality. The molecular mechanism behind the death of the fetus in relation to the homozygous NM_017419.3:c.680G>T at exon 4 (ASIC5Saudi) in the ASIC5 gene should be studied in detail. Early prenatal diagnosis of pathogenic variation like ASIC5Saudi can provide a choice for the parent to decide pregnancy termination within the allowed time among high-consanguinity population (60).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board (IRB) at Imam Abdulrahman Bin Faisal University. IRB approval number: IRB-2017-13-137 dated 07 June 2017 and extended on 14 Dec 2020. The patients/participants provided their written informed consent to participate in this study.
NHA, SA, NBA, HAA, AA-G, BR, and JB conceived and designed the research and analyzed the experiments. SA, NBA, NF, HSA, BR, and JB performed and analyzed the experiments. SA, MA, SS, and JB performed the whole-genome analysis. NHA, HAA, LA, and AA-G performed the clinical analysis. SA, HSA, and JB wrote the paper with the contributions of NHA, NBA, NF, HAA, AA-G, BR, MA, LA, and SS. All authors reviewed and approved the final manuscript.
This study was partially supported by The Deanship of Scientific Research, Imam Abdulrahman Bin Faisal University (to JB, Grant No: 2017-100-IRMC). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank the Dean, Institute for Research and Medical Consultations (IRMC), Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia, for her continuous support and encouragement. We thank the Saudi Human Genome Program, King Abdulaziz City for Science and Technology team, for their support. The authors thank Mr. Ranilo M. Tumbaga, Mr. Horace T. Pacifico, and Mrs. Jee Entusiasamo Aquino for their assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.699672/full#supplementary-material
1. Baek KH, Lee EJ, Kim YS. Recurrent pregnancy loss: the key potential mechanisms. Trends Mol Med. (2007) 13:310–7. doi: 10.1016/j.molmed.2007.05.005
2. Turki RF, Banni HA, Assidi M, Al-Qahtani MH, Abduljabbar HS, Jamel HS, et al. Analysis of chromosomal and genetic disorders in patients with recurrent miscarriages in Saudi Arabia. BMC Genom. (2014) 15:P73. doi: 10.1186/1471-2164-15-S2-P73
3. McNamee K, Dawood F, Farquharson R. Recurrent miscarriage and thrombophilia: an update. Curr Opin Obstetr Gynecol. (2012) 24:229–34. doi: 10.1097/GCO.0b013e32835585dc
4. Turki RF, Assidi M, Banni HA, Zahed HA, Karim S, Schulten HJ, et al. Associations of recurrent miscarriages with chromosomal abnormalities, thrombophilia allelic polymorphisms and/or consanguinity in Saudi Arabia. BMC Med Gen. (2016) 17:69. doi: 10.1186/s12881-016-0331-1
5. Al-Ghamdi AA, Makhashen SF. Etiology of recurrent pregnancy loss in Saudi females. Saudi J Med Med Sci. (2016) 4:187. doi: 10.4103/1658-631X.188258
6. Awartani KA, Al Shabibi MS. Description of cytogenetic abnormalities and the pregnancy outcomes of couples with recurrent pregnancy loss in a tertiary-care center in Saudi Arabia. Saudi Med J. (2018) 39:239. doi: 10.15537/smj.2018.3.21592
7. Tan Y, Yin X, Zhang S, Jiang H, Tan K, Li J. Clinical outcome of preimplantation genetic diagnosis and screening using next generation sequencing. Gigascience. (2014) 3:30. doi: 10.1186/2047-217X-3-30
8. Gaboon NE, Mohamed AR, Elsayed SM, Zaki OK, Elsayed MA. Structural chromosomal abnormalities in couples with recurrent abortion in Egypt. Turkish J Med Sci. (2015) 45:208–13. doi: 10.3906/sag-1310-5
9. Alkhuriji AF, Alhimaidi AR, Babay ZA, Wary AS. The relationship between cytokine gene polymorphism and unexplained recurrent spontaneous abortion in Saudi females. Saudi Med J. (2013) 34:484–9.
10. Al-Hassan S, Hellani A, Al-Shahrani A, Al-Deery M, Jaroudi K, Coskun S. Sperm chromosomal abnormalities in patients with unexplained recurrent abortions. Arch Androl. (2005) 51:69–76. doi: 10.1080/014850190518062
11. Ghneim HK and Alshebly MM. Biochemical markers of oxidative stress in Saudi women with recurrent miscarriage. J Korean Med Sci. (2016) 31:98–105. doi: 10.3346/jkms.2016.31.1.98
12. Ghneim HK, Al-Sheikh YA, Alshebly MM, Aboul-Soud MA. Superoxide dismutase activity and gene expression levels in Saudi women with recurrent miscarriage. Mol Med Rep. (2016) 13:2606–12. doi: 10.3892/mmr.2016.4807
13. Atia TA. Placental apoptosis in recurrent miscarriage. Kaohsiung J Med Sci. (2017) 33:449–52. doi: 10.1016/j.kjms.2017.06.012
14. Rouzi AA, Alamoudi R, Turkistani J, Almansouri N, Alkafy S, Alsenani N, et al. Miscarriage knowledge among Saudi women. Fertility Sterility. (2017) 108:e383. doi: 10.1016/j.fertnstert.2017.07.1111
15. Gader AG, Al-Mishari AA, Al-Jabbari AW, Awadalla SA, Al-Momen AM. Thrombophilia in Saudi women with recurrent fetal loss. J Appl Hematol. (2010) 1:97.
16. Al Omar SY, Mansour L, Alkhuriji AF, Alwasel S, Al-Qahtani S. Genetic association between the HLA-G 14-bp insertion/deletion polymorphism and the recurrent spontaneous abortions in Saudi Arabian women. Genet Mol Res. (2015) 14:286–93. doi: 10.4238/2015.January.23.2
17. Gowri V, Udayakumar AM, Bsiso W, Al Farsi Y, Rao K. Recurrent early pregnancy loss and consanguinity in Omani couples. Acta obstetricia et gynecologica Scandinavica. (2011) 90:1167–9. doi: 10.1111/j.1600-0412.2011.01200.x
18. Almasry SM, Elmansy RA, Elfayomy AK, Algaidi SA. Ultrastructure alteration of decidual natural killer cells in women with unexplained recurrent miscarriage: a possible association with impaired decidual vascular remodelling. J Mol Histol. (2015) 46:67–78. doi: 10.1007/s10735-014-9598-8
19. Ford HB, Schust DJ. Recurrent pregnancy loss: etiology, diagnosis, and therapy. Rev Obstetr Gynecol. (2009) 2:76.
20. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. (2010) 20:1297–303. doi: 10.1101/gr.107524.110
21. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. (2010) 26:589–95. doi: 10.1093/bioinformatics/btp698
22. Abouelhoda M, Faquih T, El-Kalioby M, Alkuraya FS. Revisiting the morbid genome of Mendelian disorders. Genome Biol. (2016) 17:235. doi: 10.1186/s13059-016-1102-1
23. Abouelhoda M, Sobahy T, El-Kalioby M, Patel N, Shamseldin H, Monies D, et al. Clinical genomics can facilitate countrywide estimation of autosomal recessive disease burden. Genet Med. (2016) 18:1244. doi: 10.1038/gim.2016.37
24. The Saudi Mendliome Group. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome Biol. (2015) 16:134.
25. Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. (2003) 31:3381–5. doi: 10.1093/nar/gkg520
26. Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins Struct Funct Bioinform. (1992) 12:345–64. doi: 10.1002/prot.340120407
27. Milburn D, Laskowski RA, Thornton JM. Sequences annotated by structure: a tool to facilitate the use of structural information in sequence analysis. Protein Eng. (1998) 11:855–9. doi: 10.1093/protein/11.10.855
28. Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. (2005) 33:W89–93. doi: 10.1093/nar/gki414
29. Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. (2001) 29:221–2. doi: 10.1093/nar/29.1.221
30. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. (1997) 18:2714–23. doi: 10.1002/elps.1150181505
31. Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. (2006) 34:52–6. doi: 10.1093/nar/gkl082
32. Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. (2005) 33):W299–302. doi: 10.1093/nar/gki370
33. Capriotti E, Fariselli P, Casadio R. I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. (2005) 33:W306–10. doi: 10.1093/nar/gki375
34. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. (2015) 31:2745–7. doi: 10.1093/bioinformatics/btv195
35. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. (2010) 31:455–61. doi: 10.1002/jcc.21334
36. Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng Design Select. (1995) 8:127–34. doi: 10.1093/protein/8.2.127
37. Carp H, Toder V, Aviram A, Daniely M, Mashiach S, Barkai G. Karyotype of the abortus in recurrent miscarriage. Fertility Sterility. (2001) 75:678–82. doi: 10.1016/S0015-0282(00)01801-X
38. Stephenson MD, Awartani KA, Robinson WP. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: a case–control study. Hum Reprod. (2002) 17:446–51. doi: 10.1093/humrep/17.2.446
39. Li TC, Makris M, Tomsu M, Tuckerman E, Laird S. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. (2002) 8:463–81. doi: 10.1093/humupd/8.5.463
40. Xu S, Liu C, Ma Y, Ji HL, Li X. Potential roles of amiloride-sensitive sodium channels in cancer development. BioMed Res Int. (2016) 2016:2190216. doi: 10.1155/2016/2190216
41. Hanukoglu I. ASIC and ENaC type sodium channels: conformational states and the structures of the ion selectivity filters. FEBS J. (2017) 284:525–45. doi: 10.1111/febs.13840
42. Boiko NY, Kreko-Pierce T, Pugh J, Stockand JD. Ataxia in the ASIC5 knockout mouse associated with decreased excitability of vestibulocerebellum unipolar brush cells. FASEB J. (2019) 33:824–13. doi: 10.1096/fasebj.2019.33.1_supplement.824.13
43. McCall MN, Uppal K, Jaffee HA, Zilliox MJ, Irizarry RA. The gene expression barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res. (2010) 39:D1011–5. doi: 10.1093/nar/gkq1259
44. Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, et al. Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. (2003) 31:142–6. doi: 10.1093/nar/gkg050
45. Boiko N, Kucher V, Wang B, Stockand JD. Restrictive expression of acid-sensing ion channel 5 (asic5) in unipolar brush cells of the vestibulocerebellum. PLoS ONE. (2014) 9:e91326. doi: 10.1371/journal.pone.0091326
46. Boiko N, Kucher V, Stockand JD. Dysfunction of acid-sensing ion channel 5 (ASIC5) in unipolar brush cells of the vestibulocerebellum causes ataxia. FASEB J. (2018) 32:750–5. doi: 10.1096/fasebj.2018.32.1_supplement.750.5
47. Lehmann-Horn F, Jurkat-Rott K. Voltage-gated ion channels and hereditary disease. Physiol Rev. (1999) 79:1317–72. doi: 10.1152/physrev.1999.79.4.1317
48. Schaefer L, Sakai H, Mattei MG, Lazdunski M, Lingueglia E. Molecular cloning, functional expression and chromosomal localization of an amiloride-sensitive Na+ channel from human small intestine. FEBS Lett. (2000) 471:205–10. doi: 10.1016/S0014-5793(00)01403-4
49. Xiong ZG, Pignataro G, Li M, Chang SY, Simon RP. Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr Opin Pharmacol. (2008) 8:25–32. doi: 10.1016/j.coph.2007.09.001
50. Bobrow CS, Soothill PW. Causes and consequences of fetal acidosis. Arch Dis Childhood Fetal Neonatal Ed. (1999) 80:F246–9. doi: 10.1136/fn.80.3.F246
51. Zha XM. Acid-sensing ion channels: trafficking and synaptic function. Mol Brain. (2013) 6:1. doi: 10.1186/1756-6606-6-1
52. Waldmann R, Champigny G, Voilley N, Lauritzen I, Lazdunski M. The mammalian degenerin MDEG, an amiloride-sensitive cation channel activated by mutations causing neurodegeneration in Caenorhabditis elegans. J Biol Chem. (1996) 271:10433–6. doi: 10.1074/jbc.271.18.10433
53. Ng E, Lind PM, Lindgren C, Ingelsson E, Mahajan A, Morris A, et al. Genome-wide association study of toxic metals and trace elements reveals novel associations. Hum Mol Genet. (2015) 24:4739–45. doi: 10.1093/hmg/ddv190
54. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:W90–7. doi: 10.1093/nar/gkw377
55. Gong Z, Hong F, Wang H, Zhang X, Chen J. An eight-mRNA signature outperforms the lncRNA-based signature in predicting prognosis of patients with glioblastoma. BMC Med Genet. (2020) 21:1–13. doi: 10.1186/s12881-020-0992-7
56. Mashimo K, Ohno Y. Effect of ethanol on gene expression of beating neonatal rat cardiomyocytes-further research with ingenuity pathway analysis software. J Nippon Med School. (2020) 88:209–19. doi: 10.1272/jnms.JNMS.2021_88-501
57. Böhme I, Schönherr R, Eberle J, Bosserhoff AK. Membrane transporters and channels in melanoma. In: Reviews of Physiology, Biochemistry and Pharmacology. Berlin, Heidelberg: Springer (2000). p. 1–106.
58. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2018) 47:D607–13. doi: 10.1093/nar/gky1131
59. Lingueglia E, Lazdunski M. Pharmacology of ASIC channels. Wiley Interdiscip Rev Membrane Transport Signal. (2013) 2:155–71. doi: 10.1002/wmts.88
Keywords: exome, recurrent pregnancy loss, whole genome sequencing, ASIC5, Saudi Arabia, molecular docking, next generation sequencing, unknown spontaneous abortion
Citation: Al Qahtani NH, AbdulAzeez S, Almandil NB, Fahad Alhur N, Alsuwat HS, Al Taifi HA, Al-Ghamdi AA, Rabindran Jermy B, Abouelhoda M, Subhani S, Al Asoom L and Borgio JF (2021) Whole-Genome Sequencing Reveals Exonic Variation of ASIC5 Gene Results in Recurrent Pregnancy Loss. Front. Med. 8:699672. doi: 10.3389/fmed.2021.699672
Received: 23 April 2021; Accepted: 21 June 2021;
Published: 30 July 2021.
Edited by:
D. Thirumal Kumar, Meenakshi Academy of Higher Education and Research, IndiaReviewed by:
Jayalakshmi Mariakuttikan, Madurai Kamaraj University, IndiaCopyright © 2021 Al Qahtani, AbdulAzeez, Almandil, Fahad Alhur, Alsuwat, Al Taifi, Al-Ghamdi, Rabindran Jermy, Abouelhoda, Subhani, Al Asoom and Borgio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Francis Borgio, ZmJhbGV4YW5kZXJAaWF1LmVkdS5zYQ==; Ym9yZ2lvbWljcm9AZ21haWwuY29t; orcid.org/0000-0001-7199-1540
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.