
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 27 April 2021
Sec. Precision Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.651473
This article is part of the Research TopicNucleic Acids-Based Cancer TheranosticsView all 29 articles
Objective: Increasing evidence suggests that microRNA (miRNA) participates in regulating tumor cell apoptosis. We aimed to observe the effect of hsa-miR-33-5p on the apoptosis of breast cancer cells and to explore its regulatory relationship with selenoprotein T (SelT).
Methods: RT-qPCR was used to examine the expression of hsa-miR-33-5p and SelT both in breast cancer tissues and cells. MCF-7 and MDA-MB-231 cells were transfected with hsa-miR-33-5p mimics or si-SelT. Then, a flow cytometry assay was carried out to examine the apoptosis of cells. Furthermore, SelT and apoptosis-related proteins including caspase-3, caspase-8, caspase-9, Bax, and Bcl-2 were detected via RT-qPCR and western blot. A luciferase reporter assay was utilized for assessing whether SelT was targeted by hsa-miR-33-5p.
Results: Downregulated hsa-miR-33-5p was found both in breast cancer tissues and cells. After its overexpression, MCF-7 cell apoptosis was significantly promoted. Furthermore, our data showed that miR-33-5p elevated apoptosis-related protein expression in MCF-7 cells. Contrary to hsa-miR-33-5p, SelT was upregulated both in breast cancer tissues and cells. SelT expression was significantly inhibited by hsa-miR-33-5p overexpression. The luciferase reporter assay confirmed that SelT was a direct target of hsa-miR-33-5p. SelT overexpression could ameliorate the increase in apoptosis induced by hsa-miR-33-5p mimics.
Conclusion: Our findings revealed that hsa-miR-33-5p, as a potential therapeutic target, could accelerate breast cancer cell apoptosis.
Breast cancer is a commonly diagnosed malignancy worldwide (1). Despite considerable progress in early detection and diagnosis, breast cancer patients' prognosis has been only slightly improved (2, 3). Moreover, its incidence in developed countries is still high, while it is increasing in developing countries due to changes in lifestyle and life expectancy (4). Hence, it is of significance to explore novel and effective therapeutic targets for breast cancer.
miRNA is a small and evolutionarily conserved non-coding RNA, with about 18–25 nucleotides in its length (5, 6). It has been widely accepted that miRNAs can bind to the 3′ untranslated region (UTR) of target genes, thereby negatively regulating their expression and causing the degradation of target mRNAs (7–9). It has been discovered that miRNAs are involved in many critical cell processes by regulating target genes at a post-transcriptional level, including cell apoptosis (10–12). There is ample evidence that aberrantly expressed miRNAs could be associated with breast cancer progression (13–15). So far, miRNAs have become promising markers for breast cancer due to the fact that they can be easily detected in tumor biopsies or body fluids (16). Dysregulated miRNAs have been recognized as early indicators and pathogenic factors of breast cancer (7, 17, 18). Furthermore, miRNA expression can predict the prognosis and progression of breast cancer (19–21). Therefore, a deeper understanding of miRNAs may provide opportunities for novel treatment strategies for breast cancer.
Previous studies found that hsa-miR-33-5p could induce osteoblast differentiation through targeting Hmga2 (22). Apoptosis is an important cellular process that is controlled by a variety of factors, including miRNAs. Abnormal apoptosis exhibits a close relationship with breast cancer occurrence, yet the function of hsa-miR-33-5p in breast cancer cell apoptosis needs to be clarified. In this study, our results suggested that hsa-miR-33-5p expression was downregulated in breast cancer. More importantly, its overexpression could promote breast cancer cell apoptosis. By further analysis, SelT could be directly targeted by hsa-miR-33-5p. Thus, our study proposed that hsa-miR-33-5p, as an underlying therapeutic target, could facilitate breast cancer cell apoptosis by SelT.
Breast cancer tissues as well as corresponding normal tissue specimens were harvested from 20 patients with breast cancer from January 2018 to December 2019 in the Linyi Cancer Hospital. All tissues were instantly stored in liquid nitrogen. No patient received chemotherapy or radiation before surgery. All the patients provided written informed consent. The study was approved by the Ethics Committee of Linyi Cancer Hospital (LYZLYY-2018-012).
The MiRWalk 2.0 database (http://mirwalk.uni-hd.de/) was used to predict the targets of hsa-miR-33-5p, which provides predicted and validated miRNA–target interactions (23). Furthermore, the immunohistochemical results of apoptosis-related proteins including Bax, Bcl2, caspase-3/8/9, and SelT in breast cancer and normal samples were downloaded from the Human Protein Atlas website (https://www.proteinatlas.org/).
Human breast cancer MCF-7 and MDA-MB-231 cells and normal breast MCF10A cells were grown in 1640 medium (Invitrogen, CA, USA) with 10% FBS at 37°C and 5% CO2 in a humid environment. hsa-miR-33-5p mimics, siRNAs against SelT, and corresponding controls were used for transfection into two cells via Lipofectamine 2000 (Invitrogen). At 48 h after transfection, the cells were collected for further analysis. The cultured cells were treated with H2O2 as a control.
Cellular apoptotic levels were examined via the annexin V-FITC/PI apoptosis detection kit (Keygene, Nanjing, China). At 48 h after transfection, the cell resuspension (100 μl) was treated with annexin V-FITC/PI lasting for 15 min in the dark. The apoptotic rates were detected by flow cytometry.
Total RNA was extracted using TRIzol (Invitrogen), followed by reverse transcription into cDNAs. The RT-qPCR was carried out on the real-time PCR system. The primer sequences of hsa-miR-33-5p and SelT were as follows: hsa-miR-33-5p, 5′-GGAMCTWYACGVAGGTG-3′ (forward), 5′-TGAAMTGCACRGAGCTTGC-3′ (reverse); SelT, 5′-TTGCTGCTTCTGCTGGTG-3′ (forward), 5′-CGTGGCGTACTGCATCTT-3′(reverse); β-actin, 5′-CGAGAAGATGACCCAGATCATG-3′ (forward), 5′-GTGAAGCTGTAGCCGCGCTCGG-3′ (reverse). β-actin was used as a control. Then, their expression levels were determined with the 2−ΔΔCT method.
Protein was extracted via RIPA lysis buffers (Beyotime, China), which was assessed with a bicinchoninic acid protein assay kit (Beyotime). The sample was separated through sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a PVDF membrane. Then, the membrane was blocked using 5% non-fat milk for 1 h, which was incubated overnight with primary antibodies at 4°C and secondary antibody (1:2,000; Abcam, USA) lasting 1 h at room temperature. The primary antibodies included anti-Bax (1:1,000; Abcam), anti-Bcl2 (1:1,000; Abcam), anti-cleaved-caspase-3 (1:1,000; Abcam), anti-pro-caspase-3 (1:1,000; Abcam), anti-cleaved-caspase-8 (1:1,000; Abcam), anti-pro-caspase-8 (1:1,000; Abcam), anti-cleaved-caspase-9 (1:1,000; Abcam), anti-pro-caspase-9 (1:1,000; Abcam), and anti-GAPDH (1:1,000; Abcam). GAPDH served as a control. Image Lab™ Software (Bio-Rad, China) was used to quantify the intensity of blots.
The wild-type luciferase vector (wt-LucSelT) containing hsa-miR-33-5p response elements in the 3′UTR of SelT, or the mutant (mut-LucSelT) vector was constructed and transfected in RKO cells with hsa-miR-33-5p mimics or its control. Luciferase reporter assay systems (Promega, USA) were utilized to quantify the firefly and Renilla luciferase activity.
A transferase-mediated dUTP nick end labeling (TUNEL) kit (Atagenix, Wuhan, China) was used to assess the apoptotic levels. The sections were treated with TUNEL solution in the dark for 60 min. Afterwards, the samples were incubated with 0.05 μg/μl of 4′,6-diamidino-2-phenylindole solution for 10 min. Anti-fluorescence quenching mounting tablets were utilized for mounting. Images were investigated under a fluorescence microscope (Olympus, Japan).
Statistical analyses were performed using GraphPad Prism 8.0. Data are expressed as mean ± standard deviation from at least three independent experiments. Comparison between different groups was analyzed using Student's t test or one-way analysis of variance. P < 0.05 was considered statistically significant.
According to RT-qPCR results, hsa-miR-33-5p expression was lower in breast cancer than normal tissue specimens (Figure 1A; p < 0.0001). Furthermore, its lower expression was observed in MCF-7 as well as MDA-MB-231 cells more than MCF10A cells (both p < 0.05; Figure 1B). These results suggested that hsa-miR-33-5p was downregulated in breast cancer tissues as well as cells. As shown in Figure 1C, hsa-miR-33-5p mimics significantly overexpressed its expression compared to control (p < 0.0001). We also found that H2O2 significantly elevated hsa-miR-33-5p expression in MCF-7 cells (Figure 1C; p < 0.0001). hsa-miR-33-5p expression was significantly higher in MCF-7 cells following transfection by hsa-miR-33-5p overexpression than H2O2 treatment (Figure 1C; p < 0.05). The above-mentioned results suggested that hsa-miR-33-5p was successfully overexpressed. The flow cytometry results suggested that hsa-miR-33-5p overexpression could promote the apoptosis of MCF-7 cells (Figures 1D,E; p < 0.001). Furthermore, compared to control, H2O2 treatment also significantly induced cell apoptosis (p < 0.001). However, there was no significant difference in cell apoptosis between cells transfected with hsa-miR-33-5p mimics and H2O2 treatment.
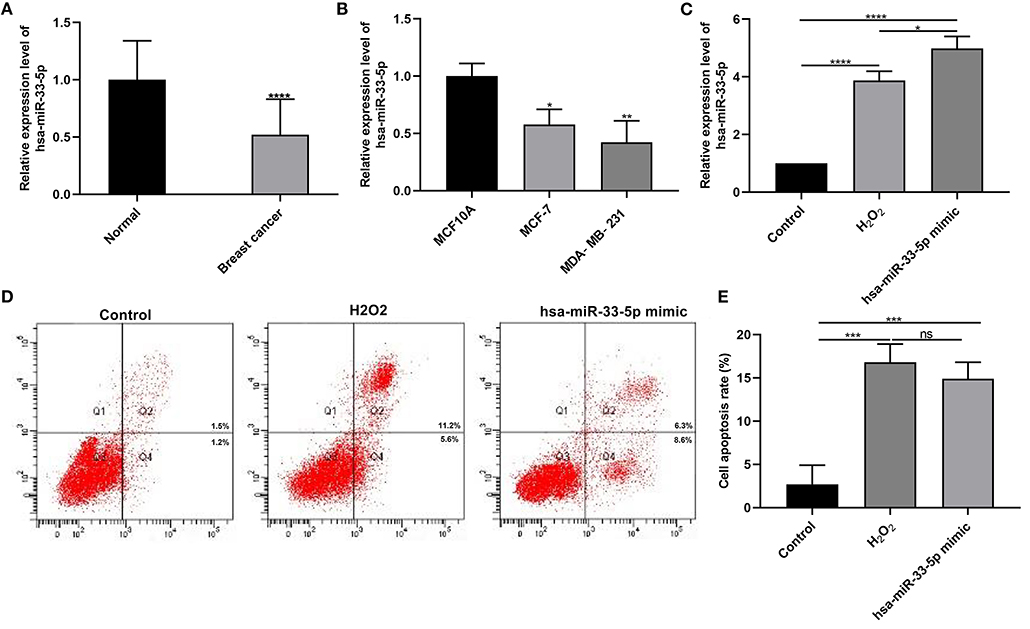
Figure 1. hsa-miR-33-5p is downregulated in breast cancer, and its overexpression promotes the apoptosis of breast cancer cells. (A,B) RT-qPCR results showing downregulated hsa-miR-33-5p both in breast cancer tissues (A) and cells (B). (C) RT-qPCR showing hsa-miR-33-5p expression in MCF-7 cells following transfection by hsa-miR-33-5p mimics. (D,E) Flow cytometry for the cell apoptosis in MCF-7 cells under transfection by hsa-miR-33-5p mimics. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant.
Apoptosis is a key cellular process in breast cancer. We detected apoptosis-related proteins in tumor tissues. Figure 2A depicts the expression and distribution of these apoptosis-related proteins including caspase-3, caspase-8, caspase-9, Bax, and Bcl-2 in breast cancer tissues according to immunohistochemistry results. Furthermore, using western blot, we found that cleaved caspase-3, cleaved caspase-8, cleaved caspase-9, and Bax proteins were significantly overexpressed in breast cancer and normal tissues (all p < 0.0001; Figures 2B,C). Meanwhile, pro-caspase-3, pro-caspase-8, pro-caspase-9, and Bcl-2 protein (all p < 0.0001) exhibited significantly low expressions in breast cancer tissues (Figures 2B,C). hsa-miR-33-5p overexpression significantly elevated the expression of cleaved caspase-3 (p < 0.01), cleaved caspase-8 (p < 0.05), cleaved caspase-9 (p < 0.0001), and Bax (p < 0.0001) compared to control (Figures 2D,E). Moreover, pro-caspase-3 (p < 0.05), pro-caspase-8 (p < 0.01), pro-caspase-9 (p < 0.05), and Bcl-2 (p < 0.0001) expressions were distinctly lowered in MCF-7 cells after transfection by overexpressed hsa-miR-33-5p (Figures 2D,E). Similar results were investigated in MCF-7 cells exposed to H2O2. Thus, hsa-miR-33-5p could mediate apoptosis-related proteins in breast cancer cells.
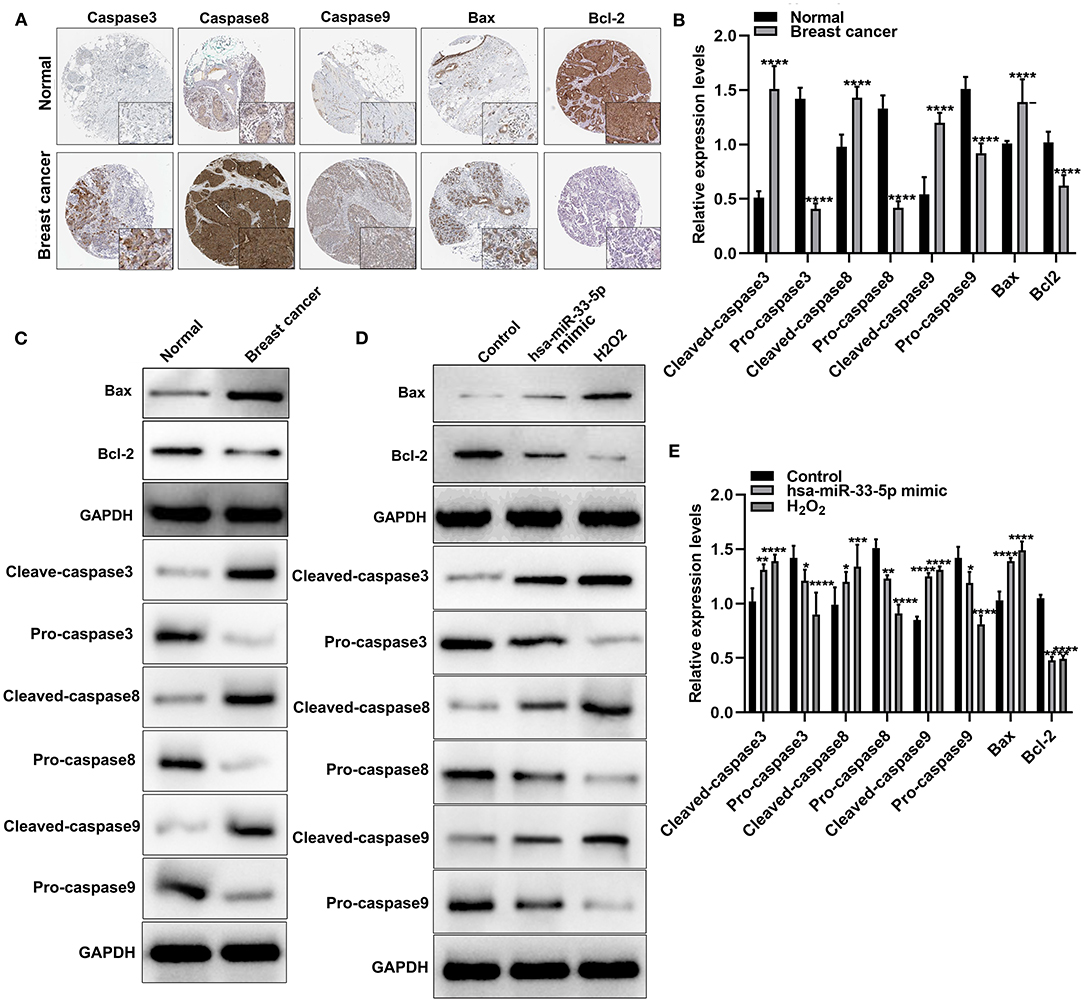
Figure 2. hsa-miR-33-5p significantly mediates apoptosis-related proteins in breast carcinoma cells. (A) Immunohistochemistry for the expression and distribution of the apoptosis-related proteins including caspase-3/8/9, Bax, and Bcl-2 in breast cancer tissues and normal tissues. (B,C) Western blot for the expression of apoptosis-related proteins including cleaved caspase-3, pro-caspase-3, cleaved caspase-8, pro-caspase-8, cleaved caspase-9, pro-caspase-9, Bax, and Bcl-2 in breast cancer tissues and normal tissues. (D,E) Western blot for the expression of cleaved caspase-3, pro-caspase-3, cleaved caspase-8, pro-caspase-8, cleaved caspase-9, pro-caspase-9, Bax, and Bcl-2 in breast cancer cells transfected with hsa-miR-33-5p mimics. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
As demonstrated by immunohistochemistry and RT-qPCR results, SelT expression had a significantly higher level in breast carcinoma than normal tissue samples (Figures 3A,B; p < 0.0001). Similarly, its upregulation was found in MCF-7 and MDA-MB-231 cells more than MCF10A cells (Figure 3C; p < 0.001). The above-mentioned results confirmed the upregulation of SelT in both breast cancer tissues and cells. As predicted, SelT could be a target of hsa-miR-33-5p. As shown in Figures 3D,E, SelT was significantly suppressed in breast cancer cells following transfection by overexpressed hsa-miR-33-5p or H2O2 treatment at the protein level (p < 0.001). However, no statistical difference in SelT expression was observed between hsa-miR-33-5p overexpression and H2O2 treatment groups. The luciferase reporter assay results confirmed that SelT was directly targeted by hsa-miR-33-5p (Figure 3F).
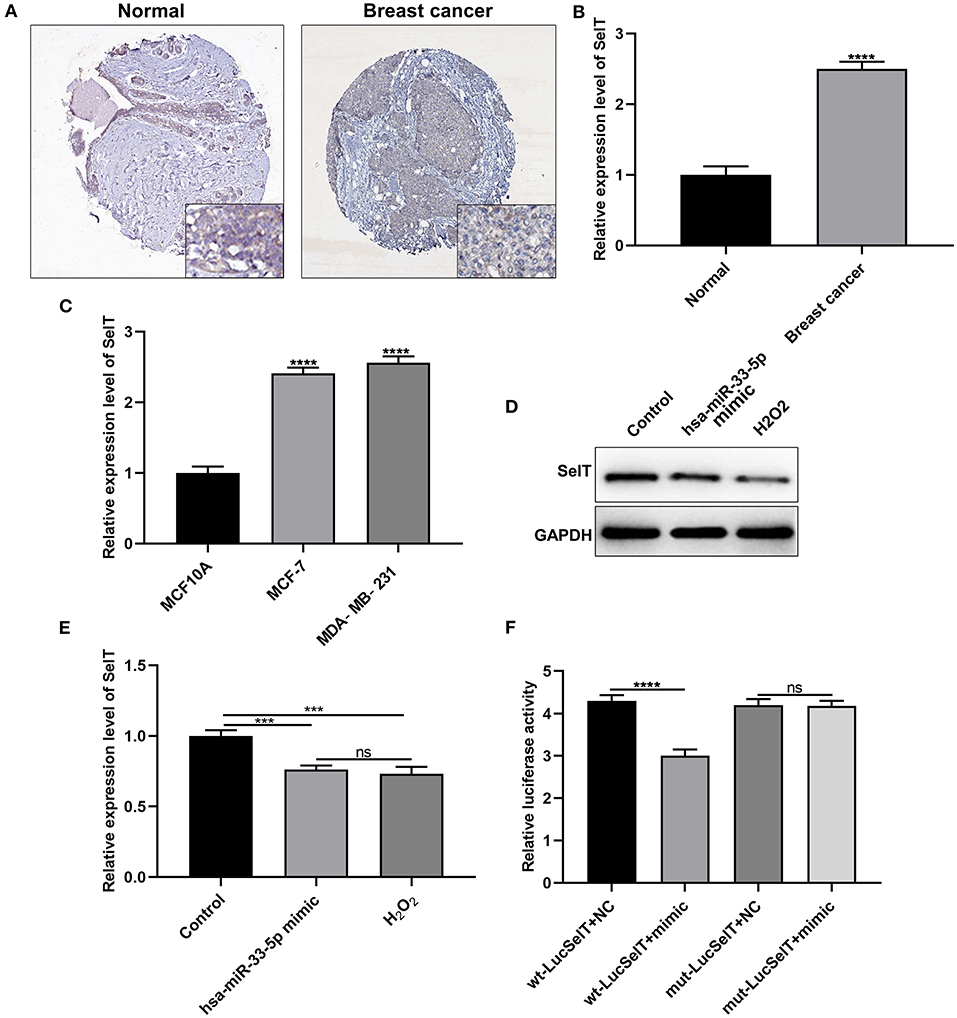
Figure 3. SelT is upregulated in breast carcinoma and is an underlying target of hsa-miR-33-5p. (A) Immunohistochemistry for the expression of SelT in breast cancer and normal tissues. (B,C) RT-qPCR results showing upregulated SelT both in breast cancer tissues (B) and cells (C). (D,E) Western blot results for the expression of SelT in breast cancer cells transfected with hsa-miR-33-5p mimics or treated with H2O2. (F) Luciferase reporter assay for hsa-miR-33-5p and SelT. ***p < 0.001; ****p < 0.0001; ns, not significant.
We further investigated whether SelT could promote the apoptosis of breast cancer cells. Two siRNAs targeting SelT were designed and transfected into breast cancer cells. As shown in Figures 4A,B, SelT expression was significantly suppressed by si-SelT#1 (p < 0.05) and si-SelT#2 (p < 0.01). Furthermore, SelT was successfully overexpressed in breast cancer cells (Figures 4C,D; p < 0.01). The flow cytometry data demonstrated that SelT knockdown markedly elevated the apoptotic levels of MCF-7 as well as MDA-MB-231 cells (Figures 4E,F). Meanwhile, SelT overexpression distinctly inhibited the apoptosis of MCF-7 as well as MDA-MB-231 cells (Figures 4G,H). The data demonstrated that inhibition of SelT may promote the apoptosis of breast cancer cells.
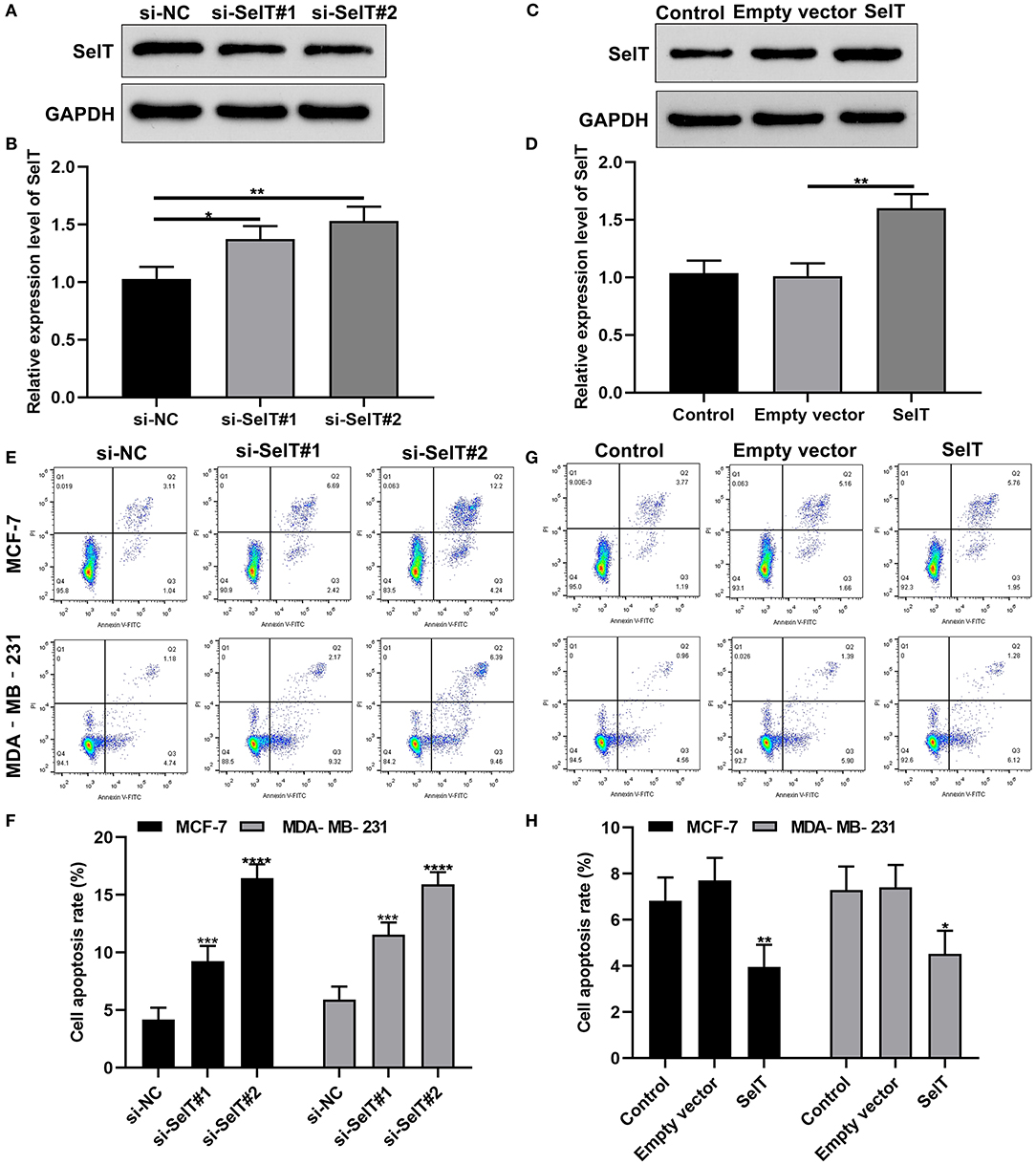
Figure 4. Inhibition of SelT promotes the apoptosis of breast cancer cells. (A,B) Western blot for the expression of SelT in breast cancer cells transfected with si-SelT#1 and si-SelT#2. (C,D) Validation of SelT expression in breast cancer cells transfected with SelT by western blot. (E,F) Flow cytometry assay for apoptotic levels of MCF-7 as well as MDA-MB-231 cells transfected by si-SelT#1 and si-SelT#2. (G,H) Assessment of breast carcinoma cell apoptosis under SelT overexpression. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Our further analysis found that hsa-miR-33-5p mimics promoted the apoptosis of MCF-7 (p < 0.0001) and MDA-MB-231 cells (p < 0.0001), which was significantly ameliorated by SelT overexpression (p < 0.0001), as shown in Figures 5A–D. Furthermore, TUNEL staining results confirmed that SelT overexpression distinctly ameliorated the increase in apoptosis induced by hsa-miR-33-5p mimics in MCF-7 (Figure 6A) and MDA-MB-231 (Figure 6B) cells. These data demonstrated that hsa-miR-33-5p may promote the apoptosis of breast cancer cells via SelT.
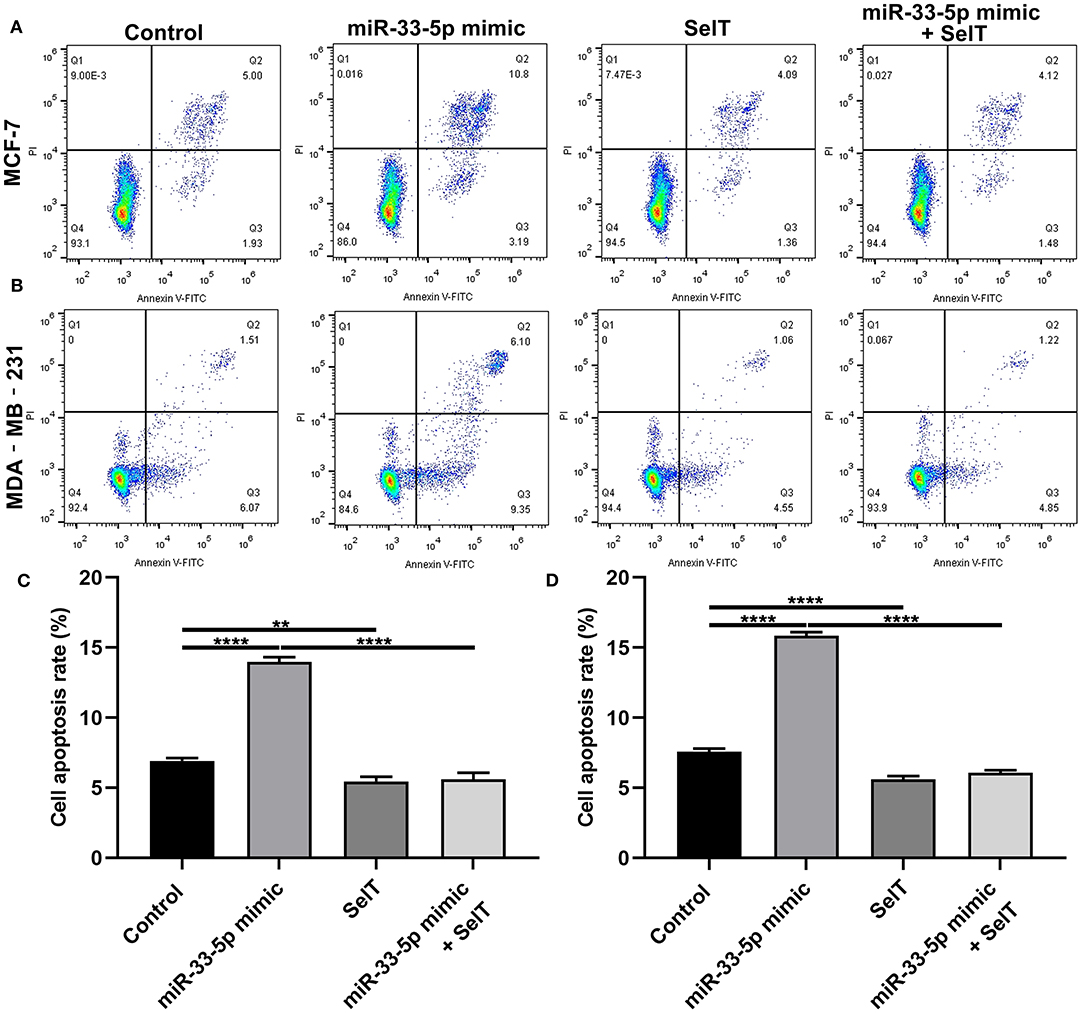
Figure 5. hsa-miR-33-5p promotes the apoptosis of breast cancer cells via SelT. (A,B) Representative images of flow cytometry assay results. (C) MCF-7 as well as (D) MDA-MB-231 cell apoptosis was determined when transfected by hsa-miR-33-5p mimics and/or SelT. **p < 0.01; ****p < 0.0001.
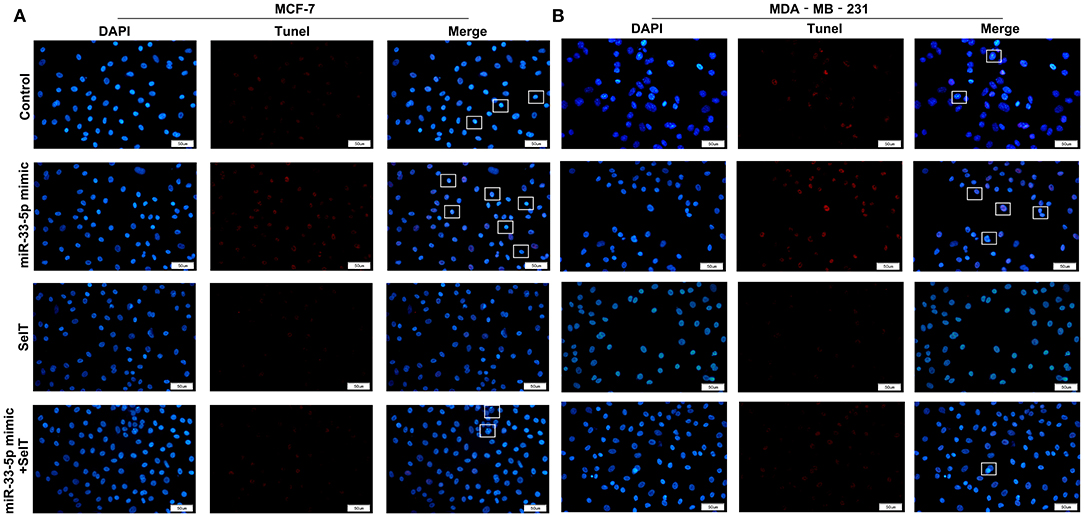
Figure 6. TUNEL staining for (A) MCF-7 as well as (B) MDA-MB-231 cell apoptosis transfected by hsa-miR-33-5p mimics and/or SelT. Bar = 50 μm. Magnification, ×400.
In these findings, we identified a novel miRNA, and found that hsa-miR-33-5p was downregulated in both breast cancer tissues and cells. Its overexpression significantly promoted the apoptosis of breast cancer cells. Furthermore, SelT was an underlying target of hsa-miR-33-5p in breast cancer. hsa-miR-33-5p could facilitate the apoptosis of breast cancer cells via SelT. Our findings deepened the understanding of molecular mechanisms in breast cancer progression and provided potential therapeutic targets.
Our findings demonstrated that hsa-miR-33-5p was dysregulated in breast cancer. So far, no studies have reported the expression and role of hsa-miR-33-5p in breast cancer. As previously reported, hsa-miR-33-5p could induce osteoblast differentiation through targeting Hmga2 (22, 24). Its knockdown could inhibit abdominal aortic aneurysm development through ABCA1 expression and activation of the PI3K/Akt pathway (25). Lowly expressed hsa-miR-33-5p is involved in inhibiting the apoptosis of murine dorsal root ganglion neurons (26). Furthermore, hsa-miR-33-5p is in association with mesangial cell apoptosis in diabetic nephropathy (27).
Resistance to apoptosis is one of the hallmarks of cancer (28). Apoptosis could maintain homeostasis via mediating senescent cell death (29). Tumor cells can resist apoptosis by upregulating anti-apoptotic proteins and/or reducing pro-apoptotic proteins (30). Apoptotic caspases mainly include initiator caspases (caspase-2/8/9/10) and executioner caspases (caspase-3/6/7) (31). Bax is an apoptotic protein, and Bcl2 is an important regulator of anti-apoptosis, both of which are involved in mitochondrial death signals (32). Thus, inducing apoptosis of tumor cells has a promising potential to eradicate cancer cells. In this study, we examined apoptosis-related proteins including caspase-3/8/9, Bax, and Bcl-2. We found that caspase-3/8/9 and Bax expressions were elevated in breast cancer tissues, while Bcl-2 expression was decreased in breast cancer tissues. These findings revealed that the apoptosis process could occur in breast cancer. As with previous findings, many miRNAs could participate in breast cancer cell apoptosis. For example, hsa-miR-106a (33), hsa-miR-205 (34), and hsa-miR-216a (35) could induce the apoptosis of breast carcinoma cells. We explored the regulatory effect of hsa-miR-33-5p in breast cancer. hsa-miR-33-5p overexpression was successfully induced in MCF-7 cells. Its overexpression significantly induced the apoptosis of breast carcinoma cells as shown in flow cytometry results. Oxidative stress is involved in the process of apoptosis. H2O2 is one of the major components of exogenous reactive oxygen species, which is considered to be a key factor in regulating tumor cell viability (36). Different cell types have different responses to H2O2-induced oxidative stress and cell viability both in a dose- and time-dependent manner (37). As previously described, in this study, H2O2 exposure significantly induced breast cancer cell apoptosis. Furthermore, H2O2 exposure promoted hsa-miR-33-5p expression in MCF-7 cells. hsa-miR-33-5p overexpression significantly elevated caspase-3, caspase-8, caspase-9, and Bax expressions, while Bcl-2 was significantly inhibited in MCF-7 cells with hsa-miR-33-5p overexpression. As expected, similar results were investigated in MCF-7 cells exposed with H2O2. The above-mentioned data confirmed that hsa-miR-33-5p expression can be induced by oxidative stress.
Upregulated SelT was found in both breast cancer tissues and cells. Its knockdown could promote the apoptosis of breast cancer cells. SelT is a recently characterized thioredoxin-like protein that is widely expressed during development (38). It is upregulated during neuroendocrine cell differentiation (39, 40). Furthermore, SelT expression is strictly regulated in time. In most adult tissues, the expression level of SelT is decreased (41, 42). Previous studies have found that SelT is dysregulated in a few cancers like gastric cancer (43) and bladder cancer (44). hsa-miR-33-5p could significantly inhibit the expression of SelT. As validated by the luciferase reporter assay, SelT was directly targeted by hsa-miR-33-5p. hsa-miR-33-5p could accelerate the apoptosis of breast cancer cells via SelT. Furthermore, the results showed that, when breast cancer cells were exposed to H2O2, SelT expression was significantly inhibited. SelT could regulate various biological processes, like apoptosis. It possesses oxidoreductase functions and is localized in the endoplasmic reticulum. A similar result has found that, when LO2 cells were exposed to H2O2, SelX expression was suppressed and cell apoptosis was induced (45). The above-mentioned findings were indicative of the fact that SelT may participate in oxidative stress response and apoptosis in breast cancer, which was consistent with a previous study (46).
In conclusion, we found that hsa-miR-33-5p could induce the apoptosis of breast cancer cells. Moreover, SelT was directly targeted by hsa-miR-33-5p in breast cancer. hsa-miR-33-5p may facilitate the apoptosis of breast cancer cells via SelT. In future studies, we will continue studying the regulatory relationships between hsa-miR-33-5p and SelT and their functions on breast carcinoma cell apoptosis in animal models.
In this study, downregulated hsa-miR-33-5p was investigated in breast cancer tissues and cells. Its overexpression could promote the apoptosis of breast cancer cells. Furthermore, SelT was targeted by hsa-miR-33-5p. These findings may offer novel therapeutic targets against breast carcinoma.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The studies involving human participants were reviewed and approved by The study was approved by the Ethics Committee of Linyi Cancer Hospital (LYZLYY-2018-012). The patients/participants provided their written informed consent to participate in this study.
WL conceived and designed the study. WZ conducted most of the experiments and data analysis and wrote the manuscript. JL participated in collecting data and helped to draft the manuscript. All the authors reviewed and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
miRNAs, microRNAs; SelT, selenoprotein T; UTR, untranslated region.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Mavaddat N, Michailidou K, Dennis J, Lush M, Fachal L, Lee A, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am J Hum Genet. (2019) 104:21–34. doi: 10.1016/j.ajhg.2018.11.002
3. Siegel RL, Jemal A, Wender RC, Gansler T, Ma J, Brawley OW. An assessment of progress in cancer control. CA Cancer J Clin. (2018) 68:329–39. doi: 10.3322/caac.21460
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. (2018) 68:7–30. doi: 10.3322/caac.21442
5. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
6. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
7. Fan CN, Ma L, Liu N. Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer. J Transl Med. (2018) 16:264. doi: 10.1186/s12967-018-1640-2
8. Pham VV, Zhang J, Liu L, Truong B, Xu T, Nguyen TT, et al. Identifying miRNA-mRNA regulatory relationships in breast cancer with invariant causal prediction. BMC Bioinformatics. (2019) 20:143. doi: 10.1186/s12859-019-2668-x
9. Wu Q, Guo L, Jiang F, Li L, Li Z, Chen F. Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER- breast cancer cell lines. J Cell Mol Med. (2015) 19:2874–87. doi: 10.1111/jcmm.12681
10. Breunig C, Pahl J, Kublbeck M, Miller M, Antonelli D, Erdem N, et al. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. (2017) 8:e2973. doi: 10.1038/cddis.2017.364
11. Sharma S, Nagpal N, Ghosh PC, Kulshreshtha R. P53-miR-191-SOX4 regulatory loop affects apoptosis in breast cancer. Rna. (2017) 23:1237–46. doi: 10.1261/rna.060657.117
12. Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. (2016) 35:448–58. doi: 10.1038/onc.2015.96
13. Aggarwal V, Priyanka K, Tuli HS. Emergence of circulating MicroRNAs in breast cancer as diagnostic and therapeutic efficacy biomarkers. Mol Diagn Ther. (2020) 24:153–73. doi: 10.1007/s40291-020-00447-w
14. Huang R, Li J, Pan F, Zhang B, Yao Y. The activation of GPER inhibits cells proliferation, invasion and EMT of triple-negative breast cancer via CD151/miR-199a-3p bio-axis. Am J Transl Res. (2020) 12:32–44.
15. Kanchan RK, Siddiqui JA, Mahapatra S, Batra SK, Nasser MW. microRNAs Orchestrate pathophysiology of breast cancer brain metastasis: advances in therapy. Mol Cancer. (2020) 19:29. doi: 10.1186/s12943-020-1140-x
16. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. (2015) 5:1122–43. doi: 10.7150/thno.11543
17. Aushev VN, Lee E, Zhu J, Gopalakrishnan K, Li Q, Teitelbaum SL, et al. Novel Predictors of breast cancer survival derived from miRNA activity analysis. Clin Cancer Res. (2018) 24:581–91. doi: 10.1158/1078-0432.Ccr-17-0996
18. Cai WL, Huang WD, Li B, Chen TR, Li ZX, Zhao CL, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer. (2018) 17:9. doi: 10.1186/s12943-017-0746-0
19. Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M, et al. Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis. (2017) 8:e3045. doi: 10.1038/cddis.2017.440
20. McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. (2015) 34:145–55. doi: 10.1007/s10555-015-9551-7
21. Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. (2017) 172:34–49. doi: 10.1016/j.pharmthera.2016.11.012
22. Wang H, Sun Z, Wang Y, Hu Z, Zhou H, Zhang L, et al. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci Rep. (2016) 6:23170. doi: 10.1038/srep23170
23. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. (2015) 12:697. doi: 10.1038/nmeth.3485
24. Wang H, Hu Z, Shi F, Dong J, Dang L, Wang Y, et al. Osteoblast-targeted delivery of miR-33-5p attenuates osteopenia development induced by mechanical unloading in mice. Cell Death Dis. (2018) 9:170. doi: 10.1038/s41419-017-0210-5
25. Zhao L, Huang J, Zhu Y, Han S, Qing K, Wang J, et al. miR-33-5p knockdown attenuates abdominal aortic aneurysm progression via promoting target adenosine triphosphate-binding cassette transporter A1 expression and activating the PI3K/Akt signaling pathway. Perfusion. (2020) 35:57–65. doi: 10.1177/0267659119850685
26. Zhang H, Wang K. Downregulation of MicroRNA-33-5p protected bupivacaine-induced apoptosis in murine dorsal root ganglion neurons through GDNF. Neurotox Res. (2019) 35:860–6. doi: 10.1007/s12640-018-9994-z
27. Tsai YC, Kuo PL, Hung WW, Wu LY, Wu PH, Chang WA, et al. Angpt2 induces mesangial cell apoptosis through the MicroRNA-33-5p-SOCS5 loop in diabetic nephropathy. Mol Ther Nucleic Acids. (2018) 13:543–55. doi: 10.1016/j.omtn.2018.10.003
28. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
29. Basu SC, Boyle P, Ma R, Agarwal A, Basu M, Moskal JR, et al. Induction of apoptosis in metastatic breast cancer cells: XV. downregulation of DNA polymerase-alpha—helicase complex (Replisomes) and glyco-genes. Adv Exp Med Biol. (2018) 1112:199–221. doi: 10.1007/978-981-13-3065-0_15
30. Thapa B, Bahadur Kc R, Uludag H. Novel targets for sensitizing breast cancer cells to TRAIL-induced apoptosis with siRNA delivery. Int J Cancer. (2018) 142:597–606. doi: 10.1002/ijc.31079
31. Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. (2002) 9:459–70. doi: 10.1016/s1097-2765(02)00482-3
32. Mirzapur P, Khazaei MR, Moradi MT, Khazaei M. Apoptosis induction in human breast cancer cell lines by synergic effect of raloxifene and resveratrol through increasing proapoptotic genes. Life Sci. (2018) 205:45–53. doi: 10.1016/j.lfs.2018.04.035
33. You F, Luan H, Sun D, Cui T, Ding P, Tang H, et al. miRNA-106a Promotes breast cancer cell proliferation, clonogenicity, migration, and invasion through inhibiting apoptosis and chemosensitivity. DNA Cell Biol. (2019) 38:198–207. doi: 10.1089/dna.2018.4282
34. Hu Y, Qiu Y, Yague E, Ji W, Liu J, Zhang J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. (2016) 7:e2291. doi: 10.1038/cddis.2016.194
35. Cui Y, Wang J, Liu S, Qu D, Jin H, Zhu L, et al. miR-216a promotes breast cancer cell apoptosis by targeting PKCalpha. Fundam Clin Pharmacol. (2019) 33:397–404. doi: 10.1111/fcp.12481
36. Orrenius S. Reactive oxygen species in mitochondria-mediated cell death. Drug Metab Rev. (2007) 39:443–55. doi: 10.1080/03602530701468516
37. Han Y-H, Zhang Z-W, Su J, Zhang B, Li S, Xu S-W. Effects of chicken selenoprotein W on H2O2-induced apoptosis in CHO-K1 cells. Biol Trace Elem Res. (2012) 147:395–402. doi: 10.1007/s12011-011-9311-7
38. Hamieh A, Cartier D, Abid H, Calas A, Burel C, Bucharles C, et al. Selenoprotein T is a novel OST subunit that regulates UPR signaling and hormone secretion. EMBO Rep. (2017) 18:1935–46. doi: 10.15252/embr.201643504
39. Boukhzar L, Hamieh A, Cartier D, Tanguy Y, Alsharif I, Castex M, et al. Selenoprotein T exerts an essential oxidoreductase activity that protects dopaminergic neurons in mouse models of parkinson's disease. Antioxid Redox Signal. (2016) 24:557–74. doi: 10.1089/ars.2015.6478
40. Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, Tanguy Y, et al. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. Faseb J. (2008) 22:1756–68. doi: 10.1096/fj.06-075820
41. Tanguy Y, Falluel-Morel A, Arthaud S, Boukhzar L, Manecka DL, Chagraoui A, et al. The PACAP-regulated gene selenoprotein T is highly induced in nervous, endocrine, and metabolic tissues during ontogenetic and regenerative processes. Endocrinology. (2011) 152:4322–35. doi: 10.1210/en.2011-1246
42. Prevost G, Arabo A, Jian L, Quelennec E, Cartier D, Hassan S, et al. The PACAP-regulated gene selenoprotein T is abundantly expressed in mouse and human beta-cells and its targeted inactivation impairs glucose tolerance. Endocrinology. (2013) 154:3796–806. doi: 10.1210/en.2013-1167
43. Lan X, Xing J, Gao H, Li S, Quan L, Jiang Y, et al. Decreased Expression of Selenoproteins as a poor prognosticator of gastric cancer in humans. Biol Trace Elem Res. (2017) 178:22–8. doi: 10.1007/s12011-016-0908-8
44. Reszka E. Selenoproteins in bladder cancer. Clin Chim Acta. (2012) 413:847–54. doi: 10.1016/j.cca.2012.01.041
45. Tang J, Cao L, Li Q, Wang L, Jia G, Liu G, et al. Selenoprotein X gene knockdown aggravated H2O2-induced apoptosis in liver LO2 cells. Biol Trace Elem Res. (2016) 173:71–8. doi: 10.1007/s12011-016-0653-z
Keywords: breast cancer, hsa-miR-33-5p, Selenoprotein T, apoptosis, therapeutic target
Citation: Zhuang W, Liu J and Li W (2021) hsa-miR-33-5p as a Therapeutic Target Promotes Apoptosis of Breast Cancer Cells via Selenoprotein T. Front. Med. 8:651473. doi: 10.3389/fmed.2021.651473
Received: 09 January 2021; Accepted: 15 March 2021;
Published: 27 April 2021.
Edited by:
Fu Wang, Xi'an Jiaotong University, ChinaReviewed by:
Chaoyun Pan, Sun Yat-sen University, ChinaCopyright © 2021 Zhuang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjin Li, bHl6bGx3akBzaW5hLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.