
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 25 June 2021
Sec. Obstetrics and Gynecological Surgery
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.645118
 Giovanni Monni1*
Giovanni Monni1* Luigi Atzori2
Luigi Atzori2 Valentina Corda1
Valentina Corda1 Francesca Dessolis1
Francesca Dessolis1 Ambra Iuculano1
Ambra Iuculano1 K. Joseph Hurt3†
K. Joseph Hurt3† Federica Murgia1,2*†
Federica Murgia1,2*†Pregnancy is a complicated and insidious state with various aspects to consider, including the well-being of the mother and child. Developing better non-invasive tests that cover a broader range of disorders with lower false-positive rates is a fundamental necessity in the prenatal medicine field, and, in this sense, the application of metabolomics could be extremely useful. Metabolomics measures and analyses the products of cellular biochemistry. As a biomarker discovery tool, the integrated holistic approach of metabolomics can yield new diagnostic or therapeutic approaches. In this review, we identify and summarize prenatal metabolomics studies and identify themes and controversies. We conducted a comprehensive search of PubMed and Google Scholar for all publications through January 2020 using combinations of the following keywords: nuclear magnetic resonance, mass spectrometry, metabolic profiling, prenatal diagnosis, pregnancy, chromosomal or aneuploidy, pre-eclampsia, fetal growth restriction, pre-term labor, and congenital defect. Metabolite detection with high throughput systems aided by advanced bioinformatics and network analysis allowed for the identification of new potential prenatal biomarkers and therapeutic targets. We took into consideration the scientific papers issued between the years 2000–2020, thus observing that the larger number of them were mainly published in the last 10 years. Initial small metabolomics studies in perinatology suggest that previously unidentified biochemical pathways and predictive biomarkers may be clinically useful. Although the scientific community is considering metabolomics with increasing attention for the study of prenatal medicine as well, more in-depth studies would be useful in order to advance toward the clinic world as the obtained results appear to be still preliminary. Employing metabolomics approaches to understand fetal and perinatal pathophysiology requires further research with larger sample sizes and rigorous testing of pilot studies using various omics and traditional hypothesis-driven experimental approaches.
The “-omics revolution” has brought promising options for high-throughput network-based analysis of DNA, RNA, proteins, and metabolites. Integrated analyses of the genome, transcriptome, proteome, and metabolome can reveal normal and pathophysiologic mechanisms that may not be detected by traditional hypothesis-driven experiments or focussed diagnostics (1). The analysis of end products of cellular or organismal biochemical processes using metabolomics approaches could revolutionize our approach to medical diagnosis and therapeutics. Metabolomics employs cutting-edge technologies to assess the presence of low molecular weight compounds, such as carbohydrates, amino acids, peptides, nucleic acids, organic acids, vitamins, and lipids, produced by cells, organs, or whole organisms (2, 3). The interaction and relationships among metabolites can be qualitatively and quantitatively characterized using powerful contemporary computational algorithms and increasingly effective informatics software (4). As a holistic characterization of physiology, metabolomics reflects genetics, environment, and the response to environmental stressors, and can reveal specific metabolic signatures due to genetics, disease, drugs, infection, nutrition, or exposures. Identifying metabolites or metabolic patterns that reflect specific disease states is a primary goal for metabolomics (5) research. In this review, we describe the techniques used in metabolomics, provide examples of metabolomics studies in prenatal care and maternal–fetal medicine, and highlight the tremendous opportunities for metabolomics applications in prenatal diagnosis.
The compounds produced by cellular metabolism are extremely diverse, involving wide variation in physicochemical properties and concentration. Their detection must be addressed by a suitable and adequate quantitative approach, and in fact, no single experimental assay can capture the full range of metabolic output (2). Therefore, investigators use multiple instruments and several analytic approaches. Two of the most common methods are metabolic profiling and metabolic fingerprinting (6). Metabolic profiling is the quantitative measurement of specific metabolites in a selected biochemical pathway or a particular class of compounds. Similarly, metabolic fingerprinting is a global screening approach that identifies metabolite patterns or “fingerprints” that are associated with known biochemical pathways, specific responses to external stimuli or endogenous signals, or disease processes. With metabolic fingerprinting, we attempt to identify discriminating metabolites that reflect specific alterations, responses, or dysfunction. Depending on the specific scientific question and the analytical approach, metabolomic analyses can be informative, discriminative, or predictive (7) (Figure 1).
Informative metabolomic analyses identify and quantify targeted or untargeted metabolites. This approach has been used to develop metabolic databases and to identify critical pathophysiologic pathways, bioactive molecules, and function or disease biomarkers. Discriminative analyses, in contrast, identify metabolic endpoint differences between samples from control and disease populations or before and after specific treatments or perturbation. This approach is typically performed using regression and other multivariate analyses to define clear diagnostic thresholds. Finally, in predictive studies, investigators use metabolic fingerprinting models based on a statistical analysis of metabolite profiles to generate predictive global algorithms that are difficult or impossible to achieve with other more focussed approaches.
Recent advances in robust high-throughput techniques such as 1H-Nuclear Magnetic Resonance spectroscopy (NMR) and Mass Spectrometry (MS) permit simultaneous measurement of many metabolites from a single biological sample (8–12) and more efficient metabolomic studies. Table 1 summarizes the key advantages and limitations of NMR and MS approaches in metabolomics research.
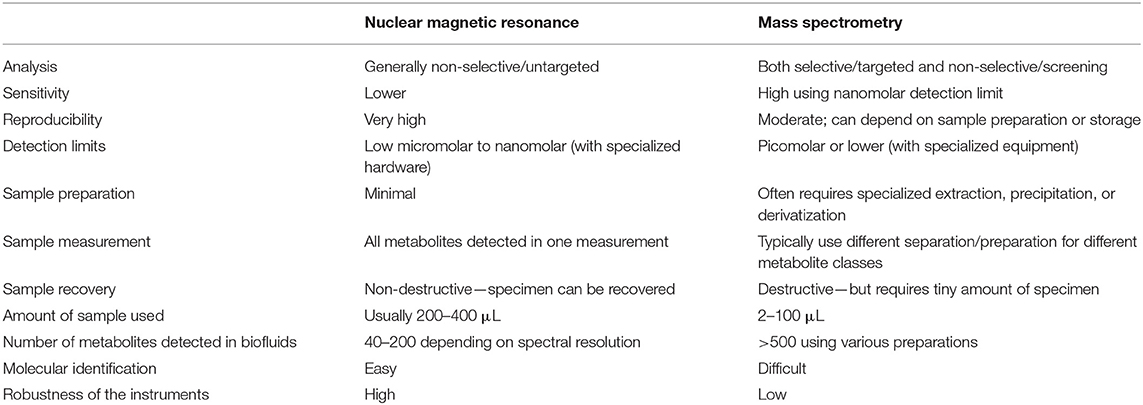
Table 1. The advantages and limitations of NMR spectroscopy and MS spectrometry as an analytical tool for metabolomics research.
Although NMR and MS are powerful techniques, the voluminous raw metabolomics data produced is only valuable after careful organization and interpretation with sophisticated contemporary bioinformatics and biostatistical tools (13, 14). Multivariate or principal component analysis transforms an assortment of metabolites into informative profiles by comparing critical elements (or components) that define health, disease, degree of disease, or other conditions and exposures. Supervised learning (e.g., machine learning) is used to transform multivariate data from metabolite profiles into patterns that are biologically relevant (15–18). The general idea behind multivariate methods is to find, if possible, distinct metabolite profiles most strongly associated with the studied phenomenon (19). Once key metabolites have been identified, metabolic network analysis can generate hypotheses for a particular condition(s) (16). Other metabolomics analysis approaches include standard methods such as parametric/non-parametric univariate tests and ANOVA and more sophisticated methodologies. The use of multiple complementary statistical methods allows the investigator to extract the most important information from a single experiment (19). Figure 2 shows the typical workflow for metabolomics experiments and analysis.
For this review of perinatal metabolomics, we conducted a comprehensive search of PubMed and Google Scholar throughout January 2020 using the following key terms: metabolomics, nuclear magnetic resonance, mass spectrometry, and metabolic profiling. We combined these with the following clinical terms: normal pregnancy, prenatal diagnosis, gestational disorders, chromosomal disorders, pre-eclampsia, fetal growth restriction, pre-term labor, and congenital anatomic defects. We identified additional articles by searching the reference lists of the identified studies. We considered peer-reviewed journal articles that appropriately described their methods and included only unique subjects, data, or analyses. We took into consideration the scientific papers issued between the years 2000–2020, thus observing that the larger number of them were mainly published in the last 10 years. Only papers dealing with the above-mentioned topics were included, those on other pathological conditions were omitted. Systematic reviews and meta-analyses were also excluded from the review together with studies focused on non-human subjects or studies published only in conference proceedings. Of 124 studies identified by the initial search, about 60 studies met the inclusion criteria. Table 2 reports the summary of each study. In particular, the data points taken from each study included the first author, year of publication, analytical technique, sample type and size, type of biomaterial examined, and results. Despite the fact that there have been a good number of pregnancy-related metabolite and metabolomics studies, the use of metabolomics specifically in prenatal diagnosis and pregnancy prediction has been more limited (63–65) although the number of pregnancy metabolomics studies is increasing (Figure 3). Next, we will review recent metabolomics publications for prenatal diagnosis and specific pregnancy pathologies.
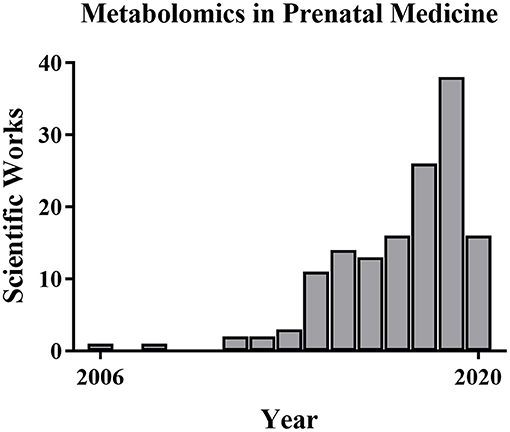
Figure 3. Metabolomics publications in prenatal medicine. The number of metabolomics publications in prenatal medicine is low but increased from 2006 to 2019 based on PubMed searches.
Contemporary fetal assessment uses both non-invasive and invasive methods. Non-invasive methods include maternal factors and history, fetal ultrasound imaging, and maternal serum analyte or cell-free fetal DNA (cfDNA) screening. Maternal blood analyte tests, such as the first trimester screen (66), sequential screen, and quadruple test (67), and maternal serum cfDNA screening carry no procedural risk for the pregnancy, but these only assess risk and cannot diagnose. In contrast, invasive diagnostic tests require fetal samples of the placenta (chorionic villus sampling) or skin cells (amniocentesis) but provide definitive results. Invasive testing incurs a small chance of procedural complications, including a 0.1–0.2% risk of fetal loss (68). The earliest possible diagnosis of fetal aneuploidy and other congenital defects is highly desired because it affords more time for decision making and reduces risks of pregnancy interruption if that is what the couple chooses.
In first trimester non-invasive screening, we estimate risk from a combination of maternal factors (e.g., age, weight, and medical considerations, such as diabetes), fetal factors (e.g., nuchal translucency [NT], nasal bone, and fetal heart rate), and feto-placental factors in maternal blood (ß-hCG and PAPP-A). The combined first trimester screen detects trisomies 21, 18, and 13 with 90, 97, and 92% sensitivity but with a set positive rate of 5% (69). The second trimester quadruple test assesses α-fetoprotein, ß-hCG, estriol, and inhibin-A and detects trisomy 21 with ~80% sensitivity and a false positive rate of 5% (67). The newest non-invasive screening test is maternal serum cfDNA which exhibits very high sensitivity and specificity for trisomies 21, 18, and 13 by examining free chromosomal DNA fragments in maternal blood that are released by normal placental apoptosis (70). Although screening tests are all helpful for alerting us about fetuses at higher risk for aneuploidy, their positive predictive value varies dramatically depending on maternal age-related risk. Many women decline invasive testing despite the minimal risk and high diagnostic accuracy of CVS or amniocentesis. Therefore, developing better non-invasive tests that cover a broader range of disorders and have lower false-positive rates is a critical need in the field (71). Omics approaches could be a reasonable solution.
High-dimensional biology techniques (i.e., multi-omics) that combine genomic, transcriptomic, proteomic, and metabolomic approaches have been applied in both normal and complicated pregnancies (72–86). Because metabolomics provides a final readout of cellular physiology, it may be the best single omics approach to characterize normal vs. pathologic pregnancies. Examining different maternal-fetal compartments at different stages of pregnancy and clearly characterizing changes throughout gestation is required in order to determine the best tests and timing for particular clinical samples. Ideally, materia for non-invasive prenatal metabolomics would be obtained from maternal blood (plasma or serum) (87), urine (88), or cervico-vaginal secretions (89). Invasive sampling can also be used to obtain amniotic fluid (90), placental biopsy (23), and cord blood (43). Samples from invasive approaches might be useful to characterize the relationship between fetal and maternal metabolites or eventually might be used for confirmatory testing as it is today. Thorough metabolomic characterization of pregnancy across gestation might suggest new diagnostic or treatment options. So far, gestational metabolomics has been used to characterize normal physiologic changes of pregnancy as well as common complications including fetal aneuploidy, pre-eclampsia, pre-term birth, fetal growth restriction (FGR), pre-term parturition, congenital fetal anomalies, and single-gene disorders. A summary of the main investigated perinatal issues and the specific identified metabolic pattern is represented in Figures 4A,B. Below, we review recent work in each of these areas.
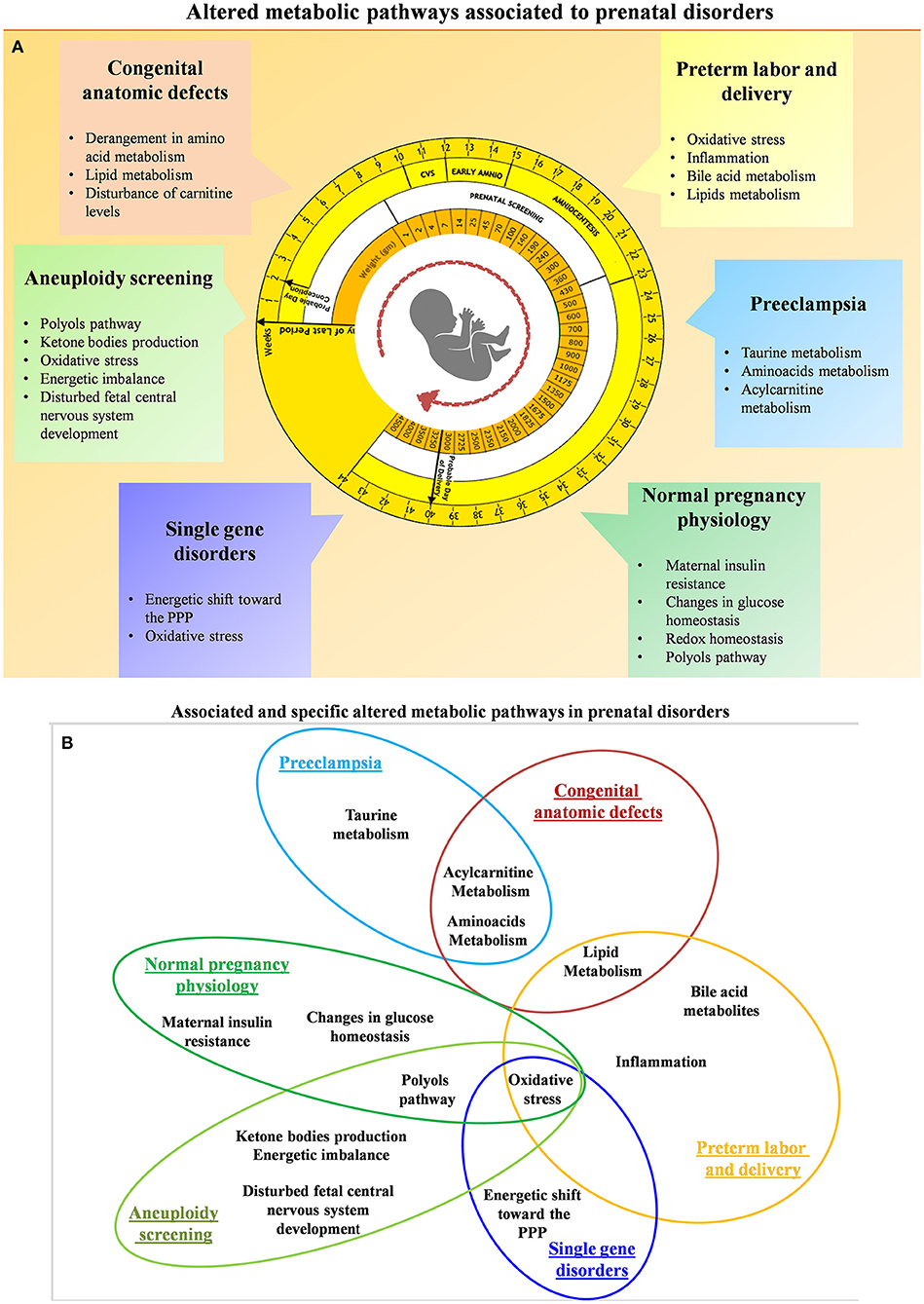
Figure 4. (A) Summary of the altered metabolic pathways associated with prenatal disorders. (B) Associated and specific altered metabolic pathways in prenatal disorders.
Pregnancy requires a wide range of adaptive physiologic changes throughout gestation in maternal, fetal, and placental function. For example, circulating maternal metabolic products such as triglycerides, cholesterol, free fatty acids, and phospholipids change dramatically during pregnancy (91) to satisfy fetal energy and catabolic needs in utero and the production of an adequate maternal milk supply post-partum (92). Maternal insulin resistance also increases significantly with gestational age in normal pregnancy to ensure adequate transfer of glucose to the fetus (93). Maternal inositols increase with gestational age, positively correlating with fetal crown-rump length. They are correlated with insulin sensitivity and could be mechanistically linked to glucose homeostasis (91).
The placenta plays a key role in regulating the metabolic milieu of pregnancy. Several studies have described an altered placental metabolic profile, due to environmental factors. For example, the oxygen tension during placental explant culture in vitro can dramatically alter metabolic signatures. The human placenta is adapted for an initial hypoxic environment (20) then switches metabolism to accommodate increased oxygenation in the second trimester when extensive spiral artery remodeling occurs (94). The intervillous oxygen tension increases from 2–3% at 8 weeks to 8.5% by 12 weeks (95) of gestation with concurrent increased placental oxidative stress. Upregulation of placental antioxidant factors maintains redox homeostasis (96), and the metabolic profile of normal placental villus explants varies with oxygen culture conditions (21). Changes in hexadecanoic acid, erythritol, and 2-deoxyribose are particularly prominent. Moreover, placental cholesterol levels were found to be increased in correlation with the CRL. The higher concentration of cholesterol may be the result of increased levels of progestational hormones. In fact, maternal blood cholesterol represents the precursor of both progesterone and estrogen (23). Other gestational age-related metabolic changes (22, 23) include placental polyol pathways which are very active in the first trimester. One hypothesis regarding elevated polyols in early pregnancy is that they are an early carbohydrate source for the placenta and embryo. Additionally, the polyol pathway may facilitate the re-oxidation of pyridine nucleotides under low oxygen conditions, helping regulate intracellular pH during periods of robust glycolysis (23). These are just a few examples of the way in which metabolomics has been used to characterize the normal physiologic changes of pregnancy.
Chromosomal abnormalities are the most frequent fetal problem diagnosed in the first trimester. Trisomy 21 (T21; Down syndrome), trisomy 18 (T18; Edwards syndrome), trisomy 13 (T13; Patau syndrome), and abnormal sex chromosomes (e.g., XO, Turner syndrome, or Monosomy X) are the most commonly screened chromosome errors. Standard routine prenatal care includes screening for these common aneuploidies (97). Although current screening tests (see above) successfully identify fetal aneuploidy in early pregnancy, the search for additional genetic biomarkers for a wider range of aneuploidies and improved sensitivity and specificity is an important goal in the science of prenatal diagnosis (24).
Several studies have examined the utility of metabolic screening for aneuploidy. For example, Bahado-Singh et al. used NMR to analyze first trimester maternal serum from T21 and control pregnancies (25). They found 11 metabolites differed between groups, including the novel biomarkers 3-hydroxybutyrate and 2-hydroxybutyrate. The first is an indispensable energy source for extrahepatic tissues such as the brain, involved in growth and myelination; the second is involved in oxidative stress defense (25). Bahado-Singh similarly studied first trimester maternal serum from normal and T18 pregnancies (26) and found that glycerol and 2-hydroxybutyrate best identified T18 fetuses. Combining discriminatory metabolites with clinical and demographic parameters, they detected T18 with 90% sensitivity and 100% specificity using the delta nuchal translucency (98) and 2-hydroxybutryate levels together. Several other metabolites appear to distinguish between T18 and T21 fetuses including threonine, trimethylamine, creatine, and formate (26). Troisi et al. published similar findings for pregnancies affected by any trisomy (combined group of T21, T18, and T13) vs. normal controls (24). A specific pattern of metabolites was altered in trisomic compared to normal fetuses, suggesting a metabolic environment of elevated oxidative stress and disturbed fetal central nervous system development. Yet another group found associations between T21 and abnormal high-density lipoprotein (HDL), methanol, and proline in the first trimester and abnormal creatinine, acetate, HDL, and low-density lipoprotein (LDL) in the second trimester (27). Overall, these studies suggest increased oxidative stress in aneuploid pregnancies and switched fuel metabolism resulting in β-oxidation and ketone body production. Aneuploid fetuses appear to utilize glucose, pyruvate, and citrate less as energy sources compared with normal pregnancies.
In addition to maternal plasma and serum, some metabolomics studies of aneuploidy have evaluated maternal urine specimens. Maternal urine metabolic signatures for chromosomal disorders generally showed altered 3-hydroxybutyrate, 2-ketoglutarate, and 1-methylhistidine. Urine metabolites specifically associated with T21 included N-methyl-2-pyridone-5-carboxamide (28). Trivedi and Iles clarified the altered cellular metabolism with T21, suggesting that metabolic profiles may improve detection of both aneuploidy and inborn errors of metabolism (29). Comparing the mass spectrometry urine metabolome of women with aneuploid or normal fetuses revealed altered levels of progesterone and dihydrouracil (29).
Our own laboratory has characterized metabolic networks from the first trimester placenta obtained via transabdominal chorionic villus sampling (TA-CVS) (23). CVS placental biopsies are ideal for metabolic analyses as they are obtained from the undisturbed placenta in situ rather than after delivery, miscarriage, or termination. We compared normal and aneuploid fetuses (T21, T18, T13) using NMR, GC-MS, and HPLC and found critical differences in energy metabolism and polyol pathways. The aneuploid placenta demonstrates excessive polyol pathway activation, decreased glutathione levels, and increased dehydroascorbate. Additionally, thorough characterization of the placental metabolome may significantly improve our ability to interpret changes in the maternal metabolic profile caused by fetal or placental dysfunction.
Pre-eclampsia (PE) is a gestational hypertensive syndrome that complicates 2–8% of pregnancies worldwide (99) and is a major cause of maternal/fetal morbidity and mortality. The current definition of PE, according to the International Society for the Study of Hypertension in Pregnancy (ISSHP) (100) and the American College of Obstetricians and Gynecologists (ACOG) (101), is a new onset of hypertension (blood pressure ≥140 mmHg systolic or ≥90 mmHg diastolic) at ≥20 weeks of gestation and proteinuria (≥300 mg/24 h or protein-to-creatinine ratio >30 mg/mmol or ≥2+ on dipstick testing) or cases without proteinuria but with severe range blood pressure (>160/110) or evidence of organ dysfunction (e.g., hematologic, renal, hepatic, and neurologic). This new definition of PE resulted in an increase in pregnancies diagnosed with PE but generally milder disease (102).
Although the exact cause has not been identified, PE is thought to be due to the interaction of maladaptive trophoblast-derived signaling factors [e.g., soluble FMS-like tyrosine kinase-1 (sFlt-1) and placental growth factor (PlGF)] (103, 104) with susceptible maternal physiology. One theory is that PE is triggered by reduced uterine perfusion leading to placental oxidative stress and apoptotic release of pro-pre-eclampsia signals. Despite decades of PE research, the only available treatment remains delivery (i.e., removal of the placenta), and the only marginally helpful prevention comes from maternal risk reduction and possibly low-dose aspirin prophylaxis (105). Combined screening using maternal factors, mean arterial pressure, uterine artery, Pulsatility Index (PI), and maternal serum PlGF has been proposed to predict about 90% of early PE (Early-PE; <34 weeks), 75% of late pre-term PE (Late Pre-term-PE; 34 to <37 weeks) and 45% of full-term PE (Term-PE; ≥37 weeks) (106). Identifying and integrating new biomarkers to predict PE prior to symptoms could facilitate diagnosis and prevention. Given the heterogeneous presentation of gestational hypertensive disorders, it is unlikely that only one or several biomarkers will predict all types of PE (e.g., gestational hypertension, PE, PE with severe features, HELLP, and eclampsia). Therefore, broad network analysis using metabolomics may be the most appropriate approach to solving this important recalcitrant clinical problem.
Several groups have examined differences in maternal metabolomics in normal and PE pregnancies. In vitro studies of villous trophoblast explants using LC-MS identified differences in culture media metabolites for normal and PE patients (30). Furthermore, the metabolic profile changes in response to different oxygen conditions varied between normal and PE placenta, suggesting not only baseline differences in PE metabolism but persistent functional differences in the response environmental stressors. Studies have not determined whether the changes in hypoxia responses in vitro preceded PE or were caused by it (30). In vivo changes due to PE have been studied in maternal urine and serum (31). Women with PE have increased urine markers of oxidative stress and renal or liver dysfunction as well as altered serum metabolites. In particular, higher total lipid content and lipoprotein levels were observed with PE (31). One study used metabolomics and Western blotting to examine placental mitochondrial funding during severe PE. Isolated placental mitochondria from severe preeclamptics showed reduced ATP levels, higher fatty acid levels, and decreased fatty acid catabolism (32).
Some of the most intriguing prenatal diagnostic studies in PE research have retrospectively assessed early metabolic changes from normal control pregnancies vs. PE cases. For instance, Bahado-Singh et al. investigated serum metabolic profiles in patients with early and late-onset PE compared with healthy controls using specific metabolic panels (33, 34). The metabolites 3-hydroxyisovalerate, arginine, and glycerol were particularly increased in Early-PE, especially when combined with uterine artery PI. An approach using metabolic, proteomic, and ultrasound parameters reached over 90% sensitivity and nearly 90% specificity for predicting PE. An important milestone in the PE study has been filed when metabolomic was combined with proteomic approaches (35) and clinical maternal features to enhance omics diagnosis from early pregnancy maternal blood samples. Significant changes in G-protein-coupled receptors, signal transduction serotonin, and glycosaminoglycan metabolisms emerged as the final result.
Similarly, Koster et al. constructed a prediction model using maternal clinical factors (e.g., maternal age, history of pre-eclampsia, other risk factors) for baseline risk and then adding protein and metabolite measures (36). More in detail, acylcarnitines of first-trimester maternal serum from women with and without PE were analyzed and correlated with the clinical factors. Interestingly, the correlation between markers selected for prediction modeling (hexanoylcarnitine, octanoylcarnitine, decenoylcarnitine, and decanoylcarnitine) showed an R > 0.8, suggesting a potential role of this class of compounds. That approach reached ~70% sensitivity and 90% specificity for Early-PE. For Late Pre-term-PE, the sensitivity and specificity were only ~30 and 90%. Moreover, these findings suggested the stearoylcarnitine as a biomarker for both EO- and LO-PE. The concentration of this biomolecule improved the prediction power of the clinical factors normally used to evaluate the baseline risk.
An important critique of PE metabolomics studies (and metabolomics diagnostic studies in general) is the wide variability in reported significant metabolites by different groups. Indeed, some studies evaluating metabolites for PE prediction have found that clinical risk factors are better than metabolite levels and that metabolite assessment does not add predictive power (37). Nonetheless, further studies may identify consistent factors we might use to create accurate, sensitive, specific prediction algorithms for PE (35).
Fetal growth restriction (FGR) is defined by a fetus that fails to meet expected growth for gestational age using estimated fetal weight below the 10th percentile. FGR occurs in 5–10% of all pregnancies (107) due to multiple causes and increases the risk of adverse perinatal outcomes. The primary challenge for FGR is that fetuses measuring below the 10th percentile for gestational age may be either constitutionally small for gestational age (SGA) or have pathologic growth restriction. Thus, there is no agreed-upon single gold standard for FGR diagnosis, and the possibility of developing metabolomics approaches to diagnose FGR pathology has generated significant interest.
Several metabolomics studies have highlighted abnormal lipid metabolism during pregnancies resulting in SGA newborns (38, 108). Interestingly, near delivery, the mother-to-newborn ratio of medium-chain fatty acids was decreased with FGR, suggesting increased energetic and structural metabolic demands of the infant (39). Differences in cholesterol synthesis were observed in FGR fetuses compared to normal fetuses (40). In particular cholesterol levels during pregnancy only increased slightly in FGR cases (2.48-fold change) while increasing substantially in normally grown controls (6.54-fold change). In neonatal urine, newborns from FGR pregnancies had increased myo-inositol, which correlated with downregulation of adipose free fatty acid release (41, 42).
Amino acid metabolism also appears to play a role in FGR, with several amino acids differing between the average for gestational age and FGR newborns (43, 44) especially ornithine (45). A role for ornithine in fetal growth was also identified in maternal serum from pregnancies affected by extreme FGR (<5th %ile) and abnormal umbilical artery Dopplers (46). Correlations between ornithine and umbilical blood flow may be related to angiogenesis via polyamine pathways. Placentae from FGR pregnancies show almost universally decreased metabolite levels (47), and generally reduced metabolite concentrations have also been detected in the maternal hair from FGR pregnancies (48). Recent mechanistic research suggests that there is a general disruption in fetal energy substrates and metabolism in FGR, but the unique metabolic adaptations of FGR vs. SGA have not been well-studied.
About 10% of all deliveries occur pre-term at <37 weeks. Pre-term birth (PTB) and the consequences of prematurity are the greatest contributors to neonatal morbidity and mortality (109, 110). Prematurity also has important long-term consequences for lifelong health and disease, and survivors of prematurity have increased risk for chronic medical conditions such as cardiovascular disease, metabolic syndrome, stroke, dyslipidemia, and neurocognitive dysfunction (111–113). Idiopathic pre-term labor (PTL) and pre-term prelabor rupture of membranes (PPROM) account for about two-thirds of pre-term deliveries while indicated deliveries (e.g., pre-eclampsia, placental abruption) account for the other third (114). PTB is the common pathophysiologic endpoint of a wide range of causes, and decades of research have failed to develop effective treatments or even provide adequate screening algorithms (115). Omics-sciences could play a role in defining normal pre-term/term physiology and the pathophysiologic pre-term processes leading to early labor and delivery (116, 117). Some have suggested that all of the great obstetrical syndromes, including PTB, represent different manifestations of various placental dysfunction or maladaptation which might be characterized effectively using metabolic assessment.
Many PTB investigators have focussed on specific markers of inflammation. Inflammation signaling is frequently associated with pre-term labor/PTB (118, 119), but a single universal marker or even several markers may not be sufficient to characterize the diverse etiologies leading to the common final pathway of labor. Analysis of urine from pregnant women before and after labor onset with GC/MS and NMR found 18 unique metabolic changes associated with labor status (49). Analysis of amniotic fluid metabolites has been of particular interest, as the fetus and placenta may be a critical cog in the pregnancy clock (50–54). One study compared amniotic fluid and maternal serum, identifying energy metabolism factors-associated PTB in the fetal compartment (pyruvate, glutamate, and glutamine) that were distinct from maternal serum metabolites that discriminated PTB (54). Another study used amniotic fluid metabolic profiling to compare women with pre-term contractions at risk for spontaneous PTB with or without intra-amniotic infection. Altered amniotic fluid carbohydrates were associated with PTB regardless of infection, while increased amino acids were present only with PTB and intrauterine infection (53). Graca et al. found decreased amino acids, citrate, and myo-inositol and increased allantoin and hexose in amniotic fluid from second trimester PTB cases (51).
Metabolic fingerprinting of pre-term labor that goes on to PTB has been attempted (55). This is a critical challenge in the field, as the majority of PTL does not proceed to PTB. Several studies found increased fatty acids associated with PTB (53). PTB was also associated with decreased levels of acetate and increased lipids (56) and a few amino acids. Interestingly, there may be important differences in PTB metabolic signatures by race and ethnicity. Menon et al. evaluated amniotic fluid from African American women with early spontaneous PTB vs. term birth. Bile acids, steroids, and xanthines were altered and the authors found 8-fold increased pantothenol levels in women who delivered early (52). To achieve the full potential of metabolomics in PTB research, careful selection of comparison groups and rigorous deep phenotyping of both maternal and fetal characteristics is needed.
It is well-known that some congenital/structural defects lead to changes in perfusion, organ function, or other factors, which could change the fetal metabolic signature and perhaps be reflected in maternal serum. Thus, even though congenital structural defects are not metabolic issues per se, we may detect some anomalies through metabolic approaches. We might also exploit metabolomics to elucidate causative metabolic mechanisms. Although NMR of amniotic fluid has been used for decades to characterize neural tube defects (57) and fetal lung or kidney maturity (58–60), more recently NMR has been paired with MS to identify other malformations. Graça et al. evaluated second-trimester amniotic fluid from normal pregnancies and from fetuses affected by congenital anatomic anomalies (61, 62). Surprisingly, maternal and fetal characteristics (e.g., maternal age and fetal sex) had no effect on the metabolic profile of normal fetuses. However, in multivariate supervised analysis, specific changes in glucose, succinate, and some amino acids and proteins were clear (62). Overall, these changes suggested a shift to glycolysis, perhaps due to hypoxic stress. Thus, it is possible that metabolic assessment could be used to screen for anatomic anomalies in combination with detailed fetal ultrasound imaging, similar to maternal serum alpha-fetoprotein for neural tube and abdominal wall defects.
Metabolomics has also been used to specifically evaluate fetal congenital heart disease (CHD) (120, 121). A few studies report metabolic screening for both structural and functional fetal heart disorders. For example, one report described metabolic changes in first trimester maternal serum for chromosomally normal fetuses with structural heart disease, showing altered phosphatidyl-choline and sphingolipids (122). Whether these metabolic changes were related to cardiac energetic processes or other end-organ compensation for altered heart function (e.g., liver dysfunction) was not determined (122). This sort of finding suggests that metabolic characterization could be used as an additional tool for CHD screening.
Common single-gene disorders are routinely evaluated with parental carrier testing and increasingly with cfDNA sequencing techniques. It is possible that some single-gene disorders, even those not affecting biochemical pathways, could be detected or at least flagged for increased scrutiny using metabolomics. That would be particularly helpful for de novo mutations. In some cases, phenotypic or genetic heterogeneity can make prenatal diagnosis difficult (e.g., β-thalassemia, Noonan's syndrome). Nonetheless, Monni et al. found that pregnancies affected by β-thalassemia exhibit significant metabolic changes (123). Comparing the metabolic profiles of placental samples obtained by TA-CVS from normal fetuses and fetuses with homozygous or heterozygous β-thalassemia identified consistent alterations in all β-thalassemia cases. We have proposed a specific metabolic fingerprint for β-thalassemia that is associated with high fetal demand for ribose 5-phosphate (for nucleotide synthesis) and nicotinamide adenine dinucleotide phosphate (for redox maintenance). It appears that fetal oxidative stress can be an important and frequent marker for a wide range of abnormal conditions, and metabolic markers of oxidative stress are readily detected with metabolomics techniques. Other heterogenous single-gene disorders may have common metabolic phenotypes as well that might be best detected by the end products of biochemical production through the analysis and quantification of metabolomics pathways.
Metabolomics is a novel and promising area of research in reproductive medicine. It can be placed in the field of precision medicine, aiming, in general, at developing personalized strategies to manage disease states by considering, at the same time, the patient's genetics, environment, lifestyle, and individual treatment responses. Considering the current prenatal screening methods, it is clear that genetics plays the most relevant role, but metabolomics can generate new insights into the biological and physio/pathological processes. In this perspective, metabolomics can offer the opportunity to find new therapeutic targets and a better understanding of pathological mechanisms.
Table 3 summarizes the advantages and limitations of metabolomics in prenatal medicine. Metabolite detection with high throughput systems coupled with advanced bioinformatics and network analysis holds promise for new prenatal biomarkers and therapeutic discoveries. This metabolomics approach can identify complex physiologic pathways that would not be detected by measuring single metabolites. Further metabolomic investigation of both normal and pathologic prenatal specimens may enhance our knowledge of pregnancy disorders and improve our ability to diagnose and treat fetal disease. Toward this goal, the implementation of large-scale metabolomics studies and secondary cohort validation will be needed. Indeed, despite the general success and the increasing number of publications in prenatal medicine as well, the impact of the metabolomics in the current clinical practice is still dim. Several aspects contribute to this statement: one of the main problems is the enormous variability (external stimuli not closely related to the disease conditions, such as diet, lifestyle in general, analytical and experimental conditions, and data analysis methods), which could influence the final result. Thus, the experimental design covers a fundamental significance and should be planned with extremely controlled conditions. In addition, based on our literature investigation, it has emerged that several clinical metabolomics research studies are affected by strong limitations (e.g., small size of the patient cohorts and incomplete patient's clinical data), and often studies with the same topics produce results that are poorly comparable. Standardization of the methods is mandatory to promote the translation of the metabolomics toward clinical practice. It is important to consider that unfortunately, validated findings have not been yet evidenced in the literature, probably by being metabolomics in prenatal medicine a pioneer new research field. For this reason, new works are still expected in this context. Clinicians who are interested in learning these new approaches and participating in such studies have much to offer the field. Hopefully, soon we will be able to offer our patients clinical metabolomics tools to more effectively characterize, diagnose, and develop treatments for a vast range of pregnancy conditions and fetal disorders.
FM and VC: conceptualization. FM, VC, and FD: methodology. LA and GM: resources. FM and AI: data curation. FM and KH: writing–original draft preparation. FM, KH, LA, and GM: writing–review & editing. GM: supervision. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Olivier M, Asmis R, Hawkins GA, Howard TD, Cox LA. The need for multi-omics biomarker signatures in precision medicine. Int J Mol Sci. (2019) 20:4781. doi: 10.3390/ijms20194781
2. Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. (2007) 26:51–78. doi: 10.1002/mas.20108
3. Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. (2011) 137:293–300. doi: 10.1039/C1AN15605E
4. Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics–a review in human disease diagnosis. Anal Chim Acta. (2010) 659:23–33. doi: 10.1016/j.aca.2009.11.042
6. Dettmer K, Hammock BD. Metabolomics–a new exciting field within the “omics” sciences. Environ Health Perspect. (2004) 112:A396–7. doi: 10.1289/ehp.112-1241997
7. Cevallos-Cevallos JM, Reyes-De-Corcuera JI, Etxeberria E, Danyluk MD, Rodrick GE. Metabolomic analysis in food science: a review. Trends Food Sci Technol. (2009) 20:557–66. doi: 10.1016/j.tifs.2009.07.002
8. Dunn WB, Ellis DavidI. Metabolomics: current analytical platforms and methodologies. TrAC Trends Analyt Chem. (2005) 24:285–94. doi: 10.1016/j.trac.2004.11.021
9. Issaq HJ, Van QN, Waybright TJ, Muschik GM, Veenstra TD. Analytical and statistical approaches to metabolomics research. J Separat Sci. (2009) 32:2183–99. doi: 10.1002/jssc.200900152
10. Wishart DS. Quantitative metabolomics using NMR. TrAC Trends Analyt Chem. (2008) 27:228–37. doi: 10.1016/j.trac.2007.12.001
11. Lei Z, Huhman DV, Sumner LW. Mass spectrometry strategies in metabolomics. J Biol Chem. (2011) 286:25435–42. doi: 10.1074/jbc.R111.238691
12. Moco S, Vervoort J, Moco S, Bino RJ, De Vos RCH, Bino R. Metabolomics technologies and metabolite identification. TrAC Trends Analyt Chem. (2007) 26:855–66. doi: 10.1016/j.trac.2007.08.003
13. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York, NY: Springer Science & Business Media (2009).
14. Oresic M, Clish CB, Davidov EJ, Verheij E, Vogels J, Havekes LM, Neumann E, et al. Phenotype characterisation using integrated gene transcript, protein and metabolite profiling. Appl Bioinformatics. (2004) 3:205–17. doi: 10.2165/00822942-200403040-00002
15. Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metabolomics. (2013) 1:92–107. doi: 10.2174/2213235X130108
16. Hollywood K, Brison DR, Goodacre R. Metabolomics: current technologies and future trends. Proteomics. (2006) 6:4716–23. doi: 10.1002/pmic.200600106
17. Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, et al. A tutorial review: metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding. Anal Chim Acta. (2015) 879:10–23. doi: 10.1016/j.aca.2015.02.012
18. Martin M, Govaerts B. Feature selection in metabolomics with PLS-derived methods. Methods. (2019) 5:1046–2023.
19. Bartel J, Krumsiek J, Theis FJ. Statistical methods for the analysis of high-throughput metabolomics data. Comput Struct Biotechnol J. (2013) 4:e201301009. doi: 10.5936/csbj.201301009
20. Jauniaux E, Cindrova-Davies T, Johns J, Dunster C, Hempstock J, Kelly FJ, et al. Distribution and transfer pathways of antioxidant molecules inside the first trimester human gestational sac. J Clin Endocrinol Metab. (2004) 89:1452–8. doi: 10.1210/jc.2003-031332
21. Heazell AEP, Brown M, Dunn WB, Worton SA, Crocker IP, Baker PN, et al. Analysis of the metabolic footprint and tissue metabolome of placental villous explants cultured at different oxygen tensions reveals novel redox biomarkers. Placenta. (2008) 29:691–8. doi: 10.1016/j.placenta.2008.05.002
22. Jauniaux E, Hempstock J, Teng C, Battaglia FC, Burton GJ. Polyol concentrations in the fluid compartments of the human conceptus during the first trimester of pregnancy: maintenance of redox potential in a low oxygen environment. J Clin Endocrinol Metab. (2005) 90:1171–5. doi: 10.1210/jc.2004-1513
23. Murgia F, Iuculano A, Peddes C, Santoru ML, Tronci L, Deiana M, et al. Metabolic fingerprinting of chorionic villous samples in normal pregnancy and chromosomal disorders. Prenat Diagnosis. (2019) 39:848–58. doi: 10.1002/pd.5461
24. Troisi J, Sarno L, Martinelli P, Di Carlo C, Landolfi A, Scala G, et al. A metabolomics-based approach for non-invasive diagnosis of chromosomal anomalies. Metabolomics. (2017) 13:140. doi: 10.1007/s11306-017-1274-z
25. Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, et al. Metabolomic analysis for first-trimester down syndrome prediction. Am J Obstet Gynecol. (2013) 208:371.e1–8. doi: 10.1016/j.ajog.2012.12.035
26. Bahado-Singh RO, Akolekar R, Chelliah A, Mandal R, Dong E, Kruger M, et al. Metabolomic analysis for first-trimester trisomy 18 detection. Am J Obstet Gynecol. (2013) 209:65.e1–9. doi: 10.1016/j.ajog.2013.03.028
27. Pinto J, Almeida LM, Martins AS, Duarte D, Marques Domingues MR, Barros AS, et al. Impact of fetal chromosomal disorders on maternal blood metabolome: toward new biomarkers? Am J Obstet Gynecol. (2015) 213:841.e1–15. doi: 10.1016/j.ajog.2015.07.032
28. Diaz SO, Barros AS, Goodfellow BJ, Duarte IF, Galhano E, Pita C, et al. Second trimester maternal urine for the diagnosis of trisomy 21 and prediction of poor pregnancy outcomes. Journal of Proteome Research. (2013) 12:2946–57. doi: 10.1021/pr4002355
29. Trivedi DK, Iles RK. Shotgun metabolomic profiles in maternal urine identify potential mass spectral markers of abnormal fetal biochemistry – dihydrouracil and progesterone in the metabolism of down syndrome. Biomed Chromatogr. (2015) 29:1173–83. doi: 10.1002/bmc.3404
30. Dunn WB, Brown M, Worton SA, Crocker IP, Broadhurst D, Horgan R, et al. Changes in the metabolic footprint of placental explant-conditioned culture medium identifies metabolic disturbances related to hypoxia and pre-eclampsia. Placenta. (2009) 30:974–80. doi: 10.1016/j.placenta.2009.08.008
31. Austdal M, Skråstad RB, Gundersen AS, Austgulen R, Iversen A-C, Bathen TF. Metabolomic biomarkers in serum and urine in women with preeclampsia. PLoS ONE. (2014) 9:e91923. doi: 10.1371/journal.pone.0091923
32. Zhou X, Han T-L, Chen H, Baker PN, Qi H, Zhang H. Impaired mitochondrial fusion, autophagy, biogenesis and dysregulated lipid metabolism is associated with preeclampsia. Exp Cell Res. (2017) 359:195–204. doi: 10.1016/j.yexcr.2017.07.029
33. Bahado-Singh RO, Syngelaki A, Mandal R, Graham SF, Akolekar R, Han B, et al. Metabolomic determination of pathogenesis of late-onset preeclampsia. J Mater Fetal Neonat Med. (2017) 30:658–64. doi: 10.1080/14767058.2016.1185411
34. Bahado-Singh RO, Syngelaki A, Akolekar R, Mandal R, Bjondahl CT, Beomsoo Han, et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am J Obstet Gynecol. (2015) 213:530.e1–10. doi: 10.1016/j.ajog.2015.06.044
35. Bahado-Singh R, Poon LC, Yilmaz A, Syngelaki A, Turkoglu O, Kumar P, et al. Integrated proteomic and metabolomic prediction of term preeclampsia. Sci Rep. (2017) 7:16189. doi: 10.1038/s41598-017-15882-9
36. Koster MPH, Vreeken RJ, Harms AC, Dane AD, Kuc S, Schielen PCJI, et al. First-Trimester serum acylcarnitine levels to predict preeclampsia: a metabolomics approach. Dis Mark. (2015) 2015:1–8. doi: 10.1155/2015/857108
37. Kuc S, Koster MPH, Pennings JLA, Hankemeier T, Berger R, Harms AC, et al. Metabolomics profiling for identification of novel potential markers in early prediction of preeclampsia. PLoS ONE. (2014) 9:e98540. doi: 10.1371/journal.pone.0098540
38. Bernard JY, Tint M-T, Aris IM, Chenc LW, Quaha PL, Tand KH, et al. Maternal plasma phosphatidylcholine polyunsaturated fatty acids during pregnancy and offspring growth and adiposity. Prostaglandins, Leukot Essent Fatty Acids. (2017) 121:21–9. doi: 10.1016/j.plefa.2017.05.006
39. Visentin S, Crotti S, Donazzolo E, D'Aronco S, Nitti D, Cosmi E, et al. Medium chain fatty acids in intrauterine growth restricted and small for gestational age pregnancies. Metabolomics. (2017) 13:54. doi: 10.1007/s11306-017-1197-8
40. Clinton CM, Bain JR, Muehlbauer MJ, Li YY, Li L, O'Neal SK, et al. Non-targeted urinary metabolomics in pregnancy and associations with fetal growth restriction. Sci Rep. (2020) 10:5307. doi: 10.1038/s41598-020-62131-7
41. Dessì A, Marincola FC, Pattumelli MG, Ciccarelli S, Corbu S, Ossicini C, et al. Investigation of the 1H-NMR based urine metabolomic profiles of IUGR, LGA and AGA newborns on the first day of life. J Matern Fetal Neonat Med. (2014) 27:13–9. doi: 10.3109/14767058.2014.955674
42. Dessì A, Atzori L, Noto A, Visser GHA, Gazzolo D, Zanardo V, et al. Metabolomics in newborns with intrauterine growth retardation (IUGR): urine reveals markers of metabolic syndrome. J Mater Fetal Neonat Med. (2011) 24:35–9. doi: 10.3109/14767058.2011.605868
43. Favretto D, Cosmi E, Ragazzi E, Visentin S, Tucci M, Fais P, et al. Cord blood metabolomic profiling in intrauterine growth restriction. Anal Bioanal Chem. (2012) 402:1109–21. doi: 10.1007/s00216-011-5540-z
44. Sanz-Cortés M, Carbajo RJ, Crispi F, Figueras F, Pineda-Lucena A, Gratacós E. Metabolomic profile of umbilical cord blood plasma from early and late intrauterine growth restricted (IUGR) neonates with and without signs of brain vasodilation. PLoS ONE. (2013) 8:e80121. doi: 10.1371/journal.pone.0080121
45. Liu J, Chen X-X, Li X-W, Fu W, Zhang W-Q. Metabolomic research on newborn infants with intrauterine growth restriction. Medicine. (2016) 95:3564. doi: 10.1097/MD.0000000000003564
46. Porter AC, Gumina DL, Armstrong M, Maclean KN, Reisdorph N, Galan HL, et al. Maternal amino acid profiles to distinguish constitutionally small versus growth-restricted fetuses defined by doppler ultrasound: a pilot study. Am J Perinatol. (2020) 37:1084–93. doi: 10.1055/s-0040-1701504
47. Bahado-Singh RO, Turkoglu O, Yilmaz A, Kumar P, Zeb A, Konda S, et al. Metabolomic identification of placental alterations in fetal growth restriction. J Mater Fetal Neonatal Med. (2020) 11, 1–10. doi: 10.1080/14767058.2020.1722632
48. Sulek K, Han T-L, Villas-Boas SG, Wishart DS, Soh SE, Kwek K, et al. Hair metabolomics: identification of fetal compromise provides proof of concept for biomarker discovery. Theranostics. (2014) 4:953–9. doi: 10.7150/thno.9265
49. Caboni P, Meloni A, Lussu M, Carta E, Barberini L, Noto A, et al. Urinary metabolomics of pregnant women at term: a combined GC/MS and NMR approach. J Matern Fetal Neonatal Med. (2014) 27:4–12. doi: 10.3109/14767058.2014.956403
50. Baraldi E, Giordano G, Stocchero M, Moschino L, Patrizia Zaramella P, Tran MR, et al. Untargeted metabolomic analysis of amniotic fluid in the prediction of preterm delivery and bronchopulmonary dysplasia. PLoS ONE. (2016) 11:e0164211. doi: 10.1371/journal.pone.0164211
51. Graça G, Duarte IF, Barros AS, Goodfellow BJ, Diaz SO, Pinto J, et al. Impact of prenatal disorders on the metabolic profile of second trimester amniotic fluid: a nuclear magnetic resonance metabonomic study. J Proteome Res. (2010) 9:6016–24. doi: 10.1021/pr100815q
52. Menon R, Jones J, Gunst PR, Kacerovsky M, Fortunato SJ, Saade GR, et al. Amniotic fluid metabolomic analysis in spontaneous preterm birth. Reprod Sci. (2014) 21:791–803. doi: 10.1177/1933719113518987
53. Romero R, Mazaki-Tovi S, Vaisbuch E, Kusanovic JP, Chaiworapongsa T, Gomez R, et al. Metabolomics in premature labor: a novel approach to identify patients at risk for preterm delivery. J Matern Fetal Neonatal Med. (2010) 23:1344–59. doi: 10.3109/14767058.2010.482618
54. Virgiliou C, Gika HG, Witting M, Bletsou AA, Athanasiadis A, Zafrakas M, et al. Amniotic fluid and maternal serum metabolic signatures in the second trimester associated with preterm delivery. J Proteome Res. (2017) 16:898–910. doi: 10.1021/acs.jproteome.6b00845
55. Lizewska B, Teul J, Kuc P, Lemancewicz A, Charkiewicz K, Goscik J, et al. Maternal plasma metabolomic profiles in spontaneous preterm birth: preliminary results. Mediat Inflamm. (2018) 2018:9362820. doi: 10.1155/2018/9362820
56. Tea I, Le Gall G, Kuester A, Guignard N, Alexandre-Gouabau MC, Darmaun D, et al. H-1-NMR-Based metabolic profiling of maternal and umbilical cord blood indicates altered materno-foetal nutrient exchange in preterm infants. PLoS ONE. (2012) 7:e029947. doi: 10.1371/journal.pone.0029947
57. Groenen PMW, Engelke UF, Wevers RA, Hendriks JCM, Eskes TKAB, Merkus HMWM, et al. High-resolution 1H NMR spectroscopy of amniotic fluids from spina bifida fetuses and controls. Euro J Obstet Gynecol Reproduct Biol. (2004) 112:16–23. doi: 10.1016/S0301-2115(03)00279-3
58. Bock JL. Metabolic profiling of amniotic fluid by proton nuclear magnetic resonance spectroscopy: correlation with fetal maturation and other clinical variables. Clin Chem. (1994) 40:56–61. doi: 10.1093/clinchem/40.1.56
59. Clifton MS, Joe BN, Zektzer AS, Kurhanewiczb J, Vigneronb DB, Coakley FV, et al. Feasibility of magnetic resonance spectroscopy for evaluating fetal lung maturity. J Pediatr Surg. (2006) 41:768–73. doi: 10.1016/j.jpedsurg.2006.02.020
60. Pearce JM, Krone JT, Pappas AA, Komoroski RA. Analysis of saturated phosphatidylcholine in amniotic fluid by 31P NMR. Magnet Reson Med. (1993) 30:476–84. doi: 10.1002/mrm.1910300410
61. Graça G, Duarte IF, Goodfellow BJ, Barros AS, Carreira IM, Couceiro AB, et al. Potential of NMR spectroscopy for the study of human amniotic fluid. Anal Chem. (2007) 79:8367–75. doi: 10.1021/ac071278d
62. Graça G, Duarte IF, Barros AS, Goodfellow BJ, Diaz S, Carreira IM, et al. 1H NMR based metabonomics of human amniotic fluid for the metabolic characterization of fetus malformations. J Proteome Res. (2009) 8:4144–50. doi: 10.1021/pr900386f
63. Bleicher SJ, O'Sullivan JB, Freinkel N. Carbohydrate metabolism in pregnancy. N Engl J Med. (1964) 271:866–72. doi: 10.1056/NEJM196410222711702
64. Heaney RP, Skillman TG. Calcium metabolism in normal human pregnancy. J Clin Endocrinol Metab. (1971) 33:661–70. doi: 10.1210/jcem-33-4-661
65. Knopp RH, Warth MR, Charles D, Childs M, Li JR, Mabuchi H, et al. Lipoprotein metabolism in pregnancy, fat transport to the fetus, and the effects of diabetes. Neonatology. (1986) 50:297–317. doi: 10.1159/000242614
66. Nicolaides KH. First-Trimester screening for chromosomal abnormalities. Semi Perinatol. (2005) 29:190–4. doi: 10.1053/j.semperi.2005.06.001
67. Nicolaides KH. Screening for fetal aneuploidies at 11 to 13 weeks. Prenat Diagn. (2011) 31:7–15. doi: 10.1002/pd.2637
68. Akolekar R, Beta J, Picciarelli G, Ogilvie C, D'Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound Obstet Gynecol. (2015) 45:16–26. doi: 10.1002/uog.14636
69. Santorum M, Wright D, Syngelaki A, Karagioti N, Nicolaides KH. Accuracy of first-trimester combined test in screening for trisomies 21, 18 and 13. Ultrasound Obstet Gynecol. (2017) 49:714–20. doi: 10.1002/uog.17283
70. Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol. (2017) 50:302–14. doi: 10.1002/uog.17484
71. Cheng W-L, Hsiao C-H, Tseng H-W, Lee T-P. Noninvasive prenatal diagnosis. Taiwan J Obstet Gynecol. (2015) 54:343–9. doi: 10.1016/j.tjog.2015.05.002
72. Buffat C, Mondon F, Rigourd V, Boubred F, Bessìeres B, Fayol L, et al. A hierarchical analysis of transcriptome alterations in intrauterine growth restriction (IUGR) reveals common pathophysiological pathways in mammals. J Pathol. (2007) 213:337–46. doi: 10.1002/path.2233
73. Hansson SR, Chen Y, Brodszki J, Chen M, Hernandez-Andrade E, Inman JM, et al. Gene expression profiling of human placentas from preeclamptic and normotensive pregnancies. Mol Hum Reprod. (2006) 12:169–79. doi: 10.1093/molehr/gal011
74. Hassan SS, Romero R, Tarca AL, Draghici S, Pineles B Bugrim A, et al. Signature pathways identified from gene expression profiles in the human uterine cervix before and after spontaneous term parturition. Am J Obstet Gynecol. (2007) 197:250.e1–7. doi: 10.1016/j.ajog.2007.07.008
75. Slonim DK, Koide K, Johnson KL, Tantravahid U, Cowanc JM, Jarrahc Z, et al. Functional genomic analysis of amniotic fluid cell-free mRNA suggests that oxidative stress is significant in down syndrome fetuses. Proc Natl Acad Sci USA. (2009) 106:9425–9. doi: 10.1073/pnas.0903909106
76. Timmons BC, Mahendroo M. Processes regulating cervical ripening differ from cervical dilation and postpartum repair: insights from gene expression studies. Reprod Sci. (2007) 14:53–62. doi: 10.1177/1933719107309587
77. Tsai M-S, Hwang S-M, Chen K-D, Lee YS, Hsu LW, Chang CJ, et al. Functional network analysis of the transcriptomes of mesenchymal stem cells derived from amniotic fluid, amniotic membrane, cord blood, and bone marrow. Stem Cells. (2007) 25:2511–23. doi: 10.1634/stemcells.2007-0023
78. Breuiller-Fouche M, Charpigny G, Germain G. Functional genomics of the pregnant uterus: from expectations to reality, a compilation of studies in the myometrium. BMC Pregnancy Childbirth. (2007) 7:S4. doi: 10.1186/1471-2393-7-S1-S4
79. Chiu RWK, Chan KCA, Gao Y, Laua VYM, Zhenga W, Leunge TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. PNAS. (2008) 105:20458–20463. doi: 10.1073/pnas.0810641105
80. Dolan S, Biermann J, Damus K. Genomics for Health in Preconception and Prenatal Periods. Journal of Nursing Scholarship. (2007) 39:4–9. doi: 10.1111/j.1547-5069.2007.00136.x
81. Tromp G, Kuivaniemi H, Romero R, Chaiworapongsa T, Kim YM, Kim MR, et al. Genome-wide expression profiling of fetal membranes reveals a deficient expression of proteinase inhibitor 3 in premature rupture of membranes. Am J Obstet Gynecol. (2004) 191:1331–8. doi: 10.1016/j.ajog.2004.07.010
82. Wilson RD. Genomics: new technology for obstetrics and gynaecology. J Obstet Gynaecol Can. (2005) 27:63–8. doi: 10.1016/S1701-2163(16)30175-X
83. Cho CKJ, Shan SJ, Winsor EJ, Diamandis EP. Proteomics analysis of human amniotic fluid. Mol Cell Proteomics. (2007) 6:1406–15 doi: 10.1074/mcp.M700090-MCP200
84. Park JS, Oh KJ, Norwitz ER, Han JS, Choi HJ, Seong HS, et al. Identification of proteomic biomarkers of preeclampsia in amniotic fluid using SELDI-TOF mass spectrometry. Reprod Sci. (2008) 15:457–68. doi: 10.1177/1933719108316909
85. Gravett MG, Thomas A, Schneider KA, Reddy AP, Dasari S, Jacob T, et al. Proteomic analysis of cervical–vaginal fluid: identification of novel biomarkers for detection of intra-amniotic infection. J Proteome Res. (2007) 6:89–96. doi: 10.1021/pr060149v
86. Kim Y-S, Kim M-S, Lee S-H, Choi BC, Lim JM, Cha KY, et al. Proteomic analysis of recurrent spontaneous abortion: Identification of an inadequately expressed set of proteins in human follicular fluid. Proteomics. (2006) 6:3445–54. doi: 10.1002/pmic.200500775
87. Luan H, Meng N, Liu P, Feng Q, Lin S, Fu J, et al. Pregnancy-Induced metabolic phenotype variations in maternal plasma. J Proteome Res. (2014) 13:1527–36. doi: 10.1021/pr401068k
88. Diaz SO, Pinto J, Graça G, Duarte IF, Barros AS, Galhano E, et al. Metabolic biomarkers of prenatal disorders: an exploratory NMR metabonomics study of second trimester maternal urine and blood plasma. J Proteome Res. (2011) 10:3732–42. doi: 10.1021/pr200352m
89. Ghartey J, Brown A, Anglim L, Elovitz M. 11: women with preterm birth have a distinct cervico-vaginal metabolome. Am J Obstetr Gynecol. (2015) 212:S9. doi: 10.1016/j.ajog.2014.10.057
90. Iuculano A, Murgia F, Peddes C, Santoru ML Tronci L, Deiana M, et al. Metabolic characterization of amniotic fluids of fetuses with enlarged nuchal translucency. J Perinat Med. (2019) 47:311–8. doi: 10.1515/jpm-2018-0314
91. Catalano PM, Roman-Drago NM, Amini SB, Sims EAH. Longitudinal changes in body composition and energy balance in lean women with normal and abnormal glucose tolerance during pregnancy. Am J Obstetr Gynecol. (1998) 179:156–65. doi: 10.1016/S0002-9378(98)70267-4
92. Di Cianni G, Ghio A, Resi V, Volpe L. Gestational diabetes mellitus: an opportunity to prevent type 2 diabetes and cardiovascular disease in young women. Womens Health. (2010) 6:97–105. doi: 10.2217/WHE.09.76
93. McLachlan KA, O'Neal D, Jenkins A, Alford FP. Do adiponectin, TNFα, leptin and CRP relate to insulin resistance in pregnancy? Studies in women with and without gestational diabetes, during and after pregnancy. Diabet Metab Res Rev. (2006) 22:131–8. doi: 10.1002/dmrr.591
94. Burton GJ, Watson AL, Hempstock J, Skepper JN, Jauniaux E. Uterine glands provide histiotrophic nutrition for the human fetus during the first trimester of pregnancy. J Clin Endocrinol Metab. (2002) 87:2954–9. doi: 10.1210/jcem.87.6.8563
95. Jauniaux E, Watson AL, Hempstock J, Bao Y-P, Skepper JN, Burton GJ. Onset of maternal arterial blood flow and placental oxidative stress: a possible factor in human early pregnancy failure. Am J Pathol. (2000) 157:2111–22. doi: 10.1016/S0002-9440(10)64849-3
96. Burton GJ, Jauniaux E. Placental oxidative stress: from miscarriage to preeclampsia. Reprod Sci. (2004) 11:342–52. doi: 10.1016/j.jsgi.2004.03.003
97. National Collaborating Centre for Women's and Children's Health. Antenatal care: routine care for the healthy pregnant woman. In: Commissioned by the National Institute for Clinical Excellence. London: Royal College of Obstetrics and Gynaecology Press (2008). p. 218–27.
98. Santiago JC, Ramos-Corpas D. Delta-NT and center-specific ultrasound nuchal translucency medians. Ultrasound Obstet Gynecol. (2007) 30:934–40. doi: 10.1002/uog.5171
99. Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semi Perinatol. (2012) 36:56–9. doi: 10.1053/j.semperi.2011.09.011
100. Brown MA, Magee LA, Kenny LC, Karumanchi SA, McCarthy F, Saito S, et al. Hypertensive disorders of pregnancy: ISSHP classification, diagnosis, and management recommendations for international practice. Hypertension. (2018) 72:24–43. doi: 10.1161/HYPERTENSIONAHA.117.10803
101. ACOG. ACOG committee opinion No. 770: uterine morcellation for presumed leiomyomas. Obstet. Gynecol. (2019) 133:1. doi: 10.1097/AOG.0000000000003126
102. Khan N, Andrade W, Castro HD, Wright A, Wright D, Nicolaides KH. Impact of new definitions of pre-eclampsia on incidence and performance of first-trimester screening. Ultrasound Obstet Gynecol. (2020) 55:50–7. doi: 10.1002/uog.21867
103. Cnossen JS, Riet GT, Mol BW, Van der post JA, Leeflang MM, Meads CA, et al. Are tests for predicting pre-eclampsia good enough to make screening viable? A review of reviews and critical appraisal. Acta Obstet Gynecol Scand. (2009) 88:758–65. doi: 10.1080/00016340903008953
104. Staff AC. Circulating predictive biomarkers in preeclampsia. Pregnancy Hypertens. (2011) 1:28–42. doi: 10.1016/j.preghy.2010.10.012
105. Roberge S, Demers S, Bujold E. Low-Dose aspirin for prevention of morbidity and mortality from preeclampsia. Ann Inter Med. (2014). Available online at: https://www.acpjournals.org/doi/abs/10.7326/L14/5020/4 (accessed September 9, 2020).
106. O'Gorman N, Wright D, Syngelaki A, Akolekar R, Wright A, Poon LC, et al. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11-13 weeks gestation. Am J Obstet Gynecol. (2016) 214:103.e1–2. doi: 10.1016/j.ajog.2015.08.034
107. Vedmedovska N, Rezeberga D, Teibe U, Melderis I, Donders GGG. Placental pathology in fetal growth restriction. Euro J Obstet Gynecol Reproduct Biol. (2011) 155:36–40. doi: 10.1016/j.ejogrb.2010.11.017
108. Leite DFB, Morillon A-C, Júnior EFM, Souza RT, Khashan AS, Baker PN, et al. Metabolomics for predicting fetal growth restriction: protocol for a systematic review and meta-analysis. BMJ Open. (2018) 8:e022743. doi: 10.1136/bmjopen-2018-022743
109. Auray-Blais C, Raiche E, Gagnon R, Berthiaume M, Pasquier JC. Metabolomics and preterm birth: what biomarkers in cervicovaginal secretions are predictive of high-risk pregnant women? Int J Mass Spectrometry. (2011) 307:33–8. doi: 10.1016/j.ijms.2011.02.009
110. Conde-Agudelo A, Romero R, Da Fonseca E, O'Brien JM, Cetingoz E, Creasy GW, et al. Vaginal progesterone is as effective as cervical cerclage to prevent preterm birth in women with a singleton gestation, previous spontaneous preterm birth, and a short cervix: updated indirect comparison meta-analysis. Am J Obstet Gynecol. (2018) 219:10–25. doi: 10.1016/j.ajog.2018.03.028
111. Crump C. Preterm birth and mortality in adulthood: a systematic review. J Perinatol. (2020) 40:833–43. doi: 10.1038/s41372-019-0563-y
112. Luu TM, Katz SL, Leeson P, Thébaud B, Nuyt AM. Preterm birth: risk factor for early-onset chronic diseases. CMAJ. (2016) 188:736–40. doi: 10.1503/cmaj.150450
113. Institute of Medicine (US) Committee on Understanding Premature Birth and Assuring Healthy Outcomes; Behrman RE Butler AS. Preterm Birth: Causes, Consequences, and Prevention. Washington, DC: National Academies Press (2007).
114. Gotsch F, Gotsch F, Romero R, Erez O, Vaisbuch E, Kusanovic JP, et al. The preterm parturition syndrome and its implications for understanding the biology, risk assessment, diagnosis, treatment and prevention of preterm birth. J Matern Fetal Neonatal Med. (2009) 22:5–23. doi: 10.1080/14767050902860690
115. Georgiou HM, Di Quinzio MKW, Permezel M, Brennecke SP. Predicting preterm labour: current status and future prospects. Dis Mark. (2015) 2015:1–9. doi: 10.1155/2015/435014
116. Kuć P, Laudański P, Kowalczuk O, Chyczewski L, Laudański T. Expression of selected genes in preterm premature rupture of fetal membranes. Acta Obstet Gynecol Scand. (2012) 91:936–43. doi: 10.1111/j.1600-0412.2012.01445.x
117. Tsiartas P, Holst RM, Wennerholm UB, Hagberg H, DM Hougaard DM, Skogstrand K, et al. Prediction of spontaneous preterm delivery in women with threatened preterm labour: a prospective cohort study of multiple proteins in maternal serum. BJOG. (2012) 119:866–73. doi: 10.1111/j.1471-0528.2012.03328.x
118. Chan RL. Biochemical markers of spontaneous preterm birth in asymptomatic women. Bio Med Res Int. (2014) 2014:1–8. doi: 10.1155/2014/164081
119. Laudanski P, Raba G, Kuc P, Lemancewicz A, Kisielewski R, Laudanski T. Assessment of the selected biochemical markers in predicting preterm labour. J Matern Fetal Neonatal Med. (2012) 25:2696–9. doi: 10.3109/14767058.2012.699116
120. Hoffman JIE, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. (2002) 39:1890–900. doi: 10.1016/S0735-1097(02)01886-7
121. Mitchell SC, Korones SB, Berendes HW. Congenital heart disease in 56,109 births incidence and natural history. Circulation. (1971) 43:323–32. doi: 10.1161/01.CIR.43.3.323
122. Bahado-Singh RO, Ertl R, Mandal R, Bjorndahl TC, Syngelaki A, Han B, et al. Metabolomic prediction of fetal congenital heart defect in the first trimester. Am J Obstet Gynecol. (2014) 211:240.e1–14. doi: 10.1016/j.ajog.2014.03.056
Keywords: congenital anatomic defects, fetal growth restriction, metabolomics, normal pregnancy, pre-eclampsia, prenatal diagnosis, prenatal medicine, pre-term labor and delivery
Citation: Monni G, Atzori L, Corda V, Dessolis F, Iuculano A, Hurt KJ and Murgia F (2021) Metabolomics in Prenatal Medicine: A Review. Front. Med. 8:645118. doi: 10.3389/fmed.2021.645118
Received: 22 December 2020; Accepted: 04 May 2021;
Published: 25 June 2021.
Edited by:
Zaleha Abdullah Mahdy, National University of Malaysia, MalaysiaReviewed by:
Rachel Sabine Kelly, Brigham and Women's Hospital and Harvard Medical School, United StatesCopyright © 2021 Monni, Atzori, Corda, Dessolis, Iuculano, Hurt and Murgia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Federica Murgia, Zi5tdXJnaWFjYWdsaWFyaUBnbWFpbC5jb20=; ZmVkZXJpY2EubXVyZ2lhQHVuaWNhLml0; Giovanni Monni, cHJlbmF0YWxtb25uaUB0aXNjYWxpLml0
†These authors have contributed equally to this work and share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.