- Institute of General Physiology, University of Ulm, Ulm, Germany
Idiopathic pulmonary fibrosis (IPF) is a progressive and fatal lung disease with limited therapeutic options. The current model suggests that chronic or repetitive “micro-injuries” of the alveolar epithelium lead to activation and proliferation of fibroblasts and excessive extracellular matrix (ECM) deposition. Disruption of alveolar type II (ATII) epithelial cell homeostasis and the characteristics of mesenchymal cell populations in IPF have received particular attention in recent years. Emerging data from single cell RNA sequencing (scRNAseq) analysis shed novel light on alterations in ATII cell progenitor dysfunction and the diversity of mesenchymal cells within the fibrotic lung. Within this minireview, we summarize the data from most recent human scRNAseq studies. We aim to collate the current knowledge on cellular plasticity and heterogeneity in the development and progression of IPF, effects of drug treatment on transcriptional changes. Finally, we provide a brief outlook on future challenges and promises for large scale sequencing studies in the development of novel therapeutics for IPF.
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive, irreversible, and usually fatal respiratory disease with limited therapeutic options (1). The current model suggests that IPF is a result of repeated epithelial cell injury and inadequate repair of the alveolar epithelium that leads to excessive fibroblast activity and lung fibrosis (2). Alveolar type 2 epithelial (ATII) cells function as progenitor cells that maintain homeostasis of the epithelium and repair the damaged epithelium after injury (2–6). Disruption of ATII cell homeostasis and loss of facultative progenitor capacity are drivers for lung fibrosis (1, 7, 8). The aberrantly activated lung epithelial cells secrete mediators that lead to fibroblast proliferation and differentiation to highly active myofibroblasts, which deposit excessive amounts of extracellular matrix (ECM) (9). This results in overall remodeling of the alveolar structure, formation of scar tissue, thickening of the alveolar septae and an increase in tissue stiffness (10, 11). The inability to resolve fibrotic processes results in ongoing activation of fibroblasts creating a vicious cycle with fatal outcome for the patient.
Disruption of ATII cell homeostasis and the characteristics of mesenchymal cell populations in IPF have therefore received particular attention in recent years. Apart from ATII cells and alveolar fibroblasts, recent studies also suggested that other cell types may have an important role in IPF initiation. This entails further, recently identified, sub-populations of specific epithelial and/or mesenchymal cells and possible contributions of other cells in the distal lung, most notably macrophages (12–15).
Overall, detailed cellular contributions to the onset and progression of IPF are still not fully elucidated. The advent of scRNA technology has increased our understanding of lung cell heterogeneity and cellular interplay in healthy and diseased lung tissue. Initial efforts were mainly conducted in mice, which also offer cell-type specific lineage tracing to study disease onset and progression (6, 16–18). However, the human lung contains many structural and cellular differences to the mouse lung and, although the alveolus is one of the most conserved regions between mouse and human lungs, it is still debated whether all cells in the human distal lung have corresponding counterparts in the murine lung (5, 8, 19). Mice appear to lack a substantial proportion of the molecularly defined human lung cell populations, including sub-populations of alveolar epithelial and mesenchymal cells (20). Hence, mice might not accurately recapitulate human disease if critical cell types are not present. In particular, mouse models of IPF, including the widely used bleomycin model, still do not recapitulate all features of IPF pathogenesis (21). It is therefore indispensable to extend our knowledge of the human lung. Recent efforts, have led to ever increasing datasets aiming at establishing a detailed cellular atlas of the human lung. These novel datasets are a valuable source to identify and map disease origins, progression and potential therapeutic targets benefiting patients (20, 22). Recently, scRNAseq approaches were also applied in studying transcriptional changes in IPF and extended our knowledge of the human IPF transcriptome (Table 1). Within this review, we summarize the data from most recent scRNAseq studies aiming to collate the current knowledge on cellular plasticity and heterogeneity in the development and progression of IPF in humans as well as effects of drug treatment on transcriptional changes. We also provide a brief outlook on future challenges and promises for large scale sequencing studies in the development of novel therapeutics for IPF.
Diversity of Lung Cell Types and Regulatory Mechanisms During Health and Disease
Epithelial Cells
Inadequate alveolar epithelial repair is believed to constitute the initial trigger for fibrotic remodeling in IPF. Within the alveolar epithelium, constituted of alveolar type I epithelial (ATI) and ATII cells, ATII cells are considered self-renewing stem cells that also regenerate ATI cells after injury (3). The contribution of ATI cells to alveolar repair is thought to be very limited (30).
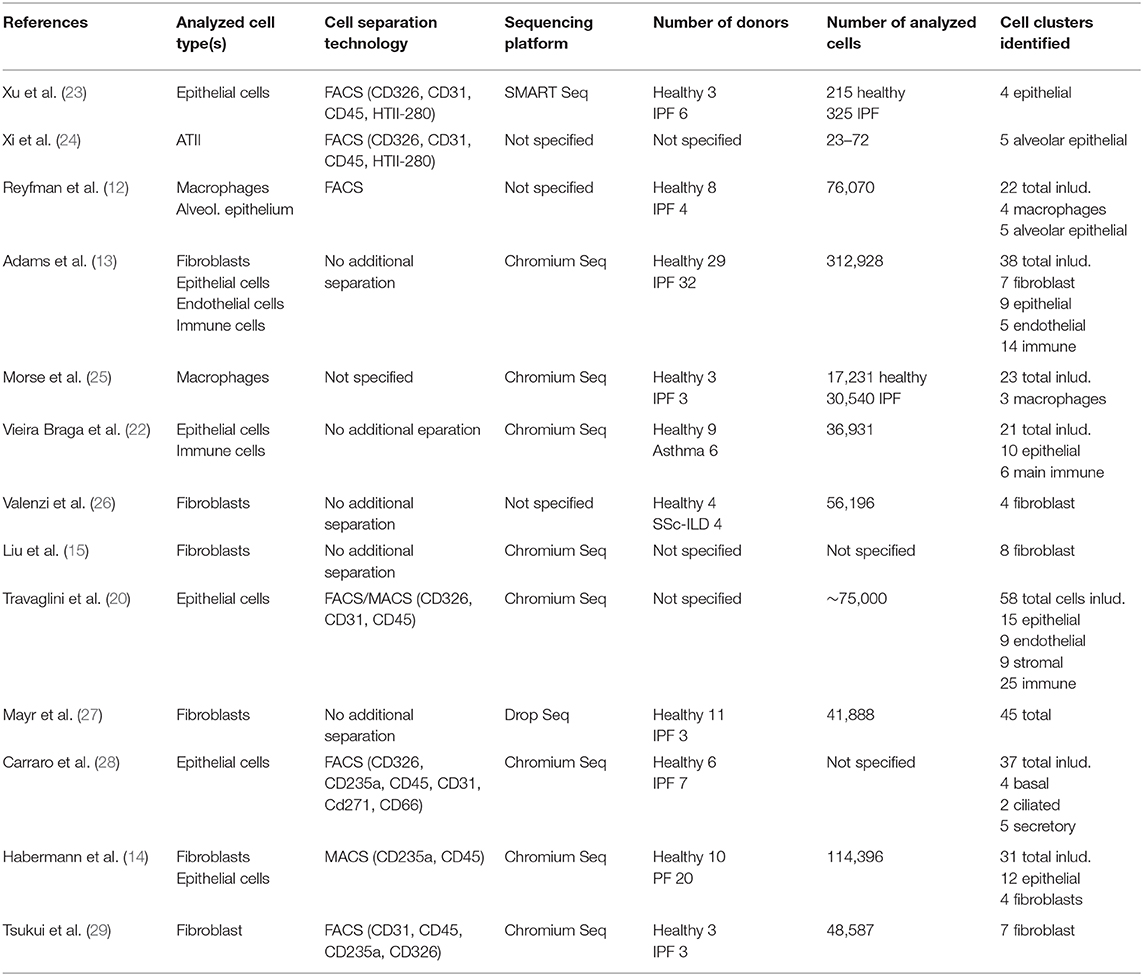
Recent scRNAseq studies identified sub-lineages within the ATII cell population in mice and humans that serve as facultative progenitor cell population to regenerate alveoli after injury. In contrast to “bulk”, quiescent ATII cells, these ATII signaling (ATII-s) cells are Wnt-active (20), and in mice, were initially characterized by expression of the transcriptional target of Wnt signaling, Axin2 (6, 18). A recent scRNAseq study directly comparing lung tissue obtained from (healthy) transplant donors and recipients with pulmonary fibrosis, however, was unable to identify this sub-population in humans, albeit finding AXIN2 expression in ATII cells. Interestingly, this study reported non-overlapping expression of Wnt ligands and AXIN2 in epithelial cells, suggesting that Wnt secretion and response were separated to specific cells. These authors also found small clusters of ATII cells that were distinct between healthy and fibrotic donors, with fibrosis-specific ATII cells expressing DMBT1, SERPINA1, and CHI3L1, genes previously associated with pulmonary fibrosis (12). The study by Reyfman et al. also identified rare cell populations including airway stem cells and senescent cells to emerge during fibrosis (12). Similarly, principal component analysis (PCA) and whole-genome unsupervised hierarchical clustering of scRNAseq-derived transcriptomes clearly separated ATII cells in a study comparing normal and fibrotic lungs. ATII cells from fibrotic lungs revealed a hypoxic sub-population with activated Notch, suppressed surfactant protein C (SFTPC), and transdifferentiation towards a KRT5+ airway basal cell-like state, whereas Wnt activity prior to Krt5 activation favors differentiation toward ATII cells, leading to normal alveolar epithelial repair (24). Atypical sub-populations of alveolar epithelial cells expressing airway-associated genes (SOX2, MUC5B, and PAX9), were also reported in explant tissue of patients undergoing transplant for end-stage IPF (23). Identification of airway stem cells within fibrotic lesions therefore matches the histological observation that epithelial cells with airway markers accumulate in fibrotic regions of human lungs, in a process termed bronchiolization (Figure 1) (31). Epithelial cells with airway markers are also found in micro-honeycomb cysts, another feature of fibrotic remodeling in human lung (32).
Figure 1. Schematic representation of cell types identified by scRNA sequencing in the distal lung of healthy and IPF donors. Vertical lines separate marker genes identified by different authors for similar cell types. Arrows depict presumed transdifferentiation pathways and respective signaling pathways. For details see main text.
Basal cells are the major stem cells in the human airways that self-renew and can give rise to all other cell types of the airways (33). scRNAseq studies have identified various clusters of basal cells in humans, that might differ in their ability to function as stem cells (20, 22, 28). Carraro et al. reported a sub-population of CD66+ secretory cell primed basal (SPB) cells, that share a molecular signature with basal cells recovered from fibrotic lung tissue (28). SPB cells from IPF were enriched in CEACAM6 expression and could be distinguished from normal SPB cells by their gene signature. IPF SPB cells expressed genes previously identified as serum biomarkers for higher risk of mortality and proposed to be involved in disease development (MUC1, MMP7, ICAM1). Two scRNAseq studies published this year, further added to the notion of aberrant basaloid cells, expansion of airway epithelial cells and reduction in alveolar epithelial cells in IPF lungs (13, 14). Habermann et al. observed a remarkable shift in epithelial cells by comparing IPF with normal lung tissue. Cells expanded in IPF lungs were airway epithelial cells (KRT5+ basal cells, FOXJ1+ ciliated cells, and secretory cells), whereas alveolar epithelial cells were recovered to a lower extend (14). They also identified a novel KRT5−/KRT15−/KRT17+/TP63+, pathologic ECM-producing epithelial cell population which was drastically increased in IPF lungs. This new cell population was in parallel described by Adams et al. (13). Both studies linked the new cell type with the co-expression of basal epithelial, mesenchymal, and senescence markers as well as developmental transcription factors. The origin of the KRT5−/KRT17+ was either traced to ATII or Club cells (14) or myofibroblast foci (13). Overall, this leads to the hypothesis that ATII as well as Club cells can differentiate into an intermediate KRT5−/KRT17+ epithelial cell subtype which can further differentiate into ATI cells, which is in accordance to other studies describing an intermediate alveolar epithelial cell population (34, 35). These reports are in line with the hypothesis that an abnormal differentiation program is initiated in the tissue microenvironment of IPF in which the proximal-peripheral patterns of cell differentiation are disrupted. This results in many respiratory epithelial cells acquiring aberrant, multilineage-like states and some individual cells sharing the characteristics of both conducting airway and alveolar epithelial cells (23). The mobilization/activation of airway stem cells may be a potential mechanism to maintain tissue integrity. However, their differentiation into airway rather than alveolar phenotypes potentially accounts for the dysplastic structures observed in fibrotic lung tissues (36). These structural defects are considered to be secondary to epithelial progenitor cell dysfunction and defective epithelial-mesenchymal signaling (28).
Mesenchymal Cells
Mesenchymal cells in the alveolus serve multiple functions to maintain alveolar integrity. They generate and remodel the ECM, co-regulate the vasculature, help maintain and restore the epithelium, act as sentries for the immune system, and have been found to have trophic interactions with alveolar epithelial cells (37). After epithelial injury, fibroblasts become activated and induce many developmental pathways that are important for proliferation, ECM deposition, and chemokine and cytokine secretion to mediate tissue repair (8, 38). Aberrant proliferation and excessive ECM deposition result in fibrotic tissue remodeling and increased tissue stiffness found in lungs from IPF patients.
Historically, mesenchymal cells were mainly divided into fibroblasts, smooth muscle cells (SMCs) and pericytes, a derivative of fibroblasts with myofibroblast characteristics, that is closely associated with capillaries (39). Recent studies, including scRNAseq efforts, have vastly increased the heterogeneity of mesenchymal cells found in the human lung (12–15, 20, 22, 26, 27).
A recent comprehensive droplet- and plate-based scRNAseq study identified 4 fibroblast clusters within alveolar tissue samples. One cluster expressing classical fibroblast markers (COL1A1, COL1A2), two clusters enriched for ACTA2, a canonical marker of myofibroblasts, and a fourth cluster representing lipofibroblasts (APOE+, PLIN2+, COL1A1+), a cell that's presence in the human lung has long been controversial (40). Myofibroblasts are of particular interest in the development and progression of IPF. Myofibroblasts help forming alveoli during development and restore tissue integrity after injury. However, they are also considered the primary drivers of ECM deposition in fibrosis, and the key effector cells in IPF combining of the synthesizing features of fibroblasts with the cytoskeletal contractile characteristics of SMCs (2, 41). Interestingly, the ACTA2+ “myofibroblast” population could be separated into two clusters. One WIF1+, FGF18+, ASPN+ cluster representing “classical” myofibroblasts localized to alveolar ducts. The other sub-population, however, showed higher expression of contractile genes (MYH11, CNN1, TAGLN) and was termed “fibromyocyte”. Both expressed a rich set of genes associated with TGF-β signaling (LTBP1, LTBP2, ASPN, DPT, TGFBR3, TGFBI, SCX, MDFI) (20).
In line, a study by Liu et al. also identified up to eight mesenchymal cell populations in lung tissue from healthy and IPF donors. This included intermediate fibroblasts, again two sub-populations of ACTA2+ myofibroblasts and a population of lipofibroblasts (TCF21+, A2M+, RARRES2+, LIMCH1+, MACF1+), and an EBF1+ sub-population. Concordantly, myofibroblasts expressed contractile genes (MYH11). This was increased in IPF. These authors also found that all mesenchymal subtypes contributed to the excessive ECM production in fibrosis and that there is little evidence for trans-differentiation of fibroblast subtypes and that fibroblast fate is determined early in lung development (15). This is supported by another recent study that suggested a continuous trajectory toward IPF archetypes co-occurring within fibroblasts and myofibroblasts, but not necessarily across them. This study identified two stromal populations in IPF and control lungs. Fibroblasts, characterized by expression of CD34, FBN1, FBLN2, VIT, and myofibroblasts consistently expressing MYLK, NEBL, MYO10, MYO1D, RYR2, and ITGA8. The IPF lung in addition contained a myofibroblast phenotype enriched with fibrillar collagens, and ACTA2 and a fibroblast phenotype that exhibited increased expression of HAS1, HAS2, FBN1, and CXCL14. The authors suggest that pathological, ACTA2-expressing IPF myofibroblasts are not a discrete cell type, but rather one extreme pole of a continuum connected to a quiescent ACTA2-negative stromal population represented in control lungs (13). A HAS1hi fibroblast population was also found in a study released at the same time and suggested to represent an invasive phenotype as it appeared restricted to the immediate subpleural regions (14) where fibrotic foci are usually found first.
Overall, these studies suggest that the heterogeneity of the fibroblast population increases in IPF lungs compared to controls, that transdifferentiation of fibroblast subtypes likely is not relevant in IPF and that fibroblasts are transcriptionally distinct from myofibroblasts. Maybe even all sub-populations may contribute to the excessive ECM production seen in fibrosis.
Other Cells
Alveolar cell injury leads to inflammation and activation of immune cells. Murine models of bleomycin-induced fibrosis suggested that (monocyte-derived) macrophages could contribute to progression of fibrosis through overreactive responses to alveolar injury (42, 43). Recent scRNAseq reports also provide evidence for the involvement of macrophages in human fibrosis. A first study defined four macrophage clusters in IPF lungs in total. Two clusters including alveolar macrophages were equally represented in normal and fibrotic lungs. The other two clusters were almost exclusively found in lungs of patients with fibrosis. In combination with immunocytochemistry, CHI3LI, MMP9, and SSP1 were suggested as markers for macrophages detected in IPF samples (12). A subsequent study found three discrete macrophage sub-populations, which were all present in normal and fibrotic lungs, one expressing monocyte markers, one highly expressing FABP4 and INHBA (FABP4hi), and one highly expressing SPP1 and MERTK (SPP1hi). Proliferation of SPP1hi macrophages was strikingly increased in IPF lungs. Colocalization and causal modeling supported the role for these highly proliferative SPP1hi macrophages in activation of IPF myofibroblasts in lung fibrosis (25). In line, a recent study also found a profibrotic macrophage archetype that gradually and sequentially shifted toward its most extreme terminus with IPF progression. Expression of SPP1 and LIPA steadily increased relatively early along the trajectory with ECM remodeling genes SPARC, GPC4, PALLD, CTSK, and MMP9 ramping up at later stages (13).
In addition, this recent study also identified changes in vascular endothelial cells in IPF lungs. A vascular endothelial cell population that expresses COL15A1 was found in the IPF lung. This is transcriptomically indistinguishable from systemically supplied bronchial vascular endothelial cells detected in control lungs. However, in IPF lungs, these cells can readily be found in affected regions in the distal IPF parenchyma whereas these cells are restricted to the peri-bronchial vasculature in the normal lung and are never seen in the lung parenchyma. The authors speculate that the ectopic presence of these cells provides the cellular molecular correlate to an old notion that the bronchial vascular network is expanded throughout the IPF lung (13).
NEW Insights From scRNAseq Studies Into Disease Progression and Biomarker Identification
scRNAseq studies have vastly increased our understanding of cell types resident in the distal lung and also highlighted differences in cell populations/states between healthy and IPF tissue (12–15, 20, 25, 28, 44, 45). Recent efforts aimed at extending these insights to a better understanding of mechanisms for the local and temporal disease progression or leveraging scRNAseq analysis to progress disease biomarker identification (27, 46). Subclassifying IPF tissue samples into three pathological defined groups of no fibrosis detectable, mild fibrosis and advanced fibrosis a recent study detected large changes in IPF patients' samples even before histological changes in the respective regions were detectable. Thirteen different gene expression tracks were identified and assigned to changes in mRNA and microRNA (regulators of transcription) expression within different cell types over the respective disease progression states (46). Combining scRNAseq of lung tissue with proteomic analysis of bronchoalveolar lavage (BAL) and plasma from healthy and fibrotic donors another study discovered that fluid proteome signatures were predictive of specific cell state changes in the lung. Markers of several pro-fibrogenic cell types were strongly increased in protein measurements of patients that were diagnosed with different forms of interstitial lung diseases (ILD) including patients with IPF when compared to non-ILD controls. These protein signatures correlated with diagnosis, lung function, smoking and injury status (27). scRNAseq data also offer the opportunity to identify transcription factors, signaling interactions and hormone/immune targets linked to disease onset and progression (20). Similarly, it offers the opportunity to identify novel diseases-related genes.
scRNAseq to Understand Effects of Drug Treatment for IPF
To date, Nintedanib and Pirfenidone are the only approved drugs for treatment of IPF (47, 48). Two recent studies employed scRNAseq to investigate the effects of drug treatment on cells from IPF patients. A comparative transcriptomic approach on lung homogenates and lung fibroblasts derived from IPF patients treated with or without pirfenidone revealed that pirfenidone exerts the beneficial effects via its action on multiple pathways in both fibroblasts and other pulmonary cells. In fibroblasts, pirfenidone therapy predominantly affected growth and cell division pathways indicating a major cellular metabolic shift. CEMIP, linked to cell migration and ECM production, was downregulated in fibroblasts and lung homogenates (49). Nintedanib treatment resulted in 157 up- and 151 downregulated genes in IPF fibroblasts. Bioinformatics analysis revealed strong protein–protein interactions within these dysregulated genes, mostly involved in the pathways of cell cycle and mitotic cell cycle. Moreover, changes in expression of several miRNAs were identified, in particular upregulation of hsa-miR-486-3p which might repress expression of DDX11, E2F1, and PLXNA4, genes linked to fibroblast proliferation and reduction of angiogenesis (50). Further studies will be required to foster our understanding of drug treatment on individual cell populations in IPF. Corticosteroids are often used to dampen the effects of acute exacerbations in IPF, however, the benefit is still debated (51) and might even be contra-indicated in combination with immunosuppressive treatment (52). scRNAseq could potentially help in identifying beneficial or adverse effects of corticosteroid treatment on individual cell populations and delineate cellular target effects.
Challenges and Future Perspectives
scRNAseq methods enable unbiased, high-throughput, and high-resolution transcriptomic analysis of individual cells. This provides an additional dimension to transcriptomic information relative to traditional methods that profile bulk populations of cells and are uniquely suited to look for heterogeneity within cell populations that might emerge during disease (53). However, scRNAseq approaches still have limitations and require cautions interpretation of data. In comparison with bulk RNAseq, the obtained data are noisier, complicating computational analysis and leading to a drop-out of weakly expressed genes (54). Likewise, identifying clusters based on delineating marker genes reported in earlier studies introduce a bias during the process of cell clustering (15). Although new analytical tools have been designed, methods resolving technical noise, considering expression variability and offering unbiased clustering of cells are still not available (55). In addition, cell isolation procedures, including up- and downstream processing, are highly variable between studies, limiting reproducibility and impacting on accuracy of the reported data sets often leading to over- or underrepresentation of specific cellular sub-populations. It will be a major challenge for the future to converge the various datasets and identify unifying marker sets to identify and specify individual cell types. In addition, the abundance of data generated in scRNAseq studies are currently lacking realistic methods of validation on the protein level due to the lack of methods for proteomics at this scale. Albeit, first attempts are under way link scRNAseq data and proteomic analysis to identify disease specific changes on the cellular level and their corresponding reflection in body fluid proteomes (27). Animal studies, despite their limitations, will also benefit in validating data from scRNAseq studies in experimental models. Some studies are already available that confirm the predicted alterations in cellular composition between healthy and fibrotic lung tissue. For example, the appearance of cells expressing airway stem cell markers in the alveolar region was associated with fibrosis in a murine bleomycin model (56).
Nevertheless, new tools enabling data analysis, visualization, and interpretation are being developed at a rapid pace and are readily available as “open source” tools (8). Within the last years, further development and optimization of new scRNAseq protocols are now enabling high throughput measurements (57–59). Novel technologies including RNA velocity and “spatial transcriptomics” can extend and complement scRNAseq studies and deepen our understanding of dynamic processes and/or spatial differences in lung development, cellular organization, and disease progression. RNA velocity, the time derivative of the gene expression state, is a high-dimensional vector that predicts the future state of individual cells (on a timescale of hours) and aids the analysis of developmental lineages and cellular dynamics (60, 61) while “spatial transcriptomics,” allows visualization and quantitative analysis of the transcriptome with high spatial resolution in individual tissue sections (62, 63). Such technologies can be particularly useful in investigating the onset and progression of IPF, a disease with great local heterogeneity (“fibrotic foci”) and rapid progression. Therefore, recent studies investigating lung cell identity and/or changes in fibrotic tissue have already started to apply such approaches (14, 29, 44, 56, 64, 65).
The rising number of sequenced cells and the number of patients per study are strengthening accuracy and resolution (Table 1). The reduction of costs and increase in throughput also facilitates scRNAseq studies to be performed at multiple time points, supporting the delineation of disease progression. Most datasets are publicly available and offer the opportunity to analyze combined datasets in the quest for discovering new therapeutic targets. In addition, exploiting these advancements in scRNAseq and combining them with complementary technologies including bulk RNASeq, imaging, and functional studies will further strengthen mechanistic insights into the onset and progression of IPF and aid in the development of new therapeutics.
Author Contributions
JN, AS, and MF wrote the manuscript. All authors agree to be accountable for the content of the work.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Projektnummer(n): 278012962 (GRK Pulmosens), 251293561 (SFB1149), and 175083951 (to MF) and a grant from the Ministry of Science, Research and the Arts of Baden-Württemberg (Az: 32-7533.-6-10/15/5) (to MF).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. (2018) 378:1811–23. doi: 10.1056/NEJMra1705751
2. Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. (2012) 122:2756–62. doi: 10.1172/JCI60323
3. Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. (2013) 123:3025–36. doi: 10.1172/JCI68782
4. Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. (2014) 507:190–4. doi: 10.1038/nature12930
5. Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. (2014) 15:123–38. doi: 10.1016/j.stem.2014.07.012
6. Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. (2018) 555:251–5. doi: 10.1038/nature25786
7. Sisson TH, Mendez M, Choi K, Subbotina N, Courey A, Cunningham A, et al. Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am J Respir Crit Care Med. (2010) 181:254–63. doi: 10.1164/rccm.200810-1615OC
8. Basil MC, Katzen J, Engler AE, Guo M, Herriges MJ, Kathiriya JJ, et al. The cellular and physiological basis for lung repair and regeneration: past, present, and future. Cell Stem Cell. (2020) 26:482–502. doi: 10.1016/j.stem.2020.03.009
9. Wolters PJ, Blackwell TS, Eickelberg O, Loyd JE, Kaminski N, Jenkins G, et al. Time for a change: is idiopathic pulmonary fibrosis still idiopathic and only fibrotic? Lancet Respir Med. (2018) 6:154–60. doi: 10.1016/S2213-2600(18)30007-9
10. Bagnato G, Harari S. Cellular interactions in the pathogenesis of interstitial lung diseases. Eur Respir Rev. (2015) 24:102–14. doi: 10.1183/09059180.00003214
11. Tschumperlin DJ, Ligresti G, Hilscher MB, Shah VH. Mechanosensing and fibrosis. J Clin Invest. (2018) 128:74–84. doi: 10.1172/JCI93561
12. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 199:1517–36. doi: 10.1164/rccm.201712-2410OC
13. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single Cell RNA-seq reveals ectopic and aberrant lung resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. (2020) 6:eaba1983. doi: 10.1101/759902
14. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Carla L, et al. Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv. (2020) 6:1972. doi.org/10.1101/753806
15. Liu X, Rowan SC, Liang J, Yao C, Huang G, Deng N, et al. Definition and signatures of lung fibroblast populations in development and fibrosis in mice and men. bioRxiv [Preprint]. (2020) 2020.07.15.203141. doi: 10.1101/2020.07.15.203141
16. Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, et al. Anatomically and functionally distinct lung mesenchymal populations marked by Lgr5 and Lgr6. Cell. (2017) 170:1149–63.e12. doi: 10.1016/j.cell.2017.07.028
17. Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. (2017) 170:1134–48.e10. doi: 10.1016/j.cell.2017.07.034
18. Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. (2018) 359:1118–23. doi: 10.1126/science.aam6603
19. Tang YW, Johnson JE, Browning PJ, Cruz-Gervis RA, Davis A, Graham BS, et al. Herpesvirus DNA is consistently detected in lungs of patients with idiopathic pulmonary fibrosis. J Clin Microbiol. (2003) 41:2633–40. doi: 10.1128/JCM.41.6.2633-2640.2003
20. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single cell RNA sequencing. Nature. (2020). doi: 10.1038/s41586-020-2922-4. [Epub ahead of print].
21. Moore C, Blumhagen RZ, Yang IV, Walts A, Powers J, Walker T, et al. Resequencing study confirms that host defense and cell senescence gene variants contribute to the risk of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 200:199–208. doi: 10.1164/rccm.201810-1891OC
22. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med. (2019) 25:1153–63. doi: 10.1038/s41591-019-0468-5
23. Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. (2016) 1:e90558. doi: 10.1172/jci.insight.90558
24. Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol. (2017) 19:904–14. doi: 10.1038/ncb3580
25. Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J. (2019) 54:1802441. doi: 10.1183/13993003.02441-2018
26. Valenzi E, Bulik M, Tabib T, Morse C, Sembrat J, Trejo Bittar H, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis. (2019) 78:1379–87. doi: 10.1136/annrheumdis-2018-214865
27. Mayr C, Simon L, Leuschner G, Ansari M, Geyer P, Angelidis I, et al. Integrated single cell analysis of human lung fibrosis resolves cellular origins of predictive protein signatures in body fluids. SSRN Electron J. (2020). doi: 10.1101/2020.01.21.20018358
28. Carraro G, Mulay A, Yao C, Mizuno T, Konda B, Petrov M, et al. Single cell reconstruction of human basal cell diversity in normal and IPF lung. Am J Respir Crit Care Med. (2020). doi: 10.1164/rccm.201904-0792oc. [Epub ahead of print].
29. Tsukui T, Sun K-H, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun. (2020) 11:1920. doi: 10.1038/s41467-020-15647-5
30. Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, et al. Plasticity of Hopx+ type I alveolar cells to regenerate type II cells in the lung. Nat Commun. (2015) 6:6727. doi: 10.1038/ncomms7727
31. Chilosi M, Poletti V, Murer B, Lestani M, Cancellieri A, Montagna L, et al. Abnormal re-epithelialization and lung remodeling in idiopathic pulmonary fibrosis: the role of ΔN-p63. Lab Investig. (2002) 82:1335–45. doi: 10.1097/01.LAB.0000032380.82232.67
32. Seibold MA, Smith RW, Urbanek C, Groshong SD, Cosgrove GP, Brown KK, et al. The idiopathic pulmonary fibrosis honeycomb Cyst contains a mucocilary pseudostratified epithelium. PLoS ONE. (2013) 8:e58658. doi: 10.1371/journal.pone.0058658
33. Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. (2009) 106:12771–5. doi: 10.1073/pnas.0906850106
34. Jansing NL, McClendon J, Henson PM, Tuder RM, Hyde DM, Zemans RL. Unbiased quantitation of alveolar type II to alveolar type i cell transdifferentiation during repair after lung injury in mice. Am J Respir Cell Mol Biol. (2017) 57:519–26. doi: 10.1165/rcmb.2017-0037MA
35. Aspal M, Zemans RL. Mechanisms of ATII-to-ATI cell differentiation during lung regeneration. Int J Mol Sci. (2020) 21:3188. doi: 10.3390/ijms21093188
36. Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. (2015) 517:621–5. doi: 10.1038/nature14112
37. Barron L, Gharib SA, Duffield JS. Lung pericytes and resident fibroblasts: busy multitaskers. Am J Pathol. (2016) 186:2519–31. doi: 10.1016/j.ajpath.2016.07.004
38. Green J, Endale M, Auer H, Perl AKT. Diversity of interstitial lung fibroblasts is regulated by platelet-derived growth factor receptor a kinase activity. Am J Respir Cell Mol Biol. (2016) 54:532–45. doi: 10.1165/rcmb.2015-0095OC
39. Weibel ER. On pericytes, particularly their existence on lung capillaries. Microvasc Res. (1974) 8:218–35. doi: 10.1016/0026-2862(74)90096-X
40. Tahedl D, Wirkes A, Tschanz SA, Ochs M, Mühlfeld C. How common is the lipid body-containing interstitial cell in the mammalian lung? Am J Physiol Lung Cell Mol Physiol. (2014) 307:386–94. doi: 10.1152/ajplung.00131.2014
41. Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. (2007) 170:1807–16. doi: 10.2353/ajpath.2007.070112
42. Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. (2017) 214:2387–404. doi: 10.1084/jem.20162152
43. McCubbrey AL, Barthel L, Mohning MP, Redente EF, Mould KJ, Thomas SM, et al. Deletion of c-FLIP from CD11bhi macrophages prevents development of bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol. (2018) 58:66–78. doi: 10.1165/rcmb.2017-0154OC
44. Peyser R, MacDonnell S, Gao Y, Cheng L, Kim Y, Kaplan T, et al. Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am J Respir Cell Mol Biol. (2019) 61:74–85. doi: 10.1165/rcmb.2018-0313OC
45. Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, Chapman HA. Distinct airway epithelial stem cells hide among club cells but mobilize to promote alveolar regeneration. Cell Stem Cell. (2020) 26:346–58.e4. doi: 10.1016/j.stem.2019.12.014
46. McDonough JE, Ahangari F, Li Q, Jain S, Verleden SE, Maya JH, et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight. (2019) 4:1–15. doi: 10.1172/jci.insight.131597
47. King TE, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. (2014) 370:2083–92. doi: 10.1056/NEJMoa1402582
48. Richeldi L, Du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. (2014) 370:2071–82. doi: 10.1056/NEJMoa1402584
49. Kwapiszewska G, Gungl A, Wilhelm J, Marsh LM, Puthenparampil HT, Sinn K, et al. Transcriptome profiling reveals the complexity of pirfenidone effects in idiopathic pulmonary fibrosis. Eur Respir J. (2018) 52:1800564. doi: 10.1183/13993003.00564-2018
50. Sheu C-C, Chang W-A, Tsai M-J, Liao S-H, Chong I-W, Kuo P-L. Gene expression changes associated with nintedanib treatment in idiopathic pulmonary fibrosis fibroblasts: a next-generation sequencing and bioinformatics study. J Clin Med. (2019) 8:308. doi: 10.3390/jcm8030308
51. Collard HR, Ryerson CJ, Corte TJ, Jenkins G, Kondoh Y, Lederer DJ, et al. Acute exacerbation of idiopathic pulmonary fibrosis an international working group report. Am J Respir Crit Care Med. (2016) 194:265–75. doi: 10.1164/rccm.201604-0801CI
52. The Idiopathic Pulmonary Fibrosis Clinical Research Network Raghu G Anstrom KJ King TE Lasky JA Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. (2012) 366:1968–77. doi: 10.1056/NEJMoa1113354
53. Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. (2015) 58:610–20. doi: 10.1016/j.molcel.2015.04.005
54. Bai YL, Baddoo M, Flemington EK, Nakhoul HN, Liu YZ. Screen technical noise in single cell RNA sequencing data. Genomics. (2020) 112:346–55. doi: 10.1016/j.ygeno.2019.02.014
55. Chen G, Ning B, Shi T. Single-cell RNA-seq technologies and related computational data analysis. Front Genet. (2019) 10:317. doi: 10.3389/fgene.2019.00317
56. Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun. (2020) 11:3559. doi: 10.1038/s41467-020-17358-3
57. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. (2015) 161:1202–14. doi: 10.1016/j.cell.2015.05.002
58. Nichterwitz S, Chen G, Aguila Benitez J, Yilmaz M, Storvall H, Cao M, et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun. (2016) 7:12139. doi: 10.1038/ncomms12139
59. Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, et al. Seq-Well: Portable, low-cost rna sequencing of single cells at high throughput. Nat Methods. (2017) 14:395–8. doi: 10.1038/nmeth.4179
60. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. (2018) 560:494–8. doi: 10.1038/s41586-018-0414-6
61. Svensson V, Pachter L. RNA Velocity: molecular kinetics from single-cell RNA-seq. Mol Cell. (2018) 72:7–9. doi: 10.1016/j.molcel.2018.09.026
62. Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. (2016) 353:78–82. doi: 10.1126/science.aaf2403
63. Salmén F, Vickovic S, Larsson L, Stenbeck L, Vallon-Christersson J, Ehinger A, et al. Multidimensional transcriptomics provides detailed information about immune cell distribution and identity in HER2+ breast tumors. bioRxiv [Preprint]. (2018) 358937. doi: 10.1101/358937
64. Riemondy KA, Jansinsg NL, Jiang P, Redente EF, Gillen AE, Fu R, et al. Single-cell RNA sequencing identifies TGF-β as a key regenerative cue following LPS-induced lung injury. JCI Insight. (2019) 5:e123637. doi: 10.1172/jci.insight.123637
Keywords: fibroblast, alveolar, lung, IPF, ATII cells, scRNA sequencing
Citation: Nemeth J, Schundner A and Frick M (2020) Insights Into Development and Progression of Idiopathic Pulmonary Fibrosis From Single Cell RNA Studies. Front. Med. 7:611728. doi: 10.3389/fmed.2020.611728
Received: 29 September 2020; Accepted: 16 November 2020;
Published: 16 December 2020.
Edited by:
Elena Lopez-Rodriguez, Charité–Universitätsmedizin Berlin, GermanyReviewed by:
Amiq Gazdhar, Bern University Hospital, SwitzerlandAlessandro Venosa, The University of Utah, United States
Copyright © 2020 Nemeth, Schundner and Frick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manfred Frick, bWFuZnJlZC5mcmlja0B1bmktdWxtLmRl
†These authors have contributed equally to this work
 Julia Nemeth
Julia Nemeth Annika Schundner
Annika Schundner Manfred Frick
Manfred Frick