- 1School of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology (POSTECH), Pohang, South Korea
- 2Department of Mechanical Engineering, Pohang University of Science and Technology (POSTECH), Pohang, South Korea
- 3Department of Convergence IT Engineering, Pohang University of Science and Technology (POSTECH), Pohang, South Korea
- 4Institute of Convergence Science, Yonsei University, Seoul, South Korea
Increasing evidence has accumulated that gut microbiome dysbiosis could be linked to neurological diseases, including both neurodegenerative and psychiatric diseases. With the high prevalence of neurological diseases, there is an urgent need to elucidate the underlying mechanisms between the microbiome, gut, and brain. However, the standardized animal models for these studies have critical disadvantages for their translation into clinical application, such as limited physiological relevance due to interspecies differences and difficulty interpreting causality from complex systemic interactions. Therefore, alternative in vitro gut–brain axis models are highly required to understand their related pathophysiology and set novel therapeutic strategies. In this review, we outline state-of-the-art biofabrication technologies for modeling in vitro human intestines. Existing 3D gut models are categorized according to their topographical and anatomical similarities to the native gut. In addition, we deliberate future research directions to develop more functional in vitro intestinal models to study the gut–brain axis in neurological diseases rather than simply recreating the morphology.
Introduction
The gut–brain axis (GBA) refers to bidirectional interactions among the brain, gut, and intestinal microbiome (1, 2) (Figure 1A). Many studies link dysregulation of the GBA to various pathologies from gastrointestinal (GI) symptoms (6) to neurological diseases including neurodegenerative diseases (7–9) and psychiatric disorders (10) (Figure 1B). In particular, the interesting modulation effect of the intestinal microbiome in GBA has been highlighted in neurological diseases such as Alzheimer's disease (11–13), Parkinson's disease (14–16), epilepsy (9, 17), autism spectrum disorders (18, 19) and anxiety or depression (20). Thus far, three major communication pathways have been identified in GBA: (a) the immune system that carries cytokines, (b) the vagus nerve that carries neuronal messages, and (c) the neuroendocrine system that carries neurotransmitters and GI hormones (21).
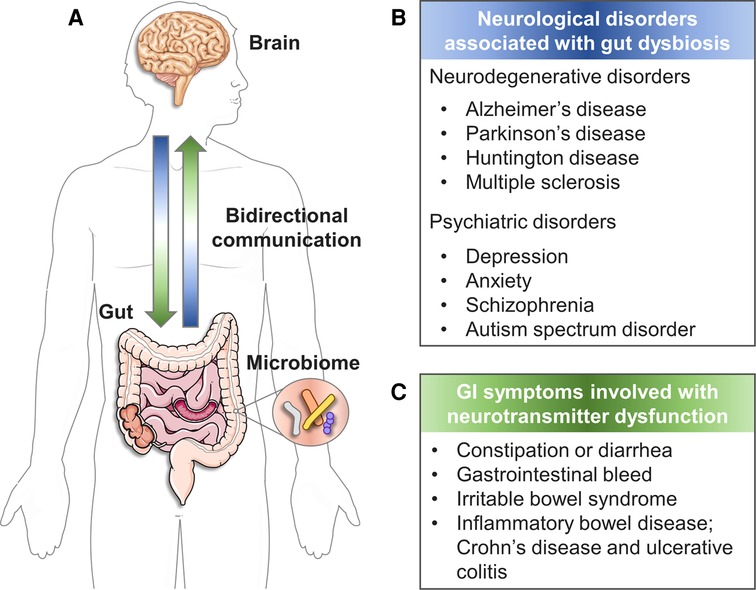
Figure 1. The bidirectional GBA. (A) The reciprocal communication between gut and brain. (B) Neurological disorders associated with gut dysbiosis (3). (C) GI symptoms involved with neurotransmitter dysfunction (4, 5). (created with Servier Medical Art; smart.servier.com).
Conventional animal models for GBA (e.g., gnotobiotic mice) have been invaluable in advancing our insights into how the microbiota and its changes impact the GI and brain (patho-) physiology, while also demonstrating causal linkages between certain microbial cohorts and disease phenotypes (22–25). However, there are critical unsolved issues when translating data from animal models to the human system. First, the interspecies differences in GI topography, microbiome profile, immune system, and brain function limit the relevance of animal models (26). In addition, it is challenging to decipher the etiology of multifactorial disorders involving the GBA due to the extremely convoluted nature of systemic interactions between multiple organs as well as the immune and nervous systems. Furthermore, disentangling the impact of specific microbiome-derived compounds from the context of the whole gut environment is a demanding task (27).
The questionable validity of animal GBA models led to a compelling need for a human-based, preclinical in vitro model that is able to dissect the intricate interplay in GBA. From an engineering viewpoint, the essence of in vitro GBA models is versatile modularity. In other words, engineers ultimately aim to (a) define the vital factors of complicated disease conditions involving multiple organs, (b) deconvolute them as independent parameters and capture these in the simplest possible configuration, and (c) couple them in combination in a scalable, well-controlled, and reproducible manner (26–28).
In this review, we attempt to suggest research directions to create in vitro GBA models in the context of neurological disease as an alternative to conventional animal models. Discussions regarding the in vitro brain and blood-brain barrier models for GBA have been extensively described elsewhere (29); therefore, here we would like to delineate the state-of-the-art in vitro 3D gut models highlighted in the field of GBA modeling so far. Major biofabrication technologies for gut modeling are classified according to their dimensions and geometrical properties. In addition, we discuss the challenges ahead toward functional gut models for in vitro GBA and strategies to surmount them.
In vitro blood-brain barrier models for GBA
Tardy progress in therapeutics development for neurological diseases has driven a need for in vitro blood-brain barrier (BBB) models. Based on their geometrical and dimensional features, these models can be categorized as: planar microphysiological systems (MPSs) based on porous membrane substrates (30–32), spheroid models (33, 34), perfusable hydrogel-laden MPS (35, 36), and perfusable microfluidic model (37) (Figure 2). Although these models have provided mechanistic knowledge about neurological diseases, they need to be optimized for future use in GBA platforms. Simplifying the multi-step, complex fabrication process of current BBB models (38, 39) will increase fabrication efficiency of multi-organ platforms, such as GBA. More realistic recapitulation of BBB anatomies (e.g., tubular architectures with curves or bifurcations in various diameters) is indispensable as it enhances the physiological relevance of neuroinflammatory responses in in vitro BBB models (40, 41) that will increase the reliability of the GBA models. Finally, free-standing BBB models would be easy to assembly with other organ modules and study the molecular transport between them. For more information about in vitro BBB models, the readers may refer to the references (42, 43).
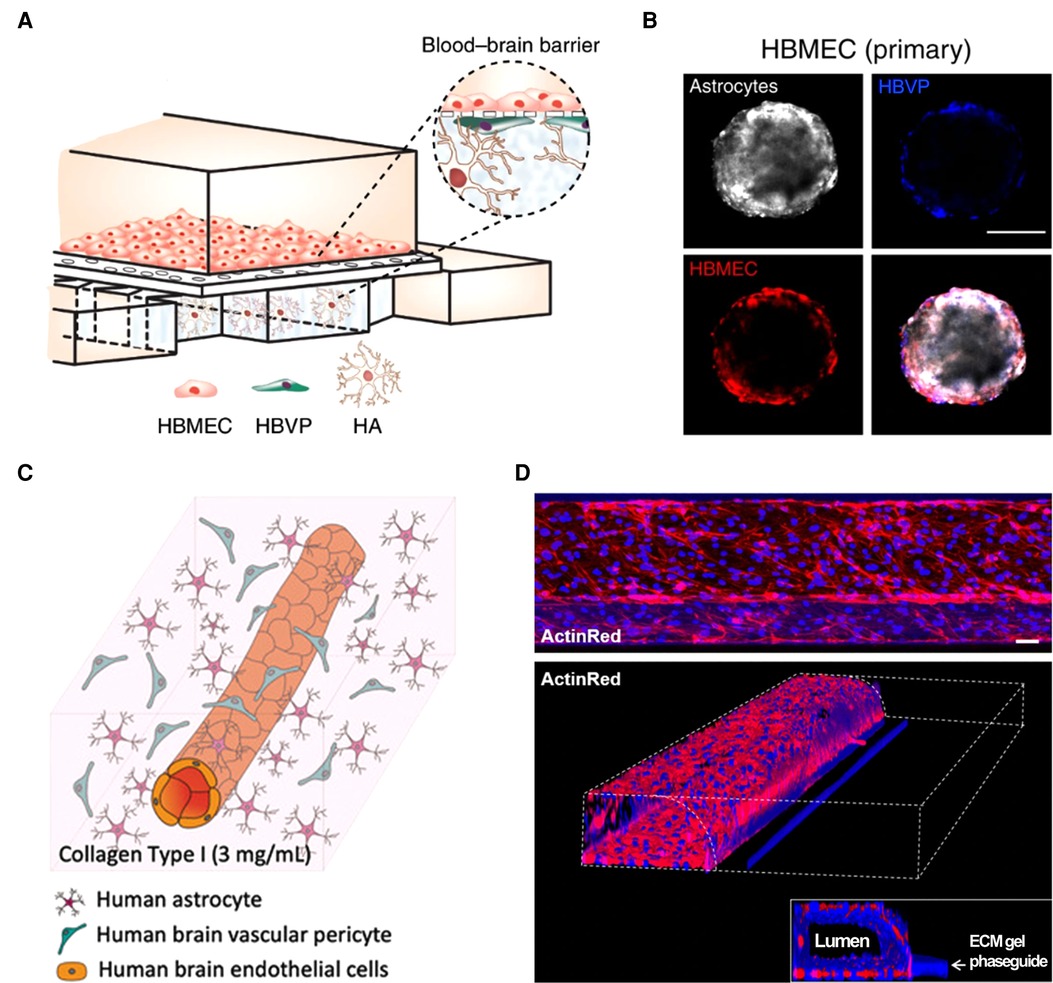
Figure 2. Current in vitro blood–brain barrier models. (A) Planar MPS based on porous membrane substrate (32). (B) Spheroid model (33). (C) Perfusable hydrogel-laden MPS (36). (D) Perfusable microfluidic model (37). All open access (CC-BY).
Current in vitro gut models with different anatomical complexity
2D models (e.g. planar cell culture) have been the mainstream approach to studying human intestinal normal- or patho-physiology and defining potential therapeutic strategies (44–46). However, these simplified 2D models cannot truly capture the complexity of intestinal tissue morphology and physiology (46–48); cells interact with their surrounding cells and heterogeneous and complex environments via elaborate biochemical signals cascade (49), i.e. cell–cell and cell–matrix interactions. Accordingly, more sophisticated 3D models that closely capture the in vivo milieu are believed to bridge the gap between conventional cell cultures and animal models (50–52). Here, we distinguished the up-to-date in vitro gut models by their level of anatomical complexity and briefly introduced biofabrication techniques and their features employed to fabricate the models.
Quasi-3D intestinal epithelium
Microphysiological systems
Kim et al. (53) opened a window into the world of human intestinal MPSs with their revolutionary work on dynamic mechanical stimulation of the intestinal epithelium. MPSs are defined as microfluidic platform devices that emulate in vivo organ physiology and function in vitro in a controlled and standardized manner (Figure 3A) (54, 55). The gut-on-a-chip developed by Kim et al. consisted of two parallel microchannels separated by a porous membrane coated with ECM and lined with human enterocyte cell line Caco-2 to reproduce the intestinal barrier. This gut-on-a-chip emulating peristalsis-like motions and luminal flow in vivo demonstrated its capability in coculture with the microbiome (62, 63), modeling gut inflammation (64, 65) and intestinal morphogenesis (66, 67) and integration with an ECM membrane (68) and intestinal organoids/enteroids (56, 67, 69) in separate reports. The organoid-mounted microfluidics could be a useful tool for studying dynamic GI hormone secretion related to digestion and response to nutrients (56). Besides, gut-on-a-chip could be a great experimental model for the real-time, non-invasive monitoring of oxygen gradient (63) or mucus production (70).
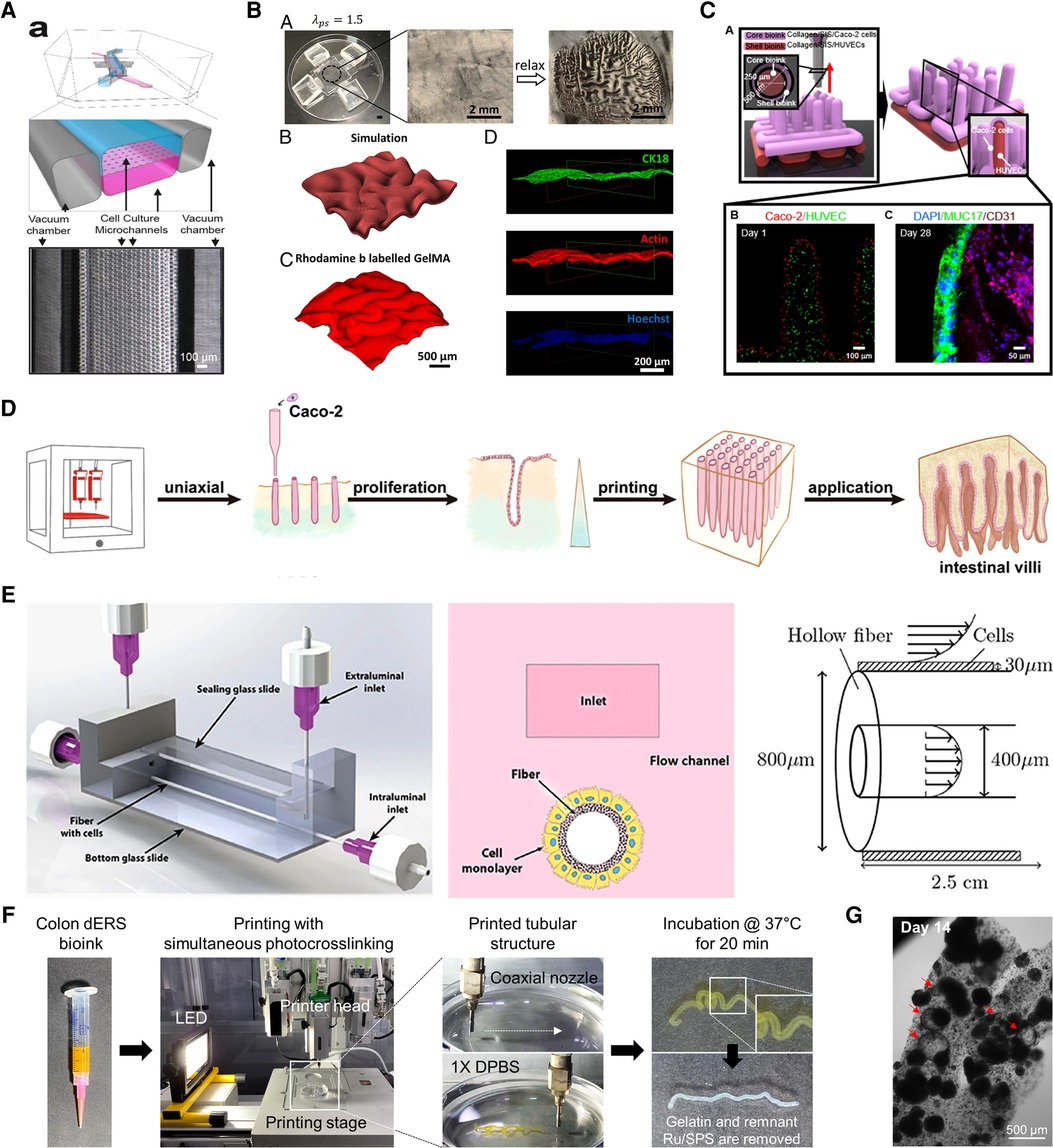
Figure 3. Biofrbrication technologies for in vitro gut models. (A) Quasi-3D microphysiological systems. Reproduced from Kasendra et al. (56). Open access (CC-BY). (B) 3D topography patterning. Reproduced from Chan et al. (57). with permission from the author. (C) 3D direct bioprinting. Reproduced from Kim et al. (58). Open access (CC-BY 4.0). (D) 3D embedded bioprinting. Adapted from Lian et al. (59) with permission from the publisher. (E) 3D hollow scaffold. Reproduced from Langerak et al. (60). Open access (CC-BY 4.0). (F,G) 3D coaxial bioprinting assisted with photocrosslinking. Reproduced from Han et al. (61) with permission from the publisher.
However, this design only includes part of the four layers of the intestinal walls, resulting in the absence of other elements that can be involved in certain disorders. (46). A fundamental limitation in the molding-and-replication-based fabrication technique is that it not only involves multiple labor-intensive steps but also impedes dimensional expansion into 3D architectures, confining them to mainly 2D or 2.5D geometries (54). Lastly, polydimethylsiloxane (PDMS) is the most favorable material for manufacturing MPSs but the PDMS surface in microfluidic channels also has potential problems in adsorbing small and hydrophobic molecules.
3D Intestinal topographies
Patterning
Capturing the 3D topography of the intestine in an in vitro culture environment can stimulate the exhibition of more realistic functions since numerous signaling gradients are present along the crypt–villus axis (71, 72). Sung et al. used PDMS replica to microfabricate a 3D hydrogel intestinal villi structure (73). Natural and synthetic hydrogels, collagen, and polyethylene glycol diacrylate respectively, were used to make the villi substrate where Caco-2 cells were seeded. The substrate was fully covered with epithelial cells three weeks after seeding and generated similar finger-like epithelial morphologies to those of the human jejunum. This kind of biomimetic scaffolds with a crypt–villus topography can impose geometric boundary conditions that resemble those in vivo to organoids or enteroids and guide their self-organization, thus improving their architecture and size (74, 75). Recently, a microfabricated array of collagen crypts and villi cultured with human enteroids effectively generated a self-renewing monolayer, crypt–villus architecture, and opposing gradients of morphogens seen in the native niche of the intestinal epithelium (76).
Another surface feature of the intestine, the circular folds in the mucosal layer, also has certain important roles. The morphological patterns seen in the mucosa considerably increase the surface area, which is beneficial for absorbing nutrients and water (77). In addition, the folded mucosa ease the passage of luminal contents because the luminal diameter of the organ can expand significantly by releasing the folds without causing high stress that may harm the mucosal layer (78–80). In nature, these wrinkle-like structures are generated due to heterogeneous growth rates during development, which generates stress mismatches between tissue layers and accordingly induces mechanical instability (81). Zhao et al. (57) reported a method to recapitulate the folding of artificial mucosa in a controllable manner using layered hydrogel systems (Figure 3B). First, a tough and stretchable hydrogel substrate was made simply by pressing the pregel solution between two layers of glass. PDMS holders stretched and fixed the substrate in a uniaxial or biaxial direction and then an epithelial cell-laden soft gelatin methacrylate (GelMA) hydrogel was attached to the prestretched substrate. As a result, the relaxation of prestretched tough hydrogel induced programmed self-folding. This system demonstrated the mesenchymal condensation process in vitro, which facilitates the understanding of mucosal folding.
Bioprinting
3D bioprinting is a groundbreaking technology that encompasses a wide range of disciplines and is one of the most attractive tools in tissue engineering (TE) (82, 83). Traditionally, this additive manufacturing process can be classified based on its distinct approach to create a solidified layer: vat polymerization (84), material extrusion (85), and material jetting (86). 3D bioprinting using hydrogel bioink with cells in TE applications is categorized into four types according to their ink-dispensing method: laser-assisted printing, stereolithography, inkjet printing, and microextrusion printing (87). Harnessing bioprinting has established a new pillar of engineering intestinal tissues in vitro (58, 59, 61, 88, 89). Lian et al. (59) described an embedded bioprinting strategy using a dual-layered support base to produce a vertical array of intestinal villi with dimensions close to the native tissue (100–200 µm in diameter and ≈2 mm in length) (Figure 3D) (90). Embedded bioprinting enables the extrusion of inks into a (sacrificial) support bath/matrix and is advantageous when using mechanically weak bioinks, which often pose a trade-off between printing fidelity and biocompatibility (91–93). The authors printed a bioink composed of gelatin and Caco-2 cells into a vertical filament shape into GelMA support, which has an upper and lower layer of different concentrations. Gelatin dissolved away at physiological temperature conditions during culture and left microchannel-like villus while still supporting the attachment and growth of encapsulated cells. Intriguingly, the heterogeneous microenvironment of GelMA created gradients in both nutrients and oxygen along the length of the hollow villus structure, simply by reconstructing the hypoxic crypt and normoxic villus bases.
Among various 3D bioprinting techniques, coaxial extrusion printing has provoked tremendous interest because of its ability to directly fabricate perfusable tubular structures by co-extruding multiple materials through a concentrically assembled core–shell nozzle (94, 95). Kim et al. (58) demonstrated a free-standing 3D villi structure composed of a small intestinal epithelium core and microvascular shell using coaxial printing (Figure 3C). To make the projective finger-like villus using collagen/decellularized small intestinal submucosa-based bioink with limited mechanical stiffness, tannic acid was adopted as a crosslinking agent. Analysis of the bioengineered villus showed enhanced cellular activities inclusive of cell viability, proliferation, and the expression of mucin and junctional protein.
3D Geometry with a hollow lumen
Although tubular structures of epithelial organs have been of great interest to TE researchers (72, 96–105), there have only been a few studies on the subject of culturing intestinal epithelial cells (IECs) on the inner surface of the hollow structure until recently. Unique advantages that such hollow tubular shapes can offer in intestinal models include (a) accessibility to both the apical and basolateral side of the intestinal epithelium, which is necessary to study trans-epithelial transport (106); (b) extending the lifespan of the intestinal tissue model via perfusion through the lumen (107); (c) intraluminal oxygen gradient, which is critical in the mutual interaction between intestinal microbiomes and altered epithelium condition (108); (d) accelerated differentiation of IECs (61, 71). In this regard, subsequent sections explore representative fabrication methods for 3D hollow tubular intestinal models.
Conventional scaffold approach
As one of the earliest works on the effect of 3D lumen configuration in intestinal models, a culture system using porous hollow fibers of polyethersulfone was applied to grow Caco-2 cells (71). Differentiation of seeded Caco-2 cells was accelerated over six days. In addition, it was investigated that the tight paracellular barriers formation and brush border enzymes expression was increased compared to the conventional Transwell culture. The shortened time required to differentiate the Caco-2 cells in the hollow tubular system is highly relevant to the rapid differentiation of enterocytes in in vivo human intestines (3–5 days) (109, 110).
Microenvironmental cues such as ECM and external forces are important factors to mimic the architecture and physiological parameters of the native intestine (Figure 3E) (60, 106). In this bioengineered intestinal tubule, Caco-2 cells were grown on a human collagen IV- and levodopa-coated hollow fiber membrane with different curvatures (106). In addition, the intestinal tubules were exposed to unidirectional shear stress for the last few days of culture. Under the dynamic condition, Caco-2 cells rapidly formed a monolayer, increasing the speed of the polarization process, inducing apical and basolateral sides, and promoting differentiation into multiple phenotypes including enterocytes, goblet, Paneth, enteroendocrine (EEC), and stem cells compared to the static condition.
Different cell types can be involved by adding layers hierarchically. Roh et al. (111) fabricated silk scaffolds with a hollow lumen space in different sizes using cylindrical molds. Human colonic organoids (colonoids) were seeded on the inner surface of the smaller scaffold and then assembled with the outer scaffold where human primary macrophages were cultured. In response to inflammation caused by Escherichia coli lipopolysaccharide, the migration of macrophages toward the epithelium was observed. Other inflammatory responses, such as increased macrophage infiltration and the production of pro-inflammatory cytokines were also verified.
Bioprinting with a high cell population
One current trend in TE is the scaffold-free approach because scaffolding material often interferes with cell-to-cell or cell-to-matrix interactions (112–114). In this context, researchers seek to reinforce cells' ability to produce a matrix by applying proper exogenous stimuli such as stiffness, mechanical stretch, and contact with ECM. These exogenous cues can initiate cellular self-assembly and self-organization when involving external forces such as centrifugation or 3D bioprinting (115). Recently, bioprinting-assisted tissue emergence (BATE) was suggested based on depositioning high-density organoid-forming stem cells directly into a highly permissive ECMs suspension of Matrigel and collagen (88). BATE permitted spatiotemporal control of the cells and the bioactive ECM liquid precursor facilitated cellular self-assembly into macrostructures following the geometrical constraints imposed by the printing process. The printed intestinal organoids robustly fused and evolved into native-like tubular tissue with budding structure, demonstrating their potential in guiding tissue morphogenesis. BATE eminently exemplified the advantage of the automated 3D bioprinting system in handling organoids/enteroids; it can handle these delicate cells in a scalable and reproducible manner (116).
Bioprinting with tube-like geometry
The first attempt to fabricate a hollow tubular in vitro human intestinal model with the coaxial printing technique was achieved by Kang et al. (89) using a colon-derived decellularized extracellular matrix (dECM)-based bioink. The tissue-specific bioink was supplemented with visible light photoinitiator ruthenium/sodium persulfate (Ru/SPS) to increase the capacity of the crosslinking speed (dERS) (117). Introducing a photoinitiator into the hydrogel bioink is ingenious because of the indispensable requirement for successful coaxial printing that bioinks should be gelated immediately after being extruded from the nozzles (94, 95, 118). Combining tissue-specific microenvironmental niche material and adequate manufacturing methods promoted tissue functionality and printing fidelity simultaneously. The tubular intestinal model printed with Caco-2-laden dERS showed luminal lining of mucin similar to in vivo, where a mucus layer covers the intestinal epithelium to house microbiomes and restrain their translocation into underlying tissues (119). Given that in vitro models with Caco-2 often fail to develop a luminal mucus layer, the recapitulation of the luminal mucus lining in the colon dERS tubular models is compelling evidence for the significance of combining suitable materials and fabrication techniques.
This approach was further developed by Han et al. (61) to enhance the intestine-specific functions of the model based on the dERS bioink (Figure 3F). The hollow tubular intestinal model was fabricated again with Caco-2 and colon dERS but some important printing parameters (e.g., initial cell density, bioink, and photoinitiator concentration) were fine-tuned. As a result, the single cells that were evenly distributed along the printed tube simultaneously aggregated to form multicellular spheroids and self-organized into lumenized cysts (Figure 3G). This transition—called lumenogenesis—is a hallmark of distinct epithelial morphogenesis that occurs under biomimetic conditions (120). In addition, the differentiation of Caco-2 cells into functional intestinal phenotypes was identified by the expression of EEC markers such as chromogranin A and lysozyme. This indicates that 3D bioprinting and tissue-specific biochemical cues hold promise for geometrical guidance and the accelerated differentiation of accommodated cells.
The necessity of neuroendocrine models
The gut contributes to the GBA as the body's largest endocrine organ. In particular, EEC cells in the intestinal epithelium produce numerous hormones and neuroactive peptides. These signaling mediators secreted from EEC cells bind to the receptors of the vagus nerve, accomplishing direct bidirectional communication (7). In other words, EEC cells have a paramount role in the neuroendocrine pathway in GBA. However, as discussed in the previous section, the most recent technologies developed to fabricate 3D in vitro gut models were focused on recreating the morphological features of the intestine rather than its endocrine function. Likewise, current in vitro multi-organ GBA models lack the intrinsic secretory property of the intestine. For example, to elicit mutual responses between the gut and brain, modulation by exogenous immune cells (27) or microbial byproducts (121) were exploited but regulation via GI hormones or neurotransmitters has not yet been successfully recapitulated.
An in vitro neuroendocrine gut model would be beneficial for studying neurological diseases, considering the crucial functions of EEC hormones and neurotransmitters in GBA. For instance, serotonin is a critical signaling regulator in GBA and about 95% of it is produced by enterochromaffin cells (one phenotype of EEC cells) in the epithelium (122). Serotonin dysfunction is heavily associated with important brain functions such as mood, sleep, and behavior (123). Unfortunately, the fabrication of an in vitro EEC model is still in its infancy. In this regard, EEC cell sources are briefly presented as an important component for an in vitro neuroendocrine model.
Candidate cell sources for EEC models
There have been enormous efforts to reconstruct the in vitro EEC function at a cellular level. For decades, two immortalized EEC cell lines of human origin, NCI-H716 and HuTo-80, have provided a starting framework to study the secretion of gut hormones in vitro. NCI-H716, a poorly differentiated adenocarcinoma of the human cecum (124), is a representative type of distal L-cells among various subtypes of EEC cells. This cell line displays endocrine features including secretory granules and chromogranin A (125) and can secrete GI hormones in response to nutrients (126). Moreover, NCI-H716 exhibits receptors for several neurotransmitters such as gastrin, serotonin, and somatostatin (127). HuTu-80 is derived from duodenal carcinoma and is the only widely available human-derived small intestinal small cell line (128). This cell line also resembles L-cells and has been utilized as a model to study the secretion of tastant-induced gut hormones (129, 130). However, it is difficult to extrapolate the physiological function of the EEC system from single-cell cultures because they lack other types of IECs that influence the production and secretion of gut hormones.
Reimann et al. (131) established a protocol to culture primary enteroendocrine cells in vitro and enabled the study of the secretory mechanisms of gut hormones at the molecular level. Primary cells refer to non-transformed ex vivo cells that are isolated from tissue specimens obtained during biopsies or surgeries (132). Studies with purified primary human EEC cells enable a better understanding of hormone secretion and the metabolic pathway of the gut (133–135). However, some critical issues remain regarding primary cells; the finite lifespan and proliferation of primary cells should be taken into consideration (136). Relatively small proportions of EEC cells (1% of the intestinal epithelium) are also a challenge when deciphering the dynamics of hormone secretion (137).
Contrary to primary IECs, intestinal enteroids/organoids maintain their viability for over a year in vitro; this indefinite proliferation feature is valuable to studying long-term intestinal illnesses. Identification of adult intestinal stem cells (ISCs) and their niche and generation of an organotypic culture system have engendered advancements in the field of intestinal epithelial study (138) (Figure 4). Interestingly, EEC cells in enteroids and organoids can be enriched by the expression of some translational factors and small molecules (140–143), which is promising for the customization of EEC cells in enteroids/organoids. Intestinal enteroids/organoids have been used in various applications including modeling intestinal development, physiology and pathophysiology, nutrition transport, and metabolism (144). The practical problems associated with enteroids/organoids models is that they are not cost-effective and are difficult to scale up to meet the size requirements of drug screening or TE approaches, where centimeter-scale material is often desired (145, 146). Besides, the microanatomy of organoids/enteroids is typically confined to spheroidal shapes, which fail to recapitulate in vivo-like crypt–villus architecture.
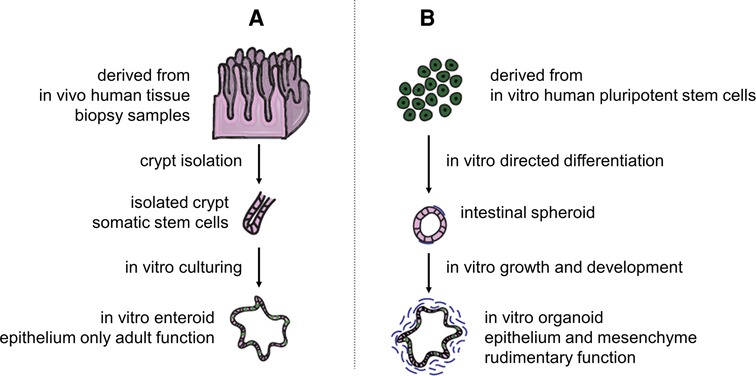
Figure 4. Difference between enteroids and organoids. (A) Enteroids are human mini-guts resembling the native intestine derived from adult ISCs of isolated crypts in intestinal biopsy samples. (B) Alternatively, intestinal organoids can be directly generated from induced pluripotent stem cells. The terminology, enteroids and organoids, was suggested to differentiate epithelial only in vitro cultures from both epithelial and mesenchymal ones, respectively. Adapted from Singh et al. (139). Open access (CC-BY 4.0).
The usefulness of microenvironmental cues in gut models
Imposing microenvironmental cues via surrounding material can maneuver the differentiation of EEC cells in vitro. For example, Bruïne et al. (127) reported that ECM has a determinant role in the endocrine differentiation of NCI-H716 cells. Culturing NCI-H716 on various extracted ECM components (e.g., colon ECM or collagen IV/heparan sulfate proteoglycan mix) enhances the adhesion of cells and induced EEC differentiation. Of note, individual ECM components, unlike complex combinations, do not induce endocrine differentiation, and the adhesion of cells onto a substrate is a prerequisite for inducing endocrine phenotype (125). Therefore, appropriate ECM environments are paramount for the differentiation of IECs.
As shown in BATE, bioprinting assisted by a proper ECM environment can achieve organoids/enteroids-derived tissue structures at centimeter scale, which is physiologically relevant for use in implantable regenerative medicine. Conventionally, the size of single organoids/enteroids is typically limited to millimeter-scale at best because the inner core becomes deficient in nutrient supply as the organoids/enteroids grow larger, resulting in necrosis (145). Furthermore, bioprinting employing multiple biomaterials is expected to assist in the integration of various organoids/enteroids and enhance their functionality in a single tissue system (147). Additionally, an enteroids polarity reversal strategy was developed based on understanding how basement membrane extract (BME) affects the epithelial polarity. Basolateral-out enteroids grown within BME showed an inversion of polarity when transferred to a BME-free suspension culture environment, enabling easy access to the apical side of the enteroids without technically demanding microinjection. Thus far, many aspects of soluble niche components important for culturing enteroids/organoids have been unraveled, whereas the role of insoluble ECM as a vital niche element remains a mystery (148). Since the IECs interact constantly with the local niche, which is comprised of both soluble factors and ECM gradients, the delicate balance between proliferating and differentiating ISCs is elicited by the dynamic microenvironment along the crypt–villus axis (149).
Compared to single ECMs (e.g. collagen or fibronectin), utilizing dECMs derived from normal or diseased tissue could contribute to mimicking a reliable microenvironment because they could reflect the structural and compositional disorganization of ECM during disease progression (150, 151). Interestingly, Alfano et al. (152) showed different intestinal models using three types of dECM substrates derived from healthy, perilesional, and colorectal carcinoma (CRC) human tissue. The specific characteristics of various cancer cells such as invasive phenotype, turnover, differentiation, and polarization were sustained, recapitulating the native tissue homeostasis and tumorigenesis more faithfully in vitro. Further investigations were taken to unveil the biochemical and mechanical features of the three dECMs and their underlying mechanisms regarding increased stiffness in the perilesional and CRC tissue (153). A recent study presented a CRC model that mimics the alteration of ECM in each tumor stage by using a dECM substrate originating from human CRC tissue of different stages (154). As the tumor stage increased, the imbalance of ECM composition was observed similar to in vivo, which induced changes in the proliferation and migration of the seeded cancer cells.
Evaluation techniques for GBA
Although In vitro BBB and gut models have seen tremendous progress in recent years, unresolved issues concerning connecting each component into a single GBA model remain. First, most of the state-of-the-art BBB and gut models are fabricated through a multi-step, complicated process that demands a lot of time and effort. In addition, as each model is advanced, they become to include various cellular compositions, making it difficult to find an optimized condition to coculture them. Last but not least, there is no defined method to track the dynamics which occur in in vitro GBA models. Many studies rely on visual assessment of cell morphology or end-point analysis since it is challenging to monitor the changes continuously without terminating the sample. However, as the BBB and gut epithelium works as physiological barriers in our body, it is necessary to quantitatively evaluate their wall tightness and integrity in vitro in a real-time and non-destructive way.
Trans-epithelial electrical resistance (TEER) is the most representative technique to measure barrier integrity (155). Recently, TEER-interfaced BBB (31, 156, 157) and gut (158) models enabled continuous and non-invasive detection of their barrier properties in situ and demonstrated the strength of sensor-implemented tissue platforms. Nevertheless, the above-mentioned models are still confined to a planar dimension and need to be expanded to 3D tissues. So far, only few studies have incorporated electronics into 3D BBB or gut tissue models (159). Moreover, electrochemical biosensors-assisted platforms for monitoring cell secretomes and behavior (160–164) would help chronological and rapid readout of multi-organ axies such as GBA.
Conclusion
This paper comprehensively reviews the necessity of EEC models in the future in terms of an in vitro tool to unravel the underlying influence of GBA in neurological diseases. The traditional GBA animal models have widened our understanding of the reciprocal interaction between the microbiome, gut, and brain. However, their intrinsic differences in tissue morphology and physiology to humans and the complex interplay in multiple organs necessitates a dismantled in vitro human GBA model. Therefore, we focused on introducing the most advanced 3D gut models and biofabrication methods so far and characterized them by their topographical and geometrical properties. Unfortunately, existing gut models are largely restricted to capturing the typical crypt–villus topography and thus miss capturing the secretory function of the intestine in response to various substances (e.g. microbial metabolites and hormones) related to GBA dysregulation. A gut model with neuroendocrine function is urgently needed and to accelerate advancements of in vitro EEC models where proper cell source, material, and fabrication technology should work in harmony (Figure 5).
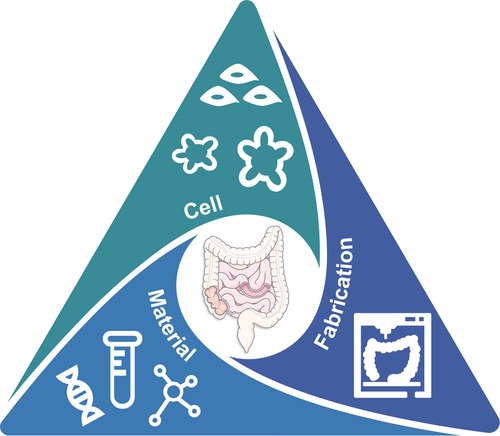
Figure 5. The TE trinity toward advanced in vitro gut model for GBA: cell, material, and fabrication.
Author contributions
JJ and HH conceptualized, reviewed, and edited the manuscript. HH wrote the manuscript and organized the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Basic Science Research Program through the National Research Foundation of South Korea (NRF) funded by the Ministry of Education (No. 2020R1A6A1A03047902). This work was also supported by the National Research Foundation of South Korea (NRF) grant funded by the Ministry of Science and ICT (No. 2021R1A2C2004981).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dinan TG, Cryan JF. The microbiome-gut-brain axis in health and disease. Gastroenterol Clin. (2017) 46:77–89. doi: 10.1016/J.GTC.2016.09.007
2. Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLOS Pathog. (2013) 9:e1003726. doi: 10.1371/JOURNAL.PPAT.1003726
3. Needham BD, Kaddurah-Daouk R, Mazmanian SK. Gut microbial molecules in behavioural and neurodegenerative conditions. Nat Rev Neurosci. (2020) 21:717–31. doi: 10.1038/s41583-020-00381-0
4. Mittal R, Debs LH, Patel AP, Nguyen D, Patel K, O’connor G, et al. Neurotransmitters: the critical modulators regulating gut-brain axis. J Cell Physiol. (2017) 232:2359–72. doi: 10.1002/jcp.25518
5. Margolis KG, Cryan JF, Mayer EA. The microbiota-gut-brain axis: from motility to mood. Gastroenterology. (2021) 160:1486–501. doi: 10.1053/J.GASTRO.2020.10.066
6. Banfi D, Moro E, Bosi A, Bistoletti M, Cerantola S, Crema F, et al. Impact of microbial metabolites on microbiota–gut–brain axis in inflammatory bowel disease. Int J Mol Sci. (2021) 22:1623. doi: 10.3390/IJMS22041623
7. Peterson CT. Dysfunction of the microbiota-gut-brain axis in neurodegenerative disease: the promise of therapeutic modulation with prebiotics, medicinal herbs, Probiotics, and Synbiotics. J Evid-Based Integr Med. (2020) 25. doi: 10.1177/2515690X20957225
8. Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J Neuroinflammation. (2019) 16:1–14. doi: 10.1186/S12974-019-1434-3
9. Fusco F, Perottoni S, Giordano C, Riva A, Iannone LF, De Caro C, et al. The microbiota-gut-brain axis and epilepsy from a multidisciplinary perspective: clinical evidence and technological solutions for improvement of in vitro preclinical models. Bioeng Transl Med. (2022) 9(2):e10296. doi: 10.1002/BTM2.10296
10. Ochoa-Repáraz J, Ramelow CC, Kasper LH. A gut feeling: the importance of the intestinal microbiota in psychiatric disorders. Front Immunol. (2020) 0:2735. doi: 10.3389/FIMMU.2020.510113
11. Lin C, Zhao S, Zhu Y, Fan Z, Wang J, Zhang B, et al. Microbiota-gut-brain axis and toll-like receptors in Alzheimer’s Disease. Comput Struct Biotechnol J. (2019) 17:1309–17. doi: 10.1016/J.CSBJ.2019.09.008
12. Doifode T, Giridharan V V, Generoso JS, Bhatti G, Collodel A, Schulz PE, et al. The impact of the microbiota-gut-brain axis on Alzheimer's Disease pathophysiology. Pharmacol Res. (2021) 164:105314. doi: 10.1016/J.PHRS.2020.105314
13. Kesika P, Suganthy N, Sivamaruthi BS, Chaiyasut C. Role of gut-brain axis, gut microbial composition, and probiotic intervention in Alzheimer's Disease. Life Sci. (2021) 264:118627. doi: 10.1016/J.LFS.2020.118627
14. Ghyselinck J, Verstrepen L, Moens F, Van Den Abbeele P, Bruggeman A, Said J, et al. Influence of probiotic bacteria on gut microbiota composition and gut wall function in an in-vitro model in patients with Parkinson's Disease. Int J Pharm X. (2021) 3:100087. doi: 10.1016/J.IJPX.2021.100087
15. Bindas AJ, Kulkarni S, Koppes RA, Koppes AN. Parkinson's disease and the gut: models of an emerging relationship. Acta Biomater. (2021) 132:325–44. doi: 10.1016/J.ACTBIO.2021.03.071
16. Dogra N, Mani RJ, Katare DP. The gut-brain axis: two ways signaling in Parkinson's Disease. Cell Mol Neurobiol. (2021) 42:315–32. doi: 10.1007/S10571-021-01066-7
17. Boeri L, Donnaloja F, Campanile M, Sardelli L, Tunesi M, Fusco F, et al. Using integrated meta-omics to appreciate the role of the gut microbiota in epilepsy. Neurobiol Dis. (2022) 164:105614. doi: 10.1016/J.NBD.2022.105614
18. Van Sadelhoff JHJ, Pardo PP, Wu J, Garssen J, Van Bergenhenegouwen J, Hogenkamp A, et al. The gut-immune-brain axis in autism spectrum disorders; a focus on amino acids. Front Endocrinol (Lausanne). (2019) 10:247. doi: 10.3389/FENDO.2019.00247/BIBTEX
19. Israelyan N, Margolis KG. Reprint of: serotonin as a link between the gut-brain-microbiome axis in autism spectrum disorders. Pharmacol Res. (2019) 140:115–20. doi: 10.1016/j.phrs.2018.12.023
20. Bear T, Dalziel J, Coad J, Roy N, Butts C, Gopal P. The microbiome-gut-brain axis and resilience to developing anxiety or depression under stress. Microorg. (2021) 9:723. doi: 10.3390/MICROORGANISMS9040723
21. Sandhu KV, Sherwin E, Schellekens H, Stanton C, Dinan TG, Cryan JF. Feeding the microbiota-gut-brain axis: diet, microbiome, and neuropsychiatry. Transl Res. (2017) 179:223–44. doi: 10.1016/J.TRSL.2016.10.002
22. Paul W, Marta C, Van de Wiele T. Resolving host–microbe interactions in the gut: the promise of in vitro models to complement in vivo research. Curr Opin Microbiol. (2018) 44:28–33. doi: 10.1016/J.MIB.2018.07.001
23. Walker AW, Parkhill J. Fighting obesity with bacteria. Science (80-). (2013) 341:1069–70. doi: 10.1126/SCIENCE.1243787/ASSET/A003E517-B8DA-4C7C-BA2E-660148E01313/ASSETS/GRAPHIC/341_1069_F1.JPEG
24. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's Disease. Cell. (2016) 167:1469–80. doi: 10.1016/J.CELL.2016.11.018
25. Douglas AE. Simple animal models for microbiome research. Nat Rev Microbiol. (2019) 17:764–75. doi: 10.1038/s41579-019-0242-1
26. Moysidou CM, Owens RM. Advances in modelling the human microbiome–gut–brain axis in vitro. Biochem Soc Trans. (2021) 49:187–201. doi: 10.1042/BST20200338
27. Trapecar M, Wogram E, Svoboda D, Communal C, Omer A, Lungjangwa T, et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci Adv. (2021) 7:eabd1707. doi: 10.1126/SCIADV.ABD1707/SUPPL_FILE/ABD1707_SM.PDF
28. Lee J, Choi JH, Kim HJ. Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments? Expert Rev Gastroenterol Hepatol. (2016) 10:883–5. doi: 10.1080/17474124.2016.1200466
29. Raimondi I, Izzo L, Tunesi M, Comar M, Albani D, Giordano C. Organ-on-a-chip in vitro models of the brain and the blood-brain barrier and their value to study the microbiota-gut-brain axis in neurodegeneration. Front Bioeng Biotechnol. (2020) 0:435. doi: 10.3389/FBIOE.2019.00435
30. Wang JD, Khafagy ES, Khanafer K, Takayama S, Elsayed MEH. Organization of endothelial cells, pericytes, and astrocytes into a 3D microfluidic in vitro model of the blood-brain barrier. Mol Pharm. (2016) 13:895–906. doi: 10.1021/ACS.MOLPHARMACEUT.5B00805/ASSET/IMAGES/LARGE/MP-2015-00805V_0008.JPEG
31. Wang YI, Abaci HE, Shuler ML. Microfluidic blood–brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng. (2017) 114:184–94. doi: 10.1002/BIT.26045
32. Ahn SI, Sei YJ, Park HJ, Kim J, Ryu Y, Choi JJ, et al. Microengineered human blood–brain barrier platform for understanding nanoparticle transport mechanisms. Nat Commun. (2020) 11:1–12. doi: 10.1038/s41467-019-13896-7
33. Cho CF, Wolfe JM, Fadzen CM, Calligaris D, Hornburg K, Chiocca EA, et al. Blood-brain-barrier spheroids as an in vitro screening platform for brain-penetrating agents. Nat Commun. (2017) 8:1–14. doi: 10.1038/ncomms15623
34. Boutin ME, Kramer LL, Livi LL, Brown T, Moore C, Hoffman-Kim D. A three-dimensional neural spheroid model for capillary-like network formation. J Neurosci Methods. (2018) 299:55–63. doi: 10.1016/J.JNEUMETH.2017.01.014
35. Zhao N, Guo Z, Kulkarni S, Norman D, Zhang S, Chung TD, et al. Engineering the human blood–brain barrier at the capillary scale using a double-templating technique. Adv Funct Mater. (2022) 32:2110289. doi: 10.1002/ADFM.202110289
36. Seo S, Nah S, Lee K, Choi N, Kim HN. Triculture model of in vitro BBB and its application to study BBB-associated chemosensitivity and drug delivery in glioblastoma. Adv Funct Mater. (2021) 32:2106860. doi: 10.1002/adfm.202106860
37. Wevers NR, Kasi DG, Gray T, Wilschut KJ, Smith B, Vught R, et al. A perfused human blood-brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS. (2018) 15:1–12. doi: 10.1186/S12987-018-0108-3/FIGURES/4
38. Sivandzade F, Cucullo L. In-vitro blood–brain barrier modeling: a review of modern and fast-advancing technologies. J Cereb Blood Flow Metab. (2018) 38:1667–81. doi: 10.1177/0271678X18788769
39. Seo S, Kim H, Sung JH, Choi N, Lee K, Kim HN. Microphysiological systems for recapitulating physiology and function of blood-brain barrier. Biomaterials. (2020) 232:119732. doi: 10.1016/J.BIOMATERIALS.2019.119732
40. Herland A, Van Der Meer AD, FitzGerald EA, Park TE, Sleeboom JJF, Ingber DE. Distinct contributions of astrocytes and pericytes to neuroinflammation identified in a 3D human blood-brain barrier on a chip. PLoS One. (2016) 11:e0150360. doi: 10.1371/JOURNAL.PONE.0150360
41. Han W, Singh NK, Kim JJ, Kim H, Kim BS, Park JY, et al. Directed differential behaviors of multipotent adult stem cells from decellularized tissue/organ extracellular matrix bioinks. Biomaterials. (2019) 224:119496. doi: 10.1016/J.BIOMATERIALS.2019.119496
42. Bhalerao A, Sivandzade F, Archie SR, Chowdhury EA, Noorani B, Cucullo L. In vitro modeling of the neurovascular unit: advances in the field. Fluids Barriers CNS. (2020) 17:1–20. doi: 10.1186/S12987-020-00183-7
43. Li QY, Lee JH, Kim HW, Jin GZ. Research models of the nanoparticle-mediated drug delivery across the blood–brain barrier. Tissue Eng Regen Med. (2021) 18:917. doi: 10.1007/S13770-021-00356-X
44. Seidlitz T, Koo BK, Stange DE. Gastric organoids—an in vitro model system for the study of gastric development and road to personalized medicine. Cell Death Differ. (2020) 28:68–83. doi: 10.1038/s41418-020-00662-2
45. Yu H, Hasan NM, In JG, Estes MK, Kovbasnjuk O, Zachos NC, et al. The contributions of human mini-intestines to the study of intestinal physiology and pathophysiology. Annu Rev Physiol. (2017) 79:291–312. doi: 10.1146/annurev-physiol-021115-105211
46. Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol. (2018) 5:659–68. doi: 10.1016/J.JCMGH.2017.12.010
47. Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. (2014) 32:760–72. doi: 10.1038/NBT.2989
48. Knight E, Przyborski S. Advances in 3D cell culture technologies enabling tissue-like structures to be created in vitro. J Anat. (2015) 227:746–56. doi: 10.1111/JOA.12257
49. Costa J, Ahluwalia A. Advances and current challenges in intestinal in vitro model engineering: a digest. Front Bioeng Biotechnol. (2019) 7:144. doi: 10.3389/FBIOE.2019.00144/BIBTEX
50. Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. (2007) 8:839–45. doi: 10.1038/nrm2236
51. Mattei G, Giusti S, Ahluwalia A. Design criteria for generating physiologically relevant in vitro models in bioreactors. Processes. (2014) 2:548–69. doi: 10.3390/PR2030548
52. Roth A, Singer T. The application of 3D cell models to support drug safety assessment: opportunities / challenges. Adv Drug Deliv Rev. (2014) 69–70:179–89. doi: 10.1016/J.ADDR.2013.12.005
53. Kim HJ, Hu D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. (2012) 12:2165–74. doi: 10.1039/C2LC40074J
54. Creff J, Malaquin L, Besson A. In vitro models of intestinal epithelium: toward bioengineered systems. J Tissue Eng. (2021) 12:2041731420985202. doi: 10.1177/2041731420985202
55. Marx U, Andersson TB, Bahinski A, Beilmann M, Beken S, Cassee FR, et al. Biology-inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX. (2016) 33:272–321. doi: 10.14573/altex.1603161.Biology-inspired
56. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Reports. (2018) 8:1–14. doi: 10.1038/s41598-018-21201-7
57. Chan HF, Zhao R, Parada GA, Meng H, Leong KW, Griffith LG, et al. Folding artificial mucosa with cell-laden hydrogels guided by mechanics models. Proc Natl Acad Sci USA. (2018) 115:7503–8. doi: 10.1073/PNAS.1802361115/SUPPL_FILE/PNAS.1802361115.SAPP.PDF
58. Kim WJ, Kim GH. An intestinal model with a finger-like villus structure fabricated using a bioprinting process and collagen/SIS-based cell-laden bioink. Theranostics. (2020) 10:2495–508. doi: 10.7150/thno.41225
59. Lian L, Zhou C, Tang G, Xie M, Wang Z, Luo Z, et al. Uniaxial and coaxial vertical embedded extrusion bioprinting. Adv Healthc Mater. (2021) 11:2102411. doi: 10.1002/ADHM.202102411
60. Langerak N, Ahmed HMM, Li Y, Middel IR, Eslami Amirabadi H, Malda J, et al. A theoretical and experimental study to optimize cell differentiation in a novel intestinal chip. Front Bioeng Biotechnol. (2020) 8:763. doi: 10.3389/FBIOE.2020.00763/BIBTEX
61. Han H, Park Y, Choi Y, Yong U, Kang B, Shin W, et al. A bioprinted tubular intestine model using a colon-specific extracellular matrix bioink. Adv Healthc Mater. (2022) 11:2101768. doi: 10.1002/ADHM.202101768
62. Kim HJ, Lee J, Choi JH, Bahinski A, Ingber DE. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. J Vis Exp. (2016) 114:3–9. doi: 10.3791/54344
63. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. (2019) 3:520–31. doi: 10.1038/s41551-019-0397-0
64. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci. (2016) 113:E7–E15. doi: 10.1073/PNAS.1522193112
65. Shin W, Kim HJ. Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci. (2018) 115:E10539–47. doi: 10.1073/PNAS.1810819115
66. Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (United Kingdom). (2013) 5:1130–40. doi: 10.1039/c3ib40126j
67. Shin W, Hinojosa CD, Ingber DE, Kim HJ. Human intestinal morphogenesis controlled by transepithelial morphogen gradient and flow-dependent physical cues in a microengineered gut-on-a-chip. iScience. (2019) 15:391–406. doi: 10.1016/J.ISCI.2019.04.037
68. Youn J, Hong H, Shin W, Kim D, Kim HJ, Kim DS. Thin and stretchable extracellular matrix (ECM) membrane reinforced by nanofiber scaffolds for developing in vitro barrier models. Biofabrication. (2022) 14:025010. doi: 10.1088/1758-5090/ac4dd7
69. Shin YC, Shin W, Koh D, Wu A, Ambrosini YM, Min S, et al. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines (Basel). (2020) 11:663. doi: 10.3390/MI11070663
70. Sontheimer-Phelps A, Chou DB, Tovaglieri A, Ferrante TC, Duckworth T, Fadel C, et al. Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell Mol Gastroenterol Hepatol. (2020) 9:507–26. doi: 10.1016/J.JCMGH.2019.11.008
71. Deng X, Zhang G, Shen C, Yin J, Meng Q. Hollow fiber culture accelerates differentiation of Caco-2 cells. Appl Microbiol Biotechnol. (2013) 97:6943–55. doi: 10.1007/S00253-013-4975-X/FIGURES/7
72. Baptista D, Teixeira L, Van Blitterswijk C, Giselbrecht S, Truckenmüller R. Overlooked? Underestimated? Effects of substrate curvature on cell behavior. Trends Biotechnol. (2019) 37:838–54. doi: 10.1016/J.TIBTECH.2019.01.006
73. Sung JH, Yu J, Luo D, Shuler ML, March JC. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip. (2011) 11:389–92. doi: 10.1039/c0lc00273a
74. Brassard JA, Lutolf MP. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. (2019) 24:860–76. doi: 10.1016/J.STEM.2019.05.005
75. Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. (2018) 19:671–87. doi: 10.1038/s41576-018-0051-9
76. Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. (2017) 128:44–55. doi: 10.1016/J.BIOMATERIALS.2017.03.005
77. DR H AMC, JR S MA. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol. (2017) 66:81–93. doi: 10.1016/J.SEMCDB.2017.01.011
78. Gregersen H. Biomechanics of the gastrointestinal tract: new perspectives in motility research and diagnostics. 2003th edition (Book Chapter). Springer (2003). p. 268. ISBN: 978-1-84996-880-5
79. Gregersen H, Kassab G, Pallencaoe E, Lee C, Chien S, Skalak R, et al. Morphometry and strain distribution in Guinea pig duodenum with reference to the zero-stress state. Am J Physiol - Gastrointest Liver Physiol. (1997) 273:G865–74. doi: 10.1152/AJPGI.1997.273.4.G865/ASSET/IMAGES/LARGE/AGIJ3101808.JPEG
80. Gregersen H, Lee TC, Chien S, Skalak R, Fung YC. Strain distribution in the layered wall of the esophagus. J Biomech Eng. (1999) 121:442–8. doi: 10.1115/1.2835072
81. Lei X, Ye D, Chen J, Tang S, Sun P, Chen L, et al. Customizable multidimensional self-wrinkling structure constructed via modulus gradient in chitosan hydrogels. Chem Mater. (2019) 31:10032–9. doi: 10.1021/ACS.CHEMMATER.9B02812/SUPPL_FILE/CM9B02812_SI_003.MP4
82. Shapira A, Dvir T. 3D Tissue and organ printing—hope and reality. Adv Sci. (2021) 8:2003751. doi: 10.1002/ADVS.202003751
83. Lee S-H, Nam H, Jeong B, Jang J. 3D printing and bioprinting technologies. In: Kim HJ, editors. Biomimetic microengineering. 1st edition. Boca Raton: CRC Press (2020). p. 191–212. doi: 10.1201/9780367814809-10
84. Ng WL, Lee JM, Zhou M, Chen YW, Lee KXA, Yeong WY, et al. Vat polymerization-based bioprinting—process, materials, applications and regulatory challenges. Biofabrication. (2020) 12:022001. doi: 10.1088/1758-5090/AB6034
85. Jiang T, Munguia-Lopez JG, Flores-Torres S, Kort-Mascort J, Kinsella JM. Extrusion bioprinting of soft materials: an emerging technique for biological model fabrication. Appl Phys Rev. (2019) 6:011310. doi: 10.1063/1.5059393
86. Li X, Liu B, Pei B, Chen J, Zhou D, Peng J, et al. Inkjet bioprinting of biomaterials. Chem Rev. (2020) 120:10793–833. doi: 10.1021/ACS.CHEMREV.0C00008/ASSET/IMAGES/LARGE/CR0C00008_0028.JPEG
87. Jang J, Park JY, Gao G, Cho DW. Biomaterials-based 3D cell printing for next-generation therapeutics and diagnostics. Biomaterials. (2018) 156:88–106. doi: 10.1016/J.BIOMATERIALS.2017.11.030
88. Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat Mater. (2020) 20:22–9. doi: 10.1038/s41563-020-00803-5
89. Kang B, Park Y, Hwang DG, Kim D, Yong U, Lim KS, et al. Facile bioprinting process for fabricating size-controllable functional microtissues using light-activated decellularized extracellular matrix-based bioinks. Adv Mater Technol. (2022) 7:2100947. doi: 10.1002/ADMT.202100947
90. Yang Y, Armour M, Kang-Hsin Wang K, al -, Cervantes-Alcalá R, Felipe Rivera F, et al. Dynamic photopolymerization produces complex microstructures on hydrogels in a moldless approach to generate a 3D intestinal tissue model. Biofabrication. (2019) 11:025007. doi: 10.1088/1758-5090/AB0478
91. Jang J, Kim TG, Kim BS, Kim SW, Kwon SM, Cho DW. Tailoring mechanical properties of decellularized extracellular matrix bioink by vitamin B2-induced photo-crosslinking. Acta Biomater. (2016) 33:88–95. doi: 10.1016/J.ACTBIO.2016.01.013
92. Jang J, Park HJ, Kim SW, Kim H, Park JY, Na SJ, et al. 3D Printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials. (2017) 112:264–74. doi: 10.1016/J.BIOMATERIALS.2016.10.026
93. Choi YJ, Jun YJ, Kim DY, Yi HG, Chae SH, Kang J, et al. A 3D cell printed muscle construct with tissue-derived bioink for the treatment of volumetric muscle loss. Biomaterials. (2019) 206:160–9. doi: 10.1016/J.BIOMATERIALS.2019.03.036
94. Gao G, Lee JH, Jang J, Lee DH, Kong J-S, Kim BS, et al. Tissue engineered bio-blood-vessels constructed using a tissue-specific bioink and 3D coaxial cell printing technique: a novel therapy for ischemic disease. Adv Funct Mater. (2017) 27:1700798. doi: 10.1002/ADFM.201700798
95. Gao G, Park W, Kim BS, Ahn M, Chae S, Cho WW, et al. Construction of a novel in vitro atherosclerotic model from geometry-tunable artery equivalents engineered via in-bath coaxial cell printing. Adv Funct Mater. (2021) 31:2008878. doi: 10.1002/ADFM.202008878
96. Alberti C. Hollow organ tissue engineering: short updating about current approaches and forecast for major research advances. G Chir. (2011) 32:345–51. PMID: 22018253
97. Saksena R, Gao C, Wicox M, de Mel A. Tubular organ epithelialisation. J Tissue Eng. (2016) 7:2041731416683950. doi: 10.1177/2041731416683950
98. Assoian RK, Bade ND, Cameron C V, Stebe KJ. Cellular sensing of micron-scale curvature: a frontier in understanding the microenvironment. Open Biol. (2019) 9:190155. doi: 10.1098/RSOB.190155
99. Del Gaudio C, Baiguera S, Ajalloueian F, Bianco A, Macchiarini P. Are synthetic scaffolds suitable for the development of clinical tissue-engineered tubular organs? J Biomed Mater Res Part A. (2014) 102:2427–47. doi: 10.1002/JBM.A.34883
100. Jeong H-J, Nam H, Jang J, Lee S-J. 3D Bioprinting strategies for the regeneration of functional tubular tissues and organs. Bioeng. (2020) 7:32. doi: 10.3390/BIOENGINEERING7020032
101. Boys AJ, Barron SL, Tilev D, Owens RM. Building scaffolds for tubular tissue engineering. Front Bioeng Biotechnol. (2020) 8:1357. doi: 10.3389/FBIOE.2020.589960/BIBTEX
102. Galliger Z, Vogt CD, Panoskaltsis-Mortari A. 3D Bioprinting for lungs and hollow organs. Transl Res. (2019) 211:19–34. doi: 10.1016/j.trsl.2019.05.001
103. Singh A, Lee D, Sopko N, Matsui H, Sabnekar P, Liu X, et al. Biomanufacturing seamless tubular and hollow collagen scaffolds with unique design features and biomechanical properties. Adv Healthc Mater. (2017) 6:1601136. doi: 10.1002/ADHM.201601136
104. Hendow EK, Guhmann P, Wright B, Sofokleous P, Parmar N, Day RM. Biomaterials for hollow organ tissue engineering. BMC Res Notes. (2016) 9:1–7. doi: 10.1186/S13069-016-0040-6/TABLES/1
105. Asnaghi MA, Jungebluth P, Raimondi MT, Dickinson SC, Rees LEN, Go T, et al. A double-chamber rotating bioreactor for the development of tissue-engineered hollow organs: from concept to clinical trial. Biomaterials. (2009) 30:5260–9. doi: 10.1016/J.BIOMATERIALS.2009.07.018
106. Jochems PGM, van Bergenhenegouwen J, van Genderen AM, Eis ST, Wilod Versprille LJF, Wichers HJ, et al. Development and validation of bioengineered intestinal tubules for translational research aimed at safety and efficacy testing of drugs and nutrients. Toxicol Vitr. (2019) 60:1–11. doi: 10.1016/J.TIV.2019.04.019
107. Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nat. (2020) 585:574–8. doi: 10.1038/s41586-020-2724-8
108. Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, Li C, et al. Robust bioengineered 3D functional human intestinal epithelium. Sci Reports. (2015) 5:1–11. doi: 10.1038/srep13708
109. Potten CS, Wilson JW, Booth C. Regulation and significance of apoptosis in the stem cells of the gastrointestinal epithelium. Stem Cells. (1997) 15:82–93. doi: 10.1002/STEM.150082
110. Potten CS. Epithelial cell growth and differentiation. II. Intestinal Apoptosis. Am J Physiol Gastrointest Liver Physiol. (1997) 273(2):G253–7. doi: 10.1152/AJPGI.1997.273.2.G253
111. Roh TT, Chen Y, Paul HT, Guo C, Kaplan DL. 3D Bioengineered tissue model of the large intestine to study inflammatory bowel disease. Biomaterials. (2019) 225:119517. doi: 10.1016/J.BIOMATERIALS.2019.119517
112. Lui C, Chin AF, Park S, Yeung E, Kwon C, Tomaselli G, et al. Mechanical stimulation enhances development of scaffold-free, 3D-printed, engineered heart tissue grafts. J Tissue Eng Regen Med. (2021) 15:503–12. doi: 10.1002/TERM.3188
113. Rogozhnikov D, Luo W, Elahipanah S, O’Brien PJ, Yousaf MN. Generation of a scaffold-free three-dimensional liver tissue via a rapid cell-to-cell click assembly process. Bioconjug Chem. (2016) 27:1991–8. doi: 10.1021/ACS.BIOCONJCHEM.6B00187/ASSET/IMAGES/MEDIUM/BC-2016-00187N_0007.GIF
114. Yan Z, Yin H, Nerlich M, Pfeifer CG, Docheva D. Boosting tendon repair: interplay of cells, growth factors and scaffold-free and gel-based carriers. J Exp Orthop. (2018) 5:1–13. doi: 10.1186/S40634-017-0117-1/TABLES/3
115. Burdis R, Kelly DJ. Biofabrication and bioprinting using cellular aggregates, microtissues and organoids for the engineering of musculoskeletal tissues. Acta Biomater. (2021) 126:1–14. doi: 10.1016/J.ACTBIO.2021.03.016
116. Rawal P, Tripathi DM, Ramakrishna S, Kaur S. Prospects for 3D bioprinting of organoids. Bio-Design Manuf. (2021) 4:627–40. doi: 10.1007/S42242-020-00124-1
117. Kim H, Kang B, Cui X, Lee SH, Lee K, Cho DW, et al. Light-activated decellularized extracellular matrix-based bioinks for volumetric tissue analogs at the centimeter scale. Adv Funct Mater. (2021) 31:2011252. doi: 10.1002/ADFM.202011252
118. Gao G, Kim H, Kim BS, Kong JS, Lee JY, Park BW, et al. Tissue-engineering of vascular grafts containing endothelium and smooth-muscle using triple-coaxial cell printing. Appl Phys Rev. (2019) 6:041402. doi: 10.1063/1.5099306
119. Virumbrales-Muñoz M, Ayuso JM, Gong MM, Humayun M, Livingston MK, Lugo-Cintrón KM, et al. Microfluidic lumen-based systems for advancing tubular organ modeling. Chem Soc Rev. (2020) 49:6402–42. doi: 10.1039/D0CS00705F
120. Cerchiari AE, Samy KE, Todhunter ME, Schlesinger E, Henise J, Rieken C, et al. Probing the luminal microenvironment of reconstituted epithelial microtissues. Sci Rep. (2016) 6:1–7. doi: 10.1038/srep33148
121. Kim MH, Kim D, Sung JH. A Gut-Brain Axis-on-a-Chip for studying transport across epithelial and endothelial barriers. J Ind Eng Chem. (2021) 101:126–34. doi: 10.1016/J.JIEC.2021.06.021
122. Alcaino C, Knutson KR, Treichel AJ, Yildiz G, Strege PR, Linden DR, et al. A population of gut epithelial enterochromaffin cells is mechanosensitive and requires Piezo2 to convert force into serotonin release. Proc Natl Acad Sci USA. (2018) 115:E7632–41. doi: 10.1073/PNAS.1804938115/-/DCSUPPLEMENTAL
123. Martin AM, Young RL, Leong L, Rogers GB, Spencer NJ, Jessup CF, et al. The diverse metabolic roles of peripheral serotonin. Endocrinology. (2017) 158:1049–63. doi: 10.1210/EN.2016-1839
124. Park JG, Frucht H, LaRocca R V, Bliss DP, Kurita Y, Chen TR, et al. Characteristics of cell lines established from human gastric carcinoma. Cancer Res. (1990) 50:2773–80. PMID: 2158397
125. De Bruine AP, Dinjens WNM, Linden EPMVD, Pijls MMJ, Moerkerk PT, Bosman FT. Extracellular matrix components induce endocrine differentiation in vitro in NCI-H716 cells. Am J Pathol. (1993) 142:773. PMID: 8456938
126. Reimer RA, Darimont C, Gremlich S, Nicolas-Métral V, Rüegg UT, Macé K. A human cellular model for studying the regulation of glucagon-like peptide-1 secretion. Endocrinology. (2001) 142:4522–8. doi: 10.1210/ENDO.142.10.8415
127. de Bruïne AP, Dinjens WNM, Pijls MMJ, Edith EPM, Rousch MJM, Moerkerk PT, et al. NCI-H716 cells as a model for endocrine differentiation in colorectal cancer. Virchows Arch B Cell Pathol Incl Mol Pathol. (1992) 62:311–20. doi: 10.1007/BF02899698
128. Brosnahan AJ, Brown DR. Porcine IPEC-J2 intestinal epithelial cells in microbiological investigations. Vet Microbiol. (2012) 156:229–37. doi: 10.1016/J.VETMIC.2011.10.017
129. Ohtsu Y, Nakagawa Y, Nagasawa M, Takeda S, Arakawa H, Kojima I. Diverse signaling systems activated by the sweet taste receptor in human GLP-1-secreting cells. Mol Cell Endocrinol. (2014) 394:70–9. doi: 10.1016/J.MCE.2014.07.004
130. Pham H, Hui H, Morvaridi S, Cai J, Zhang S, Tan J, et al. A bitter pill for type 2 diabetes? The activation of bitter taste receptor TAS2R38 can stimulate GLP-1 release from enteroendocrine L-cells. Biochem Biophys Res Commun. (2016) 475:295–300. doi: 10.1016/J.BBRC.2016.04.149
131. Reimann F, Habib AM, Tolhurst G, Parker HE, Rogers GJ, Gribble FM. Glucose sensing in L cells: a primary cell study. Cell Metab. (2008) 8:532–9. doi: 10.1016/J.CMET.2008.11.002
132. Richter M, Piwocka O, Musielak M, Piotrowski I, Suchorska WM, Trzeciak T. From donor to the lab: a fascinating journey of primary cell lines. Front Cell Dev Biol. (2021) 9:1869. doi: 10.3389/FCELL.2021.711381/BIBTEX
133. Liñán-Rico A, Ochoa-Cortes F, Zuleta-Alarcon A, Alhaj M, Tili E, Enneking J, et al. UTP - Gated signaling pathways of 5-HT release from BON cells as a model of human enterochromaffin cells. Front Pharmacol. (2017) 8:429. doi: 10.3389/FPHAR.2017.00429/BIBTEX
134. Raghupathi R, Duffield MD, Zelkas L, Meedeniya A, Brookes SJH, Sia TC, et al. Identification of unique release kinetics of serotonin from Guinea-pig and human enterochromaffin cells. J Physiol. (2013) 591:5959–75. doi: 10.1113/JPHYSIOL.2013.259796
135. Lumsden AL, Martin AM, Sun EW, Schober G, Isaacs NJ, Pezos N, et al. Sugar responses of human enterochromaffin cells depend on gut region, sex, and body mass. Nutrients. (2019) 11:234. doi: 10.3390/NU11020234
136. Gillooly JF, Hayward A, Hou C, Gordon Burleigh J. Explaining differences in the lifespan and replicative capacity of cells: a general model and comparative analysis of vertebrates. Proc R Soc B Biol Sci. (2012) 279:3976–80. doi: 10.1098/RSPB.2012.1129
137. Svendsen B, Holst JJ. Regulation of gut hormone secretion. Studies using isolated perfused intestines. Peptides. (2016) 77:47–53. doi: 10.1016/j.peptides.2015.08.001
138. Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nat. (2009) 459:262–5. doi: 10.1038/nature07935
139. Singh A, Poling HM, Spence JR, Wells JM, Helmrath MA. Gastrointestinal organoids: a next-generation tool for modeling human development. Am J Physiol - Gastrointest Liver Physiol. (2020) 319:G375–81. doi: 10.1152/AJPGI.00199.2020/ASSET/IMAGES/LARGE/ZH30082078200003.JPEG
140. Chang-Graham AL, Danhof HA, Engevik MA, Tomaro-Duchesneau C, Karandikar UC, Estes MK, et al. Human intestinal enteroids with inducible neurogenin-3 expression as a novel model of gut hormone secretion. Cell Mol Gastroenterol Hepatol. (2019) 8:209–29. doi: 10.1016/J.JCMGH.2019.04.010
141. Du A, McCracken KW, Walp ER, Terry NA, Klein TJ, Han A, et al. Arx is required for normal enteroendocrine cell development in mice and humans. Dev Biol. (2012) 365:175–88. doi: 10.1016/J.YDBIO.2012.02.024
142. Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nat. (2010) 470:105–9. doi: 10.1038/nature09691
143. Boonekamp KE, Dayton TL, Clevers H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J Mol Cell Biol. (2020) 12:562–8. doi: 10.1093/JMCB/MJAA034
144. Rallabandi HR, Yang H, Oh KB, Lee HC, Byun SJ, Lee BR. Evaluation of intestinal epithelial barrier function in inflammatory bowel diseases using murine intestinal organoids. Tissue Eng Regen Med. (2020) 17:641–50. doi: 10.1007/S13770-020-00278-0/FIGURES/3
145. Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. (2021) 6:402–20. doi: 10.1038/s41578-021-00279-y
146. Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid models of human gastrointestinal development and disease. Gastroenterology. (2016) 150:1098–112. doi: 10.1053/J.GASTRO.2015.12.042
147. Ravanbakhsh H, Karamzadeh V, Bao G, Mongeau L, Juncker D, Zhang YS, et al. Emerging technologies in multi-material bioprinting. Adv Mater. (2021) 33:2104730. doi: 10.1002/ADMA.202104730
148. Rezakhani S, Gjorevski N, Lutolf MP. Extracellular matrix requirements for gastrointestinal organoid cultures. Biomaterials. (2021) 276:121020. doi: 10.1016/J.BIOMATERIALS.2021.121020
149. Malijauskaite S, Mulvihill JJE, Grabrucker AM, McGourty K. IPSC-derived intestinal organoids and current 3D intestinal scaffolds. (2021).
150. Kim J, Jang J, Cho D-W. Controlling cancer cell behavior by improving the stiffness of gastric tissue-decellularized ECM bioink with cellulose nanoparticles. Front Bioeng Biotechnol. (2021) 0:152. doi: 10.3389/FBIOE.2021.605819
151. Park W, Bae M, Hwang M, Jang J, Cho DW, Yi HG. 3D cell-printed hypoxic cancer-on-a-chip for recapitulating pathologic progression of solid cancer. JoVE (Journal Vis Exp). (2021) 167:e61945. doi: 10.3791/61945
152. Genovese L, Zawada L, Tosoni A, Ferri A, Zerbi P, Allevi R, et al. Cellular localization, invasion, and turnover are differently influenced by healthy and tumor-derived extracellular matrix. Tissue Eng - Part A. (2014) 20:2005–18. doi: 10.1089/TEN.TEA.2013.0588/SUPPL_FILE/SUPP_FIGURE8.PDF
153. Nebuloni M, Albarello L, Andolfo A, Magagnotti C, Genovese L, Locatelli I, et al. Insight on colorectal carcinoma infiltration by studying perilesional extracellular matrix OPEN. Sci Rep. (2016) 6:22522. doi: 10.1038/srep22522
154. Li ZL, Wang ZJ, Wei GH, Yang Y, Wang XW. Changes in extracellular matrix in different stages of colorectal cancer and their effects on proliferation of cancer cells. World J Gastrointest Oncol. (2020) 12:267. doi: 10.4251/WJGO.V12.I3.267
155. Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. TEER measurement techniques for in vitro barrier model systems. J Lab Autom. (2015) 20:107–26. doi: 10.1177/2211068214561025
156. Maherally Z, Fillmore HL, Tan SL, Tan SF, Jassam SA, Quack FI, et al. Real-time acquisition of transendothelial electrical resistance in an all-human, in vitro, 3-dimensional, blood-brain barrier model exemplifies tight-junction integrity. FASEB J. (2018) 32:168–82. doi: 10.1096/FJ.201700162R
157. Liang Y, Yoon JY. In situ sensors for blood-brain barrier (BBB) on a chip. Sensors and Actuators Reports. (2021) 3:100031. doi: 10.1016/J.SNR.2021.100031
158. Shah P, Fritz J V, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human–microbe interface. Nat Commun. (2016) 7:1–15. doi: 10.1038/ncomms11535
159. Moysidou C-M, Pitsalidis C, Al-Sharabi M, Withers AM, Zeitler JA, Owens RM. 3D Bioelectronic model of the human intestine. Adv Biol. (2021) 5:2000306. doi: 10.1002/ADBI.202000306
160. Shin SR, Kilic T, Zhang YS, Avci H, Hu N, Kim D, et al. Label-free and regenerative electrochemical microfluidic biosensors for continual monitoring of cell secretomes. Adv Sci. (2017) 4:1600522. doi: 10.1002/ADVS.201600522
161. Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci. (2017) 114:E2293–302. doi: 10.1073/PNAS.1612906114
162. Jodat YA, Lotfi P, Abadi PPSS, Mun JY, Seo J, Shin EA, et al. PH-responsive DNA nanolinker conjugated hybrid materials for electrochemical microactuator and biosensor applications. ACS Appl Nano Mater. (2018) 1:6630–40. doi: 10.1021/ACSANM.8B01429/ASSET/IMAGES/LARGE/AN-2018-014299_0006.JPEG
163. Aleman J, Kilic T, Mille LS, Shin SR, Zhang YS. Microfluidic integration of regeneratable electrochemical affinity-based biosensors for continual monitoring of organ-on-a-chip devices. Nat Protoc. (2021) 16:2564–93. doi: 10.1038/s41596-021-00511-7
Keywords: gut-brain axis, neurological disease, enteroendocrine function, biofabrication, in vitro gut models
Citation: Han H and Jang J (2022) Recent advances in biofabricated gut models to understand the gut-brain axis in neurological diseases. Front. Med. Technol. 4:931411. doi: 10.3389/fmedt.2022.931411
Received: 29 April 2022; Accepted: 22 August 2022;
Published: 14 September 2022.
Edited by:
Pedro Morouço, Polytechnic Institute of Leiria, PortugalReviewed by:
Anna-Maria Pappa, Khalifa University, United Arab EmiratesWei Long Ng, Nanyang Technological University, Singapore
© 2022 Han and Jang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinah Jang amluYWhqYW5nQHBvc3RlY2guYWMua3I=
Specialty Section: This article was submitted to Regenerative Technologies, a section of the journal Frontiers in Medical Technology
 Hohyeon Han1
Hohyeon Han1 Jinah Jang
Jinah Jang