
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Med. Technol. , 12 March 2021
Sec. Pharmaceutical Innovation
Volume 3 - 2021 | https://doi.org/10.3389/fmedt.2021.640981
This article is part of the Research Topic Bridging Membrane Biophysics to Microbiology: Innovating Towards New Peptide and Peptide-based Antimicrobials View all 12 articles
Antimicrobial peptides (AMPs) have been extensively studied due to their vast natural abundance and ability to kill microbes. In an era critically lacking in new antibiotics, manipulating AMPs for therapeutic application is a promising option. However, bacterial pathogens resistant to AMPs remain problematic. To improve AMPs antimicrobial efficacy, their use in conjunction with other antimicrobials has been proposed. How might this work? AMPs kill bacteria by forming pores in bacterial membranes or by inhibiting bacterial macromolecular functions. What remains unknown is the duration for which AMPs keep bacterial pores open, and the extent to which bacteria can recover by repairing these pores. In this mini-review, we discuss various antimicrobial synergies with AMPs. Such synergies might arise if the antimicrobial agents helped to keep bacterial pores open for longer periods of time, prevented pore repair, perturbed bacterial intracellular functions at greater levels, or performed other independent bacterial killing mechanisms. We first discuss combinations of AMPs, and then focus on histones, which have antimicrobial activity and co-localize with AMPs on lipid droplets and in neutrophil extracellular traps (NETs). Recent work has demonstrated that histones can enhance AMP-induced membrane permeation. It is possible that histones, histone fragments, and histone-like peptides could amplify the antimicrobial effects of AMPs, giving rise to antimicrobial synergy. If so, clarifying these mechanisms will thus improve our overall understanding of the antimicrobial processes and potentially contribute to improved drug design.
Bacterial infections are an increasing threat to global health, due to both an increase in bacterial resistance to current therapeutics and also a decline in new antibiotic development. This results in rising numbers of untreatable health complications and deaths worldwide (1). There is thus an urgent need to identify new antibacterial strategies to effectively treat drug-resistant pathogens. The demand for such new strategies has encouraged scientists to investigate biologically-abundant antimicrobial tools that can be manipulated to kill bacteria. Repurposing and modifying known natural antimicrobial proteins may contribute to successful development of new therapeutic strategies.
Antimicrobial peptides (AMPs) have broad spectrum antimicrobial activity and are found ubiquitously in nature. They have been extensively studied as a promising option to combat multidrug-resistant bacteria. However, the rapid ability of bacteria to evolve requires new approaches to limit potential bacterial resistance to AMPs (2–4). Here, we discuss the use of AMPs in conjunction with other antimicrobials to form antimicrobial synergy, in which the combined antimicrobial effect is greater than the sum of either treatment alone. Antimicrobial synergy could potentially reduce the rise of bacterial resistance. A number of synergistic approaches using AMPs have been sought, with 300 reports made during the last 5 years as determined by PubMed. We examine and propose potential mechanisms that give rise to antimicrobial synergy with AMPs.
AMPs are ubiquitously observed in nature and are known for their physiological antimicrobial roles. They are produced by both prokaryotic and eukaryotic organisms, ranging from bacteria (5, 6), insects (7–9), amphibians (10–12), and humans (13–16). AMPs protect organisms from microbial harm and thus play vital roles in innate immunity (17) by directly or indirectly killing microbes. AMPs directly kill microbes by acting at the bacterial membrane (18, 19) or eliciting bacterial cell death via inhibition of macromolecular functions (20). AMPs indirectly kill microbes by directing cytokines to sites of infection for increased immunological responses in hosts (21). Neutrophils, the first line of innate immune defense, have dense granules that are packed with AMPs that are used to defend against microbial infections (22). When stimulated, neutrophils can also release their intracellular contents to form neutrophil extracellular traps (NETs). These web-like structures, consisting of DNA, AMPs, and other antimicrobial agents, can entrap and kill bacteria (23, 24). Similar to neutrophil elastases, AMPs have vital roles in NETs in controlling microbial threats (25). A recent report indicates that AMPs also localize to cellular lipid droplets with histones (26) and contribute to lipid-droplet based cellular immunity.
AMPs are typically small peptides, ranging from about 5 to 50 amino acids, but can be as large as over 100 amino acids (27). Most AMPs are positively charged (+2 to +9) due to their high proportions of arginine and lysine residues (28), though negatively charged AMPs do also exist (21, 29, 30). Structures of AMPs include α-helix, β-sheet, extended, and loop (31), with α-helix and β-sheet structures being the most common. More complex structures also exist, including cyclic and lasso peptides (32). AMPs are known for their amphipathic nature, typically consisting 50% of hydrophobic residues including alanine, glycine, and leucine (28, 33). The biophysical properties of AMPs contribute to their potent antimicrobial activity. Cationic (positively charged) AMPs can bind to anionic (negatively charged) lipopolysaccharide (LPS) and lipoteichoic acid (LTA), which are major components of bacterial membranes (34). The amphipathic nature of AMPs also enables them to interact with and insert into bacterial cell membranes.
Many reports attribute the antimicrobial activity of AMPs to the formation of pores within bacterial membranes, which can elicit cell damage and death. Several different classes of AMP-induced membrane pores have been proposed, including barrel-stave, toroidal, and carpet (20). In a barrel-stave model, peptide monomers form a transmembrane channel that is parallel to bacterial membrane phospholipids. A toroidal model proposes that AMPs insert into bacterial cell membranes and force membrane lipid structures to change in conformation, as opposed to pore insertion through an intact membrane like that of the barrel-stave model. The carpet model suggests that AMPs do not form transmembrane pores but instead localize to the bacterial membrane surface, where they disrupt membrane organization and integrity (35). These membrane disruptions can cause loss of bacterial membrane proton gradient, cell leakage, and eventually cell death (19). Alternative models to pore formation in membranes have also been proposed, with pore formation and cell leakage being attributed to the high concentrations of AMPs that are typically used in membrane pore formation studies (35). In particular, the entry of AMPs into bacterial cells may induce intracellular damage, including disruption of bacterial nucleic acid synthesis, protein synthesis, cell wall synthesis, and cell division (20).
LPS in Gram-negative bacterial membranes and LTA in Gram-positive cell walls contribute to overall negative charges of bacterial cell exteriors. Negatively charged membranes, which are conserved among bacteria, provide cytoplasmic rigidity and proper cationic gradients that are necessary for bacterial survival (36). However, cationic AMPs can easily bind to anionic components of bacterial membranes via electrostatic interactions to elicit cell damage. Complete bacterial resistance to AMPs is unlikely because evolving a bacterial membrane that possesses an outer neutral or positive charge simply for the purpose of avoiding AMPs would be too evolutionarily costly (37, 38). Still, many studies have shown that bacteria can have intrinsic resistance or evolve resistance to AMPs (2–4, 39, 40).
A vast array of bacterial resistance and defense mechanisms against AMPs exist, including the utilization of efflux pumps (41–43), modifications to cell membrane charge (38), expression of protective barriers around bacterial membranes (44), inhibition of antimicrobials via peptide cleavage (45, 46), and potential membrane healing and recovery post-damage (47). Both multidrug-resistant Gram-negative and Gram-positive bacteria utilize efflux pump mechanisms to actively pump AMPs back out into the extracellular environment to prevent cell damage (41, 42). In Gram-negative S. Typhimurium and P. aeruginosa, the lipid A portion of LPS is modified with the addition of 4-amino-4-deoxy-L-arabinose, which reduces the overall negative charge and thus reduces the binding affinity of positively charged AMPs, including azurocidin, polymyxin B (PMB), indolicidin, and LL-37 (48–50). In Gram-positive S. aureus, lysine is added to membrane phospholipids, reducing the overall anionic charge and affinity to defensin-like cationic AMPs (51). Colanic acid is a polysaccharide which functions as a protective capsule around many Enterobacteriaceae (52) and may prevent AMP-mediated activity. It has been suggested that these capsular polysaccharides play a role in bacterial resistance (40, 53) and virulence (54, 55). For example, capsular polysaccharides increase resistance of K. pneumoniae, S. pneumoniae, and P. aeruginosa to both PMB and human neutrophil alpha-defensin 1 (53). Additionally, increased slime production by S. epidermidis in medical catheters has been reported when bacterial capsular polysaccharides are expressed (54, 55). Bacterial species like E. coli and S. Typhimurium also release proteases to cleave and inhibit antimicrobials that threaten their survival, particularly protamine and alpha helical cationic AMPs, respectively (45, 46).
Recent work suggests that bacteria can recover from pores formed by LL-37 (47). However, the duration in which AMPs can keep bacterial pores open and the extent to which bacteria can repair these pores is unknown. It is possible that efflux pumps are used to eject AMPs out of the membrane to allow for bacterial lipid bilayers to reform. Additionally, bacterial cell wall biosynthesis may be upregulated for the purpose of membrane repair.
To optimize the use of antibiotics, it is important to mitigate potential bacterial resistance mechanisms. Many AMPs have been tested in clinical trials due their potent antimicrobial activity (56, 57). However, as with any antibiotic, using AMPs is associated with the risk of ever-evolving bacterial resistance that could negate their effects. A potential way to reduce the risk of drug-resistance to AMPs in clinical settings is to use AMPs in conjunction with other antimicrobials, focusing on combinations that lead to effective antimicrobial synergies. Synergistic combinations that have multiple targets in independent pathways could require two independent and simultaneous sets of mutations to address both challenges. Synergy could also be more lethal, decreasing the likelihood that bacteria can escape and develop resistance.
It has been suggested that bacteria are less likely to evolve resistance to antibiotic cocktails than to a single antimicrobial (58, 59). Consistent with this is the fact that multiple AMPs are released during immune responses in vivo, making it difficult for bacteria to develop resistance (60). Therefore, using AMP cocktails, especially ones that convey antimicrobial synergy, could be an effective strategy. Synergistic antibacterial combinations with AMPs could enable bacterial pores to stay open for longer durations, prevent pore repair, increase perturbation of bacterial intracellular functions, or convey other independent but complementary bacterial killing mechanisms. These mechanisms may potentially increase antimicrobial efficacy, decrease resistance, and reduce host toxicity if only low concentrations of each antimicrobial component are needed to carry out a large antimicrobial effect (61). The abundance of antimicrobial synergies discovered with AMPs presents exciting possibilities for the potential use of synergistic AMP combinations in clinical settings.
Numerous reports indicate that AMPs synergize with other AMPs. We discuss antimicrobial synergies of AMPs from organisms like insects, amphibians, and mammals, suggesting that synergistic interactions are common between AMPs within the animal kingdom.
The insect AMPs, diptericins and attacins, show synergistic killing against P. burhodogranariea in flies (62). A combination of the synthetic AMP pexiganan and bumblebee AMP melittin show S. aureus killing effects comparable to that of Vancomycin, a last line of defense antibiotic (39). Additionally, the antimicrobial activity of a bumblebee AMP, abaecin, is synergistically enhanced by the presence of a pore forming AMP, hymenoptaecin (63). In this example, hymenoptaecin forms membrane pores, potentially causing cell leakage or lytic cell death and enabling the entry of abaecin into bacterial cells. The hymenoptaecin-induced pores may increase the ability for abaecin to access and bind to DnaK, a molecular chaperone, to inhibit bacterial replication (63). Thus, the two AMPs work together to kill bacteria on both a membrane and intracellular level.
AMPs can potentially bind to other AMPs to form more potent antibacterial agents. For example, the amphibian AMPs magainin-2 and peptidyl-glycylleucine-carboxyamide (PGLa) work synergistically to inhibit E. coli growth (11). When magainin-2 and PGLa are added together, they form a “supramolecule” to quickly induce bacterial membrane pores and mediate pore stabilization (64). Moreover, it has been reported that PGLa forms an antiparallel dimer that spans the cell membrane where it binds to magainin-2 at the C-terminus (65), forming toroidal pore structures (66). These results are consistent with an additional report in which fused AMPs induce greater killing activities in S. mutans than on their own (67). These findings suggest that AMPs can bind other AMPs or other types of antimicrobials to give rise to antimicrobial synergy.
The mammalian AMP protegrin 1 has been reported to exhibit synergistic killing activity with indolicidin, LL-37, and bactenecin against P. aeruginosa and E. coli (68). Additionally, the combination of indolicidin and bactenecin gives rise to antimicrobial synergy against E. coli (68). The combinations of protegrin 1 with LL-37, bactenecin with LL-37, and protegrin 1 with bactenecin are also synergistic against E. faecalis (68). Lastly, human platelet-derived synthetic AMP combinations of PD1 through PD4 and Arg-Trp repeats RW1 through RW5 are synergistically antimicrobial in platelets (69).
AMPs can be effective when their mechanisms are complementary, such as in the case of the AMPs coleoptericin and defensin. Coleoptericin contributes to the survival of the mealworm beetle, Tenebrio molitor, but does not reduce bacterial load. In contrast, defensin does not improve host survival but reduces bacterial load (70). Their combined use both significantly increases host survival and reduces bacterial load (70). Using multiple AMPs together can thus maintain the independent functions of each AMP, resulting in a more effective treatment strategy.
While many studies demonstrate robust antimicrobial synergies with just two AMPs, synergies with three AMPs reveal even greater effects. For example, while apidaecin functions antagonistically with either pexiganan or LL 19-27 (an analog of LL-37), the triple combination of apidaecin, pexiganan, and LL 19-27 demonstrate strong synergism (58). Synergy was also observed from the combination of human β-defensin, LL-37, and lysozyme, which are produced on the skin, against S. aureus and E. coli (13). The observation of synergy between these antimicrobials is an example in which natural defense molecules have greater activity in combination rather than individually. Thus, combining natural antimicrobials could yield further discoveries of synergy.
AMPs can also synergize with antibiotics, and in some cases, overcome antibiotic resistance. The use of AMPs to increase the efficacy of already approved antibiotics appears to be a promising option to combat commonly drug-resistant pathogens. The human AMPs, LL-37 and human β-defensin 3 (HBD3), have antimicrobial synergy with the antibiotics tigecycline, moxifloxacin, piperacillin-tazobactam, and meropenem. Specifically, antibiotic killing against C. difficile is improved when both LL-37 and HBD3 are present (71). Lastly, LL 17-29 establishes antimicrobial synergy with the antibiotic chloramphenicol against highly virulent bacterial strains, including methicillin-resistant S. aureus and multidrug-resistant P. aeruginosa (59).
Combining the AMPs nisin Z, pediocin, or colistin with various antibiotics, including penicillin, ampicillin, or rifampicin, is effective in overcoming antibiotic-resistance in P. fluorescens (72). Also, the AMP melamine has synergistic killing activities when paired with ciprofloxacin, a fluoroquinolone antibiotic, against antibiotic-resistant strains of P. aeruginosa. This combination may aid in overcoming P. aeruginosa resistance to fluoroquinolone antibiotics (73). Synergistic combinations of AMPs with PMB (originally discovered as an AMP), erythromycin, and tetracycline have also been shown. In particular, variants of the AMP indolicidin synergize with the antibiotics PMB, tobramycin, gentamycin, and amikacin (74).
One of the mechanisms by which AMPs improve antibiotic function is by disrupting bacterial membranes to aid in the delivery of antibiotics into the bacterial cytoplasm, where antibiotics can act on intracellular targets. For example, the AMP arenicin-1 synergistically functions with antibiotics including ampicillin, erythromycin, and chloramphenicol to kill S. aureus, S. epidermis, P. aeruginosa, and E. coli (75). Arenicin-1 assists in the uptake of antibiotics into cells and inhibits bacterial growth via hydroxyl radical formation (75), which suggests complementary mechanisms are at play.
Histones, more commonly known for their roles in condensing eukaryotic DNA, have antibacterial properties (76, 77). However, the mechanisms by which histones kill bacteria have not previously been understood (78). Since histones are positively charged and have similar structures to that of AMPs, it has been suggested that histones and AMPs have redundant antibacterial roles (79, 80). Histones and AMPs colocalize in innate immunity components, including on cellular lipid droplets and in NETs, suggesting that they could work together to kill microbes (26, 81–83). For fish in particular, fractions of salmon histone H1 have reported antimicrobial synergy with lysozyme and a flounder AMP, pleurocidin (84). Recent work demonstrates that histones H2A and H3 can function with the pore-forming AMPs LL-37 and magainin-2 to produce antibacterial synergy against Gram-positive and Gram-negative bacteria (47). Additionally, H2A and the pore-forming antimicrobial PMB synergistically work together to completely inhibit E. coli growth over 24 hours (47). It is important to note that histones must be paired with pore forming AMPs in order for this synergistic model to be effective; histones alone have minimal antimicrobial effects at physiological conditions (47). It is possible that other histones, histone fragments, and histone-like peptides also amplify the antimicrobial effects of AMPs and give rise to antimicrobial synergy.
The mechanism of synergy between AMPs and histones is due the ability of AMPs to form pores in bacterial membranes, enabling histones to enter the bacterial cytoplasm (47, 85). Here, histones inhibit global transcription and reorganize bacterial chromosomes. Furthermore, histones enhance AMP-mediated pores that bacteria otherwise would be able to recover from, leading to reduced cell sizes and increased cytoplasmic leakage (47). The uptake of AMPs and histones into bacterial cells elicits an effective antimicrobial response consistent with a positive feedback loop (47). Importantly, if bacterial intracellular functions, like transcription and translation, are inhibited, this could reduce bacterial cell membrane integrity and repair.
Another potential effect of histones is that they may induce stress on bacterial membranes. This membrane stress could aid AMPs to more effectively form bacterial membrane pores. Altered membrane physiology, revealed through scanning electron microscopy (SEM), suggests that when bacteria are treated with only an individual AMP or histone, the membrane largely remains intact (Figure 1). However, the treatment with both AMPs and histones induces gross cell deformation and leakage of cytoplasmic contents (Figure 1). The reduced membrane integrity from the AMP and histone treatment also inhibits E. coli from maintaining their proton gradient, which is necessary for ATP production (47). Thus, membrane damage caused by synergistic combinations with AMPs may lead to lack of recovery from AMP-mediated pores, rapid loss of cytoplasmic content, failure to produce ATP, and ultimately bacterial cell death. In response to histone exposure, the rcs gene responsible for colanic acid expression is upregulated in E. coli (47). The bacterial upregulation of colanic acid, which functions as a bacterial membrane protective capsule, suggests that there is an active microbial attempt to mitigate potential membrane stress effects due to histones.
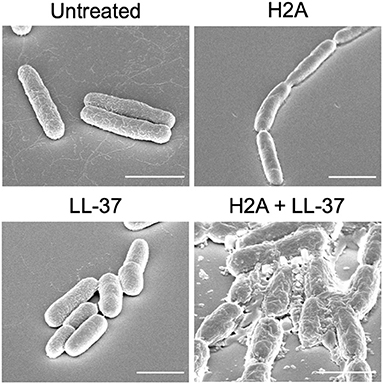
Figure 1. SEM images E. coli that are untreated or treated with H2A, LL-37, or both. E. coli that are treated with both H2A and LL-37 demonstrate extensive cellular damage. Scale bars indicate 2 μm.
AMPs also synergize with other antimicrobial agents. For example, silver nitrate and silver nanoparticles can synergize with PMB and Gramicidin S, enhancing their intracellular antimicrobial effects in Gram-negative bacteria (86). Additionally, peptoid analogs of AMPs are known to have effective and specific antimicrobial activity (87). AMPs can synergize with peptoids against Gram-negative bacteria (88). The AMP Galleria mellonella anionic peptide 2 and antimicrobial enzyme lysozyme are also synergistic against Gram-negative bacteria (89).
The combination of AMPs with current antimicrobial strategies can produce synergy through a number of distinct mechanisms (Figure 2). The introduction of antibiotics inside bacteria has often been a challenge. However, AMPs can address this challenge by forming membrane pores, thus facilitating entry of antibiotics into the cytoplasm, where the antibiotics can bind to their intracellular targets. The combination of AMPs with antibiotics could thus be an effective antibacterial strategy. This strategy could limit bacterial resistance because defense from the multifaceted attack could be significantly more difficult to achieve.
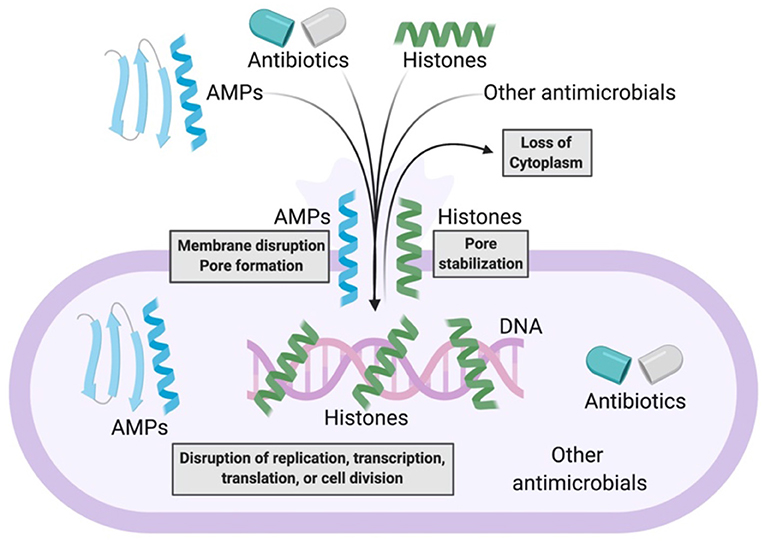
Figure 2. Model of antimicrobial synergy between AMPs and other AMPs, antibiotics, histones, and other antimicrobials. AMPs form bacterial membrane pores or disrupt bacterial membranes. This enables the entry of more AMPs, antibiotics, histones, or other antimicrobials into bacteria. As a result, there is loss of bacterial cytoplasm and disruption of bacterial macromolecular functions. Histones potentially stabilize AMP-induced pores that enable further synergistic antimicrobial activity.
If the ability for AMPs to synergize with other AMPs or antimicrobials is a conserved characteristic, then relatively low doses of each antimicrobial can be used as antibiotic treatments to exhibit large antimicrobial effects. Lower drug concentrations might also limit harmful side effects. For example, PMB is now an FDA-approved and potent last-resort antibiotic; however, PMB is also highly toxic to the nephrotic and nervous systems (90, 91). Using PMB in a synergistic antimicrobial combination, like with indolicidin or histones, would potentially require lower doses of each antimicrobial agent, potentially reducing host toxicity, while maintaining effective antimicrobial activity. Since the production of peptides can be costly, taking advantage of lower antimicrobial doses needed for synergistic treatments may also reduce production expenses. If toxicity remains an issue even with the low doses required in synergistic antimicrobial combinations, changing amino acids on AMPs has been shown to have strong effects on synergy (74). Moreover, AMPs that can synergize with preexisting AMPs in hosts could be especially potent in vivo, due to the activation of natural AMP release by the immune system. In innate immunity, humans express LL-37; therefore, synergies that arise with LL-37, like histones and protegrin 1, would be especially critical to consider for antibiotic applications.
Synergistic antimicrobial combinations are promising candidates that reduce potential bacterial resistance, overcome preexisting resistance to current antibiotics, prevent host toxicity, and increase antimicrobial efficacy. Thus, an improved understanding of mechanisms by which AMPs synergize with other antimicrobials is necessary. Moving forward, the synergistic interactions between AMPs and other antimicrobials will provide promising options to be explored in the development of new antibiotics.
LD wrote the initial draft of the manuscript. LD, SG, and AS edited the manuscript. All authors discussed the vision, direction, and scope of the manuscript.
AS was supported by NIH R21 Grant R21AI139968.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Tory Doolin for providing the SEM images. Figure 2 was created using BioRender.com.
2. El Shazely B, Yu G, Johnston PR, and Rolff J. Resistance evolution against antimicrobial peptides in staphylococcus aureus alters pharmacodynamics beyond the MIC. Front Microbiol. (2020) 11:103. doi: 10.3389/fmicb.2020.00103
3. Bauer ME, and Shafer WM. On the in vivo significance of bacterial resistance to antimicrobial peptides. Biochim Biophys Acta. (2015) 1848:3101–11. doi: 10.1016/j.bbamem.2015.02.012
4. Anaya-López JL, López-Meza JE, and Ochoa-Zarzosa A. Bacterial resistance to cationic antimicrobial peptides. Crit Rev Microbiol. (2013) 39:180–95. doi: 10.3109/1040841X.2012.699025
5. McCafferty DG, Cudic P, Yu MK, Behenna DC, and Kruger R. Synergy and duality in peptide antibiotic mechanisms. Curr Opin Chem Biol. (1999) 3:672–80. doi: 10.1016/s1367-5931(99)00025-3
6. Nes IF, Diep DB, and Holo H. Bacteriocin diversity in streptococcus and enterococcus. J Bacteriol. (2007) 189:1189–98. doi: 10.1128/JB.01254-06
7. Choi JH, Jang AY, Lin S, Lim S, Kim D, Park K, et al. Melittin, a honeybee venom-derived antimicrobial peptide, may target methicillin-resistant Staphylococcus aureus. Mol Med Rep. (2015) 12:6483–90. doi: 10.3892/mmr.2015.4275
8. Romanelli A, Moggio L, Montella RC, Campiglia P, Iannaccone M, Capuano F, et al. Peptides from royal jelly: studies on the antimicrobial activity of jelleins, jelleins analogs and synergy with temporins. J Pept Sci. (2011) 17:348–52. doi: 10.1002/psc.1316
9. Wu Q, Patočka J, and Kuča K. Insect antimicrobial peptides, a mini review. Toxins. (2018) 10:461. doi: 10.3390/toxins10110461
10. Mor A, Hani K, and Nicolas P. The vertebrate peptide antibiotics dermaseptins have overlapping structural features but target specific microorganisms. J Biol Chem. (1994) 269:31635–41.
11. Westerhoff HV, Zasloff M, Rosner JL, Hendler RW, Waal AD, Gomes AV, et al. Functional synergism of the magainins PGLa and Magainin-2 in Escherichia coli, tumor cells and liposomes. Eur J Biochem. (1995) 228:257–64. doi: 10.1111/j.1432-1033.1995.0257n.x
12. Patocka J, Nepovimova E, Klimova B, Wu Q, and Kuca K. Antimicrobial peptides: amphibian host defense peptides. Curr Med Chem. (2019) 26:5924–46. doi: 10.2174/0929867325666180713125314
13. Chen X, Niyonsaba F, Ushio H, Okuda D, Nagaoka I, Ikeda S, et al. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J Dermatol Sci. (2005) 40:123–32. doi: 10.1016/j.jdermsci.2005.03.014
14. Singh PK, Tack BF, McCray PB, and Welsh MJ. Synergistic and additive killing by antimicrobial factors found in human airway surface liquid. Am J Physiol Lung Cell Mol Physiol. (2000) 279:L799–805. doi: 10.1152/ajplung.2000.279.5.L799
15. Tollin M, Bergman P, Svenberg T, Jörnvall H, Gudmundsson GH, and Agerberth B. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides. (2003) 24:523–30. doi: 10.1016/s0196-9781(03)00114-1
16. Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals. (2014) 7:545–94. doi: 10.3390/ph7050545
17. Diamond G, Beckloff N, Weinberg A, and Kisich KO. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. (2009) 15:2377–92. doi: 10.2174/138161209788682325
18. Lehrer RI, Barton A, Daher KA, Harwig SS, Ganz T, and Selsted ME. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J Clin Invest. (1989) 84:553–61.
19. Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. (2002) 66:236–48. doi: 10.1002/bip.10260
20. Le CF, Fang CM, and Sekaran SD. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob Agents Chemother. (2017) 61:e02340–16. doi: 10.1128/AAC.02340-16
21. Lai Y, and Gallo RL. AMPed Up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. (2009) 30:131–41. doi: 10.1016/j.it.2008.12.003
22. Wetering S van, Tjabringa GS, and Hiemstra PS. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J Leukoc Biol. (2005) 77:444–50. doi: 10.1189/jlb.0604367
23. Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. (2007) 176:231–41. doi: 10.1083/jcb.200606027
24. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–5. doi: 10.1126/science.1092385
25. Rocha JDB, Nascimento MTC, Decote-Ricardo D, Côrte-Real S, Morrot A, Heise N, et al. Capsular polysaccharides from Cryptococcus neoformans modulate production of neutrophil extracellular traps (NETs) by human neutrophils. Sci Rep. (2015) 5:8008. doi: 10.1038/srep08008
26. Bosch M, Sánchez-Álvarez M, Fajardo A, Kapetanovic R, Steiner B, Dutra F, et al. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science. (2020) 370:eaay8085. doi: 10.1126/science.aay8085
27. Bahar AA, and Ren D. Antimicrobial peptides. Pharmaceuticals. (2013) 6:1543–75. doi: 10.3390/ph6121543
28. Yeaman MR, and Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. (2003) 55:27–55. doi: 10.1124/pr.55.1.2
29. Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. (2005) 3:238–50. doi: 10.1038/nrmicro1098
30. Steffen H, Rieg S, Wiedemann I, Kalbacher H, Deeg M, Sahl H-G, et al. Naturally processed dermcidin-derived peptides do not permeabilize bacterial membranes and kill microorganisms irrespective of their charge. Antimicrob Agents Chemother. (2006) 50:2608–20. doi: 10.1128/AAC.00181-06
31. Powers J-PS, and Hancock REW. The relationship between peptide structure and antibacterial activity. Peptides. (2003) 24:1681–91. doi: 10.1016/j.peptides.2003.08.023
32. Koehbach J, and Craik DJ. The vast structural diversity of antimicrobial peptides. Trends Pharmacol Sci. (2019) 40:517–28. doi: 10.1016/j.tips.2019.04.012
33. Wang G, Li X, and Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. (2009) 37:D933–7. doi: 10.1093/nar/gkn823
34. Sitaram N, and Nagaraj R. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochim Biophys Acta BBA. (1999) 1462:29–54. doi: 10.1016/S0005-2736(99)00199-6
35. Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. (2010) 5:905–17. doi: 10.1021/cb1001558
36. Farha MA, Verschoor CP, Bowdish D, and Brown ED. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chem Biol. (2013) 20:1168–78. doi: 10.1016/j.chembiol.2013.07.006
37. Yu G, Baeder DY, Regoes RR, and Rolff J. Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc R Soc B Biol Sci. (2018) 285:20172687. doi: 10.1098/rspb.2017.2687
38. Andersson DI, Hughes D, and Kubicek-Sutherland JZ. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist Updat. (2016) 26:43–57. doi: 10.1016/j.drup.2016.04.002
39. Dobson AJ, Purves J, Kamysz W, and Rolff J. Comparing Selection on S. aureus between antimicrobial peptides and common antibiotics. PLoS ONE. (2013) 8:e76521. doi: 10.1371/journal.pone.0076521
40. Campos MA, Vargas MA, Regueiro V, Llompart CM, Albertí S, and Bengoechea JA. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun. (2004) 72:7107–14. doi: 10.1128/IAI.72.12.7107-7114.2004
41. Schindler BD, and Kaatz GW. Multidrug efflux pumps of Gram-positive bacteria. Drug Resist Updat Rev Comment Antimicrob Anticancer Chemother. (2016) 27:1–13. doi: 10.1016/j.drup.2016.04.003
42. Otreebska-Machaj E, Chevalier J, Handzlik J, Szymańska E, Schabikowski J, Boyer G, et al. Efflux pump blockers in gram-negative bacteria: the new generation of hydantoin based-modulators to improve antibiotic activity. Front Microbiol. (2016) 7:622. doi: 10.3389/fmicb.2016.00622
44. Pando JM, Karlinsey JE, Lara JC, Libby SJ, and Fang FC. The Rcs-regulated colanic acid capsule maintains membrane potential in Salmonella enterica Serovar typhimurium. mBio. (2017) 8:e00808–17. doi: 10.1128/mBio.00808-17
45. Stumpe S, Schmid R, Stephens DL, Georgiou G, and Bakker EP. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J Bacteriol. (1998) 180:4002–6. doi: 10.1128/JB.180.15.4002-4006.1998
46. Guina T, Yi EC, Wang H, Hackett M, and Miller SI. A PhoP-regulated outer membrane protease of Salmonella enterica Serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol. (2000) 182:4077–86. doi: 10.1128/JB.182.14.4077-4086.2000
47. Doolin T, Amir HM, Duong L, Rosenzweig R, Urban LA, Bosch M, et al. Mammalian histones facilitate antimicrobial synergy by disrupting the bacterial proton gradient and chromosome organization. Nat Commun. (2020) 11:3888. doi: 10.1038/s41467-020-17699-z
48. Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, et al. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. (1998) 27:1171–82. doi: 10.1046/j.1365-2958.1998.00757.x
49. Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, and Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica Serovar typhimurium. Infect Immun. (2000) 68:6139–46. doi: 10.1128/iai.68.11.6139-6146.2000
50. McPhee JB, Lewenza S, and Hancock REW. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. (2003) 50:205–17. doi: 10.1046/j.1365-2958.2003.03673.x
51. Ernst CM, Staubitz P, Mishra NN, Yang S-J, Hornig G, Kalbacher H, et al. The Bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLOS Pathog. (2009) 5:e1000660. doi: 10.1371/journal.ppat.1000660
52. Grant WD, Sutherland IW, and Wilkinson JF. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J Bacteriol. (1969) 100:1187–93. doi: 10.1128/JB.100.3.1187-1193.1969
53. Llobet E, Tomás JM, and Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiol Read Engl. (2008) 154:3877–86. doi: 10.1099/mic.0.2008/022301-0
54. Christensen GD, Simpson WA, Bisno AL, and Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. (1982) 37:318–26.
55. Davenport DS, Massanari RM, Pfaller MA, Bale MJ, Streed SA, and Hierholzer WJ. Usefulness of a test for slime production as a marker for clinically significant infections with coagulase-negative staphylococci. J Infect Dis. (1986) 153:332–9. doi: 10.1093/infdis/153.2.332
56. Koo HB, and Seo J. Antimicrobial peptides under clinical investigation. Pept Sci. (2019) 111:e24122. doi: 10.1002/pep2.24122
57. Greber KE, and Dawgul M. Antimicrobial peptides under clinical trials. Curr Top Med Chem. (2017) 17:620–8. doi: 10.2174/1568026616666160713143331
58. Yu G, Baeder DY, Regoes RR, and Rolff J. Combination effects of antimicrobial peptides. Antimicrob Agents Chemother. (2016) 60:1717–24. doi: 10.1128/AAC.02434-15
59. Rajasekaran G, Kim EY, and Shin SY. LL-37-derived membrane-active FK-13 analogs possessing cell selectivity, anti-biofilm activity and synergy with chloramphenicol and anti-inflammatory activity. Biochim Biophys Acta Biomembr. (2017) 1859:722–33. doi: 10.1016/j.bbamem.2017.01.037
60. Pasupuleti M, Schmidtchen A, and Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. (2012) 32:143–71. doi: 10.3109/07388551.2011.594423
61. Xu X, Xu L, Yuan G, Wang Y, Qu Y, and Zhou M. Synergistic combination of two antimicrobial agents closing each other's mutant selection windows to prevent antimicrobial resistance. Sci Rep. (2018) 8:7237. doi: 10.1038/s41598-018-25714-z
62. Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, and Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife. (2019) 8:e44341. doi: 10.7554/eLife.44341
63. Rahnamaeian M, Cytryńska M, Zdybicka-Barabas A, Dobslaff K, Wiesner J, Twyman RM, et al. Insect antimicrobial peptides show potentiating functional interactions against Gram-negative bacteria. Proc R Soc B Biol Sci. (2015) 282:20150293. doi: 10.1098/rspb.2015.0293
64. Matsuzaki K, Mitani Y, Akada KY, Murase O, Yoneyama S, Zasloff M, et al. Mechanism of synergism between antimicrobial peptides magainin 2 and PGLa. Biochemistry. (1998) 37:15144–53. doi: 10.1021/bi9811617
65. Zerweck J, Strandberg E, Kukharenko O, Reichert J, Bürck J, Wadhwani P, et al. Molecular mechanism of synergy between the antimicrobial peptides PGLa and magainin 2. Sci Rep. (2017) 7:13153. doi: 10.1038/s41598-017-12599-7
66. Tremouilhac P, Strandberg E, Wadhwani P, and Ulrich AS. Synergistic transmembrane alignment of the antimicrobial heterodimer PGLa/magainin. J Biol Chem. (2006) 281:32089–94. doi: 10.1074/jbc.M604759200
67. He J, Eckert R, Pharm T, Simanian MD, Hu C, Yarbrough DK, et al. Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother. (2007) 51:1351–8. doi: 10.1128/AAC.01270-06
68. Yan H, and Hancock REW. Synergistic interactions between mammalian antimicrobial defense peptides. Antimicrob Agents Chemother. (2001) 45:1558–60. doi: 10.1128/AAC.45.5.1558-1560.2001
69. Mohan KVK, Rao SS, Gao Y, and Atreya CD. Enhanced antimicrobial activity of peptide-cocktails against common bacterial contaminants of ex vivo stored platelets. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. (2014) 20:O39–46. doi: 10.1111/1469-0691.12326
70. Zanchi C, Johnston PR, and Rolff J. Evolution of defence cocktails: antimicrobial peptide combinations reduce mortality and persistent infection. Mol Ecol. (2017) 26:5334–43. doi: 10.1111/mec.14267
71. Nuding S, Frasch T, Schaller M, Stange EF, and Zabel LT. Synergistic effects of antimicrobial peptides and antibiotics against clostridium difficile. Antimicrob Agents Chemother. (2014) 58:5719–25. doi: 10.1128/AAC.02542-14
72. Naghmouchi K, Le Lay C, Baah J, and Drider D. Antibiotic and antimicrobial peptide combinations: synergistic inhibition of Pseudomonas fluorescens and antibiotic-resistant variants. Res Microbiol. (2012) 163:101–8. doi: 10.1016/j.resmic.2011.11.002
73. Kampshoff F, Willcox MDP, and Dutta D. A pilot study of the synergy between two antimicrobial peptides and two common antibiotics. Antibiotics. (2019) 8:60. doi: 10.3390/antibiotics8020060
74. Ruden S, Rieder A, Chis Ster I, Schwartz T, Mikut R, and Hilpert K. Synergy pattern of short cationic antimicrobial peptides against multidrug-resistant Pseudomonas aeruginosa. Front Microbiol. (2019) 10:2740. doi: 10.3389/fmicb.2019.02740
75. Choi H, and Lee DG. Synergistic effect of antimicrobial peptide arenicin-1 in combination with antibiotics against pathogenic bacteria. Res Microbiol. (2012) 163:479–86. doi: 10.1016/j.resmic.2012.06.001
76. Miller BF, Abrams R, Dorfman A, and Klein M. Antibacterial properties of protamine and histone. Science. (1942) 96:428–30. doi: 10.1126/science.96.2497.428
77. Hirsch JG. Bactericidal action of histone. J Exp Med. (1958) 108:925–44. doi: 10.1084/jem.108.6.925
78. Doolin T, Gross S, and Siryaporn A. Physical mechanisms of bacterial killing by histones. In: Duménil S, van Teeffelen S, editors. Physical Microbiology Advances in Experimental Medicine Biology (Cham: Springer International Publishing). p. 117–33. doi: 10.1007/978-3-030-46886-6_7
79. Kawasaki H, Isaacson T, Iwamuro S, and Conlon JM. A protein with antimicrobial activity in the skin of schlegel's green tree frog Rhacophorus schlegelii (Rhacophoridae) identified as histone H2B. Biochem Biophys Res Commun. (2003) 312:1082–6. doi: 10.1016/j.bbrc.2003.11.052
80. Pavia KE, Spinella SA, and Elmore DE. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim Biophys Acta. (2012) 1818:869–76. doi: 10.1016/j.bbamem.2011.12.023
81. Cermelli S, Guo Y, Gross SP, and Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. (2006) 16:1783–95. doi: 10.1016/j.cub.2006.07.062
82. Anand P, Cermelli S, Li Z, Kassan A, Bosch M, Sigua R, et al. A novel role for lipid droplets in the organismal antibacterial response. eLife. (2012) 1:e00003. doi: 10.7554/eLife.00003
83. Brinkmann V, and Zychlinsky A. Neutrophil extracellular traps: is immunity the second function of chromatin? J Cell Biol. (2012) 198:773–83. doi: 10.1083/jcb.201203170
84. Patrzykat A, Zhang L, Mendoza V, Iwama GK, and Hancock REW. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob Agents Chemother. (2001) 45:1337–42. doi: 10.1128/AAC.45.5.1337-1342.2001
85. Duong L, Gross SP, and Siryaporn A. A novel antibacterial strategy: histone and antimicrobial peptide synergy. Microb Cell. (2020) 7:309–11. doi: 10.15698/mic2020.11.736
86. Ruden S, Hilpert K, Berditsch M, Wadhwani P, and Ulrich AS. Synergistic interaction between silver nanoparticles and membrane-permeabilizing antimicrobial peptides. Antimicrob Agents Chemother. (2009) 53:3538–40. doi: 10.1128/AAC.01106-08
87. Chongsiriwatana NP, Patch JA, Czyzewski AM, Dohm MT, Ivankin A, Gidalevitz D, et al. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci USA. (2008) 105:2794–9. doi: 10.1073/pnas.0708254105
88. Chongsiriwatana NP, Wetzler M, and Barron AE. Functional synergy between antimicrobial peptoids and peptides against gram-negative bacteria. Antimicrob Agents Chemother. (2011) 55:5399–402. doi: 10.1128/AAC.00578-11
89. Zdybicka-Barabas A, Mak P, Klys A, Skrzypiec K, Mendyk E, Fiołka MJ, et al. Synergistic action of Galleria mellonella anionic peptide 2 and lysozyme against Gram-negative bacteria. Biochim Biophys Acta BBA - Biomembr. (2012) 1818:2623–35. doi: 10.1016/j.bbamem.2012.06.008
90. Falagas ME, and Kasiakou SK. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit Care. (2006) 10:R27. doi: 10.1186/cc3995
Keywords: antimicrobial peptides, histones, antimicrobial synergism, antibiotic resistance, intracellular targeting
Citation: Duong L, Gross SP and Siryaporn A (2021) Developing Antimicrobial Synergy With AMPs. Front. Med. Technol. 3:640981. doi: 10.3389/fmedt.2021.640981
Received: 12 December 2020; Accepted: 12 February 2021;
Published: 12 March 2021.
Edited by:
Sattar Taheri-Araghi, California State University, Northridge, United StatesReviewed by:
Laura Orian, University of Padua, ItalyCopyright © 2021 Duong, Gross and Siryaporn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven P. Gross, c2dyb3NzQHVjaS5lZHU=; Albert Siryaporn, YXNpcnlhQHVjaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.