
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mater., 24 November 2022
Sec. Polymeric and Composite Materials
Volume 9 - 2022 | https://doi.org/10.3389/fmats.2022.1039247
This article is part of the Research TopicNanotechnology and Nanoscience to manage SARS-CoV-2 Variants of ConcernView all 14 articles
 Maria Victoria Hangad1,2
Maria Victoria Hangad1,2 Sarah Keshvani2
Sarah Keshvani2 Niya Kelpin3
Niya Kelpin3 Jonathan Walters-Shumka3
Jonathan Walters-Shumka3 McKayla Hood4
McKayla Hood4 Cameo Volk2
Cameo Volk2 Danika Pal2
Danika Pal2 Stephanie M. Willerth4,5,6,7*
Stephanie M. Willerth4,5,6,7*Nanomaterials have played a significant role in effectively combating the global SARS-CoV-2 pandemic that began in December 2019 through the development of vaccines as well as antiviral therapies. These versatile, tunable materials can interact and deliver a broad range of biologically relevant molecules for preventing COVID-19 infection, generating immunity against COVID-19, and treating infected patients. Application of these nanomaterials and nanotechnologies can further be investigated in conjunction with disease models of COVID-19 and this holds immense potential for accelerating vaccine or therapeutic process development further encouraging the elimination of animal model use during preclinical stages. This review examines the existing literature on COVID-19 related nanomaterial applications, including perspective on nanotechnology-based vaccines and therapeutics, and discusses how these tools can be adapted to address new SARS-CoV-2 variants of concern. We also analyze the limitations of current nanomaterial approaches to managing COVID-19 and its variants alongside the challenges posed when implementing this technology. We end by providing avenues for future developments specific to disease modelling in this ever-evolving field.
The COVID-19 pandemic, a historical event that will impact several generations, started with a pneumonia outbreak in Wuhan, China in January 2020. A new virus (SARS-CoV-2) was discovered that infected human lung epithelial cells. The global spread of SARS-CoV-2, along with the death of thousands, led to the World Health Organization to declare a global pandemic on 12 March 2020 (Ciotti et al., 2020). The ongoing spread of SARS-CoV-2 and its variants has imposed a significant burden on people around the world. Governments have implemented curfews, lockdown, physical distancing, and quarantining to prevent the disease from spreading (Miyamura, 2021). The requirements for social distancing along with the enforcement of lockdowns had a major impact on many businesses and jobs, leading to an increase in unemployment and other devastating effects on the global economy (Kawohl and Nordt, 2020). Moreover, the COVID-19 pandemic also led to psychological struggles, including high amounts of psychological stress as well as anxiety due to lack of social interactions (Banerjee, 2020). Additionally, the fear of infection causes stress during large gatherings, including events like weddings and funerals.
It is hypothesized that the origins of the disease resulted in infected animals sold in wet markets due to similarities of the SARS-CoV-2 RNA sequences to known coronaviruses found in bats (Morens et al., 2020). SARS-CoV-2 belongs to the genera Betacoronavirus, and it consists of spherical enveloped particles ranging in diameter from 70 to 100 nm covered in club-shaped glycoproteins (Mousavizadeh and Ghasemi, 2021). The particles contain a single-stranded RNA genome (Mousavizadeh and Ghasemi, 2021). SARS-CoV-2 has a spike protein containing two subunits: S1 and S2 (Choi et al., 2021). S1 enables the virus to attach to host cell surface receptors while S2 promotes the fusion of host cells with viral membranes (Choi et al., 2021) as shown in Figure 1 (Belete, 2020). The spike protein also to angiotensin-converting enzyme 2 (ACE2) receptors which enables the take up of viruses into host cells (Nugent, 2022). Because human host cells contain large amounts of ACE2, type II alveolar epithelial cellsare thought to be the main target for infection (Mondal et al., 2022). The virus is transmitted through inhalation or contact with droplets containing viral particles, and the incubation period ranges within 2–14 days of infection (Choi et al., 2021). Symptoms of SARS-CoV-2 range from mild to severe and include fever, cough, shortness of breath, vomiting, diarrhea, and abdominal pain (Ciotti et al., 2020). Other reported symptoms include severe pneumonia, acute respiratory distress syndrome (ARDS), and organ failure (Choi et al., 2021). However, these symptoms are progressively changing as new variants arise. For example, the delta variant has a higher risk for hospitalization compared to the alpha variant (Shiehzadegan et al., 2021).
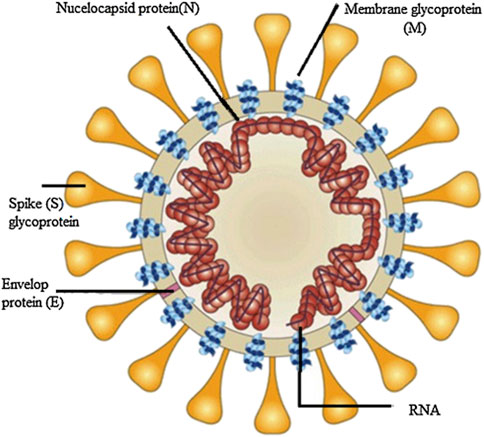
FIGURE 1. Simple Illustration of Coronavirus structure sourced from A review on Promising vaccine development progress for COVID-19 disease adapted from (Belete, 2020). Reproduced with permission of Elsevier as this work is part of their open access COVID-19 resource centre.
New variants of concern (VOC) have emerged during the pandemic as the virus has mutated. A variant of concern contains mutations that can increase transmission rates, worsen symptoms from those caused by the original virus, and/or provide viral immunity from treatments and preventative methods such as vaccines (Otto et al., 2021). Currently, these VOCs include the Alpha, Beta, Gamma, Delta, and Omicron variants (Zeng et al., 2022). These variants differ from each other due to differences in their genome sequence encoding for variations in spike protein structure (Hadj Hassine, 2022a). These subtle yet significant differences in spike protein led to changes in viral properties, such as, how contagious the variants are (transmissibility) and the types of symptoms, as well as, the severity of symptoms that can be experienced by infected individuals. Changes in viral properties can lead to issues in vaccine efficacy, as vaccines formulated for one variant may not be able to effectively mediate symptoms induced by another version of the viral pathogen.
Novel forms of nanotechnology are currently being used to treat and prevent the infection of the virus, controlling the progression of SARS-CoV-2 and easing the burdens of the pandemic. Nanotechnology uses materials with dimensions ranging from 1 to 100 nm (Hulla et al., 2015), including naturally produced particles like proteins, and synthetically produced molecules like metal nanoparticles (Mcneil, 2011). The small size, higher solubility, and multifunctionality of nanomaterials provides benefits when addressing medical issues (Mcneil, 2005). Many nanomaterials, such as quantum dots, metal nanoparticles, and nanowires (Gao and Xu, 2009), can be formulated to induce specific effects in human cells and tissues (Chakraborty et al., 2011). They can also be functionalized with different ligands for applications in drug delivery, imaging, and therapeutics. The versatile nature of nanotechnology has significant promise for managing SARS-CoV-2 through diagnostics, vaccines, and therapeutics. This review will discuss properties of nanomaterials currently being used and or are under consideration for managing the SARS-CoV-2 virus and their future application as an important tool for managing variants of concern.
In recent years, advancements in the field of nanotechnology have led to vaccines that are used to protect against current and future variants of COVID-19. Both traditional and mRNA vaccines illicit consistent immune responses towards fighting infection despite the varying mechanisms to how these responses are induced (Bullock, 2021). They both activate the adaptive immune system consisting of B-cells, antibodies, and T-cells. The adaptive immune system produces antibodies which neutralize the virus. The information of how to make these antibodies to fight against future infection is stored in memory B and T cells. This memory is what gives a vaccine recipient lasting immunity. The difference between traditional vaccines and mRNA vaccines is that traditional vaccines typically use a weakened virus or the antigen of a virus as a whole (Pardi et al., 2018) to generate immunity, whereas mRNA vaccines generate the same immunity by using the host cell machinery to synthesize the antigen of a virus encoded in a delivered mRNA strand. Here, we discuss traditional vaccine types and the innovation of mRNA vaccines against COVID-19. This section highlights nanotechnology-based formulations, specifically lipid nanoparticles (LNPs), their composition and manufacturing, and the nanomaterial-based ingredients playing a role in delivering mRNA cargo used to address SARS-CoV-2 and its variants of concern.
COVID-19 vaccines work to prevent or lessen the symptoms of an infection (Bonanni and Santos, 2011) by first eliciting an innate immune response by exposing the immune system to the COVID-19 spike protein so that if the pathogen is encountered, the presence of SARS-CoV-2 antibodies will enable a stronger immune response (Mathieu et al., 2021; Sette and Crotty, 2021). Although mRNA vaccines are widely used, non-mRNA vaccines, which include viral vector vaccines, protein subunit vaccines, and whole pathogen vaccines, are also present in the market shown to induce immunity against COVID-19 through different mechanisms.
Viral vector vaccines consist of engineered, chemically weakened viruses with SARS-CoV-2 genes. They infect immune cells to induce the production of COVID-19 proteins or display COVID-19 proteins on their surface (Samaranayake et al., 2021). Viral vector vaccines can be replicating (i.e. can make a new viral particle) or non-replicating (i.e. cannot make a new viral particle) (Samaranayake et al., 2021). Non-replicating viral vector vaccine technology has been used by Oxford-AstraZeneca to create the Vaxzevria COVID-19 vaccine (ChAdOx1 nCoV-19) and by Janssen to create the Janssen Jcovden COVID-19 Vaccine (Ad26.COV2.S) (Mathieu et al., 2021; Voysey et al., 2021; Andrews et al., 2022).
Subunit vaccines contain SARS-CoV-2 whole proteins or their protein fragments (Samaranayake et al., 2021). The Novavax Nuvaxovid COVID-19 vaccine (NVX-CoV2373) is an approved and effective protein subunit vaccine which carries a recombinant SARS-CoV-2 spike protein (Keech et al., 2020). While Novavax vaccines targeting the Beta and Delta variants of COVID-19 are in clinical trials, the production of COVID-19 variant-specific protein-subunit vaccines has been delayed compared with mRNA vaccines due to differences in vaccine development procedures (Lee and Kim, 2022).
Virus-like particle vaccines are a whole pathogen type of vaccine that mimics the structure of the SARS-CoV-2 virus that do not contain genetic material and are not infectious (Ndwandwe and Wiysonge, 2021). In Canada, the Medicago Covifenz COVID-19 vaccine is an approved plant-based virus-like particle vaccine (Hager et al., 2022). This coronavirus-like particle vaccine utilizes Nicotiana benthamiana plants to produce virus-like particles of a modified spike glycoprotein from SARS-CoV-2 strain hCoV-19/USA/CA2/2020 (Stander et al., 2022). Although there are none approved in Canada, whole-pathogen vaccines, such as live attenuated vaccines and inactivated vaccines, which utilize a weakened form of the virus to stimulate immune responses, area also in development for the SARS-CoV-2 virus (Ndwandwe and Wiysonge, 2021).
The utilization of various types of COVID-19 vaccines has been met with various levels of success as the type of vaccine influences the degree of significant reduction of infectious symptoms, transmissibility, and mortality outcomes at a global scale (Baden et al., 2021; Mathieu et al., 2021; Voysey et al., 2021; Andrews et al., 2022). Highly successful vaccines are characterized based on efficacy and are dependent on two factors: potency and administration. In order to assess vaccine efficacy, the seroconversion must be determined. It is a quantitative measure of antibody persistence which defines the level of immune response (Orenstein et al., 1985). In a 2021 review by Yan et al. (2021), vaccine trials and obtained seroconversion rates of mRNA-based vaccines as well as non-mRNA-based vaccines were summarized. While Pfizer/BioNTech’s BNT162b1/b2 mRNA lipid nanoparticle vaccine demonstrated a dose-dependent antibody response with no serious adverse events, Moderna’s mRNA-1273 demonstrated 100% seroconversion rates by day 15. On the other hand, traditional viral vaccines like Oxford-AstraZeneca’s ChAdOx1 nCoV-19 had shown to induce antibody titres five times more when comparing booster doses to single-dose groups with noted serious adverse events. Protein subunit vaccines like Novavax’s NVX-CoV2373 were demonstrated to have antibody responses such as neutralization levels and IgG GMT presence exceeding normal serum levels.
A metanalysis of vaccine efficacy in protecting against severe disease caused by COVID-19 reveals that, in adults, two doses of the Vaxzevria COVID-19 vaccine has 74% efficacy, a single dose of the Janssen Jcovden COVID-19 vaccine has 66% efficacy, two doses of the Novavax Nuvaxovid COVID-19 vaccine has 90% efficacy, and two doses of the Medicago Covifenz COVID-19 vaccine has 71% efficacy (Government of Canada 2020b; Government of Canada 2020c; Government of Canada 2020e; Government of Canada, 2022a). Two doses of the Pfizer-BioNTech Comirnaty mRNA COVID-19 vaccine possessed 95% efficacy in individuals 16 years and older and two doses of the Moderna Spikevax mRNA vaccine exhibited 94.1% efficacy in adults (Government of Canada 2022d; Government of Canada, 2022f)Of these COVID-19 vaccines, the mRNA-based vaccines, Pfizer-BioNTech Comirnaty and Moderna Spikevax, had greater efficacy than approved traditional COVID-19 vaccines. Given this data, mRNA nanoparticle-based vaccines have demonstrated to show higher success in fighting against viral pathogens like SARS-CoV-2.
Two nanoparticle-based vaccines for COVID-19 have been approved for use by Health Canada after their determination as safe and effective: the Pfizer-BioNTech Comirnaty COVID-19 vaccine (BNT162b2 mRNA) (Polack et al., 2020) and Moderna Spikevax COVID-19 vaccine (mRNA-1273 SARS-CoV-2) (Baden et al., 2021). These vaccines utilize LNP technology to deliver an mRNA transcript intramuscularly for the in vivo translation and protein synthesis of the COVID-19 spike glycoprotein (Kowalzik et al., 2021).
When delivered without a carrier, the uptake of mRNA by cells is low and considered unsuitable for therapeutic use due to the quick degradation rate of the mRNA in circulation (Kowalzik et al., 2021). Naked mRNA is negatively charged, large, and unstable, making its passive diffusion across the cell membrane challenging (Sahin et al., 2014). LNPs have arisen as powerful tools for the delivery of mRNA as they address these issues. LNPs envelop mRNA and act to protect mRNA from degradation and deliver it to the cytosol of target cells (Melamed et al., 2022).
The LNPs used in Pfizer and Moderna’s mRNA COVID-19 vaccines have four primary components: ionisable cationic lipids, helper lipids including neutral phospholipids, cholesterol, and polyethylene-glycol (PEG)-lipids (Figure 2) (Evers et al., 2018; Zhi et al., 2018; Albertsen et al., 2022). The most important part of these components is the ionizable lipids as they promote particle uptake by the desired cell population. LNPs loaded with mRNA have a core-shell structure (Albertsen et al., 2022; Pantelić et al., 2022). The nanoparticles consist of a cationic lipid shell containing the mRNA. The ionizable lipids, helper lipids, cholesterol and PEG-lipids form a lipid shell around these core molecules (Alameh et al., 2021; Hassine, 2022). Cationic ionizable lipids have a positively charged head group that make up the bulk of the LNP bilayer (Zhi et al., 2018). The cationic ionizable lipids used in LNPs generally have a pKa just below neutral pH resulting in a positive charge at acidic pH and a neutral charge at a pH above the pKa. The lipids and SM-102 and Alc-0315, used by Pfizer and Moderna, have pKas of 6.09 and 6.75 respectively.
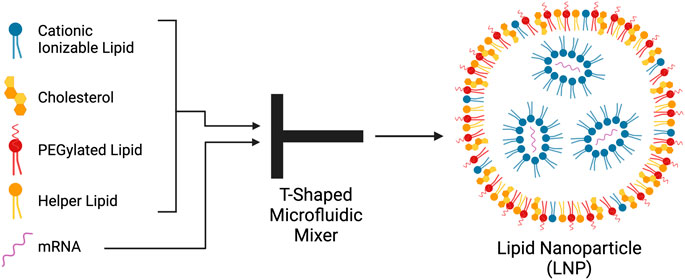
FIGURE 2. Components, structure and method of production of lipid nanoparticles (LNP). Cationic ionizable lipids cholesterol, PEGylated lipids, and helper-lipids are dissolved in ethanol and mixed with mRNA in water in a T-shaped microfluidic mixer. The lipids and mRNA self-assemble into LNPs. The negatively charged mRNA largely associates with the cationic ionizable lipids. The lipids create an outer shell surrounding the mRNA-cationic lipid complexes. Created with BioRender.com.
Production of the LNPs occurs at an acidic pH of around 4 (Cheng and Lee, 2016; Kulkarni et al., 2018). The positive charge of these lipids interacts with negatively charged nucleic acids through electrostatic interactions, increasing the encapsulation efficiency of the mRNA (Cheng and Lee, 2016; Evers et al., 2018; Kulkarni et al., 2018; Zhi et al., 2018). This interaction leads to the formation of a lipid bilayer, sandwiching condensed nucleic acids. However, at physiological pH (7.4) the ionizable cationic lipids have a neutral charge (Evers et al., 2018; Schlich et al., 2021). This effect protects against clearance of the LNPs by the immune system which would occur if permanently changed LNPs were used (Schlich et al., 2021). This property also avoids the uptake of negatively charged molecules by the LNP if a permanently charged lipid were to be used (Evers et al., 2018).
An advantage of using ionizable cationic lipids is that they promote endosomal escape within the target cells (Evers et al., 2018; Schlich et al., 2021). Escape occurs because the pH of the endosome is more acidic than the pKa of the ionizable lipid (Hu et al., 2015; Schlich et al., 2021; Pantelić et al., 2022). Ionizable cationic lipids in LNPs are neutral at physiological pH but become protonated at pH ∼6.5 in the acidic environment of the endosome (Chan et al., 2012; Suk et al., 2016). The cationic lipids then interact with the anionic endosome membrane which destabilizes the endosome membrane resulting in endosomal escape (Hafez et al., 2000; Evers et al., 2018; Buschmann et al., 2021; Schlich et al., 2021). Endosomal escape is critical to efficient nanoparticle delivery (Smith et al., 2019). LNPs that have poor endosomal escape properties remain trapped in the endo/lysosomal pathway and are consequently degraded in the acidic environment and are therefore unable to deliver their cargo (Smith et al., 2019). Mechanisms that facilitate the endosomal escape of nanoparticles include, but are not limited to, membrane fusion, osmotic pressure, nanoparticle swelling, and membrane destabilization (Smith et al., 2019).
Helper lipids increase the stability of the nanoparticle and increase the efficiency of the delivery of the mRNA to the target cells (Cheng and Lee, 2016; Evers et al., 2018). Helper lipids often have either a conical shape or a cylindrical shape, which aids in the formation of the LNP and increases the stability of the lipid bilayer of the nanoparticle. The Pfizer and Moderna vaccines both use the helper lipid 1,2-Distearoyl-sn-glycero-3-phosphocholine (DSPC) whose cylindrical geometry aids in the formation of the lipid bilayer (Cheng and Lee, 2016). Cholesterol acts as a helper lipid, stabilizing the LNP by filling the gaps between phospholipids in the lipid bilayer, and promoting membrane fusion (Cheng and Lee, 2016; Buschmann et al., 2021). Cholesterols also function within the LNPs to improve the efficiency of endosomal escape (Cheng and Lee, 2016; Evers et al., 2018). The four components of the LNP are all crucial for its proper function when forming and when in the body. The ionizable lipids help with formation and endosomal escape, the PEG lipids shield the LNP from the immune system and increase its circulation time, the helper lipids aid in formation and stability, and cholesterol aids in membrane fusion and increases the stability of the LNP.
PEG-lipids function in the nanoparticles to shield the LNP and increase its stability in the body (Suk et al., 2016; Evers et al., 2018). PEG, a hydrophilic polymer, can be attached to molecules in a process called PEGylation. PEGylation is an established method of increasing stability and circulation of drugs in the body. They shield the LNPs from opsonins, resulting in decreased phagocytosis and preventing the LNPs from aggregating during production and in the body, resulting in longer circulation time in the body (Mui et al., 2013; Evers et al., 2018). While they increase the stability of LNPs, they can also reduce uptake by target cells and endosomal escape (Mui et al., 2013; Li et al., 2014; Evers et al., 2018). PEG lipids reduce uptake by target cells because PEG is hydrophilic, therefore reducing interactions with the hydrophobic lipid bilayer of the target cell. PEG lipids also reduce ApoE-mediated fusion required for endocytosis of the LNP into hepatocytes (Akinc et al., 2010; Mui et al., 2013). The PEGylated lipids used in the Pfizer and Moderna vaccines are 2-[(polyethylene glycol)-2000]-N,N ditetradecylacetamide and 1,2-dimyristoyl-rac-glycerol, methoxypolyethylene glycol respectively (Government of Canada). The specific reasons for why these PEGylated lipids were used is proprietary information. They are likely chosen to find a balance between stability and the ability to fuse with membranes. This balance is determined by the PEGylated lipid used and its concentration within the LNPs(Evers et al., 2018; Sarode et al., 2022).
The formulation of nanotechnologies utilized for vaccines and therapeutics determine its biocompatibility within the body. Chemical composition, degradation, metabolism, and resulting toxicological behaviours are key examples of nanomaterial properties that can influence the therapeutic efficacy, or the ability of a substance to treat the disease of concern with the most minimal side effects.
Two vaccines currently in use are the Moderna and Pfizer/BioNTech vaccines (29); both showing efficacies of around 95% (Schoenmaker et al., 2021). Despite these high efficacies, analysis of antibody and T-cell responses of 122 individuals 6 months after two doses of the Pfizer vaccine revealed the median levels of S-RBG IgG dropped to 7% of their peak potencies (Naaber et al., 2021). This is indicative of short-lived immunity prompting the need for more subsequent booster doses. Aside from this, a wide variety of side effects on the range of less severe to adverse have been experienced by individuals who have received these mRNA-LNP vaccines.
Common side effects of nanomaterial-based vaccines, particularly those that utilize mRNA-LNP formulations include swelling at the location of injection, fatigue, fever, and headache (Ciotti et al., 2020; Morens et al., 2020; Anand and Stahel, 2021) A study conducted by Ndeupen et al. (2021), where empty lipid nanoparticles formulated with phosphate buffered saline was injected in mice, found intense inflammation caused by the lipids which could possibly lead to side-effects in mRNA vaccines. More adverse side effects include hypersensitivity reactions that are commonly attributed to the PEGylated (polyethylene-glycol) lipid component shown to stimulate larger complement activation (Senti et al., 2022) pre-existing antibody responses (Ju et al., 2022), and activation of allergy specific immune cells (Troelnikov et al., 2021). Although the addition of PEG has been known to play a beneficial role in nanoparticle stability and improving specificity for uptake of targeted cell types (Bigini et al., 2021), further studies have yet to thoroughly quantify the extent to which it contributes to stimulating these adverse immune responses. Experiments conducted by Tanaka et al., 2021 demonstrate how lowering the PEG-lipid content from 3% to 0.75% alters the surface density of LNPs and results in enhanced transfection of mRNA through the biological membrane (Tanaka et al., 2021). Hence, application of these PEG-related molecular modifications can be a way to optimize current LNP formulations in the SARS-CoV-2 vaccines.
To optimize the formulation of the LNPs used in COVID-19 vaccines, thousands of preliminary lipid formulations have been tested in search of sufficient lipid ratios for the mRNA carriers (Pantelić et al., 2022). There are many factors which affect the properties of the LNPs that change based on which components and ratios of them are used. Properties that frequently change among different LNP formulations are individual size and size variability within a batch (batch-to-batch variability) (Evers et al., 2018; Maeki et al., 2022). It has been shown that the size of the LNPs used in an mRNA vaccine can impact the immunogenicity of the vaccine (Yan et al., 2013; Hassett et al., 2021; Okuda et al., 2022). LNPs typically range from ∼30 to 200 nm, and within this range, the immunogenicity of the LNPs changes. For example, a study conducted by Hassett et al., in 2021 found that nanoparticles around 100 nm in size were able to recruit sufficient amounts of immune cells for participation in antigen presenting cell (APC) uptake for mRNA delivery. This study indicates that factors like immune cell recruitment and interactions with APCs can be influenced by particle size which consequently affect the degree of antigen expression as a result of mRNA-LNP delivery to host cells (Hassett et al., 2021).
Adjustment of other molecular components within LNPs can also serve to address challenges with nanoparticle stability and long-term effects of mRNA-based vaccines. In fact, a study conducted by Xu et al., 2022 confirms how the geometry and molecular chirality properties of nanoparticles conferred by its composition greatly influences the cellular uptake and lysosomal escape of nanostructures by the immune system (Xu et al., 2022) and, therefore, its therapeutic efficacy. Similarly, Alvarez-Benedicto et al., 2022 highlights the chemical properties like phospholipid head group charge, or the type of lipid DSPC or DOPE, within LNPs that mediate the destination of mRNA payloads either to the spleen or liver; they were also able to observe which phospholipid component in LNPs are more likely to contribute to endosomal escape after endocytosis (Alvarez-Benedicto et al., 2022). In terms of mRNA compatibility and stability with LNPs, a study by Packer et al., 2021 found that the presence of mRNA impurities caused by ionized lipids could disrupt mRNA translation and therefore, can negatively impact the efficacy of mRNA-lipid-based products (Packer et al., 2021). Experiments conducted by Cornebise et al., 2022 further illustrate the significant influence of lipid components of LNPs on their mRNA payload through the discovery of amino lipids that form stabilizing hydrogen bonds with the mRNA resulting in enhanced in vivo delivery (Cornebise et al., 2022). (Cornebise et al., 2022)
Because of the unstable nature of current COVID-19 vaccines and their requirement to be stored at low temperatures (Schoenmaker et al., 2021), there has been recent research into preserving the stability of the mRNA-LNPs without altering its chemical composition. One technique for LNP preservation, as employed by researchers Muramatsu et al., 2022, is freeze-drying or lyophilization where structural integrity from particle diameter, zeta potential, mRNA loading concentration, to encapsulation efficiency under several storage conditions was maintained without affecting the mRNA translation in vivo (Muramatsu et al., 2022). Despite LNPs ability to maintainin their structural integrity and to encapsulate a broad range of payloads, current efforts to optimize mRNA-LNP vaccines to address variants of concern are primarily achieved through modulating mRNA sequences as these mRNA constructs can easily be administered for critical evaluation.
LNPs are most commonly produced using rapid mixing (Evers et al., 2018; Maeki et al., 2022) In this method, the lipid particles are dissolved in an organic solvent solution, such as ethanol, and the mRNA is dissolved in an aqueous buffer solution. These solutions are rapidly mixed and the LNPs self-assemble. At commercial scales, T-shaped microfluidic devices are used in the production of the currently approved vaccines for COVID-19 as they allow for large batches to be made (Cheng and Lee, 2016; Pantelić et al., 2022) In a T-shaped microfluidic device, the ethanol and buffer mix together at the junction of the T shaped channel (Maeki et al., 2022), as shown in Figure 2. Here, the LNPs self-assemble from the lipids and the mRNA which are collected at the end of the exit channel. As previously mentioned, this takes place at an acidic pH below the pKa of the ionizable cationic lipid. The cationic ionizable lipids form electrostatic interactions with the negatively charged mRNA which aids in encapsulation efficiency. The components also form a shell around the inner ionizable lipid membrane enclosing the mRNA (Albertsen et al., 2022; Maeki et al., 2022; Pantelić et al., 2022).
LNPs produced for these mRNA vaccines are of a consistent size as it affects several aspects of cargo delivery to the targeted cells as previously mentioned. It has been found that fast mixing of the aqueaus and ethanol solutions is associated with the production of smaller nanoparticles and slow mixing is associated with larger LNPs (Maeki et al., 2022). Mixing of the ethanol and aqueous solutions so that there is a consistent ratio of both phases’ results in little variability between the sizes of LNPs while a ratio gradient refers to size variability outcomes. Microfluidic devices are tuned to have little variability in the size of LNPs and are also tuned to create the desired size of nanoparticles. Previously used dilution methods such as vortexing have large variability in the size of the LNPs produced. This is because the mixing is very] non uniform in this method resulting in variously sized LNPs. Research is being done on improving the process of making nanoparticles. One such company is Precision Nanosystems (Vancouver, B.C.). Their NanoAssemblr® technology decreases the variability in size of nanoparticles compared to a typical T shaped microfluidic device as demonstrated in a study that encapsulated silencing mRNA (siRNA) in LNPs for therapeutic purposes (Walsh et al., 2014).
Emerging variants of SARS-CoV-2 demand the production of effective vaccines to protect against them. SARS-CoV-2 variants have been associated with increased disease severity, transmissibility, and the ability to avoid detection by the adaptive immune system (Tao et al., 2021). Mutations occurring primarily in the spike protein of SARS-CoV-2 have resulted in new VOCs of the virus.
Variants of SARS-CoV-2 were first discussed as early as April 2020 (Forster et al., 2020) and these variants were first detected in late 2020 (Tao et al., 2021). Notable VOCs include the Alpha, Beta, Gamma, Delta, and Omicron variants, which differ from each other and the originalstrain of SARS-CoV-2 mainly by various mutations in the spike glycoprotein (Hadj Hassine, 2022b). A high incidence of point mutations in the N-terminal domain (NTD) and receptor binding domain (RBD) of the spike gene suggests selection for increased viral fitness via changes in the spike protein that improve host-receptor binding or the ability to evade host antibodies (McLean et al., 2022). Other viral characteristics affected by spike glycoprotein mutations include infectivity, transmissibility, pathogenicity, antigenicity, as well as immune escape shown in Figure 3 (Harvey et al., 2021). In 2020 and 2021, the Alpha, Beta, and Gamma VOCs drove worldwide surges of COVID-19 infection until the rise of the Delta variant, which displaced all other VOCs as the dominant variant (Tao et al., 2021; Viana et al., 2022). The Omicron variant of SARS-CoV-2 was first detected in November 2021 and within 3 weeks spread to 87 countries (Viana et al., 2022). Omicron, characterized by its ability to rapidly spread between vaccinated individuals, quickly proceeded to replace the Delta variant as the dominant variant (Tao et al., 2021; Viana et al., 2022). Currently these variants are being addressed through acquiring booster doses, and modifications to vaccine formulation specific to mRNA cargo, alongside enhancement of adjuvant properties through the addition of nanomaterials such as liposomes to non-mRNA vaccine types.
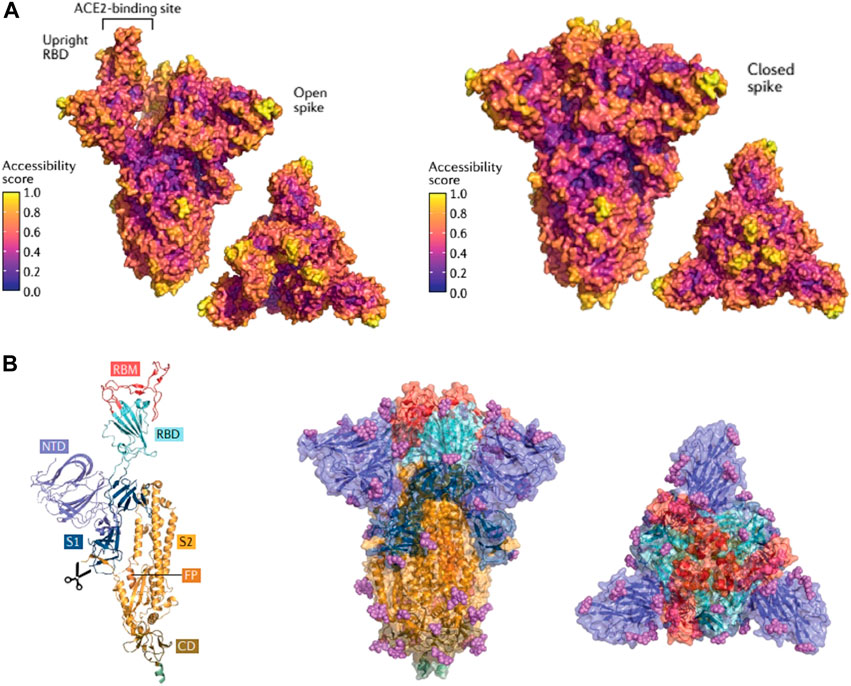
FIGURE 3. SARS-CoV-2 Spike protein structures illustrating regions specific for antibody binding and domains prone to specific amino acid mutations responsible for emerging variants of concern synthesized by researchers (Harvey et al., 2021). (A). SARS-CoV-2 Spike protein in an open conformation and closed conformation exhibiting key areas of antibody binding based on conformational epitopes. Colors are based on antibody accessibility scores where high values on the scale indicate higher antibody recognition. Lower values of antibody accessibility indicate a lower antibody recognition which strongly correlates to domains containing amino acid mutations characteristic of emerging variants of concern. (B). SARS-CoV-2 Spike Protein composition: RBM (receptor binding motif ), RBD (receptor binding domain), NTD (amino-terminal domain), S1 (subunit 1, part of NTD and RBD responsible for binding ACE2), S2 (Subunit 2, part of trimeric protein core responsible for membrane fusion). This figure is reprinted with permission from Nature Springer from (Harvey et al., 2021).
The two nanoparticle-based vaccines in use in Canada are effective against COVID-19 variants (Liu J. et al., 2022). One study showed that two doses of Pfizer’s BNT162b2 vaccine elicited the neutralization of Delta plus, Lambda, Mu, B.1.1.519, and Theta SARS-CoV-2 variants (Liu J. et al., 2022). Subsequent doses of mRNA COVID-19 vaccines, coined booster shots, have been recommended to restore wanning immunity over time and as protective tools against the loss of vaccine efficacy from emerging COVID-19 variants (Scheaffer et al., 2022). Emphasizing the importance of booster shots, studies have shown that three doses of Pfizer’s BNT162b2 vaccine and Moderna’s mRNA-1273 SARS-CoV-2 vaccine elicit improved neutralizing antibody activity and protection against the SARS-CoV-2 Omicron variant when compared to two doses (Andrews et al., 2022; Ariën et al., 2022). Variant-specific mRNA booster vaccines are also in trial, showing promising results in mouse models (Flemming, 2022). For example, one study demonstrated that immunizing mice with a booster vaccine of an Omicron-specific version of Moderna’s mRNA vaccine elicited improved protection against the Omicron variant than being boosted by the original version of the mRNA-1273 COVID-19 vaccine, demonstrating the enormous potential of altering mRNA sequences in mRNA vaccines to address COVID-19 variants (Flemming, 2022; Gagne et al., 2022). In early September 2022, Health Canada approved for adults the Moderna Spikevax Bivalent COVID-19 booster, an mRNA vaccine that includes the mRNA sequence from the original COVID-19 strain and the Omicron BA.1 variant (doi:10.1136/bmj.o2144). This booster is predicted to offer protection against further emerging subvariants of the Omicron variant (Dyer, 2022).
Booster doses may not exhibit consistent efficacy in populations Older adults may have a weaker immune response and faster declining antibodies than younger adults; recent data found that adults 80 years and older have lower antibody elicitation and titres than younger adults under the age of 60 years old when vaccinated with two doses of the Pfizer/BioNTech Comirnaty COVID-19 vaccine (Muller et al., 2021). Due to this low efficacy, older adults may have a need for a greater number of booster doses to maintain protection against severe disease as a single booster dose may not provide sufficient protection as it would in a younger adult. Distributing booster doses to communities with a high proportion of elderly individuals or immuno-deficient individuals has been a strategy used to maintain adequate protection in vulnerable populations (Galanis et al., 2022).
Further, in communities, concern of vaccine safety and side effects are critical factors in booster dose refusal and henceforth, distribution (Galanis et al., 2022). Recent analysis of voluntary reports to the Vaccine Adverse Events Reporting System (VAERS) in the United States between September 2021 and February 2022 revealed 39,286 cases of adverse reactions to a booster dose of an mRNA COVID-19 vaccine and revealed that adverse reactions are more frequent when receiving a heterologous booster (e.g., Moderna Spikevax for the first two doses and Pfizer/BioNTech Comirnaty the booster) than a homologous one (e.g. Moderna Spikevax for the first two doses and the booster dose) (Hause et al., 2022) It is possible that hesitant individuals may be more willing to receive vaccines if they were given the choice of homologous boosting. Adequate education about age group-dependent efficacy and booster side effects should remain in practise during booster dose distribution.
According to a 2021 review, Pfizer/BioNTech and Moderna’s mRNA-LNP vaccines have demonstrated a greater than 90% retention of efficacy in preventing disease towards the original D614G and alpha strain of SARS-CoV-2 but has shown a lower efficacy of approximately 80% in preventing disease alongside infection for other variants like Beta, Gamma, or Delta (Kandikattu et al., 2021). This is indicative of how mRNA-LNP vaccines induce broad based immunity. A 2022 systematic review and meta-analysis of COVID-19 vaccine effectiveness against SARS-CoV-2 variants of concern also details the same trends from clinical trial data and ongoing vaccination programs showing a consistent decrease in vaccine effectiveness for LNP based vaccines (Zeng et al., 2022).
To address the mutated RBD and NTD regions in the SARS-CoV-2 spike protein in variant strains, the robust properties of LNP based formulations are utilized to deliver specific modified mRNA sequences. The nature of the mRNA transcript used in the vaccine allows its sequence modification so that an altered mRNA transcript, specific to a COVID-19 variant, can be loaded in an LNP for delivery, which is extraordinarily advantageous for tackling COVID-19 variants (Forchette et al., 2021). For example, designed mRNA encoding for specific RBD proteins rather than the full-length S protein of SARS-CoV-2 and assessed the vaccine efficacy through cellular immune responses in mice for wild-type SARS-CoV-2 along with variants Delta and Omicron. As a result, Liu et al. (2022) experiments illustrated strong neutralizing antibody responses and significant protection from two to three doses against wild-type, Delta, and Omicron variants (Liu C. et al., 2022). This study further serves as an example to how mRNA modifications could increase specificity in immune responses through presentation of an immunogen rather than the whole antigen. Variant-specific vaccination using encapsulated mRNA in LNPs designed to include mutations in the spike protein have also been investigated (Peng et al., 2022; Zhang et al., 2022), although the resulting levels of neutralization vary, which poses a challenge when formulating a succinct, universal, one-dose vaccine against COVID-19.
mRNA-LNP vaccines offer several advantages over other vaccine types in the light of emerging COVID-19 variants by being cost-effective, and rapidly synthesized. Many LNPs are self-adjuvanting due to the nanomaterials they are composed of andexhibit adjuvanting activity, which is the ability to enhance an immune response towards an antigen (Alameh et al., 2021; Igyártó et al., 2021). An adjuvant is any ingredient that creates a stronger immune response to a vaccine. The cholesterol and phospholipid used in LNPs also occur in mammalian cell membranes, making the possibility of triggering inflammation or an innate immune response unlikely (Igyártó et al., 2021). The adjuvating properties of LNPs thus likely stem from other nanomaterial components of the LNP, such as the inflammatory properties of the cationic ionizable lipid, the mRNA, or both (Alameh et al., 2021). In mice, a LNP formulation used to deliver mRNA for immunization against COVID-19 was shown to exhibit intrinsic adjuvant activity, leading to the promotion of T follicular helper cell, germinal center B cell, long-lived plasma cells, and memory B cell responses for antibody production (Alameh et al., 2021).
Addition of nanomaterials commonly comprising LNPs can increase adjuvant properties to non-mRNA vaccine types. For example, liposome adjuvants are a cost-effective and rapidly made solution (Abhyankar et al., 2021) that may contain nanomaterials, phospholipids or cholesterol components, similar to LNPs that enhance other vaccine types like protein subunit vaccines specific for COVID-19 variants. In a mouse model of COVID-19, a spike protein vaccine containing a dual Toll-like receptor ligand nanoliposome adjuvant was shown to elicit anti-spike neutralizing antibody responses and protect mice from lethal COVID-19 infection and lung immunopathology (Abhyankar et al., 2021). Similarly, NARUVAX-C19, a recombinant spike protein vaccine candidate formulated with a nanoemulsion oil adjuvant, was shown to produce high levels of neutralizing antibodies against wildtype and delta COVID-19 variants (Tabynov et al., 2022).
Liposomes similar to LNPs can also be utilized to deliver the SARS-CoV-2 spike protein antigen as part of the nanoparticle formulation. In a separate mouse model of COVID-19, nanoparticles structurally comprised of liposomes that were decorated with either SARS-CoV-2 spike protein or its receptor binding domain on its surface exhibited effective adjuvant properties and high thermal stability (Mabrouk et al., 2021). The material formulation of the vaccine also included cobalt porphyrin-phospholipid for particle induction and the adjuvants QS-21 and PHAD, a synthetic monophosphoryl lipid A. The high thermal stability demonstrated in this paper was due to the lyophilisation or freeze-drying of the particle to preserve the nanoparticle-liposome structure alongside the additional adjuvant properties of the liposome to provide stability in vivo upon administration. The nanoparticle vaccine produced by this group exemplifies how the incorporation of additional nanomaterials or components like liposomes in vaccine formulation can make vaccines easier to store than current Pfizer and Moderna vaccines as thermostable vaccines are ideal for global distribution as well as storage in areas experiencing high case volumes of COVID-19 variants.
Due to the implausibility of fool-proof prophylactic measures, health care professionals sought treatments to combat COVID-19 post infection. Where vaccines provide acquired immunity to transmissible pathogens, therapeutics can prevent viral replication and work to bolster the body’s immune response following infection (Rehman et al., 2020). In the early pandemic, doctors pivoted towards broad-spectrum antivirals with well-studied safety profiles to test for therapeutic potential against SARS-CoV-2 (Weiss et al., 2020a). However, antiviral treatments present new challenges, such as adverse side effects from interactions with prescription medications, low bioavailability, and selectivity as highlighted by Figure 4 (Chakravarty and Vora, 2021). Thus, treatments adopted nanotechnological strategies to develop safer drug-delivery systems and improve the therapeutic index (TI) of existing drug candidates (Zhou et al., 2021).
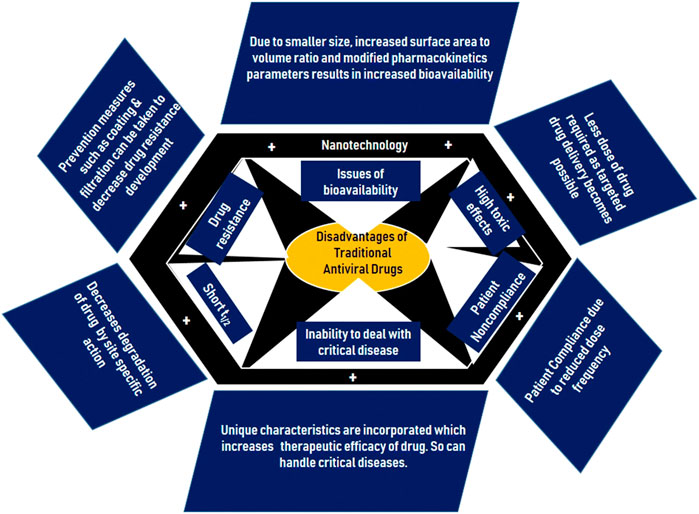
FIGURE 4. Summary of the shortfalls of traditional antivirals and methods of revision with nanoparticles (Chakravarty and Vora, 2021). Permission to use this figure is available. This figure is reprinted with permission from Springer Nature from (Chakravarty and Vora, 2021).
Nanotechnology in therapeutics is not a novel concept, however previous applications focused mainly on cancer treatments, specifically to overcome the dose-limiting toxicity of many chemotherapy agents (Sinha et al., 2006). The use of nanoparticles in tandem with currently available medications minimizes potential drug toxicity or deleterious effects through targeted drug delivery systems that ultimately increase drug bioavailability at specific sites or tissues. Furthermore, administering medications via nanoparticles does not require dilution of the compounds. Previous problems arose when following dilution: medications lost their effectiveness due to virus-compound dissociation, allowing the virus to regenerate. However, nanoparticles have the potential to inflict permanent damage to the virus (Weiss et al., 2020a; Cavalcanti and Nogueira, 2020).
Pivoting to the overall benefits of nanoparticle-based COVID-19 therapeutics, the biggest draw of such treatments is their ability to effectively deliver medications while limiting the risk of side effects by serving as drug carriers. There are two major strategies to achieve this, the first being their ability to lower the overall dose of a drug through refining the region of administration (i.e., targeting more specific regions of infected tissues). Such a tactic reduces the chance of harmful drug agents aimed at diseased cells from contacting healthy ones (Xiao et al., 2022). The second method focuses specifically on the delivery of hydrophobic medications, which often require harmful solubilizing agents. One study looking at celastrol, a compound demonstrating anticancer activity, found a way to bypass the use of toxic solubilizing agents using celastrol liposomes. The liposomal envelopment of the hydrophobic drug solved the problem of low water solubility by embedding within the lipid bilayer, while hydrophilic drugs can exist within the aqueous core of the particle (Wolfram et al., 2014) Such success could by expanded for COVID-19 therapeutics.
Limited research exists detailing the efficacy of nanotechnological therapies against emerging COVID-19 variants of concern such as Omicron, Delta, and BA.5. Antivirals such as Remdesivir (G7S-5734) are a prominent treatment choice following inhibition of SARS-CoV-2 and MERS-CoV replication in various in-vitro environments (Sheahan et al., 2017). Similarly, chloroquine and hydroxychloroquine, two antimalarials that rose exponentially in popularity at the start of the pandemic, remain an option due to their ability to prevent pneumonia exacerbation in acute COVID-19 infections, though cardiotoxicity associated with the compounds is a concern (Alhajj and Gencer, 2020). Though chloroquine was not produced specifically to treat SARS-CoV-2, a standard dose of the drug impedes viral replication (limiting COVID-19 entry into cells) and supports the immune system through activating cellular anti-inflammatory responses.
In contrast to the previously stated antivirals, Paxlovid has been a standout COVID-19 therapy for treating mild to moderate cases since gaining Emergency Use Authorization (EUA) by the United States Food and Drug Administration (USFDA) in December 2021 (Najjar-Debbiny et al., 2022). Produced by Pfizer, the drug is a combination of nirmatrelvir (a SARS-CoV-2 protease inhibitor) and ritonavir (an HIV protease inhibitor that impedes cytochrome P450 and P-glycoprotein, thus increasing the half-life of nirmatrelvir) (Islam et al., 2022; Najjar-Debbiny et al., 2022). Researchers found that when given Paxlovid within the first 5 days of SARS-CoV-2 infection, progression to acute COVID-19 and mortality decreases significantly. More importantly, the findings from Israel during the height of Omicron wave demonstrated Paxlovid has potential to treat COVID-19 subvariants (Najjar-Debbiny et al., 2022). Though Paxlovid itself is not considered a nanotechnology, it is worth noting as it was produced specifically for COVID-19 and could hold further potential as a broad-spectrum antiviral. Furthermore, it has a thorough safety profile and is easier to produce than more complex nanotechnologies, allowing for a long-term and cost-effective method to managing COVID-19 in everyday life (Wen et al., 2022). It could also be combined with other nanotechnologies to provide controlled release to treat COVID-19, reducing dose frequency and limiting the potential for over usage.
Though nanotechnology improves the efficacy (and mitigates adverse side effects) of antiviral treatments, further research and funding is needed to make them a viable option for those suffering from COVID-19. Presently available therapeutic nanomedicines are not specific to SARS-CoV-2, with most research drawing on their success in cancer, Ebola, or HIV treatments. Furthermore, a cost-effective production strategy has yet to be proposed, especially one on a large enough scale for a worldwide population (as most studies using such technologies are pre-clinical and do not require an abundance of materials) (Alhajj and Gencer, 2020).
Studies focusing on employing nanotechnology to develop COVID-19 therapeutics have found some promising evidence supporting their role in eradicating SARS-CoV-2. One of the most common strategies implemented for SARS-CoV-2 therapeutics is blocking the interaction between the viral spike protein and ACE-2 receptor. Furthermore, nanosponges and nanodecoys have been shown to effectively block the viral entry into the cells by blocking this interaction (Caldera-Crespo et al., 2022). Similarly, researchers have also employed ways to neutralize viral proteins and enzymes along with dampening the host cytokine responses to prevent tissue damage. Utilizing nanobodies, gene delivery systems involving nanocapsules, metal nanoparticles, inorganic and organic nanoparticles, and quantum dots are some of the interesting applications of nanoparticles in neutralizing viral enzymes and reducing cytokine storms in the host to reduce inflammation. Although these therapeutic approaches would have some side effects, their potential in combating SARS-CoV-2 cannot be neglected.
Therapeutic approaches against COVID-19 requires blocking the interaction between the viral spike protein with the angiotensin-converting enzyme (ACE2) receptor of the cells to prevent the virus from entering host cells (Weiss et al., 2020b). Zhang et al. developed novel cellular nanosponges by coating human cell-derived membranes onto poly (lactic-co-glycolic acid) (PLGA) nanoparticle cores to divert SARS-CoV-2 from entering their usual cell targets as illustrated in Figure 5 (Zhang et al., 2020). Nanosponges express similar receptors to the target cells allowing the virus to interact with CD147 and ACE2 receptors to enter the cells. The ACE2 receptor is highly expressed in Vero E6 cells isolated from the kidney cells of African green monkey making this cell line ideal for use as an infection model for COVID-19. The expression of host serine protease TMPRSS2 is then induced in this cell line as it is responsible in cleaving viral glycoprotein to activate the virus (Mollica et al., 2020). Experiments conducted in this in vitro study demonstrated that the entry of live SARS-CoV-2 virus into Vero E6 cells was neutralized by human lung epithelial type II cell nanosponges and human macrophage nanosponges in a concentration dependent manner. Similarly, PLGA nanoparticles have been investigated for their role in drug delivery to administer therapeutics against SARS-CoV-2. An in vitro kinetic study done by Burcu et al. focused on controlled release of Oseltamivir phosphate (OP), an antiviral model drug, when loaded onto PLGA NPs conjugated to spike-binding peptide 1(SBP1) of SARS-CoV-2 (Ucar et al., 2021). The goal was to prevent the entry of the virus by using SBP1 conjugated NPs as SBP1 peptide interacts with the receptor binding domain (RBD) of SARS-CoV-2. Thus, this approach of controlled drug release prevents COVID-19 infection. The study reported steady release rates of 53.7% and 50.4% after 72 days for OP loaded NPs and OP loaded NPs conjugated to SB1 indicating the potential of this novel controlled drug delivery system to tackle COVID-19.
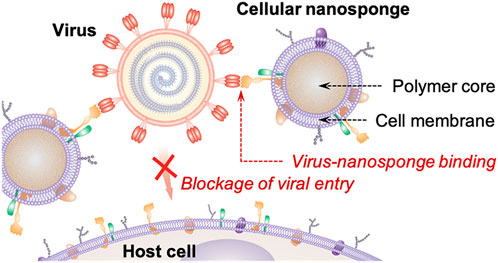
FIGURE 5. Depiction of nanosponges mimicking the target cell (host cell) membranes consisting of similar receptors (CD147 and ACE2). This inhibits the interaction of the virus with the host cell resulting in the blockage of viral entry into the host cell. This image is adapted from (Zhang et al., 2020) under the ACS Author’s Choice usage agreement. This is an unofficial adaptation of an image that appeared in an ACS publication. ACS has not endorsed the content of this adaptation or the context of its use.
The need for COVID-19 therapeutics has led to numerous studies exploring the potential of nanovesicles, nanocapsules, and nanoparticles as drug delivery systems. Immune response induced by SARS-CoV-2 leads to the production of reactive oxygen species (ROS), cytokines, and proteases in COVID-19 patients leading to hyperinflammation (Qin et al., 2020). It is imperative to treat the overproduction of ROS as it could otherwise lead to an increased viral replication advancing the cells to apoptotic stage of COVID-19 infection. Qin et al. designed nanocapsules made of 2-methacryloyloxyethyl phosphorylcholine (MPC), N-(3-aminopropyl) methacrylamide hydrochloride (APM), and N,N′-methylenebisacrylamide (BIS) monomers (called n (CAT)) that encapsulates catalase enzyme to ubiquitylate ROS (H2O2). Utilizing nanocapsules to encapsulate the enzyme led to a 16.8-fold increase in the half-life of the encapsulated enzyme in vivo. The results indicate 100% cell viability when 1,000 × 106 M H2O2 was added to human pulmonary alveolar epithelial cells (HPAEpiC) with n (CAT) when compared to 63% viability of control cells. Notably, eliminating H2O2 dampened the downstream ROS production in vivo. This led to the repression of SARS-CoV-2 replication in rhesus macaques further elucidating n (CAT)’s role in protecting tissues from oxidative injury, reinvigorating injured cells, and immunoregulation.
Another strategy employed to target COVID-19 uses decoy nanoparticles or nanodecoys that are designed to mimic cellular targets to inhibit virions. Taking advantage of ACE2 expressing lung spheroid cells (LSC), Zhenhua et al. designed LSC-membrane nanovesicles to neutralize intranasally delivered SARS-CoV-2 in cynomolgus macaques (Li et al., 2021). The study reported that the presence of sub genomic RNA (sgRNA) in bronchoalveolar lavage decreased from 6.243 log10 RNA copies ml−1 in control animals to <1.70 log10 in nanodecoy treatment groups. Similarly, immunohistochemistry (IHC) data proved a decrease in SARS nucleocapsid protein (SARS-N) in LSC-nanodecoy treatment groups in lung tissues along with a reduction in viral replication as evident from the RNAscope data, suggesting the potential of this technology in reducing the effects of COVID-19.
New COVID-19 VOCs provide challenges when designing therapeutics employing decoy strategies. Gunnels et al. identified a Beta mutant consisting of three mutations in its receptor binding domain making it resistant to FDA approved mAbs (K417N, E484K, N501Y) that inhibit its interaction with the host ACE2 (Gunnels et al., 2022). These mutations increase affinity of the spike protein for the host ACE2 by three folds. Another variant tested in the study was F486S that contains point mutation in the spike receptor binding domain decreasing its affinity for ACE2 receptor. They utilized dose response curves to analyze if the decoy extracellular vesicles containing ACE2 (obtained via ultracentrifugation (UC)) would inhibit Spike-lenti variants. The results indicated inhibition of Spike-lenti variants (Beta and F486S) via decoy UC-EVs (including design variations) that have evolved to be resistant against mAbs. These results remained the same when UC-EVs were used to inhibit Delta (L452R, T478K), Delta-plus (K417N, L452R, T478K), and Lambda (L452Q, F490S) variants.
A recent Cryo-EM study has discovered an epitope on the spike protein that is conserved in major variants of SARS-CoV-2 along with an antibody fragment VH Ab6 that can neutralize the variants by interacting with the epitope (Mannar et al., 2022). Though more sensitive to some than others, VH Ab6 confers tolerance to variants including Alpha, Beta, Gamma, Delta, Kappa, Epilson, and Omicron due to its biochemical properties, making this fragment tolerant to RBD mutations. This study opens doors for soluble mAb therapies amongst others. Interestingly, it could potentially be combined with the nanodecoy strategy mentioned above to generate VH Ab6 coated nanoparticles to neutralize SARS-CoV-2. Even though evidence does not currently exist for this particular antibody-nanodecoy strategy, there are numerous studies on developing antibody conjugated nanoparticles to treat various diseases (Juan et al., 2020). Another strategy that has become popular recently is utilizing nanobodies to neutralize the spike protein of SARS-CoV-2. Nanobodies are obtained from camelids and sharks and only consist of one variable heavy chain domain (VHH) (Mahmud et al., 2022). In contrast to human IgGs, their small size of 12–15 kDa not only allows greater permeability, but also allows for a greater interaction efficiency within the protein grooves. A recent study by Fu et al. constructed humanized nanobody library and one of the nanobodies RBD-1-2G neutralized a psuedotyped particle mimicking N501Y mutation as seen in the receptor binding domain of SARS-CoV-2 variant B.1.1.7 (Fu et al., 2022). The study utilized these nanobodies to specifically target the spike-ACE2 interactions and hence, preventviral entry.
Research studies have further emphasized the role of nanoparticles as carriers of gene delivery therapies to treat COVID-19. Gene delivery therapeutics ultimately focus on ceasing disease progression. Some of the nucleic-acid based therapeutic strategies include silencing the viral proteins post transcriptionally using RNA interfering therapies, inhibiting protein function via aptamers, and inhibiting viral replication using DNAzymes (Piyush et al., 2020). Since the genome of SARS-CoV-2 is encoded by RNA, short double stranded RNA molecules called small interfering RNA (siRNA) can be utilized to knock out the virus by silencing the genes transcriptionally or post transcriptionally (Idris et al., 2021). Idris et al. have developed a stealth lipid nanoparticle (sLNP) delivery system to screen three siRNA candidates (siUTR3, siUC7, and siHel1) that target conserved regions of SARS-CoV-2 in vivo. Their novel formulation involved a reduced use of cationic lipid particles DOTAP (∼40%) to reduce toxicity and incorporating ionizable lipid MC3 which helps in the release of siRNA. The DiD labelled DOTAP/MC3 LNP-siRNAs (∼80 nm) were found to localise in lungs, liver and spleen and outgrowth analysis from lungs after 7–8 days post infection suggested repression of SARS-CoV-2 in vivo. Such novel methods re-emphasize the immense potential of nanoparticles in COVID-19 therapeutics.
Investigations exploring the role of metal and theranostic nanoparticles, and quantum dots have reported promising findings that bring nanoparticles one step closer to clinical trials for COVID-19 treatment. Gold nanoparticles are of interest due to their biocompatibility and poor immunogenicity. Labouta et al. (2021) proposed a novel plasmonic photothermal therapy (PPT) based on theoretical discrete dipole approximation that utilizes ACE-2 functionalized gold nanorods (AuNRs) to target the virus which is followed by irradiation with near-infrared (NIR) light via flexible bronchoscopy. The study provides mathematical evidence to prove that the tip-to-tip configuration of AuNR absorbs NIR leading to the formation of strong field hotspots leading to localized heat generation to deform the virus bound to AuNRs. In addition, Auranofin is a drug that was approved for rheumatoid arthritis and it is a gold-containing triethyl phosphine (Iraci et al., 2022). A study conducted using human Huh7 cells to investigate its inhibitory impact on SARS-CoV-2 demonstrated inhibition at very low concentrations along with a reduction in cytokine production. Auranofin induces apoptosis by inhibiting essential redox enzymes leading to oxidative stress (Gil-Moles et al., 2020). Copper nanoparticles are of interest to researchers to combat SARS-CoV-2 due to its antiviral properties. Cu2+ ions are effective in producing ROS which are harmful to the viral genome as they deactivate viral enzymes. Almaki et al. utilized Co. (II) and Cu (II) complexes to produce a thiazole derivative that interacts with 6lu7 and 7bz5 proteins of SARS-CoV-2 that could be used as an antiviral treatment (Almalki et al., 2021).
Another molecular docking study on iron oxide nanoparticles (FDA approved for the treatment of anemia) found that Fe2O4 formed a stable complex with S1 subunit of receptor binding domain of the SARS-CoV-2 due to the formation of 4 hydrogen bonds (binding free energy of −10.66 kcal/mol) (Abo-zeid et al., 2020). The higher affinity of SARS-CoV-2 for IONPs inhibits its interaction with ACE2 host receptors emphasizing the potential of IONPs as antiviral therapeutic agents against this virus. Figure 6 taken from (Chenthamara et al., 2019) provides a broader perspective on the different roles of iron oxide nanoparticles for biomedical applications. Potential toxic effects of IONPs have been investigated and further research needs to be performed before its consideration as an effective therapeutic against viral pathogens like SARS-CoV-2 and as drug delivery agents (Martins et al., 2021). In 2011, an experiment was performed where the toxic effects of IONPs were tested for both in vivo (Wistar rats) and in vitro environments (Szalay et al., 2012). The experiments provided mixed results where general and organ (lung) toxicity was found in vivo, and only moderate toxicity levels were detected in vitro (Szalay et al., 2012).
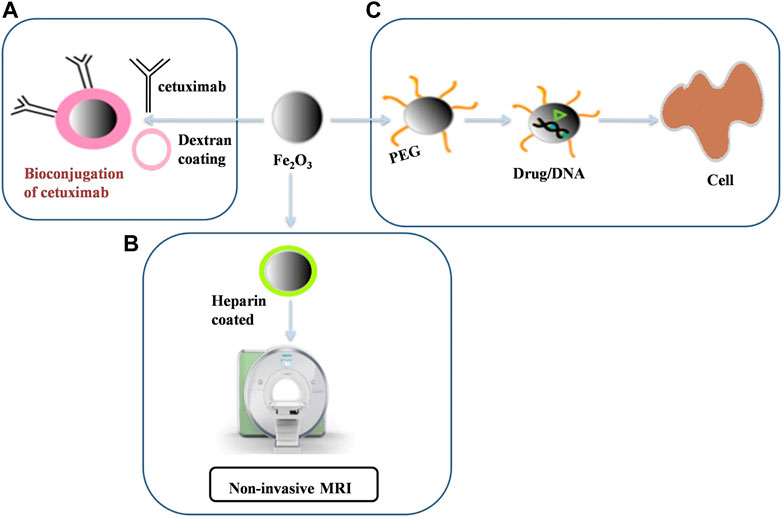
FIGURE 6. Summary of the use of iron oxide nanoparticles in (A) Conjugation of drugs like cetuximab via a PEG linker (B) Non-invasive MRIs via Heparin coated iron oxide nanoparticles and (C) Coating them with polymers like PEG for drug delivery purposes. This image was adapted from (Chenthamara et al., 2019). This image is reprinted under the terms of the Creative Commons Attribution 4.0. International License).
Like iron, zinc oxide nanoparticles have also shown potential as antiviral therapeutics against for COVID-19. Zinc is an FDA-approved substance (Hamdi et al., 2021). A recent study combining zinc oxide nanoparticles coated in triptycene organic molecules (TRP) and ellagic acid (ELG) illustrated highly effective antiviral potential for deactivating human influenza and human coronavirus (Abouaitah et al., 2021). Aside from this, there is evidence that zinc oxide nanoparticles are useful as drug delivery systems. Perera et al. (2022) study investigating the efficacy of albumin-grafted, polycaprolactone-coated, zinc oxide-loaded cloxacillin (APCL-CLOX-ZnO) nanoparticles for drug delivery revealed that 70% of the particles were able to reach the alveoli level during the in vitro lung deposition. Because SARS-CoV-2 is known for targeting respiratory organs, similar formulations may be beneficial for delivering therapeutic drugs to patients. Another study analyzing the drug releasing properties of zinc oxide nanoparticles when loaded with amoxicillin found that they could generate controlled delivery of the amoxicillin (Palanikumar et al., 2013). This study showcases zinc oxide nanoparticles and its ability to enhance the potency of treatment through targeted delivery of drugs to specific organ systems in addition to being more cost effective. Current research has yet to thoroughly elucidate the effects of zinc oxide nanoparticles on specific SARS-CoV-2 variants.
In terms of its toxicological properties, the hemolytic effects of zinc oxide nanoparticles are known to increase with higher doses. However, only 5% of hemolysis was determined in the tested concentrations according to a 2021 study (Hamdi et al., 2021). When analyzing the cellular uptake of zinc oxide nanoparticles, major reduction of cell viability was illustrated at a concentration close to 100 μg/ml of zinc oxide nanoparticles (Hamdi et al., 2021). While these studies may indicate a low toxic profile for zinc oxide nanoparticles with specific doses, there are other toxicological aspects to consider. For example, the process of preparing zinc oxide may release particles in the air that could be inhaled by those around the site. Inhalation of zinc oxide can induce a condition known as metal fume fever which carries symptoms such as coughing and fatigue (Liu et al., 2016). There is speculation that there may be similarities between metal fume fever and effects of inhaling zinc oxide nanoparticles, therefore, proper protection should be used when working with these inorganic nanomaterials (Liu et al., 2016).
Recently, the role of quantum dots (semiconductive nanoscale particles) in COVID-19 therapeutics have caught the attention of researchers. Rocha et al. proposed the use of hybrid nanoceria (CeO2) quantum dots composed of nanoceria and polymeric matrix of microcrystalline cellulose (MCC) (Rocha et al., 2022). Antioxidant properties along with a unique cubic fluorite structure with a vacant oxygen site (allowing redox cycling) makes nanoceria a nanoparticle of interest that possesses the ability to neutralize ROS. Furthermore, this nanoparticle has been known to inhibit TGF-β and NFkB signalling pathways which are both a part of COVID-19 pathogenesis as induced by the release of cytokines like IL-6, IL-8, TNF-α, and TGF- β. This group has performed spectroscopic and microscopic analysis to produce these hybrid quantum dots that could be adapted for clinical applications such as aerosol delivery to neutralize ROS in COVID-19 patients. Another interesting application of quantum dots is modifying carbon quantum dots (CQD) with boronic acid via a hydrothermal process (Loczechin et al., 2019; Xue et al., 2022)). Boronic acid inactivates the variant of human coronavirus (HCoV-229E) at an effective inhibition concentration of 52 ±8 ug/ml by interacting with the glycan units on the virus surface. Hence, modifying quantum dots increases their antiviral activity against the virus. Although metal nanoparticles provide several interesting ways to tackle SARS-CoV-2, their toxicity and appropriate dosage needs to be evaluated if these novel approaches reach clinical trials. Besides that, the other challenge associated with metal nanoparticles is keeping their formulation the same by ensuring their thermal and chemical properties remains the same. For instance, when delivering the NPs, they are exposed to external environment which might lead to chemical damage (Kumar et al., 2022) To address the potential toxic impacts of several nanoparticles, recent studies have emerged on organic nanoparticles. Several properties of organic nanoparticles such as non-toxicity, site-specificity, and biocompatibility make them attractive for COVID-19 therapeutics (Kerry et al., 2019). Lee et al. (2021) proposed a DNase-1 coated polydopamine-poly (ethylene glycol) or polydopamine-PEG to counteract tissue damage caused by cytokine storm or neutrophil activities (production of neutrophil extracellular trap or NETs) in patients infected with SARS-CoV-2. The in vivo studies performed also showed inhibition of NF- κB and cytokines.
Vaccines are typically expected to reach the market in around 5–18 years and production of these solutions for distribution or public use addsadditional months to years. During the pre-clinical stages of vaccine development, the primary objective is to assess the safety and immunogenicity in cell culture or animal disease models to determine a starting dose (Plotkin et al., 2017). Although the development of mRNA vaccines to address COVID-19 were completed in record-breaking timelines compared to traditional vaccine developments (Ball, 2021), there is still room for accelerating processes to get candidates to successfully enter phase I vaccine trials. One way to accelerate vaccine development in the context of SARS-CoV-2 and its VOCs is to design effective disease models. In doing so, potential nanotechnology-based vaccines and therapeutics can be screened in a cost-effective manner prompting for reduction in animal use, as well as customizability to screen on different patient-specific models including those in vulnerable populations like the pregnant and the elderly.
While two-dimensional (2D) tissue models and animal models allow for a characterization in cell phenotype, physiology, and function, the 2D environment does not replicate the entire complexity of interactions or behaviour of cells in vivo. 3D models can potentially mimic the natural extracellular matrix (ECM) of a specific cell type prompting for better understanding of spatial limitations, mechanical properties, and delivery of essential factors required for growth (Duval et al., 2017). Since the onset of SARS-CoV-2, research groups have developed three-dimensional (3D) models from organoids to 3D bioprinted constructs to model the effect of SARS-CoV-2 infection in several tissue types. The most common tissues utilized to investigate SARS-CoV-2 infection contain cells from the respiratory system (lung models), cardiovascular system (heart models), and nervous system (brain models). In this section, we outline current disease models of SARS-CoV-2 and highlight their potential to screen nanomaterial-based vaccines and therapeutics.
Research on 3D organoid models for SARS-CoV-2 infection often focuses on human alveolar cell types or the respiratory epithelium cell layer to detail the extent of viral infection, progression of infection, and potential immune responses. Youk et al., 2020 illustrates phenotypic changes alongside increased expression of interferon and proinflammatory genes that occur following SARS-CoV-2 infection in human lung alveolar type 2 cells self-organized in a 3D fashion (Youk et al., 2020). Similarly, Lamers et al. (2021) organoid-derived bronchioalveolar model comprising of alveolar, basal, and rare neuroendrocrine cells grown in 3D culture reveal consistent interferon responses against SARS-CoV-2 infection. Application of these 3D lung epithelium models have been utilized to study COVID-19 variants. In fact, experiments on respiratory epithelium organoids alongside designed 2D airway organoid, conducted by Chiu et al., 2022, have successfully modelled transmissibility and infectivity of the SARS-CoV-2 Omicron variant in vitro (Chiu et al., 2022). These respiratory models have also been utilized for drug screening potential therapeutics. For example, Wu et al. (2022) paper utilized type 2 alveolar epithelial cells (AT2) cultured in 3D spheres as a distal lung model to screen the ability of protease inhibitor Camostat Mesylate in blocking viral entry of SARS-CoV-2. Other respiratory models comprise of 3D synthetic hydrogel-based culture systems as an infection platform to test nanoparticle decoys mechanisms against SARS-CoV-2 (Pennarossa et al., 2021). Such models provide insight into the biology of COVID-19 and can serve a tool for drug screening.
Given evidence of SARS-CoV-2 inducing cytotoxic effects on human cardiomyocytes (Bojkova et al., 2020), and the observed rare cases of myocarditis (inflammation of the heart muscle or myocardium) or pericarditis (swelling of the pericardium) after COVID-19 vaccination (Diaz et al., 2021) researchers have worked toward modelling the effects of SARS-CoV-2 in heart tissues to elucidate the mechanisms that lead to the inflammation. One study conducted by Bailey et al., 2021 infected SARS-CoV-2 into engineered heart tissue models used human myocardial cells to highlight the correlation between cardiomyocyte infection and COVID-19 myocarditis as supported by evidence of innate immune response activation via RNA sequencing (Bailey et al., 2021). This study also serves as evidence of decreased cardiomyocyte contractility, increased sarcomere breakdown and cardiomyocyte apoptosis resulting from SARS-CoV-2 infections. Anti-inflammatory therapeutics specific to COVID-19 have yet to be tested on these platforms.
For studies that investigate the mechanisms of nervous system infection or neurotropism of SARS-CoV-2, 3D organoid models and 3D bioprinted models have been introduced. Experiments conducted by Yi et al., 2020 elucidates the infectious properties of spike protein-pseudotyped SARS-CoV-2 on ACE2 receptor expressing cortical neurons within a 3D forebrain organoid model to quantify the virus’s multiplicity of infection (MOI). As a result, the studies imply that the degree of neural infectivity is independent of viral load further validating the use of their 3D organoid models (Yi et al., 2020). Researchers have applied 3D bioprinting techniques in combination with tunable scaffold based, cell-laden, bioinks to produce 3D bioprinted neural tissue models to model neurotropism to mimic the in vivo extracellular matrix of neural tissues for enhanced infectious modelling of SARS-CoV-2.3D Bioprinting allows for precise placement of cells alongside components of its extracellular matrix to support the cells leading to better modelling of cellular behaviours in vitro. A key example of this development is exemplified by researchers de Melo et al., 2022, where they infected a 3D bioprinted neural-tissue like model consisting of murine astrocytes in a gelatin, GelMa, and fibrinogen-based bioink with mouse-adapted SARS-CoV-2. In observing their 3D bioprinted model, this paper validates the potential of 3D bioprinting techniques for disease modelling of infectious diseases especially in tissue areas that are challenging to study (de Melo et al., 2022).
In conclusion, several respiratory, cardiovascular, and nervous system disease models have been thoroughly made to SARS-CoV-2 infection yet there is a limited number of studies that have applied mRNA-LNP vaccine technologies and other nanotechnology-based therapeutics to these models. Although these models are preliminary in that they do not entirely demonstrate the adaptive immunity mechanisms and some generally utilize a single cell type as opposed to cocultures of various cell types, innate immunity mechanisms would be helpful in further characterizing nanotechnology-based behaviours within the human body.
Ultimately, the use of nanomaterials shows great promise for addressing COVID-19 through future advancements in vaccines and therapeutics. However, further research must be made to these treatments to fully optimize their potentials in healthcare. As previously stated, evidence illustrates that mRNA vaccines using lipid nanoparticles can be highly advantageous compared to conventional formulations as they are cost-effective, can be rapidly produced, and carry self-adjuvanting properties. mRNA is also flexible for sequence modifications which aids in addressing variants. Despite these benefits, there is still speculation on the longevity and physiological effects of LNPs. A previously mentioned study illustrated a substantial decrease in immunity 6 months after two doses of the Pfizer vaccine. This may be linked to mRNA impurities caused by the ionized lipids disrupting mRNA translation. This is an indicator that the implementation of annual boosters may be necessary for maintaining immunity against the virus and new variants. Given the fast rate of production, flexible sequences, and cost-effectiveness of these vaccines, they may be good candidates for large scale, annual production. In addition, adverse effects of mRNA vaccines such as fever, fatigue, and hypersensitivity may be triggered using LNPs. To illustrate, research found that LNPs caused intense inflammation in mice which may cause symptoms and that the PEGylated (polyethylene-glycol) lipid component can stimulate immune responses leading to hypersensitivity.
In terms of therapeutics, the utilization of nanoparticles alongside currently available treatments has shown promise in minimizing drug toxicity and bioavailability through drug delivery systems. There are also nanomaterials that have merit in their antiviral properties. In a study analyzing monoclonal antibody (mAb) production using modified mRNA encapsulated in LNPs (RBD mRNA-LNPs), found the mAbs produced by immunizing mice with RBD mRNA-LNPs had potent viral neutralizing abilities against the Alpha and Delta variants (Hsu et al., 2022). Unfortunately, there is very limited research detailing the efficacy of nanotechnological therapies against COVID-19 variants of concern and ways of addressing potential toxicological effects of these particles. For example, a previously stated study indicated that zinc oxide nanoparticles may work as an antiviral therapeutic against COVID-19, but for implementation into medical practice, potential toxic properties of the particles such as hemolysis occurring upon certain doses must be further studied and addressed.
Considering the known benefits and obstacles of nanoparticles in COVID-19 treatments, the medical advancements and knowledge gained through the pandemic can be significantly helpful in improving upon current medicines and aid in production of treatments against other viruses. For example, a study using chimeric spike mRNA vaccines in mice demonstrated that the vaccines were able to neutralize various SARS-like zoonotic corona viruses (Martinez et al., 2021). Considering how mRNA vaccines are currently being used for preventing COVID-19, this work proves that technology used for combating COVID-19 may be used and adjusted for other viruses. With this in mind, we can foresee future advancements in medicine utilizing nanotechnological strategies devised during the pandemic to aid in preventing and treating other illnesses.
SW conceptualized contents of the paper, gathered references, and guided the group in synthesizing the paper at each stage, including revisions of the manuscript. MVH created an outline for the paper based on SW’s guidance, wrote Section 4 entirely and parts of Section 2 along with gathering references, edited drafts, organized references, formatted final manuscript/figures. MVH organized references, wrote the introduction, conclusion, and Section 4.2; assisted with edits. NK gathered/organized references for their written contributions; Sections 2.1–2.3. JWS gathered/organized references for their written contributions; Section 2 introduction, Sections 2.2–2.3. SK gathered/organized references, wrote Section 3.2 and acknowledgments section, completed edits for their section. CV gathered/organized references, wrote Section 3.1, completed edits for their section. DP for gathered references and written portions of Section 2.1.
SW would like to acknowledge funding from the Natural Science and Engineering Research Council, the Canadian Institutes of Health Research, the New Frontiers Transformational Fund, and the Canadian Research Chairs program.
The authors would like to extend their gratitude to SW and the Willerth Lab members for all their support.
SW is the C.E.O. of Axolotl Biosciences a start-up focused on selling novel bioinks for bioprinting human tissue models.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abhyankar, M. M., Mann, B. J., Sturek, J. M., Brovero, S., Moreau, G. B., Sengar, A., et al. (2021). Development of COVID-19 vaccine using a dual Toll-like receptor ligand liposome adjuvant. npj Vaccines 6 (1), 137. doi:10.1038/s41541-021-00399-0
Abo-zeid, Y., Ismail, N. S. M., McLean, G. R., and Hamdy, N. M. (2020). A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur. J. Pharm. Sci. 153. 105465, ARTN 105465 doi:10.1016/j.ejps.2020.105465
Abouaitah, K., Allayh, A. K., Wojnarowicz, J., Shaker, Y. M., Swiderska-Sroda, A., and Lojkowski, W. (2021). Nanoformulation composed of ellagic acid and functionalized zinc oxide nanoparticles inactivates DNA and RNA viruses. Pharmaceutics 13 (12), 2174. doi:10.3390/pharmaceutics13122174
Akinc, A., Querbes, W., De, S., Qin, J., Frank-Kamenetsky, M., Jayaprakash, K. N., et al. (2010). Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther., 18, 1357–1364. doi:10.1038/mt.2010.85
Alameh, M.-G., Tombácz, I., Bettini, E., Lederer, K., Ndeupen, S., Sittplangkoon, C., et al. (2021). Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54(12), 2877–2892.e7.e2877. doi:10.1016/j.immuni.2021.11.001
Albertsen, C. H., Kulkarni, J. A., Witzigmann, D., Lind, M., Petersson, K., and Simonsen, J. B. (2022). The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 188, 114416–114416. doi:10.1016/j.addr.2022.114416
Alhajj, S., and Gencer, S. “Investigating side effects of existing drugs used in covid-19 treatment,” in Proceedings of the IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM), NEW YORK, April 2020 (IEEE), 803–810.
Almalki, S. A., Bawazeer, T. M., Asghar, B., Alharbi, A., Aljohani, M. M., Khalifa, M. E., et al. (2021). Synthesis and characterization of new thiazole-based Co(II) and Cu(II) complexes; therapeutic function of thiazole towards COVID-19 in comparing to current antivirals in treatment protocol. J. Mol. Struct. 1244, 130961. doi:10.1016/j.molstruc.2021.130961
Alvarez-Benedicto, E., Farbiak, L., Ramirez, M. M., Wang, X., Johnson, L. T., Mian, O., et al. (2022). Optimization of phospholipid chemistry for improved lipid nanoparticle (LNP) delivery of messenger RNA (mRNA). Biomater. Sci. 10 (2), 549–559. doi:10.1039/d1bm01454d
Anand, P., and Stahel, V. P. (2021). The safety of covid-19 mRNA vaccines: A review. Patient Saf. Surg. 15, 20(1). ARTN 20 doi:10.1186/s13037-021-00291-9
Andrews, N., Stowe, J., Kirsebom, F., Toffa, S., Rickeard, T., Gallagher, E., et al. (2022). Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. Overseas. Ed. 386 (16), 1532–1546. doi:10.1056/nejmoa2119451
Ariën, K. K., Heyndrickx, L., Michiels, J., Vereecken, K., Van Lent, K., Coppens, S., et al. (2022). Three doses of BNT162b2 vaccine confer neutralising antibody capacity against the SARS-CoV-2 Omicron variant. npj Vaccines 7 (1), 35. doi:10.1038/s41541-022-00459-z
Baden, L. R., El Sahly, H. M., Essink, B., Kotloff, K., Frey, S., Novak, R., et al. (2021). Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 384 (5), 403–416. doi:10.1056/nejmoa2035389
Ball, P. (2021). The lightning-fast quest for COVID vaccines - and what it means for other diseases. Nature 589 (7840), 16–18. doi:10.1038/d41586-020-03626-1
Belete, T. M. (2020). A review on Promising vaccine development progress for COVID-19 disease. Vacunas 21 (2), 121–128. doi:10.1016/j.vacun.2020.05.002
Bigini, P., Gobbi, M., Bonati, M., Clavenna, A., Zucchetti, M., Garattini, S., et al. (2021). The role and impact of polyethylene glycol on anaphylactic reactions to COVID-19 nano-vaccines. Nat. Nanotechnol. 16 (11), 1169–1171. doi:10.1038/s41565-021-01001-3
Bonanni, P., and Santos, J. I. (2011). Vaccine evolution. Perspect. Vaccinol. 1, 1–24. doi:10.1016/j.pervac.2011.05.001
Bullock, T. N. J. (2021). Fundamentals of cancer immunology and their application to cancer vaccines. Clin. Transl. Sci. 14 (1), 120–131. doi:10.1111/cts.12856
Buschmann, M. D., Carrasco, M. J., Alishetty, S., Paige, M., Alameh, M. G., and Weissman, D. (2021). Nanomaterial delivery systems for mRNA vaccines. Vaccines 9 (1), 65. doi:10.3390/vaccines9010065
Caldera-Crespo, L. A., Paidas, M. J., Roy, S., Schulman, C. I., Kenyon, N. S., Daunert, S., et al. (2022). Experimental models of COVID-19. Front. Cell. Infect. Microbiol. 11, 792584. doi:10.3389/fcimb.2021.792584
Cavalcanti, I. D. L., and Nogueira, M. (2020). Pharmaceutical nanotechnology: Which products are been designed against COVID-19? J. Nanopart. Res. 22 (9), 276. doi:10.1007/s11051-020-05010-6
Chakraborty, M., Jain, S., and Rani, V. (2011). Nanotechnology: Emerging tool for diagnostics and therapeutics. Appl. Biochem. Biotechnol. 165 (5-6), 1178–1187. doi:10.1007/s12010-011-9336-6
Chakravarty, M., and Vora, A. (2021). Nanotechnology-based antiviral therapeutics. Drug Deliv. Transl. Res. 11 (3), 748–787. doi:10.1007/s13346-020-00818-0
Chan, C.-L., Majzoub, R. N., Shirazi, R. S., Ewert, K. K., Chen, Y.-J., Liang, K. S., et al. (2012). Endosomal escape and transfection efficiency of PEGylated cationic liposome–DNA complexes prepared with an acid-labile PEG-lipid. Biomaterials 33 (19), 4928–4935. doi:10.1016/j.biomaterials.2012.03.038
Cheng, X. W., and Lee, R. J. (2016). The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 99, 129–137. doi:10.1016/j.addr.2016.01.022
Chenthamara, D., Subramaniam, S., Ramakrishnan, S. G., Krishnaswamy, S., Essa, M. M., Lin, F. H., et al. (2019). Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 23, 20(1). ARTN 20 doi:10.1186/s40824-019-0166-x
Chiu, M. C., Li, C., Liu, X. J., Yu, Y. F., Huang, J. J., Wan, Z. X., et al. (2022). A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS-CoV-2 Omicron variant. Cell Discov. 8, 57(1). ARTN 5 doi:10.1038/s41421-022-00422-1
Choi, H. M., Moon, S. Y., Yang, H. I., and Kim, K. S. (2021). Understanding viral infection mechanisms and patient symptoms for the development of COVID-19 therapeutics. Int. J. Mol. Sci. 22 (4), 1737. doi:10.3390/ijms22041737
Ciotti, M., Ciccozzi, M., Terrinoni, A., Jiang, W.-C., Wang, C.-B., and Bernardini, S. (2020). The COVID-19 pandemic. Crit. Rev. Clin. Laboratory Sci. 57 (6), 365–388. doi:10.1080/10408363.2020.1783198
Cornebise, M., Narayanan, E., Xia, Y., Acosta, E., Ci, L., Koch, H., et al. (2022). Discovery of a novel amino lipid that improves lipid nanoparticle performance through specific interactions with mRNA. Adv. Funct. Mat. 32, 2106727(8). ARTN 2106727 doi:10.1002/adfm.202106727
de Melo, B. A. G., Mundim, M. V., Lemes, R. M. R., Cruz, E. M., Ribeiro, T. N., Santiago, C. F., et al. (2022). 3D bioprinted neural-like tissue as a platform to study neurotropism of mouse-adapted SARS-CoV-2. Adv. Biol. 6 (8), 2200002. ARTN 2200002. doi:10.1002/adbi.202200002
Diaz, G. A., Parsons, G. T., Gering, S. K., Meier, A. R., Hutchinson, I. V., and Robicsek, A. (2021). Myocarditis and pericarditis after vaccination for COVID-19. Jama-Journal Am. Med. Assoc. 326 (12), 1210–1212. doi:10.1001/jama.2021.13443
Duval, K., Grover, H., Han, L. H., Mou, Y., Pegoraro, A. F., Fredberg, J., et al. (2017). Modeling physiological events in 2D vs. 3D cell culture. Physiol. (Bethesda) 32 (4), 266–277. doi:10.1152/physiol.00036.2016
Dyer, O. 2022. US and Canada to roll out the first omicron specific boosters within days. 378. [Accessed October 20, 2022].
Evers, M. J. W., Kulkarni, J. A., van der Meel, R., Cullis, P. R., Vader, P., and Schiffelers, R. M. (2018). State-of-the-Art design and rapid-mixing production techniques of lipid nanoparticles for nucleic acid delivery. Small Methods 2 (9), 1700375. doi:10.1002/smtd.201700375
Flemming, A. (2022). Are variant-specific vaccines warranted? Nat. Rev. Immunol. 22 (5), 275. doi:10.1038/s41577-022-00722-3
Forchette, L., Sebastian, W., and Liu, T. (2021). A comprehensive review of COVID-19 virology, vaccines, variants, and therapeutics. Curr. Med. Sci. 41 (6), 1037–1051. doi:10.1007/s11596-021-2395-1
Forster, P., Forster, L., Renfrew, C., and Forster, M. (2020). Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U. S. A. 117 (17), 9241–9243. doi:10.1073/pnas.2004999117
Fu, Y., Da Fonseca Rezende E Mello, J., Fleming, B. D., Renn, A., Chen, C. Z., Hu, X., et al. (2022). A humanized nanobody phage display library yields potent binders of SARS CoV-2 spike. PLOS ONE 17 (8), e0272364. doi:10.1371/journal.pone.0272364
Gagne, M., Moliva, J. I., Foulds, K. E., Andrew, S. F., Flynn, B. J., Werner, A. P., et al. (2022). mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell 185(9), 1556–1571.e18.e1518. doi:10.1016/j.cell.2022.03.038
Galanis, P., Vraka, I., Katsiroumpa, A., Siskou, O., Konstantakopoulou, O., Katsoulas, T., et al. (2022). First COVID-19 booster dose in the general population: A systematic review and meta-analysis of willingness and its predictors. Vaccines (Basel) 10 (7), 1097. doi:10.3390/vaccines10071097
Gao, J. H., and Xu, B. (2009). Corrigendum to “applications of nanomaterials inside cells” [nano today 4 (2009) 37–51]. Nano Today 4 (3), 281. doi:10.1016/j.nantod.2009.05.002
Gil-Moles, M., Basu, U., Büssing, R., Hoffmeister, H., Türck, S., Varchmin, A., et al. (2020). Gold metallodrugs to target coronavirus proteins: Inhibitory effects on the spike-ACE2 interaction and on PLpro protease activity by Auranofin and gold organometallics. Chem. Eur. J. 26 (66), 15140–15144. doi:10.1002/chem.202004112
Gunnels, T. F., Stranford, D. M., Mitrut, R. E., Kamat, N. P., and Leonard, J. N. (2022). Elucidating design principles for engineering cell-derived vesicles to inhibit SARS-CoV-2 infection. Small 18 (19), 2200125. doi:10.1002/smll.202200125
Hadj Hassine, I. (2022a). Covid-19 vaccines and variants of concern: A review. Rev. Med. Virol. 32 (4), e2313. doi:10.1002/rmv.2313
Hadj Hassine, I. (2022b). Covid-19 vaccines and variants of concern: A review. Rev. Med. Virol. 32 (4), e2313. doi:10.1002/rmv.2313
Hafez, I. M., Ansell, S., and Cullis, P. R. (2000). Tunable pH-sensitive liposomes composed of mixtures of cationic and anionic lipids. Biophysical J. 79 (3), 1438–1446. doi:10.1016/s0006-3495(00)76395-8
Hager, K. J., Pérez Marc, G., Gobeil, P., Diaz, R. S., Heizer, G., Llapur, C., et al. (2022). Efficacy and safety of a recombinant plant-based adjuvanted covid-19 vaccine. N. Engl. J. Med. Overseas. Ed. 386 (22), 2084–2096. doi:10.1056/nejmoa2201300
Hamdi, M., Abdel-Bar, H. M., Elmowafy, E., El-Khouly, A., Mansour, M., and Awad, G. A. S. (2021). Investigating the internalization and COVID-19 antiviral computational analysis of optimized nanoscale zinc oxide. ACS Omega 6 (10), 6848–6860. doi:10.1021/acsomega.0c06046
Harvey, W. T., Carabelli, A. M., Jackson, B., Gupta, R. K., Thomson, E. C., Harrison, E. M., et al. (2021). SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 19 (7), 409–424. doi:10.1038/s41579-021-00573-0
Hassett, K. J., Higgins, J., Woods, A., Levy, B., Xia, Y., Hsiao, C. J., et al. (2021). Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release 335, 237–246. doi:10.1016/j.jconrel.2021.05.021
Hassine, I. H. (2022). Covid-19 vaccines and variants of concern: A review. Rev. Med. Virol. 32, e2313(4). ARTN e2313 doi:10.1002/rmv.2313
Hause, A. M., Baggs, J., Marquez, P., Myers, T. R., Su, J. R., Blanc, P. G., et al. (2022). Safety monitoring of COVID-19 vaccine booster doses among adults - United States, september 22, 2021-february 6, 2022. MMWR. Morb. Mortal. Wkly. Rep. 71 (7), 249–254. doi:10.15585/mmwr.mm7107e1
Hsu, F.-F., Liang, K.-H., Kumari, M., Chen, W.-Y., Lin, H.-T., Cheng, C.-M., et al. (2022). An efficient approach for SARS-CoV-2 monoclonal antibody production via modified mRNA-LNP immunization. bioRxiv 2022–488878. doi:10.1101/2022.04.20.488878
Hu, Y.-B., Dammer, E. B., Ren, R.-J., and Wang, G. (2015). The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 4 (1), 18. doi:10.1186/s40035-015-0041-1
Hulla, J., Sahu, S., and Hayes, A. (2015). Nanotechnology. Hum. Exp. Toxicol. 34 (12), 1318–1321. doi:10.1177/0960327115603588
Idris, A., Davis, A., Supramaniam, A., Acharya, D., Kelly, G., Tayyar, Y., et al. (2021). A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 29 (7), 2219–2226. doi:10.1016/j.ymthe.2021.05.004
Igyártó, B. Z., Jacobsen, S., and Ndeupen, S. (2021). Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr. Opin. Virology 48, 65–72. doi:10.1016/j.coviro.2021.03.008
Iraci, N., Corsaro, C., Giofrè, S. V., Neri, G., Mezzasalma, A. M., Vacalebre, M., et al. (2022). Nanoscale technologies in the fight against COVID-19: From innovative nanomaterials to computer-aided discovery of potential antiviral plant-derived drugs. Biomolecules 12 (8), 1060. doi:10.3390/biom12081060
Islam, T., Hasan, M., Rahman, M. S., and Islam, M. R. (2022). Comparative evaluation of authorized drugs for treating Covid-19 patients. Health Sci. Rep. 5, e671(4). ARTN e671 doi:10.1002/hsr2.671
Ju, Y., Lee, W. S., Pilkington, E. H., Kelly, H. G., Li, S. Y., Selva, K. J., et al. (2022). Anti-PEG antibodies boosted in humans by SARS-CoV-2 lipid nanoparticle mRNA vaccine. Acs Nano 16, 11769–11780. doi:10.1021/acsnano.2c04543
Juan, A., Cimas, F. J., Bravo, I., Pandiella, A., Ocaña, A., and Alonso-Moreno, C. (2020). Antibody conjugation of nanoparticles as therapeutics for breast cancer treatment. Int. J. Mol. Sci. 21 (17), 6018. doi:10.3390/ijms21176018
Kandikattu, H. K., Yadavalli, C. S., Venkateshaiah, S. U., and Mishra, A. (2021). Vaccine efficacy in mutant SARS-CoV-2 variants. Int. J. Cell Biol. Physiol. 4 (1-2), 1–12.
Kawohl, W., and Nordt, C. (2020). COVID-19, unemployment, and suicide. Lancet Psychiatry 7 (5), 389–390. doi:10.1016/s2215-0366(20)30141-3
Keech, C., Albert, G., Cho, I., Robertson, A., Reed, P., Neal, S., et al. (2020). Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. Overseas. Ed. 383 (24), 2320–2332. doi:10.1056/nejmoa2026920
Kerry, R. G., Malik, S., Redda, Y. T., Sahoo, S., Patra, J. K., and Majhi, S. (2019). Nano-based approach to combat emerging viral (NIPAH virus) infection. Nanomedicine Nanotechnol. Biol. Med. 18, 196–220. doi:10.1016/j.nano.2019.03.004
Kowalzik, F., Schreiner, D., Jensen, C., Teschner, D., Gehring, S., and Zepp, F. (2021). mRNA-Based Vaccines. Vaccines 9 (4), 390. doi:10.3390/vaccines9040390
Kulkarni, J. A., Darjuan, M. M., Mercer, J. E., Chen, S., Van Der Meel, R., Thewalt, J. L., et al. (2018). On the formation and morphology of lipid nanoparticles containing ionizable cationic lipids and siRNA. ACS Nano 12 (5), 4787–4795. doi:10.1021/acsnano.8b01516
Kumar, M., Dogra, R., and Mandal, U. K. (2022). Nanomaterial-based delivery of vaccine through nasal route: Opportunities, challenges, advantages, and limitations. J. Drug Deliv. Sci. Technol. 74, 103533. doi:10.1016/j.jddst.2022.103533
Labouta, H. I., Hooshmand, N., Upreti, T., and El-Sayed, M. A. (2021). Localized plasmonic photothermal therapy as a life-saving treatment paradigm for hospitalized COVID-19 patients. Plasmonics 16 (4), 1029–1033. doi:10.1007/s11468-020-01353-x
Lamers, M. M., van der Vaart, J., Knoops, K., Riesebosch, S., Breugem, T. I., Mykytyn, A. Z., et al. (2021). An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. Embo J. 40, e105912(5). ARTN e105912 doi:10.15252/embj.2020105912
Lee, W., and Kim, S.-J. (2022). Current updates on COVID-19 vaccines and therapeutics: As of june 2022. Biotechnol. Bioprocess Eng. 13. doi:10.1007/s12257-022-0188-4
Lee, Y. Y., Park, H. H., Park, W., Kim, H., Jang, J. G., Hong, K. S., et al. (2021). Long-acting nanoparticulate DNase-1 for effective suppression of SARS-CoV-2-mediated neutrophil activities and cytokine storm. Biomaterials 267, 120389. doi:10.1016/j.biomaterials.2020.120389
Li, Y., Kröger, M., and Liu, W. K. (2014). Endocytosis of PEGylated nanoparticles accompanied by structural and free energy changes of the grafted polyethylene glycol. Biomaterials 35, 8467–8478. doi:10.1016/j.biomaterials.2014.06.032
Li, Z., Wang, Z., Dinh, P.-U. C., Zhu, D., Popowski, K. D., Lutz, H., et al. (2021). Cell-mimicking nanodecoys neutralize SARS-CoV-2 and mitigate lung injury in a non-human primate model of COVID-19. Nat. Nanotechnol. 16 (8), 942–951. doi:10.1038/s41565-021-00923-2
Liu, C., Rcheulishvili, N., Shen, Z. G., Papukashvili, D., Xie, F. F., Wang, Z. Q., et al. (2022a). Development of an LNP-encapsulated mRNA-RBD vaccine against SARS-CoV-2 and its variants. Pharmaceutics 14, 1101(5). ARTN 1101 doi:10.3390/pharmaceutics14051101
Liu, J., Feng, X., Wei, L., Chen, L., Song, B., and Shao, L. (2016). The toxicology of ion-shedding zinc oxide nanoparticles. Crit. Rev. Toxicol. 46 (4), 348–384. doi:10.3109/10408444.2015.1137864
Liu, J., Liu, Y., Xia, H., Zou, J., Weaver, S. C., Swanson, K. A., et al. (2022b). BNT162b2-elicited neutralization of delta plus, Lambda, Mu, B.1.1.519, and Theta SARS-CoV-2 variants. npj Vaccines 7 (1). doi:10.1038/s41541-022-00462-4
Loczechin, A., Seron, K., Barras, A., Giovanelli, E., Belouzard, S., Chen, Y. T., et al. (2019). Functional carbon quantum dots as medical countermeasures to human coronavirus. ACS Appl. Mat. Interfaces 11 (46), 42964–42974. doi:10.1021/acsami.9b15032
Mabrouk, M. T., Chiem, K., Rujas, E., Huang, W. C., Jahagirdar, D., Quinn, B., et al. (2021). Lyophilized, thermostable Spike or RBD immunogenic liposomes induce protective immunity against SARS-CoV-2 in mice. Sci. Adv. 7 (49), eabj1476. doi:10.1126/sciadv.abj1476
Maeki, M., Uno, S., Niwa, A., Okada, Y., and Tokeshi, M. (2022). Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Control. Release 344, 80–96. doi:10.1016/j.jconrel.2022.02.017
Mahmud, N., Anik, M. I., Hossain, M. K., Khan, M. I., Uddin, S., Ashrafuzzaman, M., et al. (2022). Advances in nanomaterial-based platforms to combat COVID-19: Diagnostics, preventions, therapeutics, and vaccine developments. ACS Appl. Bio Mat. 5 (6), 2431–2460. doi:10.1021/acsabm.2c00123
Mannar, D., Saville, J. W., Sun, Z. H., Zhu, X., Marti, M. M., Srivastava, S. S., et al. (2022). SARS-CoV-2 variants of concern: Spike protein mutational analysis and epitope for broad neutralization. Nat. Commun. 13 (1), 4696. doi:10.1038/s41467-022-32262-8
Martinez, D. R., Schafer, A., Leist, S. R., De la Cruz, G., West, A., Atochina-Vasserman, E. N., et al. (2021). Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science 373 (6558), 991–998. doi:10.1126/science.abi4506
Martins, E. S., Espindola, A., Britos, T. N., Chagas, C., Barbosa, E., Castro, C. E., et al. (2021). Potential use of DMSA-containing iron oxide nanoparticles as magnetic vehicles against the COVID-19 disease. ChemistrySelect 6 (31), 7931–7935. doi:10.1002/slct.202101900
Mathieu, E., Ritchie, H., Ortiz-Ospina, E., Roser, M., Hasell, J., Appel, C., et al. (2021). A global database of COVID-19 vaccinations. Nat. Hum. Behav. 5 (7), 947–953. doi:10.1038/s41562-021-01122-8
McLean, G., Kamil, J., Lee, B., Moore, P., Schulz, T. F., Muik, A., et al. (2022). The impact of evolving SARS-CoV-2 mutations and variants on COVID-19 vaccines. Mbio 13 (2), e0297921. doi:10.1128/mbio.02979-21
Mcneil, S. E. (2005). Nanotechnology for the biologist. J. Leukoc. Biol. 78 (3), 585–594. doi:10.1189/jlb.0205074
Mcneil, S. E. (2011). “Unique benefits of nanotechnology to drug delivery and diagnostics,” in Methods in molecular biology (China: Humana Press), 3–8.
Melamed, J. R., Hajj, K. A., Chaudhary, N., Strelkova, D., Arral, M. L., Pardi, N., et al. (2022). Lipid nanoparticle chemistry determines how nucleoside base modifications alter mRNA delivery. J. Control. Release 341, 206–214. doi:10.1016/j.jconrel.2021.11.022
Miyamura, S. (2021). Turbulence ahead: Labour and struggles in times of the covid-19 pandemic in India. Can. J. Dev. Stud./Revue Can. d'etudes. du Dev. 42 (1-2), 165–177. doi:10.1080/02255189.2021.1894415
Mollica, V., Rizzo, A., and Massari, F. (2020). The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol. 16 (27), 2029–2033. doi:10.2217/fon-2020-0571
Mondal, J., Samui, P., and Chatterjee, A. N. (2022). Dynamical demeanour of SARS-CoV-2 virus undergoing immune response mechanism in COVID-19 pandemic. Eur. Phys. J. Spec. Top. 23, 1–14. doi:10.1140/epjs/s11734-022-00437-5
Morens, D. M., Daszak, P., and Taubenberger, J. K. (2020). Escaping pandora’s box — another novel coronavirus. N. Engl. J. Med. Overseas. Ed. 382 (14), 1293–1295. doi:10.1056/nejmp2002106
Mousavizadeh, L., and Ghasemi, S. (2021). Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 54 (2), 159–163. doi:10.1016/j.jmii.2020.03.022
Mui, B. L., Tam, Y. K., Jayaraman, M., Ansell, S. M., Du, X., Tam, Y. Y., et al. (2013). Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. - Nucleic Acids 2, e139. doi:10.1038/mtna.2013.66
Muller, L., Andree, M., Moskorz, W., Drexler, I., Walotka, L., Grothmann, R., et al. (2021). Age-dependent immune response to the biontech/pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin. Infect. Dis. 73 (11), 2065–2072. doi:10.1093/cid/ciab381
Muramatsu, H., Lam, K., Bajusz, C., Laczko, D., Kariko, K., Schreiner, P., et al. (2022). Lyophilization provides long-term stability for a lipid nanoparticle-formulated , nucleoside-modified mRNA vaccine. Mol. Ther. 30 (5), 1941–1951. doi:10.1016/j.ymthe.2022.02.001
Naaber, P., Tserel, L., Kangro, K., Sepp, E., Jurjenson, V., Adamson, A., et al. (2021). Dynamics of antibody response to BNT162b2 vaccine after six months: A longitudinal prospective study. Lancet Reg. Health-Europe 10. ARTN 100208. doi:10.1016/j.lanepe.2021.100208
Najjar-Debbiny, R., Gronich, N., Weber, G., Khoury, J., Amar, M., Stein, N., et al. (2022). Effectiveness of Paxlovid in reducing severe COVID-19 and mortality in high risk patients. Clin. Infect. Dis. ciac443. doi:10.1093/cid/ciac443
Ndeupen, S., Qin, Z., Jacobsen, S., Bouteau, A., Estanbouli, H., and Igyarto, B. Z. (2021). The mRNA-LNP platform's lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. Iscience 24 (12), 103479. ARTN 103479. doi:10.1016/j.isci.2021.103479
Ndwandwe, D., and Wiysonge, C. S. (2021). COVID-19 vaccines. Curr. Opin. Immunol. 71, 111–116. doi:10.1016/j.coi.2021.07.003
Nugent, M. A. (2022). The future of the COVID-19 pandemic: How good (or bad) can the SARS-CoV2 spike protein get? Cells 11 (5). ARTN 855.
Okuda, K., Sato, Y., Iwakawa, K., Sasaki, K., Okabe, N., Maeki, M., et al. (2022). On the size-regulation of RNA-loaded lipid nanoparticles synthesized by microfluidic device. J. Control. Release 348, 648–659. doi:10.1016/j.jconrel.2022.06.017
Orenstein, W. A., Bernier, R. H., Dondero, T. J., Hinman, A. R., Marks, J. S., Bart, K. J., et al. (1985). Field evaluation of vaccine efficacy. Bull. World Health Organ. 63 (6), 1055–1068.
Otto, S. P., Day, T., Arino, J., Colijn, C., Dushoff, J., Li, M., et al. (2021). The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 31 (14), R918–R929. doi:10.1016/j.cub.2021.06.049
Packer, M., Gyawali, D., Yerabolu, R., Schariter, J., and White, P. (2021). A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 12 (1), 6777. ARTN 6777. doi:10.1038/s41467-021-26926-0
Palanikumar, L., Ramasamy, S., Hariharan, G., and Balachandran, C. (2013). Influence of particle size of nano zinc oxide on the controlled delivery of Amoxicillin. Appl. Nanosci. 3 (5), 441–451. doi:10.1007/s13204-012-0141-5
Pantelić, I., Ilić, T., Nikolić, I., and Savić, S. (2022). Lipid nanoparticles employed in mRNA-based COVID-19 vaccines: An overview of materials and processes used for development and production. Arh. za Farm. 72 (1), 20–35. doi:10.5937/arhfarm72-33660
Pardi, N., Hogan, M. J., Porter, F. W., and Weissman, D. (2018). mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 17 (4), 261–279. doi:10.1038/nrd.2017.243
Peng, L., Renauer, P. A., Okten, A., Fang, Z. H., Park, J. J., Zhou, X. Y., et al. (2022). Variant-specific vaccination induces systems immune responses and potent in vivo protection against SARS-CoV-2. Cell Rep. Med. 3 (5), 100634. ARTN 100634. doi:10.1016/j.xcrm.2022.100634
Pennarossa, G., Fazeli, A., Ledda, S., Gandolfi, F., and Brevini, T. A. L. (2021). Use of virus-mimicking nanoparticles to investigate early infection events in upper airway 3D models. Methods Mol. Biol. 2273, 131–138. doi:10.1007/978-1-0716-1246-0_8
Perera, W. P. T. D., Dissanayake, D. M. R. K., Unagolla, J. M., De Silva, R. T., Bathige, S. D. N. K., and Pahalagedara, L. R. (2022). Albumin grafted coaxial electrosparyed polycaprolactone-zinc oxide nanoparticle for sustained release and activity enhanced antibacterial drug delivery. RSC Adv. 12 (3), 1718–1727. doi:10.1039/d1ra07847j
Piyush, R., Rajarshi, K., Chatterjee, A., Khan, R., and Ray, S. (2020). Nucleic acid-based therapy for coronavirus disease 2019. Heliyon 6 (9)–e05007. doi:10.1016/j.heliyon.2020.e05007
Plotkin, S., Robinson, J. M., Cunningham, G., Iqbal, R., and Larsen, S. (2017). The complexity and cost of vaccine manufacturing - an overview. Vaccine 35 (33), 4064–4071. doi:10.1016/j.vaccine.2017.06.003
Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., et al. (2020). Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. Overseas. Ed. 383 (27), 2603–2615. doi:10.1056/NEJMoa2034577
Qin, M., Cao, Z., Wen, J., Yu, Q. S., Liu, C. Y., Wang, F., et al. (2020). An antioxidant enzyme therapeutic for COVID-19. Adv. Mat. 32 (43), 2004901. doi:10.1002/adma.202004901
Rehman, M., Tauseef, I., Aalia, B., Shah, S. H., Junaid, M., and Haleem, K. S. (2020). Therapeutic and vaccine strategies against SARS-CoV-2: Past, present and future. Future Virol. 15 (7), 471–482. doi:10.2217/fvl-2020-0137
Rocha, L. S. R., Simoes, A. Z., Macchi, C., Somoza, A., Giulietti, G., Ponce, M. A., et al. (2022). Synthesis and defect characterization of hybrid ceria nanostructures as a possible novel therapeutic material towards COVID-19 mitigation. Sci. Rep. 12 (1), 3341. doi:10.1038/s41598-022-07200-9
Scheaffer, S. M., Lee, D., Whitener, B., Ying, B., Wu, K., Liang, C. Y., et al. (2022). Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant in mice. Nat. Med. doi:10.1038/s41591-022-02092-8
Sahin, U., Karikó, K., and Türeci, Ö. (2014). mRNA-based therapeutics — developing a new class of drugs. Nat. Rev. Drug Discov. 13 (10), 759–780. doi:10.1038/nrd4278
Samaranayake, L. P., Seneviratne, C. J., and Fakhruddin, K. S. (2021). Coronavirus disease 2019 (COVID-19) vaccines: A concise review. Oral Dis. doi:10.1111/odi.13916
Sarode, A., Fan, Y., Byrnes, A. E., Hammel, M., Hura, G. L., Fu, Y., et al. (2022). Predictive high-throughput screening of PEGylated lipids in oligonucleotide-loaded lipid nanoparticles for neuronal gene silencing. Nanoscale Adv. 4 (9), 2107–2123. doi:10.1039/d1na00712b
Schlich, M., Palomba, R., Costabile, G., Mizrahy, S., Pannuzzo, M., Peer, D., et al. (2021). Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 6 (2), e10213. doi:10.1002/btm2.10213
Schoenmaker, L., Witzigmann, D., Kulkarni, J. A., Verbeke, R., Kersten, G., Jiskoot, W., et al. (2021). mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 601, 120586. ARTN 120586. doi:10.1016/j.ijpharm.2021.120586
Senti, M. E., de Jongh, C. A., Dijkxhoorn, K., Verhoef, J. J. F., Szebeni, J., Storm, G., et al. (2022). Anti-PEG antibodies compromise the integrity of PEGylated lipid-based nanoparticles via complement. J. Control. Release 341, 475–486. doi:10.1016/j.jconrel.2021.11.042
Sette, A., and Crotty, S. (2021). Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184 (4), 861–880. doi:10.1016/j.cell.2021.01.007
Sheahan, T. P., Sims, A. C., Graham, R. L., Menachery, V. D., Gralinski, L. E., Case, J. B., et al. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9 (396), eaal3653. doi:10.1126/scitranslmed.aal3653
Shiehzadegan, S., Alaghemand, N., Fox, M., and Venketaraman, V. (2021). Analysis of the delta variant B.1.617.2 COVID-19. Clin. Pract. 11 (4), 778–784. doi:10.3390/clinpract11040093
Sinha, R., Kim, G. J., Nie, S. M., and Shin, D. M. (2006). Nanotechnology in cancer therapeutics: Bioconjugated nanoparticles for drug delivery. Mol. Cancer Ther. 5 (8), 1909–1917. doi:10.1158/1535-7163.mct-06-0141
Smith, S. A., Selby, L. I., Johnston, A. P. R., and Such, G. K. (2019). The endosomal escape of nanoparticles: Toward more efficient cellular delivery. Bioconjug. Chem. 30 (2), 263–272. doi:10.1021/acs.bioconjchem.8b00732
Stander, J., Mbewana, S., and Meyers, A. E. (2022). Plant-derived human vaccines: Recent developments. BioDrugs 36, 573–589. doi:10.1007/s40259-022-00544-8
Suk, J. S., Xu, Q., Kim, N., Hanes, J., and Ensign, L. M. (2016). PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 99, 28–51. doi:10.1016/j.addr.2015.09.012
Szalay, B., Tátrai, E., Nyírő, G., Vezér, T., and Dura, G. (2012). Potential toxic effects of iron oxide nanoparticles in in vivo and in vitro experiments. J. Appl. Toxicol. 32 (6), 446–453. doi:10.1002/jat.1779
Tabynov, K., Turebekov, N., Babayeva, M., Fomin, G., Yerubayev, T., Yespolov, T., et al. (2022). An adjuvanted subunit SARS-CoV-2 spike protein vaccine provides protection against Covid-19 infection and transmission. npj Vaccines 7 (1), 24. doi:10.1038/s41541-022-00450-8
Tanaka, H., Miyama, R., Sakurai, Y., Tamagawa, S., Nakai, Y., Tange, K., et al. (2021). Improvement of mRNA delivery efficiency to a T cell line by modulating PEG-lipid content and phospholipid components of lipid nanoparticles. Pharmaceutics 13 (12), 2097. ARTN 2097. doi:10.3390/pharmaceutics13122097
Tao, K., Tzou, P. L., Nouhin, J., Gupta, R. K., De Oliveira, T., Kosakovsky Pond, S. L., et al. (2021). The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 22 (12), 757–773. doi:10.1038/s41576-021-00408-x
Troelnikov, A., Perkins, G., Yuson, C., Ahamdie, A., Balouch, S., Hurtado, P. R., et al. (2021). Basophil reactivity to BNT162b2 is mediated by PEGylated lipid nanoparticles in patients with PEG allergy. J. Allergy Clin. Immunol. 148 (1), 91–95. doi:10.1016/j.jaci.2021.04.032
Ucar, B., Acar, T., Arayici, P. P., and Derman, S. (2021). A nanotechnological approach in the current therapy of COVID-19: Model drug oseltamivir-phosphate loaded PLGA nanoparticles targeted with spike protein binder peptide of SARS-CoV-2. Nanotechnology 32 (48)–485601. doi:10.1088/1361-6528/ac1c22
Viana, R., Moyo, S., Amoako, D. G., Tegally, H., Scheepers, C., Althaus, C. L., et al. (2022). Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603 (7902), 679–686. doi:10.1038/s41586-022-04411-y
Voysey, M., Clemens, S. A. C., Madhi, S. A., Weckx, L. Y., Folegatti, P. M., Aley, P. K., et al. (2021). Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, south Africa, and the UK. Lancet 397 (10269), 99–111. doi:10.1016/s0140-6736(20)32661-1
Walsh, C., Ou, K., Belliveau, N. M., Leaver, T. J., Wild, A. W., Huft, J., et al. (2014). Microfluidic-based manufacture of siRNA-lipid nanoparticles for therapeutic applications. Methods Mol. Biol., 2nd Edition 1141, 109–120. doi:10.1007/978-1-4939-0363-4_6
Weiss, C., Carriere, M., Fusco, L., Capua, I., Regla-Nava, J. A., Pasquali, M., et al. (2020a). Toward nanotechnology-enabled approaches against the COVID-19 pandemic. Acs Nano 14 (6), 6383–6406. doi:10.1021/acsnano.0c03697
Weiss, C., Carriere, M., Fusco, L., Capua, I., Regla-Nava, J. A., Pasquali, M., et al. (2020b). Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 14 (6), 6383–6406. doi:10.1021/acsnano.0c03697
Wen, W., Chen, C., Tang, J. K., Wang, C. Y., Zhou, M. Y., Cheng, Y. R., et al. (2022). Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: A meta-analysis. Ann. Med. 54 (1), 516–523. doi:10.1080/07853890.2022.2034936
Wolfram, J., Suri, K., Huang, Y., Molinaro, R., Borsoi, C., Scott, B., et al. (2014). Evaluation of anticancer activity of celastrol liposomes in prostate cancer cells. J. Microencapsul. 31 (5), 501–507. doi:10.3109/02652048.2013.879932
Wu, T., Rabi, S. A., Michaud, W. A., Becerra, D., Gilpin, S. E., Mino-Kenudson, M., et al. (2022). Protease inhibitor Camostat Mesyalte blocks wild type SARS-CoV-2 and D614G viral entry in human engineered miniature lungs. Biomaterials 285–121509. doi:10.1016/j.biomaterials.2022.121509
Xiao, M. F., Zeng, C., Li, S. H., and Yuan, F. L. (2022). Applications of nanomaterials in COVID-19 pandemic. Rare Met. 41 (1), 1–13. doi:10.1007/s12598-021-01789-y
Xu, L. G., Wang, X. X., Wang, W. W., Sun, M. Z., Choi, W. J., Kim, J. Y., et al. (2022). Enantiomer-dependent immunological response to chiral nanoparticles. Nature 601(7893), 366-373. doi:10.1038/s41586-021-04243-2
Xue, Y. X., Liu, C. C., Andrews, G., Wang, J. Y., and Ge, Y. (2022). Recent advances in carbon quantum dots for virus detection, as well as inhibition and treatment of viral infection. Nano Converg. 9 (1), 15 ARTN 15. doi:10.1186/s40580-022-00307-9
Yan, S. Y., Gu, W. Y., and Xu, Z. P. (2013). Re-considering how particle size and other properties of antigen-adjuvant complexes impact on the immune responses. J. Colloid Interface Sci. 395, 1–10. doi:10.1016/j.jcis.2012.11.061
Yan, Z. P., Yang, M., and Lai, C. L. (2021). COVID-19 vaccines: A review of the safety and efficacy of current clinical trials. Pharm. (Basel) 14 (5), 406. doi:10.3390/ph14050406
Yi, S. A., Nam, K. H., Yun, J., Gim, D., Joe, D., Kim, Y. H., et al. (2020). Infection of brain organoids and 2D cortical neurons with SARS-CoV-2 pseudovirus. Viruses 12 (9), 1004. doi:10.3390/v12091004
Youk, J., Kim, T., Evans, K. V., Jeong, Y. I., Hur, Y., Hong, S. P., et al. (2020). Three-dimensional human alveolar stem cell culture models reveal infection response to SARS-CoV-2. Cell Stem Cell 27(6), 905-919.e10. doi:10.1016/j.stem.2020.10.004
Zeng, B., Gao, L., Zhou, Q., Yu, K., and Sun, F. (2022). Effectiveness of COVID-19 vaccines against SARS-CoV-2 variants of concern: A systematic review and meta-analysis. BMC Med. 20 (1), 200. doi:10.1186/s12916-022-02397-y
Zhang, Q., Honko, A., Zhou, J., Gong, H., Downs, S. N., Vasquez, J. H., et al. (2020). Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 20 (7), 5570–5574. doi:10.1021/acs.nanolett.0c02278
Zhang, Q., Tiwari, S. K., Wang, S., Wang, L., Li, W., Zhang, L., et al. (2022). Induction of neutralizing antibodies against SARS-CoV-2 variants by a multivalent mRNA-lipid nanoparticle vaccine encoding SARS-CoV-2/SARS-CoV Spike protein receptor-binding domains. bioRxiv. 2022.04.28.489834, doi:10.1101/2022.04.28.489834
Zhi, D., Bai, Y., Yang, J., Cui, S., Zhao, Y., Chen, H., et al. (2018). A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 253, 117–140. doi:10.1016/j.cis.2017.12.006
Keywords: nanotechnology, COVID-19, nanomaterials, vaccine development, antiviral therapeutics, disease-modelling, lipid nanoparticles, inorganic nanoparticles
Citation: Hangad MV, Keshvani S, Kelpin N, Walters-Shumka J, Hood M, Volk C, Pal D and Willerth SM (2022) Using nanomaterials to address SARS-CoV-2 variants through development of vaccines and therapeutics. Front. Mater. 9:1039247. doi: 10.3389/fmats.2022.1039247
Received: 07 September 2022; Accepted: 01 November 2022;
Published: 24 November 2022.
Edited by:
Aditya Kumar, Indian Institute of Technology Dhanbad, IndiaReviewed by:
Parikshit Moitra, The Pennsylvania State University (PSU), United StatesCopyright © 2022 Hangad, Keshvani, Kelpin, Walters-Shumka, Hood, Volk, Pal and Willerth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie M. Willerth, d2lsbGVydGhAdXZpYy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.