
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 19 March 2025
Sec. Aquatic Physiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1575833
This research identified functional genes and regulatory pathways of salinity stress in red claw crab Uca arcuata, which a globally distributed intertidal species inhabiting saline ecosystems, providing data and theoretical basis for the protection and utilization of U. arcuata in global climate change. In this study, we performed RNA sequencing on U. arcuata gill tissues exposed to salinity gradients (15‰, 25‰ control, and 35‰) to profile transcriptional responses. A total of 63.83 GB of high-quality clean reads were generated, yielding 125,462 unigenes with robust assembly metrics (N50 = 969 bp; mean length = 688 bp). The transcriptome analysis predicted 101,280 coding sequences (CDSs) and 52,706 simple sequence repeats (SSRs). Compared with the control group, the high-salinity group obtained 52 differentially expressed genes (DEGs), with 36 upregulated and 16 downregulated genes. The low-salinity group obtained 1,035 DEGs, with 780 upregulated and 255 downregulated genes. GO analysis showed a significant enrichment of DEGs in signal transduction, enzymatic activity, and binding. KEGG analysis showed that most DEGs were associated with signaling pathways and metabolism. APOA1, APOA2, GPX and GST were specific genes related to salinity adaptation. Five DEGs were randomly selected for quantitative RT-PCR validation, and the results demonstrated that the transcriptome data are highly reliable. However, we did not delve into the key functional genes and their regulatory mechanisms. Joint analysis of the genome and transcriptome of U. arcuata should conduct in the future, and comprehensively elucidate its adaptation mechanism to salinity fluctuations.
Salinity is a pivotal abiotic factor governing the survival, distribution, and physiological performance of aquatic organisms (Doney et al., 2012). Climate change, through the alteration of precipitation patterns and the disruption of ocean circulation, has significantly exacerbated the salinity fluctuations in marine ecosystems. These exacerbated salinity variations pose a substantial threat to the aquatic biodiversity, jeopardizing the survival and ecological balance of numerous aquatic species within these ecosystems (Brierley and Kingsford, 2009; Durack et al., 2012). These environmental disturbances disrupt habitat structure (Durack, 2015) and impair key physiological processes (Röthig et al., 2023). Specific impacts include compromised osmoregulation (Soria et al., 2007), disrupted energy metabolism (Verween et al., 2007), and altered hormonal balance (Arjona et al., 2007). While osmoregulatory adaptations have been extensively studied in brachyuran crabs such as Portunus pelagicus (a euryhaline species with well-characterized osmoregulatory pathways) (Romano and Zeng, 2006) and Carcinus maenas (a model organism for salinity stress responses) (Henry et al., 2002), the molecular mechanisms underlying salinity tolerance remain poorly characterized, particularly in ecologically vital species like U. arcuata. The above research can provide critical comparative insights into how U. arcuata might adapt molecularly to fluctuating salinity.
U. arcuata, also known as red claw crab, a decapod crustacean of the family Ocypodidae, thrives in tropical to temperate intertidal zones, where it excavates burrows in compact coastal sediments (Nabout et al., 2010). This species serves as a “mangrove engineer” (Kristensen, 2008; Penha-Lopes et al., 2009), enhancing ecosystem health through bioturbation-driven sediment oxygenation, where organisms like crabs or worms dig burrows, allowing oxygen to penetrate deeper layers, nutrient cycling (breaking down organic matter and redistributing nitrogen, phosphorus, and other essential elements), and pollutant mitigation (trapping heavy metals or breaking down toxins through microbial activity) (Botto et al., 2005; Holdredge et al., 2010; Lautenschlager et al., 2010). Despite its ecological significance, research on U. arcuata has historically prioritized morphology (Yamaguchi, 2001; Yamaguchi and Henmi, 2001; Yamaguchi et al., 2005) and behavior (Wada and Murata, 2000; Wada et al., 2011), likely due to their direct relevance to ecological interactions and taxonomic classification. However, its molecular responses to salinity stress remain understudied.
RNA sequencing (RNA-seq) has emerged as a powerful tool in deciphering transcriptomic adaptations—the analysis of RNA transcripts produced under specific conditions or within specific cells—to environmental stressors (Qian et al., 2014). RNA-seq has identified salinity-responsive genes and pathways in aquatic species such as Oreochromis niloticus (Cheng et al., 2022; Xu et al., 2022) and Pseudosciaena crocea (Zeng et al., 2017), as well as Charybdis japonica (Shui et al., 2022) and Scylla paramamosain (Wang et al., 2018), elucidating osmoregulatory mechanisms. However, such studies have yet to be conducted for U. arcuata. Given the sensitivity of crustacean gills to osmotic challenges (Ni et al., 2021), we conducted RNA-seq on U. arcuata gills under low-salinity (15‰), medium-salinity (25‰), and high-salinity (35‰) conditions. This study aims to (1) identify stress-responsive key genes and pathways involved in salinity adaptation, providing a molecular toolkit for targeted habitat restoration in climate-vulnerable regions, and (2) enhance understanding of climate driven habitat changes to guide adaptive management strategies, such as designing marine protected areas with optimal salinity thresholds or providing information for aquaculture practices that improve population adaptation to salinity fluctuations.
U. arcuata is not an endangered or protected species. During the sampling process, all U. arcuata individuals were chilled to induce anesthesia to minimize the suffering of animals. The Animal Care and Use Committee of Zhejiang Ocean University reviewed and approved this animal study.
From October 2023 to November 2023, 24 adult male U. arcuata were collected from the muddy intertidal zone beneath the Aoshan Bridge in Zhoushan. The specimens were transported to the laboratory and evenly distributed among three plastic water tanks for 24 hours. During this time, the water temperature and salinity in the tanks were maintained at 20°C and 25, respectively. Feeding was suspended to minimize the potential effect of other factors. After 24 h, 18 U. arcuata were evenly divided into three groups and placed in three plastic water tanks. Three groups were established: the high-salinity group (HSG), the low-salinity group (LSG), and the control group (SCG). The salinity levels for LSG, SCG, and HSG were 15, 25, and 35, respectively. According to the preliminary experiment, the duration of salinity stress was determined to be 24 hours, which can ensure the survival of all individuals. Throughout the experiment, the salinity among the tanks varied, whereas the water temperature was consistently maintained at 20°C for each group, with no significant variations in other factors. During the experiment, all groups ceased feeding, and individuals on the brink of death or already deceased were promptly removed. After the experiment, three crabs were randomly selected from each experimental group and placed on ice for freezing anesthesia. The gill tissues were then quickly dissected and immediately placed in numbered sterilized sterilized tubes. Subsequently, they were rapidly frozen in liquid nitrogen. After all samples were collected and frozen with liquid nitrogen, they were transferred to a −80°C refrigerator for storage.
After nine samples were removed from the −80°C refrigerator, total RNA was extracted from each tissue sample by using the TRIzol Reagent Kit (Invitrogen, Carlsbad, CA, USA). The RNA samples were then analyzed by 1% agarose gel electrophoresis to assess integrity and check for DNA contamination. RNA concentration (ng/μL) and purity (A260/280 ratio between 1.8 and 2.2) were measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA integrity was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA libraries were constructed with the TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA). First, under the action of reverse transcriptase, random primers were utilized to synthesize the first strand of cDNA by using mRNA as a template, followed by the synthesis of the second strand. A blend of enzymes (e.g., T4 DNA polymerase and Klenow fragment) was added to repair frayed or uneven ends of double-stranded cDNA, converting them into blunt termini compatible with downstream adapter ligation. Purification and fragment sorting were conducted on the products linked to the adapter, followed by PCR amplification to purify the final library. Finally, paired-end sequencing (150 bp) was performed on the Illumina HiSeq platform.
Raw reads from three experimental groups obtained through the sequencing platform were filtered using SeqPrep and Sickle software to obtain high-quality clean reads. Raw reads containing sequencing adapters, unknown nucleotide ratio exceeding 10%, and sequence end mass values below 20, which account for over 50% of all base quantities, were eliminated to ensure high-quality clean reads. Quality evaluation of the clean reads was performed using FastQC software to obtain their Q30, representing the percentage of bases with a quality value higher than 30 relative to the total number of reads. Clean reads that passed quality testing were utilized for subsequent transcript splicing.
Trinity software was employed to assemble the file with default parameters to obtain transcripts and unigenes. The integrity of the transcriptome assembly was assessed using BUSCO. TransDecoder software was utilized to predict the coding sequences (CDSs) of unigenes. Simple sequence repeats (SSRs) were analyzed using MISA (version 1.0) software with default parameters. The repeating units and their minimum number of nucleotide repetitions for mono-nucleotide, di-nucleotide, tri-nucleotide, tetra-nucleotide, penta-nucleotide, and hexa-nucleotide were 10, six, five, five, five, and five, respectively.
The assembled unigenes were compared against six major databases (Protein Families [Pfam], Swiss-Prot, Non-Redundant [NR] Protein Sequence, EuKaryotic Orthologous Groups [KOG], Gene Ontology [GO], and Kyoto Encyclopedia of Genes and Genomes [KEGG]) to obtain annotation information from each database.
The unigenes assembled from Trinity served as the reference by using RSEM (version 1.3.0) (Langmead, 2009), utilizing bowtie2 (Li and Dewey, 2011) with default parameters to compare the clean reads of each sample against the reference. The comparison results were compiled to determine the number of reads mapped to each transcript for each sample, which were then converted to fragments per kilobase per million base values. EdgeR and DESeq2 (Leng et al., 2013) were employed to analyze the DEGs between different groups. Subsequently, false discovery rate (FDR) and fold change (FC) values were applied to identify DEGs, with screening criteria set at FDR < 0.05 and |log2FC| ≥ 1. Volcano and MA plots were generated using R for visual analysis. Functional information of the differentially expressed genes (DEGs) was analyzed using Goatools and KOBAS (version 2.0), leveraging the GO and KEGG databases to identify key functional genes and regulatory mechanisms of U. arcuata under salinity stress.
Five DEGs from three experimental groups were randomly selected to validate the RNA-seq results by using 18S rRNA as the internal reference gene. As shown in Table 1, Specific primers were designed with Primer Premier 5.0. Initially, the complementary DNA (cDNA) samples were diluted by fivefold. Then, following the manufacturer’s instructions, the reaction system totaled 20 μL, comprising 2.0 μL of diluted cDNA template, 10 μL of ChamQ Universal Sybr qPCR Master Mix (2×), 0.4 μL of forward and reverse primers, and 7.2 μL of ddH2O. The amplification procedure was conducted using the ABI 7300 Plus real-time PCR instrument (Applied Biosystems, Carlsbad, CA, USA). It involved holding at 95°C for 3 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Three parallel experiments were performed for each cDNA template. The relative expression levels of these genes were calculated using the 2−ΔΔCT method (ΔCT = CTtarget gene − CTreference gene; ΔΔCT = ΔCTtreatment − ΔCTcontrol) (Livak and Schmittgen, 2001).
The raw RNA-seq reads of nine U. arcuata were placed in the NCBI Sequence Read Archive (SRA) database under the accession number of SRR32325637, SRR32325636, SRR32325635, SRR32325634, SRR32325633, SRR32325632, SRR32325631, SRR32325630 and SRR32325629 under BioProject PRJNA1222120. As shown in Table 2, RNA sequencing of Uca arcuata gill tissues in high-salinity group (HSG, 35‰), salinity control group (SCG, 25‰), and low-salinity group (LSG, 15‰) generated 426.85 million clean reads after rigorous quality control. Each sample yielded ≥ 6.81 Gb of high-quality data, with Q30 scores exceeding 97.36%. De novo assembly produced 223,813 transcripts (N50 = 1,407 bp; mean length = 858 bp) and 125,462 unigenes (N50 = 969 bp; mean length = 688 bp), demonstrating robust assembly continuity (Table 3). The BUSCO (version 3.1.0) analysis identified 1013 genes in the BUSCO database. Among them, 93.3% were detected for complete BUSCOs and 3.4% for missing BUSCOs, indicating good assembly quality (Table 4).
The unigenes were compared and annotated in databases such as Protein Families [Pfam], Swiss-Prot, Non-Redundant [NR] Protein Sequence, EuKaryotic Orthologous Groups [KOG], Gene Ontology [GO], and Kyoto Encyclopedia of Genes and Genomes [KEGG]. As shown in Table 5, 29.44% of unigenes were annotated in the Pfam database, representing the highest proportion. The unigenes annotated in the NR database accounted for 20.99%, and 8059 unigenes were annotated in the GO database, constituting 7.96% of the total. Additionally, 2945 unigenes were annotated in the KEGG database, accounting for 2.91% only. Meanwhile, 33,936 (33.51%) unigenes matched one or more databases.
The transcriptome information was compared with the GO database and categorized into three classifications: biological process, cellular component, and molecular function. The top 20 most annotated secondary classifications within each category were selected for plotting (Figure 1). The most significant annotated items in biological process were cellular process and metabolic process. The most significant annotated item in cellular component was cellular anatomical entity, and the most significant annotated items in molecular function were binding and catalytic activity.
KEGG annotation revealed that the unigenes were mapped to 33 pathways and categorized into five groups on the basis of their involvement in KEGG pathways (Figure 2). This classification included 652 in cellular processes, 838 in environmental information processing, 643 in genetic information processing, 1189 in metabolism, and 1499 in organismal systems. Signal transduction was the most dominant group, with 767 unigenes in 33 pathways.
In the KOG database, 11,643 genes were annotated into 25 orthologous groups (Figure 3). Among them, the largest group consisted of function unknown, Ttranslation, ribosomal structure and biogenesis, and unnamed protein, followed by transcription, intracellular trafficking and secretion, and vesicular transport.
Based on TransDecoder software, 101,280 CDSs were obtained. Additionally, 52,706 SSRs were discovered (Figure 4). The numbers of mono-nucleotide, di-nucleotide, tri-nucleotide, tetra-nucleotide, penta-nucleotide, and hexa-nucleotide were 10,620, 22,307, 9946, 1396, 241, and four, respectively.
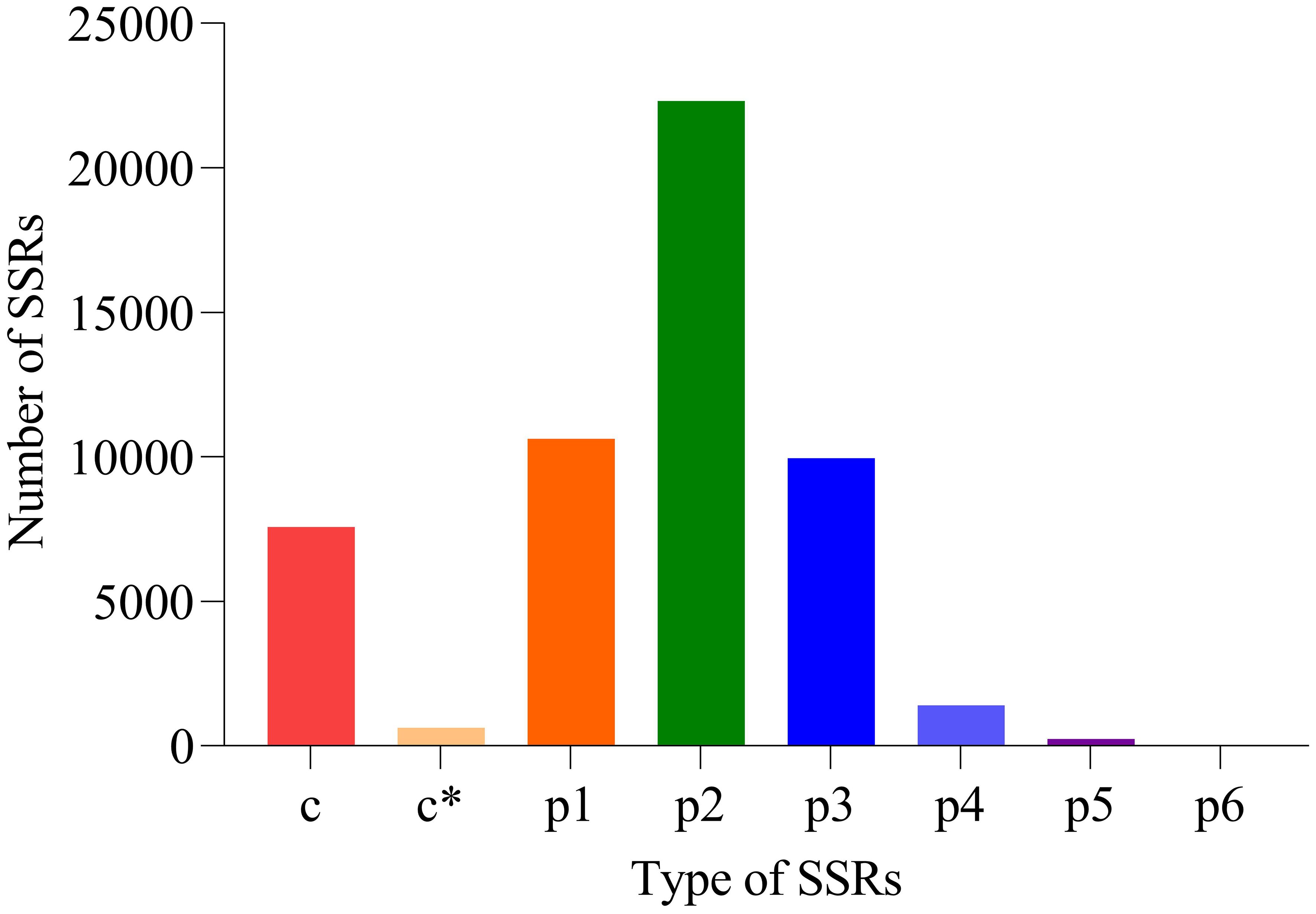
Figure 4. Results of SSRs. x-coordinate: SSR type, y-coordinate: number of SSR. p1: mono-nucleotide, p2: di-nucleotide, p3: tri-nucleotide, p4: tetra-nucleotide, p5: penta-nucleotide, p6: hexa-nucleotide and c: compound type. c* means compound type with additional annotations (such as special structure or functional markings).
A salinity of 25 was used in SCG to statistically analyze the number of differentially expressed genes (DEGs) in HSG and LSG. As shown in Figure 5, the expression levels of DEGs were visualized using volcano and MA plots. A total of 52 DEGs were identified in HSG compared with SCG, including 36 upregulated and 16 downregulated ones. In LSG versus SCG, 1035 DEGs were identified, with 780 upregulated and 255 downregulated ones. In LSG versus HSG, 660 DEGs were found, comprising 359 upregulated and 301 downregulated ones.
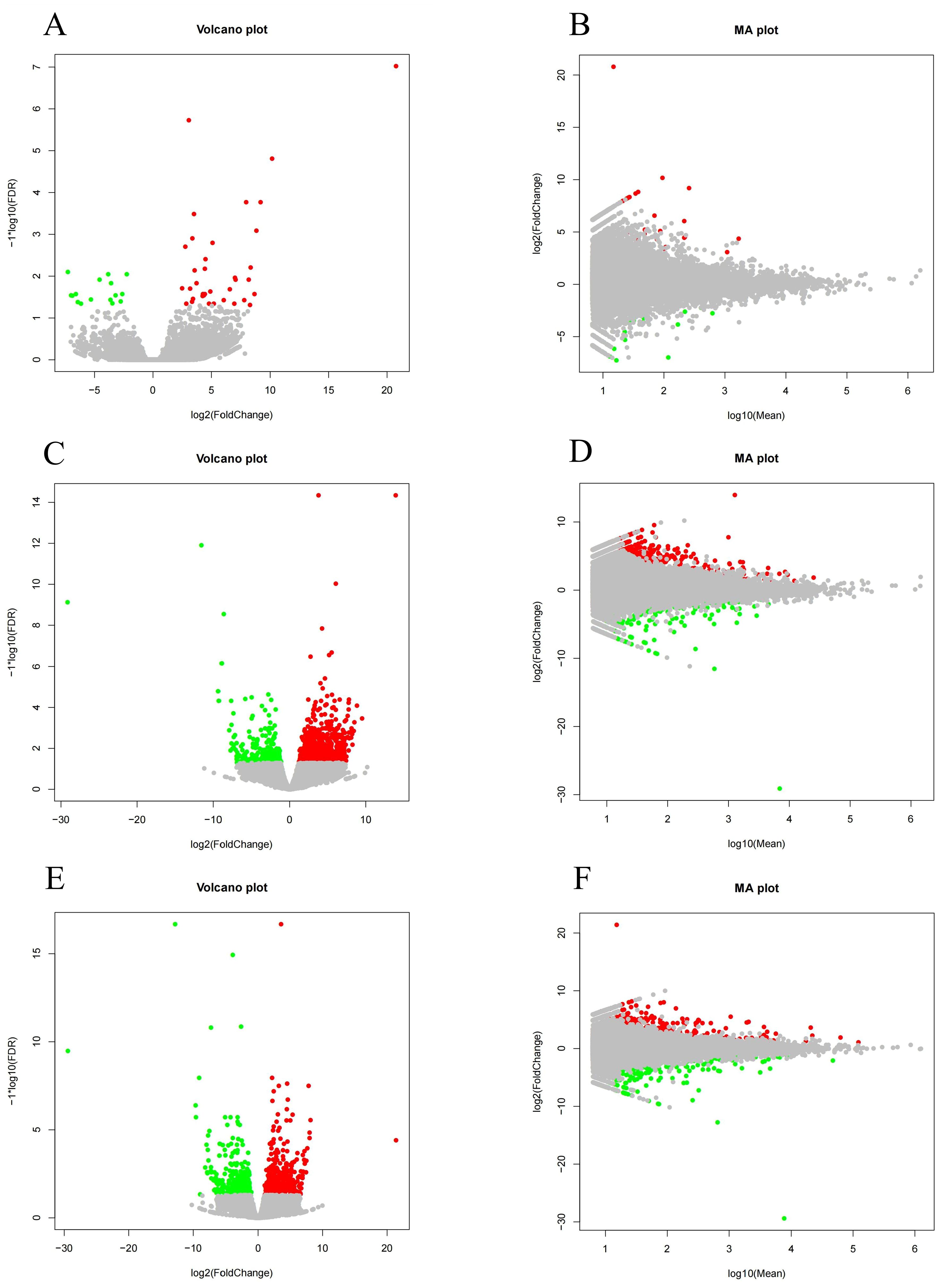
Figure 5. Volcano and MA plots of DEGs in different salinity comparison groups. (A, B) represent the DEGs between HSG and SCG; (C, D) represent the DEGs between LSG and SCG; and (E, F) represent the DEGs between LSG and HSG.
GO enrichment analysis can identify the primary biological functions and characteristics of DEGs. In this study, DEGs were categorized into three main categories in GO: biological process, cellular component, and molecular function. As shown in Figure 6, the GO analysis revealed that the significant DEGs in HSG were involved in GO terms such as phosphorelay signal transduction system (GO:0000160), G protein-coupled receptor signaling pathway (GO:0007186), and DNA integration (GO:0015074) in biological processes and G protein-coupled receptor activity (GO:0004930) in molecular function. The significant DEGs in LSG were primarily involved in the phosphorelay signal transduction system (GO:000160) in biological processes. In HSG versus LSG, the significant DEGs were involved in GO terms such as lipid transport (GO:0006869) and carboxylic acid metabolic process (GO:0019752) in biological process; extracellular space (GO:0005615) and extracellular region (GO:0005576) in cellular component; and signaling receptor binding (GO:0005102), iron ion binding (GO:000556), lipid binding (GO:0008289), oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (GO:0016705), carboxy-lyase activity (GO:0016831), heme binding (GO:0020037), and pyridoxal phosphate binding (GO:0030170) in molecular function.
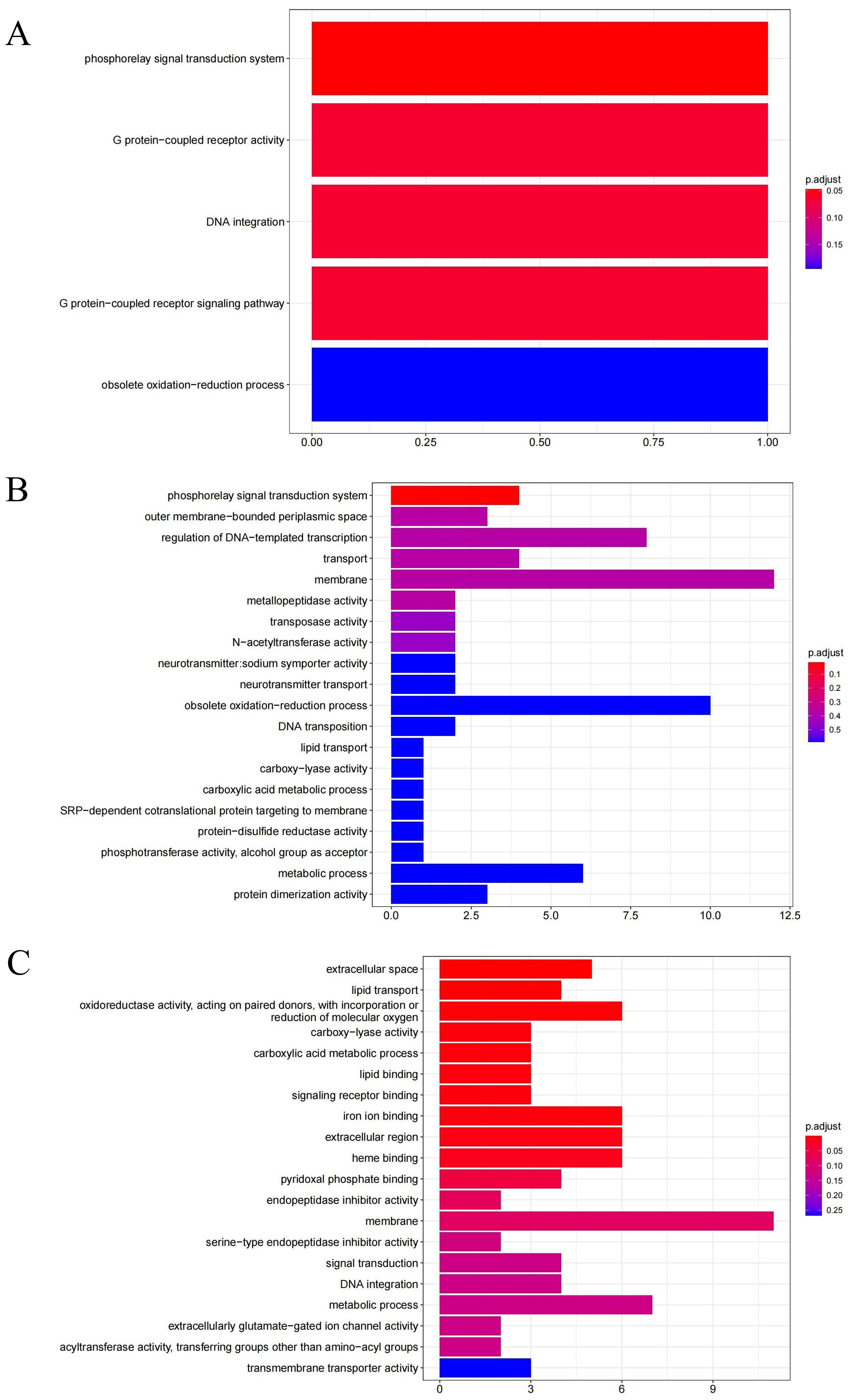
Figure 6. GO analysis of DEGs in different salinity comparison groups. (A) represent the DEGs between HSG and SCG; (B) represent the DEGs between LSG and SCG; and (C) represent the DEGs between LSG and HSG.
The functions of gene products and their potential metabolic pathways were analyzed by KEGG annotation. As shown in Figure 7, the KEGG analysis revealed that the significantly enriched pathways in HSG were primarily associated with signaling pathways, including the Fc epsilon RI signaling pathway (ko04664), the NF-kappa B signaling pathway (ko04064), the B cell receptor signaling pathway (ko04662), and the phospholipase D signaling pathway (ko04072). Additional pathways included natural killer cell mediated cytotoxicity (ko04650), arachidonic acid metabolism (ko00590), and Fc gamma R-mediated phagocytosis (ko04666). In LSG, the DEGs were significantly enriched in glutathione metabolism (ko00480). In HSG versus LSG, the DEGs were significantly enriched in the PPAR signaling pathway (ko03320), fat digestion and absorption (ko04975), complement and coagulation cascades (ko04610), and platelet activation (ko04611). Most of the enrichment pathways in the three groups were related to signaling pathways and metabolism.
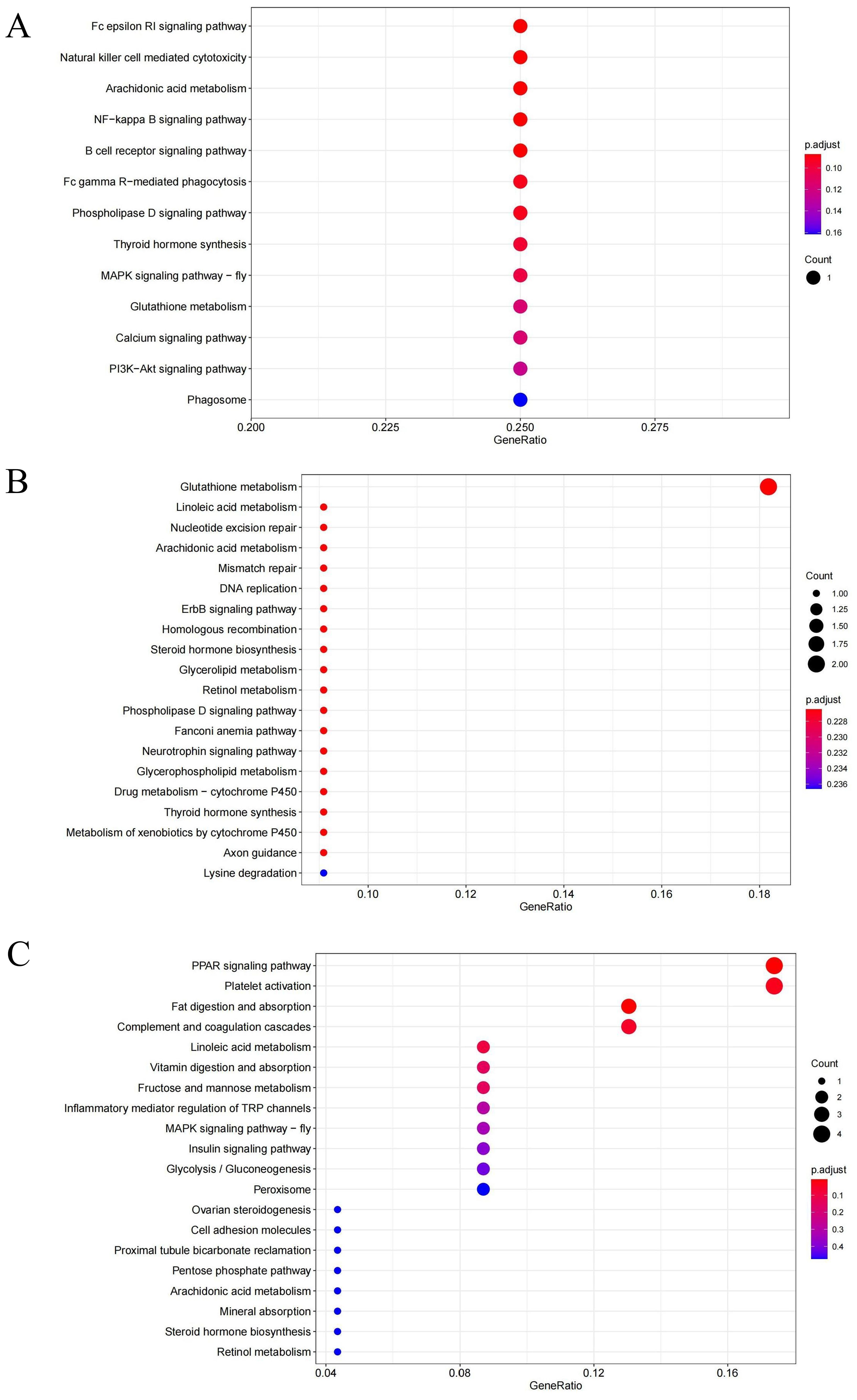
Figure 7. KEGG analysis of DEGs in different salinity comparison groups. (A) represent the DEGs between HSG and SCG; (B) represent the DEGs between LSG and SCG; and (C) represent the DEGs between LSG and HSG.
Five significant DEGs were randomly selected from the three experimental groups for validation to verify the accuracy of the transcriptome data. As shown in Figure 8, the results indicated that the expression trend of qRT-PCR results was consistent with the RNA-seq results, confirming the reliability of the latter. Furthermore, the expression trends of different genes, whether based on transcriptome or qRT-PCR, were specific at different salinities. The gene expression levels of APOA1 and APOA2 increased with increasing and decreasing salinity, whereas those of PPP1C, CYP2J, and CYP3A4 decreased.
At present, global climate change has resulted in continuous changes in ocean salinity. When salinity changes exceed the tolerance range of organisms, it can lead to mortality. Salinity is one of the crucial environmental factors that significantly affect aquatic organisms. Numerous studies have confirmed that salinity plays a key role in influencing the development and survival of crustaceans (Bœuf and Payan, 2001). Changes in salinity typically have a direct effect on the osmotic regulation ability of aquatic organisms (Ponce-Palafox et al., 2013). Salinity significantly affects the respiratory function, growth, survival, and immune defense of crabs (Sang and Fotedar, 2004). Therefore, crustaceans may be more vulnerable to the effects of salinity stress (Romano and Zeng, 2012).
Under salinity stress, marine organisms undergo cellular stress responses to changes in salinity. Cells can exert repair and protective effects, which can result in the inhibition of cell proliferation and programmed cell death (Evans and Kültz, 2020). Changes in seawater salinity directly affect the osmotic pressure regulation mechanism of aquatic animals. Once an osmotic pressure imbalance occurs, it can damage other physiological processes such as growth, development, metabolism, and immunity in aquatic animals (Shaughnessy et al., 2015). So far, no research has indicated the salinity adaptation mechanism and the effect of salinity changes on U. arcuata. Therefore, in the present study, U. arcuata was selected as the research subject to investigate its response mechanism to salinity stress.
U. arcuata was placed in high-salinity (35‰) and low-salinity (15‰) environments and sequenced using the Illumina HiSeq platform. The results showed that the quality values of clean read bases in each sample were relatively high, at least 97.36%, indicating that the sequences had high sequencing accuracy. The obtained clean reads are reliable, and they could be used for subsequent transcript splicing. The N50 value and average value of the sequence were high, indicating a good assembly effect of the transcripts, which can be further analyzed.
Analysis of the results of DEGs revealed that under salinity stress, the expression levels of many genes in U. arcuata changed significantly to cope with the stress induced by external salinity variations, respond to salinity regulation mechanisms, and ensure survival. However, the number of differentially expressed genes in low-salinity group was significantly higher than that in high-salinity group. This result is consistent with a study on Charybdis japonica (Shui et al., 2022), but we cannot simply infer that U. arcuata was under greater stress at this salinity. In addition, the number of upregulated genes in LSG was significantly higher than that in HSG. This difference may be attributed to the necessity for a larger number of genes to regulate salt tolerance mechanisms in U. arcuata under low-salt environment.
In the GO enrichment analysis, DEGs were primarily enriched in signal transduction, enzymatic activity, and binding. Due to salinity fluctuations, U. arcuata activates cellular regulatory mechanisms, such as osmotic balance control and stress response pathways to survive. When salinity changes, cells adjust ion concentrations (e.g., pumping out excess sodium via Na+/K+-ATPase) and produce protective molecules like osmolytes to prevent dehydration or swelling. These mechanisms ensure that critical functions like nutrient uptake, energy production, and cell structure stability are maintained. Without this adaptability, salinity shifts could overwhelm the organism, thereby leading to cellular dysfunction or death. This alteration ultimately helps maintain a relative balance between the organism’s internal environment and the external surroundings (Rahi et al., 2018). Crustaceans possess a mechanism for regulating osmotic pressure, and changes in salinity can disrupt this balance. At this point, the osmotic pressure regulation mechanism begins to function. When U. arcuata are in a low-salt environment, they absorb a significant amount of water through their gill cells, along with a considerable number of ions, and continuously excrete urine to maintain isotonic flow. In a high-salt environment, they lose a substantial amount of water from their bodies, and the gills expel more salt to sustain osmotic pressure balance. Moreover, ion transport enzymes in crustaceans contribute to maintaining osmotic pressure balance. Na+-K+-ATPase, which has been extensively studied, is the primary ion transport enzyme in aquatic animals. It facilitates ion absorption or emission to enhance salinity regulation in response to changes in salinity (Liu et al., 2008). Therefore, under salinity stress, U. arcuata is speculated to regulate the activity of key enzymes in its body to adapt to external salinity changes. The expression levels of key salinity-responsive enzymes, such as ion transporters (e.g., Na+/K+-ATPase, V-type H+-ATPase), osmolytes synthesis enzymes (e.g., myo-inositol oxygenase), and stress signaling regulators (e.g., NF-κB), were dynamically regulated in U. arcuata to maintain critical bodily functions. For example, ion transporters adjust cellular salt balance, osmolytes stabilize proteins and membranes under osmotic stress, and signaling genes coordinate systemic responses to salinity changes. This precise regulation of gene networks ensures physiological processes like ion homeostasis, cellular hydration, and oxidative stress mitigation remain functional, enabling survival in fluctuating salinity environments.
Most DEGs were associated with signaling pathways and metabolism. In HSG, the DEGs were significantly enriched in the Fc epsilon RI signaling pathway, NF-kappa B signaling pathway, B cell receptor signaling pathway, and phospholipase D signaling pathway. The IGH (immunoglobulin heavy chain) genes, which encode critical components of antibodies essential for recognizing and neutralizing pathogens like bacteria and viruses, showed a trend of downregulation. Immunoglobulins (Igs) are produced by B cells, which are essential components of the humoral immune response (Mashoof and Criscitiello, 2016). The basic structure of Ig consists of two identical heavy (H) chains and two identical light (L) chains (Schroeder and Cavacini, 2010). The expression levels of three Ig isotype classes, IgM, IgD, and IgT, increased following parasitic, bacterial, and fungal pathogen infections in Misgurnus anguillicaudatus (Xu et al., 2019). This downregulation suggests a potential weakening of the organism’s immune defense capacity, as fewer functional antibodies could impair its ability to combat infections. Such a decline in immune-related gene activity may leave U. arcuata more vulnerable to diseases, particularly in stressful environments where energy is diverted toward coping with osmotic challenges instead of maintaining robust immunity. The B cell receptor (BCR) plays a vital role in B cell development and antibody production. After Streptococcus agalactiae infection, CD79, which forms B cell antigen receptors with membrane immunoglobulin, was significantly upregulated in Oreochromis niloticus (Wu et al., 2019). Similarly, Cryptocaryon irritans infection led to CD79 upregulation in Epinephelus coioides (Mo et al., 2016). The expression levels of IGH genes is closely linked to the activity of BCRs (Lange et al., 2009). This may be related to the high-salt tolerance mechanism of U. arcuata, which does not necessitate upregulation of IGH in high-salt environments.
Glutathione metabolism was significantly downregulated in LSG. It plays a crucial role in regulating metabolism and promoting detoxification and antioxidation. It is also an essential metabolic process in organisms (Wu et al., 2004). The gpx (glutathione peroxidase) and GST (glutathione S-transferase) are key genes in cellular detoxification systems that protect cells against reactive oxygen species (ROS) (Choi et al., 2008). Changes in salinity can induce an increase in GPX activity in the white leg shrimp (Litopenaeus vannamei) and lead to severe oxidative stress (Liu et al., 2007). Furthermore, a correlation exists between changes in salinity and GST activity. Salinity fluctuations may be a nonbiological factor contributing to an increase in GST activity in Mediterranean mussels (Mytilus galloprovincialis) (Bebianno et al., 2007). Low-salt stress accelerates cellular senescence and places cells at increased risk of stress (Maher, 2005). The downregulation of key enzymes related to glutathione levels in these results may be associated with cellular senescence.
In HSG versus LSG, the DEGs were significantly enriched in the PPAR signaling pathway, fat digestion and absorption, complement and coagulation cascades, and platelet activation. These pathways are associated with fat metabolism and traumatic coagulation reactions. The FABP1 (fatty acid-binding protein 1, liver), FABP2 (fatty acid-binding protein 2, intestinal), APOA1 (apolipoprotein A-I), APOA2 (apolipoprotein A-II) and APOA4 (apolipoprotein A-IV) in these pathways showed a trend of upregulation, as found in Synechogobius hasta (Chen et al., 2020). Peroxisome proliferators activated receptors (PPARs) are nuclear hormone receptors activated by fatty acids and their derivatives. Essentially, they represent a class of ligand-dependent transcriptional regulatory factors that govern numerous cellular metabolic processes (Chen et al., 2017). Current research has indicated that PPAR is involved in lipid, sugar, and energy metabolisms in fish (Feng et al., 2021). PPAR can assist fish in maintaining energy balance in extreme environments (Ning et al., 2016; Xiong et al., 2022). Fat serves as a vital energy source and the primary form of energy storage (Borlongan and Benitez, 1992). Most aquatic organisms possess a strong ability to utilize fat while exhibiting poor efficiency in carbohydrate utilization, making fat the optimal energy source (Meng et al., 2019). Fatty acids are the essential building blocks of fat. They can enhance the activity of various enzymes such as Na+-K+-ATPase, promote ion absorption or release, and improve the body’s ability to adapt to fluctuations in salinity (Moya-Falcón et al., 2004; Huang et al., 2010, 2020). In the present study, PPAR levels were speculated to be significantly upregulated, enhancing fat digestion and absorption, thereby regulating lipid metabolism processes to maintain body balance. The complement system is the humoral component of the immune system, and it plays a crucial role in human immune defense by inducing hemolysis, phagocytosis, chemotaxis, and natural immune bactericidal effects. The coagulation cascade reaction is a vital process in the body that helps prevent blood vessel damage and bleeding. An association was found between complement and coagulation cascade reactions in certain physiological processes (Amara et al., 2010). Platelets in crustaceans are referred to as clotting cells. Platelets not only participate in clotting but also function as blood macrophages, forming a protective barrier against foreign substances (Yu et al., 2021). They can directly clear cell debris from the bloodstream through phagocytosis (Zhou et al., 2018). U. arcuata may prevent vascular rupture by activating the coagulation cascade reaction. It also regulates the killing effect of immune cells by activating the complement system and enhances the phagocytic ability of blood cells by activating platelets. This process boosts the body’s immune function to adapt to changes in osmotic pressure under salinity stress.
Salinity is one of the environmental factors essential for aquatic organisms. For intertidal crabs like U. arcuata, fluctuations in salinity may have a more profound impact compared to those experienced by species living exclusively in the ocean. Therefore, studying its salinity tolerance mechanism is of paramount importance. The RNA-seq analysis revealed that gill tissues of U. arcuata in high-salinity group (HSG) and low-salinity group (LSG) exhibited 52 and 1035 differentially expressed genes (DEGs), respectively. DEGs are genes with significantly altered activity levels under environmental stress. The number of DEGs in the LSG is nearly 20 times higher than that in the HSG, which may indicate that low salinity imposes a greater transcriptional burden on the organism. This implies that U. arcuata must activate or suppress a larger suite of genes to adapt to low-salinity stress, potentially disrupting biological processes such as ion transport, energy metabolism, and osmotic balance.
The GO annotation and KEGG pathway analysis indicated that U. arcuata mainly adapts to salinity fluctuations by regulating signaling pathways, enzyme activity, and metabolism. Fluctuations in salinity can profoundly influence the physiology of U. arcuata, impacting processes such as osmotic regulation, enzyme activity, and ion homeostasis. For instance, abrupt changes in salinity disrupt osmotic balance, forcing the organism to expend energy to regulate intracellular water and ion concentrations, which may divert resources from growth and development. Additionally, salinity shifts can alter the function of enzymes critical for metabolism (e.g., those involved in ATP production or nitrogen excretion), either by destabilizing their structure or modifying substrate affinity. Ion transport systems, particularly those managing sodium (Na+), potassium (K+), and chloride (Cl-), may also become dysregulated under extreme salinities, leading to cellular stress or impaired neuromuscular function. These physiological disruptions collectively compromise survival, reproduction, and long-term fitness in dynamic estuarine environments. The results of this study provide data and a theoretical basis for understanding the adaptation of U. arcuata to salinity stress, but further research is needed to explore the regulatory mechanism.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR32325637; https://www.ncbi.nlm.nih.gov/, SRR32325636; https://www.ncbi.nlm.nih.gov/, SRR32325635; https://www.ncbi.nlm.nih.gov/, SRR32325634; https://www.ncbi.nlm.nih.gov/, SRR32325633; https://www.ncbi.nlm.nih.gov/, SRR32325632; https://www.ncbi.nlm.nih.gov/, SRR32325631; https://www.ncbi.nlm.nih.gov/, SRR32325630; https://www.ncbi.nlm.nih.gov/, SRR32325629.
The animal study was approved by The Animal Care and Use Committee of Zhejiang Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
BW: Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. SS: Writing – original draft, Writing – review & editing. FZ: Writing – original draft, Writing – review & editing. ZH: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Fundamental Research Funds for Zhejiang Provincial Universities and Research Institutes (2024J004) and National Natural Science Foundation of China (32070513).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amara U., Flierl M. A., Rittirsch D., Klos A., Chen H., Acker B., et al. (2010). Molecular intercommunication between the complement and coagulation systems. J. Immunol. 185, 5628–5636. doi: 10.4049/jimmunol.0903678
Arjona F. J., Vargas-Chacoff L., Ruiz-Jarabo I., Del Río M. P. M., Mancera J. M. (2007). Osmoregulatory response of Senegalese sole (Solea Senegalensis) to changes in environmental salinity. Comp. Biochem. Phys. A. 148, 413–421. doi: 10.1016/j.cbpa.2007.05.026
Bebianno M. J., Lopes B., Guerra L., Hoarau P., Ferreira A. M. (2007). Glutathione S-tranferases and cytochrome P450 activities in Mytilus galloprovincialis from the South coast of Portugal: effect of abiotic factors. Environ. Int. 33, 550–558. doi: 10.1016/j.envint.2006.11.002
Bœuf G., Payan P. (2001). How should salinity influence fish growth? Comp. Biochem. Phys. C. 130, 411–423. doi: 10.1016/s1532-0456(01)00268-x
Borlongan I. G., Benitez L. V. (1992). Lipid and fatty acid composition of milkfish (Chanos chanos Forsskal) grown in freshwater and seawater. Aquaculture 104, 79–89. doi: 10.1016/0044-8486(92)90139-c
Botto F., Valiela I., Iribarne O., Martinetto P., Alberti J. (2005). Impact of burrowing crabs on C and N sources, control, and transformations in sediments andfood webs of SW Atlantic estuaries. Mar. Ecol. Prog. Ser. 293, 155–164. doi: 10.3354/meps293155
Brierley A. S., Kingsford M. J. (2009). Impacts of climate change on marine organisms and ecosystems. Curr. Biol. 19, R602–R614. doi: 10.1016/j.cub.2009.05.046
Chen X. N., Gao Y. L., Wu G. J., Gu J. Z., Cai Y. F., Xu J. H., et al. (2020). Molecular cloning, tissue expression, and transcriptional regulation of fabp1 and fabp2 in javelin goby (Synechogobius hasta) in response to starvation stress. Comp. Biochem. Phys. B. 250, 110484. doi: 10.1016/j.cbpb.2020.110484
Chen G. H., Luo Z., Chen F., Shi X., Song Y. F., You W. J., et al. (2017). PPARα, PPARγ and SREBP-1 pathways mediated waterborne iron (Fe)-induced reduction in hepatic lipid deposition of javelin goby Synechogobius hasta. Comp. Biochem. Phys. C. 197, 8–18. doi: 10.1016/j.cbpc.2017.04.003
Cheng Y. M., Zhao J. L., Ayisi C. L., Cao X. Y. (2022). Effects of salinity and alkalinity on fatty acids, free amino acids and related substance anabolic metabolism of Nile tilapia. Aquacul. Fish. 7, 389–395. doi: 10.1016/j.aaf.2020.06.005
Choi C. Y., An K. W., An M. I. (2008). Molecular characterization and mRNA expression of glutathione peroxidase and glutathione S-transferase during osmotic stress in olive flounder (Paralichthys olivaceus). Comp. Biochem. Phys. A. 149, 330–337. doi: 10.1016/j.cbpa.2008.01.013
Doney S. C., Ruckelshaus M., Emmett Duffy J., Barry J. P., Chan F., English C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Durack P. J. (2015). Ocean salinity and the global water cycle. Oceanography 28, 20–31. doi: 10.5670/oceanog.2015.03
Durack P. J., Wijffels S. E., Matear R. J. (2012). Ocean salinities reveal strong global water cycle intensification during 1950 to 2000. Science 336, 455–458. doi: 10.1126/science.1212222
Evans T. G., Kültz D. (2020). The cellular stress response in fish exposed to salinity fluctuations. J. Exp. Zool. Part. A. 333, 421–435. doi: 10.1002/jez.2350
Feng H. X., Zhang Y. P., Liang X. F., He S., Li L. (2021). Dietary supplementation of exogenous probiotics reduces excessive liver lipid deposition in Chinese perch (Siniperca chuatsi). Aquac. Res. 52, 5430–5440. doi: 10.1111/are.15413
Henry R. P., Garrelts E. E., McCarty M. M., Towle D. W. (2002). Differential induction of branchial carbonic anhydrase and Na+/K+ ATPase activity in the euryhaline crab, Carcinus maenas, in response to low salinity exposure. J. Exp. Zool. 292, 595–603. doi: 10.1002/jez.10075
Holdredge C., Bertness M. D., Herrmann N. C., Gedan K. B. (2010). Fiddler crab control of cordgrass primary production in sandy sediments. Mar. Ecol. Prog. Ser. 399, 253–259. doi: 10.3354/meps08331
Huang C. Y., Chao P. L., Lin H. C. (2010). Na+/K+-ATPase and vacuolar-type H+-ATPase in the gills of the aquatic air-breathing fish Trichogaster microlepis in response to salinity variation. Comp. Biochem. Phys. A. 155, 309–318. doi: 10.1016/j.cbpa.2009.11.010
Huang M., Zhou Y. G., Liu C. Y., Davis D. A., Li L., Gao Q. F., et al. (2020). Fatty acid composition, osmolality, Na+, K+-ATPase activity, cortisol content and antioxidant status of rainbow trout (Oncorhynchus mykiss) in response to various dietary levels of eicosapentaenoic acid and docosahexaenoic acid. Aquac. Res. 51, 2777–2789. doi: 10.1111/are.14617
Kristensen E. (2008). Mangrove crabs as ecosystem engineers; with emphasis on sediment processes. J. Sea. Res. 59, 30–43. doi: 10.1016/j.seares.2007.05.004
Lange M. D., Waldbieser G. C., Lobb C. J. (2009). Patterns of receptor revision in the immunoglobulin heavy chains of a teleost fish. J. Immun. 182, 5605–5622. doi: 10.4049/jimmunol.0801013
Langmead B. (2009). Ultrafast andmemory-efficient alignment of short DNA sequences to the human genome. Genome. Biol. 10, R25. doi: 10.1186/gb-2009-10-3-r25
Lautenschlager A. D., Brandis D., Storch V. (2010). Morphology and function of the reproductive system of representatives of the genus Uca. J. Morphol. 271, 1281–1299. doi: 10.1002/jmor.10869
Leng N., Dawson J. A., Thomson J. A., Ruotti V., Rissman A. I., Smits B. M., et al. (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. doi: 10.1093/bioinformatics/btt087
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC. Bioinf. 12, 1–16. doi: 10.1186/1471-2105-12-323
Liu H. Y., Pan L. Q., Fu L. (2008). Effect of salinity on hemolymph osmotic pressure, sodium concentration and Na+-K+-ATPase activity of gill of Chinese crab, Eriocheir sinensis. J. Ocean. U. China. 7, 77–82. doi: 10.1007/s11802-008-0077-2
Liu Y., Wang W. N., Wang A. L., Wang J. M., Sun R. Y. (2007). Effects of dietary vitamin E supplementation on antioxidant enzyme activities in Litopenaeus vannamei (Boone 1931) exposed to acute salinity changes. Aquaculture 265, 351–358. doi: 10.1016/j.aquaculture.2007.02.010
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maher P. (2005). The effects of stress and aging on glutathione metabolism. Ageing. Res. Rev. 4, 288–314. doi: 10.1016/j.arr.2005.02.005
Mashoof S., Criscitiello M. F. (2016). Fish immunoglobulins. Biology 5, 45. doi: 10.3390/biology5040045
Meng Y. Q., Qian K. K., Ma R., Liu X. H., Han B. Y., Wu J. H., et al. (2019). Effects of dietary lipid levels on sub-adult triploid rainbow trout (Oncorhynchus mykiss): 1. Growth performance, digestive ability, health status and expression of growth-related genes. Aquaculture 513, 734394. doi: 10.1016/j.aquaculture.2019.734394
Mo Z. Q., Yang M., Wang H. Q., Xu Y., Huang M. Z., Lao G. F., et al. (2016). Grouper (Epinephelus coioides) BCR signaling pathway was involved in response against Cryptocaryon irritans infection. Fish. Shellfish. Immun. 57, 198–205. doi: 10.1016/j.fsi.2016.08.011
Moya-Falcón C., Hvattum E., Dyrøy E., Skorve J., Stefansson S. O., Thomassen M. S., et al. (2004). Effects of 3-thia fatty acids on feed intake, growth, tissue fatty acid composition, β-oxidation and Na+, K+-ATPase activity in Atlantic salmon. Comp. Biochem. Phys. B. 139, 657–668. doi: 10.1016/j.cbpc.2004.08.009
Nabout J. C., Bini L. M., Diniz-Filho J. A. (2010). Global literature of fiddler crabs, genus Uca (Decapoda, Ocypodidae): trends and future directions. Iheringia. Ser. Zool. 100, 463–468. doi: 10.1590/s0073-47212010000400019
Ni Q., Li W. Q., Liang X. F., Liu J. L., Ge H. X., Dong Z. G. (2021). Gill transcriptome analysis reveals the molecular response to the acute low-salinity stress in Cyclina sinensis. Aquacult. Rep. 19, 100564. doi: 10.1016/j.aqrep.2020.100564
Ning L. J., He A. Y., Li J. M., Lu D. L., Jiao J. G., Li L. Y., et al. (2016). Mechanisms and metabolic regulation of PPARα activation in Nile tilapia (Oreochromis niloticus). BBA-Mol. Cell. Biol. L. 1861, 1036–1048. doi: 10.1016/j.bbalip.2016.06.005
Penha-Lopes G., Bartolini F., Limbu S., Cannicci S., Kristensen E., Paula J. (2009). Are fiddler crabs potentially useful ecosystem engineers in mangrove wastewater wetlands? Mar. pollut. Bull. 58, 1694–1703. doi: 10.1016/j.marpolbul.2009.06.015
Ponce-Palafox J. T., Ruíz-Luna A., Gómez M. G., Esparza-Leal H. M., Arredondo-Figueroa J. L., Martinez-Palacios C. A., et al. (2013). A response-surface analysis of the relative importance of the temperature, salinity and body weight on the respiratory metabolism of the white shrimp Litopenaeus vannamei (Boone 1931). Mar. Freshw. Behav. Phy. 46, 399–417. doi: 10.1080/10236244.2013.849058
Qian X., Ba Y., Zhuang Q. F., Zhong G. F. (2014). RNA-Seq technology and its application in fish transcriptomics. Omics. J. Inte. Bio. 18, 98–110. doi: 10.1089/omi.2013.0110
Rahi M. L., Moshtaghi A., Mather P. B., Hurwood D. A. (2018). Osmoregulation in decapod crustaceans: physiological and genomic perspectives. Hydrobiologia 825, 177–188. doi: 10.1007/s10750-018-3690-0
Romano N., Zeng C. S. (2006). The effects of salinity on the survival, growth and haemolymph osmolality of early juvenile blue swimmer crabs, Portunus pelagicus. Aquaculture 260, 151–162. doi: 10.1016/j.aquaculture.2006.06.019
Romano N., Zeng C. S. (2012). Osmoregulation in decapod crustaceans: implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture 334, 12–23. doi: 10.1016/j.aquaculture.2011.12.035
Röthig T., Trevathan Tackett S. M., Voolstra C. R., Ross C., Chaffron S., Durack P. J., et al. (2023). Human-induced salinity changes impact marine organisms and ecosystems. Global. Change. Biol. 29, 4731–4749. doi: 10.1111/gcb.16859
Sang H. M., Fotedar R. (2004). Growth, survival, haemolymph osmolality and organosomatic indices of the western king prawn (Penaeus latisulcatus Kishinouye 1896) reared at different salinities. Aquaculture 234, 601–614. doi: 10.1016/j.aquaculture.2004.01.008
Schroeder H. W. Jr., Cavacini L. (2010). Structure and function of immunoglobulins. J. Allergy Clin. Immun. 125, S41–S52. doi: 10.1016/j.jaci.2009.09.046
Shaughnessy C. A., Baker D. W., Brauner C. J., Morgan J. D., Bystriansky J. S. (2015). Interaction of osmoregulatory and acid–base compensation in white sturgeon (Acipenser transmontanus) during exposure to aquatic hypercarbia and elevated salinity. J. Exp. Biol. 218, 2712–2719. doi: 10.1242/jeb.125567
Shui B. N., Wang Y. J., Lou F. R., Han Z. Q. (2022). Salinity fluctuation on the genetic regulatory mechanisms of the crustacean, Charybdis japonica. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.870891
Soria G., Merino G., von Brand E. (2007). Effect of increasing salinity on physiological response in juvenile scallops Argopecten purpuratus at two rearing temperatures. Aquaculture 270, 451–463. doi: 10.1016/j.aquaculture.2007.05.018
Verween A., Vincx M., Degraer S. (2007). The effect of temperature and salinity on the survival of Mytilopsis leucophaeata larvae (Mollusca, Bivalvia): The search for environmental limits. J. Exp. Mar. Biol. Ecol. 348, 111–120. doi: 10.1016/j.jembe.2007.04.011
Wada K., Murata I. (2000). Chimney building in the fiddler crab Uca arcuata. J. Crustacean. Biol. 20, 505–509. doi: 10.1163/20021975-99990066
Wada K., Watanabe Y., Kamada M. (2011). Function of vertical claw-waving in the fiddler crab Uca arcuata. J. Crustacean. Biol. 31, 413–415. doi: 10.1651/10-3441.1
Wang H., Tang L., Wei H., Lu J., Mu C., Wang C. (2018). Transcriptomic analysis of adaptive mechanisms in response to sudden salinity drop in the mud crab, Scylla paramamosain. BMC. Genomics 19, 1–12. doi: 10.1186/s12864-018-4803-x
Wu L. T., Bian X., Kong L. H., Yin X. X., Mu L. L., Wu S. W., et al. (2019). B cell receptor accessory molecule CD79 gets involved in response against Streptococcus agalactiae infection and BCR signaling in Nile tilapia (Oreochromis niloticus). Fish. Shellfish. Immun. 87, 212–219. doi: 10.1016/j.fsi.2019.01.012
Wu G. Y., Lupton J. R., Turner N. D., Fang Y. Z., Yang S. (2004). Glutathione metabolism and its implications for health. J. Nutr. 134, 489–492. doi: 10.1093/jn/134.3.489
Xiong S. L., Wang W., Kenzior A., Olsen L., Krishnan J., Persons J., et al. (2022). Enhanced lipogenesis through Pparγ helps cavefish adapt to food scarcity. Curr. Biol. 32, 2272–2280. doi: 10.1016/j.cub.2022.03.038
Xu H., Ren M. C., Liang H. L., Ge X. P., Ji K., Huang D. Y., et al. (2022). Interactive effects of water salinity and dietary methionine levels on growth performance, whole-body composition, plasma parameters, and expression of major nutrient metabolism genes in juvenile genetically improved farmed Tilapia (Oreochromis niloticus). Aquaculture 546, 737381. doi: 10.1016/j.aquaculture.2021.737381
Xu J., Yu Y. Y., Huang Z. Y., Dong S., Luo Y. Z., Yu W., et al. (2019). Immunoglobulin (Ig) heavy chain gene locus and immune responses upon parasitic, bacterial and fungal infection in loach, Misgurnus anguillicaudatus. Fish. Shellfish. Immun. 86, 1139–1150. doi: 10.1016/j.fsi.2018.12.064
Yamaguchi T. (2001). Dimorphism of chelipeds in the fiddler crab, Uca arcuata. Crustaceana 913–923. doi: 10.1163/15685400152682665
Yamaguchi T., Henmi Y. (2001). Studies on the differentiation of handedness in the fiddler crab, Uca arcuata. Crustaceana 735–747. doi: 10.1163/156854001317015562
Yamaguchi T., Henmi Y., Ogata R. (2005). Sexual differences of the feeding claws and mouthparts of the fiddler crab, Uca arcuata (De Haan, 1833)(Brachyura, Ocypodidae). Crustaceana 1233–1263. doi: 10.1163/156854005775903528
Yu L., Shen H. J., Ren X. H., Wang A. Q., Zhu S., Zheng Y. F., et al. (2021). Multi-omics analysis reveals the interaction between the complement system and the coagulation cascade in the development of endometriosis. Sci. Rep. 11, 11926. doi: 10.1038/s41598-021-90112-x
Zeng L., Ai C. X., Wang Y. H., Zhang J. S., Wu C. W. (2017). Abrupt salinity stress induces oxidative stress via the Nrf2-Keap1 signaling pathway in large yellow croaker Pseudosciaena crocea. Fish. Physiol. Biochem. 43, 955–964. doi: 10.1007/s10695-016-0334-z
Keywords: Uca arcuata, salinity stress, transcriptional response, RNA sequencing, differentially expressed genes
Citation: Wang B, Sun S, Zhang F and Han Z (2025) Transcriptional response of salinity stress in red claw crab Uca arcuata. Front. Mar. Sci. 12:1575833. doi: 10.3389/fmars.2025.1575833
Received: 14 February 2025; Accepted: 03 March 2025;
Published: 19 March 2025.
Edited by:
Hongyu Ma, Shantou University, ChinaReviewed by:
Sofia Priyadarsani Das, National Taiwan Ocean University, TaiwanCopyright © 2025 Wang, Sun, Zhang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiqiang Han, ZDYzMzkxMjRAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.