
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 March 2025
Sec. Aquatic Physiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1573506
Studying the impact of genotype, tissue type, and their interaction on key sexual factor expression provides valuable insights into the spectrum and consequence of sex differences, particularly for maternal and paternal selection in the breeding of crustaceans. However, the interactions between genotype and tissue types, and their ability to distinguish the sexual differences of mud crab Scylla paramamosain remain unclear. Here we employed additive main effects and multiplicative interaction (AMMI), split-plot analysis (SP), and genotype main effects and genotype × environment interaction (GGE) biplot analysis to examine genotype × tissue interactions related to sexual factors expression of S. paramamosain. SP and AMMI analyses underscored genotype, tissue type, and their interaction can significantly affect sexual factor expression in both female and male crabs. AMMI analysis highlighted that genotype × tissue interactions exerted the most substantial influence on sexual factor expression in the females, while genotype alone played the most critical role in the males. GGE biplot analysis revealed positive correlations between gonad and other tissues, with the ovary and intestine showing the greatest discriminatory power for sexual factors in the females and the males, respectively. Moreover, gene fru2 exhibited relatively high and stable expression in both sexes which would be the preferred breeding index. This comprehensive statistical analysis sheds new light on the intricate relationship between genetics and phenotypes underlying sexual differences in S. paramamosain, offering valuable insights for developing tailored breeding strategies in mud crab parental selection.
Sexual dimorphism, the phenotypic differences between males and females of the same species, is a widespread phenomenon observed across the animal kingdoms (Cui et al., 2021a). Crustaceans, a diverse group of arthropods, exhibit a fascinating array of sexual dimorphism in various aspects, including morphology, behavior, and reproductive strategies (Zhang et al., 2021). One of the most noticeable manifestations of sexual dimorphism in crustaceans is the morphological distinctions between males and females (Farhadi et al., 2021a). These differences can include size, shape, and the development of specialized structures. For instance, male fiddler crabs often have an enlarged claw used in courtship displays and competitions, while female mantis shrimps may display different body shapes and coloration (Baeza et al., 2012). Additionally, male reproductive organs may include specialized appendages or gonopods for transferring sperm, while females may possess modified structures for egg production and release (Mantelatto and Martinelli, 2001). These adaptations reflect the evolutionary pressures associated with efficient reproduction. Courtship rituals, mating behaviors, and parental care duties can vary significantly between the sexes (Kobayashi and Archdale, 2020). In certain species, females may be responsible for protecting and caring for offspring, while males compete for mating opportunities through elaborate displays or aggressive behaviors (Spooner et al., 2007). Besides, varied reproductive strategies and behaviors contribute to the species’ adaptability and resilience in different environments. These different traits allow each sex to better adapt to the environment and occupy a favorable ecological niche (Bear and Monteiro, 2013).
Beyond the differences in appearance, sexual dimorphism is evident in the internal structure like the sexual organ origin from the tissue differentiation (Williams and Carroll, 2009). The cellular processes that drive sexual dimorphism are closely linked to tissue differentiation. In many species, the activation of specific signaling pathways, triggered by sex hormones, directs the development of primary and secondary sexual characteristics. This includes the differentiation of gonadal tissues, which produce hormones that further influence the development of non-reproductive tissues, such as muscle, bone, brain, and adipose tissue (Goossens et al., 2021; Uhl et al., 2022). In these non-reproductive tissues, therefore, a discernible functional dichotomy between sexes was evident. In the crustaceans, furthermore, sex-related genes of mud crab Scylla paramamosain such as Sp-Pol (Farhadi et al., 2021b), Sp-DMRT99B (Zhang et al., 2021), Sp-IAG (Jiang et al., 2020), Sp-Vih (Liao et al., 2020), and Sp-Wnt4 (Farhadi et al., 2021a) exhibited sex-biased expression. This observation suggests that certain tissues, including the gill, stomach, and hepatopancreas of S. paramamosain, display sexual dimorphism in some functions. These findings underscore the possibility that, apart from the gonads, internal tissues such as the gills, liver, and hepatopancreas of S. paramamosain may also manifest sexual dimorphism. Besides crustaceans often display secondary sexual characteristics that are associated with gender-specific genes. Nevertheless, a more in-depth meta-analysis based on these genes or molecular experimentation is imperative to conclusively establish the extent of sexual dimorphism in these organs. Studying the mechanism of this difference and clarifying the role of gene, tissue, and gene × tissue effects in the variation would be of great significance to analyze the genetic mechanism of sexual factors in depth. However, relevant research in this field has not been reported yet.
The mud crab, Scylla paramamosain, belonging to the genus Scylla, exhibits pronounced sexual dimorphism in growth rate, body size, cheliped size, and abdomen morphology (Shi et al., 2019). This sexual dimorphism extends beyond external characteristics to encompass various morphological, behavioral, and physiological traits, collectively known as secondary sexual traits. These traits, ranging from external genitalia to courtship behaviors, enable each sex to thrive in specific environmental conditions and occupy favorable ecological niches (Cui et al., 2021c). Moreover, investigations into gene expression have unveiled sex differences across organs and tissues (Cui et al., 2021b; Wan et al., 2022b; Wu et al., 2020). These findings underscore that the sexual disparities are not confined to external morphology but extend to internal organ and tissue structures. Moreover, the pronounced physiological divergence between male and female reproductive systems of S. paramamosain, particularly the distinct nutrient allocation strategies observed during gonadal maturation phase, presents a critical model for investigating evolutionary adaptations in sexual dimorphism. The elucidation of regulatory mechanisms controlling these sex-specific physiological adaptations - particularly the genotype × tissue interactions - remains fundamental to understanding reproductive fitness in ecologically significant crustacean species. In the context of aquaculture, mud crab females with mature ovaries command a premium in price and nutrition, presenting a lucrative opportunity for mono-sex culture (Waiho et al., 2020). Furthermore, given the conflict between female-biased vitellogenesis efficiency and male-specific spermatophore quality traits, the optimization of parental selection in mud crab aquaculture represents a critical bottleneck in sustainable crustacean domestication.
In this study, an additive main effects and multiplicative interaction (AMMI) (Gollob, 1968), genotype main effects and genotype × environment interaction (GGE) biplot analysis (Khan et al., 2021), and split-plot (SP) design (Jones and Nachtsheim, 2017) were used to analyze genotype × tissue interactions for sexual factors in S. paramamosain. The purpose of this study was to analyze the interactions between genotype and tissue types, and their ability to distinguish the sexual traits, and explore the intricate relationship between genetics and phenotypes underlying the sexual differentiation of S. paramamosain in-depth, and provide a reference for formulating a breeding plan for parental selection of mud crab.
The unmated male S. paramamosain weighing over 250g and female crabs prior to reproductive molting were procured for producing F1 generation following the methodology outlined by our published (Cui et al., 2021a). The offspring larvae were hatched and breed in concrete rearing tanks, they are transferred to isolated ponds to grow into adults when the larvae grow up to crablets. The F2 generation was cultured by the same process as F1 offspring. When F2 generation crablets matured into adults after approximately four months, individuals were randomly sampled. Culture conditions were ambient temperature (almost 30 °C), natural photoperiod, and salinity of approximately 30 ppt. The 30 female mud crabs Scylla paramamosain, with ovaries at stage IV weighing 250g to 300g, and 30 male crabs, with testis at stage II weighing around 200g, were selected. Meanwhile, the 30 wild female crabs and 30 male crabs with the same ovary developmental stages were captured from the inshore by the farmer. All male crabs are mixed into male crab groups and all female crabs are mixed into female crab groups.
The external abdomen morphology was obvious different between male and female S. paramamosain, while the male with narrow and straight abdomen and the female with wide and round abdomen (Shi et al., 2019). Besides, the male crab possesses the bigger cheliped than the female. Inner morphology of female S. paramamosain at stage IV (pre-maturation stage), the ovary increases in size compared to before (covering almost half of the posterolateral stomach) and changes from light yellow to bright orange (Wu et al., 2020). In the male crab, the testis at stage 2 with differentiated vas deferens, primary/secondary spermatocyte formation, but no spermatophores (Mhd et al., 2018). Upon arrival at the laboratory, they underwent an acclimation period at room temperature. The genetic gender was detected using the method we published to determine the consistency of genetic gender and morphological sex (Cui et al., 2021b). Subsequently, tissues including testes, ovaries, heart, hepatopancreas, muscle, gills, thoracic ganglion, intestines, and stomach were promptly dissected and preserved in RNA keeper solution (Vazyme Biotech Co., Ltd., Nanjing, China) at -80°C for RNA analysis.
Total RNA extraction from all samples was performed using the Animal Tissues Total RNA Extraction Kit (Tiangen Biotech Co., Ltd., Beijing, China). Subsequently, RNA integrity was assessed through 1% agarose electrophoresis. Quantification of RNA concentration and purity was conducted using a nucleic acid-protein analyzer (Eppendorf, Hamburg, Germany) and NanoDrop spectrophotometer (A260/A280 ratio ≥ 1.8; A260/A230 ratio ≥ 2.0). The extracted RNA was carefully sealed with a film and stored at -80°C for further use. For cDNA synthesis, the FastKing RT Kit (With gDNase) (TIANGEN Biotech, Beijing, China) was employed. A mixture comprising 1 μg of total RNA, 2 μL of gDNA buffer, and 8 μL RNase-free ddH2O was incubated at 42°C for 3 minutes. Subsequently, 2 μL of 5 × King RT reaction buffer, 1 μL of FastKing RT Enzyme Mix, 2 μL of FQ-RT Primer Mix, and 5 μL of RNase-free ddH2O were added to the reaction system, resulting in a total volume of 20 μL. The PCR conditions were as follows: 42°C for 15 minutes, 95°C for 3 minutes, followed by preservation at 4°C or for subsequent experiments.
Quantitative real-time polymerase chain reaction (qRT-PCR) was employed to assess mRNA expression levels across various tissues. The SYBR Green method (TIANGEN Biotech, Beijing, China) was adopted for qRT-PCR, following these conditions: initial denaturation at 95°C for 3 minutes; followed by 40 cycles of denaturation at 95°C for 5 seconds, annealing at 60°C for 10 seconds, and extension at 72°C for 15 seconds. Specific primers targeting sex-biased genes (fru1-b, fru2, fru1-a, IAG, IR1, and IR2), identified through previous research in our laboratory, along with a housekeeping gene (18S rRNA), are provided in Table 1. All data are expressed as the change with respect to the corresponding 18S Ct level. The 2–ΔΔCt method was used to analyze the expression of genes. Each sample underwent three replicates, and the gray value was expressed as mean ± standard deviation (Supplementary Table S1).
The experiment was structured as a split-plot design (Piepho and Edmondson, 2018), with wherein sex served as the main-plot factor distributed across two main plots within each of the three complete replicate blocks. A split-plot design is characterized by separate random assignments of levels of factors, where levels of some factors are assigned to larger experimental units called whole plots. Each whole plot is divided into smaller units, called split-plots, and levels of another factor are randomly assigned to split-plots (Wang et al., 2022a). Herein, tissue was designated as the sub-plot factor, with eight tissues (ganglion, gonad, stomach, heart, intestine, gill, muscle, and hepatopancreas) assigned to eight sub-plots within each main plot. Sex-related genes constituted the sub-plot factor, with six sexual genes (fru1-b, fru2, fru1-a, IAG, IR1, and IR2) allocated to individual sub-plots within each sub-plot. The split-plot analysis model is expressed as follows:
where: ymnp represents the expression of the m-th of sexual factor for the n-th tissue in the p-th complete block, μ denotes a general intercept, gmn represents the mn-th effect, ap signifies the effect of the p-th block; hmp is the main-plot error associated with the p-th block and m-th sex, assumed to be random with zero mean and variance σ2f, represents a residual sub-plot error with zero mean and variance σ2 (Wang et al., 2022b).
The main feature of the AMMI model combines analysis of variance with principal component analysis (Gollob, 1968; Wang et al., 2023). The AMMI model for the gth genotype (fru1-b, fru2, fru1-a, IAG, Dmrt99B, IR1, IR2, and wnt4) in the eth tissue (ganglion, ovary, stomach, heart, intestine, gill, muscle, testis, and hepatopancreas) defined as following:
where: yge indicates the expression of sexual factor genotype g in tissue e; μ is the grand mean; αg represents the average deviation of genotypes (the average value of each genotype minus the grand average value); βe signifies the average deviation of the tissue (the average of each tissue minus the grand average); λn denotes the eigenvalue of the nth interaction principal component axis (IPCA); γgn represents the genotype principal component score of the nth principal component; δen signifies the tissue principal component score of the nth principal component; N is the total number of principal component axes; θge denotes the residual.
GGE Biplot analysis serves to elucidate intricate interactions among various factors (Yan, 2002; Yan and Holland, 2010; Wang et al., 2023). In this study, the sex-related gene expression data from different tissue experiments were organized into a two-way table, encompassing sex factor and tissue, with each value representing the average expression of corresponding sexual factors in the respective tissue (that is, the phenotype value yge). Singular value decomposition of the first two principal components was utilized to fit the GGE Biplot model (Yan, 2002), expressed as follows:
where: yge denotes the mean expression for genotype g in tissue e; μ represents the grand mean; βe signifies the main effect of tissue e; with μ+βe representing the mean expression across all genotypes in tissue e; λ1 and λ2 denote the singular values (SV) for the first and second principal components (PC1 and PC2), respectively; γg1 and γg2 represent eigenvectors of genotype g for PC1 and PC2, respectively; δe1 and δe2 signify eigenvectors of tissue e for PC1 and PC2, respectively; θge denotes the residual associated with genotype g in tissue e.
The SP, AMMI, and GGE biplot analyses were conducted using the DPS data processing system (Tang and Feng, 2007; Wang et al., 2022a).
The results of the split-plot analysis of variance were listed in Table 2 which showed that the p values of factors tissue, gene, and gene × tissue were 0.0018, 0, and 0 for female, and were 0.0031, 0, and 0 for male, indicating that significant (P < 0.01) impacts of gene, tissue, and gene × tissue interactions on the expression of sexual factors in both female and male crabs (Table 2). The analysis of variance in the SP framework highlighted differentiated F-values for gene, tissue, and tissue × gene effects in both male and female crabs, yet the F-value observed in female crabs surpassed that of male crabs.
The AMMI analysis results demonstrated significant (P < 0.001) effects of gene, tissue, and gene × tissue interactions on the expression of sexual factors in both female and male crabs (Table 3). The analysis of variance revealed that 9.03% of the total sum of squares (SS) for sexual factor expression in females was accounted for by the genotype effect (G), while 39.77% and 50.89% of the total SS were attributed to tissue effects and genotype × tissue interactions, respectively. Additionally, PCA1 was obtained, contributing to 99.50% of the genotype × tissue interaction. For males, 56.76% of the total sum of squares (SS) for sexual factor expression was explained by the genotype effect (G), with 6.68% and 35.24% of the total SS attributed to tissue effects and genotype × tissue interactions, respectively. Similarly, PCA1 contributed to 69.36% of the genotype × tissue interaction.
GGE biplot analysis was conducted based on the mean expression of six sexual factors across eight tissues (Table 4 and Supplementary Table). The interpretation of various GGE biplot figures, including the “relationship among different tissues” (Figure 1), “which-won-where” (Figure 2), “High expression and expression stability” view (Figure 3), and “concentric circles” view (Figure 4), was derived from the conclusions drawn in Table 5. Due to the intrinsic regulatory network architecture, the convergence of expressional profiles was identified during GGE biplot analysis, where genes like IAG, IR1, and IR2 within co-expression modules exhibit synchronized dynamics. This functional coherence leads to indistinguishable clustering patterns in dimensionality-reduced visualizations, as evidenced by identical input expression value and yield mean.
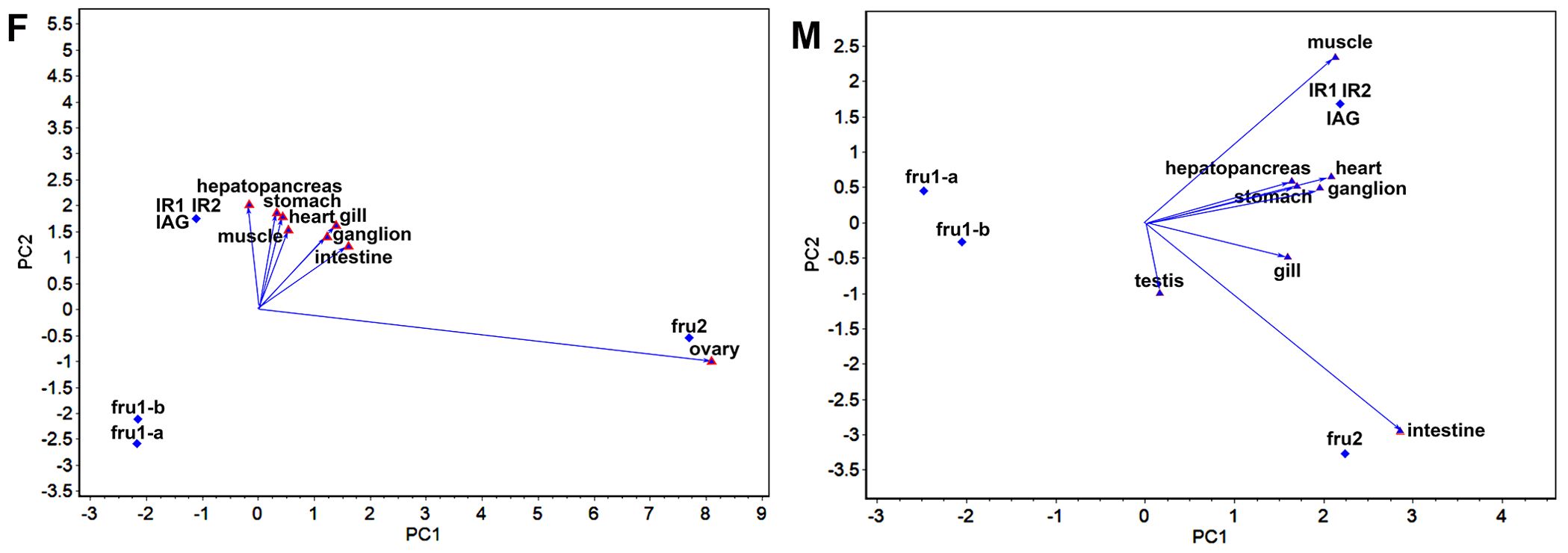
Figure 1. GGE-Biplot of the relationship among different tissues (F) and (M) indicate the GGE-Biplot of the relationship among different tissues for females and males, respectively. The ovary and testis are the crab gonads for females and males.
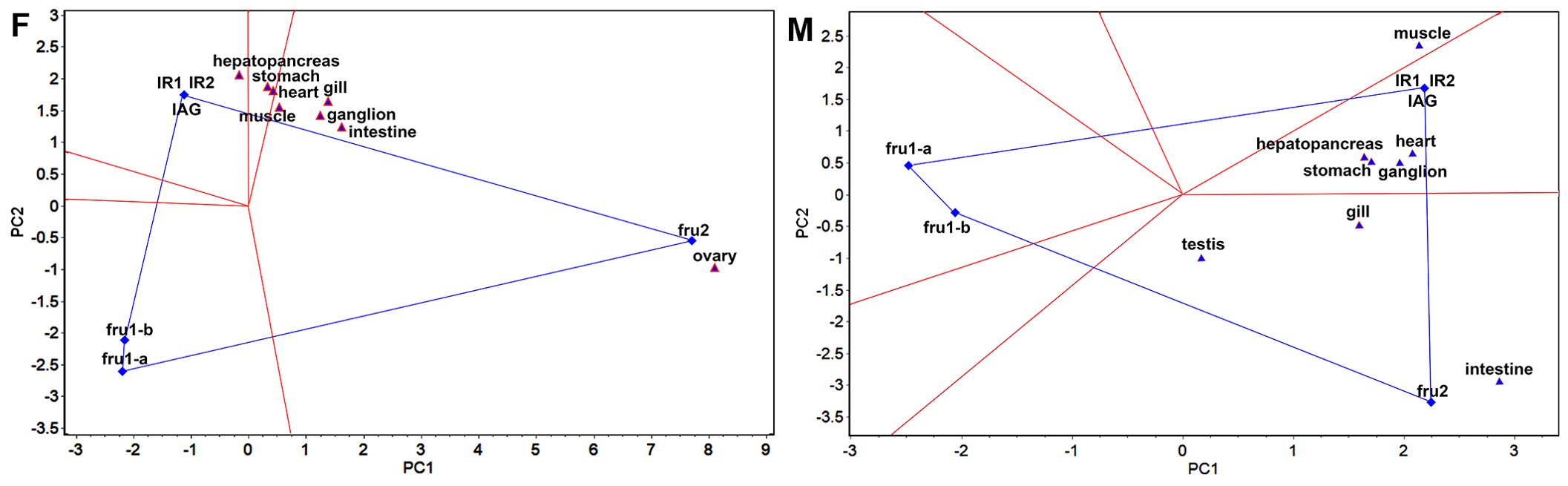
Figure 2. “Which-won-where” view of GGE-Biplot (F) and (M) indicate the “Which-won-where” view of GGE-Biplot for females and males, respectively. The ovary and testis are the crab gonads for females and males.
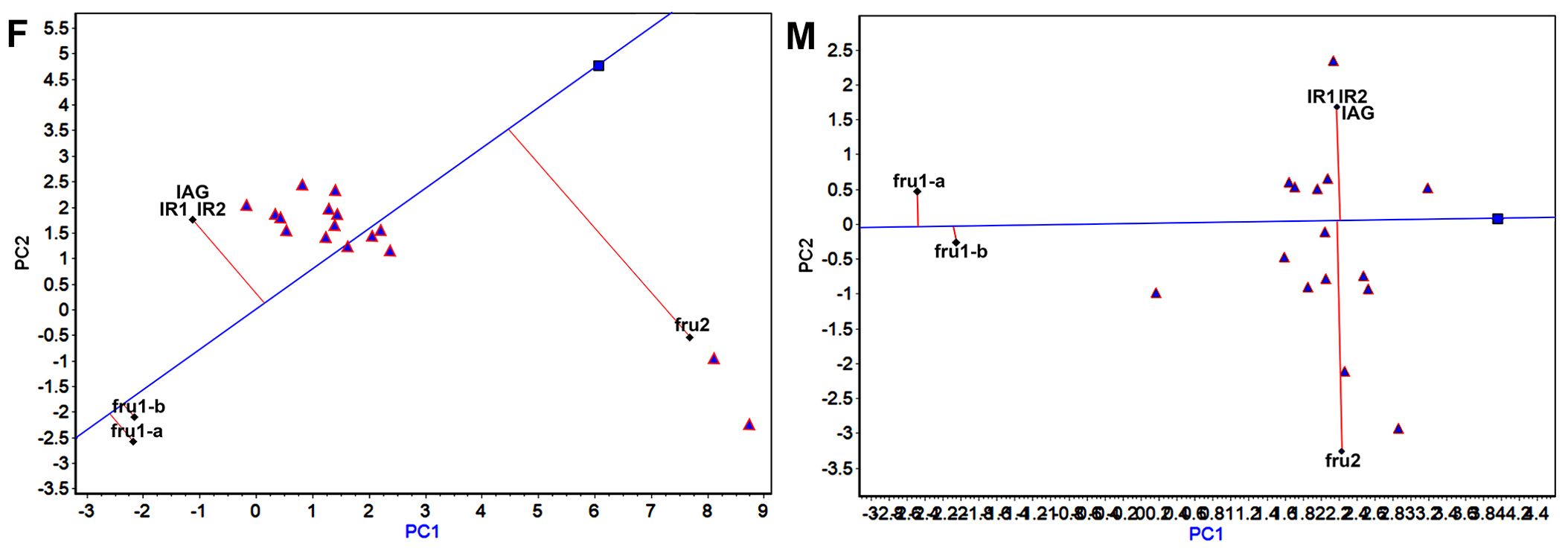
Figure 3. “High expression and expression stability” of sexual factors in GGE-Biplot (F) and (M) indicate the “High expression and expression stability” of sexual factors in GGE-Biplot for females and males, respectively.
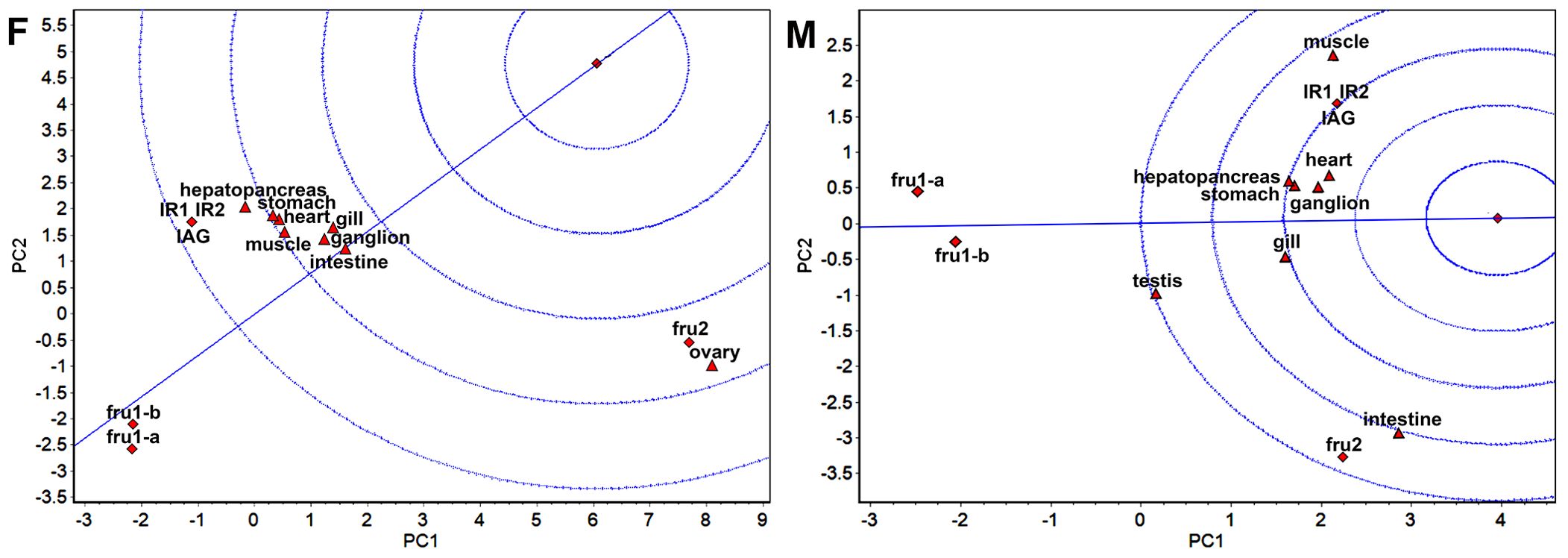
Figure 4. GGE-Biplot with concentric circles (F) and (M) indicate the concentric circles for females and males, respectively. The ovary and testis are the crab gonads for females and males.
The “relationship among different tissues” plot (Figure 1) primarily assesses the similarity of sexual factor expression across different tissues. The cosine of the angle between two-line segments represents the correlation coefficient between the corresponding tissues. An angle less than 90° indicates a positive correlation, signifying similar expression patterns of sexual factors, while an angle greater than 90° indicates a negative correlation, suggesting opposite expression patterns. An angle equal to 90° indicates no correlation. The length of the line segment signifies the tissue’s ability to distinguish sexual factors, with longer segments indicating stronger discriminatory ability. The “relationship among different tissues” plot (Figure 1) highlights distinct patterns in sexual factor expression across various tissues in female and male crabs. In female crabs, the angles between ovary and intestine, muscle, stomach, heart, ganglion, and gill, were less than 90°, indicating a positive correlation among these tissues. Conversely, the angle involving ovary and hepatopancreas exceeded 90°, suggesting a negative correlation. This suggests two distinct expression patterns of sexual factors across the eight tissues in female crabs, with ovary demonstrating the strongest discriminatory ability. In male crabs, the “relationship among different tissues” plot displayed a contrasting pattern. The angles between testis and hepatopancreas, heart, ganglion, stomach, intestine, and gill were less than 90°, indicating a positive correlation. However, the angle between testis and muscle were more than 90°, also indicating negative correlations. Furthermore, intestine and muscle exhibited the strongest discriminatory ability among the eight tissues in male crabs.
In the “which-won-where” view of the GGE biplot (Figure 2), the experimental regions are delineated based on the interaction between sexual factors and tissues, aiming to elucidate the sexual factor with the highest expression level in each region. The sexual factor positioned at the top corner of the polygon within each region signifies the sexual factor with the highest expression in that specific region. In females, the “which-won-where” view revealed that the eight tested tissue locations were consolidated into three regions, hepatopancreas belonging to one area where IR1, IR2, and IAG exhibited the highest expression, and stomach and heart belonging to one area, and muscle, gill, ganglion, intestine, and ovary belonging to one area where fru2 had the highest expression. Similarly, in males, the tested tissue locations were categorized into three distinct regions. The muscle constituted one area, the hepatopancreas, heart, stomach, and ganglion comprised one region where IR1, IR2, and IAG exhibited the highest expression, while testis, gill, and intestine formed a third region where fru2 displayed the highest expression.
In the “High expression and expression stability” view of the GGE biplot (Figure 3), the determination of sexual factors with high and stable expression is facilitated. The transverse oblique line to the right represents the approximate average expression of sexual factors across all tissues. The perpendicular line to the transverse slash indicates the tendency of sexual factor × tissue interaction. Deviation from the transverse oblique line signifies instability in expression. For female crabs, the GGE biplot revealed that the average expression of fru2 was the highest, followed by IR2, IR1, IAG, fru1-a, and fru1-b in descending order. Among these, fru2 exhibited the most unstable expression. Conversely, fru1-a, and fru1-b demonstrated high stability, with fru1-a displaying the highest stability. In male crabs, the average expression of IAG, IR2, and IR1 ranked highest, followed by fru2, with fru1-a at the lowest. The most unstable expression was observed for fru2, while fru1-b exhibited the highest stability.
In the GGE biplot with concentric circles (Figure 4), high expression and expression stability are determined based on the distance of various sexual factors from the central point. Smaller distances indicate higher and more stable expression of sexual factors (Tang and Feng, 2007). The concentric circles biplot depicted that the sexual genes fru2 exhibited high expression and stability in females. Notably, IAG, IR2, and IR1 emerged as the best in terms of expression and stability in males, followed closely by fru2.
The inheritance of the XX or XY set of sex chromosomes in humans largely determines the development of male or female sex-specific traits in adults. However, the mechanisms governing sex determination in lower invertebrates like crabs are still rudimentary compared to vertebrate mammals. Consequently, sex-biased gene expression, even in tissues not directly contributing to sexually dimorphic traits, assumes greater significance in the crab sex system (Oliva et al., 2020). Previous studies have identified several genes, including fru1-b, fru2, fru1-a, IAG, IR1, and IR2, as potential regulators of sex determination/differentiation, influenced by genetic and environmental factors. In our laboratory, fru1-a, fru1-b, and fru2 were identified and named by analyzing the transcriptome data of mud crab. It is speculated that fru1-a, fru1-b, and fru2 may significantly impact the ovary development and maturation of mud crabs, fru1-a may also participate in the development of nerve cells, and fru2 may also have a great impact on the testis development and maturation, fru1-b gene may be involved in the regulation of female crabs’ courtship behavior (Qiu, 2021). Insulin-like androgenic gland hormone (IAG) produced by androgenic gland (AG) in male crustaceans is regarded as a key regulator of sex differentiation (Huang et al., 2014). As a hormone, IAG interacts with its membrane receptor to initiate downstream signal pathways to exert its biological functions. In S. paramamosain, it suggests that the gene IR (insulin-like receptor) is involved in testicular development and plays a crucial role in transitioning from primary to secondary spermatocytes (Liu et al., 2023). Furthermore, our laboratory explored the different function of insulin-like receptor Sp-IR1 and Sp-IR2 gene in ovarian development.
Despite progress, challenges such as limited genome data, technological constraints, the complexity of the sex-determination system, and variability in master genes, hinder a comprehensive understanding of sex determination/differentiation mechanisms in crabs (Zhong et al., 2022). Given this primitive evolutionary state, internal tissues such as ganglion, gonad, stomach, heart, intestine, gill, muscle, and hepatopancreas likely exhibit sexual differences. Moreover, the same sexual factor may exhibit varied expressions across different tissues within the same organism. Therefore, analyzing the expression of key sexual factors across various internal tissues can provide insights into the sexual functions of crabs and aid in parental selection in the breeding.
A prerequisite for advancing sexual manipulation and maternal and paternal selection respectively is a comprehensive understanding of the genetic mechanisms governing sex determination and differentiation, processes that vary considerably across animal phyla (Fang et al., 2020). Studies on the genetic mechanism of crab sex determination or differentiation primarily concentrates on the genetic regulation of sex traits, including genetic interplay among these traits (Cui et al., 2021a, Cui et al., 2021b). Crabs represent prominent macro-fauna, inhabiting the transitional zones between land and sea, particularly in intertidal mangrove areas extending to shorelines (Kathiresan and Bingham, 2001). Within this diverse crab population, various crustacean species exhibit significant sexual dimorphism in growth rate, nutritional requirements, and economic importance (Wan et al., 2022a). Notably, the mud crab Scylla paramamosain has garnered substantial attention in maternal and paternal selection in the breeding process due to its economic significance and pronounced sexual dimorphism. As with most decapod crustaceans, the mud crab displays sexual dimorphism in structure, morphology, and physiology, resulting in considerable differences in economic value between females and males (Yang et al., 2017).
In this study, we employed AMMI and GGE biplot analyses to investigate genotype × tissue interactions concerning sexual factors in S. paramamosain. These analyses, namely SP, AMMI, and GGE biplots, amalgamate principal component analysis (PCA) with graphical representations to elucidate genotype × tissue interactions. The AMMI model, which integrates analysis of variance (ANOVA) and PCA, serves as an efficient tool for assessing genotype stability across multiple tissues, thereby aiding in the identification of genotypes exhibiting high expression levels tailored to specific tissues. Furthermore, the analysis of GGE biplots enhances our comprehension of genotype × tissue interactions, facilitating the identification of representative tissues, assessment of the discriminatory ability of test tissues, and identification of stable genotypes across multi-tissue trials (Shahriari et al., 2018).
The outcomes of the SP and AMMI analyses unveiled that the expression of sexual factors was significantly influenced by genotype, tissue, and genotype × tissue interactions across both female and male specimens. This underscores that the gene expressions of sex-related factors were impacted by sex as well as the specific tissues examined, highlighting sex disparities across tissues. AMMI analysis indicated that genotype × tissue interactions played a pivotal role in driving the total variation in sexual factor expression in comparison to tissue and genotype of females, while genotype is the predominant contribution to the total variation of males. Specifically, for females, 50.89% of the total sum of squares was attributed to the effect of genotype × tissue interactions, whereas for males, this figure stood at 35.24%. These results suggest that the gene expressions of sex-related factors were shaped by dynamic interplay between sexes and tissues for females, but genotype for males. The dominance of genotype × tissue interactions in female crustaceans stems from their reproductive physiology-driven evolutionary constraints, contrasting with male crabs’ behavioral selection pressures. This dichotomy arises from fundamental differences in sex-specific life history strategies and energy allocation patterns (Alava et al., 2007; Zeng et al., 2024). This report about the male crabs diverges from previous studies on traits in rice (Akter et al., 2014), Plantago species (Islam et al., 2014), and fish (Wang and Ma, 2021). In AMMI analysis of these traits, genotype predominantly accounted for 67.11%, 53.57%, and 86.11% of the total variation in grain yield, seed yield, and disease resistance, respectively. However, the female crab possesses the higher price compared to the males. Consequently, in the context of selection, the ranking of genotype × tissue interactions become crucial, as it is determined by both genotypes and tissues. In such scenarios, mitigating or eliminating the effects of tissue and genotype main effects becomes imperative. Therefore, it is imperative to consider the reciprocal interaction between sex and tissue when selecting sexual factors as a basis for breeding endeavors.
GGE biplot analysis revealed notable discrepancies in the ranking of the six sexual factors across the eight tissues both within and between sexes. These variations in tissue correlation between males and females underscore the importance of considering differences between male and female tissues when investigating gene expression patterns of sex-related factors. The GGE-Biplot of the relationship among different tissues illustrated a negative correlation between gonad and hepatopancreas in the female. This is consistent with the implications of lipid transfer from the hepatopancreas to ovaries, alongside lipid accumulation in ovaries, a concurrent decrease in total lipid content is observed in the hepatopancreas throughout vitellogenesis in mud crab and other crabs (Alava et al., 2007; Wouters et al., 2001; Zeng et al., 2024). The GGE-Biplot of the relationship among different tissues indicated a positive correlation between gonad and all tissues except for muscle in males, consistent with previous study in which the correlation between the taste component content and degree of gonad (testis) maturation was low (Chiou and Huang, 2010). Moreover, the ovary exhibited the highest discriminatory ability for the six sexual factors in female crabs, and other tissues suggested similar discriminatory ability, consistent with our knowledge of that crab ovary is the most important sexual organ (Aaqillah-Amr et al., 2018). The intestine and muscle whereas displayed the strongest discriminatory ability in male crabs, which is in accordance with that the male muscle as a major edible part has high nutritional value for amino acid composition with percentages changing differently from the gonad (Wu et al., 2019).
The “which-won-where” view indicated that the eight test tissue locations were consolidated into three regions in the both male and female, with fru2 exhibiting the highest expression in this gonad region. In the “high expression and expression stability” view, the fruitless (fru) gene exhibited both high expression and stability in both females and males. These findings is harmonious with previous study, in which the fruitless (fru) gene plays an important role in the sex determination pathway and courtship behavior of crabs (Li et al., 2017). Stable fru2 expression ensures the integrity of neural circuits governing species-specific mating rituals like claw-waving displays in crabs, and fru2 stabilizes the expression of vitellogenin (yolk protein) in females and spermatophore-binding proteins in males. Its instability may cause asynchronous gonad development or defective gametes (Kimura et al., 2005). The concentric circles biplot illustrated that these sexual genes exhibited comparable expression and stability, with exceptions noted for fru2 in females, where IAG, IR1, and IR2 showcased superior expression and stability in males. These findings are attributed to differences in the expression patterns of sex factors across various tissues, suggesting that the regulatory mechanisms governing sex differ among the eight organs examined. It is speculated that IR families, characterized by high expression and stable properties, play a pivotal role in distinguishing tissue and sex differences of the male crab (Cui et al., 2021d).
Studying the intricate relationship between genetics and phenotypes for sexual difference and clarifying the role of gene, tissue, and gene ×tissue effects in the variation would be of great significance to analyze the genetic mechanism of sexual factors in depth. AMMI, SP, and GGE were used in this study to analyze genotype × tissue interactions for sexual factors in S. paramamosain. The results of AMMI analysis revealed that genotype × tissue interaction was the major contributor to the total variation in sexual factor expression in the females while genotype alone played the most critical role in the males, which indicated that not only the sex factor but also the phenotype should be considered for the parental selection index in the breeding of mud crab. GGE biplot analysis revealed positive correlations between gonad and other tissues, with the ovary and intestine showing the greatest discriminatory power for sexual factors in the females and the males, respectively. This indicated that ovary and intestine should be preferred as the breeding indicators in maternal and paternal selection respectively. Moreover, gene fru2 exhibited relatively high and stable expression in both sexes according to the expression levels of six sexual factors in eight tissues, which indicated that, in general, fru2 would be the preferred breeding index. Therefore, this study provides new insight into the advances of the parental selection index in the breeding of mud crab.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because Crabs belong to the lower vertebrates. Written informed consent was obtained from the owners for the participation of their animals in this study.
WZ: Conceptualization, Investigation, Methodology, Writing – original draft. ZX: Investigation, Writing – review & editing. ZZ: Investigation, Writing – review & editing. WC: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the (National Natural Science Foundation of China) with grant number (42206127), (Guangdong Basic and Applied Basic Research Fund Regional Joint Fund - Youth Fund Project) with grant number (2021A1515110514), (Science and Technology Project of Guangdong Province) with grant number (STKJ2023028), and (STU Scientific Research Foundation) with grant number (NTF22034).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1573506/full#supplementary-material
Aaqillah-Amr M. A., Hidir A., Noordiyana M. N., Ikhwanuddin M. (2018). Morphological, biochemical and histological analysis of mud crab ovary and hepatopancreas at different stages of development. Anim. Reprod. Sci. 195, 274–283. doi: 10.1016/j.anireprosci.2018.06.005
Akter A., Jamil Hassan M., Umma Kulsum M., Islam M. R., Hossain K., Mamunur Rahman M. (2014). AMMI biplot analysis for stability of grain yield in hybrid rice (Oryza sativa L.). J. Rice. Res. 2, 126. doi: 10.4172/jrr.1000126
Alava V. R., Quinitio E. T., De Pedro J. B., Orosco Z. G. A., Wille M. (2007). Reproductive performance, lipids and fatty acids of mud crab Scylla serrata (Forsskål) fed dietary lipid levels. Aquac. Res. 38, 1442–1451. doi: 10.1111/j.1365-2109.2007.01722.x
Baeza J. A., Anderson J. R., Spadaro A. J., Behringer D. C. (2012). Sexual dimorphism, allometry, and size at first maturity of the Caribbean King crab, Mithrax spinosissimus, in the Florida Keys. J. Shellfish Res. 31, 909–916. doi: 10.2983/035.031.0401
Bear A., Monteiro A. (2013). Both cell-autonomous mechanisms and hormones contribute to sexual development in vertebrates and insects. BioEssays 35, 725–732. doi: 10.1002/bies.201300009
Chiou T., Huang J. (2010). Chemical constituents in the abdominal muscle of cultured mud crab Scylla serrata in relation to seasonal variation and maturation. Fish. Sci. 69, 597–604. doi: 10.1046/j.1444-2906.2003.00662.x
Cui W., Fang S., Lv L., Huang Z., Lin F., Wu Q., et al. (2021b). Evidence of sex differentiation based on morphological traits during the early development stage of mud crab Scylla paramamosain. Front. Vet. Sci. 8. doi: 10.3389/fvets.2021.712942
Cui W., Guan M., Sadek M. A., Wu F., Wu Q., Tan H., et al. (2021a). Construction of a genetic linkage map and QTL mapping for sex indicate the putative genetic pattern of the F1 hybrid Scylla (Scylla serrata ♀ × S. paramamosain ♂). Aquaculture 545, 737222. doi: 10.1016/j.aquaculture.2021.737222
Cui W., Yang Q., Zhang Y., Farhadi A., Fang H., Zheng H., et al. (2021c). Integrative transcriptome sequencing reveals the molecular difference of maturation process of ovary and testis in mud crab Scylla paramamosain. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.658091
Cui Z., Liu Y., Yuan J., Zhang X., Ventura T., Ma K. Y., et al. (2021d). The Chinese mitten crab genome provides insights into adaptive plasticity and developmental regulation. Nat. Commun. 12, 2395. doi: 10.1038/s41467-021-22604-3
Fang S., Zhang Y., Shi X., Zheng H., Li S., Zhang Y., et al. (2020). Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus) with implication of an XX/XY sex determination system. Genomics 112, 404–411. doi: 10.1016/j.ygeno.2019.03.003
Farhadi A., Fang S., Zhang Y., Cui W., Fang H., Ikhwanuddin M., et al. (2021a). The significant sex-biased expression pattern of Sp-Wnt4 provides novel insights into the ovarian development of mud crab (Scylla Paramamosain). Int. J. Biol. Macromol. 183, 490–501. doi: 10.1016/j.ijbiomac.2021.04.186
Farhadi A., Shi X., Zhang Y., Zhang Y., Li S., Zheng H., et al. (2021b). A novel imprinted gene (Sp-Pol) with sex-specific SNP locus and sex-biased expression pattern provides insights into the gonad development of mud crab (Scylla paramamosain). Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.727607
Gollob H. F. (1968). A statistical model which combines features of factor analytic and analysis of variance techniques. Psychometrika 33, 73–115. doi: 10.1007/BF02289676
Goossens G. H., Jocken J. W. E., Blaak E. E. (2021). Sexual dimorphism in cardiometabolic health: the role of adipose tissue, muscle and liver. Nat. Rev. Endocrinol. 17, 47–66. doi: 10.1038/s41574-020-00431-8
Huang X., Ye H., Huang H., Yang Y., Gong J. (2014). An insulin-like androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 204, 229–238. doi: 10.1016/j.ygcen.2014.06.002
Islam M., Anisuzzaman M., Khatun H., Sharma N., Islam M., Akter A., et al. (2014). AMMI analysis of yield performence and stability of rice genotype across different haor areas. J. Eco-friendly Agric. 7, 20–24. doi: 10.1016/S0065-2881(01)40003-4
Jiang Q., Lu B., Wang G., Ye H. (2020). Transcriptional inhibition of Sp-IAG by crustacean female sex hormone in the mud crab, Scylla paramamosain. Int. J. Mol. Sci. 21, 5300. doi: 10.3390/ijms21155300
Jones B., Nachtsheim C. J. (2017). Split-plot designs: what, why, and how. J. Qual. Technol. 41, 340–361. doi: 10.1080/00224065.2009.11917790
Kathiresan K., Bingham B. L. (2001). Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 40, 81–251. doi: 10.1016/S0065-2881(01)40003-4
Khan M. M. H., Rafii M. Y., Ramlee S. I., Jusoh M., Al Mamun M. (2021). AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trails (METs). Sci. Rep. 11, 22791. doi: 10.1038/s41598-021-01411-2
Kimura K. I., Ote M., Tazawa T., Yamamoto D. (2005). Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438, 229–233. doi: 10.1038/nature04229
Kobayashi S., Archdale M. V. (2020). Sexual traits and reproductive strategy of the leucosiid crab Pyrhila pisum. J. Mar. Biol. Assoc. UK 100, 939–948. doi: 10.1017/s0025315420000776
Li P., Liu Y., Luo D., Song C., Cui Z. (2017). Two spliced isoforms of the sex-determination gene fruitless in the Chinese mitten crab Eriocheir sinensis. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 208-209, 75–83. doi: 10.1016/j.cbpb.2017.04.008
Liao J., Zhang Z., Jia X., Zou Z., Liang K., Wang Y. (2020). Transcriptional regulation of vih by oct4 and sox9 in scylla paramamosain. Front. Endocrinol. 11. doi: 10.3389/fendo.2020.00650
Liu A., Hao S., Liu F., Huang H., Ye H. (2023). Isolation of an insulin-like receptor involved in the testicular development of the mud crab Scylla paramamosain. Int. J. Mol. Sci. 24, 13639. doi: 10.3390/ijms241713639
Mantelatto F. L. M., Martinelli J. M. (2001). Relative growth and sexual dimorphism of the South Atlantic hermit crab Loxopagurus loxochelis (Anomura, Diogenidae) from Ubatuba, Brazil. J. Nat. Hist. 35, 429–437. doi: 10.1080/002229301300009621
Mhd I., Ghazali A., Nahar S. F., Wee W., Azra M. N., Abol-Munafi A. B. (2018). Testis maturation stages of mud crab (Scylla olivacea) broodstock on different diets. Sains Malaysiana 47, 427–432. doi: 10.17576/jsm-2018-4703-01
Oliva M., Muñoz-Aguirre M., Kim-Hellmuth S., Wucher V., Gewirtz A. D. H., Cotter D. J., et al. (2020). The impact of sex on gene expression across human tissues. Science 369, eaba3066. doi: 10.1126/science.aba3066
Piepho H. P., Edmondson R. N. (2018). A tutorial on the statistical analysis of factorial experiments with qualitative and quantitative treatment factor levels. J. Agron. Crop Sci. 204, 429–455. doi: 10.1111/jac.12267
Qiu B. (2021). Cloning, expression and regulation of fruitless gene in gonadal development of mud crab (Scylla paramamosain). Shantou University. doi: 10.27295/d.cnki.gstou.2021.000557
Shahriari Z., Heidari B., Dadkhodaie A. (2018). Dissection of genotype × environment interactions for mucilage and seed yield in Plantago species: Application of AMMI and GGE biplot analyses. PloS One 13, e0196095. doi: 10.1371/journal.pone.0196095
Shi X., Lu J., Wu Q., Waiho K., Aweya J. J., Fazhan H., et al. (2019). Comparative analysis of growth performance between female and male mud crab Scylla paramamosain crablets: Evidences from a four-month successive growth experiment. Aquaculture 505, 351–362. doi: 10.1016/j.aquaculture.2019.02.062
Spooner E. H., Coleman R. A., Attrill M. J. (2007). Sex differences in body morphology and multitrophic interactions involving the foraging behaviour of the crab Carcinus maenas. Mar. Ecol. 28, 394–403. doi: 10.1111/j.1439-0485.2007.00186.x
Tang Q. Y., Feng M. G. (2007). DPS data processing system: experimental design, statistical analysis and data mining (Beijing: Science).
Uhl M., Schmeisser M. J., Schumann S. (2022). The sexual dimorphic synapse: From spine density to molecular composition. Front. Mol. Neurosci. 15. doi: 10.3389/fnmol.2022.818390
Waiho K., Fazhan H., Zhang Y., Li S., Zhang Y., Zheng H., et al. (2020). Comparative profiling of ovarian and testicular piRNAs in the mud crab Scylla paramamosain. Genomics 112, 323–331. doi: 10.1016/j.ygeno.2019.02.012
Wan H., Zhong J., Zhang Z., Sheng Y., Wang Y. (2022a). Identification and functional analysis of the doublesex gene in the mud crab Scylla paramamosain. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 266, 111150. doi: 10.1016/j.cbpa.2022.111150
Wan H., Zhong J., Zhang Z., Zou P., Wang Y. (2022b). Comparative transcriptome reveals the potential modulation mechanisms of Spfoxl-2 affecting ovarian development of Scylla paramamosain. Mar. Biotechnol. 24, 125–135. doi: 10.1007/s10126-022-10091-6
Wang X., Liu Z., Ma A. (2022a). Interpretation of the genotype by tissue interactions of four genes (AFP1, CIRP, YB-1, and HMGB1) in Takifugu rubripes under different low-temperature conditions. Front. Mol. Biosci. 9. doi: 10.3389/fmolb.2022.897935
Wang X., Ma A. (2021). Dissection of genotype x tissue interactions for immunological factors in turbot (Scophthalmus maximus) infected with Vibrio Anguillarum. Fish Shellfish Immunol. 119, 60–66. doi: 10.1016/j.fsi.2021.09.042
Wang X., Meng Z., Ma A. (2023). Genotype by temperature interaction for plasma physiological indexes in turbot (Scophthalmus maximus) under acute heat stress - Exploring a method for screening physiological biomarkers of nontoxic stress in aquatic environments. Environ. Sci. Technol. 57, 2813–2825. doi: 10.1021/acs.est.2c07958
Wang X., Zhao T., Ma A. (2022b). Genetic mechanism of tissue-specific expression of PPAR genes in turbot (Scophthalmus maximus) at different temperatures. Int. J. Mol. Sci. 23, 12205. doi: 10.3390/ijms232012205
Williams T. M., Carroll S. B. (2009). Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 10, 797–804. doi: 10.1038/nrg2687
Wouters R., Molina C., Lavens P., Calderón J. (2001). Lipid composition and vitamin content of wild female Litopenaeus vannamei in different stages of sexual maturation. Aquaculture 198, 307–323. doi: 10.1016/S0044-8486(01)00522-1
Wu H., Ge M., Zhou X., Jiang S., Lin L., Lu J. (2019). Nutritional qualities of normal and precocious adult male Chinese mitten crabs (Eriocheir sinensis). Aquac. Res. 50, 2267–2275. doi: 10.1111/are.14107
Wu Q., Waiho K., Huang Z., Li S., Zheng H., Zhang Y., et al. (2020). Growth performance and biochemical composition dynamics of ovary, hepatopancreas and muscle tissues at different ovarian maturation stages of female mud crab, Scylla paramamosain. Aquaculture 515, 734560. doi: 10.1016/j.aquaculture.2019.734560
Yan W. (2002). Singular-value partitioning in biplot analysis of multienvironment trial data. Agron. J. 94, 990–996. doi: 10.2134/agronj2002.9900
Yan W., Holland J. B. (2010). A heritability-adjusted GGE biplot for test environment evaluation. Euphytica 171, 355–369. doi: 10.1007/s10681-009-0030-5
Yang X., Ikhwanuddin M., Li X., Lin F., Wu Q., Zhang Y., et al. (2017). Comparative transcriptome analysis provides insights into differentially expressed genes and long non-coding RNAs between ovary and testis of the mud crab (Scylla paramamosain). Mar. Biotechnol. 20, 20–34. doi: 10.1007/s10126-017-9784-2
Zeng X., Li Z., Zhang Z., Shi X., Wang Y. (2024). Variations in lipid composition of ovaries and hepatopancreas during vitellogenesis in the mud crab Scylla paramamosain: Implications of lipid transfer from hepatopancreas to ovaries. Aquacult. Rep. 35, 102008. doi: 10.1016/j.aqrep.2024.102008
Zhang Y., Fang S., Lin F., Li S., Zheng H., Zhang Y., et al. (2021). Identification and characterization of gene Spdmrt99b and its sex-biased expression profile in the mud crab, Scylla paramamosain. J. Ocean. U. China. 20, 1495–1504. doi: 10.1007/s11802-021-4765-5
Keywords: genotype, tissue, genotype × tissue interactions, sexual difference, mud crab
Citation: Zhang W, Xie Z, Zhao Z and Cui W (2025) The dissection of genotype × tissue interactions in mud crab Scylla paramamosain indicates the sexual differences in parental selection. Front. Mar. Sci. 12:1573506. doi: 10.3389/fmars.2025.1573506
Received: 09 February 2025; Accepted: 03 March 2025;
Published: 21 March 2025.
Edited by:
Yafei Duan, South China Sea Fisheries Research Institute, ChinaReviewed by:
Qun Zhao, Hainan University, ChinaCopyright © 2025 Zhang, Xie, Zhao and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxiao Cui, d3hjdWlAc3R1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.