Abstract
Hibernation is a physiological strategy animals use to survive in hostile environments with extreme temperature challenges and food scarcity. During this state, significant changes occur in metabolism and cellular function, with numerous stress response pathways recalibrated to survive physiological challenges that could otherwise be fatal. Numerous studies were performed to explain the molecular mechanisms of mammalian hibernation, but detailed analyses remain scarce in reptiles. Given the limited understanding of the mechanisms regulating hibernation, we performed a comprehensive analysis of liver gene expression in the Chinese softshell turtle (Pelodiscus sinensis) comparing summer active (SA), hibernation (H), and early arousal (EA) states using RNA-sequencing. A total of 435 million high-quality reads were generated, identifying 3,508, 3,607, and 2,993 differentially expressed genes (DEGs) in the SA vs. H, H vs. EA, and EA vs. SA respectively. Gene ontology analysis revealed a shift in metabolic fuel utilization, with the down-regulation of metabolic and cellular processes during hibernation, reflecting a conserved strategy for energy conservation. The transition from hibernation to early arousal was marked by up-regulation of immune-related genes (e.g., CXCL12, ITGA4, PIGR) and endocrine regulators (e.g., CDKN1A, DLL4, IGF1R), facilitating metabolic recovery and cellular protection. Besides, we observed dynamic changes in carbohydrate and lipid metabolism, with down-regulation of hexokinase 2 (HK2) and glucose transporters during hibernation, and up-regulation of lipid metabolism genes (LSS, GPLD1) to support membrane integrity and signaling. Our findings provide insights into the molecular mechanisms underlying hibernation and arousal in ectotherms, with implications for understanding metabolic adaptations, immune regulation, and stress responses in extreme conditions.
1 Introduction
Animals living in the wild must adapt to seasonal variations in ambient temperature and photoperiod (day length), which influence food availability. Ectothermic species must undergo significant physiological adaptations to these seasonal fluctuations (Gautier et al., 2018). As winter approaches, ectothermic vertebrates employ strategies of metabolic compensation or metabolic depression to optimize their fitness in low-temperature environments (Heldmaier et al., 2004; Geiser and Turbill, 2009). Hibernation is the most extreme and efficient adaptive survival strategy employed by various animal species, including mammals and reptiles, to cope with seasonal changes. Several mammalian species have shown evidence of metabolic rate depression during hibernation (Geiser, 2004; Storey and Storey, 2004). The organism can decrease physiological activities (like oxygen consumption and heart rate), and decelerate endogenous fuel reserve depletion, allowing them to enter a hypometabolic state to prolong survival time during winter (Ruf and Geiser, 2015). Low temperature inhibits a wide array of biochemical reactions directly in vivo, but it also affects intracellular processes such as fatty acid synthesis, oxidative phosphorylation, and RNA and protein synthesis by down-regulating these processes (Niu et al., 2024). Hibernation helps preserve species and allows them to expand their distribution, which in turn promotes animal diversity and evolution. Hibernation reportedly increases the chances of survival for these animals (Heldmaier et al., 2004).
To date, large-scale genomic techniques have been used widely in mammals to investigate the molecular and genetic basis of hibernation physiology (Yan et al., 2006; Fedorov et al., 2009). Evidence indicates that many metabolism-related genes are differentially expressed during hibernation. For instance, in hibernating squirrels, genes linked to fatty acid, carbohydrate, lipid, cholesterol, amino acid metabolism, detoxification, molecular transport, and xenobiotic processing were down-regulated in the liver (Srere et al., 1992; Williams et al., 2006; Xiao et al., 2015). A comparison of differentially expressed genes in the liver of active versus hibernating bats revealed that they primarily relate to metabolic depression, shifts in energy source utilization, immune function, and stress responses (Xiao et al., 2015). The DEGs in hibernation are important for the homeostasis of metabolism and absorption of nutrients. For example, during hibernation, genes related to the urea cycle and detoxification are down-regulated in the liver of hibernating the greater horseshoe bat (Rhinolophus ferrumequinum). Key genes involved in body metabolism, specifically the metabolism of pyruvate, glucose, and amino acids, are down-regulated during hibernation. Several core clock genes in the Chinese horseshoe bat (Rhinolophus sinicus) regulate hibernation onset and termination in response to seasonal changes. Different seasonal effects on gene expression have been found in R. sinicus, for instance, which appears to be regulated by several clock genes that initiate and terminate hibernation (Wang et al., 2024). In the liver of the American black bear (Ursus americanus), genes associated with protein biosynthesis and fatty acid catabolism are up-regulated, whereas those concerning lipid biosynthesis and carbohydrate catabolism exhibit a coordinated depletion in mRNA transcription (Capraro et al., 2019). Although these studies have furthered our knowledge of mammalian liver physiology, gene expression in the livers of hibernating turtles in situ has not been examined.
The Chinese soft-shelled turtle (Pelodiscus sinensis) is a major commercial aquatic animal species in Southeast Asia, especially in China, and is considered a tonic food with high nutritional and medicinal value (Jia et al., 2005; Wu et al., 2023). The underlying cause for the high mortality rates of P. sinensis during hibernation is unresolved, despite progress having been made. Recent studies on the hibernation of P. sinensis have mainly considered physiological characteristics and antioxidant responses (Zhang et al., 2017; Tang et al., 2021; Lin et al., 2023), while some transcriptomic work has focused on sexual dimorphism and differentiation in adults (Zhang et al., 2007). The species has a period of hibernation from late October or November to March or April of the following year, affected by yearly temperature changes in its distribution range (Zhang et al., 2007; Hu et al., 2016). However, the molecular basis of these adaptations, especially in liver gene expression regulation during hibernation, is poorly characterized. To fill this gap, this study aimed to examine liver gene expression in P. sinensis in hibernation, with a focus on metabolic adjustments at the molecular level. To investigate the mechanisms of hibernation beginning and organ safety in a facultative hibernator, we conducted unbiased RNA sequencing analysis on the liver of P. sinensis during three phases: summer active (SA), hibernation (H), and early arousal (EA). The current study will provide important insights into the molecular basis of how organisms respond to stress and recover during hibernation.
2 Materials and methods
2.1 Animals
In this study, we captured P. sinensis with an average body weight of 513.04 ± 17.31 g from the wild ponds located in the southeastern area of Weinan city, GPS coordinates: N34°53′33″~34°59′13″, E110°10′23″~110°13′06″, Shanxi province. We particularly sampled six turtles in each physiological state: summer active (SA), mid-July water temperature: 30.7 °C, hibernation (H), mid-January water temperature: 3.9 °C, and early arousal (EA) in mid-March water temperature: 14.8 °C. Before sample collection, we evaluated the turtles’ health and confirmed that they were normal without abnormalities. After anaesthetizing turtles with 100 mg L-1 MS-222 (Sigma, St. Louis, MO, USA) solution, they were euthanized by cervical bleeding. After euthanasia, we collected the liver from each individual, flash-froze it in liquid nitrogen, and stored it at −80 °C until it was needed for RNA extraction. The Experimental Animal Ethics Committee at Hainan Normal University reviewed the handling of the turtles, and they adhered to appropriate guidelines No. HNECEE-2016-005. Our study also attempted to reduce turtle suffering as much as possible.
2.2 RNA extraction and sequencing
Total RNA was extracted using TRIzol® Reagent by the manufacturer’s instructions. RNA quality determination was done on 5300 Bioanalyzer (Agilent) and quantification was completed by using the ND-2000 (NanoDrop Technologies). The library was constructed using the Illumina® Stranded mRNA Prep, Ligation kit from Illumina (San Diego, CA). The cDNA library of samples (n=3) that met quality requirements (Total RNA ≥ 1 μg, concentration ≥ 35 ng/μL, OD260/280 = 1.8~2.2; OD260/230≥2.0) was generated.
2.3 Library preparation and illumina sequencing assembly
RNA purification, reverse transcription (RT), library construction and sequencing of the cDNA library were completed (n=3) at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd (Shanghai, China) according to the manufacturer’s protocol (Illumina, San Diego). The preparation of the Liver RNA-seq transcriptome library was done through the Illumina® Stranded mRNA Prep, Ligation kit from Illumina (San Diego, CA) and 1 μg of total RNA was used. In summary, Messenger RNA was isolated by the polyA selection method with oligo(dT) beads and fragmented by the fragmentation buffer. Double-strand cDNA was synthesized with a random hexamer primer (Illumina) method by using a SuperScript double-stranded cDNA synthesis kit from Invitrogen, CA. A cDNA was generated according to Illumina’s library construction method, which included end-repair, phosphorylation, and the addition of an ‘A’ base. Libraries were made with 2 % Low Range Ultra Agarose and 300 bp cDNA fragments were amplified with Phusion DNA polymerase (NEB), performing 15 PCR cycles. Following the quantitative analysis of the paired-end RNA-seq library by Qubit 4.0, the paired-end library was then sequenced using the NovaSeq X Plus (2 × 150 bp read length). The raw paired-end reads for quality control and mapping were trimmed using default parameters, followed by quality control with Fastp (Chen et al., 2018). HISAT2 (Kim et al., 2019) software was used for aligning clean reads to the reference genome by orientation mode. The mapped reads of each sample for the reference-based model were assembled by StringTie (Pertea et al., 2016).
2.4 Differential expression analysis and functional enrichment
Unigenes were annotated based on the databases of GO, SwissProt, KEGG, COG, NR, and Pfam. Differentially expressed genes (DEGs) were screened by the DESeq2 package for R based on a negative binomial distribution model. To identify the DEGs among two different samples, we calculated the expression level of each transcript, utilizing the output of transcripts per million (TPM) method. Gene abundances were quantified with RSEM (Li and Dewey, 2011). Differential expression analysis was performed using DESeq2 (Love et al., 2014) R package (1.34.0). DEGs with |Log2Fold Change| ≥ 1 and FDR < 0.05 were considered significantly differentially expressed. Functional enrichment analysis was performed to determine which DEGs were significantly enriched in GO terms (GO and KEGG) with a Bonferroni-corrected p-value < 0.05 in comparison to the whole transcriptome context.
2.5 qRT- PCR
To authenticate our measurements, we conducted qRT-PCR to examine the relative mRNA expression levels of 12 randomly selected genes in the transcriptome sequencing during hibernation, and we used the β-actin gene as the housekeeping gene (Supplementary Table S1). Primers were designed specifically and compared with sequences from different species and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). By the manufacturer’s guidelines, we performed qRT-PCR using the SYBR® PrimeScript™ Kit (Takara, Tokyo, Japan). Briefly, the PCR reactions were executed in triplicate for each sample, in 20 µL each consisting of 0.4 µL of 5× FastPfu Buffer, 2 µL of 2.5 mM dNTPs, 0.8 µL of each primer (5 µM), 0.4 µL of FastPfu polymerase, and 10 ng of template DNA. Amplification was performed using the following thermal cycler program: 5 min at 95°C; 40 cycles of 30 s at 95°C, 30 s at 55°C, and 35 s at 72°C; followed by a final extension of 8 min at 72°C. Gene relative expression levels in mRNA were evaluated by the 2−ΔΔCt method. All raw sequencing data are deposited to the NCBI Sequence Read Archive (SRA) under the accession number PRJNA1056536.
2.6 Statistical analysis
All experiments were performed in triplicate, the results were presented as the means ± SEM and analyzed using SPSS 24.0 and R software packages. Differential expression analysis was conducted using ANOVA and was visualized as a volcano plot using the “EnhancedVolcano” package (version 1.6.0). The GraphPad Prism 10.0 (GraphPad, San Diego, CA) and Origin Pro 2024b (10.15) software were used for graphing. A Venn diagram and PCoA were performed using the vegan package. GO functional enrichment analysis was performed with Goatools, while KEGG pathway analysis was carried out by employing Python’s SciPy software. A significance level of p < 0.05 was considered statistically significant.
3 Results
3.1 De novo transcriptome assembly
This study examined transcriptional changes in the liver during the hibernation–arousal cycle by employing RNA sequencing analysis of the SA period compared to H and EA. A total of 444.58 million reads and 67.13 billion bases were retrieved from the liver transcriptome of P. sinensis, with 169.33 million reads from SA, 132.94 million from H, and 142.30 million from EA. After filtering out low-quality reads and adapter sequences, we retained 435.01 million clean reads, with 164.07 million from SA, 130.84 million from H, and 140.10 million from EA. The GC contents of the clean reads were 49.44% for SA, 50.49% for H, and 49.57% for EA. The Q30 scores of the reads exceeded 94% across all groups. The data qualities from each sample are shown in Supplementary File S1; Supplementary Table S2.
Among the three groups, the comparisons of DEGs yielded the following results: SA vs. H: 3508 DEGs (1586 up-regulated, 1922 down-regulated). H vs. EA: 3607 DEGs (2283 up-regulated, 1324 down-regulated). EA vs. SA: 2993 DEGs (1836 up-regulated, 1157 down-regulated). Notably, the shift from H to EA was primarily marked by the up-regulation in gene expression during the EA period. A total of 5705 upregulated DEGs were found between SA, H, and EA, and 4119 were up-regulated in the EA stage compared to SA, indicating an overexpression of genes associated with arousal (Figure 1).
Figure 1
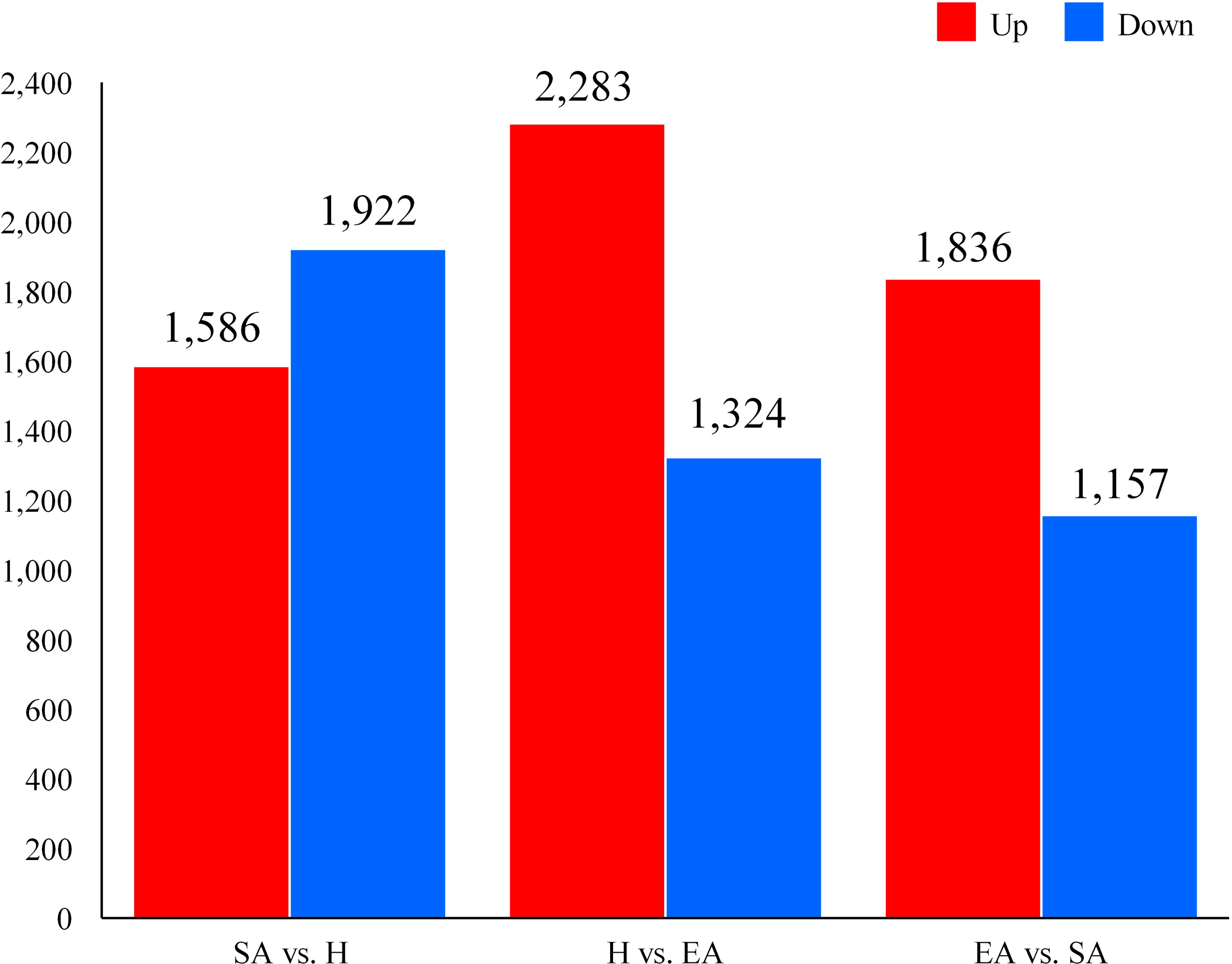
The differential expression of DEGs in the liver of Pelodiscus sinensis.
3.2 Venn diagram of DEGs and principal component analysis of samples
A sum of 13724 genes were obtained from the SA, H, and EA groups, of which 11009 (80.22%) DEGs were shared in all three groups, while 1279 genes were uniquely expressed in the H, SA, and EA groups, and 393 DEGs were shared between H vs. EA, 274 between SA vs. EA, and 769 between H vs. SA (Figure 2A). A principal component analysis (PCA) was used to characterize the compositional relationships among samples, and the results showed that SA, H, and EA were found to overlap with each other, demonstrating a good overall similarity of the biological repeats of the samples. The sum of the variability explained by the first two principal components PCs, PC1 (30.71%) and PC2 (16.59%), reached 47.30%. There were two main factors responsible for influencing P. sinensis liver changes during hibernation. Among them, the SA and EA phases overlapped, which might imply that the P. sinensis turtle shared some similar biological processes in these two phases. Additionally, the points of H and SA were adjacent, and the axis of PC1 mainly separated these two groups (Figure 2B).
Figure 2
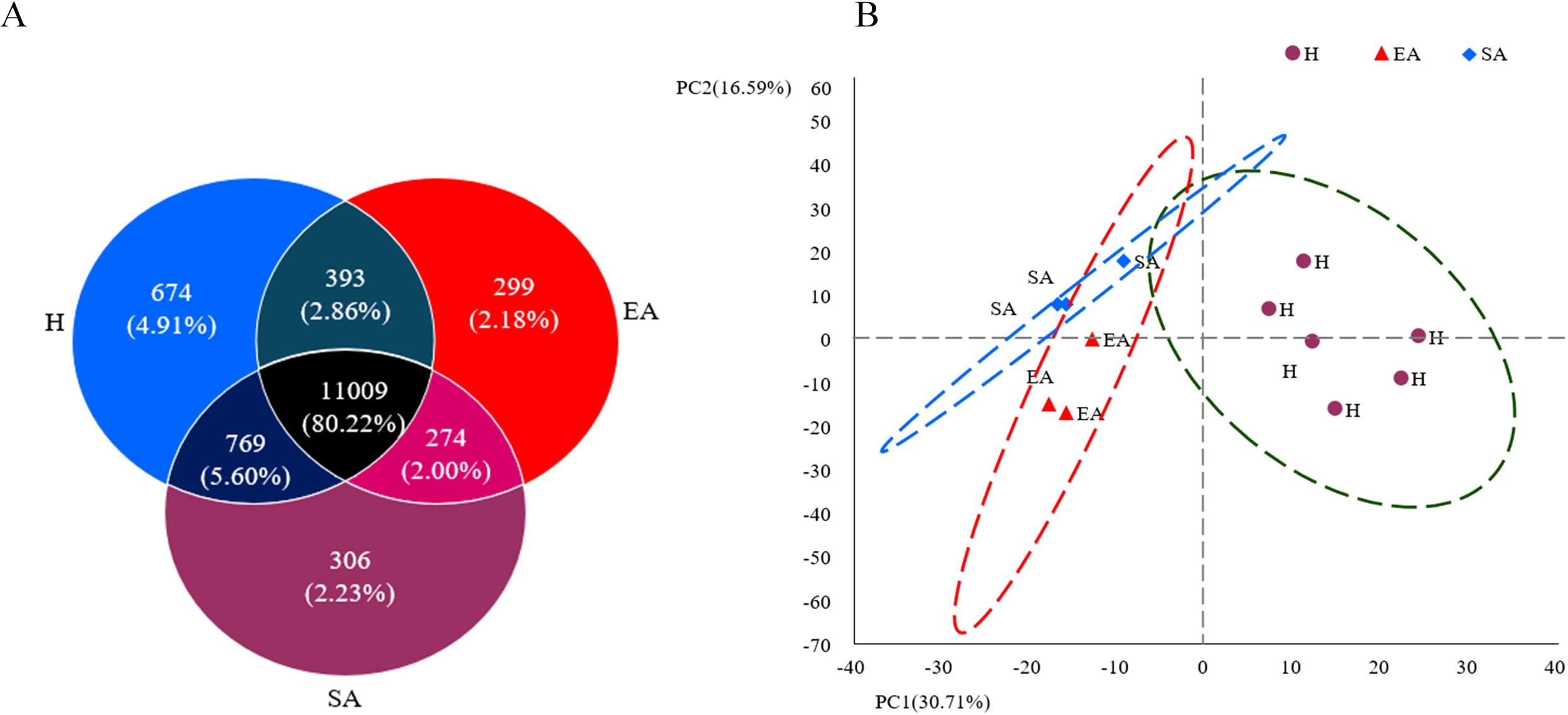
Venn diagram of DEGs and Principal component analysis of samples (A) Venn diagram (B) principal component analysis.
3.3 Gene ontology classification and annotation of DEGs
A GO functional enrichment analysis along with pathway annotation was conducted to comprehend the functions of the differentially expressed genes. We identified 146 statistically significant GO terms, including 1586 up-regulated and 1922 down-regulated DEGs in SA vs. H, 2283 up-regulated and 1324 down-regulated DEGs in H vs. EA, and 1836 up-regulated and 1157 down-regulated) DEGs in EA vs. SA group. Results were categorized into three categories: biological process, molecular function, and cellular component.
In the biological process category, the SA vs. H group showed that the proportion of down-regulated genes in metabolic processes, cellular processes, cellular component organization, and biological regulation was higher than the proportion of up-regulated genes, indicative of a repressed metabolism during hibernation. In addition, response to stimulus, the proportion of down-regulated genes was higher than the up-regulated genes in the SA vs. H group. In the category of cellular component, the proportion of up-regulated genes linked to the membrane part, cell part, and organelle part was also lower than that of down-regulated genes. Furthermore, a high percentage of down-regulated genes in the torpid liver were associated with the “transcription regulator activity” function, suggesting that transcription may be depressed during hibernation. Catalytic activity was the only molecular function category with a higher proportion of up-regulated genes than down-regulated genes (Figure 3A; Supplementary File S2).
Figure 3
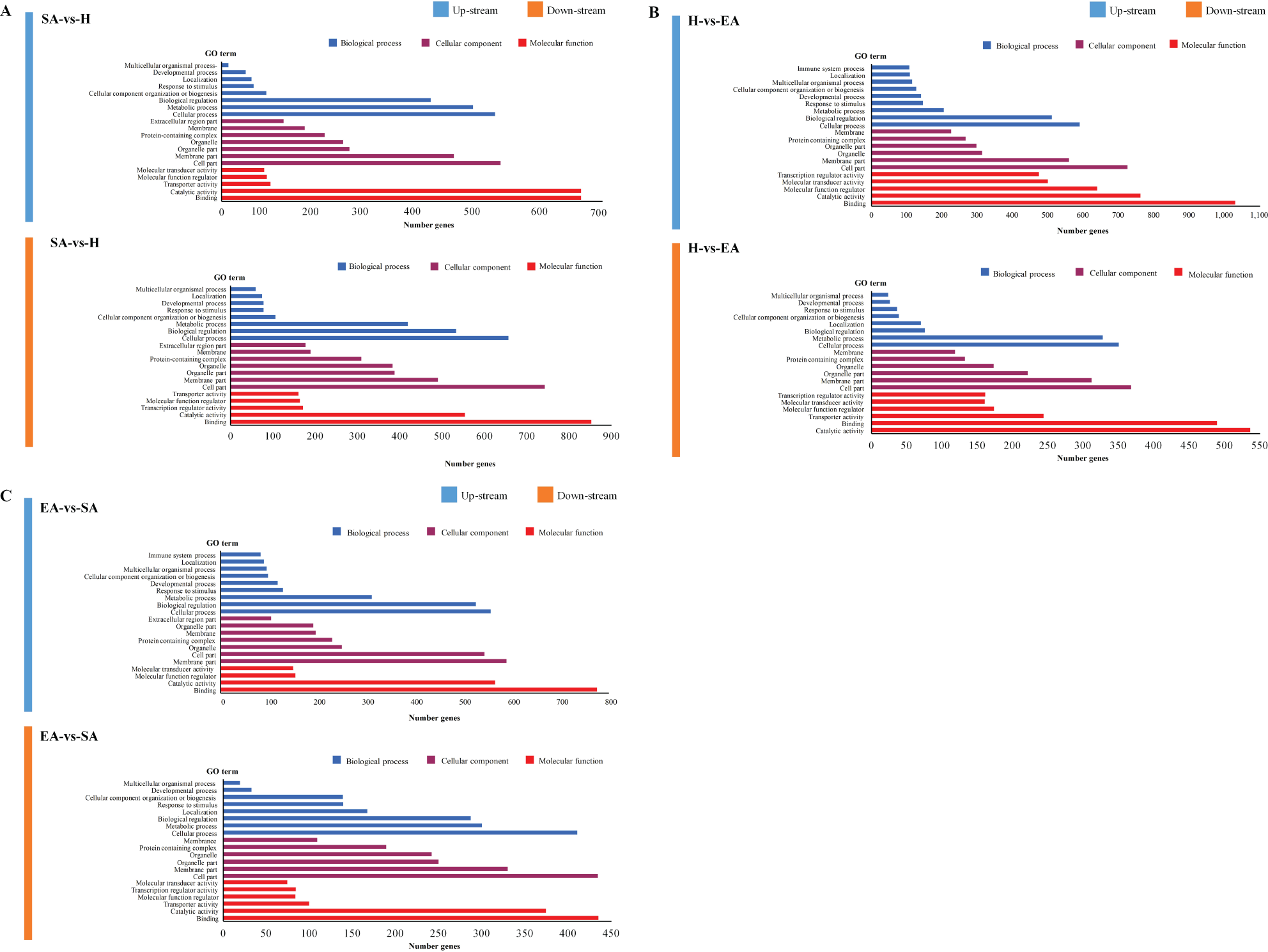
GO categories in the comparison of SA vs. H (A), H vs. EA (B), and EA vs. SA (C). All genes were divided into several functional groups within three categories: cellular component, molecular function, and biological process.
In the H vs. EA and EA vs. SA groups, the proportion of up-regulated genes involved in cellular process, and biological regulation were greater than down-regulated genes in the biological process category. Unlike the cellular process, the proportion of genes down-regulated in the metabolic process was greater than that of up-regulated genes in both the H vs. EA and EA vs. SA groups. Proportions of up-regulated genes associated with the “immune system process” “response to stimulus” “locomotion” and “developmental process” were larger than down-regulated genes during the EA and SA periods. Up-regulated terms related to cellular functions such as “cell part”, “organelle”, and “membrane part” had a higher GO percentage than down-regulated terms. There were higher numbers of differentially expressed genes associated with molecular functions including ‘binding’ and ‘catalytic activity’, as measured by GO percentages. Contrary to this, a large number of genes were down-regulated in the only H vs. EA group associated with the function of ‘binding’ and ‘catalytic activity’ (Figures 3B, C; Supplementary File S2).
3.4 GO enrichment analysis of DEGs
The data from the GO enrichment analysis indicated that in the top GO enrichment pathways among the SA vs. H group, the up-regulated DEGs were mostly enriched with the NADH dehydrogenase (ubiquinone) activity (GO: 0008137), alpha-amino acid metabolic process (GO: 1901605), while the down-regulated DEGs were found to be enriched with the series of metabolic and biosynthetic process such as negative regulation of metabolic process (GO: 0009892), negative regulation of nitrogen compound metabolic process (GO: 0051172), and positive regulation of macromolecule biosynthetic process (GO: 0010557), positive regulation of cellular biosynthetic process (GO: 0031328) (Figures 4A, B; Supplementary File S3).
Figure 4
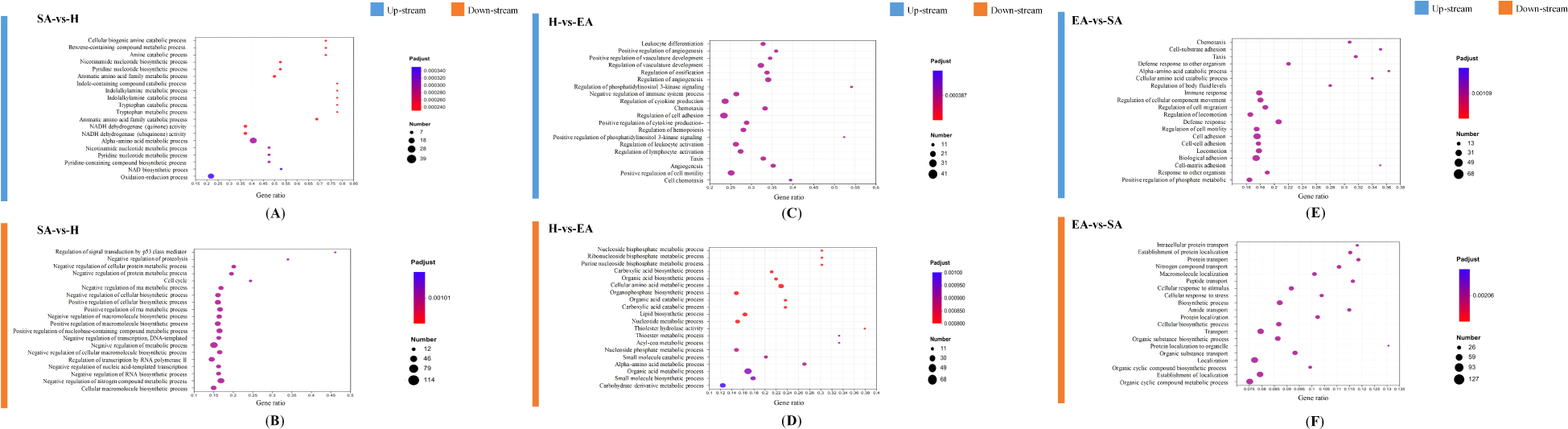
GO enrichment terms of differentially expressed genes in SA vs. H (A, B), H vs. EA (C, D), and EA vs. SA (E, F). The x-axis indicates the number of genes, and the y-axis indicates the second-level GO terms. The x-axis shows the ratio of differentially expressed genes in each term relative to the total number of genes in that term. Dot size shows the number of differentially expressed genes in that term and color is the gene set enrichment analysis test statistic with Benjamini–Hochberg adjustment for multiple testing.
In the H vs. EA group, the up-regulated DEGs were functionally linked with cell differentiation and cell development-related pathways, such as regulation of cell adhesion (GO: 0030155), regulation of vasculature development (GO: 1901342), regulation of cytokine production (GO: 0001817), and positive regulation of cell motility (GO: 2000147). The down-regulated DEGs in the H vs. EA group enriched a series of metabolism-related pathways including the cellular amino acid metabolic process (GO: 0006520), organic acid metabolic process (GO: 0006082), and carbohydrate derivative metabolic process (GO: 1901135) (Figures 4C, D; Supplementary File S3). Down-regulation of metabolic and energy derivative pathways, considering an enrichment between these pathways it is clear that metabolic and energy processes are driving the differential expression of genes throughout different stages of hibernation.
In the EA vs. SA, the up-regulated DEGs were functionally classified into immune-related and transportation pathways, including immune response (GO: 0006955), defense response (GO: 0006952), biological adhesion (GO: 0022610), and, regulation of cellular component movement (GO: 0051270), while the down-regulated DEGs mainly involved in pathways related to transport and localization including, localization (GO: 0051179), establishment of localization (GO: 0051234), transport (GO: 0006810) and organic cyclic compound metabolic process (GO: 1901360) (Figures 4E, F; Supplementary File S3). Up-regulation of enriched DEGs related to immune-related and transportation pathways indicating the turtles were prepared to come out of hibernation.
3.5 KEGG pathway enrichment analysis of DEGs and Pearson’s correlation coefficients
The Kyoto Encyclopedia of Genes and Genomes (KEGG pathway analyses) were used to determine the functions of the differentially expressed genes. Three hundred forty-three pathways were identified with a KEGG pathway annotation. The enriched KEGG level 1 pathway analyses were related to genetic information processing, metabolism, cellular processes, organismal systems, human diseases, and environmental information processing. Pathways in environmental information processing (signal transduction), organismal systems (endocrine system, immune system), and metabolism (lipid metabolism, carbohydrate metabolism) were the most significantly enriched KEGG pathways related to hibernation (Figure 5). The enrichment analysis associated with DEGs between specific hibernation stages was performed (Supplementary File S1). Pearson’s correlation coefficients were first used to test for biologically repeated correlations between samples. The generated cluster dendrogram was used to observe the overall correlation of the transcriptomes from the SA vs. H, H vs. EA, and EA vs. SA groups. Three biological replicates of liver samples from each period and the transcriptome data both exhibited good correlation (Supplementary Figure S1).
Figure 5
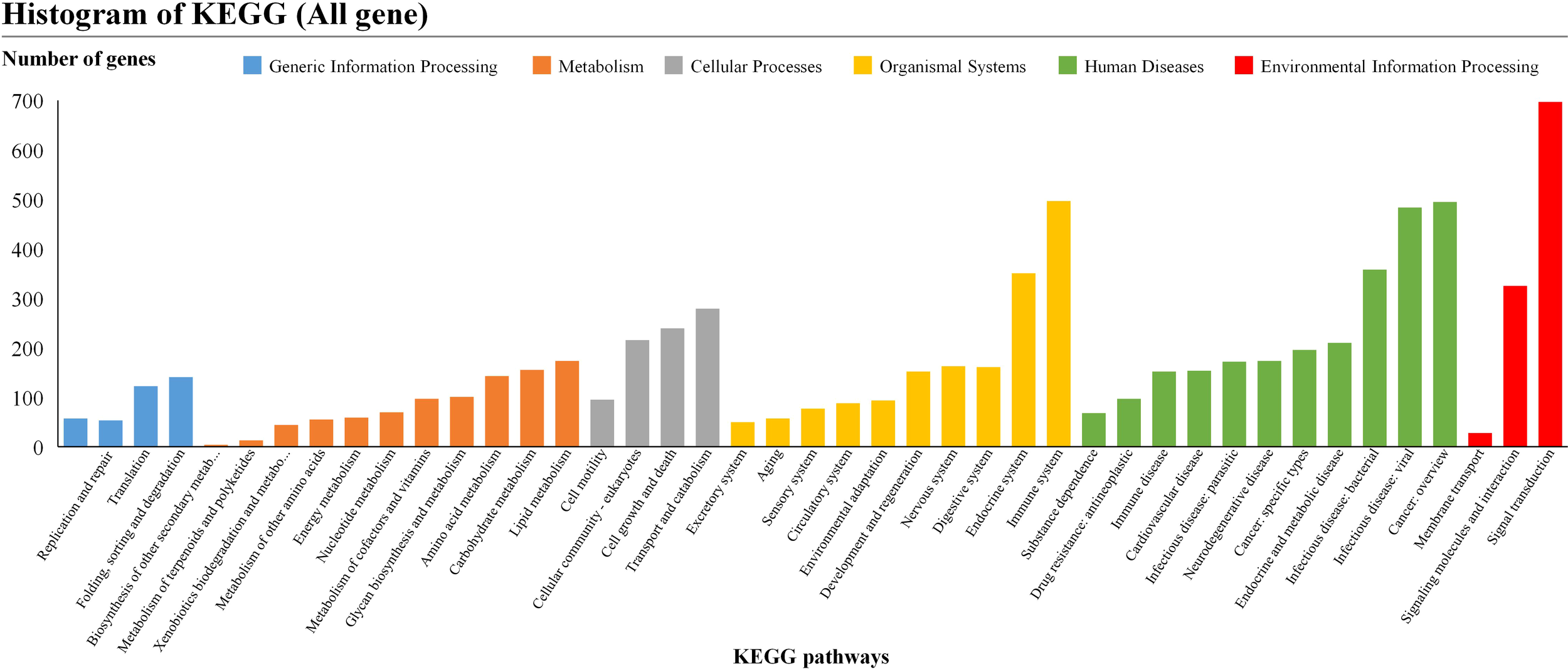
KEGG pathway enrichment analysis of differentially expressed genes. Pathways belonging to different classifications, including “Cellular Processes”, “Environmental Information Processing”, “Genetic Information Processing”, “Human Diseases” and “Metabolism”, were listed on the right of the plot.
3.6 qRT-PCR validation
The findings of RNA-seq were confirmed using qRT-PCR analysis of 12 randomly selected genes including SRD5A2, ELOVL6, GPX3, GNPNAT1, PDHA1, CAT, LOC10, MAP3K8, CEBPB, HSPA8, FOS, and SOCS. These genes are involved in the energy production metabolism and immune system metabolism. Consistent with transcriptome sequencing data, qRT-PCR analysis confirmed the precision of RNA-Seq findings (Supplementary Figure S2).
3.7 Expression difference analysis of carbohydrate, lipid, immune and endocrine-metabolism and pathway-related genes
Genes regulated in different metabolisms were examined further and categorized into four subtypes: carbohydrate metabolism, lipid metabolism, endocrine metabolism and immune metabolism (Table 1).
Table 1
| Gene Name | Description | Log2 (Fold Changes) | ||
|---|---|---|---|---|
| SA vs H | H vs EA | EA vs SA | ||
| Lipid-metabolism | ||||
| ACACA | acetyl-CoA carboxylase alpha | -2.15 | ||
| CD74 | CD74 molecule | 1.35 | ||
| GPLD1 | glycosylphosphatidylinositol specific phospholipase D1 | 1.59 | 2.11 | |
| INPP5D | inositol polyphosphate-5-phosphatase D | 1.38 | ||
| KDELR2 | KDEL endoplasmic reticulum protein retention receptor 2 | -1.35 | ||
| LSS | lanosterol synthase | 2.43 | 1.75 | |
| SERAC1 | serine active site containing 1 | -2.19 | ||
| Carbohydrate-metabolism | ||||
| HK2 | hexokinase-2 | -3.15 | ||
| LOC102447193 | phosphatidylinositol 3-kinase regulatory subunit gamma | -1.32 | ||
| LOC102447194 | solute carrier family 2, facilitated glucose transporter member 5 | 3.32 | ||
| LOC102447195 | solute carrier family 2 member 2 | 4.26 | ||
| Immune-metabolism | ||||
| CXCL12 | C-X-C motif chemokine ligand 12 | 4.35 | 3.32 | |
| FAS | Fas cell surface death receptor | 1.92 | ||
| ITGA4 | integrin subunit alpha 4 | 2.06 | 2.46 | |
| PIGR | polymeric immunoglobulin receptor, transcript variant X2 | 2.18 | 1.68 | |
| TNFRSF13C | TNF receptor superfamily member 13C | -2.87 | 4.62 | |
| CD28 | CD28 molecule | -1.68 | 2.03 | |
| ICOSLG | inducible T cell costimulator ligand | 1.92 | ||
| Endocrine-metabolism | ||||
| ADCY6 | adenylate cyclase 6 | 1.03 | -1.46 | |
| ADCY9 | adenylate cyclase 9, transcript variant X1 | -1.33 | 2.03 | |
| ATP2B4 | ATPase plasma membrane Ca2+ transporting 4, transcript variant X5 | 2.19 | ||
| CDKN1A | cyclin dependent kinase inhibitor 1A | 3.10 | 2.77 | |
| NOTCH3 | notch 3 | -2.03 | ||
| SOS2 | SOS Ras/Rho guanine nucleotide exchange factor 2 | -1.13 | 1.19 | |
| BAD | BCL2 associated agonist of cell death | -1.11 | -1.07 | |
| DLL4 | delta like canonical Notch ligand 4 | 1.62 | 1.58 | |
| GRB2 | growth factor receptor bound protein 2, transcript variant X1 | 1.36 | ||
| IGF1R | insulin like growth factor 1 receptor | 2.68 | 2.01 | |
Fold changes of differentially expressed genes regulated in different mediating pathway in the SA vs. H, H vs. EA, and EA vs. SA groups.
The sharpest response of endocrine and immune system pathway-related genes occurred in the H vs. EA and SA vs. EA groups, with the maximum number of up-regulation DEGs. As shown in Table 1. The endocrine system-related responses were most intense during hibernation, with greater up-regulation of cyclin-dependent kinase inhibitor 1A (CDKN1A), delta-like canonical Notch ligand 4 (DLL4), and insulin-like growth factor 1 receptor (IGF1R), while immune-related response includes chemokine ligand 12 (CXCL12), Fas cell surface death receptor (FAS), integrin alpha-4 (ITGA4), polymeric immunoglobulin receptor (PIGR), tumour necrosis factor receptor superfamily member 13C (TNFRSF13C) genes in the H vs. EA, and SA vs. EA group, respectively. Few endocrine and immune system-related genes were differentially expressed in the SA vs. H and H vs. EA groups (Table 1). The CDKN1A, DLL4, and IGF1R are directly involved in key regulatory processes like cell cycle control, metabolic processes, signaling, growth, and stress responses. Like CDKN1A inhibiting cyclin-dependent kinases mediate cell cycle arrest in response to stress, DLL4 is effective in Notch signaling to regulate cell differentiation, and IGF1R plays a pivotal role in growth and development through insulin-like growth factor signaling. All of which are crucial for survival during hibernation. Their up-regulation likely presents the body’s efforts to conserve energy and manage cellular stress during the hibernation period, when metabolism is slowed, and growth processes are minimal.
Moreover, the up-regulation of immunity-related genes such as CXCL12, FAS, ITGA4, PIGR, and TNFRSF13C was also observed during hibernation. The up-regulation of these immunity-related genes during hibernation suggests that, although the metabolic activity was decreased, the immune response was not completely inactive during the hypometabolic state. That is, instead of retreating into torpor, it is actively keeping a lookout for potential threats. This suggests that the organisms had activated functions to make sure their immune system was functional and could respond as soon as hibernation ended, which is essential to survive in an environment where infections or injuries could happen by the end of hibernation, regulating important immune processes such as CXCL12 and ITGA4, and other processes including apoptosis of damaged cells, mucosal defense, and B cell activation. This immune response could help to maintain a balance between energy conservation and the defense against possible threats at a time of prolonged metabolic suppression of the organism.
In the case of carbohydrate-mediated pathway-related genes solute carrier family 2 (LOC102447195), and solute carrier family 2 (LOC102447194), they had their highest response in only the SA vs. H comparison. The hexokinase 2 (HK2) and the carbohydrate-mediated pathway phosphatidylinositol 3-kinase regulatory subunit gamma (LOC102447193), were significantly down-regulated during the SA period. Among SA vs. H group down-regulated genes were, HK2 and LOC102447193. HK2 is an enzyme that phosphorylates glucose to produce glucose-6-phosphate. The down-regulation of hexokinase during hibernation is beneficial because it limits the entry of glucose into glycolysis and reduces energy consumption. This metabolic alteration helps to conserve glucose and other energy reserves, allowing the hibernating animal to maintain a low metabolic rate and survive for extended periods without food. Similarly, SLC2A family members (LOC102447194, LOC102447195) genes encode glucose transporters, and their up-regulation during hibernation suggests an adaptive response to ensure minimal but essential glucose uptake during metabolic slowdown.
In the lipid metabolism pathway, the sharpest response occurred only in LSS (lanosterol synthase) and GPLD1 (glycosylphosphatidylinositol specific phospholipase D1) related genes while ACACA (acetyl-CoA carboxylase 1), and KDELR2 (KDEL endoplasmic reticulum protein retention receptor 2) genes were significantly down-regulated in the EA vs. SA group. The SA vs. H group GPLD1 and LSS genes activity were up-regulated, these genes are associated with lipid metabolism and membrane maintenance. The LSS helps in cholesterol biosynthesis, which could be crucial for maintaining membrane integrity during metabolic suppression, while GPLD1 modulates GPI-anchored proteins, which may aid in cell signaling and stress management during low-energy conditions. Conversely, both ACACA and KDELR2 gene activities were down-regulated during the EA vs. SA period. This down-regulation of ACACA and KDELR2 facilitates the metabolic conservation of energy by limiting fatty acid synthesis and decreasing protein synthesis and cellular maintenance. Such metabolic adaptations promote the efficient utilization of stored fat as an energy source, minimize basal metabolic rate, and survival advantages during extended bouts of limited energy availability.
4 Discussion
Here, using RNA-Seq technology we examined gene expression and its regulation in a facultative hibernator, P. sinensis, an unbiased approach in the liver since its importance in metabolism, detoxification, and endocrinology. Examination of liver tissues shows transcriptomic changes that mimic the adaptive strategy used by the turtle during hibernation across three distinct phases: summer active (SA); hibernation (H); and early arousal (EA). Dynamic functional transitions of gene expression throughout hibernation stages in P. sinensis were observed by gene ontology (GO) analysis. Metabolic, cellular, and regulatory processes were suppressed during hibernation, indicating reduced physiological activity. Conversely, arousal transition was enriched in pathways related to the immune system, cellular process and biological regulation categories indicating higher phenomenon of physiological attentiveness and cellular process activity. The transition from hibernation to activity was further underscored by enrichment analyses that prominently featured the roles of immune function, and cellular differentiation and development.
The liver, a vital metabolic organ, stores surplus energy as triglycerides within lipid droplets and releases free fatty acids when energy is deficient. Analyzing differential RNA expression among the various groups highlights the dynamic nature of the transcriptome in the overwintering liver. The pathways regulated during hibernation cycles (SA vs. H, H vs. EA, and EA vs. SA) involve metabolism and transcription. The pathways described here are compatible with those previously described in seasonal hibernators’ livers using RNA-seq, and targeted metabolomics (Zaias et al., 2006; Kitao et al., 2009; Rossi et al., 2014; Faherty et al., 2018; Wang et al., 2019; Yang et al., 2019) and activity assays in facultative hibernators (Storey and Wu, 2013; Græsli et al., 2015). During hibernation, all hibernating animals shift their fuelling in hibernation (at least partly) from carbohydrates to lipids as their main fuel source (Coussement et al., 2023). Our findings shed light on the dynamics of cold-induced energy production and gene expression patterns in the liver, revealing lipidomic and transcriptional signatures in response to cold exposure. Hibernating mammals mobilize fat to produce energy while concurrently storing more fat to withstand extended periods of cold. They primarily obtain energy from the metabolism of lipids (John, 2005; Mohr et al., 2020). The rate-limiting enzyme CPT1A regulates the oxidation of fatty acids in the liver (Britton et al., 1997). In mammals, up-regulation of lipid-catabolism-linked genes results in significantly higher concentrations of triglycerides and fatty acids during hibernation than during non-hibernation (Kurtz et al., 2021). On the contrary, it has also been documented in ectotherm, wherein the expression of genes associated with glycolysis, lipid metabolism and amino acid metabolism down-regulated in hibernation of Chinese alligator (Alligator sinensis), indicating energy metabolism suppression (Lin et al., 2020). As exhibited in the current research, P. sinensis has a down-regulation of carbohydrate metabolism-associated genes like HK2, illustrating the reduced demand for glucose metabolism during hibernation. Other lipid metabolism-related factors like ACACA are down-regulated in hibernating turtles as well, indicative of a shift away from lipid biosynthesis as the organism switches to an energy conservation mode. Similar results were found in the Tibetan frog (Nanorana parkeri) and Asiatic toad (Bufo gargarizans) (Xie et al., 2023; Niu et al., 2024). Conversely, the energy production-related metabolic pathways were reactivated in the dynamic from H vs. EA. Genes for example, LSS and GPLD1, were markedly up-regulated and have also been implicated in the reinitiation of lipid metabolism in favour of energy production as metabolic processes were enhanced to prepare for the active phase. Our findings are consistent as our data shows that during the SA and EA periods, genes involved in gluconeogenesis and glycogenolysis are up-regulated. These results are parallel with those of Coussement et al. (2023) reported that genes associated with energy metabolism were up-regulated during the EA period in hamsters. Moreover, Xiao et al. (2015) reported that certain differentially expressed genes in the liver of R. ferrumequinum are associated with glycolytic pathways and lipid metabolism, fluctuating between active and inactive states. These results highlight variations in gene expression patterns among mammals, amphibians, and reptiles during hibernation, suggesting that reptiles have distinct molecular regulatory mechanisms. The variation between the groups is likely due to differences in thermoregulatory mechanisms; however, more research is needed to understand these mechanisms in detail.
Cold exposure is recognized for its regulatory influence on the immune system (Reynés et al., 2019) and, consequently, the immune response emerged as the most pronounced process (Table 1) indicating a modulatory effect of cold exposure on the expression of immune-related genes in P. sinensis. According to previous studies, hibernation has a significant impact on the immune system of ectotherms (Maniero and Carey, 1997), it includes the prolongation of both the induction and rejection of immune responses (Lin and Rowlands, 1973), suppression of bone marrow lymphocyte proliferation (Šíma et al., 1981), and dropping the proportion and degree of antibody production (Marchalonis and Cone, 1973). An example of innate immunity can be observed in desert tortoises (Gopherus agassizii) which show lesser plasma bactericidal activity in wintertime (Sandmeier et al., 2016). During hibernation low temperatures do not induce immune responses, possibly owing to the thermal sensitivity of lymphocytes and the reduction in lymphocyte numbers (Wright and Cooper, 1981). Thus, the up-regulation of immune response-associated genes, such as CXCL12, FAS, ITGA4, and TNFRSF13C, in overwintering P. sinensis is a compensatory response to a reduction in lymphatic numbers and sensitivity to cold temperatures in winter. A similar pattern of up-regulation of immune-related genes has been observed in Tibetan frogs (Nanorana parkeri), among which are TNFRSF9 (also known as CD137), TNFAIP8, TNFRSF1A, and LTB (Niu et al., 2024). Chen et al. (2019) reported similar findings in the Asian corn borer (Ostrinia furnacalis), where some immune-linked genes (including PGRPs, SPs, and AMPs) showed an up-regulation in low temperature. Previous studies revealed dynamic shifts in immune-related gene expression similar to those observed in P. sinensis, where genes like CXCL12 and FAS are up-regulated during early arousal and non-hibernation, potentially to prepare for reduced immune surveillance during torpor phases (Chow et al., 2013). However, these changes are reversible during hibernation and arousal phases, suggesting a conserved strategy across taxa to minimize immune function while preserving critical defense mechanisms during metabolic suppression.
Numerous endocrine system-related genes were up-regulated during hibernation, and the early arousal period such as CDKN1A, DDL4 and, IGF1R which showed a significant up-regulation during the H vs. EA comparison. CDKN1A is a key mediator of cell cycle arrest, apoptosis, and DNA damage response driving these events during cellular stress. In P. sinensis, the up-regulation of CDKN1A during hibernation likely reflects an adaptive mechanism to suppress cellular proliferation and prioritize energy conservation. The up-regulation of CDKN1A during hibernation in P. sinensis is likely indicative of an adaptive process to inhibit cellular proliferation and conserve energy. This role aligns with other studies, in which CDKN1A-induced cell cycle arrest promotes genomic stability and guards against oxidative damage in a stress or metabolic suppression context (Kreis et al., 2020). Notch signaling, more particularly the canonical Notch ligand 4 (DLL4), confers a critical function in modulating immune responses through T cell differentiation and immune reprogramming. In P. sinensis, the up-regulation of DLL4 during the pre-hibernation and arousal phases may represent an adaptive strategy to mediate the equilibrium between immune activation and energy saving during hibernation. The vertebrate DLL4 signalling pathway is well known to dictate the fate of immune cells, and its dominant effects in driving pro-inflammatory Th1 and Th17 responses via activation of T-cell signalling pathways have also been reported in mammals (Meng et al., 2016). In fact, in models of autoimmune disease, blocking DLL4 reduces inflammatory responses, indicating that their role is dual in terms of immune activation and regulation (Nakano et al., 2016). Like in P. sinensis modulation of DLL4 may play a role, which may reflect conservation of an immune regulation process between species to prevent unnecessary immune activation during physiological stress states such as hibernation. The increased expression of genes involved in thermogenesis, including IGF1R (insulin-like growth factor 1 receptor), is essential for regulating metabolic processes that respond to temperature fluctuations. The up-regulation of this gene during arousal implies that it is involved in the metabolic reconfigurations that are required to bring the turtle that is withdrawing from hibernation back to normal levels of activity and body temperature. This agrees with observations in mammals in which IGF1R prognosticates cell survival and apoptosis during stress situations like hibernation or fasting (Lara-Diaz et al., 2017). Such variation probably serves the organisms well in coping with immune challenges associated with hibernation, like the threat of infection and inflammation that can occur after a long, low metabolic state. Therefore, our results further confirm the temperature role in modulating metabolic pathways in ectotherms such as P. sinensis.
5 Conclusion
The liver data presented here highlight the critical importance of systemically timed sampling in the examination of differential gene expression associated with the profound physiological changes of hibernation. The findings presented here provide a reference resource, and an approach for future studies to evaluate how the physiological dynamics of hibernation can be harnessed to better understand the molecular mechanisms involved in metabolic suppression and tissue-protective pathways. During hibernation, turtles experience drastic metabolic and physiologic down-regulation to conserve energy, reduce cellular activity, and preserve immune function. It was found that the up-regulation of genes associated with immune system vigilance, cell cycle regulation, and lipid metabolism highlighted a delicate balance between energy conservation and survival under extended phases of low metabolic activity. This would allow better insight into the molecular basis of hibernation, as well as metabolic adaptation among ectotherms and has implications for understanding how other hibernating species may have similar survival strategies.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal studies were approved by the Experimental Animal Ethics Committee at Hainan Normal University, permit number HNECEE-2016-005. The studies were conducted in accordance with local legislation and institutional requirements.
Author contributions
MI: Data curation, Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Methodology, Software. XA: Software, Writing – original draft, Data curation. QZ: Software, Writing – review & editing. IK: Software, Writing – review & editing. ZA: Data curation, Writing – review & editing. TL: Methodology, Writing – review & editing, Investigation. LD: Formal analysis, Project administration, Validation, Writing – review & editing. MH: Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant numbers 32160251, 32471587).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1561403/full#supplementary-material
Supplementary Figure 1Heatmap summary of RNA-seq co-expression data for DEGs in liver. Colors indicate the relative abundance of steady-state RNA for each physiological state. Pattern names (right) in each co-expression cluster are indicated.
Supplementary Figure 2Validation of RNA-Seq results by qRT-PCR. 12 DEGs were randomly selected, and the expressions of genes at different periods were examined relative to the housekeeping gene (β-actin).
References
1
Britton C. H. Mackey D. W. Esser V. Foster D. W. Burns D. K. Yarnall D. P. et al . (1997). Fine chromosome mapping. Genomics40, 209–211. doi: 10.1006/geno.1996.4539
2
Capraro A. O’Meally D. Waters S. A. Patel H. R. Georges A. Waters P. D. (2019). Waking the sleeping dragon: gene expression profiling reveals adaptive strategies of the hibernating reptile Pogona vitticeps. BMC Genomics20, 460. doi: 10.1186/s12864-019-5750-x
3
Chen B. Niu C. Yuan L. Zhang W. (2019). Physiological responses in vitamin C system during hibernation in juvenile Chinese soft-shelled turtle Pelodiscus sinensis. J. Ocean. Limnol.37, 767–776. doi: 10.1007/s00343-019-7345-4
4
Chen S. Zhou Y. Chen Y. Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890. doi: 10.1093/bioinformatics/bty560
5
Chow H. M. Horovitz S. G. Carr W. S. Picchioni D. Coddington N. Fukunaga M. et al . (2013). Rhythmic alternating patterns of brain activity distinguish rapid eye movement sleep from other states of consciousness. Proc. Natl. Acad. Sci. U.S.A.110, 10300–10305. doi: 10.1073/pnas.1217691110
6
Coussement L. Oosterhof M. M. Guryev V. Reitsema V. A. Bruintjes J. J. Goris M. et al . (2023). Liver transcriptomic and methylomic analyses identify transcriptional mitogen-activated protein kinase regulation in facultative hibernation of Syrian hamster. Proc. R. Soc B.290, 20230368. doi: 10.1098/rspb.2023.0368
7
Faherty S. L. Villanueva-Cañas J. L. Blanco M. B. Albà M. M. Yoder A. D. (2018). Transcriptomics in the wild: Hibernation physiology in free-ranging dwarf lemurs. Mol. Ecol.27, 709–722. doi: 10.1111/mec.14483
8
Fedorov V. B. Goropashnaya A. V. Tøien Ø. Stewart N. C. Gracey A. Y. Chang C. et al . (2009). Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus). Physiol. Genomics37, 108–118. doi: 10.1152/physiolgenomics.90398.2008
9
Gautier C. Bothorel B. Ciocca D. Valour D. Gaudeau A. Dupré C. et al . (2018). Gene expression profiling during hibernation in the European hamster. Sci. Rep.8, 13167. doi: 10.1038/s41598-018-31506-2
10
Geiser F. (2004). Metabolic Rate and body temperature reduction during hibernation and daily torpor. Annu. Rev. Physiol.66, 239–274. doi: 10.1146/annurev.physiol.66.032102.115105
11
Geiser F. Turbill C. (2009). Hibernation and daily torpor minimize mammalian extinctions. Naturwissenschaften96, 1235–1240. doi: 10.1007/s00114-009-0583-0
12
Græsli A. R. Evans A. L. Fahlman Å. Bertelsen M. F. Blanc S. Arnemo J. M. (2015). Seasonal variation in haematological and biochemical variables in free-ranging subadult brown bears (Ursus arctos) in Sweden. BMC Vet. Res.11, 301. doi: 10.1186/s12917-015-0615-2
13
Heldmaier G. Ortmann S. Elvert R. (2004). Natural hypometabolism during hibernation and daily torpor in mammals. Respir. Physiol. Neurobiol.141, 317–329. doi: 10.1016/j.resp.2004.03.014
14
Hu L. Li Q. Yang P. Gandahi J. A. Arain T. S. Le Y. et al . (2016). Expression of TLR2/4 on epididymal spermatozoa of the chinese soft-shelled turtle Pelodiscus sinensis during the hibernation season. Anatomical Rec.299, 1578–1584. doi: 10.1002/ar.23463
15
Jia Y. Yang Z. Hao Y. Gao Y. (2005). Effects of animal-plant protein ratio in extruded and expanded diets on nitrogen and energy budgets of juvenile Chinese soft-shelled turtle (Pelodiscus sinensis Wiegmann). Aquac Res.36, 61–68. doi: 10.1111/j.1365-2109.2004.01184.x
16
John D. (2005). Annual lipid cycles in hibernators: integration of physiology and behavior. Annu. Rev. Nutr.25, 469–497. doi: 10.1146/annurev.nutr.25.050304.092514
17
Kim D. Paggi J. M. Park C. Bennett C. Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915. doi: 10.1038/s41587-019-0201-4
18
Kitao N. Fukui D. Hashimoto M. Osborne P. G. (2009). Overwintering strategy of wild free-ranging and enclosure-housed Japanese raccoon dogs (Nyctereutes procyonoides albus). Int. J. Biometeorol53, 159–165. doi: 10.1007/s00484-008-0199-7
19
Kreis F. Wright A. J. Hesse F. Fala M. Hu D. Brindle K. M. (2020). Measuring tumor glycolytic flux in vivo by using fast deuterium MRI. Radiology294, 289–296. doi: 10.1148/radiol.2019191242
20
Kurtz C. C. Otis J. P. Regan M. D. Carey H. V. (2021). How the gut and liver hibernate. Comp. Biochem. Physiol. Part A: Mol. Inte Physio253, 110875. doi: 10.1016/j.cbpa.2020.110875
21
Lara-Diaz V. Castilla-Cortazar I. Martín-Estal I. García-Magariño M. Aguirre G. Puche J. et al . (2017). IGF-1 modulates gene expression of proteins involved in inflammation, cytoskeleton, and liver architecture. J. Physiol. Biochem.73, 245–258. doi: 10.1007/s13105-016-0545-x
22
Li B. Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf.12, 323. doi: 10.1186/1471-2105-12-323
23
Lin J.-Q. Huang Y.-Y. Bian M.-Y. Wan Q.-H. Fang S.-G. (2020). A unique energy-saving strategy during hibernation revealed by multi-omics analysis in the Chinese alligator. Iscience23. doi: 10.1016/j.isci.2020.101202
24
Lin H. H. Rowlands J. D.T. (1973). Thermal regulation of the immune response in South American toads (Bufo marinus). Immunology24, 129.
25
Lin R. Wu J. You Z. Xu D. Li C. Wang W. et al . (2023). Induction of hibernation and changes in physiological and metabolic indices in Pelodiscus sinensis. Biology12, 720. doi: 10.3390/biology12050720
26
Love M. I. Huber W. Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550. doi: 10.1186/s13059-014-0550-8
27
Maniero G. D. Carey C. (1997). Changes in selected aspects of immune function in the leopard frog, Rana pipiens, associated with exposure to cold. J. Comp. Physiol. B.167, 256–263. doi: 10.1007/s003600050072
28
Marchalonis J. J. Cone R. E. (1973). The phylogenetic emergence of vertebrate immunity. Aust. J. Exp. Biol. Med. Sci.51. doi: 10.1038/icb.1973.44
29
Meng L. Bai Z. He S. Mochizuki K. Liu Y. Purushe J. et al . (2016). The Notch ligand DLL4 defines a capability of human dendritic cells in regulating Th1 and Th17 differentiation. J. Immunol.196, 1070–1080. doi: 10.4049/jimmunol.1501310
30
Mohr S. M. Bagriantsev S. N. Gracheva E. O. (2020). Cellular, molecular, and physiological adaptations of hibernation: the solution to environmental challenges. Annu. Rev. Cell Dev. Biol.36, 315–338. doi: 10.1146/annurev-cellbio-012820-095945
31
Nakano M. Nagaishi K. Konari N. Saito Y. Chikenji T. Mizue Y. et al . (2016). Bone marrow-derived mesenchymal stem cells improve diabetes-induced cognitive impairment by exosome transfer into damaged neurons and astrocytes. Sci. Reps6, 24805. doi: 10.1038/srep24805
32
Niu Y. Zhang X. Men S. Xu T. Zhang H. Li X. et al . (2024). Effects of hibernation on two important contractile tissues in tibetan frogs, Nanorana parkeri: a perspective from transcriptomics and metabolomics approaches. BMC Genomics25, 454. doi: 10.1186/s12864-024-10357-4
33
Pertea M. Kim D. Pertea G. M. Leek J. T. Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc.11, 1650–1667. doi: 10.1038/nprot.2016.095
34
Reynés B. van Schothorst E. M. Keijer J. Palou A. Oliver P. (2019). Effects of cold exposure revealed by global transcriptomic analysis in ferret peripheral blood mononuclear cells. Sci. Rep.9, 19985. doi: 10.1038/s41598-019-56354-6
35
Rossi G. Mangiagalli G. Paracchini G. Paltrinieri S. (2014). Hematologic and biochemical variables of hedgehogs (Erinaceus europaeus) after overwintering in rehabilitation centers. Vet. Clinl Pathol.43, 6–14. doi: 10.1111/vcp.12121
36
Ruf T. Geiser F. (2015). Daily torpor and hibernation in birds and mammals. Biol. Rev.90, 891–926. doi: 10.1111/brv.12137
37
Sandmeier F. C. Horn K. R. Tracy C. R. (2016). Temperature-independent, seasonal fluctuations in immune function of the Mojave Desert Tortoise (Gopherus agassizii). Can. J. Zool.94, 583–590. doi: 10.1139/cjz-2016-0010
38
Šíma P. Sỳkora J. Pospíšil M. (1981). Comparison of the proliferative activity of lymphoid spleen cells and the antibody response in Rana esculenta kept in 22 and 4 C. Dev. Comp. Immunol., 497–498. doi: 10.1016/B978-0-08-025922-2.50082-6
39
Srere H. K. Wang L. C. Martin S. L. (1992). Central role for differential gene expression in mammalian hibernation. Proc. Natl. Acad. Sci. U.S.A.89, 7119–7123. doi: 10.1073/pnas.89.15.7119
40
Storey K. B. Storey J. M. (2004). Metabolic rate depression in animals: transcriptional and translational controls. Biolo Revi.79, 207–233. doi: 10.1017/s1464793103006195
41
Storey K. B. Wu C.-W. (2013). Stress response and adaptation: a new molecular toolkit for the 21st century. Comp. Biochem. Physiol. A Mol. Integr. Physiol.165, 417–428. doi: 10.1016/j.cbpa.2013.01.019
42
Tang Z. Chen B. Niu C. (2021). Antioxidant defense response during hibernation and arousal in Chinese soft-shelled turtle Pelodiscus sinensis juveniles. Cryobiology99, 46–54. doi: 10.1016/j.cryobiol.2021.01.015
43
Wang Y. Han S. Li R. Cui B. Ma X. Qi X. et al . (2019). Structural characterization and immunological activity of polysaccharides from the tuber of Bletilla striata. Int. J. Biol. Macromol.122, 628–635. doi: 10.1016/j.ijbiomac.2018.10.201
44
Wang Y. Wang X. Chen Y. Du J. Xiao Y. Guo D. et al . (2024). Adapting to stress: The effects of hibernation and hibernacula temperature on the hepatic transcriptome of Rhinolophus pusillus. FASEB J.38, e23462. doi: 10.1096/fj.202301646R
45
Williams D. R. Epperson L. E. Li W. Hughes M. A. Taylor R. Rogers J. et al . (2006). Seasonally hibernating phenotype assessed through transcript screening. Physiol. Geno24, 13–22. doi: 10.1152/physiolgenomics.00301.2004
46
Wright R. K. Cooper E. L. (1981). Temperature effects on ectotherm immune responses. Dev. Comp. Immunol.5, 117–122. doi: 10.1016/0145-305X(81)90016-1
47
Wu L. Chen Q. Dong B. Han D. Zhu X. Liu H. et al . (2023). Resveratrol attenuated oxidative stress and inflammatory and mitochondrial dysfunction induced by acute ammonia exposure in gibel carp (Carassius gibelio). Eco Envi Saf.251, 114544. doi: 10.1016/j.ecoenv.2023.114544
48
Xiao Y. Wu Y. Sun K. Wang H. Zhang B. Song S. et al . (2015). Differential expression of hepatic genes of the greater horseshoe bat (Rhinolophus ferrumequinum) between the summer active and winter torpid states. PLoS One10, e0145702. doi: 10.1371/journal.pone.0145702
49
Xie R. Edwards K. M. Wille M. Wei X. Wong S.-S. Zanin M. et al . (2023). The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature622, 810–817. doi: 10.1038/s41586-023-06631-2
50
Yan J. Burman A. Nichols C. Alila L. Showe L. C. Showe M. K. et al . (2006). Detection of differential gene expression in brown adipose tissue of hibernating arctic ground squirrels with mouse microarrays. Physi Geno25, 346–353. doi: 10.1152/physiolgenomics.00260.2005
51
Yang J. Nie J. Ma X. Wei Y. Peng Y. Wei X. (2019). Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol. Cancer18, 26. doi: 10.1186/s12943-019-0954-x
52
Zaias J. Norton T. Fickel A. Spratt J. Altman N. H. Cray C. (2006). Biochemical and hematologic values for 18 clinically healthy radiated tortoises (Geochelone radiata) on St Catherines Island, Georgia. Veterinary Clin. Pathol.35, 321–325. doi: 10.1111/j.1939-165X.2006.tb00139.x
53
Zhang L. Han X. Li M. Bao H. Chen Q. (2007). Spermiogenesis in soft-shelled turtle, Pelodiscus sinensis. Anat. Rec290, 1213–1222. doi: 10.1002/ar.20587
54
Zhang W. Niu C. Chen B. Yuan L. (2017). Antioxidant responses in hibernating Chinese soft-shelled turtle Pelodiscus sinensis hatchlings. Comp. Biochy Physi A: Mol. IntegPhysi204, 9–16. doi: 10.1016/j.cbpb.2017.02.003
Summary
Keywords
Pelodiscus sinensis , liver, hibernation, transcriptomic analysis, gene expression
Citation
Iqbal MS, Ai X, Zhu Q, Khan I, Ali Z, Lan T, Ding L and Hong M (2025) Liver transcriptome analysis reveals the molecular response to hibernation challenge in the Chinese soft-shelled turtle (Pelodiscus sinensis). Front. Mar. Sci. 12:1561403. doi: 10.3389/fmars.2025.1561403
Received
15 January 2025
Accepted
05 February 2025
Published
25 February 2025
Volume
12 - 2025
Edited by
Yafei Duan, South China Sea Fisheries Research Institute, China
Reviewed by
Haiyan Liu, Hebei Normal University, China
Chuanjie Qin, Neijiang Normal University, China
Updates
Copyright
© 2025 Iqbal, Ai, Zhu, Khan, Ali, Lan, Ding and Hong.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meiling Hong, mlhong@hainnu.edu.cn; Li Ding, dingli@hainnu.edu.cn
†These authors have contributed equally to this work
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.