
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 28 February 2025
Sec. Marine Biology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1551763
 Wenguang Diao1†
Wenguang Diao1† Shengyao Zhang1†
Shengyao Zhang1† Xueying Zhu1
Xueying Zhu1 Peiqi Wu1
Peiqi Wu1 Bingyan Du1
Bingyan Du1 Zhe Han1
Zhe Han1 Yunqing Liu1*
Yunqing Liu1* Chunpeng He2*
Chunpeng He2* Zuhong Lu2*
Zuhong Lu2*Introduction: Pocillopora damicornis, a key species of stony corals, has been the subject of considerable scientific study. However, the cellular composition of P. damicornis and the roles of these cells in endosymbiosis and biomineralization remain elusive. The development of single-cell technology has provided new opportunities for researching the cellular and molecular mechanisms underlying symbiosis and mineralization. Nevertheless, the stringent environmental requirements, the complexity of the cellular components, and the paucity of high-quality reference genomes of P. damicornis have posed significant challenges for single-cell transcriptome research.
Methods: In this study, we quantified the transcriptomic expression of P. damicornis by aligning its single-cell transcriptome (scRNA-seq) data to multiple species, including Stylophora pistillata, P. damicornis, and Pocillopora verrucosa. We determined the cell types of P. damicornis by comparing its cluster-specific genes with the published cell type-specific genes of S.pistillata and conducted gene function and enrichment analyses.
Results: Unsupervised clustering analysis yielded the identification of ten distinct cell populations, including epidermis cells, gastrodermis cells, algae-hosting cells, calicoblast, cnidocytes, and immune cells. In addition, we identified 53 genes that were highly similar to known sequences in the symbiotic zooxanthellae. These genes were mainly expressed in four different cell populations, corresponding to active symbiotic populations.
Conclusion: This study identified cell types closely associated with symbiosis and calcification in P. damicornis, along with their marker genes, which are consistent with the findings in S. pistillata. These results offer insights into the cellular functions and symbiotic mechanisms of P. damicornis.
Coral reefs have been likened to tropical rainforests in the context of the oceans. These ecosystems provide a habitat for fish and play a role in the biodiversity of the ocean (Hughes et al., 2017). Coral reefs are primarily formed by the accumulation of calcium carbonate secreted by reef-building corals over extended periods of time. The survival and reef-building activities of stony corals are inseparable from the important contributions of the algae that live in symbiosis with their cells, known as Symbiodinium (Wilson and Hilferty, 1931). In the event of a disruption to the external environment, such as an increase in temperature, the symbiont undergoes disintegration, a process referred to as coral bleaching (Odum and Odum, 1955; Muscatine, 1967; Moberg and Folke, 1999; Costanza et al., 2014; Huang et al., 2019; Reimer et al., 2019). The ongoing threat to the coral ecosystem, precipitated by global warming and marine environmental pollution, has led to coral bleaching as a primary factor in the degradation of coral reefs (Swain et al., 2016). The study of the physiological characteristics and symbiotic mechanisms of corals is of great significance to the preservation of coral ecosystems. The primary methodologies employed in coral research encompass the use of isotope labeling (Clayton and Lasker, 1984), and metabolite chromatography analysis (Biel et al., 2007; Gates et al., 1995). As coral genome data continues to improve (Shinzato et al., 2011; Polato et al., 2011; Levy et al., 2021), transcriptomic analysis has become a staple of coral research. In comparison with conventional transcriptional analysis, single-cell RNA sequencing (scRNA-seq) has been shown to identify the transcriptional profile of individual cells and to identify multiple types of cells in Stylophora pistillata, including immune cells, gastrodermis cells, neurons cells, alga-hosting cells, and calicoblasts (Mansour et al., 2016; Hayward et al., 2011; Reyes-Bermudez et al., 2009; Bay et al., 2009). Nevertheless, corals have evolved over 500 million years, and there are substantial disparities between their genomes. Consequently, the analysis of single-celled organisms belonging to other stony coral families remains a significant area of research (Reimer et al., 2019).
To this end, the present study employs P. damicornis to elucidate its cell specificity. Both P. damicornis and S. pistillata are classified under the monophyletic group of the Pocilloporidae family. The family Pocilloporidae, belonging to the order Scheractinia(stony corals),is predominantly distributed in the Indo-Pacific Ocean. The family includes several genera, notably Pocillopra and Stylophora (Schmidt-Roach et al., 2014). P. damicornis exhibits a broad distributed across tropical and subtropical reefs, with notable populations in the Indo-Pacific and Eastern regions (Schmidt-Roach et al., 2014) (Barbosa et al., 2016). This study sampled cells from different parts of adult P. damicornis while retaining a small number of symbiotic algae genes to facilitate the identification of symbiotic cells.
The biological samples utilized in this study were obtained from the Xisha Islands, situated in the South China Sea (15°40’–17°10’ N, 111°–113°E). The coral samples were cultivated in a coral tank specifically designed to replicate their natural habitat conditions. All P. damicornis specimens were maintained in a RedSea® tank (redsea575, Red Sea Aquatics Ltd., London, UK) at 26°C and 1.025 salinity (Red Sea Aquatics Ltd). The temperature was maintained at 26°C and the salinity at 1.025 (as per Red Sea Aquatics Ltd. specifications). The coral cultivation system was equipped with three coral lamps (AI®, Red Sea Aquatics Ltd), a protein skimmer (regal250s, Reef Octopus), a water chiller (tk1000, TECO Ltd, Port Louis, Mauritius), two wave-making devices (VorTechTM MP40, EcoTech Marine Ltd), and a calcium reactor (Calreact 200, Reef Octopus).
To prepare the single ‐cell suspension, we first diluted a 0.02 M PBS (Solarbio, P1010‐2L, Beijing, China)to obtain a 0.035 M PBS solution with a pH of 8.0. The sample was thoroughly rinsed with this solution, and the rinsing process was repeated three times.
Next, we prepared a digestion solution by mixing a 0.1% trypsin (Sigma-Aldrich, T1426-500MG, St. Louis, MO, USA) with the 0.035 M PBS solution. The sample was then placed in this digestion solution at room temperature for 2 - 3 hours. Subsequently, the solution was filtered through a filter with a pore size of 40 μm Fisherbrand™ Sterile Cell Strainers (Thermo Fisher Scientific, 22‐363‐547, Waltham, MA, USA) and centrifuged at a centrifugal force of 350 G for 5‐6 minutes. The enzymatic reaction was terminated by adding a stop solution containing 0.4 mg of BSA.
We employed the trypan blue staining method. An automatic cell counter (TC20™, Bio‐Rad Laboratories, Inc, Hercules, CA, USA) was used to add a 0.4% trypan blue (Invitrogen, T10282, Carlsbad, CA, USA) to the cell suspension at a 1:1 ratio. To ensure the optimal single - cell capture effect, we defined the qualified criteria as a cell survival rate greater than 60% and a cell concentration ranging from 1×105 to 1×107 cells/mL. After washing and resuspension, the cell suspension was diluted to a concentration of 800 - 1200 cells/μL. The prepared cell suspension was then sent to Tianjin Novogene Company for library construction(10x Genomics, Inc, Pleasanton, CA, USA) and quality control. Finally, sequencing was carried out on the HiSeq PE150 platform(Illumina, Inc, San Diego, CA, USA).
Due to the current paucity of high-quality coral genome data and the presence of numerous gap regions, the species with relatively high-quality genome data in the family Pocilloporidae, suborder Astrocoeniina, order Scleractinia on NCBI currently include S. pistillata, P. damicornis, and Pocillopora verrucosa. This study utilizes these three species as references, in conjunction with our laboratory’s P. damicornis full-length transcriptome data (Han et al., 2022), employing the CellRanger (V8.0.1) mkref function to construct a reference genome and the count function for gene expression quantification.
To identify symbiotic zooxanthellae genes, we utilized the genome of S. pilosum (NCBI accession: GCA_964212085.1) as a reference. Through CellRanger count analysis to detect symbiodinium homologous genes in coral host cell.
The gene functions were annotated based on the previously published P. damicornis full-length transcriptome (Han et al., 2022) using Pfam (protein family; Finn et al., 2016); KOG/COG (EuKaryotic Orthologous Groups; Clusters of Orthologous Groups of proteins; Tatusov et al., 2003); KEGG (Kyoto Encyclopedia of Genes and Genomes; Kanehisa et al., 2004); and GO (Gene Ontology; Ashburner et al., 2000).
An unsupervised clustering algorithm with a resolution parameter set at 0.1 was employed for cell clustering. For the analysis of differential expression, we established thresholds of a minimum expression percentage (min.pct) of ≥ 0.01 and a log fold change (logfc.threshold) of > 0.25. Dimensionality reduction was conducted utilizing the Uniform Manifold Approximation and Projection (UMAP) algorithm, which facilitates the low-dimensional visualization of high-dimensional data.
Presently, the extant literature on marker genes for coral cell types is scant, with the study on S. pistillata constituting a rare resource. The present study employs a three-step approach to address this paucity of knowledge. First, the significantly upregulated genes that are uniquely expressed in each cluster identified in the P. damicornis dataset will be determined. Second, these cluster-specific marker genes are then cross-referenced with the cell type-specific markers reported for S. pistillata. Finally, combining the enrichment analysis result of the overlapping marker genes will allow for the inference of the corresponding cell types present in P. damicornis.
Functional enrichment analysis was conducted using the Molecular Signatures Database (MSigDB) framework (Liberzon et al., 2015). The analysis pipeline incorporated multiple resources, including GO terms for biological processes, molecular functions, and cellular components; KEGG pathways for metabolic and signaling networks; KOG for evolutionary relationships and Pfam for protein domain information. The statistical significance of the enrichment patterns was assessed using Fisher’s exact test, with a significance threshold of P < 0.05. Enriched terms were prioritized based on their statistical significance (−log10[P-value]). The resulting functional categories were then visualized using the ggplot2 package, with particular emphasis on biological processes and molecular pathways relevant to coral biology.
In this study, single-cell RNA sequencing (scRNA-seq) analysis was performed on five reference genomesfrom the family Pocilloporidae: S. pistillata, P. damicornis, P. verrucosa, and P. damicornis full-length transcriptome data, and the full-length transcriptome data of S. pistillata and P. damicornis. Among the species examined, P. verrucosa exhibited the highest number of expressed genes (32,915), followed by P. damicornis (23,077), S. pistillata (22,408), and P. damicornis (14,402) (see Table 1 for details).
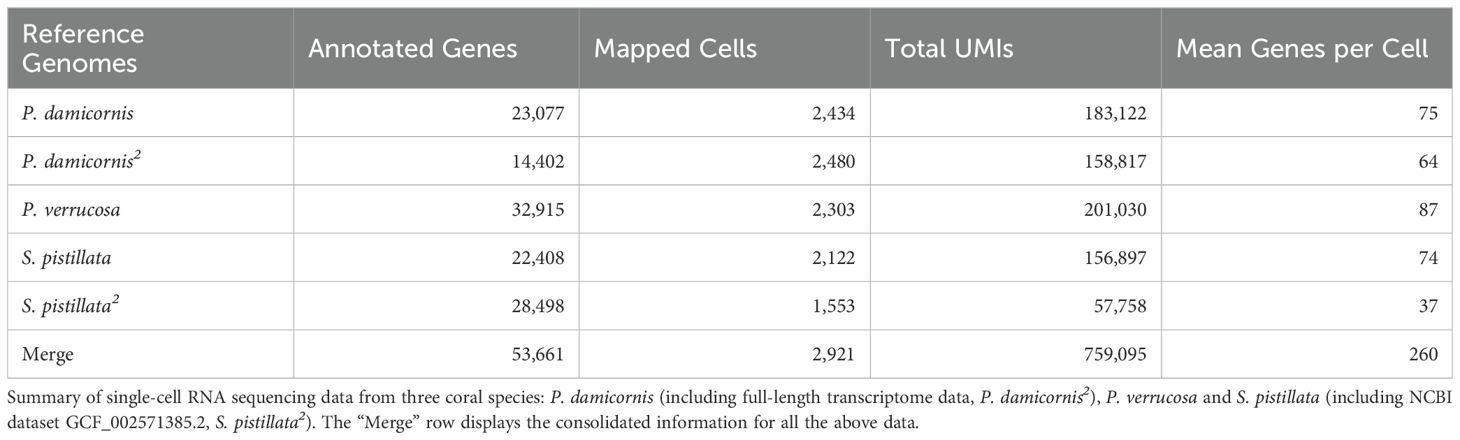
Table 1. Comparative analysis of reference genomes and integrated datasets used for single-cell transcriptome alignment.
We observed samples-specific variations in the average number of genes expressed per cell. P. verrucosa demonstrated the highest average expression levels, with 87 genes per cell. In contrast, P. damicornis, S. pistillata, and P. damicornis exhibited average expressions of 75, 74, and 74 genes per cell, respectively. Integration of transcriptomic data from all five species yielded a comprehensive dataset comprising 53,661 genes, with an average of 260 genes expressed per cell across the integrated dataset (Table 1).
UMAP visualization revealed ten distinct clusters in two-dimensional space (Figure 1A). Differential expression analysis was performed to characterize the molecular signatures of these clusters, with significance criteria set as: Absolute value of log2 fold change > 0.25, expression in > 1% of cells, and P < 0.05. The differential expression results of each cluster are shown in Table 2. The specific differentially expressed genes for each cluster are in Supplementary Table S1.

Figure 1. A study of the coral symbiotic system via single-cell transcriptome analysis. (A) Utilizing UMAP dimensionality reduction of single-cell transcriptome data, ten transcriptionally distinct cell subpopulations were identified. The utilization of distinct colors and numerical identifiers enables the clear representation of these diverse cell clusters. (B) Annotation of major cell types on the UMAP plot, including epidermis cells, gastrodermis cells, alga hosting cells, calicoblast, cnidocytes, and immune cells. (C) The expression distribution of algal symbiont genes on the UMAP plot is represented by different colors, with different colors representing different expression levels. (D) A schematic diagram of the main cell types identified is shown, with their spatial relationships within the coral symbiotic system illustrated.
Among the 53 identified Symbiodinium genes, 25 were functionally annotated (Supplementary Tables S3, S4). These genes were primarily highly expressed in clusters 4, 5, 8, and 9 (Figure 1C). These genes are mainly involved in energy metabolism, biosynthesis, stress response (including HSP family), cytoskeleton regulation, and protein synthesis and modification.
Cross-species comparison of cell-type markers between S. pistillata and P. damicornis revealed distinct cell population correspondences. Based on shared marker genes, we identified six major cell types: cluster 0 as epidermis cells, cluster 1 as gastrodermis cells, cluster 2 as algal host cells, cluster 3 as calicoblasts, cluster 6 as cnidocytes, and cluster 7 as immune cells(Figure 1B; Supplementary Table S2). This cell-type assignment was supported by overlapping marker genes between the two species (Supplementary Table S1, S8), providing molecular evidence for conserved cell populations across coral species.
The ectoderm predominantly comprises of epidermis cells (Cluster 0), which form the outermost protective barrier and contain cnidocytes (Cluster 6) for defense and predation. The endoderm, on the other hand, contains gastrodermis cells (Cluster 1) and various symbiotic cell subpopulations (Clusters 2, 4, 5, 8, and 9), which are responsible for nutrient absorption and maintaining the symbiotic relationship with zooxanthellae. The calicoblast of the basal ectoderm (Cluster 3) is responsible for bone formation, while the immune cells distributed in the mesoderm (Cluster 7) are responsible for immune defense (Figures 1B, D; Supplementary Table S3).
Gene set enrichment analysis was performed using MSigDB against KEGG and GO databases. Additionally, transcripts were categorized based on GO, KEGG, KOG/COG, and Pfam annotations, followed by Fisher’s exact test (P < 0.05 considered significant; Supplementary Table S4).
GO analysis of upregulated genes in coral epidermis cells revealed enrichment in transcription elongation, DNA binding, and ribosome biogenesis, suggesting active gene expression and protein synthesis regulation (Mayfield et al., 2014). KEGG analysis identified enriched pathways in metabolism and signal transduction (Rädecker et al., 2021). KOG and Pfam analyses highlighted specific regulatory elements, including BTG1/TOB anti-proliferative factors, Mafk bZIP transcription factor, and basic leucine zipper domains, which may modulate epidermal cell functions (Li et al., 2018; Supplementary Figure S1).
Functional enrichment analysis of upregulated genes in gastrodermis cells revealed multiple digestive-related pathways (Figure 2). These included catalytic activity regulation and cysteine peptidase activity, indicating active enzyme secretion (e.g., pepsinogen); lipid transport and lipoprotein pathways; and N-acetyltransferase activity for substrate modification. Signal transduction analysis showed enrichment in the PI3K-Akt pathway, while structural features were highlighted by enrichment in cell adhesion and actin cytoskeleton components, reflecting their epithelial barrier function (Raz-Bahat et al., 2017). The enrichment of lysosomal and protein digestion pathways further supports their role in protein processing (Coffey and De Duve, 1968; Moya et al., 2012).

Figure 2. Enrichment analysis of genes and proteins in coral gastrodermis cells. The figure shows the functional enrichment analysis results of genes and proteins categorized by biological process (BP), molecular function (MF), cellular component (CC), and KEGG pathways, as well as the enrichment fold changes of specific genes and proteins. The x-axis represents the negative logarithm transformation of the p-value (-logPV), with higher values indicating higher enrichment levels. “-logPV” represents the negative logarithm (base 10) transformation of the p-value.
GO functional enrichment analysis of coral calicoblast revealed significant pathways in energy metabolism, ion transport, and cytoskeletal regulation. Biological processes showed enrichment in ATP synthesis coupled proton transport, mitochondrial electron transport, and anion transport. Molecular functions highlighted cytochrome c oxidase activity, NADH dehydrogenase activity, and ion transport mechanisms. Cellular component analysis identified enrichment in mitochondrial structures and ion transport complexes. KEGG pathway analysis revealed enrichment in oxidative phosphorylation and related metabolic pathways (Supplementary Figure S2). KOG and Pfam analyses identified key components including carbonic anhydrases, cytoskeletal proteins, ion transporters (Na+/K+-ATPase), and regulatory domains (FerB, calponin homology, tRNA synthetase classes).
GO functional enrichment analysis of coral cnidocytes revealed significant enrichment in protein deubiquitination, calcium ion homeostasis, and protein processing pathways. Molecular function analysis identified enrichment in various enzymatic activities (metallocarboxypeptidase, ubiquitin-specific protease), calcium channels, and neurotransmitter transport. Cellular component analysis highlighted the DNA-dependent RNA polymerase III complex, suggesting active transcriptional regulation.
KEGG pathway analysis showed enrichment in protein export, endoplasmic reticulum processing, and mucin-type O-glycosylation biosynthesis, potentially involved in toxin modification (Supplementary Figure S3). KOG classification revealed enrichment in signal transduction, cytoskeleton organization, and protein modification. Pfam analysis identified domains crucial for cnidocyte function, including EF-hand calcium-binding, dynein heavy chain, myosin head, and toxin-related domains.
These findings indicate that cnidocytes possess specialized features for toxin synthesis and secretion, including protein modification systems, calcium signaling pathways, and dynamic cytoskeletal regulation (Jouiaei et al., 2015; Özbek, 2011; Beckmann and Özbek, 2012; Rachamim et al., 2015).
GO analysis revealed enrichment in protein deubiquitination, catalytic activity regulation, DNA binding, and exosome complex functions. KEGG pathways highlighted immune-related processes, including lysosomal and platelet activation pathways. KOG and Pfam analyses identified regulatory proteins such as E3 ubiquitin ligases and interferon factors, suggesting roles in coral immune regulation (Supplementary Figure S4).
As the cornerstone of the marine ecosystem, the survival of coral depends on the functional collaboration between cell groups (Levy et al., 2021). We revealed the molecular characteristics of certain cell populations in P. damicornis and their synergistic mechanisms in symbiosis, calcification and immunity through single-cell transcriptome analysis.
Epithelial cells act as the first barrier to the external environment, and they quickly respond to stress by highly expressing HSP90AB1 and Jun genes (Cziesielski et al., 2018).The cytokines they secrete may regulate the metabolic activity of adjacent gastric epithelial cells through paracrine signals. The significant upregulation of ribosomal proteins (RPL13, RPS20) and collagen (COL1A2, COL5A1) in gastric epithelial cells indicates that they are not only responsible for nutrient absorption and transport, but may also provide structural support for the symbiotic algae through extracellular matrix (ECM) remodeling (Mass et al., 2014). It is worth noting that the genes for lipid metabolism (ALDH3A2, ACSL4) in symbiotic cells are highly expressed together with the mitochondrial oxidative phosphorylation pathway (such as COX6B1, NDUFA4) in calicoblast (Drake et al., 2020). This may suggest that photosynthetic products are directly supplied via lipid carriers to ATP required for calcification.
The maintenance of the coral-alga symbiosis depends on the coordinated action of multiple cell types (Levy et al., 2021). A lipid metabolism network comprising ALDH3A2, APOD and ACSL4 in the symbiotic cells supports nutrient exchange, while the expression of ATP6V1G1 maintains an acidic pH suitable for algal photosynthesis (Barott et al., 2015). Gastric epithelial cells play a key role in regulating nutrient transport and cell proliferation by upregulating ribosomal proteins and signaling molecules such as MST1R, YAP1 and JAG2 (Rosic et al., 2014).
We found high co-expression of mesoheat shock protein (HSP90AB1) in epidermal cells with oxidative stress genes (e.g. SOD2) in symbiotic cells (Cziesielski et al., 2018). This may suggest the activation of our transcellular antioxidant network. Meanwhile the high expression of thioredoxin (Trx) in calcified cells not only supports mitochondrial energy metabolism, but may also protect calcified frontiers from oxidative damage by scavenging reactive oxygen species (ROS) (Drake et al., 2020; Ramos-Silva et al., 2013; Guo et al., 2021).This multilayered defense mechanism may be the molecular basis of coral response to climate change (DeSalvo et al., 2010; Barshis et al., 2013).
High expression of thioredoxin in calcified cells provides redox balance and energy support for biomineralization (Drake et al., 2020). Gastrodermal cells produce ATP through ETS-mediated aerobic respiration to provide energy support for the calcification process (Tambutté et al., 2011). Meanwhile, the expression of collagen proteins such as COL1A2, COL3A1, and COL5A1 in gastrodermal cells provides an extracellular matrix scaffold for the calcification process (Mass et al., 2014), ARHGEF17 and ACTB-mediated cytoskeleton promotes transport of calcified material (Zoccola et al., 2015).
The immune cells and the cnidocyte together form the defense line of the coral (Palmer, 2018; Sun et al., 2024). Immune cells express a variety of stress-related proteins that are involved in the basal defense (Rachamim et al., 2015). Cnidocyte regulate the synthesis of nematocystin through the expression of PDIA3 and RCN1 In terms of tissue defense, both immune cells and cnidocyte express specific functional genes such as FGF1 and IRF transcription factors in immune cells and ADAMTS9 and CMH1 in cnidocytes (Libro et al., 2013). The toxin and release mechanisms of cnidocyte exhibit dynamic regulatory mechanisms (Columbus-Shenkar et al., 2018).
In this study, we revealed the division of labor among multiple cell populations in corals in the maintenance of physiological symbiosis through single-cell transcriptome analysis. Epidermal, gastrodermal, and calicoblast collectively maintain coral symbiosis, calcification processes, and environmental adaptation through the synergy of metabolic networks, signaling pathways, and structural support. These findings provide new perspectives for understanding the cellular molecular basis of coral physiological functions, as well as a theoretical basis for predicting and improving coral adaptations to the environment.
Although the present study reveals the functional collaboration network among coral cell populations, there are still some shortcomings: firstly, the single-cell transcriptome analysis reflects the gene expression profiles at a specific point in time, which makes it difficult to capture the spatial and temporal characteristics of the dynamic cell-cell interactions; secondly, many of the hypothesized intercellular signaling pathways still lack functional validation. In the future, we can: 1. analyze and elucidate the dynamics of cellular interactions by combining with time series; 2. validate the key signaling pathways by combining with in vitro culture and other experiments; 3. explore the plasticity of cellular collaborative networks in response to environmental stresses; 4. verify the spatial location of coral cells by in situ hybridization experiments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WD: Data curation, Methodology, Software, Supervision, Validation, Writing – original draft. SZ: Data curation, Methodology, Software, Writing – original draft, Writing – review & editing. XZ: Methodology, Software, Visualization, Writing – review & editing. PW: Data curation, Visualization, Writing – review & editing. BD: Software, Visualization, Writing – review & editing. ZH: Data curation, Funding acquisition, Writing – review & editing. YL: Methodology, Software, Validation, Writing – original draft. HC: Conceptualization, Formal Analysis, Investigation, Project administration, Resources, Supervision, Writing – review & editing. ZL: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Project of Henan Province (242102210192, 242102210127, 242102210008).
This research is supported by the Big Data Computing Center of Southeast University and HongHe Artificial Intelligence Technology (Luoyang) Co.,Ltd., which provide computing power support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1551763/full#supplementary-material
Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: Tool for the unification of biology. Nat. Genet. 25 (1), 25–29. doi: 10.1038/75556
Barbosa M., Valentão P., Andrade P. B. (2016). Biologically active oxylipins from enzymatic and nonenzymatic routes in macroalgae. Mar. Drugs 14 (1), 23. doi: 10.3390/md14010023
Barott K. L., Venn A. A., Perez S. O., Tambutteeé S., Tresguerres M., Somero G. N. (2015). Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112 (2), 607–612. doi: 10.1073/pnas.1413483112
Barshis D. J., Ladner J. T., Oliver T. A., Seneca F. O., Traylor-Knowles N., Palumbi S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110 (4), 1387–1392. doi: 10.1073/pnas.1210224110
Bay L. K., Nielsen H. B., Jarmer H., Seneca F., van Oppen M. J. H. (2009). Transcriptomic variation in a coral reveals pathways of clonal organization. Mar. Genomics 2 (2), 119–125. doi: 10.1016/j.margen.2009.07.004
Beckmann A., Özbek S. (2012). The nematocyst: A molecular map of the cnidarian stinging organelle. Int. J. Dev. Biol. 56 (6-8), 577–582. doi: 10.1387/ijdb.113472ab
Biel K. Y., Gates R. D., Muscatine L. (2007). Effects of free amino acids on the photosynthetic carbon metabolism of symbiotic dinoflagellates. Russian J. Plant Physiol. 54, 171–183. doi: 10.1134/S1021443707020033
Clayton W. S., Lasker H. R. (1984). Host feeding regime and zooxanthellal photosynthesis in the anemone, Aiptasia pallida (Verrill). Biol. Bull. 167 (3), 590–600. doi: 10.2307/1541412
Coffey J. W., De Duve C. (1968). Digestive activity of lysosomes. I. The digestion of proteins by extracts of rat liver lysosomes. J. Biol. Chem. 243 (12), 3255–3263. doi: 10.1016/S0021-9258(18)93301-6
Columbus-Shenkar Y. Y., Sachkova M. Y., Macrander J., Fridrich A., Modepalli V., Reitzel A. M., et al. (2018). Dynamics of venom composition across a complex life cycle. Elife 7, e35014. doi: 10.7554/eLife.35014
Costanza R., de Groot R., Sutton P., van der Ploeg S., Anderson S. J., Kubiszewski I., et al. (2014). Changes in the global value of ecosystem services. Global Environ. Change 26, 152–158. doi: 10.1016/j.gloenvcha.2014.04.002
Cziesielski M. J., Liew Y. J., Cui G., Schmidt-Roach S., Campana S., Marondedze C., et al. (2018). Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proc. R. Soc. B.: Biol. Sci. 285 (1877), 20172654. doi: 10.1098/rspb.2017.2654
DeSalvo M. K., Sunagawa S., Voolstra C. R., Medina M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Mar. Ecol. Prog. Ser. 402, 97–113. doi: 10.3354/meps08372
Drake J. L., Mass T., Stolarski J., Von Euw S., van de Schootbrugge B., Falkowski P. G. (2020). How corals made rocks through the ages. Glob. Chang. Biol. 26 (1), 31–53. doi: 10.1111/gcb.14912
Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. doi: 10.1093/nar/gkv1344
Gates R. D., Hoegh-Guldberg O., Mcfall-Ngai M. J., Bil’ K. Y., Muscatine L. (1995). Free amino acids exhibit anthozoan “host factor” activity: They induce the release of photosynthate from symbiotic dinoflagellates in vitro. Proc. Natl. Acad. Sci. U.S.A. 92 (16), 7430–7434. doi: 10.1073/pnas.92.16.7430
Guo Z., Liao X., Han T., Chen J., He C., Lu Z. (2021). Full-length transcriptomics reveal the gene expression profiles of reef-building coral pocillopora damicornis and symbiont zooxanthellae. Diversity. 13 (11), 543. doi: 10.3390/d13110543
Han T., Liao X., Zhu Y., Liu Y., Lu N., Li Y., et al. (2022). Full-length transcriptome maps of reef-building coral illuminate the molecular basis of calcification, symbiosis, and circadian genes. Int. J. Mol. Sci. 23 (19), 11135. doi: 10.3390/ijms231911135
Hayward D. C., Hetherington S., Behm C. A., Grasso L. C., Forêt S., Miller D. J., et al. (2011). Differential gene expression at coral settlement and metamorphosis - A subtractive hybridization study. PloS One 6 (10), e26411. doi: 10.1371/journal.pone.0026411
Huang C., Leng D., Sun S., Zhang X. D. (2019). Re-analysis of the coral Acropora digitifera transcriptome reveals a complex lncRNAs-mRNAs interaction network implicated in Symbiodinium infection. BMC Genomics 20, 1–15. doi: 10.1186/s12864-019-5429-3
Hughes T. P., Barnes M. L., Bellwood D. R., Cinner J. E., Cumming G. S., Jackson J. B. C., et al. (2017). Coral reefs in the anthropocene. Nature 546 (7656), 82–90. doi: 10.1038/nature22901
Jouiaei M., Sunagar K., Federman Gross A., Scheib H., Alewood P. F., Moran Y., et al. (2015). Evolution of an ancient venom: Recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone. Mol. Biol. Evol. 32 (6), 1598–1610. doi: 10.1093/molbev/msv050
Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32 (suppl_1), D277–D280. doi: 10.1093/nar/gkh063
Levy S., Elek A., Grau-Bové X., Menéndez-Bravo S., Iglesias M., Tanay A., et al. (2021). A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 184 (11), 2973–2938. doi: 10.1016/j.cell.2021.04.005
Li D., Xiao L., Ge Y., Fu Y., Zhang W., Cao H., et al. (2018). High expression of Tob1 indicates poor survival outcome and promotes tumor progression via a Wnt positive feedback loop in colon cancer. Mol. Cancer 17, 1–6. doi: 10.1186/s12943-018-0907-9
Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J. P., Tamayo P. (2015). The molecular signatures database hallmark gene set collection. Cell Syst. 1 (6), 417–425. doi: 10.1016/j.cels.2015.12.004
Libro S., Kaluziak S. T., Vollmer S. V. (2013). RNA-seq profiles of immune related genes in the staghorn coral Acropora cervicornis Infected with white band disease. PloS One 8 (11), e81821. doi: 10.1371/journal.pone.0081821
Mansour T. A., Rosenthal J. J. C., Brown C. T., Roberson L. M. (2016). Transcriptome of the Caribbean stony coral Porites asteroides from three developmental stages. Gigascience 5 (1), s13742-016. doi: 10.1186/s13742-016-0138-1
Mass T., Drake J. L., Peters E. C., Jiang W., Falkowski P. G. (2014). Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc. Natl. Acad. Sci. U.S.A. 111 (35), 12728–12733. doi: 10.1073/pnas.1408621111
Mayfield A. B., Wang Y., Chen C. S., Lin C. Y., Chen S. H. (2014). Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol. Ecol. 23 (23), 5816–5830. doi: 10.1111/mec.12982
Moberg F., Folke C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econ. 29 (2), 215–233. doi: 10.1016/S0921-8009(99)00009-9
Moya A., Huisman L., Ball E. E., Hayward D. C., Grasso L. C., Chua C. M., et al. (2012). Whole Transcriptome Analysis of the Coral Acropora millepora Reveals Complex Responses to CO 2-driven Acidification during the Initiation of Calcification. Mol. Ecol. 21, 2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x
Muscatine L. (1967). Glycerol excretion by symbiotic algae from corals and Tridacna and its control by the host. Sci. (1979). 156 (3774), 516–519. doi: 10.1126/science.156.3774.516
Odum H. T., Odum E. P. (1955). Trophic structure and productivity of a windward coral reef community on eniwetok atoll. Ecol. Monogr. 25 (3), 291–320. doi: 10.2307/1943285
Özbek S. (2011). The cnidarian nematocyst: A miniature extracellular matrix within a secretory vesicle. Protoplasma 248 (4), 635–640. doi: 10.1007/s00709-010-0219-4
Palmer C. V. (2018). Immunity and the coral crisis. Commun. Biol. 1 (1), 91. doi: 10.1038/s42003-018-0097-4
Polato N. R., Vera J. C., Baums I. B. (2011). Gene discovery in the threatened elkhorn coral: 454 sequencing of the acropora palmata transcriptome. PloS One 6 (12), e28634. doi: 10.1371/journal.pone.0028634
Rachamim T., Morgenstern D., Aharonovich D., Brekhman V., Lotan T., Sher D. (2015). The dynamically evolving nematocyst content of an Anthozoan, a scyphozoan, and a hydrozoan. Mol. Biol. Evol. 32 (3), 740–753. doi: 10.1093/molbev/msu335
Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., et al. (2021). Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. U.S.A. 118 (5), e2022653118. doi: 10.1073/pnas.2022653118
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cléon I., Guichard N., et al. (2013). The skeletal proteome of the coral acropora millepora: The evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 30 (9), 2099–2112. doi: 10.1093/molbev/mst109
Raz-Bahat M., Douek J., Moiseeva E., Peters E. C., Rinkevich B. (2017). The digestive system of the stony coral Stylophora pistillata. Cell Tissue Res. 368, 311–323. doi: 10.1007/s00441-016-2555-y
Reimer J. D., Kise H., Wee H. B., Lee C. L., Soong K. (2019). Crown-of-thorns starfish outbreak at oceanic Dongsha Atoll in the northern South China Sea. Mar. Biodivers. 49, 2495–2497. doi: 10.1007/s12526-019-01021-2
Reyes-Bermudez A., DeSalvo M. K., Voolstra C. R., Sunagawa S., Szmant A. M., Iglesias-Prieto R., et al. (2009). Gene expression microarray analysis encompassing metamorphosis and the onset of calcification in the scleractinian coral Montastraea faveolata. Mar. Genomics 2 (3-4), 149–159. doi: 10.1016/j.margen.2009.07.002
Rosic N., Kaniewska P., Chan C. K. K., Ling E. Y. S., Edwards D., Dove S., et al. (2014). Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genomics 15, 1–17. doi: 10.1186/1471-2164-15-1052
Schmidt-Roach S., Miller K. J., Lundgren P., Andreakis N. (2014). With eyes wide open: A revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zool. J. Linn. Soc. 170 (1), 1–33. doi: 10.1111/zoj.12092
Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476 (7360), 320–323. doi: 10.1038/nature10249
Sun Y., Lan Y., Rädecker N., Sheng H., Diaz-Pulido G., Qian P. Y., et al. (2024). Gene expression of Pocillopora damicornis coral larvae in response to acidification and ocean warming. BMC Genomic Data 25 (1), 28. doi: 10.1186/s12863-024-01211-3
Swain T. D., Vega-Perkins J. B., Oestreich W. K., Triebold C., DuBois E., Henss J., et al. (2016). Coral bleaching response index: a new tool to standardize and compare susceptibility to thermal bleaching. Glob. Chang. Biol. 22 (7), 2475–2488. doi: 10.1111/gcb.13276
Tambutté S., Holcomb M., Ferrier-Pagès C., Reynaud S., Tambutté É., Zoccola D., et al. (2011). Coral biomineralization: From the gene to the environment. J. Exp. Mar. Biol. Ecol. 408 (1-2), 58–78. doi: 10.1016/j.jembe.2011.07.026
Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinf. 4, 1–14. doi: 10.1186/1471-2105-4-41
Wilson E. B., Hilferty M. M. (1931). The distribution of chi-square. Proc. Natl. Acad. Sci. 17 (12), 684–688. doi: 10.1073/pnas.17.12.684
Keywords: Pocillopora damicornis, ScRNA-seq, endosymbiosis, gastrodermis, epidermis, cnidocyte, calicoblast
Citation: Diao W, Zhang S, Zhu X, Wu P, Du B, Han Z, Liu Y, He C and Lu Z (2025) Single-cell transcriptome sequencing reveals the cell populations and functions associated with stony coral Pocillopora damicornis. Front. Mar. Sci. 12:1551763. doi: 10.3389/fmars.2025.1551763
Received: 26 December 2024; Accepted: 10 February 2025;
Published: 28 February 2025.
Edited by:
Ana Varela Coelho, Universidade Nova de Lisboa, PortugalCopyright © 2025 Diao, Zhang, Zhu, Wu, Du, Han, Liu, He and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunqing Liu, bGl1eXFAbGl0LmVkdS5jbg==; Chunpeng He, Y3BoZUBzZXUuZWR1LmNu; Zuhong Lu, emhsdUBzZXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.