
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 02 April 2025
Sec. Aquatic Microbiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1549732
Anthropogenic climate change is altering coastal systems globally, affecting macroalgae abundance and composition. These macroalgae host diverse microbiomes, including pathogenic bacteria. Of particular concern are Vibrio species, such as Vibrio parahaemolyticus and Vibrio vulnificus, which are linked to human disease and impact public health, the economy, and recreation in coastal areas. This study examined the presence and abundance of pathogenic Vibrio spp. across seven genera of macroalgae in a temperate estuary (Narragansett Bay, RI, USA). Using colony-forming unit (CFU) counts and multiplex qPCR, we quantified V. parahaemolyticus and V. vulnificus abundance to assess if pathogenic Vibrio abundance varied by macroalgae genus and morphology. We also examined potential environmental factors influencing pathogenic Vibrio prevalence. We demonstrate that both V. vulnificus and V. parahaemolyticus were present on all macroalgae genera, with V. vulnificus showing higher average abundance. Environmental factors like temperature, salinity, and nutrient concentrations did not strongly correlate with V. vulnificus or V. parahaemolyticus abundance, suggesting that macroalgae might offer a protective microhabitat for these pathogens. Macroalgae with opportunistic life strategies had the highest abundance of V. vulnificus and V. parahaemolyticus, highlighting their potential role as reservoirs for pathogenic Vibrio spp. Future research should explore broader environmental contexts and macroalgae–Vibrio spp. interactions to better understand and forecast pathogen dynamics.
Marine macroalgae are a diverse group of photosynthetic eukaryotic primary producers who perform vital services for ecosystems worldwide. In fact, marine macroalgae are considered the most productive and extensive organisms of all coastal vegetated ecosystems on the planet (Ji and Gao, 2021; Duarte et al., 2022). They live in every ocean and along every continent, with extensive ranges of morphology, physiological requirements, and lifestyle types (e.g., attached and slower growing vs. free floating and fast growing) (Gattuso et al., 2006; Duarte et al., 2022). The highly productive habitats formed by macroalgae support both local and global food webs and important commercial and recreational fisheries (Krumhansl and Scheibling, 2012; Pessarrodona et al., 2018; Queirós et al., 2019; Duarte et al., 2022).
Marine macroalgae abundance, composition, and distribution are changing due to human activities at global (e.g., anthropogenic climate change) and local (e.g., nutrient loading) scales. Globally, changes in ocean conditions such as acidification and warming have led to widespread replacement of slower-growing perennial macroalgae with more opportunistic species, causing fundamental changes in ecosystem function worldwide (Connell and Russell, 2010; Ji and Gao, 2021). On a local scale, nutrient-rich waters can fuel large macroalgae blooms (Valiela et al., 1997; Wiencke and Bischof, 2012; Ji and Gao, 2021). These macroalgae blooms are increasing in frequency and scale in regions across the world and are positively correlated with human-caused nutrient loading, with hotspots in western Europe, North America, Central America, and China (Joniver et al., 2021). Macroalgae blooms have significant impacts on ecosystem services and cause changes in the ecology of coastal systems spanning trophic levels (Wang et al., 2020).
Expanding macroalgae habitat and increasing macroalgae growth can be positive as humans use macroalgae in a wide variety of ways, such as in nutrition-dense food products, manufacturing, fertilizer, pharmaceuticals, and animal feed (Garcia-Vaquero and Hayes, 2016; Peñalver et al., 2020; Duarte et al., 2022). Macroalgae also represent an important global carbon sink (Pessarrodona et al., 2018) and a tool for the bioremediation of anthropogenically impacted waters (Kumar and Sudhakar, 2024). However, macroalgae blooms can lead to deleterious ecosystem consequences. For example, macroalgae can outcompete seagrasses for resources (Zribi et al., 2023), macroalgae senescence and decomposition encourages hypoxia and anoxia (Castel et al., 1996; Viaroli et al., 1996; Senga et al., 2021), and large blooms are associated with losses of ecosystem biodiversity (Ji and Gao, 2021). Another emerging potential negative impact of excess macroalgae is that they can host pathogenic bacteria (e.g., Vibrio spp.) (Noorian et al., 2023). Macroalgae are regularly colonized by diverse microbial communities, with the genera Vibrio spp. being a commonly detected member (Akrong et al., 2023). The characteristics of the macroalgae (e.g., growth style, tissue differentiation, morphology, and physiological requirements) play a major role in determining the composition of their microbial community (Littler and Littler, 1980; Steneck and Watling, 1982; Staufenberger et al., 2008; Barott et al., 2011; Lachnit et al., 2011; Lemay et al., 2021). It remains largely unknown if macroalgae could be a host for the human pathogen Vibrio spp., specifically V. vulnificus and V. parahaemolyticus. Few studies have quantified the abundance of specifically pathogenic Vibrio on macroalgae; however, those that have suggest that macroalgae may provide a habitat for pathogenic Vibrio spp. (Mahmud et al., 2007; Mahmud et al., 2008; Gonzalez et al., 2014).
Vibrio spp. are halophilic, heterotrophic bacteria that occur naturally in marine and estuarine environments worldwide. They can account for as much as 40% of the readily culturable bacteria population in estuaries (Thompson and Polz, 2014; Urakawa and Rivera, 2014). Of the over 100 species contained within the Vibrio genus, the two non-cholera species of most interest are V. vulnificus and V. parahaemolyticus. V. vulnificus has been detected across a wide range of salinities (4‰ to 37‰) and temperatures (7°C–36°C) and found to grow optimally at salinities between 10‰ and 25‰ and at a temperature of 20°C (Noorian et al., 2023). V. vulnificus is an emerging pathogen which can cause necrotizing fasciitis and primary septicemia when an open wound or an immunocompromised individual is exposed (Tison and Kelly, 1984). V. vulnificus infections account for 95% of all Vibrio spp.-related deaths in the United States and are considered pathogenic whenever present (Morris and Acheson, 2003). V. parahaemolyticus preferentially grows in warm (>15°C), low salinity <25‰ waters. V. parahaemolyticus is the most common pathogenic bacterium associated with raw or undercooked seafood consumption and typically leads to self-limiting gastroenteritis (Baker-Austin et al., 2018). Rarely, V. parahaemolyticus can also cause necrotizing fasciitis and septicemia in immunocompromised individuals (Letchumanan et al., 2014).
While not all V. parahaemolyticus strains are pathogenic, those that are possess a complex array of factors that control their virulence. Pathogenic V. parahaemolyticus strains are typically identified by the presence of two key genes: thermostable direct hemolysin (tdh) and thermostable direct-related hemolysin (trh) (Nishibuchi et al., 1992; Xu et al., 1994; Yeung and Boor, 2004; Raghunath, 2015). However, strains carrying both tdh and trh are less common in environmental isolates, with their prevalence generally lower compared to clinical strains (e.g., those isolated from hospitalized patients) (Nishibuchi et al., 1992; DePaola et al., 2000; DePaola et al., 2003). In contrast, V. vulnificus is always considered pathogenic and is commonly identified by the presence of the toxR gene, a crucial marker for its virulence (Warner and Oliver, 2008; Baker-Austin et al., 2018; U.S. Food and Drug Administration (FDA) and Center for Food Safety and Applied Nutrition, 2024).
Increases in the abundance of V. vulnificus and V. parahaemolyticus are associated with climate change, and they are undergoing a global expansion typically related to warming of coastal waters (Baker-Austin et al., 2017; Almagro-Moreno et al., 2023; Noorian et al., 2023). Reports of Vibrio infections are on the rise (Baker-Austin et al., 2010; Baker-Austin et al., 2013). In addition to increasing the abundance of Vibrio spp. in the water, elevated water temperatures may also increase the virulence and transmission of marine pathogens widely by increasing the pathogen’s metabolism and fitness (Karvonen et al., 2010; Burge et al., 2014; Lam et al., 2014; Billaud et al., 2022; Roncarati et al., 2024). The prevalence of Vibrio spp. is predicted to make a significant expansion into the higher latitudes of the northern hemisphere in the next century as temperatures rise and expand their habitable range (Baker-Austin et al., 2010; Baker-Austin et al., 2013; Almagro-Moreno et al., 2023). The increasing risk for Vibrio spp. infection has made it essential to understand the ecology, drivers, and reservoirs for pathogenic Vibrio spp. in the environment.
The purpose of this study was to assess whether the abundance and composition of pathogenic V. vulnificus and V. parahaemolyticus varied across space, time, and macroalgae genera. To do so, we quantified the abundance of these pathogenic strains on seven dominant macroalgae genera, representing all three classes of marine macroalgae, over the growing season in a temperate estuary (Narragansett Bay, RI, USA). We also measured environmental characteristics commonly linked to Vibrio spp. abundance, such as temperature, salinity, and total suspended particles. We had two hypotheses: 1) V. vulnificus and V. parahaemolyticus would be present on all macroalgae, even in small quantities, with more complex morphologies and opportunistic species exhibiting higher abundance due to their larger surface area-to-volume ratio (Ji and Gao, 2021). 2) Both V. vulnificus and V. parahaemolyticus would show a clear peak in abundance across all sites and genera, coinciding with higher temperatures, as previous research suggests these Vibrio spp. prefer warmer waters (Baker-Austin et al., 2018). To test these hypotheses, V. vulnificus and V. parahaemolyticus abundance was quantified through colony-forming unit counting, followed by species identification using qPCR and assays to assess pathogenicity.
Narragansett Bay is a well-mixed, shallow (~8 m mean depth) estuary located in the state of Rhode Island on the east coast of the United States. Narragansett Bay broadly follows a north–south gradient in salinity, nutrient loading, and productivity driven by freshwater inputs at the north end of the Bay. While typically described as a phytoplankton-based system, it also supports a robust macroalgae community of over 90 documented species (Villalard-Bohnsack and Harlin, 1992). We selected three sites for this study (Figure 1): site 1 is the northern most and shallowest site; site 2 is a small, shallow subestuary (called Greenwich Bay) that branches off along the northwest side of Narragansett Bay; and site 3 is the southernmost site, in a sheltered embayment with long-term poor water quality (Lorraine and Lucht, 2000; Oakley et al., 2012). Overall, during this study, the sites had similar physiochemical conditions, with some differences in nutrient and water column chlorophyll concentrations (Table 1). Environmental conditions in Narragansett Bay during the study period (June–October 2022) were typical for late spring through early fall. Water temperatures ranged from 21.2°C to 27.4°C, with the highest temperatures recorded in August. Salinity remained relatively stable, varying between 25‰ and 31.6‰. Dissolved inorganic nitrogen (DIN) concentrations fluctuated across sites, with the highest levels observed in August. Total suspended solids (TSS) was variable, with the highest levels in July. These environmental factors provide context for interpreting Vibrio spp. abundance patterns (please see Supplementary Table S2 for more details).
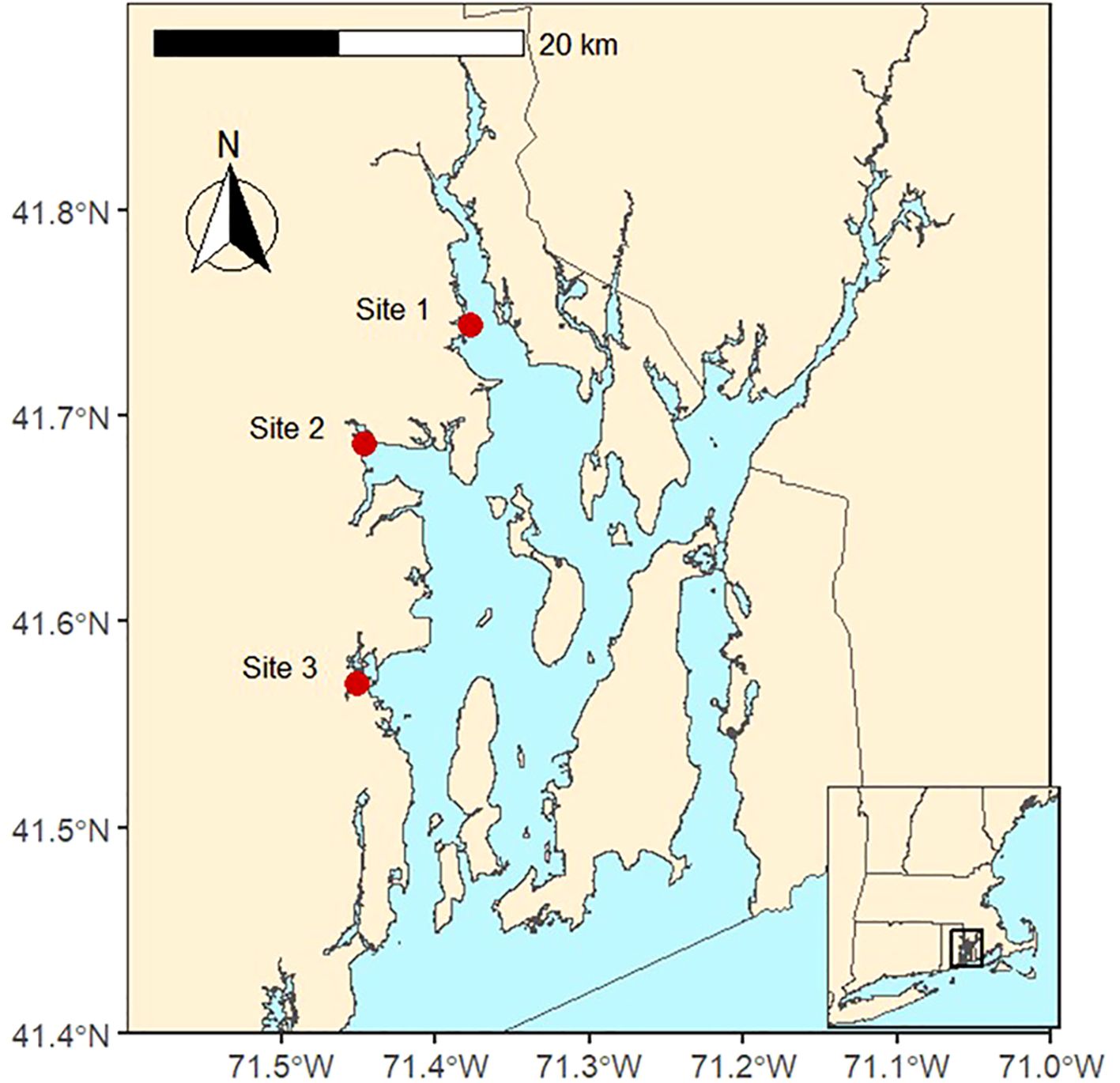
Figure 1. The three sampling locations (shown with red dots) in Narragansett Bay, Rhode Island, USA. Samples of macroalgae, water column nutrients, chlorophyll-a, and physiochemical parameters were collected at each site between June and October 2022.
We collected samples twice monthly from June 2022 to September 2022 and once in October 2022 for a total of nine sampling events at each site. We chose this sampling scheme to capture the warmest yearly temperatures, which should facilitate pathogenic Vibrio spp. growth (Baker-Austin et al., 2018).
We collected samples from the shore either off the beach (site 1; Figure 1) or off floating docks (sites 2 and 3; Figure 1). At each sampling event, triplicates of the three most abundant genera of live macroalgae were collected by hand in sterile Whirl-Pak™ bags to determine the concentration of pathogenic Vibrio spp. on the thalli. We placed the bags on ice in a dark cooler and transported them back to the laboratory at Boston University for processing within 3 h of collection. Macroalgae with visible epibiont growth (e.g., bryozoans, filamentous fouling organisms) were excluded from sampling to minimize confounding effects of epibionts on Vibrio spp. abundance.
Along with the macroalgae samples, we collected water samples for concentrations of dissolved inorganic nitrogen and phosphorus, chlorophyll-a and phaeophytin, and total suspended solids. In the field, we filtered duplicate water samples using a 60-mL acid-washed polypropylene syringe with glass fiber filters (Whatman GF/F, 0.70 micron pore size) into 30 mL of acid-washed and deionized water-leached polyethylene bottles. The filter samples were stored on ice in a dark cooler until being returned to the lab and then were stored in a freezer (−20°C) until analysis for dissolved inorganic nitrogen [DIN: ammonium (NH4+), nitrite (NO2−), nitrate (NO3−)] and dissolved inorganic phosphate (DIP) (Foster and Fulweiler, 2014). Chlorophyll-a and phaeophytin measurements were also collected in duplicate at each site using a deionized water cleaned 60 mL polypropylene syringes (Arar and Collins, 1997; Fagherazzi et al., 2014; Ray et al., 2020). Filters were stored in the dark, on ice, until being returned to the lab, and then at −80°C until analysis. During each macroalgae collection, we also measured water column temperature, salinity, and dissolved oxygen using a Hach sensor (LDO101).
Over the duration of this study, 233 macroalgae samples were collected and analyzed for the presence of pathogenic Vibrio spp. bacteria. Out of the 233 total macroalgae samples, phylum Rhodophyta accounted for 112 samples, phylum Chlorophyta for 112 samples, and phylum Phaeophyceae accounted for the rest. The majority (n = 210) of the macroalgae were opportunistic. Of the seven macroalgae genera we studied, three genera were classified as a thin blade morphology, two had a finely branched morphology, and two were coarsely branched (further sample information may be found in Supplementary Table S1) (Hurd, 2000).
Macroalgae was identified to the genus level and other characteristics (e.g., morphology type) were identified using surface area to volume (SA:V) ratios reported by (Littler and Littler, 1980). Note that while the genera of Enteromorpha and Ulva contain multiple synapomorphies and the nomenclature is altered, for the purpose of this study, we chose to keep them separated due to the large differences in morphology (and therefore SA:V) (Hayden et al., 2003; Ning et al., 2022). We processed the macroalgae samples as outlined by (Gonzalez et al., 2014). Briefly, we weighed 10 g (wet weight) of the macroalgae sample, then combined it with 100 mL of phosphate-buffered saline (PBS) and shook the sample for 5 min to dislodge the microbial biofilm on the macroalgae where Vibrio spp. may be present (Figure 2). Immediately after shaking, samples from the resulting liquid were serially diluted with PBS and then plated. Specifically, we plated all macroalgae samples on CHROMagar™ Vibrio medium (CHROMagar™, Paris, France) to determine the counts for V. vulnificus and V. parahaemolyticus (Supplementary Figure S1). To control for the variable initial water content of the macroalgae, we determined the dry weight of each sample by drying the initial 10 g of the samples in preweighed tins in a 60°C oven for a minimum of 48 h (or until constant weight) and reweighed.
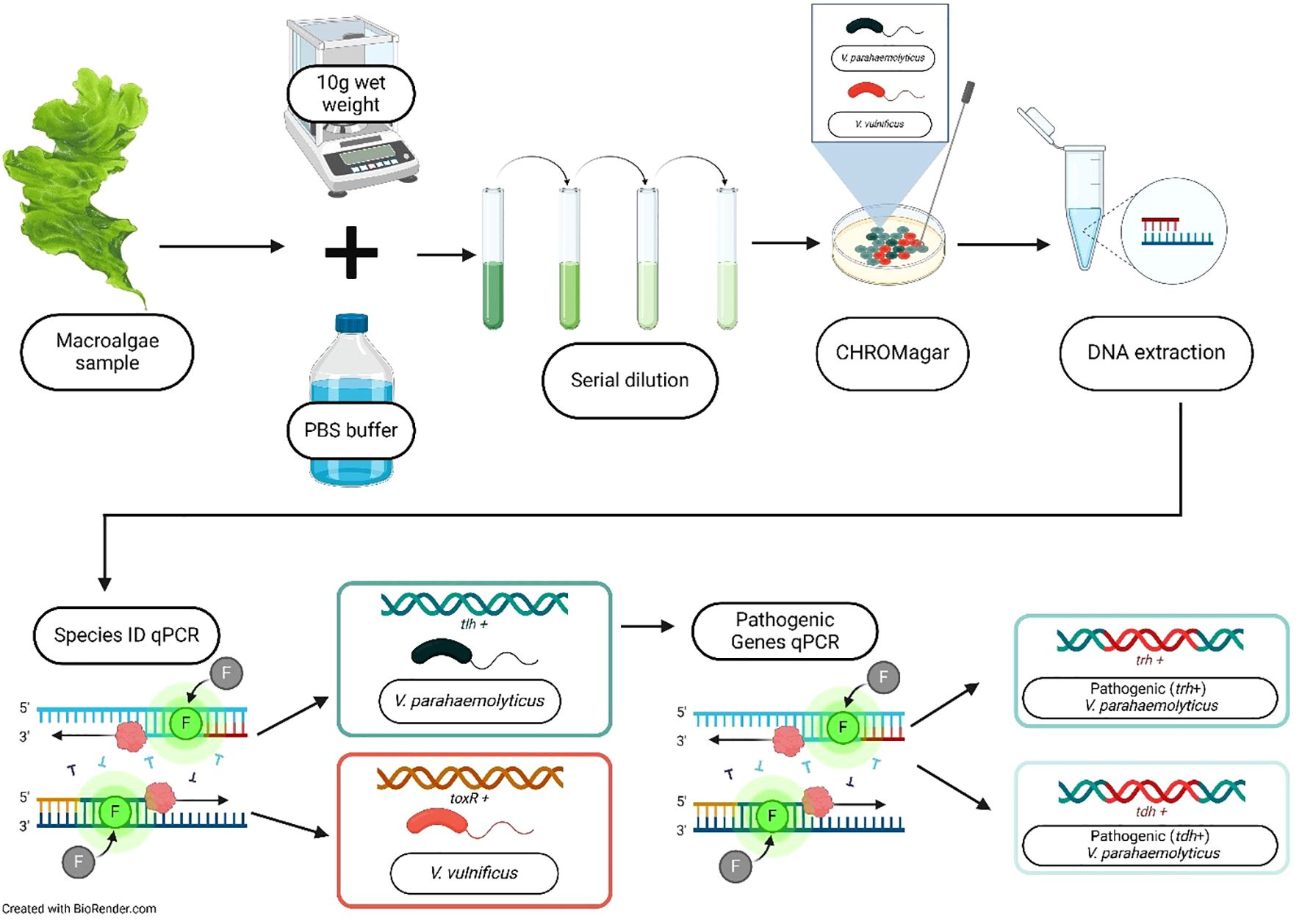
Figure 2. Flowchart of the methods for quantifying V. vulnificus and V. parahaemolyticus on macroalgae. Each 10 g (wet weight) of the macroalgae sample was shaken in 100 mL of phosphate-buffered saline (PBS) to dislodge surface-associated Vibrio spp. The mixture was serially diluted, plated on CHROMagar™, and incubated, and colony-forming units (CFUs) were counted. Presumptive V. vulnificus and V. parahaemolyticus colonies were picked for DNA extraction using the Qiagen purification kit. Multiplex qPCR was then performed to confirm species presence and quantify pathogenic genes. Created in BioRender. Geisser, A. (2025) https://BioRender.com/0fwtfx1.
We first confirmed Vibrio species type using previously published duplex qPCR assays (Scro et al., 2019). We confirmed the presence of V. parahaemolyticus by using specific primers for the gene encoding for thermostable hemolysin (tlh) that is found in all V. parahaemolyticus. V. vulnificus was confirmed by the toxR gene, which codes for the transmembrane virulence regulator gene present in this species (Warner and Oliver, 2008; Scro et al., 2019). The primers and probes for both Vibrio species and V. parahaemolyticus pathogenic gene identification in a secondary multiplex qPCR can be found in Supplementary Table S1 (Scro et al., 2019).
Next, we performed another multiplex qPCR assay on any sample positive for V. parahaemolyticus to determine the presence of pathogenic genes (Supplementary Table S3). Genes that encode for thermostable direct hemolysin (tdh) and thermostable direct hemolysin-related hemolysin (trh) are strongly representative of the virulence of V. parahaemolyticus (Nishibuchi et al., 1992; Gutierrez West et al., 2013). All V. vulnificus species were considered pathogenic, so no further genetic identification was needed.
We used standard colorimetric techniques and a high-resolution digital colorimetry on a SEAL AutoAnalyzer 3 with segmented flow injection to analyze water samples for dissolved inorganic nitrogen and phosphorus (Grasshoff et al., 1999; Solórzano, 1969; Johnson and Petty, 1983). The detection limits for these analyses were 0.080 µM for NH4+, 0.013 µM for NOx, 0.013 µM for NO3−, 0.006 µM for NO2−, and 0.010 µM for PO43−. We analyzed filters for chlorophyll-a and pheophytin using standard acetone extraction and a Turner Designs TD-700 fluorometer (Sigma-Aldrich, St. Louis, MO, USA) calibrated with pure chlorophyll-a (Solórzano, 1969; Johnson and Petty, 1983; Hansen and Koroleff, 2007; Foster and Fulweiler, 2014).
All statistical analyses were conducted in R version 4.3.3 (R Core Team, 2024). The results were considered statistically significant when p <0.05. Both V. vulnificus and V. parahaemolyticus count data were found to best fit a negative binomial distribution, using the fitdistruplus package (Delignette-Muller and Dutang, 2015). The data for both V. vulnificus and V. parahaemolyticus met all criteria for a zero-inflated negative binomial (ZINB) regression model, meaning the data had excess zeros compared to a standard Poisson or negative binomial distribution, variance greater than the mean which indicates overdispersion, and heterogeneity and justification in both the count and zero-generating facets of the process. Because standard models cannot capture the underlying structure of these data, we employed a ZINB regression model. We used the R package nyiuab/NBZIMM, built on top of the commonly used R packages nlme and MASS, to evaluate the potential environmental drivers of V. vulnificus and V. parahaemolyticus bacteria counts (Zhang et al., 2017).
To determine which predictor variables were driving pathogenic Vibrio counts on macroalgae, we created two sets of ZINB models using predictor variables of interest that did not covary (determined by first calculating a Pearson correlation, Supplementary Figure S2). These predictor variables are the environmental parameters we collected at each sampling date to correspond with Vibrio spp. abundance. Each environmental parameter was included in the model as an individual measurement corresponding to each sampling date rather than as a seasonal average. To assess which predictor variables to include in the final model, we assessed a combination of statistical significance, theoretical relevance, and ZINB model considerations. For a predictor variable to be included in the final model, the count model (negative binomial part) for each parameter needed to have a p-value (Pr(>|z|)) less than the significance level of 0.05, signifying a statistically relevant association between the predictor variable and the Vibrio spp. count. Temporal variation was inherently captured by using environmental data from each sampling event rather than aggregated seasonal values. If significant seasonal patterns were present, they would be reflected in the relationships identified by the ZINB model. Once the variables of interest were selected, we performed backwards stepwise variable elimination to find the best fit model using the function be.zeroinfl in the package mpath (Wang et al., 2015). The model for each Vibrio species that best explained the variation in the data and had the least number of parameters was selected based on the lowest Akaike’s information criterion (AIC) (Sakamoto et al., 1986). To quantify the explanatory power of the ZINB model, we calculated Nagelkerke’s adjusted R2 value (Foster et al., 2005; Martin and Hall, 2016). Due to the complex nature of ZINB models, we based our analysis and interpretation of predictor variable importance on Nagelkerke’s adjusted R2 due to this approach being scaled to resemble the R2 from linear regression more closely (Foster et al., 2005). We tested whether V. vulnificus and V. parahaemolyticus abundances were significantly different between sites, genera, class, morphology, and lifestyle type of macroalgae using pairwise least-square means tests through the emmeans package (Lenth, 2017).
V. vulnificus and V. parahaemolyticus were quantified using CFU counts, and the two species were then confirmed using qPCR multiplex assays to target genes present in each respective species. V. vulnificus is always considered pathogenic, while V. parahaemolyticus needs to be further analyzed for the presence of pathogenic genes using a qPCR multiplex assay with samples that tested positive for initial detection of the species (Scro et al., 2019). We first give background on macroalgae samples tested, then describe pathogenic Vibrio spp. abundance on macroalgae via CFUs of each species that have been confirmed using molecular methods. We do this because molecular methods paired with traditional counting methods (CFU and MPN) are more sensitive and precise in enumerating pathogenic Vibrio spp. Then, we describe the pathogenic gene abundance in the context of all samples taken, as well as further identifying the presence of pathogenic genes within V. parahaemolyticus.
We acknowledge that while CHROMagar™ Vibrio is a widely used selective medium, there is a potential for misidentification due to the metabolic overlap among Vibrio species (Yeung and Thorsen, 2016; Parveen et al., 2020). However, we followed the FDA-BAM protocol for enumeration and species confirmation, which is based on statistical likelihood rather than exhaustive molecular confirmation of all colonies. To enhance confidence in our identifications, we tested as many colonies as we could identify, beyond the minimum subset required. This approach aligns with previous studies using similar methods (Gonzalez et al., 2014; Barberi et al., 2020, Givens et al., 2014; Parveen et al., 2008; Brumfield et al., 2023). While we did not adjust CFU estimates based on PCR confirmation rates, our methodological consistency with FDA protocols and prior research suggests that our reported Vibrio spp. abundances are robust. Future work using whole-genome sequencing or MALDI-TOF could further validate species assignments and refine culture-based methods for Vibrio identification (Moussa et al., 2021). We found that pathogenic Vibrio spp. were present across all genera, with mean counts of 1.12 × 105 CFU g−1 for V. parahaemolyticus and 3.68 × 106 CFU g−1 for V. vulnificus (Figure 3). Ulva spp. had the highest count for V. vulnificus (3.8 × 107 CFU g−1), and Fucus spp. had the highest count for V. parahaemolyticus (3.3 × 106 CFU g−1).
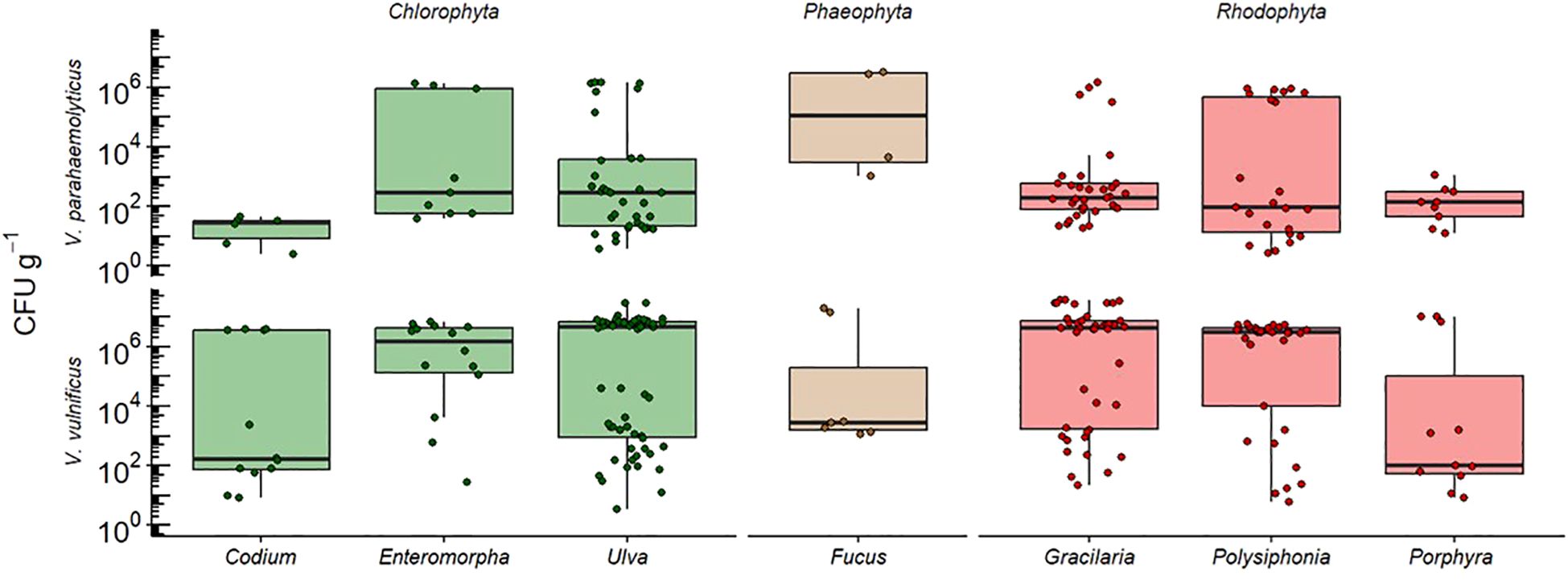
Figure 3. Counts of V. vulnificus and V. parahaemolyticus colony-forming units (CFUs) isolated from macroalgae samples during the study period. CFU counts were determined following the US FDA-BAM protocol, with presumptive identifications based on CHROMagar™ color and morphology and species confirmation through qPCR targeting the toxR (for V. vulnificus) and tlh (for V. parahaemolyticus) genes. While not every individual colony was tested, genetic confirmation was conducted on representative colonies from each sampling site and date. Whiskers represent ± SE and the median is represented by the solid black line in the box plot. There were no significant differences in bacterial abundance between macroalgae genera.
We hypothesized that V. vulnificus and V. parahaemolyticus would vary by life strategy because microbial communities may have more time to accumulate on older tissues (Goecke et al., 2010). Instead, we found that V. vulnificus and V. parahaemolyticus abundance did not vary by life strategy (Supplementary Table S3). However, there was a clear abundance of opportunistic macroalgae (e.g., Ulva) present at all sites (n = 210) compared to a lack of slower-growing genera (e.g., Fucus, n = 23), so there may be associations not captured in this study.
We hypothesized that macroalgae morphology, which is reflective of the host’s ecological niche and surface area to volume ratio, would be a significant predictor of V. parahaemolyticus and V. vulnificus abundance. We anticipated this because bacteria generally follow the well-established ecological paradigm, where increasing habitat structural complexity increases biodiversity and richness in the system (Lemay et al., 2021). We found that V. vulnificus abundances were significantly higher (p = 0.0203) on the finely branched macroalgae with complex morphology and higher surface-area-to-volume ratio compared to other morphologies. V. parahaemolyticus abundance, however, did not vary significantly by morphology type (Supplementary Table S3).
Overall, 233 macroalgae samples across seven genera were taken throughout the study period. Out of the 233 samples, 117 had no growth of presumptive Vibrio spp. bacteria on CHROMagar plates, while 116 samples tested positive for the V. parahaemolyticus species identifier gene (tlh) and 101 tested positive for the V. vulnificus identifier gene (toxR, Figure 4). These two genes are not mutually exclusive, and both species of Vibrio may be present in the sample and grow on the CHROMagar plate. The genera Ulva, Gracilaria, and Polysiphonia have higher numbers of samples that tested positive for the toxR (V. vulnificus) and tlh (V. parahaemolyticus) genes, and these were also the genera with the most total samples collected. V. vulnificus is considered pathogenic with the detection of the toxR alone, so further analysis was not necessary. However, to determine the pathogenicity of V. parahaemolyticus, we quantified pathogenic gene presence (tdh and/or trh). Any sample that tested positive for the presence of the V. parahaemolyticus species identifier gene tlh was then further tested for the presence of the two pathogenic genes trh and tdh. Of the tlh-positive samples, 101 tested positive for trh and 21 tested positive for tdh (Figure 4). These genes are also not mutually exclusive and do often co-occur. In this study, positive tdh detections always co-occurred in samples that were also positive for trh.
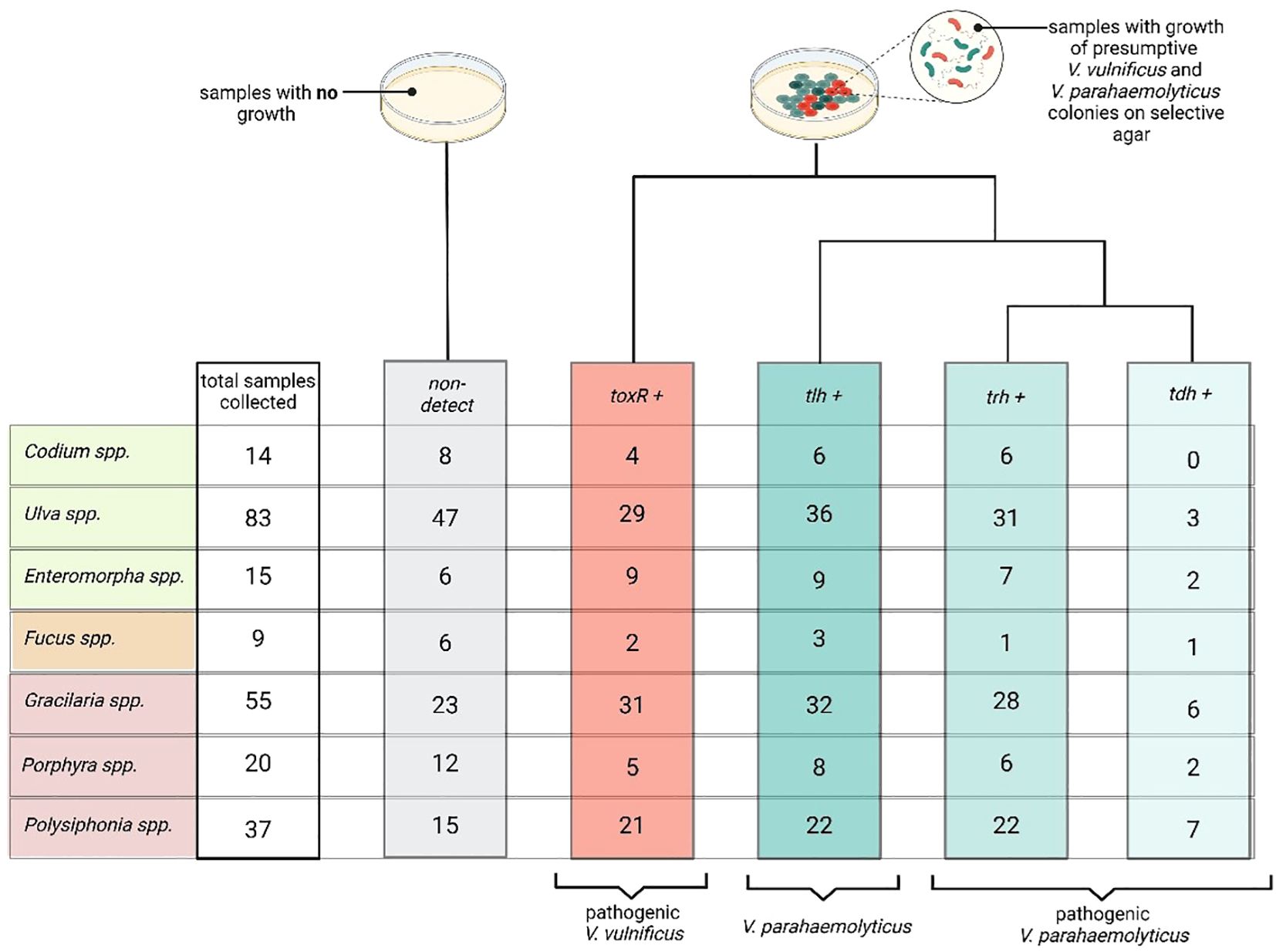
Figure 4. Summary of macroalgae samples collected, plated, and tested for Vibrio species and pathogenic genes. The first column represents the total number of macroalgae samples collected for each genus, including both positive and negative detections. The second column (gray, non-detected) indicates samples that did not grow colonies with the characteristic Vibrio morphology on CHROMagar™ and were therefore not tested further. The third column (red, toxR+) represents samples that contained colonies confirmed as V. vulnificus via qPCR detection of the toxR gene, which is species-specific and associated with pathogenicity. The fourth column (blue, tlh+) shows samples with V. parahaemolyticus, confirmed through detection of the tlh gene. The next columns display samples that tested positive for the pathogenic trh (blue, trh+) and tdh (blue, tdh+) genes, both of which indicate virulent V. parahaemolyticus. tdh was only detected in samples that were also positive for trh, suggesting the presence of highly virulent serotypes. Created in BioRender. Geisser, A. (2025) https://BioRender.com/0fwtfx1.
We hypothesized that both V. parahaemolyticus and V. vulnificus abundances would be linked to environmental parameters because previous research has established these links, particularly with salinity, temperature, and, to varying degrees, nutrient concentrations (Thompson and Polz, 2006; Baker-Austin et al., 2010; Takemura et al., 2014; Paranjpye et al., 2015; Sha et al., 2022; Kalvaitienė et al., 2023). The bulk of water column physiochemical parameters linked to Vibrio spp. abundance is inconsistent across studies and relationships dependent on taxonomic, geographic, and temporal resolution (Takemura et al., 2014). Our ZINB model identified NH4+, NOx, and salinity as the most significant predictors of V. parahaemolyticus abundance (CFU g−¹), with the lowest AIC values supporting their inclusion in the model (Table 2). In contrast, V. vulnificus abundances were best predicted by TSS, DIP, and salinity, based on AIC model selection (Table 2). These predictor variables are the environmental parameters collected at each sampling date, which were used to assess their association with Vibrio spp. abundance. Despite these being significant predictors, the overall explanatory power of the model remained limited (Nagelkerke’s R² = 0.20 for V. parahaemolyticus and 0.05 for V. vulnificus), indicating that other unmeasured factors may also be influencing Vibrio dynamics in these samples.
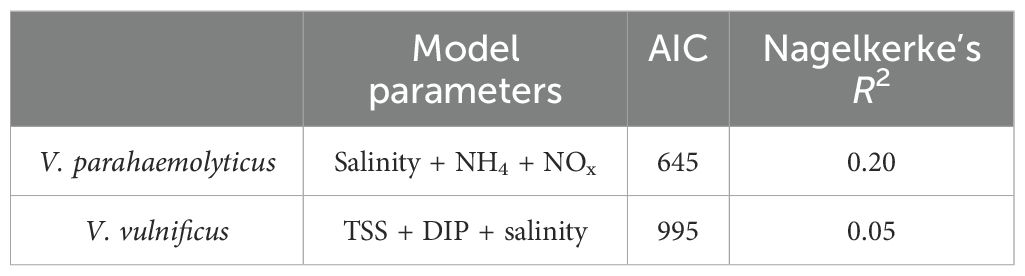
Table 2. ZINB models were created with the predictor variables determined to be statistically significant to Vibrio spp. abundance.
The final models were chosen based on AIC values, indicating the model’s quality of model fit and complexity, where a lower AIC value indicates a model with a better trade-off between goodness-of-fit and the number of parameters. Nagelkerke’s R2 was calculated to assess the explanatory power of the model as it is scaled to resemble the R2 from linear regression more closely, allowing for a clearer interpretation. For stepwise AIC values for each model, please see Supplementary Tables S4, S5.
Here, we show that V. parahaemolyticus and V. vulnificus are present on a variety of common temperate macroalgae. The abundances of V. parahaemolyticus and V. vulnificus documented in this work are on par or higher than those reported in previous studies investigating macroalgae and pathogenic Vibrio spp. For example (Gonzalez et al., 2014), report maximum abundances of 1.2 × 104 CFU g−1 of V. parahaemolyticus and 3.2 × 103 CFU g−1 of V. vulnificus on Gracilaria spp. during the growing season in Virginia, in the mid-Atlantic region of the United States. Additionally, (Barberi et al., 2020). found a maximum of 8.0 × 102 CFU g−1 of V. parahaemolyticus on Saccharina latissima during its growing season in the Gulf of Maine in the Northern Atlantic United States. Another study from Japan on Porphyra spp. and Undaria spp. reported 86 MPN g−1 of V. vulnificus and 110 MPN g−1 of V. parahaemolyticus (Mahmud et al., 2007; Mahmud et al., 2008). (MPN and CFU are considered equivalent units and thus directly comparable, with MPN being a statistics-based measurement and CFU being a direct counting measurement. MPN can have a lower limit of detection, but CFU can provide finer-scale information). Our initial hypothesis that both Vibrio spp. would vary by macroalgae morphology type was not supported for V. parahaemolyticus and very weakly supported for V. vulnificus. These data suggest that while V. vulnificus may prefer to colonize different morphological types of macroalgae, morphology is not the main driver of pathogenic Vibrio spp. abundance, at least in this study.
Another key metric used widely to describe Vibrio bacteria and their role in the environment is their pathogenicity levels and strain types. While the presence of virulence genes like tdh and trh is crucial for determining the pathogenic potential of a V. parahaemolyticus strain, these genes do not determine the serotype directly. Instead, they indicate that the strain may be more virulent or have a higher potential to cause illness. The serotype is established through serotyping methods that assess the O and K antigens, whereas the virulence genes provide additional information about the strain’s pathogenic characteristics (Iida et al., 1997; Nair et al., 2007). Samples that are positive for both V. parahaemolyticus pathogenic genes, however, may indicate the presence of the O3:K6 serotype, which has been linked to the V. parahaemolyticus pandemic clonal complex (VpCC) (Martinez-Urtaza et al., 2005; Gonzalez-Escalona et al., 2011; Martinez-Urtaza et al., 2016). Environmental isolates with the tdh and trh genes have rarely been detected in the past, with only 0%–6% testing positive for both genes found in coastal regions of the U.S., Europe, and Asia between 1995 and 2000 (Nishibuchi et al., 1992; DePaola et al., 2000). The potential spread of the predominant pandemic clone, O3:K6, highlights the evolving nature of V. parahaemolyticus virulence (Bej et al., 1999; Okura et al., 2003; Parvathi et al., 2006). The historical rarity of environmental isolates containing both virulence genes indicates that these strains are now potentially becoming more prevalent. This trend could reflect changes in environmental conditions, local human activities, climate change, or likely a combination of all these changes, necessitating further research into the factors driving these emerging genetic patterns. Similarly, V. vulnificus serotypes are based on O antigens; however, no single serotype is consistently linked to virulence the way O3:K6 is in V. parahaemolyticus (Bisharat et al., 1999). Instead, V. vulnificus is classified into three biotypes based on pathogenicity: biotype 1, responsible for most human infections; biotype 2, primarily affecting eels but occasionally infecting humans; and biotype 3, which has been linked to wound infections specifically in Israel (Tison et al., 1982; Høi et al., 1998; Warner and Oliver, 2008). Key genetic markers, such as the vcg (virulence-correlated gene) and rrn (ribosomal RNA operon) type, differentiate clinical and environmental strains, with the vcgC genotype and type B rRNA more commonly associated with human infections (Wright et al., 1981; Gulig et al., 2005; Rosche et al., 2005). As warming ocean temperatures expand its geographical range, V. vulnificus infections are increasingly reported in higher latitudes, emphasizing the need for ongoing environmental surveillance and molecular epidemiology to assess future risks (Johnson et al., 2010). The model results investigating environmental drivers of Vibrio abundance mostly align with general previous reported trends. For example, V. parahaemolyticus grows at higher salinities, with optimum growth found in salinity approximately 25‰, which is similar to the mean salinities from the sites in this study, ranging from 25‰ to 29‰ (Martinez-Urtaza et al., 2008). Nutrient concentrations, particularly nitrogen, may influence Vibrio spp. for varying reasons. The bacteria may simply be responding to an influx of nitrogen stimulating growth of host organisms, as well as using the nutrients for their own growth and metabolism (Blackwell and Oliver, 2008).
Our model identified nitrogen and TSS as factors associated with Vibrio spp. abundance, albeit with a weak correlation. While these relationships were not strong, they align with previous research suggesting that Vibrio spp. may respond to nutrient availability and suspended particles in multiple ways (Blackwell and Oliver, 2008). Elevated nitrogen levels may indirectly promote Vibrio spp. abundance by stimulating the growth of macroalgal hosts, thereby providing more surface area and organic matter for bacterial colonization (Thompson and Polz, 2006; Wang et al., 2020). Additionally, nitrogen could directly support Vibrio metabolism and growth (Thompson and Polz, 2006). Similarly, the presence of TSS may provide microhabitats for Vibrio attachment, shielding them from environmental fluctuations and predation (Venkateswaran et al., 1990; Main et al., 2015; Liang et al., 2019). However, the relatively weak association observed in this study suggests that while TSS may play a role, it is not the primary driver of Vibrio spp. abundance in these macroalgal communities. The weak correlations observed could also reflect the complexity of Vibrio spp. dynamics, where multiple interacting environmental factors influence bacterial abundance. Future research incorporating finer-scale analyses of nutrient cycling, organic matter availability, and particulate-associated Vibrio communities may help clarify these relationships. Previous studies have also reported that the concentration of suspended particles in the water column is an important predictor of pathogenic Vibrio spp. abundance (Venkateswaran et al., 1990; Fries et al., 2008). Vibrio spp. attach to particles in the water column that provide refuse from grazing, higher nutrient concentrations, and buffers from environmental fluctuations (Eiler et al., 2006). While V. vulnificus can be found across a wide range of salinities (4‰ to 37‰), it functions optimally at salinities between 10‰ and 25‰, which is consistent with the salinities in this study (Noorian et al., 2023). Although our model identified key environmental predictors of V. vulnificus abundance, the overall model performance was weak, indicating that additional unmeasured factors may be influencing V. vulnificus dynamics.
We were surprised that temperature was not a significant predictor of pathogenic Vibrio spp. in this study, as rising water temperature is linked to higher metabolism and overall increased fitness (Karvonen et al., 2010). However, our study was conducted between May and October, during which temperatures ranged from 16.3°C to 27.4°C, which is well within the known survival and growth range for Vibrio spp. It is possible that within this relatively constrained temperature window, other environmental variables, such as nutrient availability, particle attachment, or competition with other microbes, exerted a stronger influence on V. vulnificus and V. parahaemolyticus abundance. Additionally, while temperature is a well-established driver of Vibrio seasonality, its role in explaining fine-scale variation in abundance within a single season may be less pronounced. As coastal waters continue to warm, including in Narragansett Bay, the seasonal window of Vibrio prevalence may extend earlier into the spring and persist later into the fall, which could have implications for future monitoring and risk assessments.
Vibrio spp. are recognized as highly successful opportunists and generalists in various environments (Goecke et al., 2010; Samsing and Barnes, 2024). Unlike many marine bacteria, Vibrio spp. thrive even without a host association (Lovell, 2017). These bacteria are found in diverse ecosystems, from near-shore areas (Kaneko and Colwell, 1973; DePaola et al., 1994; DePaola et al., 2000; Cox and Gomez-Chiarri, 2012) to the open ocean (Martinez-Urtaza et al., 2016) and from tropical regions (Deepanjali et al., 2005; Zimmerman et al., 2007) to as far north as Alaska and Scandinavia (Høi et al., 1998; McLaughlin et al., 2005; Baker-Austin et al., 2013). Since Vibrio spp. do not rely on a host organism for survival, their presence on macroalgae suggests that they may benefit from this association. Pathogenic Vibrio spp. have been found to be more abundant on macroalgae surfaces than in the surrounding water (Mahmud et al., 2007; Mahmud et al., 2008). Additionally, higher abundances of pathogenic Vibrio spp. are found in sediments and particulate matter compared to their free-living presence in the water column (Parveen et al., 2008; Johnson et al., 2012; Gutierrez West et al., 2013; Vezzulli et al., 2013; Main et al., 2015). The absence of a strong relationship between pathogenic Vibrio spp. abundance and environmental parameters may actually provide helpful insight to understanding macroalgae as a host for V. parahaemolyticus and V. vulnificus. Similar to water column particles and sediments, the surface of macroalgae offers a micro-niche habitat conducive to bacterial survival and proliferation as it provides a buffer against environmental fluctuations (Beleneva et al., 2006; Englebert et al., 2008; Goecke et al., 2010; Saha et al., 2020). The ability of pathogenic Vibrio spp. to form biofilms on both biotic and abiotic surfaces facilitates their persistence in the environment, making macroalgae an excellent host (Baker-Austin et al., 2018). While beyond the scope of this study, it is possible that Vibrio spp. may benefit their macroalgae host by producing secondary metabolites that inhibit the colonization of other microbes, potentially enhancing the overall health of the macroalgae (Goecke et al., 2010). While we aimed to reduce potential confounding from macrofouling epibionts by excluding visibly colonized macroalgae, we recognize that microscopic epibionts may still have played a role in shaping Vibrio dynamics. Epibionts, including other microbes, small invertebrates, or protists, could have influenced Vibrio spp. through a variety of mechanisms (e.g., serving as hosts, providing additional organic matter, competing for resources, or producing antimicrobial compounds) (Egan et al., 2013; Dang and Lovell, 2016). Previous studies have shown that epiphytic microbial communities can both facilitate and inhibit Vibrio colonization, depending on species interactions and environmental conditions (Lemire et al., 2015; Liu et al., 2022; Liu et al., 2024). Future work incorporating metagenomic or microscopy-based approaches could help clarify the potential role of epibionts in structuring Vibrio populations on macroalgae.
There is limited knowledge about the abundance of V. parahaemolyticus and V. vulnificus on macroalgae. This study highlights that macroalgae in this temperate estuary are a reservoir of pathogenic V. parahaemolyticus and V. vulnificus. The environmental parameters measured here were not the key drivers of Vibrio spp. abundance. Thus, we may not have captured a large enough range of the parameters we measured, we may be missing key environmental parameters, or importantly, environmental conditions matter less when the Vibrio can attach to and be protected by the macroalgae host.
Though pathogenic Vibrio were found on all genera of macroalgae, this does not directly translate to human disease. An infectious dose of a pathogen often combined with poor immune status or previous skin lesion is required to cause disease in the host organism. Thus, the pathology of an infectious dosage in both V. parahaemolyticus and V. vulnificus is highly variable and situationally dependent (Drake et al., 2007). However, this study does demonstrate that as macroalgae continue to proliferate and our climate warms, macroalgae may be an emerging route of human exposure to pathogenic Vibrio spp. worthy of study. Future research could focus on quantifying V. parahaemolyticus and V. vulnificus abundance across a wider range of systems, seasons, and environmental parameters, as well as investigating potential species-specific chemical interactions underpinning the relationship between pathogenic Vibrio spp. and macroalgae host.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. AS: Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – review & editing. RS: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – review & editing. RF: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by a Rhode Island Sea Grant award to RF and RS.
We are thankful to the entirety of the Fulweiler lab for their support. We especially want to thank Ocean Adhikari, Brenden Blakely, Melissa Ederington Hagy, Carolyn Hagy, Lydia Jefferson, Cat Mahoney, Kwetzpallin Mexika, Helena Shenk, and Rene Yang for their help with field and lab work. We would like to thank Kyle Oliveira, Sawyer Balint, and Nia Bartolucci for their help with coding. We thank Alia Al-Haj and Nick Ray whose conversations with RF started the initial interest in this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1549732/full#supplementary-material
Akrong M. O., Anning A. K., Addico G. N. D., Hogarh J. N., Adu-Gyamfi A., deGraft-Johnson K. A. A., et al. (2023). Variations in seaweed-associated and planktonic bacterial communities along the coast of Ghana. Mar. Biol. Res. 19, 219–233. doi: 10.1080/17451000.2023.2213894
Almagro-Moreno S., Martinez-Urtaza J., Pukatzki S. (2023). “Vibrio Infections and the Twenty-First Century,” in Vibrio spp Infections. Eds. Almagro-Moreno S., Pukatzki S. (Cham: Springer International Publishing) 1404, 1–16. doi: 10.1007/978-3-031-22997-8_1
Arar E. J., Collins G. B. (1997). Method 445.0: In vitro determination of chlorophyll a and pheophytin a in marine and freshwater algae by fluorescence (United States Environmental Protection Agency). Available at: http://flrules.elaws.us/gateway/refpdf/62/D62/Ref-02922/m445_0[1].pdf.
Baker-Austin C., Oliver J. D., Alam M., Ali A., Waldor M. K., Qadri F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers. 4, 1–19. doi: 10.1038/s41572-018-0005-8
Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2, 7–18. doi: 10.1111/j.1758-2229.2009.00096.x
Baker-Austin C., Trinanes J., Gonzalez-Escalona N., Martinez-Urtaza J. (2017). Non-cholera vibrios: the microbial barometer of climate change. Trends Microbiol. 25, 76–84. doi: 10.1016/j.tim.2016.09.008
Baker-Austin C., Trinanes J. A., Taylor N. G., Hartnell R., Siitonen A., Martinez-Urtaza J. (2013). Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Climate change. 3, 73–77. doi: 10.1038/nclimate1628
Barberi O. N., Byron C. J., Burkholder K. M., St. Gelais A. T., Williams A. K. (2020). Assessment of bacterial pathogens on edible macroalgae in coastal waters. J. Appl. Phycol. 32, 683–696. doi: 10.1007/s10811-019-01993-5
Barott K. L., Rodriguez-Brito B., Janouškovec J., Marhaver K. L., Smith J. E., Keeling P., et al. (2011). Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis. Environ. Microbiol. 13, 1192–1204. doi: 10.1111/j.1462-2920.2010.02419.x
Bej A. K., Patterson D. P., Brasher C. W., Vickery M. C. L., Jones D. D., Kaysner C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiological Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Beleneva I., Zhukova N. Beleneva I. A., Zhukova N. V. (2006). Bacterial communities of brown and red algae from Peter the Great Bay, the Sea of Japan. Mikrobiologiia. 75, 410–419. doi: 10.1134/S0026261706030180
Billaud M., Seneca F., Tambutté E., Czerucka D. (2022). An increase of seawater temperature upregulates the expression of vibrio parahaemolyticus virulence factors implicated in adhesion and biofilm formation. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.840628
Bisharat N., Agmon V., Finkelstein R., Raz R., Ben-Dror G., Lerner L., et al. (1999). Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354, 1421–1424. doi: 10.1016/S0140-6736(99)02471-X
Blackwell K. D., Oliver J. D. (2008). The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina Estuaries. J. Microbiol. 46, 146–153. doi: 10.1007/s12275-007-0216-2
Burge C. A., Mark Eakin C., Friedman C. S., Froelich B., Hershberger P. K., Hofmann E. E., et al. (2014). Climate change influences on marine infectious diseases: implications for management and society. Annu. Rev. Mar. Sci. 6, 249–277. doi: 10.1146/annurev-marine-010213-135029
Castel J., Caumette P., Herbert R. (1996). Eutrophication gradients in coastal lagoons as exemplified by the Bassin d’Arcachon and the Étang du Prévost. Hydrobiologia 329, ix–xxviii. doi: 10.1007/BF00034542
Connell S. D., Russell B. D. (2010). The direct effects of increasing CO 2 and temperature on non-calcifying organisms: increasing the potential for phase shifts in kelp forests. Proc. R Soc. B. 277, 1409–1415. doi: 10.1098/rspb.2009.2069
Cox A. M., Gomez-Chiarri M. (2012). Vibrio parahaemolyticus in rhode island coastal ponds and the estuarine environment of narragansett bay. Appl. Environ. Microbiol. 78, 2996–2999. doi: 10.1128/AEM.07519-11
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/MMBR.00037-15
Deepanjali A., Kumar H. S., Karunasagar I., Karunasagar I. (2005). Seasonal Variation in Abundance of Total and Pathogenic Vibrio parahaemolyticus Bacteria in Oysters along the Southwest Coast of India. Appl. Environ. Microbiol. 71, 3575–3580. doi: 10.1128/AEM.71.7.3575-3580.2005
Delignette-Muller M. L., Dutang C. (2015). fitdistrplus: An R package for fitting distributions. J. Stat. Software 64, 1–34. doi: 10.18637/jss.v064.i04
DePaola A., Capers G. M., Alexander D. (1994). Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60, 984–988. doi: 10.1128/aem.60.3.984-988.1994
DePaola A., Kaysner C. A., Bowers J., Cook D. W. (2000). Environmental Investigations of Vibrio parahaemolyticus in Oysters after Outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66, 4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000
DePaola A., Ulaszek J., Kaysner C. A., Tenge B. J., Nordstrom J. L., Wells J., et al. (2003). Molecular, serological, and virulence characteristics of vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69, 3999–4005. doi: 10.1128/AEM.69.7.3999-4005.2003
Drake S. L., DePaola A., Jaykus L. A. (2007). An Overview of Vibrio vulnificus and Vibrio parahaemolyticus. Compr. Rev. Food Sci. Food Safety. 6, 120–144. doi: 10.1111/j.1541-4337.2007.00022.x
Duarte C. M., Gattuso J., Hancke K., Gundersen H., Filbee-Dexter K., Pedersen M. F., et al. (2022). Global estimates of the extent and production of macroalgal forests. Global Ecol. Biogeogr. 31, 1422–1439. doi: 10.1111/geb.13515
Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
Eiler A., Johansson M., Bertilsson S. (2006). Environmental influences on vibrio populations in northern temperate and boreal coastal waters (Baltic and skagerrak seas). Appl. Environ. Microbiol. 72, 6004–6011. doi: 10.1128/AEM.00917-06
Englebert E., Mcdermott C., Kleinheinz G. (2008). Effects of the Nuisance Algae, Cladophora, on Escherichia coli at Recreational Beaches in Wisconsin. Sci. total environment. 404, 10–17. doi: 10.1016/j.scitotenv.2008.05.025
Fagherazzi S., Mariotti G., Banks A. T., Morgan E. J., Fulweiler R. W. (2014). The relationships among hydrodynamics, sediment distribution, and chlorophyll in a mesotidal estuary. Estuarine Coast. Shelf Science. 144, 54–64. doi: 10.1016/j.ecss.2014.04.003
Foster S. Q., Fulweiler R. W. (2014). Spatial and historic variability of benthic nitrogen cycling in an anthropogenically impacted estuary. Front. Mar. Sci. 1. doi: 10.3389/fmars.2014.00056
Foster J. J., Yavorsky C., Barkus E. (2005). Understanding and Using Advanced Statistics: A Practical Guide for Students. (Thousand Oaks, CA, USA: Sage Publications), 1–192.
Fries J. S., Characklis G. W., Noble R. T. (2008). Sediment–water exchange of Vibrio sp. and fecal indicator bacteria: Implications for persistence and transport in the Neuse River Estuary, North Carolina, USA. Water Res. 42, 941–950. doi: 10.1016/j.watres.2007.09.006
Garcia-Vaquero M., Hayes M. (2016). Red and green macroalgae for fish and animal feed and human functional food development. Food Rev. Int. 32, 15–45. doi: 10.1080/87559129.2015.1041184
Gattuso J. P., Gentili B., Duarte C. M., Kleypas J. A., Middelburg J. J., Antoine D. (2006). Light availability in the coastal ocean: impact on the distribution of benthic photosynthetic organisms and their contribution to primary production. Biogeosciences. 3, 489–513. doi: 10.5194/bg-3-489-2006
Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Series. 409, 267–299. doi: 10.3354/meps08607
Gonzalez D. J., Gonzalez R. A., Froelich B. A., Oliver J. D., Noble R. T., McGlathery K. J. (2014). Non-native macroalga may increase concentrations of Vibrio bacteria on intertidal mudflats. Mar. Ecol. Prog. Series. 505, 29–36. doi: 10.3354/meps10771
Gonzalez-Escalona N., Strain E. A., De Jesús A. J., Jones J. L., DePaola A. (2011). Genome sequence of the clinical O4:K12 serotype vibrio parahaemolyticus strain 10329▿. J. Bacteriol. 193, 3405–3406. doi: 10.1128/JB.05044-11
Grasshoff K., Kremlingl K., Ehrhardt M. (1999). Methods of Seawater Analysis, Third, Completely Revised and Extended Edition. (Weinheim; New York; Chiester; Brisbane; Singapore; Toronto: Wiley-VCH).
Gulig P. A., Bourdage K. L., Starks A. M. (2005). Molecular pathogenesis of vibrio vulnificus. J. Microbiol. 43, 118–131.
Gutierrez West C. K., Klein S. L., Lovell C. R. (2013). High Frequency of Virulence Factor Genes tdh, trh, and tlh in Vibrio parahaemolyticus Strains Isolated from a Pristine Estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/AEM.03792-12
Hansen H. P., Koroleff F. (2007). “Determination of nutrients.” in Methods of Seawater Analysis, 3rd Edn., 159–228. doi: 10.1002/9783527613984.ch10
Hayden H. S., Blomster J., Maggs C. A., Silva P. C., Stanhope M. J., Waaland J. R. (2003). Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur. J. Phycology. 38, 277–294. doi: 10.1080/1364253031000136321
Høi L., Larsen J. L., Dalsgaard I., Dalsgaard A. (1998). Occurrence of vibrio vulnificus biotypes in danish marine environments. Appl. Environ. Microbiol. 64, 7–13. doi: 10.1128/AEM.64.1.7-13.1998
Hurd C. L. (2000). Water motion, marine macroalgal physiology, and production. J. Phycology. 36, 453–472. doi: 10.1046/j.1529-8817.2000.99139.x
Iida T., Suthienkul O., Park K. S., Tang G. Q., Yamamoto R. K., Ishibashi M., et al. (1997). Evidence for genetic linkage between the ure and trh genes in Vibrio parahaemolyticus. J. Med. Microbiol. 46, 639–645. doi: 10.1099/00222615-46-8-639
Ji Y., Gao K. (2021). Effects of climate change factors on marine macroalgae: A review. Adv. Mar. Biol. 88, 91–136. doi: 10.1016/bs.amb.2020.11.001
Johnson C. N., Bowers J. C., Griffitt K. J., Molina V., Clostio R. W., Pei S., et al. (2012). Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the Coastal and Estuarine Waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 78, 7249–7257. doi: 10.1128/AEM.01296-12
Johnson C. N., Flowers A. R., Noriea N. F., Zimmerman A. M., Bowers J. C., DePaola A., et al. (2010). Relationships between environmental factors and pathogenic vibrios in the northern gulf of Mexico. Appl. Environ. Microbiol. 76, 7076–7084. doi: 10.1128/AEM.00697-10
Johnson K. S., Petty R. L. (1983). Determination of nitrate and nitrite in seawater by flow injection analysis. Limnology Oceanography. 28, 1260–1266. doi: 10.4319/lo.1983.28.6.1260
Joniver C. F., Photiades A., Moore P. J., Winters A. L., Woolmer A., Adams J. M. (2021). The global problem of nuisance macroalgal blooms and pathways to its use in the circular economy. Algal Res. 58, 102407. doi: 10.1016/j.algal.2021.102407
Kalvaitienė G., Vaičiūtė D., Bučas M., Gyraitė G., Kataržytė M. (2023). Macrophytes and their wrack as a habitat for faecal indicator bacteria and Vibrio in coastal marine environments. Mar. pollut. Bulletin. 194, 115325. doi: 10.1016/j.marpolbul.2023.115325
Kaneko T., Colwell R. R. (1973). Ecology of vibrio parahaemolyticus in chesapeake bay. J. Bacteriol. 113, 24–32. doi: 10.1128/jb.113.1.24-32.1973
Karvonen A., Rintamäki P., Jokela J., Valtonen E. T. (2010). Increasing water temperature and disease risks in aquatic systems: climate change increases the risk of some, but not all, diseases. Int. J. parasitology. 40, 1483–1488. doi: 10.1016/j.ijpara.2010.04.015
Krumhansl K. A., Scheibling R. E. (2012). Production and fate of kelp detritus. Mar. Ecol. Prog. Series. 467, 281–302. doi: 10.3354/meps09940
Kumar S. S., Sudhakar M. S. (2024). “Algae as a Source of Bioremediation and Resource Recycling,” in Toxicity of Aquatic System and Remediation (CRC Pres), 237–254. Available at: https://www.taylorfrancis.com/chapters/edit/10.1201/9781003297901-17/algae-source-bioremediation-resource-recycling-senthil-kumar-magapu-solomon-sudhakar.
Lachnit T., Meske D., Wahl M., Harder T., Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Lam O., Wheeler J., Tang C. M. (2014). Thermal control of virulence factors in bacteria: A hot topic. Virulence. 5, 852–862. doi: 10.4161/21505594.2014.970949
Lemay M. A., Chen M. Y., Mazel F., Hind K. R., Starko S., Keeling P. J., et al. (2021). Morphological complexity affects the diversity of marine microbiomes. ISME J. 15, 1372–1386. doi: 10.1038/s41396-020-00856-z
Lemire A., Goudenège D., Versigny T., Petton B., Calteau A., Labreuche Y., et al. (2015). Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 9, 1523–1531. doi: 10.1038/ismej.2014.233
Lenth R. V. (2017). Emmeans: Estimated Marginal Means, aka Least-Squares Means doi: 10.32614/CRAN.package.emmeans
Letchumanan V., Chan K. G., Lee L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5, 705. doi: 10.3389/fmicb.2014.00705
Liang J., Liu J., Wang X., Lin H., Liu J., Zhou S., et al. (2019). Spatiotemporal dynamics of free-living and particle-associated vibrio communities in the northern chinese marginal seas. Appl. Environ. Microbiol. 85, e00217–e00219. doi: 10.1128/AEM.00217-19
Littler M. M., Littler D. S. (1980). The evolution of thallus form and survival strategies in benthic marine macroalgae: field and laboratory tests of a functional form model. Am. Naturalist. 116, 25–44. doi: 10.1086/283610
Liu Y., Wikfors G. H., Clark P., Pitchford S., Krisak M., Dixon M. S., et al. (2022). A deep dive into the epibiotic communities on aquacultured sugar kelp Saccharina latissima in Southern New England. Algal Res. 63, 102654. doi: 10.1016/j.algal.2022.102654
Liu M., Yin F., Zhao W., Tian P., Zhou Y., Jia Z., et al. (2024). Diversity of culturable bacteria from the coral reef areas in the South China sea and their agar-degrading abilities. Microorganisms. 12, 187. doi: 10.3390/microorganisms12010187
Lorraine J., Lucht J. (2000). Wickford harbor watershed assessment Vol. 00 (Kingston, Rhode Island: College of Environment and Life Sciences and Rhode Island Cooperative Extension, University of Rhode Island Technical Report).
Lovell C. R. (2017). Ecological fitness and virulence features of Vibrio parahaemolyticus in estuarine environments. Appl. Microbiol. Biotechnol. 101, 1781–1794. doi: 10.1007/s00253-017-8096-9
Mahmud Z. H., Neogi S. B., Kassu A., Mai Huong B. T., Jahid I. K., Islam M. S., et al. (2008). Occurrence, seasonality and genetic diversity of Vibrio vulnificus in coastal seaweeds and water along the Kii Channel, Japan. FEMS Microbiol. ecology. 64, 209–218. doi: 10.1111/j.1574-6941.2008.00460.x
Mahmud Z. H., Neogi S. B., Kassu A., Wada T., Islam M. S., Nair G. B., et al. (2007). Seaweeds as a reservoir for diverse Vibrio parahaemolyticus populations in Japan. Int. J. Food Microbiol. 118, 92–96. doi: 10.1016/j.ijfoodmicro.2007.05.009
Main C. R., Salvitti L. R., Whereat E. B., Coyne K. J. (2015). Community-level and species-specific associations between phytoplankton and particle-associated vibrio species in delaware’s inland bays. Appl. Environ. Microbiol. 81, 5703–5713. doi: 10.1128/AEM.00580-15
Martin J., Hall D. (2016). R 2 measures for zero-inflated regression models for count data with excess zeros. J. Stat. Comput. Simulation. 86, 1–14. doi: 10.1080/00949655.2016.1186166
Martinez-Urtaza J., Lozano-Leon A., Varela-Pet J., Trinanes J., Pazos Y., Garcia-Martin O. (2008). Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl. Environ. Microbiol. 74, 265–274. doi: 10.1128/AEM.01307-07
Martinez-Urtaza J., Powell A., Jansa J., Rey J. L. C., Montero O. P., Campello M. G., et al. (2016). Epidemiological investigation of a foodborne outbreak in Spain associated with U.S. West Coast genotypes of Vibrio parahaemolyticus. Springerplus 5, 87. doi: 10.1186/s40064-016-1728-1
Martinez-Urtaza J., Simental L., Velasco D., DePaola A., Ishibashi M., Nakaguchi Y., et al. (2005). Pandemic vibrio parahaemolyticus O3:K6, europe. Emerg. Infect. Dis. 11, 1319–1320. doi: 10.3201/eid1108.050322
McLaughlin J. B., DePaola A., Bopp C. A., Martinek K. A., Napolilli N. P., Allison C. G., et al. (2005). Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl. J. Med. 353, 1463–1470. doi: 10.1056/NEJMoa051594
Morris J. G. Jr., Acheson D. (2003). Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin. Infect. Diseases. 37, 272–280. doi: 10.1086/375600
Moussa M., Cauvin E., Le Piouffle A., Lucas O., Bidault A., Paillard C., et al. (2021). A MALDI-TOF MS database for fast identification of Vibrio spp. potentially pathogenic to marine mollusks. Appl. Microbiol. Biotechnol. 105, 2527–2539. doi: 10.1007/s00253-021-11141-0
Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Ning L., Yao Z., Zhu B. (2022). Ulva (Enteromorpha) polysaccharides and oligosaccharides: A potential functional food source from green-tide-forming macroalgae. Mar. Drugs 20, 202. doi: 10.3390/md20030202
Nishibuchi M., Fasano A., Russell R. G., Kaper J. B. (1992). Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60, 3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992
Noorian P., Hoque M. M., Espinoza-Vergara G., McDougald D. (2023). “Environmental Reservoirs of Pathogenic Vibrio spp. and Their Role in Disease: The List Keeps Expanding,” in Vibrio Spp. Infections. Eds. Almagro-Moreno S., Pukatzki S. Advances in Experimental Medicine and Biology. (Cham: Springer International Publishing) 1404, 99–126. doi: 10.1007/978-3-031-22997-8_6
Oakley B. A., Alvarez J. D., Boothroyd J. C. (2012). Benthic Geologic Habitats of Shallow Estuarine Environments: Greenwich Bay and Wickford Harbor, Narragansett Bay, Rhode Island, U.S.A. J. Coast. Res. 28, 760–773.
Okura M., Osawa R., Iguchi A., Arakawa E., Terajima J., Watanabe H. (2003). Genotypic analyses of vibrio parahaemolyticus and development of a pandemic group-specific multiplex PCR assay. J. Clin. Microbiol. 41, 4676–4682. doi: 10.1128/JCM.41.10.4676-4682.2003
Paranjpye R. N., Nilsson W. B., Liermann M., Hilborn E. D., George B. J., Li Q., et al. (2015). Environmental influences on the seasonal distribution of Vibrio parahaemolyticus in the Pacific Northwest of the USA. FEMS Microbiol. Ecol. 91, fiv121. doi: 10.1093/femsec/fiv121
Parvathi A., Kumar H. S., Bhanumathi A., Ishibashi M., Nishibuchi M., Karunasagar I., et al. (2006). Molecular characterization of thermostable direct haemolysin-related haemolysin (TRH)-positive Vibrio parahaemolyticus from oysters in Mangalore, India. Environ. Microbiol. 8, 997–1004. doi: 10.1111/j.1462-2920.2006.00990.x
Parveen S., Hettiarachchi K. A., Bowers J. C., Jones J. L., Tamplin M. L., McKay R., et al. (2008). Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int. J. Food Microbiol. 128, 354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019
Parveen S., Jacobs J., Ozbay G., Chintapenta L. K., Almuhaideb E., Meredith J., et al. (2020). Seasonal and Geographical Differences in Total and Pathogenic Vibrio parahaemolyticus and Vibrio vulnificus Levels in Seawater and Oysters from the Delaware and Chesapeake Bays Determined Using Several Methods. Appl. Environ. Microbiol. 86, e01581–e01520. doi: 10.1128/AEM.01581-20
Peñalver R., Lorenzo J. M., Ros G., Amarowicz R., Pateiro M., Nieto G. (2020). Seaweeds as a functional ingredient for a healthy diet. Mar. Drugs 18, 301. doi: 10.3390/md18060301
Pessarrodona A., Moore P. J., Sayer M. D. J., Smale D. A. (2018). Carbon assimilation and transfer through kelp forests in the NE Atlantic is diminished under a warmer ocean climate. Global Change Biol. 24, 4386–4398. doi: 10.1111/gcb.2018.24.issue-9
Queirós A. M., Stephens N., Widdicombe S., Tait K., McCoy S. J., Ingels J., et al. (2019). Connected macroalgal-sediment systems: blue carbon and food webs in the deep coastal ocean. Ecol. Monographs. 89, e01366. doi: 10.1002/ecm.1366
Raghunath P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5, 805. doi: 10.3389/fmicb.2014.00805
Ray N., Al-Haj A., Fulweiler R. (2020). Sediment biogeochemistry along an oyster aquaculture chronosequence. Mar. Ecol. Prog. Series. 646, 13–27. doi: 10.3354/meps13377
Roncarati D., Vannini A., Scarlato V. (2024). Temperature sensing and virulence regulation in pathogenic bacteria. Trends Microbiol. doi: 10.1016/j.tim.2024.07.009
Rosche T. M., Yano Y., Oliver J. D. (2005). A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49, 381–389. doi: 10.1111/j.1348-0421.2005.tb03731.x
Saha M., Dove S., Weinberger F. (2020). Chemically mediated microbial “Gardening” Capacity of a seaweed holobiont is dynamic. Microorganisms. 8, 1893. doi: 10.3390/microorganisms8121893
Sakamoto Y., Ishiguro M., Kitagawa G., Sakamoto K., Ishiguro M. (1986). Akaike Information Criterion Statistics. Mathematics and Its Applications (D. Reidel Publishing Company). (Tokyo, Dordrecht, Boston: KTK Scientific Publishers; D. Reidel ; Sold and distributed in the U.S.A. and Canada by Kluwer Academic Publishers), 290. Available at: http://www.gbv.de/dms/hbz/toc/ht002888076.pdf (Accessed May 29, 2024).
Samsing F., Barnes A. C. (2024). The rise of the opportunists: What are the drivers of the increase in infectious diseases caused by environmental and commensal bacteria? Rev. Aquaculture. doi: 10.1111/raq.12922
Scro A. K., Lundgren K. M., Smolowitz R. (2019). Multiplex qPCR detection of nonpathogenic and pathogenic Vibrio parahaemolyticus and Vibrio vulnificus from oysters cultured in Massachusetts and Rhode Island: the reliability of MPN as an indicator. J. Shellfish Res. 38, 123–130. doi: 10.2983/035.038.0112
Senga Y., Kobayashi W., Mikawa K., Kitazawa T., Lee S., Shiraki Y. (2021). Influences of green macroalgae blooms on nutrients and sulfide dynamics in hypereutrophic intertidal ecosystems. Limnology. 22, 187–196. doi: 10.1007/s10201-020-00644-w
Sha H., Li L., Lu J., Xiong J. (2022). High nutrient induces virulence in the AHPND-causing Vibrio parahaemolyticus, interpretation from the ecological assembly of shrimp gut microbiota. Fish Shellfish Immunol. 127, 758–765. doi: 10.1016/j.fsi.2022.07.016
Solórzano L. (1969). Determination of ammonia in natural waters by the phenolhypochlorite method 1 1 This research was fully supported by U.S. Atomic Energy Commission Contract No. ATS (11-1) GEN 10, P.A. 20. Limnology Oceanography 14, 799–801. doi: 10.4319/lo.1969.14.5.0799
Staufenberger T., Thiel V., Wiese J., Imhoff J. F. (2008). Phylogenetic analysis of bacteria associated with Laminaria saccharina. FEMS Microbiol. Ecology. 64, 65–77. doi: 10.1111/j.1574-6941.2008.00445.x
Steneck R. S., Watling L. (1982). Feeding capabilities and limitation of herbivorous molluscs: A functional group approach. Mar. Biol. 68, 299–319. doi: 10.1007/BF00409596
Takemura A. F., Chien D. M., Polz M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5, 38. doi: 10.3389/fmicb.2014.00038
Thompson J. R., Polz M. F. (2006). “Dynamics of Vibrio Populations and Their Role in Environmental Nutrient Cycling,” in The Biology of Vibrios (Washington, DC, USA: John Wiley & Sons, Ltd), 190–203. doi: 10.1128/9781555815714.ch13
Thompson J. R., Polz M. F. (2014). “Dynamics of Vibrio Populations and Their Role in Environmental Nutrient Cycling,” in The Biology of Vibrios. Eds. Thompson F. L., Austin B., Swings J. (Washington, DC, USA: ASM Press), 190–203. doi: 10.1128/9781555815714.ch13
Tison D. L., Kelly M. T. (1984). Vibrio species of medical importance. Diagn. Microbiol. Infect. disease. 2, 263–276. doi: 10.1016/0732-8893(84)90057-9
Tison D. L., Nishibuchi M., Greenwood J. D., Seidler R. J. (1982). Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44, 640–646. doi: 10.1128/aem.44.3.640-646.1982
Urakawa H., Rivera I. N. G. (2014). “Aquatic Environment,” in The Biology of Vibrios. Eds. Thompson F. L., Austin B., Swings J. (Washington, DC, USA: ASM Press), 173–189. doi: 10.1128/9781555815714.ch12
U.S. Food and Drug Administration (FDA) and Center for Food Safety and Applied Nutrition (2004). “Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio species.” in Bacteriological analytical manual online. (Gaithersburg, MD: Center for Food Safety and Applied Nutrition, U.S. Food and Drug Administration). Available online at: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm.
Valiela I., McClelland J., Hauxwell J., Behr P. J., Hersh D., Foreman K. (1997). Macroalgal blooms in shallow estuaries: Controls and ecophysiological and ecosystem consequences. Limnology Oceanography 42, 1105–1118. doi: 10.4319/lo.1997.42.5_part_2.1105
Venkateswaran K., Kiiyukia C., Nakanishi K., Nakano H., Matsuda O., Hashimoto H. (1990). The role of sinking particles in the overwintering process of Vibrio parahaemolyticus in a marine environment. FEMS Microbiol. Ecology. 6, 159–166. doi: 10.1111/j.1574-6968.1990.tb03936.x
Vezzulli L., Colwell R. R., Pruzzo C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65, 817–825. doi: 10.1007/s00248-012-0163-2
Viaroli P., Naldi M., Bondavalli C., Bencivelli S. (1996). Growth of the seaweed Ulva rigida C. Agardh in relation to biomass densities, internal nutrient pools and external nutrient supply in the Sacca di Goro lagoon (Northern Italy). Hydrobiologia. 329, 93–103. doi: 10.1007/BF00034550
Villalard-Bohnsack M., Harlin M. M. (1992). Seasonal Distribution and Reproductive Status of Macroalgae in Narragansett Bay and Associated Waters, Rhode Island, U.S.A (Berlin, Germany: Botanica Marina), Vol. 35. 205–214.
Wang Z., Ma S., Wang C. Y. (2015). Variable selection for zero-inflated and overdispersed data with application to health care demand in Germany. Biom J. 57, 867–884. doi: 10.1002/bimj.201400143
Wang H., Wang G., Gu W. (2020). Macroalgal blooms caused by marine nutrient changes resulting from human activities. Montero-Serra I editor. J. Appl. Ecology. 57, 766–776. doi: 10.1111/1365-2664.13587
Warner E. B., Oliver J. D. (2008). Multiplex PCR assay for detection and simultaneous differentiation of genotypes of Vibrio vulnificus biotype 1. Foodborne Pathog. Dis. 5, 691–693. doi: 10.1089/fpd.2008.0120
Wiencke C., Bischof K. (Eds.) (2012). Seaweed Biology: Novel Insights into Ecophysiology, Ecology and Utilization Vol. 219. Ecological Studies. (Berlin, Heidelberg: Springer Berlin Heidelberg). doi: 10.1007/978-3-642-28451-9
Wright A. C., Simpson L. M., Oliver J. D. (1981). Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect. Immun. 34, 503–507. doi: 10.1128/iai.34.2.503-507.1981
Xu M., Yamamoto K., Honda T., Ming X. (1994). Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176, 4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994
Yeung P. S. M., Boor K. J. (2004). Epidemiology, pathogenesis, and prevention of foodborne vibrio parahaemolyticus infections. Foodborne Pathog. Disease. 1, 74–88. doi: 10.1089/153531404323143594
Yeung M., Thorsen T. (2016). Development of a more sensitive and specific chromogenic agar medium for the detection of vibrio parahaemolyticus and other vibrio species. J. Vis. Exp. 117), 54493. doi: 10.3791/54493
Zhang X., Mallick H., Tang Z., Zhang L., Cui X., Benson A. K., et al. (2017). Negative binomial mixed models for analyzing microbiome count data. BMC Bioinf. 18, 4. doi: 10.1186/s12859-016-1441-7
Zimmerman A. M., DePaola A., Bowers J. C., Krantz J. A., Nordstrom J. L., Johnson C. N., et al. (2007). Variability of total and pathogenic Vibrio parahaemolyticus densities in northern Gulf of Mexico water and oysters. Appl. Environ. Microbiol. 73, 7589–7596. doi: 10.1128/AEM.01700-07
Keywords: Vibrio parahaemolyticus, Vibrio vulnificus, marine pathogens, qPCR, Narragansett Bay, Ulva spp., Gracilaria spp., Fucus spp.
Citation: Geisser AH, Scro AK, Smolowitz R and Fulweiler RW (2025) Macroalgae host pathogenic Vibrio spp. in a temperate estuary. Front. Mar. Sci. 12:1549732. doi: 10.3389/fmars.2025.1549732
Received: 21 December 2024; Accepted: 05 March 2025;
Published: 02 April 2025.
Edited by:
Jesus L. Romalde, University of Santiago de Compostela, SpainReviewed by:
Eric James Schott, University of Maryland, College Park, United StatesCopyright © 2025 Geisser, Scro, Smolowitz and Fulweiler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra H. Geisser, YWdlaXNzZXJAYnUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.