- 1Anhui Key Laboratory of Aquaculture and Stock Enhancement, Hefei, China
- 2Fisheries Research Institute, Anhui Academy of Agricultural Sciences, Hefei, China
- 3Hefei Animal Husbandry and Aquatic Extension Technology Center, Hefei, China
- 4Hefei Taolin Shuixiang Snakehead Ecological Aquaculture Development Co., Ltd., Hefei, China
To explore the effects of rice floating beds on fish growth and intestinal microbiota, the present study compared the fish growth performance and the microbial diversity, microbial construction, and microbial composition of water and Gibel carp (Carassius auratus gibelio) intestine in rice floating beds ponds and normal ponds. The results revealed that the Gibel carp raised in rice floating beds ponds exhibited significantly greater body length, body weight, and weight gain rates than those in normal ponds. Microbial community analysis showed enhanced richness and diversity in water and intestine of the rice floating beds group. Furthermore, the assembly processes of these microbial communities were predominantly influenced by stochastic mechanisms in rice floating beds ponds. The bacterial phyla associated with nutrient cycling, such as Cyanobacteria, Chloroflexi, and Ignavibacteria, were significantly assembled in the rice floating beds group. Overall, these results highlight the potential of rice floating bed systems as a sustainable aquaculture technique, fostering improved growth performance and microbial diversity, which are critical for the health of cultured fish.
1 Introduction
Aquaculture, a rapidly growing sector within global food production, has undergone significant transformations over the years, evolving from normal pond culture systems to more innovative and sustainable practices (Boyd et al., 2020; Thomas et al., 2021). Conventional fish farming methods, particularly those reliant on static pond environments, have been associated with several pressing environmental issues. These include the deterioration of water quality due to nutrient runoff, the overuse of harmful chemicals, and a decline in biodiversity, which collectively threaten the ecological balance within aquatic ecosystems (Diana, 2009; De Silva, 2012; Edwards, 2015). In light of these challenges, aquaculture has explored alternative approaches that prioritize ecological integrity and sustainability.
As a response to these challenges, the rice floating beds are gradually being implemented in practice, which involve growing rice on platforms that maintain water levels (Goda et al., 2024; Jiang et al., 2024). This method maximizes space utilization, improves water quality through natural filtration systems, and allows for a more diverse ecosystem. In particular, the use of floating beds has been reported to improve nutrient cycling, reduce contaminants, and enhance the biodiversity of aquatic organisms (Zhang et al., 2014; Chen et al., 2020; Xu et al., 2024). The interplay between these aquatic plants and fish species can significantly influence the composition of intestinal microbiota in fish (Ke et al., 2021). The intestinal microbiota plays a crucial role in fish health, affecting digestion, immunity, and overall well-being (Uma et al., 2020; Medina-Félix et al., 2023). Therefore, exploring the effects of floating beds on the intestinal microbiota can provide valuable insights into fish health management in aquaculture.
Numerous factors shape the intestinal microbiota of fish, including diet, environment, and interactions with other organisms (Uma et al., 2020). In particular, studies have shown that the diversity and composition of intestinal microbiota can be influenced by the availability of different dietary sources (Miyake et al., 2015; Ringø et al., 2016), which may be abundant in floating bed systems. In the floating bed ponds, the abundant root of floating plants normally attracts zooplankton, which often serve as important food sources for omnivorous–planktivorous fishes (Yamaki and Yamamuro, 2013; Wang et al., 2018). The presence of floating plant beds in the ponds not only provides alternative food sources but also creates a more complex habitat that may foster beneficial microbial communities (Zhang et al., 2022; Sopawong et al., 2024). The interaction between fish and the floating bed ecosystem could lead to changes in intestinal microbiota diversity, potentially enhancing the fish’s resistance to pathogens and improving growth performance (Ke et al., 2021).
Despite the acknowledged benefits of floating rice beds in aquaculture, limited research has been conducted on their specific effects on the microbial communities of water, and fish intestine. To investigate this, the present study employed 16S rRNA sequencing to identify the microbiota community of water and fish intestine in normal ponds, and identify the water, fish intestine, and floating rice roots microbiota community in rice floating beds ponds. By analyzing the microbial composition and diversity of fish intestine and water in floating bed systems compared to those in normal ponds, we explored how these aquatic plants influence water and intestinal microbiota, which provide novel insights into the theoretical foundation of rice-fish symbiosis.
2 Materials and methods
2.1 Experimental design and sampling
The experiment was conducted at a specialized rice-fish coculture farm located in Hefei, Anhui Province, China. A total of 3000 healthy Carassius auratus gibelio with an average body weight of 107.91 ± 7.58g were randomly put into six ponds (each measuring 30m × 10m). Each pond was designated with a label: P1, P2, P3, RP1, RP2, and RP3. Notably, P1, P2, and P3 were set as normal ponds without floating beds; RP1, RP2, and RP3 had a rice cultivation area that accounted for 15% of the total pond area. All ponds sourced their aquaculture water from a reservoir. The experiment was performed from April to August 2023, and the rice was transplanted on 1 May. During the experiment, fish were fed twice a day at 2% to 4% of body mass using a commercial diet purchased from Anhui TianBang Feed Co., Ltd. (Anhui, China). Before the rice harvest, 2 fish were randomly sampled from each pond at the end of August. The fish were anesthetized with neutralized tricaine methanesulfonate (MS-222, 200 mg L-1). After that, the body length and weight (n = 12) were measured, and the intestinal content (n = 6) was collected and stored at -20°C for testing. We selected each fish as a single sample. All animal experiments followed the guidelines and were approved by the Animal Research and Ethics Committee of the Anhui Academy of Agricultural Sciences. For water sample, we collected equal volumes of water from 30 cm below the surface at the center and four corners of each pond. The collected water was thoroughly mixed and then stored in 250 mL water sampling bags. Each pond sampled twice, and a total of 6 water samples (n = 6) were sampled from normal ponds and rice floating beds ponds, respectively. For rice roots sample, we collected rice roots from rice floating beds using sterile scissors and two sets of duplicated samples were prepared for each pond.
2.2 Microbial community analysis
2.2.1 DNA extraction and PCR amplification
The rice roots samples (0.25 ~ 0.5 g) were individually homogenized in sterile centrifuge tubes (800 μL CD1 and 5 μL RnaseA) and then were centrifuged at 14,000 g, 4°C for 8 min. The supernatant (500 μL) were subsequently transferred to a fresh sterile tube for DNA extraction. The water, rice root, and intestinal microbial genomic DNA was extracted using the FastPure Stool DNA Isolation Kit (MJYH, shanghai, China) according to manufacturer’s instructions. The quality and concentration of DNA were determined by 1.0% agarose gel electrophoresis and a NanoDrop®ND-2000 spectrophotometer (Thermo Scientific Inc., USA) and kept at -80°C prior to further use. The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R(5’-GGACTACHVGGGTWTCTAAT-3’) (Liu et al., 2016) by a T100 Thermal Cycler (BIO-RAD, USA). The PCR reaction mixture including 10 μL 2 × Pro Taq, 0.8 μL each primer (5 μM), 10 ng of template DNA, and ddH2O to a final volume of 20 μL. PCR amplification cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 30 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 4°C. All samples were amplified in triplicate. The PCR product was extracted from 2% agarose gel and purified. Then quantified using Synergy HTX (Biotek, USA).
2.2.2 Illumina sequencing
Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina NextSeq 2000 PE300 platform (Illumina, San Diego,USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
2.2.3 Amplicon sequence processing and analysis
After demultiplexing, the resulting sequences were quality-filtered with fastp (v0.19.6) and merged with FLASH (v1.2.11). Then the high-quality sequences were denoised using the DADA2 plugin in the Qiime2 (version 2020.2) pipeline with recommended parameters. DADA2 denoised sequences are usually called amplicon sequence variants (ASVs). To minimize the effects of sequencing depth on alpha and beta diversity measure, the number of sequences from each sample was rarefied to 20,000, which still yielded an average Good’s coverage of 97.90%. Taxonomic assignment of ASVs was performed using the Naive Bayes consensus taxonomy classifier implemented in Qiime2 and the SILVA 16S rRNA database (v138).
2.3 Statistical analysis
Mean ± SE values were used to present Gibel carp body length, body weight, and weight gain rates. Each dataset underwent assessment for normality and homogeneity of variance. For statistical analysis, one-way analysis of variance (ANOVA) was performed using R (v3.6.1) to determine statistical significance (P < 0.05).
Bioinformatic analysis of the intestinal microbiota was carried out using the Majorbio Cloud platform (https://cloud.majorbio.com). Based on the ASVs information, alpha diversity indices including Chao1 richness, Shannon and Simpson indexes were calculated with Mothur software (v1.30.2) (Duan et al., 2024), and were compared among samples using the Kruskal-Wallis test (P = 0.05). A Venn diagram was used to quantify the number of unique and shared OTUs in multiple groups (Liang et al., 2024).
Beta diversity based on Bray-Curtis dissimilarity metrics was evaluated by the Principal Component Analysis (PCA) using the Vegan package and was compared among groups using the Student’s T-test (P = 0.05) in R (v3.6.1). The neutral community models (NCMs) were constructed to determine the assembly mechanisms for the microbial communities in air samples using R (v3.6.1) (Burns et al., 2016). In addition, the normalized stochasticity ratio (NST) was calculated to determine the contribution of the stochastic process to the community assembly of microbes using R (v3.6.1) with the package of “NST” (Ning et al., 2019).
The linear discriminant analysis (LDA) effect size (LEfSe) (http://huttenhower.sph.harvard.edu/LEfSe) was performed to identify the significantly abundant taxa (phylum to genera) of bacteria among the different groups (LDA score > 2, P < 0.05).
3 Results
3.1 Growth performance of Gibel carps
At the end of the experiment, the body length and body length of Gibel carps in RP group were significantly higher (P < 0.05) than those in P group. At the end of the experiment, the body length and body weight of Gibel carps in the P and RP groups were both significantly higher (P < 0.05) than those at the beginning of the experiment, respectively. Besides, the body weight gain rate of Gibel carps in the RP group was significantly higher (P < 0.05) than that in the P group (Figure 1).
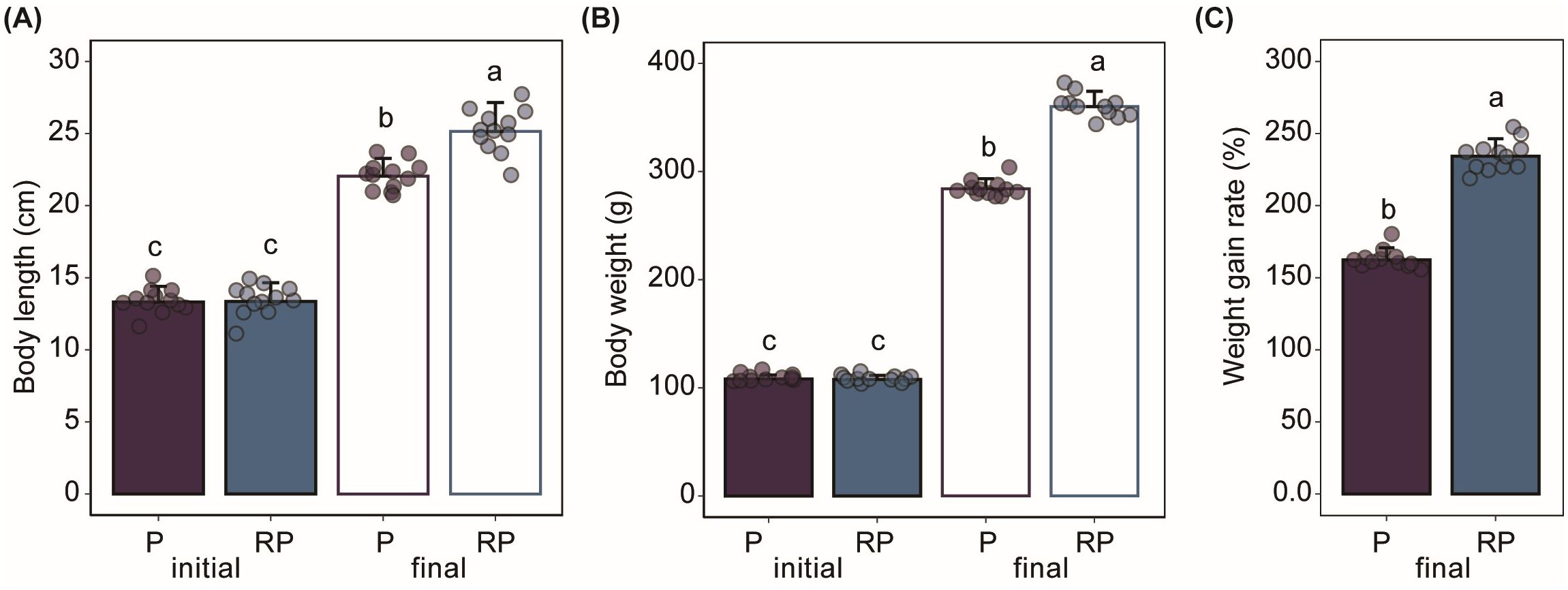
Figure 1. The differences in growth performance of Gibel carps in the normal pond group (P) and the rice floating beds pond group (RP). (A) body length. (B) body weight. (C) weight gain rate. Different letters represent significant differences between different groups (P < 0.05). Weight gain rate = (Wfinal − Winitial)/Winitial × 100%.
3.2 Microbial community changes
3.2.1 Changes in α and β diversity of microbial community
The water and rice floating beds root microbial showed significantly higher α-diversity indexes than intestinal microbiota, such as Shannon and Chao1 (P < 0.01), while the Simpson index decreased significantly of water microbial (P < 0.01) (Figures 2A–C). In addition, the Shannon index in the RIM group was significantly higher (P < 0.001) than that in the PIM group, while the Simpson index decreased significantly in the RIM group compared with the PIM group (P < 0.001) (Figures 2A, C).
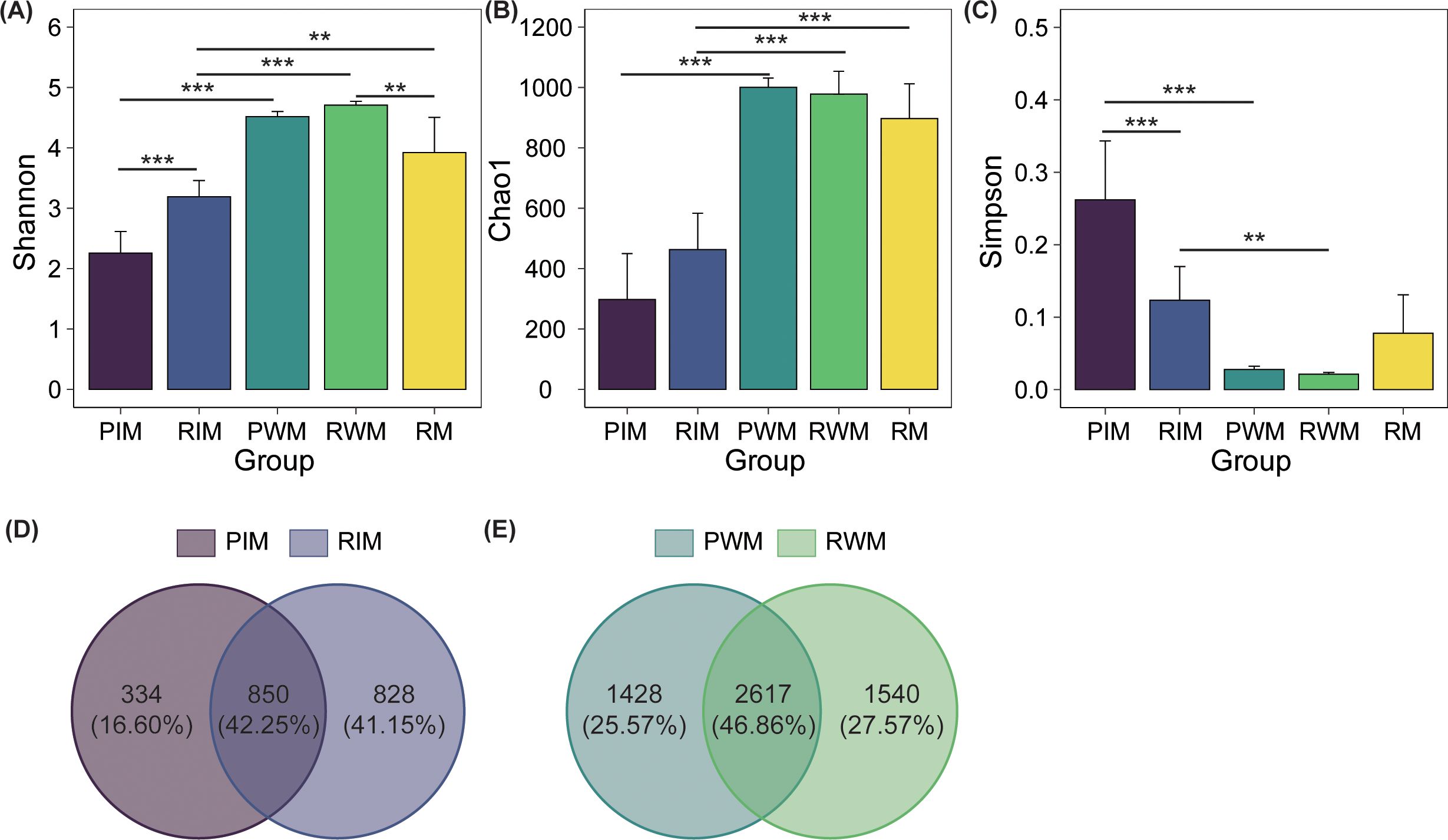
Figure 2. The α diversity and Venn diagrams of intestinal and water microbiota. (A) Shannon index. (B) Chao1 index. (C) Simpson index. (D) The Venn diagram of intestinal microbiota. (E) The Venn diagram of water microbiota. The bar chart is marked with “*” signifying a statistically significant difference (**P < 0.01, ***P < 0.001). Abbreviation codes: PIM, the intestinal microbiota in the normal pond group; RIM, the intestinal microbiota in the rice floating beds pond group; PWM, the water microbiota in the normal pond group; RWM, the water microbiota in the rice floating beds pond group; RM, the rice floating beds root microbial communities. The same as below.
For intestinal microbiota, the OTU analysis indicated that the total number of OTUs of intestinal microbiota in PIM and RIM groups was 2012. The PIM and RIM shared 850 OTUs. The unique OUT sequences in RIM (828) were higher than that in PIM (334) (Figure 2D). For water microbiota, the total number of OTUs of water microbiota in PWM and RWM groups was 5585. Specially, there were 1428 OTU sequences in the PWM groups and 1540 OTU sequences in the RWM group, with 2617 OTUs shared (Figure 2E).
The β diversity showed high variation in different groups (Figure 3A). For intestinal microbiota, the β diversity in the PIM group was significantly higher than that in the RIM group (P < 0.001). For water microbiota, the β diversity in the RWM group was significantly higher than that in the PWM group (P < 0.05). Compared with water microbiota, the intestinal microbiota in both normal ponds and rice floating beds ponds showed significantly higher β diversity (P < 0.05). In the rice floating beds pond group, the RWM showed significantly lower β diversity than the RIM and RM groups (P < 0.05). For microbiota of intestine, water, and rice root, the principal component analysis (PCA) showed the samples from different groups were separated clearly (Figure 3B).
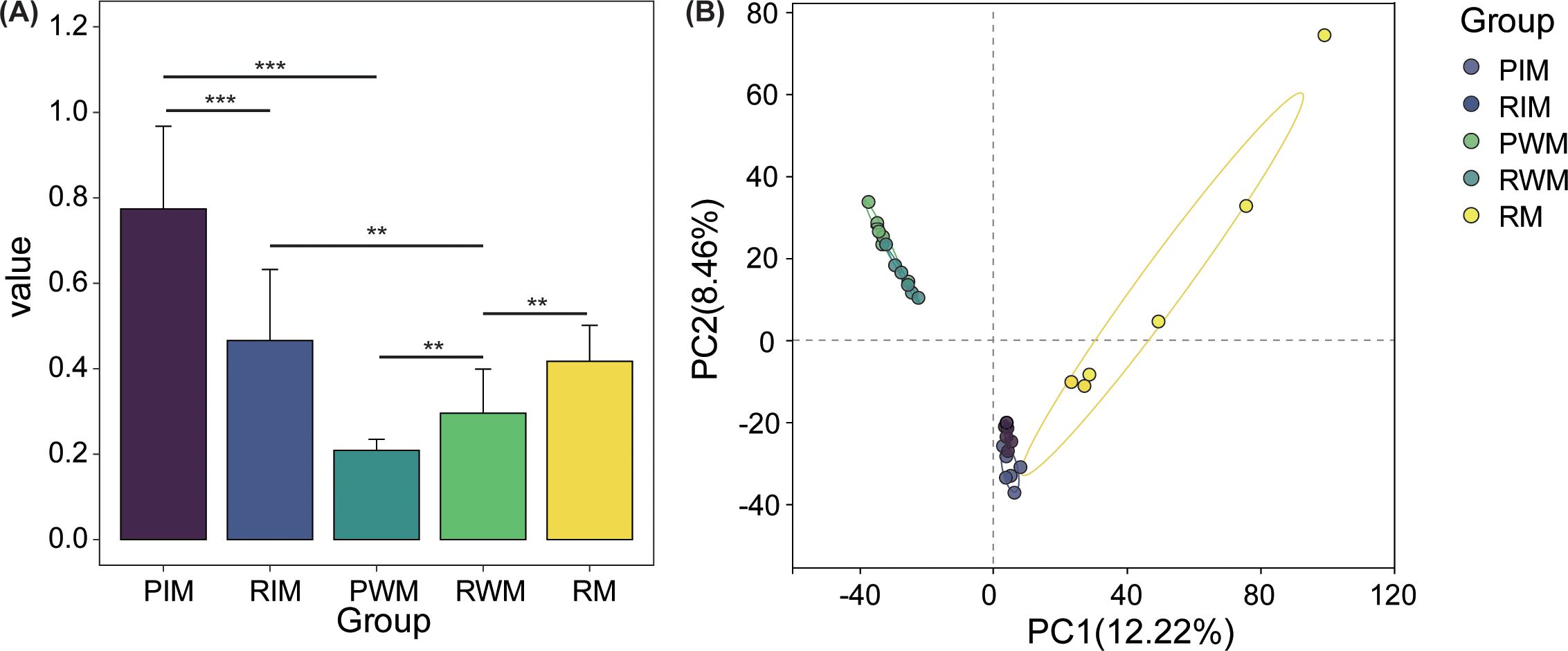
Figure 3. The β diversity and the principal component analysis (PCA) diagrams of intestinal microbiota. (A) The variations in β diversity among different groups. (B) The PCA analysis of intestinal microbiota. The bar chart displays an asterisk (*) to signify significant differences, with **P < 0.01, and ***P < 0.001. Ellipses on PCA score plots represented a confidence interval of 95%.
3.2.2 The differences in the assembly process and biomarker taxa of different microbial community
The NCM successfully estimated a large fraction of the relationship between the frequency of OTUs and their relative abundance variations (Figure 4), and explained the differences in microbial community assembly between rice floating bed ponds and normal ponds, which were 0.4376% and 0.0707%, respectively. Further, the NST revealed the relative importance of stochastic and deterministic processes for the construction of water, intestine, and root microbial communities in rice floating bed ponds and normal ponds. Most of the NST values were higher than the 50% boundary point for the microbial community in both rice floating bed ponds and normal ponds, while it was significantly lower than the 50% boundary point for the RIM group (Figure 4 bar plots). For the intestinal microbial community in the normal pond, the NST value was near 50%.
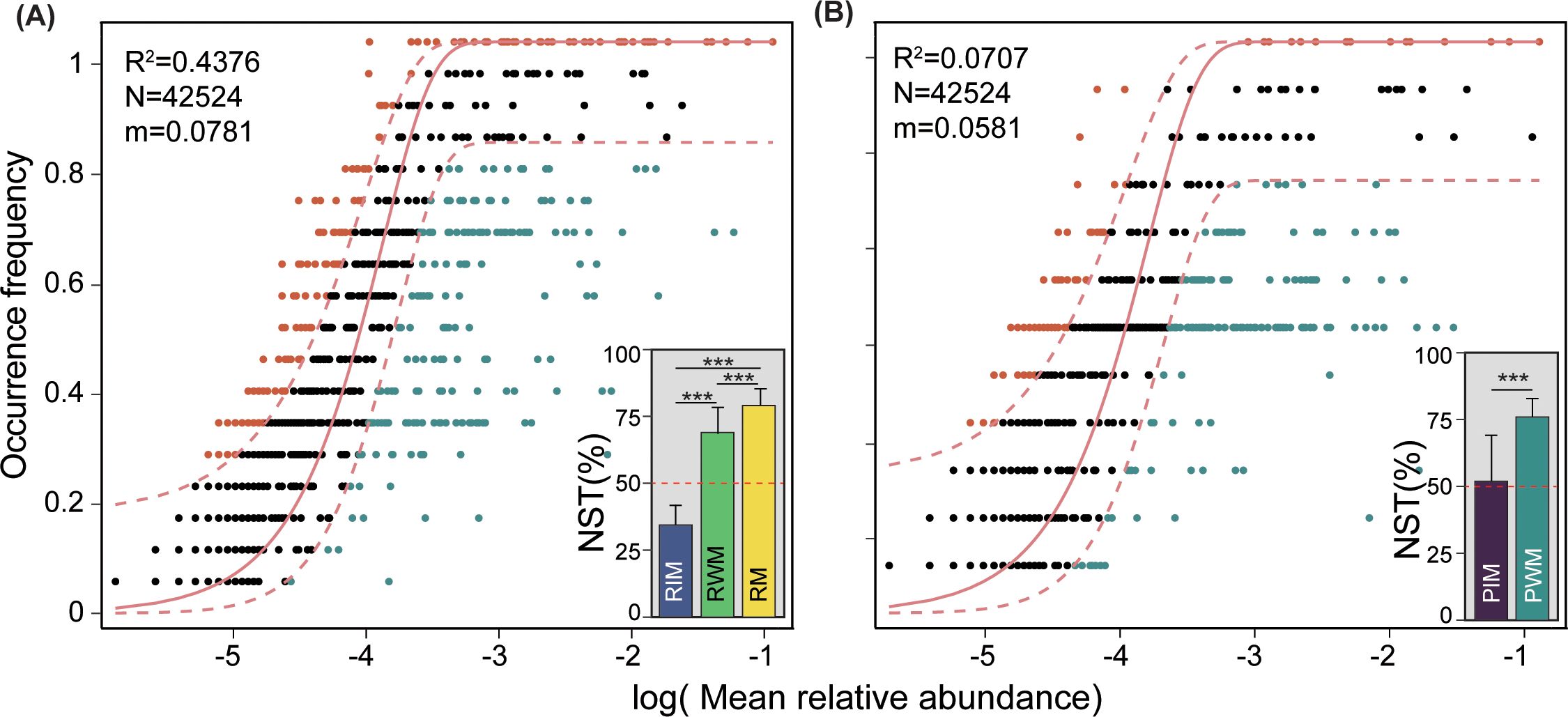
Figure 4. Community assembly process measurements. (A) The fit of the neutral community model (NCM) for RIM, RWM, and RM communities in rice floating beds pond. (B) The fit of NCM for PIM and PWM communities in normal ponds. Bar plots showing the normalized stochastic ratio (NST) represented the relative contribution of stochastic processes to the community assembly. The bar chart displays an asterisk (*) to signify significant differences, with ***P < 0.001. The NST value > 50% indicates that stochastic processes predominated in regulating the community assembly of microbes.
LEfSe analysis was used to determine biomarker taxa with the most significant differences in abundance (Figure 5). The results showed that there were more enriched taxa in the RIM group than in the PIM group. Compared with PIM, the phyla Actinobacteriota, Armatimonadota, Bdellovibrionota, Chloroflexi, Cyanobacteria, Desulfobacterota, Patescibacteria, and Planctomycetota were all significantly enriched in RIM (Figure 5A). Further, the classes Bacteroidia, Ignavibacteria, and Gammaproteobacteria were significantly higher in the RMW group and mainly contributed to the difference between the RWM and PWM groups. In the PWM group, the classes Vicinamibacteria, Subgroup_22, and Caldisericia exhibited significantly higher abundances than those in the RWM group (Figure 5B).

Figure 5. Cladogram of microbiota via LEfSe method identifying the significantly different abundant taxa. (A) LEfSe analysis of intestinal microbial communities between RIM and PIM groups. (B) LefSe analysis of water microbial communities between RWM and PWM groups. LEfSe analysis was performed based on the LDA effect size > 2 and P < 0.05. The taxonomic classification was shown from phylum to genus level from the center outward.
4 Discussion
As a compound aquaculture production system, the rice floating bed is characterized as a low-cost, solar-energy-based, and eco-friendly technology (Li et al., 2018). In this system, fish and plants provide mutual benefits, as the fish consume rice straw while their waste acts as a natural fertilizer for the rice plants (Goda et al., 2024). Based on this unique ecological interaction, by comparing the growth performance, intestinal microbial diversity, intestinal microbial construction, and intestinal microbial composition, this study revealed that the utilization of rice floating bed promoted the growth of Gibel carp, and cultivated a more diverse and complex microbial community compared with the normal pond system. These results further validate the potential advantages of floating rice beds in sustainable farming.
The results indicated that the Gibel carps raised in rice floating bed exhibited significantly enhanced body length, body weight, and weight gain rates compared to those in normal pond setups. The improved growth performance could be attributed to several interconnected factors inherent to the floating bed ecosystem, such as rich food resources, complex habitats, and efficient nutrient cycling (Tanner and Headley, 2011; Wang et al., 2020; Zhang et al., 2022). On the one hand, the plants and the microbiota attached to the plant roots could absorb nutrients from the water to support their growth and reproduction, which promoted nutrient cycling and water purification (Tanner and Headley, 2011). On the other hand, the presence of both aquatic plants and associated microorganisms provided a diverse array of nutritional resources that can enhance the availability of essential dietary components (Hossain et al., 2024).
The microbial diversity analyses revealed differences in the intestinal microbiota composition between Gibel carp in normal ponds (PIM) and those in rice floating bed ponds (RIM). The Shannon and Chao1 indices were significantly higher in the intestinal microbiota of fish raised in rice floating bed ponds, indicating a richer microbial community. The increase in diversity may provide multiple metabolic pathways and greater abilities to resist infections, thereby supporting overall fish health (Ringø et al., 2022; Medina-Félix et al., 2023). Furthermore, the larger number of unique operational taxonomic units (OTUs) found in the RIM suggested that floating rice beds created a favorable environment conducive to the establishment of microbial species, thereby leading to increased α diversity. Taken together, it could be deduced that the rice floating bed could enhance the species abundance and diversity of Gibel carp intestinal microbiota, which may contribute to regulating intestinal immunity, nutrient absorption, and host healthy status (Medina-Félix et al., 2023). Moreover, the results also indicated that the intestinal microbial communities exhibited lower α diversity and OTUs compared with those of water and root microbiota, while the β diversity of intestinal microbiota was significantly higher than that of water and root microbiota, which revealed the high variability of intestinal microbiota. This could be attributed to that the intestinal bacteria directly colonize the intestinal tract of fish from the surrounding environmental water (Li et al., 2017), and are highly affected by the culture environment, including the water temperature, salinity, pH, and feed source (Eichmiller et al., 2016; Zhao et al., 2020; Kim et al., 2021).
In addition to the differences in microbial abundance and diversity, the present study revealed the different assembly processes of microbial communities among groups. By evaluating these assemblies through a stochastic lens, the NCM offers an approach to assess the relative influence of random processes on community composition (Roguet et al., 2015; Wang et al., 2022). In the model, a high fit of the model (high R2 value) indicates a great influence by stochastic processes (Roguet et al., 2015). In the present study, we found that the R2 values for the microbial communities in the rice floating beds pond and normal pond were 0.4376 and 0.0707, respectively, indicating that stochastic processes play a higher role in the construction of bacterial communities in the rice floating beds pond than that in normal pond. This may be due to the rice floating bed enhancing the microbial diversity and accelerating their assembly towards stochastic evolution. Moreover, the values of NST serves as an auxiliary correction for determining the contribution of stochastic processes. In the present study, most of the NST values were higher than 50%, but for the RIM group, suggesting that the deterministic processes contributed more to the assembly of the intestinal microbial community in rice floating bed ponds. In general, when deterministic processes are dominant, selection via biotic or abiotic factors deeply influences the shaping of the microbial community (Liu et al., 2022). In the present study, the abundant root system under the water surface not only provided alternative food sources for Gibel carp but also created a complex microhabitat community, which may shape the intestinal microbial community of Gibel carp.
Bacterial communities are essential for maintaining the environmental health of aquaculture systems. They play a vital role in various physiological processes, including digestion, metabolism, and immune function. In this investigation, the composition of intestinal bacterial communities in the RIM group was found to be richer than that in the PIM group, with Chloroflexi and Cyanobacteria were significantly enriched in RIM group. Plant-Cyanobacteria interactions, as a beneficial symbiosis, have long been demonstrated in rice-growing areas where the most efficient nitrogen-fixing cyanobacteria are present (Nowruzi et al., 2021). Members of Chloroflexi are also frequently found in plant systems and capable of fixing inorganic CO2 and nitrite (Narsing Rao et al., 2022). In this study, the elevation of Cyanobacteria and Chloroflexi suggested a high efficiency of nutrient cycling in the rice floating beds ponds. Intriguingly, the level of certain plant-associated bacteria (e.g., Ignavibacteria) also exhibited a noticeable increase in the RWM group. The Ignavibacteria are ubiquitously abundant in paddy soils and contribute to the decomposition of complex polymers, such as cellulose, hemicellulose, and chitin (Bei et al., 2021). Therefore, a higher abundance of Ignavibacteria in the rice floating beds ponds may be associated with more plant tissue, such as rice roots, scattered in the water.
5 Conclusion
The present study evidenced that Gibel carps cultivated in rice floating bed systems exhibit significantly enhanced growth performance compared to those reared in normal pond environments. The results indicate that the rice floating bed ecosystem promotes greater body length, body weight, and weight gain rates, likely due to enriched nutrient availability and diverse habitats that facilitate efficient nutrient cycling. Furthermore, the microbial community analysis reveals noteworthy differences in the intestinal microbiota composition between the two cultivation systems. The higher α diversity and unique operational taxonomic units in the rice floating bed ponds suggest a more robust microbial ecosystem that may benefit the fish’s health. The assembly processes of these microbial communities, largely influenced by stochastic mechanisms in the rice floating beds, further underscore the ecological complexity inherent in this aquaculture practice. Notably, in rice floating beds ponds, the identification of significant bacterial phyla associated with nutrient cycling highlights the multifaceted roles of microbial communities in sustaining environmental health within aquaculture systems. Overall, these findings highlight the potential of rice floating bed systems as a sustainable aquaculture technique, fostering improved growth performance and microbial diversity, which are critical for the health of cultured fish.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was approved by Anhui Academy of Agricultural Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YJ: Conceptualization, Resources, Validation, Writing – original draft, Writing – review & editing. YS: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. BZ: Methodology, Software, Visualization, Writing – review & editing. CR: Validation, Writing – review & editing. SR: Resources, Writing – review & editing. MW: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the China Agriculture Research System (CARS-45); the Special Fund for Anhui Agriculture Research System (AARS-08); the New doctoral talents project of Anhui Academy of Agricultural Sciences (XJBS-0034); the Feixi County Special Program for Science and Technology Commissioners.
Conflict of interest
Author SR was employed by the company Hefei Taolin Shuixiang Snakehead Ecological Aquaculture Development Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bei Q., Peng J., Liesack W. (2021). Shedding light on the functional role of the Ignavibacteria in Italian rice field soil: A meta-genomic/transcriptomic analysis. Soil Biol. Biochem. 163, 108444. doi: 10.1016/j.soilbio.2021.108444
Boyd C. E., D’Abramo L. R., Glencross B. D., Huyben D. C., Juarez L. M., Lockwood G. S., et al. (2020). Achieving sustainable aquaculture: Historical and current perspectives and future needs and challenges. J. World Aquacult. Soc. 51, 578–633. doi: 10.1111/jwas.12714
Burns A. R., Stephens W. Z., Stagaman K., Wong S., Rawls J. F., Guillemin K., et al. (2016). Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664. doi: 10.1038/ismej.2015.142
Chen L., Ling H., Tan J., Shao X. (2020). Removing nutrients from crab-breeding wastewater by a floating plant–effective microorganism bed. Water 12, 3384. doi: 10.3390/w12123384
De Silva S. S. (2012). Aquaculture: a newly emergent food production sector—and perspectives of its impacts on biodiversity and conservation. Biodivers. Conserv. 21, 3187–3220. doi: 10.1007/s10531-012-0360-9
Diana J. S. (2009). Aquaculture production and biodiversity conservation. BioScience 59, 27–38. doi: 10.1525/bio.2009.59.1.7
Duan Y., Nan Y., Zhu X., Yang Y., Xing Y. (2024). The adverse impacts of ammonia stress on the homeostasis of intestinal health in Pacific white shrimp (Litopenaeus vannamei). Environ. pollut. 340, 122762. doi: 10.1016/j.envpol.2023.122762
Edwards P. (2015). Aquaculture environment interactions: Past, present and likely future trends. Aquaculture 447, 2–14. doi: 10.1016/j.aquaculture.2015.02.001
Eichmiller J. J., Hamilton M. J., Staley C., Sadowsky M. J., Sorensen P. W. (2016). Environment shapes the fecal microbiome of invasive carp species. Microbiome 4, 44. doi: 10.1186/s40168-016-0190-1
Goda A. M. A.-S., Aboseif A. M., Mohammedy E. Y., Taha M. K. S., Mansour A. I. A., Ramadan E. A., et al. (2024). Earthen pond-based floating beds for rice-fish co-culture as a novel concept for climate adaptation, water efficiency improvement, nitrogen and phosphorus management. Aquaculture 579, 740215. doi: 10.1016/j.aquaculture.2023.740215
Hossain M. M., Rahman M. H., Tina F. W., Shahjahan M. (2024). Present scenario and prospects of the use of aquatic plants in aquaculture: a review. Aquacult. Int. 32, 6791–6825. doi: 10.1007/s10499-024-01489-1
Jiang L., Yi M., Jiang Z., Wu Y., Cao J., Liu Z., et al. (2024). Effect of pond-based rice floating bed on the microbial community structure and quality of water in pond of mandarin fish fed using artificial diet. Biology 13, 549. doi: 10.3390/biology13070549
Ke X., Yi M., Li Q., Liu Z., Wang M., Cao J., et al. (2021). Effect of the herbal Houttuynia cordata floating bed on the Nile tilapia pond culturing system. Aquacult. Rep. 20, 100680. doi: 10.1016/j.aqrep.2021.100680
Kim P. S., Shin N.-R., Lee J.-B., Kim M.-S., Whon T. W., Hyun D.-W., et al. (2021). Host habitat is the major determinant of the gut microbiome of fish. Microbiome 9, 166. doi: 10.1186/s40168-021-01113-x
Li G., Tao L., Li X., Peng L., Song C., Dai L., et al. (2018). Design and performance of a novel rice hydroponic biofilter in a pond-scale aquaponic recirculating system. Ecol. Eng. 125, 1–10. doi: 10.1016/j.ecoleng.2018.10.001
Li X., Zhou L., Yu Y., Ni J., Xu W., Yan Q. (2017). Composition of Gut Microbiota in the Gibel Carp (Carassius auratus gibelio) Varies with Host Development. Microb. Ecol. 74, 239–249. doi: 10.1007/s00248-016-0924-4
Liang Y., Wang Z., Gao N., Qi X., Zeng J., Cui K., et al. (2024). Variations and interseasonal changes in the gut microbial communities of seven wild fish species in a natural lake with limited water exchange during the closed fishing season. Microorganisms 12, 800. doi: 10.3390/microorganisms12040800
Liu C., Zhao D., Ma W., Guo Y., Wang A., Wang Q., et al. (2016). Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 100, 1421–1426. doi: 10.1007/s00253-015-7039-6
Liu H., Sun F., Peng J., Shen M., Li J., Dong Y. (2022). Deterministic process dominated belowground community assembly when suffering tomato bacterial wilt disease. Agronomy 12, 1024. doi: 10.3390/agronomy12051024
Medina-Félix D., Garibay-Valdez E., Vargas-Albores F., Martínez-Porchas M. (2023). Fish disease and intestinal microbiota: A close and indivisible relationship. Rev. Aquacult. 15, 820–839. doi: 10.1111/raq.12762
Miyake S., Ngugi D. K., Stingl U. (2015). Diet strongly influences the gut microbiota of surgeonfishes. Mol. Ecol. 24, 656–672. doi: 10.1111/mec.13050
Narsing Rao M. P., Luo Z.-H., Dong Z.-Y., Li Q., Liu B.-B., Guo S.-X., et al. (2022). Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles. Environ. Res. 209, 112888. doi: 10.1016/j.envres.2022.112888
Ning D., Deng Y., Tiedje J. M., Zhou J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. U.S.A. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Nowruzi B., Bouaïcha N., Metcalf J. S., Porzani S. J., Konur O. (2021). Plant-cyanobacteria interactions: Beneficial and harmful effects of cyanobacterial bioactive compounds on soil-plant systems and subsequent risk to animal and human health. Phytochemistry 192, 112959. doi: 10.1016/j.phytochem.2021.112959
Ringø E., Harikrishnan R., Soltani M., Ghosh K. (2022). The effect of gut microbiota and probiotics on metabolism in fish and shrimp. Animals 12, 3016. doi: 10.3390/ani12213016
Ringø E., Zhou Z., Vecino J. L. G., Wadsworth S., Romero J., Krogdahl Å., et al. (2016). Effect of dietary components on the gut microbiota of aquatic animals. A never-ending story? Aquacult. Nutr. 22, 219–282. doi: 10.1111/anu.12346
Roguet A., Laigle G. S., Therial C., Bressy A., Soulignac F., Catherine A., et al. (2015). Neutral community model explains the bacterial community assembly in freshwater lakes. FEMS Microbiol. Ecol. 91, fiv125. doi: 10.1093/femsec/fiv125
Sopawong A., Yusoff F. M., Zakaria M. H., Khaw Y. S., Monir M. S., Hashim A.M. (2024). Development of a bio-green floating system (BFAS) for the improvement of water quality, fish health, and aquaculture production. Aquacult. Int. 32, 1101–1118. doi: 10.1007/s10499-023-01207-3
Tanner C. C., Headley T. R. (2011). Components of floating emergent macrophyte treatment wetlands influencing removal of stormwater pollutants. Ecol. Eng. 37, 474–486. doi: 10.1016/j.ecoleng.2010.12.012
Thomas M., Pasquet A., Aubin J., Nahon S., Lecocq T. (2021). When more is more: taking advantage of species diversity to move towards sustainable aquaculture. Biol. Rev. 96, 767–784. doi: 10.1111/brv.12677
Uma A., Subash P., Abraham T. J. (2020). Importance of gut microbiota in fish – A review. ijah 59, 181–194. doi: 10.36062/ijah.59.2SPL.2020.181-194
Wang R., Chen H., Zhu Y., Al-Masqari Z. A., Yan M., Wang G., et al. (2022). Survival status of Penaeus vannamei is associated with the homeostasis and assembly process of the intestinal bacterial community. Aquaculture 558, 738398. doi: 10.1016/j.aquaculture.2022.738398
Wang J., Fu G., Li W., Shi Y., Pang J., Wang Q., et al. (2018). The effects of two free-floating plants (Eichhornia crassipes and Pistia stratiotes) on the burrow morphology and water quality characteristics of pond loach (Misgurnus anguillicaudatus) habitat. Aquacult. Fish. 3, 22–29. doi: 10.1016/j.aaf.2017.12.001
Wang W.-H., Wang Y., Sun L.-Q., Zheng Y.-C., Zhao J.-C. (2020). Research and application status of ecological floating bed in eutrophic landscape water restoration. Sci. Total Environ. 704, 135434. doi: 10.1016/j.scitotenv.2019.135434
Xu X., Li C., Li J., Wang F., Zhou S. (2024). The enhancement of wastewater purification efficiency in ecological floating bed aquaculture through alginate oligosaccharide treatment. Aquacult. Int. 32, 9529–9546. doi: 10.1007/s10499-024-01627-9
Yamaki A., Yamamuro M. (2013). Floating-leaved and emergent vegetation as habitat for fishes in a eutrophic temperate lake without submerged vegetation. Limnology 14, 257–268. doi: 10.1007/s10201-013-0403-2
Zhang Q., Achal V., Xu Y., Xiang W.-N. (2014). Aquaculture wastewater quality improvement by water spinach (Ipomoea aquatica Forsskal) floating bed and ecological benefit assessment in ecological agriculture district. Aquacultural Eng. 60, 48–55. doi: 10.1016/j.aquaeng.2014.04.002
Zhang H., Liu H., Cao W. (2022). Nutrient removal effect and characteristics of integrated floating beds at low temperature. Environ. Res. 204, 112139. doi: 10.1016/j.envres.2021.112139
Keywords: rice floating bed, Carassius auratus gibelio, intestine, water, microbial communities
Citation: Jiang Y, Sun Y, Zhou B, Rong C, Ruan S and Wu M (2025) Enhancing aquaculture sustainability: the role of rice floating beds in improving Gibel carp growth and microbiota composition. Front. Mar. Sci. 12:1549639. doi: 10.3389/fmars.2025.1549639
Received: 07 January 2025; Accepted: 24 February 2025;
Published: 12 March 2025.
Edited by:
Marcel Martinez-Porchas, National Council of Science and Technology (CONACYT), MexicoReviewed by:
Yiran Hou, Chinese Academy of Fishery Sciences, ChinaDiana Medina Felix, State University of Sonora, Mexico
Copyright © 2025 Jiang, Sun, Zhou, Rong, Ruan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minglin Wu, bWx3MjAxOEAxMjYuY29t
†These authors have contributed equally to this work and share first authorship
 Yangyang Jiang1,2†
Yangyang Jiang1,2† Yongxu Sun
Yongxu Sun Minglin Wu
Minglin Wu