
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mar. Sci., 03 March 2025
Sec. Ocean Observation
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1531729
Placozoa are small disc-shaped animals representing one of the early branching metazoan lineages with only a dozen cell types, fast effector reactions, and complex behaviors. The simplest organization and small cell sizes limit standard microscopy applications. Here, we implement a new methodology and protocol for expansion microscopy, improving both the resolution and preservation of fragile placozoans and kin. As a result, the proposed approaches can be applicable to a diversity of microscopic animals and their larvae with broad applicability for both laboratory and cross-disciplinary field research during long-term marine expeditions.
The origin and early evolution of animals and their cell diversity are unclear (Mikhailov et al., 2009; Nielsen, 2012; 2019; Ruiz-Trillo et al., 2023). In this direction, representatives of the phylum Placozoa are essential for all reconstructions of metazoan evolution (Schierwater et al., 2009; Romanova and Moroz, 2024). These are the simplest known free-living metazoans with a relatively small set of cell types that are organized in three layers (Schulze, 1883; Grell, 1971; Rassat and Ruthmann, 1979; Smith et al., 2014; Romanova et al., 2021): the lower epithelium (lipophil, gland, epithelial cells), the middle (fiber, neural-like, crystal cells) and the upper (shiny spheres, epithelium). One of the features of the placozoan cell architecture is their relatively small size - on average 4-7 microns, with the disk-like animal thickness of 20-25 microns, and difficulty in the preservation of ultrastructural features, which provide limitations in the broad applicability of methods for convenient microscopy.
Currently, many studies in Placozoa targeting cell-type physiology include fluorescent in situ hybridization (FISH), immunohistochemical staining, and the use of highly selective cell organelle markers (Sebé-Pedrós et al., 2018; Smith et al., 2021; Najle et al., 2023). However, convenient microscopy is primarily achieving cell-type-specific resolution, often with a limited visualization of subcellular structures. This conceptual problem has been resolved by methods of expansion microscopy (ExM) (Chen et al., 2015; Cho et al., 2018; Nakamoto et al., 2022) as promising tools for increasing the dimensions of a studied object from 2x to 20x (Wassie et al., 2019; Wen et al., 2023). The ExM methods include six successive stages: 1) fixation of animals, 2) staining, 3) placing samples in a monomer solution with an initiator and a catalyst, 4) polymerization, 5) processing the polymer with digestion buffer, 6) washing the hydrogel with deionized water (Chozinski et al., 2016; Sun et al., 2021; M’Saad et al., 2022).
Here, we extend the ExM methodology to the enigmatic placozoans, enabling a reliable 2- to 5-fold increase in their body sizes and, therefore, a detailed examination of subcellular organization. Importantly, we show for the first time that stained samples can be stored in hydrogel for a extended time (2-9 weeks) till the digestion buffer stage and subsequent expansion of the sample at the final stage of preparation. This significantly expands the scope of applications of expansion microscopy since there are numerous fragile species, often collected at remote locations of the world ocean, that require cost-efficient methods of storage and fixation in the field conditions of marine expeditions. Thus, we also present a modified ExM protocol as a system for storage (at +4°С) of samples until microscopy.
We used axenic clonal cultures of the major placozoan reference species: Trichoplax adhaerens (Grell’s strain Н1, from the Red Sea). We maintained T. adhaerens culture for 11 years (2014-2024), allowing long-term observations and adjustments of culture conditions (Romanova et al., 2022; 2024).
A suspension of the green alga Tetraselmis marina (WoRMS Aphia, ID 376158) was added to the culture dishes. When the biofilm of microalgae became thinner or depleted, freshly prepared 1-2 mL suspension of T. marina could be added to the culture dishes weekly. H1 haplotype was maintained at the constant temperature of 24°С in climate chambers with 6:18 h D/N light period.
In all tests, animals were placed in 35 mm Petry dishes (n=50 specimens total) and examined under stereo microscopy (Zeiss). For experiments, we used animals ranging from 300 to 400 μm in size. Before staining, animals were maintained for 30 minutes in sterile Petri dishes with artificial seawater (ASW) and washed two to three times to remove any remains of algae. For the current study, we used 1 mM DAPI (#996-1mL, LumiProbe, RF) and 1 mM acridine orange (#O050, Paneco, RF). The samples were examined using a laser confocal Cerna-based microscope (Thorlabs, USA). Image processing was carried out using ImageJ software.
The chemistry of the protocol is based on (Chozinski et al., 2016; Sun et al., 2021; M’Saad et al., 2022), with our modifications summarized in Figure 1 and below. For staining we used such dyes as DAPI and the acridine orange, which labels nuclei and organelles, respectively.
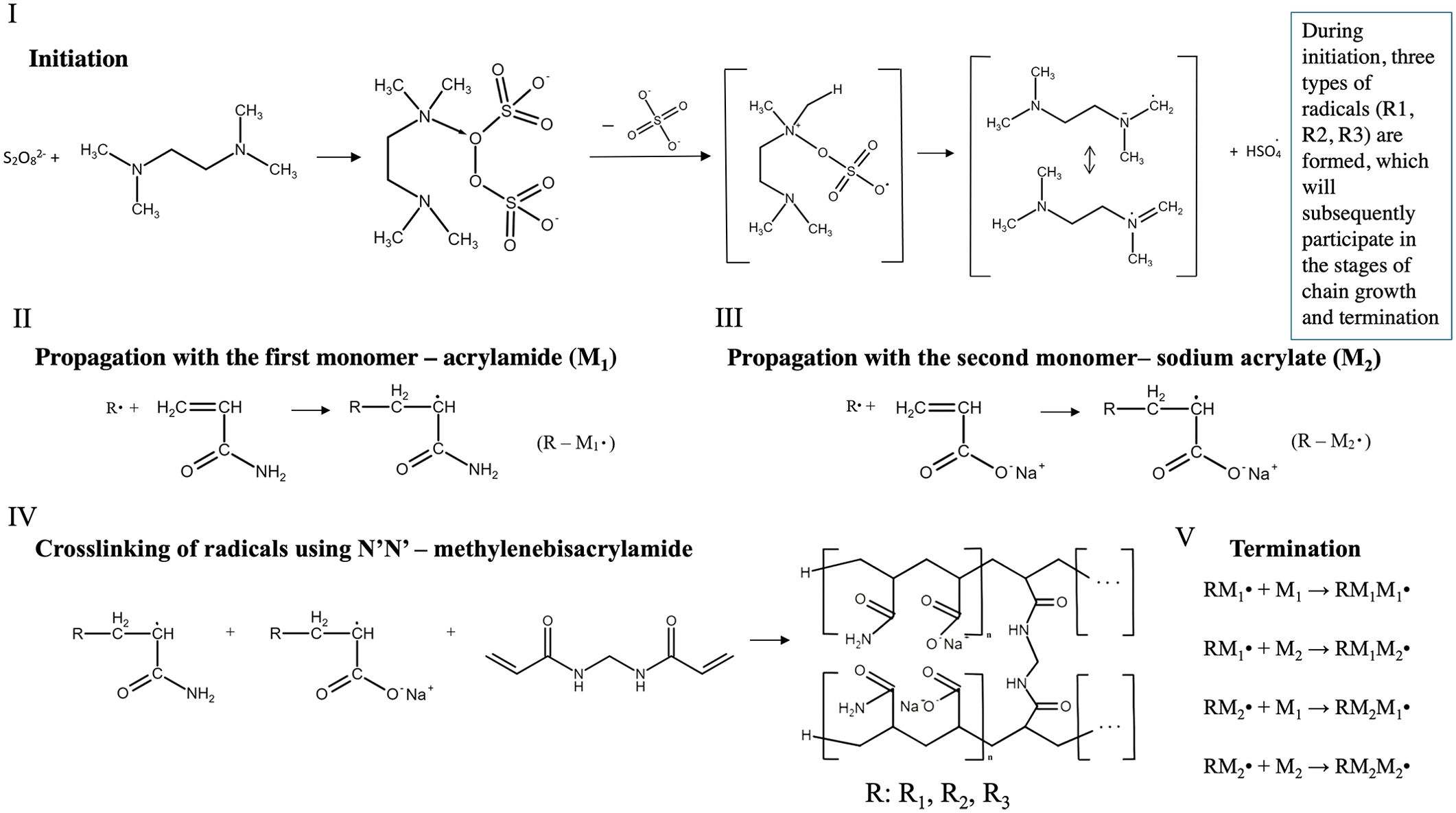
Figure 1. Principal chemical scheme of mechanism for obtaining copolymer. I) Free radical formation using the initiator persulfate anion and the initiation catalyst TEMED (Shirangi et al., 2015). II) A chain propagation involving the first monomer, acrylamide, where R• - is all possible radicals formed in the initiation stage. III) A chain propagation involving the second monomer, sodium acrylate, where R• - is all possible radicals formed in the initiation stage. IV) Cross-linking of radicals produced in the chain propagation step using N’N’-methylenebisacrylamide with an example of regular copolymer production (Menter, 2000). V) Termination with disappearance and recombination of free radicals and formation of low-active radicals. The chemical formula editor of the Department of Inorganic, Physical, and Colloidal Chemistry, Pyatigorsk Medical and Pharmaceutical Institute created the formulas. (https://physcolloid.ru/?page_id=343).
Placozoans are fragile marine animals, so we used vital staining of 30 mL DAPI and 30 mL acridine orange during 1 h incubation in 35 mm Petri dishes with 3 mL of artificial seawater or ASW (pH=8.0, RT). Then, we fixed animals with 0.25% glutaraldehyde for 10 minutes at room temperature. Gently washed with phosphate buffer solution (PBS) (0.1 M, pH=7.4) three times for 5 minutes. The monomer solution (1x PBS, 2M NaCl, 2.5% acrylamide, 0.15% methylenebisacrylamide, 8,625% sodium acrylate) and freshly prepared polymerization initiator (10% ammonium persulfate) and the catalyst (10% TEMED) we added as (Chozinski et al., 2016; Sun et al., 2021; M’Saad et al., 2022).
For mounting samples, we used 35 mm Petri dish with cover glass (22x22 mm) on the bottom. After mounting samples of placozoans: we 1) placed the half volume of monomer solution (50 µL) at the cover glass, then 2) transferred individuals (separately) with a minimum of liquid PBS (3-5 µL), and 3) covered with 50 µL of monomer solution (MS). Gently mixed MS and placed animals in the thickness of MS. All steps for mounting samples were performed on cold blue ice (+4°С); then we placed the Petri dish at +4° С, for 5 min to impregnate the sample with MS. Next, we transferred the Petri dish to the heater chamber at +37° С for 1 hour to polymerization progress.
After polymerization, we had a hardened polymer, which we incubated in 3 mL digestion buffer (50 mM Tris, 1 mM EDTA, 0.1% Triton, and 0.8 M guanidine HCl, pH=8.0) with 16 µL proteinase K at +37° С for 2 hours. The next step is replacing the solution with deionized water: we added 1 mL of deionized water, waited for 20 minutes (3 times), then repeated it for 2-3 hours, gradually reducing time intervals (Figure 2). Store hydrogel at +4° С in a refrigerator.
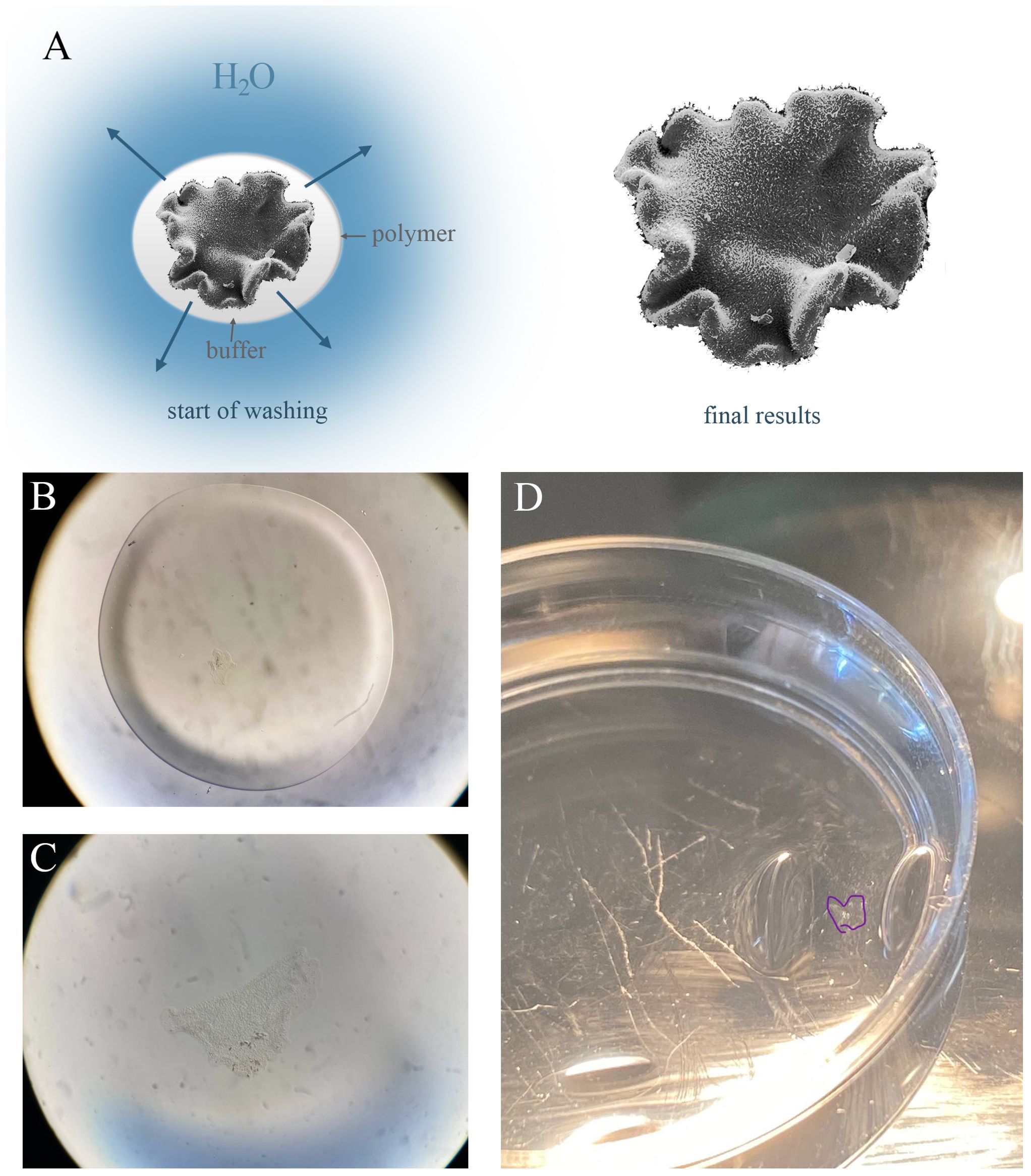
Figure 2. Expansion process during dialysis. (A) Scheme of dialysis of a biological object (Trichoplax) during dialysis. The animal was incorporated in a thin polymer (white color) with the buffer. At the start of dialysis individual has a control size. At the limit of hydrogel saturation and replacement of buffer with water molecules, the animal increases in size due to the stretching of cells. (B–D) Successive stages of hydrogel expansion: the start of washingpolymer (50 µL) with animal (B) to the intermediate stage with expansion sample (C). (B, C) – the same magnification. (D) The animal in the hydrogelis outlined in purple in a 35 mm Petri dish.
We tested the possibility of long-term storage of samples in hydrogel for 1-2 weeks (n=20) and 8-9 weeks (n=4). This is feasible due to the stop reaction at the end of polymer incubation at +37°C: after sample polymerization, the gel was placed in a phosphate buffer solution (0.1 M, pH = 8.0) for storage of the sample at +4°C. At this stage, the dialysis process is started due to a certain amount of water in the PBS solution, but the polymer expands slightly. After storage, the phosphate buffer solution must be removed from the Petri dish without exposure samples to air (or drying), and the hydrogel must be placed in a digestion buffer, where guanidine HCl and proteinase K trigger weak protein denaturation and purification of the sample from protein impurities, as well as inactivation of nucleases. Incubation is carried out according to the standard protocol for two hours, after which the buffer is replaced with deionized water for 2-3 hours. In our hands, all samples were preserved prior to subsequent microscopy.
The important reference point of the sample preparation is that after the hydrogel expansion process, we did not find any detectable morphological damage in animals. There was no separation of nuclei from the cytoplasm nor any abnormal and uncharacteristic cell structures. This indicates the sufficient preservation of membrane and cell structure during stretching the sample preparation process for ExM (Figure 3).
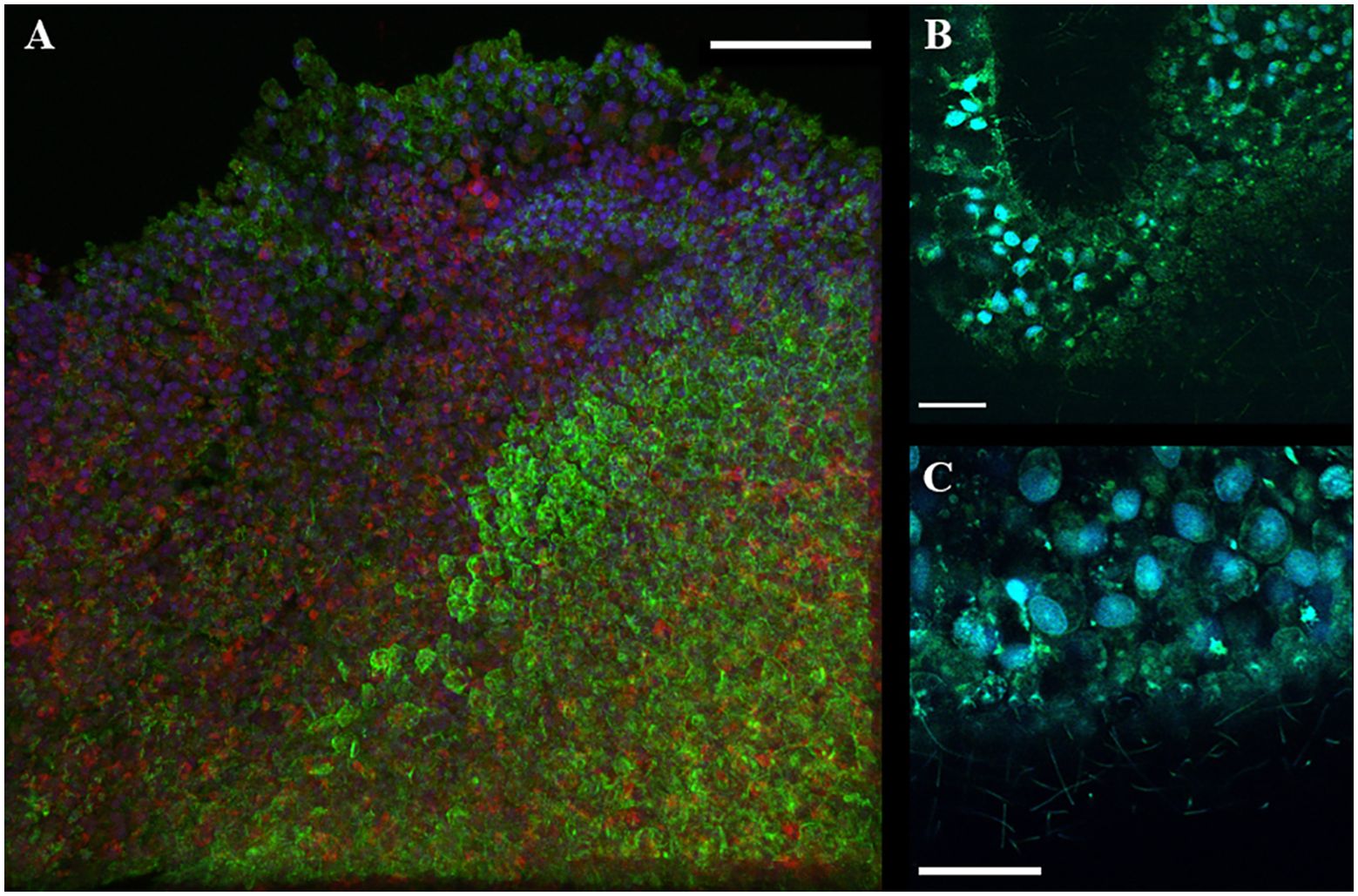
Figure 3. Confocal microscopy of ExM samples of T. adhaerens with a comparison of normal animal size. (A) Regular images on confocal microscopy with 60x objective focused on the rim area of epithelium. Staining with DAPI (blue), phalloidin (green) and mitotracker (red). (B, C) expansion microscopy with DAPI and acridine orange (blue and green channels with 40x objective) for the rim area of epithelial cells. Scale bar: (A–C) - 30 μm.
DAPI staining showed the preservation of native microscopic morphology of nuclei and their localization in the apical part for cells of both lower and upper epithelium layers as in electron microscopic studies. The middle layer fiber and crystal cells (Figure 4) also preserved their localization and microscopic morphology.
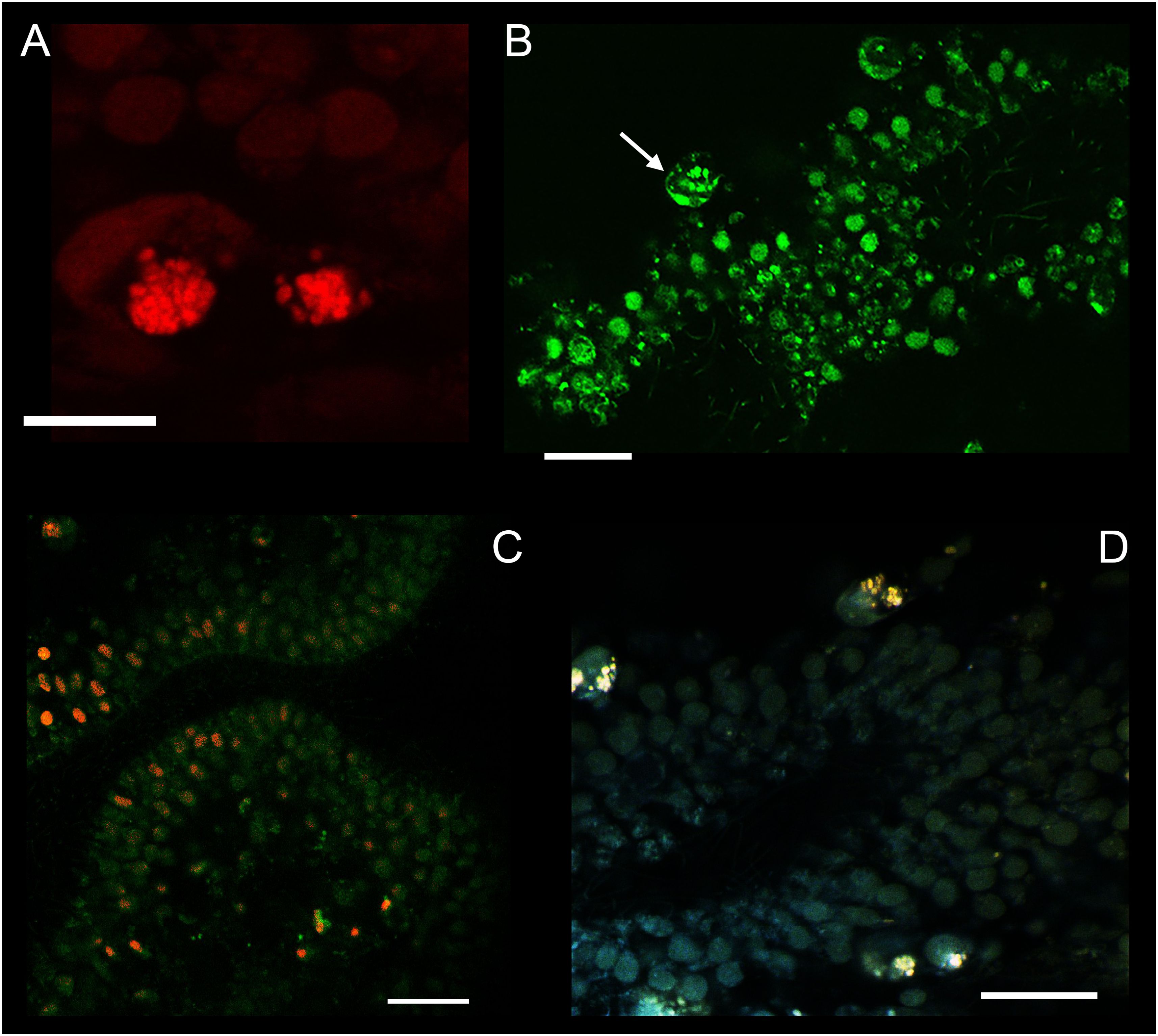
Figure 4. Confocal microscopy of ExM samples of T. adhaerens using acridine orange. (A) Two fiber cells with lysosomes in a red channel. (B) The same sample (as A section) in a green channel at different intracellular locations. (C) Colocalization of labeling (from green and red channels). AO in a green channel marked double-stranded DNA. In the red channel it revealed acidic lysosomes. (D) Colocalization of green and red channel signals for fiber cells: lysosomes have shown in green and red channels. Scale bar: (A) – 20 μm, (B–D) – 30 μm.
We used acridine orange as a vital dye at pH=8 in seawater. A lower pH level can induce injury and placozoan body compression, which can negatively affect microscopic anatomy. After fixing the animals with glutaraldehyde, we used PBS with a lower pH of 7.4 to wash the samples. The localization of acridine orange in placozoan cells was studied with emission in the green (488 nm) and red (633 nm) spectrum to assess the differences for cell population. It was shown that emission in the green spectrum detects colocalization of the signal from some cell nuclei in the lower epithelium, as well as multiple signals from the cell cytoplasm in all cells of the animal (Figure 4). With emission in the red spectrum, we localized granular structures similar to lysosomes in the cells (Figure 4A). It has been previously shown that during intravital staining with acridine orange (AO), metachromatically fluorescent granules are detected in the cytoplasm in the red region of the spectrum since AO monomers accumulate in acidic lysosomes due to the proton gradient across the membrane (Millot et al., 1997).
We show that fiber cells contain many (n>10) lysosomes in the cytoplasm (Figures 4A, B (Smith et al., 2014; Romanova et al., 2021)). It is important to note that the somas of fiber cells contain a mega cluster of mitochondria, and between them are small acidophilic structures that have not been previously identified. We suggest that the mitochondrial cluster may be associated with lysosomes due to the presumed functions of fiber cells, namely their involvement in immunity and digestion, as well as housing endosymbiotic bacteria.
The expansion microscopy significantly broadens the opportunities for detailed microscopic examinations coupled with physiological studies on the emerging model species. Although standard staining methods with a diversity of dyes, antibodies or RNA probes were used to analyze cytological structural differences between cell types (Smith et al., 2017; Ruthmann et al., 1986; Syed and Schierwater, 2002), only transmission electron microscopy provided the sufficient subcellular resolution for Trichoplax and kin (Smith et al., 2014; Romanova et al., 2021) due to relatively small cell sizes. Thus, there are growing needs for a simpler methodology, such as expansion microscopy, to monitor subcellular architecture in the context of labeling signal molecules or low-expressed mRNA (Sebé-Pedrós et al., 2018; Smith et al., 2021; Najle et al., 2023) at subcellular resolution. Here, the expansion microscopy (ExM) method was adapted for Placozoa for the first time based on the formation of a polymer hydrogel and its subsequent expansion while maintaining the morphological integrity of the samples for confocal microscopy. Specifically, we optimized conditions for Trichoplax to increase their body dimensions without observable morphological or structural disarrays.
As the second outcome, the use of similar polymerization embedding for potential long-term preserving samples in the field during expedition conditions (Moroz, 2015) is an effective solution applicable to many marine organisms and their developmental states collected in the field. For example, some species of comb jellies [e.g., representatives of Lobata or deepwater species or their larvae/juvenile (Moroz et al., 2024)] are super fragile. It would be reasonable to test whether the proposed protocol can work for other reference species in the field. We anticipate it would be possible to incubate animals in a container with a dye for 1-2 hours and gently fix the animal with manipulations in a monomer solution, following its polymerization, expansion, and preservation for weeks. If proven, the ExM protocol can be helpful for long-term land and oceanic expeditions, saving samples with subcellular resolution for months if needed. Finally, we should be careful in all 3D morphological interpretations of ExM since it was noted that expansion is not entirely uniform for different cellular components (Büttner et al., 2021); and additional control tests would be required to compare preservations of intracellular architecture using complementary methods.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
DR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YF: Data curation, Investigation, Methodology, Software, Writing – review & editing. LM: Conceptualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the Russian Science Foundation grant (23-14-00050) to DR.
The authors thank students Povernov A.A., Kololeeva V.S. and Allyamova M.T. for help in short-term maintaining cultural dishes.
All authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Büttner M., Lagerholm C. B., Waithe D., Galiani S., Schliebs W., Erdmann R., et al. (2021). Challenges of using expansion microscopy for super-resolved imaging of cellular organelles. Chembiochem 22, 686–693. doi: 10.1002/cbic.202000571
Chen F., Tillberg P. W., Boyden E. S. (2015). Expansion microscopy. Science 347, 543–548. doi: 10.1126/science.1260088
Cho I., Seo J. Y., Chang J. (2018). Expansion microscopy. J. Microscopy 271, 123–128. doi: 10.1111/jmi.2018.271.issue-2
Chozinski T. J., Halpern A. R., Okawa H., Kim H. J., Tremel G. J., Wong R. O., et al. (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488. doi: 10.1038/nmeth.3833
Grell K. G. (1971). Trichoplax adhaerens FE Schulze und die Entstehung der Metazoen. Naturwiss. Rundschau 24, 160–161.
M’Saad O., Shribak M., Bewersdorf J. (2022). Unclearing microscopy. bioRxiv 2022.2011.2029.518361. doi: 10.1101/2022.11.29.518361
Mikhailov K. V., Konstantinova A. V., Nikitin M. A., Troshin P. V., Rusin L. Y., Lyubetsky V. A., et al. (2009). The origin of Metazoa: a transition from temporal to spatial cell differentiation. Bioessays 31, 758–768. doi: 10.1002/bies.200800214
Millot C., Millot J. M., Morjani H., Desplaces A., Manfait M. (1997). Characterization of acidic vesicles in multidrug-resistant and sensitive cancer cells by acridine orange staining and confocal microspectrofluorometry. J. Histochem. Cytochem. 45, 1255–1264. doi: 10.1177/002215549704500909
Moroz L. L. (2015). Biodiversity meets neuroscience: from the sequencing ship (Ship-Seq) to deciphering parallel evolution of neural systems in Omic’s era. Integr. Comp. Biol. 55, 1005–1017. doi: 10.1093/icb/icv084
Moroz L. L., Collins R., Paulay G. (2024). Ctenophora: Illustrated Guide and Taxonomy. Methods Mol Biol. 2757, 27–102. 10.1007/978-1-0716-3642-8_2
Najle S. R., Grau-Bové X., Elek A., Navarrete C., Cianferoni D., Chiva C., et al. (2023). Stepwise emergence of the neuronal gene expression program in early animal evolution. Cell 186, 4676–4693. doi: 10.1016/j.cell.2023.08.027
Nakamoto M. L., Forró C., Zhang W., Tsai C. T., Cui B. (2022). Expansion microscopy for imaging the cell–material interface. ACS nano 16, 7559–7571. doi: 10.1021/acsnano.1c11015
Nielsen C. (2012). Animal evolution: interrelationships of the living phyla (Oxford: University Press).
Nielsen C. (2019). Early animal evolution: a morphologist's view. R. Soc. Open Sci. 6, 190638. doi: 10.1098/rsos.190638
Rassat J., Ruthmann A. (1979). Trichoplax adhaerens FE Schulze (Placozoa) in the scanning electron microscope. Zoomorphologie 93, 59–72. doi: 10.1007/BF02568675
Romanova D. Y., Moroz L. L. (2024). Brief History of Placozoa. In Ctenophores: Methods and Protocols. Moroz L. L. ed. (Springer US), pp. 103–122. doi: 10.1007/978-1-0716-3642-8_3
Romanova D. Y., Nikitin M. A., Shchenkov S. V., Moroz L. L. (2022). Expanding of life strategies in Placozoa: insights from long-term culturing of Trichoplax and Hoilungia. Front. Cell Dev. Biol. 10, 823283. doi: 10.3389/fcell.2022.823283
Romanova D. Y., Varoqueaux F., Daraspe J., Nikitin M. A., Eitel M., Fasshauer D., et al. (2021). Hidden cell diversity in placozoa: ultrastructural insights from Hoilungia hongkongensis. Cell Tissue Res. 385, 623–637. doi: 10.1007/s00441-021-03459-y
Romanova D. Y., Varoqueaux F., Eitel M., Yoshida M. A., Nikitin M. A., Moroz L. L. (2024). “Long-term culturing of placozoans (Trichoplax and Hoilungia),” in Ctenophores: Methods and Protocols (Springer US, New York, NY), 509–529.
Ruiz-Trillo I., Kin K., Casacuberta E. (2023). The origin of metazoan multicellularity: a potential microbial black swan event. Annu. Rev. Microbiol. 77, 499–516. doi: 10.1146/annurev-micro-032421-120023
Ruthmann A., Behrendt G., Wahl R. (1986). The ventral epithelium of Trichoplax adhaerens (Placozoa): Cytoskeletal structures, cell contacts and endocytosis. Zoomorphology 106, 115–122. doi: 10.1007/BF00312113
Schierwater B., de Jong D., DeSalle R. (2009). Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int. J. Biochem. Cell Biol. 41, 370–379. doi: 10.1016/j.biocel.2008.09.023
Sebé-Pedrós A., Chomsky E., Pang K., Lara-Astiaso D., Gaiti F., Mukamel Z., et al. (2018). Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2, 1176–1188. doi: 10.1038/s41559-018-0575-6
Shirangi M., Sastre Toraño J., Sellergren B., Hennink W. E., Somsen G. W., Van Nostrum C. F. (2015). Methyleneation of peptides by N, N, N, N-tetramethylethylenediamine (TEMED) under conditions used for free radical polymerization: a mechanistic study. Bioconjugate Chem. 26, 90–100. doi: 10.1021/bc500445d
Smith C. L., Abdallah S., Wong Y. Y., Le P., Harracksingh A. N., Artinian L., et al. (2017). Evolutionary insights into T-type Ca2+ channel structure, function, and ion selectivity from the Trichoplax adhaerens homologue. J. Gen. Physiol. 149, 483–510. doi: 10.1085/jgp.201611683
Smith C. L., Mayorova T. D., Winters C. A., Reese T. S., Leys S. P., Heyland A. (2021). “Microscopy studies of placozoans,” in Developmental Biology of the Sea Urchin and Other Marine Invertebrates: Methods and Protocols (NY: Springer US), 99–118.
Smith C. L., Varoqueaux F., Kittelmann M., Azzam R. N., Cooper B., Winters C. A., et al. (2014). Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Cur. Biol. 24, 1565–1572. doi: 10.1016/j.cub.2014.05.046
Sun D. E., Fan X., Shi Y., Zhang H., Huang Z., Cheng B., et al. (2021). Click-ExM enables expansion microscopy for all biomolecules. Nat. Methods 18, 107–113. doi: 10.1038/s41592-020-01005-2
Syed T., Schierwater B. (2002). The evolution of the Placozoa: a new morphological model. Senckenbergiana lethaea 82, 315–324. doi: 10.1007/BF03043791
Wassie A. T., Zhao Y., Boyden E. S. (2019). Expansion microscopy: principles and uses in biological research. Nat. Methods 16, 33–41. doi: 10.1038/s41592-018-0219-4
Keywords: placozoa, ctenophora, trichoplax, expansion microscopy, marine biology, biodiversity, evolution, cell types
Citation: Romanova DY, Frank YA and Moroz LL (2025) Expansion microscopy in Placozoa: improving resolution and preservation of fragile samples during marine expedition. Front. Mar. Sci. 12:1531729. doi: 10.3389/fmars.2025.1531729
Received: 24 November 2024; Accepted: 06 February 2025;
Published: 03 March 2025.
Edited by:
Bernd Schierwater, University of Veterinary Medicine Hannover, GermanyReviewed by:
Allen G. Collins, NOAA Fisheries, United StatesCopyright © 2025 Romanova, Frank and Moroz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daria Y. Romanova, ZGFyamFyb21hbm92YUBnbWFpbC5jb20=
†ORCID: Daria Y. Romanova, orcid.org/0000-0002-7508-3969
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.