
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 10 February 2025
Sec. Marine Pollution
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1528020
MicroRNA is an important regulatory factor at the post-transcriptional level. Previous miRNAomics analysis found that miRNA-375 was steadily upregulated in the clam Ruditapes philippinarum upon ammonia nitrogen exposure. However, we have no knowledge about its regulatory mechanism yet. In this study, the clams were challenged by the injection of miRNA-375 mimics/inhibitor in vivo. Then, a combined approach of qRT-PCR, enzyme assay, and ultrastructure observation was applied to investigate its regulatory effects on the related genes, cellular parameters, and histological structures, respectively. Results showed that increased expression of miRNA-375 interfered with the expression levels of both its target genes and ammonia toxicity-related genes, which would probably lead to oxidative stress, migration of damaged cells, apoptosis resistance, and increased possibility of tumor formation. In addition, miRNA-375 increased MDA content but decreased glutamate content and caused serious structure damage to the clam gills. Thus, increased miRNA-375 probably brings a disastrous fate to the clam R. philippinarum by inducing oxidative damage but promoting apoptosis resistance and cell migration. Overall, this study revealed for the first time the regulatory effects of miRNA-375 in the clams and gave valuable clues to understand the toxicological mechanisms of ammonia nitrogen on the marine bivalve.
With the rapid development of modern economy and intensified human activities around coastal areas in China, ammonia nitrogen (NH3-N) pollution becomes serious in the offshore environment. According to Li C. et al. (2023), the concentration of NH3-N has reached up to 0.139 mg/L at some polluted marine sites. Previously, we revealed that 0.1 mg/L NH3-N affected the clams’ gene expression and important enzyme activities involved in calcium metabolism, redox equilibrium, and glutamine metabolism, which were vital for the clams to survive (Cong et al., 2017; 2019). Thus, ammonia nitrogen under environmental concentration is a considerable stress for the healthy development of marine organisms, especially for those that inhabit at the confluence of land and sea, such as clams (Ruditapes philippinarum). However, we have not understood the related toxicological mechanism clearly, especially at the post-transcriptional regulation (such as microRNA) level.
MicroRNAs (miRNA) are a class of regulatory factors at the post-transcriptional level by degrading its target mRNAs or hindering their translation (Wheeler et al., 2009). Now, miRNAs are found to play important roles that regulate the physiological responses of marine invertebrates upon external stresses. For example, there were 55 miRNAs participating in the immune regulation of oyster Crassostrea gigas after the Vibrio splendidus challenge (Chen, 2017). In Tegillarca granosa, 16 miRNAs were responsive to cadmium pollution by regulating the expression levels of several target genes (Bao et al., 2014). However, there are only few studies on miRNAs involved in toxicological regulation upon ammonia nitrogen pollution.
In recent studies (Tian et al., 2023), miRNA expression profiles were analyzed by high-throughput sequencing in clams (R. philippinarum) under 0.1 mg/L ammonia nitrogen stress. Among the differentially expressed miRNAs, miRNA-375 was found to be significantly upregulated on the 1st day (1.63-fold) and on the 30th day (4.09-fold). Thus, miRNA-375 is considered as an important miRNA to regulate the clam physiology steadily in response to the ammonia nitrogen challenge. In the vertebrate, miR-375 is regarded as an important regulator with multiple functions in immunity, inflammation, development, and cancer formation (reviewed by Liu Y. et al., 2021). Higher expression levels of miR-375 were detected in the early stage of enzootic bovine leukosis (Murakami et al., 2024) and in the late stage of human prostate carcinogenesis (Torres-Ferreira et al., 2015), indicating that miR-375 was closely related to cancer formation and development. However, miRNA-375 also exerted tumor suppressor function in human non−small cell carcinoma (Cheng et al., 2017), asopharyngeal carcinoma (Xu et al., 2021), and small intestinal neuroendocrine tumors (Arvidsson et al., 2018). Therefore, miRNA-375 performs variable and even conflicting regulations during different cellular processes, especially in carcinogenesis development. Its positive or negative regulations on the genes probably depend on its promoter methylation and regulator circRNA (Liu L. et al., 2021). However, there is no report about the regulatory modes of miRNA-375 in the marine invertebrate yet. Thus, it is meaningful for us to investigate the regulatory functions of miRNA-375 in the clams to fulfill such gap in marine bivalves.
In the present study, miRNA-375 mimics and miRNA-375 inhibitor were used to enhance or knock down the expression level of miRNA-375 in the clams. Based on the computational prediction from the miRNAomics data, eight target genes were selected to test their expression levels by qRT-PCR. In order to clarify the regulatory roles of miRNA-375 more clearly, another six genes related to apoptosis, calcium metabolism, glutamine metabolism, and redox equilibrium involved in ammonia toxicity were also chosen to investigate their expression levels. In addition, three cellular parameters were assayed to indicate the cellular state of the clams. Because of the multiple functions of the targeted genes of miRNA-375, MDA, glutamate, and Ca2+-ATPase were selected as the tested parameters, which were all involved in the toxic responses of the clams upon NH3-N stress. Moreover, the microstructures of gill tissues were observed to directly perceive the regulatory effects of miRNA-375 on the clams. Overall, the present study aimed to investigate the regulative functions of miRNA-375 comprehensively at the gene, cell, and tissue levels, and give valuable clues to understand the complicated mechanism of clam miRNAs.
The secondary structure of miRNA-375 was predicted and drawn by the software mirdeep2 and srna-tools-cli (Friedlander et al., 2012). The characteristics of miRNA-375 hairpin structure were predicted by software miREvo (Wen et al., 2012) and mirdeep2.
Healthy clams (R. philippinarum) were obtained from a supermarket (Yantai, China) and acclimated in tanks (60 cm × 60 cm × 30 cm) fulfilled with sand-filtered seawater (31‰ salinity, pH 8.0), aerated at 18 ± 1°C for 7 days. They were fed once a day with commercial condensed Isochrysis galbana and Chlorella vulgaris Beij (density ≈ 2×1010 cells/g, 3% of the wet weight of the clams). During the acclimation period, each clam was sawed a notch in its shell near the adductor in order to facilitate the following procedure of miRNA injection as described by Zhang et al. (2014) and Chen et al. (2016). The mimics and inhibitor of miRNA-375 were synthesized by GenePharma Company (Shanghai, China) and diluted with filtered and sterile seawater.
Approximately 180 clams were divided into four groups, with 45 clams in each group. Based on the miRNA concentration used in the oysters (Chen et al., 2016) and in consideration of the average weight ratios of single clam to single oyster, 1 μM of each miRNA product (Table 1) was prepared and used to challenge the clams. Given that the clam miRNA-375 was upregulated after ammonia exposure (Tian et al., 2023), an ammonia exposure group was also set in the present study accordingly, parallel to the mimics group and the inhibitor group, and used as a positive group. The expression level of miRNA-375 after ammonia exposure was detected by qRT-PCR in a pre-experiment of the present study, which came out at approximately 1.40 ± 0.27-fold to that of the control group. It was a value close to that reported by Tian et al. (2023).
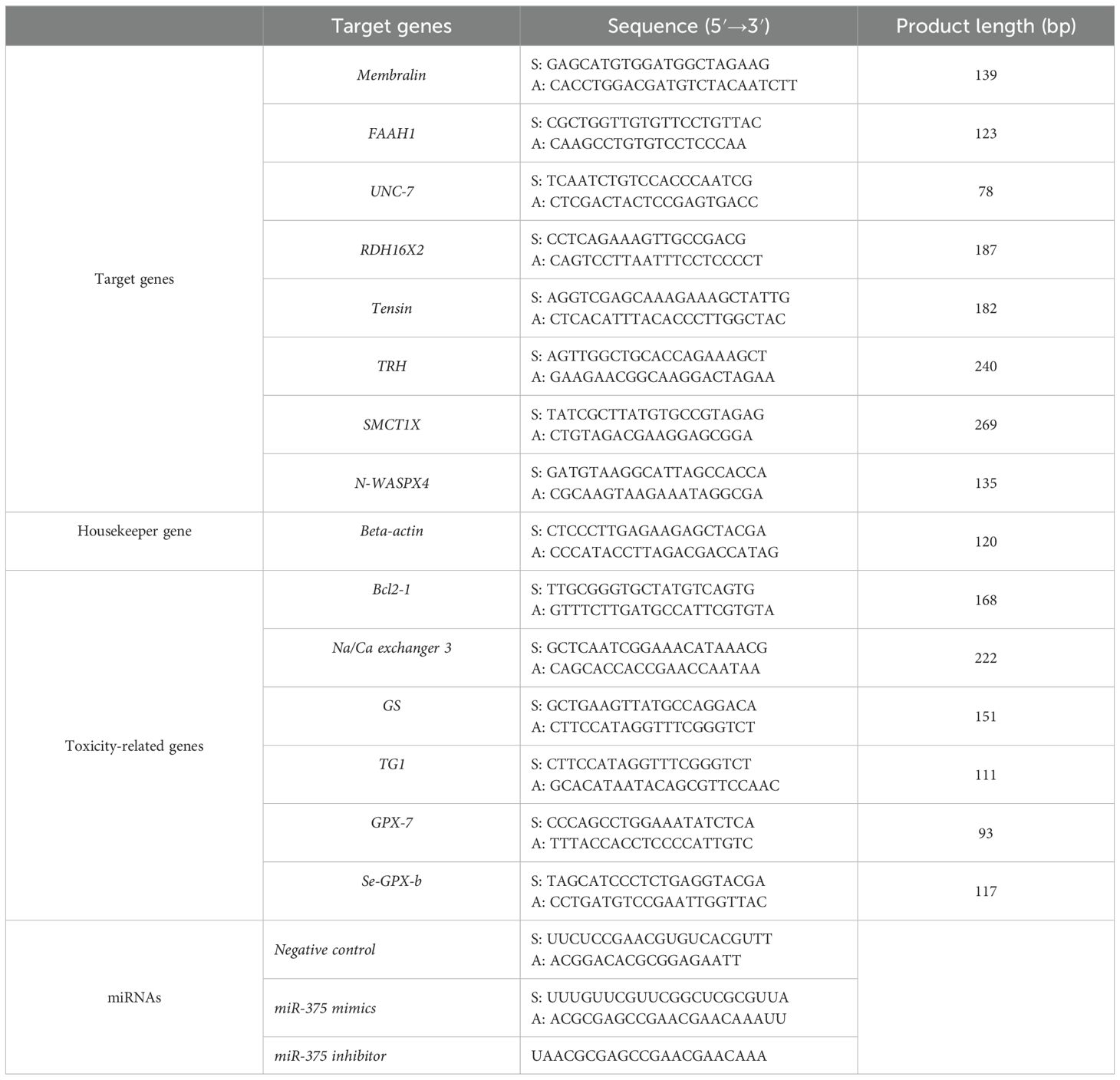
Table 1. Primers for the target genes, NH3-N toxicity-related genes, housekeeper gene, and the sequences of synthesized miRNAs.
The first group of clams were injected with 100 μL of negative control miRNA into the adductor muscle by syringe with a needle and used as the negative control group. Similarly, each clam in the second group received an injection of 100 μL of miRNA-375 mimics and used as the miRNA-375 mimics group. The clams in the third group were injected with 100 μL of miRNA-375 inhibitor and used as the miRNA-375 inhibitor group. The fourth group of clams were injected with 100 μL of seawater and then cultured in seawater added with 0.1 mg/L of NH3-N, and used as the positive group (named as NH3-N group). Each group was set to three replicate tanks, and the clam gills were sampled at 24 h post challenge because most miRNAs can perform their regulatory functions within 24 h (Tian et al., 2023; Mao et al., 2024). Five clams in each group were used for quantitative RT-PCR detection, with the sampled gills stored into Trizol. Another five clams in each group were used to assay enzyme activities, with the sampled gills stored in liquid nitrogen. In addition, three clams in each group were picked and used for transmission electron microscopy (TEM) observation.
Total RNAs were extracted from gill samples by using Trizol, and their integrity and concentration were precisely measured by an Agilent 2100 bioanalyzer. Then, mRNAs were reverse-transcribed and synthesized following the protocol of TranScript® All-in-One for qPCR and gDNA Remover and cDNA Synthesis SuperMix (TransGen Biotech, China).
The expression levels of eight target genes of miRNA-375 and six genes involved in ammonia nitrogen toxicity were tested in each group. The target genes included membralin (Membralin, XM_060695276), fatty-acid amide hydrolase 1 like (FAAH1, XM_060752551), innexin unc-7 (UNC-7, XM_060734619), retinol dehydrogenase 16 isoform X2 (RDH16X2, XM_060742233), tensin-1 like (Tensin, XM_060739587), thyrotropin-releasing hormone (TRH, XM_060711906), sodium-coupled monocarboxylate transporter1-like isoform X (SMCT1X, XM_060722458), and the neural Wiskott-Aldrich syndrome protein isoform X4 (N-WASPX4, XM_060727258). The NH3-N-toxicity related genes included B-cell lymphoma-2 1 (Bcl2-1, KC506418), Na+/Ca2+ exchanger 3 (Na/Ca exchanger 3, XM_060697531), glutamine synthetase (GS, XM_060748742), transglutaminase 1 (TG1, XP_060573211), glutathione peroxidase 7 (GPX-7, GQ384395), and selenium-dependent glutathione peroxidase (Se-GPX-b, MH085059). The primers for each gene were designed by the Primer Premier5.0 software and listed in Table 1. PerfectStart® Green qPCR SuperMix (TransGen Biotech, China) was used to amplify the target genes in Real-time PCR Systems (Bio-Rad CFX96) and calculated by the 2−ΔΔCt method.
Test kits from Jiancheng Co., Ltd (Nanjing, China) were used to detect Ca2+-ATPase activity and the contents of MDA and glutamate in gill tissues. For each sample, 25 mg of clam gill was homogenized, centrifuged, and assayed under a multifunctional microplate reader according to the kit instructions. Last, the total protein concentration in each sample was determined by the Bradford protein quantification kit (Nanjing, China) and used to normalize the above parameters (expressed as units per milligram of protein) in each sample.
The clam gills were taken by surgical scissors, quickly washed with PBS buffer solution, and cut into small pieces (1 mm×1 mm). Then, the gill fragments were stored in 2.5% glutaraldehyde and prepared according to the normal protocols for ultrathin slices described in a previous study (Cong et al., 2021). After a series of fixation, dehydration, embedding, and hardening procedures, the gills were cut into slices with 70 nm thickness and finally stained by uranyl acetate and alkaline lead citrate. Then, the slices were observed under a high-resolution TEM (JEM1200, Japan).
Each clam was used as one individual sample in the parameter tests and qRT-PCR quantification, and all the data were subjected to one-way ANOVA followed by least significant difference (LSD) analysis by using SPSS19.0. The statistical significances were defined at p < 0.05. Origin 2024 is employed to process the experimental data into box diagram.
To discriminate the differences in damage among the exposed groups well, semiquantified methods (Riba et al., 2004) were used to grade the gill lesion in each group by detecting the frequency of several kinds of histological alterations. These alterations included occurrences of necrotic organelle, empty vacuole, myeloid body, deformity in mitochondria, secondary lysosome, affected goblet cells, and deformed cilia. Based on the semi-quantitation values, lesion frequencies in each group were graded with the following criteria to assign a score: 0 (−), 0.5 (+/−), 1 (+), 2 (++), and 3 (+++). Histopathological damage was calculated as total scores/number of indices.
There were 101 bases in the miRNA-375 precursor sequence that started at U (UAGAGAAAAGUGGUCGCCAACUGACCCGAGCCGUUUGUAACAAGGCUUAAAUUACAAUGUUUUGUUCGUUCGGCUCGCGUUACAAAGGCUGACAAAGUCGU), containing a mature sequence (UUUGUUCGUUCGGCUCGCGUUA) of 22 bases in length. The characteristic hairpin structure of miRNA-375 precursor is shown in Figure 1. Several motifs were found in the miRNA-375 precursor in its stem loop structure, including three loops and two bulges.
Among the eight target genes (Figure 2A), no significant differences were detected in membralin and SMCT1X, although slight up- or downtrends were observed in their expression profiles. Significant differences were detected in the other six genes, namely, FAAH1, UNC-7, N-WASPX4, RDH16X2, Tensin, and TRH.

Figure 2. Real-time PCR quantification of gene expressions for eight target genes and six genes involved in NH3-N toxicity. (A) Target genes; (B) NH3-N toxicity-related genes. Notes: Different letters on top of the column means significant difference between the challenged group with the control group. Asterisks denote significant differences between the exposed group and the control at the same time point, with one asterisk (*) at p < 0.05, two asterisks (**) at p < 0.01, and three asterisks (***) at p < 0.001.
For FAAH1, significant increments of its mRNA expression were detected after exposures to NH3-N and miRNA-375 inhibitor (p < 0.01 and p < 0.01, respectively). It indicated that knockdown of miRNA-375 induced similar upregulation of FAAH1 transcription to that after NH3-N exposure. Slight downregulation (p > 0.05) of FAAH1 expression was detected in the miRNA-375 mimics group.
For UNC-7, significant downregulations levels were detected after exposures to NH3-N (p < 0.01) and miRNA-375 inhibitor (p < 0.05), indicating their negative regulations on UNC-7 expression. However, no significant difference was detected in the miRNA-375 mimics group.
For N-WASPX4, no significant difference was detected in the miRNA-375 mimics group and the NH3-N group, but a significant increment was observed in the miRNA-375 inhibitor group (p < 0.05), compared with the control group. It indicated that upregulation of miRNA-375 contributed to the stable expression of N-WASPX4, and an unbalanced expression would occur once miRNA-375 was knocked down.
For RDH16X2, a significant increment of its mRNA expression was detected in the miRNA-375 mimics group (p < 0.05). Although a slight increment and decrement were detected in the miRNA-375 inhibitor group and NH3-N group, respectively, there were no significant differences in each of them. It suggested that NH3-N exposure had a slight negative effect on the expression of RDH16X2, but miRNA-375 mimics had positive regulation instead.
For Tensin and TRH, significant upregulations were only detected in the miRNA-375 inhibitor group (p < 0.01 and p < 0.001, respectively). Slight but not significant variations were detected in the miRNA-375 mimics group and the NH3-N group. It indicated that miRNA-375 mimics played important roles to maintain the transcriptional stability of Tensin and TRH, and these two genes would significantly increase their expressions once miRNA-375 expression was inhibited.
The expression profiles of six genes related to NH3-N toxicity are shown in Figure 2B. Bcl-2 expression significantly increased in the miRNA-375 mimics group (p < 0.001) and the NH3-N group (p < 0.001), but decreased in the miRNA-375 inhibitor group (p < 0.001). The expression levels of Bcl-2 in the mimics group and in the NH3-N group were both significantly higher than those in the inhibitor group (p < 0.001 and p < 0.001, respectively).
The expression levels of Na/Ca exchanger 3 significantly decreased in the three stressed groups (p < 0.001, p < 0.001, and p < 0.001). However, the expression level of Na/Ca exchanger 3 in the mimics group was significantly higher than that in the inhibitor group and in the NH3-N group (p < 0.001 and p < 0.001).
Similarly, significant decrements of GS expression were detected in the miRNA-375 mimics group (p < 0.001) and miRNA-375 inhibitor group (p < 0.001), and more decrement was detected in the NH3-N group (p < 0.001).
For TG1 gene, a slight but not significant increment of expression was detected in the miRNA-375 mimics group, and significant decrements of mRNA expression levels were detected in the miRNA-375 inhibitor group (p < 0.001) and the NH3-N group (p < 0.001). The decrement was even lower in the inhibitor group than that in the NH3-N group (p < 0.001).
For Gpx-7 gene, significant decrements of expression levels were detected in all three stressed groups (p < 0.001, p < 0.001, and p < 0.001). Comparatively, the expression level of Gpx-7 was significantly higher in the mimics group than that in the inhibitor group (p < 0.05) and the NH3-N group (p < 0.05).
For Se-Gpx-b gene, similar decrements of expression levels occurred in the miRNA-375 mimics group (p < 0.001) and the NH3-N group (p < 0.001). In addition, the decrement was even lower (p < 0.001) in the inhibitor group, compared with the other two groups.
Among the three cellular parameters, MDA contents (Figure 3A) in the NH3-N group and the miRNA-375 mimics group increased significantly (p < 0.001 and p < 0.001), indicating that exposure of miRNA-375 mimics led to a similar increment of MDA content to that after ammonia exposure.

Figure 3. Contents of MDA and glutamate, and the activity of Ca2+-ATPase in the clam gills. (A) MDA; (B) glutamate; (C) Ca2+-ATPase. Different letters on top of the column means significant difference between the challenged group with the control group. Asterisks denote significant differences between the exposed group and the control at the same time point, with one asterisk (*) at p < 0.05, two asterisks (**) at p < 0.01, and three asterisks (***) at p < 0.001.
Glutamate content (Figure 3B) decreased significantly in the clams challenged by miRNA-375 mimics (p < 0.05). It suggested that miRNA-375 mimics had a negative regulation on glutamate content. However, no significant change was detected in the NH3-N group or the miRNA-375 inhibitor group.
Ca2+-ATPase activity (Figure 3C) showed a significant decrement in the NH3-N group (p < 0.01). In the miRNA-375 mimics group and miRNA-375 inhibitor group, slight but not significant increments were detected in Ca2+-ATPase activity.
The ultrastructures of clam gills are shown in Figure 4. In the negative control group (Figure 4A), most of the cells were intact and tightly packed. Cell nucleus was stained clearly. Mucus particulates were evident in the goblet cells, which scattered throughout the epithelial cells. The gill tissue had strong muscle elements surrounded by slender cilia and dense microvilli. There were quite a few of round or oval mitochondria, with complete outer membrane and neatly arranged crests. There were some Golgi apparatus, swollen endoplasmic reticulum (ER), and some secondary lysosomes.
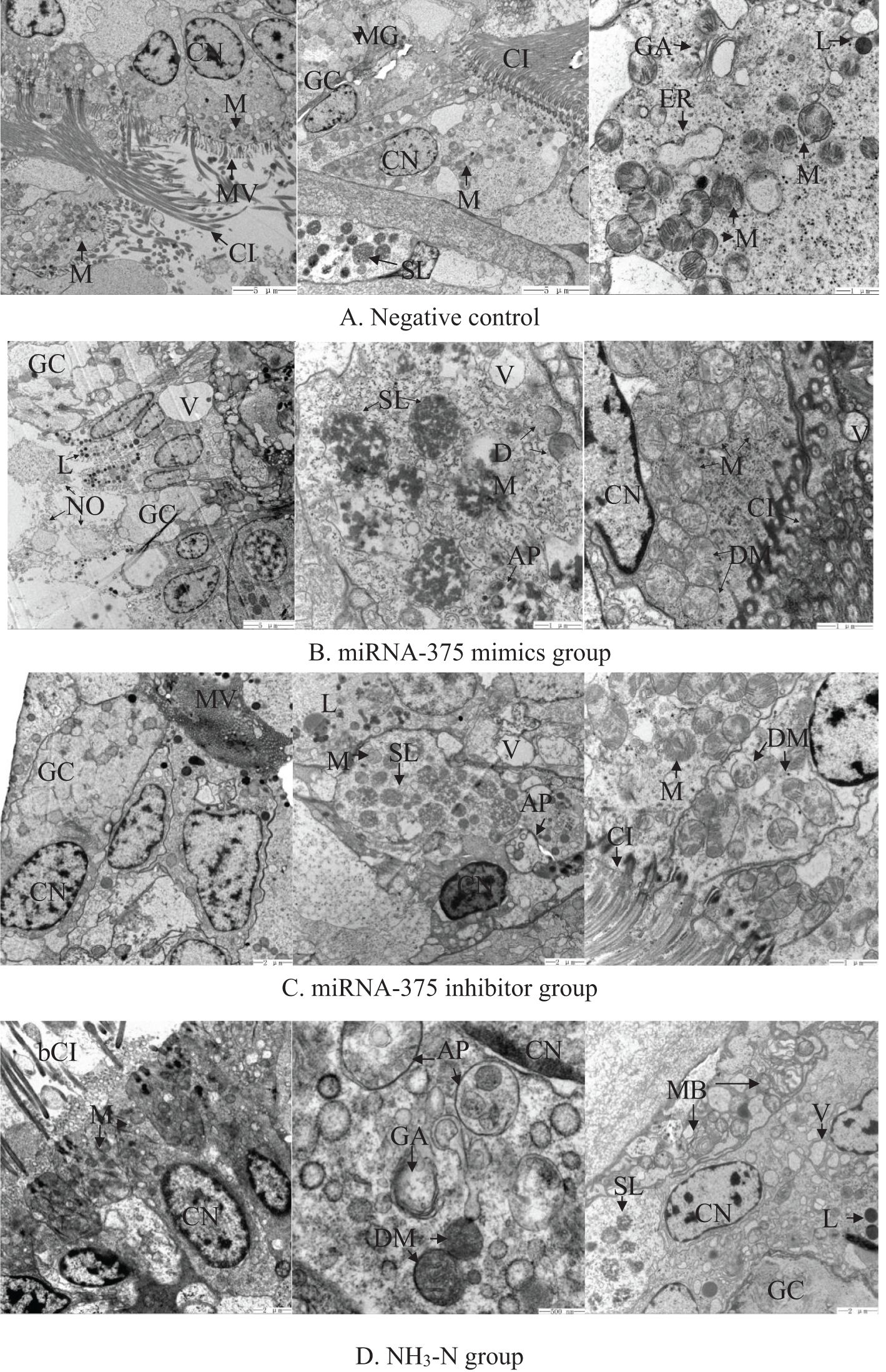
Figure 4. Electron microscopic observation of the gill tissue of clams in (A) the negative control group, (B) the miRNA-375 mimics group, (C) the miRNA-375 inhibitor group, and (D) the NH3-N group. Notes: AP, autophagosome; bCI, broken cilium; CI, cilium; CN, cell nucleus; dER, dense endoplasmic reticulum; DM, damaged mitochondria; GA, Golgi apparatus; GC, goblet cell; M, mitochondria; MB, myeloid body; ME, muscle element; MG, mucous granule; MV, microvilli; NO, necrotic organelle; PL, primary lysosome; SL, secondary lysosome; V, vacuole.
In the miRNA-375 mimics group (Figure 4B), the gill structure was seriously damaged. There were some scattered necrotic organelles, large vacuoles, and myeloid bodies. Inside the gill cells, some of the mitochondria were swollen with several deformed characteristics, including an indistinct outer membrane and broken internal cristae. Quite a few ERs were swollen. In addition, primary lysosomes, secondary lysosomes, and autophagosome appeared in the gill. Goblet cells became almost white with little mucus particles inside. Some of the microvilli became swollen.
In the miRNA-375 inhibitor group (Figure 4C), the gill structure was relatively complete, with quite a few mitochondria and slender cilia. Although the goblet cells were stained lightly, some mucus particles were still discerned. Many secondary lysosomes were observed inside the gill epithelial cells. Autophagosome was also detected in the gill. Some of the mitochondria were in contorted shapes. In addition, the cilia were arranged in good order but some microvilli were swollen.
In the NH3-N group, the gill ultrastructure was seriously damaged (Figure 4D). Part of the gill had loose and broken cilia, and the microvilli became loose as well. Cell membranes of some epithelial cells disappeared, with fragmented intracellular organelles. The whole gill was in a disordered state. There was severe deformation in mitochondrial structure, including swelling, vacuolation, blurred outer membrane, and disordered inner cristae. The mucus particles inside the goblet cells were stained shallowly. In addition, there were several autophagosomes, secondary lysosomes, phagosomes, and myeloid bodies.
Comparison of histopathological damage among different exposures by semi-quantitation (Table 2) revealed that the frequencies in the negative control, miRNA-375 mimics, mimics-375 inhibitor, and NH3-N groups were 1.14, 2.28, 1.64, and 2.42, respectively. Thus, histological damage in the miRNA-375 mimics group was close to that in the NH3-N group, and histological damage in the miRNA-375 inhibitor group was close to that in the negative group.
MiRNAs play important roles to regulate protein translation at the post-mRNA level. MiRNA-375 is a known miRNA in the vertebrate, with various roles such as apoptosis inhibitor (Li et al., 2021; Noronha et al., 2022), agent to cure lung cancer (Du et al., 2023), regulator to beta-cell development (Latreille et al., 2015), immunity inhibitor, and promoter to cell carcinogenesis (Mao et al., 2016). However, there is no study about miRNA-375 involved in the stress response, especially in the invertebrate upon ammonia exposure. Hence, the present study would provide important insights into the regulative roles of miRNA-375 on the marine bivalve.
In the present study, six of the target genes exhibited significant changes during the experiment. Firstly, miRNA-375 mimics positively regulated the mRNA expression of RDH16X2, which is responsible for retinol metabolism by converting retinol to retinoic acid and can be used as a pro-metastasis biomarker in human cancers (Kropotova et al., 2013, 2014; Li L. et al., 2023). Increased expression of RDH16X2 in the miRNA-375 mimics group implied that cancer would be promoted to migrate in the clams. Although there was no cancer reported in the clams, it might imply that some cancer-like substances would be produced and migrate in the clams with the aid of increased miRNA-375.
Additionally, miRNA-375 inhibitor positively regulated the expression levels of four genes (FAAH1, N-WASPX4, Tensin, and TRH) and negatively regulated one gene (UNC-7). Among them, FAAH1 is a fatty acid amide hydrolase and plays important roles in neural activity, energy metabolism, immunity, reproduction, and even tumor formation (Shi et al., 2024). In the present study, a significant increment of FAAH1 in the miRNA-375 inhibitor group was similar to that in the NH3-N group, implying negative regulation of miRNA-375 on FAAH1 expression. It is reported that deletion of FAAH gene contributed to enhanced oxidative stress to mice (Mukhopadhyay et al., 2011). Accordingly, downregulation of FAAH1 in the miRNA-375 mimics group would result in clams suffering from oxidative stress.
N-WASPX4 is the gene coding the neural Wiskott-Aldrich syndrome protein, which is required for the accurate condensation and separation of chromosomes during mitosis. The exposure of miRNA-375 inhibitor but not miRNA-375 mimics significantly increased the expression of N-WASPX4. It implied that miRNA-375 contributed to the stable expression of N-WASPX4 by partially downregulating its expression. Depletion of N-WASP4 can induce chromosomal misalignment and abnormal segregation, especially during invasion and migration of tumor cells (Park and Takenawa, 2011). Thus, overexpression of miRNA-375 would hinder the normal mitosis and induce genetic aberrations.
Tensin-1 is a cell adhesion molecule that regulates cell adhesion, migration, and proliferation by connecting the extracellular matrix to the intracellular cytoskeleton (Lo, 2017). In this experiment, slight downregulation was detected in the miRNA-375 mimics group. However, a significant increment of Tensin-1 expression was found in the miRNA-375 inhibitor group. It proved that miRNA-375 negatively regulated Tensin-1 expression. Decreased expression of tensin-1 implied reduced adhesive ability of cells to the cytoskeleton and increased migration ability. Thus, overexpression of miRNA-375 would promote migration of clam cells, but migration of abnormal cells would probably induce bad and even cancerous fate to the clams.
TRH is a thyroid-stimulating hormone gene, which is involved in the regulation of synthesis and secretion of thyroxine to regulate the development, growth, and cell metabolism of animals (Chiamolera and Wondisford, 2009). In addition, invertebrate TRH was involved in postembryonic growth and reproduction (Van Sinay et al., 2017). In the present study, TRH expression in the miRNA-375 group was not significantly changed; however, a significant increment was detected in the inhibitor group. Hence, it can be postulated that there were other factors that played a role in the regulation of TRH, and miRNA-375 mainly contributed to decrease TRH expression. Accordingly, increased expression of miRNA-375 would probably result in inhibited growth or reproduction in the clams. It needs further investigation in the future.
Innexin UNC-7 is an important member of invertebrate gap-junction proteins for embryonic development, morphogenesis, and physiological regulation (Phelan et al., 1998). Knockdown of miRNA-375 in the inhibitor group significantly downregulated the expression of Innexin UNC-7, similar to that in the NH3-N group. It can be inferred that miRNA-375 mimics positively regulated Innexin UNC-7 expression, such that overexpressed miRNA-375 would probably result in cell disassembly and facilitate their mobility.
In addition, the expression profiles of genes involved in apoptosis, Ca2+ metabolism, glutamate metabolism, and oxidation-reduction reaction helped to clarify the regulatory roles of clam miRNA-375 more clearly. Bcl2-1 is an apoptosis suppressor and plays important roles to inhibit apoptosis by regulating mitochondrial Ca2+ homeostasis (Morris et al., 2021). In this study, increased expression of Bcl2-1 revealed that cell apoptosis was significantly inhibited in the miRNA-375 mimics and in the NH3-N group. According to the observation result of the gill structure, gill cells were found to be damaged in the miRNA-375 mimics and in the NH3-N group, such that inhibited apoptosis by increased Bcl2-1 would probably induce apoptosis resistance to the damaged cells.
Na+/Ca2+ exchanger is an antiporter in plasma membrane and responsible for Ca2+ transportation by promoting Ca2+ efflux and Na+ influx (Rodrigues et al., 2024). Increased expression of Na+/Ca2+ exchanger can alleviate apoptosis by eliminating intracellular Ca2+ (Xia et al., 2021). Although Na+/Ca2+ exchanger 3 expression was inhibited in the three stressed groups, the relative increment in the miRNA-375 mimics group indicated that Ca2+ efflux was promoted, and accordingly, apoptosis was reduced by increased miRNA-375 expression. It was in accordance with the postulation from increased Bcl2-1 gene and probably resulted in apoptosis resistance for the clam cells.
It is reported that the NMDA-type glutamate receptor is the main target for ammonia nitrogen to induce toxicity by apoptosis (Randall and Tsui, 2002). Thus, glutamate concentration is essential to estimate the toxicity of ammonia nitrogen well. In the present study, the concentration of glutamate significantly decreased in the miRNA-375 mimics group, indicating that apoptosis caused by the NMDA receptor was inhibited greatly by increased miRNA-375. In order to prove the involvement of miRNA-375 during glutamate metabolism, the expression levels of glutamine synthetase (GS) and transglutaminase 1 (TG1) were tested after the miRNA-375 challenge, because glutamine is a precursor of glutamate (Newsholme et al., 2003) and transglutaminase is a downstream enzyme invoked by glutamate (Campisi et al., 2003). The extreme decrement of GS expression level in NH3-N exposure suggested that glutamine synthesis was heavily inhibited, in accordance with previous studies (Cong et al., 2017). Although GS expression was also significantly decreased in the miRNA-375 mimics group, a similar decrement in the miRNA-375 inhibitor group indicated that miRNA-375 was not the main regulatory factor for GS decrement. However, a significant increment of TG1 expression was detected in the miRNA-375 mimics group compared with the other two stressed groups (especially the miRNA-375 inhibitor group), indicating that miRNA-375 positively regulated TG1. It was reported that high levels of transglutaminase activities were associated with cell survival after external stimuli (Ientile et al., 2007). Thus, increased miRNA-375 would probably extend the lifespan of the damaged gill cells.
Glutathione is the main kind of antioxidant present in all of the cells, but its metabolism was affected by ammonia nitrogen exposure (Tian et al., 2023). Gpx and Se-Gpx were two antioxidant enzymes related to glutathione. In the present study, significant decrements were all found in the three stressed groups for Gpx-7 and Se-Gpx-b, in accordance with previous results (Tian et al., 2023). However, significant increments of both Gpx-7 and Se-Gpx-b were detected in the miRNA-375 mimics group compared with the inhibitor group, indicating that miRNA-375 inhibited glutathione exhaustion. Besides its role in redox, recent studies revealed that an elevated level of glutathione plays important roles in cancer transformation (Kalinina and Gavriliuk, 2020). Thus, it can be postulated that increased miRNA-375 increased the risk of tumor formation in the clams.
Combining the variation profiles of its target genes and the NH3-N toxicity-related genes, it can be concluded that the increased expression of miRNA-375 would probably cause serious results to the cells, such as oxidative stress, apoptosis resistance, migration of damaged cells, and even increased possibility of tumor formation.
However, there were also some lessons about the miRNA–mRNA interaction experiment. In the present study, eight target genes were selected based on the computational prediction from the miRNAomics data and the annotation of their biological functions in clam genome. Among them, two genes (Membralin and SMCT1X) did not change their expression upon any stress; hence, these two genes needed further validation by experiment. To our knowledge, there are several factors that affect the outcomes of the miRNA–mRNA interaction, including target gene predication by computational algorithms, genome annotation, and difference in mRNA pools. Given the universal usage of computational algorithms and a well-annotated clam genome (ASM2657151v2), the major factor may be the difference in mRNA pools, because different kinds of stimulation (e.g., immersion or injection) will produce different profiles of differentially expressed mRNAs. In a previous study, miRNA-375 was screened under an immersion challenge, but injection was applied in the present study. Thus, such difference in stimulation should be considered in a future experiment.
It is known that changes in gene expression will cause changes in protein translation, resulting in changes of downstream physiological reactions. Thus, in the present study, key parameters related to the tested genes’ functions were assayed to explore the regulatory effects of miRNA-375 on the enzyme level.
Malondialdehyde (MDA) is the end product of polyunsaturated fatty acid from peroxidation and can invoke cell damage by inducing protein polymerization (Long et al., 2021). MDA content directly marks the degree of cell damage (Arora et al., 2018). NH3-N exposure can significantly increase the contents of reactive oxygen species (ROS) (Cong et al., 2022); thus, MDA is suitable to indicate oxidative damage by NH3-N exposure. In this study, significant increments of MDA contents in the miRNA-375 mimics group implied that increased expression of miRNA-375 caused significant oxidative damage to the clams, as that after NH3-N exposure. It might be a result of high oxidative stress from the downregulation of FAAH1 gene discussed in Section 4.1.
Glutamate is an excitatory neurotransmitter and plays important roles in energy metabolism, signaling metabolism, and ammonia metabolism (Brosnan and Brosnan, 2012). It can combine free NH3 and turn into glutamine to intoxication. In addition, glutamate is the central substance to bind the NMDA-type glutamate receptor and regulate apoptosis (Randall and Tsui, 2002). In the present study, exposure to miRNA-375 mimics resulted in a significant decrease in glutamate content, while no significant change was detected in the NH3-N group. Thus, increased expression of miRNA-375 will cause more toxicity to the gill cells by free ammonia, but alleviate apoptosis of the damaged cells. Therefore, we speculated that overexpressed miRNA-375 inhibited ammonia detoxification and led to apoptosis resistance, which might be the result of increased expressions of Bcl2-1, Na+/Ca2+ exchanger 3 and TG1.
Ca2+-ATPase is a cation transporter responsible for Ca2+ metabolism, and significant changes in Ca2+-ATPase activity represented a marked disturbance of Ca2+ metabolism in mitochondria (Marchi et al., 2018). In the clams, Ca2+ concentration and Ca2+-ATPase activities decreased significantly after NH3-N exposure (Cong et al., 2019, 2022). In the present study, Ca2+-ATPase activities in the miRNA-375 mimics group and inhibitor group did not change significantly compared to that in the NH3-N group. It suggested that not only overexpression but also knocking down of miRNA-375 could regulate Ca2+-ATPase activity slightly; hence, there should be other factors that co-regulate Ca2+-ATPase in the clams.
The changes in gill tissue structure reflect the degree of damage from external adverse factors. In the negative control group, the gill cells were found in a relatively healthy state with an intact cell structure, clearly stained nucleus, goblet cells with evident particulate substance, slender and intensive cilia, ER and Golgi apparatus with complete structures, and intact mitochondria with a neatly arranged inner membrane. The appearance of secondary lysosomes might result from the clearance of negative control miRNA. No extra damage was found around the secondary lysosomes, indicating that the damage from the negative control miRNA was limited. However, different degrees of damage were detected in the other three groups.
Among the different groups, the gill structure in the NH3-N exposed group was the most heavily damaged. Broken cilia and loose microvilli suggested that gill functions of respiration and food filtering were undoubtedly affected by ammonia exposure, as elucidated in a previous study (Cong et al., 2019). Deformation in mitochondrial structure (including swelling, vacuolation, blurred outer mitochondria, and confused internal crest) suggested that mitochondrial functions, such as ATP production and calcium metabolism, were probably affected by ammonia exposure. Furthermore, mitochondrial damage also implied that more ROS were produced during the oxidative phosphorylation process, which would cause oxidative damage to the clams. This hypothesis was proved by the increased content of MDA. In addition, ammonia nitrogen exposure led to the occurrence of primary lysosomes, secondary lysosomes, and phagosomes in the gill tissue. Lysosome is known as the main organelle to maintain cell homeostasis by digesting endogenous or exogenous particles, such as cell debris or pathogenic bacteria (Yang and Wang, 2021). Accordingly, the increased number of lysosomes suggested that many of the cellular organelles were destroyed by ammonia nitrogen exposure; thus, the clams used lysosomes and phagosomes to remove the damaged tissues. However, myeloid bodies were detected in the NH3-N group. Myeloid bodies are surrounded by unit membranes with piles of threaded or reticular membranous bodies, which are abnormal lysosomes (Cornell and Leung, 2021). Hence, the occurrence of myeloid bodies indicated that the lysosomal functions in clearance and immunity would be partially inhibited by ammonia nitrogen exposure. Moreover, secretory mucus particles in goblet cells were stained lighter after ammonia nitrogen exposure, indicating that the clam immunity was probably decreased after ammonia nitrogen exposure, because goblet cells play important immune roles in mucosal surfaces (Knoop and Newberry, 2018).
Comparatively, exposure of miRNA-375 mimics caused severe histological damage to the clam gills. First of all, there were necrotic organelles and scattered cells, which meant that the gill structure was seriously damaged, making it hard for the gill cells to function normally. Furthermore, the presence of abnormal mitochondria showed that miRNA-375 mimics would not only interfere with the normal functions of mitochondria, but also produce more ROS and cause heavy oxidative damage to the cells. The significantly increased MDA content were in accordance with such inference. In some goblet cells, the mucus particles almost disappeared, which would lead to decreased immunity to the clams. The occurrence of a large number of primary lysosomes, secondary lysosomes, and autophagosomes inside the cells indicated that exposure of miRNA-375 mimics resulted in cellular debris, which needed to be removed through phagocytosis. The disordered state of gill tissue might be the result of oxidative damage from increased MDA content, as elucidated in the miRNA-375 mimics group (Section 4.2). In addition, it may be the combined results from significant changes in RDH16X2, tensin-1, and Innexin UNC-7 (Section 4.1) that would cause the damaged cells in a disordered state and facilitate them to migrate around.
In contrast, the histological damage was not so serious in the miRNA-375 inhibitor group. In general, the gill structure was relatively complete. The well-ordered cilia and intensive microvilli would enable the clam gills to perform respiration and feeding functions. The discernable mucus particles in the goblet cells would help the clams to defend against foreign pathogens. However, some deformed mitochondria would hinder the clams from attaining efficient ATP production and calcium metabolism. Moreover, occurrences of empty vacuoles and secondary lysosomes indicated that some organelles were destroyed by miRNA-375 inhibitor exposure but can be cleared by the lysosomes through phagocytosis and auto-phagocytosis.
In summary, increased expression of miRNA-375 induced significant changes in genes, resulting in oxidative damage (FAAH1, Gpx-7, and Se-Gpx-b), apoptosis resistance and Ca2+ transportation (Bcl2-1, TG1, and Na+/Ca2+ exchanger 3), genetic aberrations (N-WASP4), and cancer migration (RDH16X2, tensin-1, and Innexin UNC-7). Increased expression of miRNA-375 also resulted in significant changes in cellular parameters related to oxidative damage (MDA) and glutamate metabolism. As a result, clam gills suffered from oxidative damage and exhibited a disordered and scattered state with abnormal organelles. Thus, clam miRNA-375 probably acts as a cancer promoter but not an inhibitor.
In this study, the regulative mechanism of miRNA-375 was investigated at the gene, cell, and tissue levels, respectively. Increased expression of miRNA-375 induced oxidative damage to the clam cells, messed the tissue structure, but inhibited apoptosis and promoted migration of the damaged cells, which probably resulted from the significant changes in genes related to oxidative damage, apoptosis resistance, Ca2+ transportation, genetic aberrations, and cancer migration. Therefore, increased miRNA-375 probably brings a disastrous fate to the clams (R. philippinarum).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by Ethics committee of Yantai University. The study was conducted in accordance with the local legislation and institutional requirements.
MC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. WT: Data curation, Formal Analysis, Methodology, Project administration, Software, Validation, Writing – original draft. ZL: Conceptualization, Investigation, Methodology, Software, Validation, Writing – review & editing. YY: Investigation, Methodology, Project administration, Software, Validation, Writing – original draft. YC: Methodology, Resources, Software, Visualization, Writing – review & editing. JL: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Shandong Province Natural Science Foundation (ZR2023MD059) and the Natural Science Foundation of China (NSFC 41876135).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Arora M. K., Sarup Y., Tomar R., Singh M., Kumar P. (2018). Amelioration of diabetes-induced diabetic nephropathy by aloe vera: implication of oxidative stress and hyperlipidemia. J. Diet. Suppl. 16, 227–244. doi: 10.1080/19390211.2018.1449159
Arvidsson Y., Rehammar A., Bergström A., Andersson E., Altiparmak G., Swärd C., et al. (2018). miRNA profiling of small intestinal neuroendocrine tumors defines novel molecular subtypes and identifies miR-375 as a biomarker of patient survival. Modern Pathol. 31, 1302–1317. doi: 10.1038/s41379-018-0010-1
Bao Y., Zhang L., Dong Y., Lin Z. (2014). Identification and comparative analysis of the Tegillarca granosa haemocytes microRNA transcriptome in response to Cd using a deep sequencing approach. PloS One 9, e93619. doi: 10.1371/journal.pone.0093619
Brosnan J. T., Brosnan M. E. (2012). Glutamate: a truly functional amino acid. Amino Acids 45, 413–418. doi: 10.1007/s00726-012-1280-4
Campisi A., Caccamo D., Raciti G., Cannavò G., Macaione V., Currò M., et al. (2003). Glutamate-induced increases in transglutaminase activity in primary cultures of astroglial cells. Brain Res. 978, 1–2. doi: 10.1016/S0006-8993(03)02725-2
Chen H. (2017). miRNA-mediated modulation in the stress response of oyster Crassostrea gigas. Dissertation for Philosophy Doctor, University of Chinese Academy of Sciences, Beijing, China.
Chen H., Zhou Z., Wang H., Wang L., Wang W., Liu R., et al. (2016). An invertebrate-specific and immune-responsive microRNA augments oyster haemocyte phagocytosis by targeting CgIκB2. Sci. Rep.-UK 6, 29591. doi: 10.1038/srep29591
Cheng L., Zhan B., Luo P., Wang B. (2017). miRNA-375 regulates the cell survival and apoptosis of human non-small cell carcinoma by targeting HER2. Mol. Med. Rep. 15, 1387–1392. doi: 10.3892/mmr.2017.6112
Chiamolera M. I., Wondisford F. E. (2009). Thyrotropin-releasing hormone and the thyroid hormone feedback mechanism. Endocrinology 150, 1091–1096. doi: 10.1210/en.2008-1795
Cong M., Li Y., Xu H., Lv J., Wu H., Zhao Y. (2021). Ammonia nitrogen exposure caused structural damages to gill mitochondria of clam Ruditapes philippinarum. Ecotoxicol. Environ. Saf. 222, 112528. doi: 10.1016/j.ecoenv.2021.112528
Cong M., Wu H., Cao T., Ji C., Lv J. (2019). Effects of ammonia nitrogen on gill mitochondria in clam Ruditapes philippinarum. Environ. Toxicol. Pharmacol. 65, 46–52. doi: 10.1016/j.etap.2018.12.003
Cong M., Wu H., Yang H., Zhao J., Lv J. (2017). Gill damage and neurotoxicity of ammonia nitrogen on the clam Ruditapes philippinarum. Ecotoxicology 26, 459–469. doi: 10.1007/s10646-017-1777-4
Cong M., Xu H., Li Y., Tian W., Lv J. (2022). Modifications of calcium metabolism and apoptosis after ammonia nitrogen exposure imply a tumorous fate in clam Ruditapes philippinarum? Aquat. Toxicol. 245, 6110. doi: 10.1016/j.aquatox.2022.106110
Cornell L. D., Leung N. (2021). Myeloid bodies in acute tubular injury. Kidney Int. 99, 1027. doi: 10.1016/j.kint.2020.08.018
Du S., Qu H., Zhang Y., Zhu S., Wang Y., Zhang S., et al. (2023). MiR-375 promotes cisplatin sensitivity of lung adenocarcinoma. Pathol. Res. Pract. 249, 154765. doi: 10.1016/j.prp.2023.154765
Friedlander M. R., Mackowiak S. D., Li N., Chen W., Rajewsky N. (2012). miRDeep2 accurately identifies known and hundreds of novel microrna genes in seven animal clades. Nucleic. Acids Res. 40, 37–52. doi: 10.1093/nar/gkr688
Ientile R., Caccamo D., Griffin M. (2007). Tissue transglutaminase and the stress response. Amino Acids 33, 385–394. doi: 10.1007/s00726-007-0517-0
Kalinina E. V., Gavriliuk L. A. (2020). Glutathione synthesis in cancer cells. Biochem. Moscow 85, 895–907. doi: 10.1134/S0006297920080052
Knoop K. A., Newberry R. D. (2018). Goblet cells: multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 11, 1551–1557. doi: 10.1038/s41385-018-0039-y
Kropotova E. S., Zinov’eva O. L., Zyryanova A. F., Choinzonov E. L., Afanas’ev S. G., Cherdyntseva N. V., et al. (2013). Expression of genes involved in retinoic acid biosynthesis in human gastric cancer. Mol. Biol. 47, 280–292. doi: 10.1134/S0026893313020076
Kropotova E. S., Zinovieva O. L., Zyryanova A. F., Dybovaya V. I., Prasolov V. S., Beresten S. F., et al. (2014). Altered expression of multiple genes involved in retinoic acid biosynthesis in human colorectal cancer. Pathol. Oncol. Res. 20, 707–717. doi: 10.1007/s12253-014-9751-4
Latreille M., Herrmanns K., Renwick N., Tuschl T., Malecki M. T., McCarthy M. I., et al. (2015). miR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. J. Mol. Med. 93, 1159–1169. doi: 10.1007/s00109-015-1296-9
Li C., Wang F., Song Z., Zhou H., Gu J., Wang. H., et al. (2023). Water quality monitoring and assessment of the shellfish culturing areas in Dongying River estuary, Shandong Province. J. Fish. Res. 45, 155–162. doi: 10.14012/j.cnki.fjsc.2023.02.007
Li D., Wang T., Sun F. F., Feng J. Q., Peng J. J., Li H., et al. (2021). MicroRNA-375 represses tumor angiogenesis and reverses resistance to sorafenib in hepatocarcinoma. Cancer Gene Ther. 28, 126–140. doi: 10.1038/s41417-020-0191-x
Li L., Wu Q., Miao C., Muhammad H., Zhang L., Jiang Z., et al. (2023). 5-Azacytidine promotes HCC cell metastasis by up-regulating RDH16 expression. Eur. J. Pharmacol. 950, 175736. doi: 10.1016/j.ejphar.2023.175736
Liu Y., Wang Q., Wen J., Wu Y., Man C. (2021). MiR-375: A novel multifunctional regulator. Life Sci. 275, 119323. doi: 10.1016/j.lfs.2021.119323
Liu L., Xiao C., Sun Q. (2021). MiRNA-375 inhibits retinoblastoma progression through targeting ERBB2 and inhibiting MAPK1/MAPK3 signalling pathway. Cutaneous Ocular Toxicol. 41, 1–10. doi: 10.1080/15569527.2021.1994587
Long J., Cui Y., Wang R., Chen Y., Zhao N., Wang C., et al. (2021). Combined effects of high salinity and ammonia-N exposure on the energy metabolism, immune response, oxidative resistance and ammonia metabolism of the Pacific white shrimp Litopenaeus vannamei. Aquac. Rep. 20, 100648. doi: 10.1016/j.aqrep.2021.100648
Mao Q., Quan T., Luo B. (2016). MiR-375 targets KLF4 and impacts the proliferation of colorectal carcinoma. Tumor. Biol. 37, 463–471. doi: 10.1007/s13277-015-3809-0
Mao Z., Wang X., Zhao Y., Yang F., Qin Q., Jiang R. (2024). The role of miR-375 in migration and invasion of h.pylori-induced gastric cancer cell model. Cell Biochem. Biophys. 37, 463–471. doi: 10.1007/s12013-024-01473-9
Marchi S., Patergnani S., Missiroli S., Morciano G., Rimessi A., Wieckowski M. R., et al. (2018). Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium 69, 62–72. doi: 10.1016/j.ceca.2017.05.003
Morris J. L., Gillet G., Prudent J., Popgeorgiev N. (2021). Bcl-2 family of proteins in the control of mitochondrial calcium signalling: an old chap with new roles. Int. J. Mol. Sci. 22, 3730. doi: 10.3390/ijms22073730
Mukhopadhyay P., Horváth B., Rajesh M., Matsumoto S., Saito K., Bátkai S., et al. (2011). Fatty acid amide hydrolase is a key regulator of endocannabinoid-induced myocardial tissue injury. Free Radical Bio. Med. 50, 179–195. doi: 10.1016/j.freeradbiomed.2010.11.002
Murakami K., Matsunaga T., Matsuzaki T., Naruke Y., Miyauchi S., Kobayashi S., et al. (2024). Serum bta-miRNA-375 as a potential biomarker for the early diagnosis of enzootic bovine leukosis. PloS One 19, e0302868. doi: 10.1371/journal.pone.0302868
Newsholme P., Procopio J., Lima M. M. R., Pithon-Curi T. C., Curi R. (2003). Glutamine and glutamate—their central role in cell metabolism and function. Cell Biochem. Funct. 21, 1–9. doi: 10.1002/cbf.1003
Noronha O., Mesarosovo L., Anink J. J., Iyer A., Aronica E., Mills J. D. (2022). Differentially expressed miRNAs in age-related neurodegenerative diseases: a meta-analysis. Genes 13, 1034. doi: 10.3390/genes13061034
Park S. J., Takenawa T. (2011). Neural wiskott-aldrich syndrome protein is required for accurate chromosome congression and segregation. Mol. Cells 31, 515–522. doi: 10.1007/s10059-011-2292-8
Phelan P., Bacon J. P., Davies J. A., Stebbings L. A., Todman M. G. (1998). Innexins: a family of invertebrate gap-junction proteins. Trends Genet. 14, 348–349. doi: 10.1016/S0168-9525(98)01547-9
Randall D. J., Tsui T. K. N. (2002). Ammonia toxicity in fish. Mar. pollut. Bull. 45, 17–23. doi: 10.1016/S0025-326X(02)00227-8
Riba I., de Canales M. G., Forja J. M., DelValls T. A. (2004). Sediment quality in the Guadalquivir estuary: sublethal effects associated with the Aznalcóllar mining spill. Mar. pollut. Bull. 48, 153–163. doi: 10.1016/S0025-326X(03)00392-8
Rodrigues T., Ramos V., do-Prado-Souza L. F. L., Tersariol I. (2024). “Emerging Roles of Sodium/Calcium Exchangers in Cancer,” in Handbook of Cancer and Immunology. Ed. Rezaei N. (Springer, Cham). doi: 10.1007/978-3-030-80962-1_362-1
Shi Y., Wu S., Zhang X., Cao Y., Zhang L. (2024). Lipid metabolism-derived FAAH is a sensitive marker for the prognosis and immunotherapy of osteosarcoma patients. Heliyon. 10, e23499. doi: 10.1016/j.heliyon.2023.e23499
Tian W., Li Y., Li Z., Lv J., Cong M. (2023). Comparative analysis of microRNA expression profiles in clam Ruditapes philippinarum after ammonia nitrogen exposure. Aquat. Toxicol. 261, 106624. doi: 10.1016/j.aquatox.2023.106624
Torres-Ferreira J., Oliveira J., Gonçalves C. S., Costa B. M., Henrique R., Jerónimo C. (2015). MicroRNA-375 plays a dual role in prostate carcinogenesis. Clin. Epigenet. 7, 42. doi: 10.1186/s13148-015-0076-2
Van Sinay E., Mirabeau O., Depuydt G., Van Hiel M. B., Peymen K., Watteyne J., et al. (2017). Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. P. Natl. Acad. Sci. U.S.A. 114, E4065–E4074. doi: 10.1073/pnas.161739211
Wen M., Shen Y., Shi S., Tang T. (2012). miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinf. 13, 140. doi: 10.1186/1471-2105-13-140
Wheeler B. M., Heimberg A. M., Moy V. N., Sperling E. A., Holstein T. W., Heber S., et al. (2009). The deep evolution of metazoan microRNAs. Evol. Dev. 11, 50–68. doi: 10.1111/j.1525-142X.2008.00302
Xia Z., Wang C., Wang X., Yu H., Yao H., Shen H., et al. (2021). NCX3 alleviates ethanol-induced apoptosis of SK-N-SH cells via the elimination of intracellular calcium ions. Toxicol. Vitro 72, 105104. doi: 10.1016/j.tiv.2021.105104
Xu J., Li B., Song W., Cao L., Zhu C., Lin S. (2021). Tumor suppressor functions of miRNA-375 in nasopharyngeal carcinoma through inhibition of ubiquitin-specific protease 1 expression. Int. J. Biochem. Cell Biol. 141, 106092. doi: 10.1016/j.biocel.2021.106092
Yang C., Wang X. (2021). Lysosome biogenesis: Regulation and functions. J. Cell Biol. 220, e202102001. doi: 10.1083/jcb.202102001
Keywords: Ruditapes philippinarum, miRNA-375, NH3-N exposure, oxidative damage, apoptosis resistance
Citation: Cong M, Li Z, Tian W, Yu Y, Che Y and Lv J (2025) Increased miRNA-375 causes oxidative damage but promotes apoptosis resistance and cell migration in the clam Ruditapes philippinarum. Front. Mar. Sci. 12:1528020. doi: 10.3389/fmars.2025.1528020
Received: 14 November 2024; Accepted: 15 January 2025;
Published: 10 February 2025.
Edited by:
Xiaoshan Zhu, Hainan University, ChinaReviewed by:
Wei Xu, Texas A&M University Corpus Christi, United StatesCopyright © 2025 Cong, Li, Tian, Yu, Che and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Cong, bWNvbmdAeXR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.