- 1School of Medicine, Department of Earth and Biological Sciences Loma Linda University, Loma Linda, CA, United States
- 2Department of Natural Resources and Environment Tasmania, Hobart, TAS, Australia
- 3Centre d’Etudes Biologiques de Chizé, UMR 7372 du CNRS-La Rochelle Université, Villiers-en-Bois, France
- 4Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS, Australia
- 5CSIRO Environment, Hobart, TAS, Australia
Understanding the foraging preference of cetaceans is crucial for assessing their role as apex predators and indicators of marine ecosystem health. Using stable isotope analysis, we investigated trophic niche differentiation and foraging plasticity in 141 long-finned pilot whales (Globicephala melas edwardii) that stranded along the Tasmanian coast at three locations (Marion Bay, King Island, and Maria Island). Stranding location accounted for most of the variability in skin isotopic values (δ13C: - 17.9 ± 0.2 ‰ to –16.9 ± 0.2 ‰, Principal Coordinate 1 = 89%), likely reflecting differences in foraging habitats. In addition, isotopic niche overlap ranged from minimal (0-10% between Marion Bay and Maria Island) to moderate (between Marion Bay and King Island, and King Island and Maria Island). While sex related differences in isotopic niche space were minimal overall, there was some variability in the core niche space between males and females at Maria Island. Dietary proportions from our mixing model support a predominantly cephalopod diet for pilot whales in Tasmanian waters (91%, CI: 63-90%), with greater contributions from offshore dietary sources (68%, CI: 25-95%). The dietary variability across the three strandings highlights the foraging plasticity of pilot whales, which despite their preferences for a wide range of oceanic cephalopods, can adopt a more generalist feeding strategy when necessary. These findings provide valuable insights into the ecological role and adaptability of pilot whales in Tasmanian waters, highlighting the importance of monitoring apex predators to inform conservation and ecosystem management strategies in dynamic marine ecosystems.
Introduction
Cetaceans are apex predators that can shape the dynamics of marine ecosystems (Becker et al., 2021). Their foraging behavior can directly influence both the structure and function of their ecosystems, by impacting prey abundance, which then may initiate a cascade of trophic effects across the broader food web (Estes et al., 2011, 2016; Machovsky-Capuska and Raubenheimer, 2020). Simultaneously, cetaceans also contribute to biogeochemical cycling by facilitating nutrient distribution and supporting energy flow from lower trophic levels, helping to sustain marine productivity as both predators and prey (Baum and Worm, 2009; Strong and Frank, 2010; Roman et al., 2014; Estes et al., 2016). They are recognized as ecological sentinels due to their ability to reflect changes in the marine environment and hence the health of the ecosystem (Hazen et al., 2019; Williamson et al., 2021). The distribution and abundance of cetaceans are key indicators used to assess ecosystem health and stability, emphasizing the critical nature of studying cetacean feeding ecology to understand their roles in maintaining the balance of marine systems (Ramos and González-Solís, 2012; McCluskey et al., 2021).
There are two closely related species of pilot whales in the cetacean genus, Globicephala. Although these two species have similar morphologies, they inhabit contrasting thermal niches. The short-finned pilot whale, Globicephala macrorhynchus, is found mainly in warmer tropical waters (Soto et al., 2008; Mahaffy et al., 2015), while the long-finned pilot whale, Globicephala melas, is adapted to the temperate and subpolar regions of the northern and southern hemispheres, which defines the geographic distribution of the two G. melas subspecies (Abend and Smith, 1999; Becker et al., 2021). The southern subspecies, G. m. edwardii, has a more extensive distribution than its northern counterpart, G. m. melas, reaching as far south as the Antarctic Polar Front (Abend and Smith, 1999; Oremus et al., 2009; Mansilla et al., 2012; Santos et al., 2014; Fontaine et al., 2015; Olson, 2018). The wide latitudinal gradient of the southern species emphasizes its ecological importance and potential impact in relation to food web dynamics in the marine ecosystem.
While the Southern Hemisphere subspecies, G. m. edwardii, is widely distributed across mid- and high-latitude regions, these whales primarily inhabit deeper waters off the continental edge and shelf, frequently exploiting both pelagic and deep oceanic environments for foraging (Gales et al., 1992; Monteiro et al., 2015; Beasley et al., 2019; Becker et al., 2021). Increasing evidence also highlights their occasional use of coastal waters for foraging, demonstrating their ability to adapt to diverse habitats (Beatson et al., 2007a, b; Spitz et al., 2011). Their distribution and foraging patterns, like other cetaceans, are likely to be closely linked to the availability and distribution of their primary prey species (MacLeod et al., 2003). Thus, understanding the level of trophic flexibility is crucial to understand how this species may adapt to varying habitats across its wide-ranging distribution.
Studying the trophic dynamics of cetaceans, including long-finned pilot whales, poses significant challenges due to the difficulties in observing their foraging behavior and diet in deep oceanic habitats (Kiszka et al., 2021). However, important information has been provided through stomach content analysis in relation to their feeding preferences. Cephalopods dominate their diet, although this varies both temporally and spatially (Gales et al., 1992; Gannon et al., 1997; dos Santos and Haimovici, 2001). These studies show that their cephalopod diet can range from just a few species (Beatson et al., 2007a, b; Beatson and O’Shea, 2009) to a diverse assemblage comprising many species (Gales et al., 1992; Santos et al., 2014; Beasley et al., 2019), with occasional fish consumption in some regions (Overholtz and Waring, 1991; Gannon et al., 1997; Spitz et al., 2011). However, stomach content analysis has limitations, such as the differential retention of cephalopod beaks over fish remains, which can lead to an overestimation of their importance in the diet (Pierce et al., 2004; Bowen and Iverson, 2013).
To overcome the challenges associated with stomach content analysis, researchers have employed indirect dietary tracers to investigate the trophic ecology of cetaceans. Fatty acid signature analysis provides qualitative insights into foraging (Groß et al., 2020) and quantitative assessments of dietary intake to estimate prey consumption in whales and other marine organisms (Remili et al., 2023). Dietary fatty acids are transferred from prey to consumer with minimal modification, allowing the fatty acid composition of the consumer to reflect that of its prey (Couturier et al., 2020). Similarly, dietary metabarcoding using feces, or stomach contents from cetaceans, as well as environmental water samples from their habitat, provides a powerful method for accurately identifying prey species, even in the absence of physical remains (Berry et al., 2017; Reidy et al., 2022). Furthermore, although more challenging than the prior methods, direct observation provides valuable insights into the diet of cetaceans, particularly for species feeding in surface waters (García-Cegarra et al., 2021).
Another widely used method in foraging ecology, stable isotope analysis, is based on the natural variation of elements like carbon and nitrogen that are incorporated into the tissue of a predator through their diet (Teixeira et al., 2022). The ratios of lighter to heavier isotopes (δ¹³C for carbon, δ15N for nitrogen) provide dietary information over various time scales, depending on tissue turnover (Newsome et al., 2010). δ¹³C tracks foraging areas and primary productivity due to its minimal variation (~1 ‰) during trophic transfer, distinguishing between inshore versus offshore or pelagic versus benthic environments (DeNiro and Epstein, 1981; Kurle and Worthy, 2002; Cherel and Hobson, 2007; Newsome et al., 2010). Conversely, δ15N shows predictable enrichment with each trophic level (2-5 ‰), helping determine a predator’s trophic position and niche overlap (Minagawa and Wada, 1984; Vanderklift and Ponsard, 2003). Therefore, isotopic analysis is used as an important tool to expand our understanding of cetacean feeding ecology.
Stable isotope analysis has proven critical in delineating dietary niches and trophic plasticity among cetaceans, emphasizing how species across various feeding guilds, including both mysticetes (baleen whales) and odontocetes (toothed whales), allocate prey resources and adjust to changes in prey availability (Teixeira et al., 2022) For example, the cetacean community in the Azores showed evidence of spatial and temporal resource partitioning, thereby facilitating species coexistence (Lebon et al., 2024). Similarly, baleen whales in Icelandic waters also showed clear ecological niche partitioning, allowing them to coexist (García-Vernet et al., 2021). An examination of five cetacean species in the northwestern Mediterranean revealed exclusive trophic niches for all but two species, thus avoiding competitive exclusion. However, the overlap in niches, and hence competition, between the striped dolphin (Stenella coeruleoalba) and common dolphin (Delphinus delphis) may have altered the distribution of the species, thus mitigating resource competition (Borrell et al., 2021). Isotopic analysis also revealed that a fluctuation in the isotopic values over time of the endangered Maui dolphin (Cephalorhynchus hectori maui) may reflect a shift in prey due to environmental conditions (Ogilvy et al., 2022). Thus, isotopic analysis can provide a comprehensive understanding of the foraging preferences of cetaceans.
The present study will use isotopic analysis to determine the dietary niche and habitat of the long-finned pilot whale, (G. m. edwardii, hereafter referred to as pilot whale) off Tasmania, Australia. Tasmania is a region that experiences frequent strandings of pilot whales (Gales et al., 1992; Beasley et al., 2019). However, to our knowledge, a detailed isotopic study has not been undertaken on this species in this region, despite its importance as a predator in the ecosystem. We aim to determine regional variations in isotopic values and whether these reflect distinct isotopic niches. We will analyze skin, a metabolically active tissue that represents prior feeding over the previous 2-3 months for δ¹³C and 3-8 months for δ15N (Giménez et al., 2016; Teixeira et al., 2022). Moreover, we will document whether fish form an important part of their diet or if the diet remains dominated by cephalopods, as previously reported using stomach content analysis. In addition, we will examine whether our results are supported by those patterns observed in other regions, when considering both the variability in the composition of dietary prey as well as local foraging habitats.
Materials and methods
Sample collection
Consumers - pilot whales
A standardized stranding protocol for marine mammals (Geraci and Lounsbury, 2005), performed by the Department of Natural Resources and Environment Tasmania (NRE Tas), previously Department of Primary Industries, Parks, Water and Environment (DPIPWE), was undertaken on 141 pilot whales that mass stranded around Tasmania, Australia. Strandings occurred in November 2004 on Maria Island (n = 19) and King Island (n = 32) and in October 2005 on the beach at Marion Bay (n = 90) (Figure 1). For each pilot whale a straight-line total length (mm) from the tip of the rostrum to the deepest part of the notch in the tail fluke was recorded. Sex, as well as an age/maturity class was assigned according to length and maturity (i.e., adult, subadult and juvenile) based on established protocol (Bloch, 1993). Stomach contents were also collected from the Marion Bay and Maria Island strandings.
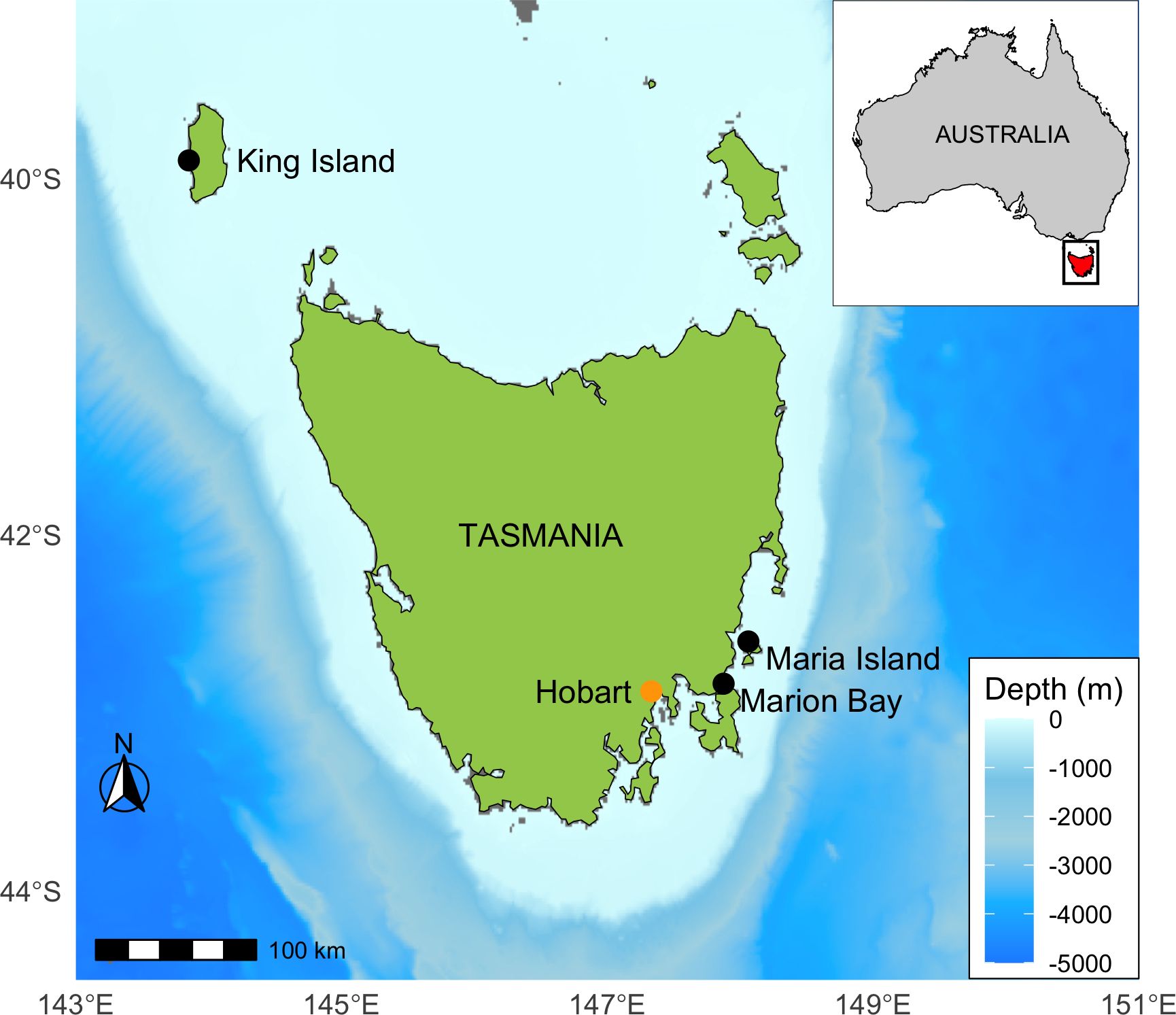
Figure 1. Locations of long-finned pilot whale, Globicephala melas edwardii, standings in 2004 and 2005 along the Tasmanian coast. Marion Bay, n = 90, Maria Island, n = 19, King Island, n= 32.
Large sample blocks of skin and blubber were immediately packed on ice and later frozen and archived in a -20°C freezer at the NRE Tas laboratory in Hobart, Tasmania. Most archived samples were taken dorsally on a body site just anterior to the dorsal fin. However, Maria Island samples were obtained from a lateral body site, except for one individual. This difference likely reflected logistical constraints during field collection. However, to minimize variability all samples were taken in a standardized manner to ensure consistency. To evaluate whether the location of skin sampling (dorsal, lateral and ventral) influenced isotopic values, samples were compared from four whales stranded at Maria Island. Sub-samples of skin (approximately 1 cm2) for all analyses were then cut from these larger blocks.
Prey – cephalopods and fish
For cephalopod prey, we focused on key cephalopod species identified from the stomach contents of pilot whales stranded in Tasmanian waters (see Table 8 in Beasley et al., 2019). For some of the cephalopod prey we used beaks obtained and archived from the stomachs of pilot whales that stranded at Bicheno (Sept, 1992), Marion Bay (Oct, 2005) and Ocean Beach (Jan, 2006). Cephalopod beaks were sorted and kept in 70% ethanol until further analysis, and subsequently identified to species level where possible (see Beasley et al., 2019). Lower rostral length (LRL) was measured with digital calipers to the nearest mm. As ommastrephids were only identified to family level, we obtained known whole specimens of ommastrephid species to use as potential prey items. The additional ommastrephids, as well as inshore cephalopods and fish used as potential prey items were sourced from local fisherman operating in Tasmanian waters in 2006 and early 2007 (Table 1).
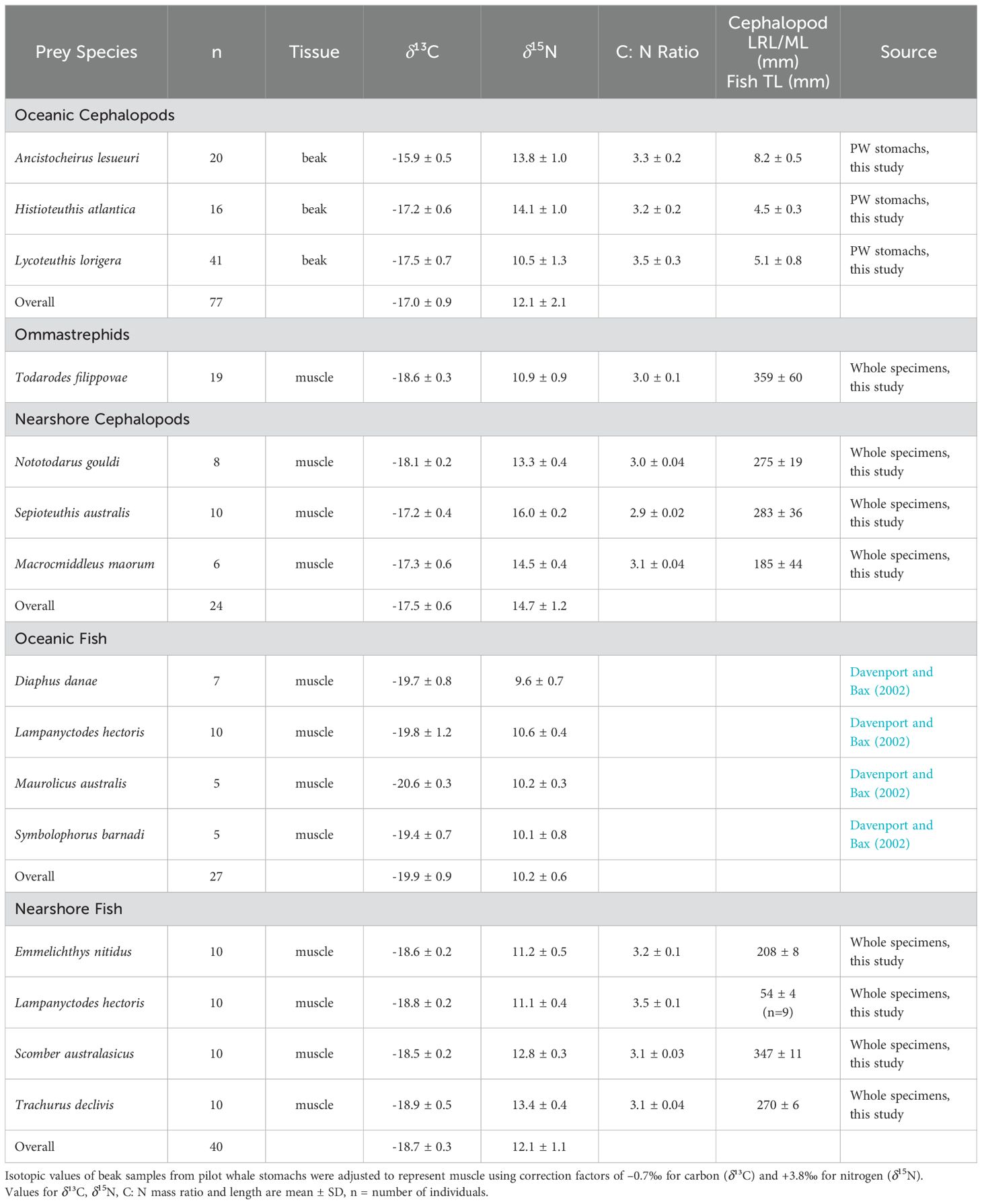
Table 1. Isotopic composition for prey species used in the MixSIAR model for long-finned pilot whales Globicephala melas edwardii.
Isotope analysis
A small section of the tip of the wing in the direction of growth was sampled from each cephalopod beak and rinsed with distilled water. The tip of the wing was selected as it signifies the most recent growth phase of the beak and therefore the most recent somatic growth (Cherel and Hobson, 2005). For isotopic analysis, beak wing tip samples were examined whole or cut in half for larger specimens. For squid species from pilot whale stomach contents, beaks were selected based on the mode of the LRL length for each prey species.
In whole prey specimens, approximately 1 cm2 pieces of anterior dorsal white muscle was dissected from behind the head for fish and mid-anterior ventral mantle for cephalopods, cleaned of any skin or mesentery, and then rinsed with distilled water. A similar sized piece of pilot whale skin was also subsampled from larger frozen samples of skin and blubber. Any remaining subcutaneous adipose tissue was removed with a scalpel and the subsequent sample rinsed in distilled water. All samples were then frozen in preparation for isotopic analysis. Just prior to isotopic analysis, samples were freeze dried and then ground to a powder with a Wig–L-Bug®. Since lipids are depleted in ¹³C, all tissue samples (except beaks) were delipidated using cyclohexane, which effectively adjusts δ¹³C values without affecting δ15N (Chouvelon et al., 2014). The ground whale skin, fish muscle or cephalopod mantle samples, along with 3 ml of cyclohexane, were left to stand overnight in small, covered test tubes under a fume hood. After delipidating overnight, samples were then centrifuged, and the supernatant removed. Samples were subsequently subjected to two more cyclohexane rinses. Between rinses the samples were allowed to stand for another 1 - 2 hours. Following delipidation, samples were left to dry uncovered under a fume hood overnight.
Relative abundance of carbon (13C/12C) and nitrogen (15N/14N) stable isotopes was determined by a Finnigan Delta Plus Advantage stable isotope-ratio mass spectrometer at the University of Victoria, British Columbia, Canada. For all isotopic analysis there was a 10 percent replication measurement for each isotopic sample run. The results of the isotopic analysis are presented in the usual δ notation relative to Vienna PDP belemnite for δ13C and atmospheric N2 (AIR) for δ15N. Replicate measurements of internal laboratory standards (DORM) indicated measurement errors of ± 0.1 ‰ and ± 0.2 ‰ for δ13C and δ15N, respectively. In addition, mean C: N mass ratios for pilot whales and prey species were within desirable ranges ≤ 3.5 (Tables 1, 2).
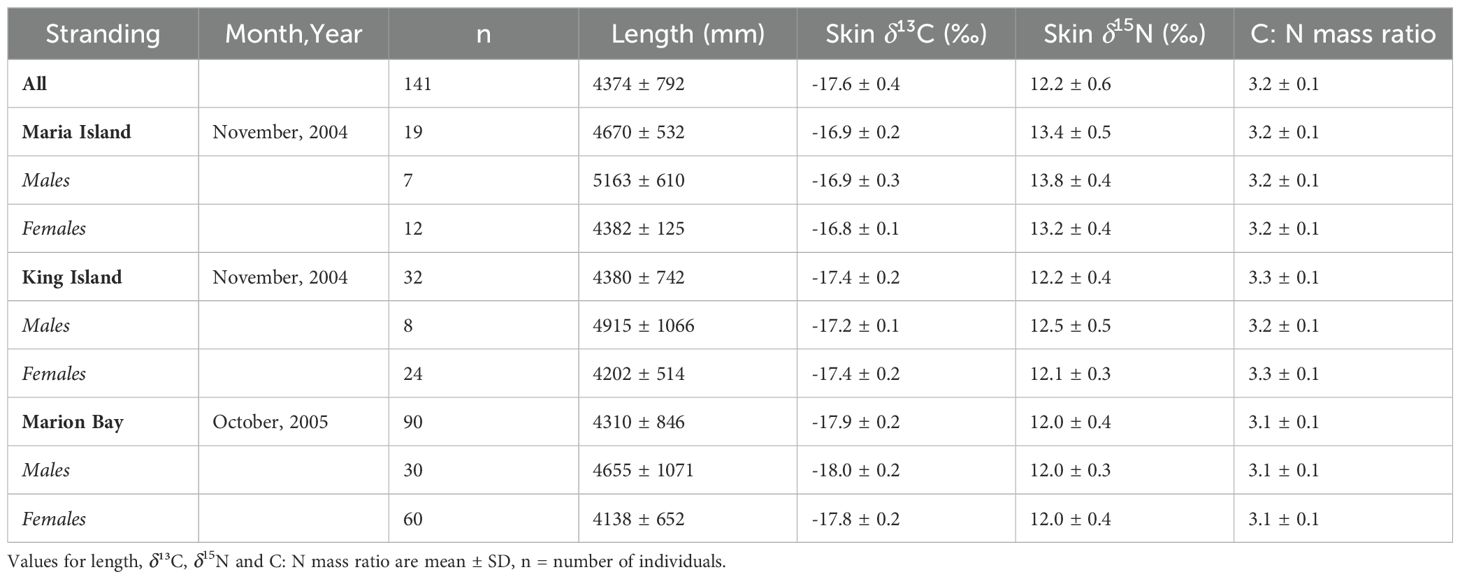
Table 2. Location of the strandings and sex of long-finned pilot whales Globicephala melas edwardii along Tasmanian shores.
Statistical analysis
Since skin samples were examined from differing body sites, a nested analysis of similarities (ANOSIM) with 9999 permutations was used to determine if there were differences between standardized body sites (dorsal, lateral, and ventral) on separate δ¹³C and δ15N values of skin from four individuals. Body site was treated as a fixed factor, and individuals as a random factor. ANOSIM was used because it is distribution free, robust for small sample sizes, and relies on permutations to determine significance. A permutational multivariate analysis of variance (PERMANOVA) model was then used to determine differences in pilot whale skin δ¹³C and δ15N values simultaneously. The non-parametric PERMANOVA model was based on 9999 permutations using a Euclidean distance matrix with sex and stranding as fixed factors. PERMDISP was used to determine variation within factors, and any significant differences in the factors were examined using pairwise comparisons. Results of the PERMANOVA were visualized using a Principal Coordinates Analysis (PCO). Spearman’s correlations were also used to assess relationships between total length and δ13C and δ15N values. These correlations were conducted across all strandings, separately for males and females across all strandings, and within each stranding for males and females separately.
Niche analysis
We employed the Stable Isotope Bayesian Ellipses in R (SIBER) package version 2.1.9 (Jackson et al., 2011) as a proxy for examining trophic diversity of the pilot whales. This method facilitated the quantification and comparison of isotopic niche widths and overlaps within and across different strandings, defined by stranding location and sex.
To quantify isotopic niches, we calculated three metrics: the Total Area (TA), the Standard Ellipse Area corrected for small sample sizes (SEAc), and the Bayesian Standard Ellipse Area (SEAB). The TA represents the whole isotopic niche, encompassing 100% of the data, but is sensitive to outliers, making it less reliable for analyzing the core niche. In comparison, the SEAc measures the core isotopic niche area, approximately 40% of the data, providing a robust estimate of central tendency while also correcting for small sample sizes. In contrast, the SEAB, which also measures the area of the ellipse, accounts for uncertainty in the data using a Bayesian framework. Accounting for uncertainty is particularly important for small or uneven sample sizes, such as the male samples for Maria Island and King Island. Using Monte Carlo resampling, the SEAB generates posterior distributions of ellipse areas. This allows estimation of the mode with 95% credible intervals, which reflect uncertainty in niche area estimates.
Furthermore, we also conducted a Bayesian analysis to evaluate the overlap and relative sizes of isotopic niches between different stranding groups and between sexes within those groups. This Bayesian approach considers the entire distribution of potential overlaps and size comparisons. Using the ‘bayesianOverlap’ function, we generated posterior distributions for the overlap, employing 4000 draws and a 95% probability interval to estimate and quantify the shared dietary isotopic space. We then calculated the bin mode of these distributions to identify the most probable range in overlap values. We also used the Bayesian posterior distribution of SEAB to assess the probability as to whether the isotopic niche of one group was larger than another. The probabilities were calculated as the proportion of posterior draws where the SEAB of one group exceeded that of another. Ellipses were also visualized using SEAB.
Dietary analysis
Dietary proportions of pilot whales were estimated by implementing the MixSIAR package version 3.1.12 (Stock and Semmens, 2016), which allows for the incorporation of prior information and uses Bayesian inference to model the contributions of various prey sources to the diet. This approach allowed us to quantify the proportional consumption of prey items between the stranding groups, utilizing δ15N and δ¹³C isotopic values of the whales (consumers) and their prey (sources).
Pilot whales consume a diverse range of cephalopods and fish. To reduce complexity caused by overlapping isotopic values among multiple species, prey items were categorized into five different groups: oceanic cephalopods (OC), oceanic fish (OF), near-shore cephalopods (NSC), near-shore fish (NSF), and ommastrephids represented by Todarodes filippovae (TF). Nearshore species included both shelf and neritic species. PERMANOVA was used to assess differences between the assigned prey groups for which we had raw isotopic data. These groups were then aggregated a posteriori to determine the proportion of cephalopods versus fish in the diet and whether the whales were more likely to feed nearshore or offshore.
Oceanic and near-shore cephalopod isotopic values were obtained from beaks retained in the pilot whale stomach contents as well as the mantle of whole specimens from local fishermen. As a result, we had both mantle (whole specimens) and beak tissues (PW stomach contents) from cephalopods for dietary analysis. Since isotopic discrimination varies according to the type of tissue (Teixeira et al., 2022), we applied a correction factor of -0.7 ‰ and 3.8 ‰ to the isotopic value of the beaks (wings) for δ¹³C and δ15N, respectively (Jackson, unpublished data), which is similar to the difference in isotopic values between beaks and muscle observed in T. filippovae (Cherel et al., 2009). Isotopic data for oceanic fish, represented by a weighted mean and standard deviation, were sourced from Davenport and Bax (2002), while near-shore fish isotopic values were derived from the muscle of specimens collected by local fishermen (see Table 1).
We first ran a MixSIAR model with uninformed priors (equal contributions from all five sources). However, to refine the estimates of the dietary composition of pilot whales, we ran two other models, which incorporated prior information based on known global dietary preferences of long-finned pilot whales (G. melas) as compiled in the literature (see Becker et al., 2021, online resource 1). This sensitivity analysis allowed us to evaluate whether our choice of TEFs influenced the dietary estimates for each stranding.
While the global dietary preferences for G. melas were obtained from Becker et al. (2021), the cephalopod and fish prey categories were slightly modified according to the known distribution of species in Tasmanian waters (i.e. whether they were oceanic cephalopods, oceanic fish, near-shore cephalopods, near-shore fish or ommastrephids). For each cephalopod or fish species, percent contribution from multiple studies were averaged, then normalized so that all contributions summed to 100%. These normalized values were then scaled to represent the five sources used in the model. The informative priors for the five sources used in the models are as follows: OC = 1.64, OF = 0.07, NSC = 2.04, NSF = 0.2 and OMM = 1.04.
The trophic enrichment factors (TEFs) between the whale’s diet and its tissue were derived from feeding experiments on bottlenose dolphins (Tursiops truncatus). These experiments provided insights into the diet-to-skin enrichment factors (Browning et al., 2014; Giménez et al., 2016). Given the close taxonomic relationship between bottlenose dolphins and long-finned pilot whales, we assumed that the TEFs would be similar for pilot whales. Therefore, we used TEFs from Giménez et al. (2016) for both the uninformed model (Model 1) and one of the informative models (Model 2) with five source groups to assess how cephalopods and fish prey contributed to the diet of pilot whales (Table 3). To address initial issues with model convergence, we also ran a third model comparison, using bottlenose dolphin TEFs from Browning et al. (2014) (see Table 3). While the dietary compositions are only reported for Models 1 and 2, Model 3 was used to compare convergence diagnostics and goodness of fit to ensure our models were robust.
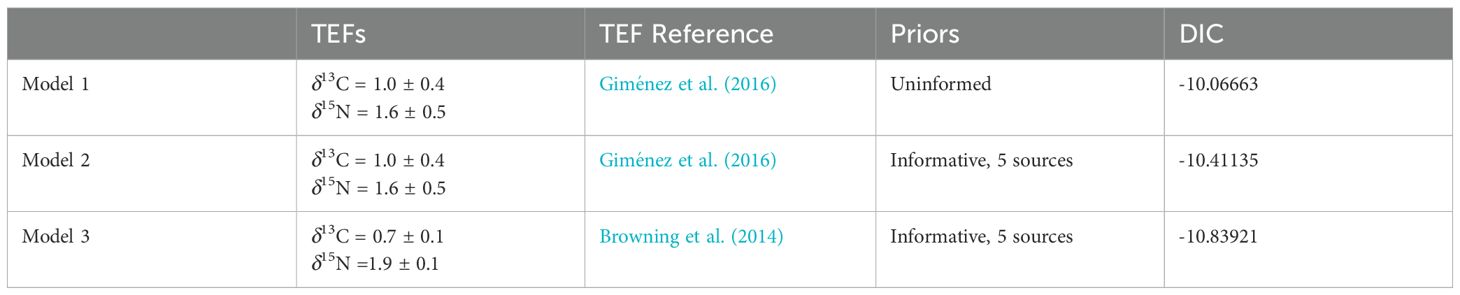
Table 3. Summary of trophic enrichment factors (TEF), type of priors used in MixSIAR models, and the resulting Deviance Information Criteria (DIC) for dietary analysis of long-finned pilot whales Globicephala melas edwardii.
Each of the three separate MixSIAR models was run with three MCMC chains, executing three million iterations, with a burn-in phase of 1.5 million iterations and a thinning interval of 500. To account for uncertainty in the models, we used both residual and process errors. The Gelman-Rubin diagnostic was used to determine if models converged, while the Deviance Information Criteria (DIC) was used to compare the goodness of fit between models.
All analyses were run in R version 4.3.3, except for PERMANOVA and ANOSIM which were undertaken using Primer v7 software (Primer-E). Values for δ¹³C, δ15N, C: N mass ratio and length are all reported as mean ± SD. SEAB is reported using the mode, along with 95% credible intervals. However, for dietary proportions we report medians, along with 95% credible intervals, which are a robust measure of central tendency but may not sum to one due to independent calculations for each dietary source.
Results
Across all strandings males (min = 2266 mm, max = 6080 mm) were larger than females (min = 2220 mm, max = 4940 mm). Skin isotope values ranged from -18.4 to -16.6 ‰ for δ13C and 11.3 to 14.3 ‰ for δ15N. Maria Island pilot whales had the most enriched isotopic value in δ¹³C and δ15N, while Marion Bay exhibited the least enriched values (Table 2).
ANOSIM results indicated weak to no significant differences in δ¹³C and δ15N values of skin between body sites (dorsal, lateral or ventral). For δ13C there was some variation (global R = 0.125, p = 12.7%) but this likely resulted from random variation, and was not significant. For δ15N the values were very similar across the three body sites, with no significant variation (global R = -0.19, p = 99.3%).
Across all strandings and within sexes, no significant correlations were found between total length and skin δ13C or δ15N values (all p > 0.05), except for Marion Bay males, where significant negative correlations were observed for δ13C (ρ = -0.54, p = 0.002), and δ15N (ρ = -0.47, p = 0.008), and for King Island females, where δ15N values also showed a negative correlation (ρ = -0.53, p = 0.007). However, the data for King Island females showed a lot of variability.
There was a significant interaction between stranding and sex (PERMANOVA, F 2,135 = 6.228, p = 0.003) on skin δ13C and δ15N values in multivariate space. Differences in the PERMANOVA was not due to within group variation of the combined factor (stranding and sex) (PERMDISP, F5,135 = 0.793, p = 0.61). Pairwise comparisons using the combined factor revealed that within each sex category (females and males), all stranding groups were significantly different from each other (p< 0.001). In addition, significant differences were observed between females and males within each stranding group (p ≤ 0.03).
Principal Coordinates Analysis (PCO) on the combined variable revealed that the first two coordinates accounted for 100% of the variation. The data along the PCO1 axis (accounting for 89.3% of the variation) were primarily separated by stranding, indicating that it captured most of the variation in the values. The remainder of the variation in data (10.7%) represented on the PCO2 axis is likely due to differences based on sex (Figure 2).
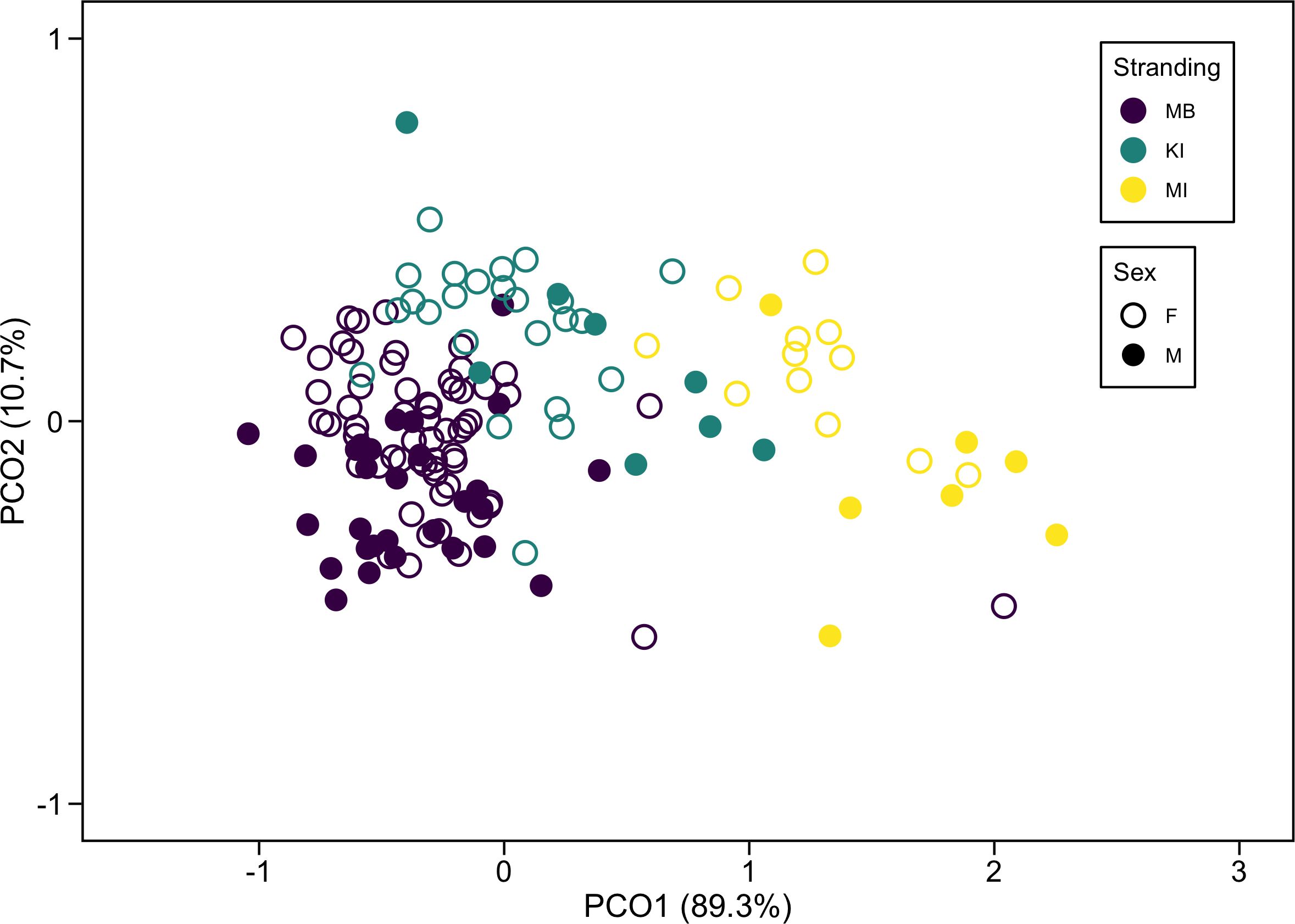
Figure 2. Principal Coordinates Analysis (PCO) of skin δ¹³C and δ15N isotopic values for Marion Bay (MB), King Island (KI), and Maria Island (MI) strandings, as well as for males and females of long-finned pilot whales (Globicephala melas edwardii) within each stranding. Values in parentheses represent the percent variance explained by each axis (PCO 1 and PCO 2).
Niche analysis
Maria Island samples had the largest Bayesian Standard Ellipse Area (SEAB) (0.3; CI 0.2 - 0.4) among the three strandings. Based on Bayesian overlap function, it showed minimal overlap with Marion Bay and King Island (bin mode range: 0-10%), indicating a distinct isotopic niche. There was an 80% and 68% probability that Maria Island had a larger SEAB than that of Marion Bay and King Island, respectively. King Island (SEAB 0.2; CI 0.2 - 0.3) had a 65% probability of a larger ellipse area than Marion Bay (SEAB 0.2; CI 0.2 - 0.3) with an overlap range of 50-60%, suggesting that at least half of the isotopic space is shared between these strandings (Table 4; Figures 3A, B).
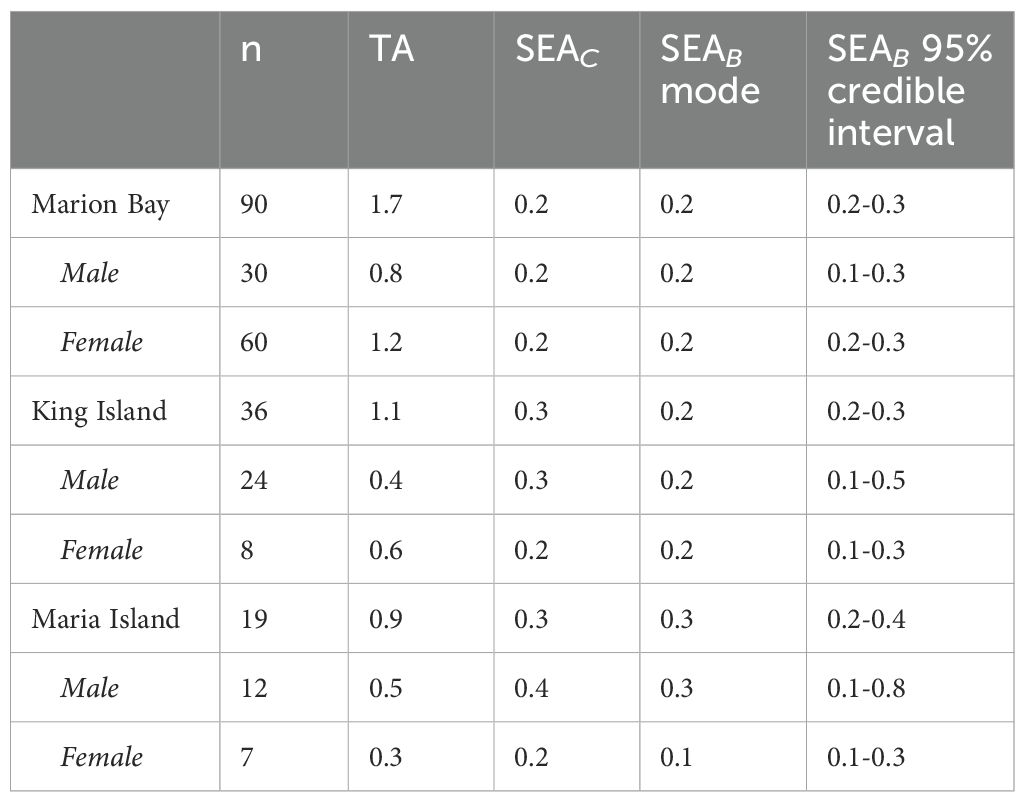
Table 4. Isotopic niche areas for each stranding, as well as for males and females within each stranding of long-finned pilot whales Globicephala melas edwardii, including Total Area (TA), Standard Ellipse Area adjusted for small sample size (SEAc), and Bayesian Standard Ellipse Area (SEAB) mode and 95% credible intervals.
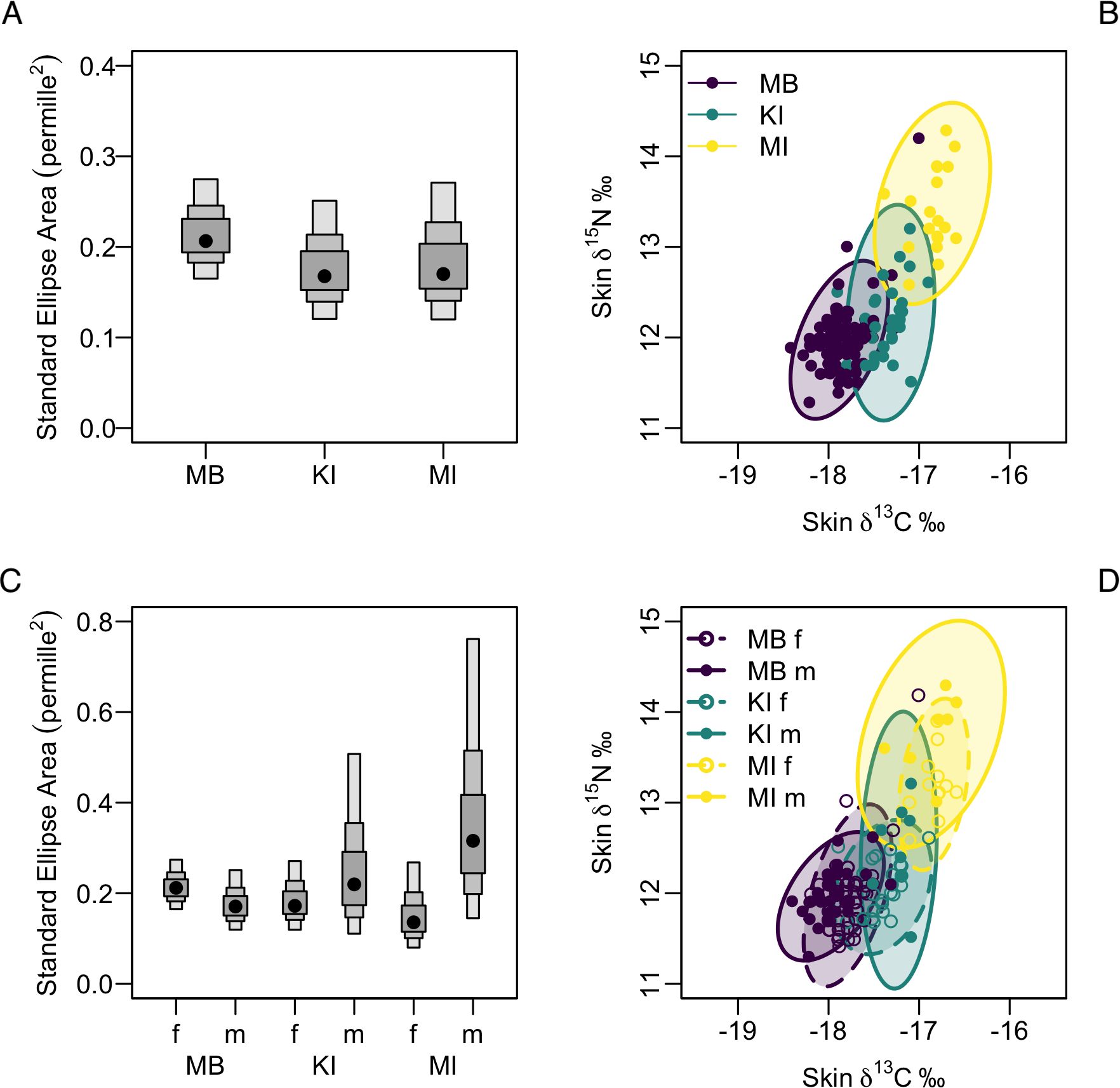
Figure 3. Density plots and isotopic niche comparisons of long-finned pilot whales (Globicephala melas edwardii) from Marion Bay (MB), King Island (KI), and Maria Island (MI). (A) Posterior distributions of Standard Ellipse Area (SEAb) for each stranding, with black dots indicating modes and shaded boxes showing 50%, 75% and 95% credible intervals. (B) Isotopic biplot with 95% SEAb ellipses for each location, illustrating core isotopic niches in δ¹³C and δ15N space. (C) SEAb distributions for males (m) and females (f) within each location, with modes and credible intervals. (D) Isotopic biplot with 95% SEAbellipses for males and females, highlighting differences in niche size and potential overlap between sexes.
The Bayesian overlap function also revealed a 70-80% overlap in the isotopic niches of Marion Bay females and males, indicating considerable similarity in their foraging habits or prey preferences. However, males exhibited a smaller isotopic niche (Total Area, TA: 0.8) than females (TA: 1.2), although both showed similar core niche metrics (SEAC and SEAB both at 0.2) indicating consistent niche utilization. Bayesian probability analysis revealed an 80% probability that the isotopic niche of females was larger than males, suggesting females had slightly greater dietary variation (Table 4; Figures 3C, D).
In contrast to Marion Bay, males from King Island and Maria Island were more likely to have larger isotopic niches than females (80% and 97%, respectively). The overlap between males and females was much lower at King Island (0-10%) and Maria Island (0-20%) compared to Marion Bay, indicating more distinct niches for females at these locations. Despite this, core niche metrics were similar between males and females across all strandings (SEAC and SEAB all at 0.2), males from King Island (CI 0.1 - 0.5) and Maria Island (CI 0.1 - 0.8) exhibited greater variability than females at both locations (CI 0.1 - 0.3), further highlighting the greater niche diversity among males (Table 4; Figures 3C, D).
Dietary analysis - MixSIAR
A PERMANOVA on four of the five aggregated prey groups (nearshore cephalopods, offshore cephalopods, ommastrephids and nearshore fish) for which we had raw isotopic data showed a difference based on prey group (F 3, 156 = 49.58, p = 0.0001). Pairwise comparisons revealed that all prey groups differed from one another (all p< 0.001). The fifth prey source (oceanic fish) was obtained from the literature (Davenport and Bax, 2002) (Table 1). A visual inspection confirmed that the consumer’s isotopic data (pilot whales) fell within the expected mixing space defined by the five sources and adjusted for the TEFs sourced from Giménez et al. (2016) (Figure 4) and Browning et al. (2014).
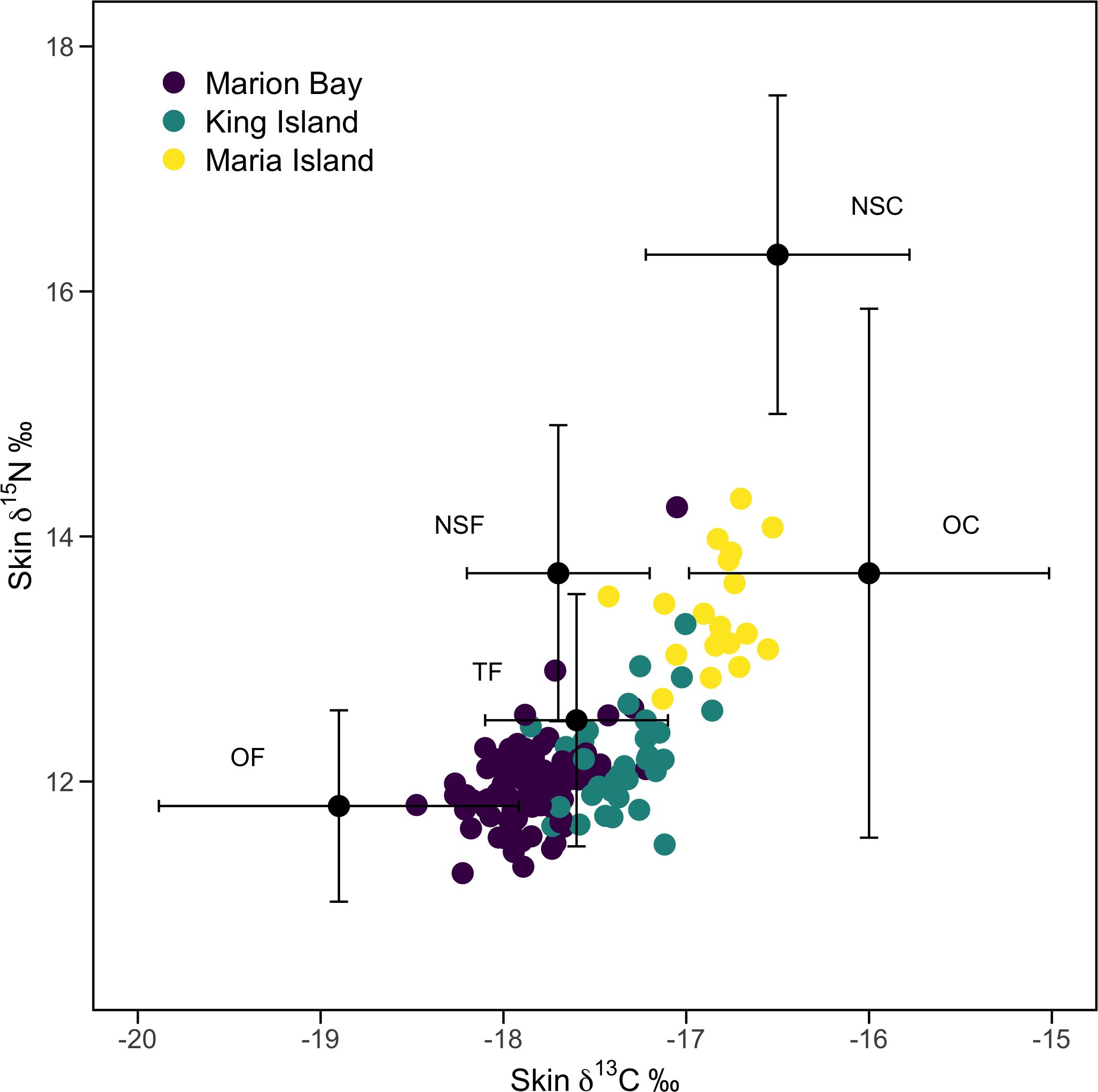
Figure 4. Biplot of skin δ¹³C and δ15N isotopic values from all long-finned pilot whales (Globicephala melas edwardii) along with mean prey values. Error bars represent ±1 SD. Near-shore cephalopods (NSC), ommastrephids [represented by Todarodes filippovae (TF)], and near-shore fish (NSF) were collected from muscle of whole animals. Oceanic cephalopods (OC) are from beaks found in pilot whale stomach contents, and offshore fish values are from Davenport and Bax (2002). Beak isotopic values were adjusted to muscle values, with all prey values corrected for trophic enrichment based on Giménez et al. (2016).
In our three MixSIAR models (see Table 3), we analyzed predicted dietary proportions using stranding as a fixed factor, as PCO analysis revealed that stranding was the most important factor accounting for variation in isotopic values. The Gelman-Rubin diagnostic for all parameters indicated strong convergence for all three models, with all below 1.05. A comparison of DIC values between Models 1 and 2, which used the same TEFs (Giménez et al., 2016) exhibited almost identical DIC values, indicating that informative priors resulted in a minimally stronger fit. Model 3, using TEFs from Browning et al. (2014) and informed priors, also converged well and fit within isotopic space.
When examining dietary contributions at each stranding using the informed priors model (Model 2), ommastrephids dominated the predicted diet of both Marion Bay (75%, CI: 59-86%) and King Island (87%, CI: 75-100%). Marion Bay included a smaller percentage of offshore fish (24%, CI: 14-37%), while King Island showed a smaller contribution from offshore cephalopods (11%, CI: 0-21%) (Figure 5). Aggregated dietary proportions revealed that cephalopods dominated the diet at King Island (96%, CI: 92-100) and Marion Bay (75%, CI: 63-86), while Marion Bay also sourced 25% (CI: 13-37%) of its diet from fish. Both strandings predominantly sourced their prey offshore (King Island: 100%, CI: 94-100%; Marion Bay: 100%, CI: 96-100%) (Figure 6). Overall, both aggregated and individual source contributions predicted from the generalist priors (Model 1) are almost identical to that of Model 2 (Supplementary Tables S1, S2).
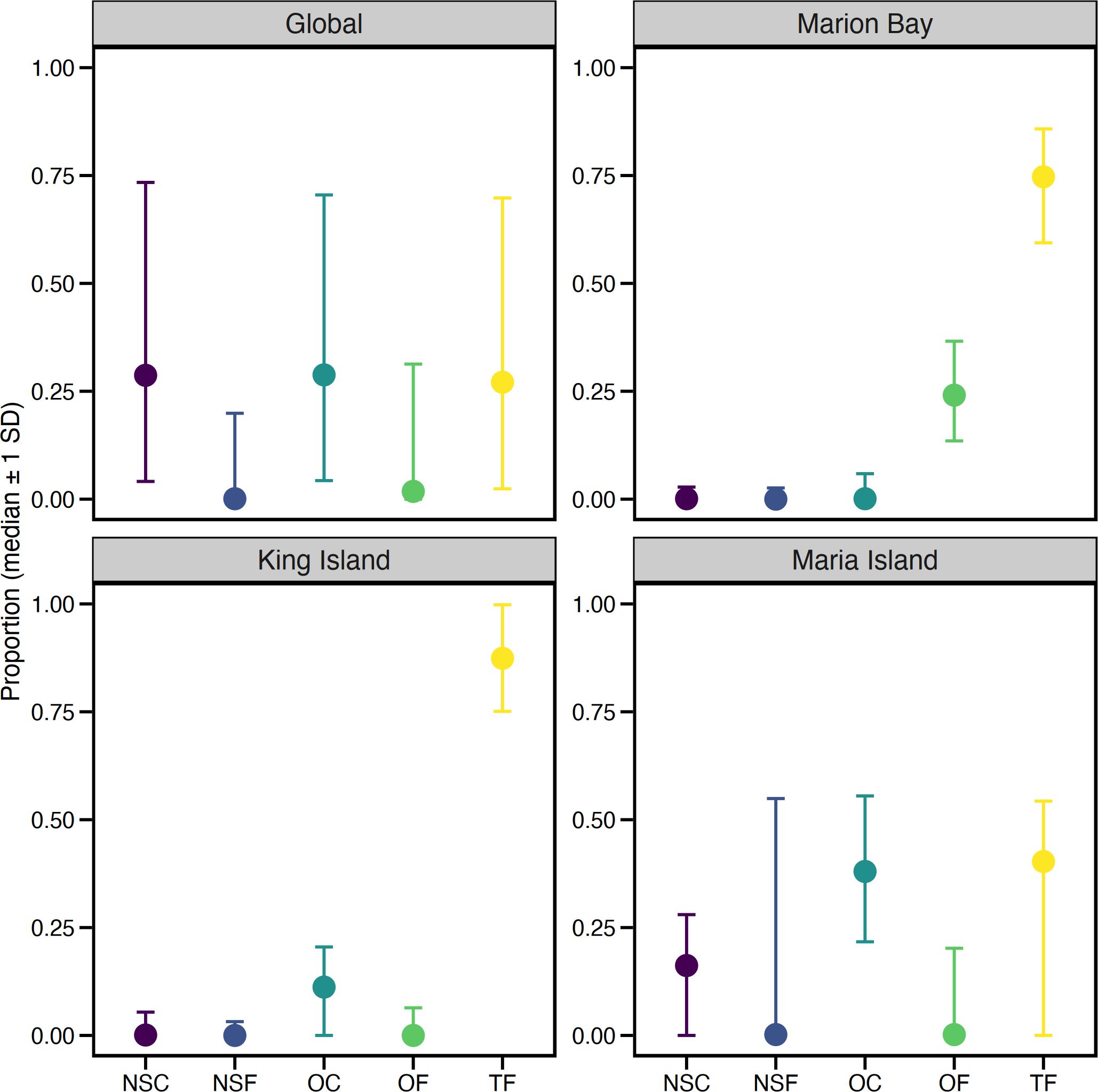
Figure 5. Results of MixSIAR diet analysis for long-finned pilot whales (Globicephala melas edwardii). Each panel represents a global estimate across all strandings and each site-specific diet proportions (Marion Bay, King Island, Maria Island). Proportions of the potential five prey groups consumed, including nearshore cephalopods (NSC), nearshore fish (NSF), offshore cephalopods (OC), offshore fish (OF), and Todarodes filippovae representing ommastrephids (TF), shown as median values with ± 1 standard deviation.
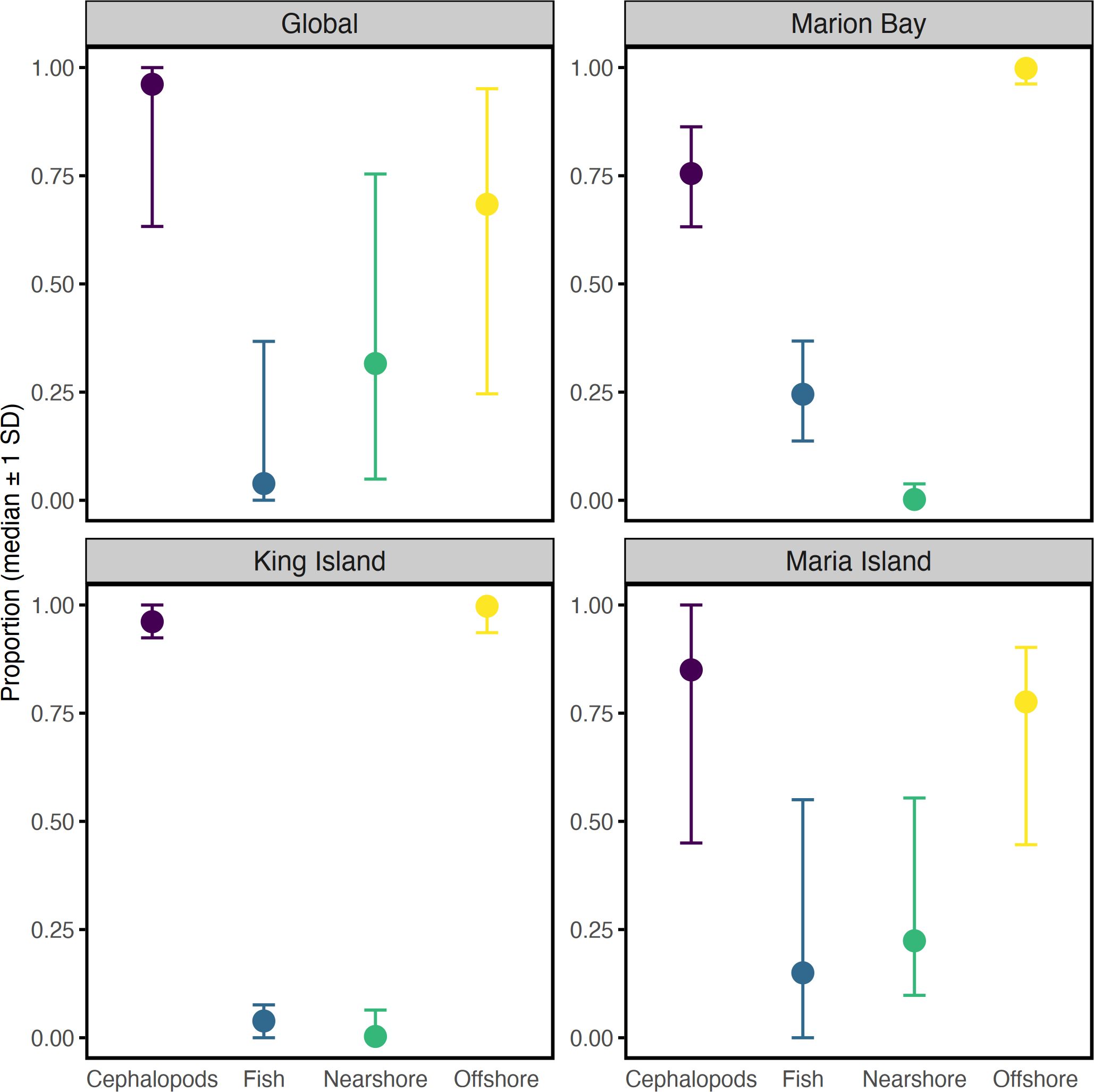
Figure 6. Results of aggregated proportions from MixSIAR diet analysis for long-finned pilot whales (Globicephala melas edwardii). Each panel represents a global estimate across all strandings and each site-specific diet proportions (Marion Bay, King Island, Maria Island) where potential prey groups are combined into broader categories: cephalopods versus fish, nearshore prey versus offshore prey, shown as median values and ±1 standard deviation. Cephalopods category includes nearshore and offshore cephalopods, and ommastrephids; fish category includes nearshore and offshore fish; nearshore category includes nearshore cephalopods and nearshore fish; offshore category includes offshore cephalopods, offshore fish and ommastrephids.
In contrast, based on the informed model, the Maria Island stranding showed greater dietary variability, with offshore cephalopods contributing 38% (CI: 22-56%), ommastrephids 40% (CI: 0-54%), and nearshore cephalopods 16% (CI: 0-55%) (Figure 5A). Aggregated dietary proportions revealed that cephalopods accounted for 85% (CI: 45-100) of the diet, while fish contributed 15% (CI: 0-55%). Compared to the other strandings, Maria Island whales sourced prey predominantly from offshore (78%, CI: 45-90%) but also used nearshore habitats (22%, CI: 10-55%) (Figure 6), suggesting greater plasticity in foraging strategies, as reflected by its distinctive mixing space. While the uninformed model also produced similar contributions at an aggregated level, individual sources revealed some variability. The model had a lower contribution of nearshore cephalopods (9%) and ommastrephids (25%) but higher contribution of nearshore fish (15%). Despite these differences, the diet of pilot whales across all strandings was dominated by cephalopods and offshore resources in both models, consistent with individual stranding events (Supplementary Tables S1, S2).
Discussion
The frequent strandings of pilot whales (Globicephala melas edwardii) in Tasmanian coastal waters provide a unique opportunity to investigate their foraging ecology, including habitat preference and dietary composition. In this study, stranding location emerged as the primary driver of variability in the skin isotopic values and niche differentiation of pilot whales in this region. The influence of stranding location on isotopic values and niche space is likely a reflection of foraging behavior and habitat use in the months prior to the stranding event. Although sex was a factor in driving isotopic variability, the effects were relatively minor in comparison to the spatial differences linked to stranding locations. This reinforces that habitat selection and foraging strategies are dominant factors influencing isotopic variability and niche partitioning in these pilot whales.
Habitat preference
Variations in δ¹³C values often define differences in foraging strategies and habitat use (Cherel and Hobson, 2007). In our study, δ¹³C values showed a small but significant gradient among the three stranding locations, reflecting spatial variation in foraging habitats of marine predators due to δ¹³C differences at the base of the food web. These δ¹³C differences are influenced by factors such as latitudinal, nearshore/offshore, or pelagic/benthic gradients (Cherel and Hobson, 2007). Nearshore or demersal habitats, which tend to be more enriched in δ¹³C, generally have higher nutrient concentrations and greater productivity than offshore or pelagic habitats (France, 1995; McMahon et al., 2013).
Baseline δ¹³C values of the marine food web can also result from interannual changes in sea surface temperature (SST) and nutrient availability (McMahon et al., 2013). While all three whale strandings occurred in similar months (October and November), the King Island and Maria Island strandings occurred in late 2004, coinciding with rising SSTs resulting from weak to moderate El Niño conditions. In contrast, the Marion Bay stranding occurred late in 2005, when sea surface temperatures had begun to decline after the El Niño event (NOAA, 2006). Elevated SSTs during an El Niño event can reduce primary productivity, leading to more depleted baseline δ¹³C values due to slower carbon fixation rates. This variability in baseline δ¹³C values ascends through the food chain, influencing isotopic composition throughout the food web, including prey and predators (Rodríguez-Malagón et al., 2021). However, the distinct δ¹³C and δ15N values observed between Maria Island and King Island, which both stranded in late 2004, suggests that spatial differences in foraging habitats, rather than any temporal variability were likely the primary driver of isotopic variation.
Spatial variability is further supported by the niche analysis, which revealed little to no overlap between the Marion Bay and Maria Island strandings, as well as the results from our mixing model. These findings suggest that pilot whales from both Marion Bay and King Island strandings primarily foraged in offshore habitats, consistent with their lower δ¹³C values. In contrast, the more enriched δ¹³C values of pilot whales from Maria Island suggest foraging in both nearshore and offshore habitats. This interpretation aligns with the PERMANOVA results, which revealed that the isotopic values differed between all locations. The slightly enriched δ15N values for Maria Island whales further support this dual habitat use, as enriched δ15N baselines are typically associated with inshore and demersal habitats (McMahon et al., 2013).
The differences in isotopic values observed in Tasmanian pilot whales also mirrors findings from New Zealand strandings. Isotopic differences were attributed to variations in primary productivity and baseline isotopic values across habitats rather than dietary shifts (Hinton et al., 2022). These findings suggest that spatially distinct foraging behavior is due to local ecological conditions and plasticity in foraging behavior, demonstrating ecological resilience. Together, these patterns highlight stranding location as a key driver in defining isotope niches and habitat preferences.
Other studies have similarly documented spatial differences in the foraging ecology of long-finned pilot whales based on stomach content analysis. Variability in diet has been documented due to seasonal, annual, or geographical differences (Gannon et al., 1997; Santos et al., 2014). For example, stranded pilot whales in New Zealand waters displayed evidence of both neritic and oceanic diets across different stranding events, though it remains unclear whether these patterns are a result of altered feeding behavior prior to stranding (Beatson et al., 2007a, b; Beatson and O’Shea, 2009).
Our isotopic dietary analysis reflects a preference for foraging in oceanic waters, with pilot whales inhabiting deeper waters off the continental shelf and edge (Gales et al., 1992; Monteiro et al., 2015). However, they exhibit plasticity in their foraging behavior by extending their foraging to nearshore and neritic waters at times, as the dietary pattern of Maria Island pilot whales suggests. Other researchers have also observed that both sub species of long-finned pilot whales, G. melas, can exploit both oceanic and neritic waters (Beatson et al., 2007a, b; Beatson and O’Shea, 2009; Spitz et al., 2011; Mèndez-Fernandez et al., 2012; Goetz et al., 2015; Becker et al., 2021). Isotopic mixing models confirmed a coastal or demersal foraging habitat for pilot whales (G. m. melas) stranded in Northwest Iberia, reinforcing findings from prior stomach content analyses (Monteiro et al., 2015). In contrast, pilot whales (G. m. melas) stranded in Scotland demonstrated a stronger preference for oceanic habitats and prey (Monteiro et al., 2015). Previous research also noted that pilot whales (G. m. edwardii) in the southwestern Atlantic Ocean primarily feed in oceanic waters, though they may sporadically exploit neritic or coastal prey when moving toward shore (Becker et al., 2021). These studies collectively highlight how regional differences in habitat use and prey distribution are key drivers of isotopic variation and differences in the foraging niche of long-finned pilot whale populations.
Sex niche variability
Some sexually dimorphic cetaceans display sex-specific foraging strategies due to differing physiological needs or habitat use (Evans and Hindell, 2004; Mendes et al., 2007; Louis et al., 2021, 2022) but evidence for such differences in pilot whales is less pronounced. In our study, sex played a lesser role than stranding site in explaining isotopic variability. This aligns with other studies that found no broad-scale differences in cephalopod prey selection between sexes. For example, a stomach content analysis of pilot whales from Tasmania, which included some of the same individuals used in this study for isotopic values from Marion Bay and Maria Island, reported no differences in the cephalopod assemblage between sexes. However, larger females generally consumed smaller Ommastrephid squid (Beasley et al., 2019). Similarly, in terms of fatty acid profiles, no difference was observed between male and female pilot whales stranded off southeast Tasmania (Walters, 2005). Similarly, research on G. melas species from the Strait of Gibraltar, the Atlantic, and New Zealand waters reported no significant sex-based differences in δ¹³C and δ15N values (de Stephanis et al., 2008; Monteiro et al., 2015; Hinton et al., 2022). By contrast, male pilot whales in Kerguelen waters exhibited slightly higher skin δ15N values than females (Fontaine et al., 2015), suggesting that finer-scale differences may be detectable between males and females.
Despite the lesser role of sex in isotopic variability in this study, finer-scale differences in isotopic values were observed. Bayesian overlap analysis documented minimal overlap between males and females from the King Island and Maria Island strandings. In contrast, the Marion Bay stranding revealed females were likely to have a larger niche than males, with niches between males and females exhibiting a 70-80% overlap. While the Marion Bay stranding shows significant dietary overlap, we also observed a negative correlation between male δ¹³C and δ15N values and body length. The negative correlation between δ¹³C and body length suggests that larger males may take deeper dives than females while foraging, as δ¹³C values are typically more depleted in offshore waters and at greater depth due to reduced primary productivity and differences in nutrient cycling (Vanderklift and Ponsard, 2003; Cherel and Hobson, 2007). Diving deeper and longer would be consistent with larger individuals having some physiological advantages, such as a greater capacity for oxygen storage for prolonged dives (Schreer et al., 2001; Soto et al., 2008). A relationship between body size and depth has also been noted in the false killer whale (Pseudorca crassidens), where larger individuals exhibit similar depth-foraging behaviors (Riccialdelli and Goodall, 2015). In contrast, the negative correlation between δ15N values and body length may reflect differences in prey selection or foraging strategies, rather than depth alone, as δ15N values of particulate organic matter generally increase with depth (Mintenbeck et al., 2007). We suggest that larger males may be targeting different prey sizes or prey types located deeper in the water column than females. This facilitates reduced intra-group competition by accessing different prey resources, particularly for such a highly social species where shared foraging is common. In contrast, the broader isotopic niche observed in male pilot whales in the King Island and Maria Island strandings might reflect the ability to exploit a more extensive foraging range or adapt to changes in prey availability, providing an ecological advantage in a dynamic marine ecosystem.
Given their highly social nature, pilot whales within the same stranding event are expected to exhibit similar foraging patterns due to their strong cohesive social bonds (de Stephanis et al., 2008; Augusto et al., 2017). Nevertheless, observed variability in isotopic niches of males and females suggests that mass strandings may include multiple social groups (de Stephanis et al., 2008; Oremus et al., 2013), complicating the assessment of their dietary habits. Pilot whales typically form stable, matrilineal groups, social units made up of closely related individuals through maternal lines. These groups support strong kin-based social bonds, shaping the pod social structure and promoting coordinated foraging strategies (Oremus et al., 2013). However, pilot whales also display flexibility in their social organization, sometimes forming large, temporary aggregations that may exploit different foraging zones and depths (de Stephanis et al., 2008; Augusto et al., 2017). This social flexibility may contribute to the variability observed in the isotopic niches between males and females, as dynamic social units and changing foraging niches may result in distinct foraging adaptations among individuals.
Pilot whale dietary composition
Determining whether fish or squid are the major dietary component in the diet of pilot whales is essential for assessing their foraging strategies. Cephalopods are an important dietary component of many cetaceans, as approximately 80% of toothed whale species frequently consume cephalopods (Clarke, 1996; Rodhouse, 2013). Our mixing model suggested that these pilot whales also primarily consumed cephalopods in the months prior to stranding, with fish supplementing their diet to a lesser extent. For example, the model suggests that 15-25% of dietary intake was fish in the Maria Island and Marion Bay strandings. Stomach content analysis from these same strandings in a prior study also corroborates our findings of a preference for cephalopod consumption (Beasley et al., 2019). At Maria Island, 63% of pilot whale stomachs examined contained prey, primarily cephalopods (39%) with a small number of individuals with fish (3%). At Marion Bay, however, 71% of stomachs contained prey, exclusively cephalopods, with no fish recorded.
These findings generally align with our isotopic results, and are further supported by evidence of a cephalopod-dominated diet observed in pilot whales from New Zealand (Beatson et al., 2007a, b; Beatson and O’Shea, 2009), and Chile (Mansilla et al., 2012). However, cephalopod hard parts are preferentially retained in whale stomachs, which may overestimate cephalopod consumption (Bowen and Iverson, 2013). Nevertheless, other dietary tracers, such as fatty acid analysis, also support a cephalopod-dominant diet. Pilot whales from Tasmania exhibited a myctophid signature in their fatty acid profile (Walters, 2005), which was attributed to secondary effects from cephalopods, such as ommastrephids that eat myctophids (Pethybridge et al., 2012, 2013). This myctophid signature was retained in squid and subsequently transferred to its predator. Isotopic studies on pilot whales from the Southwest Atlantic also suggest a cephalopod-dominant diet, although there was a small proportion of fish consumption (Becker et al., 2021). Moreover, in the Kerguelen waters, heavy metal and isotopic analyses suggest a mixed diet of fish and squid, although no quantitative distinction between them was determined (Fontaine et al., 2015).
Off Tasmania, pilot whales are reported to target a diverse cephalopod assemblage. For example, a total of 26 cephalopod species across four strandings has been identified, with a broad range of 4 to 16 taxa recovered per stranding event (Beasley et al., 2019). A diverse cephalopod diet for the long-finned pilot whale in the Northern Hemisphere (G. m. melas) has also been documented (Santos et al., 2014). In contrast, diets in other regions in the Southern Hemisphere such as New Zealand (Beatson et al., 2007a, b; Beatson and O’Shea, 2009) and Chile (Mansilla et al., 2012) are reported to be dominated by three or fewer cephalopod species. Notably, the pilot whales stranded at Maria Island also consumed just three prey species, primarily nearshore, underscoring variability in foraging strategies across different locations.
The primary cephalopod species found in the stomachs of stranded individuals in Tasmania, Ommastrephidae sp., L. lorigera, and A. lesueurii, inhabit a range of marine environments (Beasley et al., 2019). Ommastrephids, which undertake both vertical and horizontal migrations (Gilly et al., 2006; Ji and Guo, 2024) were the main prey of pilot whales in the Marion Bay and King Island strandings and made up a significant proportion of the diet consumed at Maria Island, as well as in other regions (Spitz et al., 2011; Santos et al., 2014; Monteiro et al., 2015). Two important ommastrephid species commonly found in Tasmanian waters are N. gouldi and T.filippovae. N. gouldi is predominantly a shelf to nearshore and even estuarine species (Nowara and Walker, 1998) that aggregates at the bottom during the day but disperses in the water column in the evening (Winstanley et al., 1983). Conversely, T. filippovae is an epipelagic species commonly found in slope waters (Dunning, 1993). L. lorigera and A lesueurii, commonly encountered over slopes, seamounts and submarine ridges, also stay at depth during the day but migrate to the surface at night (Beasley et al., 2019). The deep-diving capabilities of long-finned pilot whales, with records of up to 800 m, facilitate their access to mesopelagic prey that migrate vertically at night (Baird et al., 2002; Heide-Jørgensen et al., 2002). This diving ability enables them to forage on a wide array of prey species across different marine environments.
Despite the worldwide dietary preference of pilot whales for cephalopods, fish also comprise a proportion of their diet in certain regions. This may be driven by the lower energy density of cephalopods in comparison to higher-energy fish, which may lead to supplementation with fish when nutritional needs increase or when cephalopod resources are low (Overholtz and Waring, 1991; Lockyer, 2007; Spitz et al., 2011). This kind of dietary plasticity highlights their ability to modify their foraging behavior in response to the diversity and prevalence of prey, thus reinforcing their role as a cephalopod specialist that exhibits opportunistic foraging when subjected to various ecological pressures (Santos et al., 2014).
Our findings enhance the understanding of the foraging strategies of pilot whales in Tasmania; however, they are accompanied by notable limitations. A lack of species-specific trophic enrichment factors (TEFs) introduces some uncertainty into our isotopic mixing models (Kadye et al., 2020). Species, trophic level, dietary source and tissue type are all critical considerations when selecting suitable TEFs for modeling the study species (Stephens et al., 2023). In closely related species, TEF values are relatively consistent (Giménez et al., 2016; Stephens et al., 2023) As a result, we chose TEFs obtained from bottlenose dolphins (Tursiops truncatus), a closely related species within the Delphinidae family (LeDuc et al., 1999). Both pilot whales and bottlenose dolphins share similar metabolic and physiological traits due to their close taxonomic relationship, and in Tasmanian waters, they occupy comparable trophic levels and consume similar food sources, including squid and fish (Davenport and Bax, 2002). Moreover, the TEFs were experimentally derived from skin, the same tissue analyzed in this study, ensuring compatibility in isotopic assimilation rates (Giménez et al., 2016). Monteiro et al. (2015) further validated the use of TEFs from bottlenose dolphins, demonstrating a similar diet to that of stomach contents examined in pilot whales from the Northeast Atlantic Ocean. By selecting TEFs from a closely related species, we have minimized uncertainty in our estimates of dietary composition, although a species-specific TEF would further refine our analysis.
Similarly, the selection of priors can also influence predictions of dietary contributions, particularly for whales in locations where foraging strategies are more flexible. Overall, aggregated dietary compositions between the informed and generalist models were consistent, as were the individual sources for Marion Bay and King Island. However, Maria Island displayed greater variability at the individual source level, highlighting the importance of using informed priors to reduce uncertainty. More ecologically plausible dietary estimates are achieved when using informed priors that reflect global dietary knowledge of pilot whales.
Additional uncertainties may be due to the exclusion of infrequent but ecologically important prey items in the mixing model, which may introduce error into dietary composition estimates. This is particularly relevant for highly mobile species, such as pilot whales, whose wide-ranging foraging habits can result in a diet that varies significantly across locations and seasons. In addition, data collected from stranded individuals may not accurately reflect natural foraging behavior, as strandings may result in altered prey intake. However, isotopic values of skin integrate multiple feeding events over time (Teixeira et al., 2022) reducing biases associated with stranding events.
Future implications
Our study highlights the foraging plasticity of pilot whales in Tasmania, whereby they can potentially modify their foraging strategy based on the distribution or availability of prey. By confirming a dietary preference for a wide range of cephalopod species, we highlight their crucial ecological role as more generalist feeders when necessary. This ability for pilot whales to forage in both oceanic or offshore waters and nearshore waters increases their resilience to environmental changes and potential fluctuations in prey availability. Moreover, the ability to reduce intra-group competition via niche differentiation likely provides a buffer to pilot whale populations against environmental stressors, thus ensuring stable group dynamics over time. However, environmental changes, such as increasing sea surface temperatures, have resulted in many species of cetaceans demonstrating a poleward shift in their distribution or habitat (van Weelden et al., 2021). Similarly, increased ocean temperatures may also have a deleterious effect on their prey, with cephalopods likely to experience reduced survival rates, shorter development times and increased metabolic stress (Borges et al., 2023). Understanding the foraging ecology of cetaceans, including their interactions with prey, is therefore crucial, as they play an important role as sentinels of environmental change in the marine environment.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because worked on archived material not live animals.
Author contributions
CJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft. RG: Conceptualization, Data curation, Funding acquisition, Investigation, Resources, Writing – review & editing. YC: Conceptualization, Methodology, Writing – review & editing. GJ: Methodology, Resources, Writing – review & editing. PV: Conceptualization, Investigation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Data collection for this project was supported through the Princess Melikoff Trust. The ANZ Holdsworth Wildlife Research Endowment and Winifred Violet Scott Charitable Trust provided funds for the isotopic analysis in this project.
Acknowledgments
We thank the Department of Natural Resources and Environment Tasmania Marine Conservation Program, Hobart and the Tasmanian Museum and Art Gallery for their assistance and support with collection and provision of skin samples from pilot whales and associated biological information. Data collection for this project was supported through the Princess Melikoff Trust for which we are very grateful. We also thank The ANZ Holdsworth Wildlife Research Endowment and Winifred Violet Scott Charitable Trust who provided funds for the isotopic analysis in this project. A special thanks to Isabel Beasley for her assistance with beak ID and measurements, Alan Williams who identified the myctophids, and to Graeme Ewing and Simon Willcox who arranged and obtained some fish specimens for us. We are also grateful to the captain (Brian Cooksley) and crew of the Adriatic Pearl who collected squid specimens for the project. Miriam Honson assisted with some preparation with isotope samples. Vancouver Island University also provided laboratory space when needed.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1520905/full#supplementary-material
References
Abend A. G., Smith T. D. (1999). Review of distribution of the long-finned pilot whale (Globicephala melas) in the North Atlantic and Mediterranean. NOAA technical memorandum NMFS-NE. 117
Augusto J., Frasier T., Whitehead H. (2017). Social structure of long-finned pilot whales (Globicephala melas) off northern Cape Breton Island, Nova Scotia. Behavior 154, 509–540. doi: 10.1163/1568539X-00003432
Baird R. W., Borsani J. F., Hanson M. B., Tyack P. L. (2002). Diving and night-time behavior of long-finned pilot whales in the Ligurian Sea. Mar. Ecol. Prog. Ser. 237, 301–305. doi: 10.3354/meps237301
Baum J. K., Worm B. (2009). Cascading top-down effects of changing oceanic predator abundances. J. Anim. Ecol. 78, 699–714. doi: 10.1111/j.1365-2656.2009.01531.x
Beasley I., Cherel Y., Robinson S., Betty E., Hagihara R., Gales R. (2019). Stomach contents of long-finned pilot whales, Globicephala melas mass-stranded in Tasmania. PloS One 14, e0206747. doi: 10.1371/journal.pone.0206747
Beatson E., O’Shea S. (2009). Stomach contents of long-finned pilot whales, Globicephala melas, mass-stranded on Farewell Spit, Golden Bay in 2005 and 2008. New Z. J. Zool. 36, 47–58. doi: 10.1080/03014220909510139
Beatson E., O’Shea S., Ogle M. (2007a). First report on the stomach contents of long-finned pilot whales, Globicephala melas, stranded in New Zealand. New Z. J. Zool. 34, 51–56. doi: 10.1080/03014220709510063
Beatson E., O’Shea S., Stone C., Shortland T. (2007b). Notes on New Zealand mammals 6. Second report on the stomach contents of long-finned pilot whales, Globicephala melas. New Z. J. Zool. Vol 34, 359–362 doi: 10.1080/03014220709510095
Becker Y. A., Fioramonti N. E., Dellabianca N. A., Riccialdelli L. (2021). Feeding ecology of the long finned pilot whale, Globicephala melas edwardii, in the southwestern Atlantic Ocean, determined by stable isotopes analysis. Polar Biol. 44, 1655–1667. doi: 10.1007/s00300-021-02908-2
Berry T. E., Osterrieder S. K., Murray D. C., Coghlan M. L., Richardson A. J., Grealy A. K., et al. (2017). DNA metabarcoding for diet analysis and biodiversity: A case study using the endangered Australian sea lion (Neophoca cinerea). Ecol. Evol. 7, 5435–5453. doi: 10.1002/ece3.2017.7.issue-14
Bloch D. (1993). Age and growth parameters of the long-finned pilot whale off the Faroe Islands. Rep. Int. Whaling Commission Special Issue 14, 163–207.
Borges F. O., Sampaio E., Santos C. P., Rosa R. (2023). Climate-change impacts on Cephalopods: A meta-analysis. Integr. Comp. Biol. 63, 1240–1265. doi: 10.1093/icb/icad102
Borrell A., Gazo M., Aguilar A., Raga J. A., Degollada E., Gozalbes P., et al. (2021). Niche partitioning amongst northwestern Mediterranean cetaceans using stable isotopes. Prog. Oceanogr. 193, 102559. doi: 10.1016/j.pocean.2021.102559
Bowen W., Iverson S. (2013). Methods of estimating marine mammal diets: a review of validation experiments and sources of bias and uncertainty. Mar. Mammal Sci. 29, 719–754. doi: 10.1111/j.1748-7692.2012.00604.x
Browning N. E., Dold C., I-Fan J., Worthy G. A. (2014). Isotope turnover rates and diet–tissue discrimination in skin of ex situ bottlenose dolphins (Tursiops truncatus). J. Exp. Biol. 217, 214–221. doi: 10.1242/jeb.093963
Cherel Y., Fontaine C., Jackson G. D., Jackson C. H., Richard P. (2009). Tissue, ontogenic and sex-related differences in δ¹³C and δ15N values of the oceanic squid Todarodes filippovae (Cephalopoda: Ommastrephidae). Mar. Biol. 156:699–708. doi: 10.1007/s00227-008-1121-x
Cherel Y., Hobson K. A. (2005). Stable isotopes, beaks and predators: a new tool to study the trophic ecology of cephalopods, including giant and colossal squids. Proc. R. Soc. B: Biol. Sci. 272, 1601–1607. doi: 10.1098/rspb.2005.3115
Cherel Y., Hobson K. A. (2007). Geographical variation in carbon stable isotope signatures of marine predators: a tool to investigate their foraging areas in the Southern Ocean. Mar. Ecol. Prog. Ser. 329, 281–287. doi: 10.3354/meps329281
Chouvelon T., Caurant F., Cherel Y., Simon-Bouhet B., Spitz J., Bustamante P. (2014). Species-and size-related patterns in stable isotopes and mercury concentrations in fish help refine marine ecosystem indicators and provide evidence for distinct management units for hake in the Northeast Atlantic. ICES J. Mar. Sci. 71, 1073–1087. doi: 10.1093/icesjms/fst199
Clarke M. R. (1996). Cephalopods as prey. III. Cetaceans. Philos. Trans. R. Soc. London Ser. B: Biol. Sci. 351, 1053–1065. doi: 10.1098/rstb.1996.0093
Couturier L. I., Michel L. N., Amaro T., Budge S. M., Da Costa E., De Troch M., et al. (2020). State of art and best practices for fatty acid analysis in aquatic sciences. ICES J. Mar. Sci. 77, 2375–2395. doi: 10.1093/icesjms/fsaa121
Davenport S. R., Bax N. J. (2002). A trophic study of a marine ecosystem off southeastern Australia using stable isotopes of carbon and nitrogen. Can. J. Fisheries Aquat. Sci. 59, 514–530. doi: 10.1139/f02-031
DeNiro M. J., Epstein S. (1981). Influence of diet on the distribution of nitrogen isotopes in animals. Geochimica cosmochimica Acta 45, 341–351. doi: 10.1016/0016-7037(81)90244-1
de Stephanis R., García-Tíscar S., Verborgh P., Esteban-Pavo R., Pérez S., Minvielle-Sebastia L., et al. (2008). Diet of the social groups of long-finned pilot whales (Globicephala melas) in the Strait of Gibraltar. Mar. Biol. 154, 603–612. doi: 10.1007/s00227-008-0953-8
dos Santos R. A., Haimovici M. (2001). Cephalopods in the diet of marine mammals stranded or incidentally caught along southeastern and southern Brazil (21–34 S). Fisheries Res. 52, 99–112. doi: 10.1016/S0165-7836(01)00234-X
Dunning M. M. (1993). “Summer populations of Ommastrephes bartramii (Lesueur 1821) and Todarodes filippovae Adam 1975 (Cephalopoda: Ommastrephidae) from the Tasman Sea,” in Recent advances in cephalopod fisheries biology. Ed. Okutani T. (Tokyo: Tokai University Press), 97–118.
Estes J. A., Heithaus M., McCauley D. J., Rasher D. B., Worm B. (2016). Megafaunal impacts on structure and function of ocean ecosystems. Annu. Rev. Environ. Resour. 41, 83–116. doi: 10.1146/annurev-environ-110615-085622
Estes J. A., Terborgh J., Brashares J. S., Power M. E., Berger J., Bond W. J., et al. (2011). Trophic downgrading of planet Earth. science 333, 301–306. doi: 10.1126/science.1205106
Evans K., Hindell M. A. (2004). The diet of sperm whales (Physeter macrocephalus) in southern Australian waters. ICES J. Mar. Sci. 61, 1313–1329. doi: 10.1016/j.icesjms.2004.07.026
Fontaine M., Carravieri A., Simon-Bouhet B., Bustamante P., Gasco N., Bailleul F., et al. (2015). Ecological tracers and at-sea observations document the foraging ecology of southern long-finned pilot whales (Globicephala melas edwardii) in Kerguelen waters. Mar. Biol. 162, 207–219. doi: 10.1007/s00227-014-2587-3
France R. (1995). Carbon-13 enrichment in benthic compared to planktonic algae: foodweb implications. Mar. Ecol. Prog. Ser. 124, 307–312. doi: 10.3354/meps124307
Gales R., Pemberton D., Clarke M., Lu C. (1992). Stomach contents of long-finned pilot whales (Globicephala melas) and bottlenose dolphins (Tursiops truncatus) in Tasmania. Mar. Mammal Sci. 8, 405–413. doi: 10.1111/j.1748-7692.1992.tb00055.x
Gannon D. P., Read A. J., Craddock J. E., Fristrup K. M., Nicolas J. R. (1997). Feeding ecology of long-finned pilot whales Globicephala melas in the western North Atlantic. Mar. Ecol. Prog. Ser. 148, 1–10. doi: 10.3354/meps148001
García-Cegarra A., Castro C., Van Waerebeek K. (2021). Feeding of humpback whales in low latitudes of the Southeast Pacific Ocean. Neotropical Biodiversity 7, 421–430. doi: 10.1080/23766808.2021.1971041
García-Vernet R., Borrell A., Víkingsson G., Halldórsson S. D., Aguilar A. (2021). Ecological niche partitioning between baleen whales inhabiting Icelandic waters. Prog. Oceanogr. 199, 102690. doi: 10.1016/j.pocean.2021.102690
Geraci J. R., Lounsbury V. J. (2005). Marine mammals ashore: a field guide for strandings (National Aquarium in Baltimore).
Gilly W. F., Markaida U., Baxter C., Block B., Boustany A., Zeidberg L., et al. (2006). Vertical and horizontal migrations by the jumbo squid Dosidicus gigas revealed by electronic tagging. Mar. Ecol. Prog. Ser. 324, 1–17. doi: 10.3354/meps324001
Giménez J., Ramírez F., Almunia J., Forero M. G., de Stephanis R. (2016). From the pool to the sea: Applicable isotope turnover rates and diet to skin discrimination factors for bottlenose dolphins (Tursiops truncatus). J. Exp. Mar. Biol. Ecol. 475, 54–61. doi: 10.1016/j.jembe.2015.11.001
Goetz S., Read F. L., Ferreira M., Portela J. M., Santos M. B., Vingada J., et al. (2015). Cetacean occurrence, habitat preferences and potential for cetacean–fishery interactions in Iberian Atlantic waters: results from cooperative research involving local stakeholders. Aquat. Conservation: Mar. Freshw. Ecosyst. 25, 138–154. doi: 10.1002/aqc.2481
Groß J., Virtue P., Nichols P. D., Eisenmann P., Waugh C. A., Bengtson Nash S. (2020). Interannual variability in the lipid and fatty acid profiles of east Australia-migrating humpback whales (Megaptera novaeangliae) across a 10-year timeline. Sci. Rep. 10, 18274. doi: 10.1038/s41598-020-75370-5
Hazen E. L., Abrahms B., Brodie S., Carroll G., Jacox M. G., Savoca M. S., et al. (2019). Marine top predators as climate and ecosystem sentinels. Front. Ecol. Environ. 17, 565–574. doi: 10.1002/fee.v17.10
Heide-Jørgensen M. P., Bloch D., Stefansson E., Mikkelsen B., Helen Ofstad L., Dietz R. (2002). Diving behavior of long-finned pilot whales Globicephala melas around the Faroe Islands. Wildlife Biol. 8, 307–313.
Hinton B., Stockin K. A., Bury S. J., Peters K. J., Betty E. L. (2022). Isotopic niche analysis of long-finned pilot whales (Globicephala melas edwardii) in Aotearoa New Zealand waters. Biology 11, 1414. doi: 10.3390/biology11101414
Jackson A. L., Inger R., Parnell A. C., Bearhop S. (2011). Comparing isotopic niche widths among and within communities: SIBER–Stable Isotope Bayesian Ellipses in R. J. Anim. Ecol. 80, 595–602. doi: 10.1111/j.1365-2656.2011.01806.x
Ji F., Guo X. (2024). A new way to understand migration routes of oceanic squid (Ommastrephidae) from satellite data. Remote Sens. Ecol. Conserv. 10, 248–263. doi: 10.1002/rse2.v10.2
Kadye W. T., Redelinghuys S., Parnell A. C., Booth A. J. (2020). Exploring source differences on diet-tissue discrimination factors in the analysis of stable isotope mixing models. Sci. Rep. 10, 15816. doi: 10.1038/s41598-020-73019-x
Kiszka J. J., Caputo M., Méndez-Fernandez P., Fielding R. (2021). Feeding ecology of elusive Caribbean killer whales inferred from Bayesian stable isotope mixing models and whalers’ ecological knowledge. Front. Mar. Sci. 8, 648421. doi: 10.3389/fmars.2021.648421
Kurle C. M., Worthy G. A. (2002). Stable nitrogen and carbon isotope ratios in multiple tissues of the northern fur seal Callorhinus ursinus: implications for dietary and migratory reconstructions. Mar. Ecol. Prog. Ser. 236, 289–300. doi: 10.3354/meps236289
Lebon M., Colaço A., Prieto R., Cascão I., Oliveira C., Tobeña M., et al. (2024). Isotopic niches reveal the trophic structure of the cetacean community in the oceanic waters around the Azores. Front. Mar. Sci. 11, 1283357. doi: 10.3389/fmars.2024.1283357
LeDuc R., Perrin W., Dizon A. (1999). Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Mar. mammal Sci. 15, 619–648. doi: 10.1111/j.1748-7692.1999.tb00833.x
Lockyer C. (2007). All creatures great and smaller: a study in cetacean life history energetics. J. Mar. Biol. Assoc. United Kingdom 87, 1035–1045. doi: 10.1017/S0025315407054720
Louis M., Routledge J., Heide-Jørgensen M. P., Szpak P., Lorenzen E. D. (2022). Sex and size matter: foraging ecology of offshore harbor porpoises in waters around Greenland. Mar. Biol. 169, 140. doi: 10.1007/s00227-022-04123-x
Louis M., Skovrind M., Garde E., Heide-Jørgensen M. P., Szpak P., Lorenzen E. D. (2021). Population-specific sex and size variation in long-term foraging ecology of belugas and narwhals. R. Soc. Open Sci. 8, 202226. doi: 10.1098/rsos.202226
Machovsky-Capuska G. E., Raubenheimer D. (2020). The nutritional ecology of marine apex predators. Annu. Rev. Mar. Sci. 12, 361–387. doi: 10.1146/annurev-marine-010318-095411
MacLeod C. D., Santos M., Pierce G. J. (2003). Review of data on diets of beaked whales: evidence of niche separation and geographic segregation. J. Mar. Biol. Assoc. United Kingdom 83, 651–665. doi: 10.1017/S0025315403007616h
Mahaffy S. D., Baird R. W., McSweeney D. J., Webster D. L., Schorr G. S. (2015). High site fidelity, strong associations, and long-term bonds: Short-finned pilot whales off the island of Hawai ‘i. Mar. Mammal Sci. 31, 1427–1451. doi: 10.1111/mms.2015.31.issue-4
Mansilla L., Olavarría C., Vega M. A. (2012). Stomach contents of long-finned pilot whales (Globicephala melas) from southern Chile. Polar Biol. 35, 1929–1933. doi: 10.1007/s00300-012-1222-3
McCluskey S., Sprogis K., London J., Bejder L., Loneragan N. (2021). Foraging preferences of an apex marine predator revealed through stomach content and stable isotope analyses. Glob Ecol. Conserv. 25, e01396. doi: 10.1016/j.gecco.2020.e01396
McMahon K. W., Hamady L. L., Thorrold S. R. (2013). A review of ecogeochemistry approaches to estimating movements of marine animals. Limnol. oceanogr. 58, 697–714. doi: 10.4319/lo.2013.58.2.0697
Mendes S., Newton J., Reid R. J., Frantzis A., Pierce G. J. (2007). Stable isotope profiles in sperm whale teeth: variations between areas and sexes. J. Mar. Biol. Assoc. United Kingdom 87, 621–627. doi: 10.1017/S0025315407056019
Mèndez-Fernandez P., Bustamante P., Bode A., Chouvelon T., Ferreira M., Lopez A., et al. (2012). Foraging ecology of five toothed whale species in the Northwest Iberian Peninsula, inferred using carbon and nitrogen isotope ratios. J. Exp. Mar. Biol. Ecol. 413, 150–158. doi: 10.1016/j.jembe.2011.12.007
Minagawa M., Wada E. (1984). Stepwise enrichment of 15N along food chains: further evidence and the relation between δ15N and animal age. Geochimica cosmochimica Acta 48, 1135–1140. doi: 10.1016/0016-7037(84)90204-7
Mintenbeck K., Jacob U., Knust R., Arntz W., Brey T. (2007). Depth-dependence in stable isotope ratio δ15N of benthic POM consumers: the role of particle dynamics and organism trophic guild. Deep Sea Res. Part I: Oceanographic Res. Papers 54, 1015–1023. doi: 10.1016/j.dsr.2007.03.005
Monteiro S., Ferreira M., Vingada J. V., López A., Brownlow A., Méndez-Fernandez P. (2015). Application of stable isotopes to assess the feeding ecology of long-finned pilot whale (Globicephala melas) in the Northeast Atlantic Ocean. J. Exp. Mar. Biol. Ecol. 465, 56–63. doi: 10.1016/j.jembe.2015.01.007
Newsome S. D., Clementz M. T., Koch P. L. (2010). Using stable isotope biogeochemistry to study marine mammal ecology. Mar. Mammal Sci. 26, 509–572. doi: 10.1111/j.1748-7692.2009.00354.x
NOAA (2006). El Niño/Southern Oscillation for Annual 2005, published online January 2006, retrieved on January 13, 2025. Available online at: https://www.ncei.noaa.gov/access/monitoring/monthly-report/enso/200513 (Accessed January 13, 2025).
Nowara G. B., Walker T. I. (1998). Effects of time of solar day, jigging method and jigging depth on catch rates and size of Gould’s squid, Nototodarus gouldi (McCoy), in southeastern Australian waters. Fisheries Res. 34, 279–288. doi: 10.1016/S0165-7836(97)00095-7
Ogilvy C., Constantine R., Bury S. J., Carroll E. L. (2022). Diet variation in a critically endangered marine predator revealed with stable isotope analysis. R. Soc. Open Sci. 9, 220470. doi: 10.1098/rsos.220470
Olson P. A. (2018). “Pilot Whales: Globicephala melas and G. macrorhynchus,” in Encyclopedia of marine mammals, 3rd ed. (Elsevier), 701–705.
Oremus M., Gales R., Dalebout M. L., Funahashi N., Endo T., Kage T., et al. (2009). Worldwide mitochondrial DNA diversity and phylogeography of pilot whales (Globicephala spp.). Biol. J. Linn. Soc. 98, 729–744. doi: 10.1111/j.1095-8312.2009.01325.x
Oremus M., Gales R., Kettles H., Baker C. S. (2013). Genetic evidence of multiple matrilines and spatial disruption of kinship bonds in mass strandings of long-finned pilot whales, Globicephala melas. J. Heredity 104, 301–311. doi: 10.1093/jhered/est007
Overholtz W. J., Waring G. T. (1991). Diet composition of pilot whales Globicephala sp. and common dolphins Delphinus delphis in the Mid-Atlantic Bight during spring 1989. Fishery Bull. 89, 723–728.
Pethybridge H. R., Nichols P. D., Virtue P., Jackson G. D. (2013). The foraging ecology of an oceanic squid, Todarodes filippovae: The use of signature lipid profiling to monitor ecosystem change. Deep Sea Res. Part II: Topical Stud. Oceanogr. 95, 119–128. doi: 10.1016/j.dsr2.2012.07.025
Pethybridge H., Virtue P., Casper R., Yoshida T., Green C., Jackson G., et al. (2012). Seasonal variations in diet of arrow squid (Nototodarus gouldi): stomach content and signature fatty acid analysis. J. Mar. Biol. Assoc. United Kingdom 92, 187–196. doi: 10.1017/S0025315411000841
Pierce G. J., Santos M. B., Learmonth J., Mente E., Stowasser G. (2004). “Methods for dietary studies on marine mammals,” in Investigating the roles of cetaceans in marine ecosystems (The Mediterranean Science Commission, CIESM Workshop Monographs), 29–36.
Ramos R., González-Solís J. (2012). Trace me if you can: the use of intrinsic biogeochemical markers in marine top predators. Front. Ecol. Environ. 10, 258–266. doi: 10.1890/110140
Reidy R. D., Lemay M. A., Innes K. G., Clemente-Carvalho R. B., Janusson C., Dower J. F., et al. (2022). Fine-scale diversity of prey detected in humpback whale feces. Ecol. Evol. 12, e9680. doi: 10.1002/ece3.v12.12
Remili A., Dietz R., Sonne C., Samarra F. I., Rikardsen A. H., Kettemer L. E., et al. (2023). Quantitative fatty acid signature analysis reveals a high level of dietary specialization in killer whales across the North Atlantic. J. Anim. Ecol. 92, 1216–1229. doi: 10.1111/1365-2656.13920
Riccialdelli L., Goodall N. (2015). Intra-specific trophic variation in false killer whales (Pseudorca crassidens) from the southwestern South Atlantic Ocean through stable isotopes analysis. Mamm. Biol. 80, 298–302. doi: 10.1016/j.mambio.2015.01.003
Rodhouse P. G. (2013). Role of squid in the Southern Ocean pelagic ecosystem and the possible consequences of climate change. Deep Sea Res. Part II: Topical Stud. Oceanogr. 95, 129–138. doi: 10.1016/j.dsr2.2012.07.001
Rodríguez-Malagón M. A., Speakman C. N., Sutton G. J., Angel L. P., Arnould J. P. (2021). Temporal and spatial isotopic variability of marine prey species in south-eastern Australia: Potential implications for predator diet studies. PloS One 16, e0259961. doi: 10.1371/journal.pone.0259961
Roman J., Estes J. A., Morissette L., Smith C., Costa D., McCarthy J., et al. (2014). Whales as marine ecosystem engineers. Front. Ecol. Environ. 12, 377–385. doi: 10.1890/130220
Santos M. B., Monteiro S. S., Vingada J. V., Ferreira M., López A., Martínez Cedeira J. A., et al. (2014). Patterns and trends in the diet of long-finned pilot whales (Globicephala melas) in the northeast Atlantic. Mar. Mammal Sci. 30, 1–19. doi: 10.1111/mms.2013.30.issue-1
Schreer J. F., Kovacs K. M., O’Hara Hines R. (2001). Comparative diving patterns of pinnipeds and seabirds. Ecol. Monogr. 71, 137–162. doi: 10.1890/0012-9615(2001)071[0137:CDPOPA]2.0.CO;2
Soto N. A., Johnson M. P., Madsen P. T., Díaz F., Domínguez I., Brito A., et al. (2008). Cheetahs of the deep sea: deep foraging sprints in short-finned pilot whales off Tenerife (Canary Islands). J. Anim. Ecol. 77, 936–947. doi: 10.1111/j.1365-2656.2008.01393.x
Spitz J., Cherel Y., Bertin S., Kiszka J., Dewez A., Ridoux V. (2011). Prey preferences among the community of deep-diving odontocetes from the Bay of Biscay, Northeast Atlantic. Deep Sea Res. Part I: Oceanographic Res. Papers 58, 273–282. doi: 10.1016/j.dsr.2010.12.009
Stephens R. B., Shipley O. N., Moll R. J. (2023). Meta-analysis and critical review of trophic discrimination factors (Δ13C and Δ15N): Importance of tissue, trophic level and diet source. Funct. Ecol. 37, 2535–2548. doi: 10.1111/1365-2435.14403
Stock B. C., Semmens B. X. (2016). Unifying error structures in commonly used biotracer mixing models. Ecology 97, 2562–2569. doi: 10.1002/ecy.2016.97.issue-10
Strong D. R., Frank K. T. (2010). Human involvement in food webs. Annu. Rev. Environ. Resour. 35, 1–23. doi: 10.1146/annurev-environ-031809-133103
Teixeira C. R., Troina G. C., Daura-Jorge F. G., Simões-Lopes P. C., Botta S. (2022). A practical guide on stable isotope analysis for cetacean research. Mar. Mammal Sci. 38, 1200–1228. doi: 10.1111/mms.12911
Vanderklift M. A., Ponsard S. (2003). Sources of variation in consumer-diet δ 15 N enrichment: a meta-analysis. Oecologia 136, 169–182. doi: 10.1007/s00442-003-1270-z
van Weelden C., Towers J. R., Bosker T. (2021). Impacts of climate change on cetacean distribution, habitat and migration. Climate Change Ecol. 1, 100009. doi: 10.1016/j.ecochg.2021.100009
Walters A. (2005). Long-finned pilot whale (Globicephala melas): tissue lipid profiles (Hobart: University of Tasmania).
Williamson M. J., ten Doeschate M. T., Deaville R., Brownlow A. C., Taylor N. L. (2021). Cetaceans as sentinels for informing climate change policy in UK waters. Mar. Policy 131, 104634. doi: 10.1016/j.marpol.2021.104634
Keywords: cephalopods, cetacean, diet, isotopes, MixSIAR
Citation: Jackson CH, Gales R, Cherel Y, Jackson GD and Virtue P (2025) Trophic niche differentiation and foraging plasticity of long-finned pilot whales (Globicephala melas edwardii) in Tasmanian waters: insights from isotopic analysis. Front. Mar. Sci. 12:1520905. doi: 10.3389/fmars.2025.1520905
Received: 31 October 2024; Accepted: 20 January 2025;
Published: 06 February 2025.
Edited by:
Lorenzo Zane, University of Padua, ItalyCopyright © 2025 Jackson, Gales, Cherel, Jackson and Virtue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine H. Jackson, Y2hyaXN0aW5lamFja3NvbkBsbHUuZWR1
 Christine H. Jackson
Christine H. Jackson Rosemary Gales2
Rosemary Gales2 Yves Cherel
Yves Cherel George D. Jackson
George D. Jackson Patti Virtue
Patti Virtue