
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 25 March 2025
Sec. Marine Biology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1520773
This article is part of the Research Topic Physiological Processes, Disease Progression, Behavior Change and Population Dynamics of Fish in Response to External Environmental Stresses View all 3 articles
 Pengcheng Hu1,2
Pengcheng Hu1,2 Zilai Yang1,2
Zilai Yang1,2 Min Yang1,2
Min Yang1,2 Piao Sheng1,2
Piao Sheng1,2 Zhongyuan Huang1,2
Zhongyuan Huang1,2 Shuai Liu1,3
Shuai Liu1,3 Yushun Chen1,2
Yushun Chen1,2 Hong Cao1,2*
Hong Cao1,2* Xin Gao1,2*
Xin Gao1,2*Introduction: Different ecotypes exhibit distinct life histories and occupy various environmental conditions for local adaptation. Coilia nasus, an anadromous fish, resides in offshore waters of the Pacific Northwest and reproduces in connected rivers and lakes, including the middle-lower Yangtze River. Populations of C. nasus have differentiated into two ecotypes: migratory and landlocked.
Methods: This study examined the spatiotemporal distributions of these ecotypes and the environmental conditions of their habitats by analyzing environmental DNA collected from Poyang Lake and Tai Lake during 2020–2021.
Results: A total of 157 eDNA samples were obtained, with 62 yielding positive results, including 23 from Poyang Lake and 39 from Tai Lake. The migratory population exhibited seasonal movements in Poyang Lake, entering the Yangtze River-Poyang Lake channel in spring, migrating southward during summer, and returning to the channel in autumn. In contrast, the spatial distribution of the landlocked population remained relatively stable throughout the seasons. Partial least squares structural equation modeling (PLS-SEM) analysis indicated that the distribution patterns of the migratory population were not significantly associated with environmental factors. In contrast, the distribution of the landlocked population was significantly affected by lower water turbidity and reduced conductivity.
Discussion: The findings suggest that temporal variations in the spatial distribution of C. nasus in Poyang Lake primarily reflect its migratory reproductive life history. Conversely, the distribution of the landlocked C. nasus population resembles that of resident fish populations and is mainly influenced by a preference for higher water quality. The results imply that differences in life histories and adaptation to local environmental conditions may have contributed to the formation of the two ecotypes. This study also shows the effectiveness of eDNA technology in assessing fish population distribution and dynamics.
An ecotype is defined as “the product arising as a result of the genotypical response of an ecospecies to a particular habitat” (Turesson, 1922; Gregor, 1944). Ecotypic differentiation represents an early stage in the speciation process, providing important insights into adaptation mechanisms, speciation, and the maintenance of biodiversity and ecosystem functions (Seliskar et al., 2002; Todesco et al., 2020; Schwarzer and Joshi, 2017; Khalil et al., 2017; Faria and Plastino, 2022). Ecotypes develop distinct traits that enhance survival and reproductive success, allowing adaptation to specific environmental conditions (Martín-Forés et al., 2017; Stinson et al., 2017). These variations may be reflected in morphology, physiology, behavior, and population dynamics (Eriksen and Nordal, 1989; Lu and Bernatchez, 1999; Kristensen et al., 2021; Miller et al., 2011) and are largely shaped by natural selection acting on heritable traits (Thorpe et al., 2005; Sakaguchi et al., 2018). Despite considerable variation, ecotypes are not classified as separate species due to the absence of complete genetic differentiation (Hufford and Mazer, 2003; Curran et al., 2022). Differences in life history traits also contribute to ecotypic differentiation by influencing reproductive strategies, growth rates, and behavioral adaptations (Olischläger et al., 2016; May et al., 2023; Silvestro et al., 2020). However, this perspective remains insufficiently explored, with limited research and empirical evidence available to support it.
Coilia nasus, an anadromous fish, inhabits offshore waters in the Northwestern Pacific and spawns in connected rivers and lakes, including the middle-lower Yangtze River (Froese and Pauly, 2014; Ma et al., 2020). These fish form aggregations and migrate upstream toward the Yangtze River beginning in mid-February, with breeding occurring in river channels and lakes from April to July each year (Yuan, 1988; Duan et al., 2012). Historically, it was a significant commercial fish species in the middle-lower Yangtze River, with annual fisheries producing thousands of tons (Zhang et al., 2005). However, overfishing and habitat degradation in recent decades have caused a severe decline in population abundance (Duan et al., 2012). Following glacial movements during the late Pleistocene, the ancestral C. nasus populations underwent adaptive differentiation, leading to the formation of two distinct ecotypes: the migratory and the landlocked populations (Li et al., 2022; Liu et al., 2024). The migratory populations maintain a migratory life history pattern between the sea and the Yangtze River, whereas the landlocked populations complete their entire life cycle in freshwater environments. Despite notable morphological and ecological differences, genetic divergence between these ecotypes has not yet reached the subspecies level (Xu et al., 2020). Evidence from mitochondrial genetic markers and protein-coding genes (PCGs) suggests that geographical isolation caused by Pleistocene glaciation contributed to the initial divergence of C. nasus populations in the middle-lower Yangtze River (Gao et al., 2020; Liu et al., 2024). Additionally, functional analyses of mitochondrial genomes indicate that the complex environmental conditions encountered during migration facilitated adaptive divergence (Xu et al., 2020). These findings suggest that differences in life history traits may promote further divergence between the two ecotypes.
Compared to the high time and labor costs associated with traditional methods, the noninvasive environmental DNA (eDNA) approach has gained widespread adoption due to its high sensitivity, ease of operation, and reduced resource demands (Sengupta et al., 2019; McColl-Gausden et al., 2020). For instance, eDNA enables highly sensitive monitoring of Asian carp population distribution and invasion range (Jerde et al., 2013) and provides a broader geographical perspective on marine fish community diversity compared to visual surveys (Valdivia-Carrillo et al., 2021). Since the heterogeneous distribution of fish eDNA in water corresponds to actual spatial fish population distributions, researchers frequently utilize quantitative polymerase chain reaction (qPCR) technology with species-specific primers to quantitatively analyze target fish distribution and environmental variations in aquatic ecosystems (Maruyama et al., 2014; Yamamoto et al., 2016). Although qPCR effectively assesses fish biomass, its accuracy is highly dependent on primer specificity, which, if not carefully designed, can result in false-positive detections (Bohmann et al., 2014). Thus, the development and application of highly specific primers are essential for ensuring the reliability of eDNA-based research methodologies.
In the present study, a specific primer for C. nasus was designed and developed. The spatiotemporal distribution patterns of the two C. nasus ecotypes, which inhabit Poyang Lake and Tai Lake separately, were analyzed using the eDNA method to examine the effects of life history and adaptation to environmental conditions on ecotypic differentiation. The findings contribute to narrowing the knowledge gap regarding ecotype formation. Furthermore, this study enhances the development and practical application of the eDNA method in fish population assessments.
Poyang Lake (115°46’–116°45’E, 28°22’–29°45’N), located on northern Jiangxi Province, is the largest freshwater lake in the middle and lower reaches of the Yangtze River. The lake undergoes significant seasonal fluctuations in water surface area due to periodic variations in the flow of the Yangtze River’s main stream and its tributaries. During the wet season (April to September), the lake expands to about 3,900 km2, gradually shrinking to 2,792 km2 in the normal season. Between October and March of the following year, the water surface area contracts dramatically to about 1,000 km2 (Dai et al., 2015; Han et al., 2015). These abundant water resources and dynamic hydrological conditions make Poyang Lake an essential habitat for diverse fish species, particularly migratory fish in the Yangtze River basin. Tai Lake (119°54’–120°36’E, 30°56’–31°54’N), situated in southern Jiangsu Province, is the largest landlocked shallow lake in the region. Maintaining a consistent water surface area of about 2,336 km2 with stable water levels throughout the year, Tai Lake has an average depth of about 2 m, with a maximum depth not exceeding 3 m (Qin, 2015). Unlike Poyang Lake, Tai Lake is not hydrologically connected to the Yangtze River. The construction of locks and sluices in the 1970s severed its tributary connections, resulting in a closed aquatic ecosystem primarily dominated by sedentary fish species (Mao et al., 2011).
In this study, environmental DNA was seasonally sampled at 22 sites in Poyang Lake and 19 sites in Tai Lake from April 2020 to January 2021 (April-May 2020 for spring, July-August 2020 for summer, October 2020 for autumn, and December 2020-January 2021 for winter) (Figure 1). Sampling was not conducted at site P22 in spring, site P14 in summer, and sites P12, P13, and P21 in winter due to significant water level fluctuations and adverse weather conditions in Poyang Lake. Similarly, sampling at site T15 in Tai Lake was not performed in autumn and winter. The sites where sampling was not conducted were excluded in subsequent analyses. To examine the spatiotemporal distribution, sampling sites in both Poyang Lake and Tai Lake were categorized into three regions: northern, middle, and southern (Figure 1).
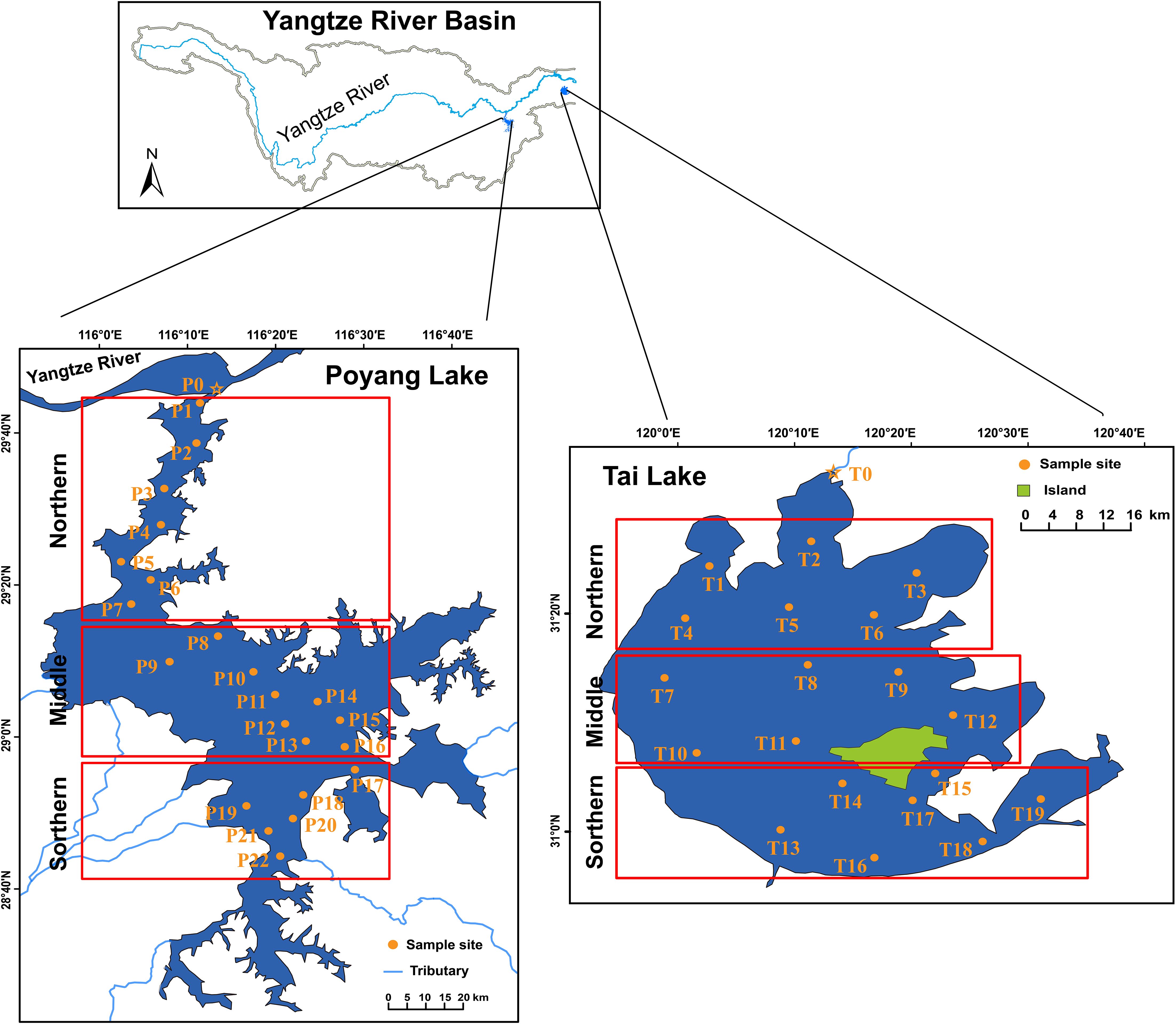
Figure 1. Sampling sites in Poyang Lake and Tai Lake. The P0 site in Poyang Lake and the T0 site in Tai Lake were designated as the starting points closest to the Yangtze River, respectively.
The geographical coordinates of each sampling site were recorded in the field using a Global Positioning System (GPS, Garmin Legend, Garmin, USA) (Supplementary Table S1). In this study, two distance variables were utilized to assess the influence of geographic factors on the spatial and temporal distribution of C. nasus ecotypes. The first variable was the straight-line distance (SD, km) from the starting point to each sampling site. Site P0 in Poyang Lake and site T0 in Tai Lake were designated as the starting points, as they are closest to the Yangtze River (Figure 1). Due to the change in topography at site P5 in Poyang Lake, the distance values for sites P6 to P22 were calculated as the sum of the straight-line distance from P0 to P5 and the straight-line distance from P5 to each subsequent site (P6–P22). The second variable was the average distance (AD, km), representing the mean distance between a given sampling site and all other sites.
The water environmental variables measured in situ included water transparency (TR, cm), water depth (WD, m), water temperature (WT, °C), dissolved oxygen (DO, mg/L), pH, and conductivity (COND, μS/cm). TR was determined using a Secchi disc, while WD was measured with a depth sounder. WT, DO, pH, and COND were recorded using the YSI ProPlus multiparameter. Additionally, except for site P22 in Poyang Lake, water samples were collected at each site and transported to the laboratory at the Institute of Hydrobiology for further analysis. Other water quality parameters, including alkalinity (ALK, mg/L), total hardness (TH, mg/L), total suspended solids (TSS, mg/L), turbidity (TUB, NTU), chloride (CL, mg/L), chemical oxygen demand (COD, mg/L), ammonia (NH4-N, mg/L), nitrite (NO2-N, mg/L), nitrate (NO3-N, mg/L), orthophosphate (PO4-P, mg/L), total phosphorus (TP, mg/L), total nitrogen (TN, mg/L), and chlorophyll a (Chla, μg/L), were analyzed in the laboratory following the methods described by Qu et al. (2020). All in-situ measurement instruments, including the depth sounder and YSI ProPlus multiparameter, were calibrated prior to each survey to ensure accuracy.
The environmental variables were classified into four categories: geographic, physical, chemical, and biological factors. Geographic factors (GF) included the straight-line distance (SD) and the average distance between a site and all other sites (AD). Physical factors (PF) encompassed ALK, COND, TH, TR, TUB, TSS, WT, and WD, representing the physical characteristics of fish habitats. Chemical factors (CF) comprised DO, pH, CL, COD, NH4-N, NO2-N, NO3-N, PO4-P, TN, and TP, which served as chemical characteristic indicators. Chlorophyll a (Chla) was considered a biological factor (BF) reflecting algae abundance.
A specific primer pair, Ca1R/F, was developed based on mitochondrial cytochrome c oxidase subunit I (COI) sequences of C. nasus using the NCBI Primer-BLAST tool to enhance the accuracy of environmental DNA (eDNA) detection (Ye et al., 2012; Duarte et al., 2023). The forward primer was designed as Ca1F (5’-TATCTCCCACGTCGTAGCCT-3’) and the reverse primer as Ca1R (5’-ACTTTAACGCCCGTTGGGAT-3’). This primer pair was 20 bp in length, targeting a 201 bp PCR product. Initial primer specificity tests were conducted using the Oligo Calc tool (Ficetola et al., 2010). To further validate primer specificity, mitochondrial DNA (mtDNA) was extracted from the dorsal muscle tissues of 12 common fish species found in the study area using a Biomarker Tissue DNA Kit. Then, DNA from each species (1 µL) was combined and divided into two groups: an experimental group containing C. nasus DNA and a control group without C. nasus DNA. The target gene was amplified using PCR gradient amplification (95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55°C~60°C for 30 s, and 72°C for 15 s, with a final extension at 72°C for 10 min) using the Ca1R/F primers. PCR reactions were performed in triplicate in 20 μL volumes, consisting of 10 μL of 2×T5 Super PCR Mix, 8 μL of ddH2O, 0.5 μL of each primer (10 μM/L), and 1 µL of template DNA. A blank control using ddH2O as the template was included in each reaction. Following amplification, products were screened using 1% agarose gel electrophoresis (AGE) to confirm successful amplification across the range of annealing temperatures. The successfully amplified products were then purified and sequenced. Finally, the BLASTn method was applied to validate sequence specificity, with a requirement of 100% similarity.
Three 2L eDNA water samples were collected at a depth of 0.5 m below the water surface using a hydrophore at each sampling site (Hänfling et al., 2016; Uchii et al., 2017) and transported to the laboratory on ice in sealed sterile bottles (Ghosal et al., 2018). The eDNA samples were filtered, extracted, and preserved following previously described methods (Yamanaka and Minamoto, 2016; Shu et al., 2020). DNA quality was assessed using 1% AGE, and the resulting eDNA solution was promptly cryopreserved at –20°C for further analysis. The collection, filtration, extraction, and cryopreservation of eDNA samples from the same batch were completed within 24 h. All collection and filtration equipment was sterilized with 10% sodium hypochlorite for 30 min and subsequently rinsed twice with ddH2O to eliminate residual disinfectant before use (Zhang et al., 2022).
A two-step process strategy was implemented to perform qPCR detection. Initially, the target gene was amplified using PCR (95°C for 3 min, followed by 40 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s) with the Ca1R/F primers. PCR reactions were conducted in triplicate in a 30 μL mixture containing 15 μL of qPCR mix, 2 μL of Mg2+ (25 mM), 0.5 μL of each primer (10 μM), 2 μL of template DNA, and 10 μL of ddH2O. Amplicons were extracted from 2% AGE, purified following the manufacturer’s instructions, and considered positive if any of the three replicates yielded a positive result. In the subsequent step, the positive amplicons were ligated into the pMD19T plasmid (size 2692 bp), followed by linearization, purification, quantification and converted into a copy number (copies/μL) by a specific formula. A standard curve was generated using a 10-fold serial dilution of above constructed standards, ranging from 10–2 to 10–6. qPCR was performed in triplicate under the following conditions: initial denaturation at 95°C for 3 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 80°C for 30 s. Each 30 μL qPCR reaction contained 15 μL of qPCR mix, 2.5 μL of Mg2+ (25 mM), 0.3 μL of each primer (10 μM), 2 μL of template DNA, 0.3 μL of Dyestuff (Eva), and 9.6 μL of ddH2O. Negative controls were prepared using ddH2O as the template. The qPCR assay was conducted on the SLAN-96P platform using an external reference method. Amplification and melting curves of both the standards and positive samples were also analyzed to evaluate the efficiency and specificity of the amplification reaction. A standard curve was then constructed by plotting the logarithm of standard plasmid concentrations against CT values to determine C. nasus DNA concentrations in positive samples, while DNA concentrations in negative samples were considered to be 0. Final DNA concentrations (copies/μL) were expressed as the mean ± standard deviation ( ± SD).
The Chi-square test was utilized to evaluate differences in eDNA detection rates between Poyang Lake and Tai Lake, as well as seasonal variations within each lake. Logistic regression analysis was applied to assess the relationship between DNA detection outcomes (positive or negative) and SD. The predicted probability of positive DNA detection, estimated through logistic regression analysis, was plotted to illustrate spatial and seasonal variations in fish occurrence. Non-parametric statistical methods were used to analyze regional and seasonal differences in DNA concentration between the two lakes. The Kruskal-Wallis test was conducted to compare multiple seasons within a single region or multiple regions within a single season. Pairwise comparisons between two seasons or two regions were performed using the Wilcoxon rank-sum test, with p-values adjusted using Holm’s correction. DNA concentration values were logarithmically transformed (log10(x + 1)) to achieve normalization and used for comparative analyses of regional and seasonal variations.
Partial least squares structural equation modeling (PLS-SEM) is an advanced multivariate analysis technique that integrates two key components: a measurement model, which defines the relationships between observed variables and their corresponding latent constructs, and a structural model, which describes the interactions among latent variables (Hair et al., 2019). This correlation-based framework facilitates the analysis of complex causal relationships by reducing multivariate dimensionality through the development of latent variables. PLS-SEM is particularly effective in addressing analytical challenges associated with non-normal data distributions and small sample sizes. In this study, this approach was applied to examine the influence of environmental variables on the distribution of the two C. nasus populations in Poyang Lake and Tai Lake. Prior to model construction, all variables were standardized to minimize the impact of differences in scale on model accuracy. Variables with correlation coefficients exceeding |0.7| were carefully selected to prevent multicollinearity issues. For measurement models, indicator loading values above 0.7 and average variance extracted (AVE) values greater than 0.5 were used as criteria for model evaluation. The coefficient of determination (R2) was used to assess the explanatory power of the structural models, while the maximum goodness of fit (GOF) determined overall model adequacy (Hair et al., 2019). Path coefficients and P-values were calculated automatically using the default parameters of the model. Statistical significance was defined as P < 0.05, with P < 0.01 and P < 0.001 indicating highly significant results. Statistical tests and logistic regression analyses were conducted using the Stats package, while PLS-SEM analysis was performed using the plspm package in R software (R Core Team, 2020).
The experimental group exhibited distinct and well-defined bands at various annealing temperatures, corresponding to the expected product sizes. In contrast, the control group displayed weak amplification, characterized by smeared bands and noticeable non-specific amplification. No bands were observed in the blank controls for either the experimental or control groups, confirming the absence of contamination and ensuring the reliability of the experimental process. BLASTn analysis verified that the forward and reverse sequenced amplicons from the experimental group (lanes A, B, H and E, F, G, respectively) matched C. nasus with 100% similarity, demonstrating the specificity and sensitivity of the custom-designed primers (Ca1R/F) for detecting free eDNA of C. nasus in water samples. Furthermore, the standard curve exhibited an R2 value of 0.998 and an E of 1.02, indicating that the qPCR system used in this study significantly improved the accuracy of detecting C. nasus DNA concentration in positive samples (Supplementary Figure S1). Therefore, subsequent analyses focused on evaluating the spatiotemporal distribution patterns of C. nasus populations using DNA positive and negative detection data and concentration values to infer fish occurrence frequency and population density.
A total of 62 positive samples were retained for analysis after excluding blank control samples that lacked correct bands. The Chi-square test revealed a significant difference in eDNA detection rates between Poyang Lake and Tai Lake, with detection rates of 27.7% (n = 23) and 52.7% (n = 39), respectively (χ2 = 10.23, P < 0.05). Across all seasons, a higher eDNA detection rate was observed in Tai Lake compared to Poyang Lake (Figure 2). In Poyang Lake, the detection rate was lowest in spring at 14.3% and highest in winter at 36.8% (Figure 2); however, no significant differences were detected among seasons (χ2 = 4.08, P > 0.05). In contrast, Tai Lake exhibited a significantly higher detection rate in winter (94.4%) compared to other seasons (χ2 = 18.69, P < 0.05), while no significant differences were observed among spring, summer, and autumn (χ2 = 2.16, P > 0.05).
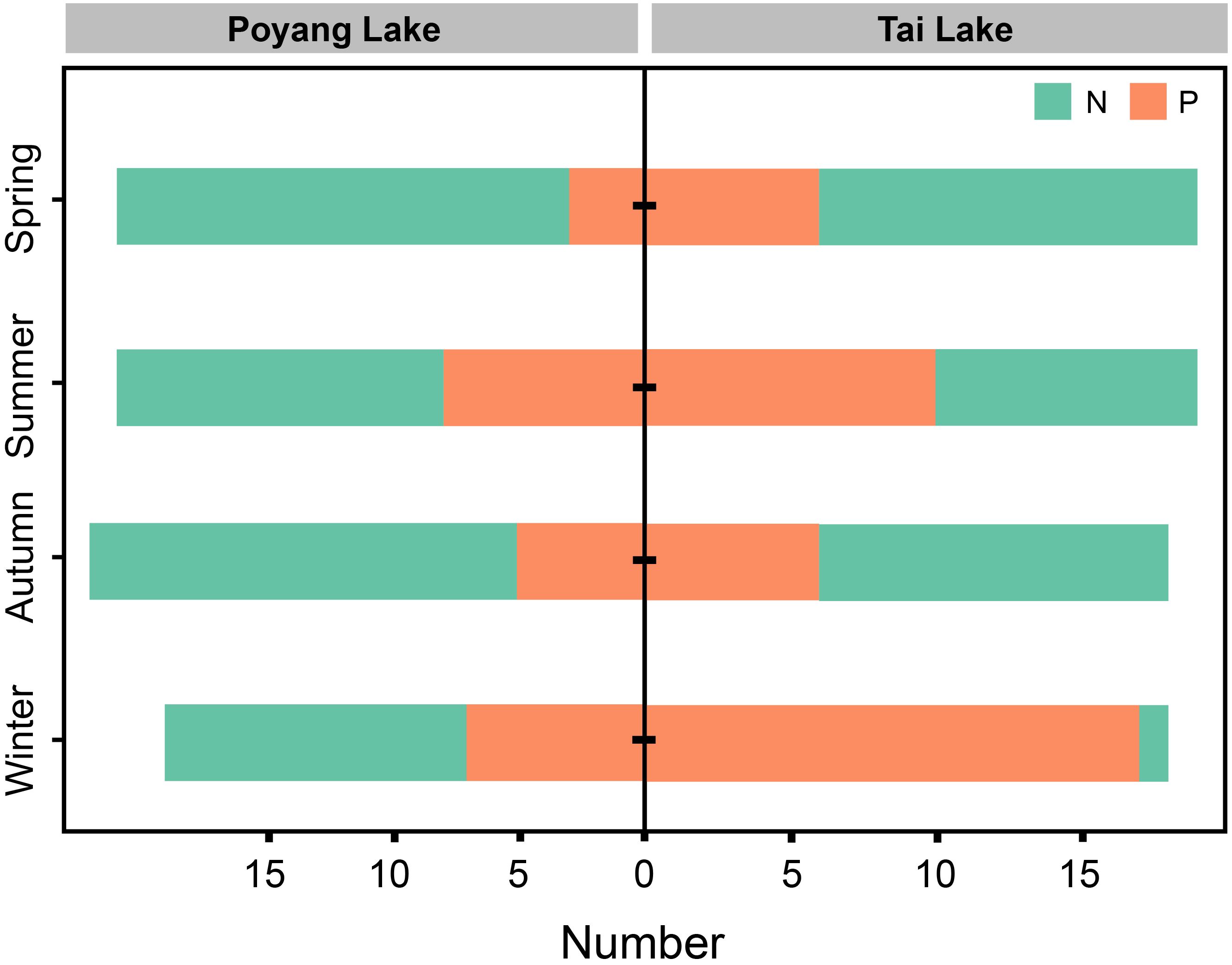
Figure 2. Number of C. nasus eDNA samples collected in Poyang Lake and Tai Lake. The green and orange bars represent negative (N) and positive (P) detections, respectively.
Logistic regression analysis indicated that the probability of DNA positive detection was not significantly associated with SD in either Poyang Lake or Tai Lake (P > 0.05). However, a statistically significant relationship between DNA positive detection probability and SD was identified during summer in Poyang Lake (P < 0.05, Figure 3). Although not statistically significant, seasonal variations in the spatial distribution patterns of DNA positive detection probability were observed in Poyang Lake. In spring, the highest probability of positive detection occurred in the lake estuary, gradually decreasing along a north-south gradient toward the southern region. In contrast, summer and spring exhibited inverse spatial patterns, with positive detection probability increasing from the lake estuary toward the southern region. The positive detection probability in the southern region during summer was higher than in autumn. However, in Tai Lake, spatial patterns of DNA positive detection probability did not show distinct seasonal variations (Figure 3).
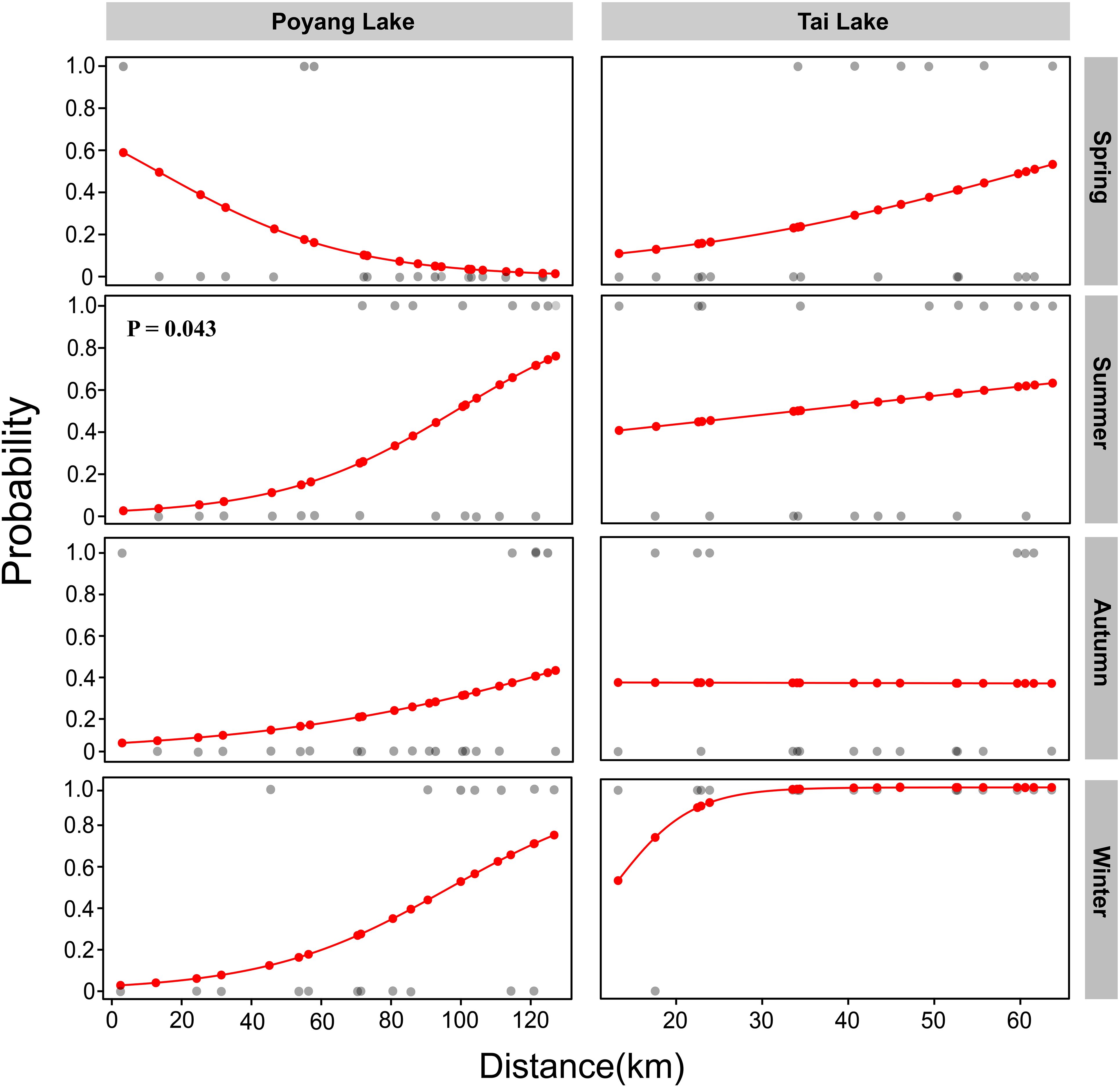
Figure 3. Spatiotemporal variations in the probability of C. nasus DNA positive detection. Grey dots represent both positive and negative DNA detection events, while red lines and dots indicate expected values derived from the logistic regression model.
The Kruskal-Wallis test indicated no statistically significant differences in DNA concentration across seasons in Poyang Lake (P > 0.05; Figure 4A). Similarly, the Wilcoxon rank-sum test did not detect significant seasonal differences in DNA concentrations (P > 0.05; Figure 4A). However, significant spatial variations were observed in spring, summer, and autumn (P < 0.05, Figure 4C). In spring, the average DNA concentration was higher in the northern region compared to 0 copies/μL in the middle and southern regions (P = 0.037, Figure 4C). In summer, DNA concentrations increased from 0 copies/μL in the northern region to 6500.1 copies/μL in the southern region (P = 0.048, Figure 4C). During autumn, the average DNA concentrations were 5818.7 copies/μL in the northern region and 2887.2 copies/μL in the southern region, while remaining 0 copies/μL in the middle region (P = 0.019, Figure 4C). In winter, DNA concentrations increased from 1793.3 copies/μL in the northern region to 3861.9 copies/μL in the southern region, though the difference was not statistically significant (P > 0.05, Figure 4C). In Tai Lake, the Kruskal-Wallis test revealed statistically significant differences in DNA concentration across seasons (P = 0.002, Figure 4B). The Wilcoxon rank-sum test further showed that DNA concentration in winter was significantly higher than in other seasons (P < 0.05, Figure 4B). No significant spatial variations in DNA concentration were detected in spring, autumn, or winter (P > 0.05; Figure 4C), but a statistically significant spatial difference was observed in summer (P = 0.027, Figure 4C).

Figure 4. Seasonal variations of C. nasus DNA concentration in Poyang Lake (A) and Tai Lake (B) and spatiotemporal distribution patterns of C. nasus DNA concentration in two lakes (C). DNA concentration values were log-transformed (log10(x + 1)) for normalization. Different lowercase letters (a and b) indicate significant differences, whereas identical lowercase letters denote no significant difference.
The Wilcoxon rank-sum test identified significant differences in sixteen environmental factors between Poyang Lake and Tai Lake, including seven physical, eight chemical, and one biological factor (P < 0.01 and P < 0.001, Table 1). With the exception of TR, WD and WT, the values of physical factors were significantly higher in Tai Lake compared to Poyang Lake (P < 0.001, Table 1). Similarly, the concentrations of several chemical factors, including CL, COD, DO, pH, and TP, were significantly greater in Tai Lake than in Poyang Lake (P < 0.01 and P < 0.001), along with Chla (Table 1). Moreover, the Kruskal-Wallis test revealed that seasonal variations in the environmental factors were statistically significant in both Poyang Lake and Tai Lake (P < 0.001), except of TR and NO2-N (P > 0.05).
After excluding environmental factors with strong multicollinearity and conducting model evaluation, PLS-SEM analysis revealed that in Poyang Lake, the models explained 2%, 66%, and 31% of the variance in Concentration, CF, and BF, respectively. The measurement model retained SD in GF, WT in PF, and DO and NO3-N in CF, achieving a GOF of 0.455. In Tai Lake, the models explained 19%, 40%, and 59% of the variance in Concentration, CF, and BF, respectively. The model included SD in GF, TUB and COND in PF, and COD and TN in CF, with a GOF of 0.457. In Poyang Lake, DNA concentration did not exhibit significant associations with any environmental factors (P > 0.05, Figure 5A). However, a significant negative correlation was observed between CF and PF (P < 0.001), consistent with its relationship with BF (P = 0.008). In Tai Lake, DNA concentration was significantly negatively correlated with PF (P = 0.002, Figure 5B), while correlations between other environmental factors and DNA concentration were not significant (P > 0.05). Unlike Poyang Lake, CF exhibited significantly positive correlations with both PF (P < 0.001) and BF (P < 0.001) in Tai Lake. Additionally, CF was negatively correlated with GF (P = 0.004, Figure 5B).
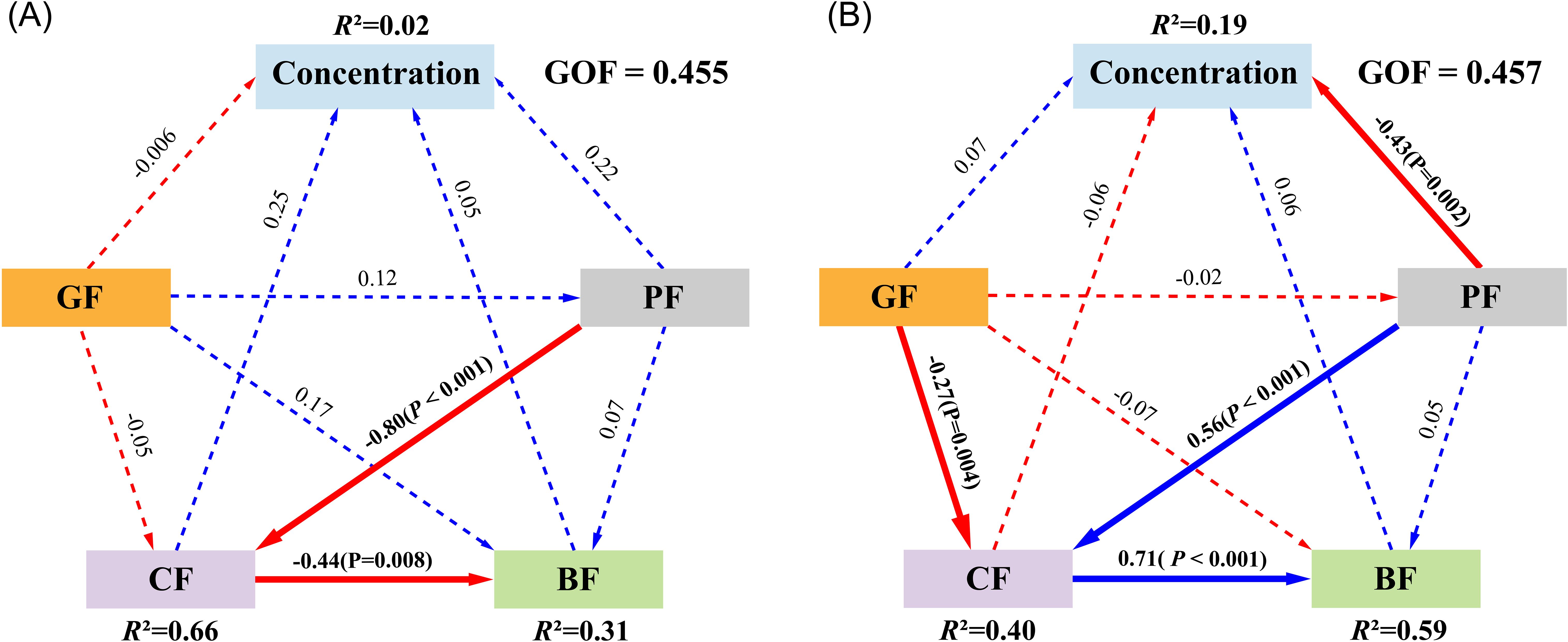
Figure 5. Partial least squares structural equation model (PLS-SEM) analysis of the effects of environmental factors on DNA concentration in Poyang Lake (A) and Tai Lake (B). Concentration refers to DNA concentration; GF represents geographic factors; PF denotes physical factors; CF corresponds to chemical factors; BF indicates biological factors. Blue and red lines represent positive and negative correlations, respectively. Solid and dotted lines denote significant and non-significant correlations, respectively. Line thickness reflects the strength of the correlation.
The findings of this study revealed distinct spatiotemporal distribution patterns between the two C. nasus ecotypes. The migratory ecotype follows a seasonal movement pattern, aggregating in the estuarine region of Poyang Lake during spring, migrating toward the southern region within the lake in summer, and returning to marine habitats in autumn and winter. This life cycle highlights the species’ strong adaptability to both freshwater and marine environments. These results are consistent with previous studies, which identified the middle and southern water areas of Poyang Lake as the primary spawning grounds for the migratory ecotype (Lu et al., 2015; Jiang et al., 2017). In contrast, the landlocked ecotype exhibits distinct behavioral patterns, lacking pronounced seasonal movements. The results speculated that the landlocked ecotype seemed to exhibite a non-selective spawning ground preference, showing no significant habitat specificity for reproductive activities. Its spatial and temporal activity patterns closely resembled those of typical lacustrine resident fish species, suggesting a stable, non-migratory life history strategy adapted to freshwater environments. Previous studies have also documented the coexistence of migratory and landlocked C. nasus ecotypes in Poyang Lake (Jiang et al., 2020; Sokta et al., 2020). Although the primers developed in this study could not differentiate between the two ecotypes, the observed spatial and seasonal distribution patterns of C. nasus in Poyang Lake were more consistent with migratory behavior, differing from the movement patterns characteristic of landlocked populations in Tai Lake.
The distinct life histories of the two ecotypes, regulated by genes encoding the S100 calcium-binding protein and the interferon regulatory factor, resulted in differences in reproductive and migratory behaviors (Liu et al., 2020). Reproductive isolation is widely recognized as a key mechanism that disrupts gene flow and contributes to ecological differentiation (Garlovsky et al., 2023; Jiménez-López et al., 2023; Rometsch et al., 2020). Currently, limited scientific evidence supports the presence of substantial gene flow between the two ecotypes. Therefore, it is speculated that differences in life history strategies, potentially leading to reproductive isolation, may play a critical role in driving ecotypic differentiation between migratory and landlocked populations of C. nasus. Reproductive isolation may result from various mechanisms, including behavioral plasticity in mating systems and ecological divergence in reproductive traits, such as nesting microhabitat preferences (Karlsson et al., 2010; Dean et al., 2021). However, the specific mechanisms underlying reproductive isolation between these ecotypes remain poorly understood and require further investigation.
The results indicated distinct environmental selection between the migratory and landlocked ecotypes. The migratory ecotype in Poyang Lake did not exhibit specific environmental preferences, whereas the landlocked ecotype in Taihu Lake demonstrated a clear preference for habitats with low turbidity and low conductivity. The effects of water turbidity on fish mobility are complex and highly variable (Rodrigues et al., 2023). Previous studies have also reported that water turbidity negatively affects both predator and prey freshwater fish, with a greater impact on predators than prey (Zanghi and Ioannou, 2025). Visual cues play a crucial role in selecting optimal foraging sites and capturing prey (Figueiredo et al., 2016). Turbid waters reduce underwater visibility (De Robertis et al., 2003), hinder visual foragers from locating food and detecting predators (Ferrari et al., 2010), and impair predation efficiency in fish (Snow et al., 2018; Figueiredo et al., 2016). These findings suggest that lower turbidity may enhance foraging efficiency in landlocked C. nasus.
The results indicated that landlocked C. nasus preferred habitats with low conductivity. Conductivity is a key water quality factor influencing the structure and distribution of fish communities (Guo et al., 2020). Increased conductivity has significant effects on a wide range of aquatic organisms, from algae to invertebrates and vertebrates (Dunlop et al., 2005). Physiologically, elevated conductivity is associated with osmotic stress, leading to the loss of sensitive species or their replacement by more tolerant species, ultimately altering community structure (Cañedo-Argüelles et al., 2013). Additionally, reduced conductivity may exacerbate the toxic effects of copper exposure in fish (Canli and Canli, 2015). However, the impact of electrical conductivity on fish growth remains debated. Dennis et al. (1995) suggested that higher conductivity could reduce fish growth, whereas Copp (2003) found no correlation between water conductivity and fish condition (K) in the River Great Ouse, U.K., catchment. The mechanisms governing fish distribution in relation to conductivity are not yet well understood and require further investigation.
Amid the rapid decline of aquatic ecosystems and increasing habitat fragmentation worldwide, traditional active monitoring methods are becoming increasingly inadequate for addressing the complexities of fish ecology (Snyder, 2003). Passive monitoring using eDNA offers a highly sensitive approach to detecting spatiotemporal variations in fish populations while minimizing habitat disturbance and destruction associated with active monitoring techniques (Bohmann et al., 2014; Tillotson et al., 2018). eDNA monitoring has demonstrated greater accuracy in estimating fish migration and abundance, as evidenced by lower data variance compared to seine fishing in assessments of Oncorhynchus tshawytscha populations (Shelton et al., 2019). Additionally, eDNA-based surveys achieved a 44% higher detection rate for endangered shark populations compared to traditional underwater video stations, significantly improving monitoring efficiency while reducing anthropogenic threats to the species (Boussarie et al., 2018). Although the application of eDNA combined with qPCR for monitoring fish populations, including the assessment of invasive, rare, and endangered species, is well-documented, the accuracy of these quantitative assessments requires careful validation (Eichmiller et al., 2015; Schmelzle and Kinziger, 2016; Plough et al., 2021).
Ensuring primer specificity and the accuracy of the standard curve is a fundamental prerequisite for achieving precise eDNA quantification, as errors in these aspects can result in false positives (Epp et al., 2011). In this study, a pair of C. nasus primers was developed and their specificity was confirmed. Melting curve analysis and the inclusion of negative controls in qPCR were also conducted, significantly reducing the occurrence of false positives during amplification and enhancing the accuracy of quantifying DNA concentration (McKee et al., 2015). Environmental factors such as flow velocity, UV exposure, pH, and external DNA contamination can also affect eDNA quantification results. Maintaining the cleanliness of sampling tools (Ruppert et al., 2019) and increasing the number of replicate samples during field sampling are essential strategies for improving sample DNA content (Davison et al., 2017). Furthermore, as eDNA currently lacks the ability to distinguish age structure and sex composition within fish populations, integration with traditional methods remains necessary to obtain more comprehensive results for fish monitoring and resource assessment. The strong concordance between eDNA and gillnet survey results suggests that eDNA can serve as a reliable complementary tool for qualitative and quantitative assessments of fish communities in large lakes (Hänfling et al., 2016). Future advancements in environmental RNA technology, based on transcriptomics theory, and the analysis of eDNA samples for specific populations (e.g., sex-reversed fish) or specific temporal periods (e.g., breeding seasons), hold promise for addressing challenges in quantitative population structure assessment (Inui et al., 2021; Marshall et al., 2021).
This study successfully utilized environmental DNA (eDNA) technology to examine the spatiotemporal distribution patterns of C. nasus populations in Poyang Lake and Tai Lake within the middle-lower Yangtze River basin. The findings clarified the distribution patterns and environmental factors influencing both migratory and landlocked ecotypes. It is speculated that the formation of these ecotypes is primarily driven by distinct life history strategies and adaptations to local environmental conditions. The migratory and landlocked ecotypes of C. nasus provide valuable models for studying ecotype formation and differentiation mechanisms. Future research should prioritize reproductive ecology, dietary preferences, and morphological characteristics to further elucidate ecotypic divergence. Additionally, this study highlights the effectiveness of eDNA technology in assessing fish population distribution and dynamics, offering critical data for the development and refinement of standardized survey methods in conservation efforts.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The research on animals was reviewed and approved by Animal Research and Ethics Committee of the Institute of Hydrobiology, Chinese Academy of Sciences.
PH: Data curation, Investigation, Writing – original draft. ZY: Formal analysis, Writing – original draft. MY: Data curation, Investigation, Writing – original draft. PS: Investigation, Writing – original draft. ZH: Formal Analysis, Writing – original draft. SL: Investigation, Writing – original draft. YC: Data curation, Methodology, Writing – original draft. HC: Conceptualization, Supervision, Writing – review & editing. XG: Conceptualization, Methodology, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Key R&D plan of Hubei Province (2023BCB039) and the Foundation of Sichuan Zumuzu River Hydropower Development Co., Ltd (Grant No. BL2022/D-77; BL2022/D-82). Sichuan Zumuzu River Hydropower Development Co., Ltd was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1520773/full#supplementary-material
Bohmann K., Evans A., Gilbert M. T. P., Carvalho G. R., Creer S., Knapp M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends. Ecol. Evol. 29, 358–367. doi: 10.1016/j.tree.2014.04.003
Boussarie G., Bakker J., Wangensteen O. S., Mariani S., Bonnin L., Juhel J.-B., et al. (2018). Environmental DNA illuminates the dark diversity of sharks. Sci. Adv. 4, eaap9661. doi: 10.1126/sciadv.aap9661
Cañedo-Argüelles M., Kefford B. J., Piscart C., Prat N., Schäfer R. B., Schulz C.-J. (2013). Salinisation of rivers: an urgent ecological issue. Environ. pollut. 173, 157–167. doi: 10.1016/j.envpol.2012.10.011
Canli E. G., Canli M. (2015). Low water conductivity increases the effects of copper on the serum parameters in fish (Oreochromis niloticus). Environ. Toxicol. Pharmacol. 39, 606–613. doi: 10.1016/j.etap.2014.12.019
Copp G. H. (2003). Is fish condition correlated with water conductivity? J. Fish Biol. 63, 263–266. doi: 10.1046/j.1095-8649.2003.00145.x
Curran E. V., Scott M. S., Olofsson J. K., Nyirenda F., Sotelo G., Bianconi M. E., et al. (2022). Hybridization boosts dispersal of two contrasted ecotypes in a grass species. Proc. R. Soc B. 289, 20212491. doi: 10.1098/rspb.2021.2491
Dai X., Wan R., Yang G. (2015). Non-stationary water-level fluctuation in China’s Poyang Lake and its interactions with Yangtze River. J. Geogr. Sci. 25, 274–288. doi: 10.1007/s11442-015-1167-x
Davison P. I., Copp G. H., Créach V., Vilizzi L., Britton J. R. (2017). Application of environmental DNA analysis to inform invasive fish eradication operations. Sci. Nat. 104, 1–7. doi: 10.1007/s00114-017-1453-9
Dean L. L., Dunstan H. R., Reddish A., MacColl A. D. C. (2021). Courtship behavior, nesting microhabitat, and assortative mating in sympatric stickleback species pairs. Ecol. Evol. 11, 1741–1755. doi: 10.1002/ece3.7164
Dennis T. E., MacAvoy S. E., Steg M. B., Bulger A. J. (1995). The association of water chemistry variables and fish condition in streams of Shenandoah National Park (USA). Water Air Soil pollut. 85, 365–370. doi: 10.1007/bf00476856
De Robertis A., Ryer C. H., Veloza A., Brodeur R. D. (2003). Differential effects of turbidity on prey consumption of piscivorous and planktivorous fish. Can. J. Fish. Aquat. Sci. 60, 1517–1526. doi: 10.1139/f03-123
Duan J., Zhang H., Liu K., Xu D., Zhang M., Shi W. (2012). An overview of coilia ectenes in Jiangsu section of the Yangtze River. Agric. Sci. Technol. 13, 1950–1954. doi: 10.16175/j.cnki.1009-4229.2012.09.047
Duarte S., Simões L., Costa F. O. (2023). Current status and topical issues on the use of eDNA-based targeted detection of rare animal species. Sci. Total. Environ. 904, 166675. doi: 10.1016/j.scitotenv.2023.166675
Dunlop J., Mcgregor G., Horrigan N. (2005). Potential impacts of salinity and turbidity in riverine ecosystems Characterisation of impacts and a discussion of regional target setting for riverine ecosystems in Queensland (Brisbane, Australia: Queensland Department of Natural Resources and Mines).
Eichmiller J. J., Miller L. M., Sorensen P. W. (2015). Optimizing techniques to capture and extract environmental DNA for detection and quantification of fish. Mol. Ecol. Resour. 16, 56–68. doi: 10.1111/1755-0998.12421
Epp L. S., Stoof-Leichsenring K. R., Trauth M. H., Tiedemann R. (2011). Molecular profiling of diatom assemblages in tropical lake sediments using taxon-specific PCR and Denaturing High-Performance Liquid Chromatography (PCR-DHPLC). Mol. Ecol. Resour. 11, 842–853. doi: 10.1111/j.1755-0998.2011.03022.x
Eriksen A. B., Nordal I. (1989). Ecotypic differentiation in relation to soil nitrogen in northern Scandinavian Cochlearia officinalis. Ecogr. 12, 31–38. doi: 10.1111/j.1600-0587.1989.tb00819.x
Faria A. V. F., Plastino E. M. (2022). Ecotypic differentiation in populations of Brazilian coast: recognizing adaptation to temperature in Gracilariopsis tenuifrons (Gracilariales, Rhodophyta). J. Appl. Phycol. 34, 2793–2805. doi: 10.1007/s10811-022-02721-2
Ferrari M. C. O., Lysak K. R., Chivers D. P. (2010). Turbidity as an ecological constraint on learned predator recognition and generalization in a prey fish. Anim. Behav. 79, 515–519. doi: 10.1016/j.anbehav.2009.12.006
Ficetola G., Coissac E., Zundel S., Riaz T., Shehzad W., Bessière J., et al. (2010). An In silico approach for the evaluation of DNA barcodes. BMC Genom. 11, 434. doi: 10.1186/1471-2164-11-434
Figueiredo B. R. S., Mormul R. P., Chapman B. B., Lolis L. A., Fiori L. F., Benedito E. (2016). Turbidity amplifies the non-lethal effects of predation and affects the foraging success of characid fish shoals. Freshw. Biol. 61, 293–300. doi: 10.1111/fwb.12703
Froese R., Pauly D. (2014). FishBase, World Wide Web Electronic Publication. Available at: http://fishbase.sinica.edu.tw/
Gao T., Ying Y., Yang Q., Song N., Xiao Y. (2020). The mitochondrial markers provide new insights into the population demographic history of coilia nasus with two ecotypes (Anadromous and freshwater). Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.576161
Garlovsky M. D., Whittington E., Albrecht T., Arenas-Castro H., Castillo D. M., Keais G. L., et al. (2023). Synthesis and scope of the role of postmating prezygotic isolation in speciation. Cold Spring Harb. Perspect. Biol. 15, a041429. doi: 10.1101/cshperspect.a041429
Ghosal R., Eichmiller J. J., Witthuhn B. A., Sorensen P. W. (2018). Attracting Common Carp to a bait site with food reveals strong positive relationships between fish density, feeding activity, environmental DNA, and sex pheromone release that could be used in invasive fish management. Ecol. Evol. 8, 6714–6727. doi: 10.1002/ece3.4169
Guo C., Chen Y., Gozlan R. E., Liu H., Lu Y., Qu X., et al. (2020). Patterns of fish communities and water quality in impounded lakes of China’s south-to-north water diversion project. Sci. Total Environ. 713, 136515. doi: 10.1016/j.scitotenv.2020.136515
Hair J. F., Risher J. J., Sarstedt M., Ringle C. M. (2019). When to use and how to report the results of PLS-SEM. EBR 31, 2–24. doi: 10.1108/ebr-11-2018-02
Han X., Chen X., Feng L. (2015). Four decades of winter wetland changes in Poyang Lake based on Landsat observations between 1973 and 2013. Remote Sens. Environ. 156, 426–437. doi: 10.1016/j.rse.2014.10.003
Hänfling B., Lawson Handley L., Read D. S., Hahn C., Li J., Nichols P., et al. (2016). Environmental DNA metabarcoding of lake fish communities reflects long-term data from established survey methods. Mol. Ecol. 25, 3101–3119. doi: 10.1111/mec.13660
Hufford K. M., Mazer S. J. (2003). Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends. Ecol. Evol. 18, 147–155. doi: 10.1016/s0169-5347(03)00002-8
Inui R., Akamatsu Y., Kono T., Saito M., Miyazono S., Nakao R. (2021). Spatiotemporal Changes of the Environmental DNA Concentrations of Amphidromous Fish Plecoglossus altivelis altivelis in the Spawning Grounds in the Takatsu River, Western Japan. Front. Ecol. Evol. 9. doi: 10.3389/fevo.2021.622149
Jerde C. L., Chadderton W. L., Mahon A. R., Renshaw M. A., Corush J., Budny M. L., et al. (2013). Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can. J. Fish. Aquat. Sci. 70, 522–526. doi: 10.1139/cjfas-2012-0478
Jiang T., Liu H., Xuan Z., Chen. X., Yang J. (2020). Classification of ecomorphotypes of Coilia nasus from the middle and lower reaches of the Yangtze River Basin. J. Lake Sci. 32, 518–527. doi: 10.18307/2020.0220
Jiang T., Yang J., Lu M. J., Liu H. B., Chen T. T., Gao Y. W. (2017). Discovery of a spawning area for anadromous Coilia nasus Temminck et Schlegel 1846 in Poyang Lake, China. J. Appl. Ichthyol. 33, 189–192. doi: 10.1111/jai.13293
Jiménez-López F. J., Arista M., Talavera M., Cerdeira Morellato L. P., Pannell J. R., Viruel J., et al. (2023). Multiple pre- and postzygotic components of reproductive isolation between two co-occurring Lysimachia species. New Phytol. 238, 874–887. doi: 10.1111/nph.18767
Karlsson K., Eroukhmanoff F., Svensson E. I. (2010). Phenotypic plasticity in response to the social environment: effects of density and sex ratio on mating behaviour following ecotype divergence. PloS One 5, e12755. doi: 10.1371/journal.pone.0012755
Khalil M. I., Gibson D. J., Baer S. G. (2017). Phylogenetic diversity reveals hidden patterns related to population source and species pools during restoration. J. Appl. Ecol. 54, 91–101. doi: 10.1111/1365-2664.12743
Kristensen M. L., Olsen E. M., Moland E., Knutsen H., Grønkjær P., Koed A., et al. (2021). Disparate movement behavior and feeding ecology in sympatric ecotypes of Atlantic cod. Ecol. Evol. 11, 11477–11490. doi: 10.1002/ece3.7939
Li Y., Chen J., Feng G., Yang J., Zhao F., Shen C., et al. (2022). Otolith microchemistry assessment: evidence of migratory coilia nasus of Yangtze River living in the Shengsi Sea area. Fishes 7, 172. doi: 10.3390/fishes7040172
Liu Y., Yang L., Lü Z., Liu J., Gong L., Liu B., et al. (2024). Allopatric divergence contributes to the population differentiation of Coilia nasus in the upper reach of Yangtze River. Ichthyol. Res. 71, 1–9. doi: 10.1007/s10228-023-00946-w
Liu D., Yang J., Tang W., Zhang X., Royster C. M., Zhang M. (2020). SINE Retrotransposon variation drives Ecotypic disparity in natural populations of Coilia nasus. Mob. DNA 11, 4. doi: 10.1186/s13100-019-0198-8
Lu G., Bernatchez L. (1999). Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): support for the ecological speciation hypothesis. Evol 53, 1491–1505. doi: 10.1111/j.1558-5646.1999.tb05413.x
Lu M., Jiang T., Liu H., Chen T., Yang J. (2015). Existence of anadromous Coilia nasus in Xinjiang River of Jiangxi Province as determined by otolith microchemistry. J. Fishery Sci. China 22, 978–985. doi: 10.3724/SP.J.1118.2015.15092
Ma F., Yin D., Fang D., Yang Y., Jiang M., You L., et al. (2020). Insights into response to food intake in anadromous Coilia nasus through stomach transcriptome analysis. Aquac. Res. 51, 2799–2812. doi: 10.1111/are.14619
Mao Z., Gu X., Zeng Q., Zhou L., Wang X., Wu L., et al. (2011). Community structure and diversity of fish in Lake Taihu. Chin. J. Ecol. 30, 2836–2842.
Marshall N. T., Vanderploeg H. A., Chaganti S. R. (2021). Environmental (e)RNA advances the reliability of eDNA by predicting its age. Sci. Rep. 11, 2769. doi: 10.1038/s41598-021-82205-4
Martín-Forés I., Avilés M., Acosta-Gallo B., Breed M. F., del Pozo A., de Miguel J. M., et al. (2017). Ecotypic differentiation and phenotypic plasticity combine to enhance the invasiveness of the most widespread daisy in Chile, Leontodon saxatilis. Sci. Rep. 7, 1546. doi: 10.1038/s41598-017-01457-1
Maruyama A., Nakamura K., Yamanaka H., Kondoh M., Minamoto T. (2014). The release rate of environmental DNA from juvenile and adult fish. PloS One 9, e114639. doi: 10.1371/journal.pone.0114639
May S. A., Shedd K. R., Rand P. S., Westley P. A. H. (2023). Tidal gradients, fine-scale homing and a potential cryptic ecotype of wild spawning pink salmon (Oncorhynchus gorbuscha). Mol. Ecol. 32, 5838–5848. doi: 10.1111/mec.17154
McColl-Gausden E. F., Weeks A. R., Coleman R. A., Robinson K. L., Song S., Raadik T. A., et al. (2020). Multispecies models reveal that eDNA metabarcoding is more sensitive than backpack electrofishing for conducting fish surveys in freshwater streams. Mol. Ecol. 30, 3111–3126. doi: 10.1111/mec.15644
McKee A. M., Spear S. F., Pierson T. W. (2015). The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biol. Conserv. 183, 70–76. doi: 10.1016/j.biocon.2014.11.031
Miller D. A., Clark W. R., Arnold S. J., Bronikowski A. M. (2011). Stochastic population dynamics in populations of western terrestrial garter snakes with divergent life histories. Ecology 92, 1658–1671. doi: 10.1890/10-1438.1
Olischläger M., Iñiguez C., Koch K., Wiencke C., Gordillo F. J. L. (2016). Increased pCO2 and temperature reveal ecotypic differences in growth and photosynthetic performance of temperate and Arctic populations of Saccharina latissima. Planta 245, 119–136. doi: 10.1007/s00425-016-2594-3
Plough L. V., Bunch A. J., Lee B. B., Fitzgerald C. L., Stence C. P., Richardson B. (2021). Development and testing of an environmental DNA (eDNA) assay for endangered Atlantic sturgeon to assess its potential as a monitoring and management tool. Environ. DNA 3, 800–814. doi: 10.1002/edn3.186
Qin B. (2015). The changing environment of Lake Taihu and its ecosystem responses. J. Freshw. Ecol. 30, 1–3. doi: 10.1080/02705060.2014.992053
Qu X., Chen Y., Liu H., Xia W., Lu Y., Gang D.-D., et al. (2020). A holistic assessment of water quality condition and spatiotemporal patterns in impounded lakes along the eastern route of China’s South-to-North water diversion project. Water Res. 185, 116275. doi: 10.1016/j.watres.2020.116275
R Core Team (2020). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. Available at: https://www.r-project.org
Rodrigues J. N., Ortega J. C. G., Petsch D. K., Padial A. A., Moi D. A., Figueiredo B. R. S. (2023). A meta-analytical review of turbidity effects on fish mobility. Rev. Fish Biol. Fish. 33, 1113–1127. doi: 10.1007/s11160-023-09785-4
Rometsch S. J., Torres-Dowdall J., Meyer A. (2020). Evolutionary dynamics of pre- and postzygotic reproductive isolation in cichlid fishes. Phil. Trans. R. Soc B 375, 20190535. doi: 10.1098/rstb.2019.0535
Ruppert K. M., Kline R. J., Rahman M. S. (2019). Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547. doi: 10.1016/j.gecco.2019.e00547
Sakaguchi S., Horie K., Ishikawa N., Nishio S., Worth J. R. P., Fukushima K., et al. (2018). Maintenance of soil ecotypes ofSolidago virgaureain close parapatry via divergent flowering time and selection against immigrants. J. Ecol. 107, 418–435. doi: 10.1111/1365-2745.13034
Schmelzle M. C., Kinziger A. P. (2016). Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol. Ecol. Resour. 16, 895–908. doi: 10.1111/1755-0998.12501
Schwarzer C., Joshi J. (2017). Parallel adaptive responses to abiotic but not biotic conditions after cryptic speciation in European peat moss Sphagnum magellanicum Brid. Perspect. Plant Ecol. Evol. Syst. 26, 14–27. doi: 10.1016/j.ppees.2017.03.001
Seliskar D. M., Gallagher J. L., Burdick D. M., Mutz L. A. (2002). The regulation of ecosystem functions by ecotypic variation in the dominant plant: a Spartina alterniflora salt-marsh case study. J. Ecol. 90, 1–11. doi: 10.1046/j.0022-0477.2001.00632.x
Sengupta M. E., Hellström M., Kariuki H. C., Olsen A., Thomsen P. F., Mejer H., et al. (2019). Environmental DNA for improved detection and environmental surveillance of schistosomiasis. Proc. Natl. Acad. Sci. U.S.A. 116, 8931–8940. doi: 10.1073/pnas.1815046116
Shelton A. O., Kelly R. P., O’Donnell J. L., Park L., Schwenke P., Greene C., et al. (2019). Environmental DNA provides quantitative estimates of a threatened salmon species. Biol. Conserv. 237, 383–391. doi: 10.1016/j.biocon.2019.07.003
Shu L., Ludwig A., Peng Z. (2020). Standards for methods utilizing environmental DNA for detection of fish species. Genes 11, 296. doi: 10.3390/genes11030296
Silvestro R., Brasseur S., Klisz M., Mencuccini M., Rossi S. (2020). Bioclimatic distance and performance of apical shoot extension: Disentangling the role of growth rate and duration in ecotypic differentiation. For. Ecol. Manage. 477, 118483. doi: 10.1016/j.foreco.2020.118483
Snow R. A., Shoup D. E., Porta M. J. (2018). Effects of turbidity on prey selection and foraging rate of hatchery-reared juvenile tiger muskellunge. N. Am. J. Fish. Manage. 38, 487–492. doi: 10.1002/nafm.10053
Snyder D. E. (2003). Invited overview: conclusions from a review of electrofishing and its harmful effects on fish. Rev. Fish Biol. Fish. 13, 445–453. doi: 10.1007/s11160-004-1095-9
Sokta L., Jiang T., Liu H., Xuan Z., Qiu C., Chen X., et al (2020). Loss of coilia nasus habitats in chinese freshwater lakes: an otolith microchemistry assessment. Heliyon. 6 (8), e04571. doi: 10.1016/j.heliyon.2020.e04571
Stinson K. A., Albertine J. M., Seidler T. G., Rogers C. A. (2017). Elevated CO2 boosts reproduction and alters selection in northern but not southern ecotypes of allergenic ragweed. Am. J. Bot. 104, 1313–1322. doi: 10.3732/ajb.1700222
Thorpe R. S., Reardon J. T., Malhotra A. (2005). Common garden and natural selection experiments support ecotypic differentiation in the Dominican anole (Anolis oculatus). Am. Nat. 165, 495–504. doi: 10.1086/428408
Tillotson M. D., Kelly R. P., Duda J. J., Hoy M., Kralj J., Quinn T. P. (2018). Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 220, 1–11. doi: 10.1016/j.biocon.2018.01.030
Todesco M., Owens G. L., Bercovich N., Légaré J.-S., Soudi S., Burge D. O., et al. (2020). Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584, 602–607. doi: 10.1038/s41586-020-2467-6
Turesson G. (1922). The genotypical response of the plant species to the habitat. Hereditas 3, 211–350. doi: 10.1111/j.1601-5223.1922.tb02734.x
Uchii K., Doi H., Yamanaka H., Minamoto T. (2017). Distinct seasonal migration patterns of Japanese native and non-native genotypes of common carp estimated by environmental DNA. Ecol. Evol. 7, 8515–8522. doi: 10.1002/ece3.3346
Valdivia-Carrillo T., Rocha-Olivares A., Reyes-Bonilla H., Domínguez-Contreras J. F., Munguia-Vega A. (2021). Integrating eDNA metabarcoding and simultaneous underwater visual surveys to describe complex fish communities in a marine biodiversity hotspot. Mol. Ecol. Resour. 21, 1558–1574. doi: 10.1111/1755-0998.13375
Xu G., Bian C., Nie Z., Li J., Wang Y., Xu D., et al. (2020). Genome and population sequencing of a chromosome-level genome assembly of the Chinese tapertail anchovy (Coilia nasus) provides novel insights into migratory adaptation. Gigascience 9, giz157. doi: 10.1093/gigascience/giz157
Yamamoto S., Minami K., Fukaya K., Takahashi K., Sawada H., Murakami H., et al. (2016). Environmental DNA as a ‘Snapshot’ of fish distribution: A case study of Japanese jack mackerel in Maizuru Bay, Sea of Japan. PloS One 11, e0149786. doi: 10.1371/journal.pone.0149786
Yamanaka H., Minamoto T. (2016). The use of environmental DNA of fishes as an efficient method of determining habitat connectivity. Ecol. Indic. 62, 147–153. doi: 10.1016/j.ecolind.2015.11.022
Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13, 1–11. doi: 10.1186/1471-2105-13-134
Yuan C. (1988). Resource and population composition changes and the reasons for Coilia ectenes in the mid-and-lower reaches of the Yangtze River. Chin. J. Zool. 23, 12–15. (in Chinese).
Zanghi C., Ioannou C. C. (2025). The impact of increasing turbidity on the predator–prey interactions of freshwater fishes. Freshw. Biol. 70, e14354. doi: 10.1111/fwb.14354
Zhang M., Xu D., Liu K., Shi W. (2005). Studies on biological characteristics and change of resource of coilia nasus schlegel in the lower reaches of the Yangtze river. Resour. Environ. Yangtze Basin 14, 694–698. (in Chinese with English abstract).
Keywords: Coilia nasus, environmental DNA, ecotypic differentiation, life history, adaptation to environment
Citation: Hu P, Yang Z, Yang M, Sheng P, Huang Z, Liu S, Chen Y, Cao H and Gao X (2025) Spatiotemporal distribution patterns of the two ecotypes of Coilia nasus in the middle-lower Yangtze River, China. Front. Mar. Sci. 12:1520773. doi: 10.3389/fmars.2025.1520773
Received: 31 October 2024; Accepted: 03 March 2025;
Published: 25 March 2025.
Edited by:
Shulian Wang, Hubei University of Technology, ChinaReviewed by:
Tingbing Zhu, Yangtze River Fisheries Research Institute, ChinaCopyright © 2025 Hu, Yang, Yang, Sheng, Huang, Liu, Chen, Cao and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Cao, cmVnYW5jYW9AaWhiLmFjLmNu; Xin Gao, Z2FveGluQGloYi5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.