- 1CellQua, Inc, Seongnam, Republic of Korea
- 2Department of Biological Sciences, Kongju National University, Gongju, Republic of Korea
- 3Gyeonggido Business and Science Accelerator, Suwon, Republic of Korea
- 4Subtropical Fisheries Research Institute, National Institute of Fisheries Science, Jeju, Republic of Korea
Introduction: This study aimed to propose the silvertip tetra (Hasemania nana) as a new experimental fish model. The silvertip tetra is a freshwater species that exhibits clear sexual dimorphism, with distinct differences in body coloration between males and females.
Methods: We analyzed the embryonic development of silvertip tetra and investigated transcriptome-level differences in gene expression between male and female brain-pituitary, caudal fin, and gonadal tissues. Additionally, we established a primary cell line derived from the caudal fin of male silvertip tetra and optimized the culture conditions for this cell line.
Results and Discussion: The optimal cell growth temperature was identified as 32°C, with a doubling time of approximately 28 hours. Successful transfection of foreign genes was confirmed by fluorescent protein expression, which was observed within 48 hours of transfection. RNA-seq analysis identified differentially expressed genes (DEGs) between sexes and tissues, particularly those involved in pigmentation, and protein interaction networks were examined to explore sex-related differences. The RNA-seq results validated by qRT-PCR suggest that the transcriptome-level gene expression patterns observed in silvertip tetra play critical roles in physiological functions and sexual dimorphism. Our findings highlight the potential of silvertip tetra as a valuable experimental model for studying pigmentation and sexual dimorphism.
1 Introduction
In biological research, animal models are essential tools not only for basic research but also for medical studies and drug development. Animal models provide the advantage of directly exploring complex biological processes that occur in vivo, thereby allowing researchers to understand new biological mechanisms and conduct preclinical studies for drug development (Vela et al., 2014; Mukherjee et al., 2022). Fish are increasingly being used across various research fields due to their lower maintenance costs compared to mammals and fewer ethical concerns in experimental settings (Harris et al., 2014; Ahn et al., 2023). Fish models are vital in life sciences research, offering several advantages such as relatively short life cycles, large numbers of offspring, and transparent embryonic development (Wakamatsu et al., 2001; Ankley and Johnson, 2004; He et al., 2014). The most widely used model, zebrafish (Danio rerio), is popular for these reasons, making it a powerful tool for genetic manipulation, developmental biology, and neuroscience research (Diekmann et al., 2004; Babin et al., 2014; He et al., 2014; Li et al., 2016).
Fish possess a greater diversity of chromatophore types compared to other vertebrates, making them an exceptional model for studying the mechanisms underlying skin coloration and pigmentation, from molecular genetics to systems biology (Luo et al., 2021). To date, fish are known to have six types of chromatophores: melanophores, xanthophores, erythrophores, iridophores, leucophores, and cyanophores (Cal et al., 2017). Based on these pigmentation differences, many fish species exhibit sexual dimorphism and sexual selection, often with males being more vibrant and females displaying simpler coloration (Mieno and Karino, 2016; Novelo et al., 2021; Luo et al., 2021). Among the well-studied small fish, the guppy (Poecilia reticulata) is recognized as a model for studying sexual dimorphism. However, it is ovoviviparous, while the Japanese medaka (Oryzias latipes), another commonly used species, is oviparous with a more distinct morphological sexual dimorphism rather than coloration-based dimorphism (Khoo et al., 1999; Otake et al., 2010). In guppies, several color patterns and fin shapes involved in sexual selection have been identified on sex chromosomes (Lindholm and Breden, 2002). These studies revealed that guppies possess an XX-XY sex determination mechanism, which has been studied through genetic investigations of body and fin coloration patterns. However, depending on the research objective, additional fish models beyond zebrafish are required, particularly when investigating traits like sexual dimorphism or pigmentation. This has created a demand for new experimental fish models that can address such research needs.
Silvertip tetra (Hasemania nana), a freshwater species native to South America, exhibits clear sexual dimorphism, with distinct differences in the body coloration of males and females (http://fishillust.com/Hasemania_nana). This characteristic makes silvertip tetra an ideal model for studying sexual differentiation and for analyzing gene expression related to pigmentation (Guiguen et al., 2018; Kottler and Schartl, 2018; Novelo et al., 2021). In addition, silvertip tetra, which is not ovoviviparous and is classified as “Least Concern” on the IUCN Red List, has a high reproductive rate and relatively short embryonic development, making it well-suited for developmental biology research. However, research on silvertip tetra remains limited, and genomic information beyond its mitochondrial genome is scarce (Xu et al., 2015). Recently, various experimental methods have been developed to replace animal testing, with cell-based assays being the most used alternative (Liebsch et al., 2011; Valerie et al., 2021). These alternative methods offer the ethical advantage of reducing animal use while also cutting experimental costs, which is why they are gaining attention from researchers worldwide (Liebsch et al., 2011). However, compared to mammalian species, fish cell lines available for distribution globally are limited. As a result, most researchers are compelled to establish cell lines on their own to conduct research. To increase the utility of experimental fish models, establishing cell lines is a critical factor (Goswami et al., 2022; Kumar et al., 2024). In this study, we established a caudal fin-derived cell line from male silvertip tetra and optimized the culture conditions for this cell line.
The aim of this research is to propose silvertip tetra as a new experimental model. As a model organism, silvertip tetra is well-suited for studies of pigmentation and sexual dimorphism, and it holds great potential for application in various biological fields. We provide detailed information on the embryonic development of silvertip tetra and analyze transcriptome-level differences in gene expression between male and female brain-pituitary, caudal fin, and gonadal tissues. Moreover, we established and optimized caudal fin-derived cell lines to provide a viable alternative to traditional animal testing methods. Through this research, we not only provide new information on silvertip tetra but also propose its potential as a valuable fish model for experimental research.
2 Materials and methods
2.1 Ethics statement
The whole process of the experiment using the fish was carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of CellQua (CQ-IACUC-2023-1-2) and Gangneung-Wonju National University (GWNU-2019-26). All methodologies of the experiment were verified by CellQua and GWNU ethical committee. Also, the authors of the present study have acquired Animal Welfare & Ethics Course certification, research ethics and compliance training program.
2.2 Fish management and tissue sampling
Silvertip tetra (H. nana) used in the experiment was in the fully matured adult stage (F1), and their average weight and length were 1.0 ± 0.038 g, 3.54 ± 0.075 cm in female (n=5), and 0.9 ± 0.019 g, 3.58 ± 0.037 cm in male (n=5), respectively (Figure 1). Fish (F0) were purchased at a local aquarium (Busan, Korea), and the F1 generation was produced by natural breeding F0 and used for experiments. Genomic DNA was extracted from the fin tissue of the silvertip tetra for species identification, and the Cytochrome c oxidase subunit I (COI) gene was analyzed (Supplementary Table S1). Embryo development was observed and recorded each stage by microscope (Motic, Xiamen, China). Fish and fertilized eggs were kept in a small-scale recirculation aquaculture system with at 24 ± 1.0°C and supplied different extruded pellets or artemia according to the growth stage. For the measure of body-size and -weight, and tissue collection, fish were anesthetized with 200 ppm of clove oil (Thermo Fisher Scientific, MA, USA). Tissue samples (brain-pituitary, caudal fin, and gonad); from females and males were kept in an ultra-freezer at -80°C for total RNA extraction. Total RNA was isolated from each tissue using Ribo EX Reagent (GeneAll, Seoul, South Korea) and purified with Hybrid-RTM Kit (GeneAll) according to the instructions of the manufacturer. The quantity and quality of the total RNA were measured using the ND-1000 nanometer (Thermo Fisher Scientific) and Bioanalyzer (Agilent, Santa Clara, CA).
2.3 Primary cell isolation and cell line establishment
Primary cells of silvertip tetra were isolated from the caudal fin tissue of an adult male. Following the manufacturer’s instructions for the explant culture-based primary cell isolation kit, pCELL-Ex kit (CellQua, Seongnam, South Korea), the cells were isolated. The isolated cells were suspended in Leibovitz’s L-15 medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% FBS (Thermo Fisher Scientific) and 1% Antibiotic-Antimycotic (GenDEPOT, Katy, TX, USA), and transferred to a T-25 cell culture flask (SPL, Pocheon, South Korea), then incubated at 26°C. The medium was replaced every three days, and the cell morphology was monitored using a fluorescence inverted microscope (Motic). When the primary cell confluency reached 70-80%, cells were subcultured using 0.25% trypsin-EDTA (Thermo Fisher Scientific). Continuous subculturing was performed for 3 to 5 days to develop a stable cell line. A total of 123 passages were performed, and the established cell line was designated as H. nana caudal fin, HNA FIN.
2.4 Proliferative characteristics of the cell line
To determine the optimal culture temperature for the cell line, passage 115 cells were used to evaluate cell growth under different temperature conditions. The cells were dissociated using 0.25% Trypsin-EDTA and seeded at a concentration of 1×104 cells/mL in a 24-well plate (Corning, Steuben, NY, USA). The cells were incubated for 5 days in L-15 medium supplemented with 10% FBS and 1% Antibiotic-Antimycotic at various temperatures: 20°C, 26°C, 32°C, and 38°C, and their viability was assessed. Cell viability was analyzed on days 3, 5, and 8 using the Quanti-Max WST-8 cell viability assay kit (Biomax, Guri, South Korea). Additionally, after culturing the cells at the optimal temperature of 32°C for 5 days, cell number was measured using the CellDrop-FL (DeNovix, Wilmington, DE, USA) to determine the doubling time.
2.5 Cryopreservation and plasmid transfection in the HNA FIN cells
The cryopreservation and post-thaw viability assay of the cell line were conducted using passage 111 HNA FIN cells at a concentration of 3×106 cells/mL. CELLBANKER 2 (ZENOGEN PHARMA, Sakamachi, Japan) was used as the cryopreservation medium. The frozen cells were stored at -80°C for varying durations: passage 52 cells were stored for 474 days, passage 115 cells for 110 days, and passage 111 cells for 10 days. After storage, the cells were quickly thawed in a 37°C water bath, and the cryopreservation medium was removed by centrifugation. The cell viability after thawing was measured using the Max-View™ Live/Dead Staining Kit (Biomax), with the number of live cells determined by fluorescence using the CellDrop-FL. To confirm the attachment and proliferation of the thawed cells, they were suspended in L-15 medium supplemented with 10% FBS and 1% Antibiotic-Antimycotic and transferred to a 90-mm dish (SPL), followed by incubation at 32°C.
To assess the transfection efficiency of foreign genes in the cell line, the pDsRed2-1 plasmid vector expressing red fluorescent protein (Takara Bio, Kusatsu, Japan) was used. An expression vector with the beta-actin 1 promoter from the Fathead Minnow (Pimephales promelas) species was inserted into passage 60 cells. The experiment was performed following the manufacturer’s instructions for the FUGENE 4K Transfection Reagent (Promega, Madison, WI, USA). The optimal transfection reagent-to-vector ratio was determined, and the ideal ratio was found to be 5:1. Fluorescence expression in the transfected cells was observed using a fluorescence inverted microscope.
2.6 Whole-transcriptome sequencing and de novo assembly
The whole-transcriptome next-generation sequencing (NGS) libraries were manufactured using Truseq stranded mRNA prep kit (Illumina, San Diego, CA, USA). NGS libraries were sequenced using NovaSeq 6000 (Illumina) on 101 x 2 stranded (paired-end) read module, and raw sequencing data were generated using base-calling software, bcl2fastq2 v2.2 and fastq quality score, ASCII Q-score (offset 33). NGS library adaptor sequences, low-quality sequences (limit=0.05), and ambiguous nucleotides (maximal two nucleotides) were removed using OmicsBox v3.3.0 (BioBam, Valencia, Spain). De novo assembly was constructed using trimmed pair-end reads since silvertip tetra genome assembly was not available for whole-transcriptome mapping reference. OmicsBox was used for generation of de novo assembly under following default parameters: word size: 25, bubble size 50, mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.5, similarity fraction: 0.8, and minimum contig length 200 bp. Coding regions of the de novo assembled reads were predicted using TransDecoder v5.5.0 with universal genetic code and default parameters (Haas et al., 2013). Transcript redundancy was reduced and clustered biological sequences of assembled sequences using CD-HIT v4.6.4 with default parameters (sequence identity type: global, sequence identity threshold: 0.95) (Fu et al., 2012). Completeness assessment was performed using the Benchmarking Universal Single-Copy Orthologs (BUSCO) v4.0.5 with default parameters in Vertebrata lineage dataset (number of species: 3354, number of BUSCOs: 67) (Seppey et al., 2019).
2.7 Functional annotation and differentially expressed genes analysis
Annotation of the de novo assembly contigs was carried out against the BLAST non-redundant protein sequences (NR) database in OmicsBox with the following conditions: E-value cut-off was 1.0E-3, the word size of BLAST parameters was 6, HSP length cut-off was 33, and HSP-Hit coverage was 0. In silico prediction was performed using InterProScan v5.45-80.0 (Finn et al., 2017), Gene Ontology (GO) (Götz et al., 2008), GO-Slim, and EggNOG-Mapper v1.0.3 with EggNOG v5.0.0 (Huerta-Cepas et al., 2019) for functional annotation. The functional annotation results were combined with those of the BLAST NR annotation to construct a whole-transcriptome reference of silvertip tetra, and the contigs from each sample were mapped to the whole-transcriptome reference assembly. Differentially expressed genes (DEGs) were investigated between females and males in each tissue (BRA, FIN, GON) with the FDR p-value below 0.05 using CLC Genomics Workbench v11.0. In addition, DEGs across various tissues were analyzed for relevant GO terms associated with pigmentation and sexual dimorphism using QuickGO (https://www.ebi.ac.uk/QuickGO/). The expression patterns of DEGs for functional analysis were classified based on fold change value based on up-regulation (fold change ≥ 2.0) and down-regulation (fold change ≤ -2.0). To analyze the expression of genes involved in pigmentation and identify their interactions, we employed STRING v11.0, utilizing the zebrafish (D. rerio) database with a minimum interaction score threshold set at > 0.5. This analysis allowed us to construct a comprehensive interaction network, incorporating multiple proteins. The resulting network was visualized using Cytoscape v3.8.1 software (Shannon et al., 2003).
2.8 Whole-transcriptome data of gene expression validation by quantitative real-time RT-PCR
qRT-PCR validation was performed on selected genes according to tissues and compared with the expression pattern of transcriptome data. Subsequent to normalize the concentration of total RNA in each tissue, cDNA was synthesized using HyperscriptTM RT Master Mix (Geneall) which included random hexamer and oligo (dT)18. Additionally, to rectify the concentration of each sample in qRT-PCR, phosphatidylinositol-binding clathrin assembly protein-like isoform x3 (PICALM) were used as an internal control gene (Lee et al., 2024). The qRT-PCR primers were designed from whole-transcriptome reference and confirmed qRT-PCR amplification efficiency using serially diluted cDNA series acquired from a single sample (Supplementary Table S1). qRT-PCR reactions were executed using QuantStudio™ 7 Flex Real-time PCR (Applied Biosystems™, Waltham, USA) with PowerUP™ SYBR™ Green Master Mix (Applied Biosystems) and gene-specific primers under the following cycling conditions: 2 steps of amplification (45 cycles), 95°C for 15 s and 60°C for 60 s. The Ct value of qRT-PCR was calculated on the delta-delta Ct method and expressed as mean ± SE and carried out by one-way ANOVA with a significance level of p < 0.05 using IBM SPSS 25.0 software (Livak and Schmittgen, 2001).
3 Result
3.1 Embryonic development of silvertip tetra
Information on embryonic development is essential in presenting the possibility as a new fish experiment model. Therefore, while securing the F1 generation of silvertip tetra, we tried to present information on the identified embryonic developmental stages and time. It took 22:10 hr from fertilization to hatching at 25°C (Table 1). Fertilized eggs are small and are characterized by non-adhesive, and transparent chorion. 10 min after fertilization, the 1-cells were observed (Figure 2A). After 5 min of 2-cells stage, the second cleavage occurred and four blastomeres were shown (Figure 2B). Blastomeres occurred up to the morula stage at 1:15 hr (Figure 2C). The early blastula (Figure 2D) was observed at 1:40 hr after fertilization and at 2:40 hr the late blastulation process was shown (Figure 2E). The movement of yolk syncytial layer (epiboly) with the closure of the blastopore, 50% epiboly, 90% epiboly, and blastopore closure of the gastrula stage with 4:10 hr, 5:40 hr, 6:40 after fertilization were showed, respectively (Figure 2F-H). 10-myotomes were observed at 9:40 hr after fertilization (Figure 2I), the heartbeat has begun, tail became sharper and longer, with melanophore on the yolk at 12:40 hr (Figure 2J). After the tail reached to the head at 14:10 hr after fertilization (Figure 2K), embryos were fully formed at 15:40 hr (Figure 2L). Before hatching, there was an increase in embryo movement, and the embryo broke the chorion at 21:50 hr (Figure 2M) and hatched at 22:10 hr after fertilization (Figure 2N).
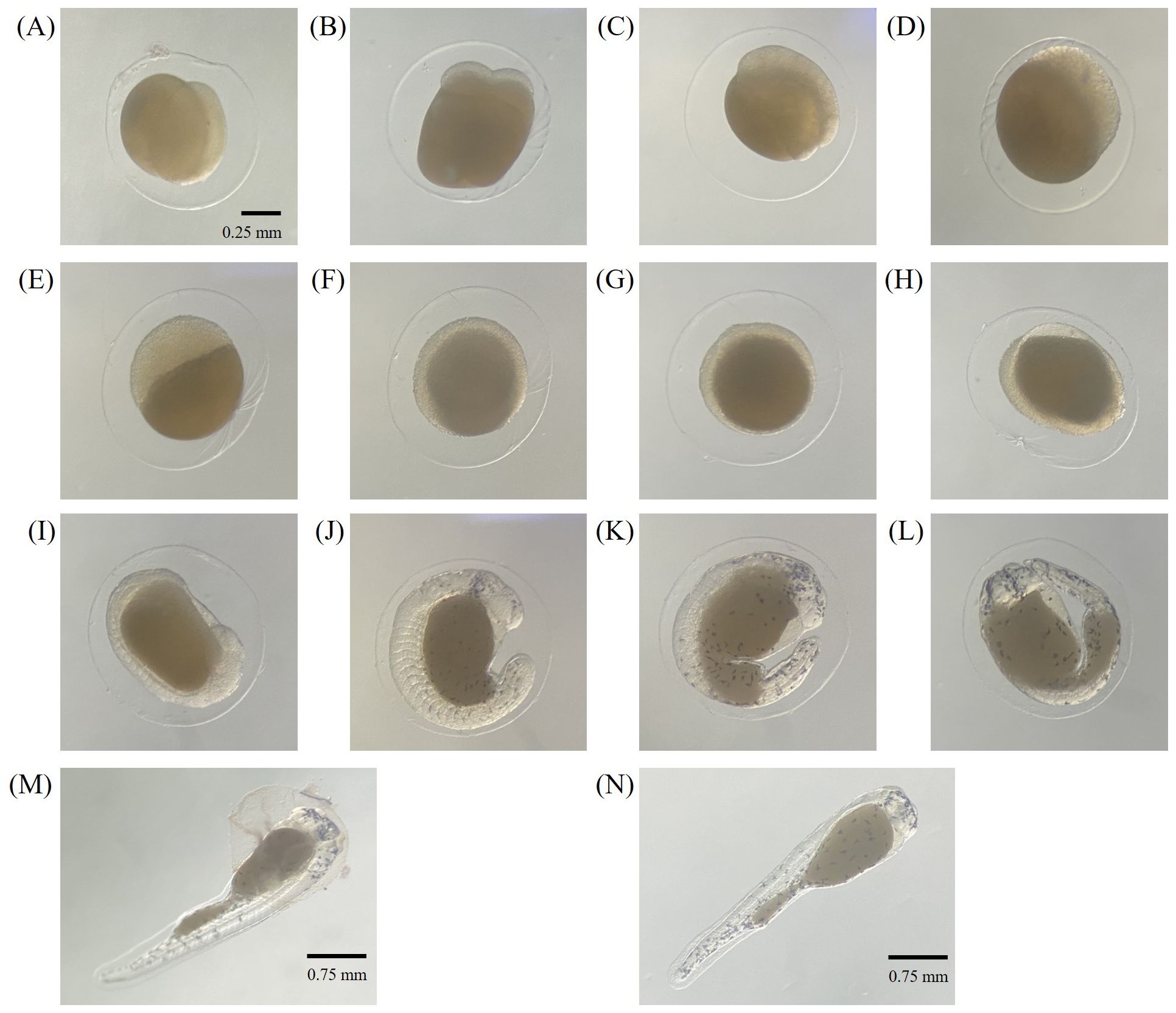
Figure 2. Embryonic developmental stages of silvertip tetra; (A) 1-cell, (B) 4-cell, (C) morula, (D) early blastula, (E) late blastula, (F) gastrula (50% epiboly), (G) gastrula (90% epiboly), (H) gastrula (blastopore closure), (I) 10 mytomes, (J) heartbeat, melanophore on the yolk, (K) tail reached to the head, (L) later development, (M) pre-hatched, (N) just hatched.
3.2 Primary cell culture and optimization of culture maintenance
Primary cells cultured using the explant culture method were observed proliferating from the tissue by day 7 after seeding. In the early stages of culture, both fibroblast-like and epithelial-like cell morphologies were observed. By day 15 after seeding, the primary cells reached approximately 70-80% confluency on the surface of the T-25 flask, and subculturing was initiated. The cell line has been continuously subcultured for over 120 passages, and the cells have been morphologically confirmed as fibroblast-like cells (Figure 3). After cryopreservation and thawing, the viability of HNA FIN cells was assessed by Live/Dead staining. The viability was 89.5 ± 2.7% for passage 52, 73.4 ± 2.5% for passage 111, and 78.8 ± 1.3% for passage 115. Additionally, after cell attachment, proliferation was confirmed, with cells reaching 70-80% confluency in the dish within 5 days of seeding. No morphological changes in the cells were observed after freezing and thawing.
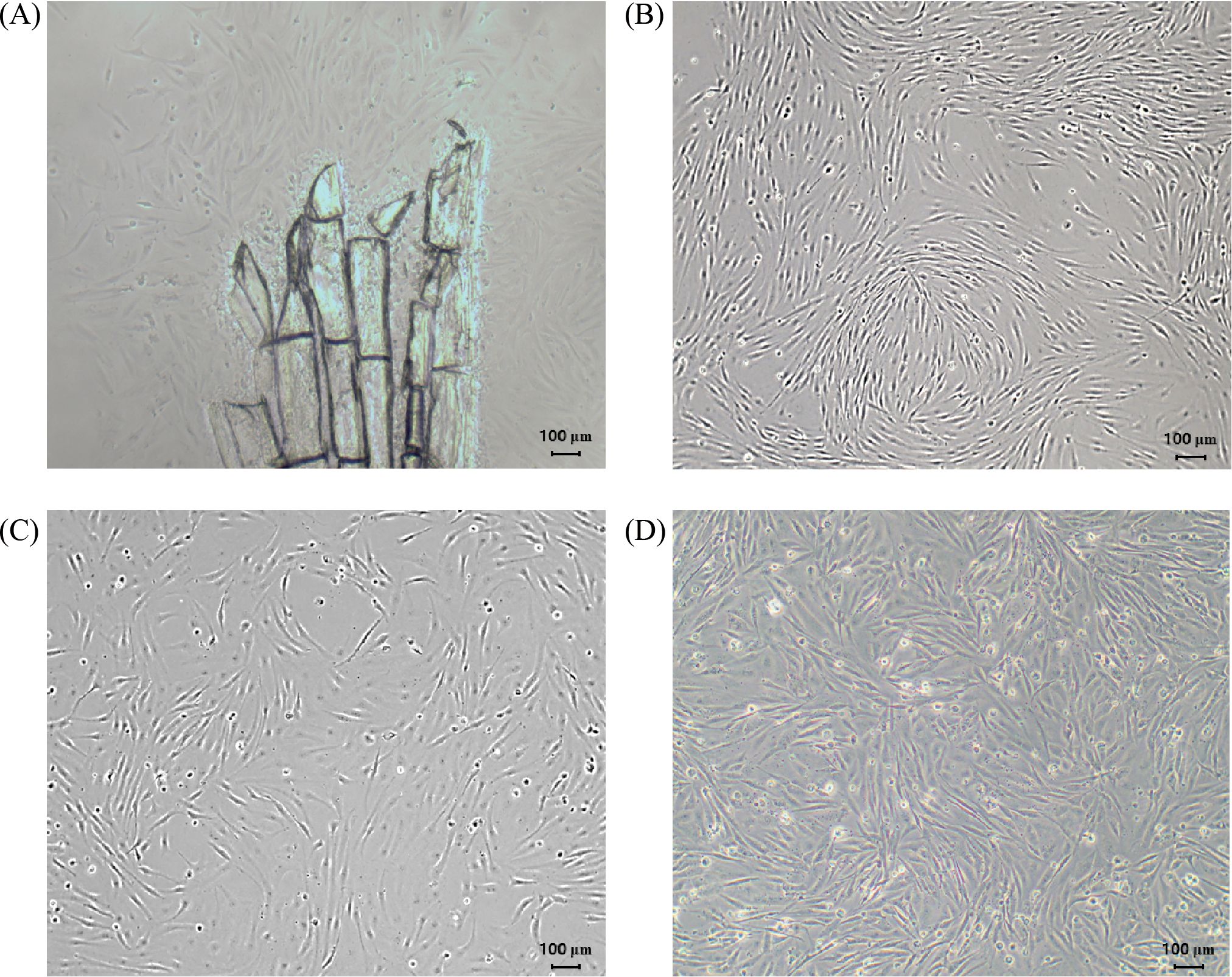
Figure 3. Morphology of caudal fin-derived HNA FIN cells from the silvertip tetra at different passages. (A) Passage 0 after 17 days. (B) Passage 40 (C) Passage 75 (D) Passage 116. Scale bar: 100 µm.
3.3 HNA FIN cells growth and transfection
To determine the optimal culture temperature for the cell line, cells were cultured at temperatures ranging from 20°C to 38°C, and the highest proliferation rate was observed at 32°C (Figure 4A). The growth rate of the cell line peaked on day 8 at 32°C. At 38°C, the cells showed a tendency to proliferate until day 3 after seeding, but the proliferation rate decreased from day 5 onward. To measure the doubling time of the cell line, the cells were cultured at the optimal temperature of 32°C for 5 days, and the cell count was used to calculate a doubling time of 28 hours (Figure 4B).
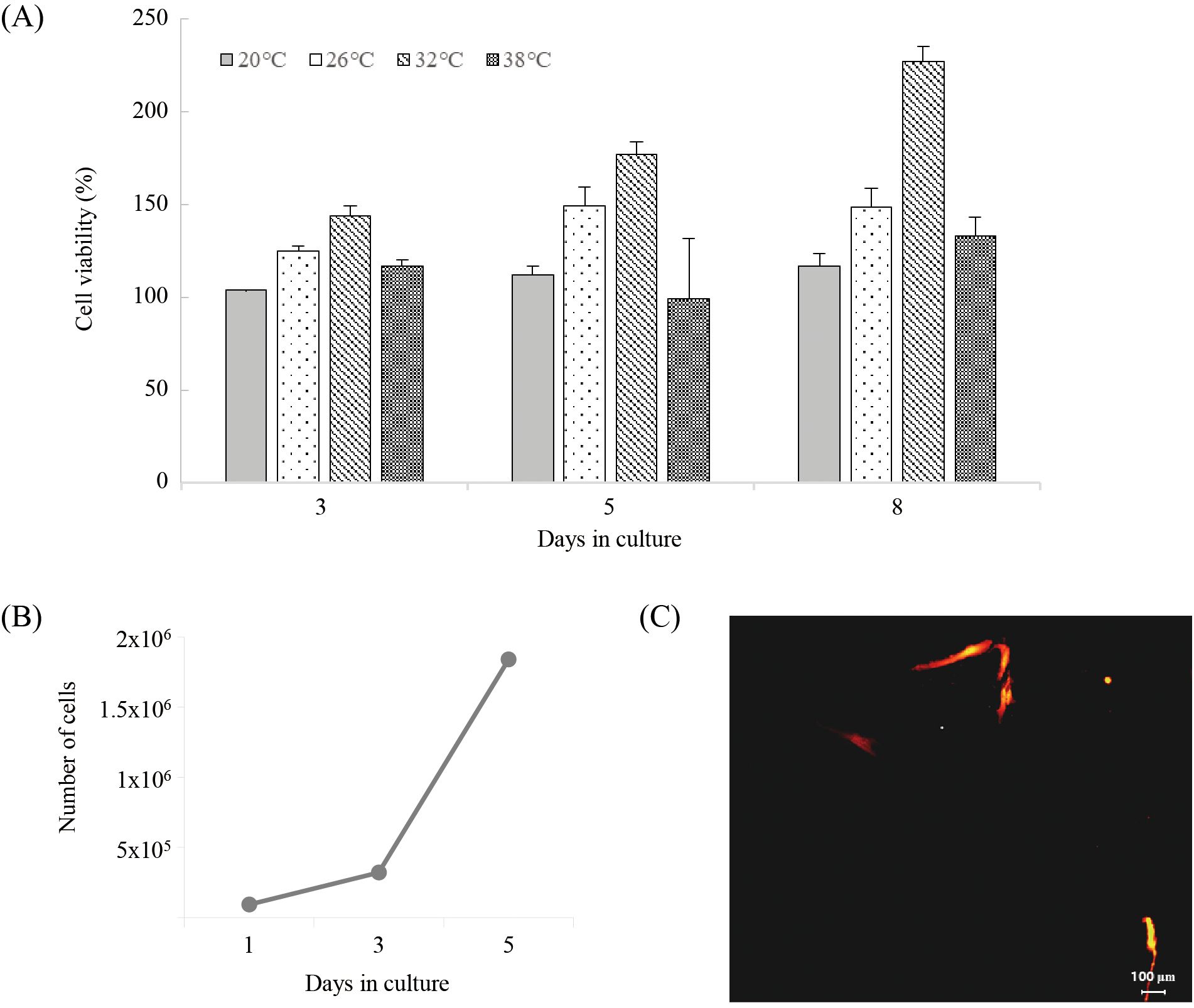
Figure 4. Cell viability of HNA FIN cells at different temperatures (20°C, 26°C, 32°C, 38°C). (B) Doubling time assay of HNA FIN cells incubated at 32°C for 1–5 days. (C) Transfection with red fluorescent protein in HNA FIN cells at passage 60.
The observation of cells expressing fluorescent protein confirmed the successful transfection of the foreign expression vector. Fluorescent protein expression was detected 48 hours after transfection, and the number of expressing cells increased over time (Figure 4C).
3.4 De novo assembly of the silvertip tetra whole-transcriptome
A total of 246,799 contigs (160,853,660 bp) was generated from females and males of silvertip tetra with the Illumina Novaseq6000 platform (Supplementary Table S2). The average length and N50 of assembled contigs was 652 bp and 1,012 bp. The result of the comparison with similar protein sequences using CD-HIT, a total of 245,900 clusters was identified at 95% identity. 17,243 contigs of the complete open reading frame (ORF), 7,509 contigs of the 5’-partial, 4,874 contigs of the 3’-partial, and 6,701 contigs of the internal ORF were predicted using TransDecoder 5.5.0. 2,779 contigs (81.63%) of the BUSCO groups have complete gene representation [single copy; 2,738 contigs ±(81.63%) and duplicated; 41 contigs (1.22%)], while 310 contigs (9.24%) were only partially recovered, and 265 contigs (7.9%) were missing in vertebrata_odb 10 lineage dataset. The high quality of the de novo assembly of silvertip tetra was verified based on the mapping rate of each tissue between 84.44% and 85.59% (Supplementary Table S3).
3.5 Functional annotation and clustering enrichment
A total of 48,876 contigs (19.8%) matched against the protein databases in BLASTx. Additional annotations were carried out against the EggNOG, GO, InterProScan, and KEGG database, resulting in a total of 37,154 contigs (15.1%), 20,465 contigs (8.3%), 54,881 contigs (22.2%), and 19,770 contigs (8.0%), respectively. 62% of the annotated genes matched the gene sequences from Astyanax mexicanus, which belongs to the same family (Characidae) as the silvertip tetra. Expression and clustering data were estimated using Principal Component Analysis (PCA) and Hierarchical clustering heat map for investigating the relationship between different gender and tissues (Figure 5A). As a result of PCA and heat map examination, each tissue-specific cluster was classified with different patterns according to gender within the tissue (Figure 5B). For gonad and caudal fin, the distribution varies along axis 1 (43.8% variability) and 2 (20.7% variability) between female and male, respectively. However, the brain-pituitary showed little difference in the distribution of axis 1 and axis 2 between female and male. In the heat map, the expression patterns of genes between female and male were similar broadly in the brain, whereas the expression patterns between genes were different as caudal fin and gonad were deviated clusters between female and male.
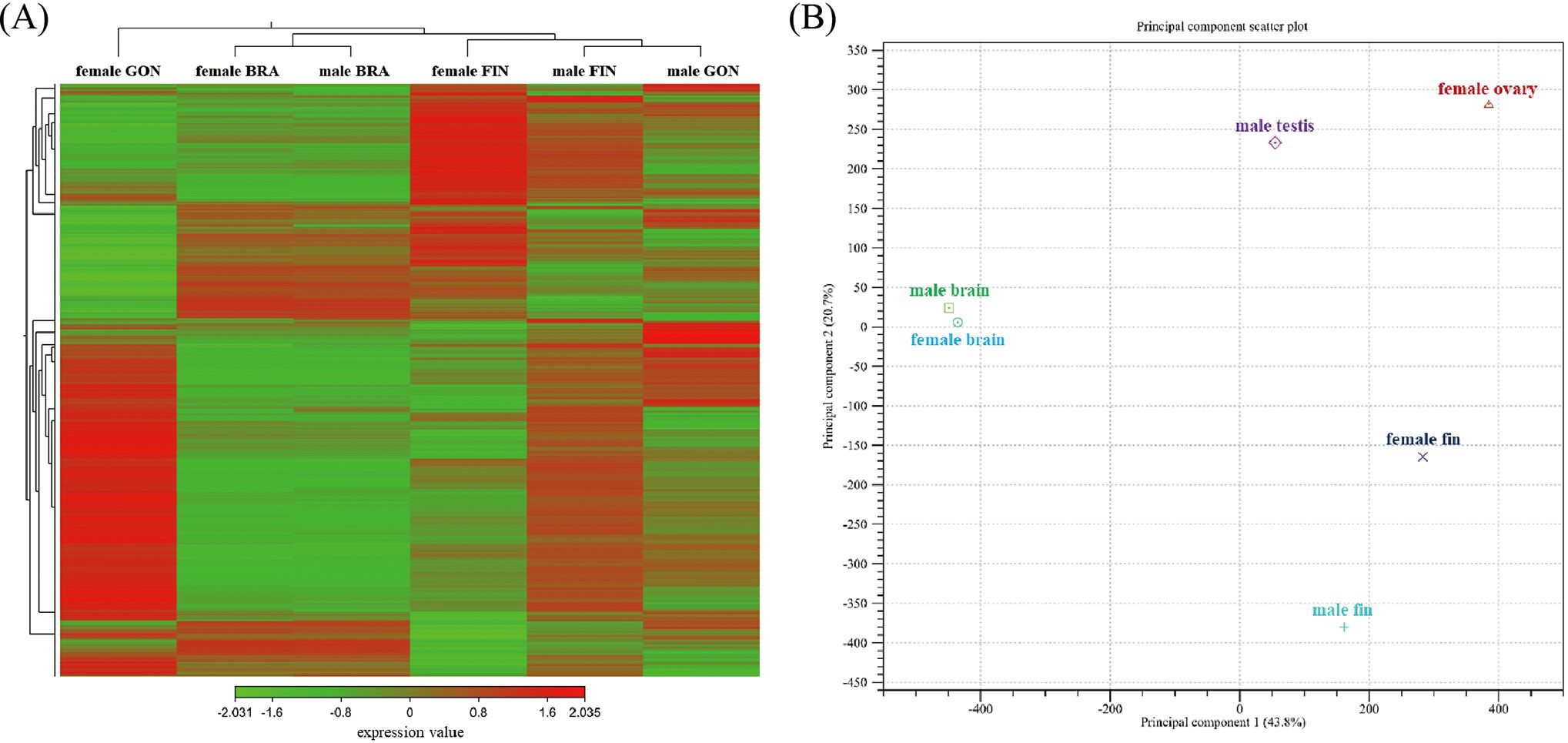
Figure 5. (A) Heatmap clustering gene expression patterns in the brain-pituitary (BRA), caudal fin (FIN), and gonad (GON) of male and female silvertip tetra. Selected genes (FDR p-value cutoff = 0.05, minimum fold change = 1.5) are represented by different saturated colors based on expression levels. Red indicates high expression, while green indicates low expression. (B) Principal component analysis (PCA) plot showing gene expression variation among tissue types and sexes. Principal component 1 (PC1) explains 43.8% of the total variance, while principal component 2 (PC2) accounts for 20.7%. Clustering is distinctly observed by tissue type, with brain, fin, and gonad forming separate groups. Within each tissue type, male and female show additional clustering patterns, reflecting sex-specific gene expression differences. Male testis and female ovary are clearly separated along PC1, suggesting strong sex-dependent gene expression divergence in gonads. Similarly, male and female fin show distinct positioning along PC1 and PC2, highlighting tissue- and sex-specific gene expression patterns.
3.6 Functional investigation of DEGs between female and male
DEGs of each tissue were investigated for functional enrichment with GO and KEGG. The distribution of three GO domains (biological process; BP, molecular function; MF, and cellular component; CC) was characterized based on level 7 classification. For DEGs of each tissue, BP domains were represented by ‘nucleic acid-templated transcription’ and ‘regulation of nucleic acid-templated transcription’. MF domains mainly were comprised by ‘nucleoside-triphosphatase activity’ and ‘zinc ion binding’. In CC domains, ‘microtubule’ among GO terms were overwhelming occupied. Furthermore, several GO terms were identified by specific tissues or gender in all three GO domains (Figure 6). Among the DEGs, several were identified as belonging to the Biological Process (BP) domain, specifically involved in pigmentation (GO:0043473), pigment metabolic process (GO:0042440), sex differentiation (GO:0007548), sex determination (GO:0007530), female sex differentiation (GO:0046660), and male sex differentiation (GO:0046661).
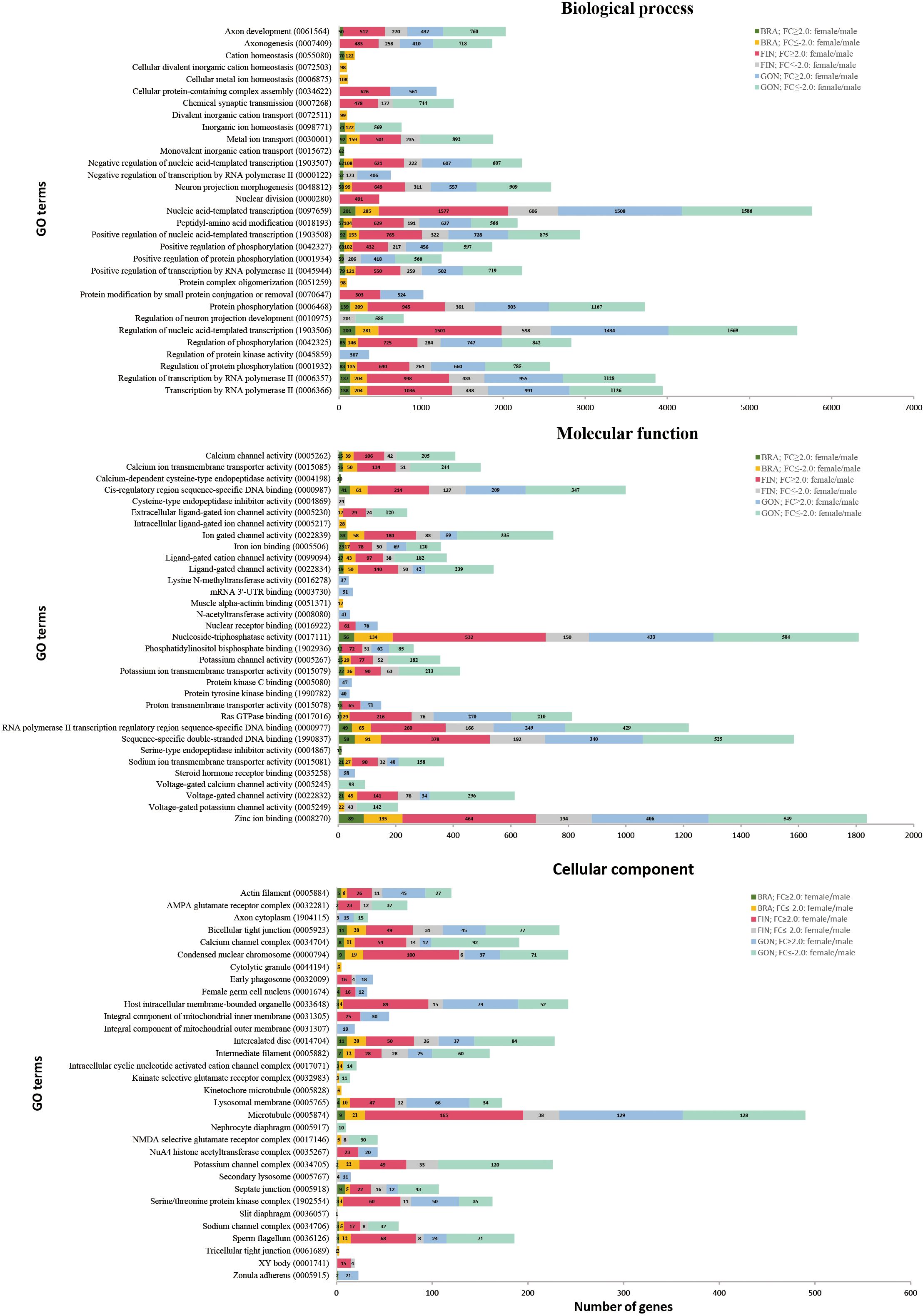
Figure 6. Gene ontology (level 7) analysis of DEGs based on tissue and gender. GO terms are categorized into three domains: biological process (BP), molecular function (MF), and cellular component (CC). The X-axis represents the number of DEGs, and the Y-axis represents GO terms.
KEGG terms were dominated by ‘metabolic pathways’ in DEGs of BRA, FIN, and GON. Additionally, some KEGG terms have been annotated for specific tissue or gender, such as GO results. In particular, ‘Rap1 signaling pathway’, ‘Ras signaling pathway’, and ‘regulation of actin cytoskeleton’ mainly involved genes that showed higher expression in males than females in BRA, FIN, and GON (Figure 7).
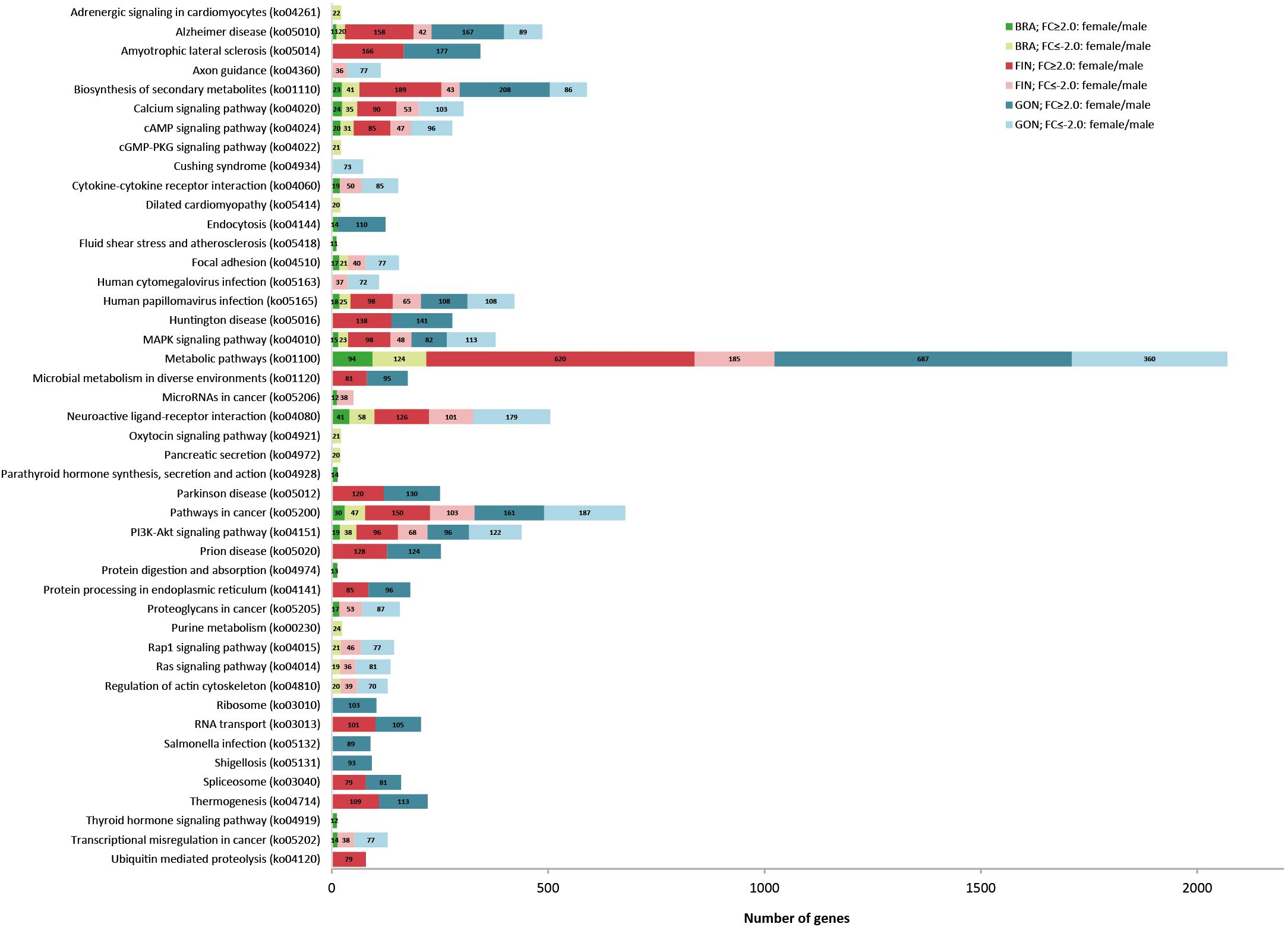
Figure 7. Comparison of the top 20 KEGG pathways with the highest DEG representation between females and males in silvertip tetra. The bar graph represents the number of DEGs with a fold change (FC) of ≥2 or ≤-2 in the BRA, FIN, and GON tissues.
3.7 Identification of DEGs between female and male
A Venn diagram was generated to investigate the types and distribution of DEGs in each tissue of females and males, with a maximum FDR p-value < 0.05 and a minimum absolute fold change > 1.5 (Figure 8A). A total of 49,605 DEGs were identified from the brain-pituitary, caudal fin, and gonad, comparing females and males. The distribution of DEGs across tissues revealed 3,715 DEGs in the brain, 28,053 in the caudal fin, and 39,931 in the gonad. Tissue-specific expression patterns were observed with 1,671 DEGs in the brain-pituitary, 7,600 in the caudal fin, and 19,159 in the gonad. Additionally, 919 DEGs were commonly expressed across the three tissues.
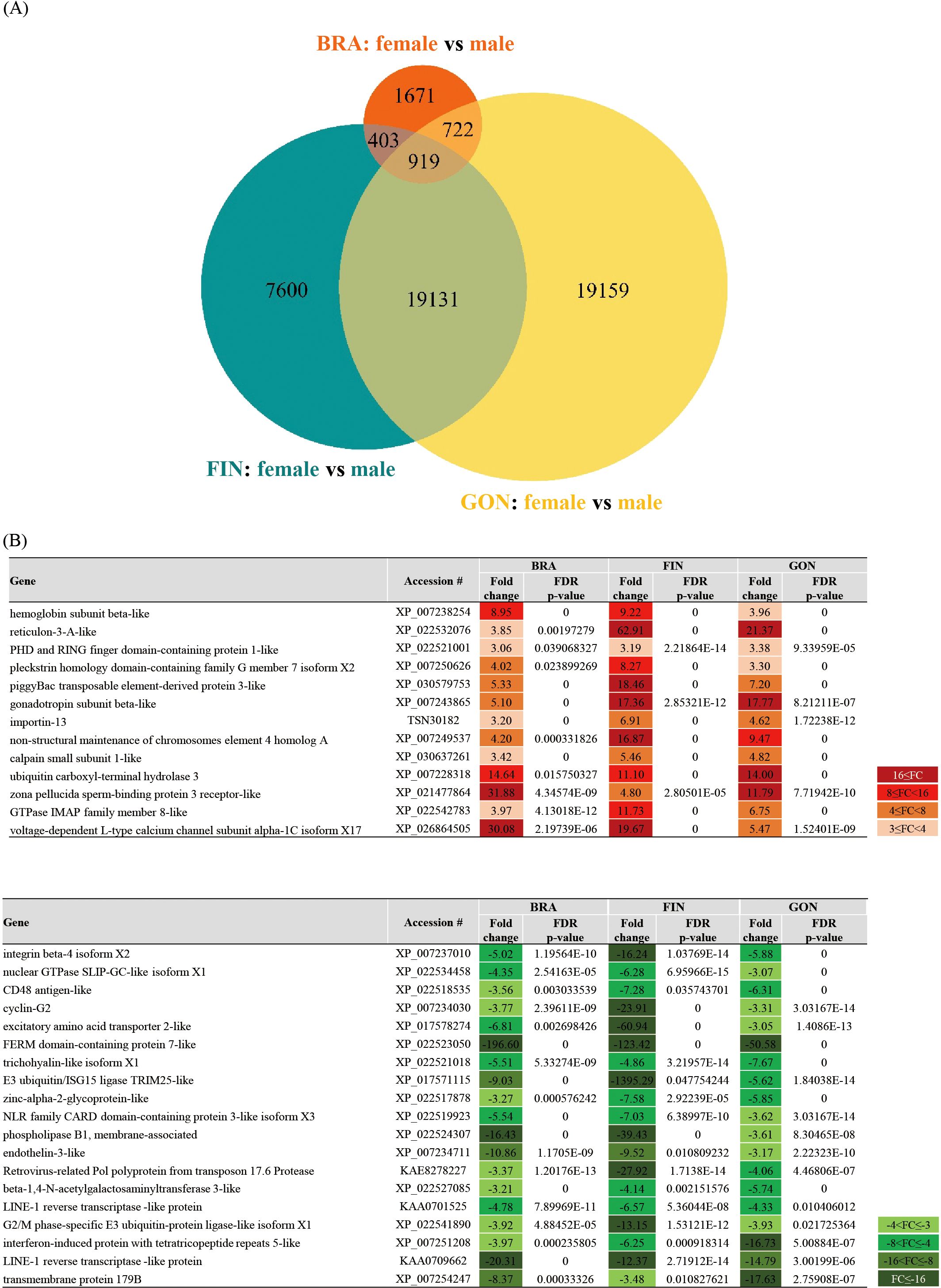
Figure 8. (A) Venn diagram showing DEGs between female and male tissues of silvertip tetra. Parameters: FDR p-value cutoff = 0.05, minimum fold change = 1.5. The orange circle represents the BRA, blue-green represents the FIN, and yellow represents the GON. (B) List of selected genes with annotations from the 919 DEGs common to all three tissues (BRA, FIN, and GON) in both females and males. The heatmap indicates expression levels with FC values≥ 3 or ≤ -3.
Among the DEGs showing common expression patterns across all three tissues, 13 DEGs in females and 19 in males exhibited a fold change ≥ 3 and an FDR p-value < 0.05, excluding undefined hypothetical proteins (Figure 8B).
In tissue-specific analyses, DEGs involved in reproduction, maturation, and growth were selected from the brain-pituitary, while DEGs involved in pigmentation and cation transport were identified in the caudal fin, and those related to gonadal development and sex differentiation were selected from the gonad.
In the brain-pituitary, the gene with higher expression in females was luteinizing hormone beta subunit (LHB), showing a fold change of 5.10. In contrast, the genes with higher expression in males were pituitary adenylate cyclase-activating polypeptide type I receptor-like isoform X4 (PACAP-R1 x4) with a fold change of -1.21, neuropeptide Y receptor type 1 (NPY1R) with -1.30, brain aromatase-like (BAA) with -1.33, insulin-like growth factor I (IGF-I) with -2.51, and prolactin-like (PRL) with -5.41 (Figure 9A).
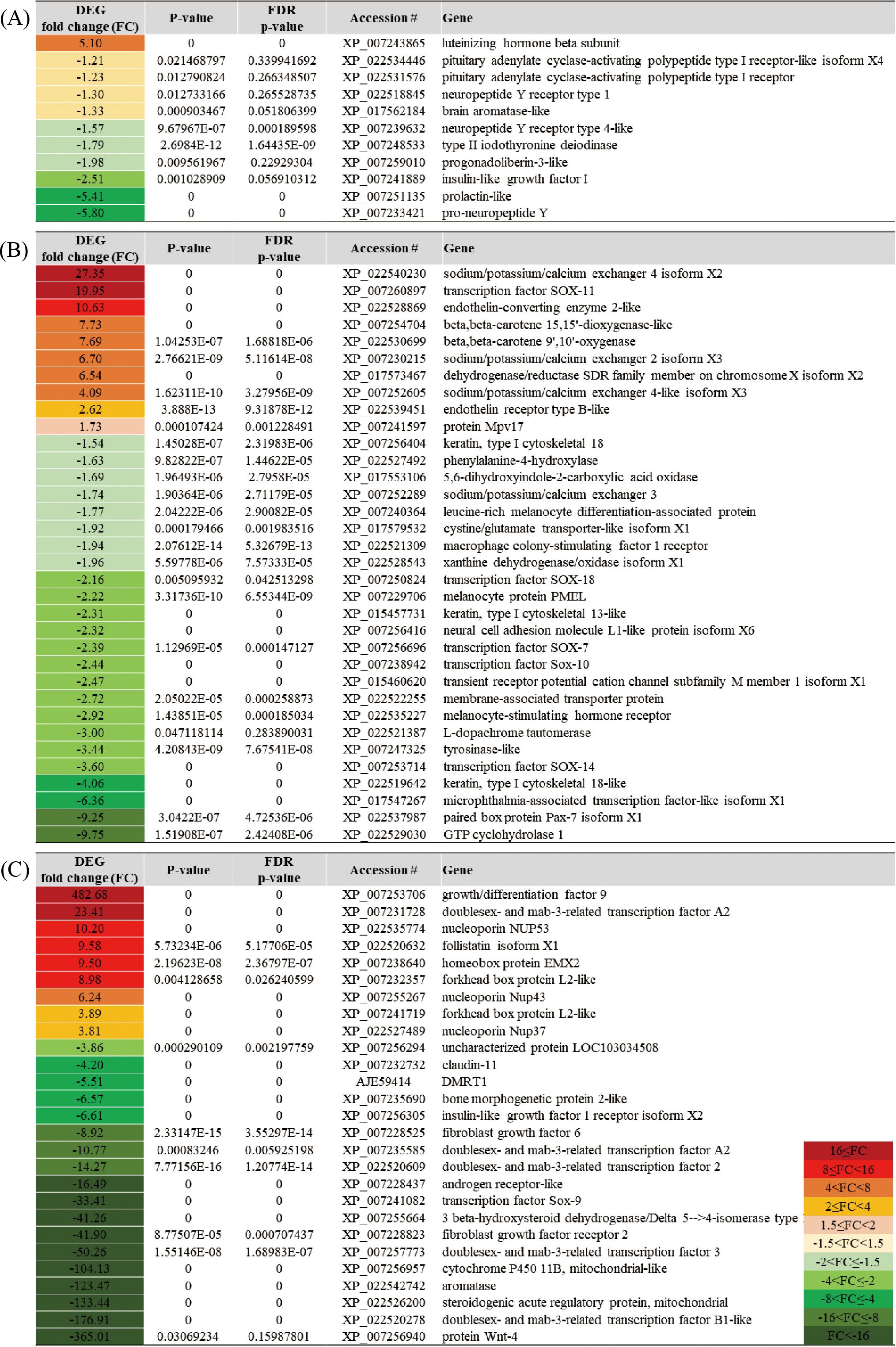
Figure 9. List of genes predominantly expressed in specific tissues: (A) BRA, (B) FIN, and (C) GON, showing differential expression patterns between females and males.
In the caudal fin, the gene with higher expression in females was sodium/potassium/calcium exchanger 4 isoform X2 (NCKX4 x2), showing a fold change of 27.35, and beta-beta-carotene 15,15’-dioxygenase-like (BCO1) with a fold change of 7.73. In contrast, the genes with higher expression in males were melanocortin receptor (MC) with a fold change of -2.22 and melanocyte-stimulating hormone receptor (MSHR) with -2.92 (Figure 9B).
In the gonad, the gene with higher expression in females was growth/differentiation factor 9 (GDF9), showing a remarkable fold change of 482.68. Other genes with higher expression in females included doublesex- and mab-3-related transcription factor A2 (DMRT2) with a fold change of 23.41, follistatin isoform X1 (FST x1) with 9.58, and homeobox protein EMX2 (EMX2) with 9.50. In contrast, the gene with higher expression in males was aromatase (CYP19A1), showing a fold change of -123.47 (Figure 9C).
3.8 protein-protein interaction in pigmentation
The interaction network of 16 proteins related to pigmentation was analyzed in the caudal fin. Among the 16 proteins, 3 were predominantly expressed in females, while the remaining 13 were predominantly expressed in males (Figure 10). The proteins with higher expression in females were identified as Endothelin-converting enzyme 2-like (ECE2), endothelin receptor type B-like (ednrb1a), and protein Mpv17 (mpv17), with fold change values of 10.63, 2.62, and 1.73, respectively. In contrast, the proteins with higher expression in males were melanocyte stimulating hormone receptor (mc1r), Tyrosinase-like (tyr), and microphtalmia-associated transcription factor-like isoform X1 (mitfa), with fold change values of -2.92, -3.44, and -6.36, respectively. Additionally, proteins such as melanocyte protein PMEL (pmela), membrane-associated transporter protein (slc45a2), transcription factor Sox-10 (sox10), and xanthine dehydrogenase/oxidase isoform x1 (xdh) exhibited higher expression in males. The protein-protein interaction (PPI) network analysis identified a total of 20 protein nodes and 60 edges. The average local clustering coefficient was 0.647, indicating a tendency for the proteins to form clusters within the network. Notably, the PPI enrichment p-value was less than 1.0e-16, strongly suggesting that these protein interactions were not random, but rather form a functionally significant protein network.
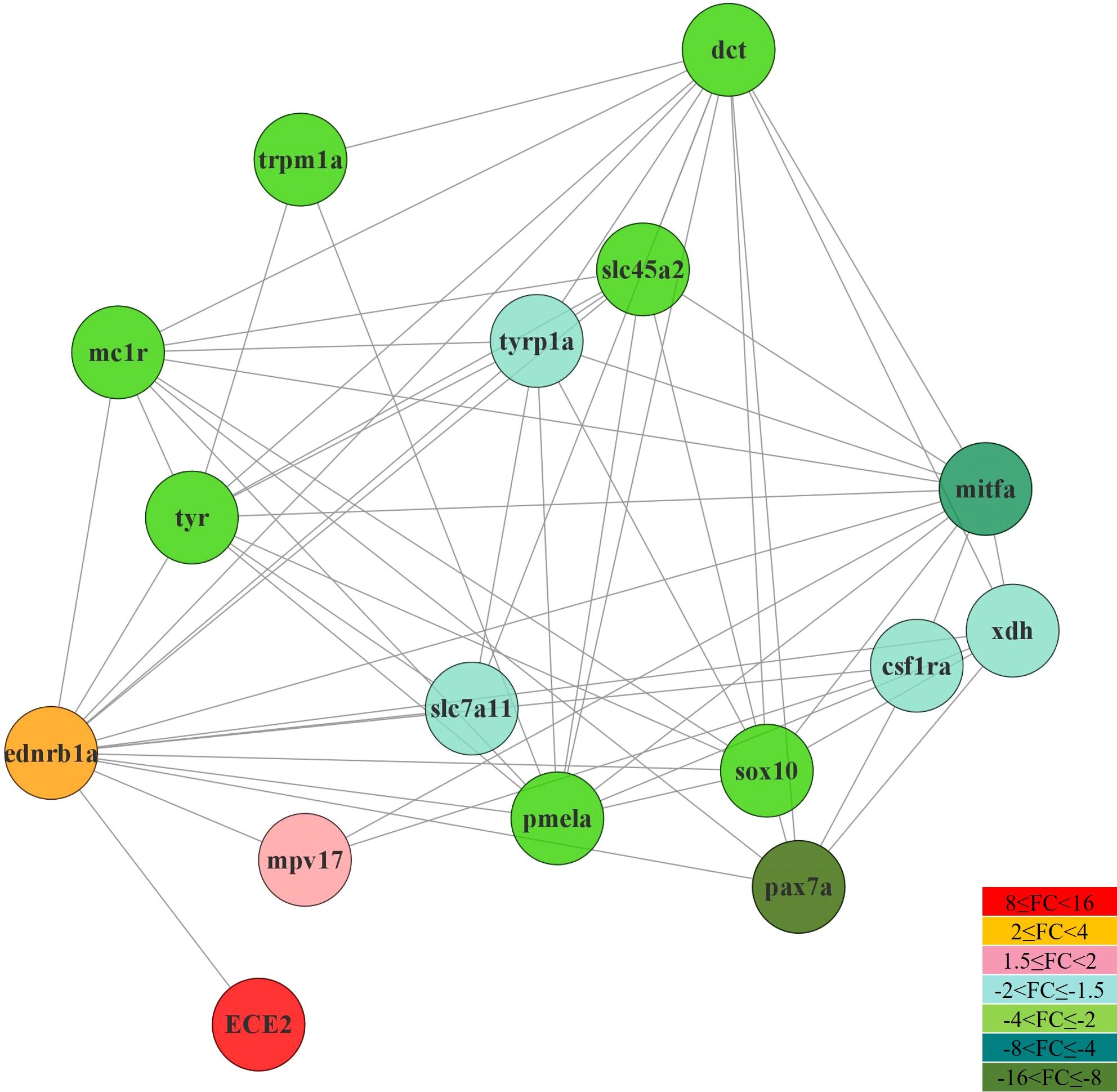
Figure 10. Interaction network and expression pattern of pigment cell-related genes in the dermis of silvertip tetra using the zebrafish (D. rerio) organism database in STRING v11.0 and Cytoscape v3.8.1.
3.9 Validation of DEGs in RNA-seq by qRT-PCR
To validate the expression patterns of DEGs, qRT-PCR was performed on five genes selected from the BRA, FIN, and GON. In the brain-pituitary, the selected genes were LHB, Type II iodothyronine deiodinase (DIO2), IGF1, PRL, and Pro-neuropeptide Y (NPY). In the caudal fin, the selected genes were BCO1, PMEL, Dehydrogenase/reductase (SDR family) member (DHRS), Sodium/calcium/potassium exchanger 2 (NCKX2), and SRY (sex determining region Y)-box 7 (SOX7). In the gonads, the selected genes were Paired box 7 (PAX7), GDF9, (sex determining region Y)-box 9 (SOX9), Doublesex and mab-3 related transcription factor 1 (DMRT1), and Forkhead box L2 (FOXL2). qRT-PCR and RNA-seq results showed similar overall expression trends, despite variations in fold change among genes. Correlation analysis between qRT-PCR data and RNA-seq data revealed a correlation coefficient (R²) of 0.9097, indicating that 90.97% of the variation between the two datasets was consistent (Figure 11).
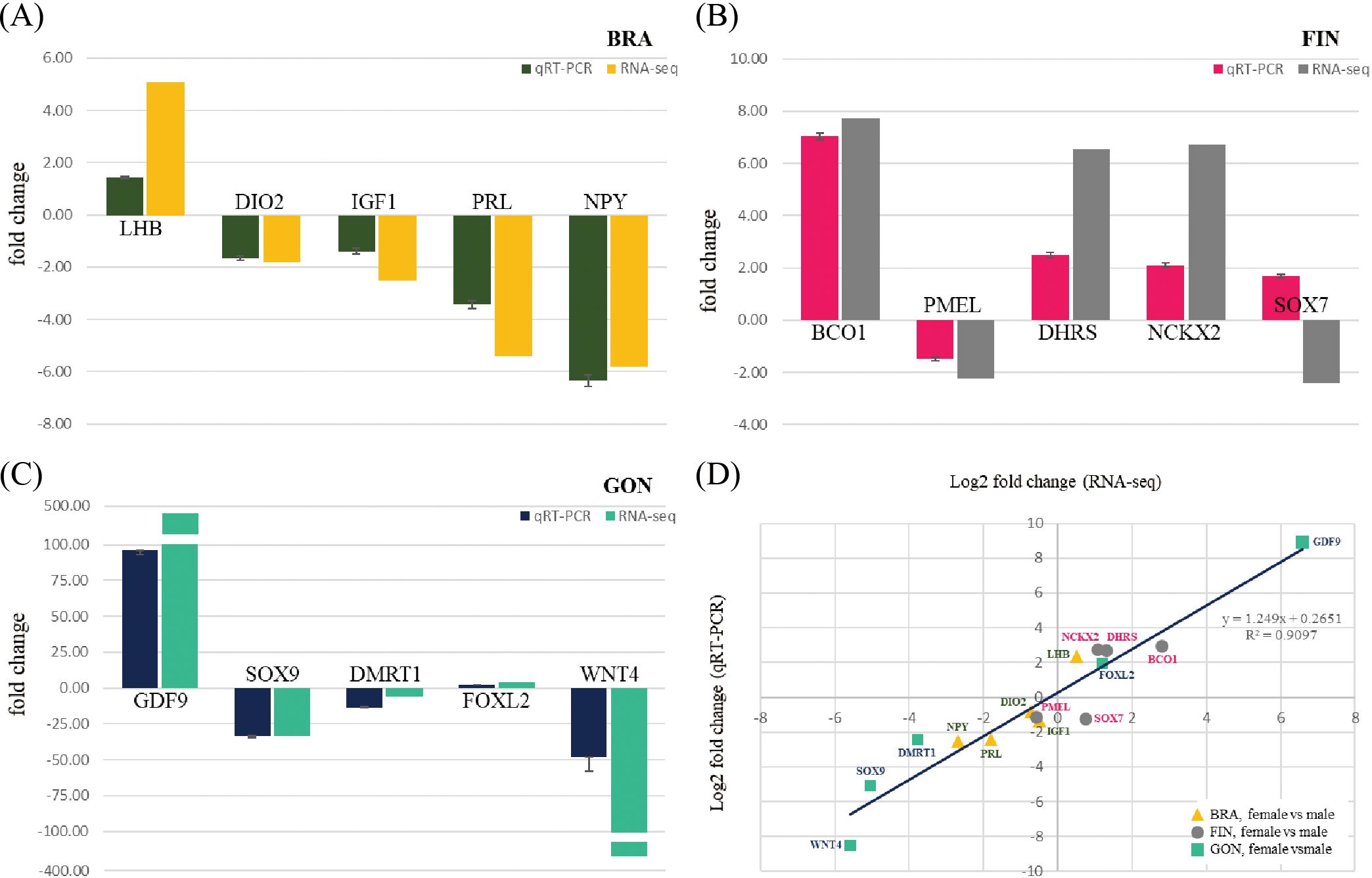
Figure 11. qRT-PCR validation of selected genes from each tissue in the whole-transcriptome assembly. Fold changes in the relative expression of selected genes from (A) BRA, (B) FIN, and (C) GON in female and male silvertip tetra. (D) Correlation between the log2 fold change from the whole-transcriptome expression value and the log2 ratio obtained by qRT-PCR analysis for 15 selected DEGs between females and males.
4 Discussion
The use of fish as experimental models has been steadily increasing (Ahn et al., 2023). Among the small fish models, zebrafish and medaka are the most widely utilized, particularly in recent studies focusing on behavioral tests under stress stimuli (Lai et al., 2023). This increased use is attributed to several advantages, including a higher rate of reproduction, easier handling techniques, sociability, and, most importantly, their evolutionary conserved genetic make-up, neural circuitry, and neuropeptide molecular structure and function shared with mammalian species (Matsui, 2017; Nakajo et al., 2020). However, despite these advantages, zebrafish and medaka present limitations for research on pigmentation and sex determination or differentiation, as distinguishing between male and female based solely on phenotype, particularly pigmentation, is challenging (Wittbrodt et al., 2002; Avdesh et al., 2012; Delcourt et al., 2018). In contrast, silvertip tetra, the fish model used in this study, offers both the advantages of a small fish model and the ease of distinguishing between sexes based on pigmentation alone. Therefore, this model shows high potential as a research model for studies on pigmentation, sex determination, and sexual differentiation.
It is known that zebrafish (D. rerio) hatch at approximately 48 hours at 28.5°C, and medaka (O. latipes) hatch at approximately 9 days at 26°C (Kimmel et al., 1995; Iwamatsu, 2004). In contrast, silvertip tetra (H. nana) hatch at around 22 hours under 25°C conditions, showing a faster hatching time compared to these two representative model species. However, just as the developmental stages of fertilized eggs in zebrafish and medaka vary with temperature, the embryonic development time of silvertip tetra may also vary depending on temperature (Schirone and Gross, 1968; Koger et al., 1999). Considering that the embryonic development process was conducted at 25°C, it is necessary to further investigate the effect of temperature changes on the development time to evaluate whether silvertip tetra exhibit consistent developmental patterns under different environmental conditions. Generally, the time required for hatching in Characidae species varies according to water temperature, but they are generally known for their short embryogenesis (Romagosa et al., 2001; Gomes et al., 2013; Park et al., 2014; Dos Santos et al., 2016; Maria et al., 2017; Santos et al., 2020). The short duration of embryogenesis in Characidae species has been reported by Reynalte-Tataje et al. (2004) and Nakaghi et al. (2014) to be associated with a large perivitelline space, small and free oocytes, as well as the need for reproductive migrations and increased chances of survival.
The HNA FIN cells exhibited a fibroblast-like morphology similar to that of small ornamental fish species such as zebrafish (D. rerio), medaka (O. latipes), guppy (P. reticulata), as well as medium to large species such as rainbow trout (Oncorhynchus mykiss), Australasian snapper (Chrysophrys auratus), and grouper (Epinephelus coioides) (Komura et al., 1988; Ossum et al., 2004; Qin et al., 2006; Gökçe and Üçüncü, 2017; Kalaiselvi Sivalingam et al., 2019; Chong et al., 2022). In terms of cell proliferation temperature, HNA FIN cells demonstrated optimal growth at a higher temperature compared to DrF cells (zebrafish), GS cells (grouper), and PSF cells (guppy) (Qin et al., 2006; Gökçe and Üçüncü, 2017; Kalaiselvi Sivalingam et al., 2019).
Rainbow trout, a cold-water species, thrives at temperatures ranging from 1-25°C, with the optimal cell proliferation temperature reported to be 21°C (Ossum et al., 2004). In contrast, zebrafish, a tropical species, inhabits temperatures between 16-28°C, with optimal cell proliferation occurring at 28°C and 30°C (Kumar et al., 2016; Sathiyanarayanan et al., 2023). Grouper, which inhabits temperatures between 24-30°C, also shows optimal cell growth at 30°C and 35°C (Wen et al., 2008). These findings suggest a correlation between the optimal environmental temperature of a fish species and its cell proliferation temperature. Although this study tested cell proliferation at 4°C intervals, further experiments with more precise temperature intervals are needed to accurately determine the optimal temperature.
Cryopreservation is known to affect cell viability depending on the cryoprotectant used (Miyamoto et al., 2012; Jaiswal and Vagga, 2022). Various cryopreservation media are used for fish cells, but DMSO and FBS are typically added to the culture medium. In this experiment, CellBanker2, a serum-free cryopreservation medium that has been used for other fish cell lines, was utilized (Kawato et al., 2017; Chen et al., 2024). Australasian snapper cell lines have been preserved with a cryopreservation medium containing 10% FBS and 20% DMSO, while turbot cell lines have been reported to use 20% FBS and 10% DMSO (Ossum et al., 2004; Chong et al., 2022). Upon thawing, both cell lines showed survival rates exceeding 80%. For zebrafish cell lines, 40% FBS and 10% DMSO were used as cryoprotectants, and the survival rate after thawing was reported to be 70-75%, which is comparable to the survival rate observed in the silvertip tetra cell line after thawing (Sathiyanarayanan et al., 2023).
The results of the PCA plot indicated that the brain-pituitary of both males and females exhibited closer gene expression relationships compared to the gonads. This can be attributed to the fact that both organs play crucial roles in regulating the reproductive axis (brain–pituitary–gonadal axis) regardless of sex (Nyuji et al., 2020). Specifically, the transcriptomic overlap between the brain-pituitary was found to be notably high, suggesting that the molecular mechanisms governing reproductive function operate similarly in both sexes. In species such as the common pandora (Pagellus erythrinus), red porgy (Pagrus pagrus), sharpsnout seabream (Diplodus puntazzo), and African cichlids, the brain and pituitary play key roles in regulating reproductive hormones, a function that is essential for both males and females, resulting in similar gene expression patterns between the sexes. On the other hand, the gonads (ovaries and testes) exhibited more pronounced differences in gene expression patterns between sexes (Böhne et al., 2014; Manousaki et al., 2014; Tsakogiannis et al., 2018). These findings reflect a common pattern observed in many fish studies, where sex-specific similarities or differences in gene expression are often tissue-dependent. An interesting observation is seen in sex-changing fish, such as the clownfish (Amphiprion bicinctus), where mature males and females exhibit gene expression relationships in the brain and gonads similar to those of other fish species. However, during sex transition, there was a marked divergence in gene expression relationships between male and female brains (Casas et al., 2016). This indicates that while the brain plays a significant role during the sex transition process, there is little difference in gene expression between the sexes when the individual is in a s Supplementary Table Sexual state. In the case of the fin, it was observed that gene expression relationships related to pigmentation were more prominent than those related to sex differences, a phenomenon that has also been mentioned in studies of albinism in cichlid fish, specifically Aulonocara baenschi (Lee and Lee, 2020).
The mature male silvertip tetra (H. nana) exhibits a vibrant orange body coloration, while the female displays a lighter yellow hue. This coloration is also reflected in their caudal fins, showing a similar pattern. Interestingly, in the caudalis ventralis region and the middle caudal fin region, a high density of melanophores is present. These areas are subject to pigment modulation in response to environmental changes, stress, and other factors. In Astatotilapia burtoni, a species of cichlid fish, increased levels of α-melanocyte-stimulating hormone (α-MSH) have been reported to cause yellowness of the body and dispersal of xanthophore pigments in both morphs (Dijkstra et al., 2017). In silvertip tetra, gene expression differences related to pigmentation were observed between males and females. In females, the expression of BCO1 and BCO2 was significantly higher. These genes are responsible for the active breakdown of carotenoids, which likely explains the lighter coloration of females compared to males. This observation is consistent with findings in salmon and rainbow trout, where beta-carotene oxygenase, a key gene involved in carotenoid metabolism, converts colored carotenoids into colorless ones (Lehnert et al., 2019; Wu et al., 2022). In addition, genes involved in ion regulation and pigment cell function, such as Sodium/potassium/calcium exchanger 2 (NCKX2) and NCKX4, as well as SOX11, which is associated with nerve development and cell differentiation in the peripheral nervous system, were more highly expressed in females than in males. This is consistent with previous studies showing that NCKX2 and SOX11 play important roles in ion channel function and their accessory subunits after peripheral nerve damage (Sandercock et al., 2019). Furthermore, studies on Boer and Macheng black crossbred goats showed that NCKX2 and NCKX4 were more highly expressed in black-coated regions than in white-coated regions, which suggests a similar trend in silvertip tetra females. The increased expression of these genes may be involved in melanosome formation (Xiong et al., 2020). Moreover, Sodium/potassium/calcium exchanger 5 (NCKX5) is known to be crucial for melanosome formation, further supporting this connection (Lamason et al., 2005; Monteiro et al., 2024). In males, several genes associated with melanophore function and pigmentation showed higher expression. These include L-dopachrome tautomerase (dct), MSHR, Paried box 7 (PAX7), Premelanosome protein (PMEL), SRY (sex determining region Y)-box 10 (SOX10), and TYR, all of which are involved in melanin synthesis and pigmentation. The expression patterns of these genes are consistent with previous studies on melanin synthesis and pigmentation (Hou et al., 2006; Harris et al., 2010; McNamara et al., 2021; Li et al., 2022). PPI network analysis revealed the functional connectivity and interactions between pigmentation-related genes. Endothelin signaling and Mpv17 play crucial roles in the survival and proliferation of iridophores (Singh and Nüsslein-Volhard, 2015). The expression of these genes was higher in females, which may be linked to their lighter body coloration compared to males.
In this study, gene expression relationships between females and males were closer in the brain-pituitary compared to the caudal fin and gonads, although certain genes exhibited sex-biased expression. The LHB, known as a glycoprotein hormone (gonadotropin) secreted by the pituitary gland, was more highly expressed in the brain-pituitary of male silvertip tetra than in females. This trend is consistent with findings in other fish species, such as the Southern catfish (Clarias gariepinus) and Catla catla (Wu et al., 2009; Rather et al., 2016). Higher expression of FST and GDF9 in female ovaries reflects their roles in ovarian function. LHB regulates ovulation in the ovaries, while FST is a key regulator of oocyte maturation and germ cell development, working alongside GDF9 to promote cellular growth in the ovaries (Li et al., 2008; Bilezikjian and Vale, 2011). Conversely, IGF-I, which exhibited higher expression in the brain-pituitary of males, is a major hormonal regulator of differentiation, growth, proliferation, and development. It has been reported to promote the expression of prolactin while inhibiting the expression of growth hormone, findings that align with the results of this study (Fruchtman et al., 2000; Kajimura et al., 2002). Additionally, the higher expression of IGF-I receptor and fibroblast growth factor in the testes of males, compared to females, further supports the connection between IGF-I expression in the brain-pituitary and its role in the gonads (De Mattos et al., 2023). Although Protein Wnt-4 (WNT4) is predominantly expressed in the ovaries during gonadal differentiation, studies have reported that mutations in WNT4 can lead to testes-biased expression (Jeays-Ward et al., 2004). Additionally, due to teleost-specific whole-genome duplication, WNT4 has evolved into various isoforms (Nicol et al., 2012). In the gonads of silvertip tetra, WNT4 was predominantly expressed in males, and sequence analysis revealed high homology with Protein Wnt-4b (WNT4b) found in other fish species. This pattern is similar to the slightly higher expression of WNT4 in the testes of rainbow trout (O. mykiss) (Nicol et al., 2012).
5 Conclusion
This study establishes the silvertip tetra (H. nana) as a promising model organism, particularly for research on pigmentation, sexual dimorphism, and gene function in fish. We demonstrated significant gene expression differences between sexes across critical tissues, such as brain-pituitary, caudal fin, and gonads, directly linked to pigmentation and sexual differentiation pathways. The transcriptomic analysis revealed key DEGs associated with pigmentation processes, including melanin synthesis and carotenoid metabolism, as well as sex differentiation, showcasing the biological relevance of silvertip tetra in studying these traits. Furthermore, successfully establishing and optimizing a caudal fin-derived cell line (HNA FIN) provides a versatile tool for molecular and cellular studies. The cell line demonstrated robust proliferation under optimized conditions and was capable of foreign gene transfection, which significantly enhances its utility in functional genomics and transgenic research. These attributes, combined with the species’ clear sexual dimorphism, short embryonic development time, and high reproductive rate, underscore its suitability for developmental biology and comparative genomic studies. Importantly, the results of this study highlight the practical applications of silvertip tetra as a cost-effective and efficient alternative to traditional model organisms, such as zebrafish and medaka, particularly in research areas where pigmentation and sex differentiation are central themes. By integrating molecular, cellular, and developmental insights, this study lays the groundwork for future exploration of silvertip tetra in a variety of biological fields, including evolutionary biology, toxicology, and aquaculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The whole-transcriptome raw data and the biological sample information used in the present study were submitted to the National Center for Biotechnology Information (NCBI). The Sequence Read Archive (SRA) raw-data can be accessed in NCBI with the following information: BioProject (PRJNA1017117) and SRA (SRX21774997, SRX21774996, SRX21774995, SRX21774994, RX21774993, SRX21774992).
Ethics statement
The animal studies were approved by Institutional Animal Care and Use Committee (IACUC) of CellQua (CQ-IACUC-2023-1-2) and Gangneung-Wonju National University (GWNU-2019-26). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
HL: Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. M-GL: Writing – review & editing. J-HC: Funding acquisition, Writing – review & editing. MK: Writing – review & editing. SL: Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (RS-2023-00254803).This study was funded by the National Institute of Fisheries Science, Ministry of Oceans and Fisheries, South Korea (R2025022).
Conflict of interest
Authors HL and SL was employed by the company CellQua, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1517938/full#supplementary-material
Supplementary Table 1 | Primer sequences for qRT-PCR validation of DEGs in the whole-transcriptome assembly and for the COI gene used in species identification are detailed below.
Supplementary Table 2 | Summary of de novo assembled contigs
Supplementary Table 3 | Information of RNA-seq raw data and reads mapped with the reference whole-transcriptome assembly
References
Ahn N., Park J., Roh S. (2023). Use of laboratory animals and issues regarding the procurement of animals for research in Korea. Lab. Anim. Res. 39, 10. doi: 10.1186/s42826-023-00161-8
Ankley G. T., Johnson R. D. (2004). Small fish models for identifying and assessing the effects of endocrine-disrupting chemicals. ILAR J. 45, 469–483. doi: 10.1093/ilar.45.4.469
Avdesh A., Chen M., Martin-Iverson M. T., Mondal A., Ong D., Rainey-Smith S., et al. (2012). Regular care and maintenance of a zebrafish (Danio rerio) laboratory: an introduction. JoVE 69, 4196. doi: 10.3791/4196
Babin P. J., Goizet C., Raldúa D. (2014). Zebrafish models of human motor neuron diseases: advantages and limitations. Prog. Neurobiol. 118, 36–58. doi: 10.1016/j.pneurobio.2014.03.001
Bilezikjian L. M., Vale W. W. (2011). The local control of the pituitary by activin signaling and modulation. Open Neuroendocrinol. J. (Online). 4, 90. doi: 10.2174/1876528901104010090
Böhne A., Sengstag T., Salzburger W. (2014). Comparative transcriptomics in East African cichlids reveals sex-and species-specific expression and new candidates for sex differentiation in fishes. Genome Biol. Evol. 6, 2567–2585. doi: 10.1093/gbe/evu200
Cal L., Suarez-Bregua P., Cerdá-Reverter J. M., Braasch I., Rotllant J. (2017). Fish pigmentation and the melanocortin system. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 211, 26–33. doi: 10.1016/j.cbpa.2017.06.001
Casas L., Saborido-Rey F., Ryu T., Michell C., Ravasi T., Irigoien X. (2016). Sex change in clownfish: molecular insights from transcriptome analysis. Sci. Rep. 6, 35461. doi: 10.1038/srep35461
Chen S. N., Huo H. J., Jin Y., Peng X. Y., Li B., Wu X. Y., et al. (2024). The infectious haemorrhagic syndrome virus (IHSV) from rice-field eel (Monopterus albus): Isolation, genome sequence, cross-infection and induced-immune response in Chinese perch (Siniperca chuatsi). Aquaculture. 583, 740561. doi: 10.1016/j.aquaculture.2024.740561
Chong G. L., Böhmert B., Lee L. E., Bols N. C., Dowd G. C. (2022). A continuous myofibroblast precursor cell line from the tail muscle of Australasian snapper (Chrysophrys auratus) that responds to transforming growth factor beta and fibroblast growth factor. In Vitro Cell. Dev. Biol.-Anim. 58, 922–935. doi: 10.1007/s11626-022-00734-2
Delcourt J., Ovidio M., Denoël M., Muller M., Pendeville H., Deneubourg J-L., et al. (2018). Individual identification and marking techniques for zebrafish. Rev. Fish Biol. Fish. 28, 839–864. doi: 10.1007/s11160-018-9537-y
De Mattos K., Pierre K. J., Tremblay J. J. (2023). Hormones and signaling pathways involved in the stimulation of leydig cell steroidogenesis. Endocrines. 4, 573–594. doi: 10.3390/endocrines4030041
Diekmann M., Hultsch V., Nagel R. (2004). On the relevance of genotoxicity for fish populations I: effects of a model genotoxicant on zebrafish (Danio rerio) in a complete life-cycle test. Aquat. Toxicol. 68, 13–26. doi: 10.1016/j.aquatox.2004.01.020
Dijkstra P. D., Maguire S. M., Harris R. M., Rodriguez A. A., DeAngelis R. S., Flores S. A., et al. (2017). The melanocortin system regulates body pigmentation and social behaviour in a colour polymorphic cichlid fish. Proc. R. Soc B Biol. Sci. 284, 20162838. doi: 10.1098/rspb.2016.283
Dos Santos M. P., Yasui G. S., Xavier P. L. P., de Macedo Adamov N. S., do Nascimento N. F., Fujimoto T., et al. (2016). Morphology of gametes, post-fertilization events and the effect of temperature on the embryonic development of Astyanax altiparanae (Teleostei, Characidae). Zygote. 24, 795–807. doi: 10.1017/S0967199416000101
Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., Bridge A. J., et al. (2017). InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Fruchtman S., Jackson L., Borski R. (2000). Insulin-like growth factor I disparately regulates prolactin and growth hormone synthesis and secretion: studies using the teleost pituitary model. Endocrinology. 141, 2886–2894. doi: 10.1210/endo.141.8.7616
Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gökçe B., Üçüncü S. (2017). The establishment of a fibroblastic cell line from caudal tissue of poecilia reticulata, peters 1859. Asian J. Biol. 3, 1–7. doi: 10.9734/AJOB/2017/34783
Gomes R. Z., Sato Y., Rizzo E., Bazzoli N. (2013). Early development of Brycon orthotaenia (Pisces: Characidae). Zygote. 21, 11. doi: 10.1017/S0967199411000311
Goswami M., Yashwanth B. S., Trudeau V., Lakra W. S. (2022). Role and relevance of fish cell lines in advanced in vitro research. Mol. Biol. Rep. 1–19. doi: 10.1007/s11033-021-06997-4
Götz S., García-Gómez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Guiguen Y., Fostier A., Herpin A. (2018). Sex determination and differentiation in fish: genetic, genomic, and endocrine aspects. Sex Control Aquacult, 35–63. doi: 10.1002/9781119127291.ch2
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D., Bowden J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Harris M. L., Baxter L. L., Loftus S. K., Pavan W. J. (2010). Sox proteins in melanocyte development and melanoma. Pigm. Cell Melanoma Res. 23, 496–513. doi: 10.1111/j.1755-148X.2010.00711.x
Harris M. P., Henke K., Hawkins M. B., Witten P. E. (2014). Fish is Fish: the use of experimental model species to reveal causes of skeletal diversity in evolution and disease. J. Appl. Ichthyol. 30, 616–629. doi: 10.1111/jai.12533
He J., Gao J., Huang C., Li C. (2014). Zebrafish models for assessing developmental and reproductive toxicity. Neurotoxicol. Teratol. 42, 35–42. doi: 10.1016/j.ntt.2014.01.006
Hou L., Arnheiter H., Pavan W. J. (2006). Interspecies difference in the regulation of melanocyte development by SOX10 and MITF. PNAS. 103, 9081–9085. doi: 10.1073/pnas.0603114103
Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Iwamatsu T. (2004). Stages of normal development in the medaka Oryzias latipes. Mech. Dev. 121, 605–618. doi: 10.1016/j.mod.2004.03.012
Jaiswal A. N., Vagga A. (2022). Cryopreservation: A review article. Cureus. 14, e31564. doi: 10.7759/cureus.31564
Jeays-Ward K., Dandonneau M., Swain A. (2004). Wnt4 is required for proper male as well as female sexual development. Dev. Biol. 276, 431–440. doi: 10.1016/j.ydbio.2004.08.049
Kajimura S., Uchida K., Yada T., Hirano T., Aida K., Grau E. G. (2002). Effects of insulin-like growth factors (IGF-I and-II) on growth hormone and prolactin release and gene expression in euryhaline tilapia, Oreochromis mossambicus. Gen. Comp. Endocrinol. 127, 223–231. doi: 10.1016/S0016-6480(02)00055-2
Kalaiselvi Sivalingam N. N., Seepoo A. M., Gani T., Selvam S., Azeez Sait S. H. (2019). Zebrafish fin-derived fibroblast cell line: A model for in vitro wound healing. J. Fish Dis. 42, 573–584. doi: 10.1111/jfd.12965
Kawato Y., Yamashita H., Yuasa K., Miwa S., Nakajima K. (2017). Development of a highly permissive cell line from spotted knifejaw (Oplegnathus punctatus) for red sea bream iridovirus. Aquaculture. 473, 291–298. doi: 10.1016/j.aquaculture.2017.02.027
Khoo G., Lim T. M., Chan W., Phang V. P. (1999). Linkage analysis and mapping of three sex-linked color pattern genes in the guppy, Poecilia reticulata. Zool. Sci. 16, 893–903. doi: 10.2108/zsj.16.893
Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Koger C. S., Teh S. J., Hinton D. E. (1999). Variations of light and temperature regimes and resulting effects on reproductive parameters in medaka (Oryzias latipes). Biol. Reprod. 61, 1287–1293. doi: 10.1095/biolreprod61.5.1287
Komura J., Mitani H., Shima A. (1988). Fish cell culture: Establishment of two fibroblast-like cell lines (OL-17 and OL-32) from fins of the medaka, Oryzias latipes. In Vitro Cell. Dev. Biol. Anim. 24, 294–298. doi: 10.1007/BF02628830
Kottler V. A., Schartl M. (2018). The colorful sex chromosomes of teleost fish. Genes. 9, 233. doi: 10.3390/genes9050233
Kumar A., Singh N., Goswami M., Srivastava J. K., Mishra A. K., Lakra W. S. (2016). Establishment and characterization of a new muscle cell line of zebrafish (Danio rerio) as an in vitro model for gene expression studies. Anim. Biotechnol. 27, 166–173. doi: 10.1080/10495398.2016.1147455
Kumar M. S., Singh V. K., Mishra A. K., Kushwaha B., Kumar R., Lal K. K. (2024). Fish cell line: depositories, web resources and future applications. Cytotechnology. 76, 1–25. doi: 10.1007/s10616-023-00601-2
Lai N. H. Y., Mohd Zahir I. A., Liew A. K. Y., Ogawa S., Parhar I., Soga T. (2023). Teleosts as behaviour test models for social stress. Front. Behav. Neurosci. 17. doi: 10.3389/fnbeh.2023.1205175
Lamason R. L., Mohideen M. P., Mest J. R., Wong A. C., Norton H. L., Aros M. C., et al. (2005). SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 310, 1782–1786. doi: 10.1126/science.11162
Lee H. J., Kim K. T., Kim M. S., Lee S. Y. (2024). Dataset for selection of stable reference genes for accurate quantitative gene expression analysis in silvertip tetra (Hasemania nana): Implications for sex differentiation and determination. Data Brief. 53, 110221. doi: 10.1016/j.dib.2024.110221
Lee S. Y., Lee H. J. (2020). Comprehensive RNA-seq analysis to evaluate the pigmentation-related genes involved in albinism of cichlid fish. Aulonocara baenschi. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00723
Lehnert S. J., Christensen K. A., Vandersteen W. E., Sakhrani D., Pitcher T. E., Heath J. W., et al. (2019). Carotenoid pigmentation in salmon: variation in expression at BCO2-l locus controls a key fitness trait affecting red coloration. Proc. R. Soc B. 286, 20191588. doi: 10.1098/rspb.2019.1588
Li Z., Li Q., Xu C., Yu H. (2022). Molecular characterization of Pax7 and its role in melanin synthesis in. Crassostrea gigas. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 260, 110720. doi: 10.1016/j.cbpb.2022.110720
Li Q., McKenzie L. J., Matzuk M. M. (2008). Revisiting oocyte–somatic cell interactions: in search of novel intrafollicular predictors and regulators of oocyte developmental competence. Mol. Hum. Reprod. 14, 673–678. doi: 10.1093/molehr/gan064
Li M., Zhao L., Page-McCaw P. S., Chen W. (2016). Zebrafish genome engineering using the CRISPR–Cas9 system. Trends Genet. 32, 815–827. doi: 10.1016/j.tig.2016.10.005
Liebsch M., Grune B., Seiler A., Butzke D., Oelgeschläger M., Pirow R., et al. (2011). Alternatives to animal testing: current status and future perspectives. Arch. Toxicol. 85, 841–858. doi: 10.1007/s00204-011-0718-x
Lindholm A., Breden F. (2002). Sex chromosomes and sexual selection in poeciliid fishes. Am. Nat. 160, S214–S224. doi: 10.1086/342898
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Luo M., Lu G., Yin H., Wang L., Atuganile M., Dong Z. (2021). Fish pigmentation and coloration: Molecular mechanisms and aquaculture perspectives. Rev. Aquacult. 13, 2395–2412. doi: 10.1111/raq.1258
Manousaki T., Tsakogiannis A., Lagnel J., Sarropoulou E., Xiang J. Z., Papandroulakis N., et al. (2014). The sex-specific transcriptome of the hermaphrodite sparid sharpsnout seabream (Diplodus puntazzo). BMC Genomics 15, 1–16. http://www.biomedcentral.com/1471-2164/15/6
Maria A. N., Ninhaus-Silveira A., Orfão L. H., Viveiros A. T. (2017). Embryonic development and larval growth of Brycon nattereri Günthe(Characidae) and its implications for captive rearing. Zygote. 25, 711–718. doi: 10.1017/S0967199417000594
Matsui H. (2017). The use of fish models to study human neurological disorders. Neurosci. Res. 120, 1–7. doi: 10.1016/j.neures.2017.02.004
McNamara M. E., Rossi V., Slater T. S., Rogers C. S., Ducrest A-L., Dubey S., et al. (2021). Decoding the evolution of melanin in vertebrates. Trends Ecol. Evol. 36, 430–443. doi: 10.1016/j.tree.2020.12.0
Mieno A., Karino K. (2016). Sexual dimorphism and dichromatism in the cyprinid fish Puntius titteya. Ichthyol. Res. 64, 250–255. doi: 10.1007/s10228-016-0559-y
Miyamoto Y., Noguchi H., Yukawa H., Oishi K., Matsushita K., Iwata H., et al. (2012). Cryopreservation of induced pluripotent stem cells. Cell Med. 3, 89–95. doi: 10.3727/215517912X639405
Monteiro J. P., Pröhl H., Lyra M. L., Brunetti A. E., de Nardin E. C., Condez T. H., et al. (2024). Expression patterns of melanin-related genes are linked to crypsis and conspicuousness in a pumpkin toadlet. Mol. Ecol. e17458. doi: 10.1111/mec.17458
Mukherjee P., Roy S., Ghosh D., Nandi S. K. (2022). Role of animal models in biomedical research: a review. Lab. Anim. Res. 38, 18. doi: 10.1186/s42826-022-00128-1
Nakaghi L. S. O., Neumann E., Faustino F., Mendes J. M. R., Braga F. M. (2014). Moments of induced spawning and embryonic development of Brycon amazonicus (Teleostei, Characidae). Zygote. 22, 549. doi: 10.1017/S0967199413000130
Nakajo H., Tsuboi T., Okamoto H. (2020). The behavioral paradigm to induce repeated social defeats in zebrafish. Neurosci. Res. 161, 24–32. doi: 10.1016/j.neures.2019.11.004
Nicol B., Guerin A., Fostier A., Guiguen Y. (2012). Ovary-predominant wnt4 expression during gonadal differentiation is not conserved in the rainbow trout (Oncorhynchus mykiss). Mol. Reprod. Dev. 79, 51–63. doi: 10.1002/mrd.21404
Novelo N. D., Gomelsky B., Coyle S. D., Kramer A. G. (2021). Evaluation of growth, sex (male proportion; sexual dimorphism), and color segregation in four cross combinations of different strains of XX female and YY male Nile Tilapia. J. World Aquacult. Soc 52, 445–456. doi: 10.1111/jwas.12742
Nyuji M., Hongo Y., Yoneda M., Nakamura M. (2020). Transcriptome characterization of BPG axis and expression profiles of ovarian steroidogenesis-related genes in the Japanese sardine. BMC Genomics 21, 1–18. doi: 10.1186/s12864-020-07080-1
Ossum C. G., Hoffmann E. K., Vijayan M. M., Holt S. E., Bols N. C. (2004). Characterization of a novel fibroblast-like cell line from rainbow trout and responses to sublethal anoxia. J. Fish Biol. 64, 1103–1116. doi: 10.1111/j.1095-8649.2004.0378.x
Otake H., Masuyama H., Mashima Y., Shinomiya A., Myosho T., Nagahama Y., et al. (2010). Heritable artificial sex chromosomes in the medaka, Oryzias latipes. Heredity. 105, 247–256. doi: 10.1038/hdy.2009.174
Park J. M., Kim N. R., Han K. H., Han J. H., Son M. H., Cho J. K. (2014). Spawning behavior, egg development, larvae and juvenile morphology of hyphessobrycon eques (Pisces: characidae) characidae fishes. Dev. Reprod. 18, 241–249. doi: 10.12717/devrep.2014.18.4.241
Qin Q. W., Wu T. H., Jia T. L., Hegde A., Zhang R. Q. (2006). Development and characterization of a new tropical marine fish cell line from grouper, Epinephelus coioides susceptible to iridovirus and nodavirus. J. Virol. Methods 131, 58–64. doi: 10.1016/j.jviromet.2005.07.009
Rather M. A., Bhat I. A., Sharma R. (2016). Identification, cDNA cloning, and characterization of luteinizing hormone beta subunit (Lhb) gene in Catla catla. Anim. Biotechnol. 27, 148–156. doi: 10.1080/10495398.2016.1140055
Reynalte-Tataje D., Zaniboni-Filho E., Esquivel J. R. (2004). Embryonic and larvae development of piracanjuba, Brycon orbignyanus Valencienne>(Pisces, Characidae). Acta Scientiarum. 26, 67–71. doi: 10.4025/actascibiolsci.v26i1.1660
Romagosa E., Narahara M. Y., Fenerich-Verani N. (2001). Stages of embryonic development of the “matrinxã”, Brycon cephalus (Pisces, Characidae). Bol. Inst. Pesca. 27, 29–32.
Sandercock D. A., Barnett M. W., Coe J. E., Downing A. C., Nirmal A. J., Di Giminiani P., et al. (2019). Transcriptomics analysis of porcine caudal dorsal root ganglia in tail amputated pigs shows long-term effects on many pain-associated genes. Front. Vet. Sci. 6, 314. doi: 10.3389/fvets.2019.00314
Santos J. A., Soares C. M., Bialetzki A. (2020). Early ontogeny of yellowtail tetra fish Astyanax lacustris (Characiformes: Characidae). Aquacult. Res. doi: 10.1111/are.14746
Sathiyanarayanan A., Yashwanth B. S., Pinto N., Thakuria D., Chaudhari A., Gireesh Babu P., et al. (2023). Establishment and characterization of a new fibroblast-like cell line from the skin of a vertebrate model, zebrafish (Danio rerio). Mol. Biol. Rep. 50, 19–29. doi: 10.21203/rs.3.rs-1890769/v1
Schirone R. C., Gross L. (1968). Effect of temperature on early embryological development of the zebrafish. Brachydanio rerio. J. Exp. Zool. 169, 43–52. doi: 10.1002/jez.1401690106
Seppey M., Manni M., Zdobnov E. M. (2019). BUSCO: assessing genome assembly and annotation completeness. Gene Prediction: Methods Protoc. 227–245. doi: 10.1007/978-1-4939-9173-0_14
Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Singh A. P., Nüsslein-Volhard C. (2015). Zebrafish stripes as a model for vertebrate colour pattern formation. Curr. Biol. 25, R81–R92. doi: 10.1016/j.cub.2014.11.013
Tsakogiannis A., Manousaki T., Lagnel J., Sterioti A., Pavlidis M., Papandroulakis N., et al. (2018). The transcriptomic signature of different sexes in two protogynous hermaphrodites: Insights into the molecular network underlying sex phenotype in fish. Sci. Rep. 8, 3564. doi: 10.1038/s41598-018-21992-9
Valerie Z., Adelaide D., David A. B., Sofia B. L., Elisabet B., Stephanie B., et al. (2021). Non-animal methods in science and regulation.
Vela J. M., Maldonado R., Hamon M. (2014). The 3Ns of preclinical animal models in biomedical research. In Vivo Models Drug Discovery. (Weinheim: Wiley-VCH). p. 1–26. doi: 10.1002/9783527679348.ch01
Wakamatsu Y., Pristyazhnyuk S., Kinoshita M., Tanaka M., Ozato K. (2001). The see-through medaka: a fish model that is transparent throughout life. PNAS. 98, 10046–10050. doi: 10.1073/pnas.18120429
Wen C. M., Lee C. W., Wang C. S., Cheng Y. H., Huang H. Y. (2008). Development of two cell lines from Epinephelus coioides brain tissue for characterization of betanodavirus and megalocytivirus infectivity and propagation. Aquaculture. 278, 14–21. doi: 10.1016/j.aquaculture.2008.03.020
Wittbrodt J., Shima A., Schartl M. (2002). Medaka-a model organism from the far East. Nat. Rev. Genet. 3, 53–64. doi: 10.1038/nrg704
Wu S., Huang J., Li Y., Zhao L., Liu Z. (2022). Analysis of yellow mutant rainbow trout transcriptomes at different developmental stages reveals dynamic regulation of skin pigmentation genes. Sci. Rep. 12, 256. doi: 10.1038/s41598-021-04255-y
Wu F., Zhang X., Zhang W., Huang B., Liu Z., Hu C., et al. (2009). Expression of three gonadotropin subunits in Southern catfish gonad and their possible roles during early gonadal development. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 153, 44–48. doi: 10.1016/j.cbpa.2008.12.013
Xiong Q., Tao H., Zhang N., Zhang L., Wang G., Li X., et al. (2020). Skin transcriptome profiles associated with black-and white-coated regions in Boer and Macheng black crossbred goats. Genomics. 112, 1853–1860. doi: 10.1016/j.ygeno.2019.10.019
Keywords: silvertip tetra (Hasemania nana), sexual dimorphism, experimental model, cell line, gene expression, transcriptome analysis
Citation: Lee HJ, Lee M-G, Cho J-H, Kim MS and Lee SY (2025) Comprehensive transcriptome profiling of silvertip tetra (Hasemania nana), a new freshwater fish model for gender classification based on color and establishment of a caudal fin-derived cell line. Front. Mar. Sci. 12:1517938. doi: 10.3389/fmars.2025.1517938
Received: 27 October 2024; Accepted: 24 January 2025;
Published: 14 February 2025.
Edited by:
Taewoo Ryu, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Ting Xue, Fujian Normal University, ChinaMurali Sanjeev Kumar, National Bureau of Fish Genetic Resources (ICAR), India
Copyright © 2025 Lee, Lee, Cho, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Yoon Lee, c2FuZ3lvb24wNDIzQGNlbGxxdWEuaW8=
 Hwa Jin Lee
Hwa Jin Lee Mi-Gi Lee
Mi-Gi Lee Jeong-Hyeon Cho
Jeong-Hyeon Cho Min Sun Kim
Min Sun Kim Sang Yoon Lee
Sang Yoon Lee