Abstract
This study investigates the practicality and potential impact of three-dimensional (3D) scanning technology to assess marine mammal strandings. Mobile phones were used to evaluate the technology and its capability to accurately reconstruct topographic features of 11 distinct marine mammal species, across diverse environmental conditions. This process was validated by initially measuring an inflatable whale to an accuracy of greater than 99%, with most morphometrics collected being within 3% of conventional tape rule measurements. Our findings demonstrate the adaptability of the technology in remote environments, particularly for large whale strandings, while showcasing its utility to record morphometrics and enhance necropsy documentation. The study underscores the transformative role of 3D scanning in marine mammal postmortem examinations and conservation efforts, offering avenues for improved research, education, and management practices. It emphasizes the importance of accessible technology in engaging communities and advancing wildlife conservation efforts globally.
1 Introduction
As sentinels of broader ocean health (Bossart, 2011), marine mammals reflect the intricate balance of marine ecosystems and play a crucial role in providing insights into complex and dynamic environmental changes (Simmonds and Isaac, 2007; MacLeod, 2009; Derville et al., 2019; Burek et al., 2008), pollution levels (Williams et al., 2023), and other factors (Bowen, 1997; Rhodes-Reese et al., 2021). The study of marine mammals such as large whales, dolphins, and other pelagic creatures often poses significant challenges due to the time spent submerged and vast and remote oceanic habitats (Nowacek et al., 2016). Necropsies, the postmortem examination of deceased animals, offer invaluable opportunities to assess marine mammal health, shedding light on both individual and population-level factors influencing their wellbeing (Küker et al., 2018). Necropsies not only provide insights into the cause of death but also offer unique glimpses into other aspects of marine mammal health. Tracking emerging diseases (Waltzek et al., 2012; Leguia et al., 2023; VanWormer et al., 2019), assessing variations in body condition (Castrillon and Bengtson Nash, 2020), and identifying anthropogenic trauma (Schoeman et al., 2020; Cassoff et al., 2011) are critical observations that can be derived from necropsies and ancillary diagnostic studies. Results from these examinations facilitate temporospatial monitoring of stranding events (Wund et al., 2023), identify trends in mortality rates and proximate causes of death (Bogomolni et al., 2010), and assess the impacts of environmental stressors on marine mammal populations (Carrier-Belleau et al., 2021).
Best practices in conducting necropsies worldwide continue to improve the information collected around marine mammal strandings and overall health. In remote areas where access to specialized equipment, personnel, and expertise is limited, it is difficult to extract a consistent level of detail from these examinations in comparison to resourced areas (Fitton et al., 2021). In regions like British Columbia, Canada, where expansive marine mammal populations thrive amidst hundreds of kilometers of remote coastline, the need for innovative approaches to necropsy procedures becomes increasingly relevant. Despite these challenges, extensive data have been compiled on stranding trends (Raverty et al., 2024; Barbieri et al., 2013), anthropogenic threats (Storlund et al., 2024), emerging diseases (Berhane et al., 2022; Teman et al., 2021; Rosenberg et al., 2016), and contaminant levels (Lee et al., 2023a, 2023b) of marine mammals in this region. The opportunity to improve the quantity and quality of data derived from postmortem examinations is critical and can be further developed by available imaging instruments and procedures (Tsui et al., 2020). Improved documentation and sample management are other enhancements for necropsy procedures (Brownlie and Munro, 2016).
Novel complementary methods enhance necropsy data collection and analysis and can be realized in part by the application of rapidly developing three-dimensional (3D) reconstruction sensors and technology (Bois et al., 2021; Farahani et al., 2017). Continued advancements in this technology will further improve the accessibility and usefulness of 3D reconstruction tools. By providing detailed visualizations of marine mammal anatomy and pathology, these virtual reconstructions have the potential to enhance public engagement, interest, and education (Au and Lee, 2017). Virtual reality (VR) environments in particular offer immersive experiences (Cipresso et al., 2018) that will potentially allow users to explore reconstructed marine mammal specimens in unprecedented detail, fostering greater awareness and appreciation for these creatures and the challenges they face.
Models of marine mammals have been used successfully to estimate body size and morphology, often to understand animal health and bioenergetics. Digital modeling combining photographs and select morphometric measurements have been successfully applied to create 3D models of pilot whales (Adamczak et al., 2019), finless porpoises (Zhang et al., 2023), and humpback whales (Hirtle et al., 2022), as well as many other animals (Irschick et al., 2022). These models are typically used as a proxy for live animals and animated for biomechanical or nutritional analysis. Often these models are validated using stranded animals, which further justifies the need for continued and improved collection of morphometric data during necropsy of marine mammal species. Digital imaging has also been combined with 3D printing to create high-definition 3D printed specimen replicas of a killer and blue whale for use in a marine science center (Mills et al., 2022). In Alaska, a humpback whale was 3D scanned using an iPad and unmanned aerial vehicle (UAV) photogrammetry to create a “Virtual Necropsy” educational tool (Chenoweth et al., 2022). The use of accessible remote sensing tools is invaluable for both education and in documentation of stranded animals, although the technology must continue to be tested, improved, and implemented to provide long-term value for marine mammal conservation. This includes being tested in time-constrained situations, non-ideal environmental conditions, and by inexperienced users with varied specimens and species.
In this paper, we present a novel rapid approach for 3D reconstruction to complement conventional necropsy techniques in various necropsy conditions and species. The primary purpose of our work is to showcase the current state of the imaging technology with marine mammal strandings, and that it can be utilized by community partners to improve information collected during a necropsy, particularly in challenging and resource-poor environments. This includes testing the feasibility of collecting 3D data in the typical (often poor) environmental conditions where marine mammals are beach cast or live stranded in British Columbia. This is particularly important for large whale strandings, or in cases where carcass relocation from remote areas is not feasible. In these cases, limited external examinations and sample collection must be conducted in place. Our methodology was tested by collecting data from a diverse range of marine mammal specimens, including large whales, dolphins, porpoises, pinnipeds, and sea otters, with a focus on accessible and user-friendly handheld devices equipped with LiDAR sensors. This approach was pursued in conjunction with a validation of the method using an inflatable pilot whale. Ancillary data included UAV photogrammetry of large whales to provide a comparison to LiDAR. In cases of large cetacean strandings with comprehensive necropsies, individual datasets of organ systems were opportunistically collected. These scans were processed and created into detailed virtual models for morphometric assessments, and/or as a visualization and educational tool. These scans were also implemented in a web application and VR environment as an example of marine mammal stranding data visualization for multiple species. Our study demonstrates the potential of 3D reconstruction technology with easily available sensors (cell phones) to enhance the study of marine mammal health and morphometrics. Detailed visualizations of marine mammal specimens and continued collection of these data afford researchers, community members, and educators with valuable tools to better document and understand marine mammal stranding events. We highlight the importance of innovation in necropsy procedures for advancing our understanding of marine mammal health and informing conservation efforts particularly in remote and challenging environments.
2 Materials and methods
2.1 Study area
The province of British Columbia, located on the western seaboard of Canada, is renowned for its diverse and dramatic physical geography along its extensive coastline. Spanning over 27,000 km, the coastline features a complex network of fjords, inlets, and islands, including the prominent Vancouver Island and Haida Gwaii (Taylor et al., 1997). The coastal waters are characterized by a mixture of cold, nutrient-rich currents and deep ocean upwellings that support a diverse and vibrant marine ecosystem including many marine mammals. There are 31 species of marine mammals recorded in British Columbian waters, including 25 cetaceans (whales, dolphins, and porpoises) and 6 species of carnivores (seals, sea lions, and sea otter) (Ford, 2014).
Marine mammal strandings in British Columbia are monitored province-wide by the British Columbia Marine Mammal Response Network (BCMMRN) coordinated by the Department of Fisheries and Oceans Canada (DFO). Necropsies for this study were conducted in cooperation with Indigenous Community Members, Fisheries Officers, Canadian Coast Guard (CCG), research scientists, non-government organizations, and the Provincial Animal Health Centre in Abbotsford, British Columbia.
In May 2022, data collection for strandings that were conducive to a postmortem examination and 3D reconstruction within the territorial waters of British Columbia was initiated. In this case series, 11 species and 13 specimens were examined, including 8 cetacean and 3 carnivore species.
2.2 3D model reconstruction background
While in-depth descriptions of the background theory for the methods used can be found elsewhere (see Gomes et al., 2014; Kang et al., 2020; Ma and Liu, 2018; Zhou et al., 2024), given the novelty of the application, a short overview is presented here.
Light Detection and Ranging (LiDAR) is a remote sensing technology that employs laser pulses to measure the distance between the instrument and a target surface or object. Distance is determined through a measure of the time it takes for the pulses generated by the instrument to travel to the surface and be reflected to the sensor. This process generates precise, high-resolution data about the physical environment. The resulting output, typically a 3D point cloud, provides a detailed representation of surfaces and objects, making LiDAR a versatile tool for applications ranging from topographic mapping to structural analysis (Wandinger, 2005).
LiDAR systems are available in various types, including terrestrial, RPAS, airborne, and handheld devices, each suited to specific applications. In recent years, LiDAR technology has been miniaturized and integrated into consumer-grade mobile devices, such as the latest iPhones and iPads (Vacca, 2023). Third-party applications make use of these iOS integrated devices to generate 3D point clouds and solid meshes with minimal user training needed. They are also relatively low-cost and more accessible than survey-graded systems. Although these built-in systems lack the range and resolution of professional-grade devices, they are sufficient for many applications, including conservation (Abbas et al., 2024), small-scale mapping (Abdel-Majeed et al., 2024), and preliminary structural analysis (Tondo et al., 2023). Handheld and mobile LiDAR devices are particularly efficient for data collection in diverse environments, from controlled indoor spaces to rugged field settings (Bauwens et al., 2016; Chio and Hou, 2021; Desai et al., 2021). Advanced techniques, such as simultaneous localization and mapping (SLAM) algorithms, also enhance the functionality of these systems, enabling real-time processing of spatial data, even in dynamic or GPS-deprived environments (Al-Tawil et al., 2024).
While professional LiDAR systems can be costly, with tripod-based survey devices typically ranging in excess of US$20,000, iOS mobile devices with integrated LiDAR provide a lower-cost alternative (Yen et al., 2011). For instance, an iPhone equipped with LiDAR can be purchased for under US$1,500 (Apple, Cupertino, California), offering researchers and professionals an affordable way to perform 3D scanning and mapping tasks.
In contrast to LiDAR, structure-from-Motion Multi-View Stereo (SfM-MVS) photogrammetry is a technique used to create 3D models from 2D photographs (Eltner and Sofia, 2020). By capturing a series of overlapping photographs from different angles, SfM software can identify common points within the images and reconstruct their 3D positions. This process involves camera calibration, feature detection, and matching, followed by the generation of a sparse point cloud that represents the object’s or scene’s geometry (Fathi and Brilakis, 2011). Further refinement through dense point cloud reconstruction is achieved through algorithms such as MVS (Strecha et al., 2012).
A 3D point cloud is a collection of discrete points in 3D space, each representing a specific location on the surface of an object or scene. These points collectively describe the object’s geometry but lack information about the surface’s continuity (Liu et al., 2019). In contrast, a 3D mesh provides a more structured representation, consisting of vertices, edges, and faces that form a continuous surface (Rassineux, 1997). Figure 1 shows what the two different formats of point cloud (a) and 3D mesh model (b) represent visually for reference on an inflatable test whale.
Figure 1

Point cloud representation of inflatable test whale collected from LiDAR (A) and the resultant 3D mesh model (B).
2.3 Marine mammal specimens examined
Over the study period, 13 marine mammal specimens composed of 11 species were collected or examined for this study. A summary of the stranding information and 3D data collection for each case is provided in Supplementary Table 1, and the geographic location of each specimen is shown in Supplementary Figure 1. In addition to the specimens listed in Supplementary Table 1, LiDAR and SfM data from an inflatable test whale were also collected to validate the process of data collection for beach cast animals.
2.3.1 Validation of reference measurements
In July 2022, 15 LiDAR datasets of an inflatable pilot whale were collected (British Divers Marine Life Rescue, East Sussex, UK), typically used for live stranding training. These data were collected with ideal clear sky conditions at the DFO warehouse on Annacis Island, British Columbia. Multiple software settings were tested and scanning sequences were completed to determine what may work ideally in the field. A 5.00-m tape measure was set up as a ground reference to assess the accuracy of the reconstructed model. Three UAV flights to collect photogrammetry data were also performed (Supplementary Table 2).
Standard photographs and morphometric measurements were collected for each stranding in accordance to conventional marine mammal necropsy procedure (Pugliares et al., 2007). This was performed opportunistically with the maximum possible number of samples and measurements taken given the surrounding conditions and time limitations, resources, and personnel available for the postmortem exam.
2.3.2 Large whale specimens collected
In September 2024, a 20.07-m female adult fin whale (Balaenoptera physalus) was discovered dead and floating in Prince Rupert, B.C. The animal was towed to a site on Digby Island, where it was secured up the shoreline at high tide. As the tide receded, the whale became accessible for a full necropsy, conducted on 3 September 2024. The animal was positioned in dorsal recumbency, and for human safety concerns, the right lateral aspect of the torso from the thoracic inlet to the peduncle was not accessible. Dissection of the head and neck confirmed acute subcutaneous hemorrhage and edema, consistent with a “suspect” vessel strike. The animal presented in good body and moderate postmortem condition, with widespread skin sloughing, as well as the cervix and a 1-m segment of the colon exteriorized. During this period, two 3D datasets of the animal were collected: one prior to the necropsy and one following its completion.
In May 2023, a 12.30-m female gray whale (Eschrichtius robustus) and calf were entrapped in a tidal lagoon at the end of Jarvis Inlet, approximately 10 km northwest of Port Hardy, B.C. Despite rescue attempts, only the calf escaped from the lagoon. The mother was found floating dead outside of the lagoon a few days after the final rescue efforts in late August 2023. At necropsy, the animal presented in poor body and postmortem condition. Generalized emaciation was attributed to entrapment in the lagoon, lack of appropriate prey, and sustained lactation for 4 months. The calf was assessed as well nourished. After detection outside the lagoon, the animal was towed to a nearby inlet and winched as close as possible to shoreline at high tide. As the tide receded, the animal was accessible, and two 3D datasets of the animal were completed: one pre- and one post-necropsy. Heavy rain during the postmortem examination precluded UAV flights collecting photogrammetry data; however, a video of moving the whale to the necropsy site was collected.
In June 2022, a 9.9-m male gray whale (E. robustus) presented stranded on the west side of Nootka Island. The animal was in fair to poor postmortem condition (decomposition code 3.5 (Bogomolni et al., 2010)) and moderate body condition. The abdomen was moderately distended and firm. The CCG was able to bring three personnel to attend this necropsy via CCG helicopter. The animal was on shore at low tide allowing for examination during a half tidal cycle. The timeline limited the external examination of the carcass but allowed for a limited “windowed” approach, where rectangular sections of the abdomen and thorax were opened to access organs for examination and sampling. To collect photogrammetry data and obtain video of the pathologists sampling the animal, two UAV flights (Supplementary Table 2) were completed, and two 3D datasets of the animal were completed: one pre-necropsy with the animal as it was stranded, and one post-necropsy.
In May 2022, a 12.2-m female humpback whale (Megaptera novaeangliae) was stranded on Langara Island, Haida Gwaii. The animal was in moderate body and fair postmortem condition (decomposition code 3.5). The opportunity was available for three personnel to attend this remote necropsy. The whale was floating in a narrow tidal gorge, precluding any meaningful necropsy, but the decision was made to anchor the animal to shore as far up the inlet as possible at high tide. At low tide the following day, the animal was on shore and more exposed and the necropsy was completed during the half tidal cycle before the animal refloated. Two UAV flights were completed: one to collect photogrammetry data and the other to record video of the pathologist sampling the animal. One 3D dataset of the animal post-necropsy was collected, which included stepping on the animal itself as the surrounding terrain was inaccessible. The animal’s fluke and a portion of the tail was submerged during the examination, precluding that portion of the animal from being included in the 3D dataset.
2.3.3 Dolphin specimens collected
In March 2024, a 6.00-m pregnant adult female and calf killer whale (Orcinus orca) were entrapped in a lagoon, ultimately resulting in a live stranding event near Zeballos, BC. Despite efforts to refloat the animal, the adult female died later in the day. Multiple personnel were available to assist with this necropsy, with pre-necropsy 3D datasets collected on both the adult and unborn fetus. Logistic and personnel challenges unfortunately precluded additional scans from being conducted.
In July 2023, a 2.23-m juvenile Risso’s dolphin (Grampus griseus) was stranded on East Beach, Haida Gwaii. The animal was in poor body condition and very poor postmortem condition (decomposition code 4). The animal was subsequently frozen, and a necropsy was conducted in August 2023 in Daajing Giids as part of a training course for First Nations community partners and Fisheries Officers in marine mammal conservation. 3D datasets of the animal were collected pre-necropsy.
In August 2023, a 1.95-m adult female striped dolphin (Stenella coeruleoalba) live stranded near Ucluelet, B.C. The animal was agonal and attempts to refloat and release the animal were unsuccessful. The dolphin was frozen and, in October 2023, had a full postmortem exam at Annacis Island. This analysis included 3D datasets of the body before and after necropsy, as well as individual excised organs and organ systems that were suitable for scanning.
2.3.4 Porpoise specimens collected
In May 2022 a 2.04-m adult male Dall’s porpoise (Phocoenoides dalli) was found floating dead in open water near Victoria, B.C. The animal was in fair-poor body condition with insect larvae around its mouth and prominent abdominal distention. The animal was frozen in Victoria and transported to Annacis Island for a full necropsy in July 2023. This included collecting 3D datasets of the body before and after necropsy as well as images of excised organs.
In May 2023, a 1.50-m adult male harbor porpoise (Phocoena phocoena) was stranded on Salt Spring Island, B.C. in moderate body condition. The animal was frozen and stored at Annacis Island until a full necropsy was performed in August 2023. Datasets of the animal before and after necropsy in addition to individual excised organ systems were collected.
In July 2023 a 0.83-m harbor porpoise calf was live stranded in Tsawwassen, B.C. Efforts to transport the animal to the Vancouver Aquarium’s marine mammal rescue facility were unsuccessful. The animal died in transit. The animal was frozen at Annacis Island, and a full necropsy was performed in August 2023. 3D datasets of the animal before and after necropsy in addition to excised organ systems were collected.
2.3.5 Carnivore specimens collected
In January 2021, an 0.80-m juvenile female northern fur seal (Callorhinus ursinus) was stranded near Port Hardy, B.C. in poor postmortem and fair body condition. The animal was frozen and transported to Annacis Island, where a necropsy was performed in July 2023. 3D datasets of the animal before and after necropsy in addition to excised organ systems were collected. In December 2023, a 0.79-m harbor seal (Phoca vitulina) calf was reported abandoned in Vancouver, B.C. The animal was collected by the Vancouver Aquarium’s marine mammal rescue team but died in transit to the facility. The animal was frozen, and a full necropsy was conducted in January 2024 at Annacis Island. 3D datasets of the animal before and after necropsy in addition to organ systems were collected. Lastly, in March 2022 a 1.14-m subadult male sea otter (Enhydra lutris) was stranded near Tofino, B.C. in fair body and moderate postmortem condition. The animal was frozen and transferred to Annacis Island for necropsy in July 2023. 3D datasets of the animal before and after necropsy in addition to individual organ systems were collected.
2.4 LiDAR data acquisition and processing
LiDAR data were collected using an iPhone 12 Pro (Apple, Cupertino, CA) using the free Scaniverse application (Niantic, San Francisco, CA). The LiDAR sensor is available on the Pro and Pro Max models of the iPhone 12 and later, in addition to iPad Pro models from 2020.
The acquisition was achieved by walking around the animal with the iPhone in hand, carefully covering the entire extent of the animal and ensuring to not exceed the adjustable LiDAR range (i.e., 0.3–5 m) in distance from the surface of the animal while scanning. This range was adjusted as needed per animal, with the larger whales generally requiring a farther range in order to enable the data collector to step around the animal and not lose contact with the animal surface. The details of the range used for each scan is contained in Supplementary Figure 1. For large whales, data collection took up to 10 min, where with the smaller marine mammals, this process was completed in a maximum of 2–3 min.
The Scaniverse app collects both LiDAR and photographs from the phone’s main camera. Processing through the application generates a realistic triangular 3D mesh as the primary output. The scanning systems on iOS device applications use a form of SLAM that continuously tracks the sensor’s position and orientation in three dimensions over time. SLAM relies on optical data overlaps, utilizing previously observed features to establish relative coordinates and maintain accurate image registration (Lehtola et al., 2021). The Scaniverse interface and an example workflow for marine mammal data collection and subsequent export capabilities using the Scaniverse application are depicted in Figure 2.
Figure 2
Workflow using the Scaniverse application for the purpose of collecting marine mammal scans. On the left is a written explanation of the processing steps. On the right is the same steps in pictorial form as would be viewed in the Scaniverse application.
2.5 Structure-from-motion photogrammetry acquisition and processing
For large whale necropsy conditions that allowed for collection of UAV photogrammetry, a Mavic Professional [Da-Jiang Innovations (DJI), Shenzhen, China] UAV was used. Data collection consisted of capturing photographs of the exposed surfaces of stranded whales and the surrounding area. The UAV was flown manually as opposed to the widely used preprogrammed orthogonal flight path transects as no previous understanding of the stranding sites was available to create a detailed flight plan. During the manual flights, the best effort was made to create orthogonal flight lines with high overlap (~80%) in a cross-hatched pattern. See Supplementary Table 2 for flight details. Pix4DMapper (Pix4D, Prilly, Switzerland) was used to implement the SfM-MVS workflow. The resultant 3D mesh output for these large whale cases was used primarily to provide additional context for the large whale strandings.
One important consideration in the creation of realistic models is the determination of scale. The LiDAR system measures distances accurately and therefore evaluates the appropriate scale of the models within Scaniverse without the need for input of a known reference measurement in the scene. In the case of UAV photogrammetry here, the geotagging of each image used to create the model provides this distance measurement within a scene that is required to produce accurate scale. For non-georeferenced images, a known reference measurement is required to add these scale constraints to the model.
2.6 Data processing workflow and visualization
The acquired marine mammal datasets were generally categorized into two main groups, each corresponding to distinct use cases. The first is the application of 3D data collection in the instance of remote or inaccessible necropsies that benefit from the ease of use of these sensors and software. This is the case where trained regional partners are most likely to use this technology to collect 3D reconstruction information in the field where the animal is too large (for large whales) or too remote to be transported for a full postmortem examination or for pathologists to travel to the area. In this scenario, the mesh output of the Scaniverse workflow from a handheld device (Figure 2) provided an acceptable reconstruction without postprocessing. When possible, the use of UAV photogrammetry for large whales was implemented, to provide more environmental context and auxiliary data to the handheld LiDAR collection alone. This procedure could only be done if appropriate personnel and equipment were available. The second case is more exploratory in using this technology in the context of animals that were stranded or were transported to resourced areas where a full necropsy can be conducted, with all organ systems sampled and excised with a 3D dataset collected. Both data collection protocols followed a similar workflow, which is outlined in depth in Supplementary Figure 2.
Following processing and inspection of the resulting model, the meshes were exported to both.obj (geometry definition file format) and.las (RGB 3-dimensional point cloud data) files from the Scaniverse application to further refine in post-processing.
Post-processing the Scaniverse output was done by the free open-source 3D point cloud and mesh processing software CloudCompare Version 2.13. The two main tools used were to crop and segment data as well as merge multiple scans of the same specimen. For the cases where UAV photogrammetry data were collected, Pix4DMapper Version 4.8 was used to process the UAV data into meshes that could then be combined with the Scaniverse mesh outputs in CloudCompare.
2.7 Morphometric measurements
The key morphometric quantities of interest for cetaceans during postmortem exams and photogrammetry (and in general) are straightline length (from rostral limit of upper jaw), appendage lengths, girth, and blubber thickness. There are 12 straightline length measurements, 4 appendage measurements, 4 girth measurements, 2 throat pleat measurements (if applicable), and 9 blubber measurements typically recorded for complete morphometric characterization during postmortem exams (Supplementary Figure 3). For carnivores, morphometrics typically include straightline and curvilinear length (tip of snout to tip of tail), two appendage measurements, three girth measurements, and three blubber measurements (if applicable) (Rowles et al., 2001; Burns and Gillespie, 2003; IJsseldijk et al., 2019).
In this case series, digital morphometric measurements (Euclidean distance) were made in Scaniverse using a straightline length or using a polyline measurement for curvilinear measurements of length or girth. For all carcasses, the multiple digital measurements are compared against the manually collected morphometric data as a proof of concept. One important consideration in this work is that there is no way to verify the accuracy of the manual measurements; experienced personnel were primarily tasked with these measurements, but there is an unquantifiable source of error intrinsic to manual measurements that must be mentioned. To minimize the impact of this, only qualified personnel with appropriate necropsy experience were taking measurements in these cases, with the veterinary pathologist supervising if unable to take the measurement themselves.
2.8 Web visualization and virtual reality
The generated 3D reconstructions of the specimens and auxiliary information were stored in an accessible web platform for visualization and further investigation. The exported. obj meshes were uploaded to a server using the open-source 3D Heritage Online Presenter (3DHOP) framework (Visual Computing Laboratory - ISTI-CNR, Pisa, Italy) for interactive web presentations of high-resolution 3D models. These models can be evaluated through this medium for their quality, reproducibility, and reusability.
Uploading and incorporating the models into VR was implemented using the ENGAGE VR (Immersive VR Education, Ireland) platform. This platform allows for the uploading of.glb files, which, unlike.obj, provide a packaged single file containing all textures and shaders of a model. Within the platform, a presentation was created to view the output of these scans in lifelike sizes, providing an opportunity to view what happens during a necropsy through VR for multiple species types.
3 Results
3.1 Validation of model geometry with inflatable whale
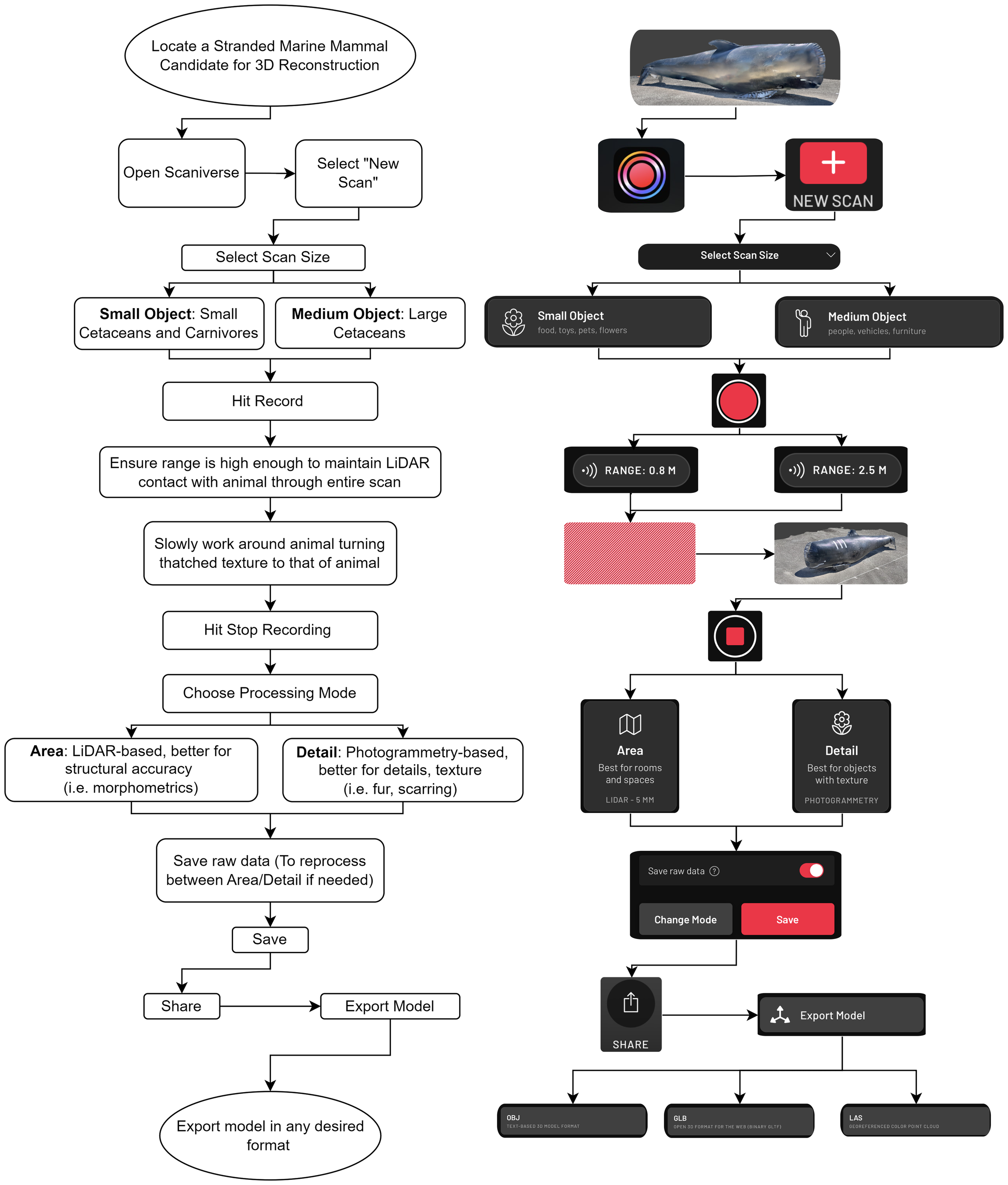
The 5.00-m measuring tape next to the inflatable whale was used as a reference and measured over 15 scan iterations within the Scaniverse app to be 4.99 ± 0.03 m. This demonstrates that barring any obvious distortions, errors in scanning, or errors in using the measurement tool within Scaniverse, measurements achieved through reconstructions have approximately a 1% uncertainty. These three error sources must be closely monitored to obtain accurate measurements from a 3D reconstruction. An example of the reference measurement as well as three example morphometric measurements of the inflatable whale including dorsal fin height, flipper anterior length, and half tail fluke width are shown in Figure 3. The proportions and overall shape of the inflatable whale are not entirely comparable to a real animal; however, the reconstruction can clearly distinguish the structure of the fins, as well as the body shape.
Figure 3
3D model (generated from Scaniverse) of the inflatable pilot whale used for live stranding training. (A) Reference assessment against known length (5.0 m), and straightline measurement of the whale at 4.40 m. (B) Side profile of whale for reference and display shape of inflatable. (C) Simulated measurement of anterior flipper length. (D) Simulated measurement of dorsal fin height. (E) Simulated measurement of (half) tail fluke width.
During testing, it was found that for most cases, using “detail mode” (photogrammetry) for processing within Scaniverse occasionally resulted in large artifacts (e.g., two tails) especially for the large whales, while in “area mode”, the structural data provided by LiDAR allow for the best representation of true form in all cases. For smaller animals and excised organ systems, detail mode was able to better represent the surface texture, which is important for diagnostics.
3.2 Large whales
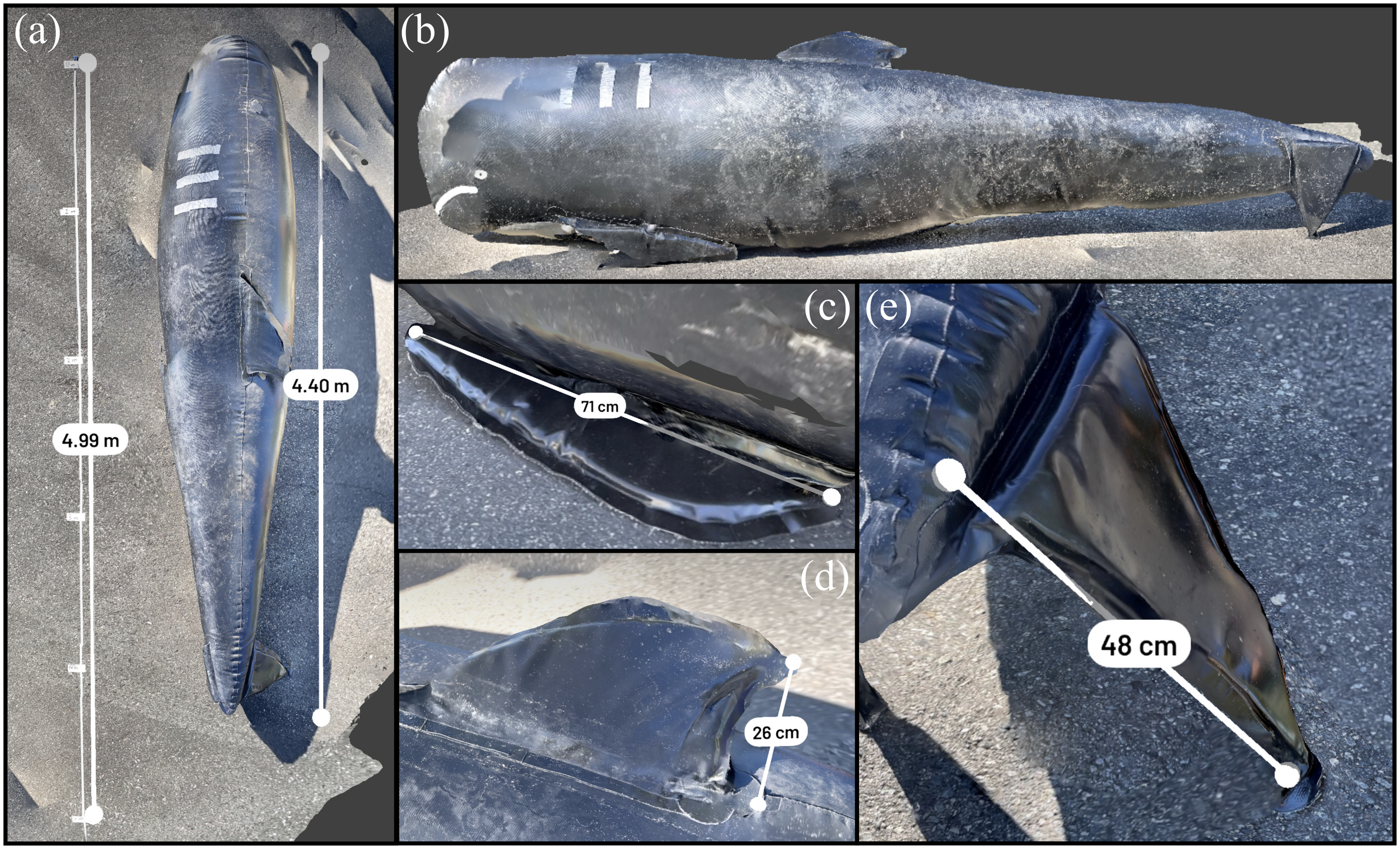
The fin whale that presented dead in Prince Rupert had ample subcutaneous and visceral adipose stores and the animal is well muscled as seen in Figure 4. Pre-necropsy morphometrics via 3D data showed a measured straightline length of 19.45 m, which differed by 3.14% from the straightline length determined from the manual measurement (20.07 m) seen in Figure 4A. The tail fluke width differed by 2.25% (Figure 4C), while the anterior flipper length differed by 0.43% (Figure 4D). The dorsal fin height differed by 2.60% (Figure 4E). Externally, the right dorsolateral aspect of the head and neck shows localized swelling and asymmetry, consistent with acute subcutaneous hemorrhage and edema caused by the suspected vessel strike. Multifocal skin erosions and occasional ulcerations appear as irregular surface textures and depressions on the body. The reduced blubber thickness in the ventral third, indicative of moderate atrophy, is evident, particularly in cross-sectional views. Post-necropsy in Figure 4B, the internal trauma to the head and neck is clearly visible, with areas of hemorrhage and edema in the underlying musculature appearing as disrupted and discolored tissue. Additionally, muscle degeneration and necrosis in the skeletal musculature are apparent as irregular and separated muscle fibers with fluid accumulation. Together, the 3D datasets capture the extent of trauma and pathological findings consistent with a suspected vessel strike. The gross necropsy findings of edema (and hemorrhage) are borne microscopically and also consistent with cause of death due to a traumatic incident.
Figure 4
3D reconstruction of fin whale stranded on cargo ship recovered near Prince Rupert. (A) Straightline length calculated in Scaniverse. (B) Animal after necropsy. (C) Tail fluke width measurement. (D) Anterior flipper measurement. (E) Dorsal fin measurement.
The gray whale that was reported dead after a 4-month entrapment in a lagoon was severely emaciated but presented with some postmortem bloating. Pre-necropsy morphometrics via 3D data in addition to a post-necropsy representation of the animal that displays several internal organs are shown in Figure 5. With full exposure of the animal at necropsy, an accurate straightline length can be determined directly from Scaniverse. The measured straightline length (12.35 m) was <1% different from the value measured manually in the field (12.30 m). The axillary girth, doubled at half, was different by a factor of 1.4%. This can partially be explained by the discrepancy when using a polyline measurement in Scaniverse as opposed to a tape measure that can adequately lie flush to the animal. In addition, postmortem bloat, liquefaction of the blubber, posture of the carcass on the beach, and other factors may have contributed to this discrepancy. In this case, polyline measurements at less than 10 cm between vertices were used. The tail fluke measurement had a discrepancy of 0.4 m or approximately 10% if using a straightline fluke length, reduced to 2% using a six-part polyline measurement. This is an important consideration in this case and other strandings where the animal’s fluke was in a curved shape, meaning a straightline length as shown in Figure 5B will underestimate this value if the morphometric measurement was done with a tape measure flush to the fluke length as was done here. A 13% discrepancy can be seen with the anterior flipper measurement as a small portion of the pectoral was submerged, another consideration when compiling morphometrics from 3D data. The animal had marked reductions in subcutaneous and visceral adipose stores and was poorly muscled. There was salmon discoloration of the blubber. This emaciation manifests in a “peanut head” appearance, or a marked bilaterally symmetric narrowing of the caudal aspect of the skull, which is readily apparent in the 3D representation of the animal (Figures 5D, E). The salmon discoloration of the blubber has been observed in other gray whales, and it is difficult to infer whether this may be related to dietary changes (abundant carotenoids), endogenous pigment production, or some other process. The animal also had generalized sloughing of the skin, attributed to postmortem change.
Figure 5

3D reconstruction of gray whale stranded near Jarvis Inlet after being trapped in a tidal lagoon for 4 months. (A) Straightline length calculated in Scaniverse. (B) Tail fluke measurement (underestimated if straightline length used). (C) Anterior flipper measurement, underestimated by 10% due to water cutoff. (D) From mid dorsal looking towards head. (E) Front dorsal towards tail obvious “peanut head” as a result of extreme emaciation before death captured in the 3D reconstruction. (F) Animal after necropsy.
The gray whale stranded on Nootka island was bloated, and upon incision of the abdominal musculature, there was marked deflation of the abdominal cavity. Figure 6 displays the UAV 3D model output obtained during this necropsy and 3D dataset collection conducted before and after necropsy. The animal’s total length was estimated at 9.9 m from nadir UAV photographs; the fluke of the animal was still submerged at low tide. The analysis included a correcting factor given the estimated 20° decline of the tail in the water. An anterior flipper measurement from the pre-necropsy LiDAR in Scaniverse was collected to be 1.44 m, within 2% of the manual measurement of 1.47 m. In the post-necropsy scan, it is apparent that, upon incision, a moderate amount of brown–red fluid was drained from the animal as a result of advanced autolysis. The small intestines and colon were distended with gas, which was well characterized by the 3D reconstruction. The post-necropsy dataset of this animal did not characterize the head adequately. This is due to the LiDAR collection losing contact (out of range) halfway through the scan. The processing software cannot register where the data are collected from and must re-register from a new reference point. This resulted in this scan essentially having two lower halves of the animal separated by approximately 15°, which was rectified in postprocessing via segmentation of the scan at the point of deviation and subsequent re-registration through the use of equivalent reference points that are present in both segments (Xu et al., 2023). Despite the visual success shown in Figure 6D, this scan is not suitable for use in morphometric analysis, only a visual assessment.
Figure 6
3D reconstruction of gray whale stranded on Nootka Island. (A) UAV photogrammetry output before necropsy. (B) LiDAR output before necropsy. (C) UAV photogrammetry output after necropsy. (D) LiDAR output after necropsy.
The stranded humpback whale in Haida Gwaii was in dorsal recumbency and the UAV photogrammetry and 3D dataset are presented in Figure 7. The UAV photogrammetry shown before and after necropsy especially outlines the steep tidal gorge the animal was in that precluded much of the normal necropsy procedure for safety concerns. LiDAR data were collected after necropsy and showed that on incision of the musculature, multiple tense loops of gas that inflated the small intestine were extruded. The flukes and distal third of the peduncle were submerged. The oral cavity was also only partially exposed from the water. Morphometric measurements were collected solely through assessing the 3D reconstruction and UAV images due to the difficult environment, although some measurements such as that of the anterior flippers could be verified via a measuring tape. With some areas of the animal inaccessible to the scanner being underwater, the straightline length could not be calculated directly in Scaniverse. The animal’s length was estimated at 12.2 m from these scans and associated photographs in postprocessing. Other morphometrics were collected from Scaniverse, including an example anterior fluke measurement at 2.47 m, consistent with the manual measurement (2.50 m) to within 2%. The 3D reconstruction adequately displays the animal’s moderate body condition, prominent mammae, and open abdominal cavity, including a moderate amount of ingesta primarily consisting of herring bones.
Figure 7
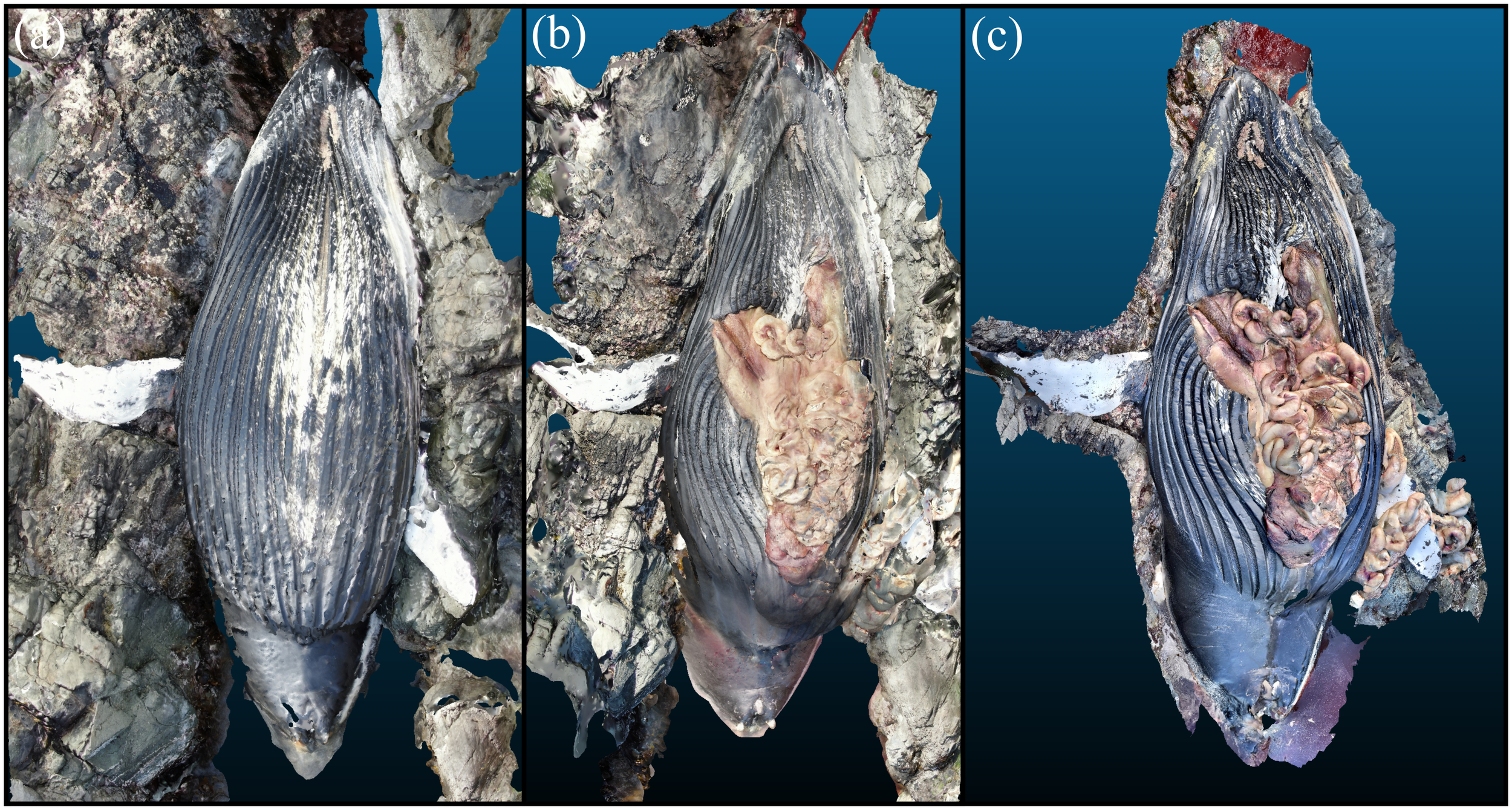
3D reconstruction of humpback whale in Haida Gwaii. (A) UAV photogrammetry output before necropsy. (B) UAV photogrammetry output after necropsy. (C) LiDAR output after necropsy.
3.3 Dolphins and porpoises
Stranded killer whales are rare in British Columbia relative to the large whales (Barbieri et al., 2013) and each event provides a unique opportunity to showcase 3D reconstructions of this species. For the adult pregnant female transient that was stranded near Zeballos, this was especially true. This animal initially was live stranded but subsequently died despite efforts to refloat her, and a postmortem examination was undertaken within 36 h. Figure 8 displays the pre-necropsy scan of the near-term pregnant killer whale. Extremely wet and humid conditions caused specular anomalies of the scanner operator and other surrounding areas to be present on the surface of the whale in the 3D reconstruction. This did not impact the LiDAR information obtained from the scan (and thus ability to collect morphometrics). The straightline length is measured at 6.02 m with a 6.00-m tape measurement in the field. The straightline length of the fetus was measured to within 1% of the field morphometric measurement, with the 1.50-m measurement in Figure 8E comparable to the 1.53 m measured in the field.
Figure 8
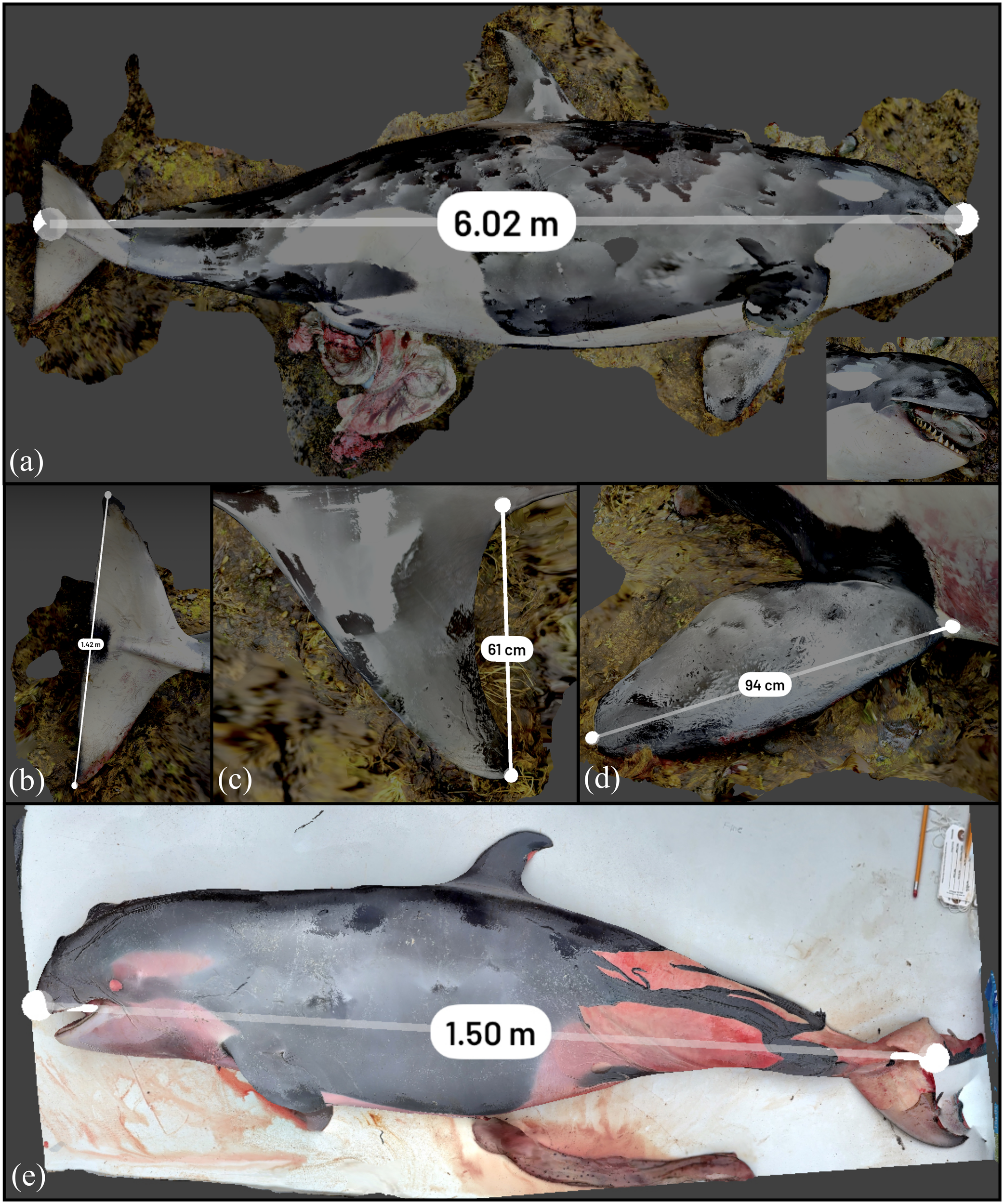
3D reconstruction of killer whale stranded near Zeballos. (A) Straightline length calculated in Scaniverse. (B) Tail fluke measurement (underestimated if straightline length used). (C) Dorsal fin measurement. (D) Anterior flipper measurement. (E) Unborn fetus removed from the whale.
The Risso’s dolphin specimen was found with extensive scavenging and poor postmortem condition, with extensive skin sloughing. The animal was 2.21 m in Scaniverse compared to 2.23 m during the necropsy with a difference of less than 1%. The pre-necropsy scan of the animal is shown in Figure 9. The suboptimal nutrition of this animal coupled with the lack of ingesta and blubber atrophy found during the necropsy is consistent with a negative energy balance that would have been severe enough to cause the loss of this animal. There were no other apparent internal or external lesions.
Figure 9
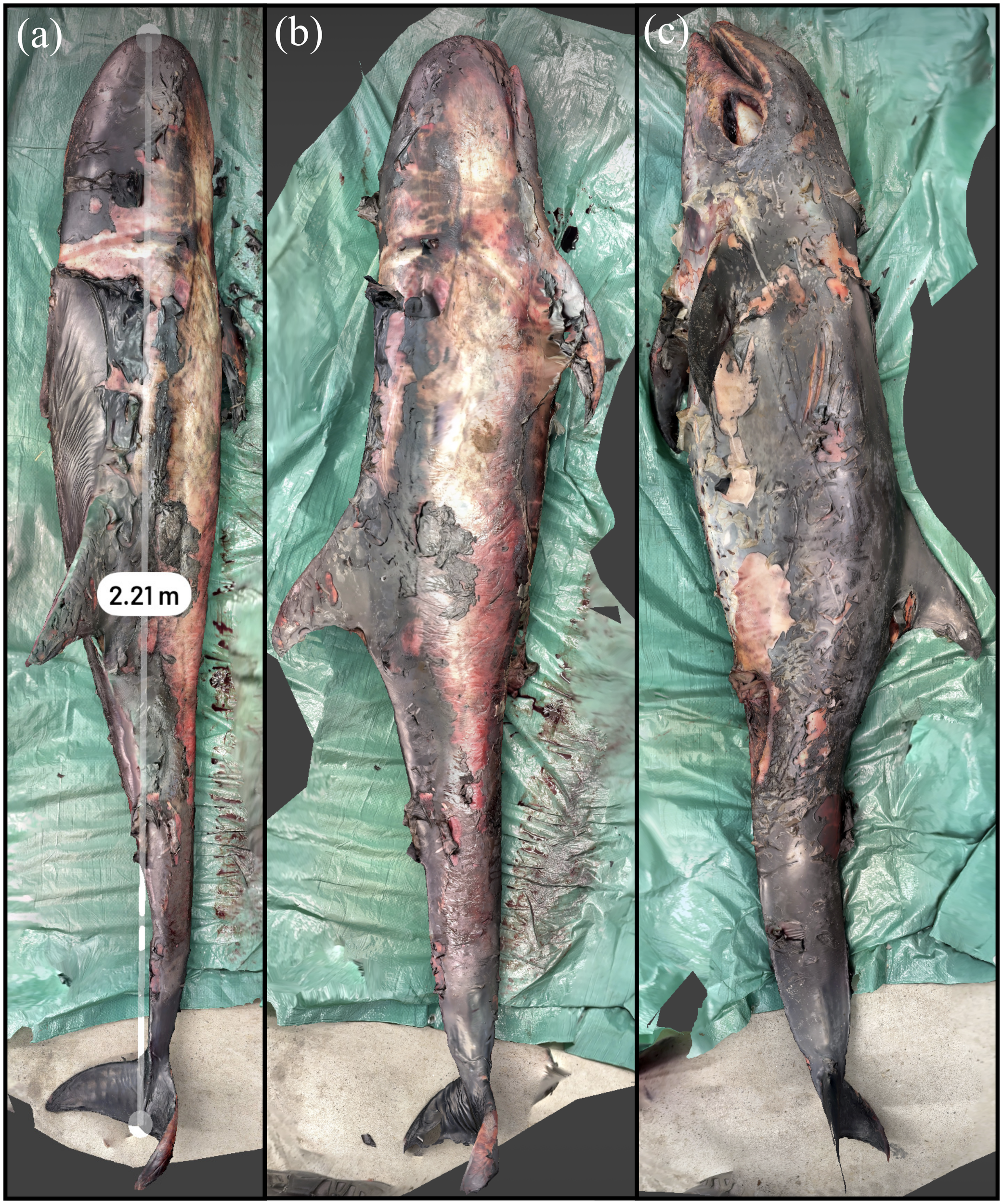
3D reconstruction of a Risso’s dolphin with necropsy conducted as part of a training exercise with community partners. (A) Measurement of straightline length of the animal. (B) Animal lying on the left side. (C) Animal lying on the right side.
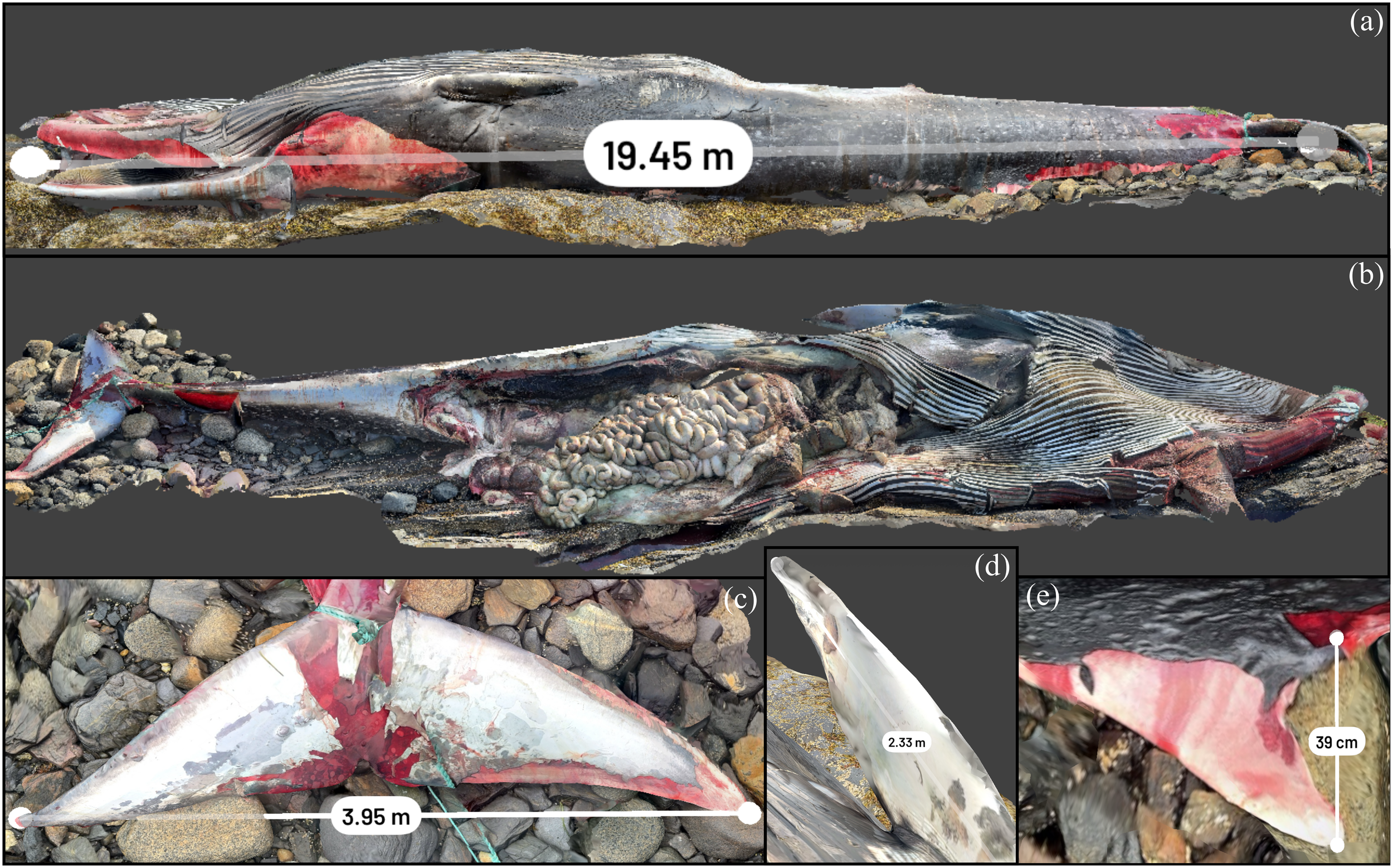
The live stranded striped dolphin is shown through its 3D reconstruction in Figure 10. The animal had superficial abrasions on the leading edge of the dorsal fin and edges of the tail flukes, possibly as a result of the live stranding event. The straightline length of the animal was measured by Scaniverse at 1.91 m, with a field measurement of 1.95 m, an approximately 2% discrepancy. This animal and the remaining carcasses included in this case series had complete necropsies and tissues were scanned in situ and excised as was logistically feasible. The following strandings with full necropsies were less time- and resource-constrained and allowed for more focused systematic visualization of the stranded animal and associated internal anatomy. This procedure facilitated review of the stranded animal as a 3D dataset to compare against different specimens or measure/visually assess areas, including that of organ systems. The sequential removal of all tissues and organs during the full necropsy is displayed for select organs. The topographic features and unique structure of the organs and stomach, pluck including goosebeak and thoracic viscera, as well as the kidney were well depicted by the scan and are displayed in Figure 10. There were no significant lesions in the adipose tissue, peripheral vasculature, penis, urinary bladder, trachea, kidney, thyroid gland, adrenal gland, diaphragm, or liver.
Figure 10
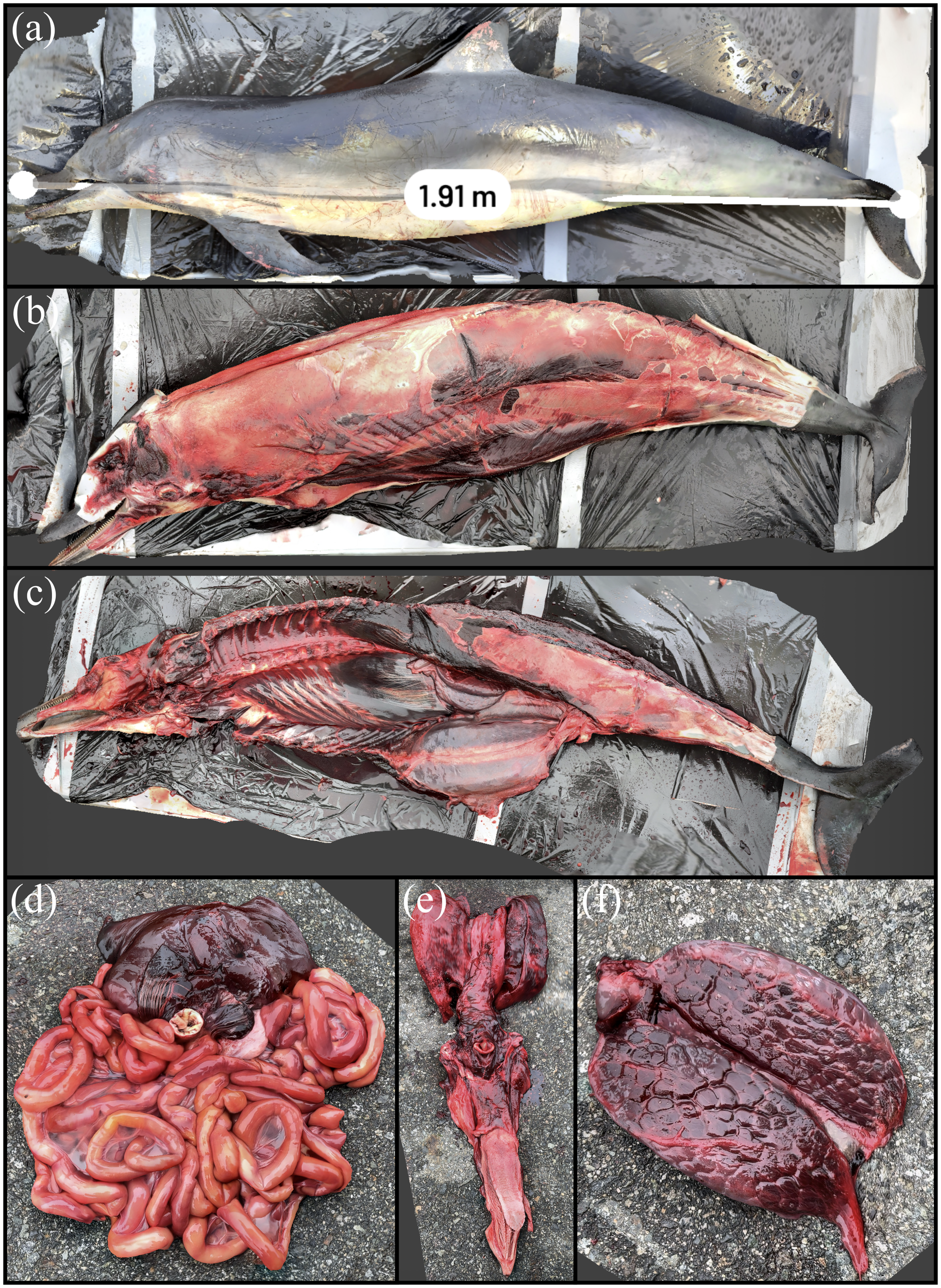
3D reconstruction of a striped dolphin that was live stranded in Ucluelet and was euthanized. (A) Measurement of straightline length of the animal. (B) Removal of external blubber layer. (C) Carcass with organ systems removed. (D) Digestive system. (E) Pluck including goosebeak. (F) Reniculate (lobed) kidney seen in marine mammals.
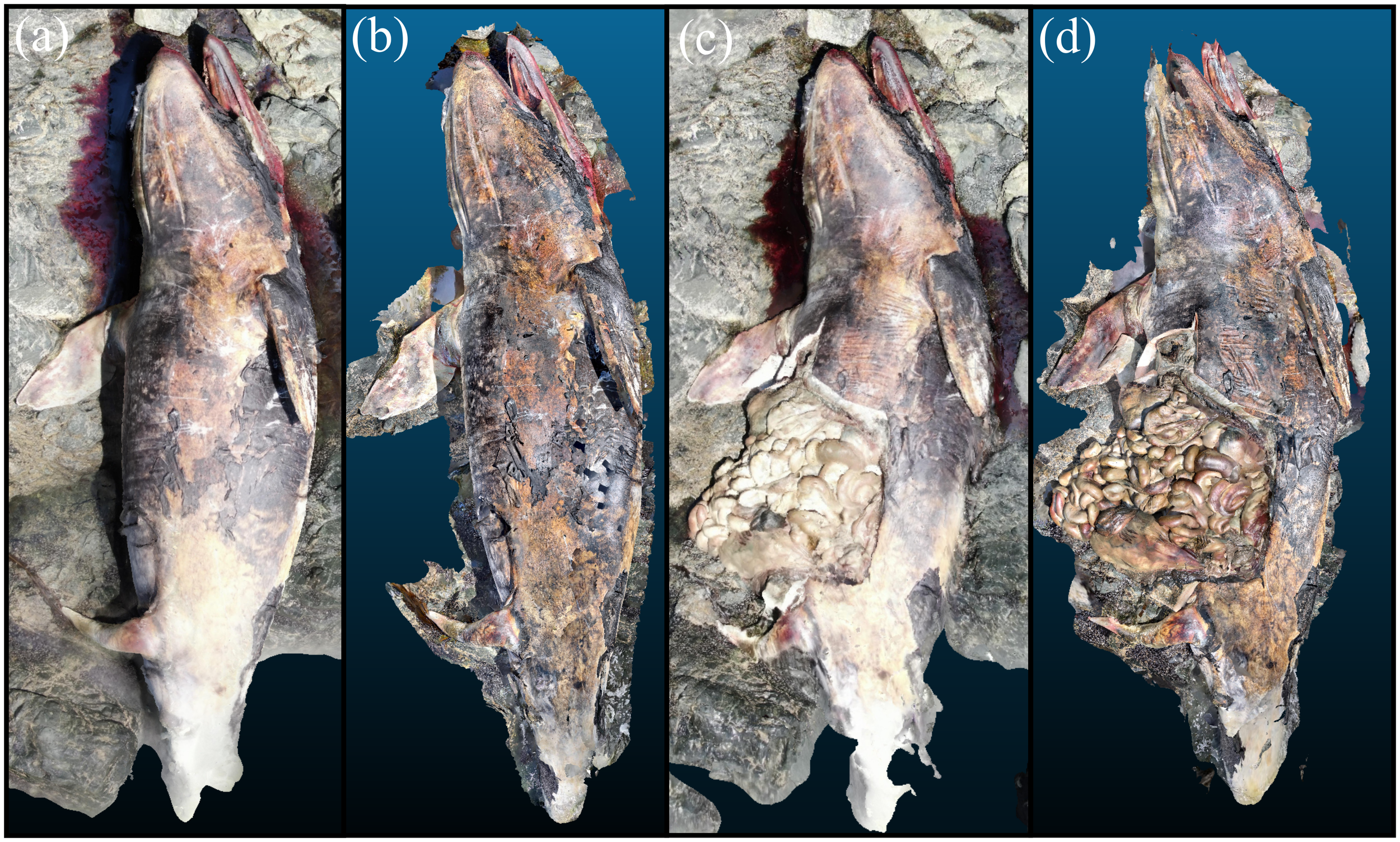
The stranded Dall’s porpoise is shown via its reconstruction in Figure 11. The animal was measured to be 1.97 m straightline length by Scaniverse, a difference of 3.5% from the manual measurement of 2.04 m. The animal had reduced subcutaneous and visceral adipose stores and was fairly muscled. There were no apparent internal or external lesions that would have contributed to the loss of this animal. All organs were sampled with select scans of organ systems shown in Figure 11.
Figure 11
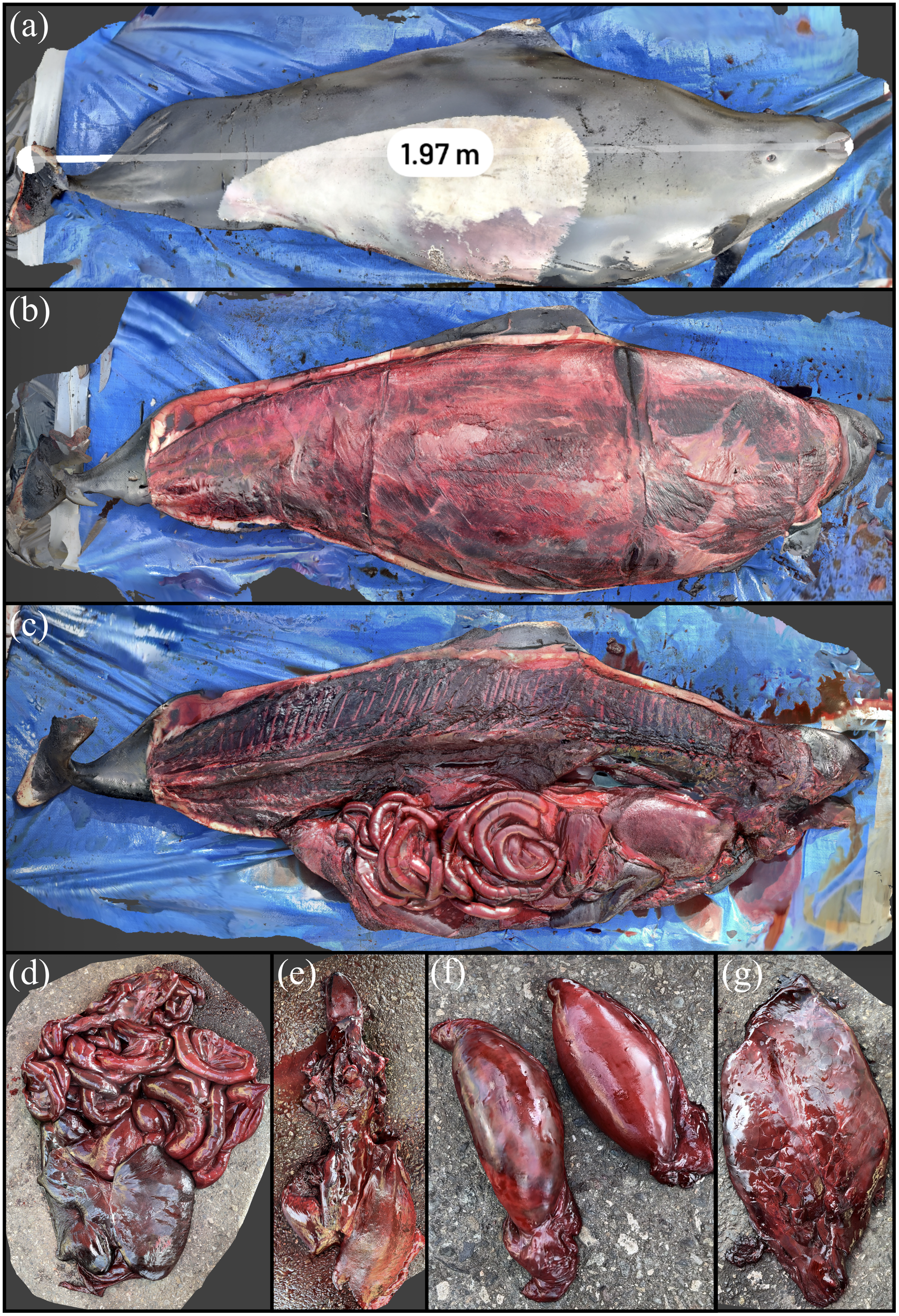
3D reconstruction of a Dall’s porpoise. (A) Measurement of straightline length of the animal. (B) Removal of external blubber layer. (C) Carcass with organ systems visible. (D) Digestive system. (E) Pluck including goosebeak. (F) Testes. (G) Reniculate (lobed) kidney seen in marine mammals.
Two harbor porpoise reconstructions are displayed in Figure 12. The adult was moderately fleshed with possible otitis interna. Incised cutaneous wounds are visible on this animal and suggestive of a possible vessel strike interaction. These wounds are visible on the scan and can be subsequently analyzed by various characteristics of the injury (Byard et al., 2012). The animal was measured with a straightline length of 1.46 m, within 3% of the manual measurement of 1.50 m. All organs were sampled with select organ systems shown in Figures 12a–g. The harbor porpoise calf was in good postmortem and body condition with no apparent internal or external lesions. The animal was measured at a straightline length of 0.81 m, within 2% of the manual measurement of 0.83 m. Selected scans of the calf’s overall body condition and organ systems are shown in Figures 12A–E.
Figure 12
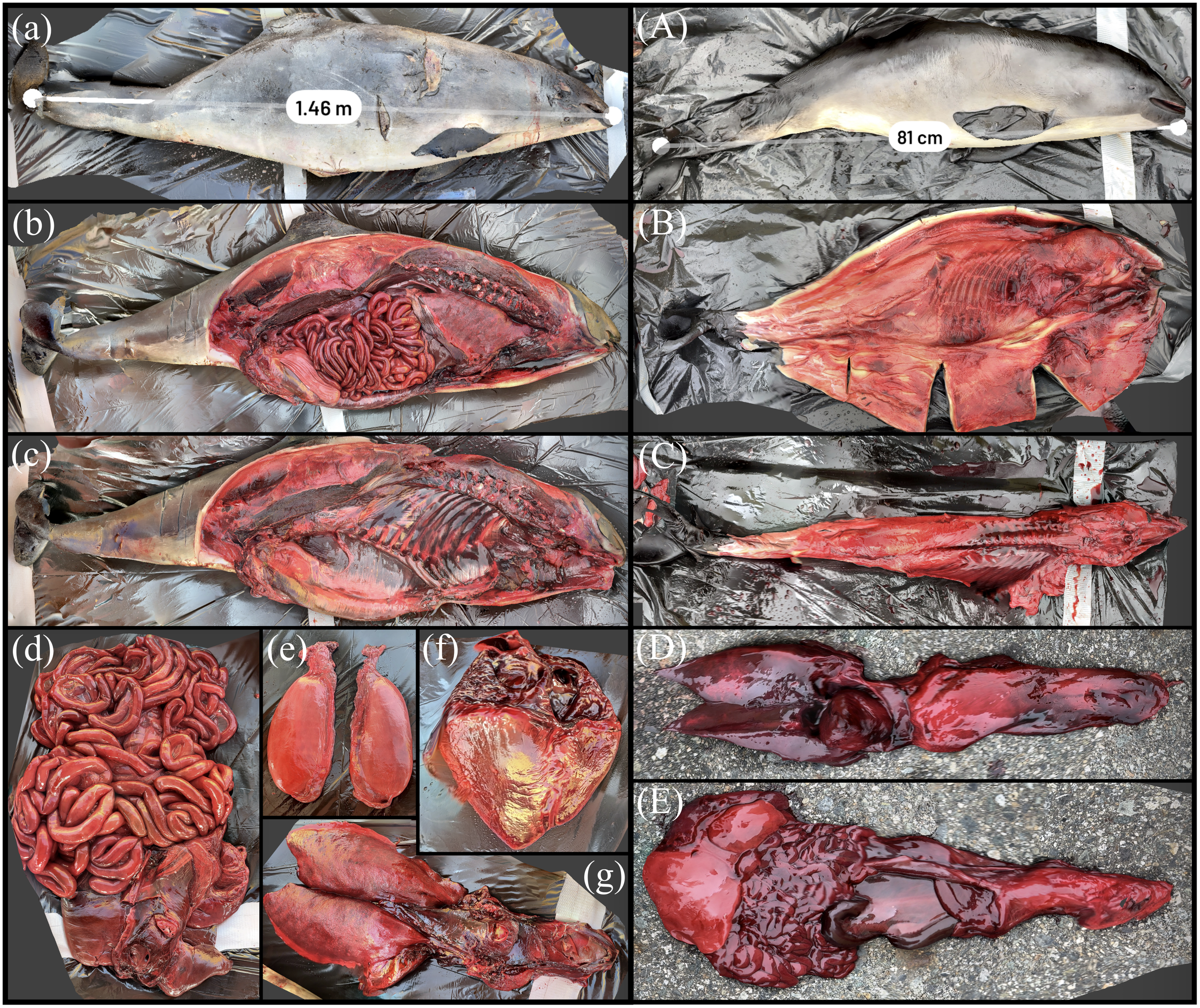
3D reconstruction of an adult and calf harbor porpoise. (a) Measurement of straightline length of the adult harbor porpoise. (b) Carcass with organ systems visible. (c) Carcass with organ systems removed. (d) Digestive system. (e) Testes. (f) Heart. (g) Pluck including gooseback. (A) Measurement of straightline length of harbor porpoise calf. (B) Carcass with blubber flensed. (C) Carcass with organ systems removed. (D) Pluck. (E) Digestive system.
3.4 Sea otter, northern fur seal, and harbor seal
All carnivore full-body reconstructions are displayed in Figure 13. The use of “detail” mode in Scaniverse improved the resolution of the fur texture in some scans. The sea otter was autolyzed, with a pendulous fluid filled abdomen. The abdomen was fluctuant with a prominent fluid line on ballottement. The reconstruction confirmed moderate abdominal distention with fluid accumulation in Figure 13a. The straightline length of the animal was 1.09 m using the reconstruction, a difference of 4% from the manual measurement during the examination of 1.14 m. The subcutis is a dull pale green brown and tacky, and the liver is moderately enlarged. Oysters in the stomach contents can be viewed in a scan of the digestive system, along with scans of various organs presented in the web application (Supplementary Table 3). Unfortunately, postmortem change hampered a gross examination of the animal although the suspected cause of death is trauma to the head. The northern fur seal specimen was also in a state of advanced autolysis, although was fairly fleshed (Figure 13A). There is mild swelling of the head and vulva, and the lungs are mottled pale to dark red. The animal was measured at 0.77 m, approximately a 3% difference from that of the manual measurement at 0.80 m. The reconstructed harbor seal pup is presented in Figure 13α; the reconstruction measurement was 0.78 m with a difference of approximately 1% from the manual morphometric measurement of 0.79 m.
Figure 13
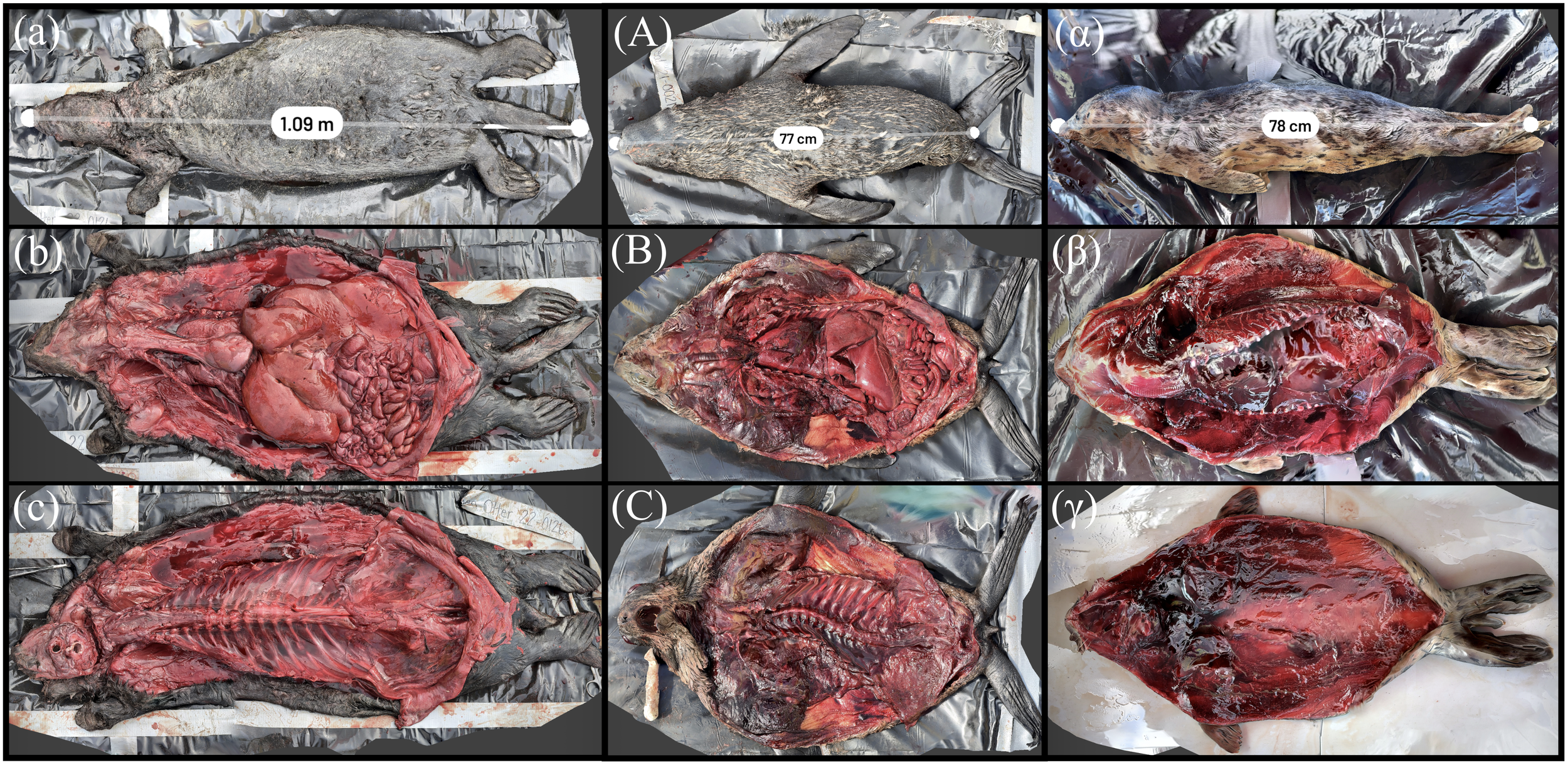
3D reconstruction of an adult sea otter, juvenile fur seal, and harbor seal pup. (a) Measurement of straightline length of adult sea otter (b) Sea otter carcass with organ systems visible. (c) Sea otter carcass with organ systems removed. (A) Measurement of straightline length of the juvenile fur seal. (B) Fur seal carcass with organ systems visible. (C) Fur seal carcass with organ systems removed. (α) Measurement of straightline length of harbor seal pup. (β) Harbor seal carcass with organ systems visible. (γ) Harbor seal carcass with organ systems removed.
A summary of each specimen and associated measurement types, manual and model morphometrics, and the percentage difference between the two is compiled in Table 1.
Table 1
| Species | Measurement type | Manual morphometric measurement (m) | Model morphometric measurement (m) | Percentage difference (%) |
|---|---|---|---|---|
| Inflatable whale | Straightline length | 4.41 | 4.40 | 0.23 |
| Anterior flipper length | 0.72 | 0.71 | 1.40 | |
| Dorsal fin height | 0.27 | 0.26 | 3.77 | |
| Tail fluke width (half) | 0.50 | 0.48 | 4.08 | |
| Fin whale | Straightline length | 20.07 | 19.45 | 3.14 |
| Dorsal fin height | 0.38 | 0.39 | 2.60 | |
| Tail fluke width | 4.04 | 3.95 | 2.25 | |
| Anterior flipper length | 2.34 | 2.33 | 0.43 | |
| Gray whale (Tsibass) | Straightline length | 12.30 | 12.35 | 0.41 |
| Axillary girth (doubled at half) | 5.64 | 5.60 | 0.71 | |
| Tail fluke width | 3.17 | 3.10 | 2.23 | |
| Anterior flipper length | 2.27 | 1.98 | 13.65 | |
| Gray whale (Nootka) | Anterior flipper length | 1.47 | 1.44 | 2.06 |
| Humpback whale | Anterior flipper length | 2.50 | 2.47 | 1.21 |
| Killer whale | Straightline length | 6.00 | 6.02 | 0.33 |
| Tail fluke width | 1.44 | 1.42 | 1.40 | |
| Dorsal fin height | 0.63 | 0.61 | 3.23 | |
| Anterior flipper length | 0.95 | 0.94 | 1.06 | |
| Killer whale fetus | Straightline length | 1.53 | 1.50 | 1.98 |
| Risso’s dolphin | Straightline length | 2.23 | 2.21 | 0.90 |
| Striped dolphin | Straightline length | 1.95 | 1.91 | 2.07 |
| Dall’s porpoise | Straightline length | 2.04 | 1.97 | 3.49 |
| Harbor porpoise (adult) | Straightline length | 1.50 | 1.46 | 2.70 |
| Harbor porpoise (calf) | Straightline length | 0.83 | 0.81 | 2.44 |
| Sea otter | Straightline length | 1.14 | 1.09 | 4.48 |
| Northern fur seal | Straightline length | 0.80 | 0.77 | 3.82 |
| Harbor seal | Straightline length | 0.79 | 0.78 | 1.27 |
Morphometrics of each specimen achieved manually and through measurement of the models.
3.5 Web application and virtual reality
All the scans presented here in addition to those of other organ systems or stages of the necropsy are in Supplementary Table 3, where the scans can be visualized, explored, and zoomed into. These manipulations are a major advantage of 3D data versus a series of two-dimensional photographs. Future strandings where LiDAR or photogrammetry data are collected will continue to be added to this dataset, which is intended for public access.
The VR implementation of these scans was achieved through a short presentation in ENGAGE VR, which highlights the importance of marine mammal necropsies and the possibility of VR in improving the science and education of necropsies. The users within the VR environment can walk around and experience several perspectives of necropsies at close to life size while a video of the stranding response and pathologists dissecting the whale is displayed above. The application provides a unique sense of the scale of necropsies to those that have never witnessed them in this context. An example screenshot of navigating the VR environment is shown in Figure 14.
Figure 14

Navigation and example of the VR environment where students can investigate what conditions are like during a marine mammal necropsy. (A) The humpback whale shown in VR with the video outlining the necropsy playing above. (B) The gray whale from Nootka Island relative to a person navigating the VR environment.
4 Discussion
While all marine mammal 3D datasets were well visually reconstructed in the field, challenges such as water immersion affecting straightline measurements for large whales were noted (Figures 6, 7), prompting the need for complementary data integration methods like UAV photographs for large whale strandings for accurate measurements. For larger specimens (e.g., the fin whale in Figure 4 at 20 m), deviations in the manual measurement due to bending and warping of the tape measure over the larger distance are magnified and provide a potential explanation for differences between measurements obtained from the models and those gathered manually. Polyline measurements were utilized for curvilinear metrics such as girth or flukes that rested curved and showed good agreement to manual tape measurements (Figure 5). Specific excised organ system scans for smaller carcasses including the striped dolphin, porpoises, and carnivores facilitated unique data collection opportunities, enhancing documentation and the ability to revisit interpretation of necropsy results, particularly concerning internal and external lesions (e.g., previous sign of vessel strike interaction in Figure 12).
The integration of 3D reconstruction technology into necropsy procedures offers several benefits that were conducted with minimal reduction in the capacity to perform thorough postmortem examinations in this case series. This is despite the fact that many of the remote necropsies were conducted by a small complement of three to four personnel. This technology enables the collection of comprehensive data beyond what is achievable through photographs and morphometric data sheets alone. The detailed topographic and anatomic information obtained from 3D data facilitates more accurate assessments of causes of mortality and pathology when the physical specimen is not accessible, while also serving as datasets for morphometric analysis and virtual reconstructions. 3D reconstruction technology enables researchers to visualize, consult, and analyze remotely stranded specimens in ways that were previously impossible, opening new avenues for research and education. This enhanced documentation of a specimen’s anatomy is useful in creating unique additions to permanent records of necropsy findings and simplifying data sharing with other researchers or regions for collaborative analyses. These 3D reconstructions can also be easily overlaid and compared between multiple specimens, enabling the identification of patterns, variations, and anomalies across different individuals or species. Where access to specialized equipment and facilities may be limited, handheld devices equipped with LiDAR sensors such as that of current mobile phone models (iPhone) offer a cost-effective and portable solution to conduct necropsies and collect ancillary data. Although all scans can be subsequently processed after data collection, the more accurate and representative the initial scans are, the more likely community partners will be motivated to continue using and finding value in them. This reduces the effort and barriers to making the adoption of this technology standard for postmortem exams.
The findings of this study suggest that the application of this technology is accessible to most users when the outlined instructions are followed diligently. The highest value of this technology is currently in scenarios where conventional necropsies are limited or novel species may be investigated, such as shown with large whales in Figures 4–7. Two primary use cases emerge: supplementing limited external examinations and complementing internal complete necropsies. The first case is essential for morphometric analysis where traditional measurements are not feasible, and the identification of external lesions, particularly signs of trauma. The second case enables later assessment of internal lesions and organs. While all species are viable subjects, animals assessed in rainy conditions or with high specular reflection, such as the killer whale case here, may exhibit visual anomalies during the creation of the 3D mesh. The combination of LiDAR and photogrammetry during scans collected by Scaniverse is crucial for creating lifelike representations. LiDAR data are critical for accurate morphometric measurements.
The advent of scanning technology in handheld phones has made this technology easily accessible in a broad range of applications primarily in mapping and geosciences (Tavani et al., 2022; Günen et al., 2023; Luetzenburg et al., 2021), and in fields such as human body measurement (Mikalai et al., 2022). The use of iOS-based LiDAR has been studied for repeatability and bias against reference values collected via manual measurements of a vehicle and filing cabinet (Heinrichs and Yang, 2021). These scans are analogous in size to a large whale and smaller cetaceans/carnivores, respectively, with their results also producing centimeter-level deviations from manual measurements as seen in these cases for marine mammals. These systems have also been used for cultural heritage applications, where different iOS scanning applications were tested against each other and distance metrics were compared (Teppati Losè et al., 2022). This study outlines that although all iOS LiDAR sensors are comparable, the choice of scanning application impacts the outcome of the scan by dictating parameters such as scanning point density. In this work, only Scaniverse was used and evaluated, although other applications such as 3DScanner, Polycam, and SiteScape perform similar functions. In one study comparing these applications, point clouds against high-precision terrestrial LiDAR scanners (Askar and Sternberg, 2023), it was shown that Scaniverse has a far lower density of points than any other application, which makes on-site field processing far more rapid in Scaniverse. This is especially useful in the use cases outlined in this study for remote and inaccessible environments where quickly analyzing the scan output is important to ensure the scan was captured properly. This does come at the expense of a less-dense point cloud, although in our examinations, the meshes output by Scaniverse were within an acceptable margin of error for morphometric assessments, given these measurements in the field are subject to their own errors, especially in the large whale cases.
Previous studies that have investigated 3D reconstructions of stranded marine mammals primarily emphasize using this technology either for creating a digital collection of fossils or specimens for museums (Merella et al., 2023; Franci and Berta, 2018; Niven et al., 2009), or for input into models that can estimate free-ranging animals’ body mass and volume for objectives such as bioenergetic analysis and swimming dynamics (Zhang et al., 2023; Irschick et al., 2022). The former cases have created extremely detailed reconstructions that can be 3D printed and re-articulated into true representations of skeletons. The later purpose is rapidly developing and allows for accurate determinations of many aspects of living marine mammals and advance the ability to study and understand these species in the wild. The only identified uses of this technology in assessing stranded marine mammals in the field is the case of the “Virtual Necropsy” conducted in Alaska (Chenoweth et al., 2022) as well as a fin whale reconstruction in Italy (Del Pizzo et al., 2021). The former appears to be the first time that this technology was used to generate a 3D reconstruction of a necropsy over the course of a decomposing animal. This work garnered broad interest and showcased the value of these scans for use in education, research, and public interest. In the fin whale case, the use of mobile phone photogrammetry to reconstruct a large fin whale was also successful. In this work, we provide a complement and extension to this kind of data collection by providing scans from a variety of necropsy situations and species types and showcasing their utility as a documentation and visualization tool as this technology progresses.
The continued advancement of 3D reconstruction technology in the field of marine mammal strandings has significant implications for overall research and conservation efforts in this field. The morphometric and structural data obtained from 3D data offer invaluable reference and validation datasets for constructing accurate models of free-ranging cetaceans, crucial for bioenergetics and other physiological studies. These data also immortalize these animals, enabling revisitation and improved documentation of rare events such as killer whale strandings. This is essential for monitoring causes of death and population health over time and providing context for future stranding events of all species. The collection of 3D data across populations also simplifies comparative studies in body condition, offering more nuanced methods of comparison beyond girth and straightline length. The accessibility of this handheld technology allows non-experts and community members in remote or inaccessible regions to perform scans and engage actively in marine mammal conservation. Beyond British Columbia, this technology provides an opportunity for stranding programs globally with lower resources to collect more comprehensive data on marine mammal strandings by simply using their phone and downloading a 3D scanning application. An example of this is the recent Unusual Mortality Event (UME) for gray whales, where areas with lesser capacity to respond to strandings such as Mexico reported almost half (44.9%) of all strandings in the Eastern North Pacific relative to a small 4.2% in British Columbia by comparison (Raverty et al., 2024). The utilization of simple accessible tools such as that presented here could augment the limited data collection from stranding hotspot location in lower-income nations with limited capacity to respond and collect comprehensive stranding information going forward. Preserving these data for analysis by experts that cannot attend a necropsy is another use case of this technology as progressive decomposition from delayed examination limits the utility of a traditional assessment. Progressive decomposition also compromises the utility of measurements for validation of free-living animals as the animal decomposes, bloats, and changes shape (Christiansen et al., 2019). The fresher the specimen, the more likely both external and internal factors can be observed and determined to contribute to cause of death. These scan products when combined with rapidly improving tools such as VR also present the potential to improve training the new generation of responders and volunteers in necropsy procedures, enhancing their preparedness for stranding events and improving examination efficiency and outcomes. The integration of website and VR visualizations not only facilitates data compilation and dissemination but also ensures its longevity and accessibility to diverse user groups, including students, researchers, and conservation practitioners. The continued collection and use of data from 3D reconstructed marine mammals contributes to a growing integration of technology into wildlife conservation and ecological studies. Further work with this dataset will combine these reconstructions with that of medical imaging including computed tomography (CT) and magnetic resonance imaging (MRI) (Dennison et al., 2012) to showcase the ability of imaging products to improve the ability to diagnose, document, and understand marine mammal health.
5 Conclusion
This study investigates the potential application of 3D scanning technology within the domain of marine mammal strandings, to assess its applicability, practicality, and consequential impact. By leveraging accessible LiDAR scanners present in mobile phones, we explored the technology’s capacity to reconstruct marine mammal carcasses with accuracy and consistency across varied postmortem conditions, geographic regions, and species. Our investigation demonstrated the utility of 3D reconstructions in precisely measuring morphometrics in field settings, offering valuable supplementation to traditional field measurements and enhancing documentation practices. In most cases, discrepancies between manual morphometric measurements and that of the 3D reconstruction were less than 3%. For larger specimens (e.g., the fin whale in Figure 4 at a colossal 20 m), deviations in the manual measurement due to bending and warping of the tape measure over the larger distance are magnified and provide a potential explanation for differences between measurements obtained from the models and those gathered manually. The adaptability of this technology extends to educational realms, where it can be employed to train responders and volunteers in necropsy procedures, thereby improving stranding event preparedness, procedures, and outcomes. The integration of VR and web visualizations serves to disseminate these data effectively across diverse user groups and stakeholders in marine mammal health and management. Our research underscores the potential impact 3D scanning technology can have on marine mammal and ecological studies, offering a pathway for enhanced research, education, and conservation efforts.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
BC: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. MK: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – review & editing. JA-M: Funding acquisition, Writing – review & editing. OL: Methodology, Writing – review & editing. PC: Investigation, Writing – review & editing. TL: Investigation, Methodology, Writing – review & editing. SR: Conceptualization, Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We would like to thank the McCall MacBain Scholarships, the McGill Department of Geography Rathlyn GIS Award, the Natural Sciences and Engineering Research Council of Canada (NSERC) as well as the Canadian Wildlife Health Cooperative and Department of Fisheries and Oceans Canada, Science for their support in this work.
Acknowledgments
We acknowledge the Canadian Coast Guard and the Department of Fisheries and Oceans Canada for their support in facilitating transport and assistance with several of the necropsies. We also thank the local partners from each region for their assistance in the more remote necropsies as well as for collecting carcasses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1485788/full#supplementary-material
References
1
Abbas S. F. Abed F. M. (2024). “Evaluating the accuracy of iPhone LiDAR sensor for building façades conservation,” in Recent research on geotechnical engineering, remote sensing, geophysics and earthquake seismology. Eds. BezzeghoudM.ErgülerZ. A.Rodrigo-CominoJ.JatM. K.KalatehjariR.BishtD. S.BiswasA.et al (Springer Nature Switzerland, Cham), 141–144. doi: 10.1007/978-3-031-48715-6_31
2
Abdel-Majeed H. M. Shaker I. F. Abdel-Wahab A. M. Awad A. A. L. D. I. (2024). Indoor mapping accuracy comparison between the apple devices’ LiDAR sensor and terrestrial laser scanner. HBRC J.20, 915–315. doi: 10.1080/16874048.2024.2408839
3
Adamczak S. K. McLellan A. P. W.A. Thorne L. H. (2019). Using 3D models to improve estimates of marine mammal size and external morphology. Front. Mar. Sci.6. doi: 10.3389/fmars.2019.00334
4
Al-Tawil B. Hempel T. Abdelrahman A. Al-Hamadi. A. (2024). A review of visual SLAM for robotics: evolution, properties, and future applications. Front. Robotics AI.11. doi: 10.3389/frobt.2024.1347985
5
Askar C. Sternberg H. (2023). Use of smartphone LiDAR technology for low-cost 3D building documentation with iPhone 13 pro: A comparative analysis of mobile scanning applications. Geomatics3, 563–795. doi: 10.3390/geomatics3040030
6
Au E. H. Lee J. J. (2017). Virtual reality in education: A tool for learning in the experience age. Int. J. Innovation Educ.4, 2155. doi: 10.1504/IJIIE.2017.091481
7
Barbieri M. M. Raverty S. Bradley Hanson M. Venn-Watson S. Ford J. K.B. Gaydos J. K. (2013). Spatial and temporal analysis of killer whale (Orcinus orca) strandings in the north pacific ocean and the benefits of a coordinated stranding response protocol. Mar. Mammal Sci.29, E448–E625. doi: 10.1111/mms.12044
8
Bauwens S. Bartholomeus H. Calders K. Lejeune P. (2016). Forest inventory with terrestrial LiDAR: A comparison of static and hand-Held mobile laser scanning. Forests7, 1275. doi: 10.3390/f7060127
9
Berhane Y. Joseph T. Lung O. Embury-Hyatt C. Xu W. Cottrell P. et al . (2022). Isolation and characterization of novel reassortant influenza A(H10N7) virus in a harbor seal, British Columbia, Canada. Emerging Infect. Dis.28, 1480–1845. doi: 10.3201/eid2807.212302
10
Bogomolni A. L. Pugliares K. R. Sharp S. M. Patchett K. Harry C. T. LaRocque J. M. et al . (2010). Mortality trends of stranded marine mammals on Cape Cod and southeastern Massachusetts, USA 2000 to 2006. Dis. Aquat. Organisms88, 143–555. doi: 10.3354/dao02146
11
Bois M. C. Morris J. M. Boland J. M. Larson N. L. Scharrer E. F. Aubry M.-C. et al . (2021). Three-dimensional surface imaging and printing in anatomic pathology. J. Pathol. Inf.12, 22. doi: 10.4103/jpi.jpi_8_21
12
Bossart G. D. (2011). Marine mammals as sentinel species for oceans and human health. Veterinary Pathol.48, 676–690. doi: 10.1177/0300985810388525
13
Bowen W. (1997). Role of marine mammals in aquatic ecosystems. Mar. Ecology-Progress Ser. - Mar. ECOL-PROGR Ser.158, 267–274. doi: 10.3354/meps158267
14
Brownlie H.W.B. Munro R. (2016). The veterinary forensic necropsy: A review of procedures and protocols. Veterinary Pathol.53, 919–285. doi: 10.1177/0300985816655851
15
Burek K. A. Gulland F. M.D. O’Hara T. M. (2008). Effects of climate change on arctic marine mammal health. Ecol. Appl.18, S126–S134. doi: 10.1890/06-0553.1
16
Burns W. Gillespie A. (2003). “Agreement on the conservation of cetaceans of the black sea, mediterranean sea and contiguous atlantic area (ACCOBAMS) (1996),” in The future of cetaceans in a changing world. (Transnational Publishers), 421–442. doi: 10.1163/9789004480599_020
17
Byard R. W. Winskog C. MaChado A. Boardman W. (2012). The assessment of lethal propeller strike injuries in sea mammals. J. Forensic Legal Med.19, 158–615. doi: 10.1016/j.jflm.2011.12.017
18
Carrier-Belleau C. Drolet D. McKindsey C. W. Archambault P. (2021). Environmental stressors, complex interactions and marine benthic communities’ Responses. Sci. Rep.11, 41945. doi: 10.1038/s41598-021-83533-1
19
Cassoff R. M. Moore K. M. McLellan W. A. Barco S. G. Rotstein D. S. Moore M. J. (2011). Lethal entanglement in baleen whales. Dis. Aquat. Organisms96, 175–855. doi: 10.3354/dao02385
20
Castrillon J. Nash S. B. (2020). Evaluating cetacean body condition; a review of traditional approaches and new developments. Ecol. Evol.10, 6144–6625. doi: 10.1002/ece3.6301
21
Chenoweth E. M. Houston J. Huntington K. B. Straley J. M. (2022). A virtual necropsy: applications of 3D scanning for marine mammal pathology and education. Animals12, 5275. doi: 10.3390/ani12040527
22
Chio S.-H. Hou K.-W. (2021). Application of a hand-held LiDAR scanner for the urban cadastral detail survey in digitized cadastral area of Taiwan urban city. Remote Sens.13, 49815. doi: 10.3390/rs13244981
23
Christiansen F. Sironi M. Moore M. J. Martino M. D. Ricciardi M. Warick H. A. et al . (2019). Estimating body mass of free-living whales using aerial photogrammetry and 3D volumetrics. Methods Ecol. Evol.10, 2034–2445. doi: 10.1111/2041-210X.13298
24
Cipresso P. Giglioli I. A. C. Alcañiz Raya M. Riva G. (2018). The past, present, and future of virtual and augmented reality research: A network and cluster analysis of the literature. Front. Psychol.9. doi: 10.3389/fpsyg.2018.02086
25
Del Pizzo S. Troisi S. Testa R. L. Sciancalepore G. Pedrotti D. Pietroluongo G. (2021). “Photogrammetry applications on stranded cetaceans: fin whale case study and preliminary results,” 2021 International Workshop on Metrology for the Sea; Learning to Measure Sea Health Parameters (MetroSea), Reggio Calabria, Italy, 2021, pp. 63–67. doi: 10.1109/MetroSea52177.2021.9611588
26
Dennison S. Fahlman A. Moore M. (2012). The use of diagnostic imaging for identifying abnormal gas accumulations in cetaceans and pinnipeds. Front. Physiol.3. doi: 10.3389/fphys.2012.00181
27
Derville S. Torres L. G. Albertson R. Andrews O. Baker C.S. Carzon P. et al . (2019). Whales in warming water: assessing breeding habitat diversity and adaptability in oceania’s changing climate. Global Change Biol.25, 1466–1481. doi: 10.1111/gcb.14563
28
Desai J. Liu J. Hainje R. Oleksy R. Habib A. Bullock D. (2021). Assessing vehicle profiling accuracy of handheld LiDAR compared to terrestrial laser scanning for crash scene reconstruction. Sensors21, 80765. doi: 10.3390/s21238076
29
Eltner A. Sofia G. (2020). Structure from motion photogrammetric technique. In. Developments Earth Surface Processes23, 1–24. doi: 10.1016/B978-0-444-64177-9.00001-1
30
Farahani N. Braun A. Jutt D. Huffman T. Reder N. Liu Z. et al . (2017). Three-dimensional imaging and scanning: current and future applications for pathology. J. Pathol. Inf.8, 36. doi: 10.4103/jpi.jpi_32_17
31
Fathi H. Brilakis I. (2011). Automated sparse 3D point cloud generation of infrastructure using its distinctive visual features. Special Section: Adv. Challenges Computing Civil Building Eng.25, 760–705. doi: 10.1016/j.aei.2011.06.001
32
Fitton J. M. Addo K. A. Jayson-Quashigah P.-N. Nagy G. J. Gutiérrez O. Panario D. et al . (2021). Challenges to climate change adaptation in coastal small towns: examples from Ghana, Uruguay, Finland, Denmark, and alaska. Ocean Coast. Manage.212, 105787. doi: 10.1016/j.ocecoaman.2021.105787
33
Ford J. K. B. (2014). Marine mammals of british columbia (British Columbia: Royal BC Museum Victoria).
34
Franci G. Berta A. (2018). Relative growth of the skull of the common minke whale (Balaenoptera acutorostrata) using a 3D laser surface scanner. Aquat. Mammals43, 529–375. doi: 10.1578/AM.44.5.2018.529
35
Gomes L. Bellon O. R. P. Silva L. (2014). 3D reconstruction methods for digital preservation of cultural heritage: A survey. Depth Image Anal.50, 3–14. doi: 10.1016/j.patrec.2014.03.023
36
Günen M. A. Erkan İ. Aliyazıcıoğlu Ş. Kumaş C. (2023). Investigation of geometric object and indoor mapping capacity of apple iPhone 12 pro LiDAR. Mersin Photogrammetry J.5, 82–895. doi: 10.53093/mephoj.1354998
37
Heinrichs B. E. Yang M. (2021). Bias and repeatability of measurements from 3D scans made using iOS-based LiDAR. In. SAE Int. J. Adv. Curr. Practices Mobility3, 2219–2226. doi: 10.4271/2021-01-0891
38
Hirtle N. O. Stepanuk J. E.F. Heywood E. I. Christiansen F. Thorne L. H. (2022). Integrating 3D models with morphometric measurements to improve volumetric estimates in marine mammals. Methods Ecol. Evol.13, 2478–2905. doi: 10.1111/2041-210X.13962
39
IJsseldijk L. L. Brownlow A. C. Mazzariol S. (2019). European best practice on cetacean post-mortem investigation and tissue sampling. Research reports or papers. ACCOBAMS/ASCOBANS. doi: 10.31219/osf.io/zh4ra
40
Irschick D. J. Christiansen F. Hammerschlag N. Martin J. Madsen P. T. Wyneken J. et al . (2022). 3D visualization processes for recreating and studying organismal form. iScience25, 104867. doi: 10.1016/j.isci.2022.104867
41
Kang Z. Yang J. Yang Z. Cheng S. (2020). A review of techniques for 3D reconstruction of indoor environments. ISPRS Int. J. Geo-Information9, 3305. doi: 10.3390/ijgi9050330
42
Küker S. Faverjon C. Furrer L. Berezowski J. Posthaus H. Rinaldi F. et al . (2018). The value of necropsy reports for animal health surveillance. BMC Veterinary Res.14, 1915. doi: 10.1186/s12917-018-1505-1
43
Lee K. Alava J. J. Cottrell P. Cottrell L. Grace R. Zysk I. et al . (2023a). Emerging contaminants and new POPs (PFAS and HBCDD) in endangered southern resident and bigg’s (Transient) killer whales (Orcinus orca). In Utero Maternal Transfer pollut. Manage. Implications. Environ. Sci. Technol.57, 360–745. doi: 10.1021/acs.est.2c04126
44
Lee K. Raverty S. Cottrell P. Zoveidadianpour Z. Cottrell B. Price D. et al . (2023b). Polycyclic aromatic hydrocarbon (PAH) source identification and a maternal transfer case study in threatened killer whales (Orcinus orca) of british columbia, Canada. Sci. Rep.13, 225805. doi: 10.1038/s41598-023-45306-w
45
Leguia M. Garcia-Glaessner A. Muñoz-Saavedra B. Juarez D. Barrera P. Calvo-Mac C. et al . (2023). Highly pathogenic avian influenza A (H5N1) in marine mammals and seabirds in Peru. Nat. Commun.14, 5489. doi: 10.1038/s41467-023-41182-0
46
Lehtola V. V. Nikoohemat S. Nüchter A. (2021). “Indoor 3D: overview on scanning and reconstruction methods,” in Handbook of big geospatial data. Eds. WernerM.ChiangY.-Y. (Springer International Publishing, Cham), 55–97. doi: 10.1007/978-3-030-55462-0_3
47
Liu W. Sun J. Li W. Hu T. Wang P. (2019). Deep learning on point clouds and its application: A survey. Sensors19 (19), 4188. doi: 10.3390/s19194188
48
Luetzenburg G. Kroon A. Bjørk A. A. (2021). Evaluation of the apple iPhone 12 pro LiDAR for an application in geosciences. Sci. Rep.11, 222215. doi: 10.1038/s41598-021-01763-9
49
Ma Z. Liu S. (2018). A review of 3D reconstruction techniques in civil engineering and their applications. Advanced Eng. Inf.37, 163–174. doi: 10.1016/j.aei.2018.05.005
50
MacLeod C. D. (2009). Global climate change, range changes and potential implications for the conservation of marine cetaceans: A review and synthesis. Endangered Species Res.7, 125–136. doi: 10.3354/esr00197
51
Merella M. Farina S. Scaglia P. Caneve G. Bernardini G. Pieri A. et al . (2023). Structured-Light 3D scanning as a tool for creating a digital collection of modern and fossil cetacean skeletons (Natural History Museum, University of Pisa). Heritage6, 6762–6765. doi: 10.3390/heritage6100353
52
Mikalai Z. Andrey D. Hawas H. S. Tеtiana Н. Oleksandr S. (2022). Human body measurement with the iPhone 12 pro LiDAR scanner. AIP Conf. Proc.2430, 0900095. doi: 10.1063/5.0078310
53
Mills N. Semans S. Flannery M. Squire C. Grimes S. Jacobsen J. (2022). A tale of two whales: applying digital imaging and 3D printing to cetacean research and education. Front. Ecol. Environ.20, 422–305. doi: 10.1002/fee.2508
54
Niven L. Steele T. E. Finke H. Gernat T. Hublin J.-J. (2009). Virtual skeletons: using a structured light scanner to create a 3D faunal comparative collection. J. Archaeological Sci.36, 2018–2235. doi: 10.1016/j.jas.2009.05.021
55
Nowacek D. P. Christiansen F. Bejder L. Goldbogen J. A. Friedlaender A. S. (2016). Studying cetacean behaviour: new technological approaches and conservation applications. Anim. Behav.120, 235–244. doi: 10.1016/j.anbehav.2016.07.019
56
Pugliares K. R. Bogomolni A. Touhey K. M. Herzig S. M. Harry C. T. (2007). Marine mammal necropsy: an introductory guide for stranding responders and field biologists. Report. (Woods Hole, MA, USA: Woods Hole Oceanographic Institution). doi: 10.1575/1912/1823
57
Rassineux A. (1997). 3D mesh adaptation. Optimization of tetrahedral meshes by advancing front technique. Comput. Methods Appl. Mechanics Eng.141, 335–354. doi: 10.1016/S0045-7825(96)01116-4
58
Raverty S. Duignan P. Greig D. Huggins J. L. Huntington K. B. Garner M. et al . (2024). Gray Whale (Eschrichtius Robustus) Post-Mortem Findings from December 2018 through 2021 during the Unusual Mortality Event in the Eastern North Pacific. Edited by Vitor Hugo Rodrigues Paiva. PloS One19, e0295861. doi: 10.1371/journal.pone.0295861
59
Rhodes-Reese M. Clay D. Cunningham C. Moriles-Miller J. Reese C. Roman J. et al . (2021). Examining the role of marine mammals and seabirds in southeast alaska’s marine ecosystem dynamics. Front. Mar. Sci.8. doi: 10.3389/fmars.2021.720277
60
Rosenberg J. Haulena M. Hoang L. Morshed M. Zabek E. Raverty S. (2016). Cryptococcus gattii type VGIIa infection in harbor seals (Phoca vitulina) in british columbia, Canada. J. Wildlife Dis.52 (3), 677–681. doi: 10.7589/2015-11-299
61
Rowles T. K. Dolah F. M.V. Hohn A. A. (2001). “Gross necropsy and specimen collection protocols,” in CRC handbook of marine mammal medicine, 2nd (Boca Raton, FL: CRC Press).
62
Schoeman RenéeP. Patterson-Abrolat C. Plön S. (2020). A global review of vessel collisions with marine animals. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.00292
63
Simmonds M. P. Isaac S. J. (2007). The impacts of climate change on marine mammals: early signs of significant problems. Oryx41, 19–265. doi: 10.1017/S0030605307001524
64
Storlund R. L. Cottrell P. E. Cottrell B. Roth M. Lehnhart T. Snyman H. et al . (2024). Aquaculture related humpback whale entanglements in coastal waters of british columbia from 2008–2021. PloS One19, e02977685. doi: 10.1371/journal.pone.0297768
65
Strecha C. Bronstein A. Bronstein M. Fua P. (2012). LDAHash: improved matching with smaller descriptors. IEEE Trans. Pattern Anal. Mach. Intell.34, 66–78. doi: 10.1109/TPAMI.2011.103
66
Tavani S. Billi A. Corradetti A. Mercuri M. Bosman A. Cuffaro M. et al . (2022). Smartphone assisted fieldwork: towards the digital transition of geoscience fieldwork using LiDAR-equipped iPhones. Earth-Science Rev.227, 103969. doi: 10.1016/j.earscirev.2022.103969
67
Taylor B. Taylor E. BC Environment, and Canada (1997). Responding to global climate change in british columbia and yukon. Victoria, B.C: Environment Canada, Pacific and Yukon Region, Aquatic and Atmospheric Sciences Division, 1997.
68
Teman S. Gaydos J. Norman S. Huggins J. Lambourn D. Calambokidis J. et al . (2021). Epizootiology of a cryptococcus gattii outbreak in porpoises and dolphins from the salish sea. Dis. Aquat. Organisms146, 129–143. doi: 10.3354/dao03630
69
Teppati Losè L. Spreafico A. Chiabrando F. Tonolo F. G. (2022). Apple LiDAR sensor for 3D surveying: tests and results in the cultural heritage domain. Remote Sens.14, 41575. doi: 10.3390/rs14174157
70
Tondo G. R. Riley C. Morgenthal G. (2023). Characterization of the iPhone LiDAR-Based sensing system for vibration measurement and modal analysis. Sensors23, 78325. doi: 10.3390/s23187832
71
Tsui H. C.L. Kot B. C.W. Chung T. Y.T. Chan D. K.P. (2020). Virtopsy as a revolutionary tool for cetacean stranding programs: implementation and management. Front. Mar. Sci.7. doi: 10.3389/fmars.2020.542015
72
Vacca G. (2023). 3D survey with apple LiDAR sensor—Test and assessment for architectural and cultural heritage. (Basel, Switzerland: MDPI). doi: 10.3390/heritage6020080
73
VanWormer E. Mazet J. Hall A. Gill V. A. Boveng P. L. London J. M. et al . (2019). Viral emergence in marine mammals in the north pacific may be linked to arctic sea ice reduction. Sci. Rep.9, 15569. doi: 10.1038/s41598-019-51699-4
74
Waltzek T. B. Cortés-Hinojosa G. Wellehan J. F. X. Jr. Gray G. C. (2012). Marine mammal zoonoses: A review of disease manifestations. Zoonoses Public Health59, 521–355. doi: 10.1111/j.1863-2378.2012.01492.x
75
Wandinger U. (2005). “Introduction to LiDAR,” in LiDAR: range-resolved optical remote sensing of the atmosphere (Springer Science+Business Media Inc., NY: Springer), 1–18.
76
Williams R. S. Brownlow A. Baillie A. Barbe J. L. Barnett J. Davison N. J. et al . (2023). Spatiotemporal trends spanning three decades show toxic levels of chemical contaminants in marine mammals. Environ. Sci. Technol.57, 20736–20749. doi: 10.1021/acs.est.3c01881
77
Wund S. Méheust E. Dars C. Dabin W. Demaret F. Guichard B. et al . (2023). Strengthening the health surveillance of marine mammals in the waters of metropolitan France by monitoring strandings. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1116819
78
Xu N. Qin R. Song S. (2023). Point cloud registration for LiDAR and photogrammetric data: A critical synthesis and performance analysis on classic and deep learning algorithms. ISPRS Open J. Photogrammetry Remote Sens.8, 100032. doi: 10.1016/j.ophoto.2023.100032
79
Yen K. S. Ravani B. Lasky T. Advanced Highway Maintenance and Construction Technology Research Center (Calif.) (2011). LiDAR for data efficiency. WA-RD. Available online at: https://rosap.ntl.bts.gov/view/dot/23658 (Accessed June, 6, 2024).
80
Zhang C. Zhou H. Christiansen F. Hao Y. Wang K. Kou Z. et al . (2023). Marine mammal morphometrics: 3D modeling and estimation validation. Front. Mar. Sci.10. doi: 10.3389/fmars.2023.1105629
81
Zhou L. Wu G. Zuo Y. Chen X. Hu H. (2024). A comprehensive review of vision-based 3D reconstruction methods. Sensors24, 23145. doi: 10.3390/s24072314
Summary
Keywords
3D scan, LiDAR, marine mammals, necropsy, morphometrics, photogrammetry, remote sensing, visualizations
Citation
Cottrell B, Kalacska M, Arroyo-Mora JP, Lucanus O, Cottrell P, Lehnhart T and Raverty S (2025) 3D reconstructions of stranded marine mammals via easily accessible remote sensing tools for use in morphometrics and visualizations. Front. Mar. Sci. 12:1485788. doi: 10.3389/fmars.2025.1485788
Received
24 August 2024
Accepted
09 January 2025
Published
05 March 2025
Volume
12 - 2025
Edited by
Takafumi Hirata, Hokkaido University, Japan
Reviewed by
Shaowei Zhang, Chinese Academy of Sciences (CAS), China
Pavel Gol’din, National Academy of Sciences of Ukraine (NAN Ukraine), Ukraine
Updates
Copyright
© 2025 Cottrell, Kalacska, Arroyo-Mora, Lucanus, Cottrell, Lehnhart and Raverty.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen Raverty, stephen.raverty@gov.bc.ca
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.