
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mar. Sci., 26 February 2025
Sec. Aquatic Microbiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1482946
Marine biofilms are globally ubiquitous surface-associated microbial communities that have gained increasing attention due to their distinctive structure and functions. The aim of this study is to provide a comprehensive overview of the current scientific understanding, with a specific focus on naturally occurring biofilms that develop on diverse marine abiotic surfaces, including microplastics, seafloor sediments, subsurface particles, and submerged artificial structures susceptible to biocorrosion and biofouling induced by marine biofilms. This article presents recent advancements and discoveries concerning the diversity, structure, function, and dynamics of these surface-associated microbial communities in the marine environment, highlighting their ecological and biogeochemical dimensions, while also serving as an inspiration for further investigations into marine biofilms.
Microorganisms can live either as free cells or in a consortium, as a biofilm, consisting of the same or different species. Biofilms are surface-associated microbial communities encased in the self-secreted extracellular matrix (Costerton et al., 1995) (Figure 1). This matrix, known as the extracellular polymeric substances (EPSs), mainly consisting of exopolysaccharides, secreted proteins, and extracellular DNA (eDNA), is believed to maintain the structural integrity of the biofilm by holding cells together as “molecular glue.” In aquatic environments, EPSs provide protection for biofilm cells against harsh environmental conditions and shear forces (Stoodley et al., 2002; Yu et al., 2015). It is widely acknowledged that biofilms represent one of the most successful and prevalent forms of life in natural habitats and industrial and hospital settings. It has been estimated that approximately 40%–80% of prokaryotes have the ability to form biofilms (Flemming and Wuertz, 2019).
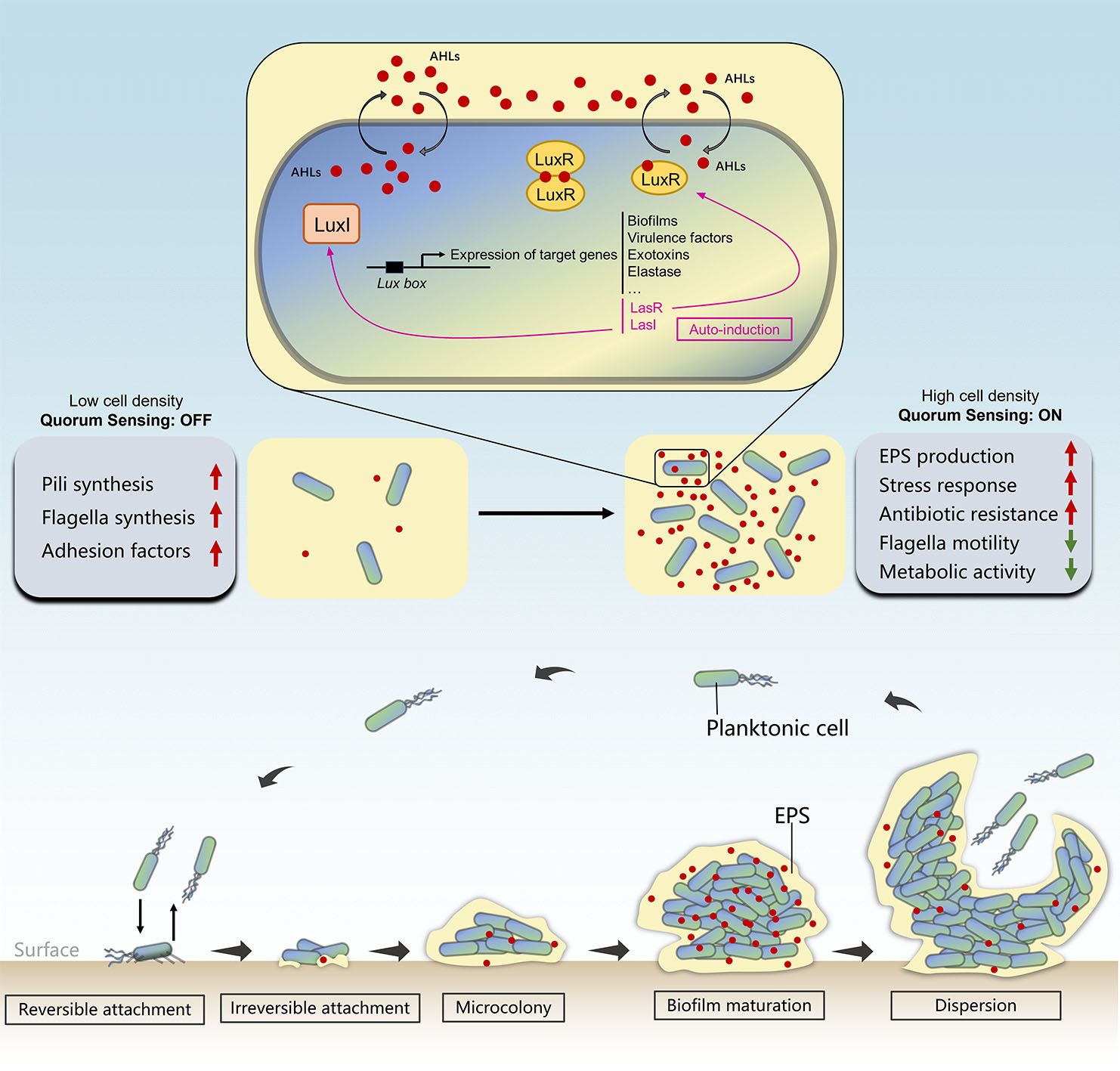
Figure 1. Typical stages of biofilm formation and the regulatory role of quorum sensing that takes part in biofilm development.
The transition from free-swimming planktonic cells to biofilm-making sessile aggregates is a multi-step process called biofilm formation. Biofilm formation follows a five-stage multicellular cycle (Ma et al., 2009) (Figures 1, 2A). In the initial step, free-floating microbial cells loosely and reversibly attached to a surface mediated by cell surface structures such as flagella and pili. The intracellular signaling molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) is essential for the initial stage of biofilm formation, as it inhibits flagella-mediated swimming motility and promotes the production of biofilm matrix (Römling et al., 2013). The Pil-Chp surface-sensing system located in microbial surfaces increases the concentration of c-di-GMP with each attachment/detachment event. Therefore, the formation of biofilms begins with the conversion of surface-naive planktonic cells (bacteria that have not yet encountered surfaces and have a low concentration of c-di-GMP) to surface-sentient planktonic cells (bacteria that have encountered surfaces and have a high concentration of c-di-GMP), resulting in the irreversible attachment of cells to surfaces (Armbruster and Parsek, 2018). Following this, attached microorganisms begin to multiply and aggregate within the self-produced EPS matrix in the presence of a high concentration of c-di-GMP. Flagella and type IV pili-mediated motilities play essential roles in microbe-surface interactions and cell–cell aggregations, respectively, leading to the formation of microcolonies (Rabin et al., 2015). The subsequent biofilm maturation can result in the development of “mushroom”-shaped structure with multilayered cells (Figure 2B), depending on the species involved. EPS is essential for the maturation of biofilms, as it facilitates microbial attachment to surfaces; stabilizes the 3-D structure of the biofilm; groups cells together; protects from a variety of stressors, such as the host immune system response, antimicrobials, oxidative damage, and metallic cations; and encapsulates signaling molecules that are necessary for quorum sensing (QS), metabolic products, and enzymes (Toyofuku et al., 2016). During the final dispersion stage, biofilm ruptures either actively (motility and EPS degradation-dependent dispersion) or passively (physical causes such as liquid flow-dependent dispersion), and microbes are released as planktonic cells to colonize new sites (Chandki et al., 2011). External factors such as pH, temperature, gravitational forces, Brownian movements, hydrodynamic forces, signal molecules, and the nature of the inhabited surfaces all influence this complex formation process (Zhao et al., 2017). For instance, bacteria generally possess a net negative charge due to the carboxyl, amino, and phosphate groups on their cell wall surfaces, leading to more adhesion on positively charged surfaces (Kovačević et al., 2016; Guo et al., 2018). Moreover, according to a study by Hou et al. (2018), shear flow promotes biofilm formation by stimulating S. aureus’ EPS production and EPS-matrix strength. Their research supported earlier hypotheses regarding pressure-induced EPS production (Hou et al., 2018).
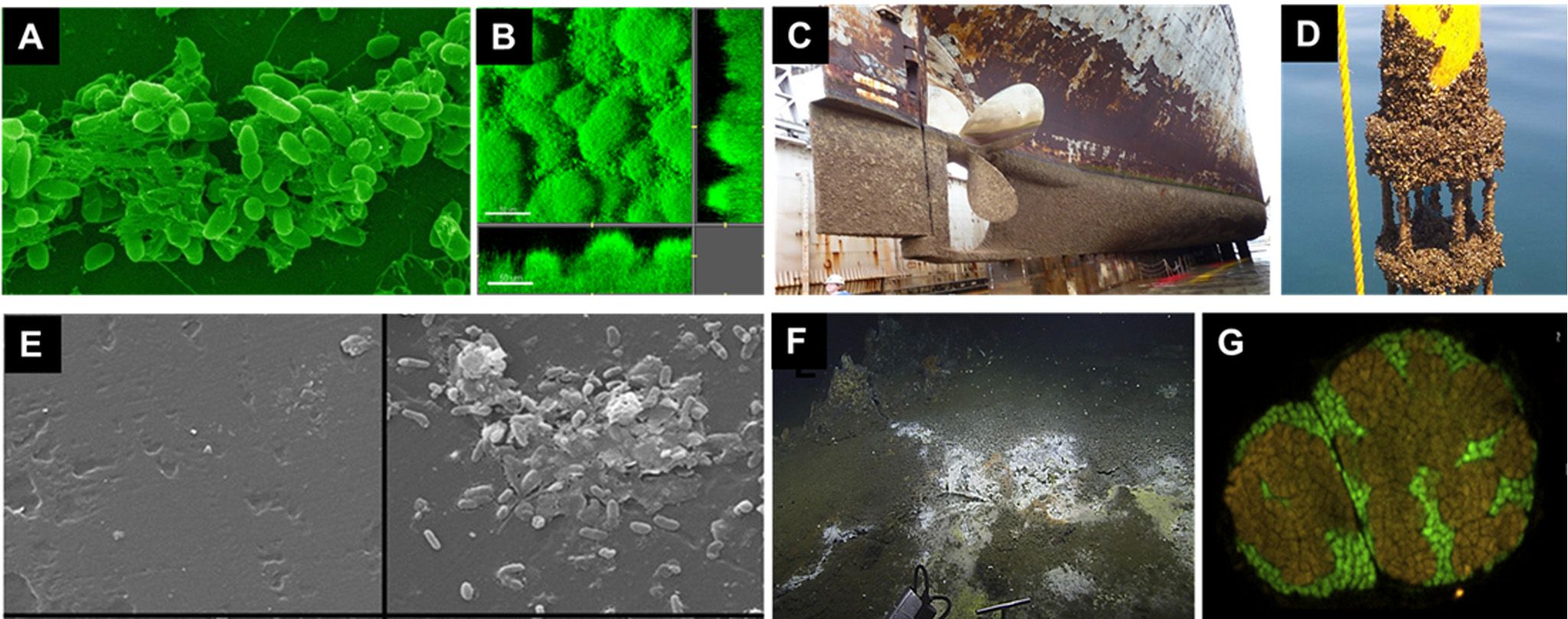
Figure 2. Photos of biofilms and marine corrosion formed on different surfaces. (A) Scanning electron micrographs of aggregates of Pseudomonas aeruginosa cells encased by the self-secreted EPS (Alain, 2021). (B) Confocal laser scanning microscopic images of Pseudomonas aeruginosa biofilm that takes the form of mushroom-like structures (Haagensen et al., 2015). (C) Biocorrosion on the surfaces of the ship hull (Lewis, 2018). (D) Current measurement instrument biofouling with zebra mussels. (E) Scanning electron micrographs showing prokaryotic attachment on a microplastic surface sampled either immediately (left) or after 14 days of inoculation (right) (Harrison et al., 2014). (F) White bacterial mat on sediment located in an area where hydrothermal fluids are seeping through cracks in the seafloor (Fahy et al., 2017). (G) Aggregates of ANME and SRB cells in the subsurface sediments of the Sonora Margin cold seeps, Guaymas Basin (Vigneron et al., 2014).
Despite their self-sufficiency, microorganisms communicate and coordinate with each other to accomplish the biofilm formation; the mechanism of this cell-to-cell communication process is referred to as “quorum sensing” (Fuqua et al., 1996). A typical QS system consists of microbial groups, signal molecules, and behavioral genes. It enables microbial cells to sense population density by monitoring the accumulation of a specific signal molecules (termed as the autoinducers, AIs) secreted and released by community members. The accumulation of these signal molecules in the surrounding environment is only adequate to activate the response with a sufficient quorum size. Once a minimal signal concentration is reached, signal molecules, known as the autoinducers, interacts with receptor proteins, leading to coordinated behavior through changes in gene expression. In this communication system, signal molecules serve as “a language” to help microorganisms to “make decisions” whether to conduct coordinative group behaviors such as biofilm formation, virulence production, enzymes secretion, and antibiotic release (Ruparell et al., 2016). Key processes involved in biofilm formation including initial bacteria–surface interactions, surface attachment, biofilm initiation, biofilm maturation, and biofilm differentiation are all believed to be regulated through this cell-density-dependent manner (Ng and Bassler, 2009). Chemical signals can be categorized into several groups. A majority of them were discovered through research on infection-causing bacteria. N-Acylated homoserine lactones (AHLs), commonly found in a number of Gram-negative bacteria, represent the first described class of QS signals. In this context, AHLs synthesized by synthase LuxI can freely diffuse across the membrane into the surrounding environment. Once their concentration reaches a certain threshold level, they bind to the receptor protein LuxR. Subsequently, LuxR dimerizes and gains the ability to act as a transcription factor by binding to the Lux box in DNA. Consequently, genes involved in biofilm formation, exotoxins production, etc. are activated while also promoting expression of both LuxI and LuxR (auto-induction) (Camilli and Bassler, 2006) (Figure 1). Two N-acyl-homoserine lactone (AHL)-based QS systems, las and rhl, and one alkylquinolone (AQ)-mediated QS have been discovered in P. aeruginosa (Jimenez et al., 2012). Different QS systems function hierarchically, regulating the virulence factors and biofilm formation of P. aeruginosa (Sharma et al., 2024). Complexes formed by the two QS systems induce expression of various virulence factors such as protease, elastase, alkaline protease, and HCN production (Brint and Ohman, 1995; Pessi and Haas, 2000; Miller and Bassler, 2001). QS promotes the production of biofilms by releasing extracellular DNA (eDNA), which is necessary for adhesion, cell-to-cell attachment, biofilm formation, stability, and defense against detergents and antibiotics (Das and Manefield, 2013). Moreover, Gram-positive systems utilize secreted oligopeptides and two-component systems, which are composed of cytoplasmic transcription factors and membrane-bound sensor kinase receptors that regulate gene expression (Novick and Geisinger, 2008). Genetic competence in B. subtilis and S. pneumoniae, virulence response and the production of antimicrobial peptides in S. aureus, and numerous other processes are reported to be regulated by QS in Gram-positive bacteria with similar fundamental objectives (Haque et al., 2019).
The synthesis of extracellular polymeric substances is crucial for biofilm development. The production of biofilms characterizes many chronic illnesses, prompting extensive research into how bacterial biopolymers influence both pathogenesis and biofilm formation. Bacterial biopolymers, along with their production and biological roles, present targets for the development of innovative antibacterial agents (Qvortrup et al., 2019). In addition, the enhanced tolerance to harmful contaminants and increased breakdown capacities of bacterial biofilms are often ascribed to the EPS matrix (Mishra et al., 2022). Through ion exchange, precipitation, binding, emulsification, solubilization, and complexation, EPS layers interact with harmful environmental contaminants (Shukla et al., 2017). Various functional groups of EPS, including carboxyl, amide, phosphoryl, and hydroxyl, participate in the elimination of toxic substances from contaminated environments. Biofilm EPS is involved in several remedial processes, including the sorption and degradation of dyes and pesticides, the emulsification of petroleum hydrocarbons, the binding and solubilization of polycyclic aromatic hydrocarbons (PAHs), and the sequestration of heavy metals (Mahto et al., 2022). Therefore, bacterial biofilm and EPS offer an appealing approach for decontaminating highly contaminated areas.
On the other hand, substantial research has concentrated on utilizing the unique material features of bacterial polymers for industrial applications in medical and technical fields. On polysaccharides, the presence of hydrophilic groups—such as hydroxy and carboxyl groups—confer great water-binding capacity and enable intermolecular contacts and crosslinks (for example, polymer–polymer, polymer–drug, and polymer–host tissue and cell interactions). Porous hydrogels formed by polysaccharides can be employed for the controlled release of anticancer drugs (Li and Mooney, 2016), drug delivery, tissue engineering (Miao et al., 2018), immobilization of enzymes (Mohan et al., 2015), therapeutic cell entrapment, and protection of transplanted cells from the host immune system (Mitrousis et al., 2018). Polyamides or poly(amino acid) chains produced by bacteria can serve as capsules or biofilm matrix or as storage material (Yu et al., 2016) (Zhang and Yang, 2019). The biodegradability, non-toxicity, and modifiability of bacterial polyamides have made them viable alternatives to chemically produced polymers for use in pharmaceutical, cosmetic, biomedical, and industrial formulations (Lee et al., 2019). Polyhydroxyalkanoates (PHAs) are bacterially produced bioplastics that can be chemically modified, bioengineered, and processed into low-value commodity bioplastics or high-value medical materials (e.g., tissue engineering scaffolds, drug carriers, sutures, and particle vaccinations) (Kai and Loh, 2014). The synthesis of polyphosphate (PolyP) is an evolutionarily ancient capability of bacteria. PolyPs not only serve as a reservoir for phosphate but also supply chemical energy for biosynthetic pathways, act as a buffer against alkalinity, function as a metal-chelating agent, and play a role in channel complexes for DNA uptake. Because of their superior energy storage properties, industry has increasingly investigated polyPs to drive energy-consuming enzyme-catalyzed reactions (Yoo et al., 2018). In regenerative medicine, they are also regarded as morphogenetically active biomaterials for bone regeneration and cartilage repair (Müller et al., 2017). Unlike other biopolymers, such as polysaccharides and polyesters, extracellular DNA, polypeptides, and proteins are fascinating programmable biomaterial platforms because of their genetic programmability and simplicity of engineering (Nguyen et al., 2018). The development of engineered living materials—that is, living cells that are designed to autonomously self-assemble entire materials with novel and tunable properties for a variety of applications, including microbial electrosynthesis, biosensors, electronic monitoring devices, and bioremediation—has attracted a lot of attention recently due to the straightforward genetic programmability of these polymers (Gilbert and Ellis, 2019).
In marine environments, a wide variety of surfaces can be colonized by diverse microorganisms, including bacteria, archaea, diatoms, fungi, flagellates, ciliates, and multicellular eukaryotes, leading to the subsequent formation of highly complex biofilms. These surfaces include both biotic surfaces, such as algae and marine living animals, and abiotic surfaces like different types of particles, aggregates, immersed constructs, vessel surfaces, and inert or bio-reactive mineral substrata (Figure 3).
The association with these surfaces provides microorganisms with numerous ecological advantages including increased access to nutritional resources, improved organism interactions, and enhanced environmental stability. These characteristics are particularly crucial in marine habitats, where nutrients are usually a restrictive factor of growth and ambient circumstances are highly dynamic and occasionally unfavorable (Dang and Lovell, 2016). The composition of the microbial community in marine biofilms is distinct from that of freshwater and plankton (Battin et al., 2016; Catão CP et al., 2021). For example, in freshwater biofilms, Pseudomonadota typically form the dominating phylum, while in marine biofilms, their abundance is lower (Battin et al., 2016). Furthermore, SAR11, Prochlorococcus, and Synechococcus are prevalent in seawater; however, pennate diatoms, Sphingomonadaceae of Alphaproteobacteria, Alteromonadaceae of Gammaproteobacteria, and Bacteroides are widespread bacterial species in marine biofilms (Harrison et al., 2018). The chemical composition of marine biofilms, such as EPS and metabolites, alters during community succession; qualitative and quantitative changes in the chemical profiles of marine biofilm extracts were observed at different stages of development (Chung et al., 2010). Moreover, the formation of marine biofilms and the associated microbial metabolic activities can also lead to deleterious outcomes such as biofouling, biocorrosion, the persistence of drug-resistant bacteria, and medical infections, resulting in significant costs and health threats (Watnick and Kolter, 2000; Cottingham et al., 2003).
Marine biofilms are primarily composed of prokaryotes in oceans (Antunes et al., 2019). Globally, more than 25,000 operational taxonomic units (OTUs) of the 16S rRNA genes of marine biofilm prokaryotes have been clustered (97% identity) (Zhang W. et al., 2019). According to an empirical study, marine biofilms are estimated to harbor a diverse assemblage of over 25,000 species, with the majority of strains exhibiting a sequence similarity exceeding 97% based on the analysis of 16 S rRNA gene (Konstantinidis and Tiedje, 2005). As the most diverse and dynamic species, microorganisms are widely recognized for their crucial role in marine environments by driving biogeochemical cycles (Paerl and Pinckney, 1996; Hawley et al., 2017) and providing substances and energy to higher trophic levels (Azam et al., 1983; de Carvalho, 2012). Moreover, they are the primary residents on marine surfaces, forming ubiquitous biofilms whose configuration, dynamics, and function may be determined by early colonizers (Dang et al., 2008, 2011). According to an empirical study, marine biofilms are estimated to harbor over 25,000 species. These prokaryotic communities encompass a diverse range of phyla including Proteobacteria, Acidobacteriota, Actinomycetota, and Crenarchaeota, among which Proteobacteria stands out as the predominant group (Zhang et al., 2019). The microbial community composition of marine biofilms is distinct, as demonstrated by a metagenomic survey that revealed 7,300 OTUs exclusively found in marine biofilms (Zhang et al., 2019). Although viral predation is restricted in biofilms due to the survival advantage of biofilm microorganisms over their planktonic counterparts (De Carvalho, 2018), it has been demonstrated that viruses confined within a biofilm matrix can remain active and infect colonizing cells, as evidenced by T7 phages (Bond et al., 2021). In a recent study, a total of 2,446 connections between viruses and prokaryotes were identified within 84 marine biofilms. The predominant connections observed were between bacteriophages in the Uroviricota phylum and bacteria belonging to the Proteobacteria, Cyanobacteriota, and Bacteroidota taxa (Zhou et al., 2023).
Our understanding of the social behaviors and interactions of microorganisms in natural biofilms is limited. According to the hypothesis of species sorting, certain microbial species may assemble into a community as a result of selective pressures exerted by local abiotic and biotic environmental factors (Zhang et al., 2014). Moreover, the spatial architecture of biofilms is primarily shaped by microbial interactions among neighboring cells. Consequently, the cooperative and competitive dynamics within marine biofilms are significantly influenced by the spatial distribution of microbial cells (Nadell et al., 2016). The findings of a recent study on signal transduction in natural biofilm development indicate that signaling molecules have the potential to modify the composition of marine biofilms (Wang et al., 2020). The expression levels of signaling-related genes, including several QS gene families (e.g., QS in Vibrio, QS in Yersinia, QS regulation in Pseudomonas, AI-2 transporters, and AHL inducers), two-component regulator families (e.g., two-component regulatory systems in Campylobacter, mazE-mazF system, and oxygen and light sensor PpaA-PpsR), virulence (e.g., Streptococcus pyogenes virulence regulators), and unknown regulatory genes associated with biofilm formation (e.g., biofilm formation in Staphylococcus) were markedly elevated in biofilm samples compared to planktonic cells, as demonstrated through an analysis of signal transduction systems within 101 marine biofilms formed on diverse surfaces and across multiple oceanic regions (Wang et al., 2022). In addition, the taxonomic affiliation of signal transduction genes in marine biofilms was found to be distinct from that observed in seawater samples, with the potential for inter-phyla interactions between bacteria residing in marine biofilms and those present in the seawater (Wang et al., 2022). The interaction and communication among microorganisms within marine biofilms and their response to environmental changes remain poorly understood due to the limited investigation conducted on natural marine biofilms.
Given the significant impacts of marine biofilms on the marine industry and biogeochemical cycles, it is imperative to gain a comprehensive understanding of the key participants, assembly mechanisms, and functional roles. Biofilms formed on biotic surfaces differ from those formed on abiotic surfaces due to differences in selective forces. The focus of this review is only on the microbial diversity, functions, microbial interactions, spatial and temporal variations in microbial community structures of marine biofilms formed on abiotic natural or manufactured surfaces, and their impacts on maritime industries.
Marine biofilm formation on manmade surfaces and involved microbial metabolic processes can have macroscale negative effects including biofouling, biocorrosion, persistence, and transmission of harmful or pathogenic bacteria and virulence determinants. The following is a summary of the main types of adverse biofilms formed on artificial marine surfaces, including major characteristics, community composition, influence factor, ecological roles, and impacts on human lives.
Oceans serve as a repository for plastic particles, hosting an estimated 5.25 trillion pieces of plastic debris within the marine system. Of these, 229,000 tons float on the surface, while 4 billion microplastics per square kilometer are globally distributed in the deep sea (Parker, 2015). Plastics, therefore, provide a distinctive and enduring habitat that can be colonized by microbes and transported over vast distances. This community of plastic-debris-associated microorganisms is generally referred to as “the plastisphere” (Zettler et al., 2013) (Figure 2E). In terms of open ocean waters, the known biomass of plastisphere has been previously estimated to be 0.01%–0.2% of total microbial biomass (Amaral-Zettler et al., 2020).
Several studies suggest that microbial diversity and richness may be highly influenced by environmental factors and niche partitioning (Dussud et al., 2018; Frere et al., 2018). Biogeographical and environmental parameters such as salinity and nutrient content have a strong influence on the structure of marine microplastic biofilms (Amaral-Zettler et al., 2015; Oberbeckmann et al., 2018). The microbial colonization of MP surfaces is also significantly influenced by the unique structural characteristics of the colonizing microorganisms and the properties of the microplastics (Pompilio et al., 2008; Gong et al., 2019; Feng et al., 2020). Electrostatic interactions governed by surface charge in plastics and microbial communities result in chemical adsorption, while the adhesion of microbial cells to substrates with extracellular polymeric substances (EPS) leads to physical adsorption (Kor and Mehdinia, 2020; Kumar et al., 2019). Bacteria possess a negative charge and attach more rapidly to surfaces with a positive charge. Compared to other microplastics, polyethylene and polystyrene are less favorable to bacterial attachment due to their negative charges (He et al., 2022). The polymer type of microplastics has been extensively investigated (Lozano et al., 2021; McGivney et al., 2020; Meng et al., 2022). The varying composition of microplastics regulates their explicit buoyancy and superficial rugosities, which influence biofilm formation, microbial colonization, and the capacity for long-distance dispersion of microorganisms (Hossain et al., 2019). A monitoring investigation of biofilms on four distinct microplastics revealed that polyolefins had the highest total suspended solids and organic matter content due to their low surface energy (Artham et al., 2009). Xie et al. indicated that the predominant bacteria on the surfaces of four microplastics were associated with particular groups on the microplastic molecules (Xie et al., 2021). It is intriguing to note that the chemical components of the plastic debris collected from a Mediterranean Sea beach affected the functions expressed by the microbial communities rather than their structure (Delacuvellerie et al., 2022). Moreover, Sooriyakumar et al. determined that surface roughness influences the types of microorganisms that inhabit the plastic surface (Sooriyakumar et al., 2022). Aged microplastics exhibited an enhanced microbial community composition, attributed to their high surface area, roughness, and polarity (Rai et al., 2021). Goldstein et al. (2014) demonstrated a positive correlation between the diversity of fouling microorganisms in the North Pacific and the size of microplastic fragments (Goldstein et al., 2014). Carson et al. (2013) observed that an increase in the size of MP fragments corresponded with a rise in the abundance of microbial taxa, while diversity remained unchanged (Carson et al., 2013). According to Gong et al., microplastics with varying particle sizes had surface biofilms with different microbial-community compositions (Gong et al., 2023). Yao et al. proposed that the maintenance of biomass in the biofilm may be compromised by the more incompact biofilm formed on the surface that larger microplastics (Yao et al., 2019). Smaller microplastics have a larger surface area and potential surface chemical interactions, which could have an impact on the adsorption capacity (Horton et al., 2017). Furthermore, the particle size and surface modification of plastics greatly influenced their potential toxic effects on aquatic biofilms (Miao et al., 2019).
A core bacterial community comprising Cyanobacteriota, Bacteriodota, and Proteobacteria typically represents the dominant groups of microbial assemblages inhabiting microplastics in various marine ecosystems (Oberbeckmann et al., 2014; Dussud et al., 2018; Jiang et al., 2018; Xu et al., 2019; Li et al., 2020; Zhang et al., 2021). Among these groups, Bacteroidia and Alpha-, and Gamma-proteobacteria are the most abundant classes (Delacuvellerie et al., 2019; Kirstein et al., 2019; Xu et al., 2019; Dudek et al., 2020). Additionally, archaeal communities could also potentially be present in plastic-associated biofilms. In a study, Crenarchaeota were found in all deep ocean-collected micro- and mesoplastic biofilms (Woodall et al., 2018). Moreover, using next-generation sequencing, De Tender et al. (2017) investigated the biofilm ecosystems on polyethylene microplastics cultured in marine environments. Their findings revealed a variety of microorganisms belonging to the Ascomycota and Basidiomycota phyla, with a lesser presence of Zygomycota (De Tender et al., 2017).
Paints used to coat surfaces in aquatic environments often contain biocides to prevent biofouling, and as these coatings degrade, antifouling paint particles (APPs) end up in aquatic, and especially marine, sediments. Paint particles in the marine environment are often overlooked in microplastic pools (Turner, 2021). APPs release biocides and other chemicals that reduce the formation of biofilms, inhibiting the proliferation of organisms on submerged surfaces like ship hulls. Consequently, although microplastics are rapidly inhabited by microbial communities similar to those found on other inert substrates such as glass, rock, or wood (Wright et al., 2020), antifouling surfaces offer a niche that favors specific, albeit occasionally diverse, bacterial populations that exhibit resistance to active biocides (Chen et al., 2013; Flach et al., 2017). Tagg et al. (2019) investigated and compared biofilm communities on microplastics and paint particles collected from sediment grabs in the coastal Baltic. Researchers discovered that alkyd- and epoxy-based paints, presumably containing antifouling components, sustained unique and more stable communities than biofilms on polypropylene and polyvinyl chloride microplastics and on natural particles. The prevalence of the Desulfobacteraceae family on certain paint particles indicates that their presence in sediment may influence the sulfur metabolism cycle (Tagg et al., 2019). A recent study revealed that sediment microcosms contaminated with spiking APPs exhibit a distinct and consistent alteration in their microbial community, 71 indicator taxa associated with antifouling presence and 454 associated with antifouling absence were identified (Tagg et al., 2024).
Recently, the potential impacts of microplastics on the ecological functions in coastal environments have attracted increasing attention. For instance, the leaching of dissolved organic carbon from microplastics has been shown to significantly influence the oceanic carbon cycle by increasing microbial activity (Romera-Castillo et al., 2018). The nitrification and denitrification of salt marsh sediments were facilitated by polyurethane foam and poly(lactic acid) (PLA) microplastics treatments, as demonstrated by Seeley et al.; conversely, poly(vinyl chloride) (PVC) inhibited both processes (Seeley et al., 2020). In addition, biodegradable microplastics were observed to promote the microbial-mediated reduction of sulfate to chromium-reducible sulfides in coastal sediments, demonstrating their substantial effects on the coastal biogeochemical S cycle (Wang et al., 2023).
It is reasonable to assume that QS has the potential to influence the formation and composition of the plastisphere, despite the fact that the pertinent research has not yet been conducted. Rhodobacteraceae were found to comprise 16.4% of the bacterial biofilms of marine microplastics in the Yangtze River Estuary, China (Jiang et al., 2018) and to occupy core populations in the plastisphere (Debroas et al., 2017; Dussud et al., 2018). According to Zhang et al. (2014), a significant percentage of Rhodobacteraceae in the plastisphere was ascribed to the conserved and ubiquitous QS signaling molecules involved in a number of metabolic processes, particularly flagellum movement and biofilm formation on plastic particles (Zan et al., 2014). QS may facilitate the attachment and aggregation of Rhodobacteraceae to the surface of marine microplastics and the inhibition of other bacteria, such as Gammaproteobacteria. Furthermore, QS bacteria isolated from marine microplastics demonstrated a strong capacity to form biofilms and showed the ideal conditions for epiphytic growth. Consequently, Oceanicola sp. strain D3, exhibiting QS capability, was isolated from a PVC biofilm (Li et al., 2019). It is noteworthy that bacteriostasis, algae inhibition, and the degradation ability of dimethylsulfoniopropionate (DMSP) by QS bacteria could influence the pollution of microplastics in the marine environment (Gagné, 2017). Additionally, QS has the potential to serve as an innovative approach to investigate the degradation of marine microplastic biofilms. For instance, AHLs have the potential to alter the activity of extracellular hydrolases (Jatt et al., 2015). Consequently, a QS system that utilizes AHLs could play a significant role in the degradation of microplastic biofilms. Furthermore, QS bacteria may serve as effective indicators for monitoring MP contamination in the marine environment due to their rapid sensitivity to organic substances and dominant colonization.
It is believed that microbial communities present in microplastics play a potential role in the degradation of plastic polymers (Jacquin et al., 2019). Due to their hydrocarbon-clastic nature, microbial biofilms could also change the physical properties of microplastics, such as size and buoyancy, therefore allowing them to utilize microplastics as a source of energy by breaking down petroleum derivatives and complex biopolymers (Zettler et al., 2013; Ogonowski et al., 2018). Alteromonadaceae and Burkholderiales, Erythrobacter spp., and Alcanivorax borkumensis in microplastic biofilms were demonstrated to be capable of degrading poly(3-hydroxybutyrate-co-3-hydroxyhexanoate (PHBH), hydrocarbons, and low-density polyethylene, respectively (Morohoshi et al., 2018; Curren and Leong, 2019; Delacuvellerie et al., 2019). Moreover, the ratio of microplastics microbiological communities to free microbes in the oceans is expected to increase over time, thereby potentially impacting environmental ecological dynamics due to the presence of dense biofilms on these microplastics. The formerly less active or inactive species of the marine ecosystem can be selectively enriched on the newly introduced surface plastics in oceans. In addition, the leachate from microplastics can significantly enhance the levels of dissolved organic matter, thereby elevating microbial biomass in the ocean (Romera-Castillo et al., 2018). Wang et al. discovered that biofilms formed on microplastics act as viable carriers for aquatic contaminants, facilitating their trophic transmission across the food web to higher organisms (Wang et al., 2021).
The growing concern about microplastics arises from their potential to harm organisms and aquatic environments (Amaral-Zettler et al., 2020). An especially significant issue is that microplastics might act as substrates, facilitating the proliferation and dissemination of diseases (Barros and Seena, 2021). Some pathogenic bacteria were more prevalent on microplastics than on natural substrates (Sabatino et al., 2024; Stevenson et al., 2024); some pathogens were exclusively detected on microplastics (Hu et al., 2021). Opportunistic pathogens, including Vibrio (Kesy et al., 2021), Acinetobacter (Shi et al., 2021), and Mycobacterium (Zhao et al., 2021), have a substantial tendency to adhere to microplastics, as demonstrated by the most recent research. Moreover, microplastics play an important role in the proliferation, harboring, and dissemination of ARGs, hence exacerbating the difficulty of managing and treating infections (Liu et al., 2021).
Moreover, in comparison to planktonic cells, horizontal gene transfer (HGT) is believed to occur more frequently and be more widespread in microplastic biofilms through type IV secretion systems and conjugation (Arias-Andres et al., 2018). Although the underlying mechanisms are currently unknown, a high frequency of exchange of antibiotic resistance genes (ARGs) has been observed, which is postulated to contribute to the dissemination of antimicrobial resistance (AMR) (Arias-Andres et al., 2018; Donlan, 2000). The functionality of biofilms and the prevalence and types of gene exchange taking place on marine microplastic particles are both intriguing areas that continue to be subjects of ongoing research.
Marine corrosion is a complex process that refers to the electrochemical degradation of materials, typically metals, under the influence of prolonged contact of marine structures with seawater (Lawal et al., 2024). The seawater itself acts as an electrolyte with high corrosive properties. Chemical corrosion is predominantly initiated by pure chemical activity. Corrosion occurs when metal materials are exposed to the marine environment and interact directly with it. Gases such as SO3, SO2, and CO2 can dissolve in the liquid layer interacting with the metal surface, resulting in an acidic environment that erodes the metal (Sun et al., 2016). Additionally, the oceanic environment is intricate due to the influence of marine organisms and their metabolites, which collectively contribute to corrosion (Li et al., 2019). Upon submergence in seawater, surfaces quickly become colonized by microorganisms, leading to the formation of highly heterogeneous and dynamic three-dimensional structures. This deleterious biofilm formation has a significant impact on maritime engineering, resulting in considerable damage and financial losses worldwide (Enning and Garrelfs, 2014).
MIC is the corrosion of materials that is promoted directly by the living activities of microbes or indirectly by their metabolites, while marine biofouling is the result of the unfavorable settlement and accumulation of marine microorganisms and macro-foulers on submerged surfaces of materials (Li et al., 2019). Thus, MIC is a corrosion process that takes place at the microscopic level, involving interactions between microorganisms and material surfaces, while biofouling is a macro-level process characterized by the deposition and accumulation of fouling substances on surfaces. Both MIC and biofouling are closely associated with biofilms that are formed by marine microorganisms that are sorted by the initial film (KoChina et al., 2022). Organisms associated with MIC are simply a variety of microbes. Organisms associated with biofouling comprise different microorganisms, plants, and animals. Microorganisms are the cause of biofouling creation because they generate appropriate settling sites and circumstances and nutrients that attract additional organisms (Li et al., 2019).
Metal corrosion could be accelerated either directly or indirectly by microbiological activities or their metabolites, known as microbiologically influenced corrosion (Liu et al., 2017) (Figure 2C). A prominent example of this phenomenon occurs in the oil and gas industry, including storage and transportation processes. While microbes play a crucial role in oil bioremediation, the microbial contamination in oil and natural gas facilities is undesirable because they can metabolize hydrocarbons, alter sulfur content, and affect oil density and viscosity. The anaerobic conditions common in the oil sector, together with the abundance of microbe substrates, such as hydrocarbons and organosulfur compounds, stimulate the biofilm formation, which causes microbial-influenced corrosion (Vigneron et al., 2016; Abu Bakar et al., 2017; Li et al., 2017).
Biofilm formation and development, which are influenced by microbial metabolisms and corrosion, play a critical role in MIC. Several studies have revealed that different stages in biofilm formation exert varying impacts on corrosion. The formation of heterogeneous biofilms due to the detachment of unstable ones can result in localized corrosion, thereby accelerating the overall corrosion rate (Xia et al., 2015; Lenhart et al., 2014).
There are two primary theories explaining the mechanism by which microbial biofilms induce corrosion. According to the oxygen concentration cell theory, when heterogeneous biofilms form on a material surface, regions covered by dense biofilms experience oxygen deprivation due to prevention of biofilm formation and consumption by aerobic bacteria. Consequently, these areas serve as anodic sites in the corrosion process. Conversely, locations without biofilm or covered by thin biofilms act as cathodic sites for electron consumption and oxygen reaction (Jia et al., 2019). On the other hand, the bio-catalytic cathodic sulfate reaction (BCSR) theory posits that sulfate-reducing bacteria (SRB)-formed biofilms function as barriers to mass transfer, hindering carbon source diffusion. As a result of limited external carbon and electron availability, starved SRB near the metal surface utilize iron as an electron donor leading to iron corrosion. The released electrons are subsequently utilized by SRB for sulfate reduction (Skovhus et al., 2017). Dinh et al. (2004) found that unique marine deltaproteobacterial SRB enriched and isolated using metallic iron as the only electron donor reduced sulfate faster than traditional strains and were more corrosive. Furthermore, they generated a large amount of H2 rather than scavenging it, probably due to an initial excess of iron-derived reducing power (Dinh et al., 2004). Accordingly, direct electron intake from iron has been proposed as a kinetically more effective way to employ Fe0 for sulfate respiration than through the slowly generated abiotic H2, i.e., through a faster bypass (Dinh et al., 2004). Under laboratory conditions, sulfate- and iron-reducing bacteria were found in different layers of anaerobic biofilm on carbon steel in seawater, and electrochemical impedance spectroscopy (EIS) capacitance changes could indicate corrosion product alterations. The primary mechanism of corrosion inhibition is the formation of green rust, which is induced by biofilms. The change in capacitance in EIS can be used to indicate the change in corrosion products (Duan et al., 2008). It is further discovered that two SRB strains, Desulfopila corrodens strain IS4 and Desulfovibrio ferrophilus strain IS5, which were isolated from marine sediment, rapidly reduced sulfate while oxidizing metallic iron (as the only electron donor) (Enning et al., 2012). Moreover, recent research has shown that outer-membrane cytochromes (OMCs) are extensively conserved in sediment oxidized sulfur species (OSS)-respiring bacteria and allow cells to directly utilize electrons from insoluble minerals through extracellular electron transport (Deng et al., 2018).
Assorted bacteria, including sulfur-oxidizing bacteria (SOB), SRB belonging to the Delta-proteobacteria, non-cultivable iron-oxidizing bacteria (IOB) belonging to the Zeta-proteobacteria, iron-reducing bacteria (IRB), slime-producing bacteria (SPB), and acid-producing bacteria (APB), are known to be involved in bio-corrosion (Enning and Garrelfs, 2014). The growth of biofilm was found to be essential for electron uptake on a cathodic surface by Desulfovibrio ferrophilus IS5, an iron-corroding strain. However, the mechanism of electron uptake from the cathode may differ from that involved in iron corrosion (McCully and Spormann, 2020). Unlike hydrogenase-positive bacteria that typically utilize H2 as an electron carrier, anaerobic microbial iron corrosion involves a more direct acquisition of electrons from metallic iron rather than through hydrogen consumption (Enning et al., 2012). The corrosion process typically involves sulfate reduction, nitrate reduction, metal reduction, sulfur oxidation, metal oxidation, and fermentation processes. In natural biofilms, a diverse range of species initiates a cascade of metabolic activities that result in more severe corrosion compared to the effects caused by a single species (Kip and Van Veen, 2015). SRB and IOB often cooperate within biofilms formed on metal substrates, mediating this process as the main culprits. IOB depletes oxygen, creating a favorable environment for the growth of anaerobic SRBs, which subsequently promotes corrosion by the matrix (Dong et al., 2011; Belkaid et al., 2011; Jones and Amy, 2002). The coexistence of iron-oxidizing bacteria (which directly contribute to corrosion) and iron-reducing bacteria facilitates the continuous progression of the corrosion process (Videla and Herrera, 2009). The formation and maintenance of biofilm structures are crucial in the process of marine biocorrosion. The biofilms contain high concentrations of organic acids, which exhibit corrosive properties towards metals and concrete (Procópio, 2019). Moreover, by selectively chelating cations, the EPS of biofilms can serve as efficient electron transporters between biofilms and metals (Ma et al., 2020). Bacteria account for over 98% of the microbial population assessed by qPCR in samples from three anaerobic biofilms inside a severely corroded steel pipe at an offshore oil facility in the Gulf of Mexico; however, archaea species were also detected (Vigneron et al., 2016). The findings of various studies have consistently shown that sulfur oxidizers and methanogens are the predominant archaeal groups associated with marine microbial corrosion (Usher et al., 2014; Zhang et al., 2019).
Biofilm formation on metal surfaces, however, can hinder corrosion through various mechanisms, such as bacterial aerobic respiration that neutralizes corrosive agents, the creation of protective films, and the inhibition of corrosion-inducing bacteria growth via antimicrobial secretion (Videla and Herrera, 2009). Adequate evidence exists in the literature that molecules that disrupt quorum sensing have been employed to mitigate biocorrosion. These molecules function by either inhibiting the release of signaling proteins or degrading them. These quorum quenchers (QQ) or QS inhibitors (QSI), which were recommended as a means of preventing the corrosion caused by multiple microorganisms, consist of a variety of synthetic and natural biocides, such as surfactin, magnesium peroxide, capsaicin, penicillic acid, gramicidin, patulin, cinnamaldehyde, vanillin, hexadecanoic acid, isonaamidine, phenolics, saponins, quinones, tannins, terpenoids, AHL acylases, and AHL lactonases (Scarascia et al., 2016). There is still much to be explored regarding biofilm growth and surface corrosion in the maritime environment.
The undesired settlement and aggregation of marine microbes, plants, and animals on submerged material surfaces are referred to as marine biofouling (Lindholdt et al., 2015). Marine substrates such as aquaculture equipment, oil and gas installations, and ship hulls often exhibit diverse accumulations of micro- and macro-foulers (Blackwood et al., 2017). On a single ship’s hull, for instance, there exists various materials and structures that provide distinct fouling niches favored by different organisms. The formation of marine biofouling biofilms increases frictional resistance leading to higher fuel consumption. Additionally, it initiates or accelerates the corrosion of metals and concrete, which elevates the risk of failure in marine facilities and equipment (Blackwood et al., 2017) (Figure 2D).
Marine biofouling is a microbial biofilm-related process that encompasses the following sequential steps, as depicted in Figure 4. First, surfaces undergo rapid physicochemical alterations within seconds to minutes due to the deposition of an organic conditioning film composed of various organic molecules. Subsequently, bacteria adhere to this film and develop into a biofilm as primary colonizers. The biofilm communities colonizing artificial surfaces in Mauritian coastal waters were predominantly composed of Proteobacteria, specifically Gamma-proteobacteria (Rampadarath et al., 2017). Third, secondary colonizers such as larvae, diatoms, and microalgae spores accumulate on these surfaces to exploit the nutrients provided by the pre-existing biofilm matrix. Finally, macro-planktonic communities such as mussels and barnacles subsequently settle and proliferate on material surfaces over time (Roberts et al., 1991). As pivotal initiators of biofouling processes, microbial biofilms create favorable settlement sites with optimal conditions while also serving as nutrient sources for attracting new species. The production of macro-foulers may be regulated by bacterial activities, while the presence of macro-foulers can offer protection for bacteria and their biofilms against eradication.
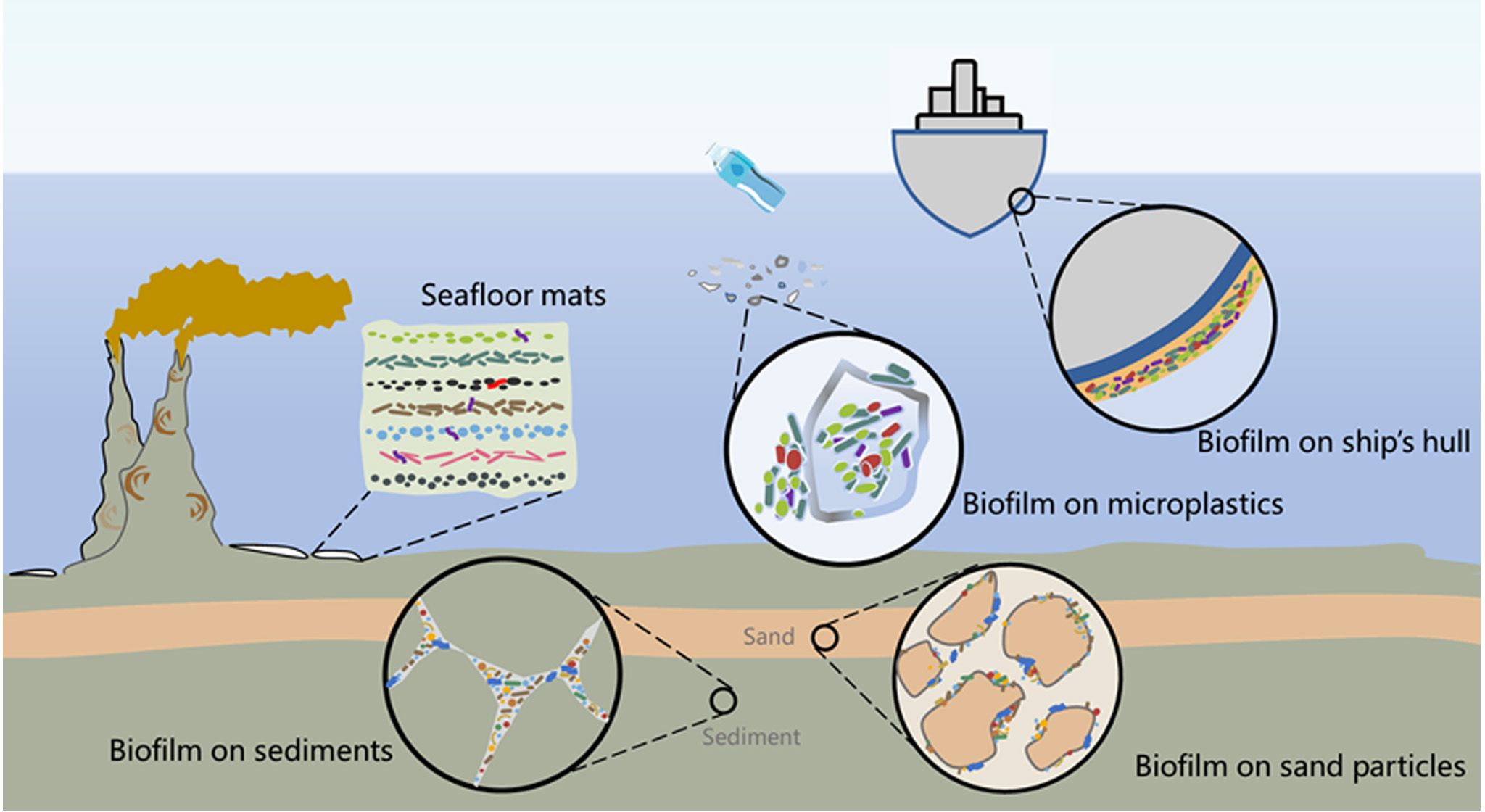
Figure 4. Marine biofilms on various abiotic surfaces in the ocean. Microbial coverage is present on various surfaces in marine environments, including seafloor microbial mats that are formed by multiple biofilms of microorganisms embedded in a matrix of exopolysaccharides, in a vertical fashion, multispecies biofilm formation on the surface of microplastic particles, MIC biofilm formed on submerged ship hulls, microbial biofilms in fractured suboceanic sediments, and biofilms coating sand particles.
Marine biofilms play a crucial role in facilitating the settlement and morphogenesis of macro-fouling organisms (Agostini et al., 2021). Model studies have demonstrated that specific bacterial strains can induce morphogenesis in marine invertebrates by extracellular phage-like structures or the released vesicles (Dobretsov and Rittschof, 2020). The morphogenic development of brown and green algae, in contrast, requires the co-cultivation of distinct strains (Tapia et al., 2016; Wichard, 2023). This symbiotic relationship appears to be validated by model testing of anti-fouling strategies. A study investigating various cementitious materials for biofouling prevention demonstrated that surfaces with biofilms exhibiting lower total cell counts exhibited a correspondingly reduced total biomass of macrofouling organisms (Hayek et al., 2021). Similarly, a study on the development of Shewanella marisflavi biofilms and their impact on mussel settlement revealed that enzymatic inhibition of total bacterial protein count significantly reduced both the bacterial cell count and the settlement of mussels (Hayek et al., 2021).
Marine biofouling is a significant international challenge that has a substantial impact on global economies and ecosystems. It manifests as the widespread and costly colonization of submerged surfaces by marine organisms, and the global transportation of invasive, harmful species (Byers et al., 2023). The utilization of antifouling coatings has emerged as the predominant method for preventing fouling, owing to their efficacy, cost-effectiveness, and ease of maintenance (Liu et al., 2023). Currently, long-term effective anti-fouling coatings are non-passive. These systems predominantly depend on the exfoliation of surface layers (self-polishing), which consistently release deleterious metallic ions and accompanying booster biocides into the surrounding environment. In spite of the stringent environmental regulations in Europe, the 11 antifouling biocides that have been approved continue to pose a danger to the marine environment (de Campos et al., 2022). While these coatings can greatly reduce adherent organisms on exposed surfaces, their toxic effects on nearby marine ecosystems and indiscriminate targeting of all proximate marine species have depleted essential marine-based human food sources (Ali et al., 2024).
The Galvanic Anode Cathodic Protection system (GACP) is extensively employed to protect submerged metallic structures from corrosion. Due to their low mass consumption, high efficiency, negative corrosion potential, and, of course, affordability, aluminum (Al), zinc (Zn), and magnesium (Mg) are the main metals that compose galvanic anodes (Levallois et al., 2023). Because the galvanic anode has a lower electrochemical potential than the metal to be protected, the metal oxidizes and releases toxicologically significant components into the marine environment as ions or oxyhydroxides (Reese et al., 2020). In addition, electroactive bacteria are naturally present in marine environments. These bacteria spontaneously colonize anodes to form biofilms and maintain electrooxidation processes on their surface (Flemming et al., 2016). As more immerged metallic structures are and will be protected by galvanic anodes, such as the growing number of offshore wind farms, the potential effects of metals released by galvanic anodes on marine ecosystems are critical. Nevertheless, the coatings industry appears to be very conservative, primarily as a result of economic and technical constraints. Passive technology development is now beyond critical. The majority of current research on the synthesis of alternative passive anti-fouling solutions has concentrated on natural-occurring interfacial derivations (Ali et al., 2024). The development of environmentally benign, high-performance, and broad-spectrum surfaces has been facilitated by evolutionary processes in nature. Current research is focused on the adaptation of these innovative structures (Ali et al., 2024). Furthermore, an ambitious and comprehensive regulatory framework is essential to facilitate the necessary green technological revolution while balancing economic sustainability, global trade interests, conservation initiatives, and inspiring future innovations.
Despite the functional importance of mediating biogeochemical cycles, research on biofilms in their natural settings, particularly at the sediment–water interface, has received limited attention. Microbial mats are a specialized type of biofilm that typically consist of one to several cell layers and vary in thickness from a few millimeters to a centimeter (Bolhuis and Stal, 2011). Another defining feature of microbial mats is that the vast majority of them is autotrophic; in other words, they utilize inorganic carbons as carbon sources photosynthetically or chemosynthetically (Zammit et al., 2021; Nakagawa and Takai, 2008).
The seafloor mats are vertically stratified benthic microbial communities that thrive at the seawater–seabed interface. Depending on the environmental circumstances, these mats can cover entire basins or span a few thousand square meters (Valentine et al., 2016). Functional groups of marine microorganisms are densely compacted into a thin mat laterally where varying amounts of minerals such as silicates and carbonates may also be embedded (Stal et al., 1984). The formation of mat is primarily driven by microbial activities, interactions between microorganisms and their grazers, and the viral shunt (i.e., viral lysis that converts microbial biomass into a pool of dissolved organic matter; Rastelli et al., 2017). The development of the mat is facilitated by the mat-associated microbiota, which produces EPS and encases organic sheaths (Flood et al., 2021). The hyphae of fungi can enhance the stability of microbial mats and serve a structural function by traversing the physicochemical gradients present in the layers (Pasulka et al., 2019; Velázquez et al., 2016). Seafloor mat residents, specialized consortia of bacteria and archaea, interact closely with each other in spatial and temporal physiological couplings (Allen et al., 2009; Spring et al., 2015). Diverse metabolic activities can occur in close proximity due to rapid nutrient cycling across microgradients and potential niche diversification within mat layers. As a result, these conditions create an excellent setting for cross-genera communication and the formation of unique community symbioses.
The processes of denitrification, metal reduction, and sulfate reduction are prevalent and essential in microbial mats. Photosynthesis is the primary energy source for microbial mats at shallow depths. The Cyanobacteria in shallow-water mats utilize solar energy to assimilate inorganic carbon, synthesize sugars, and release oxygen; many of them also possess the ability to fix N2, resulting in intricate patterns of nitrogenase activity (Prieto-Barajas et al., 2018). The SRBs also play a crucial role in microbial mats by facilitating the conversion of sulfates into sulfur and the oxidation of organic materials (Bolhuis et al., 2014). At deeper depth, marine microbial mats are typically supported by chemosynthetic processes (Kato et al., 2018). Microbial mats formed on the benthic–planktonic interface of deep-sea vents and cold seeps are predominantly self-sustaining, as they support most of the major biogeochemical cycles. Seafloor-associated bacteria that establish close symbiotic relationships with metazoans form the foundation of the food chain, resulting in highly specialized and diversified metazoan vent communities (Dubilier et al., 2008).
In the oligotrophic deep sea, hydrothermal vents and seeps function as oases of life with exceptional biodiversity and productivity, owing to the elevated concentration of metals contained in the expelled fluids that can be utilized by chemoautotrophs. Commonly, SRBs from the Delta-proteobacteria are consistently found in hydrothermal sites (Houghton et al., 2007; Frank et al., 2013). Additionally, SOBs typically dominate the chemosynthetic microbial communities in many hydrothermal vents (Huegler et al., 2010; Flores et al., 2011; Jaeschke et al., 2012). At cold seeps, microbial mats support a diverse community of methane- and sulfur-oxidizing bacteria (Crepeau et al., 2011). Mat-forming, sulfur-oxidizing Gamma-proteobacteria and Epsilon-proteobacteria (e.g., Sulfurovum and Sulfurimonas), particularly filamentous forms such as Beggiatoa and Thiothrix, are responsible for the visually noticeable white-colored microbial mats (Crepeau et al., 2011; Yamamoto and Takai, 2011; Kato et al., 2012) (Figure 2F). Depending on the environmental condition, Beggiatoa oxidize sulfide to elemental sulfur and usually further to sulfate-utilizing oxygen or nitrate as a terminal electron acceptor in cold seeps (Grünke et al., 2011). The reverse tricarboxylic acid (rTCA) cycle is utilized by other microbial mat formers, such as Sulfurovum (Campylobacterota), for carbon fixation. These organisms exhibit remarkable metabolic adaptability, including the ability to oxidize hydrogen and sulfur while reducing oxygen, nitrates, and sulfur compounds (Stokke et al., 2015).
The production of a variety of extractable QS signals by mats has been demonstrated (Charlesworth et al., 2019; Decho et al., 2009). It has been proposed that QS could be in charge of the coordination of metabolic processes in these closely linked ecosystems reflecting strongly coupled microbial interactions (Prescott and Decho, 2020). Particularly, QS-mediated S transformations may play a particularly significant role in microbial mats. Sivakumar et al. (2019) identified a relationship between QS and sulfate reduction in two SRB: the marine bacterium Desulfobacterium corrodens and the soil bacterium Desulfovibrio vulgaris. Their results showed that QSI addition dramatically reduced the growth rates, biofilm formation, and sulfate reduction capacities of these strains, even at subinhibitory concentrations (Sivakumar et al., 2019).
Microbial mats play an important role in regulating the erosive reaction of sediment particles to hydrodynamic forces (Paterson et al., 2000; Gerbersdorf et al., 2008; Vardy et al., 2007). Upon settling at the sediment surfaces, microbial-secreted EPS glue the sediment particles together in the matrix (Decho, 1990; Flemming, 2011), effectively covering and shielding excessive portions of the sedimentary surfaces from erosion (Noffke and Paterson, 2008), a process known as “bio-stabilization” (Paterson, 1989). Furthermore, mineral precipitation is closely associated with the development of microbial mats (Sanz-Montero and Rodriguez-Aranda, 2013). Initial mats can further evolve into solid, typically reef-like structures like stromatolites, by retaining sediment particles and facilitating mineral precipitation (“biomineralization”) under favorable conditions (Noffke and Paterson, 2008; Phoenix and Konhauser, 2008). When the by-products of microbial reactions disrupt the chemical balance of the micro-environment, substances such as carbonate, iron sulfide, iron oxide, or silica precipitate. For example, microbial mats can facilitate biological carbonate precipitation through processes like photosynthesis, biological oxidation, reduction, or hydrolysis (Grotzinger and Knoll, 1999). In addition, when mineral elements (such as iron) are concentrated by complexing (binding) with organic molecules produced by mats communities, mineral precipitation can be triggered (Baskar et al., 2008). Moreover, in certain seafloor environments, such as thermal springs, abiotic precipitation can occur. In such cases, mat microbes may act as templates for precipitation when fluid saturation level is sufficiently high for spontaneous precipitation (Dupraz et al., 2009). As stromatolite analogs, microbial mats are widely recognized as the oldest ecosystem on Earth, with their fossils dating back nearly 3.5 billion years (Margulis et al., 1980).
Over geological timeframes, the deep biosphere serves as a vital component of biogeochemical fluxes and processes in the Earth’s system (Parkes et al., 2014). Despite typically low level of activity, with generation durations spanning thousands of years, comprehensive calculations indicate that subsurface sediments contribute significantly to sediment activity, establishing their global biogeochemical relevance (Colwell and D’Hondt, 2013). It has been confirmed that the deep marine biosphere is actively populated by microbial cells, which gradually form clusters (Teske, 2005). The deep oceanic subsurface represents a habitat predominantly dominated by biofilms, particularly in regions exhibiting high biogeochemical activity.
The estimated range of the habitable deep suboceanic zone extends from 0.5 km to 5 km crust depth (from 1 million year old to 180 million year old), based on modeled 120°C isotherms (with 120°C as the temperature limit for microbial life) (Bar-On et al., 2018; Heberling et al., 2010). Despite concerns about its validity, lipid analysis reveals that the deep suboceanic biosphere harbors approximately 90% of the suboceanic biomass, with archaea accounting for up to 35% of the biomass in marine subsurface sediments (Danovaro et al., 2015). The basaltic layer with significant porosity serves as the main layer that provides space for microbial residence and activities (Schrenk et al., 2010). Crustal communities act as a conduit for critical elements transfer from the mantle to surface water (Menez et al., 2012).
The acquisition of samples and separation of sessile cells pose challenges in studying subsurface biofilms, with key aspects of their ecology remaining unclear. Homogenization of sediment samples, followed by separation and enrichment of microbial cells contained within, which are subsequently counted using fluorescence microscopy (Morono and Inagaki, 2016), has been established as a direct approach for the quantification of sessile cells. However, both direct and indirect approaches fail to distinguish between single cells, aggregates, and mature biofilms.
Under energy constraints, adhesion to surfaces has been regarded as a survival strategy (Marshall, 1985). Substrates such as fatty acids can adhere to the particles surfaces and accumulate locally (Lever et al., 2015), providing a plausible explanation. Sediment particles serve as excellent colonization substrates for biofilms, offering nutrients and different types of electron acceptors and donors (Battin et al., 2016). The marine sediments harbor the largest methane reservoir on Earth (Liu et al., 2022). Sessile bacteria in the subsurface reduce carbon dioxide or low-molecular-weight organic compounds, forming the biotic proportion (Thorseth et al., 2001). Boetius et al. demonstrated that marine microbial consortia performed anaerobic oxidation of methane in anoxic sediments (Boetius et al., 2000). The oxidation primarily takes place in biofilms formed by anaerobic methanotrophic archaea (ANME), generally together with SRB partners (Knittel and Boetius, 2009; Yang et al., 2021) (Figure 2G).
The sedimentary biofilm microbiota is affected by physical and chemical parameters in its surrounding environments, which in turn exerts influences on the intricate nature of sediments. Therefore, a thorough understanding of both deep-sea microbial life and marine sediment dynamics relies heavily on comprehending the critical process of microbial biofilm formation on particle surfaces. Several studies conducted on natural sediments have demonstrated that sand and silt particles are preferred habitats, while clay particles are rarely colonized due to their small size (Weise and Rheinheimer, 1978; Deflaun and Mayer, 1983). Additionally, the degree of roundness of particles plays a significant role in microbial colonization: as particle roundness (and age) increases, microbial colonization decreases. Moreover, it is observed that microbial biofilms tend to form within low-relief areas on particles, such as crevices, depressions, and surface fissures where cells are better protected from external mechanical damage (Meadows and Anderson, 1966; Weise and Rheinheimer, 1978). Furthermore, subsurface sediment layers can be stabilized by biofilm-associated activity during fluid eruption events. In pelagic environments, these pervasive biofilms can alter the characteristics of eroded sediment flocs including size, shape, density, and settling velocity. Consequently, sediment transport and deposition processes are influenced (Droppo, 2001, 2004).
Various marine surfaces harbor diverse, distinct microbial communities that remain poorly explored. In this review, the recent advancements in understanding marine surface colonization and biofilm formation on multiple abiotic surfaces are synthesized and discussed (Figure 4). However, there are still significant knowledge gaps regarding community diversity, metabolites, ecological functions, and their response and influence on the changing marine environment. Testing and applying new bioinformatics pipelines with reduced cost and errors, techniques with high spatial and temporal resolution targeting cell phenotype, metabolism, and response to environmental fluctuations, and sensitive in situ measurements are essential for investigating natural marine biofilms. Integrating ecological findings combined with metabolic network reconstructions and biogeochemical modeling would provide valuable insights into higher-level properties of the biofilm-associated populations such as cooperative and other socio-microbial functions and biogeochemical roles.
SY: Conceptualization, Resources, Visualization, Writing – original draft, Writing – review & editing, Investigation, Methodology, Software. XL: Writing – review & editing. HL: Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by China Geological Survey (grant number DD20221703 and DD20230063) and Guangdong Major project of Basic and Applied Basic Research Program (grant number 2020B0301030003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abu Bakar A., Ali M. K. F. M., Noor N. M., Yahaya N., Ismail M., Abdullah A. (2017). Bio-corrosion of carbon steel by sulfate reducing bacteria consortium in oil and gas pipelines. J. Mechaic. Eng. Sci. 11, 2592–2600. doi: 10.15282/jmes.11.2.2017.3.0237
Agostini V. O., Muxagata E., Pinho G. L. L., Pessi I. S., Macedo A. J. (2021). Bacteria-invertebrate interactions as an asset in developing new antifouling coatings for man-made aquatic surfaces. Environ. pollut. 271, 116284. doi: 10.1016/j.envpol.2020.116284
Alain F. (2021). Research-Professor Alain Filloux. Available online at: https://www.imperial.ac.uk/people/a.filloux/research.html (Accessed May 11, 2022).
Ali A., Culliton D., Fahad S., Ali Z., Kang E.-T., Xu L. (2024). Nature-inspired anti-fouling strategies for combating marine biofouling. Prog. Organ. Coat. 189, 108349. doi: 10.1016/j.porgcoat.2024.108349
Allen M. A., Goh F., Burns B. P., Neilan B. A. (2009). Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology 7, 82–96. doi: 10.1111/j.1472-4669.2008.00187.x
Amaral-Zettler L. A., Zettler E. R., Mincer T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Amaral-Zettler L. A., Zettler E. R., Slikas B., Boyd G. D., Melvin D. W., Morrall C. E., et al. (2015). The biogeography of the Plastisphere: implications for policy. Front. Ecol. Environ. 13, 541–546. doi: 10.1890/150017
Antunes J., Leão P., Vasconcelos V. (2019). Marine biofilms: diversity of communities and of chemical cues. Environ. Microbiol. Rep. 11, 287–305. doi: 10.1111/1758-2229.12694
Arias-Andres M., Kluemper U., Rojas-Jimenez K., Grossart H.-P. (2018). Microplastic pollution increases gene exchange in aquatic ecosystems. Environ. Pollut. 237, 253–261. doi: 10.1016/j.envpol.2018.02.058
Armbruster C. R., Parsek M. R. (2018). New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. 115, 4317–4319. doi: 10.1073/pnas.1804084115
Artham T., Sudhakar M., Venkatesan R., Madhavan Nair C., Murty K. V. G. K., Doble M. (2009). Biofouling and stability of synthetic polymers in sea water. Int. Biodeterior. Biodegr. 63, 884–890. doi: 10.1016/j.ibiod.2009.03.003
Azam F., Fenchel T., Field J. G., Gray J. S., Meyerreil L. A., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Bar-On Y. M., Phillips R., Milo R. (2018). The biomass distribution on Earth. Proc. Natl. Acad. Sci. United States America 115, 6506–6511. doi: 10.1073/pnas.1711842115
Barros J., Seena S. (2021). Plastisphere in freshwaters: An emerging concern. Environ. pollut. 290, 118123. doi: 10.1016/j.envpol.2021.118123
Baskar S., Baskar R., Lee N., Kaushik A., Theophilus P. K. (2008). Precipitation of iron in microbial mats of the spring waters of Borra Caves, Vishakapatnam, India: some geomicrobiological aspects. Environ. Geol. 56, 237–243. doi: 10.1007/s00254-007-1159-y
Battin T. J., Besemer K., Bengtsson M. M., Romani A. M., Packmann A. I. (2016). The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 14, 251–263. doi: 10.1038/nrmicro.2016.15
Belkaid S., Ladjouzi M. A., Hamdani S. (2011). Effect of biofilm on naval steel corrosion in natural seawater. J. Solid State Electrochem. 15, 525–537. doi: 10.1007/s10008-010-1118-5
Blackwood D. J., Lim C. S., Teo S. L. M., Hu X., Pang J. (2017). Macrofouling induced localized corrosion of stainless steel in Singapore seawater. Corrosion Sci. 129, 152–160. doi: 10.1016/j.corsci.2017.10.008
Boetius A., Ravenschlag K., Schubert C. J., Rickert D., Widdel F., Gieseke A., et al. (2000). A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407, 623–626. doi: 10.1038/35036572
Bolhuis H., Cretoiu M. S., Stal L. J. (2014). Molecular ecology of microbial mats. FEMS Microbiol. Ecol. 90, 335–350. doi: 10.1111/1574-6941.12408
Bolhuis H., Stal L. J. (2011). Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing. Isme J. 5, 1701–1712. doi: 10.1038/Ismej.2011.52
Bond M. C., Vidakovic L., Singh P. K., Drescher K., Nadell C. D. (2021). Matrix-trapped viruses can prevent invasion of bacterial biofilms by colonizing cells. Elife 10. doi: 10.7554/eLife.65355
Brint J. M., Ohman D. E. (1995). Synthesis of multiple exoproducts in pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177, 7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995
Byers J. E., Blaze J. A., Dodd A. C., Hall H. L., Gribben P. E. (2023). Exotic asphyxiation: interactions between invasive species and hypoxia. Biol. Rev. 98, 150–167. doi: 10.1111/brv.12900
Camilli A., Bassler B. L. (2006). Bacterial small-molecule signaling pathways. Science 311, 1113–1116. doi: 10.1126/science.1121357
Carson H. S., Nerheim M. S., Carroll K. A., Eriksen M. (2013). The plastic-associated microorganisms of the North Pacific Gyre. Mar. pollut. Bull. 75, 126–132. doi: 10.1016/j.marpolbul.2013.07.054
Catão Cp E., Pollet T., Garnier C., Barry-Martinet R., Rehel K., Linossier I., et al. (2021). Temperate and tropical coastal waters share relatively similar microbial biofilm communities while free-living or particle-attached communities are distinct. Mol. Ecol. 30, 2891–2904. doi: 10.1111/mec.15929
Chandki R., Banthia P., Banthia R. (2011). Biofilms: A microbial home. J. Indian Soc. Periodontol. 15, 111–114. doi: 10.4103/0972-124x.84377
Charlesworth J. C., Watters C., Wong H. L., Visscher P. T., Burns B. P. (2019). Isolation of novel quorum-sensing active bacteria from microbial mats in Shark Bay Australia. FEMS Microbiol. Ecol. 95. doi: 10.1093/femsec/fiz035
Chen C.-L., Maki J. S., Rittschof D., Teo S. L.-M. (2013). Early marine bacterial biofilm on a copper-based antifouling paint. Int. Biodeterior. Biodegr. 83, 71–76. doi: 10.1016/j.ibiod.2013.04.012
Chung H. C., Lee O. O., Huang Y.-L., Mok S. Y., Kolter R., Qian P.-Y. (2010). Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 4, 817–828. doi: 10.1038/ismej.2009.157
Colwell F. S., D’Hondt S. (2013). Nature and extent of the deep biosphere. Rev. Mineral. Geochem. 75, 547–574. doi: 10.2138/rmg.2013.75.17
Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappinscott H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Cottingham K. L., Chiavelli D. A., Taylor R. K. (2003). Environmental microbe and human pathogen: the ecology and microbiology of Vibrio cholerae. Front. Ecol. Environ. 1, 80–86. doi: 10.2307/3868034
Crepeau V., Bonavita M.-A. C., Lesongeur F., Randrianalivelo H., Sarradin P.-M., Sarrazin J., et al. (2011). Diversity and function in microbial mats from the Lucky Strike hydrothermal vent field. FEMS Microbiol. Ecol. 76, 524–540. doi: 10.1111/j.1574-6941.2011.01070.x
Curren E., Leong S. C. Y. (2019). Profiles of bacterial assemblages from microplastics of tropical coastal environments. Sci. Total Environ. 655, 313–320. doi: 10.1016/j.scitotenv.2018.11.250
Dang H., Chen R., Wang L., Shao S., Dai L., Ye Y., et al. (2011). Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ. Microbiol. 13, 3059–3074. doi: 10.1111/j.1462-2920.2011.02583.x
Dang H., Li T., Chen M., Huang G. (2008). Cross-Ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74, 52–60. doi: 10.1128/aem.01400-07
Dang H., Lovell C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80, 91–138. doi: 10.1128/mmbr.00037-15
Danovaro R., Corinaldesi C., Rastelli E., Dell’Anno A. (2015). Towards a better quantitative assessment of the relevance of deep-sea viruses, Bacteria and Archaea in the functioning of the ocean seafloor. Aquat. Microbial. Ecol. 75, 81–90. doi: 10.3354/ame01747
Das T., Manefield M. (2013). Phenazine production enhances extracellular DNA release via hydrogen peroxide generation in Pseudomonas aeruginosa. Commun. Integr. Biol. 6, e23570. doi: 10.4161/cib.23570
Debroas D., Mone A., Ter Halle A. (2017). Plastics in the North Atlantic garbage patch: A boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 599-600, 1222–1232. doi: 10.1016/j.scitotenv.2017.05.059
de Campos B. G., Figueiredo J., Perina F., Abessa D., Loureiro S., Martins R. (2022). Occurrence, effects and environmental risk of antifouling biocides (EU PT21): Are marine ecosystems threatened? Crit. Rev. Environ. Sci. Technol. 52, 3179–3210. doi: 10.1080/10643389.2021.1910003
de Carvalho C. C. C. R. (2012). Biofilms: new ideas for an old problem. Recent patents Biotechnol. 6, 13–22. doi: 10.2174/187220812799789163
De Carvalho C. C. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00126
Decho A. W. (1990). Microbial exopolymer secretions in ocean environments - their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. 28, 73–153.
Decho A. W., Visscher P. T., Ferry J., Kawaguchi T., He L., Przekop K. M., et al. (2009). Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (AHLs) and abundance changes that may relate to diel pH. Environ. Microbiol. 11, 409–420. doi: 10.1111/j.1462-2920.2008.01780.x
Deflaun M. F., Mayer L. M. (1983). Relationships between bacteria and grain surfaces in intertidal sediments. Limnol. Oceanogr. 28, 873–881. doi: 10.4319/lo.1983.28.5.0873
Delacuvellerie A., Cyriaque V., Gobert S., Benali S., Wattiez R. (2019). The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 380. doi: 10.1016/j.jhazmat.2019.120899
Delacuvellerie A., Géron A., Gobert S., Wattiez R. (2022). New insights into the functioning and structure of the PE and PP plastispheres from the Mediterranean Sea. Environ. pollut. 295, 118678. doi: 10.1016/j.envpol.2021.118678
Deng X., Dohmae N., Nealson K. H., Hashimoto K., Okamoto A. (2018). Multi-heme cytochromes provide a pathway for survival in energy-limited environments. Sci. Adv. 4, eaao5682. doi: 10.1126/sciadv.aao5682
De Tender C., Devriese L. I., Haegeman A., Maes S., Vangeyte J., Cattrijsse A., et al. (2017). Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ. Sci. Technol. 51, 7350–7360. doi: 10.1021/acs.est.7b00697
Dinh H. T., Kuever J., Mußmann M., Hassel A. W., Stratmann M., Widdel F. (2004). Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832. doi: 10.1038/nature02321
Dobretsov S., Rittschof D. (2020). Love at first taste: induction of larval settlement by marine microbes. Int. J. Mol. Sci. 21, 731. doi: 10.3390/ijms21030731
Dong Z. H., Liu T., Liu H. F. (2011). Influence of EPS isolated from thermophilic sulphate-reducing bacteria on carbon steel corrosion. Biofouling 27, 487–495. doi: 10.1080/08927014.2011.584369
Donlan R. M. (2000). Role of biofilms in antimicrobial resistance. ASAIO J. 46, S47–S52. doi: 10.1097/00002480-200011000-00037
Droppo I. G. (2001). Rethinking what constitutes suspended sediment. Hydrol. Process. 15, 1551–1564. doi: 10.1002/hyp.228
Droppo I. G. (2004). Structural controls on floc strength and transport. Can. J. Civil Eng. 31, 569–578. doi: 10.1139/l04-015
Duan J., Wu S., Zhang X., Huang G., Du M., Hou B. (2008). Corrosion of carbon steel influenced by anaerobic biofilm in natural seawater. Electro. Acta 54, 22–28. doi: 10.1016/j.electacta.2008.04.085
Dubilier N., Bergin C., Lott C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6, 725–740. doi: 10.1038/nrmicro1992
Dudek K. L., Cruz B. N., Polidoro B., Neuer S. (2020). Microbial colonization of microplastics in the Caribbean Sea. Limnol. Oceanogr. Lett. 5, 5–17. doi: 10.1002/lol2.10141
Dupraz C., Reid R. P., Braissant O., Decho A. W., Norman R. S., Visscher P. T. (2009). Processes of carbonate precipitation in modern microbial mats. Earth-Sci. Rev. 96, 141–162. doi: 10.1016/j.earscirev.2008.10.005
Dussud C., Meistertzheim A. L., Conan P., Pujo-Pay M., George M., Fabre P., et al. (2018). Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. pollut. 236, 807–816. doi: 10.1016/j.envpol.2017.12.027
Enning D., Garrelfs J. (2014). Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl. Environ. Microbiol. 80, 1226–1236. doi: 10.1128/aem.02848-13
Enning D., Venzlaff H., Garrelfs J., Dinh H. T., Meyer V., Mayrhofer K., et al. (2012). Marine sulfate-reducing bacteria cause serious corrosion of iron under electroconductive biogenic mineral crust. Environ. Microbiol. 14, 1772–1787. doi: 10.1111/j.1462-2920.2012.02778.x
Fahy E., Runyan J., Arriola G. A. (2017). "Rockin" Welcome Mats: Ciliate and Bacteria Mats of Hydrothermal Vents. New London: NAUTILUS LIVE: Ocean exploration trust. Available online at: https://nautiluslive.org/album/2017/10/29/rockin-welcome-mats-ciliate-and-bacteria-mats-hydrothermal-vents (Accessed April 30, 2024).
Feng L., He L., Jiang S., Chen J., Zhou C., Qian Z.-J., et al. (2020). Investigating the composition and distribution of microplastics surface biofilms in coral areas. Chemosphere 252, 126565. doi: 10.1016/j.chemosphere.2020.126565
Flach C.-F., Pal C., Svensson C. J., Kristiansson E., Östman M., Bengtsson-Palme J., et al. (2017). Does antifouling paint select for antibiotic resistance? Sci. Total Environ. 590-591, 461–468. doi: 10.1016/j.scitotenv.2017.01.213
Flemming H.-C. (2011). The perfect slime. Colloids Surf. B-Biointerf. 86, 251–259. doi: 10.1016/j.colsurfb.2011.04.025
Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Flemming H.-C., Wuertz S. (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260. doi: 10.1038/s41579-019-0158-9
Flood B. E., Louw D. C., van der Plas A. K., Bailey J. V. (2021). Giant sulfur bacteria (Beggiatoaceae) from sediments underlying the Benguela upwelling system host diverse microbiomes. PloS One 16, e0258124. doi: 10.1371/journal.pone.0258124
Flores G. E., Campbell J. H., Kirshtein J. D., Meneghin J., Podar M., Steinberg J. I., et al. (2011). Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 13, 2158–2171. doi: 10.1111/j.1462-2920.2011.02463.x
Frank K. L., Rogers D. R., Olins H. C., Vidoudez C., Girguis P. R. (2013). Characterizing the distribution and rates of microbial sulfate reduction at Middle Valley hydrothermal vents. Isme J. 7, 1391–1401. doi: 10.1038/ismej.2013.17
Frere L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. (2018). Microplastic bacterial communities in the Bay of Brest: Influence of polymer type and size. Environ. pollut. 242, 614–625. doi: 10.1016/j.envpol.2018.07.023
Fuqua C., Winans S. C., Greenberg E. P. (1996). ). Census and consensus in bacterial ecosystems: The LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50, 727–751. doi: 10.1146/annurev.micro.50.1.727
Gagné F. (2017). Toxicity and disruption of quorum sensing in Aliivibrio fisheri by environmental chemicals: Impacts of selected contaminants and microplastics. J. Xenobiot 7, 7101. doi: 10.4081/xeno.2017.7101
Gerbersdorf S. U., Manz W., Paterson D. M. (2008). The engineering potential of natural benthic bacterial assemblages in terms of the erosion resistance of sediments. FEMS Microbiol. Ecol. 66, 282–294. doi: 10.1111/j.1574-6941.2008.00586.x
Gilbert C., Ellis T. (2019). Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 8, 1–15. doi: 10.1021/acssynbio.8b00423
Goldstein M. C., Carson H. S., Eriksen M. (2014). Relationship of diversity and habitat area in North Pacific plastic-associated rafting communities. Mar. Biol. 161, 1441–1453. doi: 10.1007/s00227-014-2432-8
Gong X., Ge Z., Ma Z., Li Y., Huang D., Zhang J. (2023). Effect of different size microplastic particles on the construction of algal-bacterial biofilms and microbial communities. J. Environ. Manage. 343, 118246. doi: 10.1016/j.jenvman.2023.118246
Gong M., Yang G., Zhuang L., Zeng E. Y. (2019). Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ. pollut. 252, 94–102. doi: 10.1016/j.envpol.2019.05.090
Grotzinger J. P., Knoll A. H. (1999). Stromatolites in Precambrian carbonates: Evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Planet. Sci. 27, 313–358. doi: 10.1146/annurev.earth.27.1.313
Grünke S., Felden J., Lichtschlag A., Girnth A. C., de Beer D., Wenzhöfer F., et al. (2011). Niche differentiation among mat-forming, sulfide-oxidizing bacteria at cold seeps of the Nile Deep Sea Fan (Eastern Mediterranean Sea). Geobiology 9, 330–348. doi: 10.1111/j.1472-4669.2011.00281.x
Guo S., Kwek M. Y., Toh Z. Q., Pranantyo D., Kang E. T., Loh X. J., et al. (2018). Tailoring polyelectrolyte architecture to promote cell growth and inhibit bacterial adhesion. ACS Appl. Mater Interf. 10, 7882–7891. doi: 10.1021/acsami.8b00666
Haagensen J. A., Verotta D., Huang L., Spormann A., Yang K. (2015). New in vitro model to study the effect of human simulated antibiotic concentrations on bacterial biofilms. Antimicrob. Agents Chemother. 59, 4074–4081. doi: 10.1128/aac.05037-14
Haque S., Yadav D. K., Bisht S. C., Yadav N., Singh V., Dubey K. K., et al. (2019). Quorum sensing pathways in Gram-positive and -negative bacteria: potential of their interruption in abating drug resistance. J. Chemother. 31, 161–187. doi: 10.1080/1120009X.2019.1599175
Harrison J. P., Hoellein T. J., Sapp M., Tagg A. S., Ju-Nam Y., Ojeda J. J. (2018). “Microplastic-associated biofilms: a comparison of freshwater and marine environments," in The Handbook of Environmental Chemistry, ed. Wagner M., Lambert S. (Cham: Springer), 58, 181–201. doi: 10.1007/978-3-319-61615-5_9
Harrison J. P., Schratzberger M., Sapp M., Osborn A. M. (2014). Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 14, 232. doi: 10.1186/s12866-014-0232-4
Hawley A. K., Nobu M. K., Wright J. J., Durno W. E., Morgan-Lang C., Sage B., et al. (2017). Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 8, 1507. doi: 10.1038/s41467-017-01376-9
Hayek M., Salgues M., Souche J.-C., Cunge E., Giraudel C., Paireau O. (2021). Influence of the intrinsic characteristics of cementitious materials on biofouling in the marine environment. Sustainability 13, 2625. doi: 10.3390/su13052625
He S., Jia M., Xiang Y., Song B., Xiong W., Cao J., et al. (2022). Biofilm on microplastics in aqueous environment: Physicochemical properties and environmental implications. J. Hazard. Mater. 424, 127286. doi: 10.1016/j.jhazmat.2021.127286
Heberling C., Lowell R. P., Liu L., Fisk M. R. (2010). Extent of the microbial biosphere in the oceanic crust. Geochem. Geophys. Geosys. 11, Q08803. doi: 10.1029/2009gc002968
Horton A. A., Walton A., Spurgeon D. J., Lahive E., Svendsen C. (2017). Microplastics in freshwater and terrestrial environments: Evaluating the current understanding to identify the knowledge gaps and future research priorities. Sci. Total Environ. 586, 127–141. doi: 10.1016/j.scitotenv.2017.01.190
Hossain M. R., Jiang M., Wei Q., Leff L. G. (2019). Microplastic surface properties affect bacterial colonization in freshwater. J. Basic Microbiol. 59, 54–61. doi: 10.1002/jobm.201800174
Hou J., Veeregowda D. H., Belt-Gritter B., Busscher H. J., Mei H. (2018). Extracellular Polymeric Matrix Production and Relaxation under Fluid Shear and Mechanical Pressure in Staphylococcus aureus Biofilms. Appl. Environ. Microbiol. 84, e01516–e01517. doi: 10.1128/AEM.01516-17
Houghton J. L., Seyfried W. E. Jr., Banta A. B., Reysenbach A. L. (2007). Continuous enrichment culturing of thermophiles under sulfate and nitrate-reducing conditions and at deep-sea hydrostatic pressures. Extremophiles 11, 371–382. doi: 10.1007/s00792-006-0049-7
Hu H., Jin D., Yang Y., Zhang J., Ma C., Qiu Z. (2021). Distinct profile of bacterial community and antibiotic resistance genes on microplastics in Ganjiang River at the watershed level. Environ. Res. 200, 111363. doi: 10.1016/j.envres.2021.111363
Huegler M., Gaertner A., Imhoff J. F. (2010). Functional genes as markers for sulfur cycling and CO2 fixation in microbial communities of hydrothermal vents of the Logatchev field. FEMS Microbiol. Ecol. 73, 526–537. doi: 10.1111/j.1574-6941.2010.00919.x
Jacquin J., Cheng J., Odobel C., Pandin C., Conan P., Pujo-Pay M., et al. (2019). Microbial ecotoxicology of marine plastic debris: A review on colonization and biodegradation by the “Plastisphere. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00865
Jaeschke A., Jorgensen S. L., Bernasconi S. M., Pedersen R. B., Thorseth I. H., Frueh-Green G. L. (2012). Microbial diversity of Loki’s Castle black smokers at the Arctic Mid-Ocean Ridge. Geobiology 10, 548–561. doi: 10.1111/gbi.12009
Jatt A. N., Tang K., Liu J., Zhang Z., Zhang X. H. (2015). Quorum sensing in marine snow and its possible influence on production of extracellular hydrolytic enzymes in marine snow bacterium Pantoea ananatis B9. FEMS Microbiol. Ecol. 91, 1–13. doi: 10.1093/femsec/fiu030
Jia R., Unsal T., Xu D., Lekbach Y., Gu T. (2019). Microbiologically influenced corrosion and current mitigation strategies: A state of the art review. Int. Biodeterior. Biodegr. 137, 42–58. doi: 10.1016/j.ibiod.2018.11.007
Jiang P., Zhao S., Zhu L., Li D. (2018). Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Sci. Total Environ. 624, 48–54. doi: 10.1016/j.scitotenv.2017.12.105
Jimenez P. N., Koch G., Thompson J. A., Xavier K. B., Cool R. H., Quax W. J. (2012). The multiple signaling systems regulating virulence in pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/mmbr.05007-11
Jones D. A., Amy P. S. (2002). A thermodynamic interpretation of microbiologically influenced corrosion. Corrosion 58, 638–645. doi: 10.5006/1.3287692
Kai D., Loh X. J. (2014). Polyhydroxyalkanoates: chemical modifications toward biomedical applications. ACS Sustain. Chem. Eng. 2, 106–119. doi: 10.1021/sc400340p
Kato S., Nakamura K., Toki T., Ishibashi J.-i., Tsunogai U., Hirota A., et al. (2012). Iron-based microbial ecosystem on and below the seafloor: a case study of hydrothermal fields of the Southern Mariana Trough. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00089
Kato S., Shibuya T., Takaki Y., Hirai M., Nunoura T., Suzuki K. (2018). Genome-enabled metabolic reconstruction of dominant chemosynthetic colonizers in deep-sea massive sulfide deposits. Environ. Microbiol. 20, 862–877. doi: 10.1111/1462-2920.14032
Kesy K., Labrenz M., Scales B. S., Kreikemeyer B., Oberbeckmann (2021). S. Vibrio colonization is highly dynamic in early microplastic-associated biofilms as well as on field-collected microplastics. Microorganisms 9, 76. Available online at: https://www.mdpi.com/2076-2607/9/1/76 (Accessed December 26, 2023).
Kip N., Van Veen J. A. (2015). The dual role of microbes in corrosion. ISME J. 9, 542–551. doi: 10.1038/ismej.2014.169
Kirstein I. V., Wichels A., Gullans E., Krohne G., Gerdts G. (2019). The Plastisphere-Uncovering tightly attached plastic “specific” microorganisms. PloS One 14, e0215859. doi: 10.1371/journal.pone.0215859
Knittel K., Boetius A. (2009). Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63, 311–334. doi: 10.1146/annurev.micro.61.080706.093130
KoChina T. A., Kondratenko Y. A., Shilova O. A., Vlasov D. Y. (2022). Biocorrosion, biofouling, and advanced methods of controlling them. Prot. Metals Phys. Chem. Surf. 58, 129–150. doi: 10.1134/S2070205122010129
Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kor K., Mehdinia A. (2020). Neustonic microplastic pollution in the Persian Gulf. Mar. pollut. Bull. 150, 110665. doi: 10.1016/j.marpolbul.2019.110665
Kovačević D., Pratnekar R., Godič Torkar K., Salopek J., Dražić G., Abram A., et al. (2016). Influence of polyelectrolyte multilayer properties on bacterial adhesion capacity. Polymers 8, 345. doi: 10.3390/polym8100345
Kumar M., Kumar M., Pandey A., Thakur I. S. (2019). Genomic analysis of carbon dioxide sequestering bacterium for exopolysaccharides production. Sci. Rep. 9, 4270. doi: 10.1038/s41598-019-41052-0
Lawal S. L., Afolalu S. A., Jen T. C., Akinlabi E. T. (2024). Corrosion control and its application in marine environment - A review. Solid State Phenom. 355, 61–73. doi: 10.4028/p-634sdI
Lee S. Y., Kim H. U., Chae T. U., Cho J. S., Kim J. W., Shin J. H., et al. (2019). A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33. doi: 10.1038/s41929-018-0212-4
Lenhart T. R., Duncan K. E., Beech I. B., Sunner J. A., Smith W., Bonifay V., et al. (2014). Identification and characterization of microbial biofilm communities associated with corroded oil pipeline surfaces. Biofouling 30, 823–835. doi: 10.1080/08927014.2014.931379
Levallois A., Vivier B., Caplat C., Goux D., Orvain F., Lebel J.-M., et al. (2023). Aluminium-based galvanic anode impacts the photosynthesis of microphytobenthos and supports the bioaccumulation of metals released. Aquat. Toxicol. 258, 106501. doi: 10.1016/j.aquatox.2023.106501
Lever M. A., Rogers K. L., Lloyd K. G., Overmann J., Schink B., Thauer R. K., et al. (2015). Life under extreme energy limitation: a synthesis of laboratory-and field-based investigations. FEMS Microbiol. Rev. 39, 688–728. doi: 10.1093/femsre/fuv020
Lewis A. J. (2018). Marine bio-fouling and invasive species with John Lewis. Available online at: https://www.3cr.org.au/radioblue/episode-201808261130/marine-bio-fouling-and-invasive-species-john-lewis (Accessed October 30, 2021).
Li J. J., Huang W., Jiang R. J., Han X. B., Zhang D. D., Zhang C. F. (2020). Are bacterial communities associated with microplastics influenced by marine habitats? Sci. Total Environ. 733, 139400. doi: 10.1016/j.scitotenv.2020.139400
Li J., Mooney D. J. (2016). Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 1, 16071. doi: 10.1038/natrevmats.2016.71
Li Q., Xu X., He C., Zheng L., Gao W., Sun C., et al. (2019). Complete genome sequence of a quorum-sensing bacterium, oceanicola sp. Strain D3, isolated from a microplastic surface in coastal water of Qingdao, China. Microbiol. Resour. Announc. 8. doi: 10.1128/mra.01022-19
Li X.-X., Yang T., Mbadinga S. M., Liu J.-F., Yang S.-Z., Gu J.-D., et al. (2017). Responses of microbial community composition to temperature gradient and carbon steel corrosion in production water of petroleum reservoir. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02379
Lindholdt A., Dam-Johansen K., Olsen S. M., Yebra D. M., Kiil S. (2015). Effects of biofouling development on drag forces of hull coatings for ocean-going ships: a review. J. Coat. Technol. Res. 12, 415–444. doi: 10.1007/s11998-014-9651-2
Liu H., Gu T., Asif M., Zhang G., Liu H. (2017). The corrosion behavior and mechanism of carbon steel induced by extracellular polymeric substances of iron-oxidizing bacteria. Corrosion Sci. 114, 102–111. doi: 10.1016/j.corsci.2016.10.025
Liu Y., Liu W., Yang X., Wang J., Lin H., Yang Y. (2021). Microplastics are a hotspot for antibiotic resistance genes: Progress and perspective. Sci. Total Environ. 773, 145643. doi: 10.1016/j.scitotenv.2021.145643
Liu D., Shu H., Zhou J., Bai X., Cao P. (2023). Research progress on new environmentally friendly antifouling coatings in marine settings: A review. Biomimetics 8, 200. doi: 10.3390/biomimetics8020200
Liu S., Yu S., Lu X., Yang H., Li Y., Xu X., et al. (2022). Microbial communities associated with thermogenic gas hydrate-bearing marine sediments in Qiongdongnan Basin, South China Sea. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1032851
Lozano Y. M., Lehnert T., Linck L. T., Lehmann A., Rillig M. C. (2021). Microplastic shape, polymer type, and concentration affect soil properties and plant biomass. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.616645
Ma L., Conover M., Lu H., Parsek M. R., Bayles K., Wozniak D. J. (2009). Assembly and development of the pseudomonas aeruginosa biofilm matrix. PloS Pathog. 5, e1000354. doi: 10.1371/journal.ppat.1000354
Ma Y., Zhang Y., Zhang R., Guan F., Hou B., Duan J. (2020). Microbiologically influenced corrosion of marine steels within the interaction between steel and biofilms: a brief view. Appl. Microbiol. Biotechnol. 104, 515–525. doi: 10.1007/s00253-019-10184-8
Mahto K. U., Vandana, Priyadarshanee M., Samantaray D. P., Das S. (2022). Bacterial biofilm and extracellular polymeric substances in the treatment of environmental pollutants: Beyond the protective role in survivability. J. Clean. Product. 379, 134759. doi: 10.1016/j.jclepro.2022.134759
Margulis L., Barghoorn E. S., Ashendorf D., Banerjee S., Chase D., Francis S., et al. (1980). The microbial community in the layered sediments at Laguna Figueroa, Baja California, Mexico: Does it have Precambrian analogs? Precambrian Res. 11, 93–123. doi: 10.1016/0301-9268(80)90042-x
Marshall K. C. (1985). Bacterial adhesion in oligotrophic habitats. Microbiol. Sci. 2, 321–2, 325-6.
McCully A. L., Spormann A. M. (2020). Direct cathodic electron uptake coupled to sulfate reduction by Desulfovibrio ferrophilus IS5 biofilms. Environ. Microbiol. 22, 4794–4807. doi: 10.1111/1462-2920.15235
McGivney E., Cederholm L., Barth A., Hakkarainen M., Hamacher-Barth E., Ogonowski M., et al. (2020). Rapid physicochemical changes in microplastic induced by biofilm formation. Front. Bioeng. Biotechnol. 8. doi: 10.3389/fbioe.2020.00205
Meadows P. S., Anderson J. G. (1966). Micro-organisms attached to marine and freshwater sand grains. Nature 212, 1059–1060. doi: 10.1038/2121059a0
Menez B., Pasini V., Brunelli D. (2012). Life in the hydrated suboceanic mantle. Nat. Geosci. 5, 133–137. doi: 10.1038/ngeo1359
Meng F., Yang X., Riksen M., Geissen V. (2022). Effect of different polymers of microplastics on soil organic carbon and nitrogen – A mesocosm experiment. Environ. Res. 204, 111938. doi: 10.1016/j.envres.2021.111938
Miao L., Hou J., You G., Liu Z., Liu S., Li T., et al. (2019). Acute effects of nanoplastics and microplastics on periphytic biofilms depending on particle size, concentration and surface modification. Environ. pollut. 255, 113300. doi: 10.1016/j.envpol.2019.113300
Miao T., Wang J., Zeng Y., Liu G., Chen X. (2018). Polysaccharide-based controlled release systems for therapeutics delivery and tissue engineering: from bench to bedside. Adv. Sci. 5, 1700513. doi: 10.1002/advs.201700513
Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Mishra S., Huang Y., Li J., Wu X., Zhou Z., Lei Q., et al. (2022). Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere 294, 133609. doi: 10.1016/j.chemosphere.2022.133609
Mitrousis N., Fokina A., Shoichet M. S. (2018). Biomaterials for cell transplantation. Nat. Rev. Mater. 3, 441–456. doi: 10.1038/s41578-018-0057-0
Mohan T., Rathner R., Reishofer D., Koller M., Elschner T., Spirk S., et al. (2015). Designing hydrophobically modified polysaccharide derivatives for highly efficient enzyme immobilization. Biomacromolecules 16, 2403–2411. doi: 10.1021/acs.biomac.5b00638
Morohoshi T., Ogata K., Okura T., Sato S. (2018). Molecular characterization of the bacterial community in biofilms for degradation of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) films in seawater. Microbes Environ. 33, 19–25. doi: 10.1264/jsme2.ME17052
Morono Y., Inagaki F. (2016). Chapter Three-Analysis of Low-Biomass Microbial Communities in the Deep Biosphere. Adv. Appl. Microbial. 95, 149–178. doi: 10.1016/bs.aambs.2016.04.001
Müller W. E. G., Tolba E., Ackermann M., Neufurth M., Wang S., Feng Q., et al. (2017). Fabrication of amorphous strontium polyphosphate microparticles that induce mineralization of bone cells in vitro and in vivo. Acta Biomater. 50, 89–101. doi: 10.1016/j.actbio.2016.12.045
Nadell C. D., Drescher K., Foster K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Nakagawa S., Takai K. (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol. Ecol. 65, 1–14. doi: 10.1111/j.1574-6941.2008.00502.x
Ng W.-L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Nguyen P. Q., Courchesne N. D., Duraj-Thatte A., Praveschotinunt P., Joshi N. S. (2018). Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 30, e1704847. doi: 10.1002/adma.201704847
Noffke N., Paterson D. M. (2008). Microbial interactions with physical sediment dynamics, and their significance for the interpretation of Earth’s biological history. Geobiology 6, 1–4. doi: 10.5555/20083033746
Novick R. P., Geisinger E. (2008). Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564. doi: 10.1146/annurev.genet.42.110807.091640
Oberbeckmann S., Kreikemeyer B., Labrenz M. (2018). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02709
Oberbeckmann S., Loeder M. G. J., Gerdts G., Osborn A. M. (2014). Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 90, 478–492. doi: 10.1111/1574-6941.12409
Ogonowski M., Motiei A., Ininbergs K., Hell E., Gerdes Z., Udekwu K. I., et al. (2018). Evidence for selective bacterial community structuring on microplastics. Environ. Microbiol. 20, 2796–2808. doi: 10.1111/1462-2920.14120
Paerl H. W., Pinckney J. L. (1996). A mini-review of microbial consortia: Their roles in aquatic production and biogeochemical cycling. Microbial. Ecol. 31, 225–247. doi: 10.1007/BF00171569
Parker L. (2015). Ocean Trash: 5.25 Trillion Pieces and Counting, but Big Questions Remain. Washington: National Geographic. Available online at: https://education.nationalgeographic.org/resource/ocean-trash-525-trillion-pieces-and-counting-big-questions-remain/ (Accessed March 22, 2024).
Parkes R. J., Cragg B., Roussel E., Webster G., Weightman A., Sass H. (2014). A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: geosphere interactions. Mar. Geol. 352, 409–425. doi: 10.1016/j.margeo.2014.02.009
Pasulka A., Hu S. K., Countway P. D., Coyne K. J., Cary S. C., Heidelberg K. B., et al. (2019). SSU-rRNA gene sequencing survey of benthic microbial eukaryotes from guaymas basin hydrothermal vent. J. Eukar. Microbiol. 66, 637–653. doi: 10.1111/jeu.12711
Paterson D. M. (1989). Short-term changes in the erodibility of intertidal cohesive sediments related to the migratory behavior of epipelic diatoms. Limnol. Oceanogr. 34, 223–234. doi: 10.4319/lo.1989.34.1.0223
Paterson D. M., Tolhurst T. J., Kelly J. A., Honeywill C., de Deckere E., Huet V., et al. (2000). Variations in sediment properties, Skeffling mudflat, Humber Estuary, UK. Continent. Shelf Res. 20, 1373–1396. doi: 10.1016/s0278-4343(00)00028-5
Pessi G., Haas D. (2000). Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in pseudomonas aeruginosa. J. Bacteriol 182, 6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000
Phoenix V. R., Konhauser K. O. (2008). Benefits of bacterial biomineralization. Geobiology 6, 303–308. doi: 10.1111/j.1472-4669.2008.00147.x
Pompilio A., Piccolomini R., Picciani C., D’Antonio D., Savini V., Di Bonaventura G. (2008). Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 287, 41–47. doi: 10.1111/j.1574-6968.2008.01292.x
Prescott R. D., Decho A. W. (2020). Flexibility and adaptability of quorum sensing in nature. Trends Microbiol. 28, 436–444. doi: 10.1016/j.tim.2019.12.004
Prieto-Barajas C. M., Valencia-Cantero E., Santoyo G. (2018). Microbial mat ecosystems: structure types, functional diversity, and biotechnological application. Electron. J. Biotechnol. 31, 48–56. doi: 10.1016/j.ejbt.2017.11.001
Procópio L. (2019). The role of biofilms in the corrosion of steel in marine environments. World J. Microbiol. Biotechnol. 35, 73. doi: 10.1007/s11274-019-2647-4
Qvortrup K., Hultqvist L. D., Nilsson M., Jakobsen T. H., Jansen C. U., Uhd J., et al. (2019). Small molecule anti-biofilm agents developed on the basis of mechanistic understanding of biofilm formation. Front. Chem. 7. doi: 10.3389/fchem.2019.00742
Rabin N., Zheng Y., Opoku-Temeng C., Du Y., Bonsu E., Sintim H. O. (2015). Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 7, 493–512. doi: 10.4155/fmc.15.6
Rai P. K., Lee J., Brown R. J. C., Kim K.-H. (2021). Micro- and nano-plastic pollution: Behavior, microbial ecology, and remediation technologies. J. Clean. Product. 291, 125240. doi: 10.1016/j.jclepro.2020.125240
Rampadarath S., Bandhoa K., Puchooa D., Jeewon R., Bal S. (2017). Early bacterial biofilm colonizers in the coastal waters of Mauritius. Electron. J. Biotechnol. 29, 13–21. doi: 10.1016/j.ejbt.2017.06.006
Rastelli E., Corinaldesi C., Dell’Anno A., Tangherlini M., Martorelli E., Ingrassia M., et al. (2017). High potential for temperate viruses to drive carbon cycling in chemoautotrophy-dominated shallow-water hydrothermal vents. Environ. Microbiol. 19, 4432–4446. doi: 10.1111/1462-2920.13890
Reese A., Voigt N., Zimmermann T., Irrgeher J., Pröfrock D. (2020). Characterization of alloying components in galvanic anodes as potential environmental tracers for heavy metal emissions from offshore wind structures. Chemosphere 257, 127182. doi: 10.1016/j.chemosphere.2020.127182
Roberts D., Rittschof D., Holm E., Schmidt A. R. (1991). Factors influencing initial larval settlement-temporal, spatial and surface molecular-components. J. Exp. Mar. Biol. Ecol. 150, 203–221. doi: 10.1016/0022-0981(91)90068-8
Romera-Castillo C., Pinto M., Langer T. M., Álvarez-Salgado X. A., Herndl G. J. (2018). Dissolved organic carbon leaching from plastics stimulates microbial activity in the ocean. Nat. Commun. 9, 1430. doi: 10.1038/s41467-018-03798-5
Römling U., Galperin Michael Y., Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/mmbr.00043-12
Ruparell A., Dubern J. F., Ortori C. A., Harrison F., Halliday N. M., Emtage A., et al. (2016). The fitness burden imposed by synthesising quorum sensing signals. Sci. Rep. 6, 33101. doi: 10.1038/srep33101
Sabatino R., Zullo R., Di Cesare A., Piscia R., Musazzi S., Corno G., et al. (2024). Traditional and biodegradable plastics host distinct and potentially more hazardous microbes when compared to both natural materials and planktonic community. J. Hazard. Mater. 465, 133166. doi: 10.1016/j.jhazmat.2023.133166
Sanz-Montero M. E., Rodriguez-Aranda J. P. (2013). The role of microbial mats in the movement of stones on playa lake surfaces. Sediment. Geol. 298, 53–64. doi: 10.1016/j.sedgeo.2013.10.006
Scarascia G., Wang T., Hong P. Y. (2016). Quorum sensing and the use of quorum quenchers as natural biocides to inhibit sulfate-reducing bacteria. Antibio. (Basel) 5, 39. doi: 10.3390/antibiotics5040039
Schrenk M. O., Huber J. A., Edwards K. J. (2010). Microbial provinces in the subseafloor. Annu. Rev. Mar. Sci. 2, 279–304. doi: 10.1146/annurev-marine-120308-081000
Seeley M. E., Song B., Passie R., Hale R. C. (2020). Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat. Commun. 11, 2372. doi: 10.1038/s41467-020-16235-3
Sharma S., Kumar S., Kumar P., Tripathi V. N. (2024). Quorum sensing in Gram-negative pathogens, a fresh look. Microbe 4, 100108. doi: 10.1016/j.microb.2024.100108
Shi J., Wu D., Su Y., Xie B. (2021). Selective enrichment of antibiotic resistance genes and pathogens on polystyrene microplastics in landfill leachate. Sci. Total Environ. 765, 142775. doi: 10.1016/j.scitotenv.2020.142775
Shukla S. K., Mangwani N., Karley D., Rao T. S. (2017). “Bacterial biofilms and genetic regulation for metal detoxification,” in Handbook of metal-microbe interactions and bioremediation (Boca Raton, FL: CRC press), 317–332.
Sivakumar K., Scarascia G., Zaouri N., Wang T., Kaksonen A. H., Hong P. Y. (2019). Salinity-mediated increment in sulfate reduction, biofilm formation, and quorum sensing: A potential connection between quorum sensing and sulfate reduction? Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00188
Skovhus T. L., Eckert R. B., Rodrigues E. (2017). Management and control of microbiologically influenced corrosion (MIC) in the oil and gas industry-Overview and a North Sea case study. J. Biotechnol. 256, 31–45. doi: 10.1016/j.jbiotec.2017.07.003
Sooriyakumar P., Bolan N., Kumar M., Singh L., Yu Y., Li Y., et al. (2022). Biofilm formation and its implications on the properties and fate of microplastics in aquatic environments: A review. J. Hazard. Mater. Adv. 6, 100077. doi: 10.1016/j.hazadv.2022.100077
Spring S., Brinkmann N., Murrja M., Sproeer C., Reitner J., Klenk H.-P. (2015). High diversity of culturable prokaryotes in a lithifying hypersaline microbial mat. Geomicrobiol. J. 32, 332–346. doi: 10.1080/01490451.2014.913095
Stal L. J., Grossberger S., Krumbein W. E. (1984). Nitrogen-fixation associated with the cyanobacterial mat of a marine laminated microbial ecosystem. Mar. Biol. 82, 217–224. doi: 10.1007/bf00392402
Stevenson E. M., Rushby-Jones O., Buckling A., Cole M., Lindeque P. K., Murray A. K. (2024). Selective colonization of microplastics, wood and glass by antimicrobial-resistant and pathogenic bacteria. Microbiology 170. doi: 10.1099/mic.0.001506
Stokke R., Dahle H., Roalkvam I., Wissuwa J., Daae F. L., Tooming-Klunderud A., et al. (2015). Functional interactions among filamentous Epsilonproteobacteria and Bacteroidetes in a deep-sea hydrothermal vent biofilm. Environ. Microbiol. 17, 4063–4077. doi: 10.1111/1462-2920.12970
Stoodley P., Sauer K., Davies D. G., Costerton J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Sun C., Sun J., Wang Y., Lin X., Li X., Cheng X., et al. (2016). Synergistic effect of O2, H2S and SO2 impurities on the corrosion behavior of X65 steel in water-saturated supercritical CO2 system. Corrosion Sci. 107, 193–203. doi: 10.1016/j.corsci.2016.02.032
Tagg A. S., Oberbeckmann S., Fischer D., Kreikemeyer B., Labrenz M. (2019). Paint particles are a distinct and variable substrate for marine bacteria. Mar. pollut. Bull. 146, 117–124. doi: 10.1016/j.marpolbul.2019.06.013
Tagg A. S., Sperlea T., Hassenrück C., Kreikemeyer B., Fischer D., Labrenz M. (2024). Microplastic-antifouling paint particle contamination alters microbial communities in surrounding marine sediment. Sci. Total Environ. 926, 171863. doi: 10.1016/j.scitotenv.2024.171863
Tapia J. E., González B., Goulitquer S., Potin P., Correa J. A. (2016). Microbiota influences morphology and reproduction of the brown alga Ectocarpus sp. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00197
Teske A. P. (2005). The deep subsurface biosphere is alive and well. Trends Microbiol. 13, 402–404. doi: 10.1016/j.tim.2005.07.004
Thorseth I. H., Torsvik T., Torsvik V., Daae F. L., Pedersen R. B. (2001). Diversity of life in ocean floor basalt. Earth Planet. Sci. Lett. 194, 31–37. doi: 10.1016/s0012-821x(01)00537-4
Toyofuku M., Inaba T., Kiyokawa T., Obana N., Yawata Y., Nomura N. (2016). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. doi: 10.1080/09168451.2015.1058701
Turner A. (2021). Paint particles in the marine environment: An overlooked component of microplastics. Water Res. X 12, 100110. doi: 10.1016/j.wroa.2021.100110
Usher K., Kaksonen A., MacLeod I. (2014). Marine rust tubercles harbour iron corroding archaea and sulphate reducing bacteria. Corrosion Sci. 83, 189–197. doi: 10.1016/j.corsci.2014.02.014
Valentine D. L., Fisher G. B., Pizarro O., Kaiser C. L., Yoerger D., Breier J. A., et al. (2016). Autonomous marine robotic technology reveals an expansive benthic bacterial community relevant to regional nitrogen biogeochemistry. Environ. Sci. Technol. 50, 11057–11065. doi: 10.1021/acs.est.6b03584
Vardy S., Saunders J. E., Tolhurst T. J., Davies P. A., Paterson D. M. (2007). Calibration of the high-pressure cohesive strength meter (CSM). Continent. Shelf Res. 27, 1190–1199. doi: 10.1016/j.csr.2006.01.022
Velázquez D., López-Bueno A., Aguirre de Cárcer D., De los Ríos A., Alcamí A., Quesada A. (2016). Ecosystem function decays by fungal outbreaks in Antarctic microbial mats. Sci. Rep. 6, 22954. doi: 10.1038/srep22954
Videla H. A., Herrera L. K. (2009). Understanding microbial inhibition of corrosion. A comprehensive overview. Int. Biodeterior. Biodegr. 63, 896–900. doi: 10.1016/j.ibiod.2009.02.002
Vigneron A., Alsop E. B., Chambers B., Lomans B. P., Head I. M., Tsesmetzis N. (2016). Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl. Environ. Microbiol. 82, 2545–2554. doi: 10.1128/aem.03842-15
Vigneron A., Cruaud P., Pignet P., Caprais J.-C., Gayet N., Cambon-Bonavita M.-A., et al. (2014). Bacterial communities and syntrophic associations involved in anaerobic oxidation of methane process of the Sonora Margin cold seeps, Guaymas Basin. Environ. Microbiol. 16, 2777–2790. doi: 10.1111/1462-2920.12324
Wang R., Ding W., Long L., Lan Y., Tong H., Saha S., et al. (2020). Exploring the influence of signal molecules on marine biofilms development. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.571400
Wang J., Guo X., Xue J. (2021). Biofilm-developed microplastics As Vectors of Pollutants in Aquatic Environments. Environ. Sci. Technol. 55, 12780–12790. doi: 10.1021/acs.est.1c04466
Wang H., Yang Q., Li D., Wu J., Yang S., Deng Y., et al. (2023). Stable isotopic and metagenomic analyses reveal microbial-mediated effects of microplastics on sulfur cycling in coastal sediments. Environ. Sci. Technol. 57, 1167–1176. doi: 10.1021/acs.est.2c06546
Wang R., Zhang W., Ding W., Liang Z., Long L., Wong W. C., et al. (2022). Profiling signal transduction in global marine biofilms. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.768926
Watnick P., Kolter R. (2000). Biofilm, city of microbes. J. Bacteriol. 182, 2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000
Weise W., Rheinheimer G. (1978). Scanning electron-microscopy and epifluorescence investigation of bacterial colonization of marine sand sediments. Microbial. Ecol. 4, 175–188. doi: 10.1007/bf02015075
Wichard T. (2023). From model organism to application: Bacteria-induced growth and development of the green seaweed Ulva and the potential of microbe leveraging in algal aquaculture. Semin. Cell Dev. Biol. 134, 69–78. doi: 10.1016/j.semcdb.2022.04.007
Woodall L. C., Jungblut A. D., Hopkins K., Hall A., Robinson L. F., Gwinnett C., et al. (2018). Deep-sea anthropogenic macrodebris harbours rich and diverse communities of bacteria and archaea. PloS One 13, e0206220. doi: 10.1371/journal.pone.0206220
Wright R. J., Erni-Cassola G., Zadjelovic V., Latva M., Christie-Oleza J. A. (2020). Marine plastic debris: A new surface for microbial colonization. Environ. Sci. Technol. 54, 11657–11672. doi: 10.1021/acs.est.0c02305
Xia J., Yang C., Xu D., Sun D., Nan L., Sun Z., et al. (2015). Laboratory investigation of the microbiologically influenced corrosion (MIC) resistance of a novel Cu-bearing 2205 duplex stainless steel in the presence of an aerobic marine Pseudomonas aeruginosa biofilm. Biofouling 31, 481–492. doi: 10.1080/08927014.2015.1062089
Xie H., Chen J., Feng L., He L., Zhou C., Hong P., et al. (2021). Chemotaxis-selective colonization of mangrove rhizosphere microbes on nine different microplastics. Sci. Total Environ. 752, 142223. doi: 10.1016/j.scitotenv.2020.142223
Xu X. Y., Wang S., Gao F. L., Li J. X., Zheng L., Sun C. J., et al. (2019). Marine microplastic-associated bacterial community succession in response to geography, exposure time, and plastic type in China’s coastal seawaters. Mar. pollut. Bull. 145, 278–286. doi: 10.1016/j.marpolbul.2019.05.036
Yamamoto M., Takai K. (2011). Sulfur metabolisms in epsilon- and gamma-Proteobacteria in deep-sea hydrothermal fields. Front. Microbiol. 2. doi: 10.3389/fmicb.2011.00192
Yang H., Yu S., Lu H. (2021). Iron-coupled anaerobic oxidation of methane in marine sediments: A review. J. Mar. Sci. Eng. 9, 875. doi: 10.3390/jmse9080875
Yao S., Lyu S., An Y., Lu J., Gjermansen C., Schramm A. (2019). Microalgae–bacteria symbiosis in microalgal growth and biofuel production: a review. J. Appl. Microbiol. 126, 359–368. doi: 10.1111/jam.14095
Yoo N. G., Dogra S., Meinen B. A., Tse E., Haefliger J., Southworth D. R., et al. (2018). Polyphosphate stabilizes protein unfolding intermediates as soluble amyloid-like oligomers. J. Mol. Biol. 430, 4195–4208. doi: 10.1016/j.jmb.2018.08.016
Yu S., Su T., Wu H., Liu S., Wang D., Zhao T., et al. (2015). PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 25, 1352–1367. doi: 10.1038/cr.2015.129
Yu Y., Yan F., Chen Y., Jin C., Guo J. H., Chai Y. (2016). Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of bacillus subtilis. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01811
Zammit G., Schembri S., Fenech M. (2021). Phototrophic biofilms and microbial mats from the marine littoral of the central Mediterranean. Acta Botanica Croatica 80, 112–116. doi: 10.37427/botcro-2020-031
Zan J., Liu Y., Fuqua C., Hill R. T. (2014). Acyl-homoserine lactone quorum sensing in the roseobacter clade. Int. J. Mol. Sci. 15, 654–669. doi: 10.3390/ijms15010654
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “Plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang W., Ding W., Li Y. X., Tam C., Bougouffa S., Wang R., et al. (2019). Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun. 10, 517. doi: 10.1038/s41467-019-08463-z
Zhang W. P., Wang Y., Tian R. M., Bougouffa S., Yang B., Cao H. L., et al. (2014). Species sorting during biofilm assembly by artificial substrates deployed in a cold seep system. Sci. Rep. 4, 6647. doi: 10.1038/srep06647
Zhang H., Yang C. (2019). Arginine and nitrogen mobilization in Cyanobacteria. Mol. Microbiol. 111, 863–867. doi: 10.1111/mmi.14204
Zhang B., Yang X., Liu L. C., Chen L., Teng J., Zhu X. P., et al. (2021). Spatial and seasonal variations in biofilm formation on microplastics in coastal waters. Sci. Total Environ. 770, 145303. doi: 10.1016/j.scitotenv.2021.145303
Zhao Y., Gao J., Wang Z., Dai H., Wang Y. (2021). Responses of bacterial communities and resistance genes on microplastics to antibiotics and heavy metals in sewage environment. J. Hazard. Mater. 402, 123550. doi: 10.1016/j.jhazmat.2020.123550
Zhao X., Zhao F., Wang J., Zhong N. (2017). Biofilm formation and control strategies of foodborne pathogens: food safety perspectives. RSC Adv. 7, 36670–36683. doi: 10.1039/c7ra02497e
Keywords: marine biofilms, microplastic biofilms, biofouling, biocorrosion, subsurface mats, sedimentary biofilms
Citation: Yu S, Lu X and Lu H (2025) Marine microbial biofilms on diverse abiotic surfaces. Front. Mar. Sci. 12:1482946. doi: 10.3389/fmars.2025.1482946
Received: 19 August 2024; Accepted: 31 January 2025;
Published: 26 February 2025.
Edited by:
Jin Zhou, Tsinghua University, ChinaReviewed by:
Yan-Hua Zeng, Hainan University, ChinaCopyright © 2025 Yu, Lu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shan Yu, c2hhbnl1QHBrdS5lZHUuY24=
‡ORCID: Xindi Lu, orcid.org/0000-0003-0094-4397
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.