- 1School of Pharmacy, Jiangsu University, Zhenjiang, China
- 2Shanghai Key Laboratory for Molecular Engineering of Chiral Drugs, School of Pharmacy, Shanghai Jiao Tong University, Shanghai, China
- 3Guangdong Key Laboratory for Research and Development of Natural Drugs, School of Pharmacy, Guangdong Medical University, Dongguan, China
The microbial diversity in oceans is considerable, widely distributed in seawater, marine sediments, and marine organisms. Compared with terrestrial resources in traditional natural product research, the living environments of marine microorganisms are starkly different. The drastic differences in survival conditions, such as high salinity, oligotrophic conditions, lack of light, and limited oxygen, determine that microorganisms exhibit distinctive characteristics in metabolism, survival modes, and adaptive mechanisms. These factors contribute to significant distinctions in secondary metabolic pathways and enzymatic reaction mechanisms between marine and terrestrial microorganisms. In this review, we summarized a total of 72 novel natural products with antibacterial activity, published in 2024, which are derived from marine-derived fungi. These products (polyketides, alkaloids, terpenoids, and peptides) are emphasized in terms of their structures and biological activities. This article aims to provide useful information for the research and development of novel antibiotics.
1 Introduction
Since the late 1990s, with the increasing exploration of natural resources, discovering new biologically active natural products has become increasingly challenging (Atanasov et al., 2021). Traditional strategies for the isolation and identification of natural products have led to the repeated isolation of a large number of known compounds, slowing down the process of discovering structurally novel active compounds (Zhang et al., 2020; Pirintsos et al., 2022; Young et al., 2022). Meanwhile, the emergence of drug resistance in bacteria has made it urgent for humanity to seek new natural products with novel structures, unique biological activities, and mechanisms of action as lead compounds for new drug development (Schneider, 2021; Vaou et al., 2021).
Compared to terrestrial biological resources studied in traditional natural product research, the living environments of marine organisms are strikingly different. The drastic differences in survival conditions, such as high pressure, high salinity, oligotrophic conditions, lack of light, limited oxygen, and special habitats (submarine hydrothermal vents, deep-sea trenches), determine that marine organisms exhibit significant characteristics in metabolism, survival strategies, information transfer, and adaptive mechanisms (Hai et al., 2021; Lu et al., 2021; Srinivasan et al., 2021). These factors contribute to marine organisms having almost entirely distinct secondary metabolic pathways and enzymatic reaction mechanisms compared to terrestrial organisms. Notable drugs derived from marine sources include the antiviral drug vidarabine, the anticancer drugs cytosine arabinoside and eribulin, the analgesic conotoxin, the lipid-lowering drug ethyl ester of eicosapentaenoic acid (EPA), and the “warhead” of antibody-drug conjugates (ADCs), dolastatin (Alves et al., 2020). These drugs all possess unique chemical structures, and the exploration of lead molecules with significant pharmacological activities from marine organisms is gradually demonstrating significant research value and application potential.
Marine microorganisms, as an important group within marine organisms, have always been one of the research hotspots in marine natural products (Stincone and Brandelli, 2020; Crawford et al., 2021; Shams Ul Hassan et al., 2021; Gomez-Banderas, 2022; Hassan et al., 2022; Voser et al., 2022; Hassan et al., 2024). Thanks to the rapid development of high-throughput sequencing technology, more and more microbial genomes have been sequenced in recent years. Bioinformatics analysis has revealed that microbial genomes typically contain multiple secondary metabolite biosynthetic gene clusters. Limited by conventional laboratory cultivation conditions, the variety of compounds we have discovered so far is far less than the number of compounds that microorganisms are capable of producing, and a large number of potential secondary metabolites remain undiscovered (Scherlach and Hertweck, 2021; Albarano et al., 2020; Peng et al., 2021). Activating the expression of these silent gene clusters and seeking the active secondary metabolites they contain will become an important source for discovering novel drug precursors for anti-inflammatory, antibacterial, antitumor, antiviral, and enzyme inhibitor applications, opening up new avenues for fundamental research in drug discovery (Stuart et al., 2020; Zhu et al., 2022; El-Hawary et al., 2023; Tsipinana et al., 2023).
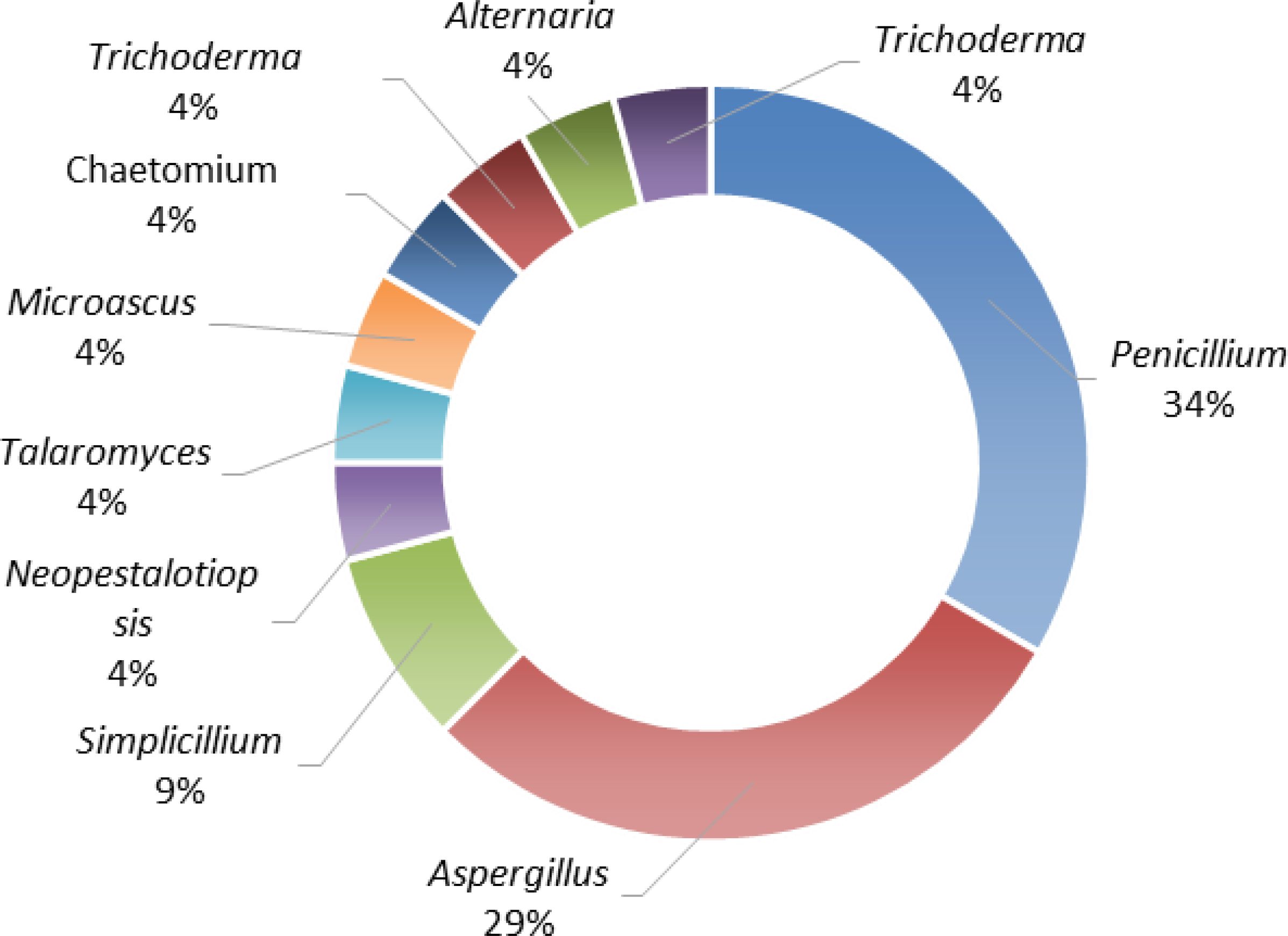
Based on data from Web of Science, PubMed, Elsevier, the American Chemical Society (ACS), and Google Scholar, this article selects studies that report novel compounds and provide minimum inhibitory concentration values (MICs), comprehensively summarizing the sources, structures, and biological activity progress of 56 newly isolated antibacterial natural products derived from marine fungi in 2024. Known compounds and compounds with other activities were not included in the statistics. According to their structural characteristics, these natural products are classified into four major categories, including polyketides (41.7%, 30/72), alkaloids (19.4%, 14/72), terpenoids (12.5%, 9/72), peptides (22.2%, 16/72), and miscellaneous (4.2%, 3/72) (Figure 1). Among these biological samples, 33.3% (8/24) originate from Penicillium fungi, indicating the great potential of Penicillium fungi secondary metabolites in the search for novel antibiotics. Meanwhile, the significance of Aspergillus fungi (29.2%, 7/24) in the exploration of novel antibiotics cannot be overlooked (Figure 2). Particularly noteworthy are the impressive discoveries by Professor Shu-Hua Qi from South China Sea Institute of Oceanology, whose 16-epiascomylactam B (38) exhibited strong antibacterial activity (Yao et al., 2024). Tables 1–5 list the names, isolation sources, categories, and activity levels (MIC) of the antibacterial active compounds.
2 Polyketides
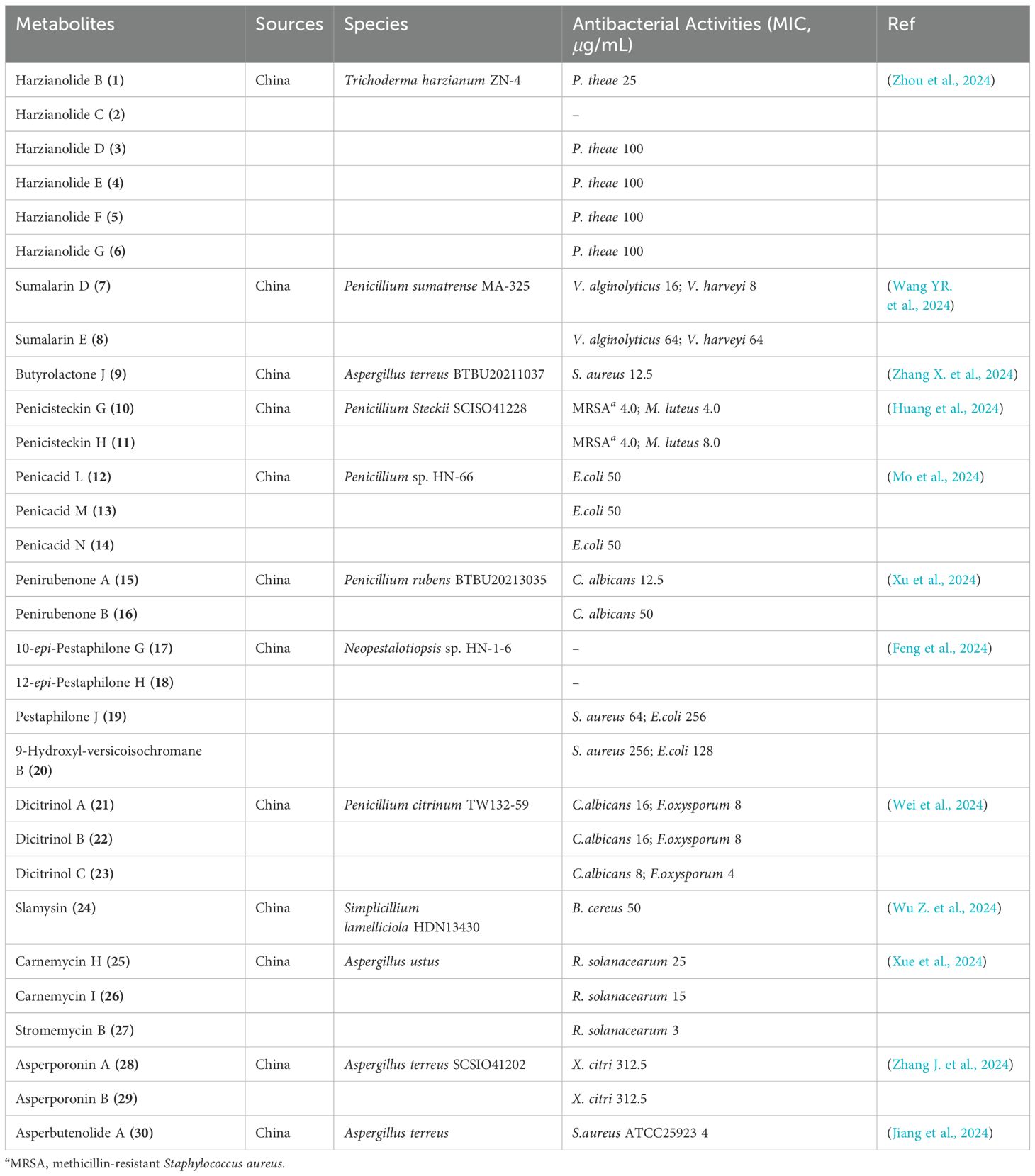
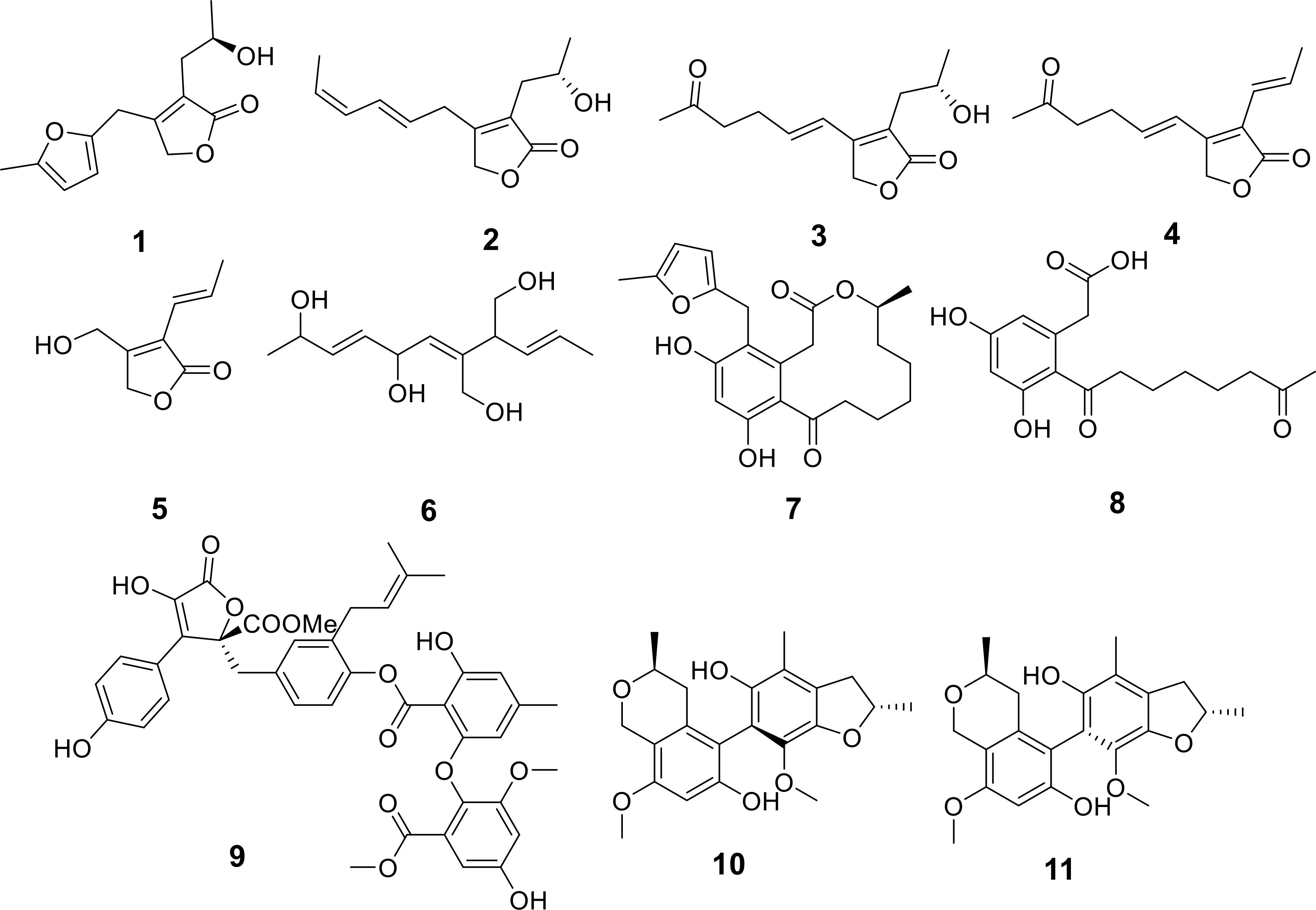
From the EtOAc extract of liquid fermentation cultures of Trichoderma harzianum ZN-4, sourced from sediments in the Zhoushan coastal region, five previously unreported γ-butyrolactone harzianolides named B-F (1-5), along with their precursor harzianolide G (6), were isolated and characterized (Figure 3) (Zhou et al., 2024). In biological testing, Compound 1 exhibited moderate inhibitory effects on the phytopathogenic fungus P. theae, with a minimum inhibitory concentration (MIC) of 25 μg/mL. Compounds 3-6 displayed weaker activity, each having an MIC of 100 μg/mL against the same fungus (Table 1). Additionally, two new curvularin derivatives, sumalarins D and E (7, 8), were isolated and identified from the mangrove-associated fungus Penicillium sumatrense MA-325 (Figure 3) (Wang YR. et al., 2024). These compounds demonstrated activity against aquatic pathogenic bacteria, V. alginolyticus and V. harveyi, with MIC values spanning from 8 to 64 μg/mL, respectively (Table 1).
A novel compound, designated as butyrolactone J (9), has been extracted from the secondary metabolites produced by the marine-derived fungal strain Aspergillus terreus BTBU20211037 (Zhang X. et al., 2024). This compound displayed inhibitory activity against Staphylococcus aureus ATCC 25923, with a minimal inhibitory concentration (MIC) of 12.5 μg/mL (Table 1). Meanwhile, a pair of uncharacterized atropo-diastereomeric dimers, named penicisteckins G (10) and H (11), were isolated from the marine coral-associated fungus Penicillium steckii SCISO41228 (Figure 3) (Huang et al., 2024). Both compounds exhibited moderate antibacterial activity against a range of pathogenic strains tested, particularly against methicillin-resistant Staphylococcus aureus (MRSA) and Micrococcus luteus, with MIC values of 4.0 μg/mL (Table 1).
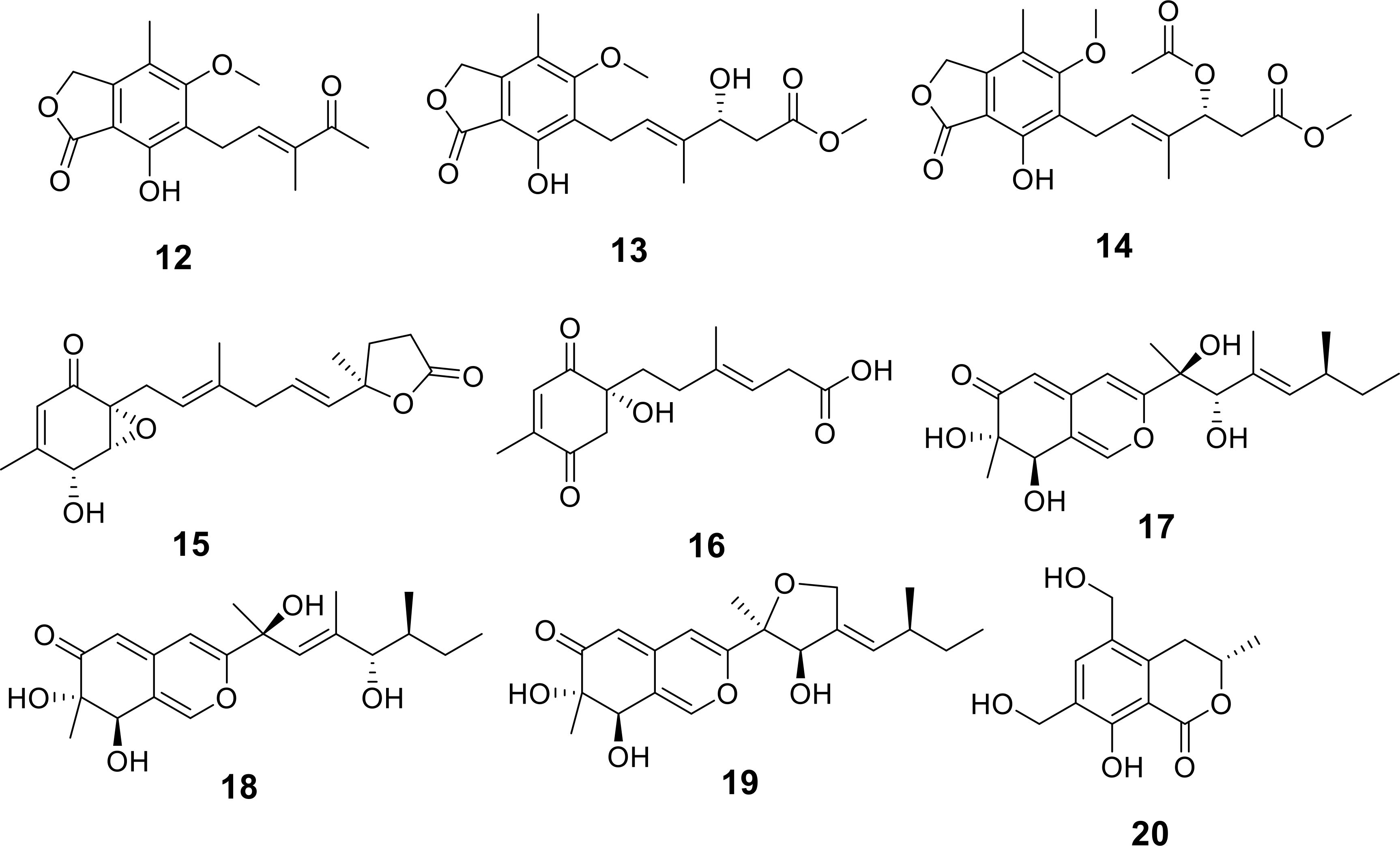
Three newly discovered mycophenolic acid derivatives, penicacids L-N (12-14), were obtained from a fungal isolate, Penicillium sp. HN-66, sourced from marine sediments in the South China Sea (Figure 4) (Mo et al., 2024). Bioassay results indicated that these compounds exhibited weak inhibitory activity against E. coli ATCC25922, with minimum inhibitory concentrations (MICs) of 50 μg/mL (Table 1). Additionally, two novel polyketide derivatives, penirubenones A and B (15 and 16), were isolated from the marine-derived fungus Penicillium rubens BTBU20213035 (Xu et al., 2024). When combined with 0.0625 µg/mL rapamycin, compounds 15 and 16 demonstrated synergistic antifungal activity against Candida albicans at concentrations of 12.5 and 50 µg/mL, respectively (Table 1). Furthermore, four new compounds, three azaphilones (17-19) and one dihydroisocoumarin (20) were isolated from the sea-mud-derived fungus Neopestalotiopsis sp. HN-1-6, collected from the Beibu Gulf of China (Figure 4) (Feng et al., 2024). Among these, compounds 19 and 20 showed antibacterial activity against Staphylococcus aureus and Escherichia coli, with MICs ranging from 64 µg/mL to 256 µg/mL (Table 1).
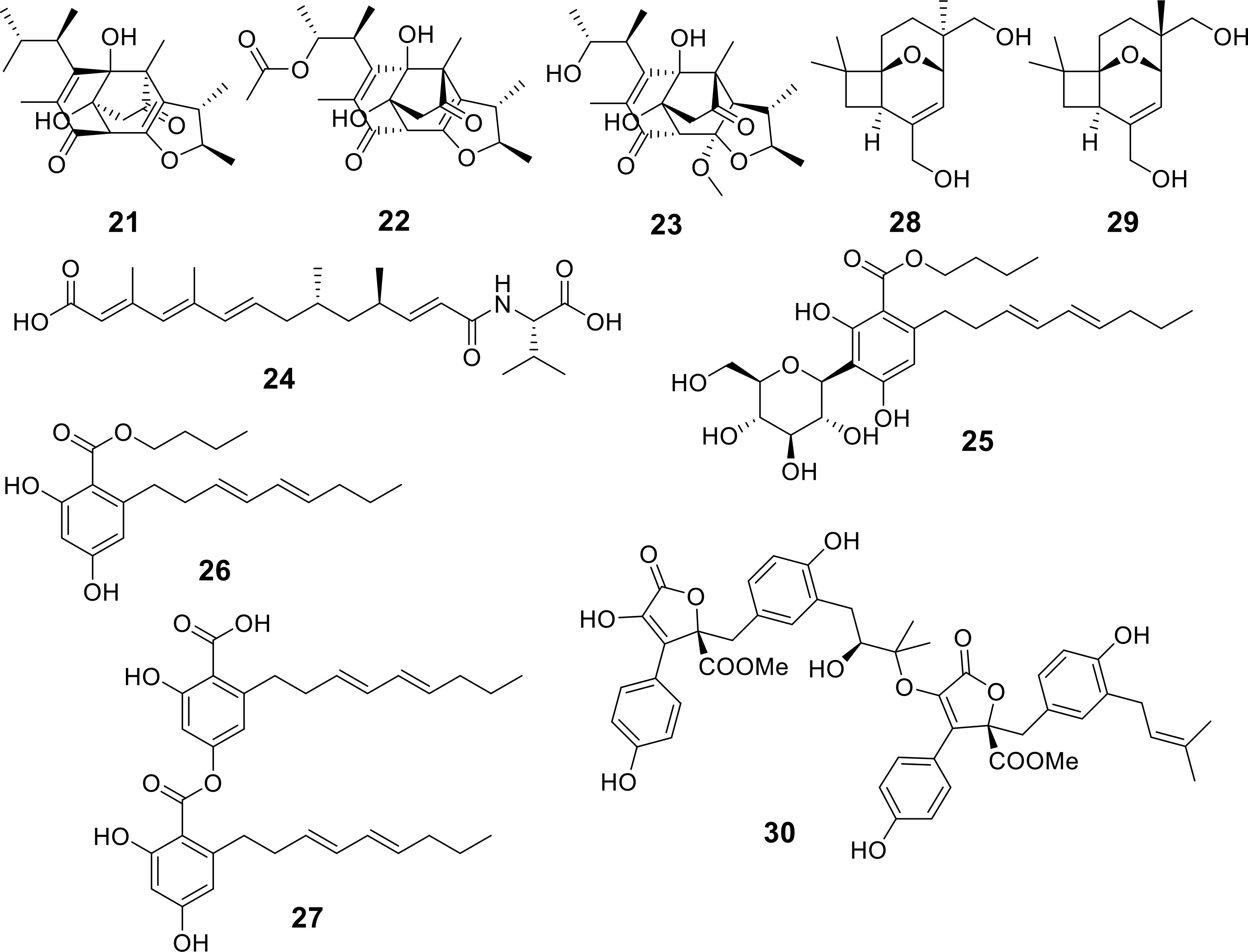
Three novel and unusual citrinin derivatives, featuring a distinctive 6/5/7/5 core structure, named dicitrinols A-C (21-23), were isolated through the fermentation process of the hydrothermal vent-associated fungus Penicillium citrinum TW132-59 (Figure 5) (Wei et al., 2024). These compounds exhibited moderate antifungal activity against Candida albicans and Fusarium oxysporum, with MIC values ranging from 4 to 16 μg/mL (Table 1). Additionally, a new compound called slamysin (24) (Figure 5) was discovered from a cryptic cytochalasin-like gene cluster (sla) within the antarctic-derived Simplicillium lamelliciola HDN13430. This compound is characterized by an N-acylated amino acid structure and demonstrated weak anti-Bacillus cereus activity with an MIC of 50 µg/mL (Wu Z. et al., 2024).
Through biological activity-guided screening, three previously undescribed compounds, carnemycins H-I (25, 26) and stromemycin B (27) were isolated from the secondary metabolites of a marine-derived Aspergillus ustus (Figure 5) (Xue et al., 2024). Among them, compound 27 exhibited excellent inhibitory activity against R. solanacearum, with an MIC value of 3 μg/mL (Table 1). Furthermore, asperporonin A (28) and asperporonin B (29) were identified as novel compounds possessing a highly unusual structural skeleton from a bioassay-guided isolation of the deep-sea fungus Aspergillus terreus SCSIO41202 (Zhang J. et al., 2024). These compounds showed weak activity against Xanthomonas citri at a concentration of 312.5 µg/mL (Table 1). An unusual aromatic butenolide, asperbutenolide A (30), with antimicrobial properties (S. aureus ATCC25923 4 µg/mL) from the marine fungus Aspergillus terreus SCAU011 (Jiang et al., 2024). Highlight the potential application valuation of asperbutenolide A as a new antibacterial agent.
3 Alkaloids
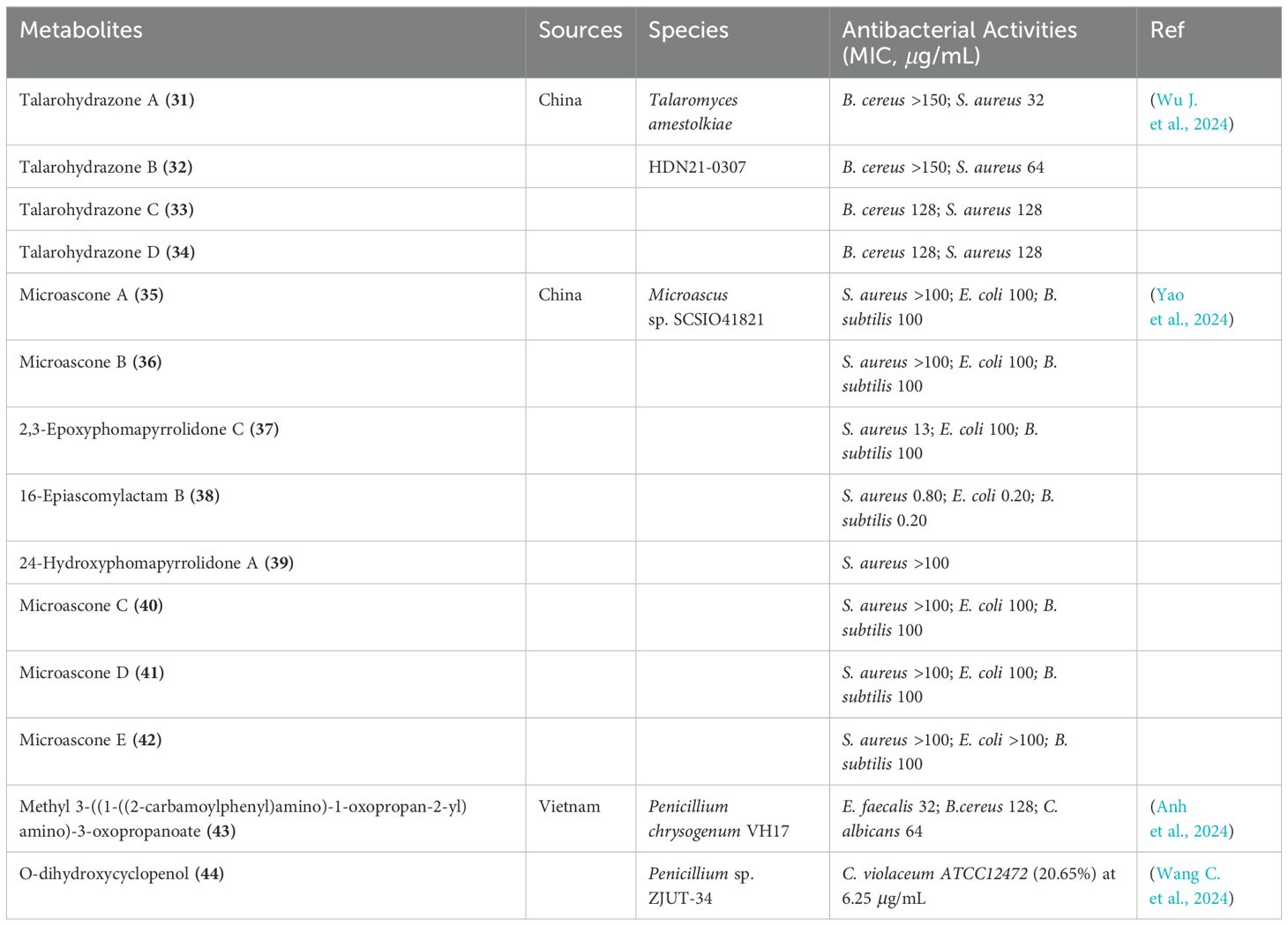
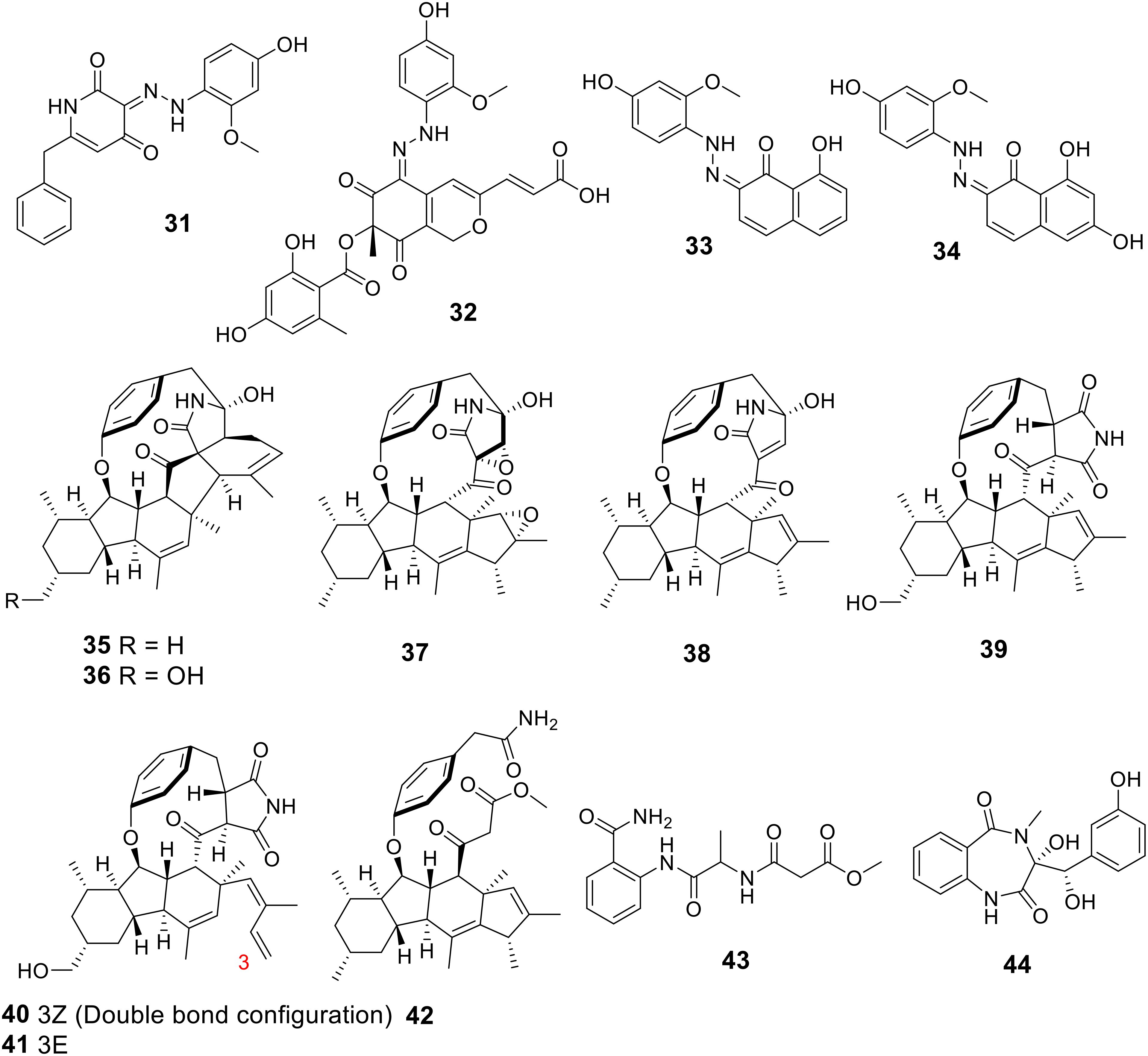
Using the one strain-many compounds (OSMAC) approach, four uncommon phenylhydrazone alkaloids, named talarohydrazones A-D (31-34), were isolated from the deep-sea cold seep-derived fungus Talaromyces amestolkiae HDN21-0307 (Figure 6) (Wu J. et al., 2024). These compounds exhibited weak antibacterial activity against Staphylococcus aureus, with MIC values falling between 32 and 128 μg/mL (Table 2). Additionally, eight novel decahydrofluorene-class alkaloids were isolated from the marine-derived fungus Microascus sp. SCSIO41821, including microascones A and B (35 and 36), 2,3-epoxyphomapyrrolidone C (37), 14,16-epiascomylactam B (38), 24-hydroxyphomapyrrolidone A (39), and microascones C-E (40-42) (Yao et al., 2024). Notably, compound 38 demonstrated potent antibacterial activity against various tested pathogens, with MIC values ranging from 0.20 to 0.80 μg/mL (Table 2). Furthermore, a new compound, methyl 3-((1-((2-carbamoylphenyl)amino)-1-oxopropan-2-yl)amino)-3-oxopropanoate (43), was isolated from the methanol extract of the marine-derived fungus Penicillium chrysogenum VH17 (Anh et al., 2024). Compound 43 showed antimicrobial activity against several reference microorganisms, with MIC values spanning from 32 to 128 μg/mL (Table 2). The marine fungus Penicillium sp. ZJUT-34, cultivated on a rice medium, yielded a novel alkaloid known as O-dihydroxycyclopenol (44). This compound exhibited a concentration-dependent inhibitory effect on the formation of violacein in C. violaceum ATCC12472 without affecting its growth. Specifically, at concentrations of 100, 50, 25, 12.5, and 6.25 μg/mL, compound 44 demonstrated inhibition rates of 42.03%, 39.77%, 34.69%, 29.41%, and 20.65%, respectively (Wang C. et al., 2024).
4 Terpenoids
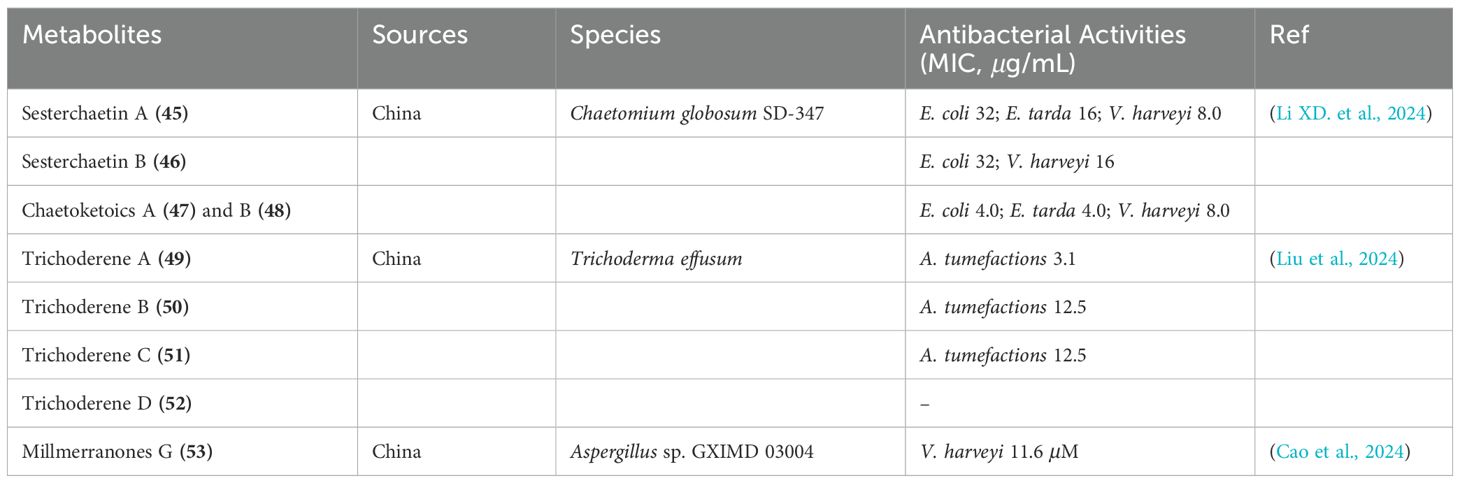
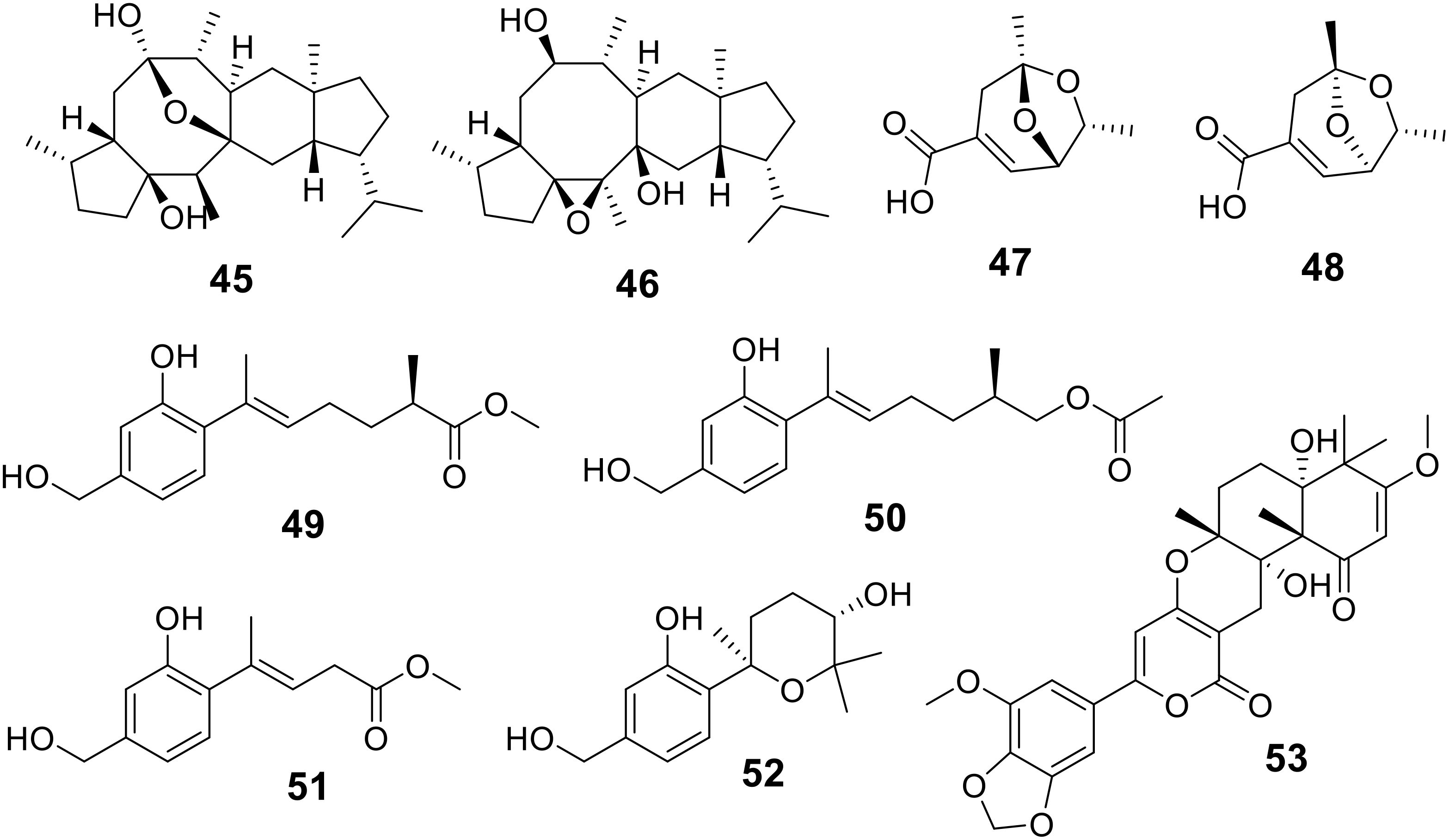
Xiao-Dong Li et al. characterized two novel sesterterpenoids, named sesterchaetins A and B (45 and 46), as well as two new diepoxide polyketides, chaetoketoics A and B (47 and 48), from the culture extract of Chaetomium globosum SD-347, a fungal strain originating from deep-sea sediment (Li XD. et al., 2024). The sesterchaetins demonstrated significant inhibitory activity against the aquatic pathogen Vibrio harveyi, with MIC values of 8.0 and 16 μg/mL, respectively. The chaetoketoics exhibited notable antimicrobial activity against E. coli and E. tarda, both with an MIC value of 4.0 μg/mL (Table 3).
Furthermore, four new sesquiterpene derivatives, trichoderenes A-D (49-52), were isolated from the marine-derived fungus Trichoderma effusum (Liu et al., 2024). These compounds were tested for their antimicrobial activity against A. tumefactions, and compounds 49-51 showed inhibitory activity with MIC values of 3.1, 12.5, and 12.5 μg/mL, respectively (Table 3). Additionally, a new meroterpene derivative, millmerranones G (53), was identified from the mangrove-derived fungus Aspergillus sp. GXIMD 03004, which was isolated from the leaves of the mangrove plant Acanthus ilicifolius L. collected in Beibu Gulf, China (Cao et al., 2024). Compound 53 displayed weak activity against Vibrio harveyi, with an MIC value of 11.6 μM (Table 3; Figure 7).
5 Peptides
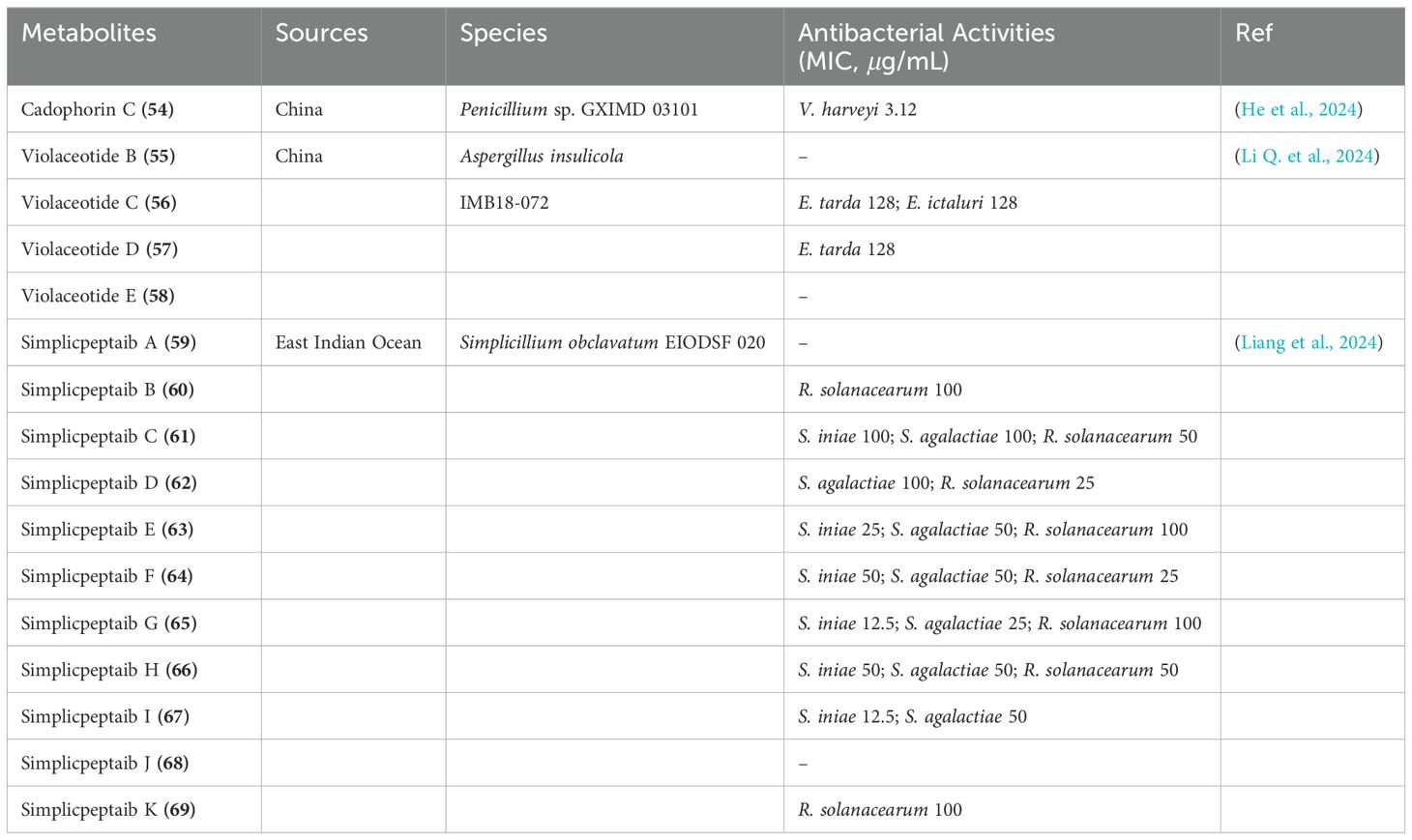
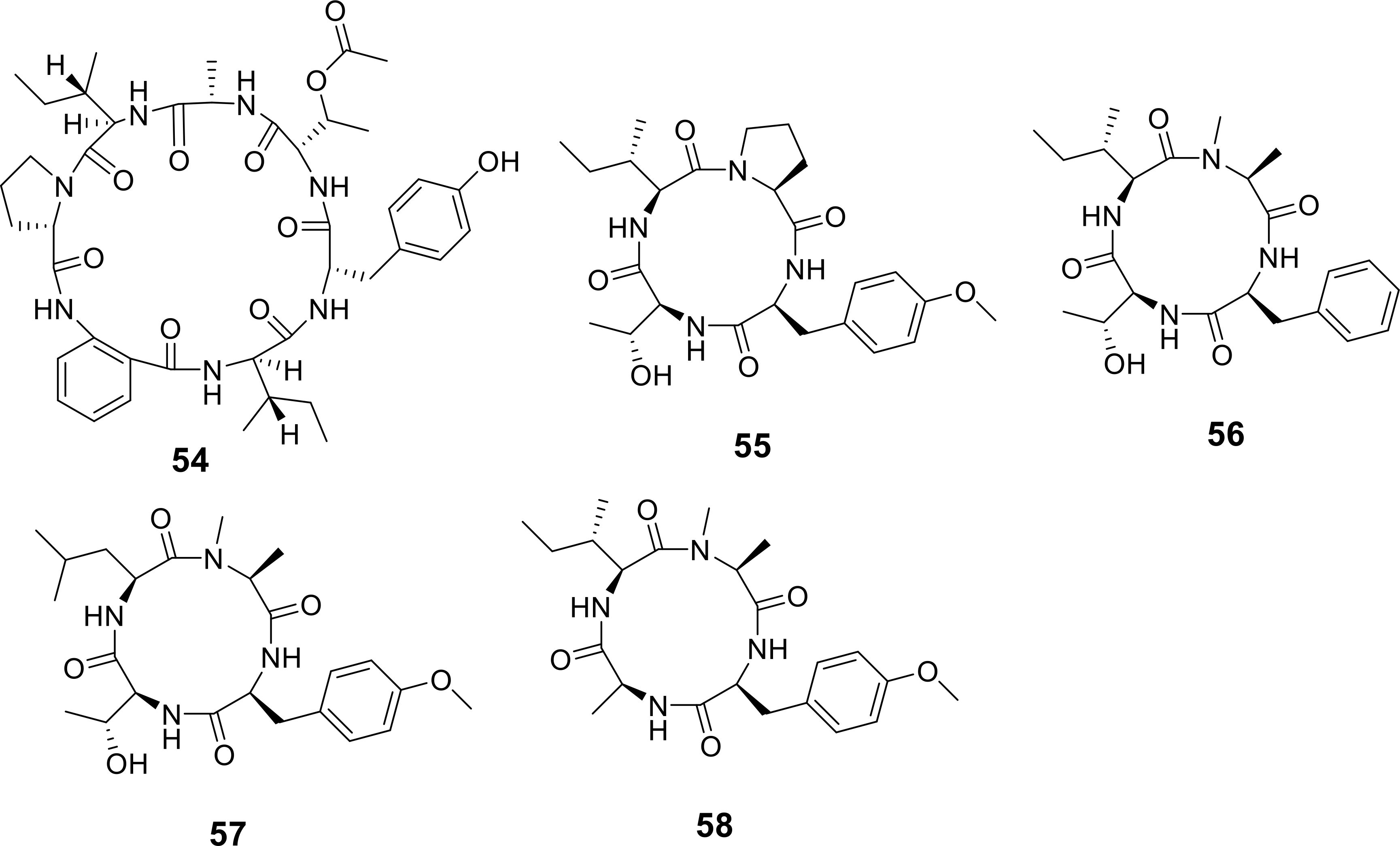
A novel cyclic heptapeptide, cadophorin C (54), was obtained from the mangrove-derived fungus Penicillium sp. GXIMD 03101, which was isolated from the mangrove species Acanthus ilicifolius L. (Figure 8) (He et al., 2024). Antibacterial testing revealed that compound 54 exhibited weak activity against the aquatic pathogen Vibrio harveyi, with an MIC value of 3.12 μg/mL (Table 4). Additionally, four new cyclic tetrapeptides, named violaceotides B-E (55-58), were discovered from the culture extract of the sponge-associated Aspergillus insulicola IMB18-072 after cocultivation with the marine-derived Alternaria angustiovoidea IMB20-805 (Figure 8) (Li Q. et al., 2024). Among these compounds, 56 and 57 demonstrated selective antimicrobial activity against the aquatic pathogenic bacteria Edwardsiella tarda and E. ictaluri, with MIC values of 128 μg/mL (Table 4).
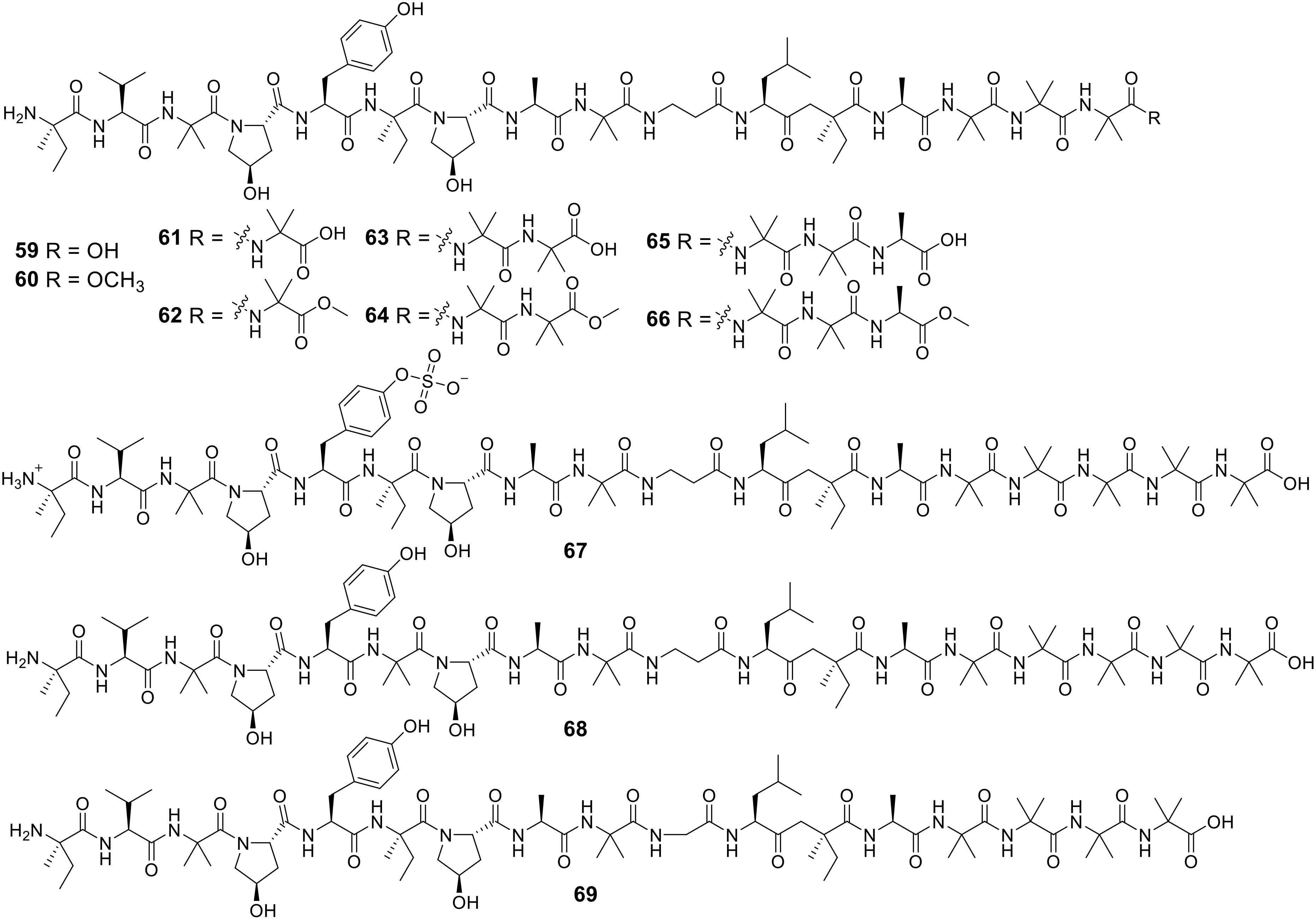
To discover novel antibacterial agents, researchers employed a bioassay-guided method to explore the secondary metabolites of Simplicillium obclavatum EIODSF 020, a fungus sourced from the deep sea, which exhibits antibacterial properties against pathogens affecting plants and fish. This approach resulted in the identification of 11 novel peptaibiotics, named simplicpeptaibs A-K (59-69) (Figure 9) (Liang et al., 2024). Notably, compounds 62, 64, 65, and 67 demonstrated potent activity against Ralstonia solanacearum, a tobacco pathogen, as well as Streptococcus iniae and Streptococcus agalactiae, pathogens that affect tilapia. The minimum inhibitory concentrations (MICs) of these compounds ranged from 12.5 to 100 μg/mL (Table 4).
Additionally, two novel cyclic depsipeptides, namely Rakicidin J and Rakicidin K, were extracted from the culture broth of the marine actinomycete Micromonospora chalcea FIM-R150103. Both compounds exhibited moderate antibacterial properties against ten strains of Gram-positive bacteria (Methicillin resistant S. aureus, C. difficile, etc.), with MIC values falling between 4 and over 32 μg/mL (Chen et al., 2024).
6 Miscellaneous
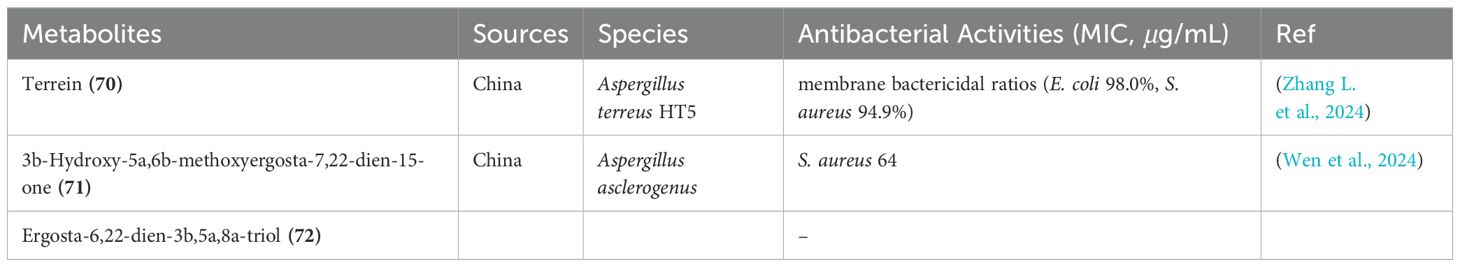
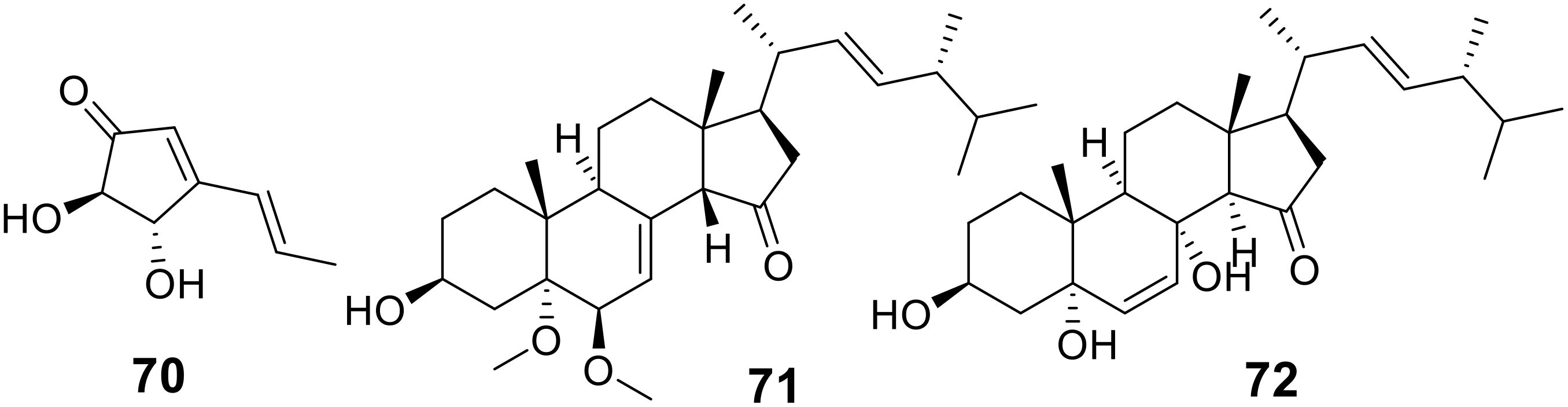
Terrein (70), a secondary metabolite initially sourced from the marine Aspergillus terres HT5 strain (Figure 10), demonstrated remarkable antibacterial properties. When integrated into a reverse osmosis membrane, it effectively eliminated 98.0% of E. coli and 94.9% of S. aureus, as shown in Table 5 (Zhang L. et al., 2024). Furthermore, two ergostane-type steroids containing oxygen, including a novel compound named 3b-hydroxy-5a,6b-methoxyergosta-7,22-dien-15-one (71), and a previously identified analogue, ergosta-6,22-dien-3b,5a,8a-triol (72), were extracted from crude samples of a marine sponge-associated fungus Aspergillus sp. (Figure 10) (Wen et al., 2024). Notably, compound 71 displayed antibacterial activity against S. aureus with a minimum inhibitory concentration of 64 μg/mL (Table 5).
7 Conclusions
The increasingly severe issue of global drug resistance has prompted vigorous efforts to search for new antibacterial agents. Marine natural products play a crucial role in drug discovery and serve as the premise for the early development of generic drugs (Hussain et al., 2021; Surendhiran et al., 2021). The marine medicinal organism resources are abundant, with good biodiversity and large variety reserves (Garcia-Perez et al., 2023; Xu et al., 2023; Zhang Y. et al., 2024). Efficient development of these resources is the focus of marine drug development. Efforts should be made in multiple aspects simultaneously, including talent cultivation, scientific research, and industrialization research. While strengthening basic research, it is even more important to facilitate the entry of active compounds into clinical trials, achieve drug marketing and sales, drive economic development, and form a development model with a certain scale. Meanwhile, cooperation among enterprises, universities, and scientific research institutes should be strengthened to attract more government policy support and funding, thereby promoting the rapid development of the marine pharmaceutical economy and achieving greater leaps in marine drug development.
This review provides a detailed discussion of 72 compounds with antibacterial activity reported in 2024, originating from marine fungi, covering polyketides (Table 1), alkaloids (Table 2), terpenoids (Table 3), peptides (Table 4) and miscellaneous (Table 5). The article introduces the sources, chemical structures, biological activities of these compounds. In summary, the continuous emergence of drug-resistant pathogens poses a significant threat to human health. Currently, the effectiveness of most traditional antibiotics is waning, while secondary metabolites from marine microorganisms offer promising resources for exploring natural antimicrobials with unique structures, potent activities, and specific mechanisms of action. The marine environment serves as a valuable source of new natural products, potentially providing crucial leads for the discovery and development of novel antibiotics in the future. Natural products from marine fungi hold promise in inspiring medicinal chemists to search for antimicrobials superior to existing drugs. Furthermore, in recent years, novel structures and metabolic (biosynthetic) pathways have attracted many researchers, and the bioactivities of these compounds have also sparked interest in the pharmaceutical development community. Overall, research on antibacterial natural drugs derived from marine fungi currently focuses mainly on isolation and structural elucidation, with a very limited number of subsequent pharmacological studies. More in-depth and thorough research efforts are needed to strengthen this promising field. Developing novel antibacterial agents from secondary metabolites of marine fungi and their synthetic derivatives deserves special attention.
Author contributions
CP: Data curation, Project administration, Resources, Visualization, Writing – review & editing. SH: Data curation, Methodology, Writing – original draft, Writing – review & editing. IM: Data curation, Formal Analysis, Methodology, Writing – review & editing. HJ: Data curation, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NSFC (No. 82404465), the Senior Talent Foundation of Jiangsu University (5501290012). The work was also supported by the Chugai foundation for innovative drug discovery science: C-FINDs (2025-CF-01). The work was supported by NSFC (81973191), project supported by the Modern Plateau Plant Medicine Research Project of Shanghai Jiao Tong University (SA1700208).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albarano L., Esposito R., Ruocco N., Costantini M. (2020). Genome mining as new challenge in natural products discovery. Mar. Drugs 18, 199. doi: 10.3390/md18040199
Alves E., Dias M., Lopes D., Almeida A., Domingues M. D. R., Rey F. (2020). Antimicrobial lipids from plants and marine organisms: an overview of the current state-of-the-art and future prospects. Antibiotics 9, 441. doi: 10.3390/antibiotics9080441
Anh N. M., Minh L. T. H., Linh N. T., Dao P. T., Quynh D. T., Huong D. T. M., et al. (2024). Secondary metabolites from marine fungus Penicillium chrysogenum VH17 and their antimicrobial and cytotoxic potential. Biosci. Biotechnol. Biochem. 88, 1254–1260. doi: 10.1093/bbb/zbae113
Atanasov A. G., Zotchev S. B., Dirsch V. M., International natural product sciences taskforce, Supuran C. T. (2021). Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 20, 200–216. doi: 10.1038/s41573-020-00114-z
Cao G. P., Huang G. Q., Chen G. Y., Yi X. X., Wang X. S., Wei F. J., et al. (2024). a meroterpene isolated from a mangrove-derived fungus Aspergillus sp. GXIMD 03004. Natural Product. Res. doi: 10.1080/14786419.2024.2402460
Chen L., Xie L., Zhou J., Fang D., Jiang H., Liu W., et al. (2024). Rakicidin J and K, two cytotoxic and antibacterial cyclic depsipeptides from the marine bacterium Micromonospora chalcea. Natural Product. Res., 1–7. doi: 10.1080/14786419.2024.2335354
Crawford J. M., Tang G. L., Herzon S. B. (2021). Natural products: an era of discovery in organic chemistry. J. Organic. Chem. 86, 10943–10945. doi: 10.1021/acs.joc.1c01753
El-Hawary S. S., Hassan M. H. A., Hudhud A. O., Abdelmohsen U. R., Mohammed R. (2023). Elicitation for activation of the actinomycete genome’s cryptic secondary metabolite gene clusters. RSC. Adv. 13, 5778–5795. doi: 10.1039/d2ra08222e
Feng T., Wu R., Wang Y., Wang P., Zhou L., Wang C., et al. (2024). Proangiogenic azaphilones from the marine-derived fungus Neopestalotiopsis sp. HN-1-6. Mar. Drugs 22, 241. doi: 10.3390/md22060241
Garcia-Perez P., Cassani L., Garcia-Oliveira P., Xiao J., Simal-Gandara J., Prieto M. A., et al. (2023). Algal nutraceuticals: a perspective on metabolic diversity, current food applications, and prospects in the field of metabolomics. Food Chem. 409, 135295. doi: 10.1016/j.foodchem.2022.135295
Gomez-Banderas J. (2022). Marine natural products: a promising source of environmentally friendly antifouling agents for the maritime industries. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.858757
Hai Y., Wei M. Y., Wang C. Y., Gu Y. C., Shao C. L. (2021). The intriguing chemistry and biology of sulfur-containing natural products from marine microorganisms (1987-2020). Mar. Life Sci. Technol. 3, 488–518. doi: 10.1007/s42995-021-00101-2
Hassan S. S. U., Abdel-Daim M. M., Behl T., Bungau S. (2022). Natural products for chronic diseases: a ray of hope. Molecules 27, 5573. doi: 10.3390/molecules27175573
Hassan S. S. U., Wu J., Li T., Ye X., Rehman A., Yan S., et al. (2024). Unlocking marine microbial treasures: new PBP2a-targeted antibiotics elicited by metals and enhanced by RSM-driven transcriptomics and chemoinformatics. Microbial. Cell Factories. 23, 303. doi: 10.1186/s12934-024-02573-0
He J. L., Jiang X. D., Gan Y. M., Qu X. J., Yi X. X., Liu Y. H., et al. (2024). Cadophorin C, a new cyclic heptapeptide isolated from a mangrove-derived fungus Penicillium sp. GXIMD 03101. Natural Product. Res. doi: 10.1080/14786419.2024.2425796
Huang X., Cai J., Xiao J., Zhou X., Liu Y., Li Y., et al. (2024). Antimicrobial isochroman-derived atropo-diastereomeric dimers from Penicillium Steckii SCISO 41228. Chem. Biodivers., e202402152. doi: 10.1002/cbdv.202402152
Hussain H., Mamadalieva N. Z., Ali I., Green I. R., Wang D., Zou L., et al. (2021). Fungal glycosides: Structure and biological function. Trends Food Sci. Technol. 110, 611–651. doi: 10.1016/j.tifs.2021.02.029
Jiang Y., Wang J., Zhang H., Tian X., Liang Z., Xu X., et al. (2024). Biological activity and sterilization mechanism of marine fungi-derived aromatic butenolide asperbutenolide A against Staphylococcus aureus. Chem. Biodivers. 21, e202301826. doi: 10.1002/cbdv.202301826
Li Q., Li S., Li S., Hao X., Wang A., Si S., et al. (2024). Antimicrobial and anti-inflammatory cyclic tetrapeptides from the co-cultures of two marine-derived fungi. J. Natural Products. 87, 365–370. doi: 10.1021/acs.jnatprod.3c01115
Li X. D., Li X. M., Wang B. G., Li X. (2024). Antimicrobial sesterterpenoids with a unique 5/8/6/5 tetracyclic carbon-ring-system and diepoxide polyketides from a deep sea-sediment-sourced fungus Chaetomium globosum SD-347. Organic. Biomol. Chem. 22, 3979–3985. doi: 10.1039/d4ob00449c
Liang X., Yang J. F., Huang Z. H., Ma X., Yan Y., Qi S. H. (2024). New antibacterial peptaibiotics against plant and fish pathogens from the deep-sea-derived fungus Simplicillium obclavatum EIODSF 020. J. Agric. Food Chem. 72, 6402–6413. doi: 10.1021/acs.jafc.4c00493
Liu Y., Qi L., Xu M., Li W., Liu N., He X., et al. (2024). Anti-agrobacterium tumefactions sesquiterpene derivatives from the marine-derived fungus Trichoderma effusum. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1446283
Lu W. Y., Li H. J., Li Q. Y., Wu Y. C. (2021). Application of marine natural products in drug research. Bioorganic. Med. Chem. 35, 116058. doi: 10.1016/j.bmc.2021.116058
Mo T., Qin Y., Zhang Y., Liang Y., Li X., Li W., et al. (2024). Three new antibacterial mycophenolic acid derivatives from the marine-derived fungus Penicillium sp. HN-66. Chem. Biodivers., e202401657. doi: 10.1002/cbdv.202401657
Peng X. Y., Wu J. T., Shao C. L., Li Z. Y., Chen M., Wang C. Y. (2021). Co-culture: stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar. Life Sci. Technol. 3, 363–374. doi: 10.1007/s42995-020-00077-5
Pirintsos S., Panagiotopoulos A., Bariotakis M., Daskalakis V., Lionis C., Sourvinos G., et al. (2022). From traditional ethnopharmacology to modern natural drug discovery: a methodology discussion and specific examples. Molecules 27, 4060. doi: 10.3390/molecules27134060
Scherlach K., Hertweck C. (2021). Mining and unearthing hidden biosynthetic potential. Nat. Commun. 12, 3864. doi: 10.1038/s41467-021-24133-5
Schneider Y. K. (2021). Bacterial natural product drug discovery for new antibiotics: strategies for tackling the problem of antibiotic resistance by efficient bioprospecting. Antibiotics 10, 842. doi: 10.3390/antibiotics10070842
Shams Ul Hassan S., Ishaq M., Zhang W. D., Jin H. Z. (2021). An overview of the mechanisms of marine fungi-derived anti-inflammatory and anti-tumor agents and their novel role in drug targeting. Curr. Pharm. Design. 27, 2605–2614. doi: 10.2174/1381612826666200728142244
Srinivasan R., Kannappan A., Shi C., Lin X. (2021). Marine bacterial secondary metabolites: a treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs 19, 530. doi: 10.3390/md19100530
Stincone P., Brandelli A. (2020). Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 40, 306–319. doi: 10.1080/07388551.2019.1710457
Stuart K. A., Welsh K., Walker M. C., Edrada-Ebel R. (2020). Metabolomic tools used in marine natural product drug discovery. Expert Opin. Drug Discovery 15, 499–522. doi: 10.1080/17460441.2020.1722636
Surendhiran D., Li C., Cui H., Lin L. (2021). Marine algae as efficacious bioresources housing antimicrobial compounds for preserving foods-A review. Int. J. Food Microbiol. 358, 109416. doi: 10.1016/j.ijfoodmicro.2021.109416
Tsipinana S., Husseiny S., Alayande K. A., Raslan M., Amoo S., Adeleke R. (2023). Contribution of endophytes towards improving plant bioactive metabolites: a rescue option against red-taping of medicinal plants. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1248319
Vaou N., Stavropoulou E., Voidarou C., Tsigalou C., Bezirtzoglou E. (2021). Towards advances in medicinal plant antimicrobial activity: a review study on challenges and future perspectives. Microorganisms 9, 2041. doi: 10.3390/microorganisms9102041
Voser T. M., Campbell M. D., Carroll A. R. (2022). How different are marine microbial natural products compared to their terrestrial counterparts? Natural Product. Rep. 39, 7–19. doi: 10.1039/d1np00051a
Wang C., Zhou Y., Yang L., Hu H., Chen J., Ying Y., et al. (2024). Discovery of 2,5-diketopiperazine alkaloids with quorum sensing inhibitory activity from the marine fungus Penicillium sp. ZJUT-34. Natural Product. Res. 38, 3605–3612. doi: 10.1080/14786419.2023.2258441
Wang Y. R., Dong Y. L., Li X. M., Shi X. S., Li H. L., Meng L. H., et al. (2024). Curvularin derivatives from the marine mangrove derived fungus Penicillium sumatrense MA-325. Phytochemistry 220, 114000. doi: 10.1016/j.phytochem.2024.114000
Wei J., Chen X., Wu B. (2024). Dicitrinols A-C: citrinin derivatives from hydrothermal vent-associated fungus Penicillium citrinum TW132-59. J. Organic. Chem. 89, 15264–15270. doi: 10.1021/acs.joc.4c02067
Wen H. M., Zhang Y. W., Feng F. J., Huang G. B., Lv Y. H., Zhang Z. Y., et al. (2024). Antibacterial oxygenated ergostane-type steroids produced by the marine sponge-derived fungus Aspergillus sp. J. Asian Natural Products. Res. 26, 548–554. doi: 10.1080/10286020.2023.2259317
Wu Z., Wang W., Li J., Ma C., Chen L., Che Q., et al. (2024). Evolution-based discovery of polyketide acylated valine from a cytochalasin-like gene cluster in Simplicillium lamelliciola HDN13430. J. Natural Products. 87, 1222–1229. doi: 10.1021/acs.jnatprod.3c01202
Wu J., Wang W., Yang Y., Shah M., Peng J., Zhou L., et al. (2024). Phenylhydrazone alkaloids from the deep-sea cold seep derived fungus Talaromyces amestolkiae HDN21-0307. J. Natural Products. 87, 1407–1415. doi: 10.1021/acs.jnatprod.4c00132
Xu X., Dong Y., Yang J., Wang L., Ma L., Song F., et al. (2024). Secondary metabolites from marine-derived fungus Penicillium rubens BTBU20213035. J. Fungi. 10, 424. doi: 10.3390/jof10060424
Xu M., Godana E. A., Dhanasekaran S., Zhang X., Yang Q., Zhao L., et al. (2023). Comparative proteome and transcriptome analyses of the response of postharvest pears to Penicillium expansum infection. Postharvest. Biol. Technol. 196, 112182. doi: 10.1016/j.postharvbio.2022.112182
Xue J., Guo X., Xu G., Chen X., Jiao L., Tang X. (2024). Discovery, identification, and mode of action of phenolics from marine-derived fungus Aspergillus ustus as antibacterial wilt agents. J. Agric. Food Chem. 72, 2989–2996. doi: 10.1021/acs.jafc.3c07826
Yao F. H., Liang X., Shen W. B., Lu X. H., Li G. C., Qi S. H. (2024). Microascones, decahydrofluorene-class alkaloids from the marine-derived fungus Microascus sp. SCSIO 41821. J. Natural Products. 87, 810–819. doi: 10.1021/acs.jnatprod.3c00984
Young R. J., Flitsch S. L., Grigalunas M., Leeson P. D., Quinn R. J., Turner N. J., et al. (2022). The time and place for nature in drug discovery. JACS. Au. 2, 2400–2416. doi: 10.1021/jacsau.2c00415
Zhang J., Gao L., Lin H., Liang Y., You M., Ding L., et al. (2024). Discovery of antibacterial compounds against Xanthomonas citri subsp. citri from a marine fungus Aspergillus terreus SCSIO 41202 and the mode of action. J. Agric. Food Chem. 72, 12596–12606. doi: 10.1021/acs.jafc.4c02769
Zhang Y., Hao R., Chen J., Li S., Huang K., Cao H., et al. (2024). Health benefits of saponins and its mechanisms: perspectives from absorption, metabolism, and interaction with gut. Crit. Rev. Food Sci. Nutr. 64, 9311–9332. doi: 10.1080/10408398.2023.2212063
Zhang X., Jia R., Yang J., Song Z., Xu X., Song F. (2024). A butyrolactone derivative from marine-derived fungal strain Aspergillus terreus BTBU20211037. Natural Product. Res. doi: 10.1080/14786419.2024.2422515
Zhang L., Song J., Kong L., Yuan T., Li W., Zhang W., et al. (2020). The strategies and techniques of drug discovery from natural products. Pharmacol. Ther. 216, 107686. doi: 10.1016/j.pharmthera.2020.107686
Zhang L., Yang Q., Jiang Y., Yu L., Song N., Zhao D., et al. (2024). Antifouling and antibacterial bioactive metabolites of marine fungus (terrein)/polyamide thin-film composite reverse osmosis membranes for desalination applications. Desalination 572, 117140. doi: 10.1016/j.desal.2023.117140
Zhou C., Ge Y., Lan D., Zhao M., Wu B. (2024). Harzianolides B-G: undescribed butenolides isolated from the fungus Trichoderma harzianum ZN-4. Fitoterapia 176, 106039. doi: 10.1016/j.fitote.2024.106039
Keywords: marine fungi, antibacterial activity, novel natural products, alkaloid, polyketide
Citation: Pan C, Hassan SSu, Muhammad I and Jin H (2025) Marine fungi as a goldmine for novel antibiotics: a 2024 perspective. Front. Mar. Sci. 11:1538136. doi: 10.3389/fmars.2024.1538136
Received: 02 December 2024; Accepted: 27 December 2024;
Published: 21 January 2025.
Edited by:
Syed Qamar Abbas, Sarhad University of Science & Information Technology (SUIT), PakistanReviewed by:
Heba H. Abdel-Kader, National Institute of Oceanography and Fisheries (NIOF), EgyptIqra Chandio, Zhengzhou University, China
Muhammad Usman, Western University, Canada
Copyright © 2025 Pan, Hassan, Muhammad and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengqian Pan, Y3FwYW5AdWpzLmVkdS5jbg==; Huizi Jin, a2ltaHpAc2p0dS5lZHUuY24=
†These authors have contributed equally to this work
 Chengqian Pan
Chengqian Pan Syed Shams ul Hassan
Syed Shams ul Hassan Ishaq Muhammad
Ishaq Muhammad Huizi Jin
Huizi Jin